Abstract
Human papillomaviruses (HPVs) cause cellular hyperproliferation-associated abnormalities including cervical cancer. The HPV genome encodes two major viral oncoproteins, E6 and E7, which recruit various host proteins by direct interaction for proteasomal degradation. Recently, we reported the structure of HPV18 E7 conserved region 3 (CR3) bound to the protein tyrosine phosphatase (PTP) domain of PTPN14, a well-defined tumor suppressor, and found that this intermolecular interaction plays a key role in E7-driven transformation and tumorigenesis. In this study, we carried out a molecular analysis of the interaction between CR3 of HPV18 E7 and the PTP domain of PTPN21, a PTP protein that shares high sequence homology with PTPN14 but is putatively oncogenic rather than tumor-suppressive. Through the combined use of biochemical tools, we verified that HPV18 E7 and PTPN21 form a 2:2 complex, with a dissociation constant of 5 nM and a nearly identical binding manner with the HPV18 E7 and PTPN14 complex. Nevertheless, despite the structural similarities, the biological consequences of the E7 interaction were found to differ between the two PTP proteins. Unlike PTPN14, PTPN21 did not appear to be subjected to proteasomal degradation in HPV18-positive HeLa cervical cancer cells. Moreover, knockdown of PTPN21 led to retardation of the migration/invasion of HeLa cells and HPV18 E7-expressing HaCaT keratinocytes, which reflects its protumor activity. In conclusion, the associations of the viral oncoprotein E7 with PTPN14 and PTPN21 are similar at the molecular level but play different physiological roles.
Keywords: CR3, E7, HPV18, human papillomavirus, PTPN21
INTRODUCTION
Human papillomaviruses (HPVs) are double-stranded DNA viruses that are known to be the causative agents of various cellular hyperproliferation-associated abnormalities (Munoz et al., 2003; Tommasino, 2014). More than 200 genotypes have been identified; among these, at least 14 are classified as “high-risk” types based on their pathogenicity in the host, including HPV16, HPV18, HPV31, and HPV45. They are well-defined carcinogens that cause several types of tumors, such as cervical cancer and head and neck squamous cell carcinoma (Kobayashi et al., 2018; Tumban, 2019; Wang et al., 2018). High-risk HPV genomes encode two oncogenic proteins that are responsible for HPV-driven tumorigenesis and malignancy: E6 and E7 (Mittal and Banks, 2017). HPV E6 recognizes p53, a key tumor suppressor protein, and provokes its proteasomal degradation by recruiting the E6AP ubiquitin ligase (Martinez-Zapien et al., 2016; Werness et al., 1990). Another oncogenic protein, HPV E7, contains three conserved regions (CRs). CR1 and CR2 are mainly responsible for targeting and promoting the degradation of pocket protein family members, such as retinoblastoma protein, a well-known cell cycle key regulator (Huh et al., 2007; Lee et al., 1998; Munger et al., 1989). CR3, which covers the C-terminal half of the E7 protein, adopts a homodimeric domain structure (Liu et al., 2006; Ohlenschlager et al., 2006) and regulates multiple viral processes involved in high-risk HPV-mediated tumorigenesis by interacting with several host factors (Avvakumov et al., 2003; Berezutskaya and Bagchi, 1997; Bodily et al., 2011; Brehm et al., 1999; Poirson et al., 2017; Todorovic et al., 2012). We recently determined the crystal structure of HPV18 E7 CR3 bound to the protein tyrosine phosphatase (PTP) domain of PTPN14, and this was the first report of the E7 CR3-mediated complex structure (Yun et al., 2019).
PTPN14 or PTPD2 is a cytosolic protein that contains an N-terminal four-point-one, ezrin-radixin-moesin (FERM) domain, central proline-rich motifs, and a C-terminal catalytic PTP domain. Given that PTPN14 is an important antitumor protein that cooperates with the p53 transcription factor and negatively regulates Hippo signaling-involved cell proliferation (Huang et al., 2013; Liu et al., 2013; Mello et al., 2017; Michaloglou et al., 2013; Wilson et al., 2014), it is not surprising that this protein is targeted by HPV E7 via direct binding, which leads to its proteasomal degradation (Hatterschide et al., 2019; Szalmas et al., 2017; White et al., 2016; Yun et al., 2019). Our previous study also revealed that PTPN21 or PTPD1 can directly interact with CR3 of HPV18 E7 (Yun et al., 2019), consistent with previous systematic proteomic analyses (Rozenblatt-Rosen et al., 2012; White et al., 2012). However, unlike PTPN14, which is defined as a tumor suppressor and particularly controls the Hippo signaling pathway (Huang et al., 2013; Liu et al., 2013; Mello et al., 2017; Michaloglou et al., 2013; Wilson et al., 2014), PTPN21 is known to be upregulated in several types of cancer cells and is associated with EGFR activation and intracellular trafficking, implying that this protein might be potentially oncogenic rather than tumor-suppressive (Carlucci et al., 2010; Plani-Lam et al., 2016; Roda-Navarro and Bastiaens, 2014; Wu et al., 2010). While the association between p53 and PTPN21 remains elusive, PTPN21 was reported to inhibit nuclear factor-κB transcriptional activity (Cho et al., 2017), which is known to play a pivotal role in immune responses and be negatively regulated by p53 (Carra et al., 2020; Jang et al., 2020; Pal et al., 2014; Schneider et al., 2010). Therefore, the biological roles and consequences of the HPV E7‒PTPN21 interaction in HPV-driven tumorigenesis might differ from those of the HPV E7‒PTPN14 interaction and warrant further investigation.
In this study, we elucidated the structural and physiological aspects of the human PTPN21 PTP domain and HPV18 E7 CR3 complex. We found that while the recombinant PTPN21 PTP domain shares high similarity with that of PTPN14 at the molecular level, the two PTP proteins differ from each other in terms of protein expression patterns and consequential cellular effects when expressed in HeLa cervical cancer cells and HaCaT keratinocytes. Therefore, these data expand our understanding of the protein interactions across species that are mediated by the HPV oncoprotein E7.
