Abstract
The combined development of new technologies for neuronal recordings and the development of novel sensors for recording both cellular activity and neurotransmitter binding has ushered in a new era for the field of neuroscience. Among these new technologies is fiber photometry, a technique wherein an implanted fiber optic is used to record signals from genetically encoded fluorescent sensors in bulk tissue. Fiber photometry has been widely adapted due to its cost-effectiveness, ability to examine the activity of neurons with specific anatomical or genetic identities, and the ability to use these highly modular systems to record from one or more sensors or brain sites in both superficial and deep-brain structures. Despite these many benefits, one major hurdle for laboratories adopting this technique is the steep learning curve associated with the analysis of fiber photometry data. This has been further complicated by a lack of standardization in analysis pipelines. In the present communication, we present pMAT, a ‘photometry modular analysis tool’ that allows users to accomplish common analysis routines through the use of a graphical user interface. This tool can be deployed in MATLAB and edited by more advanced users, but is also available as an independently deployable, open-source application.
Keywords: Fiber Photometry, Calcium Imaging, Neural Recording, Open Source
1.0. Introduction
Fiber photometry is a tool that allows for the recording of bulk fluorescent signals from a growing number of sensors including those for detecting levels of cellular activity, through the use of sensitive changes in intracellular calcium or the binding of neurotransmitters, such as GCaMP61, jGCaMP72, jRCAMP1a,b or jRGECO1a3. In addition, a number of sensors have recently been developed for detecting the binding of a wide range of specific neurotransmitters (Table 1)4–10. These sensors may be virally delivered or genetically encoded and virally activated.
Table 1. Genetically Encoded Sensors.
A list of genetically encoded fluorescent sensors, including the most up-to date information regarding whether these have been used effectively for fiber photometry calcium imaging.
| Sensor Name | Validated for Fiber Photometry | Relevant Citations | Lab/Individual contributing validation data or validating citation |
|---|---|---|---|
| GCaMP6 (f,m,s) | Y | Gunaydin et al., 2014 | Barker Lab (David Barker) |
| jGCaMP7 (b,s,f,c) | Y | Dana et al., 2019 | Rothwell Lab (Marc Pisansky) |
| jRCAMP1a,b or jRGECO1a | Y | Dana et al., 2016; Kim et al., 2016 | Lerner Lab (Jillian Seiler) |
| dLight-1.1 | Y | Patriarchi et al., 2018 | Lerner Lab (Jillian Seiler); Papaleo Lab (Daniel Dautan) |
| iGluSnFR | Y | Marvin et al., 2018; McGovern et al., 2020 | Root Lab (Dillon McGovern) |
| iGABASnFR | Y | Marvin et al., 2019; McGovern et al., 2020 | Root Lab (Dillon McGovern) |
| iAChSnFr | Y | Borden et al., 2020; McGovern et al., 2020 | Root Lab (Dillon McGovern) |
| iSeroSnFr | TBD | Unger et al., 2019 | Lin Tian (Chunyang Dong) |
| GRABDA | Y | Sun et al., 2018 | Cheer Lab (Kate Peters) |
| GRABNE | Y | Feng et al., 2019 | Feng et al., 2019 |
| GRAB-5HT | Y | Wan et al., 2020 | Wan et al., 2020 |
| iATPSnFR | TBD | Lobas et al., 2019 | |
| iGlucoSnFR | TBD | Keller et al. 2019 |
A major advantage of fiber photometry is its ease of use and capability to conduct high-throughput experiments. Recordings are accomplished via lightweight fiber optics that provide little interference in most behavioral apparatuses. Moreover, the inclusion of an internal control channel (i.e., an ‘isosbestic control’ channel)11 provides a clever method for subtracting movement artifacts that are generated during the task, as well as aid in the correction of other changes such as photobleaching or LED stability, among others. Fiber photometry can reliably record activity at cell bodies12 as well as axon terminals13 and has proven especially useful for repeated or longitudinal recordings14, 15 or for recording from deep-brain structures16 Lastly, fiber photometry provides the ability to conduct circuit-specific recordings. This can be accomplished by injecting viral vectors (e.g., GCaMP) at the soma for a target group of cells while placing the fiber optic over distally located axon terminals13 or by injecting retrograde viruses (e.g., retrograde adeno-associates or herpes simplex viruses) at the site of axon terminals and placing the fiber optic over the cell bodies from which those terminals originate (Barker et al. unpublished data).
Following a landmark paper by Gunaydin and colleagues17, there has been an exponential growth in the use of fiber photometry within the neuroscience field (Figure 1). Accordingly, there has been a concomitant increase in the number of open-source e.g.,18 or commercially available systems for the acquisition of fiber photometry data, including recent movements towards wireless systems. Nonetheless, the analysis of fiber photometry data is most often accomplished via custom programming scripts, creating a barrier for many laboratories as they struggle to tackle the analysis of the large datasets that can be amassed using the technique. Additionally, efforts to standardize methods of analysis for fiber photometry have remained stagnant, while multiple widely-used analysis packages exist for other imaging methods such as single-cell or two-photon calcium imaging. Thus, the goal of the present manuscript was to develop an open-source analysis suite capable of accomplishing basic tasks required for the visualization and analysis of fiber photometry data. The present Photometry Modular Analysis Tool or ‘pMAT’ can be downloaded and deployed as an executable file (‘.exe’) that runs independent of MATLAB and requires no previous coding experience to run. Alternatively, pMAT can be downloaded for use within MATLAB, either by individuals with no previous coding experience, or by advanced users that wish to contribute to future development. In addition to alleviating the programming barrier to entry, the analysis suite provides a starting point for the standardization of analysis protocols and can be expanded to support analysis for most of the common data acquisition systems currently on the market.
Figure 1. Growth in references to ‘fiber photometry’ since its inception.
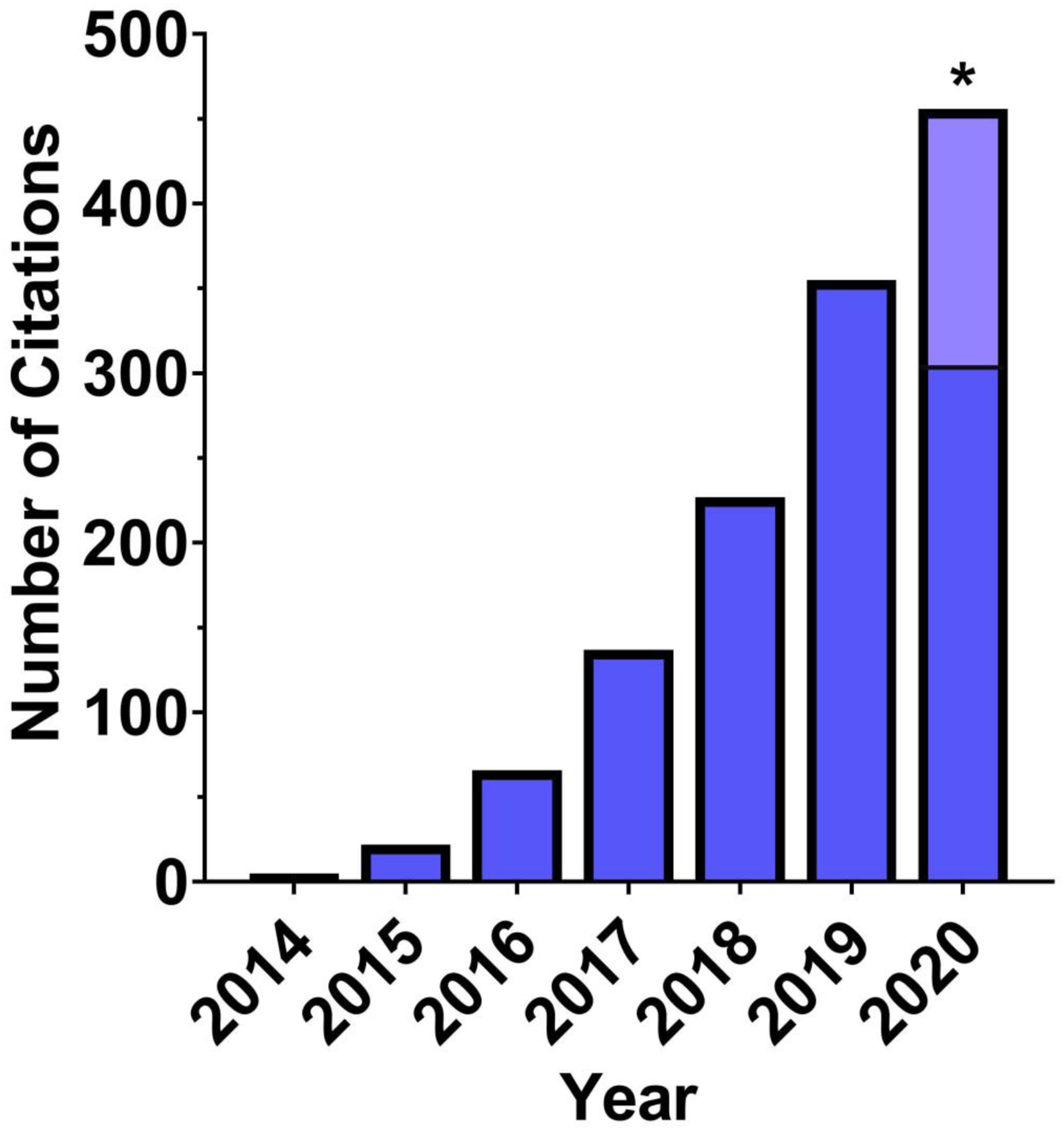
The usage and mentions for fiber photometry has increased rapidly since 2014 and continues to grow yearly at an exponential pace. The data here represents the number of Google Scholar mentions for ‘fiber photometry’, which includes papers using the technique as well as review articles and technical papers. * Projected 2020 papers, light blue; current, dark blue.
