Abstract
Cancer is a disease characterized by altered metabolism, and there has been renewed interest in understanding the metabolism of tumors. Even though nutrient availability is a critical determinant of tumor metabolism, there has been little systematic study of the nutrients directly available to cancer cells in the tumor microenvironment. Previous work characterizing the metabolites present in the tumor interstitial fluid has been restricted to the measurement of a small number of nutrients such as glucose and lactate in a limited number of samples. Here we adapt a centrifugation-based method of tumor interstitial fluid isolation readily applicable to a number of sample types and a mass spectrometry-based method for the absolute quantitation of many metabolites in interstitial fluid samples. In this method, tumor interstitial fluid (TIF) is analyzed by liquid chromatography-mass spectrometry (LC/MS) using both isotope dilution and external standard calibration to derive absolute concentrations of targeted metabolites present in interstitial fluid. The use of isotope dilution allows for accurate absolute quantitation of metabolites, as other methods of quantitation are inadequate for determining nutrient concentrations in biological fluids due to matrix effects that alter the apparent concentration of metabolites depending on the composition of the fluid in which they are contained. This method therefore can be applied to measure the absolute concentrations of many metabolites in interstitial fluid from diverse tumor types, as well as most other biological fluids, allowing for characterization of nutrient levels in the microenvironment of solid tumors.
Keywords: Nutrients, Metabolomics, Microenvironment, Interstitial fluid, Cancer metabolism, Mass spectrometry
Background
Cell division requires the duplication of the biomass of the mother cell prior to division. As a result, growing cells must be able to utilize the nutrients available in their environments to synthesize the macromolecules required to divide. To sustain cancerous proliferation, tumors often exhibit altered metabolism (DeBerardinis and Chandel, 2016). In many cases, tumor metabolism is driven by cell-intrinsic processes such as oncogenic activation ( Cairns et al., 2011 ; Nagarajan et al., 2016 ). However, recent work has highlighted the importance of cell-extrinsic factors in dictating cancer cell metabolism (Anastasiou, 2017; Bi et al., 2018 ; Muir et al., 2018 ). The importance of the extracellular environment in shaping cancer metabolism is perhaps unsurprising, as the nutrient environment in which a cancer cell exists constrains which metabolic reactions are possible within that cell. Since cell-extrinsic metabolite levels can play a role in determining the behavior of tumor cells, it is critical to examine tumor cell metabolism under physiological conditions. However, our understanding of the metabolic composition of the tumor microenvironment is lacking.
The nutrient environment that a cancer cell has access to is predominantly composed of interstitial fluid (Wiig and Swartz, 2012). Understanding the nutrient content of tumor interstitial fluid would provide insight into the metabolic constraints imposed upon tumor cells by their environment. There exist multiple methodologies for isolating interstitial fluid from normal organs and from tumors ( Wiig et al., 2010 ). However, early attempts to measure the nutrient content of interstitial fluid were limited by their inability to measure multiple metabolites, and consequently our knowledge of nutrient availability in tumors is restricted to a few metabolites in a limited number of animal tumor models (Burgess and Sylven, 1962; Gullino et al., 1964 ). The advent of mass spectrometry has allowed for detection of many metabolites simultaneously. However, despite technological advances, metabolomics studies are complicated by the fact that components present in biological fluids can suppress or enhance the detection of specific metabolites. These discrepancies in detection of metabolites between different biological fluids are termed “matrix effects,” and are a major confounding factor in comparing metabolite concentrations between different biological fluids and in accurately quantitating metabolites in those fluids ( Panuwet et al., 2016 ; Sullivan et al., 2019 ).
Here we demonstrate a method for centrifugation-based isolation of tumor interstitial fluid and the subsequent absolute quantitation of numerous metabolites within that fluid using stable isotope dilution, a technique in which stable isotope-labeled internal standards for metabolites of interest are added to experimental samples. These stable isotope internal standards are subject to the same matrix effects as the corresponding metabolite in the sample and can be distinguished by their increased mass compared to the metabolites in the sample. To measure many metabolites simultaneously, we first quantitate the concentrations of 13C metabolites from an extract of polar metabolites from yeast that are cultured with 13C isotopically labeled glucose as the sole carbon source. This quantitated yeast extract is then used as an internal standard that allows for reliable quantification of targeted metabolites in biological samples while minimizing systematic error from matrix effects. This approach provides a robust method to quantitate polar metabolites in biological fluids and complements similar existing isotope dilution based methods, such as the commercially available Biocrates AbsoluteIDQ kits ( Gieger et al., 2008 ) that primarily quantify non-polar lipids in biological samples.
The absolute quantitation of metabolite levels enabled by this protocol can allow for direct comparison of interstitial fluid composition in diverse tumor types, providing the opportunity to systematically interrogate nutrient availability in animal models of diverse cancers and human tumor samples. Further, the absolute quantification of metabolite levels in interstitial fluid allows for the generation of tissue culture media that mimics physiological conditions found in a tumor, thus expanding the range of in vitro/ ex vivo experiments that can be carried out under physiological nutrient conditions. Most broadly, this protocol provides a method to absolutely quantify many metabolites simultaneously in complex biological fluids, which can be used to study the metabolic composition of any biological material.
Materials and Reagents
50 ml conical tube (Falcon, catalog number: 14 959 49A)
Lab tape (Thermo Fisher, catalog number: 15-901-20H)
EDTA-coated plasma collection tubes (Sarstedt, catalog number: 41.1395.105)
1 ml 25G TB syringe (Becton Dickinson, catalog number: 309625)
20 μM mesh nylon filter (Spectrum Labs, catalog number: 148134)
Whatman paper (Thermo Fisher, catalog number: 88600)
Petri dishes (Corning, catalog number: 07 202 011)
Eppendorf tubes (Corning, catalog number: 14 222 168)
LC/MS sample vials (Thermo Fisher, catalog number: C4000-11)
LC/MS vial caps (Thermo Fisher, catalog number: C5000-54B)
Glassware (bottles, graduated cylinders, conical flasks) reserved for LCMS (not washed by central glass washing services, as this can introduce surfactant and other contaminants into your system)
Pipettes (Gilson, catalog number: F167380)
Mouse surgical kit (Thermo Fisher, catalog number: 14516249)
