Abstract
We describe here a detailed, refined protocol for the generation of citrulline-specific monoclonal antibodies from single human B cells from rheumatoid arthritis (RA) patients. This protocol provides a detailed guide of the procedure starting from single B cells of your choice and followed by amplification of the variable region of immunoglobulin genes by RT-PCR, subsequent immunoglobulin gene cloning, recombinant IgG1 monoclonal antibody (mAb) production and quality controls. The produced mAbs can be used for further studies including reactivity towards candidate antigens and functionality both in vitro and in vivo. This protocol can be used to generate antigen-specific mAbs from B cells derived from different tissues and compartments, including peripheral blood, synovial fluid, digested biopsies, bone marrow aspirations, and bronchoalveolar lavage fluid. Notably, although examples are given on how to identify citrulline-specific autoantibodies the general methods can also be applied to other reactivities.
Keywords: Autoantibodies, B cells, Antigen-specific monoclonal antibodies, Citrulline, Rheumatoid arthritis
Background
Rheumatoid arthritis (RA) is an autoimmune disease affecting 0.5% to 1% of adults worldwide and can if left untreated lead to joint destruction and disability (Neovius et al., 2011). Anti-citrullinated protein autoantibodies (ACPA) are a hallmark of RA and can predict a more aggressive disease course in patients that belong to the positive subgroup. This autoreactivity is directed towards citrullination, an enzyme mediated post-translational modification changing the positively charged arginine to a neutral citrulline. Interestingly, serum ACPA IgG often precede the onset of disease (Rantapää-Dahlqvist et al., 2003; Nielen et al., 2004) and while recent discoveries have started to decipher their roles in the pathogenesis and suggested that APCA may directly mediate pathogenic effects such as pain, osteoclast differentiation, and monocyte pro-inflammatory response (Harre et al., 2012; Sokolove et al., 2014; Wigerblad et al., 2016; Krishnamurthy et al., 2016; Krishnamurthy et al., 2019), much remains to be elucidated. The generation of citrulline-specific monoclonal antibodies (mAbs) from single B cells obtained from RA patient material provides a valuable tool to better understand ACPA-mediated functionality, as well as the B-cell biology and the Immunoglobulin (Ig) gene repertoire during disease. However, when generating monoclonal antibodies from autoimmune patients, there are several technical challenges and important aspects to consider, especially regarding analysis of the citrulline fine-specificity of these produced mAbs.
Hence, here we describe a detailed, refined protocol for the generation of citrulline-specific mAbs from single B cells from RA patients. The protocol provides a detailed guide of the procedure starting from single-cell sorted B cells e.g., by flow cytometry, amplification of the variable region of immunoglobulin genes by RT-PCR, and subsequent immunoglobulin gene cloning and full recombinant IgG1 monoclonal antibody (mAb) production. This protocol is derived from strategies developed some years ago in the Nussenzweig’s laboratory and that has been widely applied for generation of human monoclonal antibodies (Wardemann et al., 2003; Tiller et al., 2008). However, this protocol also includes more detailed methods for quality controls, IgG production, purification, storage and specificity-screenings. These are things that we have found to be critical to avoid false positivity in assays and to be able to separate true strong antigen-specific reactivity from the polyreactivity commonly seen in autoimmune patients that can lead to incorrect conclusions (Amara et al., 2019). Since aggregation or poor-quality IgG can give false positivity in ELISA, we strongly recommend to avoid IgG preparations with low concentrations and that any positivity found in initial exploratory screenings is confirmed in larger scale IgG expression batches that are more rigorously quality control tested. Furthermore, although this collection of methods is providing in-house ELISA protocols for detection of citrulline reactivity we recommend using several methods in parallel for confirmation of reactivity (e.g., antigen-arrays, multiplex bead arrays, and/or commercial CCP2/CCP3 clinical ELISA kits). All screenings should use an IgG concentration of 5 μg/ml IgG or lower to avoid any false positivity and any identified positive clones should subsequently be titrated from 5 μg/ml to 50 ng/ml (or lower) to confirm binding. While the frequency of polyreactive B cells clones are relatively high in the B cell repertoire, especially in memory B cells carrying low SHM, our data suggest that the true antigen-specific clones are in contrast relatively rare also in the tissue.
Therefore, it may be favorable to use an enrichment strategy for antigen-specific B cells or defined B cells subsets (e.g., antigen-tetramer sorting) rather than a broad repertoire screening. However, these methods are not within the scope of this protocol. Notably, a majority of the high-binding ACPA clones identified so far (Lloyd et al., 2018; Titcombe et al., 2018; Steen et al., 2019) carry high level of somatic hypermutations and N-linked glycosylation sites in the variable regions and these features may also have an indicative value when identifying new positive mAbs.
With this method, it is possible for an experienced researcher to produce milligrams of human recombinant mAbs within a few weeks, providing a valuable tool for downstream functional studies both in vitro and in vivo. The capacity to produce and characterize antigen-specific mAbs is extremely useful for elucidating the interactions of particular antibodies and antigens and exploring B-cell immunology.
Materials and Reagents
Common list
Adhesive PCR plate foils
Adhesive PCR plate seals
Filtered pipette tips
Graduated sterile pipettes
Parafilm
Reagent reservoir, 100 ml sterile polystyrene
1.5 and 2 ml microtubes
15 ml conical tubes
50 ml conical tubes
96-well PCR plates
96-well PCR plates, skirted
Bacto-Tryptone (Thermo Fisher Scientific, catalog number: 16299741)
CaCl2 (Sigma-Aldrich, catalog number: C1016)
DNase Away surface Decontaminant (Thermo Fisher Scientific, catalog number: 7010PK)
Dulbecco’s Phosphate Buffered Saline (DPBS) (Sigma-Aldrich, catalog number: D8662)
Ethanol 99.5% Analytical Grade (Thermo Fisher Scientific, catalog number: 12634800)
GelRed Nucleic Acid gel stain (Biotium, catalog number: 41003)
Glycine (Sigma-Aldrich, catalog number: 50046)
High-quality deoxynucleotide triphosphates (dNTPs) (Promega, catalog number: U1335)
Isopropanol PA (VWR, catalog number: 83442)
Milli-Q water
NaCl (Sigma-Aldrich, catalog number: S9888)
NaCO3 (Merck millipore, catalog number: 1063920500)
NaHCO3 (Merck millipore, catalog number: 1063290500)
Nuclease-free water (Qiagen, catalog number: 129114)
NucleoSpin 96-PCR cleanup kit (Macherey-Nagel, catalog number: 7406584)
RNase away surface Decontaminant (Thermo Fisher Scientific, catalog number: 7000TS1)
SDS (Sigma-Aldrich, catalog number: L3771)
Triton X-100 (Sigma-Aldrich, catalog number: 93442)
Trizma base (Sigma-Aldrich, catalog number: T6066)
Trizma hydrochloride (Sigma-Aldrich, catalog number: T3253)
Tween 20 (Sigma-Aldrich, catalog number: P9416)
UltraPure Agarose (Thermo Fisher Scientific, catalog number: 16500500)
Yeast extract (Thermo Fisher Scientific, catalog number: 15815488)
100 bp DNA ladder (Thermo Fisher Scientific, catalog number: 15628019)
100 ml sterile polystyrene basin
10x TBE (powder) (Thermo Fisher Scientific, catalog number: AM9864)
98% Sulphuric acid (VWR, catalog number: 1.00748.2500)
-
B-cell receptor single-cell cloning
Collection of single-cell sorted cells
40 μm cell strainer (Corning, catalog number: 431750)
DTT (Thermo Fisher Scientific, catalog number: Y00147)
Gibco PBS (10x), pH 7.2 (Fisher Scientific, catalog number: 11514526)
RNAsin ribonuclease inhibitors (Promega, catalog number: N2511)
Sorted patient B cells
70% Ethanol (in nuclease-free water)
Single-cell reverse transcription
Nonidet NP40 or TERGITOL Solution (Sigma-Aldrich, catalog number: NP40S)
Random hexamer primer (RHP) mix (Sigma-Aldrich, catalog number: 11034731001)
RNAsin ribonuclease Inhibitors (Promega, catalog number: N2511)
Superscript III reverse transcriptase kit (Thermo Fisher Scientific, catalog number: 18080085)
70% Ethanol (in nuclease-free water)
Immunoglobulin variable region gene amplification
HotStartTaq plus DNA polymerase Kit (Qiagen, catalog number: 203609)
Sequencing and PCR primers (see Appendix)
70% Ethanol (in nuclease-free water)
Ig gene-specific PCR
Gene-specific primers (see Appendix)
HotStartTaq plus DNA polymerase Kit (Qiagen, catalog number: 203609)
70% Ethanol (in nuclease-free water)
PCR products digestion with restriction enzymes
AgeI-HF (NEB, catalog number: R3552L)
BsiWI-HF (NEB, catalog number: R3553S)
SalI-HF (NEB, catalog number: R3138S)
XhoI-HF (NEB, catalog number: R0146S)
70% Ethanol (in Milli-Q water)
Ligation of PCR products to cloning expression vectors
Human IgH linearized plasmid vector with cloning sites for AgeI and SalI (Gift from Hedda Wardemann and Eric Meffre)
Human Igκ linearized plasmid vector with cloning sites for AgeI and BsiWI (Gift from Hedda Wardemann and Eric Meffre)
Human Igλ linearized plasmid vector with cloning sites for AgeI and XhoI (Gift from Hedda Wardemann and Eric Meffre)
QIAquick gel extraction kit (Qiagen, catalog number: 28704)
Quick ligase Kit (NEB, catalog number: M2200L)
70% Ethanol (in Milli-Q water)
Transformation of DH5α cells
Plastic Petri dishes (100 mm x 15 mm) (Fisher Scientific, catalog number: S33580A)
Ampicillin (Sigma-Aldrich, catalog number: A5354)
Bacteriological agar (Sigma-Aldrich, catalog number: A5306)
Disposable bacteria cell spreaders and inoculating loops
Glycerol (Sigma-Aldrich, catalog number: G5516)
Subcloning efficiency DH5α competent cells (Thermo Fisher Scientific, catalog number: 18265017)
70% Ethanol (in Milli-Q water)
Insert-check PCR
Agar plates
GoTaq G2 DNA polymerase (Promega, catalog number: M7848)
Insert check PCR primers (available in Appendix)
Sequencing primers (available in Appendix)
70% Ethanol (in Milli-Q water)
Plasmid miniprep
Ampicillin (Sigma-Aldrich, catalog number: A5354)
LB broth (Sigma-Aldrich, catalog number: L3552)
QIAprep spin miniprep Kit (Qiagen, catalog number: 27106)
70% Ethanol (in Milli-Q water)
-
Exploratory screening of monoclonal antibodies (mAbs)
Small scale production of mAbs (30 ml cultures)
Corning erlenmeyer culture flasks (125 ml) (Sigma-Aldrich, catalog number: CLS431143)