MATERIALS AND METHODS
Preparation of recombinant protein constructs
Each of the DNA fragments encoding the human PTPN21 (residues 873-1174, referred to as PTPPTPN21) and human PTPN14 (residues 886-1187, referred to as PTPPTPN14) PTP domains were cloned into the modified pPRoEX HTa plasmid (Invitrogen, USA) with an N-terminal (His)10-linked green fluorescent protein tag and the pET21a plasmid (Novagen, USA), respectively. Each of the E7 DNA fragments encoding CR3s of HPV18 (residues 54-105, referred to as CR318E7), HPV16 (residues 45-98), HPV1a (residues 39-93), and HPV11 (residues 45-98) were cloned into the modified pET28a vector (Novagen), with an N-terminal (His)6-linked maltose-binding protein tag. The PTPPTPN21 and CR318E7 constructs were used as templates for the preparation of a mutant PTPN21 protein containing F1032S·G1043Q·E1083A substitutions, referred to as PTP(SQA)PTPN21, and a mutant HPV18 E7 CR3 protein containing R84A·L91A substitutions, referred to as CR3(AA)18E7.
Recombinant protein production
The recombinant proteins used in this study were produced in the Escherichia coli BL21 (DE3) RIL strain (Novagen), which was cultured in Luria-Bertani media. Cells were incubated at 37°C until reaching an optical density of 0.6 at 600 nm, and then cultured for 16 h at 18°C following 0.2 mM isopropyl β-D-thiogalactopyranoside induction of protein expression. Cell lysates were subjected to purification steps separately for single protein purification or after proper mixing for protein complex purification. The purification was carried out using a Ni-NTA affinity chromatography column (Qiagen, Germany), HiTrap Q HP anion exchange chromatography column (GE Healthcare, USA), and HiLoad 26/600 Superdex 75 prep grade size-exclusion chromatography column (GE Healthcare). The (His)10-linked green fluorescent protein, tagged to the PTPN21 proteins, and the (His)6-linked maltose binding protein, tagged to the E7 proteins, were digested using TEV protease after Ni-NTA affinity chromatography and removed from the samples during the purification process. The protein samples were finally equilibrated with a buffer that consisted of 20 mM Tris-HCl (pH 7.5), 200 mM NaCl, and 1 mM dithiothreitol.
Small angle X-ray scattering (SAXS) experiments
SAXS data were collected on the beamline 4C at the Pohang Accelerator Laboratory at sample-to-detector distances of 1,000 and 4,000 mm (Kim et al., 2017). Measurements were conducted at 4°C with protein samples prepared with 1, 3, and 5 mg/ml, in triplicate. Data were obtained in ten successive frames of 10 s at a flow rate of 0.3 μl/s, using a Microlab 600 advanced syringe pump. An X-ray beam with a wavelength of 0.734 Å was used. Two-dimensional SAXS patterns were averaged and normalized by background subtraction using scattering intensities from the buffer solution. I(q), the scattering intensity data as a function of q, were obtained by radial averaging using the equation q = 4πsinθ/λ (θ, half of the scattering angle; λ, wavelength; 0.01 Å-1 < q < 0.6 Å-1). We used the CRYSOL (Svergun et al., 1995) and GNOM (Semenyuk and Svergun, 1991) programs to calculate the SAXS curves and distance distribution function P(r). Molecular envelope reconstructions were performed using the ab-initio shape determination program GASBOR (Svergun et al., 2001). The final refined models were obtained from 30 independently generated models that were compared and selected using the DAMAVER program. The PyMOL program was used for surface rendering. Superimposition of structural diagrams onto reconstructed dummy atoms was archived using the SUPCOMB program (Kozin and Svergun, 2001).
Isothermal titration calorimetry (ITC)
Protein samples containing 100 μM PTPPTPN21/PTP(SQA)PTPN21 samples or 5 μM CR318E7/CR3(AA)18E7 were prepared in a solution containing 50 mM Tris-HCl (pH 7.5), 200 mM NaCl, and 10 mM β-mercaptoethanol. We carried out vacuum degasification for 20 min prior to all experiments. The measurements were performed on a VP-ITC microcalorimetry system (MicroCal, UK) at 25°C and analyzed using Origin software (OriginLab, USA) as described previously (Yun et al., 2019).
Size-exclusion chromatography-multi angle light scattering (SEC-MALS)
The SEC-MALS analysis was carried out using a SuperdexTM 75 10/300 GL column (GE Healthcare) with 5 mg/ml protein samples in a buffer that contained 50 mM Tris-HCl (pH 7.5), 200 mM NaCl, and 1 mM dithiothreitol. Optilab T-rEX (Wyatt Technology Corporation, USA) combined with high-performance liquid chromatography (Shimadzu, Japan) and DAWN HELEOS-II (Wyatt Technology Corporation) was used to record the differential refractive index spectra. ASTRA 6 software (Wyatt Technology Corporation) was used to calculate the weight-average molar mass.
Plasmid preparation for the cellular assays
Plasmids p6640 MSCV-P C-FlagHA 16E7-Kozak (#35018), p6641 MSCV-P C-FlagHA 18E7-Kozak (#35019), p6644 MSCV-P C-FlagHA 45E7-Kozak (#35135), MSCV-C 11E7 (#37912), and MSCV-C 6b (#37917) were purchased from Addgene (USA). They were used for transient transfection and as templates for construct preparation. Wild-type (WT) HA-PTPN21 was generously provided by Prof. Sayeon Cho (Chung-Ang University, Korea) and used as a template to generate a binding-defective mutant construct by site-directed mutagenesis (Cho et al., 2017). The constructs were then subcloned into the pcDNA3-Flag plasmid (Addgene).
Cell culture and transfection
HeLa and C33a cervical cancer cells, HaCaT human keratinocytes, and 293T cells were cultured at 37°C and 5% CO2 in Dulbecco’s modified Eagle’s medium (DMEM; Corning, USA) containing 1% antibiotic-antimycotic (Gibco, USA) and 10% fetal bovine serum (GE Healthcare). Transient plasmid DNA transfection was conducted using Lipofectamine 3000 (Invitrogen) for HeLa and C33a cells and polyethylenimine (Sigma-Aldrich, USA) for 293T cells, according to the manufacturer’s instructions. HaCaT cells stably expressing HPV18 E7 were prepared as previously reported (Yun et al., 2019). For the delivery of siRNAs, HaCaT and HeLa cells were seeded and transfected into a 6-well culture plate with 20 nM SMARTpool ON-TARGET plus PTPN21 (SO-2868072G) and a non-targeting pool (D-001810-10; Dharmacon, USA) using LipoRNAiMAX (Invitrogen). siRNA-treated cells were harvested 48 h post-transfection for reverse transcription-polymerase chain reaction (RT-PCR) analysis.
