Abstract
Plasmodesmata (PD) are intercellular channels between walled plant cells that enable the transportation of materials between adjacent cells, which are important for plant growth and development. The permeability of PD must be tightly regulated. Assays to determine the permeability of PD are crucial for related studies on the regulation of PD development and permeability. Here we describe an assay for the determination of PD permeability via the observation and quantification of GFP diffusion and cell-to-cell transport of CMV MP-GFP in Arabidopsis leaves.
Keywords: Plasmodesmata, PD permeability, GFP, CMV MP-GFP, Particle bombardment
Background
Plasmodesmata (PD) are plant-specific channels between cells, which are important for plant growth and development as well as the interaction between plants and the surrounding environment ( Maule et al., 2011 ; Lee, 2015; Cheval and Faulkner, 2018). Water and small molecules can pass through PD freely. Other macromolecules, e.g., proteins, RNA, and some pathogens, can also move via PD. However, the mobility of those molecules between adjacent cells is tightly regulated, although the mechanism underlying this transport is not well understood ( Sevilem et al., 2015 ). Currently, there are several approaches reported for measuring the permeability of PD in plant tissues. One of the methods is to visualize the diffusion of fluorescent dextrans or other fluorescent probes from the targeted cells into neighboring cells ( Ding et al., 1996 ). This approach allows the researchers to select fluorescent dextrans of different sizes which will facilitate the study of PD permeability. In addition, this method also allows researchers to perform the coinjection of fluorescent dextrans with special drugs or proteins that allows the colleagues to assay the effect of those drugs or proteins on the permeability of PD. However, this method requires a rigorous experimental system and a highly experienced operator to perform the experiments. The second method is Drop-ANd-See (DANS) which uses the 5(6)-carboxy fluorescein diacetate (CFDA) for the rapid assessment of PD permeability ( Cui et al., 2015 ). DANS is a simple and rapid approach, but it is impossible to trace the spread of CFDA at the single cell resolution. The researchers in this field also used the green fluorescent protein (GFP) (Crawford and Zambryski, 2001; Liarzi and Epel, 2005) and Cucumber mosaic virus (CMV) Movement Protein (MP)-GFP (CMV MP-GFP) (Iglesias and Meins, 2000) as probes to assay the permeability of PD. The expression of GFP or CMV MP-GFP was achieved by delivering plasmids expressing them into plant cells via particle bombardment. This method enables the researchers to visualize the diffusion of GFP from targeted cells into adjacent cells or cell-to-cell movement of CMV MP-GFP. Here, we introduce the detailed method of GFP diffusion and cell-to-cell movement of CMV MP-GFP, which is adapted from our recently published paper ( Diao et al., 2018 ).
Materials and Reagents
9 cm glass dish (NORMAX, catalog number: 5058546)
Pipette tips (USA Scientific, catalog numbers: 1112-1720, 1110-1200)
Eppendorf tubes (Fisher Scientific, FisherbrandTM, catalog number: 05-408-129)
Macrocarriers (Bio-Rad, catalog number: 1652335)
1,100 psi rupture discs (Bio-Rad, catalog number: 1652329)
Stopping screens (Bio-Rad, catalog number: 1652336)
coverslip (THOMAS SCIENTIFIC, catalog number: 6672A46)
Arabidopsis thaliana plants
Tungsten M10 or M17 microcarriers (Bio-Rad, catalog numbers: 1652266 or 1652267)
TIANprep Midi Plasmid Kit (TIANGEN, catalog number: DP103-03)
Spermidine (Sigma-Aldrich, catalog number: S2626)
Calcium chloride dihydrate (CaCl2·2H2O) (Sigma-Aldrich, catalog number: C7902)
Plasmid pdGN ( Lee et al., 2005 )
Plasmid CMVMP-pdGN ( Diao et al., 2018 )
Bio-Rad 1.0 μm Gold Microcarriers (Bio-Rad, catalog number: 165-2263)
ethanol (AMRESCO, catalog number: E193)
50% sterile glycerin (Sigma-Aldrich, catalog number: G5516)
KNO3 (Sigma-Aldrich, catalog number: P6083)
NH4NO3 (AMRESCO, catalog number: 94629)
MgSO47H2O (Sigma-Aldrich, catalog number: M1880)
KH2PO4 (Sigma-Aldrich, catalog number: P5655)
MnSO4H2O (Macklin, catalog number: M813652)
ZnSO47H2O (Macklin, catalog number: Z820817)
H3BO4 (Sigma-Aldrich, catalog number: B6768)
KI (Sigma-Aldrich, catalog number: V900056)
Na2MoO42H2O (Sigma-Aldrich, catalog number: M1651)
CuSO45H2O (Macklin, catalog number: C805354)
CoCl26H2O (Aladdin, catalog number: C8396-01)
Glycine (Sigma-Aldrich, catalog number: V900144)
VB1 (Sigma-Aldrich, catalog number: V-014)
VB6 (Sigma-Aldrich, catalog number: V-018)
Nicotinic Acid (Sigma-Aldrich, catalog number:V900424)
Inositol (Sigma-Aldrich, catalog number: I5125)
FeSO47H2O (Macklin, catalog number: I809845)
Na2EDTA2H2O (AMRESCO, catalog number: 0105)
Sucrose (Sigma-Aldrich, catalog number: V900116)
Agar (Sigma-Aldrich, catalog number: A1296)
MS medium (see Recipes)
Gold suspension solution (see Recipes)
Equipment
Ophthalmic scissors
Ophthalmic tweezers
Centrifuge (Eppendorf, model: 5417R)
PDS-1000/HeTM Biolistic Particle Delivery System (Bio-Rad, model: 1652257)
Plant LED Incubator
Zeiss LSM 510 META or an equivalent confocal microscope
Vortex (Corning, model: Corning® LSETM Vortex Mixer)
NanoDrop 2000 (Gene Company Limited)
-20 °C freezer
Software
ImageJ (https://imagej.nih.gov/ij/ version 1.51)
IBM SPSS Statistics (version 25)
Procedure
Prepare petri dish containing 20 ml of MS medium with 0.8% agar.
