Abstract
Loop-mediated isothermal amplification (LAMP) has been widely used in the detection of pathogens. However, there are usually numerous variants in one viral pathogen and primers employed in LAMP can hardly match all these variants. The mismatches between the primers and the viral genomes, especially those at the 3′-end of the primers, hinder LAMP reactions, leading to failure of the detection. Here, we present a mismatch-tolerant RT-LAMP protocol, which utilizes the 3′-5′ exonuclease activity of the Q5 high-fidelity DNA polymerase to remove potential mismatched bases at the 3′-end of the primers during LAMP amplification. Using HIV-1 as a proof-of-principle, we showed that this protocol could represent a promising tool for accurate detection of genetically unstable viruses in laboratory, hospital and field.
Keywords: Mismatch-tolerant RT-LAMP, Highly variable viruses, Mutant, HIV-1, Visual detection, High-fidelity DNA polymerase
Background
Emerging and re-emerging infectious diseases are serious threats to global public health (Mehand et al., 2018). Many outbreaks and epidemics have been caused by viruses, such as HIV-1, HCV, MERS-CoV, Ebola virus, A/H7N9 influenza virus, and more recently Zika virus. These viral diseases have led to high morbidity and mortality that disproportionally impacted on low-income countries (Van Doorn, 2017; Fenollar and Mediannikov, 2018; Waldman and Balskus, 2018). Rapid and accurate diagnosis of viral pathogens is crucial for the prevention and control of viral infectious diseases (Mehand et al., 2018).
Isothermal amplification techniques represent a promising direction for the development of point-of-care testing (POCT) diagnostic tools especially in the low-income countries or resource-limited settings (de Paz et al., 2014). Loop-mediated isothermal amplification (LAMP) is the most widely used isothermal amplification technology in biomedical research (Notomi et al., 2000). Its principle is auto-cycling strand displacement DNA amplification reaction using Bst DNA polymerase with high strand displacement activity under isothermal condition. LAMP generally uses three pairs of primers, and two inner primers (FIP and BIP) are responsible for initiating the self-primed DNA synthesis of the dumbbell form DNA. However, the biggest challenge for the detection of viruses using LAMP is the high genetic diversity of some viral genomes, which exist in the forms of genotypes, subtypes, and/or quasispecies (Sanjuan et al., 2010; Domingo and Perales, 2018). These diverse forms can easily cause mismatches with primers during their amplification, thereby resulting in a low sensitivity of detection and a limited spectrum of detection (Zhang et al., 2017; Li et al., 2019; Zhou et al., 2019). It is virtually impossible to detect all variants or serotypes in one LAMP assay as the conserved regions in the genomes are usually too short to completely match the long (approximately 40 nt) inner primers (FIP and BIP) of the assay. Therefore, an underestimate of viral load or even failure of detection is common using RT-LAMP method, especially for highly variable RNA viruses (Waldman and Balskus, 2018; Zhou et al., 2019). This may be the most important reason that limits the commercial application of LAMP in the diagnosis of viral infectious diseases (Wong et al., 2018). To overcome this problem, we recently developed a mismatch-tolerant RT-LAMP method that contains a minuscule amount of high-fidelity DNA polymerase and utilizes its 3′-5′ exonuclease activity to remove potential mismatched bases at the 3′-end of the primers during amplification (Zhou et al., 2019). The new method was demonstrated to be especially suited for the detection of highly variable viruses (Zhou et al., 2019). In this paper, we provide a detailed protocol of the mismatch-tolerant RT-LAMP method using HIV-1 detection as an example (Table 1).
Table 1. Primers used for the HIV-1 RT-LAMP assay .
