Abstract
The essential peptidoglycan (PG) layer surrounds the cytoplasmic membrane in nearly all bacteria. It is needed to maintain the shape of the cell and protect it from lysis due to high turgor. Growth of the PG layer is a complex process that involves the activities of PG synthases and hydrolases during elongation and cell division. PG growth sites can be labeled by the recently developed fluorescent D-amino acid (FDAA) probes in a range of different bacteria. FDAAs are incorporated into PG by dd-transpeptidases (Penicillin-binding proteins, PBPs) or, if present, ld-transpeptidase (LDTs). Long-pulse in situ labeling of E. coli cells with the FDAA 7-hydroxycoumarincarbonylamino-D-alanine (HADA) is expected to result in a uniform label at the side wall of cells and enhanced label at cell division sites due to the intense PG synthesis. However, we observed reduced label at mid-cell when labeling E. coli cells with HADA. We reasoned that probe incorporated at cell division sites may be removed by PG hydrolases and modified the labeling protocol to better preserve PG-incorporated HADA for fluorescence microscopy. Here, we report the optimized HADA-labeling protocol by which cells retain an enhanced HADA signal at the division septum.
Keywords: Fluorescence D-amino acid (FDAA), HADA, Peptidoglycan synthesis, Transpeptidase, Labeling
Background
The peptidoglycan (PG) sacculus is a mesh-like, essential macromolecule that encases the cytoplasmic membrane in most bacteria. It is composed of glycan chains made of alternating β-1, 4-linked N-acetylglucosamine and N-acetylmuramic acid residues connected via short peptides and is required to maintain the shape and osmotic stability of a bacterial cell ( Typas et al., 2011 ). Growing and dividing bacteria synthesize new PG and incorporate it into their PG sacculus, and these processes are targeted by important antibiotics such as the β-lactams and glycopeptides. To visualize the growth sites on the PG sacculus Miguel de Pedro developed an elegant method in which D-cysteine (D-Cys) is first homogeneously incorporated into PG of growing bacteria (by then unknown enzymes), followed by a chase period in the absence of D-Cys. PG sacculi were isolated, followed by the selective biotinylation of D-Cys residues and the visualization of biotin with nano-gold labeled antibodies by electron microscopy ( de Pedro et al., 1997 ). PG growth sites were characterized by regions of reduced label (when D-Cys became 'diluted' upon incorporation of new, label free PG during the chase) or without label (a zone of exclusively new PG). Although this method was crucial to determine the modes of PG segregation in model species such as Escherichia coli ( de Pedro et al., 1997 ) and Caulobacter crescentus ( Aaron et al., 2007 ), it was relatively cumbersome and used mainly in specialist laboratories. Over the last years, Erkin Kuru, Michael VanNieuwenhze and Yves Brun developed an ever-increasing palette of fluorescent D-amino acid (FDAA) probes that covalently label PG in cells and can therefore be used to visualize PG by fluorescence microscopy ( Kuru et al., 2012 and 2015; Hsu et al., 2017 ). New 'rotor probes' of this series, rFDAA, produce fluorescence only upon incorporation into PG, enabling real-time visualization of probe incorporation and making it unnecessary to wash cells after the labeling procedure ( Hsu et al., 2019 ). Presumably FDAAs are incorporated into PG by PBPs and LDTs, as are other D-amino acids ( Cava et al., 2011 ; Lupoli et al., 2011 ). FDAAs efficiently label sites of active PG synthesis and have therefore been successfully used to track PG synthesis sites in different bacterial species ( Kuru et al., 2012 ; Radkov et al., 2018 ). They are easy to use and reliably label the PG in many bacteria and they have contributed to major discoveries in a variety of bacterial species ( Radkov et al., 2018 ), for example, the discovery of PG in Chlamydia trachomatis and its visualization ( Liechti et al., 2014 and 2016), the tracking of septal PG synthesis dynamics in B. subtilis (Bisson- Filho et al., 2017 ) and the visualization of preseptal PG synthesis in E. coli ( Pazos et al., 2018 ).