-
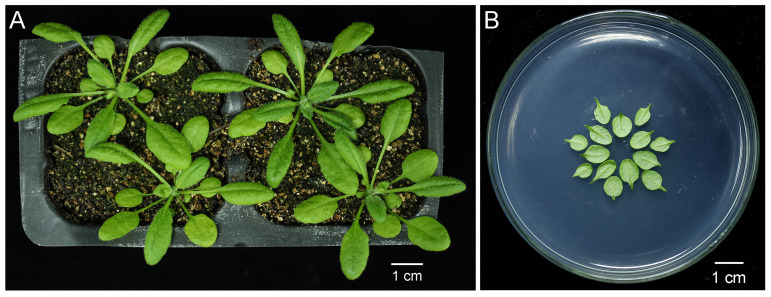
Arabidopsis plants grown under the 16 h/8 h light/dark cycle for 3-4 weeks (Figure 1A). Cut rosette leaves of Arabidopsis plants that had not yet bolted by ophthalmic scissors and place in a petri dish with the abaxial side facing upward as shown in Figure 1B.
Note: Cells of young leaves work better than old leaves. Leaves from different genotypes should be placed on MS medium equally divided (Figure 1B).
-
Prepare pdGN plasmid from E-coli ( Lee et al., 2005 ). Isolated the plasmid using A TIANprep Midi Plasmid Kit. The concentration of plasmid should be at least 1 µg/µl.
Note: The concentration of the plasmid is important for the next step. The pdGN-35S:HDEL-mCherry plasmid should also be prepared when we need to indicate the cells targeted by particle bombardment (Batoko et al., 2000) .
-
Prepare bullets of biolistic bombardment. Vortex 10 µl gold suspension solution (Recipe 2) in a microcentrifuge tube with 2 µg plasmid DNA for 1 min. Add 20 µl fresh 0.1 M spermidine and vortex again for 1 min. Add 50 µl 2.5 M CaCl2 and continue vortexing for another 3 min. Allow the mixture to settle for 5 min on ice and centrifuge the mixture for 10 s, at 2,348 × g. Discard the supernatant. Wash the pellet with 1 ml of 70% ethanol and then with 1 ml of 100% ethanol. Mix and centrifuge gently. Finally, resuspend the pellet in 15 µl 100% ethanol.
Notes:
The maximum total volume of plasmid DNA should be ≤ 10 µl. When both pDGN plasmids and CMV MP-pDGN plasmids need to be expressed simultaneously, 2 µl of each and gold particles are mixed together in a single test tube.
The 1 M spermidine stock can be stored at -20 °C for at least 3 months.
Vortex the mixture gently and transfer it to a macrocarrier using a pipette with a 10-200 µl tip. Air dry until the ethanol has evaporated, assemble all of the parts necessary for performing a particle bombardment according to the manufacturer’s instructions (Figure 2). Set the pressure to 1,100 psi and use 1,550 psi rupture discs. Place the MS plate with Arabidopsis leaves 9 cm below the macrocarrier (level 3) in the bombardment chamber (Figure 2). Set the vacuum level in the bombardment chamber to 28 psi and bombard.
Incubate the bombarded leaves for 24 h or 48 h at 23 °C in the dark.