RT-PCR analysis
Total RNA was extracted using the TRIzol reagent (Ambion, USA), and cDNA was obtained using M-MLV reverse transcriptase (Promega, USA) according to the manufacturer’s protocol. A total of 1 μg RNA was reverse-transcribed using Oligo dT primers for amplification. For quantitative RT-PCR, PTPN21 primers were used in a 2X Real-Time PCR Smart Mix (SolGent, Korea) and EvaGreen (SolGent), which was run on a real-time PCR (Bio-Rad, USA). β-Actin was used for normalization. The following primer sequences were used: PTPN21 (forward), 5′-TGAAACACAAGAATGGAAGGC-3′; PTPN21 (reverse), 5′-TCCTCTTTATTTGCCAGCTCT-3′.
Immunoblotting and immunoprecipitation
For immunoblotting, phosphate-buffered saline-resuspended cell lysates were mixed with 2X SDS sample buffer in a 1:1 ratio and boiled for 10 min. For immunoprecipitation, cells were lysed in a buffer containing 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1 mM ethylenediaminetetraacetic acid, 0.5% Triton X-100, and a protease inhibitor cocktail (Roche, Switzerland). Proteins from cell extracts were sequentially immunoprecipitated with anti-Flag M2 agarose (A2220; Sigma-Aldrich) overnight at 4°C and Protein G-Sepharose (GE Healthcare) for 2 h at 4°C. Samples were mixed with the 2X SDS sample buffer in a 1:1 ratio and boiled for 5 min. Immunoprecipitated proteins were analyzed by western blotting using anti-HA (12013819001; Sigma-Aldrich), anti-Flag (F3165; Sigma-Aldrich), and anti-β-actin (A300-491A; Bethyl Laboratories, USA). Proteins were detected using enhanced chemiluminescence reagents (GE Healthcare) according to the manufacturer’s instructions. MG132 was purchased from A.G. Scientific (M-1157).
Cell proliferation assays
For cell growth assays, HeLa and HaCaT cells were transfected with genes coding for PTPN21 constructs for 24 h or with PTPN21-targeting or non-targeting siRNA for 48 h and seeded in a 48-well plate, at 2,000 cells per well. Cells were cultured in 200 μl DMEM supplemented with 1% antibiotic-antimycotic solution and 10% fetal bovine serum, without medium change. The CellTiter-Glo luminescent cell viability assay kit (Promega) was used to analyze cell proliferation by measuring luminescence after a 10-min incubation with a luminometer Wallac VictorTM X3 (Perkin Elmer, USA).
Migration and invasion assay
Costar Transwell® chambers containing 8.0 μm polycarbonate membrane (CLS3422; Corning) were used to analyze cell migration and invasion. HeLa and HaCaT cells were collected and suspended in 100 μl of serum-free DMEM at a density of 5 × 103 cells/ml and 1.5 × 105 cells/ml, respectively, to analyze migration. HeLa cells were also collected and suspended in 100 μl of the same medium, at a density of 5 × 105 cells/ml, to analyze invasion. Medium supplemented with 10% FBS was subsequently added to the wells. Prior to seeding for the invasion assay, a thin layer of Matrigel (354234; Corning) was added to the membrane matrix, and then 100 μl of prepared cells were added on top of the inserts and Matrigel-coated inserts. The chamber plate was incubated at 37°C and 5% CO2 for 24 h. Invaded cells were fixed with 4% paraformaldehyde (Biosesang, Korea) in phosphate-buffered saline for 30 min, and stained with 0.5% crystal violet (Tokyo Chemical Industry, Japan) for 20 min. Cells were observed and imaged using a microscope Eclipse Ts2 (Nikon, Japan).
RESULTS
HPV18 E7 interacts similarly with PTPN21 and PTPN14
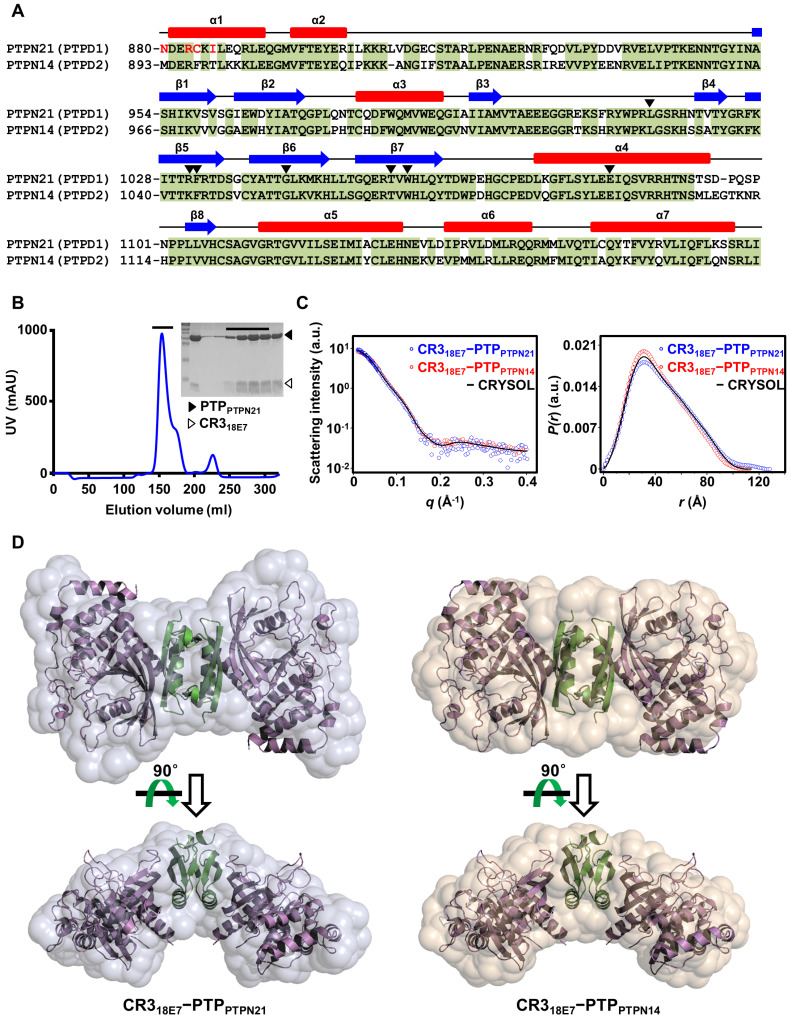