We recently showed that copper(II) inhibits LD-TPases in vivo and in vitro ( Peters et al., 2018 ). Within this study we performed long-pulse in situ labeling of E. coli cells with the FDAA 7-hydroxycoumarincarbonylamino-D-alanine (HADA), which was previously shown to uniformly label cells ( Kuru et al., 2012 and 2015). Indeed, using the published procedure wild-type cells showed homogeneous label along the lateral wall but, unexpectedly, constricting cells showed a reduced signal at the septum. No signal could be detected in a strain lacking all six LD-TPases ( Peters et al., 2018 ). During cell growth PG synthesis takes place at the division septum due to the activities of PG synthases (PBP1B and PBP3) ( de Pedro et al., 1997 ; Bertsche et al., 2006 ). We reasoned that the HADA signal at mid-cell was low despite the enhanced PG synthesis because the incorporated HADA might be removed by PG hydrolases during the time the cells were harvested and washed at neutral pH, resulting in the loss of label at the division septum. Indeed, labeling of chlamydial PG with fluorescent dipeptide required the inactivation of DD-carboxypeptidases by amplicillin ( Liechti et al., 2014 ). Here we optimized the long-pulse HADA labeling protocol to reduce the potential loss of HADA label by PG hydrolases. Specifically, we stopped cell growth and label incorporation by adding sodium citrate buffer pH 2.25 to the growing cells and washed the cells once with sodium citrate buffer at pH 3.0, followed by two washing steps with phosphate-buffered saline (PBS, pH 7.4), prior to processing the samples for fluorescence microscopy. With this procedure cells of wild-type and several mutants were homogeneously labeled at the side-wall and had enhanced signal at the septum of constricting cells. We conclude that the rapid incubation and washes of cells at acidic pH preserves the HADA label by preventing the removal of incorporated HADA by PG hydrolases (Amanuma and Strominger, 1980; Stefanova et al., 2002 ), resulting in improved signal detection at cell division sites.
Materials and Reagents
Sterile Petri dishes 92 × 16 mm with cams (SARSTEDT, catalog number: 82.1473)
Pipette tips 10 µl, 200 µl and 1,000 µl (STARLAB, catalog numbers: S1111-3700, S1113-1700, S1111-6701)
1.5 ml micro-tubes (SARSTEDT, catalog number: 72.690.001)
Sterile serological pipettes (5, 10 and 25 ml) (SARSTEDT, catalog numbers: 86.1253.001, 86.1254.001 and 86.1685.001)
Sterile 50 ml CELLSTAR tubes with blue screw cap (Greiner BIO-ONE, catalog number: 227261)
Concentric Luer-Lok Syringe 50 ml (BD Plastipak, BD Luer Lok, catalog number: 10636531)
0.22 µm syringe filter (PES membrane, sterile) (STARLAB, catalog number: E4780-1226)
Diagnostic microscopic slides (12 wells, 5.2 mm numbered) (Thermo Scientific, catalog number: 10028210)
Microscope slides SuperFrost plus (25 × 75 × 1 mm) (R. Langenbrinck Labor + Medizintechnik, catalog number: 03-0060)
Cover glasses, square (22 × 22 mm, thickness 1.5) (VWR, catalog number: 631-0125)
Semi-micro cuvettes (polystyrene, 10 × 4 × 45 mm) (SARSTEDT, catalog number: 67.742)
Milli-Q quality water (ddH2O) and distilled water
Peptone from casein, enzymatic digest (Sigma-Aldrich, catalog number: 82303)
