Defects in Rab prenylation in the rgtb1 mutant influence the transport of auxin from ovules to the maternal plant and lead to developmental arrest of the female gametophyte in Arabidopsis.
Keywords: Arabidopsis, auxin transport, female gametophyte, funiculus, ovule, PIN1, PIN3, Rab, rab geranylgeranyl transferase
Abstract
Eukaryotic cells rely on the accuracy and efficiency of vesicular traffic. In plants, disturbances in vesicular trafficking are well studied in quickly dividing root meristem cells or polar growing root hairs and pollen tubes. The development of the female gametophyte, a unique haploid reproductive structure located in the ovule, has received far less attention in studies of vesicular transport. Key molecules providing the specificity of vesicle formation and its subsequent recognition and fusion with the acceptor membrane are Rab proteins. Rabs are anchored to membranes by covalently linked geranylgeranyl group(s) that are added by the Rab geranylgeranyl transferase (RGT) enzyme. Here we show that Arabidopsis plants carrying mutations in the gene encoding the β-subunit of RGT (rgtb1) exhibit severely disrupted female gametogenesis and this effect is of sporophytic origin. Mutations in rgtb1 lead to internalization of the PIN1 and PIN3 proteins from the basal membranes to vesicles in provascular cells of the funiculus. Decreased transport of auxin out of the ovule is accompanied by auxin accumulation in tissue surrounding the growing gametophyte. In addition, female gametophyte development arrests at the uni- or binuclear stage in a significant portion of the rgtb1 ovules. These observations suggest that communication between the sporophyte and the developing female gametophyte relies on Rab-dependent vesicular traffic of the PIN1 and PIN3 transporters and auxin efflux out of the ovule.
Introduction
Rab proteins are key components of the vesicular traffic machinery found in all eukaryotes. They reside on the cytosol-facing leaflet of lipid bilayers of organellar membranes. Their interaction with effector proteins enables the recognition and loading of cargo into membrane vesicles, vesicle budding from donor membranes, their movement on the cytoskeleton, and finally recognition, docking, and fusion with acceptor compartment membranes (Pfeffer, 2017). In Arabidopsis thaliana, there are 55 Rab genes (Rutherford and Moore, 2002; Shi et al., 2016). Much effort has been directed towards understanding the in vivo significance of the GTP/GDP cycle of Rabs (Novick, 2016; Pfeffer, 2017). Less studied is yet another level of Rab activity regulation by lipid modifications on the C-terminal tail. Rabs undergo post-translational modification with two geranylgeranyl moieties on cysteine residues close to the protein C-terminus. This modification enables stable anchoring of Rabs to the membranes, which results in a 10 times higher affinity for the membranes (Silvius and l’Heureux, 1994; Shahinian and Silvius, 1995). The enzyme catalyzing the prenylation of Rab proteins is rab geranylgeranyl transferase (RGT), a complex of catalytic RGTA and lipid substrate-binding RGTB subunits and the accesory Rab escort protein (REP) (Seabra et al., 1992; Thoma et al., 2001). Single geranylgeranylated or unmodified Rab proteins are mistargeted and non-functional (Gomes et al., 2003).
In Arabidopsis, RGTA is encoded by two genes—one probably coding for a non-functional protein—REP is encoded by a single gene, and RGTB is encoded by two functional genes, RGTB1 and RGTB2 (Hala et al., 2010; Shi et al., 2016). Total depletion of Rab prenylation by disruption of both RGTB genes is lethal, causing pollen sterility (Gutkowska et al., 2015). Disruption of the RGTB1 gene alone is non-lethal; however, the plants are severely affected (Hala et al., 2010) in a way that can be interpreted as the result of defective auxin gradient formation in the organs; however, the role of auxin in rgtb1 plants has not been reported.
Auxin, the major plant growth hormone, is synthesized in the shoot apex and in leaf primordia, and is transported to other organs via vascular tissues or cell–cell polar transport (Paque and Weijers, 2016; Mroue et al., 2018). The latter is performed by a set of auxin efflux and influx facilitators, including the PIN and AUX/LAX proteins. PINs are the most important auxin efflux transporters. Their polar localization affects the direction of auxin movement in plant tissues and the formation of auxin gradients during organ growth and differentiation (Petrasek and Friml, 2009; Luo and Zhou, 2018). Polar, asymmetric distribution of PINs on the plasma membrane is regulated by their constant recycling to the endosomes and back by the intracellular vesicle transport machinery (Tanaka et al., 2013; Adamowski and Friml, 2015). The transport of PINs from the trans-Golgi network (TGN) to the plasma membrane depends on the activity of small GTPases from the ADP-ribosylation factor (ARF) and RabA families and their regulators (ARF-GEFs and Rab-GEFs) (Geldner et al., 2003; Feraru et al., 2012; Tanaka et al., 2014).
Auxin-dependent processes play an essential role in plant reproduction (Pagnussat et al., 2009; Ceccato et al., 2013; Lituiev et al., 2013; Panoli et al., 2015, Shirley et al., 2019) and the ovule’s response to fertilization (Figueiredo et al., 2015, 2016; Larsson et al., 2017; Robert et al., 2018). A well-coordinated spatiotemporal network of auxin production, transport, and signaling is critical for synchronized development of the sporophytic part of the ovule and female gametophyte (FG; Robert et al., 2015).
The plant ovule is a fundamental organ for reproduction, and is the site of megaspore formation (megasporogenesis), FG formation (megagametogenesis), fertilization, and embryo and endosperm development (Shirley et al., 2019). In Arabidopsis, ~50 ovules are formed inside the pistil of each flower. During female germline development, a single archesporial cell differentiates and undergoes meiosis in each ovule (Pinto et al., 2019). Out of the four post-meiotic spores, only one survives, and develops to form a seven-celled FG, also called the embryo sac (Webb and Gunning, 1990; Christensen et al., 1997). Concurrently, the surrounding sporophytic tissues grow to form integuments (Schneitz et al., 1995). The ovule remains connected with the maternal plant via the funiculus—a stalk filled with vascular tissue. The funiculus enables direct transport of hormones and nutrients to and from the developing ovule (Khan et al., 2015). Two sperm cells delivered by a pollen tube fertilize the central cell and the egg cell. After double fertilization (Zhou and Dresselhaus, 2019), the zygote develops into the embryo, while the fertilized central cell develops into a nutritional tissue, the endosperm (Brown et al., 1999; Faure et al., 2002).
In this study, we aimed to address the role of vesicular transport in mediating interactions between sporophytic and gametophytic tissues within the ovule: specifically, does vesicular transport deficiency in the sporophyte influence the fate of the developing FG and what transport-related mechanisms are involved? To this end, we chose to investigate the rgtb1 mutant which, unlike many other transport-related mutants, is not embryo-lethal. Rather, the rgtb1 mutant produces viable sporophytes, in addition to defective FGs at reasonably high frequency.
Our findings show that interfering with vesicle traffic affects PIN1 and to a lesser extent PIN3 recycling from the endosomes to the basal membranes of the funiculus, and prevents auxin efflux from the ovule. In particular, PIN1 internalization during meiosis/early FG development leads to increased auxin concentration in the ovule and the arrest of FG development at the functional megaspore (FM)/FG2 stage. In some rgtb1 ovules, the presence of RGTB2 activity is apparently sufficient to rescue the RGTB1 deficiency. This highlights the importance of RGTB1-mediated vesicular transport in sporophytic tissues during ovule development and FG formation.
Materials and methods
Plant material
Plant lines used were: Arabidopsis wild type (WT) Col-0, rgtb1-1 (SALK 015871), and rgtb1-2 (SALK 125416) as in Hala et al. (2010) and Gutkowska et al. (2015). PIN1–green fluorescent protein (GFP), was from the Nottingham Arabidopsis Stock Centre (NASC), number N9362 (Benkova et al., 2003), PIN3–GFP (Zadnikova et al., 2010), was a gift from Dr Katerina Schwarzerova (UEB CAS, Prague), DII-Venus, was NASC number N799173 (Brunoud et al., 2012), and pDR5rev:3×Venus, was NASC number N799364 (Heisler et al., 2005). rgtb1-1 and rgtb1-2 heterozygous plants were crossed to fluorescent marker lines, and homozygous plants were identified in the F2 generation by phenotyping and genotyping with appropriate primer pairs. Due to low fertility, the rgtb1 lines were maintained as segregating populations. F2 and further generations were used for microscopic analysis. For reciprocal inheritance crosses, heterozygous rgtb1 plants were chosen by PCR genotyping. The progeny of each cross was grown in soil for 1 month and PCR genotyped; 170–450 plants of each cross were analyzed.
For flower and ovule observations, plants were grown in soil in long-day conditions (16 h light, 8 h darkness). Homozygous plants were chosen by means of their characteristic dwarf phenotype.
Scanning electron microscopy
For SEM observations, flowers were fixed in 3% glutaraldehyde in 25 mM phosphate buffer (pH 7.2) overnight, rinsed, dehydrated and critical-point dried, coated with a thin gold layer, and examined using a LEO 1430VP scanning electron microscope (Carl Zeiss, Germany).
Sample clearing
Flower buds were fixed in acetic acid:ethanol 1:3 solution and cleared in chloral hydrate solution [66.7% chloral hydrate (w/w), 8.3% glycerol (w/w)] or cedar oil as described (Rojek et al., 2018). Ovules were examined under a Nikon Eclipse E800 epifluorescence microscope equipped with differential interference contrast (DIC) optics and a Nikon DS-5Mc CCD camera (PRECOPTIC Co.).
Fluorescence analysis of ovules
For fluorescence analysis, the ovules were mounted in 7% glucose. For FM® 4-64 dye application (Invitrogen), dissected ovules were incubated in 4 µM FM4-64 diluted in 7% glucose, on glass slides for 1–2 h in the dark and observed using confocal laser scanning microscopy (CLSM; Leica TCS SP8X). To avoid differences in fluorescence intensity due to transcript silencing in consecutive generations, sister (progeny of the same mother plant) WT and rgtb1 plants were used for imaging. Specimens were imaged using an epifluorescence microscope (Nikon Eclipse E800) or CLSM. Detection of GFP and yellow fluorescent protein (YFP; Venus) under the epifluorescence microscope was achieved with Epi-Fl Filter Block B-1E (EX 470–490, DM 505, BA 520–560). A filter Block G-2A (EX 510–560, DM 575, BA 590) was used for co-localization of non-specific fluorescent signal and cuticule-like components. For the sake of image clarity, a uniform procedure was applied: ovules at stages younger than FG4 were imaged at ×100 magnification and older ovules starting from FG4 were imaged at ×40 magnification. Roots in control experiments were obtained from 7-to 10-day-old seedlings grown vertically on 1/2 Murashige and Skoog (MS) medium without sugar, and 1.5% agar.
Under CLSM, the GFP excitation wavelength was set to 489 nm and emission was detected at 505–547 nm; for YFP (Venus), excitation was at 514 nm and emission was at 524–566 nm; and for FM4-64, excitation was at 558 nm and emission was at 674–766 nm. CLSM imaging was performed with a ×63 oil immersion objective.
All figures were prepared in Adobe Photoshop (Elements 11 and CS6 versions).
Statistical analysis
All calculations were performed in GraphPad Prism 5.0 software. Mean values and standard error of the mean were calculated and data were compared with unpaired Student t-test with a two-tailed hypothesis. Graphs were prepared using the same software. In every case, several (at least three) independent plant cultivations, each of at least 10 plants per genotype, were performed to generate data. A Fisher exact test against the H0 hypothesis that the allele transmissions are equal was applied in the case of reciprocal genetic cross analysis.
Transcriptomic analysis
RNASEQ reads from the experiment SRP075604 were downloaded from publicly available databases (Klepikova et al., 2015, 2016). Reads were mapped to the Arabidopsis genome and gene expression was calculated [in transcripts per million (TPM)] using CLC Genomics workbench ver9.5.2 (www.clcbio.com). For the purpose of this study, only selected stages and tissues were analyzed in detail. Normalized gene expression values for the Col WT nucellus, FG, and whole ovule were extracted from microarray data reported previously (Tucker et al., 2012a). Presence/absence tags were used to assess whether genes were expressed; values below an arbitrary expression value of 10 generally indicate that the transcript is undetectable.
mRNA in situ hybridization
Inflorescences from Col-0 WT plants were fixed in FAA (50% ethanol, 5% acetic acid, 4% paraformaldehyde, and 0.025% Tween-20) and embedded in paraffin as described previously (Tucker et al., 2003). To generate the probe, an RGTB1 fragment was amplified from genomic DNA using the following oligonucleotides with T7 adaptors: RGTB1_ASF (5'-TGGTCAAACAATATGGCCG), RGTB1_ASR (5'-TAATACGACTCACTATAGAGCAGCAACACAACTTCGTT), RGTB1_SF (AGCAGCAACACAACTTCGTT), and RGTB1_SR (TAATACGACTCACTATAGTGGTCAAACAATATGGCCG). Digoxigenin (DIG)-labeled probes were transcribed with T7 polymerase using the DIG-labeling kit (Roche). In situ hybridization was performed using an InsituPro VSi robot (Intavis), following a standard protocol (Javelle et al., 2011).