We recently determined the crystal structure of CR318E7 bound to PTPPTPN14 and demonstrated that each homodimeric CR318E7 protomer binds a single PTPPTPN14 molecule and that the two PTPPTPN14 molecules in a 2:2 complex do not mutually interact with each other (Yun et al., 2019). Our previous study also verified the molecular association of CR318E7 with PTPPTPN21 using biochemical analyses (Yun et al., 2019). PTPPTPN21 exhibits high amino acid sequence homology (71% identity and 88% similarity) with PTPPTPN14 (Fig. 1A). Moreover, PTPPTPN14 residues, which are known to play an important role in binding CR318E7, are highly conserved in PTPPTPN21 (Fig. 1A). Therefore, we hypothesized that CR318E7 recognizes PTPPTPN21 similarly as it recruits PTPPTPN14 by constituting a 2:2 complex. To verify this hypothesis, we prepared recombinant CR318E7 bound to PTPPTPN21 or PTPPTPN14 proteins (Fig. 1B) and conducted a SAXS analysis (Table 1). We first confirmed that the experimentally observed scattering intensity curves and distance distribution function P(r) of the two complexes matched well with the theoretical values obtained from a calculation that used the crystal structure of CR318E7 bound to PTPPTPN14 (PDB code 6IWD; Fig. 1C). Moreover, the X-ray crystallography-determined CR318E7−PTPPTPN14 structure could be incorporated into the SAXS analysis-based molecular envelopes of the same protein complex (Fig. 1D, right) and the CR318E7−PTPPTPN21 complex (Fig. 1D, left). These data indicate that the homodimeric CR318E7 constitutes the 2:2 complex together with two PTPPTPN21 molecules similarly as it does with two PTPPTPN14 molecules.
Fig. 1. Comparison between PTPPTPN21 and PTPPTPN14.
(A) Sequence alignment. Conserved residues are shaded in green, and the secondary structures of PTPPTPN14 are shown at the top. Residues that are significantly involved in binding CR318E7 are marked by arrowheads. (B) Preparation of the recombinant CR318E7−PTPPTPN21 complex protein eluted together from the size-exclusion chromatography column. Purification details are described in the Materials and Methods section. (C and D) SAXS analysis. (C) X-ray scattering profiles (left) and pair-distance distribution function (right) in aqueous solution measured at 4°C. The circles represent the experimental data, whereas the black lines indicate the theoretical SAXS curve calculated from the CR318E7−PTPPTPN14 crystal structure. (D) The CR318E7−PTPPTPN14 crystal structure, shown as ribbon drawings, was docked into two reconstructed SAXS envelopes. Additional details are described in the Materials and Methods section.
Table 1.
Structural parameters obtained from the SAXS data
| Model/Sample | Rg,Gc (Å) | Rg,p(r)d (Å) | Dmaxe (Å) | MMcalculatedf (kDa) | MMSAXg (kDa) |
|---|---|---|---|---|---|
| CR318E7−PTPPTPN14a | 34.32 ± 0.01 | 34.71 ± 0.20 | 114.0 | 83.0 | - |
| CR318E7−PTPPTPN14b | 32.31 ± 0.62 | 33.35 ± 0.72 | 113.0 | 83.0 | 78.9 |
| CR318E7−PTPPTPN21b | 34.82 ± 0.54 | 35.77 ± 0.65 | 128.0 | 83.0 | 79.0 |
aModel obtained from the PDB code 6IWD.
bSamples subjected to SAXS.
c Rg,G (radius of gyration) was obtained from the scattering data by the Guinier analysis.
d Rg,p(r) (radius of gyration) was obtained from the p(r) function by the program GNOM.
e Dmax (maximum dimension) was obtained from the p(r) function by the program GNOM.
f MMcalculated (molecular mass) was obtained from the amino acid sequence of protein.
g MMSAXS (molecular mass) was obtained from the SAXS experiments using BSA as a standard.