Peptone from soybean, enzymatic digest (Fluka Analytical, catalog number: 90765)
Sodium chloride (NaCl) (VWR, catalog number: 27810.295)
Di-Potassium hydrogen phosphate (K2HPO4) (VWR, catalog number: 26931.263)
D-Glucose (anhydrous) (Melford, catalog number: G1400)
Bacto Agar (BD Diagnostics, catalog number: 214030)
Bacto Tryptone (BD Diagnostics, catalog number: 211699)
Yeast extract powder (Ohly KAT, catalog number: FllOHLYKAT)
FDAA 7-hydroxycoumarincarbonylamino-D-alanine (HADA) provided by Michael S. VanNieuwenhze, Indiana University ( Kuru et al., 2015 )
Dimethylsulfoxide anhydrous (DMSO) (Invitrogen, catalog number: D12345)
Citric acid (anhydrous) (Sigma-Aldrich, catalog number: C2404)
Sodium hydroxide (NaOH) (VWR, catalog number: 28245.298)
10× PBS buffer with 0.017 M KH2PO4, 0.05 M Na2HPO4, 1.5 M NaCl, pH 7.4 (phosphate saline) (Lonza Walkersville INC, AccuGENE, catalog number: 51226)
16% Paraformaldehyde (formaldehyde) aqueous solution (Electron Microscopy Science, catalog number: 15710-S)
SB Molecular Biology Grade Agarose (Severn Biotech LtD., catalog number: 30 10 50)
Tryptic soy broth (TSB) and Tryptic soy agar (TSA) (see Recipes)
Miller Luria-Bertani (LB) medium and LB agar (see Recipes)
HADA stock solution (see Recipes)
10× Sodium citrate buffer, pH 2.25 (see Recipes)
1× sodium citrate buffer, pH 3.0 (see Recipes)
1× Phosphate buffered saline pH 7.4 (PBS) (see Recipes)
3% Paraformaldehyde solution (see Recipes)
Agar slides for microscopic imaging (see Recipes)
Equipment
Sterile, flexible and disposable Inoculating loops (VWR, catalog number: 612935P)
Glass beaker, 600 ml and 1,000 ml (PYREX, catalog numbers: 1000/18D and 1000/22D, respectively)
Premium single Cell Analytical Balance (Kern, model: ABT 120-4NM)
IKA RH basic 2 magnetic stirrer (IKA, catalog number: 0003339002)
pH meter (Cole-Parmer, Jenway, model: 3510, catalog number: 351001)
Pipettes (Gilson, catalog numbers: F167300 and F167500)
Ultra Low Temperature Freezer VIPTM Series -86 °C (SANYO, catalog number: MDF-U73V)
Comfort TP 1410 table-high, under worktop fridge (Liebherr, catalog number: 4016803034339)
Vortex (IKA, model: Minishaker MS2)
Microwave MS106 0.6 FT (Matsui)
50 L Laboratory Incubator with Digital Control (GenLab incubator: INC/50/DIG)
Microcentrifuge accuSpin Micro 17R, refrigerated (Fisher Scientific, catalog number: 11526873)
Nikon Eclipse Ti microscope (Nikon Plan Fluor × 100/1.30 Oil Ph3 DLL objective) equipped with a Photometrics/Cool SNAP HQ2CCD camera using the phase contrast and DAPI channel (filter set: Chroma 49000, excitation at 350/50 nm, emission 460/50 nm)
Autoclave
Software
ImageJ (Rasband W.S./U. S. NIH, Bethesda, Maryland, USA/https://imagej.nih.gov/ij/)
MetaMorph® Version 7.0 (Microscopy Automation and Image Analysis Software/Molecular DevicesLLC/https://www.moleculardevices.com/products/cellular-imaging-systems/acquisition-and-analysis-software/metamorph-microscopy)
Procedure
-
Prepare the following solutions in advance
TSB (or LB) and TSB (LB) agar plates (see Recipes).
50 mM HADA stock solution (see Recipes).
-
10 and 1x sodium citrate buffers, 1x PBS and 3% paraformaldehyde (see Recipes).
Note: Filter and cool 10x and 1x sodium citrate buffers, 1x PBS and 3% paraformaldehyde solution on ice prior to use.