2.0. Recording Parameters for Testing
For all internal recordings using to validate the software pipeline, GCaMP6 was excited at two wavelengths, a 490nm, calcium-dependent signal and 405 nm isosbestic control11, by amplitude modulated signals from two light-emitting diodes reflected off dichroic mirrors and coupled into a 400μm 0.48NA optic fiber. The isosbestic control channel (henceforth termed, ‘control’) represents the point of GCaMP excitation at which emissions from the calcium bound and unbound state have equal intensities. Thus, the control channel represents GCaMP emissions that are independent of calcium, but still susceptible to artifacts related to movement, fiber bending, and all other physical alterations. Signals emitted from GCaMP6 and its isosbestic control channel then returned through the same optic fiber and were acquired using a femtowatt photoreceiver (Model 2151; Newport), digitized at 1kHz, and then recorded by a real-time signal processor (RZ5D) from Tucker Davis Technologies (TDT) running the Synapse software suite. Behavioral inputs were collected using the digital input and output ports (DIO) using either a DIG-726TTL-G card (Med-Associates Inc.) that was connected to the TDT system with a custom-soldered DB-25 cable or using an SG-231 28V DC to TTL adapter (Med-Associates Inc) connected to the TDT system using a BNC connection. Both setups have been tested for temporal precision to verify the synchrony of behavioral data and fiber photometry data. The temporal prevision was always of <1 ms (~250 μs, on average).
3.0. Installation of the pMAT suite
3.1. Installation of pMAT as an independent program.
To run pMAT as an independent ‘.exe’ file, one of the pMAT ‘independent deployment’ folders should be downloaded from https://github.com/djamesbarker/pMAT. The folder should be unzipped and saved to your computer. Open the pMAT deployment folder and click the application named ‘pmat installer’. The default path is ‘C:\Program Files\pmat-BarkerLab’. This process will install both the pMAT software and the free MATLAB runtime environment, which allows you to use the program as an executable program without the need for a MATLAB license. At the time of this manuscript, the MATLAB 2020a runtime (i.e., runtime 9.8) is being used for deployment.
3.2. Installation of pMAT suite in MATLAB.
For installation in MATLAB, pMAT should be downloaded from https://github.com/djamesbarker/pMAT and saved to a Windows PC. It is recommended that users create a directory called “C:\pmat-BarkerLab\”, as this is the location that will be used for saving pMAT settings files. Once downloaded, it is important to open MATLAB and use the pathtool command (type ‘pathtool’ into the command window) to add and save the pMAT folder and all of its subfolders to the MATLAB path. pMAT is then run by typing pmat into the MATLAB command window. pMAT was originally developed with MATLAB 2019a, so we recommend using this version or newer.
3.3. Tutorials and sample data.
Video tutorials for the pMAT suite along with sample data in comma separated value (.CSV) or Tucker Davis Technologies (TDT) formats can be viewed and downloaded by visiting https://github.com/djamesbarker/pMAT. These files are useful for seeing the appropriate formatting needed to work with pMAT and can also be used to familiarize yourself with the software. As pMAT extends to support additional systems and file-types, these examples will continue to be updated. We recommend storing these files in a folder that is easy to access (e.g., ‘C:\pmat-BarkerLab’).
4.0. Features of the pMAT suite
4.1. Data Loading and Control Module.
The pMAT suite was initially designed around the Tucker Davis Technologies (TDT) recording platform, as many of the earliest fiber photometry systems were run on this hardware and software, which provides synchrony between behavioral data and neural signals. The first step in loading data is to import data. pMAT allows you to process either single files or whole folders full of files using ‘batch processing’. If selecting a single file, you will be prompted to select the specific folder containing your data. If selecting batch processing, an entire folder (containing many TDT data subfolders) can be selected for processing. When establishing your analysis pipeline, we recommend that users name their signal channel as ‘490’ in the TDT system and their control channel as ‘405’. These names will be automatically recognized in pMAT. Users with differing nomenclature will be prompted to select their signal and control channels from an automatically populated list.
Loading a single file allows the user to interact with the other modules in the pMAT suite. Each section can be run independently using buttons within the module, or the entire suite can be set up at once and executed using the ‘Run all Selections’ button within the control module. A similar approach is taken for batch processing. The user interacts with the first file to set up the parameters for batch processing and then runs all of the files in the folder by pressing the ‘Run All Selections’ button (Figure 2A).
Figure 2. Overview of the pMAT suite.