Results
rgtb1 produces deformed flowers
The general features of rgtb1-1 and rgtb1-2 plants were described previously (Hala et al., 2010), with both alleles showing similar phenotypes. rgtb1 flowers are smaller than those of the WT, and the perianths never open (Fig. 1A–F versus G–L). In mature rgtb1 flowers, a relatively long pistil is surrounded by a normal number of small sepals, petals, and anthers (Fig. 1G). The flower phenotypes of rgtb1 and the lack of typical anthesis make it difficult to establish the actual stage of flower and gametophyte development. To overcome this, we present the comparison of WT versus rgtb1 flowers based on pistil and stigma maturity according to Smyth et al. (1990) and Christensen et al. (1997) which we use throughout this work (Fig. 1B–F, H–L).
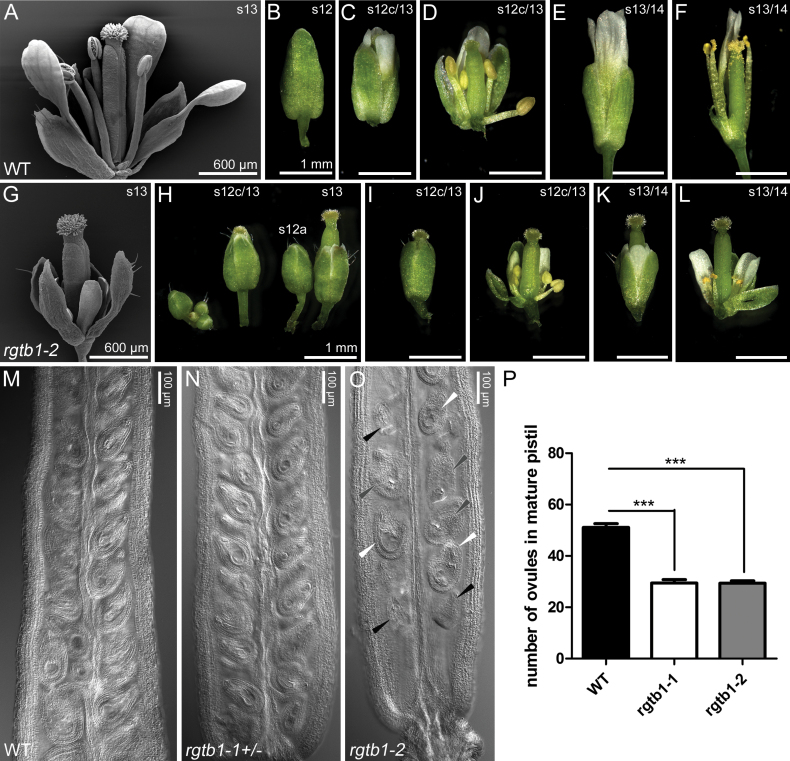
Fig. 1.
The rgtb1 mutation disturbs flower development and reduces fertility. Selected developmental stages of WT (Col-0, A–F) and rgtb1-2 (G–L) plants. Delay of stamen and vegetative organ growth in comparison with the pistil (G), precocious bud opening, and loss of typical anthesis at stage 13 (H–K). Problems with pollen release (L). (M and N) Ovules in WT (M) and rgtb1+/– (N) flowers around anthesis. (O) Empty ovules of normal size (black arrowheads) or collapsed ovules (white arrowheads) accompany normal mature ovules in rgtb1–/– flowers around anthesis. (P) Number of ovules in mature pistils (from flowers around anthesis). Number of pistils counted: WT n=25, rgtb1-1 n=24, rgtb1-2 n=36. Bars represent the mean ±SEM compared with the unpaired Student t-test. Results are highly significant (P<0.001). (A, G) SEM; (B–F, H–L) stereomicroscopy. Scale bar=1 mm in (B) and (H) which are shown at scale with the photographs in (C–F) and (I–L). Photograph of a cleared pistil under DIC on a microscope (M–O). (This figure is available in color at JXB online.)
The discrepancy between pistil and anther size was proposed to account for the low fertility of rgtb1 mutants (Hala et al., 2010) together with pollen coat defects and pollen germination deficiency (Gutkowska et al., 2015). However, closer inspection of rgtb1–/– flowers revealed a lower number of ovules per mature ovary than in the WT or rgtb1+/–, and many of the ovules, despite normal size, were deformed (Fig. 1M–P). The observations of ovule deformation were new in the context of rgtb1 mutations, hence we decided to study the reason for the reduced fertility on the female side.
rgtb1 ovule defects are sporophytic in origin
The fact that rgtb1 homozygous mutants are viable indicates that any gametophytic effect of RGTB1 mutation is not completely penetrant. On the other hand, rgtb1–/– plants are nearly infertile. In order to provide evidence that infertility in rgtb1 is of sporophytic origin, we compared the transmission efficiency through the male and female gametes in heterozygous rgtb1 plants by backcrossing them to the WT, as both pollen donors and acceptors (Table 1). Although transmission through the male gamete was decreased to ~60–70% in rgtb1 mutants, no significant change in transmission efficiency through the female gamete was observed. This result suggests that any defects in FG development and/or fertilization in rgtb1 mutants are dependent on RGT activity in sporophytic tissues. Consistent with this, in rgtb1–/– mother plants, we observed deformed and non-viable ovules, while in rgtb1+/– mother plants, apparently normal ovules containing a rgtb1– FG were functional and capable of normal genetic transmission of the allele to the progeny.
Table 1.
The rgtb1 is a recessive sporophytic mutation
| Pollen acceptor/pollen donor | Expected rgtb1 +/–:WT ratio | Obtained rgtb1 +/–:WT ratio | Transmission of the rgtb1 allele | P-value (Fisher exact test) |
|---|---|---|---|---|
| WT×rgtb1-1+/– | 1:1 | 58:114 | 0.34 | 0.0031** |
| rgtb1-1+/–×WT | 1:1 | 107:130 | 0.45 | NS |
| WT× rgtb1-2+/– | 1:1 | 150:270 | 0.36 | <0.0001*** |
| rgtb1-2+/–×WT | 1:1 | 229:218 | 0.51 | NS |
Functional megaspore arrest leads to defective female gametogenesis in the rgtb1 mutant
We examined female sporogenesis in rgtb1 homozygous plants in comparison with WT plants. No differences were observed in megaspore mother cell (MMC) differentiation, meiotic division, or FM differentiation (Supplementary Fig. S1 at JXB online; >600 ovules were observed for each genotype).
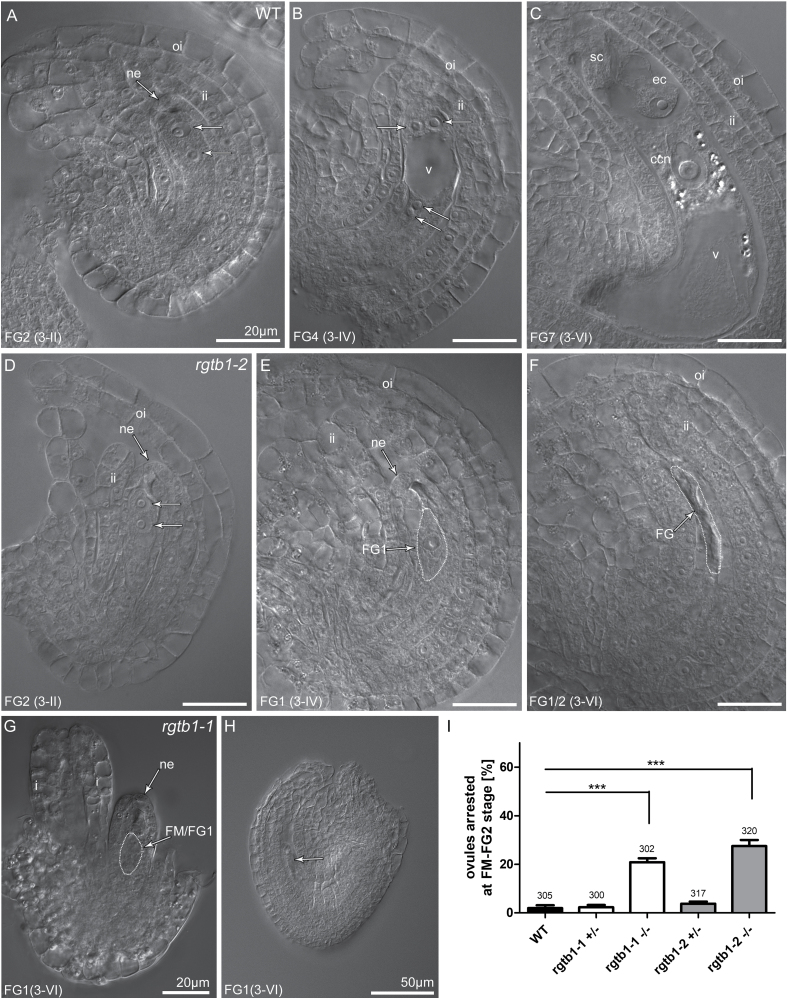
Abnormalities in rgtb1 ovules were first detected after the FM stage in comparison with the WT (Fig. 2A–C versus D–H; I). Approximately 21–27% of ovules in rgtb1-1 and rgtb1-2 homozygous plants, respectively, arrested at this stage (Fig. 2E, G, H) or immediately after the first mitotic division—at the bi-nucleate stage (Fig. 2F)—in comparison with only 2–4% of ovules in WT or heterozygous plants (~300 ovules were counted for each genotype). This observation confirmed the result of the reciprocal crosses, which suggested that sporophytic defects are responsible for abnormal ovule development (Table 1). Deformation of the integuments was also observed in rgtb1–/– plants (Fig. 2D, G). The inner or outer integument was not present, and instead the FG cells protruded or the integuments grew asymmetrically.
Fig. 2.
The rgtb1 mutation affects ovule and female gametophyte (FG) development. Stages of ovule development in WT (A–C), rgtb1-1 (G and H), and rgtb1-2 (D–F) ovules from flowers at developmental stage 3-I to 3-VI (according to Schneitz et al., 1995; Christensen et al., 1997). (A–C) WT plants; stages of FG development correlate with ovule development. Starting from the 3-I stage (corresponding to the FG2 stage of the ovule in the WT), a developmental arrest is observed in a large portion of rgtb1 ovules at the FM or FG1 stage. Normal development of ovules at the 3-II/FG2 flower stage (A versus D), developmental arrest of the FG in rgtb1 at the 3-IV/FG4 flower stage (B versus E), and the 3-VI/FG6 flower stage (C versus F–H) with normal (E, F, H) or (D, G) abnormal development of integuments. (I) Fraction of ovules arrested at the FM/FG2 stage of gametogenesis (%). In each case, >300 ovules were counted, while the exact number is given above the corresponding bar. Bars represent the mean ±SEM. Data were compared with unpaired Student t-test; ***indicates a P-value <0.001. (A–H) DIC microscopy. Abbreviations: ne, nucellar epidermis; ii, inner integument; oi, outer integument; ec, egg cell; sc, synergid cell; ccn, central cell nucleus; v, central vacuole; FM, functional megaspore; FG, female gametophyte. Nuclei in FGs are marked by arrows (A, B, D). Scale bar=20 μm for all images.
These abnormal phenotypes are reminiscent of ovules showing deficiencies in auxin biosynthesis and flux (Schruff et al., 2006). For example, hypomorphic pin1-5 mutants show similar defects in the ovule to those observed in rgtb1 (Ceccato et al., 2013). Because auxin transport by PIN proteins is Rab vesicle dependent, and PIN1 and PIN3 are the main auxin efflux proteins in the developing ovule, we decided to study PIN1 and PIN3 protein localization in rgtb1 ovules.
PIN1–GFP and PIN3–GFP are internalized from basal membranes in the funiculus provascular cells of rgtb1 ovules
We crossed rgtb1 plants to the pPIN1:PIN1-GFP (Benkova et al., 2003) and pPIN3:PIN3-GFP (Zadnikova et al., 2010) lines, markers for auxin efflux in multiple organs including the ovule, and analyzed homozygous progeny of the cross. Localization of PIN1–GFP in root tissues of the seedlings was indistinguishable between WT and rgtb1 plants (Supplementary Fig. S2A, B versus C–F), while PIN3–GFP marked fewer cells in the quiescent center (QC) of the root meristem of rgtb1-1 than in the WT (Supplementary Fig. S2G–I versus J–L).
