Abstract
Cyanobacteria represent a frequently used model organism for the study of oxygenic photosynthesis. They belong to prokaryotic microorganisms but their photosynthetic apparatus is quite similar to that found in algal and plant chloroplasts. The key players in light reactions of photosynthesis are Photosystem I and Photosystem II complexes (PSI and PSII, resp.), large membrane complexes of proteins, pigments and other cofactors embedded in specialized photosynthetic membranes named thylakoids. For the study of these complexes a mild method for the isolation of the thylakoids, their subsequent solubilization and analysis is essential. The presented protocol describes such a method which utilizes breaking the cyanobacterial cells using glass beads in an optimized buffer. This is followed by their solubilization using dodecyl-maltoside and analysis using optimized clear-native gel electrophoresis which preserves the native oligomerization state of both complexes and allows the estimation of their content.
Keywords: Cyanobacteria, Photosynthesis, Thylakoids, Photosystem, Clear-native gel, Electrophoresis
Background
Thylakoids are specialized membranes of algal and plant chloroplasts and cyanobacteria. The thylakoid lipid bilayer shares characteristic features with prokaryotic bacterial membranes but it is enriched in glycolipids, namely galactolipids, while the level of acidic lipids is relatively low and does not exceed 20% of the overall lipid content. Thylakoids contain a large number of proteins and their complexes and over 40% of them are involved in light-dependent reactions of photosynthesis. The next largest functional group includes proteins involved in protein targeting, processing and folding, oxidative stress response and translation. The most integral membrane proteins of thylakoids form large protein complexes which play an important role in the harvesting of light and its photochemical conversion. The key complexes performing this conversion are PSI and PSII. They both perform the light-driven primary charge separation resulting in the thylakoid-associated flow of electrons reducing NADP+ and generating a pH gradient used for the synthesis of ATP. Both complexes are subject to intensive research focused mainly on the elucidation of their function and underlying structural features as well as the mechanism of their assembly. For this research the isolation of native membranes and their detailed analysis as concerns the content, assembly and oligomerization state of both photosystems is essential. The existing and frequently used protocols for the isolation of cyanobacterial thylakoids either do not sufficiently preserve the native structure of fragile membrane complexes like Photosystem II dimers (e.g., Komenda and Barber, 1995), or the obtained preparations contain high concentrations of compounds (for instance 1 M glycine betaine) which are not compatible with the clear native gel electrophoresis (e.g., Gombos et al., 1994 ). The described protocol for thylakoid preparation is optimized from both points of view. The analysis of cyanobacterial membranes by clear native PAGE (CN PAGE) using a protocol modified from Wittig et al. (2007) gives direct information about the quantity and oligomeric (or assembly) state of multisubunit pigmented photosystem complexes with unprecedented speed and resolution. The isolation, solubilization, and analysis of membranes does not disrupt weak non-covalent interactions between pigments and proteins and therefore the gel with separated individual complexes can be directly scanned. Unlike denaturing SDS gels, no fixation, staining and destaining, or, blotting and immunodetection is needed for distinguishing between PSII and PSI (see Figure 2). Green PSI complexes do not show red fluorescence at room temperature and are present as dominant green bands in the gel while PSII complexes fluoresce and their green bands are hard to distinguish on the simple transparency scan. Thus, PSI quantification is done using the simple transparency scan of green non-fluorescent bands while PSII quantification is done using the red fluorescence scan of fluorescent bands.