Blood bank saline (Azer Scientific, catalog number: 16005-092)
70% Ethanol (Thermo Fisher, catalog number: 04-355-305)
Acetonitrile LC/MS Optima 4 L (Fisher Scientific, catalog number: A955-4)
Methanol LC/MS Optima 4 L (Fisher Scientific, catalog number: A456-4)
Pierce formic acid (99%, LC/MS grade, Life Technologies, catalog number: 28905)
Water LC/MS Optima 4 L (Fisher Scientific, catalog number: W64)
Ammonium carbonate (Sigma-Aldrich, catalog number: 379999)
Ammonium hydroxide solution (28%, Sigma-Aldrich, catalog number: 338818)
Metabolomics amino acid standard mix (Cambridge Isotope Laboratories, Inc., catalog number: MSK-A2-1.2)
13C isotopically labeled yeast metabolite extract (Cambridge Isotope Laboratories, Inc., catalog number: ISO1)
13C3 lactate (Sigma-Aldrich, catalog number: 485926)
13C3 glycerol (Cambridge Isotope Laboratory, catalog number: CLM-1510)
13C6 15N2 cystine (Cambridge Isotope Laboratory, catalog number: CNLM-4244)
2H9 choline (Cambridge Isotope Laboratory, catalog number: DLM-549)
13C4 3-hydroxybutyrate (Cambridge Isotope Laboratory, catalog number: CLM-3853)
13C6 glucose (Cambridge Isotope Laboratory, catalog number: CLM-1396)
13C2 15N taurine (Cambridge Isotope Laboratory, catalog number: CNLM-10253)
2H3 creatinine (Cambridge Isotope Laboratory, catalog number: DLM-3653)
13C5 hypoxanthine (Cambridge Isotope Laboratory, catalog number: CLM-8042)
13C3 serine (Cambridge Isotope Laboratory, catalog number: CLM-1574)
13C2 glycine (Cambridge Isotope Laboratory, catalog number: CLM-1017)
-
Chemical standard library pool 1
Metabolite name Manufacturer Part# Alanine Sigma A7469 Arginine Sigma A6969 Asparagine Sigma A7094 Aspartate Sigma A7219 Carnitine Sigma C0283 Citrulline Sigma C7629 Cystine Sigma C7602 Glutamate Sigma G8415 Glutamine Sigma G3126 Glycine Sigma G7126 Histidine Sigma 53319 Hydroxyproline Sigma H54409 Isoleucine Sigma I7403 Leucine Sigma L8912 Lysine Sigma L8662 Methionine Sigma M5308 Ornithine Sigma 75469 Phenylalanine Sigma P5482 Proline Sigma P5607 Serine Sigma S4311 Taurine Sigma T0625 Threonine Sigma T8441 Tryptophan Sigma T8941 Tyrosine Sigma T8566 Valine Sigma V0513 Lactate Sigma L7022 Glucose Sigma G7528 -
Chemical standard library pool 2
Metabolite name Manufacturer Part# 2-hydroxybutyric acid Sigma 220116 2-aminobutyric acid Sigma 162663-25G AMP Sigma A1752 Argininosuccinate Sigma A5707-50MG Betaine Sigma 61962 Biotin Sigma B4639 Carnosine Sigma C9625-5G Choline Sigma C7017 CMP Sigma C1006 Creatine Sigma C0780-50G Cytidine Sigma C4654 dTMP Sigma T7004-100MG Fructose Sigma F0127 Glucose-1-phosphate Sigma G6750 Glutathione Sigma G4251 GMP Sigma G8377 IMP Sigma 57510-5G O-phosphoethanolamine Sigma P0503-1G Pyridoxal Sigma P9130-500MG Thiamine Sigma T4625 trans-Urocanate Cayman 16228 UMP Sigma U6375-1G Xanthine Sigma X7375-10G -
Chemical standard library pool 3
Metabolite name Manufacturer Part# 3-hydroxybutyric acid Sigma H6501 Acetylalanine Sigma A4625-1G Acetylaspartate Sigma 00920-5G Acetylcarnitine Sigma A6706 Acetylglutamine Sigma A9125-25G ADP Sigma A5285 Allantoin Sigma 05670-25G CDP Abcam ab146214-100 mg CDP-choline Alfa J64161-10 g Coenzyme A Sigma C4282 Creatinine Sigma C4255-10MG gamma-aminobutyric acid Sigma A2129-10G GDP Sigma G7127 Glutathione disulfide Sigma G4376 Glycerate Sigma 367494 Hypoxanthine Sigma H9377 myo-Inositol Sigma I5125 NAD+ Sigma N1511 p-aminobenzoate Sigma A9878 Phosphocholine Sigma P0378-5G Sorbitol Sigma W302902 UDP Sigma 94330-100MG UDP-glucose Sigma U4625-100MG -
Chemical standard library pool 4
Metabolite name Manufacturer Part# Phenylacetylglutamine Cayman 16724-25mg Acetylglutamate Sigma 855642 Acetylglycine Sigma A16300-5G Acetylmethionine Sigma 01310-5G Asymmetric dimethylarginine Cayman 80230 ATP Sigma A2383 CTP Sigma C1506 dATP Sigma D6500 dCTP Sigma D4635 Deoxycytidine Sigma D3897 Folic acid Sigma F8758 GTP Sigma G8877 Hypotaurine Sigma H1384-100MG Methionine sulfoxide Sigma M1126-1G Methylthioadenosine Sigma D5011-25MG Phosphocreatine Sigma P7936-1G Pyridoxine Sigma P9755 Ribose-5-phosphate Sigma 83875 SAH Sigma A9384-25MG Thymidine Sigma T9250 Trimethyllysine Sigma T1660-25MG Uridine Sigma U3003 UTP Sigma U6625 -
Chemical standard library pool 5
Metabolite name Manufacturer Part# 3-phosphoglycerate Sigma P8877 cis-aconitic acid Sigma A3412-1G Citrate Mallinckrodt 754 DHAP Sigma 51269 Fructose-1,6-bisphosphate Sigma F6803 Fumarate Sigma 240745 Glucose-6-phosphate Sigma G7879 Glycerol-3-phosphate Cayman 20729-100 mg Guanidinoacetate Sigma G11608 Kynurenine Sigma K8625 Malate Sigma 2288 NADP+ Sigma N0505 Niacinamide Sigma 72340 2-oxoglutarate Sigma 75890-25G Phosphoenolpyruvate Sigma P3637 Pyruvate Sigma P5280 Succinate Sigma S3674 Uracil Sigma U0750 -
Chemical standard library pool 6
Metabolite name Manufacturer Part# 3-hydroxyisobutyric acid Adipogen CDX-H0085-M250 2-hydroxyglutarate Sigma H8378 Aminoadipate Sigma A0637 beta-alanine Sigma 14064 Carbamoylaspartate Alfa A17166-10 g Cystathionine Cayman 16061-50 mg Cysteic acid Santa Cruz sc-485621 FAD Sigma F6625 Glycerophosphocholine Sigma G5291-50MG Inosine Sigma I4125 Orotate Sigma O2875 Pantothenate Sigma P5155 Phosphoserine Fluka 79710 Riboflavin Sigma R9504 UDP-GlcNAc Sigma U4375 Uric acid Sigma U2625-25G -
Chemical standard library pool 7
Metabolite name Manufacturer Part# Itaconic acid Sigma I29204 Homocysteine TCI H0159 2-oxobutyric acid Sigma K401 2-hydroxybutyric acid Sigma 220116 Ascorbate Sigma A7506 Sarcosine Sigma 131776-100G Dimethylglycine Sigma D1156-5G N6-acetyllysine Sigma A4021-1G Pipecolate Sigma P45850-25G Indolelactate Sigma I5508-250MG-A Picolinate Sigma P42800-5G 3-methyl-2-oxobutyrate Sigma 198994-5G 3-methyl-2-oxopentanoic acid Sigma 198978-5G Formyl-methionine Sigma F3377-250MG 2-aminobutyric acid Sigma A2536-1G Homocitrulline Santa Cruz sc-269298-100 mg gamma-glutamyl-alanine Sigma 483834-500MG Mannose Sigma M6020-25G Cysteine-glycine (dipeptide) Sigma C0166-100MG Mobile Phase A (see Recipes)
Mobile Phase B and Needle Wash (see Recipes)
Rear Seal Wash (see Recipes)
80% methanol containing 13C-15N labeled amino acid mix (see Recipes)
Extraction Buffer with isotopically labeled internal standards (EB) (see Recipes)
Chemical standard library preparation (see Recipes)
Equipment
Sorvall Legend X1R Refrigerated Centrifuge (Thermo Fisher, catalog number: 75004260)
Sorvall Legend Micro 21 Refrigerated Centrifuge (Thermo Fisher, catalog number: 75002447)
Mixer Mill (Retsch, catalog number: MM301)
50 ml Mixing Jar (Retsch, catalog number: 01.462.0216)
5 mM Grinding Balls (Retsch, catalog number: 05.368.0034)
LP Vortex Mixer (Thermo Fisher, catalog number: 11676331)
Analytical balance (Mettler Toledo, catalog number: AL54)
Dionex Ultimate 3000 UHPLC equipped with RS Binary Pump, RS column oven and RS autosampler (Thermo Fisher Scientific, San Jose, CA)
QExactive hybrid quadrupole-Orbitrap benchtop mass spectrometer equipped with an Ion Max ion source and HESI-II probe (Thermo Fisher Scientific, San Jose, CA)
SeQuant® ZIC®-pHILIC 5 μm 150 x 2.1 mm polymeric analytical PEEK HPLC column (Millipore Sigma, catalog number: 1504600001)
SeQuant® ZIC®-pHILIC 5 μm 20 x 2.1 mm PEEK Guard Kit with coupler (3 pc) (Millipore Sigma, catalog number: 15043800001)
Software
Thermo Scientific Xcalibur 4.1 SP1 (Thermo Fisher Scientific)
Microsoft Excel
Procedure
-
Study design
-
How many biological replicates are recommended?