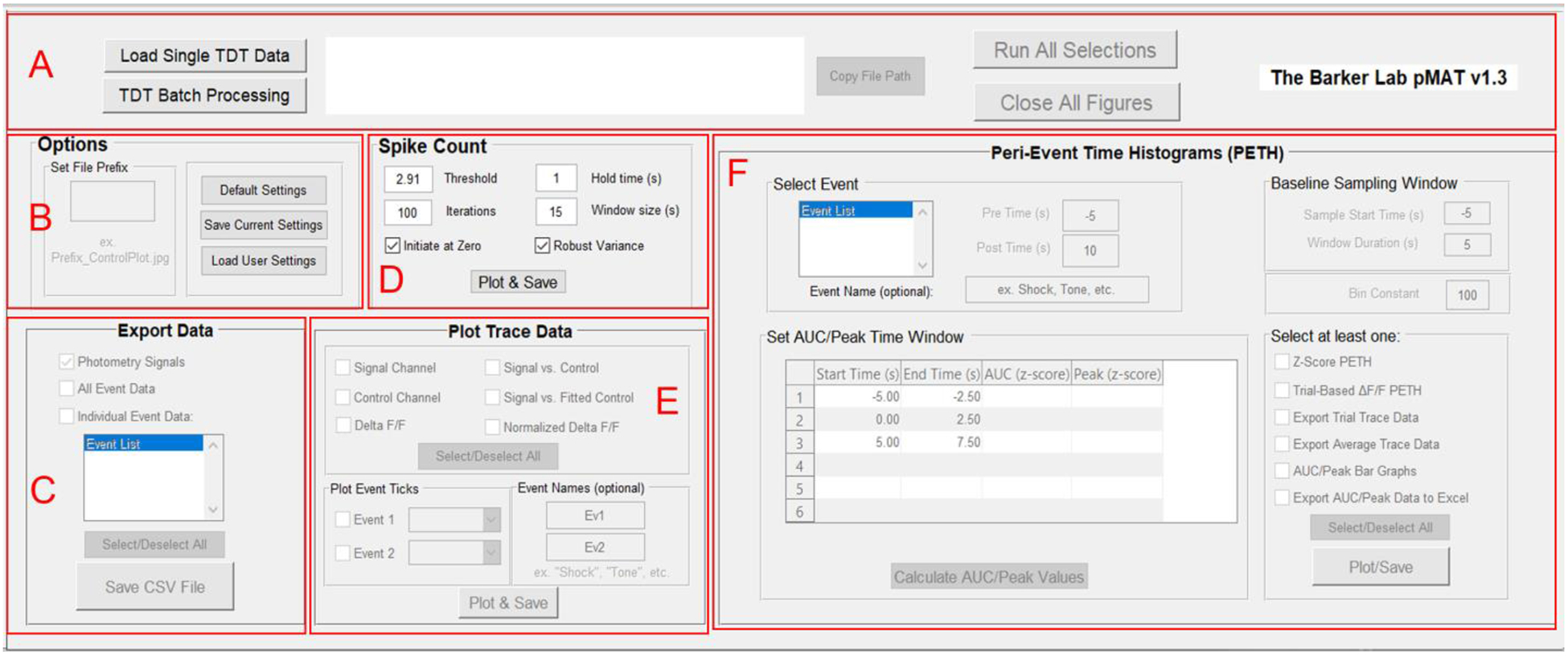
A) The control module allows for loading single files or batch processing and provides an interface of buttons to run selections across all modules or to close all of the figures generated when running the code.
B) The Options module is used for setting file prefixes as well as saving and loading pMAT options.
C) The Export Data module provides options for exporting signal, control, or event data into comma separated value (‘.CSV’) files that can be imported into other programs such as Microsoft Excel or R.
D) The Spike Count module allows users to conduct an iterative smoothing and reliable count of spikes or transients in their data.
E) The Plot Trace Data module allows for the plotting of signal and control channels, as well as the stepwise evaluation of the signal correction routines used to generate the ΔF/F. The module also allows for the visualization of events across time on the ΔF/F plots.
F) The Peri-Event Time Histogram module allows for the evaluation of the signal around an event of interest and provides the tools for the plotting and exporting of relevant data for the peri-event traces or summary statistics.
4.2. Start/End Buffer.
Upon loading a file, the user will be prompted to buffer the file by cutting out the first x seconds from the beginning and end of the recording. A time buffer is especially practical at the beginning of each session. In most commercial systems, LEDs are turned on at the start of the session, leading to a dramatic peak in the signal as the LEDs turn on and achieve maximum brightness. In addition, photobleaching is exponential and often greatest in the first few seconds of the recording. These sharp changes in signal can lead to issues with the scaling and plotting of data, among other things. Therefore, we recommend removing a buffer of 2–5 second (default 2 seconds) from the beginning and end of the recording file.
4.3. Loading Data from other systems.
Data from other systems can be loaded into the pMAT suite via the ‘Import Data’ menu. These data should be formatted in a Comma-separated value or ‘CSV’ format, with individual files for the fiber photometry recording data and event data. Fiber photometry data should be in a 3-column format that can include headers. The first column should include timestamps for the recording data, the second column should include the signal channel data, and the third column should include the control channel data. The event file should also follow a 3-column format, with the first column containing an event name (text/string), the second column containing the start/onset time for the event and the third column containing the stop/offset time for the event. CSV files are useful, as they can be generated in Microsoft Excel, Google Sheets, or equivalents (i.e., using the ‘Save As’ option) and are a universal data format that can be read by many applications. Ultimately this format accommodates many other commercial recording systems, although specific import tools are also planned in future releases.
4.4. Appending additional timestamps.
Once a file has been loaded, users can append additional event data (e.g., video scored events) from a ‘.CSV’ file using ‘Append Event Data’ selection from the ‘Import Data’ menu. The event file should follow a 3-column format, with the first column containing an event name (text/string), the second column containing the start/onset time for the event and the third column containing the stop/offset time for the event.
4.5. Options Module.
Settings for the state of the pMAT suite can be saved by the user for future use, or for collaboration across laboratories (Figure 2B). Settings that can be saved include file prefixes, checkbox statuses, list box selections, popup selections, as well as all user input text boxes (pre/post times, baseline sampling window and bin constant). The user can also set a file prefix to help organize data files (e.g., by adding an experiment name, dosing group, etc.) or to help differentiate files (e.g., by adding a version number). Upon closing, pMAT will automatically prompt users by asking whether they wish to save these settings (Figure 3A–D).
Figure 3. The pMAT options and Export Modules.
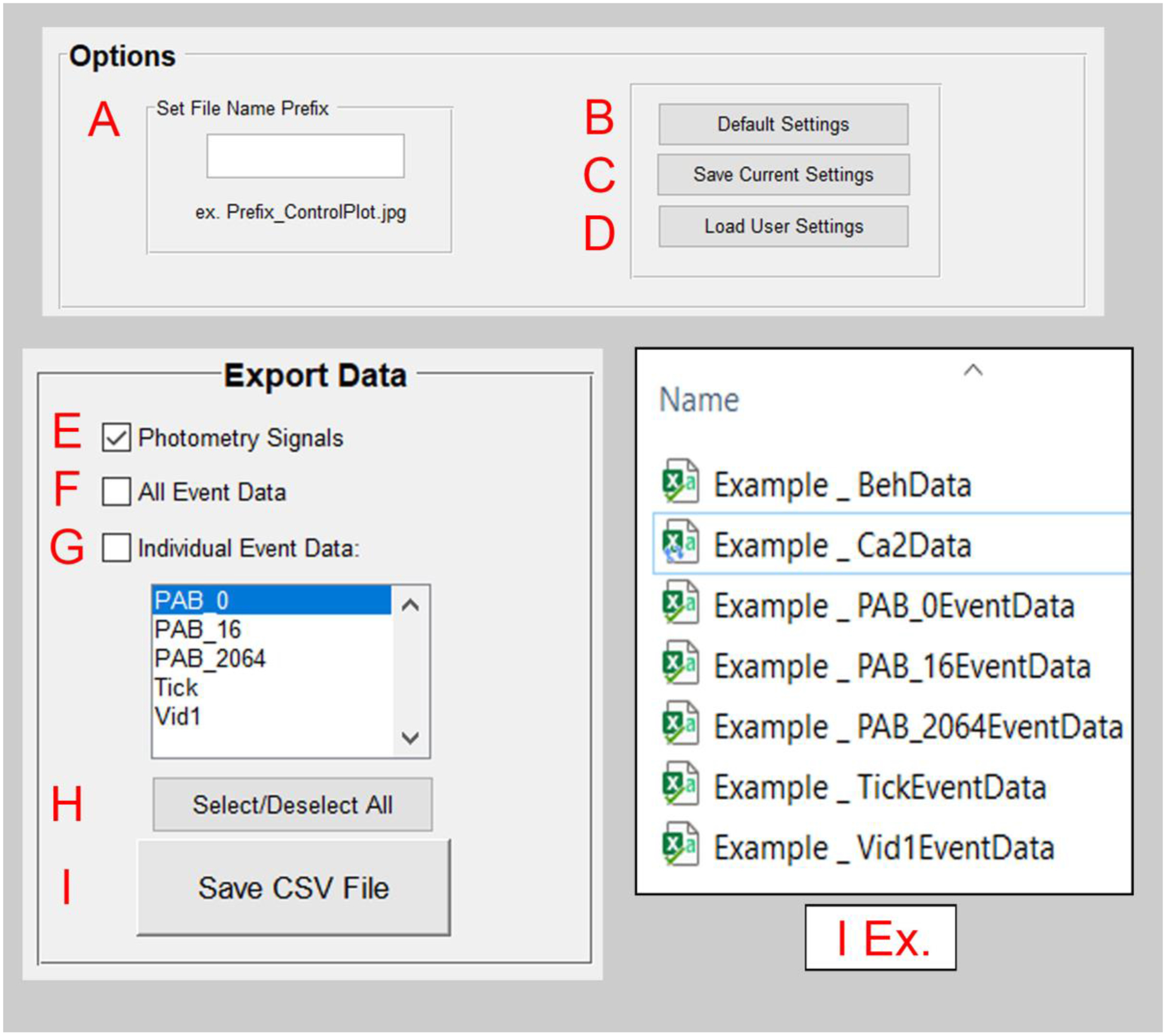
A) Within the options module, a file prefix can be set to help with the organization of files.