0.22 μm vacuum filter units (Corning, catalog number: 431096)
0.22 μm syringe filters
Expi293 cells (Thermo Fisher Scientific, catalog number: A14528)
Expi293 expression medium (Thermo Fisher Scientific, catalog number: A1435101)
OptiPRO serum free medium (sfm) (Thermo Fisher Scientific, catalog number: 12309019)
PEI-MAX 40K transfection grade linear polyethylenimine hydrochloride (Polysciences, catalog number: 24765-1)
Erythrosin B (Sigma-Aldrich, catalog number: 198269)
70% Ethanol (in Milli-Q water)
Purification of mAbs
Bio-spin disposable chromatography columns (Bio-Rad, catalog number: 732-6008)
Amicon Ultra 4 ml 50 kDa MWCO centrifugation units (Millipore, catalog number: UFC805008)
Zeba Spin Desalting Columns 7K MWCO 5 ml (Thermo Fisher Scientific, catalog number: 89892)
0.22 μm low protein binding syringe filters (Millipore Millex-GV, catalog number:SLGV013SL)
Protein G sepharose 4 fast flow antibody purification resin (GE Lifesciences, catalog number: 17061805)
70% Ethanol (in Milli-Q water)
0.1 M glycine (pH 3.0) (see Recipes)
1 M Tris (pH 8.0) (see Recipes)
IgG concentration assessment from purified mAbs
Corning® 96-well half area clear flat bottom polystyrene high bind microplate (Corning, catalog number: 3690)
3,3′,5,5′-Tetramethylbenzidine (TMB) substrate (Sigma-Aldrich, catalog number: T0440)
70% Ethanol (in Milli-Q water)
Bovine serum albumin (BSA) (Sigma-Aldrich, catalog number: A7906)
Goat F(ab’)2 anti-human IgG Fab (Jackson Immunoresearch, catalog number: 109-006-097)
HRP conjugated goat F(ab’)2 anti-human IgG Fc gamma specific (Jackson Immunoresearch, catalog number: 109-036-008)
Human IgG whole molecule (Jackson Immunoresearch, catalog number: 009-000-003)
0.5 M H2SO4 (see Recipes)
SDS-polyacrylamide gel electrophoresis
Bolt 4-12% Bis-Tris Plus protein gels (Thermo Fisher Scientific, catalog number: NW04122BOX)
Bolt lithium dodecyl sulfate (LDS) sample Buffer (4x) (Thermo Fisher Scientific, catalog number: B007)
Bolt methyl ester sulfonate (MES)-sodium dodecyl sulfate (SDS) running buffer (Thermo Fisher Scientific, catalog number: B000202)
Bolt sample reducing agent (10x) (Thermo Fisher Scientific, catalog number: B009)
SeeBlue Plus2 pre-stained protein standard (Thermo Fisher Scientific, catalog number: LC5925)
SimplyBlue SafeStain (Thermo Fisher Scientific, catalog number: LC6065)
Polyreactivity and control ELISAs
Polyreactivity ELISA- Bovine serum albumin (BSA) (Sigma-Aldrich, catalog number: A7906)
- Deoxyribonucleic acid sodium salt, from salmon testes (Sigma-Aldrich, catalog number: D1626)
- HRP conjugated F(ab)2 anti-human IgG Fc gamma specific (Jackson Immunoresearch, catalog number: 109-036-008)
- Insulin solution human (Sigma-Aldrich, catalog number: 19278-5ML)
- Lipopolydsaccharides from Escherichia coli 055:B5 (Sigma-Aldrich, catalog number: L6525-1MG)
- Nunc-ImmunoTM MicroWellTM 96-Well plates, high binding (Nunc, catalog number: 442404)
- TMB substrate (Sigma-Aldrich, catalog number: T0440)
- 0.5 M H2SO4 (see Recipes)
Anti-Bovine serum albumin (BSA)/plastic and rabbit IgG ELISA- Bovine serum albumin (BSA) (Sigma-Aldrich, catalog number: A7906)
- Chromopure rabbit IgG (Jackson Immunoresearch, catalog number: 011-000-003)
- HRP conjugated rabbit F(ab’)2 anti-human IgG Fc gamma specific (Jackson Immunoresearch, catalog number: 309-036-008)
- Malondialdehyde (MDA) modified BSA (Academy Bio-Medical, catalog number: 20P-MD-BS102)
- Molecular biology grade BSA (NEB, catalog number: B9000S)
- TMB substrate (Biolegend, catalog number: 421101)
- 0.5 M H2SO4 (see Recipes)
Monoclonal antibody screening for citrulline specificities
Anti-alpha enolase 1 and histone 4 ELISA- Corning® 96-well half area clear flat bottom polystyrene high bind microplate (Corning, catalog number: 3690)
- Bovine serum albumin (BSA) (Sigma-Aldrich, catalog number: A7906)
- Carbonate buffer (see Recipes)
- HRP conjugated goat F(ab’)2 anti-human IgG Fc gamma specific (Jackson Immunoresearch, catalog number: 109-036-008)
- Peptides (Table 7)
- Reference (pool of ACPA + RA serum samples)
- RIA buffer (see Recipes)
- TMB substrate (Sigma-Aldrich, catalog number: T0440)
- 0.5 M H2SO4 (see Recipes)
Anti-Cit-fibrinogen, Cit-vimentin and Cit-filaggrin peptide ELISA- Pierce Streptavidin Coated High Capacity Plates (Thermo Fisher Scientific, catalog number: 15500)
- Biotinylated peptides (Table 7)
- Bovine serum albumin (BSA) (Sigma-Aldrich, catalog number: A2153)
- HRP conjugated F(ab)2 anti-human IgG Fc gamma specific (Jackson Immunoresearch, catalog number: 109-036-008)
- Reference (pool of ACPA + RA serum samples)
- TMB substrate (Sigma-Aldrich, catalog number: T0440)
- RIA buffer (see Recipes)
- TBE buffer (see Recipes)
- 0.5 M H2SO4 (see Recipes)
Anti-Cyclic Citrullinated Peptides 2nd generation (CCP2) ELISA- IMMUNOSCAN CCPlus clinical assay (Euro Diagnostica, catalog number: RA-96PLUS)
-
Large scale expression of monoclonal antibodies for in-depth functionality assays
Plasmid Maxipreps
LB agar plates
225 ml conical centrifuge tubes with graduations (VWR, catalog number: 89218-286)
Ampicillin (Sigma-Aldrich, catalog number: A5354)
PureLinkTM HiPure Plasmid DNA Maxiprep Kit (Thermo Fisher Scientific, catalog number: K210017)
70% Ethanol (in Milli-Q water)
LB medium (see Recipes)
Large scale production of mAbs (400-3,200 ml cell suspensions)
Corning Erlenmeyer culture flasks (1 L) (Sigma-Aldrich, catalog number: CLS431147)
225 ml conical centrifuge tubes with graduations (VWR, catalog number: 89218-286)
0.22 μm syringe filter
Expi293 cells (Thermo Fisher Scientific, catalog number: A14528)
Expi293 Expression Medium (Thermo Fisher Scientific, catalog number: A1435101)
OptiPRO serum free medium (sfm) (Thermo Fisher Scientific, catalog number: 12309019)
PEI-MAX 40K Transfection Grade Linear Polyethylenimine Hydrochloride (Polysciences, catalog number: 24765-1)
Erythrosin B (Sigma-Aldrich, catalog number: 198269)
70% Ethanol (in Milli-Q water)
IgG concentration assessment from mAbs supernatant
Refer to “IgG concentration assessment from purified mAbs” in Materials and Reagents B
Purification of mAbs from large scale expressions
Slide-A-Lyzer G2 Dialysis cassettes, 10K MWCO 30-70 ml (Thermo Fisher Scientific, catalog number: 88254)
Zeba Spin Desalting Columns 7K MWCO 5 ml (Thermo Fisher Scientific, catalog number: 89892)
Column PD-10, Empty (GE Healthcare, catalog number: 17-0435-01)
LabMate PD-10 Buffer Reservoir (GE Healthcare, catalog number: 18-3216-03)
0.22 μm low protein binding syringe filters (Millipore Millex-GV, catalog number: SLGV013SL)
Protein G Sepharose 4 Fast Flow antibody purification resin (GE Lifesciences, catalog number: 17061805)
0.1 M glycine (pH 3.0) (see Recipes)
1 M Tris (pH 8.0) (see Recipes)
IgG concentration assessment from purified mAbs
Refer to “IgG concentration assessment from purified mAbs” in Materials and Reagents B
SDS-PAGE
Refer to “SDS-polyacrylamide gel electrophoresis” in Materials and Reagents B
Test for IgG aggregation
0.22 μm bottle-top vacuum filters (Corning, catalog number: 431096)
20% ethanol (in Milli-Q water)
70% Ethanol (in Milli-Q water)
Endotoxin test
No specific Materials and Reagents for this section of the protocol.
Citrulline-Specificity ELISAs with titration curves
Refer to “Monoclonal antibody screening for citrulline specificities” in Materials and Reagents B
CCP2 ELISA
Refer to “anti-Cyclic Citrullinated Peptides 2nd generation (CCP2) ELISA” in Materials and Reagents B
Table 7. Citrullinated peptides for screening.
| Antigens | Peptides | Concentration used for coating |
|---|---|---|
| α-enolase | CEP-1 | 2.5 μg/ml |
| α-enolase | REP-1 | |
| Vimentin | biotin-cit-vim (aa60-75) | 1 μg/ml |
| Vimentin | biotin-arg-vim (aa60-75) | |
| Fibrinogen α | biotin-cit-fib 573 (aa563-583) | 2 μg/ml |
| Fibrinogen α | biotin-arg-fib 573 (aa563-583) | |
| Fibrinogen α | biotin-cit-fib 591 (aa580-600) | 2 μg/ml |
| Fibrinogen α | biotin-arg-fib 591 (aa580-600) | |
| Fibrinogen β | biotin-cit-fib (aa36-52) | 1 μg/ml |
| Fibrinogen β | biotin-arg-fib (aa36-52) | |
| Filaggrin | biotin-cit-Fil (aa307-324) | 2 μg/ml |
| Filaggrin | biotin-arg-Fil (aa307-324) |
Equipment
Common list
Centrifuge suitable for 1.5 and 2 ml microtubes (Sigma Laborzentrifugen GmbH, catalog number: 10020)
Centrifuge suitable for 225 ml tubes, 50 ml tubes, and 96-well PCR plates (Sigma Laborzentrifugen GmbH, catalog number: 10475)
ELISA plate reader (Molecular Devices, catalog number: PLUS 384)
ELISA plate washer (Tecan, catalog number: 16029011)
Heating block suitable for 1.5 and 2 ml microtubes
Microwave (Whirlpool)
Pipette controller (Thermo Fisher Scientific, catalog number: 10759754)
Precision scale (Sartorius, catalog number: AY511)
Protein gel electrophoresis tank (Bio-Rad laboratories, catalog number: 1656001)
Vacuum Manifold (Promega, catalog number: A2291)
Vacuum Manifold (Qiagen, catalog number: 19413)
Vacuum pump (Promega, catalog number: A6722)
Vortex (Thermo Fisher Scientific, catalog number: 15547335)
-
B-cell receptor single-cell cloning
Agarose electrophoresis gel tank (Thermo Fisher Scientific, catalog number: D3-14)
Electrophoresis power supply (VWR, catalog number: 700-0448)
Gel-doc Electrophoresis Apparatus (Bio-Rad laboratories, catalog number: 1708195)
Hot plate suitable for 96-well PCR plates (Bioer Technology, catalog number: CHB-202)
Multi-tubes vortex (VWR, catalog number: 58816-116)
PCR cooler suitable for 96-well PCR plates (Thermo Fisher Scientific, catalog number: 10192281)
PCR thermal cycler suitable for 96-well PCR plates (Applied Biosystems, catalog number: 435965)
Sequencing service (e.g., internal or external provider)
37 °C incubator (Heraeus, catalog number: 26066010)
37 °C shaking incubator (Infors HT, catalog number: S-000123637)
-
Exploratory screening of monoclonal antibodies (mAbs)
Bürker counting chamber (Thermo Fisher Scientific, catalog number: 10628431)
CO2 shaking incubator (Infors HT, catalog number: S-000117022)
Light microscope (Zeiss, catalog number: 451487)
Orbital tube rotator (Abdos life sciences, catalog number: E11280)
-
Large scale expression of monoclonal antibodies for in-depth functionality assays
Äkta Explorer 10 instrument (or equivalent; GE Healthcare Life Sciences)
Enrich SEC 650 column 10 x 300 mm, 24 ml (Bio-Rad, catalog number: 7801650)
Tube Roller mixer (Sigma-Aldrich, catalog number: Z675148)
Software
International ImmunoGeneTics information system (IMGT/V-QUEST), a free sequence alignment software for the immunoglobulin (IG) and T cell receptor (TR) nucleotide sequences of the variable regions and domains. This integrated alignment tool compares the germline and rearranged input sequences with the IMGT reference sequences (IMGT/V-QUEST reference directory).