The number of biological replicates needed for studies will depend on the variability between samples. For animal studies, where a large number of variables can be controlled (i.e., tumor genetics, tumor size, animal genetics, animal diet, time of interstitial fluid isolation), variability will likely be smaller than for human samples. Additionally, the number of replicates required will depend on the intended purpose of the experiment to be performed. For example, if the intended purpose is to determine if there is a nutritional difference in the interstitial fluid between two tumor types, it is important to determine an effect size between the groups in addition to variability between samples in order to estimate sample sizes needed. We recommend generating pilot data or utilizing previously published data on TIF composition differences ( Sullivan et al., 2019 ) to estimate effect sizes and utilizing power analysis software in Metaboanalyst ( Chong et al., 2018 ) or other statistical analysis software to estimate the number of biological replicates needed.
Additionally, when determining the number of biological replicates needed, it is important to note that not every tumor will necessarily yield interstitial fluid. In our experience, roughly 75% of murine pancreatic adenocarcinomas yielded tumor interstitial fluid (TIF) with volumes ranging from 5 to 180 μl of fluid ( Sullivan et al., 2019 ). Therefore, additional samples may be required to achieve the required number of replicates determined from power analysis.
We recommend harvesting TIF at the same time from all animals involved in a study. Circadian rhythm and food intake can alter plasma metabolite levels and therefore TIF metabolite levels, introducing additional variability ( Dallmann et al., 2012 ; Abbondante et al., 2016 ). If TIF must be harvested from animals on different days, we recommend harvesting TIF at the same time during the day.
We recommend two people work together to isolate TIF and cardiac blood to increase the speed of TIF harvest to prevent alterations in TIF composition due to prolonged periods of ischemia occurring between euthanasia and TIF harvest. In our own experiments, dissection was completed in ~2 min. and we found limited evidence of ischemia altering tumor metabolite levels ( Sullivan et al., 2019 ).
We have successfully isolated TIF using the protocol described in Procedure B from multiple genetically engineered and implantation mouse models of breast, lung, prostate, pancreas and melanoma cancers and others have isolated TIF from murine models of melanoma and breast cancer ( Ho et al., 2015 ; Eil et al., 2016 ; Spinelli et al., 2017 ; Zhang et al., 2017 ) using similar protocols. Additionally, similar protocols have been used to isolate TIF from renal ( Siska et al., 2017 ) and ovarian carcinomas (Haslene- Hox et al., 2011 ). Thus, we anticipate the TIF isolation protocol can be used successfully for a variety of tumor types, of mouse and human origin.
The analysis described in Procedure C uses 7 separate chemical standard pools as described in ( Sullivan et al., 2019 ) that enable the quantification of 149 metabolites commonly measured in biofluids ( Lawton et al., 2008 ; Evans et al., 2009 ; Mazzone et al., 2016 ; Cantor et al., 2017 ). However, depending on experimental goals, the full analysis utilizing all 7 standard pools may not be required. Subsets of the chemical standard pools can be used that cover analytes of interest if the full analysis is not needed. Note though that individual metabolites in the standard pools provided in this protocol have been carefully selected so as to avoid metabolites with the same m/z (isomeric and isobaric species) being in the same pool. In addition, metabolites that could be generated by in-source fragmentation from larger metabolites have been separated.
The description of the liquid chromatography-mass spectrometry analysis in this protocol (Procedure D) is a rough guideline for experienced operators of such instruments to perform the analysis described. Successful mass spectrometry analysis of the samples will require a trained UHPLC and Thermo Scientific hybrid quadrupole-Oribtrap mass spectrometer operator.
-
-
Isolation of TIF and plasma from tumor bearing animals
-
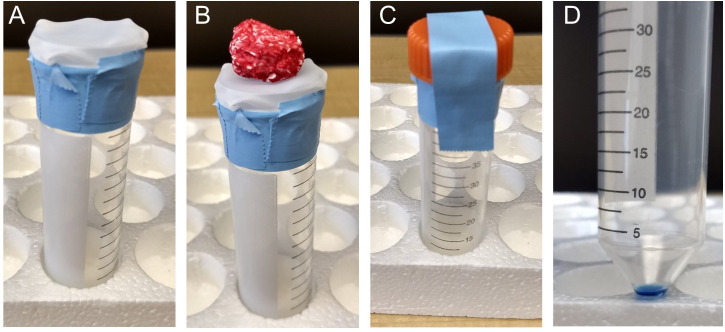
Prepare a TIF isolation tube (Figure 1 A).
Take a nylon filter and place it over the top of a 50 ml conical tube.
Tape the filter down using lab tape. Make sure the filter is affixed somewhat loosely to the top of the conical tube, such that the tumor can push the filter down slightly into the tube.
-
Prepare materials in advance to allow for rapid mouse dissection.
Put pre-chilled (4 °C) saline (~25 ml) into a Petri dish for washing the tumor.
Make a ~4 cm square piece of Whatman paper for drying the tumors after the saline rinse.
Have a 25G TB syringe ready for cardiac blood collection.
Label and pre-chill one EDTA-coated plasma collection tubes on ice for 10 min prior to mouse dissection.
Euthanize the mouse by cervical dislocation.
Spray around the incision site with 70% ethanol to prevent contamination of samples with hair.
-
Quickly and cleanly dissect the tumor away from the animal. The exact procedure for the dissection will depend on the anatomical location of the tumor. One person should continue with the following steps while the other person can start on plasma isolation (Step B6).
Place the tumor into the saline containing Petri dish to rinse the tumor.
-
Blot the tumor dry on Whatman paper. Take care at this step to ensure all saline is blotted from the tumor, so that saline does not contaminate the TIF.
If concerned about saline contamination during TIF isolation, we suggest the following additional step: add a non-natural metabolite such as norvaline to the saline. Continue with the remainder of the protocol as normal. Subsequently, when analyzing TIF samples, determine if the non-natural metabolite is present in the TIF sample. If the non-natural metabolite is detected this indicates saline contamination, whereas if the metabolite is not detected saline contamination is unlikely.
Optional: Weigh the tumor if this information is needed. Tumor volume can also be determined by caliper measurements if needed.
Place the tumor on top of the nylon mesh filter that is affixed to the conical tube (Figure 1B). Place the 50 ml conical lid over the tumor and tape the lid in place (Figure 1C).
Centrifuge the tumor at 4 °C for 10 min at 106 × g using a refrigerated clinical centrifuge (e.g., Sorvall Legend X1R).
-
Remove the 50 ml conical tube. If the isolation worked, there will be 10-50 μl of fluid in the bottom of the conical tube (Figure 1D). Keep the tube on ice.
Note: Human tumors have been found to yield 5-150 μl of fluid per gram of tumor (Haslene-Hox et al., 2011). Similarly, we isolated 10-50 μl of fluid from murine pancreatic tumors weighing ~0.3-2.5 g (Sullivan et al., 2019). There is not an exact correlation between tumor size and TIF volume, as many factors likely influence interstitial volume. However, larger tumors are more likely to yield TIF in larger volumes. Tumors with large fluid filled cysts can give hundreds of μl of fluid per gram of tumor. Disregard these from analysis as it is unclear if the cystic fluid is representative of interstitial fluid.
-
Remove the isolate fluid to a fresh Eppendorf tube. The fluid can be extracted directly for LC/MS analysis or frozen and stored at -80 °C for future analysis.
We have analyzed TIF samples both before and after 2 months of storage at -80 °C (avoiding freeze-thaw cycles) and detected similar metabolite concentrations after storage ( Sullivan et al., 2019 ). Thus, TIF samples can be stored for at least 2 months without freeze-thaw cycles prior to analysis.
-
The tumor from which TIF was isolated can be used for additional analysis using other appropriate protocols.