B) The ‘Default Settings’ button reverts all settings back to their original state.
C) The ‘Save Current Settings’ button allows for users to save settings for the state of the pMAT suite. Users will also be prompted to save their settings when exiting the program.
D) The ‘Load User Settings’ button allows users to restore previously saved settings, allowing for the reproducibility of routines.
E) The ‘Photometry Signals’ check box will store a ‘.CSV’ file with the signal and control channel data as well as their corresponding timestamps, once the ‘Save CSV File’ button (I) is pressed.
F) The ‘All Event Data’ check box will store a ‘.CSV’ file containing the tags for all behavioral events as well as their corresponding timestamps, once the ‘Save CSV File’ button (I) is pressed.
G) The ‘Individual Event Data’ check box will store a ‘.CSV’ file for each event that has been selected within the window below, once the ‘Save CSV File’ button (I) is pressed. Multiple events can be highlighted for export simultaneously.
H) Users can select or deselect all checkboxes simultaneously.
I) The ‘Save CSV File’ button executes the command to save data corresponding to the selected checkboxes in E-G.
4.6. Data Export Module.
At the most basic level, the pMAT suite allows the user to quickly export data in a universal ‘.CSV’ format that is compatible with Microsoft Excel, MATLAB, R, and Python, among others (Figure 2C). Three types of data can be exported: 1) a file containing calcium signals, control signals, and their corresponding timestamps, 2) a file containing all event data and timestamps, which correspond to all transistor-transistor logic (TTL) inputs to the recording hardware for behavioral or experimental events, and 3) selected event data and their corresponding timestamps. Each of these files are saved in the “Data” folder created within the corresponding data folder. Currently, the storage location of data is predetermined in order to facilitate the batch processing of data (Figure 3E–I). Exported data can be processed by other programs and even re-imported into pMAT as needed (see sections 4.3 and 4.4).
4.7. Plot Trace Data Module.
Traces across whole experimental sessions are constructed using a multi-step process. Each of these steps can each be individually visualized and evaluated by users with the tools provided in the ‘Plot Trace Data’ module of the GUI (Figure 2E). First, data from the signal and isosbestic control channels are extracted. Second, the channels are individually smoothed using a Lowess, local linear regression method to smooth the signal and reduce high-frequency noise (signal channel and control channel plots; Figure 4 A–C). As signal and control channel emissions generally have different levels of power (‘Signal vs. Control’ plot), the next step is to normalize the scale of the channels. This is accomplished by first using a least-squares regression to find the relationship between the signal and control channels (MATLAB polyfit function):
| eq.1. |
| eq.2. |
Where N is the number of coefficients, x is the control channel, and y is the signal channel. Next, the resulting slope and intercept are then used to generate a scaled control channel (‘Signal vs. Fitted Control’ plot; Figure 4D):
| eq.3. |
Finally, the delta F/F (ΔF/F) is generated by subtracting the fitted control channel from the signal channel. This subtraction is used to eliminate movement or other common artifacts (Figure 4E):
| eq.4. |
For the fully processed ΔF/F plots, the user also has the option to represent the whole session trace as a normalized z-score (z-score of eq. 4) of the trace and to include reference ticks showing the timing of up to two behavioral events on top of the whole-session trace (Figure 4 G–H).
Figure 4. The pMAT trace plotting module.
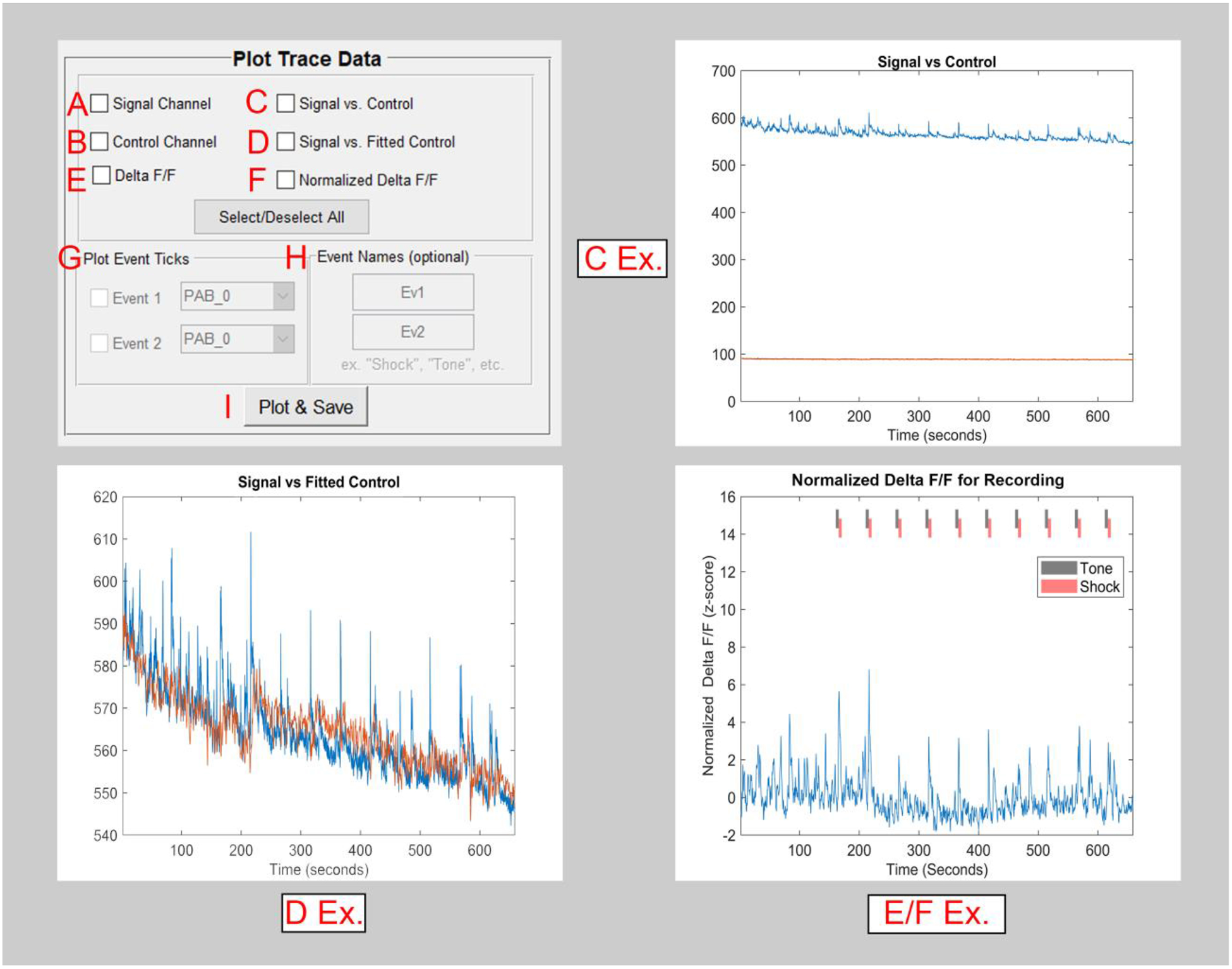
A) The checkbox for the ‘Signal Channel’ will independently plot data from a recorded sensor.