Modeling and verification of the E7−PTPN21 intermolecular interaction
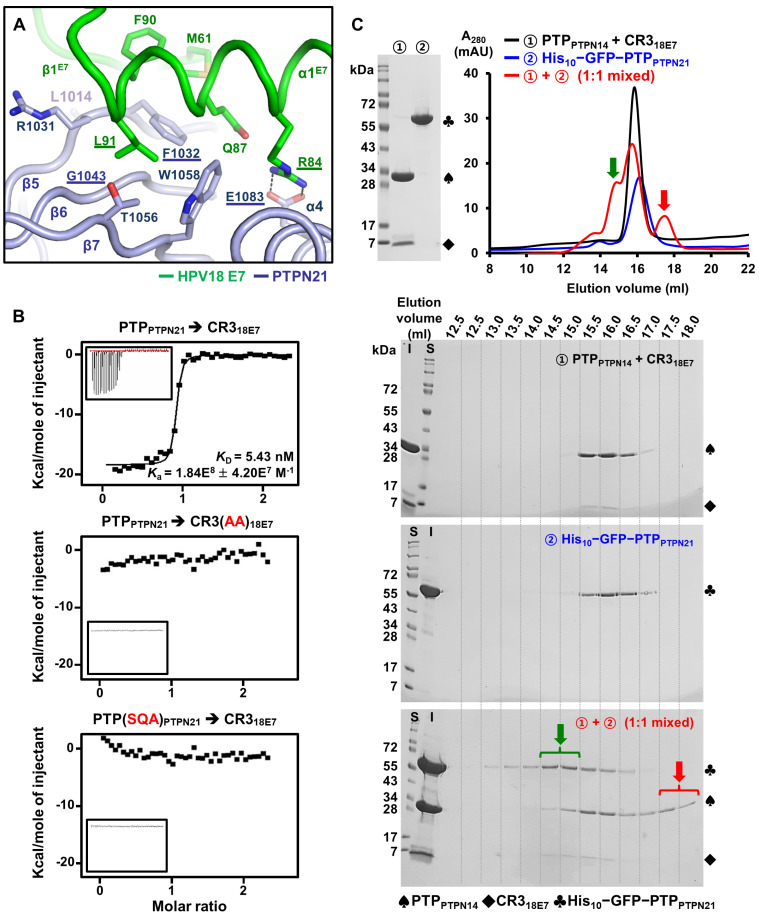
As PTPPTPN14 and PTPPTPN21 share amino acid sequence (Fig. 1A) and CR318E7-binding mode (Figs. 1C and 1D) similarities, we modeled intermolecular CR318E7−PTPPTPN21 contacts based on the CR318E7−PTPPTPN14 crystal structure. In this model, the complex formation between the two proteins is mediated by hydrophobic interactions that involve Leu1014, Arg1031, Phe1032, Gly1043, and Trp1058 of PTPN21 and Met61, Phe90, and Leu91 of HPV18 E7 (Fig. 2A). Glu1083 of PTPN21 is expected to form an electrostatic interaction with Arg84 of HPV18 E7, which might also contribute to complex formation (Fig. 2A). Therefore, based on this complex model, we selected Phe1032, Gly1043, and Glu1083 of PTPN21 and Arg84 and Leu91 of HPV18 E7 as potential key residues for intermolecular interaction studies between the two proteins. The ITC measurements confirmed that the high-affinity binding between PTPPTPN21 and CR318E7, with a dissociation constant of 5.43 nM, was completely abrogated by introducing the F1032S∙G1043Q∙E1083A (referred to as SQA in this study) triple mutation in PTPN21 or the R84A∙L91A (referred to as AA in this study) double mutation in HPV18 E7 (Fig. 2B). These findings imply the relevance of the homologous structure-based modeling of CR318E7−PTPPTPN21.
Fig. 2. Modeling and validation of the CR318E7−PTPPTPN21 interaction.
(A) The intermolecular interaction of CR318E7 with PTPPTPN21 was modeled based on the CR318E7−PTPPTPN14 crystal structure and the SAXS analysis, as shown in Fig. 1D. Residues that are assumed to contribute significantly to complex formation are shown in sticks and are labeled. Residues selected for preparing binding-defective mutants are underlined. The electrostatic interaction between Glu1083 of PTPN21 and Arg84 of HPV18 E7 is represented by dashed lines. (B) The ITC analysis was carried out using WT or mutant proteins. The PTPPTPN21 proteins (0.2 mM) were titrated into the indicated CR318E7 constructs (10 μM). (C) Competition assay. (Top) The recombinant CR318E7−PTPPTPN14 protein complex and His10−GFP−PTPPTPN21 were prepared and subjected to a HiLoad 26/600 Superdex 200 prep grade column separately or after 16 h incubation at a 1:1 molar ratio. (Bottom) Eluted fractions from each size-exclusion chromatography experiment were analyzed and visualized by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Coomassie staining. S, size marker; I, input.
Given the structurally and biochemical similarity between CR318E7−PTPPTPN14 and CR318E7−PTPPTPN21, we hypothesized that PTPPTPN14 and PTPPTPN21 are competitive with each other for binding CR318E7. To address this hypothesis, recombinant CR318E7−PTPPTPN14 protein complex was incubated with PTPPTPN21 and then subjected to size-exclusion chromatography. We determined that administration of PTPPTPN21 into the CR318E7−PTPPTPN14 complex resulted in the isolation of PTPPTPN14 from the complex (Fig. 2C; marked by a red arrow) and co-elution of CR318E7 together with PTPPTPN21 (Fig. 2C; marked by a green arrow). These results demonstrate that PTPPTPN21 is able to replace PTPPTPN14 from the CR318E7-bound complex, and that the two PTPs recognize CR318E7 in a competitive manner.
E7s from various HPV genotypes interact with PTPN21
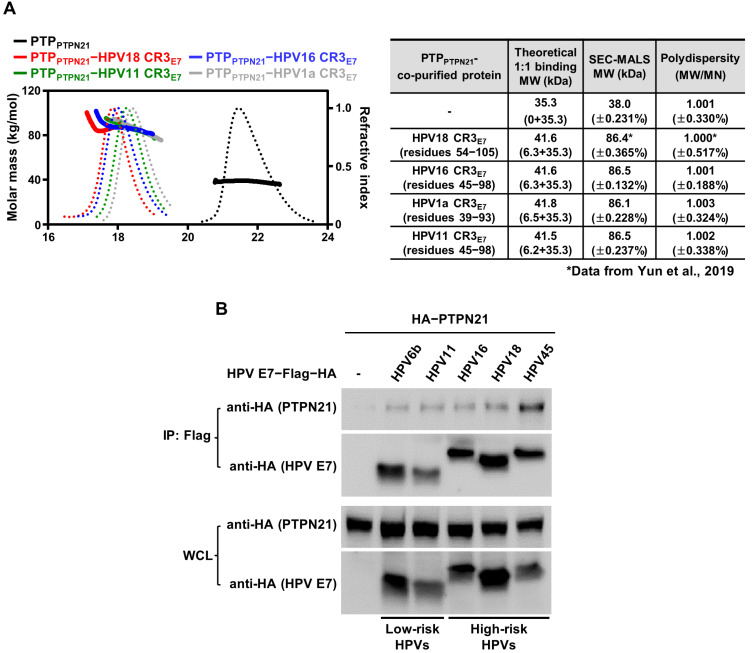
Given the molecular similarity between PTPPTPN14 and PTPPTPN21, we assumed that PTPPTPN21 associates with E7s from a wide range of HPV genotypes, as PTPPTPN14 does (Poirson et al., 2017; Szalmas et al., 2017; White et al., 2016; Yun et al., 2019). To verify this hypothesis, we first carried out SEC-MALS using recombinant PTPPTPN21 proteins in HPV E7-bound or unbound forms. As shown in Fig. 3A, PTPN21 (~38 kDa) was determined to form a stable complex with E7s (estimated to be 13 kDa as a homodimer) from HPV1a, HPV11, HPV16, and HPV18 with molecular weights of ~86 kDa, which corresponds to a 2:2 complex. Next, we conducted a coimmunoprecipitation assay using PTPN21 and E7 proteins transiently expressed in 293T cells. PTPN21 was coimmunoprecipitated with E7 from both high-risk HPVs, including HPV16, HPV18, and HPV45, and low-risk HPVs, including HPV6b and HPV11 (Fig. 3B). Collectively, these results demonstrate that E7 proteins across a wide range of HPV genotypes are able to interact with PTPN21, as they do with PTPN14.
