Abstract
Tissues are comprised of different cell types whose interactions elicit distinct gene expression patterns that regulate tissue formation, regeneration, homeostasis and repair. Analysis of these gene expression patterns require methods that can capture as closely as possible the transcriptomes of cells of interest in their tissue microenvironment. Current technologies designed to study in situ transcriptomics are limited by their low sensitivity that require cell types to represent more than 1% of the total tissue, making it challenging to transcriptionally profile rare cell populations rapidly isolated from their native microenvironment. To address this problem, we developed fluorouracil-tagged RNA sequencing (Flura-seq) that utilizes cytosine deaminase (CD) to convert the non-natural pyrimidine fluorocytosine to fluorouracil. Expression of S. cerevisiae CD and exposure to fluorocytosine generates fluorouracil and metabolically labels newly synthesized RNAs specifically in cells of interest. Fluorouracil-tagged RNAs can then be immunopurified and used for downstream analysis. Here, we describe the detailed protocol to perform Flura-seq both in vitro and in vivo. The robustness, simplicity and lack of toxicity of Flura-seq make this tool broadly applicable to many studies in developmental, regenerative, and cancer biology.
Keywords: In situ transcriptomics , Cytosine deaminase, Uracil phosphoribosyl transferase, Fluorouracil, Nascent RNA
Background
In an organ, cells interact with tissue microenvironment that includes organ-specific resident cells, immune cells, perivascular niches, extracellular matrix, cytokines, metabolites, and an oxygen concentration range. These interactions dictate the expression of specific set of genes in the cells that often have functional significances. The transcriptional responses associated with these interactions are usually dynamic and can only be observed if the interactions are preserved. Thus, it is critical to maintain the tissue microenvironment to ensure the capture of true transcriptional state of cells under the physiological conditions.
Current techniques to study cell-type specific transcriptomes have limitations that preclude their effective application in studying rare and under-represented cell populations. Single-cell RNA sequencing (scRNA-seq) with or without an intervening fluorescence activated cell sorting (FACS) step, requires extensive physical and enzymatic processing of the tissue, which not only disrupts the effects of the host microenvironment on cells but also exerts stress on these cells, severely compromising the ability to discern the impact of the host stroma from the transcriptome of the isolated cells. As many as 7,500 genes have been reported to be altered by more than two-fold during the FACS processing of muscle stem cells for transcriptomic analysis ( Machado et al., 2017 ). In situ transcriptomic profiling obviate these problems but lack the necessary sensitivity for cell populations that represent less than 1% of the tissue. For example, translating ribosome affinity purification and mRNA sequencing (TRAP-Seq) ( Heiman et al., 2008 ) is not suitable to analyze cells that constitute less than 1% of the total population ( Bertin et al., 2015 ; Obenauf et al., 2015 ). Direct enzyme based metabolic tagging of RNA with thiouracil (TU) and ethynyl cytosine (EC) in the cells of interest are limited in sensitivity and specificity due to collateral tagging of RNA in cells lacking the enzymes, and requires additional in vitro biotinylation steps ( Cleary et al., 2005 ; Miller et al., 2009 ; Gay et al., 2013 and 2014; Hida et al., 2017 ). TU tagging has a sensitivity limit of 5% ( Gay et al., 2013 ). Thiol (SH)-linked alkylation of the metabolic labeling of RNA in tissue (SLAM-ITseq) eliminates the noise associated with the purification of RNAs that are not thiol tagged in TU-tagging method ( Matsushima et al., 2018 ), but undesired TU tagging through endogenous enzymes in cells lacking UPRT expression remains a limitation. We described the development of Flura-seq, a cytosine deaminase (CD)-based method for in situ transcriptomic profiling of rare cell populations that represent as little as 0.003% of an organ ( Basnet et al., 2019 ). Flura-seq requires exogenous expression of CD, a key enzyme of the pyrimidine salvage pathway in fungi and prokaryotes ( Mullen et al., 1992 ) and UPRT from T. gondii. CD is absent in mammalian cells, which instead use cytidine deaminase for the same purpose ( Mullen et al., 1992 ). In addition to converting cytosine to uracil, CD can also convert 5-fluorocytosine (5-FC), a non-natural pyrimidine, to 5-flourouracil (5-FU). 5-FU is endogenously converted to fluorouridine triphosphate (F-UTP), which is then incorporated into RNA. The co-expression of UPRT limits the labeling of RNA in the cells expressing CD ( Basnet et al., 2019 ). Flura-seq is applicable to in vitro co-culture experiments as well as in vivo experiments where cells of interest, expressing CD-UPRT, are present in the intact tissue. Flura-seq is technically very simple that only requires very basic molecular biology skills, and the whole procedures can be completed within 24 hours. Here, we describe the detailed protocol for labeling and isolation of RNA for Flura-seq for both in vitro and in vivo experiments.