B) The checkbox for the ‘Control Channel’ will independently plot data from an isosbestic or autofluorescence control.
C) The signal and control channels can be plotted simultaneously in order to compare the various properties of these signals (e.g., rate of photobleaching).
D) The fitted control is overlaid onto the signal channel (D Ex.) in order to help visualize the scaling that has occurred and the corrections that will be made.
E) The Δ F/F is generated using the signal and fitted control. This shows the final stage of the calculated correction that occurs using the control channel.
F) A normalized (z-score) version of the Δ F/F is generated.
G) The event checkboxes allow for the visualization of events across the whole-session trace data. (e.g., the Tone and Shock ticks in the E/F Ex.)
H) Events can be given a custom name, as desired.
I) All checkboxes selected in the Plot and Trace Data module are run using the Plot and Save button.
4.8. Spike Count: Overview.
We sought to create an automatic method to quantify the number of spikes or ‘transients’ present within a fiber photometry recording. Counting these kinds of events has been valuable for differentiating and understanding neuronal activity19, 20. Nonetheless, the drift in optical signals due to photobleaching makes the accurate detection of spikes particularly challenging. To address these problems and improve upon existing threshold-based methods, we created an algorithm to first correct for photobleaching iteratively and then quantify spikes from the corrected signal. Detrending of the signal occurs by calculating the ΔF/F for small chunks of data across the entire dataset (Figure 5 D; user-defined by ‘Window Size’), centering and normalizing these windows, and then repeating this process by calculating these same values for many temporally offset windows across the same data. These iterations are then averaged to produce a final normalized ΔF/F signal. The number of iterations for this process is user-defined and defaults to 100 iterations (Figure 5C), which provides an effective balance between optimal detrending and data processing time.
Figure 5. The pMAT Spike Count Module.
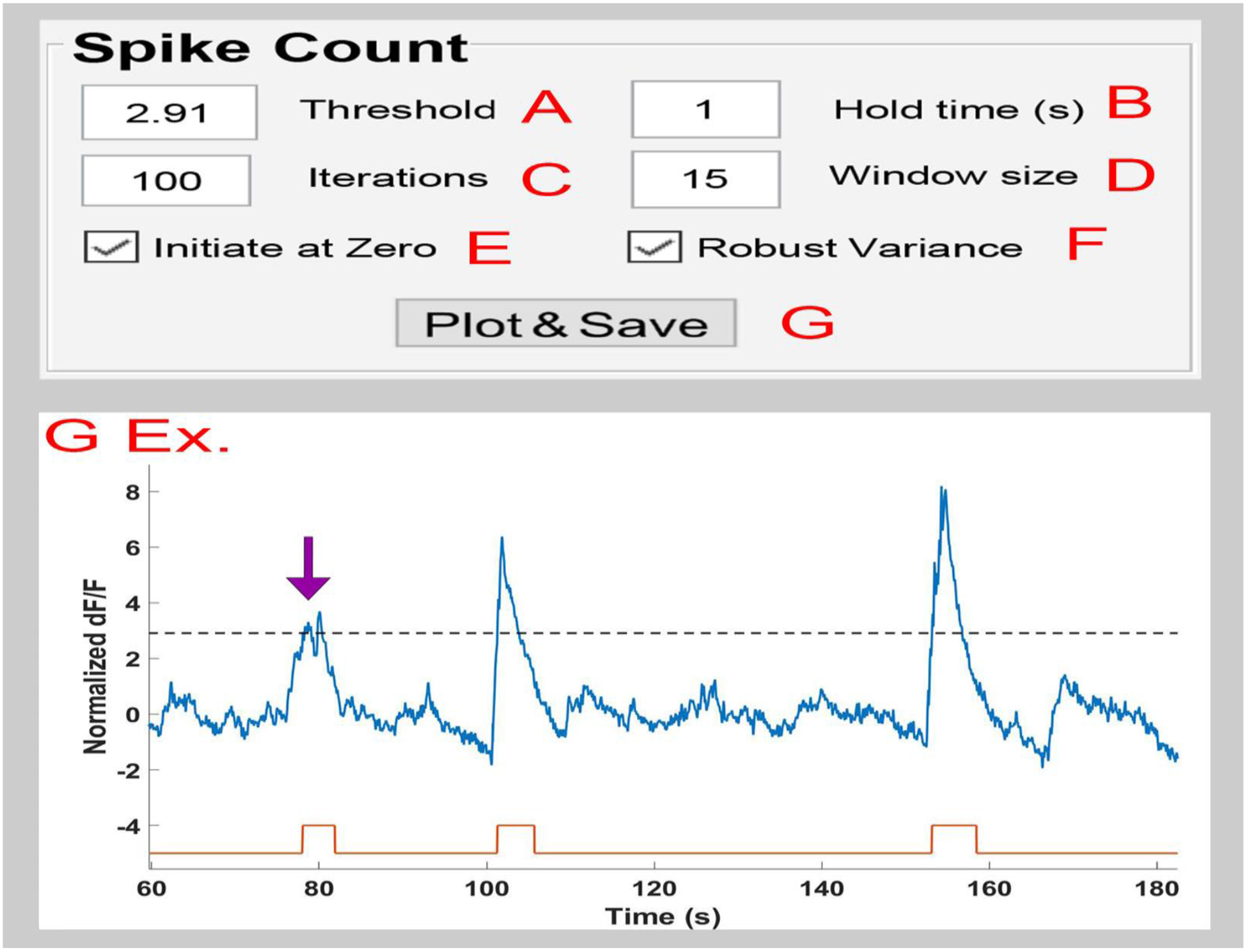
A) The ‘Threshold’ value sets the detection limit for counting spikes. This is represented by the dotted line in the returned plot (G Ex).
B) The ‘Hold Time’ value indicates the minimum amount of time that must exist between two events before counting them as separate. If multiple spikes occur during the hold time, they will be combined into a single event (purple arrow, G Ex.)
C) The ‘Iteration’ value determines the number of times that iterative de-bleaching will occur before averaging the results.
D) The ‘Window Size’ determines the bins of time used to center and normalize the data while creating the moving average.
E) The ‘Initialize at Zero’ checkbox indicates that spikes should be counted from the first zero value on the y-axis before a threshold crossing until the first zero value on the y-axis following the end of the spike.
F) The ‘Robust Variance’ checkbox employs a robust z-score approach using median absolute deviations when checked, and standard deviation when unchecked.
G) The ‘Plot & Save’ button initiates the spike detection using the settings from A-F and returns a plot showing the Δ F/F (blue trace), spike threshold (dotted line), and the start (black tick) and end (red tick) for each spike that was detected.
After the final detrending, events that surpass a user-defined threshold are counted as spikes (Figure 5A). Users are also given the option to group complex peaks or rapidly occurring events by setting the minimum amount of separation that must exist between two events (Figure 5B; ‘Hold Time’) and given the option to further define the spike duration by selecting whether spikes should start at the threshold crossing or at point where the data crosses zero on the y-axis (Figure 5E; ‘Initiate at Zero’ option). These data are then automatically saved as a plot as well as a list of spike times (Figure 5G).
4.9. Peri-Event Histograms Module: Overview.
Peri-event time histograms/heatmaps (PETHs) are a valuable tool for examining the stability of a response over repeated trials. To calculate PETHs in the pMAT suite, a user-defined event window is set around each event that begins x seconds before the event and ends y seconds after a selected event (Figure 6A). In addition, a baseline window is defined for a user-defined period of time preceding each event window (Figure 6B). For both the baseline window and event window, the ΔF/F is then calculated in the same manner as described above for the trace data (Section 4.4), albeit on a much shorter timescale. In addition, the data are centered following the ΔF/F calculation to set the first point in each trial to zero. These data are visualized using the “Trial-Based ΔF/F PETH” or “z-score PETH” feature (Figure 6D–E).