Note: We have successfully used tumors from which TIF was isolated for immunohistochemical, immunoblotting and flow cytometric analyses.
-
Use the 25G TB syringe to isolate blood from the mouse heart by cardiac puncture. Cardiac puncture can be a difficult technique to perform. Detailed protocols with video documentation of cardiac puncture blood isolation have been previously published (Schroeder, 2019). If unfamiliar with this technique, we recommend utilizing these resources for more detailed information on isolating blood in this manner.
Dissect open the thoracic cavity.
Insert the syringe into the ventricle.
Slowly withdraw blood to prevent collapse of the heart.
Remove the needle from the syringe to prevent cell lysis when expelling the cells from the syringe.
Expel the blood into the EDTA-coated plasma collection tube.
Keep plasma on ice.
Spin the EDTA-coated plasma collection tubes at 845 × g in benchtop centrifuge (e.g., Sorvall Legend Micro 21) for 15 min at 4 °C.
Remove the plasma from the pelleted blood cells and put into fresh Eppendorf tube.
-
Extract this plasma directly for LC/MS analysis or freeze and store at -80 °C for future analysis.
Note: Previous studies have found that plasma samples can be stored at -80 °C (without freeze-thaw cycles) for up to 30 months without significant alterations in the levels of many metabolites (Stevens et al., 2019). Thus, plasma samples can be stored for many months prior to analysis.
-
-
Extraction of metabolites from TIF and plasma
Prepare libraries of pooled chemical standards that include the metabolites to be quantified. See Recipes section and Tables 5-11 for details on how libraries were compounded in ( Sullivan et al., 2019 ).
-
Prepare metabolite extraction buffer (EB) (Recipe 5) with appropriate isotopically labeled internal standards. See Recipes section for details on making EB with isotopic standards as described in ( Sullivan et al., 2019 ).
Note: Make enough EB for the number of samples and standards you have plus an additional 10%, so as not to run out of EB before extracting metabolites from every sample. 45 μl of EB is needed for each sample and standard pool dilution. Isotopically labeled metabolite standards in the EB are not indefinitely stable. Prepare EB fresh prior to each experiment.
Prepare dilutions of chemical standard libraries in HPLC grade water. The highest concentration should be 5 mM.
Next, make dilution series of each standard pool in HPLC grade water as listed below in Step C5. Keep these on ice prior to extraction. Note that multiple dilutions of the chemical standard libraries are required for construction of standard curves relating known metabolite concentration to LC/MS response, which is necessary for downstream analysis of metabolite concentrations. Below we recommend a scheme to generate 8-point standard curves covering physiological concentrations of metabolites, but variations are possible.
-
The 5 mM pool will not be always in solution for each pool, therefore vortex vigorously and immediately pipette from this mixture in order to prevent error from settling particles:
Take 20 μl of 5 mM stock and dilute into 80 μl HPLC grade water to yield a 1 mM solution.
Take 30 μl of 1 mM stock and dilute into 70 μl HPLC grade water to yield a 300 μM solution.
Take 33.33 μl of 300 μM stock and dilute into 66.67 μl HPLC grade water to yield a 100 μM solution.
Take 30 μl of 100 µM stock and dilute into 70 μl HPLC grade water to yield a 30 μM solution.
Take 33.33 μl of 30 μM stock and dilute into 66.67 μl HPLC grade water to yield a 10 μM solution.
Take 30 μl of 10 μM stock and dilute into 70 μl HPLC grade water to yield a 3 µM solution.
Take 33.33 μl of 3 μM stock and dilute into 66.67 μl HPLC grade water to yield a 1 μM solution.
Thaw the TIF and plasma samples on ice.
Add 5 μl of each TIF sample, plasma sample and chemical standard library dilution (Recipe 6) to a fresh Eppendorf tube. Keep on ice.
Add 45 μl of EB to each sample/standard. Keep on ice.
Vortex all the samples for 10 min at maximum speed at 4 °C.
Spin down all samples for 10 min at 21,000 × g at 4 °C.
-
Take 20 μl of the mixtures from the Eppendorf tube and add to an LC/MS sample vial. Cap the vial.
Note: A minimum of 15 μl is needed in the LC/MS vial to ensure correct and accurate injection by the autosampler. The vials described in Materials and Reagents contain fused inserts. Vials without inserts will require larger volumes.
Freeze the remaining sample in the Eppendorf tubes and store at -80 °C. This sample can be run later if the initial LC/MS is not successful.
-
Liquid chromatography-mass spectrometry analysis of extracted metabolites
Start off with a clean system (use appropriate LC cleaning methods in place in your lab).
-
Calculate the amount of Mobile Phase A (Recipe 1) required and prepare fresh on the day of analysis. Store this for no more than one week.
Note: Depending on your system, you will use ~2 ml per injection and require an additional 50-100 ml in the bottle. Do not forget to make enough Mobile Phase A for additional injection types such as solvent blanks and system suitability tests that must be run in addition to the samples.
Calculate the amount of Mobile Phase B (Recipe 2) required and prepare more if needed. Depending on your system, you will need ~2 ml per injection plus an additional 50-100 ml in the bottle.
Check the level of rear seal wash (Recipe 3) and top up if needed.
If using an UHPLC system that has a separate needle wash, fill this with acetonitrile.
Connect a SeQuant® ZIC®-pHILIC 5 μm 150 x 2.1 mm analytical column to the Guard column using the connector supplied in the Guard kit.
Connect the column and guard to your UHPLC system using standard techniques.
Set the column oven temperature to 25 °C.
Set the autosampler temperature to 4 °C.
-
Set initial conditions: set the flow rate to 0.150 ml/min with 80% B. Record initial pressure value.
Note: ZIC-pHILIC columns cannot tolerate such high back pressures and injection volumes as typical reverse phase columns. Keep an eye on the back pressure and do not let it exceed the maximum pressure recommended by the manufacturer. It is good practice to set a maximum pressure in your method that is below that set by the manufacturer to avoid damage to the column.
Equilibrate the column with starting conditions (80% B) for 30 min prior to running anything on the system.
Check the mass calibration on the mass spectrometer. If the mass has not been calibrated within the last week, or if it fails the mass check, recalibrate using the standard calibration mixes recommended by the manufacturer. In addition, perform a custom low-mass calibration by spiking glycine and aspartate into the calibration mix, or as recommended by the manufacturer.
Ensure your entire LCMS system performance is acceptable by running system suitability tests, such as injecting a mixture of amino acids onto your column and into the MS. Check for signal intensity as well as peak shape and separation.
Use the conditions shown in Table 1 below for the UHPLC gradient:
-
Operate the mass spectrometer in full-scan, polarity-switching mode, with a scan range of 70-1000 m/z. Include an additional narrow-range scan from 220 to 700 m/z in negative mode to improve detection of nucleotides. Use the parameters shown below in Tables 2 and 3 for the MS:
Table 2. MS source parameters.
Parameter Setting Spray voltage 3.0 kV (pos); 3.1 kV (neg) Heated capillary 275 °C HESI probe 350 °C Sheath gas 40 units Aux gas 15 units Sweep gas 1 unit Table 3. MS scan parameters.
Parameter Setting Resolution 70,000 AGC target 1E6 Max IT 20 ms Note that in this particular study, we had previously collected MS/MS data for each metabolite being quantified, and used this to confirm retention times using a library of chemical standards. If adding new metabolites to your standard pools, collect MS/MS data on the standard itself, as well as on a pooled biological sample to help confirm peak identification.
-
Write your sequence (sample run order) using Thermo Scientific Xcalibur Sequence Setup View.
Note: Given the extremely low volume of samples used in this method, the sequence differs from typical sequences which will include column conditioning injections, as well as quality control pooled samples. As multiple standard curves are run and each sample includes 13C-labeled internal standards, we chose to forgo using precious sample to create QC pools and instead determine linearity and consistency of metabolite detection using the standard curves and the 13C internal standards.
Start off by injecting several water blanks to ensure system is clean from carry over and contaminants.
Include solvent blanks, using the 75/25/0.1 acetonitrile/methanol/formic acid mix that was used to make the extraction mix.
