Abstract
In our recently published paper, we highlight that during normal aging of C. elegans age-dependent aggregates of proteins form and lead to functional decline of tissues. The protocol described here details the isolation of two proteins from C. elegans in their aggregated amyloid-like form, casein kinase I isoform alpha (KIN-19) and Ras-like GTP-binding protein rhoA (RHO-1). We used nickel beads to isolate His-tagged KIN-19 and RHO-1, and thus permitting the isolation of both small and large aggregated or fibrillary forms of the proteins. We characterized their morphology by transmission electron microscopy. We further expressed RFP-tagged proteins and stained them with a fluorescent molecule, thioflavin T, which identifies β-sheet structures, and which is a defining feature of amyloid fibrils. We further applied structured illumination microscopy to determine the level of colocalization between RFP and thioflavin T.
Keywords: His-tag, RFP-tag, Amyloid, Aggregates, Structured illumination microscopy, Transmission electron microscopy, RHO-1, KIN-19
Background
During aging, there is often a dysregulation in protein homeostasis and therefore an accumulation of damaged or non-functional proteins. Protein damage can occur by reactive oxygen species or post-translational modifications, along with mutations and transcriptional changes that naturally occur with age. One of the most studied forms of protein malfunction during aging are protein aggregates referred to as amyloids. These amyloids are found in many age-associated diseases such as Alzheimer’s disease and Parkinson’s disease. Soluble monomeric protein converts into an insoluble state where structure alters and proteins aggregate to form β-sheet rich fibrils. It is not clear whether the body is sequestering damaged or misfolded proteins into these amyloid fibrils when protein homeostasis is dysregulated or whether the formation of amyloid fibrils further dysregulates protein homeostasis, or both. Screening for proteins most likely to become insoluble upon aging Caenorhabditis elegans (C. elegans) yielded two target proteins, casein kinase I isoform alpha (KIN-19) and Ras-like GTP-binding protein rhoA (RHO-1) ( David et al., 2010 ). These two proteins were further studied to investigate their aggregation propensity during the ageing of C. elegans, whether they formed amyloid-like structures and whether they correlated with a functional decline ( Huang et al., 2019 ). In this protocol we first express the amyloid-like proteins KIN-19 and RHO-1 with a 6xHis-tag, this permits us to isolate the proteins with nickel beads. There can often be fibrous structures within small organisms such as C. elegans, but isolating His-tagged proteins ascertains that we isolate the protein of interest. Use of nickel beads rather than a nickel chromatography column allowed us to isolate large structures such as fibrils and aggregates which can get stuck in nickel columns. Subsequent imaging of eluted His-tagged proteins on a transmission electron microscope (TEM) permitted us to investigate the morphology of potential amyloid fibrils due to the sub-nanometer resolution provided by the TEM. However, a defining feature of amyloid fibrils is the presence of highly structured β-sheets which cannot be determined using TEM. Thus, in order to determine the presence of β-sheet structures in our proteins of interest we used structure illumination microscopy (SIM) which has ~200 nm resolution. We expressed red fluorescent protein (RFP)-tagged proteins in C. elegans and concomitantly stained them with thioflavin-T (ThT), a molecule which fluoresces when it intercalates into β-sheet structures. We then consequently investigated the colocalization of RFP and ThT fluorescence.
