Temperature response of stem elongation in wheat grown under field conditions indicated that temperature response is highly heritable and linked to the flowering pathway.
Keywords: Development, field phenotyping, GWAS, LIDAR, physiology, plant height, temperature response, wheat
Abstract
In wheat, temperature affects the timing and intensity of stem elongation. Genetic variation for this process is therefore important for adaptation. This study investigates the genetic response to temperature fluctuations during stem elongation and its relationship to phenology and height. Canopy height of 315 wheat genotypes (GABI wheat panel) was scanned twice weekly in the field phenotyping platform (FIP) of ETH Zurich using a LIDAR. Temperature response was modelled using linear regressions between stem elongation and mean temperature in each measurement interval. This led to a temperature-responsive (slope) and a temperature-irresponsive (intercept) component. The temperature response was highly heritable (H2=0.81) and positively related to a later start and end of stem elongation as well as final height. Genome-wide association mapping revealed three temperature-responsive and four temperature-irresponsive quantitative trait loci (QTLs). Furthermore, putative candidate genes for temperature-responsive QTLs were frequently related to the flowering pathway in Arabidopsis thaliana, whereas temperature-irresponsive QTLs corresponded to growth and reduced height genes. In combination with Rht and Ppd alleles, these loci, together with the loci for the timing of stem elongation, accounted for 71% of the variability in height. This demonstrates how high-throughput field phenotyping combined with environmental covariates can contribute to a smarter selection of climate-resilient crops.
Introduction
Temperature is a major abiotic factor affecting plant growth and development. As a consequence of global warming, wheat production could decrease by 6% for each degree Celsius of global temperature increase (Asseng et al., 2015). While heat stress during critical stages can drastically reduce yield (Gibson and Paulsen, 1999; Farooq et al., 2011), warm temperatures can decrease yield by accelerating development and thereby shortening critical periods for yield formation (Fischer, 1985; Slafer and Rawson, 1994). Despite the clear effect of temperature on growth and phenology, little is known about the genotype-specific response pattern to varying temperature conditions during crop development and its genetic control. We therefore aimed to quantify the genotype-specific temperature responsiveness of European winter wheat during the stem elongation (SE) phase.
SE is a critical phase for yield formation in wheat. It occurs between the phenological stages of terminal spikelet initiation and anthesis (Slafer et al., 2015). The start of SE coincides with the transition from vegetative to reproductive development, when the apex meristem differentiates from producing leaf primordia to producing spikelet primordia (Trevaskis et al., 2007; Kamran et al., 2014). During SE, florets are initiated at the spikelets until booting (Kirby, 1988; Slafer et al., 2015). An increased duration of SE increases the number of fertile florets due to longer spike growth and higher dry matter partitioning to the spike (González et al., 2003). This in turn increases the number of grains per spike and therefore yield (Fischer, 1985). Modifying the timing of the critical phenological stages (transition to early reproductive phase and flowering), and thus SE duration, has been proposed as a way to increase wheat yield (Slafer et al., 1996; Miralles and Slafer, 2007; Whitechurch et al., 2007) or at least to mitigate adverse climate change effects on yield, for example by enhancing earliness to escape heat during flowering (Chapman et al., 2012; Hernandez-Ochoa et al., 2019). The recent warming trend causes a faster advancement in phenology. For example, flowering time occurred earlier in Germany throughout the past decade, which is attributable to both increased temperature and selection for early flowering (Rezaei et al., 2018).
Final height is also an important yield determinant. During the ‘Green Revolution’ wheat yields increased by the introduction of reduced height (Rht) genes. The resulting dwarf and semi-dwarf varieties benefit from improved resource allocation from the stem to the spike and reduced lodging, allowing more intensive nitrogen application (Hedden, 2003). Gibberellin (GA)-insensitive Rht genes (Rht-A1, Rht-B1, and Rht-D1) were shown to limit cell wall extensibility which decreases growth rates (Keyes et al., 1989) without affecting development (Youssefian et al., 1992). Moreover, the allele Rht-B1c (Wu et al., 2011) and the GA-sensitive Rht12 dwarfing gene (Chen et al., 2013) delay heading.
The main abiotic factors affecting the timing of floral initiation and flowering are temperature and photoperiod, with temperature affecting both vernalization and general rate of development (Slafer et al., 2015). These developmental transitions are controlled by major genes involved in the flowering pathway, namely vernalization (Vrn), photoperiod (Ppd), and earliness per se (Eps) genes (Slafer et al., 2015). The Ppd and Vrn genes define photoperiod and vernalization requirements which jointly enable the transition to generative development and define time to flowering. On the other hand, Eps genes fine-tune the timing of floral transition and flowering, after vernalization and photoperiod requirements are fulfilled (Zikhali and Griffiths, 2015). While vernalization and photoperiod response are well known, the role of temperature per se remains less clear. Temperature affects all developmental phases, and warmer ambient temperatures generally accelerate growth and development in crops (Slafer and Rawson, 1994, 1995a,c; Atkinson and Porter, 1996; Fischer, 2011; Slafer et al., 2015). However, it is unclear if temperature response governs growth rate and development independently. If so, the question remains as to whether there is enough genetic variability in temperature response to be used in a breeding context (Parent and Tardieu, 2012).
Genotypic variation for growth response to temperature was reported for wheat leaf elongation rate (Nagelmüller et al., 2016), as well as for canopy cover growth (Grieder et al., 2015). Kiss et al. (2017) reported significant genotype×temperature interactions in the timing of SE as well as temperature-dependent differences in the expression of Vrn and Ppd genes under controlled conditions. Under field conditions, the response of stem elongation to temperature has not yet been investigated at high temporal resolution.
In recent years, new high-throughput phenotyping technologies have enabled the monitoring of plant height with high accuracy and frequency in the field (Bendig et al., 2013; Friedli et al., 2016; Holman et al., 2016; Aasen and Bareth, 2018; Hund et al., 2019). We have previously demonstrated that the ETH field phenotyping platform (FIP; Kirchgessner et al., 2016) can be used to accurately track the development of canopy height in a large set of wheat genotypes using terrestrial laser scanning (Kronenberg et al., 2017). Considerable genotypic variation was detected for the start and end of SE which correlated positively with final canopy height (Kronenberg et al., 2017).
While many temperature-independent factors affecting plant height are known, the influences of temperature-dependent elongation and timing of the elongation phase is less clear. We hypothesize that apart from temperature-independent factors, there is a genotype-specific response to ambient temperature which affects growth as well as the timing of developmental stages. To address this, we aimed to dissect the process towards final height into the following components: (i) temperature-independent elongation; (ii) temperature-dependent elongation; and (iii) the duration of the elongation phase determined by the start and end of the process. To achieve this, we present a method to assess and measure these three processes under field conditions by means of high-frequency, high-throughput phenotyping of canopy height development. The resulting data were combined with genetic markers to identify quantitative trait loci (QTLs) controlling the aforementioned processes.
Materials and methods
Experimental set-up, phenotyping procedures, and extracted traits
Field experiments were conducted in the FIP at the ETH research station in Lindau-Eschikon, Switzerland (47.449°N, 8.682°E, 520 m a.s.l.; soil type: eutric cambisol). We used a set of ~330 winter wheat genotypes (335–352 depending on the experiment) comprising current European elite cultivars (GABI wheat; Kollers et al., 2013), supplemented with 30 Swiss varieties. These were monitored over three growing seasons in 2015, 2016, and 2017. Briefly, the field experiments were conducted in an augmented design with two replications per genotype using micro plots with a size of 1.4×1.1 m. In the 2017 growing season, the experiment was repeated again, with minor changes in genotypic composition. This resulted in 328 genotypes present across all three experiments. Details about the experimental set-up for the growing seasons 2015 and 2016 are described in Kronenberg et al. (2017).
Canopy height was measured twice weekly from the beginning of shooting (BBCH 31) using a light detection and ranging (LIDAR) scanner (FARO R Focus3D S 120; Faro Technologies Inc., Lake Mary, USA) mounted on the FIP (Kirchgessner et al., 2016). Height measurements were concluded when no further height increase was observed in any of the genotypes. Canopy height data were extracted from the LIDAR data as described in Kronenberg et al. (2017).
The start, end, and duration of SE as well as final canopy height (FH) were extracted from the height data following Kronenberg et al. (2017): FH was defined as the point after which no increase in height was observed in several consecutive measurements. Normalized canopy height was calculated as the percentage of FH on each day of measurement for every plot and then linearly interpolated between measurement points. Growing degree-days after sowing until 15% FH (GDD15) and 95% FH (GDD95) were used as proxy traits for the start and end of SE, respectively. SE duration was recorded in thermal time (GDDSE) as well as in calendar days (timeSE), as the difference between GDD95 and GDD15 (Kronenberg et al., 2017). Heading date was recorded as GDD (headingGDD) when 50% of the spikes were fully emerged from the flag leaf sheath (BBCH 59; Lancashire et al., 1991). Heading data for 2015 could not be evaluated due to insufficient data availability. Therefore, a third year of heading data was gathered in 2018, when the experiment was repeated again as described in Anderegg et al. (2020).
In order to investigate short-term growth response to temperature, average daily stem elongation rates (SERs) were calculated for each plot as the difference (∆) in canopy height (CH) between consecultive time points (t):
| (1) |
Extracting growth response to temperature
Temperature response was modelled by regressing average daily SER against average temperature of the respective interval for each plot within the respective year following
| (2) |
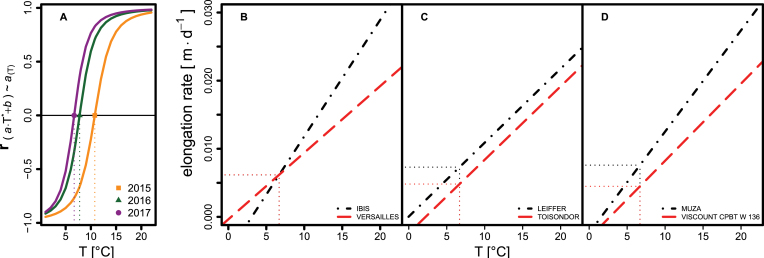
where T is the ambient temperature, a is the coefficient of the linear regression (i.e. growth response to ambient temperature; slpSER~T), and ε denotes the residual error. bTcrit is the model intercept at the temperature at which the correlation between intercept (intSER~T) and slope is zero (see below). Per definition, the intercept of a linear model would be calculated at T=0 °C—far outside the range of observed temperatures. In the observed data, the intercept at T=0 °C correlated strongly negatively with the slope (Fig. 1A) and thus did not add much additional information concerning the performance of the evaluated genotypes. Likewise, an intercept at 20 °C, at the upper range of the observed data, correlated strongly positively with the slope (Fig. 1A). Grieder et al. (2015) performed a similar analysis for the canopy cover development during winter and found a similar, strongly negative correlation between temperature response (slope) and growth at 0 °C (intercept). We sequentially calculated the intercept at 0.01 °C increments between 1 °C and 22 °C for each plot within a year. Subsequently, we calculated the Pearson correlation coefficient between the common slope of these models and each of the different intercepts (Fig. 1A). Based on this sequence, we empirically determined the critical temperature value (Tcrit) at which the correlation between slope and intercept was zero (Fig. 1A). Hence, Tcrit is defined as the temperature at which intercept and slope are independent. Due to this independence, the value of the intercept at Tcrit can be interpreted as the intrinsic growth component independent of temperature response herein referred to as ‘vigour’. Following this, two genotypes can show the same vigour but differ markedly in temperature response (Fig. 1B), have the same temperature response but differ in vigour (Fig. 1C), or differ for both temperature response and vigour (Fig. 1D).
Fig. 1.
Illustration and interpretation for the parameters of the applied temperature response model (Equation 2). (A) Distribution of Pearson correlation coefficients between the intercept and slope of the linear model for individual years, depending on the temperature at which the intercept is calculated. Dotted vertical lines indicate the critical temperature (Tcrit) for individual years used to calculate the intercept. (B–D) Illustration of the relationship between intercept and slope on contrasting genotypes (dashed and dash-dotted lines). (B) The same vigour but differing in temperature response. (C) Both have the same temperature response but differ in vigour. (D) Genotypes differ in vigour as well as in temperature response. Horizontal and vertical dotted lines indicate vigour and Tcrit, respectively. The two contrasting genotypes per example (B–D) were selected from the 2017 data based on their vales for slope and intercept.