Fig. 3. Interaction of PTPN21 with E7s from diverse HPV genotypes.
(A) SEC-MALS analysis. PTPPTPN21 forms a complex with each of the CR3 domains of E7s from the indicated HPV genotypes when co-purified. Dotted lines, refractive indexes; Solid lines, molar masses in kg/mol. MW, weight-average molar mass; MN, number-average molar mass. (B) Full-length PTPN21 and each of the indicated E7 proteins were transiently expressed in 293T cells, and their interactions were analyzed by coimmunoprecipitation. WCL, whole cell lysate.
PTPN21 is not subjected to proteasomal degradation in HeLa cells
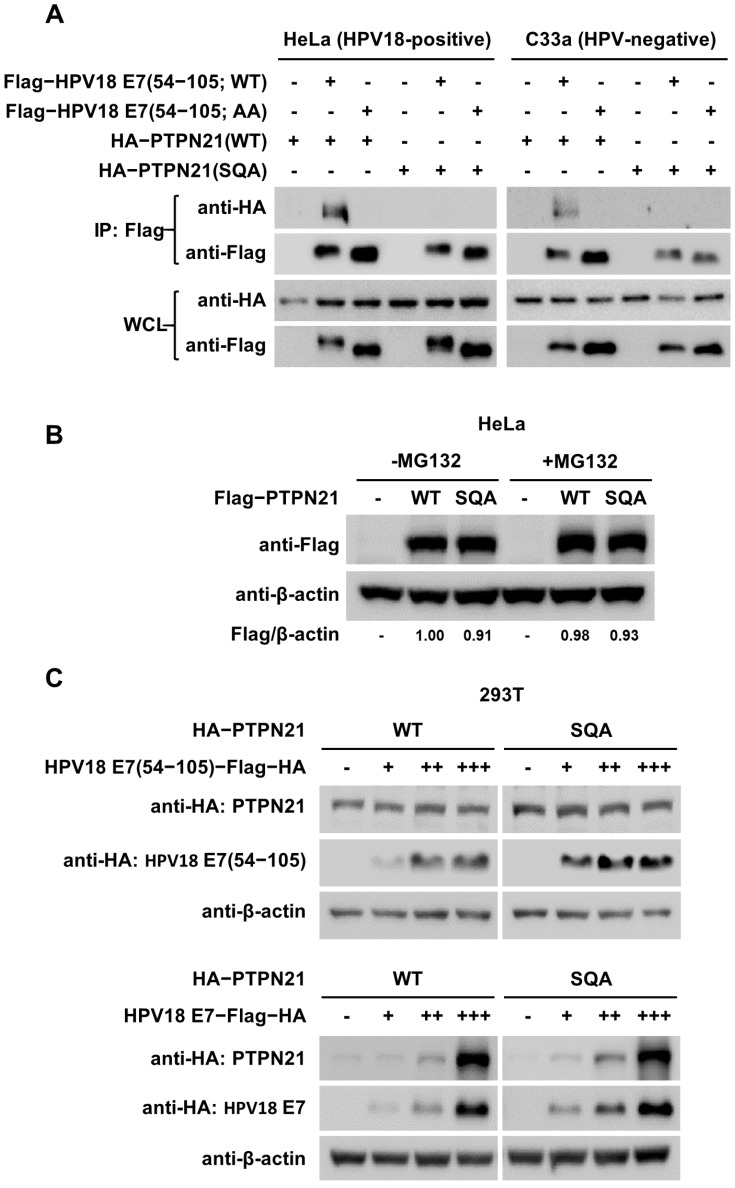
In our previous study, the interaction between PTPN14 and HPV18 E7 was verified to induce PTPN14 degradation in HPV18-positive HeLa cervical cancer cells, which was delayed by the introduction of binding-defective mutations or proteasome inhibitor MG132 treatment (Yun et al., 2019). We investigated whether the interaction between PTPN21 and HPV18 E7 results in similar biological consequences. CR318E7 and full-length PTPN21 were transiently expressed in HPV18-positive HeLa and HPV-negative C33a cervical cancer cells, in WT and binding-defective mutant forms. The immunoprecipitation results verified that the intermolecular interaction was detected between the WT proteins, but not when binding-defective mutations were introduced into PTPN21 or HPV18 E7, in both HeLa and C33a cells (Fig. 4A). Remarkably, we found that the protein levels of the WT and SQA mutant PTPN21 constructs differed from those of the PTPN14 constructs when they were expressed in HeLa cells. The cellular amount of E7 binding-defective SQA mutant PTPN14 was significantly higher than that of WT, and this protein was sensitive to proteasome inhibitor MG132 treatment (Yun et al., 2019). Contrastively, the WT and binding-defective PTPN21 protein levels were comparable to each other and hardly affected by the treatment of the proteasome inhibitor MG132 when expressed in HeLa cells (Fig. 4B). Consistently, introduction of HPV18 E7 binding-defective mutations did not cause any noticeable difference of the protein level patterns of PTPN21 when expressed in HPV-free 293T cells with increasing transfection doses of full-length or the CR3 domain of HPV18 E7 (Fig. 4C). These results suggest that PTPN21 was not targeted by the E7-induced proteasomal degradation system in HPV18-positive cervical cancer cells, unlike PTPN14, despite the high similarity in their E7-binding mode (Fig. 1).