-
Growing the cells
Defrost glycerol stock solutions of the bacterial strains on ice. Streak the strains BW25113 (Datsenko and Wanner, 2000), D456 (Edwards and Donachie, 1993) and CS703-1 ( Meberg et al., 2001 ) on TSB agar plates using an inoculation loop and incubate the plates overnight at 37 °C.
Inoculate 25 ml of TSB (or LB) media with a single bacterial colony and incubate overnight at 37 °C.
Dilute the overnight cultures to an OD578 of 0.1 in TSB (or LB) media and grow at 37 °C until they reach an exponential OD578 of 0.4.
-
Labeling the cells
Dilute the cells to an OD578 of 0.1 in a final volume of 500 μl of pre-warmed (37 °C) TSB (or LB) in a sterile 1.5 ml micro-tube.
Add 2.5 µl of the 50 mM stock solution of fluorescent derivative of D-alanine (HADA) to the sample to achieve a final concentration of 250 μM and incubate with shaking for 30 min at 37 °C.
-
Stop bacterial growth and washing steps to remove the excess of HADA
Add one-tenth of the final volume (50 µl) of ice cold 10x sodium citrate buffer (pH 2.25) to the growing cultures. Place the samples on ice until the centrifuge (step 2) is ready to use.
Centrifuge the samples for 2 min at 16,200 × g at 4 °C.
Remove the supernatant carefully and resuspend the cell pellet in 1.5 ml ice cold 1x sodium citrate buffer (pH 3.0) by gentle pipetting.
Centrifuge the samples for 2 min at 16,200 × g at 4 °C. Remove the supernatant carefully and resuspend the cells in 1.5 ml ice cold 1x PBS (pH 7.4) by gentle pipetting and centrifuge the samples again.
-
Repeat the washing step with ice cold 1x PBS (pH 7.4) for another time. Remove the supernatants of the samples carefully without touching the cell pellets.
Note: It was reported that HADA is virtually non-fluorescent in acidic pH or non-buffered water and cells imaged at a pH above 7.0 showed the maximum brightness of HADA labeling (Kuru et al., 2015). Therefore, the last two washing steps with 1× PBS (pH 7.4) are crucial.
-
Cell fixation
Resuspend the cell pellets in 12.5 µl 1× PBS and 12.5 μl of 3% paraformaldehyde (see Recipes) for cell fixation.
Mix well by pipetting and store on ice or 4 °C until microscopic analysis.
Note: Microscopic analysis of the labeled cells should be performed immediately.
-
Microscopic analysis
Pipette 3 μl of the fixed cells in a well of the prepared microscopic agarose slide (see Recipes) and place a square shaped cover glass carefully on top and press gently.
Analyze the labeled cells with a Nikon Eclipse Ti microscope (Nikon Plan Fluor × 100/1.30 Oil Ph3 DLL objective) equipped with a Photometrics/Cool SNAP HQ2CCD camera using the phase contrast and DAPI channel (filter set: Chroma 49000, excitation at 350/50 nm, emission 460/50 nm).
-
Use exposure times of 100 ms for phase contrast and 1 s for fluorescence images.