Materials and Reagents
Microlance needle 30G × 0.5” (Fisher Scientific Lt, catalog number: 10442014)
1.5 ml microcentrifuge tubes (Fisher Brand, catalog number: FB74031)
1.5 ml plastic pestles (StarLab, catalog number: I1415-539)
400 mesh copper grids with carbon coat (EM Resolutions, catalog number: C400cu100)
Transfer membrane PVDF 0.45 μm (Merck Millipore, catalog number: IVPH08100)
HisTrap FF Crude column–extract the nickel beads (Merck, catalog number: GE11-0004-58)
Nunc® Lab-Tek® II Chamber SlideTM system (Merck, catalog number: C7057-1PAK)
0.22 μm filter (Sartorius, catalog number: SAT16532K)
C. elegans uqIs22 [Pkin19::kin19::hisavi + Pkin19::birAtagrfp] also called DCD242 in ( Huang et al., 2019 )
C. elegans uqIS19 [Pmyo2::rho1::hisavi + Pmyo2::birAtagRFP] also called DCD243 in ( Huang et al., 2019 )
C. elegans uqIs9 [Pmyo-2::rho-1::tagrfp + Ptph-1::gfp] also called DCD13 in ( Huang et al., 2019 )
C. elegans fem-1(hc17ts) IV (Caenorhabditis genetic center, catalog number: BA17)
Anti-6x His tag® [4D11] primary antibody (Abcam, catalog number: ab5000)
Goat anti-Mouse IgG (H + L) Secondary Antibody, Alexa Fluor® 647 conjugate (Merck, catalog number: A21235)
Amersham ECL Mouse IgG, HRP-linked whole Ab (from sheep) (GE Healthcare, catalog number: NA931-1ML)
Bovine serum albumin (BSA) fraction V (Merck, catalog number: 9048-46-8)
PBS tablets, Dulbeco A ,1 tablet per 100 ml H2O (Oxoid, catalog number: BR0014G)
Radioimmunoprecipitation assay (RIPA) buffer (Merck, catalog number: R0278-50ML)
Protease inhibitor tablets, Complete, Mini EDTA-free (Roche, catalog number: 11836170001)
Imidazole (Merck, catalog number: I5513)
HCl (Merck, catalog number: 84435)
Thioflavin T (Abcam, catalog number: ab120751) -important to use within shelf life
4-12% Bis-Tris polyacrylamide gels (Invitrogen, catalog number: NP0323BOX)
Supersignal West PicoPLUS Chemiluminescent Substrate (Thermo Fisher Scientific, catalog number: 34577)
NuPAGETM LDS buffer (4x) (Thermo Fisher Scientific, catalog number: NP0007)
NuPAGETM MES SDS Running buffer (20x) (Thermo Fisher Scientific, catalog number: NP0002)
NuPAGETM Transfer Buffer (20x) (Thermo Fisher Scientific, catalog number: NP0006)
Methanol (Alfa Aesar, catalog number: L123255.0F)
Tween® 20 (Merck, catalog number: P1379)
0.1% (w/v) Poly-L-Lysine in H2O (Merck, catalog number: P8920-100ML)
Equipment
Sonicator (SANYO, model: Soniprep 150)
Benchtop cooled microcentrifuge (Thermo Scientific, model: Heraeus Fresco 21)
Fixed speed rotator (Stuart, model: SB2)
Microbalance (Sartorius, model: Secura 26-1S)
pH meter (Mettler Toledo, model: Education line and inLab® Routine Pro probe)
Dri-block Heat Block (Techne, model: DB-2TC)
XCell SureLock® Mini-Cell and XCell IITM Blot module for running SDS-PAGE gels and transfer (Thermo Fisher Scientific)
Power Pac (Bio-Rad, model: Power Pac 300)
Roller mixer (Stuart, model: SRT1)
G:Box Chemi imaging system (Syngene, model: XX6)
Transmission electron microscope (TEM) (Thermo Fisher Scientific, model: Tecnai G2 80-200keV)
Glow discharge system with argon gas (Quorum Technologies, Model: EMITECH K100X)
Structured illumination microscope (SIM) (Custom built) ( Muller et al., 2016 )
Software
Fiji open source image processing program (https://fiji.sc)
Procedure
This protocol describes the preparation of worms for imaging techniques. For protocols on the number of worms to grow, growth conditions, collection and freezing of worms please refer to William B. W. (1988) and Groh et al. (2017) .
-
Lysis of C. elegans
Defrost pellets of C. elegans expressing KIN-19::Hisavi and expressing RHO-1::Hisavi, these should ideally have been frozen in PBS with protease inhibitors (one protease inhibitor tablet per 10 ml of PBS, PBS is made with 1 tablet in 100 ml H2O).
To reduce C. elegans protein adherence to the microcentrifuge tubes, all tubes used in the experiments need to be incubated with 10% BSA in PBS for 10 min to coat the tubes.
Using a pipette with the end cut off giving a wide pore to allow easy mixing of the suspension, resuspend the worm pellet mixture in a 1:1 ratio with radioimmunoprecipitation assay (RIPA) buffer with protease inhibitor tablets to a volume of 250 μl.
In a 1.7 ml microcentrifuge tube sonicate the suspension at an amplitude of 10 microns for 10 s on, 30 s off, 10 s on, on ice or in a cold room and further homogenise it manually with a plastic pestle for 10 strokes.
Centrifuge the tubes for 1 min at 800 × g at 4 °C. The pellet contains the unwanted cuticle fragments and unlysed worms, carefully remove the supernatant.
Then further centrifuge the supernatant at 21,000 × g for 15 min at 4 °C. Carefully transfer the supernatant and resuspend the remaining pellet in 250 μl of PBS with protease inhibitors.
Push and pull the resuspended pellet through a 30 G needle three times.