UNICORN software version 5.2 (GE Healthcare Life Science).
Procedure
Work for the recombinant antibody pipeline (Figure 1).
Figure 1. Human monoclonal antibody production pipeline.
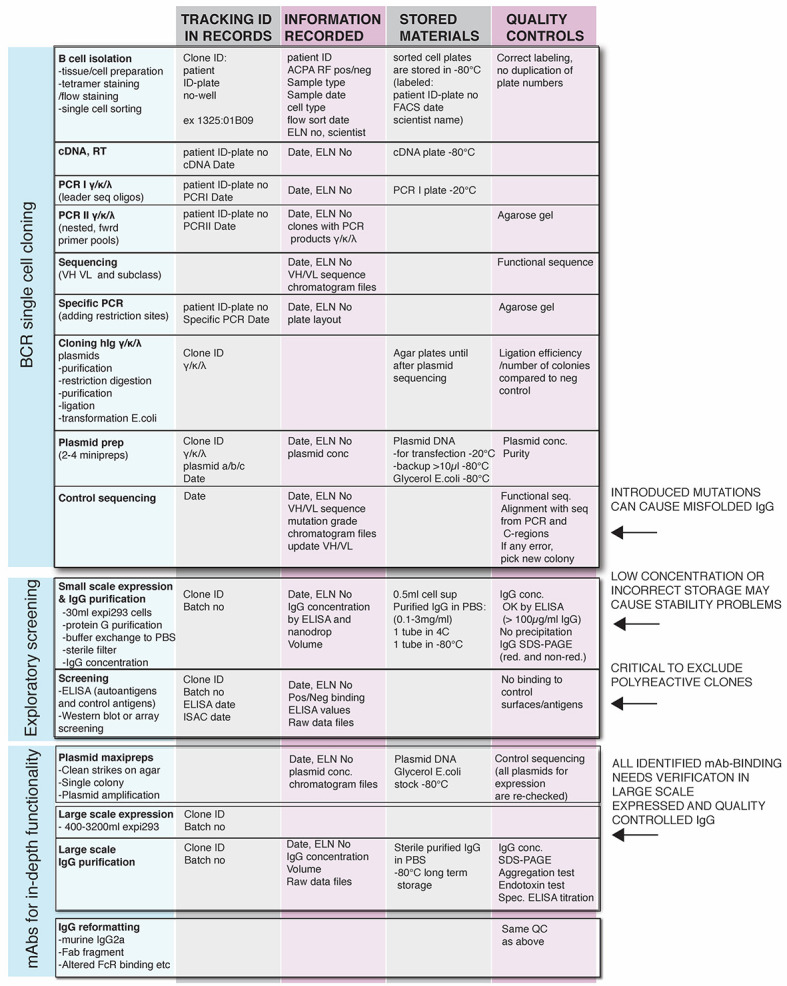
ELN: Electronic laboratory notebook.
-
B cell receptor single-cell cloning
-
Collection of single-cell sorted cells (approx. 5 h)
Process cells and tissues and sort B cells according to your validated flow cytometry methodology.
Sort single B cells into 96-well PCR plates containing 10 μl of sorting buffer (Table 1).
Immediately cover each plate with adhesive PCR plate foil, spin down the plate and place it on dry ice until the cell sorting is finished. If not immediately used for the steps described below, store the plates in -80 °C freezer until further use.
-
Single-cell reverse transcription (approx. 3 h)
Thaw plate(s) of single-sorted B cells on ice and spin down before removing the adhesive PCR plate foil.
Carefully add 6 μl of Random hexamer primers (RHP) mix (Table 2) to each well. Cover the plate with adhesive PCR plate seal and spin down.
Transfer the plate on a hot plate for 1 min at 68 °C and put back on ice or onto the PCR-cooler.
Carefully remove the adhesive PCR plate seal and add 14 μl RT mix (Table 2) to each well.
Cover the plate with a new adhesive PCR plate seal and perform the reverse transcription at 42 °C for 5 min, 25 °C for 10 min, 50 °C for 60 min, and 95 °C for 5 min.
The cDNA can be immediately used or stored at -80 °C before performing the PCRs.
Notes:
Before starting all steps in this section, it is important to clean the PCR hood or the laboratory bench and laboratory pipettes with ethanol, RNase away, and DNase away decontamination solutions.
Single-cell lysis and cDNA synthesis are performed in the original plate that was used for cell sorting, which minimizes the risk for cross-contamination.
When you calculate the volume of each reagent that you need, add 10% of reagent volume overage (e.g., for a full 96-well PCR plate, calculate for 105 wells), so that all the wells are filled properly.
High attention when working with single cells must be considered; even talking over the plate can cause contamination. It is also very important to cautiously remove the adhesive PCR plate seals to avoid splashes.
Store cDNA at -80 °C until further use.
-
Immunoglobulin variable region gene amplification (approx. 2 days)
Prepare heavy chain, lambda and kappa chain PCR master mixes for the first PCR in 3 separate tubes as described in Table 3. The targeted gene segments are all indicated in the name of the primer in Appendix.
Carefully add 38 μl of each PCR master mix 1 to the corresponding well and plate (heavy, kappa, or lambda) and then carefully add 3 μl of cDNA to each well. Cover with adhesive PCR plate seal and spin down.
Perform the first PCR at 95 °C for 15 min; for 50 cycles at 95 °C for 30 s, 58 °C (IgH/Igκ) or 60 °C (Igλ) for 30 s, 72 °C for 55 s; 72 °C for 10 min.
Prepare PCR master mixes for the second PCR, as shown in Table 3.
Carefully add 37 μl of PCR master mix 2 to each well of the plate and then carefully add 3.5 μl of first PCR product to each well. Cover with adhesive PCR plate seal.
Perform the second PCR at 95 °C for 15 min; for 50 cycles at 95 °C for 30 s, 58 °C (IgH/Igκ) or 60 °C (Igλ) for 30 s, 72 °C for 45 s; 72 °C for 10 min.
Check the size of the second PCR products by running a 2% agarose gel, and visualize the DNA bands using a UV transilluminator. Expected sizes of the second PCR are 510 bp for Igκ, 405 bp for Igλ, and 450 bp for IgH (Figure 2).
Purify second PCR products using a PCR cleanup kit following the manufacturer’s instruction.
Sequence the purified second PCR products using the respective primers describes in Appendix.
Notes:
When you calculate the volume of each reagent that you need, add 10% of reagent volume overage (e.g., for a full 96-well PCR plate, calculate for 105 wells), so that all the wells are filled properly.
To ensure that the master mix is not contaminated, prepare each master mix with enough volume to have one buffer control.
Heavy chain, lambda and kappa chain PCRs are always performed in separate plates.
PCR products can be immediately used or stored at -20 °C until further use.
-
Ig gene-specific PCR (approx. 4 h)
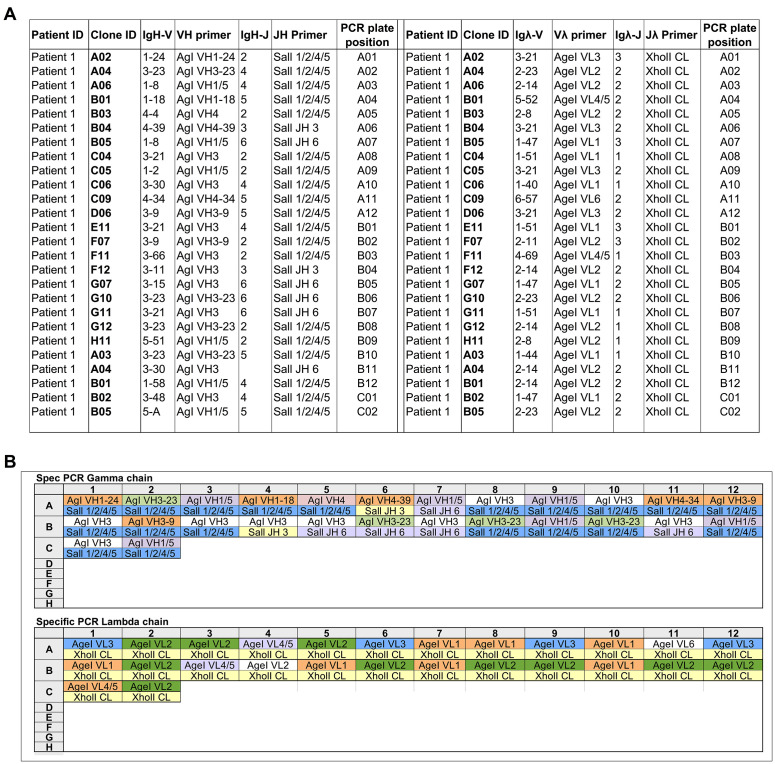
Use the data generated from the IMGT/V-QUEST to identify for each PCR product (IgH, Igκ, or Igλ) the closest germline IgV and IgJ gene and to define each gene-specific primer for each well using primers described in the Appendix (Figure 3A).
Prepare a pipetting scheme indicating the correct primer pairs for each well and plate (heavy, kappa, or lambda) (Figure 3B).
Dilute gene-specific primers to a working concentration of 5 nmol/ml in nuclease-free water.
In a 96-well plate carefully add 2 μl of the matched 5′ and 3′ primers for each individual PCR product into the same well.
Cover the plate with adhesive PCR plate seal and spin down for 10 s.
In the same plate with the primers, add 34 μl of the first PCR mix (as described in Table 3) in each well.
Add 4 μl of unpurified first PCR product into the correct well corresponding to the matched primers.
Seal the plate with adhesive PCR plate and spin down for 10 s.
Perform the second round of PCR described in Step A3f.
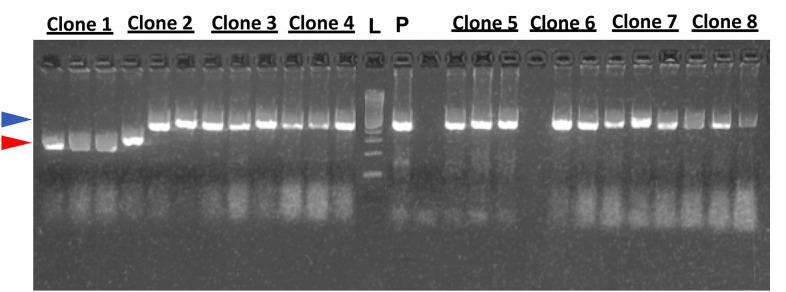
Check the size of the gene-specific PCR products as described in Step A3g (Figure 4).