Fig. 4. Immunoprecipitation and immunoblotting analysis of PTPN21.
(A) The indicated CR318E7 and full-length PTPN21 constructs were transiently expressed in HeLa (left) and C33a (right) cells and analyzed by the coimmunoprecipitation assay. WCL, whole cell lysate. (B) The protein levels of two types of PTPN21 constructs expressed in HeLa cells were analyzed with or without MG132 treatment for 8 h. The relative protein amounts normalized to β-actin were calculated using Vilber Lourmat software. (C) The two PTPN21 constructs were transfected (0.5 μg/ml) and expressed in 293T cells with increasing transfection doses (+, 0.5 μg/ml; ++, 1.0 μg/ml; +++, 1.5 μg/ml) of the indicated HPV18 E7 constructs, and their protein levels were measured and compared.
PTPN21 potentially functions as an oncoprotein rather than a tumor-suppressor
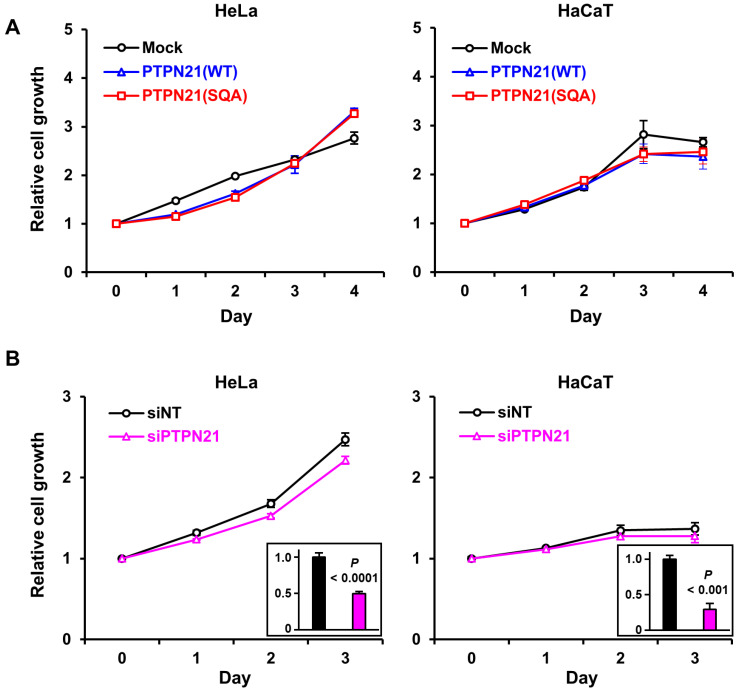
PTPN14 is a well-known tumor suppressor, and our previous study demonstrated that E7 binding-defective mutations remarkably enhance the ability of PTPN14 to delay HeLa cell proliferation, migration, and invasion (Yun et al., 2019). On the other hand, the physiological function of PTPN21 in cervical cancer development is not yet clearly understood, although PTPN21 was reported to be upregulated in several particular cancer types, such as B-cell non-Hodgkin’s gastric lymphoma and bladder cancer (Carlucci et al., 2010; Plani-Lam et al., 2016). Unlike PTPN14, PTPN21 exhibited a proteasomal degradation-evasive character, and E7-binding capability did not notably affect its protein level (Fig. 4). Therefore, we investigated whether PTPN21 could influence cell proliferation, migration, and invasion in cervical cancer cells. First, we examined how the transient expression of the full-length WT and SQA mutant PTPN21 affects the proliferation of HeLa cervical cancer cells and nontumorigenic HaCaT keratinocytes. We found that PTPN21 expression did not restrict cell proliferation in either cell type, irrespective of protein form (Fig. 5A). Consistently, siRNA-mediated suppression of PTPN21 expression hardly affected the growth of HeLa and HaCaT cells (Fig. 5B), which indicates that PTPN21 is not a proliferation regulator of these cell types.
Fig. 5. PTPN21 does not significantly affect cell proliferation.
The growth of HeLa and HaCaT cells transiently expressing the indicated PTPN21 constructs (A) or treated with the indicated siRNA (B) were compared day-to-day. Relative mRNA expression levels are represented by the graphs in black rectangles. The P values were obtained from Student’s t-tests.
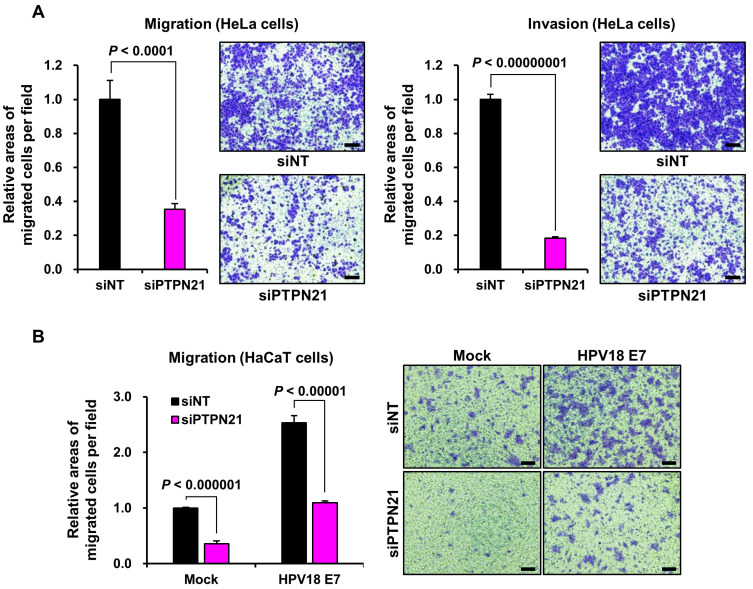
We next analyzed the effects of PTPN21 ablation on HeLa cell migration/invasion and HPV18 E7-expressing or control HaCaT cell migration. Fig. 6A shows that HeLa cell migration and invasion were critically attenuated by the genetic knockdown of PTPN21. Moreover, HaCaT cell migration was significantly repressed with the siPTPN21 treatment, in control cells and in cells stably expressing HPV18 E7 (Fig. 6B). These results collectively imply that PTPN21 positively controls cell migration and invasion. Taken together, this protein, which was bound to HPV18 E7 (Figs. 1-3) and evades proteasomal degradation (Fig. 4), might function as an oncoprotein rather than a tumor-suppressor in HPV18-positive HeLa cervical cancer cells and in keratinocytes that are vulnerable to HPV infection.