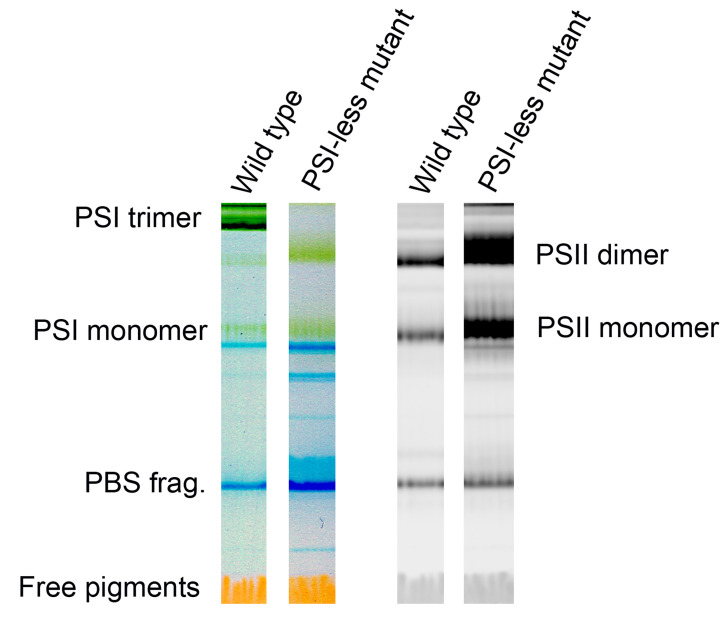
Figure 2. Representative color scans (left pair) and chlorophyll fluorescence images (right pair) of the gel with separated Photosystem I and Photosystem II complexes from the cyanobacterium Synechocystis PCC 6803 GT-P .
( Tichý et al., 2016 ) and the Photosystem I-less mutant ( Shen et al., 1993 ). Left two lanes show color scans, right ones Chl fluorescence scans with dominant bands of Photosystem II dimer and monomer. Blue bands (PBS frag.) represent fragments of phycobilisomes, the cyanobacterial antennae, fluorescence of which is not elicited by blue light.
Materials and Reagents
Pipette tips
2 ml screw cap polypropylene micro vials (Biospec Products, catalog number: 3205)
Hamilton® microsyringe series 700, 0.25 ml (Sigma, catalog number: 24538-U)
Hamilton needle, 51 mm (Sigma, catalog number: 21746)
Binder Clip (Deli 8584, Luma trading)
Magnetic stirring bars with diameter 5 mm and length 20 mm
Transparent plastic film
1.5 ml spectroscopic cuvettes (Fisherbrand PS, catalog number: 7755.0302)
The glucose tolerant strain of the cyanobacterium Synechocystis sp. PCC6803 (GT-P, Tichý et al., 2016 )
Acrylamide for molecular biology (AA) (AppliChem Panreac, catalog number: A3812,1000)
Bisacrylamide for molecular biology (BIS) (AppliChem Panreac, catalog number: A3636,0250)
6-aminohexanoic acid (ACA) (AppliChem Panreac, catalog number: A2266,500)
Bis-Tris (AppliChem Panreac, catalog number: A1025.1000)
MES (AppliChem Panreac, catalog number: A0689.0250)
Tricine (AppliChem Panreac, catalog number: A1085,1000)
N,N,N',N'-tetramethylethylenediamine (TEMED) (Sigma, catalog number: T928-25ml)
Ammonium persulfate (APS) (Sigma, catalog number: T3678-25g)
Dithiothreitol (DTT) (AppliChem Panreac, catalog number: A1101,0025)
SDS (Sigma, catalog number: 7172-100g)
Glass beads with a diameter of 150-212 μm (Sigma, catalog number: G1145)
n-Dodecyl-β-D-maltoside (DM) (AppliChem Panreac, catalog number: A0819,0005)
Sodium deoxycholate (Sigma, catalog number: 30970-25g)
Gel Filtration Markers Kit for Protein Molecular Weights (Sigma, catalog number: MWGF1000-1KT)
CaCl2 (analytical grade)
MgCl2 (analytical grade)
Methanol (analytical grade)
Sodium hydroxide (analytical grade)
Hydrochloric acid (analytical grade)
Glycerol (HPLC grade)
Milli-Q de-ionized water (Merck Millipore, Ultrapure [Type 1] water)
Buffer B (see Recipes)
Gel buffer 6x (pH 7.6) (see Recipes)
Acrylamide solution (AB) (see Recipes)
Upper (Cathode) buffer 10x (see Recipes)
Upper (Cathode) buffer 1x (see Recipes)
Lower (Anode) buffer 10x (see Recipes)
Lower (Anode) buffer 1x (see Recipes)
Resolving gel and stacking gel (see Recipes)
Equipment
Automatic pipettes (P1000, P200 and P10)
Mini-beadbeater (Sigma, catalog number: Z249289)
Magnetic stirrer Color Squid White (Thermo Fisher Scientific, catalog number: 6110.1005)
GE gradient maker (Sigma, catalog number: GESG50)
Vortex (Velp Scientifica, model: RX3)
Centrifuge (Hettich, model: Micro22 R)
UV-VIS spectrophotometer (Shimadzu, model: UV3000)
Cooled microcentrifuge (Eppendorf, model: 5415R)
PAGE apparatus (Bio-Rad, PROTEAN® II xi Cell)
Power Supply (Bio-Rad, PowerPacTM HV High-Voltage)
Cooled thermostat (Labio, model: CTB 06C)
LAS 4000 camera (Fuji; for chlorophyll fluorescence scanning used LED blue light source 460 nm for excitation and R670 filter for emission)
Scanner HP ScanJet G4050 (HP, model: G4050)