Using fluorescence microscopy in combination with DIC, we precisely revealed the tissue- and stage-specific altered expression of PIN1–GFP in rgtb1 ovules. In WT ovules containing an MMC and at the conclusion of megasporogenesis, PIN1–GFP showed polar localization in the most distal nucellar epidermal cells. This asymmetric membrane localization of PIN1–GFP was comparable between the WT and rgtb1 (Fig. 3A, B versus G, H, M, N). PIN1–GFP expression gradually decreased in the WT and rgtb1 nucellus, starting from the first mitotic division of the FG, and it was undetectable in ovules collected from mature WT and rgtb1 flowers (Fig. 3C–F versus I–L, O–T); at that stage, PIN1–GFP was restricted to the chalaza (a medial domain connecting the funiculus to the rest of the ovule) and the funiculus.
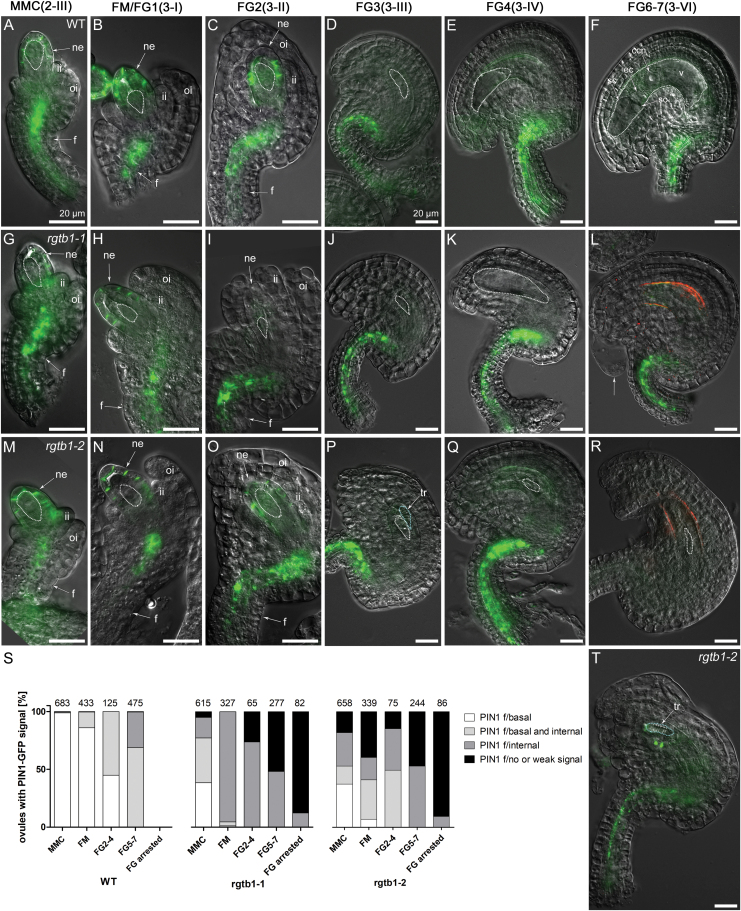
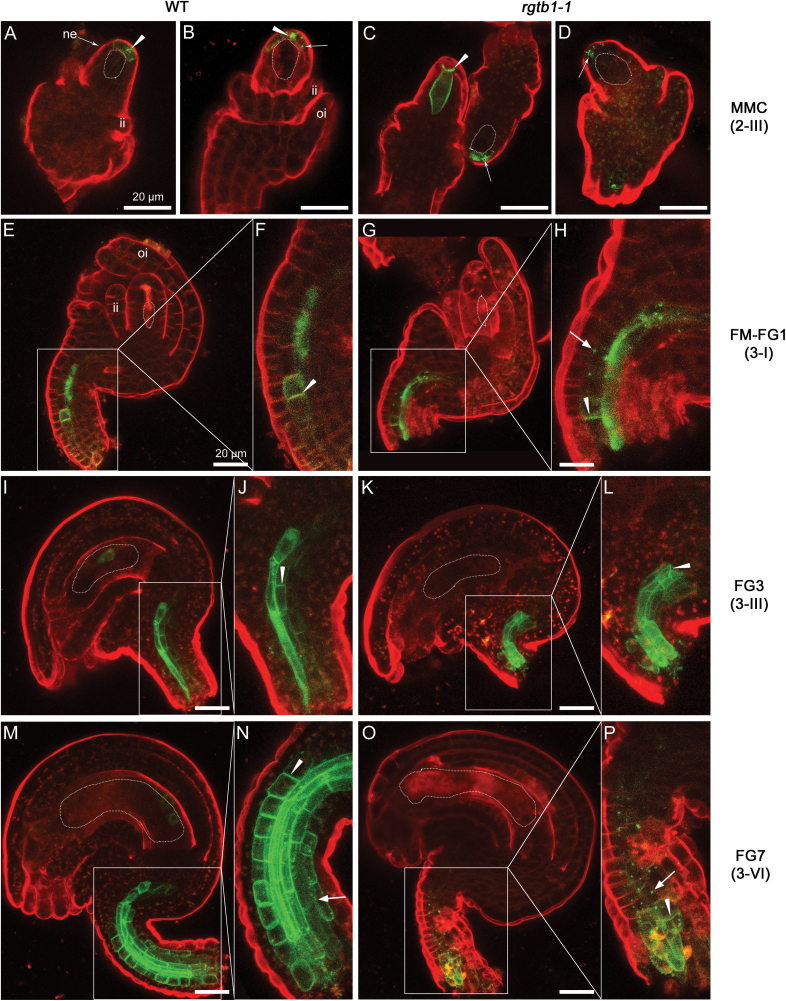
Fig. 3.
PIN1 is localized on polar membranes at the tip of the rgtb1 ovule nucellus, but mislocalized in provascular cells of the funiculus. pPIN1:PIN1-GFP expression in WT (A–F), rgtb1-1 (G–L), and rgtb1-2 (M–R, T) ovules from stages 2-III to 3-VI (according to Schneitz et al., 1995). During stages 2-III/MMC to 3-II/FG2, PIN1–GFP is polarly localized at the tip of the nucellus in WT and rgtb1 ovules (A–C versus G–I, M–O). Decreased PIN1 expression in the nucellus starts from stage 3-III/FG3, leading to an absence of nucellar signal in all ovules at later stages (D–F versus J–L, P–R). Basal polar signal in the WT funiculus provasculature at the MMC stage becomes gradually internalized at ovule maturity (examples of basal polar signal A–C and partial internalization D–F). (S) Quantification of ovules expressing PIN1–GFP according to the genotype and developmental stage. Nucellar signal in WT and rgtb1 ovules was uniform; therefore, only the differences of funiculi-localized signal were considered. The range of phenotypes of PIN1–GFP localization in rgtb1 with funiculus provasculature signal were divided into four classes: basal polar localized (white, examples in M, N); partially internalized (light gray, examples in L, Q); intracellular (dark gray, examples in (G, H–K, O–P); absent or very weak (black, example on R, T). FM/FG2-arrested ovules isolated from older ovaries were considered in a separate class. The number of ovules counted is given above each bar. (T) Example of an rgtb1-2 ovule developmentally impaired with underdeveloped integuments, lacking the FG and with a tracheary element-like structure adjacent to the aborted FG. (A–R, T) Epifluorescence microscopy; DIC and PIN1–GFP signal. The white dotted line highlights the MMC, FM, or FG, as appropriate for the image. Abbreviations: I, integuments; ii, inner integument; oi, outer integument; ne, nucellar epidermis; f, funiculus, ec, egg cell; sc, synergid cell; ccn, central cell nucleus; v, central vacuole; tr, aborted tracheary element; mmc, megaspore mother cell; FM, functional megaspore; FG, female gametophyte. Scale bar=20 μm on all images. (This figure is available in color at JXB online.)
In funiculi, the localization of PIN1–GFP appeared different between the WT and rgtb1 mutants. Therefore, we used CSLM in ovules stained with FM4-64 styryl dye to assess PIN1–GFP subcellular localization in that tissue (Fig. 4). Based on images obtained by fluorescence microscopy with DIC imaging (Fig. 3) and CLSM (Fig. 4), we calculated the frequency of ovules presenting PIN1–GFP localized only on the basal plasma membrane of the funiculus cells, partially internalized from the basal polar membrane, only endosomal, and lacking the PIN1–GFP signal. In the WT at the MMC stage, PIN1–GFP showed basal polar localization in all ovules (Fig. 4A, B). During subsequent development, PIN1–GFP signal was gradually internalized, with >50% of ovules at the FG4 stage showing at least partial endosomal localization (Fig. 4M, N). In the WT, all mature ovules had PIN1–GFP signal predominantly internalized (Figs 3S, 4S, T).
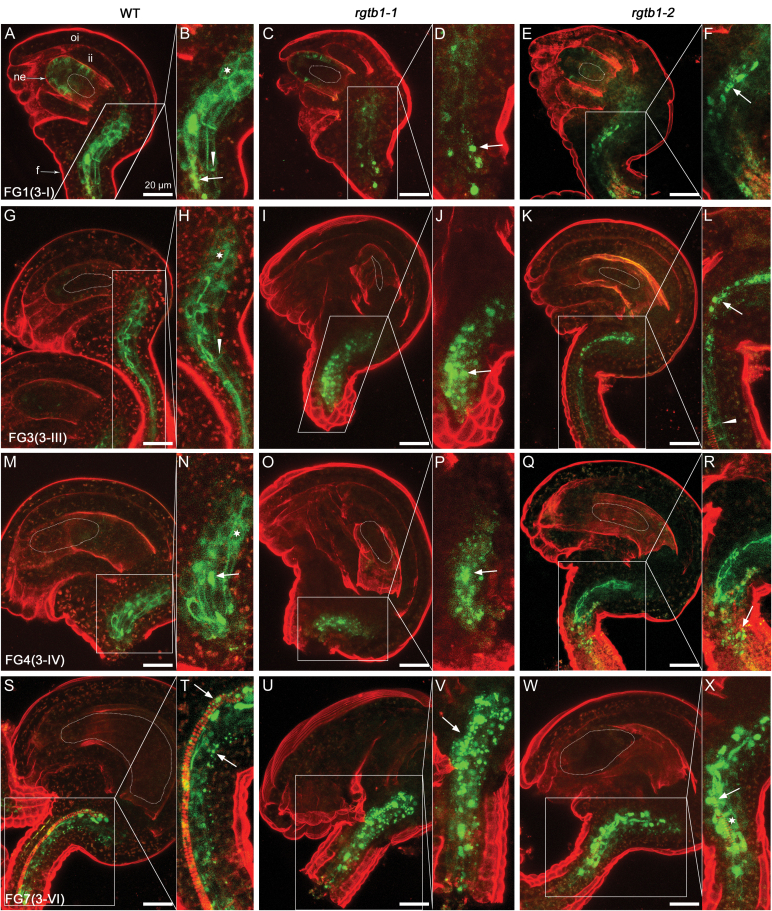
Fig. 4.
PIN1–GFP is internalized in the funiculus provasculature of rgtb1 ovules. pPIN1:PIN1–GFP expression in WT (A, B, G, H, M, N, S, T), rgtb1-1 (C, D, I, J, O, P, U, V), and rgtb1-2 (E, F, K, L,Q, R, W, X) ovules from developmental stage 3-I to 3-VI (according to Schneitz et al., 1995). (A and B) PIN1–GFP is basal polarly localized in provascular cells of the funiculus in the young WT ovules (at the FG1 stage) and with progression of development becomes partly internalized (G and H, at the FG3 stage; M and N, at the FG4 stage) and is internalized in mature ovules (S and T). In a large fraction of rgtb1 ovules, the PIN1–GFP signal is internalized, at least partially, throughout the whole of development, starting from the unicellular stage until mature FG (rgtb1-1 images C, D, I, J, O, P, U, V; rgtb1-2 images E, F, K, L, Q, R, W, X). (A–X) Confocal laser scanning microscopy, PIN1–GFP signal and FM4-64 dye fluorescence. Dotted lines highlight the MMC, FM, or FG, as appropriate for the image. Abbreviations: ne, nucellar epidermis; f, funiculus; ii, inner integument. Basal polar localization of PIN1–GFP is marked by arrowheads, intracellular localization is marked by arrows, and an asterisk marks cells with apolar PIN1–GFP localization. The left narrow image represents a magnification of the area boxed in the right image. Scale bar=20 μm for all images. (This figure is available in color at JXB online.)