Statistical analysis
The data were processed stepwise as follows: (i) correction for design factors and spatial trends; (ii) application of linear models to each plot to determine growth response to temperature; and (iii) prediction of adjusted means and calculation of the heritability for all traits across years.
The spatial correction of canopy height per measurement time point was done using the R package SpATS (Rodríguez-Álvarez et al., 2018) following:
| (3) |
where Y is the phenotypic value for the a plot in the jth row in the kth column planted with the ith genotype, f(r,c) is a smoothed bivariate surface defined over rows and ranges of a virtual grid, Gi is the effect of the ith genotype (i=1, …, n; n=335–352 depending on the year), Rj is the effect of the jth row in the virtual grid, and Ck is the effect of the kth range. With the number of genotypes, the number of ranges/rows varied across years (17/21 in 2015, 18/20 in 2016, and 18/21 in 2017, respectively). The replications were arranged diagonally in this grid with a gap of five rows and ranges between them. Thus, for example in 2017, replication 1 was at rows 1–21 and ranges 1–18; replication 2 was at rows 24–41 and ranges 27–47 of the virtual grid.
The function f(r,c) describing the bivariate surface can be decomposed in a nested-type ANOVA structure as described by Rodríguez-Álvarez et al. (2018). The number of spline points was set to two-thirds of the total number of rows and ranges in the virtual grid, respectively.
From this model, the predicted genotypic best linear unbiased estimates (BLUEs) plus residual error were kept as spatially corrected plot values. Thus, these new plot values were corrected for spatial effect as well as for the random row and range effects, and used for the subsequent dynamic model. For a visualization of the applied SpATS correction, see Supplementary Fig. S1 at JXB online.
Genotypic BLUEs across the three seasons were calculated for all traits using the R package asreml-4 (Butler, 2018) following:
| (4) |
where ypij is the spatialy corrected plot value of the respective trait (FH, GDD15, GDD95, GDDSE, timeSE, intSER~T, or slpSER~T), μ is the overall mean, Gi the fixed effect of the genotypes common in all three years (i=1,…,328), Yj is the fixed effect of the year (j=2015,…,2017), GYij is the random genotype-by-year interaction, and ε pij is the residual error.
In order to estimate best linear unbiased predictors (BLUPs) and heritability (H2) across years, Gi in Equation 3 was set as a random term and heritability was calculated following (Falconer and Mackay, 1996) using:
| (5) |
where H2 is the broad sense heritability, σ 2G is the genotypic variance, σ 2GY is the genotype×year interaction variance, and σ 2ε is the residual variance. For heading data, only one replicate per year was available. Plot-corrected values were extracted using SpATS (Rodríguez-Álvarez et al., 2018), and heritability across 3 years was calculated by omitting the GY term in Equation 4 and dividing the residual variance by three, based on the three available year–site replications.
Genotypic BLUEs across 3 years were used for subsequent correlation analysis and genome-wide association study (GWAS). All statistical analyses were performed in the R environment (R Core Team, 2015).In order to investigate the relationship between FH, temperature response, and vigour, and to test for confounding Rht or Ppd effects on temperature response, FH was modelled using the linear model
| (6) |
where yi is FH of the ith genotype, µ is the model intercept, β 1–6 are the main effect estimates of x=slpSER~T, intSER~T, GDDSE, Rht-B1, Rht-D1, or Ppd-D1, respectively. β 1,2–β 5,6 are all two-way interaction effects (n=15) and ε i is the residual error. Genotypic data for Rht-B1, Rht-D1, and Ppd-D1 alleles were available for 301 genotypes obtained from Kollers et al. (2014). There, genotyping of the Rht-1 alleles was performed using PCR markers (Ellis et al., 2002), while Ppd-D1 alleles were genotyped by the presence or absence of a 2 kb insertion using specific primers (Beales et al., 2007; Kollers et al., 2014)
Association study
GWAS was performed on the different traits to compare the phenotypic correlations with the underlying genetic architecture of the traits. As a positive control, FH data from Germany and France reported by Zanke et al. (2014b) were also compared and analysed.
Genotyping data were made previously by the GABI wheat consortium represented by the Leibniz Institute of Plant Genetics and Crop Plant Research (IPK; Zanke et al., 2014a) using the 90K illumina SNP-chip (Cavanagh et al., 2013; Wang et al., 2014). Monomorphic single nucleotide polymorphisms (SNPs) were discarded. The remaining markers were mapped to the IWGSC reference genome (International Wheat Genome Sequencing Consortium, 2018) by BLASTN search using an E-value threshold <1e–30. The genome position with the lowest E-value was assigned as the respective marker location. Markers that could not be unequivocally positioned were dropped. After filtering SNPs with a minor allele frequency and missing genotype rate <0.05, a total of 13 450 SNP markers and 315 genotypes remained in the set. The reference genome position of Rht, Ppd, Vrn, and putative Eps genes was determined with BLASTN search as described above using published GenBank sequences (Supplementary Table S1).
To mitigate against multiple testing, relatedness, and population structure, three different methods were used to calculate marker–trait associations (MTAs) between phenotypic BLUPs and SNP markers. (i) We used a mixed linear model (MLM) including principal components among marker alleles as fixed effects and kinship as random effect to account for population structure (Zhang et al., 2010). This approach was chosen to stringently prevent type I errors. The MLM GWAS was performed using the R Package GAPIT (v.2, Tang et al., 2016). Kinship was estimated according to VanRaden (2008). (ii) In a generalized linear model (GLM) framework implemented in PLINK (Purcell et al., 2007), association analysis was performed using SNP haplotype blocks consisting of adjacent SNP triplets. Using haplotype blocks takes the surrounding region of a given SNP into account, thus increasing the power to detect rare variants (Purcell et al., 2007). (iii) Finally, the FarmCPU method (Liu et al., 2016) was used, which is also implemented in GAPIT. FarmCPU tests individual markers with multiple associated markers as covariates in a fixed effect model. Associated markers are iteratively used in a random effect model to estimate kinship. Confounding between testing markers and kinship is thus removed while controlling type I error, leading to increased power (Liu et al., 2016).
For all methods, a Bonferroni correction was applied to the pointwise significance threshold of α=0.05, to avoid false positives. Hence, only markers above –log10(P-value) >5.43 were considered significant.
Linkage disequilibrium (LD) among markers was estimated using the squared correlation coefficient (r2) calculated with the R package SNPrelate (Zheng et al., 2012). A threshold of r2=0.2 (Gaut and Long, 2003) was applied to calculate the chromosome-specific distance threshold of LD decay. Putative candidate genes were identified by searching the IWGSC annotation of the reference genome (International Wheat Genome Sequencing Consortium, 2018) for genes associated with growth and development within the LD distance threshold around the respective MTA.
Results
Phenotypic results
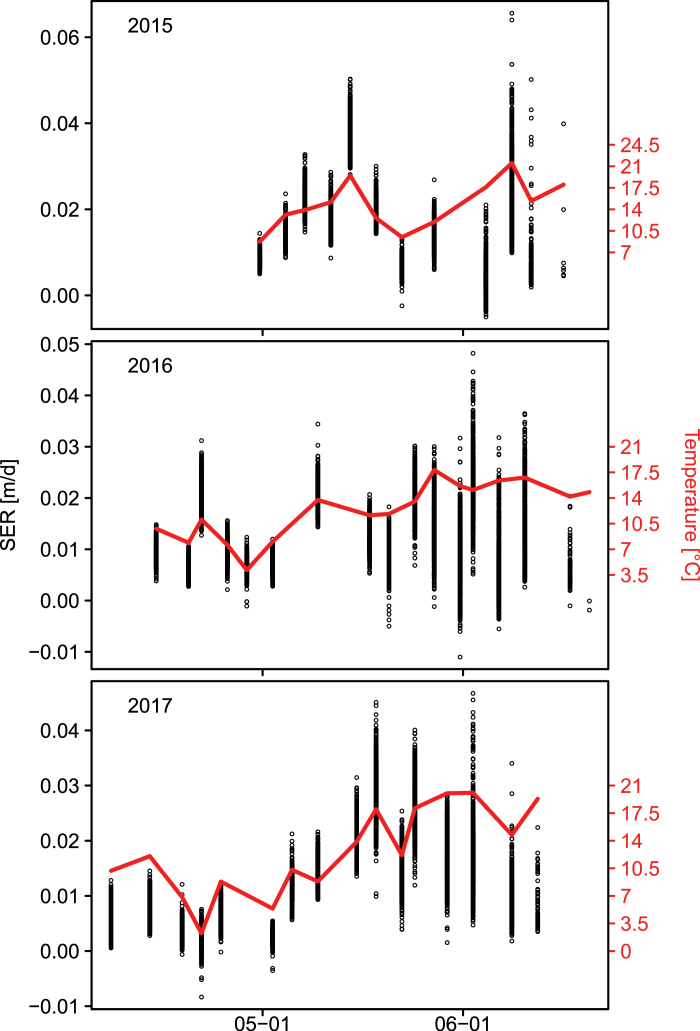
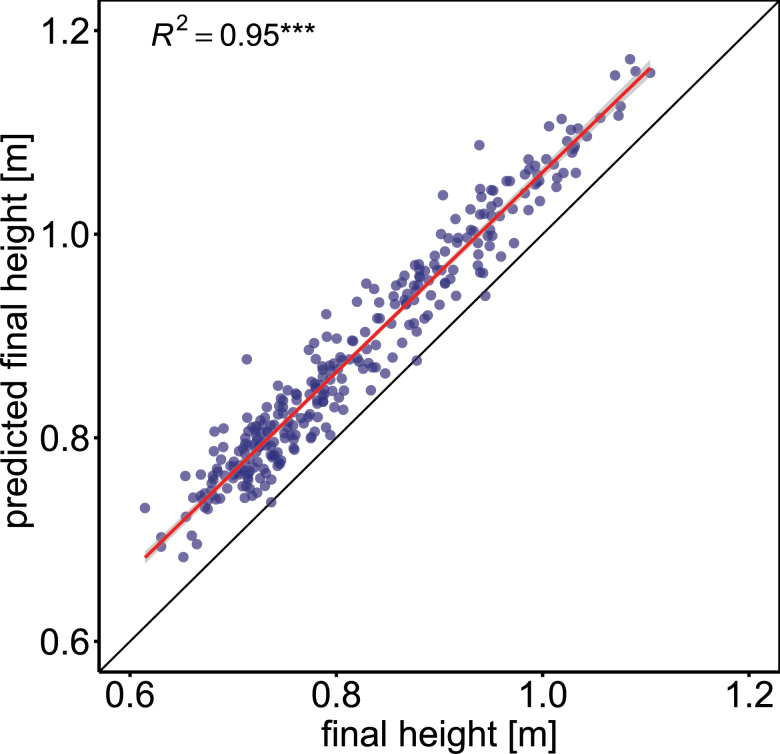
We measured the canopy height of 710–756 plots per year, containing 335–352 wheat genotypes, for three consecutive years. In each season, measurements were made between 17 and 22 times during SE. Plot-based growth rates within single years extracted from these data indicate a clear relationship between growth and temperature for the period of SE, as depicted in Fig. 2. Towards the end of the measurement period in June, there was a larger deviation, which was also reflected in the quality of plot-based linear model fits of SER versus temperature (see Equation 2), summarized in Supplementary Fig. S2. For the 2015 and especially the 2016 experiment, R2 values were low and except for the 2017 experiment, the parameter estimates were not statistically significant (Supplementary Fig. S2a). Inspection of the best and worst model fits, however, shows that failure of fitting the model for single plots was levelled out by the replications within genotypes (Supplementary Fig. S2b). The weak model fits therefore did not affect the genotype ranking of adjusted means across replications. ANOVA revealed significant (P<0.001) genotypic effects for both slpSER~T and intSER~T across 3 years. Both traits showed high heritabilities across years (H2=0.81 for slpSER~T and H2=0.77 for intSER~T; Table 1). Using the BLUEs of slpSER~T, intSER~T, and temperature sum for SE (GDDSE), FH could be predicted with high accuracy across different years (0.85≤R2≤0.89) by training a linear model on the BLUEs of one year and predicting it on the BLUEs of another independent year. In order to account for possible confounding Rht and Ppd effects, the allelic status of Rht-B1, Rht-D1, and Ppd-D1 was included as contrasts in the model (Table 2). Of the 301 genotypes with available data, 7% and 58% carried the dwarfing alleles Rht-B1b and Rht-D1b, respectively, and 13% carried the photoperiod-insensitive allele Ppd-D1a. Training the model on the 3 year BLUEs resulted in a prediction accuracy of single years between R2=0.94 and R2=0.95 (Fig. 3). Type II ANOVA revealed significant effects for slpSER~T, intSER~T, GDDSE, Rht-B1, and Rht-D1. A significant (P<0.05) interaction effect was found between Rht-D1 and Ppd-D1. Furthermore, weak interactions (P<0.1) were found for Rht-B1:Ppd-D1, intSER~T:Rht-B1, and intSER~T:Rht-D1 (Table 2). High heritabilities across 3 years (0.54≤H2≤0.98; Table 1) were also found for the other traits: FH, start of SE, end of SE, SE duration, and heading. All traits showed moderate genotype×year interaction effects which were smaller (except for SE duration) than the genotypic effects across years (Table 1).