Purify gene-specific PCR products using a PCR cleanup kit following the manufacturer’s instruction.
Notes:
It’s important to clean pipettes and the PCR hood with ethanol, RNAse away, and DNAse away decontamination solution.
It’s important to work on ice or a PCR cooler.
PCR products can be used immediately or stored at -20 °C until further use.
-
PCR products digestion with restriction enzymes (approx. 3 h)
Thaw purified PCR products from Step A4 and spin down.
-
Prepare digestion mix 1 for IgH, Igκ, and Igλ as described in Table 4.
§Enzymes used are Sall and AgeI for Heavy chain; AgeI for Kappa chain; Xhol and AgeI for Lambda chain.
Vortex mixes gently and spin down and carefully add 10 μl of the digestion mix 1 to each tube.
Add 30 μl of purified PCR products to each tube, vortex and spin down.
Incubate for 15 min at 37 °C in the heating block.
Prepare digestion mix 2 for Igκ as described in Table 5.
Spin down the tube.
Carefully add 10 μl of each mix to each tube, vortex gently and spin down.
Incubate for 1 h at 55 °C in the heating block.
Perform PCR clean-up for the digested product as described above.
Notes:
PCR products can be immediately used or stored at -20 °C until further use.
It’s absolutely needed to get rid of any of ethanol before the elution because ligase enzyme can be inhibited by minute traces of ethanol.
-
Ligation of PCR products to cloning expression vectors (approx. 5 h)
Linearize the vectors using enzymes (AgeI and Sall for IgH vector; AgeI and BsiWI for Igκ vector; Xhol and AgeI for Igλ vector) and purify the cleaved plasmids using Qiagen gel purification following the manufacturer’s protocol.
Prepare the ligation mix as described in Table 6 and add 28 μl to each tube.
Carefully add 20 μl of digested DNA to each tube.
Add 2 μl of Quick ligase to each tube.
Incubate for 15 min at 25 °C in the heating block.
Put the tubes back on ice.
Notes:
For better ligation efficiency, the quantity of the expression vectors used should be around 60 ng/μl. A three-fold molar excess of insert DNA to vector should be used.
The expression vectors are high copy number plasmids.
High purity of cleaved vectors and PCR products is required for optimal results.
To avoid self-ligation of the vector, ligase should always be added last.
-
Transformation of DH5α cells (approx. 18 h with o/n incubation)
Thaw chemically competent bacteria on ice. Mix gently and carefully transfer 40 μl of cells into the tube on ice.
Carefully add 5 μl of ligation product to the tube/well.
Mix gently and incubate for 30 min on ice.
Heat chock bacteria cells at 42 °C for 30 s and place the tubes back on ice for 2 min.
Add 150 μl LB medium (without ampicillin) to the tube.
Incubate the tubes for 30 min at 37 °C with shaking at 225 rpm.
Take LB-Ampicillin agar plates out of the cold room to warm and label them with the corresponding clone name.
Spread around 150 μl of the bacteria cells onto the agar plate.
Incubate plates overnight at 37 °C upside down.
Cover agar plates with parafilm and save them at 4 °C.
Note: Store agar plates at 4 °C until plasmids are sequenced and verified.
-
Insert-check PCR (approx. 3 h)
Prepare a PCR master mix for the insert-check PCR, as shown in Table 3. Using the primers: Ab sense/IgG int for heavy; Ab sense/xholI for lambda; and Ab sense/Cκ494 for Kappa.
Pick two to four colonies with 10 μl pipette tips from each plate, spread the colony on a backup LB agar plate, and then place the same pipette tips, each in a different well of a 96-well PCR plate containing the insert-check PCR mix.
Perform the second PCR at 95 °C for 15 min; for 25 cycles at 95 °C for 30 s, 58 °C for 30 s, 72 °C for 60 s; 72 °C for 10 min.
Check the size of the PCR products by running a 2% agarose gel, and visualize the DNA bands using a UV transilluminator. Expected sizes: 700 bp for Igκ, 590 bp for Igλ, and 650 bp for IgH (Figure 5).
-
Plasmid miniprep (approx. 2 days with o/n incubation)
For each sample, pick 4 insert positives colonies from the backup plate using a 200 μl pipette tip and inoculate 4 ml LB medium containing 100 μg/ml ampicillin in 15 ml culture tubes.
Incubate the tubes overnight at 37 °C with shaking at 225 rpm.
After incubation, pellet bacteria by centrifuging the culture tubes for 10 min at 3360 × g. Discard the supernatant.
Purify plasmids using a miniprep kit following the manufacturer’s instruction.
Sequence the plasmid DNA using the (Ab sense primer) to confirm the identity with the second PCR product.
Notes:
Before starting the plasmid miniprep make glycerol stocks of each culture by mixing 300 μl of 1:1 sterile LB/glycerol and 400 μl of the inoculate to a 2 ml tubes, mix well and freeze at -80 °C.
It’s necessary to check the complete sequence in order to verify that the PCR product is ligated in-frame within the plasmid without any introduced errors. It’s also important to check the signal peptide and the constant region sequences are correct. Sometimes miniprep sequences may not be high enough quality to generate complete sequences and it is important to repeat the procedure.
-
-
Exploratory screening of monoclonal antibodies (mAbs)
-
Small scale production of mAbs (approx. 8 days with incubation)
Mix 1.27 μg of total plasmid DNA for each 1 ml of cell suspension transfected. Half with heavy chain (0.635 μg/ml cell suspension) and half with light chain (0.635 μg/ml cell suspension). We recommend 30 ml cultures with 19 μg light chain and 19 μg heavy chain DNA for the initial screening.
Weigh in PEI-MAX and dissolve it in OptiPRO serum free media (sfm) to make the working solution (100 μg/ml) and sterile filter the solution through a 0.22 μm filter, you need 0.05 ml working solution per ml of cell suspension to transfect.
Adjust the concentration of the Expi293 cells to 2.5 x 106 cells/ml and aliquot cell suspensions to labeled cell culture shaker flasks and put the flasks into the incubator (8% CO2, 37 °C, and 125 rpm on an orbital shaker).
Add working solution of PEI-MAX in OptiPRO sfm to the tubes with DNA plasmids (0.05 ml/ml cell suspension). Mix by brief vortexing and incubate for exactly 10 min at room temperature.
Add the mix DNA plasmids/PEI-MAX in OptiPRO sfm media drop-wise to the cell suspension.
Put the flasks back into the incubator as soon as possible.
Incubate at 37 °C, 125 rpm for 6 days.
After incubation, pour the cell suspension into 50 ml tubes.
Centrifuge the cells for 30 min at 840 × g at room temperature.
Decant the supernatant by slowly pouring into new 50 ml tubes. The cell pellet should not be disrupted.
Centrifuge the supernatant for 20 min at 10,000 x g, room temperature.
Filter IgG through a 0.22 μm syringe filter.
Notes:
The day before transfection, dilute the Expi293 cells in the EXPI293 expression medium to 1.8 x 106 cells/ml.
The most critical for the expression is that the Expi293 cells are in a good growth phase and with high viability.
Always sterile filter all reagents and plasmids preparations before transfection.
Always use freshly prepared PEI-MAX working solutions.
During the 6 days incubation, there is no need to passage the cells or change the medium.
Filter the supernatant under sterile conditions.
lgG in the supernatant are stable at 4 °C for a couple of weeks, but purification is recommended as soon as possible.
-
Purification of mAbs (approx. 2 days)
Vortex Protein G Sepharose 4 Fast Flow beads and shake bottle vigorously. Add enough beads for 100 μl per sample to a 50 ml tube containing DPBS.
Wash beads by centrifuging for 10 min at 840 × g, room temperature. Take off supernatant using a pipette, then centrifuge again using the same program. Resuspend Protein G in DPBS to achieve a total volume of 400 μl per sample.
Add 400 μl of Protein G into each tube containing the supernatant.
Incubate the tubes containing supernatants and Protein G for at least 14 h at 4 °C under 25 rpm rotation.
After incubation, centrifuge supernatants for 20 min at 840 × g, room temperature. In the meantime, prepare a 2 ml microtube containing 100 μl 1 M Tris (pH 8.0).
Discard supernatant without disrupting pellet. Leave about 1 ml of the supernatant at the bottom with the beads.
Resuspend the beads in the remaining supernatant, and transfer to the empty Bio-spin chromatography column.
Wash the Protein G column three times with 2 ml DPBS each.
Blot the bottom of the Bio-spin chromatography column on a paper tissue to get rid of excess flow through droplets.
Add 1 ml of filtered 0.1 M glycine (pH 3.0) to the protein G resin in the Bio-spin chromatography column to elute the IgG into a 2 ml Eppendorf tube. Immediately mix with the neutralization buffer (1 M Tris, pH 8.0).
Change buffer to PBS using Zeba Spin 5 ml centrifugation desalting columns.
Remove the Zeba spin column’s bottom closure and place the column into a 15 ml collection tube and centrifuge at 1,000 × g, discard flow-through and replace the column into the collection tube.
Add 2.5 ml sterile DPBS to the column. Centrifuge at 1,000 × g and discard flow-through. Repeat this step two additional times.
Blot the bottom of the column to remove excess liquid. Transfer the device to a new collection tube. Apply the eluted IgG fraction on top of the resin of the Zepa spin column.
Centrifuge and retain flow-through that contains the buffer exchanged IgG. Discard spin column.
After buffer exchange, measure the protein concentration using 280 nm absorbance by nanodrop, if the concentration is less than 100 μg/ml, use Amicon Ultra centrifugation units (4 ml MWCO 50 kDa) to concentrate the IgG to 100-200 μl following the manufacturer’s instruction.
After buffer exchange and possibly IgG concentration, filter IgG preparations through a 0.22 μm low protein binding filter under sterile conditions.
Aliquot the antibodies as following: one aliquot that can be used for ELISAs and SDS-PAGE gel and kept sterile at 4 °C; and one aliquot that should be frozen and stored at -80 °C for long term storage.
Notes:
Do not use brake on any of the centrifugations involving the protein G beads, even slight breaking at the end of the spin can cause the beads to fluff, making it difficult to cleanly remove the supernatant.
Before starting confirm that elution glycine buffer mixing with the 1 M Tris will give pH 7. If not, adjust the volume of Tris used for neutralization.
The time the antibodies are at low pH should be minimized so work fast. When you start the antibody purification, make sure that after elution, immediately buffer exchange to PBS and sterile filter through a 0.22 μm filter under the hood before storage.
-
IgG concentration assessment from purified mAbs (approx. 1 day)
Coat ½ area plates with F(ab’)2 antihuman IgG Fab (2 μg/ml in DPBS), 50 μl/well overnight at 4 °C.
Wash plates 3 x with DPBS 0.05% Tween 20.
Block with 100 μl/well of 1% BSA in DPBS and incubate for 1-2 h at room temperature.
Wash plates 3 x with DPBS 0.05% Tween 20.
Add 50 μl/well of the standard (human IgG1 kappa 200 ng/ml in the first well after that a 1/2 dilution in eight steps down to 1.5 ng/ml, diluted in DPBS with 0.1% BSA).
Add 50 μl/well of purified mAbs diluted in DPBS with 0.1% BSA. Use the preliminary concentration by nanodrop 280 nm absorbance for determining the approximate dilution factor that will be required.
Incubate for 1-2 h at room temperature.
Wash plates 3 x with DPBS 0.05% Tween 20.