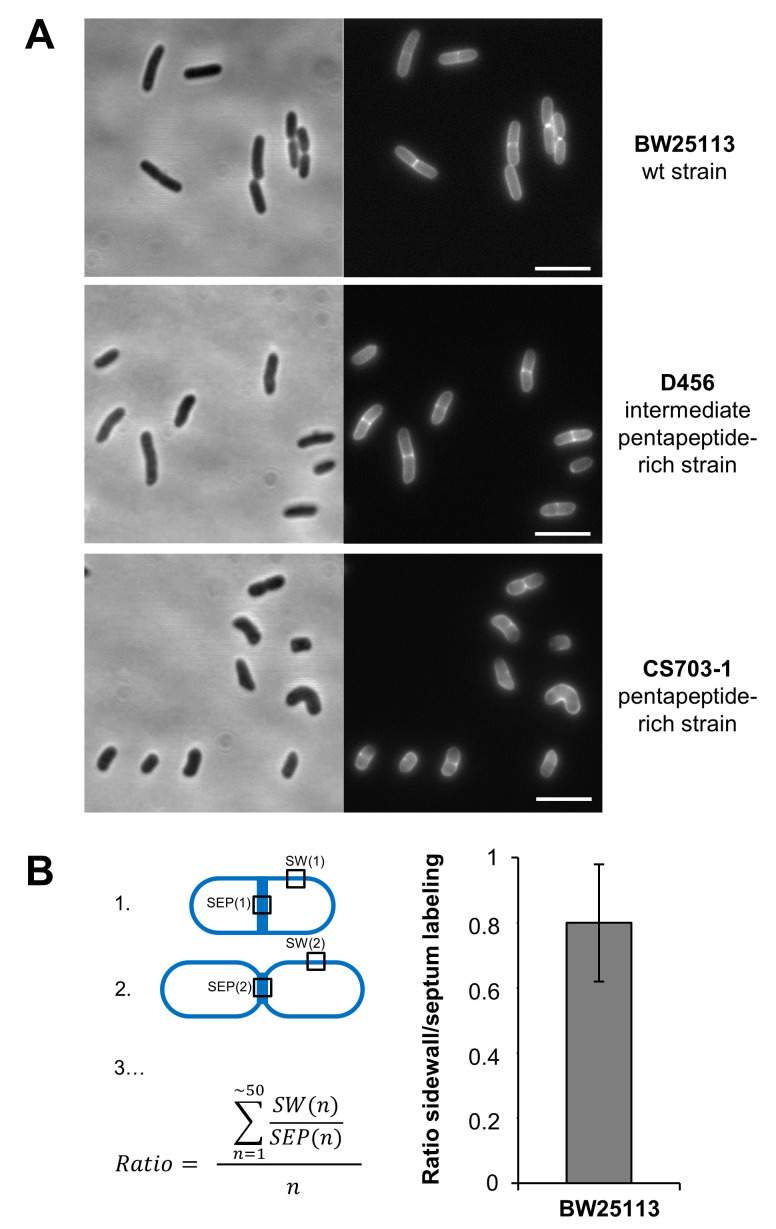
Note: Using this optimized protocol, all analyzed cells were completely labeled, a stronger signal at the septum of constricting cells was revealed (see Figure 1). The addition of 10x sodium citrate buffer to the growing cells, the initial wash with 1x sodium citrate buffer at pH 3.0, and the rapid processing of the samples on ice improved the removal of unincorporated HADA and led to a decreased background fluorescence signal and improved signal detection.
Figure 1. PG labeling of E. coli cells using the fluorescent D-amino acid HADA.
A. BW25113 (Datsenko and Wanner, 2000), D456 (Edwards and Donachie, 1993) and CS703-1 ( Meberg et al., 2011 ) cells were incubated with 250 µM HADA for 30 min (long pulse labeling). One-tenth of the final volume of 10x sodium citrate buffer (pH 2.25) was added to the cell cultures prior to short washing steps performed at 4 °C. First the cells were washed once with 1x sodium citrate buffer (pH 3.0) followed by two washing steps with phosphate buffer saline (PBS, pH 7.4). The cells were fixed with 3% paraformaldehyde and visualized by phase contrast (left) and fluorescence microscopy using the DAPI channel (right). Scale bars, 2 μm. B. Optional quantification of the sidewall (SW) and septum (SEP) fluorescence (depending on research question). As an example, the density of the fluorescence signals at the sidewall and septum in BW25113 (wt) were quantified in ~50 cells using Image J. The mean and standard deviation of the ratio of sidewall over septum signal density was calculated according to the shown formula and represented in a diagram.