-
Isolation of aggregated RHO-1 and KIN-19 by nickel beads
-
By cutting the end of a HisTrap FF Crude column remove 20 µl of precharged Nickel Sepharose 6 Fast Flow beads and incubate them with the resuspended pellet overnight at 4 °C on a fixed speed rotator at 20 rpm.
Note: Any nickel bead will likely work.
The next day centrifuge the tubes at 800 × g for 1 min remove the supernatant (containing unbound proteins) to leave the His-tagged proteins, fibrils and beads.
To remove the His-tagged aggregates and fibrils from the beads add 60 µl of 500 mM imidazole in PBS pH 8.0 (adjust pH with HCl) and incubate overnight at 4 °C on a fixed speed rotator at 20 rpm.
To isolate eluted aggregates and fibrils, centrifuge the beads at 800 × g for 1 min and collect the supernatant containing the aggregates and fibrils which are then used for imaging experiments. Note: We tried shorter times for His-tagged protein/fibril adherence to the beads and for elution, but this was not as successful as the overnight incubations. Unsuccessful elution protocol: incubate precharged Nickel Sepharose 6 Fast Flow beads with resuspended worm pellets for 15 min on a vortex mixer (Stuart, SA8) at 400 rpm. Then centrifuge the beads to pellet them at 800 × g for 1 min and remove the supernatant. Add 60 µl of 500 mM imidazole in PBS pH 8 (adjust pH with HCl) to the beads and incubated for 3 min on a vortex mixer at 400 rpm, but this was not long enough to successfully remove fibrils, shown by Western Blot and TEM in Figures 1A and 1B.
-
-
Confirming protein capture and release with nickel beads by Western blot
We tested the capture of protein aggregates and fibrils to the nickel beads and their successful elution by Western blot. Incubate 5 µl of crude worm pellet + 5 µl PBS, 3 µl of nickel beads after incubation + 7 µl PBS, 10 µl of supernatant after incubation and 10 µl of eluted aggregates with 4x LDS buffer at 100 °C in a heat block for 5 min. Use 10 µl of the sample to separate by 4-12% Bis-Tris SDS-PAGE gel and then transfer to a PVDF membrane for Western blotting.
Block the membrane with 5% BSA in PBS with 0.05% Tween-20 for 30 min to reduce background signal.
Incubate the membrane with anti-6xHis tag primary antibody at 1:1000 with PBS and 0.05% Tween-20 on a roller for 1 h. Then wash the membrane three times in PBS and 0.05% Tween-20 for 5 min each time.
Incubate the secondary antibody, anti-mouse-HRP, with the membrane at 1:1000 with PBS and 0.05% Tween-20 on a roller for 1 h. Wash the membrane five times in PBS and 0.05% Tween-20.
Incubate the membrane with Supersignal West PicoPLUS Chemiluminescent Substrate for 5 min before imaging using the G:Box Chemi imager (Figure 1A).
-
Imaging isolated protein aggregates by TEM
Glow discharge the carbon coated copper grids using argon gas on a Quorum Technology K100X at 25mAmps for 2 min.
Add 10 µl of protein sample to the grid and incubated for 1 min.
Wash the grid twice for 5 min in dH2O by making a 200 μl droplet of dH2O and placing the grid in the droplet.
Incubate the grid once in 10 μl 2% uranyl acetate for negative staining for 30 s before drying it by blotting with filter paper.
A Tecnai G2 80-200kv TEM at the Cambridge Advanced Imaging Centre was used for imaging the grids (Figures 1B-1D).
-
The Tecnai imaging software allows you to do point-to-point measurements of fibril widths and lengths, the diameters of the RHO-1 and KIN-19 fibrils were ~15-20 nm. Point-to-point measurements can be used to determine periodicity/helicality of fibrils, although periodicity was not observed in the RHO-1 or KIN-19 fibrils.
Note: We attempted to immunolabel the fibrils with the anti-6x His tag primary antibody and a secondary antibody conjugated to 10 nm gold beads, but the fibrils had been washed off the grids at the end of the staining process. It might be useful to not glow discharge the grids to keep it hydrophobic, allowing the fibrils to adhere better.