Fig. 2.
Relationship between the stem elongation rate (SER) and temperature. Plot-based SER raw data (n>700 per year) of >330 genotypes (black dots) as well as temperature (solid red line) are plotted against calendar time for the years 2015–2017.
Table 1.
Variance components and heritabilities for all investigated traits
| Trait | Variance component | Estimate | SE | z ratio | % Total variance | Heritability |
|---|---|---|---|---|---|---|
| intSER~T | Gen_ID | 7.024E-07 | 7.219E-08 | 9.730 | 49.01 | 0.77 |
| Gen_ID:Year | 5.224E-07 | 3.491E-08 | 14.963 | 36.45 | ||
| units!R | 2.084E-07 | 8.920E-09 | 23.365 | 14.54 | ||
| slpSER~T | Gen_ID | 7.348E-08 | 7.184E-09 | 10.229 | 55.00 | 0.81 |
| Gen_ID:Year | 4.516E-08 | 2.925E-09 | 15.440 | 33.80 | ||
| units!R | 1.495E-08 | 6.396E-10 | 23.372 | 11.19 | ||
| FH | Gen_ID | 1.226E-02 | 9.798E-04 | 12.511 | 92.24 | 0.98 |
| Gen_ID:Year | 5.890E-04 | 4.568E-05 | 12.893 | 4.43 | ||
| units!R | 4.417E-04 | 1.890E-05 | 23.371 | 3.32 | ||
| GDD15 | Gen_ID | 1.226E+03 | 1.171E+02 | 10.471 | 56.64 | 0.82 |
| Gen_ID:Year | 6.241E+02 | 4.370E+01 | 14.283 | 28.84 | ||
| units!R | 3.144E+02 | 1.345E+01 | 23.365 | 14.53 | ||
| GDD95 | Gen_ID | 1.190E+03 | 1.120E+02 | 10.624 | 56.84 | 0.84 |
| Gen_ID:Year | 4.953E+02 | 3.958E+01 | 12.515 | 23.66 | ||
| units!R | 4.081E+02 | 1.747E+01 | 23.365 | 19.50 | ||
| timeSE | Gen_ID | 5.844E+00 | 8.095E-01 | 7.219 | 27.84 | 0.59 |
| Gen_ID:Year | 9.481E+00 | 6.911E-01 | 13.717 | 45.16 | ||
| units!R | 5.668E+00 | 2.425E-01 | 23.370 | 27.00 | ||
| GDDSE | Gen_ID | 5.665E+02 | 8.544E+01 | 6.631 | 24.14 | 0.54 |
| Gen_ID:Year | 1.067E+03 | 8.007E+01 | 13.326 | 45.46 | ||
| units!R | 7.134E+02 | 3.052E+01 | 23.375 | 30.40 | ||
| headingGDD | Gen_ID | 1.742E+03 | 1.481E+02 | 11.764 | 80.24 | 0.92 |
| units!R | 4.290E+02 | 2.380E+01 | 18.028 | 19.76 |
Table 2.
Type II ANOVA table for the linear modela used to predict final canopy height based on temperature response (slpSER~T), vigour (intSER~T), and stem elongation duration (GDDSE)
| Predictor | Sum of squares | df | F-value | Pr(>F) | |
|---|---|---|---|---|---|
| slpSER~T | 7.63E-01 | 1 | 2.43E+03 | 5.08E-140 | *** |
| intSER~T | 5.91E-01 | 1 | 1.88E+03 | 2.57E-126 | *** |
| GDDSE | 1.29E-01 | 1 | 4.12E+02 | 5.74E-57 | *** |
| Rht-B1 | 3.16E-03 | 1 | 1.01E+01 | 1.69E-03 | ** |
| Rht-D1 | 3.53E-03 | 1 | 1.12E+01 | 9.13E-04 | *** |
| Ppd-D1 | 1.48E-06 | 1 | 4.71E-03 | 9.45E-01 | |
| slpSER~T: intSER~T | 2.56E-04 | 1 | 8.16E-01 | 3.67E-01 | |
| slpSER~T:GDDSE | 2.31E-05 | 1 | 7.36E-02 | 7.86E-01 | |
| intSER~T:GDDSE | 9.76E-06 | 1 | 3.11E-02 | 8.60E-01 | |
| Rht-B1:Ppd-D1 | 8.94E-04 | 1 | 2.85E+00 | 9.26E-02 | ° |
| Rht-D1:Ppd-D1 | 1.42E-03 | 1 | 4.51E+00 | 3.45E-02 | * |
| slpSER~T:Rht-B1 | 4.01E-06 | 1 | 1.28E-02 | 9.10E-01 | |
| slpSER~T:Rht-D1 | 1.43E-06 | 1 | 4.56E-03 | 9.46E-01 | |
| slpSER~T:Ppd-D1 | 5.53E-04 | 1 | 1.76E+00 | 1.85E-01 | |
| intSER~T:Rht-B1 | 9.01E-04 | 1 | 2.87E+00 | 9.13E-02 | ° |
| intSER~T:Rht-D1 | 7.02E-04 | 1 | 2.23E+00 | 1.36E-01 | |
| intSER~T:Ppd-D1 | 7.70E-04 | 1 | 2.45E+00 | 1.19E-01 | |
| GDDSE:Rht-B1 | 1.71E-05 | 1 | 5.46E-02 | 8.15E-01 | |
| GDDSE:Rht-D1 | 7.56E-04 | 1 | 2.41E+00 | 1.22E-01 | |
| GDDSE:Ppd-D1 | 4.48E-05 | 1 | 1.43E-01 | 7.06E-01 | |
| Residuals | 8.79E-02 | 280 |
Rht-B1, Rht-D1, band Ppd-D1 alleles and all two-way interactions (the interaction effect of Rht-B1 and Rht-D1 was dropped due to singularity) were included to test for possible confounding of temperature response with final height or photoperiod. The model was applied on the 3 year BLUEs of all genotypes with available allelic data of the respective genes (n=301).
Asterisks and dots indicate the significance of the respective predictor (***P<0.001, **P<0.01, *P<0.05, °P<0.1).
a See Equation 6.
Fig. 3.
Prediction of final height based on temperature response (slpSER~T), vigour (intSER~T), and stem elongation duration (GDDSE). Rht-B1, Rht-D1, and Ppd-D1 alleles and all two-way interactions were included to test for possible confounding of temperature response with final height or photoperiod (see Equation 6 and Table 2). The model was applied on the 3 year BLUEs of all genotypes with available allelic data of the respective genes (n=301).
Phenology, temperature response, and final height were positively correlated
To evaluate the relationships between the traits measured, Pearson correlation coefficients were calculated for each trait pair. If not indicated otherwise, the reported correlations were highly significant (P<0.001)
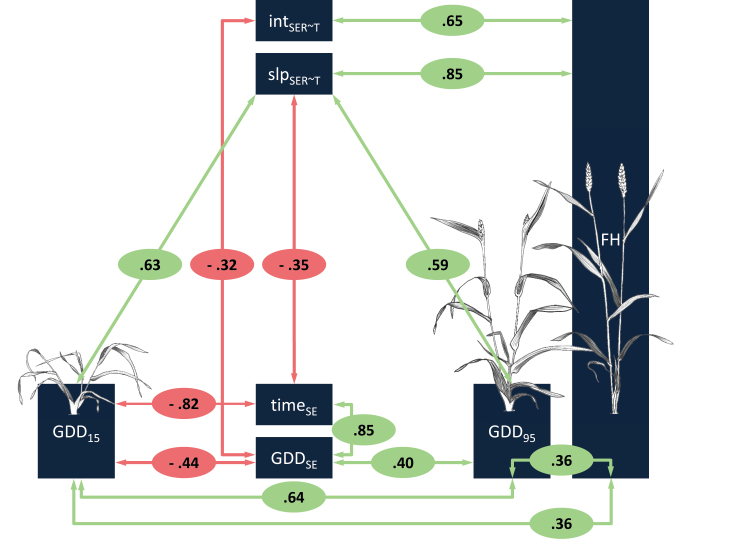
Positive correlations were found among GDD15, GDD95, and FH (0.36≤r≤0.64, Fig. 4), indicating that taller genotypes were generally later in their development towards FH. Temperature response (slpSER~T) and vigour (intSER~T) also showed a strong, positive relationship to FH (r=0.85 and r=0.65, respectively). However, only temperature response correlated with GDD15 and GDD95 (r=0.63 and r=59, respectively), whereas vigour did not (r≤0.26, Supplementary Fig. S3).
Fig. 4.
Key correlations among investigated traits. Pearson correlation coefficients between respective traits are given in red and green circles, where red denotes a negative correlation and green denotes a positive correlation. Weak correlations (r<0.3) are shown in the complete correlation matrix Supplementary Fig. S3. Illustrations of GDD15, GDD95, and FH were taken from Schürch et al. (2018).
As expected, SE duration in thermal time (GDDSE) was negatively correlated with GDD15 (r= –0.44) and positively correlated with GDD95 (r=0.4). However, GDDSE did not correlate with FH (r= –0.01, P=0.874) or temperature response (r=0.006, P=0.285), although GDDSE negatively correlated with vigour (r= –0.32). In contrast, SE duration in calendar days (timeSE) was negatively correlated with temperature response (r= –0.35) and GDD15 (r= –0.82), indicating a longer SE phase for earlier genotypes. Heading showed strong positive correlations with GDD15 (r=0.61) and GDD95 (r=0.71), and a weak correlation to temperature response (r=0.29). Furthermore, heading correlated negatively with intSER~T (r= –0.41) and showed no correlation to FH (r=0, P=0.934). Other weak correlations (r<0.3), that are not discussed, are shown in Supplementary Fig. S3.
Linkage disequilibrium and population structure
Prior to MTA analysis, we evaluated population structure and LD. Principal component analysis (PCA) of the marker genotypes revealed no distinct substructure in the investigated population. The biplot of the first two principal components showed no apparent clusters, with the first component explaining 8% and the second component explaining 3.3% of the variation in the population (Supplementary Fig. S4). This is consistent with prior work using the same population (Kollers et al., 2013; Yates et al., 2019). On average across all chromosomes, LD decayed below an r2 of 0.2 at a distance of 9 Mb. There was, however, considerable variation in this threshold among the single chromosomes (Supplementary Table S2).
Association study
Genome-wide association results differed markedly depending on the applied model. Using an MLM with kinship matrix and PCA as covariates resulted in no significant MTA for any trait (Supplementary Fig. S5). In contrast, the GLM using the haplotype method on temperature response yielded 2958 significant MTAs for α<0.05 and 1852 MTAs for α<0.001, respectively (Supplementary Fig. S6). However, investigation of the respective QQ-plots showed large P-value inflation in the haplotype method whereas the P-values were slightly deflated when using the MLM approach (Supplementary Figs S5, S6). In contrast, with FamCPU, the QQ-plots (Fig. 5) showed no P-value inflation, except for some markers. This pattern is expected, if population structure is appropriately controlled. Therefore, FarmCPU was chosen to be the most appropriate method for the given data, despite identifying fewer significant MTAs.
