Abstract
Shigella flexneri is an intracellular bacterial pathogen that gains access to the gut epithelium using a specialized Type III Secretion System (T3SS). Various determinants mediating this invasive infection have been experimentally verified using the classical gentamicin protection assay presented here. In this assay epithelial cell lines are infected by bacteria in vitro and the extracellular bacteria are killed by gentamicin. The internalized bacteria, which are protected from the bactericidal action of gentamicin, are recovered by lysing the epithelial cells and enumerated by determining the colonies formed on solid medium. Various techniques based on light microscopy, such as immunofluorescence and bacteria expressing fluorescent proteins, are also used for studying intracellular bacteria. However, these techniques are not only labor intensive and require sophisticated equipment, but mostly are also not quantitative. Despite being an easy quantitative method to study invasiveness of bacteria, the gentamicin protection assay cannot distinguish between the survival and multiplication of the internalized bacteria over longer incubation periods. To alleviate the complications created by multiplication and dissemination of internalized bacteria, complementary assays like plaque formation assays are required. This protocol presents an easy and cost-effective method to determine the invasiveness and the capacity to establish an infection of Shigella under different conditions.
Keywords: Shigella, Intracellular pathogen, Invasion, Type III Secretion System, Intestinal epithelial cells
Background
Shigella infects about 160 million people leading to about 600,000 deaths every year (Reference 8). The clinical manifestations of Shigella infection, or shigellosis, arise only after the bacterium enters the epithelium, where it multiplies and spreads to adjacent cells causing cell death and tissue necrosis. Thus, the entry into the epithelial cells is a critical step in the infectious life cycle of Shigella (Ashida et al., 2015). Most of the determinants mediating this step have been ascribed to a large virulence plasmid that encodes a T3SS responsible for injecting bacterial proteins into the host cell via a needle-like projection (Puhar and Sansonetti, 2014).
The gentamicin protection assay is a classical method that is used to assess the invasiveness of Shigella and has led to the identification of a number of mediators of invasion by mutational analysis and comparisons with the wild-type bacteria. In a typical experiment, the bacteria are allowed to infect the intestinal cells in a synchronized manner followed by removal of external bacteria by gentamicin treatment. The invasiveness is assessed by determining the number of surviving bacteria (protected from gentamicin being intracellular) from the lysates of infected cells. A lower number of surviving bacteria with respect to the wild-type indicates a defect in invasion or intracellular survival. If short infection times are used after the gentamicin treatment (typically 1h), the gentamicin protection assay allows to compare the bacterial capacity to enter host cells only. However, if longer infection times are used after gentamicin treatment (2 h or more), the assay will also reflect the bacterial capacity to survive and multiply within the target cell besides the capacity to invade. Hence, if the gentamicin protection assay is employed to study the capacity to survive and multiply by allowing long infection times, these experiments should always be complemented with tests carried out at short infection times to ensure correct interpretation of the results. The gentamicin protection assay, however, is not suitable to assess the extent of intercellular spread of bacteria throughout the monolayer, which is conveniently determined by a plaque formation assay. Although the invasiveness of bacterial pathogens can also be studied by fluorescence microscopy-based techniques such as immunofluorescence and FACS; it requires the use of costly reagents like labeled probes, special reporter strains and sophisticated equipment. The gentamicin protection assay is not only relatively easy to perform and cheap, it is fairly rapid and less labor intensive making it amenable to higher throughput.
The protocol presented here reprises the classical gentamicin protection assay, which has been optimized for use with Shigella flexneri and TC7 intestinal epithelial cells but can be modified to be used with other intracellular pathogens and other host cell types.