Dilute the secondary antibody (HRP conjugated F(ab)2 anti-human IgG Fc gamma spec) 1:20,000 in DPBS 0.05% Tween 20.
Add 50 μl/well of the secondary antibody and incubate for 1 h at room temperature.
Wash plates 3 x with DPBS 0.05% Tween 20.
Add 50 μl/well of the TMB substrate and incubate for 7-10 min in the dark.
Stop reaction by adding 50 μl/well of 0.5 M H2SO4.
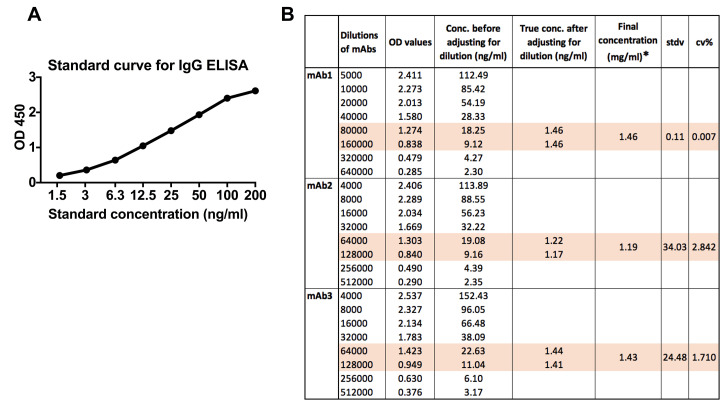
Read plate at 450 nm, reference filter 650 nm (Figure 6).
Notes:
It is a very sensitive ELISA so be careful when you do the dilutions. Never use less than 5 μl for diluting samples and the standard.
Always reserve two wells for the blank.
-
SDS-polyacrylamide gel electrophoresis (approx. 3 h)
For each reducing condition prepare and label tubes containing 5 μl of LDS sample Buffer (4x) and 2 μl of Sample Reducing Agent (10x) for reducing conditions.
Add 3 μg of each antibody into the corresponding tube. Adjust the final volume to 20 μl with nuclease-free water (if necessary). Run all IgG under both reducing and non-reducing conditions.
Place the samples in the heating block at 70 °C for 10 min.
Remove the samples from the heating block and spin them down shortly.
Place the gel in the tank and fill up the chamber with the 1x MES SDS Running Buffer.
Add the samples and a ladder (e.g., See Blue Plus2 prestained ladder) to the gel.
Let the electrophoresis go for approximately 40 min (depending on voltage settings).
Take the gel out and separate the two plastic plates surrounding it.
Use the gel knife to cut around the gel.
Rinse the gel in water twice each for 5 min.
Heat the gel in Simply Blue Safe Stain on the microwave for 30 s twice.
Rinse the gel with water twice each for 5 min.
Let the gel be in the water for at least 1 h, preferably overnight.
Note: The resulting bands for heavy chain will be between 50 and 60 kDa and the light chain will be between 25 and 30 kDa (Figure 7).
-
Polyreactivity and control ELISAs (approx. 1 day)
Polyreactivity ELISA
Coat plates with 1 μg/ml of double-stranded DNA, insulin, and LPS, 50 μl/well in DPBS overnight at 4 °C.
Wash plates 3 x with DPBS 0.05% Tween 20.
Block with 100 μl/well of 1% BSA in DPBS and incubate for at least 1 h at room temperature.
Wash plates 3 x with DPBS 0.05% Tween 20.
Add 50 μl/well of monoclonal antibodies (5 μg/ml) in the first well after that a 1/3 dilution in DPBS.
Incubate for 1 h at room temperature.
Wash plates 3 x with DPBS 0.05% Tween 20.
Dilute the secondary antibody (HRP conjugated F(ab)2 anti-human IgG Fc gamma spec) 1:20,000 in DPBS 0.05% Tween 20.
Add 50 μl/well of the secondary antibody and incubate for 1 h at room temperature.
Wash plates 3 x with DPBS 0.05% Tween 20.
Add 100 μl/well of the TMB substrate and incubate for 7-10 min in the dark.
Stop reaction by adding 100 μl/well of 0.5 M H2SO4
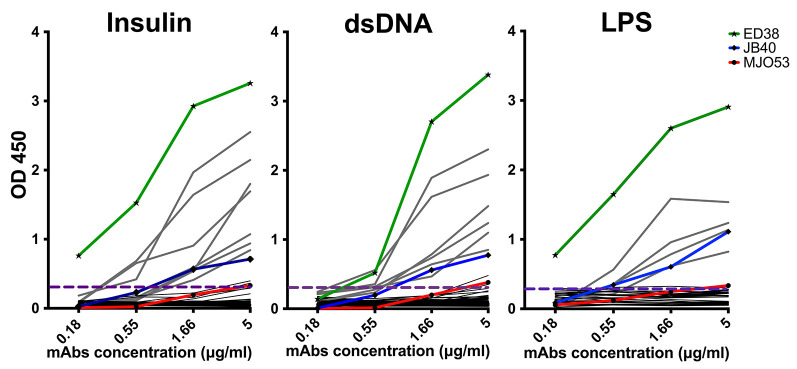
Read plate at 450 nm, reference filter 650 nm (Figure 8).
Notes:
Three different control antibodies can be included in the polyreactivity ELISAs; MGO53 (nonreactive), JB40 (weak reactive), and ED38 (high reactive) (Tiller et al., 2008; Agmon-Levin et al., 2014).
Antibodies are considered polyreactive when they recognized at least two out of the three analyzed antigens.
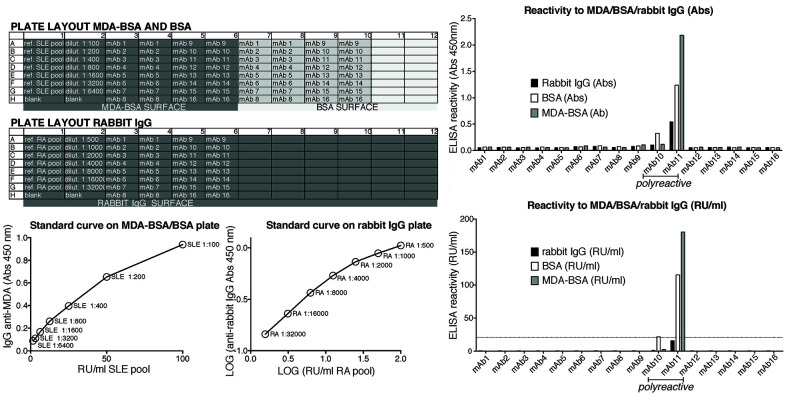
Anti-Bovine serum albumin (BSA)/plastic, anti-malondialdehyde modified BSA (MDA_BSA) and IgG anti-rabbit IgG ELISAs
Coat one ½ area plate with 3 μg/ml of BSA and MDA-BSA (six columns of each antigen on the same plate) and one plate with rabbit IgG, 5 μl/ml in DPBS overnight at 4 °C.
Wash plates 1 x with DPBS 0.05% Tween 20.
Block with 150 μl/well of 3% BSA in DPBS and incubate for 1 h at room temperature.
Wash plates 1 x with DPBS 0.05% Tween 20.
Add 50 μl/well of monoclonal antibodies (5 μg/ml) diluted in 1% BSA in DPBS. Include an RA serum pool on rabbit IgG coated wells as a control to generate a two-fold dilution series starting at around 1:1,000 dilution on every plate.
Incubate for 1.5 h at room temperature.
Wash 3 x with DPBS 0.05% Tween 20.
Dilute the secondary antibody [HRP conjugated rabbit F(ab’)2 anti-human IgG Fc gamma spec] 1:10,000 in 1% BSA in DPBS.
Add 50 μl/well of the secondary antibody and incubate for 1 h at room temperature.
Wash 4 x with PBS 0.05% Tween 20.
Add 50 μl per/well of the TMB substrate and incubate for 10 min in the dark.
Stop reaction by adding 50 μl/well of 0.5 M H2SO4.
Read plate at 450 nm (Figure 9).
Notes:
It’s important to test all the monoclonal antibodies for BSA binding in parallel to other antigens in the same plate, to ensure that we keep a reasonable incubation time and a standard curve.
The BSA assay primarily captures polyreactive antibodies binding to the plastic surface.
The MDA-BSA assay can be used for identification of MDA-modified protein autoantibodies but clones that are positive for both MDA and BSA (or rabbit IgG) are considered polyreactive.
The rabbit IgG ELISA captures antibodies with rheumatoid factor activity but could also give false positivity for unstructured or aggregated IgG.
Monoclonal antibodies that show binding in the BSA assay or any other control antigen are not considered as citrulline-specific antibodies.
-
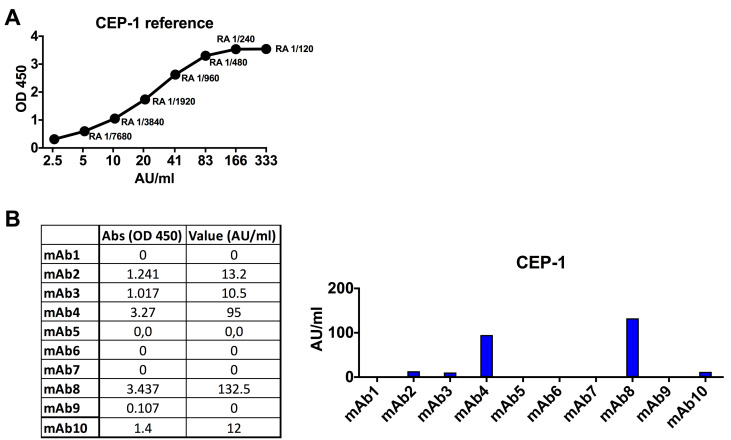
Monoclonal antibody screening for citrulline specificities (approx. 1 day)
Anti-alpha enolase 1 ELISA
Coat ½ area plates with citrullinated and arginine peptides (Table 7) in carbonate buffer pH 9.6, 50 μl/well overnight at 4 °C.
Wash plates 3 x with DPBS 0.05% Tween 20.
Block with 100 μl/well of 1% BSA in DPBS and incubate for 1-2 h at room temperature.
Wash plates 3 x with DPBS 0.05% Tween 20.
Include a positive RA serum pool as a standard to generate a two-fold dilution series starting at around 1:250 dilution on every plate.
Add 50 μl/well in duplicate of monoclonal antibodies (5 μg/ml) diluted in RIA buffer to both arginine and citrulline coated plates.
Incubate for 1-2 h at room temperature.
Wash plates 3 x with DPBS 0.05% Tween 20.
Dilute the secondary antibody (HRP conjugated F(ab’)2 anti-human IgG Fc gamma spec) 1:10,000 in RIA buffer.
Add 50 μl/well of the secondary antibody and incubate for 1 h at room temperature.
Wash plates 3 x with DPBS 0.05% Tween 20.
Add 50 μl/well of the TMB substrate and incubate for 10 min in the dark.
Stop reaction by adding 50 μl/well of 0.5 M H2SO4.
Read plate at 450 nm, reference filter 650 nm (Figure 10).
Notes:
All reactive samples were analyzed in at least 3 independent experiments.
For each plate, you should have a standard curve of a pool of ACPA + RA serum samples as a control and for standardization of reactivity.
Citrulline-reactive mAbs are re-run starting at 5 μg/ml followed by a 2-fold dilution series down to 0.15 ng/ml to generate dilution curves.