In rgtb1, similar to what is described in Fig. 2 for ovule development, we observed a range of different PIN1–GFP phenotypes in the funiculus at any developmental stage. In striking contrast to the WT, already before meiosis, PIN1–GFP started to be internalized in 36–50% of rgtb1 funiculi (Fig. 3G). At the same stage, ~30% of rgtb1 ovules showed normal WT-like signal and few lost the signal from the funiculi (Fig. 3M). At the FM/FG1 stage, nearly all rgtb1 ovules had PIN1–GFP signal internalized or even absent, depending on the genotype (Fig. 3S), while at this stage in the WT only 10% of ovules showed evidence of PIN1–GFP internalization. Examples of rgtb1 ovules with the endosomal signal at the FM/FG1 stage are shown in Fig. 4C–F. At later stages of development, the signal of PIN1–GFP in rgtb1 funiculi was always internal or absent, and the fractions of ovules that lacked the PIN1–GFP in funiculi increased during development, reaching 15–25% at FG2–FG4 stages and 50% at the FG6/7 stages (Figs 3S, 4I–L, O–R, U–X). In the WT at maturity, the PIN1–GFP signal was also internalized, but was present in all analyzed ovules.
Note that rgtb1 pPIN1:PIN1-GFP showed the same frequency of ovules arrested at the FM/FG1/FG2 stage as the mutant rgtb1. Sister ovules coming from the same ovary showed either normal or precocious PIN1–GFP internalization and were distributed randomly.
Both in WT and in rgtb1-1 ovules, PIN3–GFP signal became detectable in a few cells at the tip of the nucellus (Fig. 5A, B versus C, D), on the membranes that contact directly with the MMC, in agreement with earlier studies (Ceccato et al., 2013). No signal of PIN3–GFP was detected at the chalazal or funiculi at the MMC stage in both genotypes (Fig. 5A–D), in contrast to a well-established signal for PIN1–GFP (Fig. 3A, G, M, S). Before meiosis, WT and rgtb1-1 ovules showed a similar pattern of PIN3–GFP (Fig. 5A, B versus C, D). After meiosis, PIN3–GFP fluorescence disappeared from the micropylar pole of the ovule, and became visible in the funiculus provasculature. In the WT, PIN3–GFP was localized on the plasma membrane, but not strictly in a basal polar manner (Fig. 5E, F), while in rgtb1-1 it was partly internalized in nearly half of the ovules (Fig. 5G, H). This pattern was preserved in later stages of development (Fig. 5I, J for the WT versus K, L for the mutant). Finally, in mature WT ovules, PIN3–GFP localization became polarized in funiculus provascular cells (Fig. 5M, N), similar to earlier reports (Larsson et al., 2017). In all defective FM/FG2-arrested ovules isolated from mature ovaries of rgtb1-1, the PIN3–GFP signal remained at least partly intracellular (Fig. 5O, P).
Fig. 5.
PIN3–GFP is partly internalized in the funiculus provasculature of rgtb1 ovules. pPIN3:PIN3-GFP expression in WT (A, B, E, F, I, J, M, N) and rgtb1-1 (C, D, G, H, K, L, O, P) ovules from developmental stage 2-III to 3-VI (according to Schneitz et al., 1995). PIN3–GFP localizes to MMC-adjacent membranes of the nucellus at the tip of the ovule in the WT and rgtb1-1 (A and B versus C and D); at later stages, the nucellar signal disappears in both genotypes (E–P). PIN3–GFP is also basal–polar localized in provascular cells of the funiculus in WT plants starting from the FM stage (E, F for the FM/FG1 stage; I, J for the FG3 stage; M, N for mature ovules). In rgtb1-1, PIN3–GFP is expressed in the same regions of the funiculus but is internalized to a larger extent (G, H versus E, F; K, L versus I, J; O, P versus M, N). (A–P) Confocal laser scanning microscopy, PIN1–GFP signal and FM4-64 dye fluorescence. Dotted lines highlight the MMC, FM, or FG, as appropriate for the image. Abbreviations: ne, nucellar epidermis; f, funiculus; ii, inner integument; oi, outer integument. Basal polar localization of PIN3–GFP is marked by arrowheads, and intracellular localization is marked by arrows. The left narrow image represents magnification of the area boxed in the right image. Scale bar=20 μm for all images. (This figure is available in color at JXB online.)
Auxin accumulates in arrested ovules of the rgtb1 mutant
In order to analyze auxin accumulation in the developing ovule, we used two auxin reporters that rely on different principles of operation. The first was pDR5rev:3×Venus-N7 (Heisler et al., 2005). This construct measures the auxin signaling output by inducing the synthesis of a 3×Venus fluorescent protein bearing a nuclear localization signal (NLS). The protein is transcribed from the Cauliflower mosaic virus (CaMV) 35S minimal promoter preceded by a regulatory element that binds the ARF protein–auxin complex. Hence the cellular response to active auxin can be measured. The second construct was 35S:DIIS-Venus-NLS, which gives results complementary to those from the pDR5rev:3×Venus construct (Brunoud et al., 2012). DIIS-Venus measures the auxin signaling input. It consists of the coding sequence of the naturally existing Aux/IAA domain, which is quickly degraded by the proteasome upon auxin binding, fused to a nuclear-localized version of the Venus protein. The construct is transcribed from the CaMV35S promoter in almost every plant cell; in the presence of auxin, the reporter protein is degraded and no signal is detected. In the case of cells in which auxin is transiently present (is only transported through), the readout from pDR5rev:3×Venus-N7 may be (nearly) negative while the readout from DIIS-Venus or the related R2D2 construct is positive (Robert et al., 2015).
In control experiments in both WT and rgtb1 plants, the pDR5rev:3×Venus fluorescent signal was present in the QC of the root apical meristem, in the root epidermis, and in vascular strands (Supplementary Fig. S2M–O), while the DIIS-Venus reporter fluorescence was present only in cell nuclei of elongated epidermal cells in the root hair formation zone (Supplementary Fig. S2P–R).
Again, we decided to use DIC contrast imaging on whole-mount ovules to clearly distinguish stages of development of the FG. During early ovule development, the activity of the auxin reporter pDR5rev:3×Venus was similar in both WT and rgtb1 plants. From the MMC stage until the FG3 stage, signal was detected in the most distal epidermal nucellar cells (Fig. 6A–C versus F–H, K–M). While the pDR5rev:3×Venus signal mirrors the PIN1–GFP- and PIN3–GFP-expressing cells at the tip of the nucellus, the high expression of PIN proteins at the chalaza and funiculus is surprisingly not accompanied by the auxin reporter pDR5rev:3×Venus. This was similar in WT and rgtb1 plants at early stages of FG development, and is consistent with previous observations (Bencivenga et al., 2012; reviewed in Robert et al., 2015).
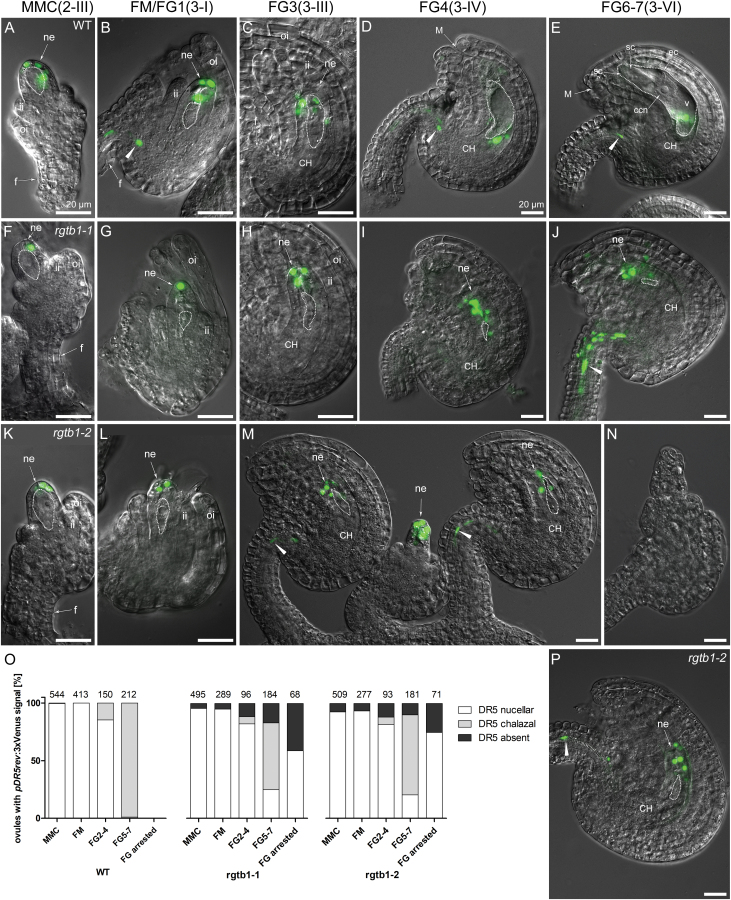
Fig. 6.
Developmentally impaired rgtb1 ovules show mislocalized response of the DR5rev:3×Venus auxin reporter. Auxin nuclear reporter DR5rev:3×Venus expression in WT (A–E), rgtb1-1 (F–J), and rgtb1-2 (K–N, P) ovules from developmental stage 2-II to 3-VI (according to Schneitz et al., 1995). DR5rev:3×Venus signal at the tip of the nucellus of WT and rgtb1 ovules at the 2-II/MMC stage (A versus F, K) until stage 3-III/FG3 (B, C versus G, H, L, M). In WT ovules from stage 3-IV/FG4 until maturity, DR5rev:3×Venus signal is localized at the chalazal part of the ovule (D, E; DR5 signal from the chalazal nucellus layer overlaps the FG layer) and the funiculus provasculature. In many rgtb1 cases, DR5rev:3×Venus signal remains localized in the micropylar pole of the ovule (for stage FG4, D versus I, M; for stage FG6, E versus J, P) and usually is also present in the funiculus. (M) rgtb1-2 ovules showing normal DR5rev:3×Venus signal localization at stage 3-III/FG3 and, in the middle, a developmentally arrested ovule expressing DR5rev:3×Venus at the tip of the nucellus. (N) Ovule arrested at an early stage of development showing no signal of DR5rev:3×Venus. (O) Fractions of ovules at different developmental stages from plants showing DR5rev:3×Venus reporter activity. Ovules were divided into three classes based on the presence of DR5rev:3×Venus signal at the top of the micropylar nucellus (white, examples in A–C, F––J, K–M, P), at the chalazal pole of the ovule in the endothelium (gray, examples in D, E), or with the signal absent (black, example in N). Ovules were counted under fluorescent and confocal microscopes. The number of ovules counted is given above each bar. (A–N, P) Epifluorescence microscopy; merged images of DIC and the DR5rev:3×Venus signal. The white dotted line highlights the MMC, FM, or FG, as appropriate for the image. Abbreviations: ii, inner integument; oi, outer integument; f, funiculus; ne, nucellar epidermis; CH, chalaza, M, micropyle; ec, egg cell; sc, synergid cell; ccn, central cell nucleus; v, central vacuole. Arrowheads point to the DR5rev:3×Venus signal in the funiculus. Scale bar=20 μm for all images. (This figure is available in color at JXB online.)
From the FG4 stage onwards, the nucellar tissue surrounding the FG continued to degenerate and cells with reporter activity were found in the chalazal pole, but always outside the FG in WT ovules, even at maturity (Fig. 6D, E). Generally, throughout FG development, the pDR5rev:3×Venus signal decreased, but persisted in the funiculus. At the time of second mitosis in the FG, 16% of WT ovules showed an auxin response at the chalazal end of the ovule (Fig. 6D) whilst a similar fraction of rgtb1 ovules completely lacked signal. During later stages of development, 21–25% of rgtb1 ovules were arrested at FM/FG2 and showed pDR5rev:3×Venus activity in the epidermal nucellar cells, a feature characteristic of earlier stages of development (Fig. 6D, E versus J, P). Conversely, pDR5rev:3×Venus activity was not detected at all in the most severely affected rgtb1 ovules at any of the analyzed stages (Fig. 6N, 5–17%). Still, >50% of the ovules isolated from mature rgtb1 ovaries expressed DR5rev:3×Venus in a WT-like manner, namely at the chalazal pole and in the funiculus (Fig. 6O). As was the case for PIN1–GFP, ovules arrested at the FM/FG2 stage with the micropylar auxin maximum and ovules progressing normally through development were randomly distributed in each ovary (Fig. 6M).
To complement the data obtained for the positive auxin reporter pDR5rev:3×Venus, we also utilized the DIIS-Venus negative reporter. In WT ovules, fluorescent signal was detected in the early stages of development, from the MMC to the first mitotic division (Fig. 7A–C), complementing the results obtained by pDR5rev:3×Venus. Around ovule maturity, DIIS-Venus signal was present in the outer integuments and the chalaza, and also more weakly in the outer cell files of the funiculus (Fig. 7D). No fluorescent signal was observed inside the embryo sac, as expected due to the CaMV35S promoter specificity that drives the DIIS-Venus expression.