Materials and Reagents
Conical flask (Duran, catalog number: 2121628)
Microfuge tubes (Eppendorf, Safe-Lock tubes, catalog numbers: 0030120086, 0030120094)
Centrifuge tubes (Sarstedt, catalog numbers: 62.554.502, 62.547.254)
Culture tubes (TPP, catalog number: 91016)
Pipette tips (VWR, catalog numbers: 89041-404, 89041-412, 89041-400)
Serological pipettes (VWR, catalog numbers: 612-3702, 612-3700, 612-3698)
6-well tissue culture test plates (TPP, catalog number: 92406)
TC7 cells (human cell line, a derivative of Caco-2 colon adenocarcinoma cells) (Chantret et al., 1994)
Shigella flexneri M90T (Sansonetti et al., 1982)
Dulbecco's Modified Eagle Medium (DMEM) (Gibco, catalog number: 21885-025)
Penicillin-Streptomycin solution (10,000 U/ml) (Gibco, catalog number: 15140122)
Minimum Essential Medium Non-Essential Amino Acids solution (100x) (Gibco, catalog number: 11140-035)
Dulbecco’s Phosphate Buffered Saline (PBS) solution (Gibco, catalog number: 14190-144)
Fetal Bovine Serum (Gibco, catalog number: 10500056)
HEPES 1 M solution (Gibco, catalog number: 15630-080)
Trypsin-EDTA (0.05%), phenol red (Gibco, catalog number: 25300-054)
Trypan Blue Stain (0.4%) (Thermo Scientific, catalog number: T10282)
Agarose (VWR Life Science, catalog number: 35-1020)
Tryptic Soy Broth (TSB) ready to use powder (Merck, catalog number: 105459)
Tryptic Soy Agar (TSA) ready to use powder (Merck, catalog number: 105458)
Ethanol (VWR chemicals, catalog number: 20821.558)
Congo red (Sigma, catalog number: C6277)
Sodium deoxycholate (Sigma, catalog number: 30970)
Gentamicin sulfate (Sigma, catalog number: G1264)
Sterile disposable petriplates (Sigma, catalog number: P5606-400EA)
Congo red solution (see Recipes)
Growth medium (see Recipes)
Infection buffer (see Recipes)
Sodium Deoxycholate solution (see Recipes)
Gentamicin solution (see Recipes)
Equipment
Biosafety cabinet (Thermo Scientific, HERAsafe KS 18)
Spectrophotometer (Amersham, Utrospec 2100 pro)
Pipette controller (VWR, Accurpette)
CO2 Incubator (Thermo Scientific, Heracell VIOS 160i)
Benchtop centrifuge (VWR, Micro Star 17R)
Centrifuge (Eppendorf, 5810R with Rotor A-4-81 for plates)
Water bath (Grant, JBA 12)
Pipettes (Eppendorf, Research plus)
Inverted microscope (Motic, AE2000 Binocular)
Cell counter (Thermo Scientific, Countess II FL)
Cell counter slides (Thermo Scientific, Countess Cell Counting Chamber Slides, C10228)
Orbital shaker (Edmund Bühler, Swip SM25)
Vacuum pump (VWR, Mini diaphragm vacuum pump VP 86)
Software
Prism 7 (GraphPad, https://www.graphpad.com)
Procedure
Note: The experiment should be carried out in a biosafety level 2 lab.
-
Preparation of bacteria
Refer to Procedure A of the Protocol–Plaque Assay to Determine Invasion and Intercellular Dissemination of Shigella flexneri in TC7 human Intestinal Epithelial Cells (Sharma and Puhar, 2019).
-
Preparation of TC7 cells
Refer to Procedure B of the Protocol–Plaque Assay to Determine Invasion and Intercellular Dissemination of Shigella flexneri in TC7 human Intestinal Epithelial Cells (Sharma and Puhar, 2019).
-
Infection and cell lysis
Day 3
On Day 3, when the bacteria reach OD600nm = 0.3-0.4, pipette 1-1.5 ml culture into a microcentrifuge tube.
Spin the tube at 3,000 × g for 5 min. Aspirate the supernatant. Add an equal volume of PBS at room temperature and resuspend the pellet by tapping the microcentrifuge tube or by gentle vortexing.
-
Repeat Step C2 at least twice and finally resuspend the pellet in infection buffer (see Recipes) at room temperature. Record the OD600nm of the bacterial suspension.
The recommended multiplicity of infection (MOI) for this experiment is 5. Since there are 1 x 106 cells in each well, the number of bacteria required per well is 5 x 106.
The number of Shigella cells at OD600nm = 1 is 0.5 x 109/ml; hence at OD = z, the number of bacterial cells = z x 0.5 x 109. Calculate the volume of bacterial suspension required for infecting each well using the following formula:
Volume of bacterial suspension required (in ml) = 5 x 106/z x 0.5 x 109
Aspirate the medium from the cells and add 2 ml of sterile PBS at room temperature to each well.
Swirl the plate gently and carefully aspirate all the liquid. After at least 2 washings, add 2 ml of infection buffer (see Recipes) at room temperature to each well.
Add the desired volume of bacterial suspension (typically about 10 μl, calculated in Step C3) to each well of the 6-well plate. Centrifuge the plate at 180 × g for 10 min at room temperature.
Incubate the plate at 37 °C, 10% CO2, for the desired infection time, typically 1 h. When Shigella comes in contact with the cells at the incubation temperature of 37 °C (35 °C and above) the secretion of effector proteins through the T3SS is triggered which mediates the bacterial invasion.
After incubation, wash the wells with sterile PBS three times as in Steps C4 and C5.