All identified citrulline-reactive mAbs should be confirmed using large scale expression and quality-controlled IgG batches (verified to be pure and of high quality on SDS-PAGE and without aggregation or endotoxin).
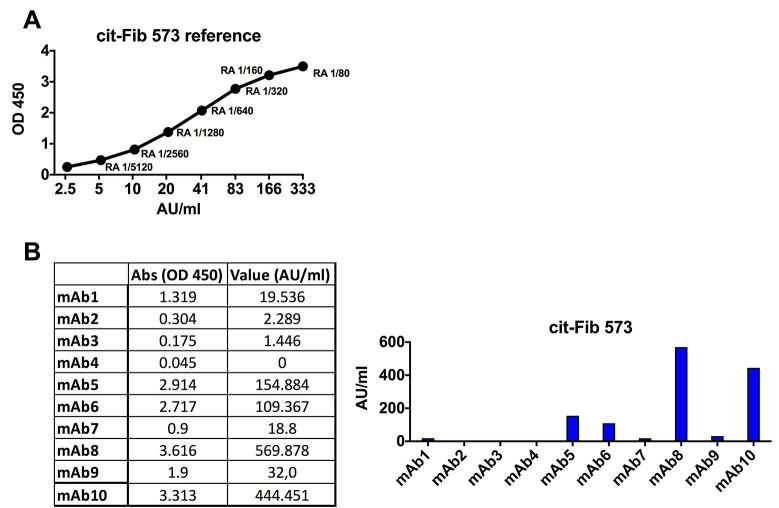
Anti-Cit-fibrinogen, Cit-vimentin and Cit-filaggrin peptide ELISA
Wash the streptavidin coated plates 2 x with DPBS 0.05% Tween 20.
Coat plates with 50 μl/well of citrullinated and arginine peptides (Table 7) in TBE buffer for 1 h at room temperature.
Wash plates 3 x with DPBS 0.05% Tween 20.
Include a positive RA serum pool as a control to generate a two-fold dilution series starting at around 1:250 dilution on every plate.
Add 50 μl/well in duplicate of monoclonal antibodies (5 μg/ml) diluted in RIA buffer to both arginine and citrulline coated plates.
Incubate for 1-2 h at room temperature.
Wash plates 3 x with DPBS 0.05% Tween 20.
Dilute the secondary antibody (HRP conjugated F(ab)2 anti-human IgG Fc gamma specific) 1:10,000 in RIA buffer.
Add 50 μl/well of the secondary antibody and incubate for 1 h at room temperature.
Wash plates 3 x with DPBS 0.05% Tween 20.
Add 100 μl/well of the TMB substrate and incubate for 10-15 min in the dark.
Stop reaction by adding 100 μl/well of 0.5 M H2SO4.
Read the plate at 450 nm, reference filter 650 nm (Figure 11).
Notes:
All reactive samples should be analyzed in at least three independent experiments.
For each plate, you should use the standard curve of a pool of ACPA + RA serum samples as a control and for standardization of reactivity.
Citrulline-reactive mAbs are re-run at a concentration of 5 μg/ml then diluted by factor of two down to 0.15 ng/ml to generate dilution curves.
All identified citrulline-reactive mAbs should be confirmed using large scale expression and quality-controlled IgG batches (verified to have high purity and of high quality on SDS-PAG and without aggregation or endotoxin).
-
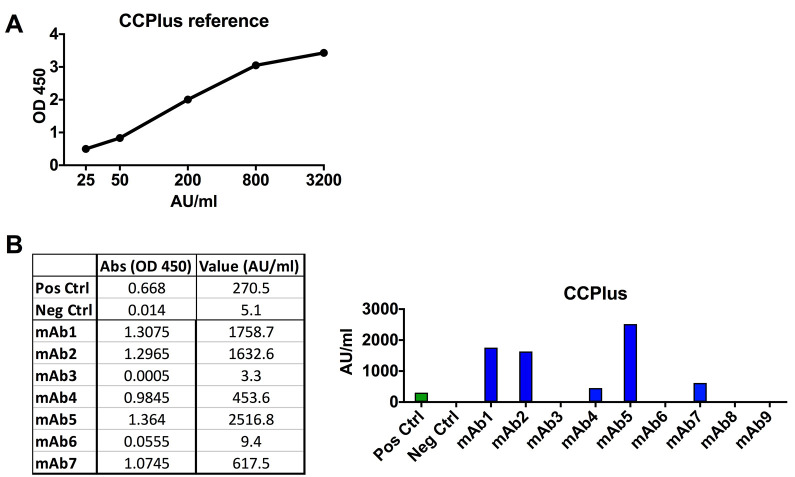
Anti-CCP2 ELISA (approx. 3 h)
Perform anti-CCP2 ELISA using IMMUNOSCAN CCPlus clinical assay for IgG at 5 μg/ml following the manufacturer’s instructions (Figure 12).
-
-
Large scale expression of monoclonal antibodies for in-depth functionality assays
-
Plasmid Maxipreps (approx. 3 days with incubations)
Bacteria from glycerol stocks of the heavy and light chains of mAbs are clean streaked on LB-agar plates containing 100 μg/ml ampicillin and incubated overnight at 37 °C. One colony from each chain of the mAb is picked and inoculated into 5 ml of LB medium containing 100 μg/ml ampicillin, after 6-8 h the cultures are expanded to 500 ml of LB medium containing 100 μg/ml ampicillin and incubated overnight at 37 °C with shaking at 225 rpm.
After incubation, make glycerol stocks as earlier described and pellet bacteria by centrifuging at 4,000 × g for 10 min and discard the supernatant.
Apply 30 ml Equilibration Buffer (EQ1) directly into the Filtration Cartridge, which is inserted in the PureLink HiPure Maxi Column. Allow the solution in the column to drain by gravity flow.
Add 10 ml Resuspension Buffer (R3) with RNase A to the cell pellet and gently shake the pellet until the cell suspension is homogenous.
Add 10 ml Lysis Buffer (L7). Mix gently by inverting the capped tube until the mixture is homogeneous. Do not vortex.
Incubate the tube at room temperature for 5 min. Do not exceed 5 min.
Add 10 ml Precipitation Buffer (N3). Mix immediately by inverting the tube until the mixture is homogeneous. Do not vortex.
Transfer the precipitated lysate into the column. Allow the lysate to filter through the column by gravity flow. Add 10 ml of wash buffer (W8) to the column and let it pass through the column by gravity flow.
Discard the inner filtration cartridge. Wash the column with 50 ml of Wash Buffer (W8). Discard the flow through after the buffer drains.
Place a sterile 50 ml tube under the HiPure Filter Column. Add 15 ml Elution Buffer (E4) to the column. Allow the solution to drain by gravity flow. Discard the column. The elution tube contains the purified DNA.
Precipitate the DNA using the DNA precipitation units that can be purchased with the kit or alternatively using a centrifuge: Add 10.5 ml isopropanol to the eluate. Mix well and centrifuge the tube at 12,000 × g for 30 min at 4 °C. Discard the supernatant and add 5 ml 70% ethanol (in Milli-Q water) to the pellet. Centrifuge the tube at 12,000 × g for 10 min at 4 °C. Remove the supernatant and air-dry the pellet.
Add 500 μl of TE Buffer to the purified DNA.
Measure the concentration of the DNA plasmid.
Store plasmid DNA at -20 °C.
Sequence the plasmid before continuing to the large-scale expression (Ab sense primer).
Notes:
The PureLink kit can be exchanged for another high-purity plasmid preparation kit. Adjust kit size from midi to maxi or mega depending on the amount of plasmid needed for expression.
Verify that the Resuspension Buffer (R3) contains RNase A, and no precipitate has formed in the Lysis Buffer (L7).
Sequence the maxiprep plasmids and ensure that the quality of the sequences is high. The maxipreps will be the starting point for all subsequent studies.
-
Large scale production of mAbs (400-3,200 ml cell suspensions) (approx. 8 days with incubation)
Increase the media volume during the 2-3 passages before transfection to achieve the correct volume of cell suspension needed.
The day before transfection, adjust the Expi293 cells to a concentration of 1.8 x 106 cells/ml in EXPI293 expression medium.
Mix 1.27 μg of total plasmid DNA for each ml of cell suspension transfected. Half with heavy chain (0.635 μg/ml cell suspension) and half with light chain (0.635 μg/ml cell suspension). For large scale production, we express 400-3,200 ml cultures.
Weigh in PEI-MAX and dissolve it in OptiPRO sfm media to make the working solution (100 μg/ml), sterile filter the solution through a 0.22 μm filter, you need 0.05 ml working solution per ml of cell suspension to transfect.
Adjust the cell concentration to 2.5 x 106 cells/ml in EXPI293 expression medium. Aliquot cell suspensions to its respective labeled flasks and put the flasks into the incubator (8% CO2, 37 °C, and 125 rpm in an orbital shaker).
Add working solution of PEI-MAX in OptiPRO sfm media to the DNA plasmids. Vortex and incubate for 10 min at room temperature.
Add the mix DNA plasmids/PEI-MAX in OptiPRO sfm media drop-wise to the cell suspension.
Put the flasks back into the incubator as soon as possible.
Incubate at 37 °C, 125 rpm for 6 days.
After incubation, pour the cell suspension into 225 ml conical centrifugation flasks.
Centrifuge the cells for 30 min at 840 × g at room temperature.
Decant the supernatant by slowly pouring into 50 ml tubes. The cell pellet should not be disrupted.
Centrifuge the supernatant for 20 min at 10,000 x g to remove cell debris, room temperature.
Filter it through a 0.22 μm filter to a new sterile bottle.
Notes:
Always use freshly prepared PEI-MAX working solutions.
It is important to add the PEI-MAX/plasmid-mix dropwise and at the same time mix the cell suspension carefully by swirling the flask. Put the flask with transfected cells as soon as possible back into the incubator.
Filter the supernatant under sterile conditions.
The supernatant can be stored at 4 °C for months.
-
IgG concentration assessment from mAbs supernatant (approx. 1 day)
Follow the previously described protocol to assess the IgG concentration in cell supernatants before continuing the purification step.
-
Purification of mAbs from large scale expressions (approx. 3 days)
Vortex Protein G Sepharose 4 Fast Flow beads and shake bottle vigorously. Add enough Protein G for each sample to a 50 ml tube containing DPBS.
Calculate the approximate total amount of IgG in the supernatant using the concentration from IgG ELISA and use the amount to calculate how much Protein G to use. Protein G has a binding capacity up to 20 mg hIgG/ml of resin.
Wash beads by centrifuging for 10 min at 840 × g, room temperature. Take off supernatant using a pipette, add the same volume of DPBS as previous, then centrifuge again using the same program.
Resuspend beads in appropriate volume DPBS or sample supernatant and add the beads to the antibody-containing supernatant.
Put flasks containing cell culture supernatant and Protein G beads on the roller (60 rpm) and incubate overnight (at least 14 h) at 4 °C.
After incubation, keep the bottles with supernatant and beads on the bench at 4 °C for a while to let the beads sink to the bottom of the flask.
Let almost all the supernatant pass through the column. Collect and save the flow through fraction at 4 °C to assess remaining IgG. Leave a small amount of supernatant together with the beads.
Add the rest of the supernatant together with the protein G beads to the column.
Add some PBS to the bead-containing bottle to be able to transfer the remaining beads over to the column.
Wash three times each with 30 ml DPBS.
Blot the bottom of the column to get rid of excess flow through droplets and place the column on top of a 50 ml tube.