Follow with a system suitability test (SST) injection. We use 80% methanol containing 13C-15N labeled amino acid mix (Recipe 4).
Add the samples to your sequence and follow with the standard curves, starting with the lowest concentration and working up to the highest concentration for each curve. Separate each curve with solvent blank and check for carry-over.
Insert additional SST injections every 8-10 samples. These will be used as QCs to ensure no loss of signal over time.
Set the injection volume to 2 μl for each injection type.
Set the instrument method to the appropriate method.
Save the sequence file.
Randomize sample running order to decrease the chance of signal loss over time. Export the sequence as a .csv file. Open in Microsoft Excel, cut and paste the samples into a new tab, leaving behind the blanks, STTs and standard curves. Add an additional column and use the = rand () function to create random numbers for each of the samples. Now sort the samples from smallest to largest using the random number values. Cut and paste back into the previous sequence containing the blanks, SSTs and the standard curves. Save the .csv file with “random” in the file name. Import the new randomized sequence back into Xcalibur Sequence View and save using “random” in the file name.
-
Place your sample vials in the autosampler in the vial positions according to the sequence.
Use the non-randomized sequence to check vial positions for your samples.
Ensure the solvent blank vials and SST vials contain enough volume for multiple injections.
-
Run the sequence.
Note: For examples of expected outputs from the LC/MS analysis of TIF and plasma samples, LC/MS data from (Sullivan et al., 2019) is available at https://www.metabolomicsworkbench.org/data/DRCCMetadata.php?Mode=Project&ProjectID=PR000750.
Figure 1. Collecting TIF using nylon mesh filters attached to conical tubes.
A. The filter is loosely affixed to the top of the conical tube using laboratory tape, so that the filter will sag slightly into the conical tube when a sample is placed on top of the filter. B. A sample on top of the filter and conical tube. C. After adding the sample to the filter, the lid of the conical tube is placed on top, but not screwed onto the conical. Instead, it is taped using laboratory tape in place. D. ~30 μl of tumor interstitial fluid (colored blue to here for contrast) collected in the conical tube after centrifugation.
Table 1. LC parameters.
| Time (min) | Flow rate (ml/min) | %B |
|---|---|---|
| 0.00 | 0.150 | 80 |
| 20.0 | 0.150 | 20 |
| 20.5 | 0.150 | 80 |
| 28.0 | 0.150 | 80 |
Data analysis
-
Identify metabolite peaks. This protocol will describe peak identification in Thermo Scientific Xcalibur, but could be adapted with any other peak identification method.
-
Generate a processing method file that will be used to identify peaks for each metabolite of interest:
Create a new processing method.
-
Load a .raw file containing LC/MS data derived from an external standard sample that contains the metabolite of interest as well as a 13C internal standard for that metabolite.
Note: Typically using an external standard sample that is in the middle of the standard curve works best; sometimes high concentrations points on the standard curve have poor quality peaks.
-
Calculate the exact mass of the metabolite of interest.
Notes:
Exact mass is determined by summing the masses of the most abundant isotopes of each element in a compound. For instance, the exact mass of CO2 would be the summed masses of a carbon-12 atom (12.000) + two oxygen-16 atoms (15.995 + 15.995) = 43.990. This exact mass of a compound will differ from its molecular weight; the molecular weight of an element is derived by averaging the masses of each of the isotopes of that element, weighted by the abundance of each isotope in nature.
If using Thermo Scientific Xcalibur, calculate the exact mass using the Isotope Simulation tab in the QualBrowser module. Enter the chemical formula, ensure the “adduct” check box in unchecked and select “New”. Ensure that the software global settings are set to a mass tolerance of 5 ppm and mass precision is set to 5 decimal places. Ideally, the peak integration software that you are using should calculate this for you.
-
Calculate the mass to charge ratio (m/z) of the metabolite of interest in positive and negative ionization mode.
Notes:
For analysis of small polar metabolites, the most common ions will be those that have gained or lost a single proton and most molecules will have a charge state of 1.
If using Thermo Scientific Xcalibur, calculate the m/z using the Isotope Simulator in the QualBrowser module by entering the formula, checking the “adduct” box and selecting a charge of either +1 or -1, depending on whether you are calculating the m/z in positive or negative mode, respectively.
There are a variety of online tools available that will provide exact mass information, as well as calculate m/z for a variety of different adducts. The most comprehensive is the Metlin data base (Guijas et al., 2018): https://metlin.scripps.edu/landing_page.php?pgcontent=mainPage.
-
In the processing method, select either positive ionization mode or negative ionization mode depending on whether you will be searching for a positively charged ion or a negatively charged ion.
Note: Some metabolites are more easily detected in positive or negative mode. A list of recommendations for which mode to use for a variety of metabolites is located in Supplementary File 1 of (Sullivan et al., 2019). If no recommendations are available, empirically determine which method gives better detection by trying both.
-
Identify and validate the retention time of each metabolite:
Search for the exact mass of the ion of interest.
Note the retention time of any peaks that match the exact mass of the ion of interest within 5 ppm.
-
Open a .raw file of a different external standard sample with the metabolite of interest at a lower concentration.
Note which peaks that match the exact mass of the ion of interest decrease in area.
Repeat with each of the external standard samples that contain the metabolite of interest, checking which peak areas track with the expected amount of the metabolite.
Refer to MS/MS data to confirm peak identification.
Open a .raw file that does not contain the metabolite of interest. Ensure that any candidate peaks are not present in this .raw file.
-
Search for the exact mass of the 13C labeled version of the ion of interest.
Note: This peak should be approximately the same area in all samples.
Check that the retention time of the 13C labeled standard peak exactly matches that of the candidate peak.
Repeat for all metabolites of interest.
-
Assign 13C labeled standards as internal standards for their corresponding 12C metabolites.
For metabolites with no 13C internal standard, assign a 13C metabolite with a similar retention time as the internal standard.
-
Use the processing method to pick and validate peaks for all metabolites in all LC/MS data files (both experimental samples and external standards):
-
Once all peaks have been automatically picked, manually inspect every peak for each metabolite and for each sample to ensure that all peaks have been correctly identified. Some common examples of errors that occur with automatic peak picking algorithms:
Incorrect peak was picked: this can occur for isobaric compounds with similar retention times, such as leucine and isoleucine.
Peak was not fully picked from baseline to baseline.
Peak was picked but overlaps with a second peak: this occasionally happens where biological samples have an overlapping peak that was not present in the external standards. If this is the case, this metabolite should not be quantitated using this LC/MS method and an alternative method of chromatographic separation should be identified.
Export the ratio of peak areas for the sample versus the 13C internal standard to Microsoft Excel or your data processing software of choice.
-
-
-
Determine the relationship between relative peak area and concentration of metabolite in external standards.
Calculate the exact concentration of each metabolite in each point on the external standard curve based on the amount that was weighed out.
Generate a graph of metabolite concentration in each external standard sample versus the relative peak area of the metabolite.
-
Check if the relative peak area of the metabolite increases linearly with concentration:
Fit a linear regression to the graph.
-
The R2 value for the linear regression should be ≥ 0.995.
Metabolites often respond non-linearly at high concentrations. If the standard curve has points that are much higher than the concentrations present in experimental samples, the highest points on the standard curve can be removed. Just ensure that the relative peak areas for all samples fall within the linear range of the standard curve.
Non-linear metabolites should be excluded from quantitative analysis, as this lack of linearity will prevent accurate quantitation by isotope dilution.
-
Determine the concentrations of the internal standards that were added to all samples.
Solve for the concentration of 13C internal standard present in each external standard sample using the following relationship:
Note: This relationship can be used to calculate the concentration of the 13C internal standard in each of the external standard samples; the same concentration should be present in each. To derive the most accurate value for the concentration of the 13C internal standard, average the concentrations derived from the external standard points most similar in concentration to the experimental samples.
Calculate the concentration of each metabolite in the experimental samples by isotope dilution. Solve for the concentration of the 12C metabolite using the same relationship defined in step C.