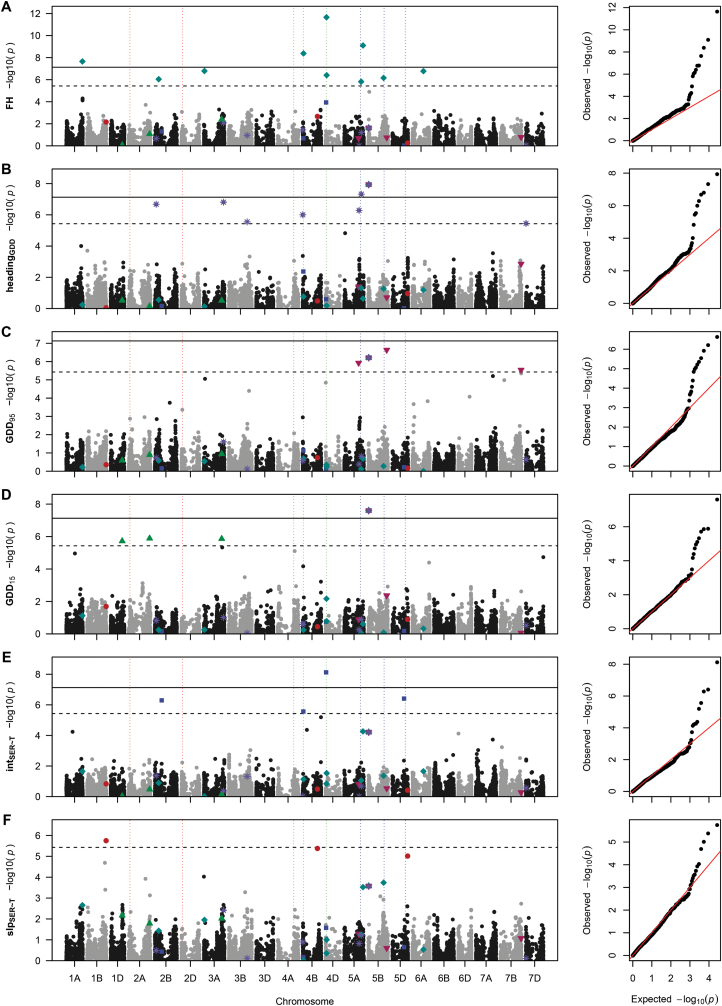
Fig. 5.
Manhattan plots and quantile–quantile plots depicting the GWAS results using FarmCPU for final height (FH, A), growing degree days until heading (headingGDD, B); end (GDD95, C) and start (GDD15, D) of stem elongation; vigour-related intercept (intSER~T, E); and temperature-related slope (slpSER~T, F) of stem elongation in response to temperature. Horizontal lines mark the Bonferroni-corrected significance threshold for P<0.05 (dashed line) and P<0.001 (solid line). Vertical dotted lines mark the positions of Ppd-1 on chromosomes 2A and 2D (red), Rht-1 on chromosomes 4A–4D (green), and Vrn-1 on chromosomes 5A–5D. Significant marker–trait associations for slpSER~T (red dots), intSER~T (blue squares), GDD15 (green upright triangles), GDD95 (magenta downward-facing triangles), heading (violet asterisks), and FH (turquoise diamonds) are highlighted in all Manhattan plots.
As a positive control, we compared our FH data and associated markers with data from Zanke et al. (2014b) who used the same population and SNP chip in field experiments in France and Germany. FH correlated strongly between the two studies (r=0.95), which is in accordance with the high heritability of the trait. In this study, we found 10 significant MTAs for FH (Table 3; Fig. 5). Zanke et al. (2014b) reported 280 significant MTAs for FH across several environments. Of these, only marker RAC875_rep_c105718_585 on chromosome 4D overlapped with the MTAs found in this study. However, by considering flanking markers, we found that of the remaining nine significant MTAs for FH, four were in LD with MTAs found by Zanke et al. (2014b; Supplementary Table S3). The significant MTA found for FH in this study are near known genes controlling FH. For example, Tdurum_contig64772_417 is 4 Mb upstream of Rht-B1 and RAC875_rep_c105718_585 is 7 Mb downstream of Rht-D1 on their respective group 4 chromosomes.
Table 3.
Marker–trait associations for slpSER~T, intSER~T, GDD15, GDD95 headingGDD, and FH, including P-value, minor allele frequency (MAF), and allelic effect estimate
| Trait | SNP | Chr | Position | P-value | MAF | Effect |
|---|---|---|---|---|---|---|
| slpSER~T | wsnp_Ex_c1597_3045682 | 1B | 688 283 256 | 1.76E-06 | 0.19 | –6.05E-05 |
| CAP7_c10839_300 | 4B | 533 724 424 | 4.17E-06 | 0.24 | –5.07E-05 | |
| IAAV7104 | 5D | 553 678 522 | 9.67E-06 | 0.13 | –6.02E-05 | |
| intSER~T | RAC875_s109189_188 | 2B | 248 149 774 | 5.08E-07 | 0.42 | 1.73E-04 |
| Ku_c63300_1309 | 4B | 21 556 672 | 2.73E-06 | 0.10 | –2.99E-04 | |
| Kukri_rep_c68594_530 | 4D | 12 773 259 | 7.47E-09 | 0.40 | –2.30E-04 | |
| Kukri_c6477_696 | 5D | 423 502 809 | 3.89E-07 | 0.21 | –2.03E-04 | |
| GDD15 | wsnp_Ex_c12447_19847242 | 1D | 416456386 | 1.91E-06 | 0.46 | 6.89E+00 |
| Tdurum_contig47508_250 | 2A | 754 339 235 | 1.31E-06 | 0.21 | 9.41E+00 | |
| Kukri_c55381_67 | 3A | 648 868 234 | 1.38E-06 | 0.17 | –1.00E+01 | |
| Excalibur_c74858_243 | 5B | 13 190 663 | 2.50E-08 | 0.47 | –7.88E+00 | |
| GDD95 | Excalibur_c49597_579 | 5A | 521 934 666 | 1.19E-06 | 0.42 | –6.58E+00 |
| Excalibur_c74858_243 | 5B | 13 190 663 | 6.15E-07 | 0.47 | –6.14E+00 | |
| Tdurum_contig44115_561 | 5B | 669 897 388 | 2.31E-07 | 0.13 | –1.02E+01 | |
| RAC875_c38693_319 | 7B | 740 056 880 | 2.87E-06 | 0.20 | 7.51E+00 | |
| headingGDD | RAC875_c12766_461 | 2B | 47 430 682 | 2.10E-07 | 0.39 | –7.77E+00 |
| Kukri_rep_c106620_208 | 3A | 714 300 397 | 1.55E-07 | 0.08 | 1.47E+01 | |
| BS00022611_51 | 3B | 659 787 924 | 2.83E-06 | 0.12 | 8.75E+00 | |
| IAAV7221 | 4B | 2 036 611 | 9.96E-07 | 0.07 | –1.29E+01 | |
| BS00000365_51 | 5A | 538 000 573 | 5.19E-07 | 0.44 | –7.65E+00 | |
| IACX2540 | 5A | 619 684 943 | 4.73E-08 | 0.35 | 9.39E+00 | |
| Excalibur_c74858_243 | 5B | 13 190 663 | 1.16E-08 | 0.47 | –8.67E+00 | |
| Excalibur_c46904_84 | 7D | 5 198 912 | 3.55E-06 | 0.13 | –9.46E+00 | |
| FH | Excalibur_c85499_232 | 1A | 582 219 427 | 2.23E-08 | 0.11 | 2.52E-02 |
| wsnp_Ku_c11665_18999583 | 2B | 139 070 721 | 9.07E-07 | 0.13 | 2.08E-02 | |
| Kukri_c49280_230 | 3A | 20 134 735 | 1.63E-07 | 0.08 | 2.90E-02 | |
| Tdurum_contig64772_417 | 4B | 26 491 482 | 4.18E-09 | 0.07 | 3.56E-02 | |
| RAC875_rep_c105718_585 | 4D | 25 989 162 | 2.20E-12 | 0.38 | –2.56E-02 | |
| BS00036421_51 | 4D | 32 347 318 | 3.96E-07 | 0.37 | –1.58E-02 | |
| RAC875_c8231_1578 | 5A | 613 588 253 | 1.52E-06 | 0.43 | 1.44E-02 | |
| wsnp_Ku_rep_c71232_70948744 | 5A | 679 663 586 | 7.93E-10 | 0.47 | –2.21E-02 | |
| BS00109560_51 | 5B | 556 182 591 | 6.91E-07 | 0.46 | –1.56E-02 | |
| BS00022120_51 | 6A | 396 301 470 | 1.68E-07 | 0.24 | –2.01E-02 |
Temperature response loci are independent of vigour loci
For slpSER~T, we detected one significant (LOD=5.75) MTA on chromosome 1B (wsnp_Ex_c1597_3045682) and two almost significant (LOD=5.38 LOD=5.01) MTAs on chromosomes 4B (CAP7_c10839_300) and 5D (IAAV7104), respectively (Fig. 5). All associated markers for slpSER~T yielded small but significant allelic effects ranging from –0.061 mm °C–1 d–1 to –0.051 mm °C–1 d–1 (Table 3). The GWAS for intSER~T yielded four significant MTAs on chromosomes 2B, 4B, 4D, and 5D, respectively (Table 3; Fig. 5). Start and end of SE yielded four MTAs each, and heading yielded eight MTAs (Table 3; Fig. 5).
Comparing the GWAS results for temperature response, vigour, FH, GDD15, GDD95, and heading revealed no common QTLs between slpSER~T and any other trait. Only one marker (Excalibur_c74858_243) was significantly associated with both GDD15 and GDD95, as well as heading. The lack of overlap of MTAs between temperature response, vigour, and timing of critical stages indicates that they are genetically independent. However, there is a genetic connection between vigour and FH on the one hand and between the start and end of SE and heading on the other.
To identify potential causative genes underlying the QTLs, we searched the reference genome annotation around the respective QTL intervals. For temperature response, we found an increased presence of genes or gene homologues involved in the flowering pathway, namely EARLY FLOWERING 3, FRIGIDA, and CONSTANS (Table 4). Around the QTLs associated with vigour, the annotation showed genes associated with growth (i.e. GRAS, CLAVATA, BSU1, and ARGONAUTE) as well as developmental progress (i.e. Tesmin/TSO1-like CXC domain, BEL1, and AGAMOUS) (Table 5). Importantly, we found GAI-like protein 1 6 Mb upstream of marker Kukri_rep_c68594_530, which we identified as Rht-D1 by blasting the Rht-D1 sequence (GeneBank ID AJ242531.1) against the annotated reference genome. Genes putatively underlying the QTLs for heading, GDD15, and GDD95 are listed in Supplementary Tables S4–S6. As expected, genes or gene homologues associated with the flowering pathway were found in the vicinity of the MTAs for heading. The common QTLs for heading, GDD15, and GDD95 on chromosome 5B (Excalibur_c74858_243) were found to be 6.6 Mb upstream of FLOWERING LOCUS T (Supplementary Tables S4–S6). Other flowering-associated genes found near the heading QTLs were CONSTANS, FRIGIDA, and a FLOWERING LOCUS C-associated gene (Supplementary Table S4). Moreover, a number of putative response regulators as well as genes putatively involved in light control of development (i.e. FAR1-RELATED SEQUENCE; Lin and Wang, 2004) were found near the heading QTLs (Supplementary Table S4). The remaining QTLs for GDD95 were near genes associated with developmental progress and flowering, such as AGAMOUS, MEI2-like 1, HAPPLESS 2, and BEL1 (Supplementary Table S5). Genes near the remaining QTLs for GDD15 were associated with developmental progress (i.e. FLOWERING LOCUS T, BEL1, TERMINAL EAR1-like, and FAR1-RELATED SEQUENCE) as well as growth (i.e. CLAVATA and DELLA; Supplementary Table S6).