-
Mount the bombarded leaves on a glass slide with a coverslip to image with a Zeiss LSM 510 META or an equivalent confocal microscope. The GFP signal is excited by a 488 nm argon laser and emission is captured in the range of 505–545 nm; the mCherry signal is excited by a 543 nm HeNe laser and the emission is captured in the range of 590–625 nm. Use a 20x/0.25 Fluar objective lens to scan the GFP cell clusters. Twenty percent laser intensity is used to image every fluorescent cell clearly.
Note: Acquire at least 60 images from each genotype for statistical analysis.
Figure 1. Preparation of the leaves of 4-week-old Arabidopsis plants for particle bombardment.
A. Arabidopsis plants after growing under the 16 h/8 h light/dark cycle for 4 weeks. scale bar = 1 cm. B. Rosette leaves that had not yet bolted were cut from Arabidopsis plants shown in (A) by ophthalmic scissors and placed concentrically with abaxial side facing upward on the surface of solid Murashige & Skoog medium. scale bar = 1 cm.
Figure 2. Side view of PDS-1000/HeTM Biolistic Particle Delivery System.

Before performing particle bombardment, the petri dish containing Arabidopsis leaves was moved into the PDS-1000/HeTM Biolistic Particle Delivery System (left). Part 1 and part 2 of the system were zoomed in at right. a is 1,100 psi rupture disk. b is microparticles carrier disk, which carries 1.0 μm Gold Microcarriers. c is stopping screen.
Data analysis
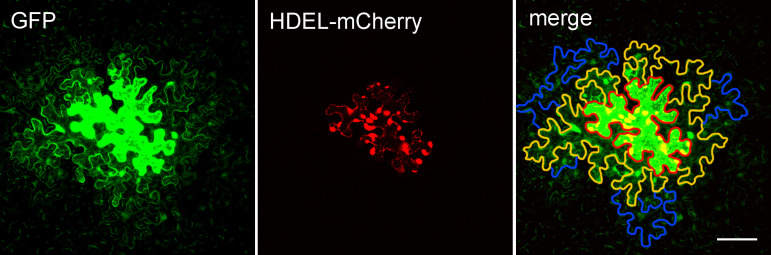
Open each image in ImageJ and find the cell expressing GFP (generally the cell with strongest fluorescence) which is defined as layer 0 (the edge of the cell is marked with red line in Figure 3). GFP protein diffuses from the layer 0 cell into other cells around it via PD. The cells that share a common cell wall with layer 0 cells were defined as layer 1 (the edge of cells is marked with orange lines in Figure 3). Cells that share a common cell wall with layer 1 cells, but not with layer 0 cells, were defined as layer 2 cells (the edge of cells is marked with blue lines marked in Figure 3). Cells that expressed GFP but showed no diffusion were not counted to avoid the situation in which damage was caused by the bombardment.
Count the number of cell layers in the cell clusters. As the bombardment will cause damage for cells or induce the non-uniform expression of GFP that prevent the subsequent analysis, we normally capture at least 60 images from more than 6 plants per set but only pick data from three plants with heathy cells and uniform expression of GFP for the statistical analysis. Repeat the experiment at least three times.
-
The IBM SPSS Statistics (version 25) software was used to perform the statistical analysis. First, the normality of the datasets was assessed by Shapiro-Wilk tests (Shapiro and Wilk, 1965) the data were not normally distributed by Shapiro-Wilk tests, the Mann-Whitney U-test (Fay and Proschan, 2010) was applied for the subsequent statistical analyses.
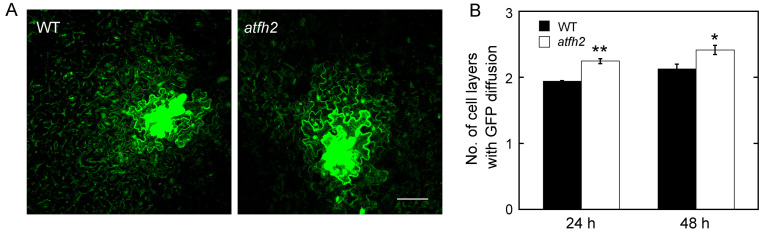
Figure 4. AtFH2 redundantly regulate PD permeability.
A. Images of GFP diffusion in leaf epidermal pavement cells of WT and atfh2 plants. scale bar = 50 µm. B. Quantification of the number of GFP diffusion layers in leaves of WT and atfh2 plants. Values represent mean ± SE. *P < 0.05, and **P < 0.01 by Mann-Whitney U-test.
Figure 3. Arabidopsis leaf epidermal cells expressing pdGN-35:GFP and pdGN-35S:HDEL-mCherry.
HDEL-mCherry is non-mobile, and was used to indicate the bombarded cell. The edge of the layer 0 cell is marked by a red line. The edges of the layer 1 cells and layer 2 cells are marked by orange and blue lines, respectively. scale bar = 50 µm.