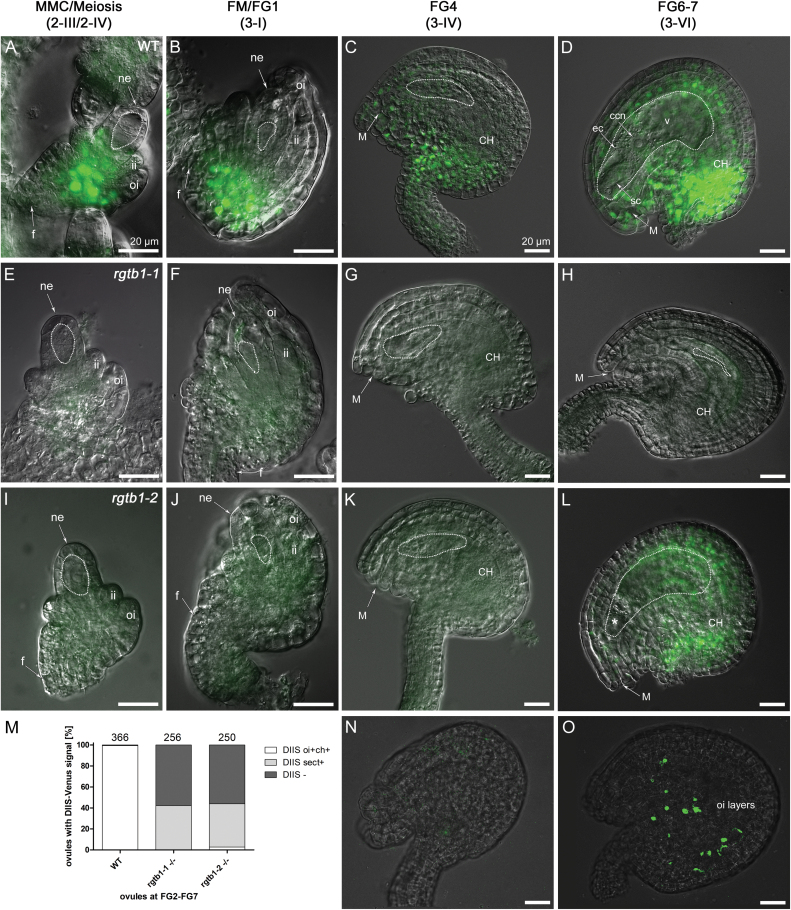
Fig. 7.
The 35S:DIIS-Venus auxin sensor signal is decreased in rgtb1 ovules. The CaMV35S:DIIS-Venus signal during ovule development in WT (A–D) and rgtb1 (E–L, N, O) ovules. (A) Meiosis, WT; the DIIS-Venus signal is in the chalaza. At FM and early FG stages, the DIIS-Venus fluorescence is also present in the outer integument (B, C). Around maturity, DIIS-Venus is detected in the integuments, the chalazal pole, and in the outer cell files of the funiculus (D). At no stage was DIIS-Venus fluorescence seen within the WT FG (D). DIIS-Venus signal was lacking in all young rgtb1 ovules (E, F, I, J) and in 60% of ovules around maturity, both with an arrested (H, N) and with a normal FG (G, K, L). At maturity, <3% of the rgtb1-2 ovules exhibit a WT-like localized signal (L) and a 40% sectorial signal in scattered cells of the outer integument and chalaza (O). (M) Graph summarizing DIIS-Venus signal localization in ovules at all analyzed stages of development. The white bar shows a typical signal at the chalaza and in the outer integument; examples in (A–D). The gray bar shows signal present only in scattered cells of the chalaza and outer integument; example in (O). The black bar shows no signal or a very weak signal; examples in (E–K, N). The exact number of ovules counted is given above each bar. Epifluorescence microscopy; merged images of DIC and the 35S:DIIS-Venus signal. The white dotted line highlights the MMC, FM, or FG, as appropriate for the image. Abbreviations: ne, nucellar epidermis; ii, inner integument; oi, outer integument; f, funiculus; M, micropyle; CH, chalaza; ec, egg cell; sc, synergid cell; ccn, central cell nucleus; v, central vacuole. Scale bar=20 μm for all images. (This figure is available in color at JXB online.)
The DIIS-Venus sensor signal was not detected in young rgtb1 ovules (Fig. 7E–G, I–K versus A–C) and was hardly detected around maturity; <3% of ovules showed a WT-like DIIS-Venus signal at the FG6/7 stage (Fig. 7L). Overall, at maturity, 42% of rgtb1 ovules had DIIS-Venus signal distributed unequally (sectorial) in single cells of the outer integument and chalaza (Fig. 7O). Moreover, the rgtb1 ovules arrested at FM/FG2 never showed DIIS-Venus signal (Fig. 7H, N). The DIIS-Venus signal localization in rgtb1 is summarized on a graph (Fig. 7M). The lack of DIIS-Venus signal in arrested ovules supported the DR5rev:3×Venus results where nucellar auxin accumulation was still maintained in young rgtb1 ovules and those unable to complete megagametogenesis (compare Fig. 6).
It is important to note that in the case of both DIIS-Venus and pDR5rev:3×Venus reporters, only plants showing reporter fluorescence in other sporophytic tissues and at least some ovules were considered for observation and counting. Again, as was the case for PIN1–GFP and DR5rev:3×Venus, the ovules showing positive and negative signal for the DIIS-Venus reporter were distributed randomly in rgtb1 ovaries.
Multiple members of the RGT complex are expressed in ovules around meiosis
The results described above are consistent with rgtb1 mutants, showing decreased efficiency of PIN1 protein recycling and auxin responses. This might be due to hypoprenylation of at least some Rab proteins in the rgtb1 mutants, to the complete absence of RGT activity in ovules, or to a different Rab protein specificity of a remaining RGTB2 enzyme isoform at the FM stage. To address these possibilities, gene expression profiles were analyzed.
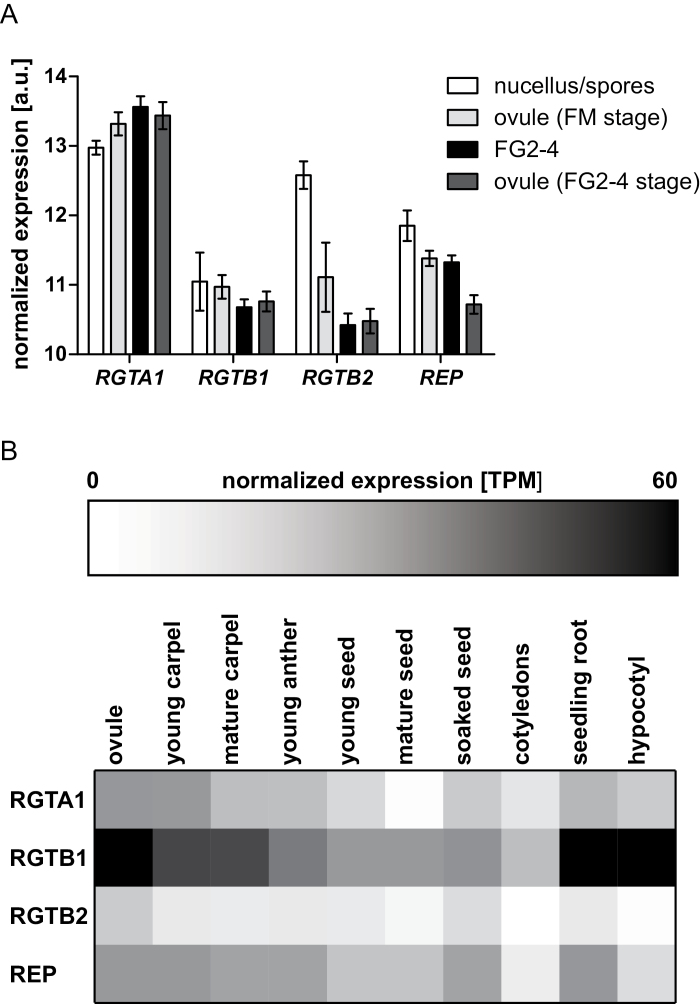
The expression level of the RGTA1, REP, RGTB1, and RGTB2 genes was examined in whole carpels, mature ovules, and seeds at different stages using published RNA-sequencing datasets (Klepikova et al., 2015, 2016; Fig. 8). RGTA1 and REP genes are expressed at relatively low levels in carpels, mature ovules, and seeds, as well as in all other studied tissues (Fig. 8B). RGTB1 expression is generally uniform, but always higher than that of RGTA1 and REP, while RGTB2 expression is barely detectable. Expression datasets from young microdissected ovules were also analyzed, including nucellar cells at the time of meiosis/FM differentiation, FG2–4 female gametophytes, and whole ovules at the respective stages (Tucker et al., 2012a). The data indicate that all members of the RGT complex are expressed in young ovules (Fig. 8A). The RGTA1, RGTB2, and REP genes show significant differences in expression between tissues; RGTB2 and REP are up-regulated in nucellar cells relative to the whole ovule [fold change (FC)=2.8, P-value <0.01; and FC=1.4, P-value <0.05, respectively], while RGTA1 is slightly more abundant in the whole ovule (FC=0.8, P<0.05) compared with its tip. REP is also up-regulated in the developing FG relative to the whole ovule (FC=1.5, P<0.05). Interestingly, in ovules around meiosis, the level of expression of RGTB2 is similar to or higher than the levels of expression of RGTB1 (Fig. 8A) in striking contrast to other tissues, where RGTB2 transcript is always less abundant (Fig. 8B).
Fig. 8.
Expression profiles of RGT genes in ovules and other tissues. (A) Expression of RGT complex-encoding genes in WT ovules during megasporogenesis and development of 2–4 nucleate female gametophytes (FG2-4). Nucellar tissue and FG2-4 samples were laser microdissected and analyzed by hybridization to Affymetrix arrays as described in Tucker et al. (2012a). The rest of the ovule tissues from dissected samples were collected separately. Mean normalized gene expression values for the Col WT nucellus, female gametophyte, and whole ovule ±SD are presented. (B) RNASEQ reads from the experiment SRP075604 were downloaded from publicly available databases (Klepikova et al., 2015, 2016). Reads were mapped to the Arabidopsis genome, and gene expression was calculated and presented as a heat map for selected plant organs.
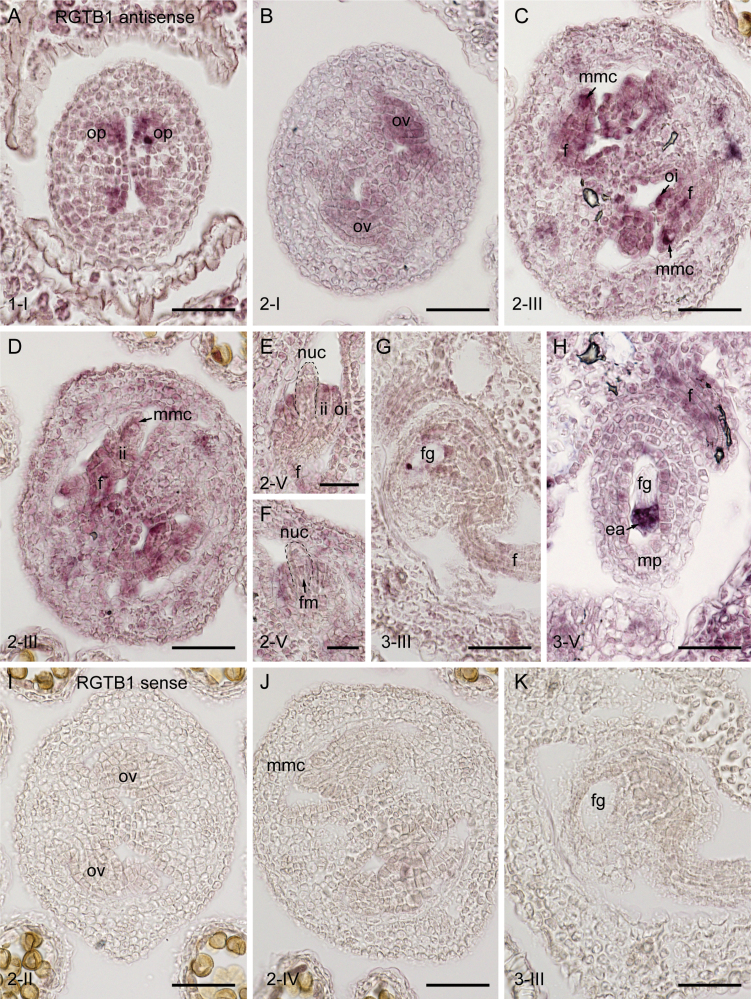
To further delineate the spatial distribution of RGTB1 mRNA in ovules, mRNA in situ hybridization was utilized. At early stages of ovule development, RGTB1 mRNA was evenly distributed in the ovule primordia and ovule proper (Fig. 9A, B). During MMC expansion, RGTB1 was detected throughout the ovule but was particularly strong in the archesporial cell/young MMC (Fig. 9C). In contrast, during meiosis, signal was strong in the developing integuments but was depleted in the nucellus and MMC (Fig. 9D). During subsequent stages, signal was not obvious in the developing FM or FG (Fig. 9E–G), but was detected in most sporophytic ovule tissues including the funiculus (Fig. 9E–G). As the ovule approached maturity, signal was maintained in the funiculus and was particularly abundant in the egg apparatus (Fig. 9H). These results indicate that RGTB1 mRNA partially overlaps with the sites of PIN accumulation, but is not abundant in the MMC or nucellus from meiosis until gametophyte cellularization.