-
Preparation of C. elegans protein extracts for imaging by SIM
We initially tried to image RHO-1::His aggregates that were fluorescently labelled with antibodies, using the Anti-6x His tag primary antibody and the Goat anti-Mouse Alexa Fluor® 647 conjugate secondary antibody, but the extensive washing involved in antibody staining meant many aggregates were washed off the chamber slides and very few remained for imaging (Figure 2A). To obtain more aggregates and fibrils for fluorescence imaging, we instead used RHO-1::tagRFP worms which were already fluorescently labeled, and control worms which did not contain any amyloid aggregates as confirmed by the absence of ThT staining, fem-1(-). Process the worms as per Protocol A, Lysis of C.elegans, and use the crude worm pellet for imaging using SIM.
Coat Nunc Lab-Tek II Chamber Slide wells with 0.1% poly-L-Lysine for 30 min at RT before incubating 200 μl of the resuspended crude worm pellets in the wells along with 50 µM of ThT in PBS for 1 h at RT, keep the chamber slide under foil to keep in the dark (ThT powder added to PBS and filtered through a 0.22 μm filter).
-
Remove the non-adhered pellet by pipetting and wash three times with 200 μl PBS by pipette before imaging (Figure 2B).
Note: The ThT stock solution must be within its recommended usage period and freshly prepared for optimal binding and fluorescence.
-
Imaging protein aggregates by SIM
To visualize amyloids from worm extracts, we used our custom-built SIM providing a spatial resolution approaching 90 nm at frame rates reaching 22 Hz3.
Objective-60x water.
Exposure time: 25 ms for each channel.
-
Wavelengths used:
561 channel for RFP-Rho1
488 channel for ThT
647 channel for immuno-labeling with secondary antibody conjugated to AF647 dye.
Hardware control and image reconstruction were performed with software written in LabView and Matlab (Ströhl and Kaminski., 2015). For visualization, Fiji was used.
Figure 1. Representative Western blot and TEM images of successful and unsuccessful binding and elution of aggregated His-tagged proteins to nickel beads.

A. Western blot using an α-6xHis primary antibody to show representative successful (lanes under blue line) elution of His-tagged RHO-1 and unsuccessful (lanes under black line) elution of KIN-19 from nickel beads. The α-6xHis has identified 6xHis-tagged RHO-1 and KIN-19 of varying sizes indicating the presence of aggregates as well as degraded protein. Negative-stain transmission electron micrographs showing (B) unsuccessful elution of KIN-19::His protein from nickel beads and (C) successful capture and elution of RHO-1::His (RHO-1 fibrils indicated by a blue arrow) and (D) KIN-19::His (KIN-19 fibrils indicated by a blue arrow). Grids are negatively stained using 2% uranyl acetate. Scale bar = 100 nm.
Figure 2. SIM imaging to confirm the presence of RHO-1 positive amyloid structures.
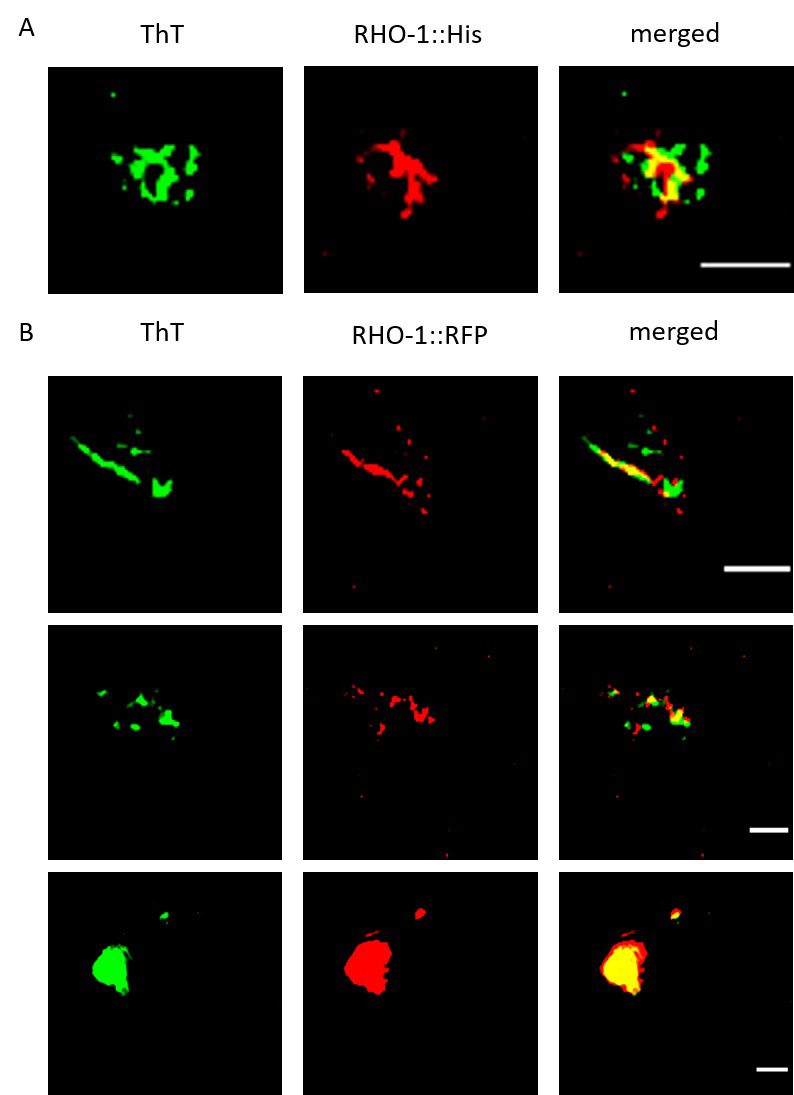
Extracts from C. elegans worms were stained for amyloid structures using ThT and examined for colocalization with RHO-1. A. SIM showed colocalization of ThT positive structures and His-tag immunostained RHO-1 proteins. B. We also observed colocalization of ThT positive structures with RHO-1::RFP and observed both fibrillar and inclusion body-like structures. Scale bar: 1 μm.