pH meter (Sartorius, PB-11)
FIM-150 flake ice maker (Biobase)
Software
ImageJ bundled with Java 1.8.0_112 (https://imagej.nih.gov/ij/download.html)
Procedure
The protocol requires 20 ml of Synechocystis PCC 6803 grown at 30 °C on an orbital shaker (about 60 rpm) under cool white fluorescent light of 35 μmol m-2 s-1 in liquid BG11 ( Rippka et al., 1979 ) with an optical density of ~0.4-0.6 at 750 nm (see Notes). Collect the cyanobacterial cells by centrifugation at 2,300 × g for 10 min at ~4 °C, resuspend the cell pellet in 1.5 ml of Buffer B and transfer the resuspended cells into a 2 ml screw cap microtube. From this point keep the tubes on ice.
Centrifuge the tube in the microcentrifuge at 9,300 × g for 2 min at 4 °C, discard the supernatant and resuspend the pellet in 0.15 ml of Buffer B and add the same volume of glass beads (measured separately in a graduated 0.5 ml microtube and added to the 2 ml microtube with the sample).
Break the cells with three cycles of 20 s beating (max. speed) and 1 min cooling in a Mini-Beadbeater.
Add 0.2 ml of Buffer B, vortex the tube for 2 s and immediately transfer the supernatant into a new pre-cooled 2 ml screw cap microtube, repeat five times.
Centrifuge the tube at 400 × g for 1 min at 4 °C to remove unbroken cells and glass beads.
Transfer the supernatant into a new pre-cooled 2 ml screw cap microvial and centrifuge at 16,000 × g for 20 min at 4 °C.
Remove supernatant and resuspend the pellet in 0.2 ml of Buffer B using a stainless-steel piston from a 0.25 ml microsyringe (length 120 mm, diameter 2 mm): hold the tube with the pellet after removal of the supernatant in one and piston in the second hand, insert the piston into the tube down to the bottom and vortex for 15 s with inserted piston, then add 0.2 ml of the buffer and vortex again with piston inside for 10 s.
Measure the chlorophyll (Chl) content in the suspension. Mix 13.5 µl of suspension with 1.5 µl of 10% DM, centrifuge at 16,000 × g for 5 min, take 10 µl of the supernatant and add to 990 µl of methanol in a screw cap microvial, vortex, centrifuge at 16,000 × g for 5 min, take the supernatant and measure absorbance at 720 and 666 nm, C (μg Chl/ml) = (A666 - A720) x 1,269 (Wellburn, 1994).
-
Prepare the resolving gel (see Video 1).
First assemble the gel cassettes from glass plates and 1 mm spacer according to the manufacturer’s instructions.
Then mix all components well (except for APS!) for each 4% and 14% resolving gel solution using magnetic stirrers and stirring bars (components of each solution are shown in Recipe 8, one column is one solution, the second and third column); then add APS to both solutions under constant mild stirring.
Pour the 4% solution together with stirring bar into the right compartment of the gradient maker located on the top of the magnetic stirrer. The gradient maker has an inlet attached to the silicone tube leading to the peristaltic pump and has a closed connecting channel to the second compartment. The outlet of the peristaltic pump is connected to the hypodermic needle attached to the upper part of the glass gel cassette using a binder clip.
Carefully open the connecting channel just enough to fill it with the 4% solution (if more solution is released, pipette it back to the right compartment) and close it again, and add now the 14% solution into the second (left) compartment of the gradient maker.
