Abstract
Mitochondria are double-membraned organelles responsible for several functions in the cell including energy production, calcium signaling, and cellular metabolism. An equilibrium between fission and fusion events of mitochondria is required for their proper functioning. Mitochondrial morphologies have been quantified in yeast using image processing modules such as MitoGraph and MitoLoc. However, the dynamics of mitochondrial fission and fusion have not been analyzed in these methods. Here, we present a method for measuring mitochondrial morphologies, as well as estimation of fission and fusion frequencies of mitochondria in individual fission yeast cells whose mitochondria are fluorescently-tagged or stained. The latter relies on counting of individual mitochondria upon signal filtering in each frame of a time-lapse. Taken together, we present a simple protocol for analyzing mitochondrial dynamics, which can easily be adopted to other model systems.
Keywords: Mitochondria, Mitochondrial dynamics, Mitochondrial fission, Mitochondrial fusion, Live-cell imaging, Fluorescence microscopy, Cell biology, Schizosaccharomyces pombe
Background
Mitochondria are essential organelles present in the form of tubular networks within cells and constantly undergo fission and fusion events throughout the cell cycle. The rate of fission and fusion events determine the morphology of the mitochondria and this is a crucial determinant of their function (Mishra and Chan, 2016). Several factors contribute to the changes in morphology of the mitochondria. For instance, mitochondrial fission can be induced by stress, metabolic state, aging and in certain diseases (Sprenger and Langer, 2019). On the contrary, mitochondrial fusion can be induced to enhance cellular respiration in starved conditions and to dilute the effects of damaged mitochondrial DNA that may be present inside the cell (Westermann, 2010). Additionally, mitochondrial dynamics can drastically vary between cell types depending on the metabolic need of the given cell type. Since mitochondrial dynamics and thereby form are direct readouts for mitochondrial and cellular function, it is imperative to accurately measure these properties.
Early quantification efforts involved the use of qualitative descriptors to assess diverse mitochondrial morphologies. In recent times, computational image processing algorithms are being used to robustly quantify the differences in mitochondrial morphologies. For instance, the image processing module ‘MitoLoc’ uses the 3D reconstruction and analysis software, ‘MitoMap’ (a plugin for ImageJ) to define an object using Otsu thresholding and then calculates mitochondrial fragmentation, condensation and tubularity using mitochondrial shape descriptors such as mitochondrial size, position and integrity to differentiate the diverse mitochondrial phenotypes in budding yeast ( Vowinckel et al., 2015 ). MitoLoc calculates the fragmentation of mitochondria using a fragmentation index f, which is defined as the sum of relative fragment volumes that individually constitute less than 20% of the total mitochondrial volume. Another open-source image analysis platform named ‘MitoGraph’ has been employed to quantify mitochondrial morphology in both yeast and mammalian cells ( Viana et al., 2015 ; Harwig et al., 2018 ).
MitoGraph is an automated image processing software which performs graph theory-based quantitative analysis of mitochondrial networks by calculating the volumes of three-dimensional organelles in living cells. For both of these modules to provide accurate morphological data, it is imperative to utilize the best practices for imaging mitochondria by using the most optimal mitochondrial markers as well as high resolution imaging techniques. However, both of these modules do not provide quantitative measurements for mitochondrial fission and fusion dynamics. Here, we detail a simple method for quantifying both mitochondrial morphologies, as well as fission and fusion dynamics.