Fig. 9.
In situ hybridization of RGTB1 in ovule tissues. (A–H) Antisense RGTB1 probe. (I–K) Sense RGTB1 probe. During early stages of ovule development, RGTB1 transcript was detected in a range of tissues including (A) ovule primordia, (B) all tissues of the young ovule, and (C) subsequently in the megaspore mother cell, chalaza, integuments, and funiculus. (D) During meiosis, signal diminished in the MMC but remained present in the chalaza, integuments, and funiculus. (E, F) During functional megaspore selection, RGTB1 was expressed weakly or was absent in the nucellus/FM but strongly in the integuments. (G) Signal was detected in the female gametophyte during later stages of cell specification, and was abundant in the egg apparatus. Signal remained in sporophytic tissues such as the integuments and funiculus. (I–K) Sense probes showed no background hybridization. op, ovule primordia; mmc, megaspore mother cell; oi, outer integument; ii, inner integument; f, funiculus; nuc, nucellus; fg, female gametophyte; ea, egg apparatus; mp, micropyle; ov, ovule; fm, functional megaspore, Scale bar=50 µm in A–D and G–K, and 25 µm in E and F. (This figure is available in color at JXB online.)
Rab family genes are expressed in young Arabidopsis ovules
Identification of the hypoprenylated Rab proteins in rgtb1 mutants could provide further mechanistic information regarding the key Rab proteins required for ovule development. Unfortunately we were unable to find differences in Rab geranylgeranylation in WT versus rgtb1 flowers by proteomic methods (Supplementary Table S1). In parallel, examination of the RNA sequencing (Supplementary Fig. S3B) and laser capture microdissection (Supplementary Fig. S3A) datasets suggests that multiple Rab-encoding genes are expressed in ovules at meiosis, during FG development, and at anthesis. All RabD genes are expressed, with RabD2b showing the most prominent expression level among all the Rabs at anthesis. In total, only 11 of the 55 Rab-encoding genes are not expressed in ovules. Notably, in post-meiotic ovules when the rgtb1 mutant phenotype is first detected, the most abundant Rab transcripts are RabE1d and RabH1b followed by RabB1c; high expression of RabA4a, RabA1b, and RabA5b is also observed. Five Rab genes are significantly up-regulated in the nucellus compared with the rest of the ovule, namely RabE1d, RabH1b, RabF2b, RabA1a, and RabA6a. In contrast, 10 genes are more abundant in other parts of the ovule compared with the tip, and these include RabA2a, RabD2b, RabA5a, RabA2b, RabA5c, RabD2c, RabG3c, RabG3d, and RabC2b. Taken together, these results indicate that multiple Rab genes are expressed in the developing ovule at meiosis and these may be targets for hypoprenylation in rgtb1. The pattern of Rab gene expression changes during the time of ovule development and seems to differ in gametophytic versus maternal sporophytic tissues.
Discussion
Earlier observations of rgtb1 mutants suggested that many of their phenotypes may result from disturbed auxin homeostasis (Hala et al., 2010). The most straightforward hypothesis linking defects in vesicular transport to a reduced auxin response is the disturbance of auxin carrier recycling, which potentially leads to defective formation of instructive auxin gradients in developing tissues and organs. Out of several auxin carriers, the efflux PIN proteins are well described to depend on vesicular traffic and basal/apical sorting (Luschnig and Vert, 2014). At least three PIN proteins are expressed in the developing ovule: PIN1, PIN3, and PIN6 (Larsson et al., 2017). Of these three proteins, PIN1 is particularly important in early stages of ovule development, followed by PIN3 (Ceccato et al., 2013). Others (including PIN6) take part in later events leading to correct formation of vascular bundles in the funiculus and auxin flux in and out of the mature FG and early embryo (Larsson et al., 2017, Robert et al., 2018). This prompted us to study the interplay of Rab-dependent vesicular traffic, PIN1, PIN3, and auxin at the early stages of ovule development, particularly during initiation of the female germline.
Expression of RGTB2 may contribute to the progression of meiosis and female megaspore differentiation in rgtb1
Transcriptomic analysis suggests that genes coding for RGTA1 and REP subunits of the RGT complex are expressed at comparable levels in most plant tissues. The RGTB subunits are expressed in most organs in a ratio of ~1:10–1:20 RGTB2 to RGTB1. The prominent exception is developing and mature pollen, where the expression of RGTB2 equals or even exceeds that of RGTB1 (Hala et al., 2010). Here we show that RGTB1 mRNA is expressed in a range of ovule tissues including the young MMC, integuments, and funiculus. However, expression was weak or absent in the nucellus and FM. In silico evidence suggests that RGTB2 is substantially expressed in the vicinity of the developing nucellus, FM, and FG, possibly even exceeding RGTB1. We speculate that additive tissue-specific expression of RGTB1 and RGTB2 in the ovule is required for normal germline development.
The hierarchy of Rab prenylation (Kohnke et al., 2013) in Arabidopsis WT or rgtb1 mutants was not addressed experimentally in previous studies (Hala et al., 2010; Gutkowska et al., 2015), hence it is difficult to determine which particular Rab is responsible for the manifestation of rgtb1 ovule phenotypes. The enzymatic activity of RGT in rgtb1 is decreased to 25% of that of the WT, at least for some substrates such as Rab A2a (Hala et al., 2010). Biochemical studies, although not completely quantitative, may also indicate that the RGTB2 subunit has higher enzymatic activity for some Rab proteins than RGTB1 (Shi et al., 2016). Hence, we also predict that there may be a compensatory effect of RGTB2 in rgtb1 mutants, at least in the tissues or stages where RGTB2 expression is relatively high, such as pollen and the young ovule. RGTB2 activity may facilitate completion of meiosis and FM specification in at least some rgtb1 ovules, due to prenylation of important housekeeping Rabs. At the same time, other Rabs, being less abundant or having lower affinity for the RGTB2/RGTA1/REP complex, remain unprenylated and are easily degraded.
The timing of developmental arrest in rgtb1 ovules is also interesting. Nucellus tissues isolated by laser capture microdissection at the meiosis/FM stage show high transcription of the RGTB2 gene, while our in situ hybridization experiments suggest that RGTB1 is weak or absent in the nucellus. Hence, RGTB2 may compensate for the lack of RGTB1 transcript in the nucellus. Indeed both PIN1 and PIN3 localization and auxin accumulation during megasporogenesis were indistinguishable in the WT and rgtb1. At the FG2/FG4 stage, RGTB2 transcript levels are low, while RGTB1 is detected in the integuments, chalaza, and funiculus. At this developmental stage, FG arrest had already initiated in many rgtb1 ovules, concurrent with PIN1 and PIN3 mislocalization from the basal membranes and an auxin concentration increase in the nucellus. This may suggest that the pool of Rabs prenylated (possibly by RGTB2) around meiosis was degraded and the number of Rab molecules still membrane bound and active is not sufficient to sustain the normal recycling of the endosomes. Meiosis, FM specification, and first mitotic division in Arabidopsis take >36 h (Schneitz et al., 1995). Prenylated Rab protein levels on the cell membranes significantly drop after chemical inhibition of RGT after 48 h (Kazmierczak et al., 2017), and the onset of acute systemic phenotypes in the absence of RGT activity was described to be at 4 d in vertebrate embryos (Moosajee et al., 2009). This fits well with the timing of early FG development of Arabidopsis. Another possibility is that at the FM/FG2 stage, a Rab protein not expressed at earlier stages in the ovule is crucial for PIN1 recycling. Even if this Rab is synthesized correctly, the lack of geranylgeranyl modification renders it inactive, and PIN(s) recycling is compromised.
Hypoprenylation of more than one Rab may explain the rgtb1 ovule phenotype
In this work, we summarize data highlighting the expression of Rab-encoding genes in gametophytic and sporophytic tissues of the ovule. A number of Rab genes were up-regulated in nucellus tissue during megasporogenesis, at the same time that polarized PIN1 and PIN3 accumulate in nucellar epidermal cells. Interesting examples are members of the A and E families, known to perform cargo transport to the plasma membrane (Zheng et al., 2005; Chow et al., 2008; Camacho et al., 2009; Asaoka et al., 2013; Drdova et al., 2013; Kirchhelle et al., 2016). This gives a hint as to which Rabs may be involved in FG differentiation and development, although direct proof requires experimental confirmation.
Additional indications as to which Rab(s) may be involved in generative processes in rgtb1 come from studies on individual/multiple Rab mutants. In particular, the rabd2a/b/c mutant is embryo lethal (Pinheiro et al., 2009) and approximately half of the ovules in the rabd2b/c double mutant are not fertilized (Peng et al., 2011). Rab D2b is the most abundant Rab transcript detected in the ovule at anthesis, and provided the highest score in the proteomic analysis of the flowers, indicating that RabD hypoprenylation in rgtb1 may be a cause of some ovule phenotypes. Mutations in a common GEF for Arabidopsis RabF proteins, VPS9, causes embryo lethality, but no obvious seed deformations are detected (Goh et al., 2007).
Neither RabD proteins nor RabF proteins have been associated with PIN1 and auxin localization, but the RabA subgroup may perform these functions. Studies highlight potential roles for RabA1 (Koh et al., 2009; Feraru et al., 2012; Naramoto et al., 2014) or RabA5 (Drdova et al., 2013) in this pathway, both of which are abundant in the young ovule. RabA1b and other members of the family take part in endosomal PIN1 protein recycling through VHA-a1-positive endosomes (Feraru et al., 2012; Berson et al., 2014). What is more, RabA1e-positive endosomes and PIN1 both show characteristic co-aggregation in protophloem cells in a sphingolipid biosynthesis mutant (Markham et al., 2011). Interestingly, high VAN4 (RabA1 GEF) promoter activity is detected in procambium cells of leaves, shoots, and ovule funiculi (Naramoto et al., 2014), marking the high demand for active RabA1 in vasculature-forming tissues. RabA1a and RabF2b expression was enriched in nucellus tissue during megasporogeneis.
PIN1 and PIN3 internalization and incorrect auxin distribution in rgtb1 ovules correlate with developmental arrest of the female gametophyte
The recycling of the main auxin efflux protein PIN1 and also PIN3 was affected in rgtb1 ovules, leading to an aberrant pDR5rev:3×Venus expression, indicative of increased auxin concentration in sporophytic tissues surrounding the developing germline. In parallel, development of the germline was blocked in a significant fraction of rgtb1 ovules, and these arrested ovules indeed showed unusually high and delocalized auxin maxima compared with adjoining ‘normal’ ovules. The symptoms of defective development in rgtb1 ovules were primarily detected after the first mitotic division of the FM. Absence of the PIN1–GFP signal in the ovule provasculature was previously noted in another transport-related mutant, hapless-13 (Wang et al., 2016). FG arrest at the uni- to binuclear stage in rgtb1 may be explained by a lack of PIN1 on the basal membranes of the provasculature cells in the chalazal part of the nucellus and in the funiculus, similar to that reported for the pin1-5 mutant (Ceccato et al., 2013).
Previous studies in Arabidopsis reported accumulation of auxin (via pDR5rev markers) in nucellus cell layers proximal to the FM (Pagnussat et al., 2009), but not inside it. During subsequent stages of gametogenesis, auxin is not detected in the FG but reaches high concentrations in adjoining sporophytic cells. As FG development proceeds towards maturity, the auxin maximum diminishes and shifts from the micropylar to the chalazal pole of the ovule (Lituiev et al., 2013). In many rgtb1 ovules, pDR5rev:3×Venus signal remained strong at the micropylar pole at maturity, and this coincided with developmental arrest. It also coincided with mislocalized PIN1 and PIN3 in the funiculus, consistent with no correct path for auxin efflux. This implies that in WT plants auxin is produced in the young ovule in sporophytic tissues around the growing FG and exported out of the ovule to the mother plant through the funiculus, with the aid of PIN1 and PIN3 (Larsson et al., 2017). This possibility is consistent with recent studies detailing analysis of auxin biosynthesis and transport in the Arabidopsis ovule (Panoli et al., 2015; Larsson et al., 2017; reviewed in Shirley et al., 2019). In the case of rgtb1, the excess auxin in the nucellus may inhibit the progression of the FG towards maturity, which is consistent with the sporophytic nature of the germline abortion phenotype. Convincing complementary results were also obtained by the use of the DIIS auxin reporter. Alternatively, failed FG development inhibits progression of nucellar degradation, resulting in persistence of cells that accumulate auxin but are unable to transport it. These effects would have to be indirect and non-cell-autonomous, since we never observed auxin accumulation inside the FG.