Data analysis
ImageJ was used to crop the images to prepare figures and MetaMorph (v7) was used to quantify the fluorescence signals at the septum and at the sidewalls of ~50 cells from at least two independent experiments. The fluorescence intensity per unit of area at septal and sidewall positions can be quantified, the background subtracted, and the ratio of the sidewall over the septum signal density calculated. The values should be given as mean ± SD; the Student’s t-test should be used to determine the significance of the results ( Peters et al., 2018 ).
Notes
Experiments were performed in TSB and LB media without changing the reproducibility of the results.
Recipes
-
Tryptic soy broth (TSB) and Tryptic soy agar (TSA)
To prepare 1 L of TSB weigh the following reagents:
17 g Peptone from casein
3 g Peptone from soybean
5 g Sodium chloride
2.5 g Di-Potassium hydrogen phosphate (K2HPO4)
2.5 g D-Glucose (anhydrous)
Dissolve all ingredients in a final volume of 1 L distilled water under gentle stirring using a magnetic stirrer
Adjust the pH to 7.3 and autoclave the solution for 15 min at 121 °C
In order to prepare TSA add 15 g/L of agar to 1 L of TSB before autoclaving, cool the solution to a temperature of 48 °C after autoclaving and pour the agarose plates under sterile conditions
-
Miller Luria-Bertani (LB) medium and LB agar
To prepare 1 L of LB, weigh the following reagents:
10 g Bacto Tryptone
10 g Sodium chloride
5 g Yeast extract
Dissolve all ingredients in a final volume of 1 L distilled water under gentle stirring using a magnetic stirrer
Adjust the pH to 7.3 and autoclave the solution for 30 min at 121 °C
In order to prepare LB agar add 15 g/L of agar to 1 L of LB medium before autoclaving, cool the solution to a temperature of 48 °C after autoclaving and pour the agarose plates under sterile conditions
-
HADA stock solution
Dilute 2.1 mg of powdered HADA (328.7 g/mol) in 127.77 µl DMSO to create a 50 mM stock solution
Mix the stock solution well by pipetting and aliquot it in 10 µl portions in sterile 1.5 ml micro-tubes
Sterilization of stock solutions is not required. Store at -80 °C and protected from light
Thaw the stock solution rapidly at room temperature shortly before use
-
Avoid excessive cycles of thawing and refreezing, or long exposure at room temperature and light
Note: Dry powders of FDAAs are stable at -20°C or lower temperature for several months (Kuru et al., 2015). In our studies we stored HADA at -80°C, either as dry powder or as stock solution in DMSO for several months and without signs of decomposition or altered labeling efficacy.
-
10x Sodium citrate buffer, pH 2.25 (500 ml)
805 mM citric acid
410 mM NaOH
1.19 M NaCl
Weigh 77.5 g citric acid (anhydrous) and 34.9 g of sodium chloride
Add 206 ml of NaOH solution (c = 1 mol/L) and top the volume up to 500 ml with MilliQ water
Dissolve completely by gentle stirring and check the pH with a pH meter. It should be 2.25
-
1x sodium citrate buffer, pH 3.0 (500 ml)
80.5 mM citric acid
41 mM NaOH
119 mM NaCl
Weigh 7.74 g citric acid (anhydrous) and 3.49 g sodium chloride
Add 206 ml NaOH solution (c = 0.1 mol/L) and top the volume up to 500 ml with MilliQ water
Dissolve completely by gentle stirring and check the pH with a pH meter. It should be 3.0
-
1x Phosphate buffered saline pH 7.4 (PBS)
To prepare 50 ml of 1x PBS:
Pipette 5 ml of commercially available 10x PBS solution into a sterile Greiner tube
-
Add 45 ml of Milli Q water and mix well
Note: Filter the prepared solutions (10x and 1x sodium citrate buffer and the 1× PBS) into sterile 50 ml Greiner tubes. Use sterile single use 50 ml syringes and 0.22 µm syringe filters (PES membrane) for filtration. Cool the solutions on ice prior to use.