Add 2 ml of gentamicin solution (see Recipes) to each well and incubate the plate for one more hour at 37 °C and 10% CO2. If intracellular bacterial survival and multiplication is assessed, this incubation time can be extended.
After incubation with gentamicin, wash the wells thrice with PBS (as in Steps C4 and C5).
Aspirate the PBS from the wells and add 1 ml of sodium deoxycholate solution (see Recipes) to lyse the TC7 cells (the bacteria resist deoxycholate).
Using a 1 ml pipette, scrape the cells off the surface and pipette up and down to lyse the cells effectively and homogenize the lysate.
-
Plating
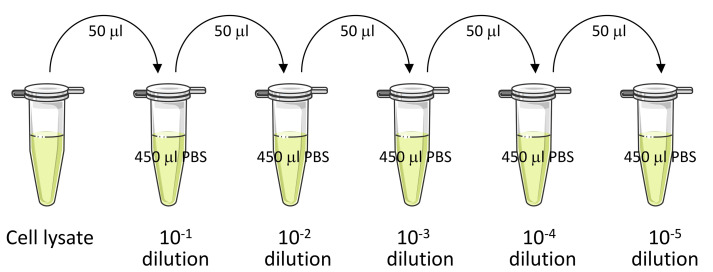
Prepare log serial dilutions of this lysate in sterile PBS as follows.
Dispense 450 μl of sterile PBS in 1.5 ml microcentrifuge tubes.
Add 50 μl of cell lysate to one tube and mark it 10-1 dilution (as in Figure 1). Vortex the tube for a few seconds and transfer 50 μl of suspension from this tube to the next tube containing 450 μl of sterile PBS. Mark this new tube as 10-2 dilution. Repeat this procedure to obtain further dilutions of the lysate (Figure 1).
For shorter infection time points, a lower dilution will provide significant results. For example, if infection time is 1 h, 10-1 dilution is enough; but if the infection time is increased to 2 h, 10-2 dilution would be more appropriate owing to bacterial multiplication. Similarly, consider higher dilutions for even longer infection times.
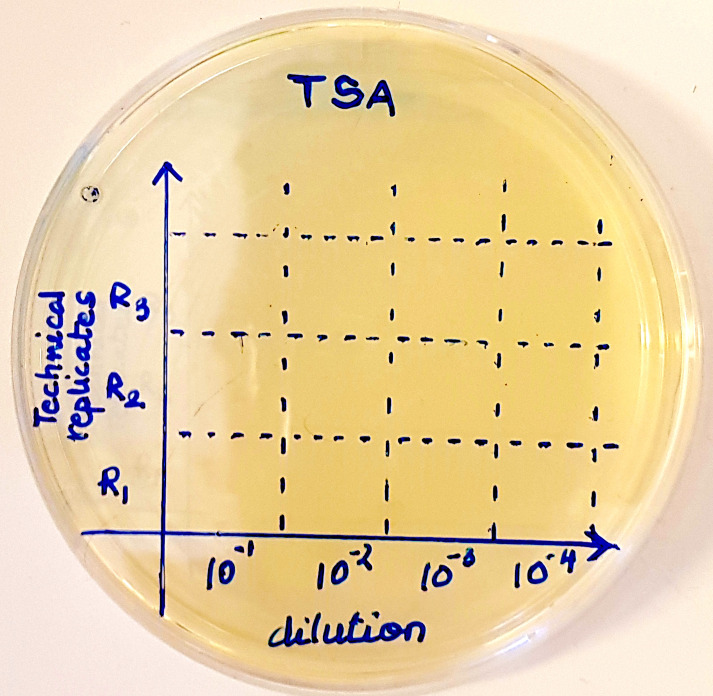
Mark sectors on the agar plates (on the bottom of the plate) as in Figure 2.
Put a 10 μl drop of diluted suspension (10-1/10-2/10-3/10-4 as labeled on the plate in Figure 2) on one of the sectors on the TSA plate. Let the drop dry.
Put at least three drops of each dilution (R1, R2 and R3 in Figure 2; representing technical replicates).
On another plate, mark sectors in a similar way (on the bottom of the plate) and put drops of suspension prepared from the lysate of a duplicate well. This plate serves as another technical replicate.
Incubate the plates overnight at 37 °C.
Count the colonies in the drop. Only consider the dilutions in which the number of colonies is within the countable range (3-30 colonies).
In parallel, make log serial dilutions of the starting bacterial suspension and spot 10 μl drops on TSA plates in a similar way as described above.
Incubate the plates overnight at 37 °C, along with the other plates, and count the colonies in the drop similar to Step D10. This colony count of the inoculum serves as a verification of the calculation performed in Step C3.