Data analysis
-
Image reconstruction
Raw images acquired by SIM were reconstructed using the Fiji plugin LAG-SIM based on fairSIM (Figure 2, Young et al., 2016 )
-
Parameter setting in LAG fairSIM-dialog
Slice for parameters: 1
Correction illumination: yes
Filter type: RL-in, RL-out
Wiener Parameter: 2.05
Apodisation cutoff: 2.9
Richardson-Lucy steps: 6
OTF attenuation: yes
Attenuation strength: 0.995
Attenuation FWHM: 1.2
Register images: yes (if acquiring multiple channels)
To determine size or colocalization between the two channels, software such as particle analysis plugin ( Schindelin et al., 2012 ) and the Coloc 2 plugin ( Manders et al., 1993 ), respectively, in the image analysis software FIJI can be used
Acknowledgments
G.S.K.S. acknowledges funding from the Wellcome Trust, the UK Medical Research Council (MRC), Alzheimer Research UK (ARUK), and Infinitus China Ltd. This protocol was based on the research paper ( Huang et al., 2019 ) in collaboration with Dr Della David.
Competing interests
None.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1. David D, Ollikainen C., N. Trinidad J, C. Cary M, P. Burlingame A, L. and Kenyon C. (2010). Widespread protein aggregation as an inherent part of aging in C. elegans . PLoS Biol 8(8): e1000450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Groh N., Gallotta I., Lechler M. C., Huang C., Jung R. and David D. C.(2017). Methods to study changes in inherent protein aggregation with age in Caenorhabditis elegans . J Vis Exp (129). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Huang C., Wagner-Valladolid S., Stephens A. D., Jung R., Poudel C., Sinnige T., Lechler M. C., Schlorit N., Lu M., Laine R. F., Michel C. H., Vendruscolo M., Kaminski C. F., Kaminski Schierle G. S. and David D. C.(2019). Intrinsically aggregation-prone proteins form amyloid-like aggregates and contribute to tissue aging in Caenorhabditis elegans . Elife 8: e43059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Manders E. M. M., Verbeek F. J. and Aten J. A.(1993) Measurement of co‐localization of objects in dual-colour confocal images. J. Microsc. 169(3): 375-382. [DOI] [PubMed] [Google Scholar]
- 5. Muller M., Monkemoller V., Hennig S., Hubner W. and Huser T.(2016). Open-source image reconstruction of super-resolution structured illumination microscopy data in ImageJ. Nat Commun 7: 10980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J. Y., White D. J., Hartenstein V., Eliceiri K., Tomancak P. and Cardona A.(2012). Fiji: an open-source platform for biological-image analysis. Nat Methods 9(7): 676-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ströhl F. and Kaminski C. F.(2015). A joint Richardson-Lucy deconvolution algorithm for the reconstruction of multifocal structured illumination microscopy data. Methods Appl Fluoresc 3(1): 014002. [DOI] [PubMed] [Google Scholar]
- 8. Young L. J., Strohl F. and Kaminski C. F.(2016). A guide to structured illumination tirf microscopy at high speed with multiple colors. J Vis Exp (111). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. William B. W.(1988). The nematode Caenorhabditis elegans Cold Spring Harbor: Cold Spring Harbor Laboratory Press. ISBN: 978-087969433-3. [Google Scholar]