Calculate the volume of elution buffer 0.1 M glycine (pH 3.0) based on the approximate total IgG in the supernatant to achieve a concentration of approximately 1.5 mg/ml in the eluted antibody suspension. Based on the elution volume calculate the volume of neutralization buffer (1 M Tris (pH 8.0)) (approx. 100 μl Tris/ml glycine). Neutralize the eluted IgG immediately and mix by turning the tube.
Change buffer to PBS for storage using either Zeba Spin desalting columns (see small scale purification) or Slide-a-lyzer dialysis cassettes depending on the volume of the eluted mAbs. For dialysis, dehydrate the cassettes for 2 min and apply eluted IgG, dialyze to 3-4 L of DPBS (diluted from 10x) 2 h at RT, transfer to new DPBS (3-4 L), followed by overnight dialysis at 4 °C (total dialysis volume should be 300x the volume of the dialysis sample).
Filter the purified antibody through a 0.22 μm filter.
Notes:
The time the antibodies are at low pH should be minimized as much as possible.
When you start the antibody purification, make sure that after elution, immediately buffer exchange to PBS and sterile filter under the hood.
Protein G Sepharose 4 Fast Flow is a first choice for general-purpose capture of antibodies at laboratory scale. It has a binding capacity of approximately 20 mg/ml of resin.
Before you start antibody purification, mix 100 μl 1 M Tris and 1 ml 0.1 M Glycine and check pH with a filter paper (should be pH 7), if the pH is too high or too low, the amount of 1 M Tris should be adjusted.
Long-time storage of large antibody preparations should be at -80 °C.
-
IgG concentration assessment from purified mAbs (approx. 1 day)
Follow the previously described protocol to assess the IgG concentration in cell supernatants before continuing to the purification step.
-
SDS-PAGE (approx. 3 h)
Follow the previously described protocol to assess the IgG purity and quality on SDS-PAGE.
-
Test for IgG aggregation (approx. 1 day)
Equilibrate the size exclusion column (Bio-Rad Enrich SEC 650) on the Äkta FPLC system with 30 ml of Milli-Q water followed by 50 ml of DPBS at a flow rate of 0.5 ml/min.
Clean the inside of the injection syringe with Milli-Q water.
Draw 10 μg of the IgG1 monoclonal antibody into the injection syringe (maximum 100 μl), Place the syringe in the holder and insert it into the injection hole.
Inject the sample with a slow and continuous movement.
Run the sample at a flow rate of 1.0 ml/min. Samples are injected on the column after 20 ml of UV stabilization. The program stops after totally 60 ml; monomeric IgG has a retention time of approx. 12.5 ml.
After the sample is finished, the column is washed with approximately 30 ml of Milli-Q water then 30 ml of 20% ethanol (in Milli-Q water) at a flow rate of 0.5 ml/min.
Notes:
All samples and buffers should be filtered through a 0.22 μm filter before being injected into the ÄKTA, if the sample volume is too low to filter, the sample should be centrifuged for 3 min at 13500 x g.
Leave the syringe in the inject-position until the run is finished.
To avoid bacterial growth, contamination, and poor column performance, always use freshly prepared buffers.
Always watch out for bubbles.
-
Endotoxin test (approx. 3 h)
All larger preparation of monoclonal antibodies that are used for functional studies should be tested for LPS contamination using Limulus Amebocyte Lysate (LAL) assay.
-
Citrulline-Specificity ELISAs with titration curves (approx. 1 day)
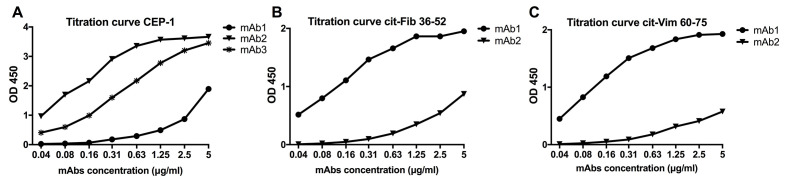
Repeat all ELISAs for citrullinated peptides for all large-scale IgG batches to confirm binding and perform titration experiments starting at 5 μg/ml (Figure 13).
-
CCP2 ELISA (approx. 4 h)
Refer to Step B7
-
Table 1. Reagent preparation for cells sorting.
| Buffer | Reagent | Concentration | 1x | 96x + 10% |
|---|---|---|---|---|
| Sorting buffer | Nuclease-free water | - | 8.00 | 840.00 |
| PBS | [10x] | 0.50 | 52.50 | |
| DTT | [100 mM] | 1.00 | 105.00 | |
| RNAsin | [40 U/μl] | 0.50 | 52.50 | |
| total | 10.00 | 1050.00 |
Table 2. Reagent preparation for reverse transcription.
| Buffers | Reagent | Concentration | 1x | 96x + 10% |
|---|---|---|---|---|
RHP mix |
Nuclease-free water | - | 4.70 | 493.50 |
| RHP | [300 ng/μl] | 1.00 | 105.00 | |
| NP-40 | [10%] | 1.00 | 105.00 | |
| RNAsin | [40 U/μl] | 0.30 | 31.50 | |
| total | 6.00 | 735.00 | ||
RT mix |
Nuclease-free water | - | 4.00 | 420.00 |
| First strand buffer | [5x] | 6.00 | 630.00 | |
| DTT | [100 mM] | 2.00 | 210.00 | |
| dNTPs | [25 mM each] | 1.00 | 105.00 | |
| RNAsin | [40 U/μl] | 0.40 | 42.00 | |
| SuperScript III | [200 U/μl] | 0.60 | 63.00 | |
| total | 14.00 | 1470.00 |
Table 3. PCR mix for 1st and 2nd PCR.
| PCR step | Reagent | Concentration | 1x | 96x + 10% |
|---|---|---|---|---|
| 1st PCR | Nuclease -free water | - | 32.16 | 3376.80 |
| PCR Buffer | [10x] | 4.00 | 420.00 | |
| Forward primer mix | [50 μM each] | 0.13 | 13.65 | |
| Reverse primer mix | [50 μM each] | 0.13 | 13.65 | |
| dNTPs | [25 mM each] | 0.40 | 42.00 | |
| HotStar Taq | [5 U/μl] | 0.18 | 18.90 | |
| subtotal mix | 37.00 | 3885.00 | ||
| cDNA template | 3.00 | |||
| Total | 40.00 | |||
| 2nd PCR | Nuclease-free water | - | 31.66 | 3324.30 |
| PCR Buffer | [10x] | 4.00 | 420.00 | |
| Forward primer mix | [50 μM each] | 0.13 | 13.65 | |
| Reverse primer mix | [50 μM each] | 0.13 | 13.65 | |
| dNTPs | [25 mM each] | 0.40 | 42.00 | |
| HotStar Taq | [5 U/μl] | 0.18 | 18.90 | |
| subtotal mix | 36.50 | 3832.50 | ||
| DNA template | 3.50 | |||
| Total | 40.00 |
Figure 2. Representative agarose gel picture of RT-PCR products of IgH (450 bp), Igλ (405 bp), and Igκ (510 bp) variable region genes amplified from single human B cells.
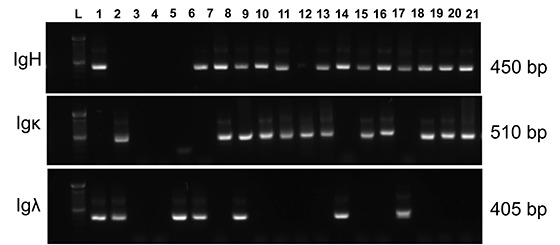
Lane L: 100 bp DNA ladder.
Figure 3. Example showing how to use the data generated from the IMGT/V-QUEST to identify the closest germline IgV and IgJ gene (A) and to define gene-specific primers for each sample (B).
Figure 4. Representative agarose gel picture of gene-specific products of IgH variable region genes.
Lane L: 100 bp DNA ladder.
Table 4. Restriction digestion mix 1.
| Reagent | Concentration | 1x |
|---|---|---|
| Nuclease-free water | - | 5.00 |
| Smart cut Buffer | [10x] | 4.99 |
| Enzymes§ | [20 U/μl each] | 0.50 |
Table 5. Digestion mix 2.
| Reagent | Concentration | 1x |
|---|---|---|
| Nuclease-free water | - | 4.00 |
| NEB3 Buffer | [10x] | 5.00 |
| BSA | [20 μg/μl] | 0.50 |
| Enzyme (BsiWI) | [20 U/μl] | 0.50 |
Table 6. Ligation mix.
| Reagent | Volume |
|---|---|
| Water | 2.00 μl |
| Quick Ligase Buffer (2x) | 25.00 μl |
| Vector | 1.00 μl |
Figure 5. Representative agarose gel picture of insert check PCR products of IgH variable region genes.
Lane L: 100 bp DNA ladder. P: positive control. Blue arrow: expected size of positive colonies. Red arrow: expected size of negative colonies.
Figure 6. Example of IgG ELISA data.
A. Standard curve for the IgG ELISA. B. Table showing absorbance data and calculation of IgG concentration. Briefly, a linear line is fitted to the standard curve and used to extrapolate concentration values from the absorbances for the different diluted samples. Only absorbance within the linear range of the standard curve are will give an accurate calculation (here highlighted in orange). The final concentration of the IgG stock is calculated by multiplication with the dilution factor. Calculation can be made manually or by using the ELISA reader software. *. The true conc. values represent the average concentration of the two values of the conc. before adjusting for dilution.
Figure 7. Example of SDS-PAGE gel picture of purified mAbs.
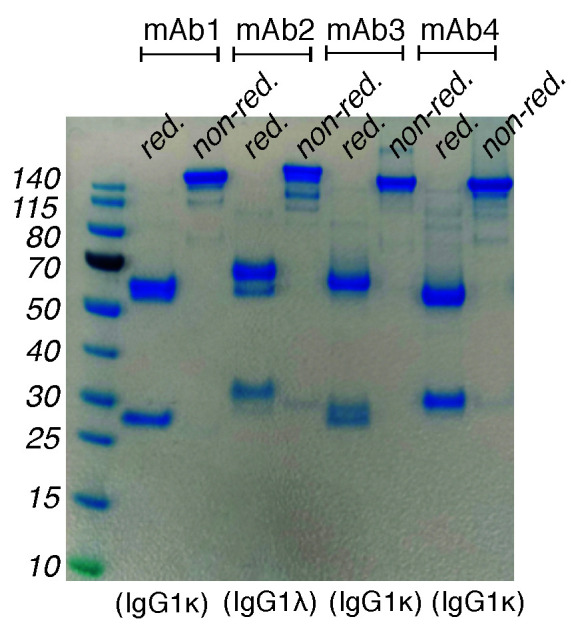
Purified antibodies run on a 4-12% Bis-Tris Plus protein gel using MES-SDS running buffer under reducing (red.) and non-reducing (non-red.) conditions. The heavy and light chain bands differ slightly in MW from antibody to antibody but typically fall between 50 and 60 kDa for the heavy chain and 25 and 30 kDa for the light chains. Notably, some ACPA mAb carry N-linked glycosylation in the VH or VL regions that will result in a size shift or double bands on the reduced gels. Proper evaluation of the presence of glycosylation can be made by parallel treatment with endoglycosidase (PNGase F compared to EndoS; data not shown). Protein bands are visualized by Coomassie blue staining.
Figure 8. Graphs showing example of polyreactivity ELISA as tested by dsDNA, LPS and insulin reactivity.