Materials and Reagents
5 ml Polystyrene Round-Bottom tube (Corning, catalog number: 352058)
60 mm Tissue culture plates (Corning, catalog number: 353004)
Oligo (dT)25 magnetic beads (New England Biolabs, catalog number: S1419S)
Protein G Dynabeads (Thermo Fisher Scientific, catalog number: 10004D)
6-8 weeks old athymic nude female mice (Envigo, catalog number: 069)
Doxycycline diet (625 mg/kg, Envigo, catalog number: TD.07383)
BrdU antibody (Abcam, catalog number: ab6326)
CD-UPRT (Addgene, plasmid number: 126677)
rtTA3 (Addgene, plasmid number: 26730)
Doxycycline (Sigma-Aldrich, catalog number: D9891)
5-Fluorocytosine (5-FC) (Sigma-Aldrich, catalog number: F7129)
5-Bromo-2’-deoxyuridine (BrdU) (Millipore Sigma, catalog number: B5002)
Thymine (Sigma-Aldrich, catalog number: T0376)
RNeasy MinElute Cleanup kit (Qiagen, catalog number: 74204)
cDNA kit-First Strand Transcriptor (Roche, catalog number: 043790-12001)
RNeasy Mini Kit (Qiagen, catalog number: 74106)
Ethylenediaminetetraacetic acid solution (EDTA) (Sigma-Aldrich, catalog number: 03690)
Tween 20 (Sigma Millipore, catalog number: P1379)
Lithium dodecyl sulfate (LiDS) (Sigma Millipore, catalog number: L4632)
Lithium chloride (LiCl) (Sigma Millipore, catalog number: 203637)
Ultrapure BSA (BSA) (Thermo Fisher Scientific, catalog number: AM2616)
Glycogen (Thermo Fisher Scientific, catalog number: R0551)
RNAlater (Sigma-Aldrich, catalog number: R0901)
20x UltraPure SSPE Buffer (Thermo Fisher Scientific, catalog number: 15591043)
PBS
Tris base
Tris-HCl
Ethanol (Decon Labs, catalog number: 2716)
DTT (Sigma-Aldrich, catalog number: 3483-12-3)
Cell lysis buffer (see Recipes)
mRNA Wash Buffer I (see Recipes)
mRNA Wash Buffer II (see Recipes)
mRNA Wash Buffer III (see Recipes)
Elution Buffer (see Recipes)
SSPET buffer (see Recipes)
TE buffer (see Recipes)
Equipment
PRO 200 grinder from PRO Scientific Inc (Homogenizer) (PRO Scientific, catalog number: 01-01200)
Magnetic rack (ThermoFisher Scientific, catalog number: 12321D)
Thermo scientific Rotator (ThermoFisher Scientific, catalog number: 88881001)
Eppendorf Thermomixer (ThermoFisher Scientific, catalog number: 05-400-200)
Centrifuge
Procedure
-
Making CD-UPRT expressing stable cells
1.Generate CD-UPRT expressing cell line of interest (for example, MDA-MB-231) by transducing cells with CD-UPRT (Addgene) and rtTA3 (Addgene) lentivirus using standard protocol.