Recipes
-
MS medium (1 L)
76 g/L KNO3
66 g/L NH4NO3
14.8 g/L MgSO47H2O
6.8 g/L KH2PO4
13.28 g/L CaCl2
1.69 g/L MnSO4H2O
8.6 g/L ZnSO47H2O
6.3 g/L H3BO4
0.83 g/L KI
0.25 g/L Na2MoO42H2O
0.025 g/L CuSO45H2O
0.025 g/LCoCl26H2O
0.2 g/L Glycine
0.1 g/L VB1
0.05 g/L VB6
0.05 g/L Nicotinic Acid
10 g/L Inositol
2.78 g/L FeSO47H2O
3.73 g/L Na2EDTA2H2O
10 g/L Sucrose
6 g/L Agar
-
Gold suspension solution
Place 30 mg of gold microcarriers into a 1 ml microcentrifuge tube
Add 1 ml of 70% ethanol and vortex for 3 min
Allow the mixture to settle for 15 min and centrifuge the mixture for 5 s
Remove the supernatant and add 1 ml sterile water and repeat the previous step two times
Remove the supernatant and add 500 µl 50% sterile glycerin
Vortex the mixture gently and store at -20 °C
Acknowledgments
This work was supported by grants from national natural science foundation of China (31471266; 31671390).
Competing interests
The authors declared that they have no conflicts of interest to this work.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1. Batoko H., Zheng H. Q., Hawes C. and Moore I.(2000). A Rab1 GTPase is required for transport between the endoplasmic reticulum and golgi apparatus and for normal golgi movement in plants. Plant Cell 12(11): 2201-2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cheval C. and Faulkner C.(2018). Plasmodesmal regulation during plant-pathogen interactions. New Phytol 217(1): 62-67. [DOI] [PubMed] [Google Scholar]
- 3. Crawford K. M. and Zambryski P. C.(2001). Non-targeted and targeted protein movement through plasmodesmata in leaves in different developmental and physiological states. Plant Physiol 125(4): 1802-1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cui W., Wang X. and Lee J. Y.(2015). Drop-ANd-See: a simple, real-time, and noninvasive technique for assaying plasmodesmal permeability. Methods Mol Biol 1217: 149-156. [DOI] [PubMed] [Google Scholar]
- 5. Diao M., Ren S., Wang Q., Qian L., Shen J., Liu Y. and Huang S.(2018). Arabidopsis formin 2 regulates cell-to-cell trafficking by capping and stabilizing actin filaments at plasmodesmata . Elife 7: e36316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ding B., Kwon M.-O. and Warnberg L.(1996). Evidence that actin filaments are involved in controlling the permeability of plasmodesmata in tobacco mesophyll. Plant J 10(1): 157-164. [Google Scholar]
- 7. Fay M. P. and Proschan M. A.(2010). Wilcoxon-Mann-Whitney or t-test? On assumptions for hypothesis tests and multiple interpretations of decision rules . Stat Surv 4: 1-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Iglesias V. A. and Meins F., Jr (2000). Movement of plant viruses is delayed in a β-1,3-glucanase-deficient mutant showing a reduced plasmodesmatal size exclusion limit and enhanced callose deposition. Plant J 21(2): 157-166. [DOI] [PubMed] [Google Scholar]
- 9. Lee J. Y.(2015). Plasmodesmata: a signaling hub at the cellular boundary. Curr Opin Plant Biol 27:133-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lee J. Y., Taoka K., Yoo B. C., Ben-Nissan G., Kim D. J. and Lucas W. J.(2005). Plasmodesmal-associated protein kinase in tobacco and Arabidopsis recognizes a subset of non-cell-autonomous proteins . Plant Cell 17(10): 2817-2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liarzi O. and Epel B. L.(2005). Development of a quantitative tool for measuring changes in the coefficient of conductivity of plasmodesmata induced by developmental, biotic, and abiotic signals. Protoplasma 225(1-2): 67-76. [DOI] [PubMed] [Google Scholar]
- 12. Maule A. J., Benitez-Alfonso Y. and Faulkner C.(2011). Plasmodesmata- membrane tunnels with attitude. Curr Opin Plant Biol 14(6): 683-690. [DOI] [PubMed] [Google Scholar]
- 13. Sevilem I., Yadav S. R. and Helariutta Y.(2015). Plasmodesmata: channels for intercellular signaling during plant growth and development. Methods Mol Biol 1217: 3-24. [DOI] [PubMed] [Google Scholar]
- 14. Shapiro S. S. and Wilk M. B.(1965). An analysis of variance test for normality(complete samples). Biometrika 4): 591-611. [Google Scholar]