Table 4.
Selected putative candidate genes for temperature response (slpSER~T) from the IWGSC reference genome annotation
| Chr | SNP [Position] | r.start | r.end | Gene | Description | Distance |
|---|---|---|---|---|---|---|
| Chr1B | wsnp_Ex_c1597_3045682 [688 283 256] | 688 282 509 | 688 286 431 | TraesCS1B01G480600 | Winged-helix DNA-binding transcription factor family protein | 747 |
| 688 352 414 | 688 354 696 | TraesCS1B01G480700 | HMG-Y-related protein A | –69 158 | ||
| 687 710 716 | 687 719 885 | TraesCS1B01G480100 | Argonaute | 572 540 | ||
| 687 128 952 | 687 135 442 | TraesCS1B01G479200 | Zinc finger protein CONSTANS | 1 154 304 | ||
| 687 078 233 | 687 084 562 | TraesCS1B01G479000 | Zinc finger protein CONSTANS | 1 205 023 | ||
| 686 928 468 | 686 931 886 | TraesCS1B01G478700 | Zinc finger protein CONSTANS | 1 354 788 | ||
| 686 749 516 | 686 755 405 | TraesCS1B01G478100 | WD-repeat protein, putative | 1 533 740 | ||
| 685 645 287 | 685 649 392 | TraesCS1B01G477400 | Early flowering 3 | 2 637 969 | ||
| Chr4B | CAP7_c10839_300 [533 724 424] | 537 474 959 | 537 479 867 | TraesCS4B01G266000 | Protein FRIGIDA | –3 750 535 |
| 541 363 317 | 541 365 139 | TraesCS4B01G267700 | Protein upstream of flc | –7 638 893 | ||
| 542 582 729 | 542 583 265 | TraesCS4B01G268300 | MADS transcription factor | –8 858 305 | ||
| Chr5D | IAAV7104 [553 678 522] | 554 357 761 | 554 360 305 | TraesCS5D01G544800 | FRIGIDA-like protein, putative | –679 239 |
| 554 467 487 | 554 472 596 | TraesCS5D01G545100 | Transducin/WD-like repeat- protein | –788 965 | ||
| 556 226 523 | 556 234 480 | TraesCS5D01G548800 | Transducin/WD-like repeat- protein | –2 548 001 |
Table 5.
Selected putative candiate genes for vigour (intSER~T) of temperature response from the IWGSC reference genome annotation
| Chr | SNP [position] | r.start | r.end | Gene | Description | Distance |
|---|---|---|---|---|---|---|
| Chr2B | RAC875_s109189_188 [248 149 774] | 243 569 388 | 243 571 100 | TraesCS2B01G239400 | GRAS transcription factor | 4 580 386 |
| Chr4B | Ku_c63300_1309 [21 556 672] | 21 187 173 | 21 192 244 | TraesCS4B01G028500 | Tesmin/TSO1-like CXC domain- containing protein | 369 499 |
| 20 005 649 | 20 008 978 | TraesCS4B01G026600 | Argonaute family protein | 1 551 023 | ||
| 19 740 974 | 19 744 058 | TraesCS4B01G026200 | WD40 repeat-like protein | 1 815 698 | ||
| 23 404 428 | 23 408 188 | TraesCS4B01G031300 | BHLH family protein, putative, expressed | –1 847 756 | ||
| 23 818 506 | 23 822 972 | TraesCS4B01G032000 | Protein UPSTREAM OF FLC | –2 261 834 | ||
| 18 162 363 | 18 165 744 | TraesCS4B01G025500 | Homeobox protein BEL1 like | 3 394 309 | ||
| 18 091 908 | 18 093 975 | TraesCS4B01G025400 | BEL1-like homeodomain protein | 3 464 764 | ||
| 17 229 197 | 17 236 874 | TraesCS4B01G024000 | Argonaute protein | 4 327 475 | ||
| 17 017 132 | 17 019 148 | TraesCS4B01G023300 | AGAMOUS-like MADS-box transcription factor | 4 539 540 | ||
| 26 335 682 | 26 336 740 | TraesCS4B01G036600 | BRI1 suppressor 1 (BSU1)-like 3 | –4 779 010 | ||
| 26 824 399 | 26 827 490 | TraesCS4B01G037200 | WD-repeat protein, putative | –5 267 727 | ||
| 15 427 017 | 15 431 870 | TraesCS4B01G021500 | Basic helix–loop–helix (bHLH) DNA-binding superfamily protein | 6 129 655 | ||
| 15 259 656 | 15 263 139 | TraesCS4B01G021200 | Basic helix–loop–helix (bHLH) DNA-binding superfamily protein | 6 297 016 | ||
| 15 146 117 | 15 150 854 | TraesCS4B01G021100 | Basic helix loop helix (BHLH) DNA-binding family protein | 6 410 555 | ||
| 14 710 395 | 14 711 057 | TraesCS4B01G020800 | Protein FAR1-RELATED SEQUENCE 5 | 6 846 277 | ||
| 28 413 432 | 28 414 112 | TraesCS4B01G041000 | Sensitive to freezing 6 | –6 856 760 | ||
| 29 673 211 | 29 674 674 | TraesCS4B01G042500 | Fantastic four-like protein | –8 116 539 | ||
| Chr4D | Kukri_rep_c68594_530 [12 773 259] | 12 700 119 | 12 703 878 | TraesCS4D01G028900 | BHLH family protein, putative, expressed | 73 140 |
| 13 096 296 | 13 096 966 | TraesCS4D01G029600 | CLAVATA3/ESR (CLE)-related protein 25 | –323 037 | ||
| 13 196 859 | 13 200 535 | TraesCS4D01G029700 | Protein UPSTREAM OF FLC | –423 600 | ||
| 11 364 404 | 11 369 466 | TraesCS4D01G026100 | Tesmin/TSO1-like CXC domain- containing protein | 1 408 855 | ||
| 10 746 363 | 10 750 251 | TraesCS4D01G024300 | Argonaute protein | 2 026 896 | ||
| 10 684 336 | 10 690 389 | TraesCS4D01G024100 | Argonaute family protein | 2 088 923 | ||
| 10 254 979 | 10 257 683 | TraesCS4D01G023600 | WD40 repeat-like protein | 2 518 280 | ||
| 15 768 990 | 15 772 059 | TraesCS4D01G034500 | WD-repeat protein, putative | –2 995 731 | ||
| 9 495 616 | 9 501 619 | TraesCS4D01G022600 | Homeobox protein BEL1 like | 3 277 643 | ||
| 9 443 778 | 9 445 575 | TraesCS4D01G022500 | BEL1-like homeodomain protein 1 | 3 329 481 | ||
| 9 069 403 | 9 071 423 | TraesCS4D01G021100 | MADS-box transcription factor | 3 703 856 | ||
| 16 584 271 | 16 584 948 | TraesCS4D01G038400 | Sensitive to freezing 6 | –3 811 012 | ||
| 8 777 205 | 8 779 670 | TraesCS4D01G020300 | Growth-regulating factor | 3 996 054 | ||
| 8 149 046 | 8 151 425 | TraesCS4D01G019200 | Basic helix–loop–helix (bHLH) DNA-binding superfamily protein | 4 624 213 | ||
| 8 135 666 | 8 137 454 | TraesCS4D01G019100 | Basic helix–loop–helix (bHLH) DNA-binding superfamily protein | 4 637 593 | ||
| 8 010 719 | 8 012 446 | TraesCS4D01G018800 | Basic helix–loop–helix (bHLH) DNA-binding superfamily protein | 4 762 540 | ||
| 7 992 104 | 7 995 445 | TraesCS4D01G018700 | Basic helix–loop–helix (bHLH) DNA-binding superfamily protein | 4 781 155 | ||
| 17 765 786 | 17 767 021 | TraesCS4D01G039900 | Fantastic four-like protein | –4 992 527 | ||
| 18 781 062 | 18 782 933 | TraesCS4D01G040400 | GAI-like protein 1 (Rht-D1) | –6 007 803 | ||
| 6 703 246 | 6 703 509 | TraesCS4D01G015200 | SAUR-like auxin-responsive protein family | 6 070 013 | ||
| 6 699 039 | 6 699 458 | TraesCS4D01G015100 | SAUR-like auxin-responsive protein family | 6 074 220 | ||
| 6 682 318 | 6 682 602 | TraesCS4D01G015000 | SAUR-like auxin-responsive protein family | 6 090 941 | ||
| 6 663 820 | 6 664 131 | TraesCS4D01G014900 | SAUR-like auxin-responsive protein family | 6 109 439 | ||
| 6 461 624 | 6 462 688 | TraesCS4D01G013800 | BRI1 suppressor 1 (BSU1)-like 3 | 6 311 635 | ||
| 19 169 377 | 19 171 147 | TraesCS4D01G040600 | Protein FAR1-RELATED SEQUENCE 5 | –6 396 118 | ||
| 6 017 847 | 6 023 948 | TraesCS4D01G012800 | Protein FAR1-RELATED SEQUENCE 5 | 6 755 412 | ||
| 4 128 933 | 4 133 919 | TraesCS4D01G008400 | WD-repeat protein, putative | 8 644 326 | ||
| 21 775 252 | 21 776 785 | TraesCS4D01G046200 | CONSTANS-like zinc finger protein | –9 001 993 | ||
| Chr5D | Kukri_c6477_696 [423 502 809] | 423 858 756 | 423 860 766 | TraesCS5D01G334100 | Armadillo repeat only | –355 947 |
| 421 503 514 | 421 504 332 | TraesCS5D01G329500 | HVA22-like protein | 1 999 295 | ||
| 426 296 827 | 426 301 957 | TraesCS5D01G337800 | WD-repeat protein, putative | –2 794 018 | ||
| 429 289 426 | 429 292 023 | TraesCS5D01G341000 | CONSTANS-like zinc finger protein | –5 786 617 | ||
| 416 787 868 | 416 788 986 | TraesCS5D01G325300 | Protein Mei2 | 6 714 941 | ||
| 416 625 946 | 416 628 639 | TraesCS5D01G325200 | Protein Mei2 | 6 876 863 | ||
| 415 622 032 | 415 622 615 | TraesCS5D01G323500 | Auxin-responsive protein | 7 880 777 |
Vigour, temperature response, and the timing of SE affect final height
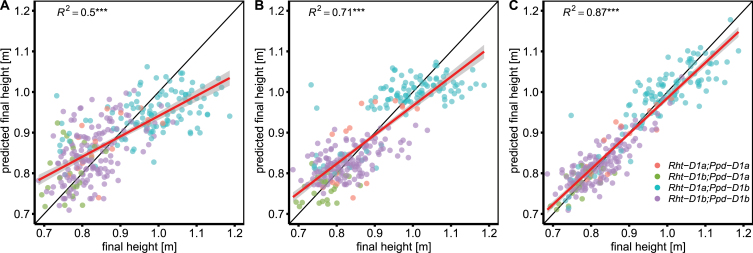
The phenotypic correlations show a strong connection between temperature response, vigour, and FH as well as weaker connections between GDD15, GDD95, and FH. In order to examine this interdependency on a genetic level, we used a linear model to predict FH with the SNP alleles of the QTLs for slpSER~T, intSER~T, GDD15, and GDD95 as predictors. The model was able to predict FH with an accuracy R2=0.5; however, clusters in the data showed clear effects of Rht-D1 and Ppd-D1 alleles (Fig. 6A). Adding Rht-B1, Rht-D1, and Ppd-D1 alleles as predictors increased the prediction accuracy to R2=0.71 (Fig. 6B). There were significant contributions by QTLs of all three traits; however, their effects were small compared with the obvious effects of Rht-B1, Rht-D1, and Ppd-D1 (Table 6). Including all two-way interaction effects among the QTLs, Rht-1 and Ppd-1 increased the prediction accuracy to R2=0.87 (Fig. 6C), indicating a fine-tuning effect of temperature response, vigour, and timing of SE on FH.
Fig. 6.