-
Labeling of RNA in vitro
Day 1
-
1
Plate 1,000 cells (for example, MDA-MB-231) expressing Doxycycline inducible CD-UPRT (cells of interest) with 1 million 4T1 cells (or other cells (optional)) in 60 mm plates in 3 ml volume (Doxycycline inducible system is optional).
Day 2
-
2
Add 1 μg/ml Doxycycline.
Day 3
-
3
24 h post Doxycycline treatment, add 5-FC and thymine to 250 μM (50 mM stock) and 125 μM (25 mM stock) final concentration, respectively. 5-FC and thymine stock solutions are made in PBS.
-
4
Harvest cells after 2-12 h using cell lysis buffer. Wash the cells 1x with 3 ml PBS before adding 1 ml of cell lysis buffer on the plate. Scrape the cells in 1.5 ml eppendorf tube.
-
1
-
Labeling of RNA in mice
Day 1
-
1
Prepare 500,000/ml CD-UPRT expressing stable cells (described above in A.1) in PBS.
-
2
Inject 100 μl (50,000 cells) in mouse through tail vein.
Day 28
-
3
Change the mice diet to Doxycycline diet, 3 days before 5-FC injection, if cells are expressing Doxycycline inducible CD-UPRT. Skip this step if cells are constitutively expressing CD-UPRT.
Day 31
-
4
Inject the mice with 250 mg/kg 5-FC in PBS (12.5 μg/ml stock) intraperitoneally, and 125 mg/kg thymine in PBS (6.25 μg/ml stock) subcutaneously.
-
5
After 4-12 h, euthanize the mice by CO2 asphyxiation, harvest the organs, and keep the organ in RNAlater buffer.
-
6
Put the organ of interest (for example, lungs) in 2 ml cell lysis buffer (Recipe 1) in 5 ml Polystyrene Round-Bottom tube, and homogenize the organ with a tissue grinder.
-
7
Divide the lysate equally into two tubes and add 3 ml of cell lysis buffer in each tube (The lysate can be flash frozen in liquid nitrogen for future use).
-
1
-
Isolation of mRNA
Add 1x volume of cell lysis buffer (4 ml) in the lysate from C.7, rotate at room temperature (RT) for 5 min. For in vitro cells, add appropriate volume of lysis buffer (1 ml for confluent 60 mm plates), and scrape the cells.
Put the lysate into 1.5 ml Eppendorf tube/s, and centrifuge at 16, 000 × g for 5 min at RT, and keep supernatant.
-
Prepare oligo dT25 beads (≥ 100 μl beads/~25 mg of lungs/brain).
Place 250 μl beads/Eppendorf tube (1.5 ml) in magnetic rack.
Remove the storage buffer.
Wash the beads 1x with 1 ml cell lysis buffer by pipetting up and down five times.
Take out the supernatant (1 ml/250 μl beads) from D.2 and add it to the freshly prepared oligo dT25 beads.
Incubate at RT for 10 min in a rotator.
Discard the supernatant.
Wash the beads 2x with 1 ml mRNA wash buffer I (Recipe 2) at RT by pipetting up and down seven times.
Wash the beads 2x with 1 ml mRNA wash buffer II (Recipe 3) at RT by pipetting up and down seven times.
Wash the beads 1x with 1 ml mRNA wash buffer III (Recipe 4) at RT by pipetting up and down seven times.
Wash the beads 1x with 1 ml TE buffer (Recipe 7) at RT by pipetting up and down seven times.
Add 50 μl of Elution buffer (Recipe 5), and elute the RNA in Eppendorf Thermomixer at 85 °C for 2 min shaking at 750 rpm.
Place the beads in magnetic rack and keep the Eluate. The mRNA can be stored at -80 °C for future use.