Materials and Reagents
Glass-bottom confocal dish, 35 x 10 mm, PP, Sterile (SPL, catalog number: 100350)
Reusable Bottle Top Filter (Tarsons, catalog number: 50060)
Membrane filter with 0.22 µm pore size (Millipore, catalog number: GSWP04700)
Kimwipes disposable wipers (Sigma-Aldrich, catalog number: Z188956)
Schizosaccharomyces pombe cells expressing fluorescently-labeled mitochondria or fission yeast cells stained with MitoTracker dye. Wild-type S. pombe cells can be obtained from National BioResource Project, Japan (https://nbrp.jp)
Lectin from Bandeiraea simplicifolia (Sigma-Aldrich, catalog number: L3759), store at 2-8 °C
Adenine (HiMedia, catalog number: TC082), store below 30 °C
Histidine (HiMedia, catalog number: GRM050), store below 30 °C
Leucine (HiMedia, catalog number: GRM054), store below 30 °C
Uracil (HiMedia, catalog number: GRM264), store below 30 °C
L- Lysine (HiMedia, catalog number: RM3028), store below 30 °C
MitoTrackerTM Orange CMTMRos (Thermo Fisher, catalog number: M7510), store at -5 to -30 °C
MgCl2·6H2O (HiMedia, catalog number: MB040), store below 30 °C
CaCl2·2H2O (HiMedia, catalog number: MB034), store below 30 °C
KCl (HiMedia, catalog number: GRM3934), store below 30 °C
NaCl (HiMedia, catalog number: MB023), store below 30 °C
Na2SO4 (HiMedia, catalog number: MB209), store below 30 °C
Pantothenic acid (HiMedia, catalog number: CMS178), store at 2-8 °C
Nicotinic acid (HiMedia, catalog number: CMS177), store below 30 °C
Inositol (HiMedia, catalog number: GRM102), store below 30 °C
Biotin (HiMedia, catalog number: TC096), store at 2-8 °C
Boric acid (HiMedia, catalog number: MB007), store below 30 °C
MnSO4 (HiMedia, catalog number: GRM1381), store below 30 °C
ZnSO4·7H2O (HiMedia, catalog number: MB233), store below 30 °C
FeCl2·6H2O (HiMedia, catalog number: GRM6353), store below 30 °C
Molybdic acid (Sigma-Aldrich, 232084), store at room temperature
KI (HiMedia, catalog number: GRM252), store below 30 °C
CuSO4·5H2O (HiMedia, catalog number: GRM630), store below 30 °C
Citric acid (HiMedia, catalog number: GRM1023), store below 30 °C
Yeast extract (HiMedia, catalog number: RM027), store below 30 °C
Glucose (Sigma-Aldrich, catalog number: G8270), store at room temperature
Potassium hydrogen phthalate (HiMedia, catalog number: GRM2405), store below 30 °C
Na2HPO4 (HiMedia, catalog number: MB024), store below 30 °C
KH2PO4 (Sigma-Aldrich, catalog number: P5655), store at room temperature
NH4Cl (HiMedia, catalog number: TC092), store below 30 °C
Agar powder, bacteriological (Himedia, catalog number: GRM026), store below 30 °C
NaOH
Phosphate buffered saline (see Recipes)
-
EMM: Edinburgh Minimal Medium with appropriate supplements (see Recipes)
50x Salt stock
1,000x Vitamin stock
10,000x Mineral stock
YE: Yeast Extract with supplements (see Recipes)
Equipment
Orbital shaking Incubator (Ascension Innovations, model: AI-100)
-
Laser scanning (point/line) or spinning disk confocal microscope
We used the confocal mode of an InCell Analyzer 6000 (GE Healthcare) Imaging system with 60x/0.7 N.A. objective, fitted with an sCMOS 5.5MP camera having an x-y pixel separation of 108, and 405, 488, 561 and 640 nm laser lines
Standard vertical autoclave (Equitron, model: 7431STWL.AFE.113)
Laminar airflow chamber
Software
IN Cell Analyzer Acquisition Software v.4.6 (GE Healthcare) or any other confocal microscopy system-specific software
Fiji/ImageJ (http://fiji.sc/Fiji, Schindelin et al., 2012 )
MATLAB software v. 2015b or above (MathWorks, https://in.mathworks.com)
Procedure
-
Preparation of S. pombe cells for imaging
Culture the yeast cells overnight by adding a loopful of cells (that have grown for at least a day in solid YE media) into 1 ml of liquid YE with appropriate supplements. Grow the cells at 30 °C for 12 h at 200 rpm to OD600 of 0.4-0.9.
Subculture 200 μl from the overnight culture into 1 ml of fresh medium and grow at 30 °C for ~2 h prior to imaging at 200 rpm.
Cover the surface of a 35 mm (No. 1) glass bottom dish with 25% NaOH solution for ~2 h to make the surface hydrophilic. Wash the dish twice with sterilized water and wipe the surface with Kimwipes.
Drop 2 μl of lectin (2 mg/ml in PBS) onto the dish and spread evenly using an inoculation loop.
Allow the lectin to dry completely inside the laminar airflow at room temperature for 10-15 min.