| Types | Primer | Sequence (5′-3′) | Sources |
|---|---|---|---|
| Outer primers | AceIN-F3 | CCMMTTTGGAAAGGACCAGC | This work |
| AceIN-B3b | AACATACATATGRTGYTTTACTA | (Ocwieja et al., 2015) | |
| AceIN-B3a | TCTTTGAAAYATACATATGRTG | ||
| Inner primers | AceIN-FIPf | CTTGGCACTACYTTTATGTCACTAAARCTYCTCTGGAAAGGTG | |
| AceIN-FIPe | CTTGGTACTACYTTTATGTCACTAAARCTACTCTGGAAAGGTG | ||
| AceIN-BIP | GGAYTATGGAAAACAGATGGCAGCCATGTTCTAATCYTCATCCTG | ||
| Loop primers | AceIN-LF | TCTTGTATTACTACTGCCCCTT | |
| AceIN-LB | GTGMTGATTGTGTGGCARGTAG | This work |
Materials and Reagents
Axygen® MicroVolume Extended-Length Filtered Pipet Tips (Axygen, catalog number: TXLF10)
Axygen® Universal Fit 100 μl Filtered Pipet Tips (Axygen, catalog number: TF100RS)
Axygen® Universal Fit 200 μl Filtered Pipet Tips (Axygen, catalog number: TF200RS)
Axygen® Universal Fit 1,000 μl Filtered Pipet Tips (Axygen, catalog number: TF1000LRS)
Axygen 1.5 ml Snaplock Microtubes (Axygen, catalog number: MCT150CS)
Axygen 0.2 ml PCR® Tubes (Axygen, catalog number: PCR02C)
LightCycler® 480 Multiwell Plate 96,white (Roche, catalog number: 4729692001)
LightCycler® 8-Tube Strips, white (Roche, catalog number: 6612601001)
DreamTaqTM Green PCR Master Mix (2x) (Thermo Fisher Scientific, catalog number: k1081)
pUC57-IN (containing partial fragment of HIV-1 integrase gene: AF033819.3. the sequence is 5’-ACGGTTAGGGCCGCCTGTTGGTGGGCGGGAATCAAGCAGGAATTTGGAATTCCCTACAATCCCCAAAGTCAAGGAGTAGTAGAATCTATGAATAAAGAATTAAAGAAAATTATAGGACAGGTAAGAGATCAGGCTGAACATCTTAAGACAGCAGTACAAATGGCAGTATTCATCCACAATTTTAAAAGAAAAGGGGGGATTGGGGGGTACAGTGCAGGGGAAAGAATAGTAGACATAATAGCAACAGACATACAAACTAAAGAATTACAAAAACAAATTACAAAAATTCAAAATTTTCGGGTTTATTACAGGGACAGCAGAAATCCACTTTGGAAAGGACCAGCAAAGCTCCTCTGGAAAGGTGAAGGGGCAGTAGTAATACAAGATAATAGTGACATAAAAGTAGTGCCAAGAAGAAAAGCAAAGATCATTAGGGATTATGGAAAACAGATGGCAGGTGATGATTGTGTGGCAAGTAGACAGGATGAGGATTAGAACATGGAAAAGTTTAGTAAAACACCATATGTATGTTTCAGGGAAAGCTAGGGGATGGTTTTATAGACATCACTATGAAAGCCCTATCGGATCCCGGGCCCGTCGACTG-3′) (Synthesized by Shanghai BioSune Biotechnology Co., Ltd.)
QubitTM RNA HS Assay Kit RNA (Life Technologies, catalog number: Q32855)
WarmStart RTx Reverse Transcriptase (NEB, catalog number: M0380L)
Q5® High-Fidelity DNA Polymerase (NEB, catalog number: M0491L)
Bst 2.0 DNA Polymerase (NEB, catalog number: M0537L)
Fast Mutagenesis System (Transgen, catalog number: FM111)
QIAgen Viral RNA Mini Kit (Qiagen, catalog number: 52906)
QIAquick® Gel Extraction Kit (Qiagen, catalog number: GC-28706)
HiScribe T7 High Yield RNA Synthesis Kit (NEB, catalog number: E2040S)
WarmStart® Colorimetric LAMP 2x Master Mix (DNA & RNA) (with cresol red) (NEB, catalog number: M1800S)
Isothermal Amplification Buffer Pack (NEB, catalog number: B0537S)
Magnesium Sulfate (MgSO4) Solution (NEB, catalog number: B1003S)
dNTP Set, 100 mM Solutions (Thermo Fisher, catalog number: R0186)
SYTOTM 9 Green Fluorescent Nucleic Acid Stain (Invitrogen, catalog number: S34854)
Nuclease-Free Water (not DEPC-Treated) (AmbionTM, catalog number: AM9938)
Agarose (Biowest, catalog number: BY-R0100)
Ultra-pure water (Genview, catalog number: GU3313-500)
GelRed Nucleic Acid Straining Dye (10000x) (TOROIVD, catalog number: RSD100-25)
Plasma samples (Previous samples from our laboratory)
Yeast extract (Oxoid, catalog number: LP0021)