Figure 6. The pMAT peri-event time histogram module.
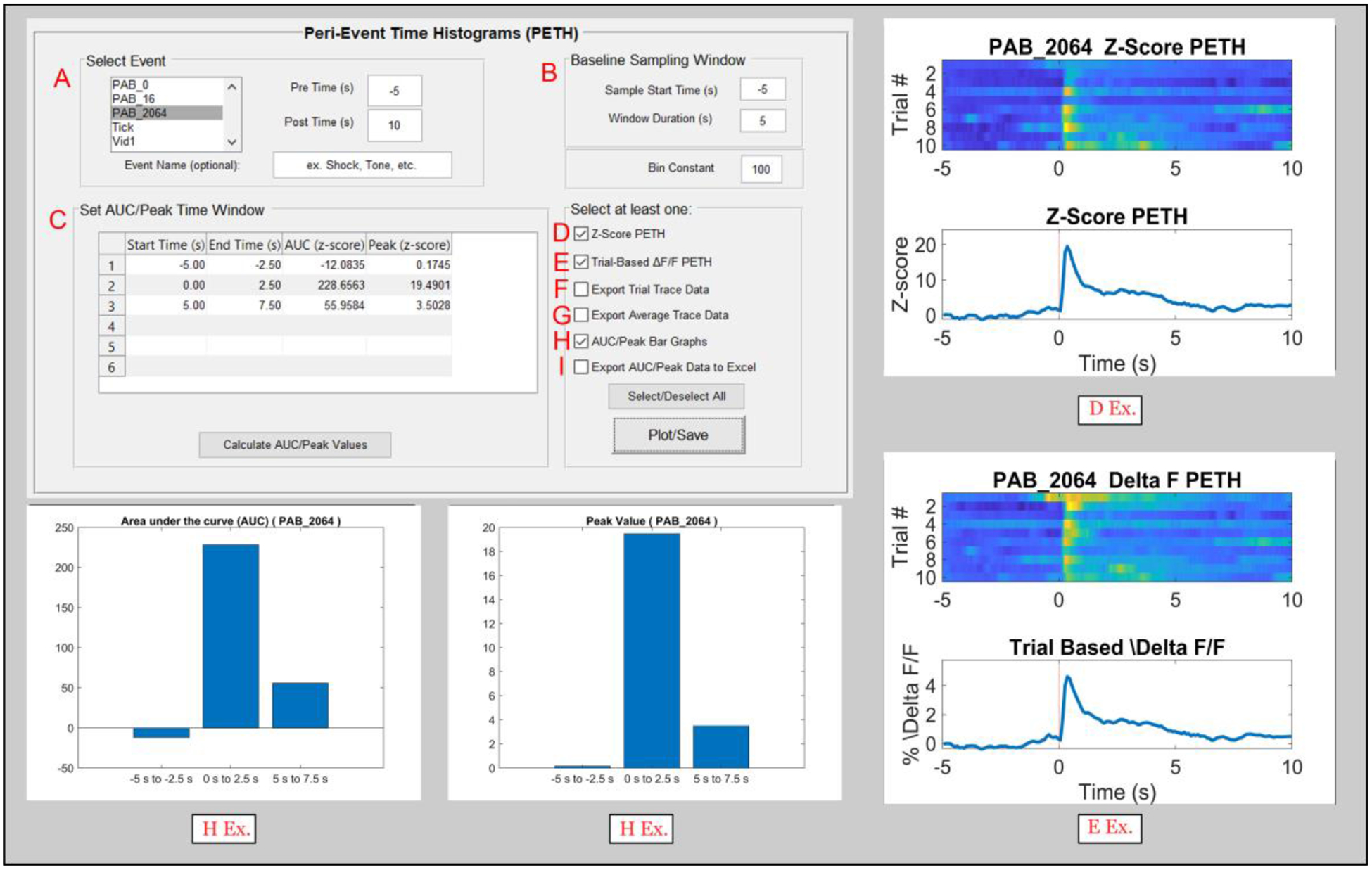
A) An event of interest can be selected to create a peri-event heatmap and histogram (PETH; D Ex. And E Ex.). A window of event before (Pre Time) and after (Post Time) can also be defined.
B) The user can then define a Baseline Sampling Window to act as a reference point for normalizing (robust z-score) data. In addition, a bin size for the event and baseline data can be defined.
C) The area under the curve (AUC) and ‘Peak’ or maximum value of a trace are two commonly used metrics collected for fiber photometry data. Users can input up to 6 values for the start and end times of specific windows and the AUC and Peak will be calculated for each of these windows. These values can also be plotted using the checkbox in H.
D) Selecting the ‘Z-Score PETH’ (peri event histogram and heatmap) will plot a normalized (robust z-score) heatmap and histogram aligned to the event selected in A. Warmer colors on the heatmap (top) represent higher fluorescent signals at that timepoint, while the line plot on the bottom represents the average for all trials corresponding to the event of interest (D Ex.).
E) The ‘Trial Based Δ F/F PETH’ will plot the absolute Δ F/F values on a peri event heatmap and histogram (E Ex.).
F) Data for individual trials of the Z-Score PETH (D; top) can be exported to a ‘.CSV’ file and saved for plotting or processing elsewhere.
G) Data for the average trace of the Z-Score PETH (D; bottom) can also be exported to a ‘.CSV’ file for plotting or processing elsewhere.
H) The checkbox for H produces two graphs, showing the AUC and Peak values defined by the user in the cells provided in C.
I) The AUC and Peak values can also be exported into a ‘.CSV’ file. These are commonly collated across experimental subjects or groups for statistical analysis.
The combination of this shortened correction window and zeroing procedure are designed to correct for changes in the stability of the signal that occur over long timescales due to photobleaching of fluorophores, photobleaching of fiber optic cables, and changes in the stability of other components for fiber photometry systems (i.e., LEDs and other electronics). The challenge of signals that weaken or change across long recordings is not unique to fiber photometry and has been effectively solved in electrochemistry experiments for some time using a similar approach21. Signals are then converted into a robust z-score using the following formula:
| eq.5. |
Traces for both the average trace, or the traces from individual trials can then be exported for further analysis or for plotting elsewhere (Figure 6 F–G).
4.10. Peri-Event Histograms Module: Area under the curve and peak calculations.
From the robust z-score, the area under the curve and peak values can be extracted from the PETH data for up to six different user-defined windows. These windows are defined by editing the cells in the “Set AUC/Peak Time Window” matrix (Figure 6C).
4.11.
The peak is calculated as the maximum value that occurs within each user-defined window. The area under the curve is calculated for the portion of the curve that falls within each user defined window using the trapezoidal method for integral calculation (MATLAB trapz function). These data can be plotted as a bar graph for examination and also exported in a ‘.CSV’ format for further analysis (Figure 6H–I).
4.12. Sensor Compatibility.
We are currently in an era where novel sensors are being developed at a rapid pace. Upon collecting external data for validating pMAT, we recognized that a comprehensive list of sensors that are compatible with fiber photometry does not yet exist. Many but not all of these sensors have been tested using fiber photometry in their original publications. With this in mind, we sought the help of numerous labs in order to provide an up-to-date list of the tested and un-tested sensors currently being used with fiber photometry. We also used this opportunity to evaluate these sensors within the pMAT suite, to ensure its broad compatibility with other sensors as well and experimental designs. From this we determined that all currently available sensors are compatible with fiber photometry and with this analysis pipeline. The list of sensors and relevant publications are found in Table 1, with plots for each sensor around behavioral events of interest in Figure 7. The specific behavioral events examined vary by the interests of the contributing laboratories. Nonetheless, behaviorally relevant increases, decreases, or biphasic responses can be seen in each peri-event histogram.