-
Purification of 5-FU tagged mRNA
Prepare 70 μl of protein G beads per sample at RT. Place the beads in magnetic rack, and remove the storage buffer. Upto 350 μl (for 5 samples) of beads can be transferred to one 1.5 ml Eppendorf tube.
Wash the beads with 1 ml of 0.5x SSPET buffer (Recipe 6) at RT by pipetting up and down five times.
Block the beads with 1 ml of 0.5x SSPET buffer containing 10 μg/ml BSA and 20 μg/ml glycogen for 1 h at 4 °C.
Wash the beads 1× with 1 ml of 0.5x SSPET buffer at RT by pipetting up and down five times.
Add 35 μl of beads in 750 μl of 0.5x SSPET buffer containing BrdU antibody (1 μg for RT-PCR, and 5 μg for RNA-seq) in 1.5 ml Eppendorf tube. Rotate at 4 °C overnight.
Wash the beads 1× with 1 ml of 0.5x SSPET buffer at RT by pipetting up and down five times.
Add 35 μl of beads in mRNA (400 μl in this case) from Step D12 diluted with the same volume (400 μl) of 1x SSPET buffer. Total volume will be about 835 μl.
Rotate for 2 h using ThermoScientific Rotator at RT.
Wash 2× with 1 ml of 0.5x SSPET buffer at RT by pipetting up and down seven times.
Wash 2× with 1 ml of 1x SSPET buffer at RT by pipetting up and down seven times.
Wash 1× with 1 ml of TE buffer at RT by pipetting up and down seven times.
Elute the bound mRNA with 200 μl of 100 μg/ml BrdU in TE buffer in Eppendorf Thermomixer at 900 rpm for 45 min at RT.
Keep the supernatant (5-FU tagged mRNA).
Mix the 200 μl supernatant with 700 μl of RLT buffer from RNeasy kit.
Add 500 μl of 100% ethanol, and mix well.
Pass the solution through RNeasy column.
Wash 1× with 500 μl of RPE buffer from RNeasy kit.
Wash 1× with 500 μl of 80% ethanol.
Spin at 13,000 × g for 2 min at RT.
Elute the RNA with 200 μl of RNase free water.
Add 600 μl of 0.5x SSPET to the elute.
Wash the Protein G Dynabeads incubated with BrdU antibody as described in Step E6.
Re-immunoprecipitate 5-FU tagged mRNA as described in Steps E7-E13.
Purify RNA as described in Steps E14-E19 using RNeasy MiniElute Cleanup kit, and elute the RNA with 12.5 μl RNase free water.
Proceed with cDNA synthesis for RT-PCR or RNA-seq.
-
Labeling and Purification of 5-Fluorouracil (5-FU) tagged mRNA from co-cultured cells and from mouse lungs in xenograft model
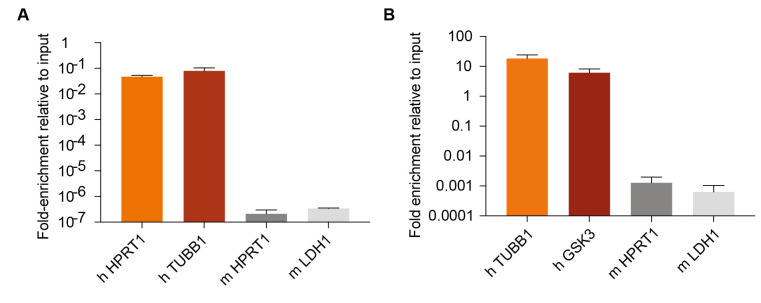
1,000 human MDA-MB-231 cells transduced with CD-UPRT (Addgene) and rtTA3 (Addgene) (MDA231-CDUPRT) were co-cultured with 1 million mouse 4T1 cells. RNAs were labeled as described in Section B, and 5-FU tagged mRNAs were purified as described in Section D and E. Enrichment of 5-FU tagged mRNAs of representative human and mouse genes relative to non immuno-purified mRNA were measured by RT-PCR (Figure 1A).