Tryptone (Oxoid, catalog number: LP0042)
Agar (Shangxiang, catalog number: 120420)
Sodium chloride (HuShi, catalog number: 10019318)
Ampicillin trihydrate (Solarbio, catalog number: A7490-5)
50x TAE buffer (Meilunbio, catalog number: MA0004)
2000 DNA Marker (Yeasen, catalog number: 10501ES60)
5000 DNA Marker (Yeasen, catalog number: 10504ES60)
TIANprep Mini Plasmid Kit (Tiangn, catalog number: DP103-03)
Liquid LB medium (see Recipes)
100 mg/ml ampicillin solution (see Recipes)
Ampicillin-resistant solid medium (see Recipes)
Ampicillin-resistant liquid LB medium (see Recipes)
2% agarose gel (see Recipes)
1% agarose gel (see Recipes)
Equipment
0.5-10 μl Eppendorf Research® plus Adjustable Volume Pipettes (Eppendorf, catalog number: I32693E)
10-100 μl Eppendorf Research® plus Adjustable Volume Pipettes (Eppendorf, catalog number: 251596Z)
20-200 μl Eppendorf Research® plus Adjustable Volume Pipettes (Eppendorf, catalog number: 4830359)
100-1,000 μl Eppendorf Research® plus Adjustable Volume Pipettes (Eppendorf, catalog number: 4847859)
NanoDropTM 2000 Spectrophotometer (Thermo Fisher, model: NanoDropTM 2000, catalog number: ND-2000)
LightCycler® 96 System (Roche, catalog number: 05815916001)
Bio-Rad CFX96TM Real-Time PCR System (Bio-Rad, catalog number: 785BR18555)
Eppendorf Centrifuge 5417R (Eppendorf, model: 5417R)
Eppendorf Mastercycler nexus (Eppendorf, catalog number:6325ZK904949)
Tanon Gel Image System (Tanon, model: 2500)
Tanon EPS 300 (Tanon, model: 300)
Tanon electrophoresis tank (Tanon, model: 400)
Software
LightCycler® 96 System software (Roche)
Bio-Rad CFX Manager 3.1 software (Bio-Rad)
Procedure
-
Preparation of HIV-1 wild type and mutant RNA standards
-
Construct two mutant pUC57-IN plasmids using the fast mutagenesis system according to the following steps. Primers for construction of the mutant plasmids are in Table 2.
Prepare the PCR reaction mixes (Table 3), PCR cycling condition: Enzyme activation and pre-denaturation at 94 °C for 3 min, 25 cycles of denaturation at 94 °C for 20 s, annealing at 55 °C for 20 s and extension at 72 °C for 1 min, followed by extending at 72 °C for 10 min.
Electrophoresis detection: measure 10 μl PCR product using 1% agarose gel electrophoresis at a constant voltage (140 V) for about 30 min.
PCR product digestion: add 1 μl of DMT enzyme to the remaining PCR product, mix and incubate for 1 h at 37 °C.
Add 5 μl of DMT digestion product to 50 μl of competent cells, mix and place on ice for 30 min. Then place the mixture at 42 °C for 45 s, and on ice for 2 min.
Add 250 μl of room temperature LB medium (without antibiotics) to the mixture, 200 rpm, 37 °C for 1 h.
Spread 100 μl of the bacterial solution evenly on a plate containing 100 μg/ml ampicillin and incubate overnight in a 37 °C incubator.
Pick the mono-clones into 3 ml LB medium containing 100 μg/ml ampicillin, for shaking culture (200 rpm), at 37 °C for 10-12 h.
Extract the plasmid DNA for Sanger sequencing (Shanghai Platinum Company) and verify the presence of the mutation. Store the mutated plasmids at -20 °C for subsequent experiments.
-
Amplify the HIV-1 integrase segment using T7 promotor-containing primer pair (HIV T7-F: TAATACGACTCACTATAGACGGTTAGGGCCGCCTGT and HIV R: CAGTCGACGGGCCCGGGA) with the wild-type and mutant HIV-1 plasmids as templates.