Remove 8.5 ml of the 14% solution with a pipette and add it directly to the bottom of the glass cassette for gel polymerization and immediately start pumping the remaining solution with a rate of about 10 ml per minute into the glass cassette and in parallel open the connecting channel between the compartments of the maker. Adjust mild stirring of the solution (2 turns per second).
After pouring the gradient, remove the needle, overlay the solution in the gel cassette with 0.5 ml of distilled water using a 0.25 ml syringe and wash the gradient maker and tubings with Milli-Q water. Watch the interface between the water and 4% solution until a clear border is formed and then remove the water with the syringe.
Prepare the stacking gel. Add APS to the 10 ml 4% solution (right column in Recipe 8), pour to the top of the polymerized resolving gel, immerse the 10 well comb avoiding air bubbles and wait for polymerization; afterwards remove the comb and wash the wells with Milli-Q water.
Attach the prepared gels into the cooling core, check if the connection is tight and not leaking with distilled water, pour out water and insert the core with gels into the electrophoresis chamber with 2 litres of 1x lower buffer, attach the silicon tubes of the cooling core to the thermostat, which is set to 4 °C and pour 200 ml of 1x upper buffer into the upper reservoir of the cooling core.
For gel analysis take the samples containing 5 μg Chl, add a corresponding volume of Buffer B buffer to reach the same volume for all samples (usually 30 μl), add 1/10 of the volume of 10% DM and centrifuge immediately at 16,000 × g for 20 min at 4 °C.
Take 25 μl of the solubilized membranes and load into the gel wells.
For estimation of the molecular size of complexes, Gel Filtration Markers Kit for Protein Molecular Weights can be used, load 30 μl of the recommended mixture.
Close the lid, attach it to the power supply and run in the constant current mode at 15 mA for 1.5-2 h with an upper voltage limit of 1,000 V (see Figure 1; during the run voltage gradually increases up to 1,000 V, after reaching this limit, the current goes down).
After the pigmented front reaches a distance of about 8 cm from the bottom of the wells, switch off the power supply, remove the lid, detach the silicone cooling tubes and take out the cooling core with the gels from the electrophoretic chamber.
Remove the gel from the glass cassette and wrap it into a transparent plastic film.
Transfer it into the scanner to acquire the color image (300 dpi resolution, 48-bit color depth).
Transfer the gel into the LAS4000 camera to take a 16 bit greyscale image in tiff format either in transparency mode to quantify green bands of PSI (Digitize DIA method, the lowest tray position, 3 s exposure, high resolution mode) or in epifluorescence mode using the blue LED excitation light (wavelength 460 nm) eliciting the chlorophyll (but not phycobiliprotein) fluorescence while the emitted light is passed through an R670 filter (long pass filter with the edge at 670 nm) to quantify PSII (Fluorescence method, the lowest tray position, 3 s exposure, high-resolution mode) (see Figure 2).
Quantify bands using ImageJ according to the user guide (https://imagej.nih.gov/ij/docs/guide/user-guide.pdf). The bands are first selected as regions of interest (ROIs) using the Area selection tools in Toolbar Tools, the background is subtracted using the processing tools and the intensity of the bands analyzed using the intensity statistics in the analysis tools.
Video 1. Pouring the resolving gel.

Figure 1. The setup of the electrophoretic apparatus (lower right corner) with cooling thermostat (left) and power supply (upper right corner).

Notes
Apart from the cyanobacterium Synechocystis PCC 6803 the described protocol has already been successfully tested for other cyanobacterial strains such as Synechococcus elongatus PCC 7942 and Synechococcus sp. PCC 7002. For the green algae Chlamydomonas reinhardtii, Chlorella sorokiniana and Scenedesmus quadricauda the protocol is identical, only larger glass beads with a diameter of 0.5 mm (Sigma) should be used for breaking the cells. The procedure for CN PAGE can also be used for isolated thylakoid membranes from plant chloroplasts.
Recipes
-
Buffer B (50 ml stock)
25 mM MES/NaOH buffer (pH 6.5)
10 mM CaCl2
10 mM MgCl2
25% glycerol
Divide into 10 ml aliquots and store frozen at -25 °C (stable for at least 6 months)
-
Gel buffer 6x (pH 7.0, 100 ml)
300 mM Bis-Tris/HCl buffer
Adjust to pH 7.0 with 4 M HCl
Store at 4 °C (stable for at least 6 months at 4 °C)
Note: Usually 100 ml is prepared.