Meanwhile, wash cells once by spinning them down and resuspending them in 1 ml of EMM liquid medium.
If cells are already expressing fluorescently-labeled mitochondria (e.g., Cox4-GFP/RFP), proceed directly to Step A10, else follow Steps A7-A9 below to stain mitochondria with MitoTracker dye.
Dissolve MitoTracker Orange/DeepRed in DMSO at a concentration of 1 mM and dilute it in liquid EMM to a stock solution of 1 µM. This stock solution can be stored at -20 °C.
On the day of the experiment, freshly prepare 200 nM of MitoTracker Orange/DeepRed from the stock solution by diluting with EMM. Incubate cells with 1 ml of 200 nM MitoTracker Orange/DeepRed solution for 15-30 min at 30 °C.
After incubation, wash the cells thrice in 1 ml of liquid EMM to eliminate background fluorescence from unincorporated dye, and finally resuspend cells in 1 ml of liquid EMM medium.
Transfer 200 μl of cells resuspended in liquid EMM to the lectin-coated glass bottom dish.
Incubate the cell suspension on the glass bottom dish for 10 min at room temperature.
Wash out unbound cells and add 200 μl of liquid EMM to the dish.
Image at room temperature (~25 °C).
-
Obtaining time-lapse images to observe mitochondrial dynamics
Transfer cells to the stage of the confocal microscope.
Visualize mitochondria labeled with GFP or RFP using the 488 nm and 561 nm laser lines along with 525/20 and 605/52 nm bandpass emission filters respectively. Alternatively, visualize mitochondria stained with the MitoTracker Orange using the 561 nm laser line along with the 605/52 nm bandpass emission filter. The signal-to-noise ratio must be at least 1.5.
Capture mitochondrial dynamics by obtaining time-lapse images that are 12 s apart, for a total time of ~4 min, with Z-stacks for each timepoint with 0.5 µm step-size encompassing the full cell (~5 steps).
Save the time-lapse images in an appropriate format, such as TIFF.
Data analysis
-
Identification of mitochondria and measurement of mitochondrial parameters in Fiji/ImageJ
Open time-lapse images obtained using Fiji/ImageJ.
Ensure that the pixel to μm conversion for the image files is accurate by checking ‘Set scale’ in Fiji/ImageJ.
Crop individual cells in the field of view.
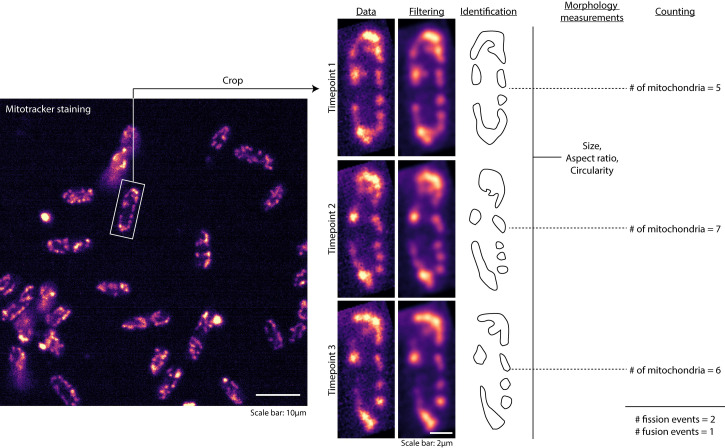
Apply ‘Mean’ filter with an appropriate radius which is sufficient to reduce background noise, while still maintaining separation between individual mitochondria. In Figure 1, we used a radius of 2 pixels.
Identify mitochondria in the images either manually or based on intensity thresholding (Figure 1).
If identifying mitochondria manually, draw regions of interest (ROIs) around mitochondria in each frame using the freehand selection tool.
In the ‘Set measurements’ function of Fiji/ImageJ, check the following options: Shape descriptors, Fit ellipse and Stack Position.
If identifying mitochondria using intensity thresholding, use the ‘Analyze particles’ function to display mitochondrial measurements, else run the ‘Measure’ function on each timepoint of the time-lapse images.