5 × 104 MDA231-CDUPRT cells were injected in the mouse lungs through tail vein. After 28 days, CD UPRT expression was induced by feeding mice with doxycycline diet for 3 days, and RNA was labeled as described in Section C, and 5-FU tagged mRNA was purified as described in Section D and E. Enrichment of 5-FU tagged mRNAs of representative human and mouse genes relative to non immuno-purified mRNA were measured by RT-PCR (Figure 1B).
Figure 1. Labeling and isolation of 5-FU tagged RNA by cytosine deaminase expression.
A. 1,000 human MDA-MB-231 cells expressing CD-UPRT were co-cultured with 1 million mouse 4T1 cells, and 5-FU tagged mRNA from human cells were isolated as described in the protocol, and enrichment of representative human genes and mouse genes relative to their corresponding 2% inputs (non immuno-purified mRNA) are shown (n = 3, ± SEM). B. 50,000 MDA-MB-231 cells expressing CD-UPRT were injected to the lungs through tail vein, and after 28 days, 5-FU tagged mRNA was isolated as described in the protocol, and the enrichment of representative human and mouse genes relative to 1% inputs (non immune-purified mRNAs) were measured by RT-PCR (n = 6, ± SEM). “h” indicates human and “m” indicates mouse.
Notes
For in vitro experiments, confluency of cells does not affect 5-FU tagging (Step B1) and the method is likely to be applicable to all cell types (adherent as well as suspension cells). We have successfully applied the method in MDA-MB-231, HCC1954, H2087, 293T and 4T1 cells.
Thymine does not dissolve completely in PBS. Filter the solution before use (Step B3).
Longer incubation of 5-FC and thymine increases labeling of RNA, and yields higher signal-to-noise ratio. 12 h of labeling is recommended for very small number of cells (< 0.01% or < 1,000 cells). For conditions consisting of more than 1 × 105 cells, 4 h of RNA labeling is sufficient. (Steps B4 and C4). In vivo experiments have been successfully performed with 4 h and 12 h of labeling, and in vitro experiments have been performed with 2-12 h of labeling.
In mice experiments, doxycycline can also be administered through drinking water (Step C3).
This protocol has been successfully used in mouse lungs, brain and mammary fat pad. Other organs have not been tested yet.
For in vivo experiments, the growth curve of different cell lines can vary. Hence, it is important to pick the time for 5-FC injection based on desired size of the tumors. For example, mouse cancer cell lines grow much faster than MDA-MB-231 cells, so earlier time point might be suitable for mouse cancer cell lines (Step C4).
The strain, sex and age of the mice can vary for different cell lines. For example, different mice strain might be suitable for syngeneic experiments.
Recipes
-
Cell lysis buffer
20 mM Tris-HCl pH 7.5
500 mM LiCl
1% LiDS
1 mM EDTA
5 mM DTT (DTT is added freshly)
-
mRNA Wash Buffer I
20 mM Tris-HCl pH 7.5
500 mM LiCl
0.1% LiDS
1 mM EDTA
5 mM DTT (DTT is added freshly)
-
mRNA Wash Buffer II
20 mM Tris-HCl pH 7.5
500 mM LiCl
1 mM EDTA
-
mRNA Wash Buffer III
20 mM Tris-HCl pH 7.5
200 mM LiCl
1 mM EDTA
-
Elution Buffer
20 mM Tris-HCl pH 7.5
1 mM EDTA
-
SSPET buffer
1× SSPE (Sodium Chloride-Sodium Phosphate-EDTA)
0.05% Tween 20
-
TE buffer
10 mM Tris pH 7.5
1 mM EDTA
Acknowledgments
H.B. was supported by a Damon Runyon Postdoctoral Fellowship. This work was supported by NIH grants P01-CA094060 (J.M.), P30-CA008748 (MSKCC), and a DOD Innovator award W81XWH-12-0074 (J.M.). The protocol is derived from Basnet et al., 2019 .