In a 0.2 ml PCR® tube with cap, a PCR reaction mix includes 25 μl 2x DreamTaqTM Green PCR Master Mix, 2 μl of 10 μM of each primer (HIV T7-F and HIV R primer) and 2 μl HIV-1 plasmid in a final volume of 50 μl.
The cycling condition is enzyme activation and pre-denaturation at 95 °C for 3 min, followed by 40 cycles of denaturation at 95 °C for 30 s, annealing at 55 °C for 30 s and extension at 72 °C for 1 min, fully extending at 72 °C for 10 min.
Measure the PCR product using 2% agarose gel electrophoresis at a constant voltage (140 V) for about 30 min.
After electrophoresis, cut the specific PCR product from the gel under UV control and purify according to the manufacturer′s instruction using the QIAquick® Gel Extraction Kit.
Obtain RNAs through in vitro transcription with the purified DNA as templates using the HiScribe T7 High Yield RNA Synthesis Kit.
-
Quantify the obtained RNA using NanoDropTM 2000 Spectrophotometer, and calculate the RNA copy number using the following formula:
Dilute RNA to 104 copies/ul, aliquot to 40 μl in a 0.2 ml PCR® tube with cap and store at -80 °C until use.
-
-
Plasma sample viral RNA extraction
Extract viral RNA from 140 μl of plasma using the QIAgen Viral RNA Mini Kit following the manufacturer′s instruction.
Elute the extracted RNA in 50 μl of nuclease-free water, aliquot to 25 μl per tube, and store at -80 °C until use.
-
Qualitative detection of HIV-1 by the real-time mismatch-tolerant HIV-1 RT-LAMP assay
-
Preparation of mixed primer sets (Table 4)
Prepare the mixed primer set 1 by adding 5 μl 100 μm each of primers AceIN-F3, AceIN-B3a, and AceIN-B3b into 85 μl nuclease-free water; the mixed primer set 2 includes 20 μl 100 μm each of primers AceIN-FIPf and AceIN-FIPe, 40 μl 100 μm AceIN-BIP, and 20 μl nuclease-free water; the mixed primer set 3 includes 20 μl 100 μm each of primers AceIN-LF and AceIN-LB, and 60 μl nuclease-free water.
-
Preparation of the real-time RT-LAMP mix (Table 5)
In a 96-well PCR plate or 8-tube PCR strip, each 25 μl reaction mix includes 3 μl RNA template (RNA standard or RNA extracts from clinical sample), 2.5 μl 10x isothermal amplification buffer, 1 μl 100mM MgSO4, 3.5 μl 10 mM dNTPs, 0.075 μl 2 units of Q5 high-fidelity DNA polymerase, 1 μl 8 units of Bst 2.0 DNA polymerase, 0.5 μl 15 units of warmstart RTx reverse transcriptase, 1 μl 1 mM SYTO 9, and 1 μl each mixed primer set (mixed primer set 1-3 in Table 4).
RT-LAMP cycling condition: Perform the reaction at 62 °C for 1 min by 60 cycles. Collect fluorescence signal at each cycle.
-
-
Colorimetric RT-LAMP detection (Table 5)
-
Preparation of the colorimetric RT-LAMP mix (Table 5)
In an 8-tube PCR strip, each 25 μl reaction mix includes 12.5 μl warmstart colorimetric LAMP 2x master mix, 1 μl each mixed primer set (mixed primer sets 1-3 in Table 4), 0.075 μl 2 units of Q5 high-fidelity DNA polymerase, and 3 μl of RNA template (RNA standard or RNA extracts from clinical sample).
Colorimetric RT-LAMP cycling condition: Perform the reaction at 62 °C for 50 min. Observe the color change at 20-, 30-, 40-, and 50-min time points by naked eyes.
-
Table 2. Primers for construction of two HIV-1 mutant plasmids.
| Primer | Sequence (5′-3′) |
|---|---|
| AceIN-F3 Mu1-G F | CTTTGGAAAGGACCAGGAAAGCTCCTC |
| AceIN-F3 Mu1-G R | CCTGGTCCTTTCCAAAGTGGATTTCTG |
| AceIN-F3 Mu2-A F | CTTTGGAAAGGACCAGAAAAGCTCCTC |
| AceIN-F3 Mu2-A R | TCTGGTCCTTTCCAAAGTGGATTTCTG |
Note: Introduced mutant is shown in Red font. F: forward primer; R: reverse primer; Mu: mutant.