-
Calculate the semi-quantitative concentration of all other analytes using the external standard curves. These values are considered semi-quantitative as they are subject to matrix effects arising from the biological samples being compared to external standards dissolved in water. These matrix effects can be substantial ( Sullivan et al., 2019 ).
Calculate the slope and intercept of the linear regression calculated in step B3a.
Use this slope and intercept to calculate the semi-quantitative value approximating the concentration of the metabolite.
-
Manually evaluate the concentrations derived from this calculation:
Check that the value of each metabolite is zero in the external standard samples that do not contain that metabolite.
Any data that shows a negative concentration should be removed.
-
Perform statistical analysis of the data using Metaboanalyst (https://www.metaboanalyst.ca/MetaboAnalyst/faces/home.xhtml) ( Chong et al., 2018 ) or another program for statistical analysis.
Auto-scale the data (mean-center and divide by the standard deviation of each concentration).
To broadly compare if there are differences in the metabolites present in two sample types, perform principal component analysis or hierarchical clustering.
To identify specific metabolites that differ in concentration between sample types, generate a volcano plot in which a raw P-value of 0.01 and a fold change of 1.5 are used to identify significantly altered metabolites.
Recipes
-
Mobile Phase A
20 mM ammonium carbonate
0.1% ammonium hydroxide (pH 9.4-9.6)
Optima LC/MS water
-
Mobile Phase B and Needle Wash
Optima LC/MS acetonitrile
-
Rear Seal Wash
10% Optima LC/MS methanol
Optima LC/MS water
-
80% methanol (containing 13C-15N amino acid standard mix) (200 ml)
160 ml Optima LC/MS methanol
40 ml Optima LC/MS water
40 μl Metabolomics Amino Acid Standard Mix
-
Extraction Buffer with isotopically labeled internal standards (EB) (Table 4)
Notes:
This is the recipe containing isotopically labeled internal standards for analysis of metabolites as in (Sullivan et al., 2019). If analysis of other metabolites using stable isotope internal standards is desired, purchase or synthesize the desired isotopically labeled metabolite and add it to the Extraction Buffer. Quantification with isotopically labeled standards performs best when the abundance of the isotopically labeled metabolite is similar to the unlabeled metabolite to be quantified. Therefore, when adding isotopically labeled internal standards, add the isotope such that it will be at roughly a similar abundance as the unlabeled metabolite when the sample is diluted in the Extraction Buffer.
This recipe is for 180 samples at 45 μl per sample for a total of 8,100 μl in volume. Adjust the volumes accordingly as needed for the number of samples you intend to analyze. After adding all components, vortex briefly to ensure EB is well mixed, and store on ice while in use. Make fresh prior to each experiment. Remember to include extra 75/25/0.1 acetonitrile/methanol/formic acid for use as solvent blanks in your calculations.
-
Chemical standard library preparation
Below are recipes for the preparation of chemical standard libraries described in ( Sullivan et al., 2019 ). To prepare these chemical libraries, purchase the chemicals from the listed supplier and weigh them out as indicated, placing each metabolite into a 50 ml mixing mill jar. Mix the combined metabolites using a Mixer Mill MM301 with five 5 mm diameter stainless steel grinding balls. Perform 6 cycles of 1 min mixing at 25 Hz followed by 3 min resting. Store the now mixed chemical standard library powder stocks at -20 °C prior to use. For use, resuspend each mixed chemical library in HPLC grade water at 5 mM concentration as indicated for each library below.
Custom chemical standard libraries can be produced by acquiring desired pure chemical standards and mixing the pure chemical standards in equimolar amounts. When generating libraries, it is important to ensure that each library will not contain metabolites that have the same exact mass, as it is not then possible to determine the correct retention time for both metabolites when compounded into the same library. Consider putting these metabolites into separate pooled libraries (Tables 5-11).
Table 5. Chemical standard library pool 1.
Metabolite name Molecular weightof metaboliteMolecular weightof chemical standardAmount to weigh (mg) Alanine 89.09 89.09 429.99 Arginine 174.2 210.66 1016.75 Asparagine 132.12 150.13 724.60 Aspartate 133.11 133.11 642.45 Carnitine 161.199 197.66 954.00 Citrulline 175.2 175.2 845.60 Cystine 240.3 240.3 1159.80 Glutamate 147.13 147.13 710.12 Glutamine 146.14 146.14 705.34 Glycine 75.066 75.066 362.30 Histidine 155.1546 155.1546 748.85 Hydroxyproline 131.13 131.13 632.89 Isoleucine 131.1729 131.1729 633.10 Leucine 131.17 131.1729 633.10 Lysine 146.19 182.65 881.56 Methionine 149.21 149.21 720.16 Ornithine 132.16 168.62 813.84 Phenylalanine 165.19 165.19 797.28 Proline 115.13 115.13 555.67 Serine 105.09 105.09 507.21 Taurine 125.15 125.15 604.03 Threonine 119.119 119.119 574.92 Tryptophan 204.225 204.225 985.69 Tyrosine 181.19 181.19 874.51 Valine 117.151 117.151 565.42 Lactate 90.09 112.06 540.85 Glucose 180.1559 180.1559 869.52 Note: Dissolve this pool at 20.19 mg/ml for 5 mM solution.Table 6. Chemical standard library pool 2.
Metabolite name Molecular weight of metabolite Molecular weight of chemical standard Amount to weigh (mg) 2-hydroxybutyric acid 104.1 126.09 12.61 2-aminobutyric acid 103.12 103.12 10.31 AMP 347.2212 347.22 34.72 Argininosuccinate 290.273 334.24 33.42 Betaine 117.1463 117.15 11.71 Biotin 244.31 244.31 24.43 Carnosine 226.2324 226.23 22.62 Choline 104.1708 139.62 13.96 CMP 323.1965 367.16 36.72 Creatine 131.133 131.13 13.11 Cytidine 243.2166 243.22 24.32 dTMP 320.1926 366.17 36.62 Fructose 180.16 180.16 18.02 Glucose-1-phosphate 260.135 336.32 33.63 Glutathione 307.3235 307.32 30.73 GMP 363.22 407.18 40.72 IMP 348.206 392.17 39.22 O-phosphoethanolamine 141.063 141.06 14.11 Pyridoxal 167.16 203.62 20.36 Thiamine 265.35 337.23 33.72 trans-Urocanate 137.118 137.12 13.71 UMP 324.1813 368.15 36.82 Xanthine 152.11 152.11 15.21 Note: Dissolve this pool at 28.54 mg/ml for 5 mM solution.Table 7. Chemical standard library pool 3.
Metabolite name Molecular weight of metabolite Molecular weight of chemical standard Amount to weigh (mg) 3-hydroxybutyric acid 104.1045 126.09 252.18 Acetylalanine 131.1299 131.13 262.26 Acetylaspartate 175.139 175.14 350.28 Acetylcarnitine 203.2356 239.70 479.40 Acetylglutamine 188.183 188.18 376.36 ADP 427.203 501.32 1002.64 Allantoin 158.121 158.12 316.24 CDP 403.177 403.20 806.40 CDP-choline 489.332 510.31 1020.62 Coenzyme A 767.535 767.53 1535.06 Creatinine 113.12 113.12 226.24 gamma-aminobutyric acid 103.12 103.12 206.24 GDP 443.201 443.20 886.40 Glutathione disulfide 612.631 612.63 1225.26 Glycerate 106.0773 286.25 572.50 Hypoxanthine 136.1115 136.11 272.22 myo-Inositol 180.16 180.16 360.32 NAD+ 663.43 663.43 1326.86 p-aminobenzoate 137.138 137.14 274.28 Phosphocholine 184.152 329.73 659.46 Sorbitol 182.17 182.17 364.34 UDP 404.1612 448.12 896.24 UDP-glucose 566.302 610.27 1220.54 Note: Dissolve this pool at 37.23 mg/ml for 5 mM solution.Table 8. Chemical standard library pool 4.