-
3% Paraformaldehyde solution
To prepare 1,000 µl of 3% paraformaldehyde for cell fixation:
Pipette 187.5 µl of 16% paraformaldehyde solution in a sterile 1.5 ml micro-tube and add 812.5 µl filtered 1× PBS solution
Vortex and cool down on ice prior use
-
Agar slides for microscopic imaging
To prepare an agarose solution with a final concentration of 1% w/v:
Dissolve 1 g agarose in 100 ml 1× PBS by microwaving
Bring the temperature of the solution to ~60 °C
Spot ~150 μl agarose solution in every second of the 12 wells on a diagnostic microscopic slide
Place a microscope slide on top and press gently down in order to get a flat, even and thin agarose pad
Let the pad solidify before you carefully remove the top microscope slide
Air-dry the agarose pad for ~1 min prior to use
Note: The cells do not settle on the agarose pad, if the agarose slide is not dried properly. This can make imaging of the cells more difficult.
Acknowledgments
This work reports some of the methods used to demonstrate that copper inhibits peptidoglycan LD-transpeptidases suppressing β-lactam resistance due to bypass of penicillin-binding proteins ( Peters et al., 2018 ). This work was funded by Wellcome Trust Grant 101824/Z/13/Z; the United Kingdom Medical Research Council within the Antimicrobial Resistance Cross-Council Initiative Collaborative Grant MR/N002679/1 (to W.V.) and NIH Grant GM113172 (to M.S.V.).
Competing interests
The authors declare that they have no conflict of interest or competing interest.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1. Aaron M., Charbon G., Lam H., Schwarz H., Vollmer W. and Jacobs-Wagner C.(2007). The tubulin homologue FtsZ contributes to cell elongation by guiding cell wall precursor synthesis in Caulobacter crescentus . Mol Microbiol 64(4): 938-952. [DOI] [PubMed] [Google Scholar]
- 2. Amanuma H. and Strominger J. L.(1980). Purification and properties of penicillin-binding proteins 5 and 6 from Escherichia coli membranes . J Biol Chem 255(23): 11173-11180. [PubMed] [Google Scholar]
- 3. Bertsche U., Kast T., Wolf B., Fraipont C., Aarsman M. E., Kannenberg K., von Rechenberg M., Nguyen-Distèche M., T. den Blaauwen, Höltje J. V. and Vollmer W.(2006). Interaction between two murein(peptidoglycan) synthases, PBP3 and PBP1B, in Escherichia coli . Mol Microbiol 61(3): 675-690. [DOI] [PubMed] [Google Scholar]
- 4. Bisson-Filho A. W., Hsu Y. P., Squyres G. R., Kuru E., Wu F., Jukes C., Sun Y., Dekker C., Holden S., VanNieuwenhze M. S., Brun Y. V. and Garner E. C.(2017). Treadmilling by FtsZ filaments drives peptidoglycan synthesis and bacterial cell division. Science 355(6326): 739-743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cava F., de Pedro M. A., Lam H., Davis B. M. and Waldor M. K.(2011). Distinct pathways for modification of the bacterial cell wall by non-canonical D-amino acids. EMBO J 30(16): 3442-3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Datsenko K. A. and Wanner B. L.(2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products . Proc Natl Acad Sci U S A 97(12): 6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. de Pedro M. A., Quintela J. C., Holtje J. V. and Schwarz H.(1997). Murein segregation in Escherichia coli. J Bacteriol 179(9): 2823-2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Edwards D. H. and Donachie W. D.(1993). Construction of a triple deletion of penicillin-binding proteins 4, 5, 6 in Escherichia coli. In: Bacterial Growth and Lysis. De Pedro, M. A., Höltje, J. V. and Löffelhardt, W.(Eds.). Boston, Springer: 369-374. Doi: 10.1007/978-1-4757-9359-8_44. [DOI] [Google Scholar]