Table 3. PCR reaction mixes of the construction of two HIV-1 mutants.
| Component | Volume (μl) |
|---|---|
| pUC57-IN (1-10 ng) | 2 |
| Mu F (10 μM) | 1 |
| Mu R (10 μM) | 1 |
| 2×TransStart FastPfu PCR Supermix | 25 |
| Nuclease-free water | 21 |
| total | 50 |
Note: F: forward primer; R: reverse primer; Mu: mutant.
Table 4. Mixed primer sets preparation of the mismatch-tolerant RT-LAMP assay.
| HIV-1 RT-LAMP assay | Regular LAMP assay | |||
|---|---|---|---|---|
| Mixed primer set | Volume (μl) | Primers | Volume (μl) | |
| Mixed primer set 1 (25×) | 100 μM AceIN-F3 | 5 | 100 μM F3 | 5 |
| 100 μM AceIN-B3a | 5 | 100 μM B3 | 5 | |
| 100 μM AceIN-B3b | 5 | Nuclease-free water | 90 | |
| Nuclease-free water | 85 | |||
| total | 100 | 100 | ||
| Mixed primer set 2 (25×) | 100 μM AceIN-FIPf | 20 | 100 μM FIP | 40 |
| 100 μM AceIN-FIPe | 20 | 100 μM BIP | 40 | |
| 100 μM AceIN-BIP | 40 | Nuclease-free water | 20 | |
| Nuclease-free water | 20 | |||
| total | 100 | 100 | ||
| Mixed primer set 3 (25×) | 100 μM AceIN-LF | 20 | 100 μM Loop F | 20 |
| 100 μM AceIN-LB | 20 | 100 μM Loop B | 20 | |
| Nuclease-free water | 60 | Nuclease-free water | 60 | |
| total | 100 | total | 100 | |
Note: The regular assay is recommended when no degenerate primers is used.
Table 5. Real-time and colorimetric reaction mixes of the mismatch-tolerant RT-LAMP assay.
| Component | Real-time RT-LAMP (μl) | Colorimetric RT-LAMP (μl) |
| 10× Isothermal Amplification Buffer | 2.5 | - |
| WarmStart Colorimetric LAMP 2x Master Mix | - | 12.5 |
| MgSO4 (100 mM) | 1 | - |
| dNTPs (10 mM) | 3.5 | - |
| Bst 2.0 DNA polymerase (8 U) | 1 | - |
| Warmstart RTx reverse transcriptase (15 U) | 0.5 | - |
| Q5 high-fidelity DNA polymerase (0.2 U/μl) | 0.75 | 0.75 |
| SYTO 9 (1×) | 1 | - |
| Mixed primer set 1 (25×) | 1 | 1 |
| Mixed primer set 2 (25×) | 1 | 1 |
| Mixed primer set 3 (25×) | 1 | 1 |
| RNA template | 3 | 3 |
| Nuclease-free water | 8.75 | 5.75 |
| Total | 25 | 25 |
Note: Obtain 0.2 U/μl Q5 high-fidelity DNA polymerase by ten-fold dilution of 2 U/μl Q5 enzyme stock.