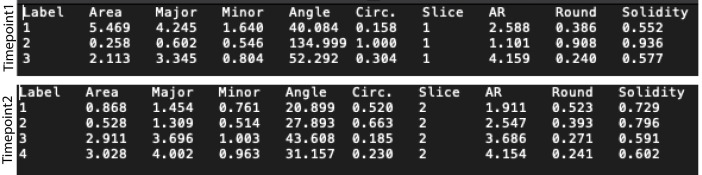
Save the measurements in appropriate format such as .txt or .csv (Figure 2).
-
Plotting of mitochondrial morphology parameters and estimation of mitochondrial fission/fusion frequencies using MATLAB
Import the data saved in .txt or .csv format for each yeast cell into MATLAB.
Plot the mitochondrial size distribution after collating the data under the column ‘Major’ of the data files across all timepoints from all the cells. This refers to the length of the ‘Major’ axis of an ellipse fitted to the mitochondria. Use the same dataset to obtain the mean and standard deviation of the mitochondrial sizes across cells.
Plot the circularity and aspect ratio of mitochondria using the data under the column ‘Circ.’ and ‘AR’ respectively of the data files across all timepoints from all the cells. Again, obtain the mean and standard deviations of the circularity and aspect ratios across cells using the same data set.
Write a script/function to count the number of mitochondria in each frame of the time-lapse movie by enumerating the number of occurrences of each stack position in the first column of the data file.
Compare the number of mitochondria in consecutive timepoints to obtain fission and fusion events. For instance, if the number of mitochondria in timepoint 1 is 5 and that in timepoint 2 is 7, then the number of fission events is 7 - 5 = 2. 2 ÷ 12 s = 0.167/s. Conversely, if the number of mitochondria in timepoint 1 is 5 and that in timepoint 2 is 4, then the number of fusion events is 5 – 4 = 1 (Figure 1).
Count the total number of fission and fusion events per cell for the entire time-lapse movie to obtain an estimate of fission and fusion frequencies. For example, if the total number of fission and fusion events for a cell in a 228 s time-lapse movie are 8 and 9 respectively, then the fission and fusion frequencies for the cell are 0.035/s and 0.039/s respectively. Obtain the mean and standard deviation of fission and fusion frequencies of all cells.
-
Statistical analysis of data
The data for fission and fusion frequencies obtained as described above can be represented in box plots with the central line indicating the median (see Mehta et al., 2019 ). To obtain the 95% confidence interval of the median, perform one-way ANOVA (‘anova1’ in MATLAB) or Kruskal-Wallis Test (‘kruskalwallis’ in MATLAB). The former is used when data is normally distributed and the latter when data are non-normally distributed. The normality of the dataset is tested using ‘chi2gof’ function in MATLAB. Significant difference (P < 0.05) between control and test groups is then be tested using Tukey’s Honestly Significant Difference procedure (‘multcompare’ in MATLAB).
Figure 1. Analysis of mitochondrial dynamics.
Workflow of analysis of the time-lapse images obtained, including cropping, image filtering, identification of mitochondria, measurement of morphology and counting.
Figure 2. Snapshots of txt files obtained after identification of mitochondria in a single cell for 2 timepoints (‘Slice’ 1 and 2) of a time-lapse movie.
Three mitochondria were identified in Timepoint 1 and four in Timepoint 2. The columns ‘Major’, ‘Circ.’ and ‘AR’ are used for quantification of mitochondrial morphology.
Recipes
-
Phosphate buffered saline (10x)
Amount Component Final concentration 80 g/L NaCl 0.137 M 2 g/L KCl 2.7 mM 14.4 g/L Na2HPO4 0.01 M 2.7 g/L KH2PO4 1.8 mM Adjust the pH to 7.2
Filter sterilize the solution by passing it through a bottle top filter containing a 0.22 µm pore size membrane filter and store at room temperature
-
EMM: Edinburgh Minimal Medium with appropriate supplements
Note: This recipe is taken from http://www-bcf.usc.edu/~forsburg/media.html.