Metabolite name Molecular weight of metabolite Molecular weight of chemical standard Amount to weigh (mg) Phenylacetylglutamine 264.3 264.3 17.17 Acetylglutamate 189.1659 189.1659 12.29 Acetylglycine 117.1033 117.1033 7.61 Acetylmethionine 191.245 191.245 12.43 Asymmetric dimethylarginine 202.25 275.2 17.88 ATP 507.18 551.14 35.82 CTP 483.1563 527.12 34.26 dATP 491.2 535.15 34.78 dCTP 467.2 511.12 33.22 Deoxycytidine 227.2172 227.2172 14.76 Folic acid 441.3975 441.3975 28.69 GTP 523.2 523.18 34.00 Hypotaurine 109.1475 109.1475 7.09 Methionine sulfoxide 165.21 165.21 10.73 Methylthioadenosine 297.3335 297.3335 19.32 Phosphocreatine 211.114 255.08 16.58 Pyridoxine 169.18 205.64 13.36 Ribose-5-phosphate 230.11 310.1 20.15 SAH 384.4 384.41 24.98 Thymidine 242.2286 242.2286 15.74 Trimethyllysine 189.279 224.73 14.60 Uridine 244.2014 244.2014 15.87 UTP 484.1411 559.09 36.34 Note: Dissolve this pool at 36.75 mg/ml for 5 mM solution.Table 9. Chemical standard library pool 5.
Metabolite name Molecular weight of metabolite Molecular weight of chemical standard Amount to weigh (mg) 3-phosphoglycerate 186.06 230.02 57.51 cis-aconitic acid 174.108 174.11 43.53 Citrate 192.124 294.10 73.53 DHAP 170.06 180.19 45.05 Fructose-1,6-bisphosphate 340.1157 406.06 101.52 Fumarate 116.07 116.07 29.02 Glucose-6-phosphate 260.135 282.12 70.53 Glycerol-3-phosphate 172.0737 370.40 92.60 Guanidinoacetate 117.1066 117.11 29.28 Kynurenine 208.2139 208.21 52.05 Malate 134.0874 134.09 33.52 NADP+ 744.413 765.39 191.35 Niacinamide 122.12 122.12 30.53 2-oxoglutarate 146.11 146.11 36.53 Phosphoenolpyruvate 168.042 267.22 66.81 Pyruvate 88.06 110.04 27.51 Succinate 118.09 118.09 29.52 Uracil 112.0868 112.09 28.02 Note: Dissolve this pool at 20.77 mg/ml for 5 mM solution.Table 10. Chemical standard library pool 6.
Metabolite name Molecular weight of metabolite Molecular weight of chemical standard Amount to weigh (mg) 3-hydroxyisobutyric acid 104.1045 126.09 22.07 2-hydroxyglutarate 148.114 192.10 33.62 Aminoadipate 161.156 161.16 28.20 beta-alanine 89.093 89.09 15.59 Carbamoylaspartate 176.128 176.13 30.82 Cystathionine 222.263 222.26 38.90 Cysteic acid 169.16 169.16 29.60 FAD 785.5497 829.51 145.16 Glycerophosphocholine 258.231 257.22 45.01 Inosine 268.229 268.23 46.94 Orotate 156.1 194.19 33.98 Pantothenate 219.23 238.27 41.70 Phosphoserine 185.07 185.07 32.39 Riboflavin 376.369 376.37 65.86 UDP-GlcNAc 607.3537 651.32 113.98 Uric acid 168.1103 168.11 29.42 Note: Dissolve this pool at 21.52 mg/ml for 5 mM solution.Table 11. Chemical standard library pool 7.
Metabolite name Molecular weight of metabolite Molecular weight of chemical standard Amount to weigh (mg) Itaconic acid 130.0987 130.10 52.04 Homocysteine 135.185 135.19 54.07 2-oxobutyric acid 102.0886 102.09 40.84 2-hydroxybutyric acid 104.1045 126.09 50.44 Ascorbate 176.1241 198.11 79.24 Sarcosine 89.0932 89.09 35.64 Dimethylglycine 103.1198 103.12 41.25 N6-acetyllysine 188.2242 188.22 75.29 Pipecolate 129.157 129.16 51.66 Indolelactate 205.2099 205.21 82.08 Picolinate 123.1094 123.11 49.24 3-methyl-2-oxobutyrate 116.1152 138.10 55.24 3-methyl-2-oxopentanoic acid 130.1418 152.12 60.85 Formyl-methionine 177.221 177.22 70.89 2-aminobutyric acid 103.1198 103.12 41.25 Homocitrulline 189.2123 189.21 75.68 gamma-glutamyl-alanine 217.2224 218.21 87.28 Mannose 180.16 180.16 72.06 Cysteine-glycine (dipeptide) 178.21 178.21 71.28 Note: Dissolve this pool at 14.33 mg/ml for 5 mM solution.
Table 4. Extraction Buffer (EB) composition.
| Component | Volume added | Final concentration | |
|---|---|---|---|
| HPLC grade acetonitrile | 5771.25 μl | 71.25% | |
| HPLC grade methanol | 1923.75 μl | 23.75% | |
| HPLC grade formic acid | 15.39 μl | 1.9% | |
|
~15 mg of isotopically labeled yeast extract (ISO1) dissolved in 1.5 ml of HPLC grade water. Note: After adding water to the yeast extract, dissolve the yeast extract by vortexing and/or rocking the yeast extract and water at 4 °C for approximately 30 min. Solution can be stored at -80 °C although some metabolites will degrade over time (see manufacturer’s instructions). |
405 μl | 5% | |
| 2 mM solution of 2H9 choline prepared in HPLC grade water (stored at -20 °C) | 4.03 μl | 1 μM | |
| 50 mM solution of 13C4 3-hydroxybutyrate prepared in HPLC grade water (stored at -20 °C) | 0.81 μl | 5 μM | |
| 200 μM solution of 13C6 15N2 cystine prepared in HPLC grade water (stored at -20 °C) | 81 μl | 2 μM | |
| 100 mM solution of 13C3 lactate prepared in HPLC grade water (stored at -20 °C) | 16.2 μl | 200 μM | |
| 57.3 mM solution of 13C6 glucose prepared in HPLC grade water (stored at -20 °C) | 7.05 μl | 50 μM | |
| 100 mM solution of 13C3 serine prepared in HPLC grade water (stored at -20 °C) | 1.62 μl | 20 μM | |
| 750 mM solution of 13C2 glycine prepared in HPLC grade water (stored at -20 °C) | 1.62 μl | 150 μM | |
| 2 mM solution of 13C5 hypoxanthine prepared in HPLC grade water (stored at -20 °C) | 2.02 μl | 0.5 μM | |
| 200 mM solution of 13C2 15N taurine prepared in HPLC grade water (stored at -20 °C) | 2.02 μl | 50 μM | |
| 60 mM solution of 13C3 glycerol prepared in HPLC grade water (stored at -20 °C) | 2.02 μl | 15 μM | |
| 4 mM solution of 2H3 creatinine prepared in HPLC grade water (stored at -20 °C) | 2.02 μl | 1 μM | |
Acknowledgments
This protocol is based on our previously published study ( Sullivan et al., 2019 ). This work has been supported by a grant to AM from the NIH (F32CA213810). MRS was supported by T32GM007287 and acknowledges additional support from an MIT Koch Institute Graduate Fellowship.
Competing interests
The authors report no competing interests. CAL is a paid consultant for ReviveMed.