Data analysis
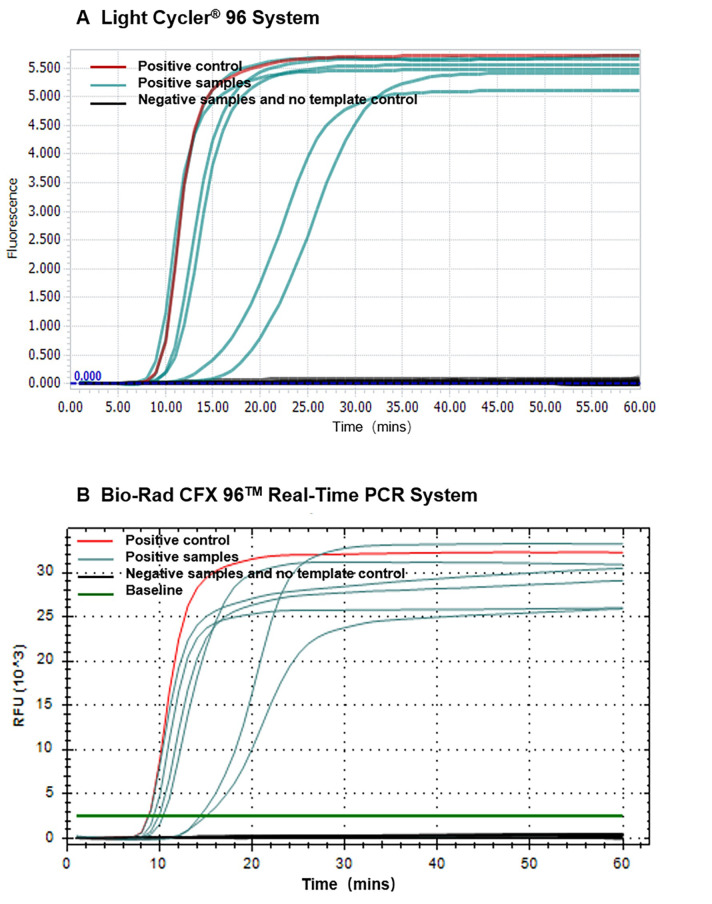
The result of the real-time mismatch-tolerant RT-LAMP assay can be seen with LightCycler® 96 system software, Bio-Rad CFX manager 3.1 software (Figure 1) or softwares implemented in other real-time PCR machines. Positive results (with the presence of HIV-1 templates) show clear "S" type amplification curves, and negative results (without HIV-1 template) have no amplification curve. The Ct values (time to appearance of the amplification curve) are negatively related to the amount of template input.
-
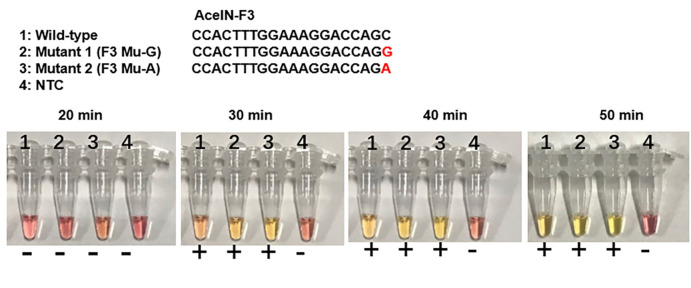
In the Colorimetric RT-LAMP detection assay, the results can be judged by naked eyes. A clear color change from burgundy to orange or yellow is considered as HIV-1 positive at 30-50 min which is dependent on the set cut-off (Figure 2).
Notes
Figure 1. Amplification curves of the real-time mismatch-tolerant HIV-1 RT-LAMP.
Amplification curves using LightCycler® 96 system (A) or Bio-Rad CFX96 TM Real-Time PCR system (B). HIV-1 RNA standard used as a positive control is shown in red, clinical samples from HIV-positive subjects are shown in cyan, and samples from HIV-negative subjects or that do not have template are shown in black.
Figure 2. Colorimetric RT-LAMP detection of HIV-1.
The color change from burgundy to orange or yellow is considered as positive (+). NTC: no template control. The mutated base is marked in red. AceIN-F3 is an outer forward primer.
The LAMP reaction is very sensitive. Attention should be paid to avoid contamination during the operation, and a stringent laboratory compartmentalization is strongly recommended for LAMP and other amplification assays.
To avoid potential contamination, agarose gel electrophoresis of LAMP products is not encouraged.
Because only a small amount of Q5 high-fidelity DNA polymerase is used in each reaction, the enzyme stock can be diluted to a lower concentration (e.g., 0.2 U/μl) to reduce pipetting error.
Other high-fidelity DNA polymerases can also be used instead of Q5 enzyme in the reaction. The recommended optimal concentration of high-fidelity DNA polymerase is between 0.1 and 0.3 units per 25 μl LAMP reaction.
The exact amount of Q5 high-fidelity DNA polymerase per 25 μl LAMP reaction needs to be optimized for detection of each specific virus.
If there are no degenerate primers to be used, the primer mix can be prepared based on the standard assay (Table 2).
The amount of RNA template per 25 μl LAMP reaction can be adjusted between the range of 0-11.75 μl for the real-time version, and 0-8.75 μl for the colorimetric version (Table 3).