Figure 7. Examples of sensors compatible with fiber photometry calcium imaging.
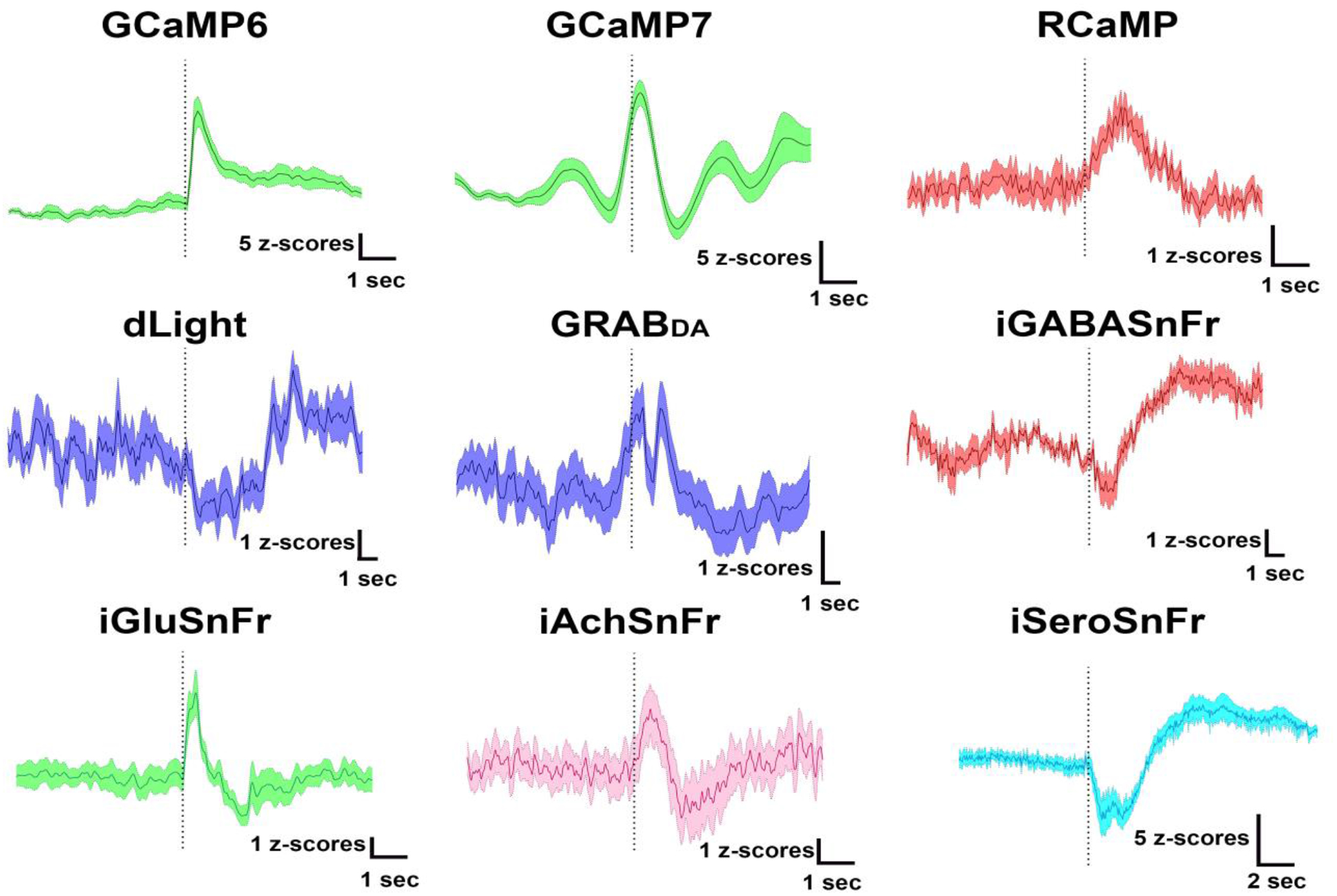
The mosaic figure above shows example peri-event traces from sensors used to detect cellular activity (GCaMP6, GCaMP7, or RCaMP) or to detect neurotransmitter binding for dopamine (dLight, GRABDA), GABA (iGABASnFr), glutamate (iGluSnFr), acetylcholine (iAchSnFr), or serotonin (iSEroSnFr). The traces were collected under different experimental conditions (e.g., reward delivery or footshock), but for each trace the fiber photometry data has been aligned to a relevant behavioral event (dotted line). As seen by the variability of responses, each of the currently available sensors is not only compatible with fiber photometry calcium imaging, but can also demonstrate behaviorally relevant increases, decreases, or even biphasic responses.
4.13. Future development goals.
The first iteration of pMAT is meant to support the most basic processing with the goal of getting the first iteration of the tool to end-users. However, the second iteration of the pMAT suite is already underway. The current plans for this iteration include a) increased compatibility for additional recordings or data formats, b) the ability to interface with other open source software platforms(e.g., DeepLabCut and SimBA) c) the ability to detrend bleached data without using regression modeling techniques d) the ability to generate multi-event histograms for events with fixed or variable intervals, and f) increased compatibility with tools from the TDT Synapse software, such as behavioral notes.
4.14. Contributing to the improvement of pMAT.
Users without coding experience can be incredibly valuable contributors to the improvement of pMAT. We encourage these users to note any problems that they have and to write a description of these ‘bugs’ or problems under the ‘Issues’ section of the pMAT GitHub (https://github.com/djamesbarker/pMAT). Users with experience with MATLAB are also encouraged to contribute to the future modifications or bug fixes to the pMAT suite. Any user wishing to contribute a change should familiarize themselves with GitHub best practices, which generally include creating a copy (i.e., ‘fork’) of the original code, and then returning a request for changes to the original owners (i.e., a ‘pull request’) so that they can evaluate the code and integrate valid changes into a future release of the code.
5.0. Discussion
Fiber photometry allows us to examine levels of neuronal activity, neurotransmitter signaling, and even the interplay between the two that were previously inaccessible in awake, freely moving animals. The ability to conduct fiber photometry recordings has been the product of advances in recording technologies17 as well as advances in fluorescent proteins, which have enabled the development of an entire library of sensors in colors that exist both within and outside of the visible spectrum22.
5.1-. Pros and cons of fiber photometry.
There are many advantages to the implementation of fiber photometry, including the ability to examine the activity of neurons with specific anatomical and/or genetic identities12–14, 16, 19, 23 as well as the capacity to record from multiple cell types or brain regions simultaneously3, 24–26. Fiber photometry systems are also relatively simple when compared to other imaging techniques27, allowing for high-throughput experiments in a wide array of behavioral tasks12–14, 16, 19, 28. Finally, the availability of both open source18 and turn-key systems, as well as the relatively low cost of these systems, has made the technique incredibly accessible for most laboratories.
The advantages of fiber photometry must be considered carefully during experimental design and weighed against a number of notable disadvantages. Indeed, fiber photometry captures a relatively homogeneous picture of neuronal signaling, which appears biased towards increases in neuronal activity29, although decreases can be observed when cellular activity is homogeneous e.g.,28 Figure 6. Moreover, fiber photometry carries poor spatial resolution when compared single-cell calcium recordings via miniaturized microscopes or two-photon microscopes27. Finally, fiber photometry recordings collect data from a fairly small fields of tissue30, as the collection of these signals is subject to both the dispersion light from low-powered LEDs and the emission of relatively weak signals from calcium and neurotransmitter sensors. Thus, the quality of recordings can be highly dependent on both the proximity of the implant and the density of neurons. This becomes especially important when recording from axon terminals, where the quality of signals are routinely lower than when recording from cell bodies13, 24.
5.2-. A role for common approaches in rigor and reproducibility for fiber photometry experiments.