Ethics
This study was performed in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals. All animal experiments were performed using protocols (#1115-110-18) that were approved by the MIT Committee on Animal Care (IACUC). All surgeries were performed using isoflurane anesthesia administered by vaporizer and every effort was made to minimize suffering.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1. Abbondante S., Eckel-Mahan K. L., Ceglia N. J., Baldi P. and Sassone-Corsi P.(2016). Comparative circadian metabolomics reveal differential effects of nutritional challenge in the serum and liver. J Biol Chem 291(6): 2812-2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Anastasiou D.(2017). Tumour microenvironment factors shaping the cancer metabolism landscape. Br J Cancer 116(3): 277-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bi J., Wu S., Zhang W. and Mischel P. S.(2018). Targeting cancer's metabolic co-dependencies: A landscape shaped by genotype and tissue context. Biochim Biophys Acta Rev Cancer 1870(1): 76-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Burgess E. A. and Sylven B.(1962). Glucose, lactate, and lactic dehydrogenase activity in normal interstitial fluid and that of solid mouse tumors. Cancer Res 22: 581-588. [PubMed] [Google Scholar]
- 5. Cairns R. A., Harris I. S. and Mak T. W.(2011). Regulation of cancer cell metabolism. Nat Rev Cancer 11(2): 85-95. [DOI] [PubMed] [Google Scholar]
- 6. Cantor J. R., Abu-Remaileh M., Kanarek N., Freinkman E., Gao X., Louissaint A. Jr., Lewis C. A., and Sabatini D.M.(2017). Physiologic Medium Rewires Cellular Metabolism and Reveals Uric Acid as an Endogenous Inhibitor of UMP Synthase. Cell 169(2): 258-272 e217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chong J., Soufan O., Li C., Caraus I., Li S., Bourque G., Wishart D. S. and Xia J.(2018). MetaboAnalyst 4.0: towards more transparent and integrative metabolomics analysis. Nucleic Acids Res 46(W1): W486-W494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dallmann R., Viola A. U., Tarokh L., Cajochen C. and Brown S. A.(2012). The human circadian metabolome. Proc Natl Acad Sci U S A 109(7): 2625-2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. DeBerardinis R. J. and Chandel N. S.(2016). Fundamentals of cancer metabolism. Sci Adv 2(5): e1600200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Eil R., Vodnala S. K., Clever D., Klebanoff C. A., Sukumar M., Pan J. H., Palmer D. C., Gros A., Yamamoto T. N., Patel S. J., Guittard G. C., Yu Z., Carbonaro V., Okkenhaug K., Schrump D. S., Linehan W M., Roychoudhuri R. and Restifo N. P.(2016). Ionic immune suppression within the tumour microenvironment limits T cell effector function. Nature 537: 539-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Evans A. M., DeHaven C. D., Barrett T., Mitchell M. and Milgram E.(2009). Integrated, nontargeted ultrahigh performance liquid chromatography/electrospray ionization tandem mass spectrometry platform for the identification and relative quantification of the small-molecule complement of biological systems. Anal Chem 81(16): 6656-6667. [DOI] [PubMed] [Google Scholar]
- 12. Gieger C., Geistlinger L., Altmaier E., Hrabé de Angelis M., Kronenberg F., Meitinger T., Mewes H. W., Wichmann H. E., Weinberger K. M., Adamski J., Illig T. and Suhre K.(2008). Genetics meets metabolomics: a genome-wide association study of metabolite profiles in human serum. PLoS Genet 4(11): e1000282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gullino P. M., Clark S. H. and Grantham F. H.(1964). The interstitial fluid of solid tumors. Cancer Res 24: 780-794. [PubMed] [Google Scholar]
- 14. Guijas C., Montenegro-Burke J. R., Domingo-Almenara X., Palermo A., Warth B., Hermann G., Koellensperger G., Huan T., Uritboonthai W., Aisporna A. E., Wolan D. W., Spilker M. E., Benton H. P. and Siuzdak G.(2018). METLIN: a technology platform for identifying knowns and unknowns. Anal Chem 90(5): 3156-3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Haslene-Hox H., Oveland E., Berg K. C., Kolmannskog O., Woie K., Salvesen H. B., Tenstad O. and Wiig H.(2011). A new method for isolation of interstitial fluid from human solid tumors applied to proteomic analysis of ovarian carcinoma tissue. PLoS One 6(4): e19217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ho P. C., Bihuniak J. D., Macintyre A. N., Staron M., Liu X., Amezquita R., Tsui Y. C., Cui G., Micevic G., Perales J. C., Kleinstein S. H., Abel E. D., Insogna K. L., Feske S., Locasale J. W., Bosenberg M. W., Rathmell J. C., Kaech S. M.(2015). Phosphoenolpyruvate is a metabolic checkpoint of anti-tumor T cell responses. Cell 162(6): 1217-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lawton K. A., Berger A., Mitchell M., Milgram K. E., Evans A. M., Guo L., Hanson R. W., Kalhan S. C., Ryals J. A. and Milburn M. V.(2008). Analysis of the adult human plasma metabolome. Pharmacogenomics 9(4): 383-397. [DOI] [PubMed] [Google Scholar]
- 18. Mazzone P. J., Wang X. F., Beukemann M., Zhang Q., Seeley M., Mohney R., Holt T. and Pappan K. L.(2016). Metabolite Profiles of the Serum of Patients with Non-Small Cell Carcinoma. J Thorac Oncol 11(1): 72-78 [DOI] [PubMed] [Google Scholar]
- 19. Muir A., Danai L. V. and Vander Heiden M. G.(2018). Microenvironmental regulation of cancer cell metabolism: implications for experimental design and translational studies. Dis Model Mech 11(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nagarajan A., Malvi P. and Wajapeyee N.(2016). Oncogene-directed alterations in cancer cell metabolism. Trends Cancer 2(7): 365-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Panuwet P., Hunter R. E. Jr. D'Souza P. E., Chen X., Radford S. A., Cohen J. R., Marder M. E., Kartavenka K., Ryan P. B. and Barr D. B.(2016). Biological matrix effects in quantitative tandem mass spectrometry-based analytical methods: advancing biomonitoring. Crit Rev Anal Chem 46(2): 93-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schroeder K. S. V. A.(2019). Blood Withdrawal I. JoVE. [Google Scholar]
- 23. Siska P. J., Beckermann K. E., Mason F. M., Andrejeva G., Greenplate A. R., Sendor A. B., Chiang Y. J., Corona A. L., Gemta L. F., Vincent B. G., Wang R. C., Kim B., Hong J., Chen C. L., Bullock T. N., Irish J. M., Rathmell W. K. and Rathmell J. C.(2017). Mitochondrial dysregulation and glycolytic insufficiency functionally impair CD8 T cells infiltrating human renal cell carcinoma. JCI Insight 2(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Spinelli J. B., Yoon H., Ringel A. E., Jeanfavre S., Clish C. B. and Haigis M. C.(2017). Metabolic recycling of ammonia via glutamate dehydrogenase supports breast cancer biomass. Science 358(6365): 941-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Stevens V. L., Hoover E., Wang Y. and Zanetti K. A.(2019). Pre-analytical factors that affect metabolite stability in human urine, plasma, and serum: A review. Metabolites 9(8):156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sullivan M. R., Danai L. V., Lewis C. A., Chan S. H., Gui D. Y., Kunchok T., Dennstedt E. A., Vander Heiden M. G. and Muir A.(2019). Quantification of microenvironmental metabolites in murine cancers reveals determinants of tumor nutrient availability. Elife 8: e44235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wiig H. and Swartz M. A.(2012). Interstitial fluid and lymph formation and transport: physiological regulation and roles in inflammation and cancer. Physiol Rev 92(3): 1005-1060. [DOI] [PubMed] [Google Scholar]
- 28. Wiig H., Tenstad O., Iversen P. O., Kalluri R. and Bjerkvig R.(2010). Interstitial fluid: the overlooked component of the tumor microenvironment? Fibrogenesis Tissue Repair 3: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang Y., Kurupati R., Liu L., Zhou X. Y., Zhang G., Hudaihed A., Filisio F., Giles-Davis W., Xu X., Karakousis G. C., Schuchter L. M., Xu W., Amaravadi R., Xiao M., Sadek N., Krepler C., Herlyn M., Freeman G. J., Rabinowitz J. D. and Ertl H. C. J.(2017). Enhancing CD8+ T cell fatty acid catabolism within a metabolically challenging tumor microenvironment increases the efficacy of melanoma immunotherapy . Cancer Cell 32(3): 377-391. [DOI] [PMC free article] [PubMed] [Google Scholar]