Recipes
-
Liquid LB medium
10 g of tryptone,5 g of yeast extract and 10 g of sodium chloride in 1 L of ultra-pure water
Mix and autoclave it
-
100 mg/ml ampicillin solution
1 g of ampicillin powder in 5 ml of ultra-pure water
After it was dissolved, add ultra-pure water to make it to 10 ml
Dispense the 100 mg/ml ampicillin solution into a 1.5 ml EP tube and store at -20 °C until use
-
Ampicillin-resistant solid medium
10 g of tryptone, 5 g of yeast extract, 10 g of sodium chloride and 15 g of agar in 1 L of ultra-pure water
Mix and autoclave it
Add 1 ml of 100 mg/ml ampicillin solution and mix it
Pour about 10 ml on each plate, and store at 4 °C after solidification
-
Ampicillin-resistant liquid LB medium
Add 50 μl of 100 mg/ml ampicillin solution in 50 ml liquid LB medium and mix it
-
2% agarose gel
2 g of agarose in 100 ml of 1x TAE and 10 μl 10000x GelRed
-
1% agarose gel
1 g of agarose in 100 ml of 1x TAE and 10 μl 10000x GelRed
Acknowledgments
This protocol was modified from our original method published previously (Zhou et al., 2019). This study was supported by grants from the National Natural Science Foundation of China (81672033 and U1302224), the National Science and Technology Major Project of China (2017ZX10103009-002), the “One Belt One Road” Project (153831KYSB20170043) of Chinese Academy of Sciences, and the 133 projects of Institute Pasteur of Shanghai, CAS. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Competing interests
The authors declare no competing interests.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1.de Paz H. D., Brotons P. and Muñoz-Almagro C.(2014). Molecular isothermal techniques for combating infectious diseases: towards low-cost point-of-care diagnostics. Expert Rev Mol Diagn 14(7): 827-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Domingo E. and Perales C.(2018). Quasispecies and virus. Eur Biophys J 47(4): 443-457. [DOI] [PubMed] [Google Scholar]
- 3.Fenollar F. and Mediannikov O.(2018). Emerging infectious diseases in Africa in the 21st century. New Microbes New Infect 26: S10-S18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li Y., Wan Z., Hu Y., Zhou Y., Chen Q. and Zhang C.(2019). A mismatch-tolerant RT-quantitative PCR: application to broad-spectrum detection of respiratory syncytial virus. Biotechniques 66(5): 225-230. [DOI] [PubMed] [Google Scholar]
- 5.Mehand M. S., Al-Shorbaji F., Millett P. and Murgue B.(2018). The WHO R&D Blueprint: 2018 review of emerging infectious diseases requiring urgent research and development efforts. Antiviral Res 159: 63-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Notomi T., Okayama H., Masubuchi H., Yonekawa T., Watanabe K., Amino N. and Hase T.(2000). Loop-mediated isothermal amplification of DNA. Nucleic Acids Res 28(12): E63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ocwieja K. E., Sherrill-Mix S., Liu C., Song J., Bau H. and Bushman F. D.(2015). A reverse transcription loop-mediated isothermal amplification assay optimized to detect multiple HIV subtypes. PLoS One 10(2): e0117852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanjuan R., Nebot M. R., Chirico N., Mansky L. M. and Belshaw R.(2010). Viral mutation rates. J Virol 84(19): 9733-9748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van Doorn H. R.(2017). Emerging infectious diseases. Medicine 45: 798-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Waldman A. J. and Balskus E. P.(2018). The human microbiota, infectious disease, and global health: challenges and opportunities. ACS Infect Dis 4(1): 14-26. [DOI] [PubMed] [Google Scholar]
- 11.Wong Y. P., Othman S., Lau Y. L., Radu S., and Chee H. Y.(2018). Loop-mediated isothermal amplification(LAMP): a versatile technique for detection of micro-organisms. J Appl Microbiol 124(3): 626-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang M., Liu K., Hu Y., Lin Y., Li Y., Zhong P., Jin X., Zhu X. and Zhang C.(2017). A novel quantitative PCR mediated by high-fidelity DNA polymerase. Sci Rep 7(1): 10365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou Y., Wan Z., Yang S., Li Y., Li M., Wang B., Hu Y., Xia X., Jin X., Yu N. and Zhang C.(2019). A mismatch-tolerant reverse transcription loop-mediated isothermal amplification method and its application on simultaneous detection of all four serotype of dengue viruses. Front Microbiol 10: 1056. [DOI] [PMC free article] [PubMed] [Google Scholar]