Figure 1. Schematic representation of logarithmic serial dilution of the cell lysate.
Figure 2. Sectors on a TSA plate for putting drops of up to four different dilutions in triplicate.
Data analysis
-
From the raw colony count, determine the CFU/ml using the following formula:
CFU/ml = No. of colonies x dilution factor x 100
For example, if 15 colonies were counted in 10-2 dilution, CFU/ml would be calculated using the aforementioned formula as follows:
CFU/ml = 15 (No. of colonies) x 100 (dilution factor, from 10-2) x 100 = 150,000 or 1.5 x 105
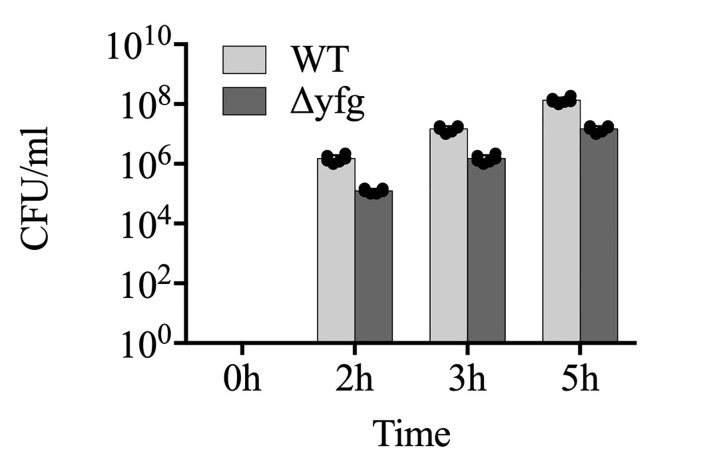
Using Prism software plot the number of bacteria expressed in CFU/ml against time for every strain to be analyzed and perform statistical analysis using the same software (Figure 3). For comparing only two sample values at a fixed time point, use Student’s t-test. However, for studying the statistical significance of multiple samples across multiple time points, use two-way ANOVA with Tukey’s post-hoc test.
Figure 3. An example plot of the number of intracellular bacteria in CFU/ml at different infection times for two different strains.
Each dot represents an independent value obtained from a replicate.
Notes
To prepare glycerol stocks of Shigella flexneri, dilute the overnight culture 200 times in fresh TSB and incubate at 37 °C with 200 rpm shaking. Monitor the OD600 of the culture periodically. At OD600 = 0.6, withdraw 1 ml of culture in a 2 ml cryovial and add sterile glycerol to a concentration of 15-50% (v/v). Mix gently and store the tube at -80 °C. The frozen stocks are stable for years at -80 °C, however, multiple freeze-thaw cycles may reduce the shelf life.
Carefully select only the red colonies on Congo red agar for the experiment. The off-white colonies have lost the virulence plasmid and are invasion-deficient. If plates are not used immediately after overnight culture, but kept at 4 °C (which is possible for up to 3 weeks), white colonies need to be marked because they will turn red due to non-specific binding of Congo red. Red staining of invasive colonies only develops during growth above 35 °C.
Congo red is able to induce secretion via the Type III Secretion System (T3SS) in Shigella. The dye binds to the bacterial colonies that have an active T3SS making them appear red in color. It is a simple and quick screening method to differentiate virulent Shigella colonies from the avirulent ones (due to curing of the virulence plasmid). Unfortunately, the mechanism of action of Congo red is not understood.
The TC7 cells are cultured and maintained in presence of Penicillin and Streptomycin (100 U/ml). However, infections are carried out in infection buffer that do not contain any antibiotics (which may inhibit the bacterial growth). This also highlights the importance of washing the cell monolayers before starting the infections.
The 6-well plates are centrifuged after addition of bacteria to aid binding, as Shigella lacks adhesion factors.
Keep all the buffers and reagents that are required at initiation of invasion and thereafter, warmed up to 37 °C before using. The secretion of T3SS effectors mediating invasion is temperature dependent. It is active above 35 °C, but inactive at room temperature. In order to synchronize the invasion, it is important to keep the bacteria in buffers at room temperature before invasion.
The medium containing gentamicin should be prepared on the day of use.
Even during longer incubations, gentamicin won’t be able to penetrate the cells and kill the intracellular bacteria. The intracellular bacteria remain ‘protected’ all the times.
Deoxycholate is a detergent and therefore TC7 cell lysates should be made with little pipetting to avoid bubbles.
Use a dedicated incubator for infections.
When using the pipette controller, gently dispense the solutions with gravity (“G” setting in Accurpipette) or at the lowest flow speed into the 6-well plate to avoid detaching the cell monolayer.