Small differences in auxin concentration in sister rgtb1 ovules cause dramatic changes in fate
Interestingly, a diverse range of phenotypes were identified in rgtb1 mutants, ranging from the complete cessation of FG development to formation of nearly normal mature ovules. Curiously, normal and affected ovules neighbor each other in one ovary; the situation where consecutive ovules are all affected or all normal is quite rare. We considered what the reason might be for unequal effects of the rgtb1 mutation on sister ovules in the same ovary. PIN proteins are typically redundant in function, and the lack of one may induce expression of another (Vieten et al., 2005).Furthermore, their expression (but not proper localization or recycling) is regulated by auxin presence in a positive feedback loop by AUX/IAA repressors, ARF and PLT transcription factors (reviewed in Habets and Offringa, 2014). If the auxin concentration is above a certain threshold, the hormone down-regulates PINs post-transcriptionally (Vieten et al., 2005) or directly regulates the number of PIN proteins present at the membrane (Paciorek et al., 2005; Robert et al., 2010). This complex network of positive and negative feedback loops of auxin biosynthesis and transport enables fine-tuning of plant response to external and internal cues. The aberrant PIN1 localization (vesicles instead of basal membrane) in rgtb1 mutants is consistent with decreased PIN recycling ability.
Deficiency of RGTB1 in rgtb1 mutants leads to reduced Rab protein prenylation (Hala et al., 2010). RGTB1 (or RGTB2 in the case of the rgtb1 mutants) may contribute to tissue development in a dose-dependent manner, and that is why phenotypes appear occasionally in neighboring ovules. We hypothesize that in some ovules, the number of prenylated and membrane-attached Rabs is above the threshold to recycle the PIN1 efficiently to the basal membranes and pump auxin out of the ovule. In neighboring ovules, even slight changes in Rab abundance may not sustain enough recycling and the concentration of PIN on the membrane falls below this threshold. As a consequence, auxin becomes trapped in the nucellus, which leads to the observed persistence of auxin maxima around the FG, and inhibition of developmental progression. A similar case of inhibitory auxin action was previously reported for meristemoid cell differentiation into mature stomata mediated by PIN3 (Le et al., 2014) and also in a pin1-5 mutant, where the lack of PIN1 on basal membranes of the funiculus comes from lower production of this protein in the cells (Ceccato et al., 2013). PIN and auxin regulatory networks are complex and the decisions on cell specification (e.g. to differentiate into a functional FG or stomata guard cell) appear to be made in any of the ovules/meristemoids independently. The reduced PIN1-mediated efflux in rgtb1 mutants may prevent the establishment of gradients and/or transcriptional profiles that are required for the transition to gametogenesis.
Sporophytic tissues of rgtb1 ovules do not develop correctly
Defects in ovules of rgtb1 mutants were not restricted to the nucellus and germline, since the integuments, which later give rise to seed coat, occasionally showed abnormal features. Similar phenotypes were previously reported in Arabidopsis ovules misexpressing auxin-dependent transcription factors (Wu et al., 2006; Kelley et al., 2012; Simonini et al., 2016) or Hieracium ovules treated with the polar auxin transport inhibitor 1-N-naphthylphthalamic acid (NPA) (Tucker et al., 2012b). Also the size and shape of rgtb1 integument cells were often disturbed. Integuments with milder deformations apparently enabled normal growth of the FG.
To summarize, this study provides a comprehensive analysis of FG development in a vesicular transport-deficient plant, highlighting the key role of sporophytic vesicle transport in development of the haploid generation. Although further work is required to identify the particular Rab protein responsible for the observed rgtb1 female gametogenesis-related phenotypes, we describe the influence of transport downturn on localization of PIN1 and PIN3 auxin efflux proteins and auxin gradient formation in the ovule. We suggest that this will encourage further, detailed studies on Rabs and involvement of their interactors in ovule development.
Supplementary data
Fig. S1. Female sporogenesis in rgtb1 mutants.
Fig. S2. Localization of PIN1–GFP, PIN3–GFP, and auxin sensors in rgtb1 seedling roots.
Fig. S3. Expression of Rab-encoding genes in the ovule.
Table S1. Proteomic analysis of Rab proteins isolated from WT and rgtb1 flowers.
The following supplementary data are available at JXB online.
Acknowledgements
The authors would like to thank Professor Ewa Swiezewska (IBB PAS) for support and comments on the manuscript, Dr Agata Malinowska (MS Facility IBB PAS) for help with MS data analysis, Dr Marta Hoffman-Sommer (IBB PAS) and Dr Martin Potocky (UEB CAS) for critically reading the manuscript, and Dr Michal Hala (Charles University) for sharing the PIN3-GFP×rgtb1-1 line. We also thank Chao Ma and Dr Xiujuan Yang for technical assistance with the in situ procedure. The work was financially supported by grant no. UMO-2016/21/D/NZ3/02615 from the National Science Centre of Poland to MG and by University of Gdansk grant nos DS530-L160-D243 and 531-D030-D243-20 to JR. MRT is supported by a grant from the Australian Research Council (DP180104092). SCP is supported by Fundação para a Ciência e Tecnologia (SFRH/BD/137304/2018).
Author contributions
JR conceived the research plan, performed the microscopic experiments, analyzed the data, prepared the figures, and revised the manuscript; MRT analyzed the data, prepared the figures, and wrote and revised the manuscript; SCP performed the in situ hybridization experiment and prepared the figures; MR and ML performed confocal microscopy observations; JN performed SEM observations; HS and GS helped with plant line breeding and genotyping; JB provided scientific support and comments; MG conceived the research plan, performed the experiments, analyzed the data, and wrote and revised the manuscript. MG agrees to serve as the author responsible for contact and ensures communication. All authors read and approved the final manuscript.
Data availability
All data supporting the findings of this study are available within the paper and within its supplementary data published online.
References
- Adamowski M, Friml J. 2015. PIN-dependent auxin transport: action, regulation, and evolution. The Plant Cell 27, 20–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asaoka R, Uemura T, Ito J, Fujimoto M, Ito E, Ueda T, Nakano A. 2013. Arabidopsis RABA1 GTPases are involved in transport between the trans-Golgi network and the plasma membrane, and are required for salinity stress tolerance. The Plant Journal 73, 240–249. [DOI] [PubMed] [Google Scholar]
- Bencivenga S, Simonini S, Benková E, Colombo L. 2012. The transcription factors BEL1 and SPL are required for cytokinin and auxin signaling during ovule development in Arabidopsis. The Plant Cell 24, 2886–2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benková E, Michniewicz M, Sauer M, Teichmann T, Seifertová D, Jürgens G, Friml J. 2003. Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115, 591–602. [DOI] [PubMed] [Google Scholar]
- Berson T, von Wangenheim D, Takáč T, Šamajová O, Rosero A, Ovečka M, Komis G, Stelzer EH, Šamaj J. 2014. Trans-Golgi network localized small GTPase RabA1d is involved in cell plate formation and oscillatory root hair growth. BMC Plant Biology 14, 252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown RC, Lemmon BE. 1999. Development of endosperm in Arabidopsis thaliana. Sexual Plant Reproduction 12, 32–42. [Google Scholar]
- Brunoud G, Wells DM, Oliva M, et al. . 2012. A novel sensor to map auxin response and distribution at high spatio-temporal resolution. Nature 482, 103–106. [DOI] [PubMed] [Google Scholar]
- Camacho L, Smertenko AP, Pérez-Gómez J, Hussey PJ, Moore I. 2009. Arabidopsis Rab-E GTPases exhibit a novel interaction with a plasma-membrane phosphatidylinositol-4-phosphate 5-kinase. Journal of Cell Science 122, 4383–4392. [DOI] [PubMed] [Google Scholar]
- Ceccato L, Masiero S, Sinha Roy D, Bencivenga S, Roig-Villanova I, Ditengou FA, Palme K, Simon R, Colombo L. 2013. Maternal control of PIN1 is required for female gametophyte development in Arabidopsis. PLoS One 8, e66148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow CM, Neto H, Foucart C, Moore I. 2008. Rab-A2 and Rab-A3 GTPases define a trans-golgi endosomal membrane domain in Arabidopsis that contributes substantially to the cell plate. The Plant Cell 20, 101–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen CA, King EJ. 1997. Megagametogenesis in Arabidopsis wild type and the Gf mutant. Sexual Plant Reproduction 10, 49–64. [Google Scholar]
- Drdová EJ, Synek L, Pečenková T, Hála M, Kulich I, Fowler JE, Murphy AS, Zárský V. 2013. The exocyst complex contributes to PIN auxin efflux carrier recycling and polar auxin transport in Arabidopsis. The Plant Journal 73, 709–719. [DOI] [PubMed] [Google Scholar]
- Faure JE, Rotman N, Fortuné P, Dumas C. 2002. Fertilization in Arabidopsis thaliana wild type: developmental stages and time course. The Plant Journal 30, 481–488. [DOI] [PubMed] [Google Scholar]
- Feraru E, Feraru MI, Asaoka R, Paciorek T, De Rycke R, Tanaka H, Nakano A, Friml J. 2012. BEX5/RabA1b regulates trans-Golgi network-to-plasma membrane protein trafficking in Arabidopsis. The Plant Cell 24, 3074–3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueiredo DD, Batista RA, Roszak PJ, Hennig L, Kohler C. 2016. Auxin production in the endosperm drives seed coat development in Arabidopsis. eLife 5, e20542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueiredo DD, Batista RA, Roszak PJ, Köhler C. 2015. Auxin production couples endosperm development to fertilization. Nature Plants 1, 15184. [DOI] [PubMed] [Google Scholar]
- Geldner N, Anders N, Wolters H, Keicher J, Kornberger W, Muller P, Delbarre A, Ueda T, Nakano A, Jürgens G. 2003. The Arabidopsis GNOM ARF-GEF mediates endosomal recycling, auxin transport, and auxin-dependent plant growth. Cell 112, 219–230. [DOI] [PubMed] [Google Scholar]
- Goh T, Uchida W, Arakawa S, Ito E, Dainobu T, Ebine K, Takeuchi M, Sato K, Ueda T, Nakano A. 2007. VPS9a, the common activator for two distinct types of Rab5 GTPases, is essential for the development of Arabidopsis thaliana. The Plant Cell 19, 3504–3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes AQ, Ali BR, Ramalho JS, Godfrey RF, Barral DC, Hume AN, Seabra MC. 2003. Membrane targeting of Rab GTPases is influenced by the prenylation motif. Molecular Biology of the Cell 14, 1882–1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutkowska M, Wnuk M, Nowakowska J, Lichocka M, Stronkowski MM, Swiezewska E. 2015. Rab geranylgeranyl transferase β subunit is essential for male fertility and tip growth in Arabidopsis. Journal of Experimental Botany 66, 213–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habets ME, Offringa R. 2014. PIN-driven polar auxin transport in plant developmental plasticity: a key target for environmental and endogenous signals. New Phytologist 203, 362–377. [DOI] [PubMed] [Google Scholar]
- Hála M, Soukupová H, Synek L, Zárský V. 2010. Arabidopsis RAB geranylgeranyl transferase beta-subunit mutant is constitutively photomorphogenic, and has shoot growth and gravitropic defects. The Plant Journal 62, 615–627. [DOI] [PubMed] [Google Scholar]
- Heisler MG, Ohno C, Das P, Sieber P, Reddy GV, Long JA, Meyerowitz EM. 2005. Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Current Biology 15, 1899–1911. [DOI] [PubMed] [Google Scholar]
- Javelle M, Marco CF, Timmermans M. 2011. In situ hybridization for the precise localization of transcripts in plants. Journal of Visualized Experiments 57, e3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaźmierczak A, Kusy D, Niinivehmas SP, Gmach J, Joachimiak Ł, Pentikäinen OT, Gendaszewska-Darmach E, Błażewska KM. 2017. Identification of the privileged position in the imidazo[1,2-a]pyridine ring of phosphonocarboxylates for development of Rab geranylgeranyl transferase (RGGT) inhibitors. Journal of Medicinal Chemistry 60, 8781–8800. [DOI] [PubMed] [Google Scholar]