Prediction of final height based on QTLs and Rht-B1, Rht-D1, and Ppd-D1 alleles. (A) The SNP alleles of significantly associated QTLs for temperature response (slpSER~T), vigour (intSER~T), and start (GDD15) and end (GDD95) of stem elongation were used in a linear model without considering interaction effects. (B) Rht-B1, Rht-D1, and Ppd-D1 alleles were added to the model used in (A). (C) All two-way interaction effects among SNP alleles and Rht-B1, Rht-D1, and Ppd-D1 alleles were included in the model. The models were applied to the 3 year BLUEs of all genotypes with available genotypic data (n=300) on all predictors. Colours indicate the allelic status regarding Rht-D1 and Ppd-D1 of the respective genotypes as depicted in (C).
Table 6.
Type II analysis of variance for the prediction of final height using QTLs and Rht-B1, Rht-D1, and Ppd-D1 alleles
| Predictor [trait.QTL.chromosome] | SNP | Sum of squares | Df | F-value | Pr(>F) | |
|---|---|---|---|---|---|---|
| slpSER~T.1.1B | wsnp_Ex_c1597_3045682 | 4.20E-03 | 1 | 1.08E+00 | 2.99E-01 | |
| slpSER~T.2.4B | CAP7_c10839_300 | 7.97E-03 | 1 | 2.05E+00 | 1.53E-01 | |
| slpSER~T.3.5D | IAAV7104 | 2.81E-02 | 1 | 7.23E+00 | 7.58E-03 | ** |
| intSER~T.1.2B | RAC875_s109189_188 | 1.72E-03 | 1 | 4.42E-01 | 5.07E-01 | |
| intSER~T.2.4B | Ku_c63300_1309 | 1.64E-02 | 1 | 4.23E+00 | 4.07E-02 | * |
| intSER~T.3.4D | Kukri_rep_c68594_530 | 7.47E-03 | 1 | 1.93E+00 | 1.66E-01 | |
| intSER~T.4.5D | Kukri_c6477_696 | 6.06E-03 | 1 | 1.56E+00 | 2.13E-01 | |
| GDD15.1.1D | wsnp_Ex_c12447_19847242 | 1.41E-02 | 1 | 3.64E+00 | 5.74E-02 | ° |
| GDD15.2.2A | Tdurum_contig47508_250 | 4.97E-02 | 1 | 1.28E+01 | 4.07E-04 | *** |
| GDD15.3.3A | Kukri_c55381_67 | 1.91E-02 | 1 | 4.91E+00 | 2.75E-02 | * |
| GDD15.4|GDD95.2|heading.7.5B | Excalibur_c74858_243 | 7.64E-08 | 1 | 1.97E-05 | 9.96E-01 | |
| GDD95.1.5A | Excalibur_c49597_579 | 4.04E-02 | 1 | 1.04E+01 | 1.40E-03 | ** |
| GDD95.3.5B | Tdurum_contig44115_561 | 1.03E-03 | 1 | 2.65E-01 | 6.07E-01 | |
| GDD95.4.7B | RAC875_c38693_319 | 4.28E-03 | 1 | 1.10E+00 | 2.95E-01 | |
| Rht-B1 | 3.32E-01 | 1 | 8.55E+01 | 6.02E-18 | *** | |
| Rht-D1 | 6.71E-01 | 1 | 1.73E+02 | 3.79E-31 | *** | |
| Ppd-D1 | 5.87E-02 | 1 | 1.51E+01 | 1.26E-04 | *** | |
| Residuals | 1.09E+00 | 282 |
SNP alleles of the QTLs for temperature response (slpSER~T), vigour (intSER~T), start (GDD15), and end (GDD95) of stem elongation as well as Rht-B1, Rht-D1, and Ppd-D1 alleles were used as predictors in a linear model (FH=ΣQTLslpSER~T+ΣQTLintSER~T+ΣGDD15+ΣQTLGDD95+Rht-B1+Rht-D1+Ppd-D1) without interaction effects (see Fig. 6B). The model was applied to the 3 year BLUEs of all genotypes with available genotypic data (n=300) on all predictors.
Asterisks and dots indicate the significance of the respective predictor (***P<0.001, **P<0.01, *P<0.05, °P<0.1).
Discussion
In this study, we present a method to measure temperature response during stem elongation of wheat using high-throughput phenotyping of canopy height in the field. We found a genotype-specific response of wheat to change in ambient temperature which was correlated with the timing of the developmental key stages. We decomposed this growth dynamic into a genotype-specific vigour component and temperature response component using regression models. We further related these parameters to plant height and the timing of developmental key stages.
Linear regression models were used to describe wheat growth response to temperature for leaf elongation (Nagelmüller et al., 2016), canopy cover (Grieder et al., 2015), as well as SER (Slafer and Rawson, 1995a). Others proposed the use of a more complex, Arrhenius type of peak function to account for decreasing growth rates at supra-optimal temperatures (Parent and Tardieu, 2012). However such models are mainly applicable when the temperatures experienced by the crop exceed the temperature optimum. Wheat has its temperature optimum at ~27 °C (Parent and Tardieu, 2012). Temperatures in the measured growth intervals during SE did not exceed 25 °C and, given the temporal resolution of the data, a simple linear model is justified (Parent et al., 2019).
The results of the correlation analysis show a clear connection between FH and temperature response (slpSER~T) as well as between FH and vigour (intSER~T). This is consistent with our hypothesis that FH can be described as a function of temperature-independent growth processes and as a function of temperature response during SE. Importantly, among all components, the temperature response was a significant driver of FH and also had a strong influence on the timing. Temperature response delayed the beginning of SE, leading to a later start and end of the whole phase. This finding might appear counter-intuitive: given the assumption that plants develop faster under higher ambient temperatures, a more responsive genotype should develop faster compared with a less responsive one. Slafer and Rawson (1995b) reported an accelerated development towards floral transition with increasing temperatures up to 19 °C, whereas higher temperatures slowed development. In that respect, a more responsive genotype would experience a stronger delay of floral transition under warm temperatures.
In terms of their correlation to FH, the effects of the timing of start and end of SE are less distinct. FH was more a function of faster growth than of a longer duration of growth, especially since genotypes with a strong temperature response had a shorter duration of SE. However, the timing of the start and end of SE was linked with temperature response. Based on this result and the correlations, it would appear that temperature response influences FH directly as well as indirectly by mediating the start and end of SE. Surprisingly, we found no correlation between heading and FH despite the positive correlation of both traits with GDD95. A correlation between heading date and FH would therefore be expected. Previous studies reported pleiotropic effects between plant height and heading time (Griffiths et al., 2010; Mo et al., 2018).
The question of whether these trait correlations are due to pleiotropic effects will substantially impact the breeding strategy (Chen and Lübberstedt, 2010). If the relationship between phenology, FH, and temperature response were to be to a large degree pleiotropic, these traits could not be independently selected. Alternative explanations are linkage and population structure. The GABI wheat panel is made of wheat varieties from different regions of Europe. As the examined traits are major drivers of adaptation to the different regions of Europe, we anticipate a very strong selection for both temperature response and timing of critical stages. Even if there is no apparent population structure at neutral markers, there may be a strong population structure at selected loci with a strong effect on local adaptation. Our phenotypic results showed a significant interaction effect between Ppd-D1 and Rht-D1 on FH, indicating either a co-selection or a pleiotropic effect of Ppd-D1. Pleiotropic effects between height and flowering time are known for maize and rice. For example, the DWARF8 gene of maize encoding a DELLA protein is associated with height and flowering time (Lawit et al., 2010) and strongly associated with climate adaptation (Camus-Kulandaivelu et al., 2006). The rice GHD7 locus has a strong effect on number of days to heading, number of grains per panicle, plant height, and stem growth (Xue et al., 2008). In wheat, the dwarf gene Rht-12 was shown to have a delaying effect on heading (Worland et al., 1994; Chen et al., 2013) as well as an additive interaction effect with Ppd-D1 on plant height (Chen et al., 2018). Furthermore, it was shown that the tall Rht-D1a and the photoperiod-sensitive Ppd-D1b allele positively affect leaf area and spike length throughout SE (Guo et al., 2018). To further examine the relationship among the different traits, we consider the following GWAS analysis using stringent correction of population structure.
The GWAS results indicate an independent genetic control of FH, temperature response, and the timing of critical stages, whereas vigour and FH as well as heading time, and start and end of SE appear to be partly linked. Yet, FH could be predicted with surprising accuracy using the QTLs for temperature response, vigour, and start and end of SE, which reflects the correlations found in the phenotypic data.
Previous studies investigating the control of developmental key stages in wheat with respect to temperature generally adopted the concept that after fulfilment of photoperiod and vernalization, Eps genes act as fine-tuning factors independent of environmental stimuli (Kamran et al., 2014; Zikhali and Griffiths, 2015). Increasing temperature, apart from vernalization, is thought to generally quicken growth and development independent of the cultivar (Slafer and Rawson, 1995b; Porter and Gawith, 1999; Slafer et al., 2015). A genotype-specific temperature effect on the duration of different phases was not considered (Takahashi and Yasuda, 1971; Slafer and Rawson 1995c). It was, however, reported that photoperiod effects vary depending on temperature (Slafer and Rawson, 1995c). Under long days, Hemming et al. (2012) reported faster development and fewer fertile florets under high compared with low temperatures. Temperature-dependent effects were also found for different Eps QTLs (Slafer and Rawson, 1995c; Gororo et al., 2001). It has previously been suggested that Eps effects could be associated with interaction effects between genotype and temperature fluctuations (Slafer and Rawson, 1995c; van Beem et al., 2005).
The mechanisms of ambient temperature sensing and of its effects on growth and development are not yet well understood (Sanchez-Bermejo and Balasubramanian, 2016). However, important findings regarding ambient temperature effects on flowering time as well as on hypocotyl elongation have come from Arabidopsis thaliana (Wigge, 2013). With respect to these two traits, Sanchez-Bermejo and Balasubramanian (2016) reported distinct genotypic differences in temperature sensitivity. According to their results, the flowering pathway genes FRIGIDA (FRI), FLOWERING LOCUS C (FLC), and FLOWERING LOCUS T (FT) are major candidate genes for ambient temperature-mediated differences in flowering time (Sanchez-Bermejo and Balasubramanian, 2016). In the present study, we found FRI homologues near two of the three QTLs for temperature response. FRI and FLC act as the main vernalization genes in A. thaliana (Johanson et al., 2000; Amasino and Michaels, 2010). In wheat, these genes are not yet well described. However, FLC orthologues were found to act as flowering repressors regulated by vernalization in monocots (Sharma et al., 2017).
The most promising candidate gene for temperature response found near the QTLs on chromosome 1B is EARLY FLOWERING 3 (ELF3). In Arabidopsis, ELF3 was found to be a core part of the circadian clock involved in ambient temperature response (Thines and Harmon, 2010). In barley, ELF3 was shown to be involved in the control of temperature-dependent expression of flowering time genes (Ejaz and von Korff, 2017). A mutant ELF3 accelerated floral development under high ambient temperatures while maintaining the number of seeds (Ejaz and von Korff, 2017). Furthermore, ELF3 has been reported as a candidate gene for Eps1 in Triticum monococcum (Alvarez et al., 2016) as well as in wheat (Zikhali et al., 2016). A recent study in wheat showed an interaction between Eps-D1 and ambient temperature which corresponded to different expression of ELF3 (Ochagavía et al., 2019). In this study, we directly measured growth response to temperature during SE and found a significant MTA near ELF3 on chromosome 1B. Following Ochagavía et al. (2019), this indicates that growth response to temperature is connected to Eps-B1 which is a homologue to Eps-D1. Furthermore, the temperature×Eps-D1 interaction effects on heading reported by Ochagavía et al. (2019) are in agreement with the correlations found among growth response to temperature, GDD15, GDD95, and heading in the present study.
One important aspect we could not address in this study is the interaction of genotype-specific temperature response with vernalization and photoperiod (Slafer and Rawson, 1995c; Gol et al., 2017; Kiss et al., 2017). Due to the climate conditions in Switzerland, we expect fulfilment of vernalization requirement in all genotypes. However, due to the broad geographic origin of the investigated genotypes, the relationship between temperature response and the timing of SE might be confounded by different photoperiod requirements. Nevertheless, the correlations between earliness and temperature response are in agreement with Ochagavia et al. (2019). It also remains unclear whether and to what extent temperature response varies across different developmental phases and how temperature response relates to other environmental stimuli such as vapour pressure deficit or radiation. Nevertheless, the results of this study present valuable information towards a better understanding of temperature response in wheat and may be of great importance for breeding. Temperature response could provide a breeding avenue for local adaptation as well as the control of plant height.