Perhaps one of the largest barriers for conducting fiber photometry experiments has been the intensive analysis required to obtain data for statistical comparisons. This single barrier complicates the process for researchers that are new to the technique and can make fiber photometry less approachable or even create problems with the rigor and reproducibility of the data. While multiple laboratories and commercial companies have offered guidance or analysis scripts for fiber photometry, the pMAT suite provides the first tool for analyzing fiber photometry data that does not require basic programming skills. Thus, pMAT is a first step towards alleviating this burden and providing an intuitive and accessible way to analyze fiber photometry data.
One of the most important features of the pMAT suite are tools for data quality control. The Plot Trace Data module allows users to independently examine their signal channels or control channels, while also allowing users to examine the stepwise process needed to produce the ΔF/F trace. These tools allow users to compare their signals with the known parameters of the sensor being recorded. For example, GCaMP signals should contain a sharp rise, followed by a slower, scalloped decay; the time constants for the rise and fall depend on the variant of GCAMP. Additionally, users can calculate the signal-to-noise ratio by examining the height of the signal during moments of quiescence as compared to the height of large transients. We recommend a minimum value of 2:1 for signal-to-noise, but ideally these values would be higher. Recorded signals should also be validated histologically by examining the expression of viral vectors and fiber placements. Moreover, experimenters should consider the use of appropriate controls in each experimental preparation. Examples of appropriate controls would be groups expressing control fluorophores in place of the active sensors, the use of neutral cues or events, combining sensors with technologies to inhibit signals of interest, or even the inclusion of positive controls such as optical or electrical stimulation in order to evoke responses. Lastly, the use of peri-event time histograms in pMAT is critically important for the validation of neural signaling, allowing users to demonstrate that both the presence and timing of sensor responses can be replicated across experimental trials.
Having an open-source tool with a well-documented description of the analytical approaches is a first step towards standardizing analysis pipelines. Moreover, the independent deployment of pMAT obviates the need for additional software, making it more widely accessible and budget-friendly. Among the tools available are those to export data for dissemination or plotting, to rapidly check the quality of signals following experimental sessions, and to evaluate the quality of movement controls in a stepwise manner. Moreover, pMAT will provide experimenters with the tools to plot sensor responses that are time-locked to behavioral events and to collect summary data that can be used for statistical analysis. Finally, the modular design of this tool allows for the rapid integration of additional tools to accommodate sensor-specific analyses or advances in the analytical approaches used by the field.
Highlights.
pMAT is an open-source tool for analyzing fiber photometry data
A point-and-click interface obviates the need for coding experience
Compatible with many fluorescent sensors and fiber photometry acquisition systems
Uses a modular design to allow community development of future tools
Supports experimental rigor and reproducibility through a standardized approach
Acknowledgements:
We thank Myles Billard and Mark Hanus from Tucker Davis Technologies for their critical feedback on the pMAT tool. We also thank Dr. David Root, Dillon McGovern, Dr. Kate Peters, Dr. Joseph Cheer, Jillian Seiler, Dr. Talia Lerner, Dr. Daniel Dautan, Chunyang Dong and Dr. Lin Tian for providing sample data for validation of the various sensors presented in the present manuscript. This work was supported by a NIDA K99/R00 Pathway to independence award (DA043572) and by pilot grants from the New Jersey Alliance for Clinical and Translational Sciences as well as the Rutgers Brain Health Institute. Funding sources were not involved in study design, data collection, and interpretation, or in the decision to submit this research for publication.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest: The authors declare that they do not have any conflicts of interest (financial or otherwise) related to the data presented in this manuscript.
References
- 1.Chen T-W, et al. , Ultrasensitive fluorescent proteins for imaging neuronal activity. 2013. 499(7458): p. 295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dana H, et al. , High-performance calcium sensors for imaging activity in neuronal populations and microcompartments. 2019. 16(7): p. 649–657. [DOI] [PubMed] [Google Scholar]
- 3.Dana H, et al. , Sensitive red protein calcium indicators for imaging neural activity. 2016. 5: p. e12727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borden PM, et al. , A fast genetically encoded fluorescent sensor for faithful in vivo acetylcholine detection in mice, fish, worms and flies. 2020. [Google Scholar]
- 5.Marvin JS, et al. , Stability, affinity, and chromatic variants of the glutamate sensor iGluSnFR. 2018. 15(11): p. 936–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marvin JS, et al. , A genetically encoded fluorescent sensor for in vivo imaging of GABA. 2019: p. 1. [DOI] [PubMed] [Google Scholar]
- 7.Patriarchi T, et al. , Ultrafast neuronal imaging of dopamine dynamics with designed genetically encoded sensors. 2018. 360(6396). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Unger E, et al. , Directed evolution of a selective and sensitive serotonin biosensor via machine learning. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun F, et al. , A genetically encoded fluorescent sensor enables rapid and specific detection of dopamine in flies, fish, and mice. 2018. 174(2): p. 481–496. e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feng J, et al. , A genetically encoded fluorescent sensor for rapid and specific in vivo detection of norepinephrine. 2019. 102(4): p. 745–761. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lerner TN, et al. , Intact-brain analyses reveal distinct information carried by SNc dopamine subcircuits. 2015. 162(3): p. 635–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barbano MF, et al. , VTA Glutamatergic Neurons Mediate Innate Defensive Behaviors. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barker DJ, et al. , Lateral preoptic control of the lateral habenula through convergent glutamate and GABA transmission. 2017. 21(7): p. 1757–1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pignatelli M, et al. , Cooperative synaptic and intrinsic plasticity in a disynaptic limbic circuit drive stress-induced anhedonia and passive coping in mice. 2020: p. 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y, et al. , Long-term Fiber photometry for Neuroscience studies. 2019. 35(3): p. 425–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Root DH, et al. , Distinct signaling by VTA glutamate, GABA, and combinatorial glutamate- GABA neurons in motivated behavior. Cell Reports, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gunaydin LA, et al. , Natural neural projection dynamics underlying social behavior. 2014. 157(7): p. 1535–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Akam T and Walton M.E.J.S.r., pyPhotometry: Open source Python based hardware and software for fiber photometry data acquisition. 2019. 9(1): p. 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Calipari ES, et al. , In vivo imaging identifies temporal signature of D1 and D2 medium spiny neurons in cocaine reward. 2016. 113(10): p. 2726–2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bavley CC, et al. , Cocaine-and stress-primed reinstatement of drug-associated memories elicit differential behavioral and frontostriatal circuit activity patterns via recruitment of L-type Ca 2+ channels. 2019: p. 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rodeberg NT, et al. , Hitchhiker’s guide to voltammetry: acute and chronic electrodes for in vivo fast-scan cyclic voltammetry. 2017. 8(2): p. 221–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O’Banion CP and Yasuda R.J.C.O.i.N, Fluorescent sensors for neuronal signaling. 2020. 63: p. 31–41. [DOI] [PubMed] [Google Scholar]
- 23.Fenno LE, et al. , Comprehensive dual-and triple-feature intersectional single-vector delivery of diverse functional payloads to cells of behaving mammals. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim CK, et al. , Simultaneous fast measurement of circuit dynamics at multiple sites across the mammalian brain. 2016. 13(4): p. 325–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meng C, et al. , Spectrally resolved fiber photometry for multi-component analysis of brain circuits. 2018. 98(4): p. 707–717. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sych Y, et al. , High-density multi-fiber photometry for studying large-scale brain circuit dynamics. 2019. 16(6): p. 553. [DOI] [PubMed] [Google Scholar]
- 27.Siciliano CA and Tye KMJA, Leveraging calcium imaging to illuminate circuit dysfunction in addiction. 2019. 74: p. 47–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhong W, et al. , Learning and stress shape the reward response patterns of serotonin neurons. 2017. 37(37): p. 8863–8875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.London TD, et al. , Coordinated ramping of dorsal striatal pathways preceding food approach and consumption. 2018. 38(14): p. 3547–3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pisanello M, et al. , The Three-dimensional signal collection field for fiber photometry in brain tissue. 2019. 13: p. 82. [DOI] [PMC free article] [PubMed] [Google Scholar]