Grey lines represent positive mAbs. Black lines represent negative mAbs. Dashed horizontal lines show a cutoff of OD 450 for positive reactivity. ED38: high-positive polyreactive control antibody (green line); JB40: low-positive polyreactive control antibody (blue line); mGO53: negative control antibody (red line).
Figure 9. Example of IgG anti- (BSA)/plastic, -MDA-BSA, and -rabbit IgG ELISA data.
. The figure shows ELISA plate setup for screening mAbs binding to BSA/plastic, MDA-BSA or rabbit IgG. In this example, a positive systemic lupus erythematosus (SLE) patient serum pool is used as a positive control reference for IgG anti-MDA reactivity (here dilution 1:100 is set as 100 Reactivity Units [RU]/ml) and a positive RF IgG containing RA patient serum pool is used as a positive control reference sample on the rabbit IgG surface (here dilution 1:500 is set as 100 RU/ml). Qualification of the mAb reactivity for comparison between ELISA runs can be made by fitting a linear or logarithmic line to the standard curves and extrapolating a relative reactivity in RU/ml from the absorbance values.
Figure 10. Example of anti-CEP-1 ELISA data.
A. Standard curve for the CEP-1 ELISA using a positive RA reference serum sample (here dilution 1:120 is set to 333 Activity Units (AU)/ml). B. Data showing ACPA-positive (blue bars) and negative mAbs for CEP-1 ELISA at 5 μg/ml IgG. Qualification of the mAb reactivity for comparison between ELISA runs can be made by fitting a linear line to the standard curve and extrapolating a relative reactivity in AU/ml from the absorbance values.
Figure 11. Example of anti cit-Fib 573 (aa563-583) ELISA data.
A. Standard curve for the cit-Fib 573 ELISA using a positive RA reference serum [here dilution 1:80 is set to 333 Activity Units (AU)/ml]. B. Data showing ACPA-positive (blue bars) and negative mAbs for cit-Fib 573 ELISA at 5 μg/ml IgG. Qualification of the mAb reactivity for comparison between ELISA runs can be made by fitting a linear line to the standard curve and extrapolating a relative reactivity in AU/ml from the absorbance values.
Figure 12. Example of anti-CCP2 ELISA data.
A. Standard curve for the CCPlus ELISA. B. Data showing ACPA-positive (blue bars) and negative mAbs for the CCPlus ELISA. Qualification of the mAb CCP-reactivity at 5 μg/ml in AU/ml is made following the manufacturer’s instructions for serum samples.
Figure 13. Example of titration curves of binding of citrulline-specific mAbs in ELISA to CEP-1 (A), Cit-Fib 36-52 (B), and Cit-Vim 60-75 (C) peptide surfaces.
Recipes
-
RIA buffer (pH 7.6)
10 g BSA
20.4 g NaCl (350 mM)
10 ml 1 M Tris HCl (pH 7.6)
10 ml Triton X-100
10 ml 10% SDS
Adjust pH
Milli-Q water (up to 1,000 ml)
Note: Store this buffer at -20 °C until use.
-
Carbonate buffer (pH 9.6)
1.59 g NaCO3
2.93 g NaHCO3
Adjust pH
Milli-Q water (up to 1,000 ml)
Note: Store this solution at room temperature.
-
10x TBS (pH 8.0)
24.2 g Tris base
80 g NaCl
Adjust pH
Milli-Q water (up to 1000 ml)
Note: Store this solution at room temperature.
-
TBE buffer
1x TBS (prepare from 10x TBS with Milli-Q water)
0.05% Tween 20
0.1% BSA
-
0.5 M H2SO4
Dilute 27.2 ml of 98% Sulphuric acid in 1,000 ml distilled water
Note: Store this solution at room temperature.
-
0.1 M glycine (pH 3.0)
7.5 g Glycine
Adjust pH
Milli-Q water (up to 1,000 ml)
0.22 μm filter and store this solution at room temperature
-
1 M Tris (pH 8.0)
121 g Tris base
Adjust pH
Milli-Q water (up to 1,000 ml)
Note: Store this solution at room temperature.
-
LB medium
10 g Bacto-Tryptone
5 g NaCl
5 g Yeast extract
Milli-Q water (up to 1,000 ml)
Note: Autoclave and store this solution at room temperature.
Acknowledgments
This work was supported by the Knut and Alice Wallenberg Foundation, the European Research Council (grant 250167), the Innovative Medicines Initiative-supported BTCure program (grant 115142-2), The Swedish Rheumatism Association, King Gustaf V's 80-year Foundation, and the Swedish Medical Research Council.
Competing interests
The authors have no conflict of interest.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
Supplementary Data.
References
- 1.Agmon-Levin N., Damoiseaux J., Kallenberg C., Sack U., Witte T., Herold M., Bossuyt X., Musset L., Cervera R., Plaza-Lopez A., Dias C., Sousa M. J., Radice A., Eriksson C., Hultgren O., Viander M., Khamashta M., Regenass S., Andrade L. E., Wiik A., Tincani A., Ronnelid J., Bloch D. B., Fritzler M. J., Chan E. K., I. Garcia-De La Torre, Konstantinov K. N., Lahita R., Wilson M., Vainio O., Fabien N., Sinico R. A., Meroni P. and Shoenfeld Y.(2014). International recommendations for the assessment of autoantibodies to cellular antigens referred to as anti-nuclear antibodies. Ann Rheum Dis 73(1): 17-23.24126457 [Google Scholar]
- 2.Amara K., Steen J., Murray F., Morbach H., Fernandez-Rodriguez B. M., Joshua V., Engstrom M., Snir O., Israelsson L., Catrina A. I., Wardemann H., Corti D., Meffre E., Klareskog L. and Malmstrom V.(2019). Retraction: Monoclonal IgG antibodies generated from joint-derived B cells of RA patients have a strong bias toward citrullinated autoantigen recognition. J Exp Med 216(1): 245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harre U., Georgess D., Bang H., Bozec A., Axmann R., Ossipova E., Jakobsson P. J., Baum W., Nimmerjahn F., Szarka E., Sarmay G., Krumbholz G., Neumann E., Toes R., Scherer H. U., Catrina A. I., Klareskog L., Jurdic P. and Schett G.(2012). Induction of osteoclastogenesis and bone loss by human autoantibodies against citrullinated vimentin. J Clin Invest 122(5): 1791-1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krishnamurthy A., Joshua V., Haj Hensvold A., Jin T., Sun M., Vivar N., Ytterberg A. J., Engstrom M., Fernandes-Cerqueira C., Amara K., Magnusson M., Wigerblad G., Kato J., Jimenez-Andrade J. M., Tyson K., Rapecki S., Lundberg K., Catrina S. B., Jakobsson P. J., Svensson C., Malmstrom V., Klareskog L., Wahamaa H. and Catrina A. I.(2016). Identification of a novel chemokine-dependent molecular mechanism underlying rheumatoid arthritis-associated autoantibody-mediated bone loss. Ann Rheum Dis 75(4): 721-729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krishnamurthy A., Ytterberg A. J., Sun M., Sakuraba K., Steen J., Joshua V., Tarasova N. K., Malmstrom V., Wahamaa H., Rethi B. and Catrina A. I.(2019). Citrullination Controls Dendritic Cell Transdifferentiation into Osteoclasts. J Immunol 202(11): 3143-3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lloyd K. A., Steen J., Amara K., Titcombe P. J., Israelsson L., Lundström S. L., Zhou D., Zubarev R. A., Reed E., Piccoli L., Gabay C., Lanzavecchia A., Baeten D., Lundberg K., Mueller D. L., Klareskog L., Malmström V. and Grönwall C.(2018). Variable domain N-linked glycosylation and negative surface charge are key features of monoclonal ACPA: Implications for B-cell selection. Eur J Immunol 48(6): 1030-1045. [DOI] [PubMed] [Google Scholar]
- 7.Neovius, M., Simard, J. F., Askling, J. and group, A. s.(2011). Nationwide prevalence of rheumatoid arthritis and penetration of disease-modifying drugs in Sweden. Ann Rheum Dis 70(4): 624-629. [DOI] [PubMed] [Google Scholar]
- 8.Nielen M. M., van Schaardenburg D., Reesink H. W., van de Stadt R. J., van der Horst-Bruinsma I. E., de Koning M. H., Habibuw M. R., Vandenbroucke J. P. and Dijkmans B. A.(2004). Specific autoantibodies precede the symptoms of rheumatoid arthritis: a study of serial measurements in blood donors. Arthritis Rheum 50(2): 380-386. [DOI] [PubMed] [Google Scholar]
- 9.Rantapaa-Dahlqvist S., de Jong B. A., Berglin E., Hallmans G., Wadell G., Stenlund H., Sundin U. and van Venrooij W. J.(2003). Antibodies against cyclic citrullinated peptide and IgA rheumatoid factor predict the development of rheumatoid arthritis. Arthritis Rheum 48(10): 2741-2749. [DOI] [PubMed] [Google Scholar]
- 10.Sokolove J., Johnson D. S., Lahey L. J., Wagner C. A., Cheng D., Thiele G. M., Michaud K., Sayles H., Reimold A. M., Caplan L., Cannon G. W., Kerr G., Mikuls T. R. and Robinson W. H.(2014). Rheumatoid factor as a potentiator of anti-citrullinated protein antibody-mediated inflammation in rheumatoid arthritis. Arthritis Rheumatol 66(4): 813-821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Steen J., Forsström B., Sahlström P., Odowd V., Israelsson L., Krishnamurthy A., Badreh S., Mathsson Alm L., Compson J., Ramsköld D., Ndlovu W., Rapecki S., Hansson M., Titcombe P. J., Bang H., Mueller D. L., Catrina A. I., Grönwall C., Skriner K., Nilsson P., Lightwood D., Klareskog L. and Malmstrom V.(2019). Recognition of amino acid motifs, rather than specific proteins, by human plasma cell-derived monoclonal antibodies to posttranslationally modified proteins in rheumatoid arthritis. Arthritis Rheumatol 71(2): 196-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tiller T., Meffre E., Yurasov S., Tsuiji M., Nussenzweig M. C. and Wardemann H.(2008). Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. J Immunol Methods 329(1-2): 112-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Titcombe P. J., Wigerblad G., Sippl N., Zhang N., Shmagel A. K., Sahlström P., Zhang Y., Barsness L. O., Ghodke-Puranik Y., Baharpoor A., Hansson M., Israelsson L., Skriner K., Niewold T. B., Klareskog L., Svensson C. I., Amara K., Malmström V. and Mueller D. L.(2018). Pathogenic citrulline-multispecific B cell receptor clades in rheumatoid arthritis. Arthritis Rheumatol 70(12): 1933-1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wardemann H., Yurasov S., Schaefer A., Young J. W., Meffre E. and Nussenzweig M. C.(2003). Predominant autoantibody production by early human B cell precursors. Science 301(5638): 1374-1377. [DOI] [PubMed] [Google Scholar]
- 15.Wigerblad G., Bas D. B., Fernades-Cerqueira C., Krishnamurthy A., Nandakumar K. S., Rogoz K., Kato J., Sandor K., Su J., Jimenez-Andrade J. M., Finn A., Bersellini Farinotti A., Amara K., Lundberg K., Holmdahl R., Jakobsson P. J., Malmstrom V., Catrina A. I., Klareskog L. and Svensson C. I.(2016). Autoantibodies to citrullinated proteins induce joint pain independent of inflammation via a chemokine-dependent mechanism. Ann Rheum Dis 75(4): 730-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.