Depending on the strain, medium with appropriate supplements is used. For imaging, liquid EMM is most ideal (Forsburg and Rhind, 2006)
Amount Component Final concentration 3 g/L potassium hydrogen phthallate 14.7 mM 2.2 g/L Na2HPO4 15.5 mM 5 g/L NH4Cl 93.5 mM 20 g/L glucose 2% w/v 20 ml/L salts 1 ml/L vitamins 0.1 ml/L minerals Minimal supplemented with amino acids: EMM + 225 mg/L supplements (ade, leu, his, lys, ura...) as required (each amino acid at 225 mg/L)
We make amino acid stock solutions at 7.5 mg/ml (3.75 for uracil) and autoclave the solution for 40 min at 121 °C with 15 psi of pressure
Stock solutions
-
50x Salt stock
Amount Component Final concentration 52.5 g/L MgCl2·6H2O 0.26 M 0.735 g/L CaCl2·2H2O 4.99 mM 50 g/L KCl 0.67 M 2 g/L Na2SO4 14.1 mM Filter sterilize the solution by passing it through a bottle top filter containing a 0.22 µm pore size membrane filter and store at 4 °C
-
1,000x Vitamin stock
Amount Component Final concentration 1 g/L pantothenic acid 4.20 mM 10 g/L nicotinic acid 81.2 mM 10 g/L inositol 55.5 mM 10 mg/L biotin 40.8 μM Filter sterilize the solution by passing it through a bottle top filter containing a 0.22 µm pore size membrane filter and store at 4 °C
-
10,000x Mineral stock
Amount Component Final concentration 5 g/L boric acid 80.9 mM 4 g/L MnSO4 23.7 mM 4 g/L ZnSO4·7H2O 13.9 mM 2 g/L FeCl2·6H2O 7.40 mM 0.4 g/L molybdic acid 2.47 mM 1 g/L KI 6.02 mM 0.4 g/L CuSO4·5H2O 1.60 mM 10 g/L Citric acid 47.6 mM Filter sterilize the solution by passing it through a bottle top filter containing a 0.22 µm pore size membrane filter and store at 4 °C
-
-
YE: Yeast Extract with supplements
Amount Component Final concentration 5 g/L Yeast extract 0.5% w/v 30 g/L Glucose 3.0% w/v Supplements: 225 mg/L adenine, histidine, leucine, uracil and lysine hydrochloride
Autoclave the solution for 40 min at 121 °C with 15 psi of pressure. To make solid medium, add 2% (w/v) Difco Bacto Agar prior to autoclaving
Acknowledgments
This research was supported by the Department of Science and Technology (India)-INSPIRE Faculty Award, the Department of Biotechnology (India) Innovative Young Biotechnologist Award, and the Science and Engineering Research Board (SERB, India) Early Career Research Award awarded to V.A. This protocol was adapted from Mehta et al., 2019 .
Competing interests
The authors declare that they have no competing interests with the contents of this article.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1. Forsburg S. L. and Rhind N.(2006). Basic methods for fission yeast. Yeast 23(3): 173-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Harwig M. C., Viana M. P., Egner J. M., Harwig J. J., Widlansky M. E., Rafelski S. M. and Hill R. B.(2018). Methods for imaging mammalian mitochondrial morphology: A prospective on MitoGraph. Anal Biochem 552: 81-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mehta K., Chacko L. A., Chug M. K., Jhunjhunwala S. and Ananthanarayanan V.(2019). Association of mitochondria with microtubules inhibits mitochondrial fission by precluding assembly of the fission protein Dnm1. J Biol Chem 294(10): 3385-3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mishra P. and Chan D. C.(2016). Metabolic regulation of mitochondrial dynamics. J Cell Biol 212(4): 379-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J. Y., White D. J., Hartenstein V., Eliceiri K., Tomancak P. and Cardona A.(2012). Fiji: an open-source platform for biological-image analysis. Nat Methods 9(7): 676-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sprenger H. G. and Langer T.(2019). The good and the bad of mitochondrial breakups. Trends Cell Biol 29(11): 888-900. [DOI] [PubMed] [Google Scholar]
- 7. Viana M. P., Lim S. and Rafelski S. M.(2015). Quantifying mitochondrial content in living cells. Methods Cell Biol 125: 77-93. [DOI] [PubMed] [Google Scholar]
- 8. Vowinckel J., Hartl J., Butler R. and Ralser M.(2015). MitoLoc: A method for the simultaneous quantification of mitochondrial network morphology and membrane potential in single cells. Mitochondrion 24: 77-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Westermann B.(2010). Mitochondrial fusion and fission in cell life and death. Nat Rev Mol Cell Biol 11(12): 872-884. [DOI] [PubMed] [Google Scholar]