With the recent advancements in unmanned aerial vehicle (UAV)-based phenotyping techniques, the growth of canopy cover and canopy height can be measured using image segmentation and structure from motion approaches (Bareth et al., 2016; Aasen and Bareth, 2018; Roth et al., 2018). Thus, temperature response can be investigated during the development of the vegetative canopy cover (Grieder et al., 2015) and during the generative height development as demonstrated here. It can also be assessed in indoor platforms (e.g. Parent and Tardieu, 2012) and the field using a leaf length tracker (Nagelmüller et al., 2016) measuring short-term responses of leaf growth to diurnal changes in temperature. Combining this information may greatly improve our understanding about the genetic variation in growth response to temperature.
Together, the results of this study indicate that temperature response may be exploitable as a breeding trait to adjust phenology towards specific environments, through either phenotypic or marker-assisted selection. Furthermore, a better understanding of temperature response may enhance the capability of crop models to predict crop performance under future climate change scenarios.
Conclusion and outlook
Modern phenotyping platforms hold great promise to map the genetic factors driving the response of developmental processes to environmental stimuli. To the best of our knowledge, this is the first experiment dissecting the SE process into its underlying components: temperature-dependent elongation, temperature-independent vigour, and duration of elongation. The independent loci detected for these traits suggest that it is possible to select them independently. The detected loci may be used to fine-tune height and the beginning and end of SE as they explain a substantial part of the overall genotypic variation. With increases in automation, growth processes may be monitored in the field on a daily basis or even multiple times per day. This will increase the precision in assessing genotype responses to the fluctuation in meteorological conditions and will allow quantification of the relationship of these responses to yield. Remote sensing by means of UAVs in combination with photogrammetric algorithms will allow measurement of these traits in breeding nurseries. We believe that this is paving the way for a more informed selection to climate adaptation within individual growing seasons.
Supplementary data
The following supplementary data are available at JXB online.
Fig. S1. Correction of canopy height for spatial as well as random row and range effects.
Fig. S2. Summary of plot-based linear model fits of stem elongation rate versus temperature.
Fig. S3. Pearson correlation coefficients among 3 year BLUEs of all investigated traits.
Fig. S4. Principal component analysis among marker genotypes.
Fig. S5. Manhattan plots and quantile–quantile plots depicting the GWAS results using the MLM approach.
Fig. S6. Manhattan plots and quantile–quantile plots depicting the GWAS results using the GLM approach.
Table S1. Genes of interest related to floral transition and flowering.
Table S2. Chromosome-wise distance thresholds for LD decay <r2=0.2
Table S3. Corresponding marker–trait associations for final canopy height with respect to Zanke et al. (2016).
Table S4. Selected putative candidate genes for headingGDD from the IWGSC reference genome annotation.
Table S5. Selected putative candidate genes for GDD95 from the IWGSC reference genome annotation.
Table S6. Selected putative candidate genes for GDD15 from the IWGSC reference genome annotation.
Table S7. Three-year BLUEs of the investigated traits FH, headingGDD, GDD15, GDD95, GDDSE, timeSE, slpSER~T, and intSER~T.
Acknowledgements
We sincerely thank Hansueli Zellweger for managing and nursing our field experiments. We further thank the members of the ETH crop science and the ETH molecular plant breeding groups, especially Michelle Nay and Beat Keller, for many fruitful discussions. We also thank Martina Binder for performing the correlation analysis between the FH data of this study and the data from Zanke et al. (2014b) in the framework of her MSc Thesis. We would like to thank Marion Röder (IPK Gatersleben) for supply of the GABI wheat panel including genetic information. Finally, we thank the anonymous reviewers for their helpful comments and suggestions. This work was supported by the Swiss National Foundation (SNF) in the framework of the project PhenoCOOL (project no. 169542).
Author contributions
LK conducted the laser scans, did all statistical analyses, and drafted the manuscript; SY assisted with the GWAS and candidate gene evaluation; MB assisted with the spatial correction; NK developed the analysis pipeline for the laser scans and contributed to measurements; AW drafted the grant application and supervised the overall concept; AH made the experimental design, developed the phenotyping models and assisted with the statistical analysis. All authors contributed to the drafting of the manuscript.
Data availability
Processed phenotypic data are available as Supplementary data. Unprocessed data and analysis scripts are available from the authors upon reasonable request.
References
- Aasen H, Bareth G. 2018. Spectral and 3D nonspectral approaches to crop trait estimation using ground and UAV sensing. In: Thenkabail PS, Lyon JG, Huete A, eds. Hyperspectral remote sensing of vegetation. Biophysical and biochemical characterization and plant species studies. Boca Raton, FL: CRC Press, 103–131. [Google Scholar]
- Alvarez MA, Tranquilli G, Lewis S, Kippes N, Dubcovsky J. 2016. Genetic and physical mapping of the earliness per se locus Eps-A (m) 1 in Triticum monococcum identifies EARLY FLOWERING 3 (ELF3) as a candidate gene. Functional & Integrative Genomics 16, 365–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amasino RM, Michaels SD. 2010. The timing of flowering. Plant Physiology 154, 516–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderegg J, Yu K, Aasen H, Walter A, Liebisch F, Hund A. 2020. Spectral vegetation indices to track senescence dynamics in diverse wheat germplasm. Frontiers in Plant Science 10, 1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asseng S, Ewert F, Martre P, et al. . 2015. Rising temperatures reduce global wheat production. Nature Climate Change 5, 143–147. [Google Scholar]
- Atkinson D, Porter JR. 1996. Temperature, plant development and crop yields. Trends in Plant Science 1, 119–124. [Google Scholar]
- Bareth G, Bendig J, Tilly N, Hoffmeister D, Aasen H, Bolten A. 2016. A comparison of UAV- and TLS-derived plant height for crop monitoring: using polygon grids for the analysis of Crop Surface Models (CSMs). Photogrammetrie, Fernerkundung, Geoinformation 2016, 85–94. [Google Scholar]
- Beales J, Turner A, Griffiths S, Snape JW, Laurie DA. 2007. A pseudo-response regulator is misexpressed in the photoperiod insensitive Ppd-D1a mutant of wheat (Triticum aestivum L.). Theoretical and Applied Genetics 115, 721–733. [DOI] [PubMed] [Google Scholar]
- Bendig J, Bolten A, Bareth G. 2013. UAV-based imaging for multi-temporal, very high resolution crop surface models to monitor crop growth variability. Photogrammetrie, Fernerkundung, Geoinformation 2013, 551–562. [Google Scholar]
- Butler D. 2018. https://asreml.kb.vsni.co.uk/knowledge-base/asreml/ asreml – fit the linear mixed model. [Google Scholar]
- Camus-Kulandaivelu L, Veyrieras JB, Madur D, Combes V, Fourmann M, Barraud S, Dubreuil P, Gouesnard B, Manicacci D, Charcosset A. 2006. Maize adaptation to temperate climate: relationship between population structure and polymorphism in the Dwarf8 gene. Genetics 172, 2449–2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh CR, Chao S, Wang S, et al. . 2013. Genome-wide comparative diversity uncovers multiple targets of selection for improvement in hexaploid wheat landraces and cultivars. Proceedings of the National Academy of Sciences, USA 110, 8057–8062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman SC, Chakraborty S, Dreccer MF, Howden SM. 2012. Plant adaptation to climate change—opportunities and priorities in breeding. Crop and Pasture Science 63, 251–268. [Google Scholar]
- Chen L, Du Y, Lu Q, et al. . 2018. The photoperiod-insensitive allele Ppd-D1a promotes earlier flowering in Rht12 dwarf plants of bread wheat. Frontiers in Plant Science 9, 1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Phillips AL, Condon AG, Parry MAJ, Hu Y-G. 2013. GA-responsive dwarfing gene Rht12 affects the developmental and agronomic traits in common bread wheat. PLoS One 8, e62285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Lübberstedt T. 2010. Molecular basis of trait correlations. Trends in Plant Science 15, 454–461. [DOI] [PubMed] [Google Scholar]
- Ejaz M, von Korff M. 2017. The genetic control of reproductive development under high ambient temperature. Plant Physiology 173, 294–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis M, Spielmeyer W, Gale K, Rebetzke G, Richards R. 2002. ‘Perfect’ markers for the Rht-B1b and Rht-D1b dwarfing genes in wheat. Theoretical and Applied Genetics 105, 1038–1042. [DOI] [PubMed] [Google Scholar]
- Falconer DS, Mackay TFC. 1996. Introduction to quantitative genetics. Essex, UK: Benjamin Cummings. [Google Scholar]
- Farooq M, Bramley H, Palta JA, Siddique KHM. 2011. Heat stress in wheat during reproductive and grain-filling phases. Critical Reviews in Plant Sciences 30, 491–507. [Google Scholar]
- Fischer RA 1985. Number of kernels in wheat crops and the influence of solar radiation and temperature. Journal of Agricultural Science 105, 447–461. [Google Scholar]
- Fischer RA 2011. Wheat physiology: a review of recent developments. Crop and Pasture Science 62, 95–114. [Google Scholar]
- Friedli M, Kirchgessner N, Grieder C, Liebisch F, Mannale M, Walter A. 2016. Terrestrial 3D laser scanning to track the increase in canopy height of both monocot and dicot crop species under field conditions. Plant Methods 12, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaut BS, Long AD. 2003. The lowdown on linkage disequilibrium. The Plant Cell 15, 1502–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson LR, Paulsen GM. 1999. Yield components of wheat grown under high temperature stress during reproductive growth. Crop Science 39, 1841–1846. [Google Scholar]
- Gol L, Tomé F, von Korff M. 2017. Floral transitions in wheat and barley: interactions between photoperiod, abiotic stresses, and nutrient status. Journal of Experimental Botany 68, 1399–1410. [DOI] [PubMed] [Google Scholar]
- González FG, Slafer GA, Miralles DJ. 2003. Floret development and spike growth as affected by photoperiod during stem elongation in wheat. Field Crops Research 81, 29–38. [Google Scholar]
- Gororo NN, Flood RG, Eastwood RF, Eagles HA. 2001. Photoperiod and vernalization responses in Triticum turgidum×T. tauschii synthetic hexaploid wheats. Annals of Botany 88, 947–952. [Google Scholar]
- Grieder C, Hund A, Walter A. 2015. Image based phenotyping during winter: a powerful tool to assess wheat genetic variation in growth response to temperature. Functional Plant Biology 42, 387–396. [DOI] [PubMed] [Google Scholar]
- Griffiths S, Simmonds J, Leverington M, Wang Y, Fish L, Sayers L, Alibert L, Orford S, Wingen L, Snape J. 2010. Meta-QTL analysis of the genetic control of crop height in elite European winter wheat germplasm. Molecular Breeding 29, 159–171. [DOI] [PubMed] [Google Scholar]
- Guo Z, Liu G, Röder MS, Reif JC, Ganal MW, Schnurbusch T. 2018. Genome-wide association analyses of plant growth traits during the stem elongation phase in wheat. Plant Biotechnology Journal 16, 2042–2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedden P 2003. The genes of the Green Revolution. Trends in Genetics 19, 5–9. [DOI] [PubMed] [Google Scholar]
- Hemming MN, Walford SA, Fieg S, Dennis ES, Trevaskis B. 2012. Identification of high-temperature-responsive genes in cereals. Plant Physiology 158, 1439–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Ochoa IM, Luz Pequeno DN, Reynolds M, et al. . 2019. Adapting irrigated and rainfed wheat to climate change in semi-arid environments: management, breeding options and land use change. European Journal of Agronomy 109, 125915. [Google Scholar]