- Kelley DR, Arreola A, Gallagher TL, Gasser CS. 2012. ETTIN (ARF3) physically interacts with KANADI proteins to form a functional complex essential for integument development and polarity determination in Arabidopsis. Development 139, 1105–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan D, Millar JL, Girard IJ, et al. . 2015. Transcriptome atlas of the Arabidopsis funiculus—a study of maternal seed subregions. The Plant Journal 82, 41–53. [DOI] [PubMed] [Google Scholar]
- Kirchhelle C, Chow CM, Foucart C, et al. . 2016. The specification of geometric edges by a plant rab GTPase is an essential cell-patterning principle during organogenesis in arabidopsis. Developmental Cell 36, 386–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klepikova AV, Kasianov AS, Gerasimov ES, Logacheva MD, Penin AA. 2016. A high resolution map of the Arabidopsis thaliana developmental transcriptome based on RNA-seq profiling. The Plant Journal 88, 1058–1070. [DOI] [PubMed] [Google Scholar]
- Klepikova AV, Logacheva MD, Dmitriev SE, Penin AA. 2015. RNA-seq analysis of an apical meristem time series reveals a critical point in Arabidopsis thaliana flower initiation. BMC Genomics 16, 466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh EJ, Kwon YR, Kim KI, Hong SW, Lee H. 2009. Altered ARA2 (RABA1a) expression in Arabidopsis reveals the involvement of a Rab/YPT family member in auxin-mediated responses. Plant Molecular Biology 70, 113–122. [DOI] [PubMed] [Google Scholar]
- Köhnke M, Delon C, Hastie ML, Nguyen UT, Wu YW, Waldmann H, Goody RS, Gorman JJ, Alexandrov K. 2013. Rab GTPase prenylation hierarchy and its potential role in choroideremia disease. PLoS One 8, e81758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson E, Vivian-Smith A, Offringa R, Sundberg E. 2017. Auxin homeostasis in arabidopsis ovules is anther-dependent at maturation and changes dynamically upon fertilization. Frontiers in Plant Science 8, 1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le J, Liu XG, Yang KZ, et al. . 2014. Auxin transport and activity regulate stomatal patterning and development. Nature Communications 5, 3090. [DOI] [PubMed] [Google Scholar]
- Lituiev DS, Krohn NG, Müller B, Jackson D, Hellriegel B, Dresselhaus T, Grossniklaus U. 2013. Theoretical and experimental evidence indicates that there is no detectable auxin gradient in the angiosperm female gametophyte. Development 140, 4544–4553. [DOI] [PubMed] [Google Scholar]
- Luo J, Zhou JJ, Zhang JZ. 2018. Aux/IAA gene family in plants: molecular structure, regulation, and function. International Journal of Molecular Science 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luschnig C, Vert G. 2014. The dynamics of plant plasma membrane proteins: PINs and beyond. Development 141, 2924–2938. [DOI] [PubMed] [Google Scholar]
- Markham JE, Molino D, Gissot L, Bellec Y, Hématy K, Marion J, Belcram K, Palauqui JC, Satiat-Jeunemaître B, Faure JD. 2011. Sphingolipids containing very-long-chain fatty acids define a secretory pathway for specific polar plasma membrane protein targeting in Arabidopsis. The Plant Cell 23, 2362–2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moosajee M, Tulloch M, Baron RA, Gregory-Evans CY, Pereira-Leal JB, Seabra MC. 2009. Single choroideremia gene in nonmammalian vertebrates explains early embryonic lethality of the zebrafish model of choroideremia. Investigative Ophthalmology & Visual Science 50, 3009–3016. [DOI] [PubMed] [Google Scholar]
- Mroue S, Simeunovic A, Robert HS. 2018. Auxin production as an integrator of environmental cues for developmental growth regulation. Journal of Experimental Botany 69, 201–212. [DOI] [PubMed] [Google Scholar]
- Naramoto S, Nodzyłski T, Dainobu T, Takatsuka H, Okada T, Friml J, Fukuda H. 2014. VAN4 encodes a putative TRS120 that is required for normal cell growth and vein development in Arabidopsis. Plant & Cell Physiology 55, 750–763. [DOI] [PubMed] [Google Scholar]
- Novick P 2016. Regulation of membrane traffic by Rab GEF and GAP cascades. Small GTPases 7, 252–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paciorek T, Zazímalová E, Ruthardt N, et al. . 2005. Auxin inhibits endocytosis and promotes its own efflux from cells. Nature 435, 1251–1256. [DOI] [PubMed] [Google Scholar]
- Pagnussat GC, Alandete-Saez M, Bowman JL, Sundaresan V. 2009. Auxin-dependent patterning and gamete specification in the Arabidopsis female gametophyte. Science 324, 1684–1689. [DOI] [PubMed] [Google Scholar]
- Panoli A, Martin MV, Alandete-Saez M, Simon M, Neff C, Swarup R, Bellido A, Yuan L, Pagnussat GC, Sundaresan V. 2015. Auxin import and local auxin biosynthesis are required for mitotic divisions, cell expansion and cell specification during female gametophyte development in Arabidopsis thaliana. PLoS One 10, e0126164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paque S, Weijers D. 2016. Q&A: Auxin: the plant molecule that influences almost anything. BMC Biology 14, 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Ilarslan H, Wurtele ES, Bassham DC. 2011. AtRabD2b and AtRabD2c have overlapping functions in pollen development and pollen tube growth. BMC Plant Biology 11, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrasek J, Friml J. 2009. Auxin transport routes in plant development. Development 136, 2675–2688. [DOI] [PubMed] [Google Scholar]
- Pfeffer SR 2017. Rab GTPases: master regulators that establish the secretory and endocytic pathways. Molecular Biology of the Cell 28, 712–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinheiro H, Samalova M, Geldner N, Chory J, Martinez A, Moore I. 2009. Genetic evidence that the higher plant Rab-D1 and Rab-D2 GTPases exhibit distinct but overlapping interactions in the early secretory pathway. Journal of Cell Science 122, 3749–3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto SC, Mendes MA, Coimbra S, Tucker MR. 2019. Revisiting the female germline and its expanding toolbox. Trends in Plant Science 24, 455–467. [DOI] [PubMed] [Google Scholar]
- Robert HS, Grunewald W, Sauer M, Cannoot B, Soriano M, Swarup R, Weijers D, Bennett M, Boutilier K, Friml J. 2015. Plant embryogenesis requires AUX/LAX-mediated auxin influx. Development 142, 702–711. [DOI] [PubMed] [Google Scholar]
- Robert HS, Park C, Gutièrrez CL, et al. . 2018. Maternal auxin supply contributes to early embryo patterning in Arabidopsis. Nature Plants 4, 548–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert S, Kleine-Vehn J, Barbez E, et al. . 2010. ABP1 mediates auxin inhibition of clathrin-dependent endocytosis in Arabidopsis. Cell 143, 111–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojek J, Kapusta M, Kozieradzka-Kiszkurno M, Majcher D, Górniak M, Sliwinska E, Sharbel TF, Bohdanowicz J. 2018. Establishing the cell biology of apomictic reproduction in diploid Boechera stricta (Brassicaceae). Annals of Botany 122, 513–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford S, Moore I. 2002. The Arabidopsis Rab GTPase family: another enigma variation. Current Opinion in Plant Biology 5, 518–528. [DOI] [PubMed] [Google Scholar]
- Schneitz K, Hulskamp M. 1995. Wild-type ovule development in Arabidopsis thaliana: a light microscope study of cleared whole-mount tissue. The Plant Journal 7, 731–749. [Google Scholar]
- Schruff MC, Spielman M, Tiwari S, Adams S, Fenby N, Scott RJ. 2006. The AUXIN RESPONSE FACTOR 2 gene of Arabidopsis links auxin signalling, cell division, and the size of seeds and other organs. Development 133, 251–261. [DOI] [PubMed] [Google Scholar]
- Seabra MC, Goldstein JL, Sudhof TC, Brown MS. 1992. Rab geranylgeranyl transferase. A multisubunit enzyme that prenylates GTP-binding proteins terminating in Cys-X-Cys or Cys-Cys. Journal of Biological Chemistry 267, 14497–14503. [PubMed] [Google Scholar]
- Shahinian S, Silvius JR. 1995. Doubly-lipid-modified protein sequence motifs exhibit long-lived anchorage to lipid bilayer membranes. Biochemistry 34, 3813–3822. [DOI] [PubMed] [Google Scholar]
- Shi W, Zeng Q, Kunkel BN, Running MP. 2016. Arabidopsis Rab geranylgeranyltransferases demonstrate redundancy and broad substrate specificity in vitro. Journal of Biological Chemistry 291, 1398–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirley NJ, Aubert MK, Wilkinson LG, Bird DC, Lora J, Yang X, Tucker MR. 2019. Translating auxin responses into ovules, seeds and yield: insight from Arabidopsis and the cereals. Journal of Integrative Plant Biology 61, 310–336. [DOI] [PubMed] [Google Scholar]
- Silvius JR, l’Heureux F. 1994. Fluorimetric evaluation of the affinities of isoprenylated peptides for lipid bilayers. Biochemistry 33, 3014–3022. [DOI] [PubMed] [Google Scholar]
- Simonini S, Deb J, Moubayidin L, Stephenson P, Valluru M, Freire-Rios A, Sorefan K, Weijers D, Friml J, Østergaard L. 2016. A noncanonical auxin-sensing mechanism is required for organ morphogenesis in Arabidopsis. Genes & Development 30, 2286–2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth DR, Bowman JL, Meyerowitz EM. 1990. Early flower development in Arabidopsis. The Plant Cell 2, 755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H, Kitakura S, Rakusová H, Uemura T, Feraru MI, De Rycke R, Robert S, Kakimoto T, Friml J. 2013. Cell polarity and patterning by PIN trafficking through early endosomal compartments in Arabidopsis thaliana. PLoS Genetics 9, e1003540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H, Nodzylski T, Kitakura S, Feraru MI, Sasabe M, Ishikawa T, Kleine-Vehn J, Kakimoto T, Friml J. 2014. BEX1/ARF1A1C is required for BFA-sensitive recycling of PIN auxin transporters and auxin-mediated development in Arabidopsis. Plant & Cell Physiology 55, 737–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoma NH, Iakovenko A, Kalinin A, Waldmann H, Goody RS, Alexandrov K. 2001. Allosteric regulation of substrate binding and product release in geranylgeranyltransferase type II. Biochemistry 40, 268–274. [DOI] [PubMed] [Google Scholar]
- Tucker MR, Araujo AG, Paech NA, Hecht V, Schmidt ED, Rossel J, Vries SC, Koltunow AM. 2003. Sexual and apomictic reproduction in Hieracium subgenus pilosella are closely interrelated developmental pathways. The Plant Cell 15, 524–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker MR, Okada T, Hu Y, Scholefield A, Taylor JM, Koltunow AM. 2012a Somatic small RNA pathways promote the mitotic events of megagametogenesis during female reproductive development in Arabidopsis. Development 139, 1399–1404. [DOI] [PubMed] [Google Scholar]
- Tucker MR, Okada T, Johnson SD, Takaiwa F, Koltunow AM. 2012b Sporophytic ovule tissues modulate the initiation and progression of apomixis in Hieracium. Journal of Experimental Botany 63, 3229–3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieten A, Vanneste S, Wisniewska J, Benková E, Benjamins R, Beeckman T, Luschnig C, Friml J. 2005. Functional redundancy of PIN proteins is accompanied by auxin-dependent cross-regulation of PIN expression. Development 132, 4521–4531. [DOI] [PubMed] [Google Scholar]
- Wang JG, Feng C, Liu HH, Ge FR, Li S, Li HJ, Zhang Y. 2016. HAPLESS13-mediated trafficking of STRUBBELIG is critical for ovule development in Arabidopsis. PLoS Genetics 12, e1006269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb MC, Gunning BES. 1990. Embryo sac development in Arabidopsis thaliana—I. Megasporogenesis, including the microtubular cytoskeleton. Sexual Plant Reproduction 3, 244–256. [Google Scholar]
- Wu MF, Tian Q, Reed JW. 2006. Arabidopsis microRNA167 controls patterns of ARF6 and ARF8 expression, and regulates both female and male reproduction. Development 133, 4211–4218. [DOI] [PubMed] [Google Scholar]
- Zádníková P, Petrásek J, Marhavy P, et al. . 2010. Role of PIN-mediated auxin efflux in apical hook development of Arabidopsis thaliana. Development 137, 607–617. [DOI] [PubMed] [Google Scholar]
- Zheng H, Camacho L, Wee E, Batoko H, Legen J, Leaver CJ, Malhó R, Hussey PJ, Moore I. 2005. A Rab-E GTPase mutant acts downstream of the Rab-D subclass in biosynthetic membrane traffic to the plasma membrane in tobacco leaf epidermis. The Plant Cell 17, 2020–2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou LZ, Dresselhaus T. 2019. Friend or foe: signaling mechanisms during double fertilization in flowering seed plants. Current Topics in Developmental Biology 131, 453–496. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the findings of this study are available within the paper and within its supplementary data published online.