Competing interests
H.B and J.M have filed for patent for Flura-seq method (PCT/US18/22092). J.M serves in the scientific advisory board and owns company stock in Scholar Rock.
Ethics
Mouse experiments were performed following the protocol (99-09-032) approved by the MSKCC Institutional Animal Care and Use Committee (IACUC) that is valid until 06/19/2020.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1. Basnet H., Tian L., Ganesh K., Huang Y. H., Macalinao D. G., Brogi E., Finley L. W. and Massagué J.(2019). Flura-seq identifies organ-specific metabolic adaptations during early metastatic colonization. Elife 8: e43627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bertin B., Renaud Y., Aradhya R., Jagla K. and Junion G.(2015). TRAP-rc, translating ribosome affinity purification from rare cell populations of Drosophila embryos . J Vis Exp (103). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cleary M. D., Meiering C. D., Jan E., Guymon R. and Boothroyd J. C.(2005). Biosynthetic labeling of RNA with uracil phosphoribosyltransferase allows cell-specific microarray analysis of mRNA synthesis and decay. Nat Biotechnol 23(2): 232-237. [DOI] [PubMed] [Google Scholar]
- 4. Gay L., Karfilis K. V., Miller M. R., Doe C. Q. and Stankunas K.(2014). Applying thiouracil tagging to mouse transcriptome analysis. Nat Protoc 9(2): 410-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gay L., Miller M. R., Ventura P. B., Devasthali V., Vue Z., Thompson H. L., Temple S., Zong H., Cleary M. D., Stankunas K. and Doe C. Q.(2013). Mouse TU tagging: a chemical/genetic intersectional method for purifying cell type-specific nascent RNA. Genes Dev 27(1): 98-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Heiman M., Schaefer A., Gong S., Peterson J. D., Day M., Ramsey K. E., Suárez-Fariñas M., Schwarz C., Stephan D. A., Surmeier D. J., Greengard P. and Heintz N.(2008). A translational profiling approach for the molecular characterization of CNS cell types. Cell 135(4): 738-748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hida N., Aboukilila M. Y., Burow D. A., Paul R., Greenberg M. M., Fazio M., Beasley S., Spitale R. C. and Cleary M. D.(2017). EC-tagging allows cell type-specific RNA analysis. Nucleic Acids Res 45(15): e138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Machado L., Esteves de Lima J., Fabre O., Proux C., Legendre R., Szegedi A., Varet H., Ingerslev L. R., Barres R., Relaix F. and Mourikis P.(2017). In situ fixation redefines quiescence and early activation of skeletal muscle stem cells . Cell Rep 21(7): 1982-1993. [DOI] [PubMed] [Google Scholar]
- 9. Matsushima W., Herzog V. A., Neumann T., Gapp K., Zuber J., Ameres S. L. and Miska E. A.(2018). SLAM-ITseq: sequencing cell type-specific transcriptomes without cell sorting. Development 145(13). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Miller M. R., Robinson K. J., Cleary M. D. and Doe C. Q.(2009). TU-tagging: cell type-specific RNA isolation from intact complex tissues. Nat Methods 6(6): 439-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mullen C. A., Kilstrup M., and Blaese R. M.(1992). Transfer of the bacterial gene for cytosine deaminase to mammalian cells confers lethal sensitivity to 5-fluorocytosine: a negative selection system. Proc Natl Acad Sci U S A 89(1): 33-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Obenauf A. C., Zou Y., Ji A. L., Vanharanta S., Shu W., Shi H., Kong X., Bosenberg M. C., Wiesner T., Rosen N., Lo R. S. and Massagué J.(2015). Therapy-induced tumour secretomes promote resistance and tumour progression. Nature 520(7547): 368-372. [DOI] [PMC free article] [PubMed] [Google Scholar]