- Holman F, Riche A, Michalski A, Castle M, Wooster M, Hawkesford M. 2016. High throughput field phenotyping of wheat plant height and growth rate in field plot trials using UAV based remote sensing. Remote Sensing 8, 1031. [Google Scholar]
- Hund A, Kronenberg L, Anderegg J, Yu K, Walter A. 2019. Non-invasive field phenotyping of cereal development. In: Ordon F, Friedt W, eds. Advances in breeding techniques for cereal crops. Cambridge, UK: Buleigh Dodds Science Publishing. [Google Scholar]
- International Wheat Genome Sequencing Consortium 2018. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361, eaar7191. [DOI] [PubMed] [Google Scholar]
- Johanson U, West J, Lister C, Michaels S, Amasino R, Dean C. 2000. Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science 290, 344–347. [DOI] [PubMed] [Google Scholar]
- Kamran A, Iqbal M, Spaner D. 2014. Flowering time in wheat (Triticum aestivum L.): a key factor for global adaptability. Euphytica 197, 1–26. [Google Scholar]
- Keyes GJ, Paolillo DJ, Sorrells ME. 1989. The effects of dwarfing genes Rht1 and Rht2 on cellular dimensions and rate of leaf elongation in wheat. Annals of Botany 64, 683–690. [Google Scholar]
- Kirby EJM 1988. Analysis of leaf, stem and ear growth in wheat from terminal spikelet stage to anthesis. Field Crops Research 18, 127–140. [Google Scholar]
- Kirchgessner N, Liebisch F, Yu K, Pfeifer J, Friedli M, Hund A, Walter A. 2016. The ETH field phenotyping platform FIP: a cable-suspended multi-sensor system. Functional Plant Biology 44, 154–168. [DOI] [PubMed] [Google Scholar]
- Kiss T, Dixon LE, Soltész A, et al. . 2017. Effects of ambient temperature in association with photoperiod on phenology and on the expressions of major plant developmental genes in wheat (Triticum aestivum L.). Plant, Cell & Environment 40, 1629–1642. [DOI] [PubMed] [Google Scholar]
- Kollers S, Rodemann B, Ling J, et al. . 2013. Whole genome association mapping of Fusarium head blight resistance in European winter wheat (Triticum aestivum L.). PLoS One 8, e57500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollers S, Rodemann B, Ling J, et al. . 2014. Genome-wide association mapping of tan spot resistance (Pyrenophora tritici-repentis) in European winter wheat. Molecular Breeding 34, 363–371. [Google Scholar]
- Kronenberg L, Yu K, Walter A, Hund A. 2017. Monitoring the dynamics of wheat stem elongation: genotypes differ at critical stages. Euphytica 213, 157. [Google Scholar]
- Lancashire P, Bleiholder H, Vandenboom T, Langeluddeke P, Stauss R, Weber E, Witzenberger A. 1991. A uniform decimal code for growth-stages of crops and weeds. Annals of Applied Biology 119, 561–601. [Google Scholar]
- Lawit SJ, Wych HM, Xu D, Kundu S, Tomes DT. 2010. Maize DELLA proteins dwarf plant8 and dwarf plant9 as modulators of plant development. Plant & Cell Physiology 51, 1854–1868. [DOI] [PubMed] [Google Scholar]
- Lin R, Wang H. 2004. Arabidopsis FHY3/FAR1 gene family and distinct roles of its members in light control of Arabidopsis development. Plant Physiology 136, 4010–4022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Huang M, Fan B, Buckler ES, Zhang Z. 2016. Iterative usage of fixed and random effect models for powerful and efficient genome-wide association studies. PLoS Genetics 12, e1005767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miralles DJ, Slafer GA. 2007. Sink limitations to yield in wheat: how could it be reduced? Journal of Agricultural Science 145, 139–149. [Google Scholar]
- Mo Y, Vanzetti LS, Hale I, Spagnolo EJ, Guidobaldi F, Al-Oboudi J, Odle N, Pearce S, Helguera M, Dubcovsky J. 2018. Identification and characterization of Rht25, a locus on chromosome arm 6AS affecting wheat plant height, heading time, and spike development. Theoretical and Applied Genetics 131, 2021–2035. [DOI] [PubMed] [Google Scholar]
- Nagelmüller S, Kirchgessner N, Yates S, Hiltpold M, Walter A. 2016. Leaf length tracker: a novel approach to analyse leaf elongation close to the thermal limit of growth in the field. Journal of Experimental Botany 67, 1897–1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochagavía H, Prieto P, Zikhali M, Griffiths S, Slafer GA. 2019. Earliness per se by temperature interaction on wheat development. Scientific Reports 9, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent B, Millet EJ, Tardieu F. 2019. The use of thermal time in plant studies has a sound theoretical basis provided that confounding effects are avoided. Journal of Experimental Botany 70, 2359–2370. [DOI] [PubMed] [Google Scholar]
- Parent B, Tardieu F. 2012. Temperature responses of developmental processes have not been affected by breeding in different ecological areas for 17 crop species. New Phytologist 194, 760–774. [DOI] [PubMed] [Google Scholar]
- Porter JR, Gawith M. 1999. Temperatures and the growth and development of wheat: a review. European Journal of Agronomy 10, 23–36. [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, et al. . 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. American Journal of Human Genetics 81, 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2015. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- Rezaei EE, Siebert S, Hüging H, Ewert F. 2018. Climate change effect on wheat phenology depends on cultivar change. Scientific Reports 8, 4891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Álvarez MX, Boer MP, van Eeuwijk FA, Eilers PHC. 2018. Correcting for spatial heterogeneity in plant breeding experiments with P-splines. Spatial Statistics 23, 52–71. [Google Scholar]
- Roth L, Aasen H, Walter A, Liebisch F. 2018. Extracting leaf area index using viewing geometry effects—a new perspective on high-resolution unmanned aerial system photography. ISPRS Journal of Photogrammetry and Remote Sensing 141, 161–175. [Google Scholar]
- Sanchez-Bermejo E, Balasubramanian S. 2016. Natural variation involving deletion alleles of FRIGIDA modulate temperature-sensitive flowering responses in Arabidopsis thaliana. Plant, Cell & Environment 39, 1353–1365. [DOI] [PubMed] [Google Scholar]
- Schürch C, Kronenberg L, Hund A. 2018. Wheat developmental stages. Available at: https://commons.wikimedia.org/wiki/File:Wheat_developmental_stages.jpg (accessed 6 July 2018). [Google Scholar]
- Sharma N, Ruelens P, D’hauw M, Maggen T, Dochy N, Torfs S, Kaufmann K, Rohde A, Geuten K. 2017. A flowering locus C homolog is a vernalization-regulated repressor in Brachypodium and is cold regulated in wheat. Plant Physiology 173, 1301–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slafer GA, Calderini DF, Miralles DJ. 1996. Yield components and compensations in wheat: opportunities for further increasing yield potential. In: Reynolds MP, Rajaram S, McNab A, eds. Increasing yield potential in wheat: breaking the barriers: Proceedings of a Workshop held in Ciudad Obregón, Sonora, Mexico. Mexico: CIMMYT, 101–133. [Google Scholar]
- Slafer GA, Kantolic AG, Appendino ML, Tranquilli G, Miralles DJ, Savin R. 2015. Genetic and environmental effects on crop development determining adaptation and yield. In: Sadras V, Calderini D, eds. Crop physiology, 2nd edn San Diego: Academic Press, 285–319. [Google Scholar]
- Slafer GA, Rawson HM. 1994. Sensitivity of wheat phasic development to major environmental factors: a re-examination of some assumptions made by physiologists and modellers. Functional Plant Biology 21, 393–426. [Google Scholar]
- Slafer GA, Rawson HM. 1995a Rates and cardinal temperatures for processes of development in wheat: effects of temperature and thermal amplitude. Functional Plant Biology 22, 913–926. [Google Scholar]
- Slafer GA, Rawson HM. 1995b Intrinsic earliness and basic development rate assessed for their response to temperature in wheat. Euphytica 83, 175–183. [Google Scholar]
- Slafer GA, Rawson HM. 1995c Photoperiod×temperature interactions in contrasting wheat genotypes: time to heading and final leaf number. Field Crops Research 44, 73–83. [Google Scholar]
- Takahashi R, Yasuda S. 1971. Genetics of earliness and growth habit in barley. In: Nilan RA, ed. Barley Genetics II. Proceedings of the Second International Barley Genetics Symposium. Pullman, WA: Washington State University Press, 388–408. [Google Scholar]
- Tang Y, Liu X, Wang J, et al. . 2016. GAPIT version 2: an enhanced integrated tool for genomic association and prediction. The Plant Genome 9, doi: 10.3835/plantgenome2015.11.0120. [DOI] [PubMed] [Google Scholar]
- Thines B, Harmon FG. 2010. Ambient temperature response establishes ELF3 as a required component of the core Arabidopsis circadian clock. Proceedings of the National Academy of Sciences, USA 107, 3257–3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevaskis B, Hemming MN, Dennis ES, Peacock WJ. 2007. The molecular basis of vernalization-induced flowering in cereals. Trends in Plant Science 12, 352–357. [DOI] [PubMed] [Google Scholar]
- van Beem J, Mohler V, Lukman R, van Ginkel M, William M, Crossa J, Worland AJ. 2005. Analysis of genetic factors influencing the developmental rate of globally important CIMMYT wheat cultivars. Crop Science 45, 2113–2119. [Google Scholar]
- VanRaden PM 2008. Efficient methods to compute genomic predictions. Journal of Dairy Science 91, 4414–4423. [DOI] [PubMed] [Google Scholar]
- Wang S, Wong D, Forrest K, et al. . 2014. Characterization of polyploid wheat genomic diversity using a high-density 90 000 single nucleotide polymorphism array. Plant Biotechnology Journal 12, 787–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitechurch EM, Slafer GA, Miralles DJ. 2007. Variability in the duration of stem elongation in wheat and barley genotypes. Journal of Agronomy and Crop Science 193, 138–145. [Google Scholar]
- Wigge PA 2013. Ambient temperature signalling in plants. Current Opinion in Plant Biology 16, 661–666. [DOI] [PubMed] [Google Scholar]
- Worland AJ, Sayers EJ, Börner A. 1994. The genetics and breeding potential of Rht12, a dominant dwarfing gene in wheat. Plant Breeding 113, 187–196. [Google Scholar]
- Wu J, Kong X, Wan J, et al. . 2011. Dominant and pleiotropic effects of a GAI gene in wheat results from a lack of interaction between DELLA and GID1. Plant Physiology 157, 2120–2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue W, Xing Y, Weng X, et al. . 2008. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nature Genetics 40, 761–767. [DOI] [PubMed] [Google Scholar]
- Yates S, Mikaberidze A, Krattinger SG, et al. . 2019. Precision phenotyping reveals novel loci for quantitative resistance to Septoria tritici blotch. Plant Phenomics 2019, doi.org/10.34133/2019/3285904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youssefian S, Kirby EJM, Gale MD. 1992. Pleiotropic effects of the GA-insensitive Rht dwarfing genes in wheat. 1. Effects on development of the ear, stem and leaves. Field Crops Research 28, 179–190. [Google Scholar]
- Zanke C, Ling J, Plieske J, et al. . 2014. a Genetic architecture of main effect QTL for heading date in European winter wheat. Frontiers in Plant Science 5, 217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanke CD, Ling J, Plieske J, et al. . 2014. b Whole genome association mapping of plant height in winter wheat (Triticum aestivum L.). PLoS One 9, e113287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Ersoz E, Lai CQ, et al. . 2010. Mixed linear model approach adapted for genome-wide association studies. Nature Genetics 42, 355–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Levine D, Shen J, Gogarten SM, Laurie C, Weir BS. 2012. A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics 28, 3326–3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zikhali M, Griffiths S. 2015. The effect of Earliness per se (Eps) genes on flowering time in bread wheat. In: Ogihara Y, Takumi S, Handa H, eds, Advances in wheat genetics: from genome to field. Tokyo: Springer, 339–345. [Google Scholar]
- Zikhali M, Wingen LU, Griffiths S. 2016. Delimitation of the Earliness per se D1 (Eps-D1) flowering gene to a subtelomeric chromosomal deletion in bread wheat (Triticum aestivum). Journal of Experimental Botany 67, 287–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Processed phenotypic data are available as Supplementary data. Unprocessed data and analysis scripts are available from the authors upon reasonable request.