Be careful while swirling the plate, washing the cells and during centrifugation, because the confluent cell monolayer detaches easily.
The TC7 cells can survive for about 6 h in a medium devoid of serum. Therefore, additional serum is not required for infection times up to about 6 h. However, for longer incubation times, serum may be added after gentamicin treatment.
For further reading and details on basic methods that have not been detailed in this manuscript (like bacterial inoculation, pouring the plates, etc.) the reader is suggested to go through the following books: A Laboratory Manual (Cappuccino and Welsh, 2017) and Culture of Animal Cells: A Manual of Basic Technique and Specialized Applications (Freshney, 2010).
Recipes
-
Congo red solution
1% (w/v) Congo red in water
Weigh 1 g of Congo red dye and dissolve it in 70 ml of water. Bring the volume of the solution to 100 ml and filter sterilize the solution, using 0.2 μm membrane filters. The Congo red solution is stable for a long time at room temperature
-
Growth medium
DMEM
1x amino acid solution
100 U/ml Pen-Strep solution
10% heat inactivated FBS
To 500 ml DMEM, add 5 ml of amino acid solution, 5 ml of Pen-Strep solution and 50 ml of FBS (inactivated by incubation at 55 °C for 30 min)
Note: The growth medium must be stored at 4 °C and can be used for 3-4 weeks.
-
Infection buffer
DMEM (serum free)
20 mM HEPES, pH 7.4
To 50 ml of serum free DMEM, add 1 ml of 1 M sterile HEPES pH 7.4
Note: Infection buffer is stable at 4 °C for months.
-
Sodium Deoxycholate solution
0.5% sodium deoxycholate in PBS
To 40 ml of PBS, add 200 mg of sodium deoxycholate and filter-sterilize
Note: The solution is stable at room temperature for several months.
-
Gentamicin solution
50 μg/ml gentamicin in infection buffer
To 15 ml of infection buffer, add 25 μl of gentamicin from a stock solution of 30 mg/ml (prepared in water)
Note: The stock solution of gentamicin must be stored at -20 °C in small aliquots. Although the frozen stock solution is stable for several months, repeated freeze-thaw cycles may render the antibiotic inefficient. The working gentamicin solution should not be stored and always be freshly prepared.
Acknowledgments
We thank members of the Puhar lab for critical reading. A.S. is the recipient of a stipend from The MIMS Excellence by Choice Postdoctoral Programme under the patronage of Emmanuelle Charpentier. The programme is financed by the Kempe Foundations and the Knut and Alice Wallenberg Foundation. A.P. acknowledges generous funding from MIMS, UCMR, Umeå University and the Wallenberg Academy Fellow programme. This protocol was used in Puhar et al. (2013).
Competing interests
The authors declare no conflict of interest.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1.Ashida H., Mimuro H. and Sasakawa C.(2015). Shigella manipulates host immune responses by delivering effector proteins with specific roles. Front Immunol 6: 219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cappuccino J. G. and Welsh C.T.(2017). Microbiology: A Laboratory Manual, 11th EditionISBN: 978-0134298672. [Google Scholar]
- 3.Chantret I., Rodolosse A., Barbat A., Dussaulx E., Brot-Laroche E., Zweibaum A. and Rousset M.(1994). Differential expression of sucrase-isomaltase in clones isolated from early and late passages of the cell line Caco-2: evidence for glucose-dependent negative regulation. J Cell Sci 1): 213-225. [DOI] [PubMed] [Google Scholar]
- 4.Freshney R. I.(2010). Culture of Animal Cells: A Manual of Basic Technique and Specialized Applications. Sixth Edition ISBN: 9780470528129. [Google Scholar]
- 5.Puhar A. and Sansonetti P. J.(2014). Type III secretion system. Curr Biol 24(17): R784-791. [DOI] [PubMed] [Google Scholar]
- 6.Puhar A., Tronchere H., Payrastre B., Nhieu G. T. and Sansonetti P. J.(2013). A Shigella effector dampens inflammation by regulating epithelial release of danger signal ATP through production of the lipid mediator PtdIns5P. Immunity 39(6): 1121-1131. [DOI] [PubMed] [Google Scholar]
- 7.Sansonetti P. J., Kopecko D. J. and Formal S. B.(1982). Involvement of a plasmid in the invasive ability of Shigella flexneri. Infect Immun 35(3): 852-860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shigellosis–Chapter 3–2018 Yellow Book | Travelers’ Health | CDC. Available at: https://wwwnc.cdc.gov/travel/yellowbook/2018/infectious-diseases-related-to-travel/shigellosis. [Google Scholar]