-
Acrylamide solution (AB)
50% AA
1.33% BIS
Milli-Q water
Store at 4 °C (stable for at least 3 months)
Note: Usually 500 ml is prepared.
-
Upper (Cathode) buffer 10x
0.5 M tricine
150 mM Bis-Tris/HCl
Adjust pH to 7.0 using 4 M HCl
Store at 4 °C (stable for at least 6 months at 4 °C)
Note: Usually 500 ml is prepared.
-
Upper (Cathode) buffer 1x (200 ml for one PAGE)
Add 20 ml of upper buffer 10x to 180 ml of Milli-Q water and 1 ml of 10% sodium deoxycholate to reach 0.05% final concentration
Add 0.4 ml of 10% DM to reach 0.02% final concentration.
Note: Prepare fresh before running.
-
Lower (Anode) buffer 10x
0.5 M Bis-Tris/HCl
Adjust pH to 7.0 with 4 M HCl
Store at 4 °C (stable for at least 6 months at 4 °C)
Note: Usually 500 ml is prepared.
-
Lower (Anode) buffer 1x (2 L)
Dilute lower (Anode) buffer 10x to lower (Anode) buffer 1x with Milli-Q water
Note: This solution is stable for at least 3 months at 4 °C and can be reused three times.
-
Resolving gel and stacking gel
Mix individual components (the first column from left) to get 4% (second column) and 14% (third column) solution for the preparation of the resolving gradient gel and 4% solution for the preparation of the stacking gel (fourth column) according to the following table:
4% (resolving gel) 14% (resolving gel) 4% (stacking gel) Glycerol 0 3.2 g 0 g Water 6.5 ml 6.1 ml 7.4 ml Buffer 1.4 ml 2.8 ml 1.7 ml AB 0.68 ml 4.8 ml 0.8 ml 10% DM 43 μl 86 μl 50 μl TEMED 3.5 μl 7.0 μl 10 μl 10% APS 19 μl 38 μl 80 μl Volume 8.5 ml 17 ml 10 ml
Acknowledgments
This work was supported by the Czech Ministry of Education, Youth and Sports of the Czech Republic (project LO1416) and bilateral projects between the Academies of Sciences of the Czech Republic and Hungary (MTA-16-11).
Competing interests
We declare no conflict of interests.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1. Gombos Z., Wada H. and Murata N.(1994). The recovery of photosynthesis from low-temperature photoinhibition is accelerated by the unsaturation of membrane lipids: a mechanism of chilling tolerance. Proc Natl Acad Sci U S A 91(19): 8787-8791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Komenda J. and Barber J.(1995). Comparison of psbO and psbH deletion mutants of Synechocystis PCC 6803 indicates that degradation of D1 protein is regulated by the QB site and dependent on protein synthesis . Biochemistry 34(29): 9625-9631. [DOI] [PubMed] [Google Scholar]
- 3. Rippka R., Deruelles J., Waterbury J. B., Herdman M. and Stanier R. Y.(1979). Generic assignments, strain histories and properties of pure cultures of cyanobacteria. Microbiology 111: 1-61. [Google Scholar]
- 4. Shen G., Boussiba S. and Vermaas W. F.(1993). Synechocystis sp PCC 6803 strains lacking photosystem I and phycobilisome function . Plant Cell 5(12): 1853-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tichý M., Beckova M., Kopecna J., Noda J., Sobotka R. and Komenda J.(2016). Strain of Synechocystis PCC 6803 with aberrant assembly of photosystem II contains tandem duplication of a large chromosomal region . Front Plant Sci 7: 648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wellburn A. R.(1994). The spectral determination of chlorophylls a and b, as well as total carotenoids, using various solvents with spectrophotometers of different resolution. J Plant Physiol 144(3): 307-313. [Google Scholar]
- 7. Wittig I., Karas M. and Schagger H.(2007). High resolution clear native electrophoresis for in-gel functional assays and fluorescence studies of membrane protein complexes. Mol Cell Proteomics 6(7): 1215-1225. [DOI] [PubMed] [Google Scholar]