- 9. Hsu Y. P., Hall E., Booher G., Murphy B., Radkov A. D., Yablonowski J., Mulcahey C., Alvarez L., Cava F., Brun Y. V., Kuru E. and VanNieuwenhze M. S.(2019). Fluorogenic D-amino acids enable real-time monitoring of peptidoglycan biosynthesis and high-throughput transpeptidation assays. Nat Chem 11(4): 335-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hsu Y. P., Rittichier J., Kuru E., Yablonowski J., Pasciak E., Tekkam S., Hall E., Murphy B., Lee T. K., Garner E. C., Huang K. C., Brun Y. V. and VanNieuwenhze M. S.(2017). Full color palette of fluorescent d-amino acids for in situ labeling of bacterial cell walls . Chem Sci 8(9): 6313-6321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kuru E., Hughes H. V., Brown P. J., Hall E., Tekkam S., Cava F., de Pedro M. A., Brun Y. V. and VanNieuwenhze M. S.(2012). In situ probing of newly synthesized peptidoglycan in live bacteria with fluorescent D-amino acids . Angew Chem Int Ed Engl 51(50): 12519-12523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kuru E., Tekkam S., Hall E., Brun Y. V. and Van Nieuwenhze M. S.(2015). Synthesis of fluorescent D-amino acids and their use for probing peptidoglycan synthesis and bacterial growth in situ . Nat Protoc 10(1): 33-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liechti G. W., Kuru E., Hall E., Kalinda A., Brun Y. V., VanNieuwenhze M. and Maurelli A. T.(2014). A new metabolic cell-wall labelling method reveals peptidoglycan in Chlamydia trachomatis . Nature 506(7489): 507-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Liechti G., Kuru E., Packiam M., Hsu Y. P., Tekkam S., Hall E., Rittichier J. T., VanNieuwenhze M., Brun Y. V. and Maurelli A. T.(2016). Pathogenic chlamydia lack a classical sacculus but synthesize a narrow, mid-cell peptidoglycan ring, regulated by MreB, for cell division. PLoS Pathog 12(5): e1005590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lupoli T. J., Tsukamoto H., Doud E. H., Wang T. S., Walker S. and Kahne D.(2011). Transpeptidase-mediated incorporation of D-amino acids into bacterial peptidoglycan. J Am Chem Soc 133(28): 10748-10751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Meberg B. M., Sailer F. C., Nelson D. E. and Young K. D.(2001). Reconstruction of Escherichia coli mrcA(PBP 1a) mutants lacking multiple combinations of penicillin binding proteins . J Bacteriol 183(20): 6148-6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pazos M., Peters K., Casanova M., Palacios P., VanNieuwenhze M., Breukink E., Vicente M. and Vollmer W.(2018). Z-ring membrane anchors associate with cell wall synthases to initiate bacterial cell division. Nat Commun 9(1): 5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Peters K., Pazos M., Edoo Z., Hugonnet J. E., Martorana A. M., Polissi A., VanNieuwenhze M. S., Arthur M. and Vollmer W.(2018). Copper inhibits peptidoglycan LD-transpeptidases suppressing β-lactam resistance due to bypass of penicillin-binding proteins. Proc Natl Acad Sci U S A 115(42): 10786-10791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Radkov A. D., Hsu Y. P., Booher G. and VanNieuwenhze M. S.(2018). Imaging bacterial cell wall biosynthesis. Annu Rev Biochem 87: 991-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Stefanova M. E., Davies C., Nicholas R. A. and Gutheil W. G.(2002). pH, inhibitor, and substrate specificity studies on Escherichia coli penicillin-binding protein 5 . Biochim Biophys Acta 1597(2): 292-300. [DOI] [PubMed] [Google Scholar]
- 21. Typas A., Banzhaf M., Gross C. A. and Vollmer W.(2011). From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat Rev Microbiol 10(2): 123-136. [DOI] [PMC free article] [PubMed] [Google Scholar]