Fig. 6. Suppression of the PTPN21 expression delays cell migration and invasion.
The effects of the siRNA treatment against PTPN21 on HeLa cell motility and invasiveness (A) or motility of HaCaT cells stably expressing the control vector or HPV18 E7 (B) were compared. The P values were obtained from Student’s t-tests. Representative cells stained with crystal violet are shown together. Scale bars = 50 μm.
DISCUSSION
The recognition of human proteins by E6- or E7-associated direct interaction and subsequent proteasomal degradation of the recruited host proteins are among the main strategies of HPV for inducing the transformation and immortalization of HPV-infected cells. Based on the determination of the complex crystal structure accompanied by biochemical and cellular analysis, we previously verified that tumor-suppressive PTPN14 is targeted by HPV18 E7 for degradation through a potent direct interaction (Yun et al., 2019). Our previous report also showed that PTPN21 is another binding partner associated with HPV18 E7 (Yun et al., 2019). In this subsequent study, we elucidated the molecular aspects of this novel interaction, which was accomplished by combining biochemical tools, including SAXS (Fig. 1), structure-based modeling (Fig. 2A), and SEC-MALS (Fig. 3). These results collectively demonstrated that PTPPTPN21 forms a 2:2 complex with CR318E7, and defined the binding interface between the two proteins, which was confirmed by binding affinity measurements (Fig. 2B) and immunoprecipitation assays (Fig. 4A) carried out using WT and binding-defective mutant constructs. Despite the high similarity between PTPN14 and PTPN21 at the molecular level, we noticed that the biological consequences of HPV E7-binding differ between the two PTPs. First, while the E7 binding-defective mutant PTPN14 protein level was more than four-times higher than that of WT when expressed in HPV18-positive HeLa cells (Yun et al., 2019), these values were comparable in the case of the PTPN21 constructs (Figs. 4B and 4C). Second, MG132 protease inhibitor treatment led to a more than three-fold increase in WT PTPN14 protein levels in HeLa cells, but it did not induce such a prominent change in PTPN21 expression (Fig. 4B). Third, whereas PTPN14 functioned as a negative regulator of HeLa cell proliferation, migration, and invasion, and E7-binding defective mutations reinforced its tumor-suppressive activities, neither the ectopic expression nor genetic knockdown of PTPN21 relevantly affected HeLa or HaCaT cell proliferation (Fig. 5). Instead, the cellular phenotypes directly associated with tumor development, such as cell migration and invasion, were significantly attenuated by the repression of PTPN21 expression in HeLa and HPV18 E7-expressing HaCaT cells, which strongly suggest that PTPN21 is putatively oncogenic rather than tumor-suppressive, as implied previously (Carlucci et al., 2008; 2010; Plani-Lam et al., 2016; Roda-Navarro and Bastiaens, 2014; Wu et al., 2010). In conclusion, PTPN21 does not appear to be the target of the HPV E7-dependent host protein degradation system, despite its high structural homology with PTPN14 in the E7 binding mode (Fig. 1) and potent binding affinity with HPV18 E7 (Fig. 2).
What might be the biological consequence of the interaction between PTPN21 and HPV18 E7? One possibility is that HPV might control the localization or host protein association of PTPN21 through direct binding to E7, and thus take advantage of its oncogenic properties. Another possibility is that PTPN21 might be a collateral binding partner of HPV E7 due to the high structural similarity between PTPPTPN14 and PTPPTPN21. At present, the precise biological function of this interaction during viral infection and E7-mediated tumorigenesis remains unclear. Another unresolved issue is how PTPN21 evades proteasomal degradation despite its potent interaction with HPV18 E7. PTPN14 degradation is known to be mediated by the E3 ubiquitin-protein ligase UBR4/p600, which is recruited through direct interaction with the N-terminal region of HPV E7 (Szalmas et al., 2017; White et al., 2016). Given that PTPN14, but not PTPN21, was subjected to proteasomal degradation (Fig. 4), we suggest that PTPN21 might somehow hinder intermolecular interactions between the N-terminal region of E7 and UBR4/p600. Interestingly, we found that expression of full-length HPV18 E7, but not that of the CR3 domain, stabilizes both WT and CR3 domain-binding defective mutant PTPN21 (Fig. 4C), in which the N-terminal regions of these proteins are presumed to be involved. We thus examined whether PTPN21 has another region that associates with HPV18 E7 and found an unidentified interaction between the PTPN21 N-terminal FERM domain and full-length HPV18 E7 using immunoprecipitation (unpublished data). The relevance and biological importance of this novel interaction are our next objectives to determine.
In this study, we analyzed the molecular basis of how HPV18 E7 associates with human PTPN21 and revealed the physiological aspects of this interaction in HeLa cervical cancer cells and HaCaT keratinocytes. Our study expands our understanding of the diverse host protein recognition mediated by the HPV-encoded oncoprotein E7. We also anticipate that our observations will eventually serve as a rational basis for therapeutic approaches to treat HPV-implicated malignant diseases.
ACKNOWLEDGMENTS
SAXS measurements were performed at the beamline 4C at the Pohang Accelerator Laboratory in Korea. We appreciate Dr. Eunha Hwang (Korea Basic Science Institute, Korea) for help with the SEC-MALS measurements and Dr. Jungwon Hwang and Dr. Myung Hee Kim (Korea Research Institute of Bioscience and Biotechnology, Korea) for help with the ITC measurements. This study was supported by the National Research Foundation of Korea (NRF_2020R1C1C1008451 and NRF_2019M3E5D6063955 to B.K., NRF_2017M3A9G5083321 to S.C.L., and NRF_2019R1C1C1002831 to E.W.L.) and the KRIBB Research Initiative Programs (to B.K.), which were funded by the Ministry of Science and ICT (MSIT) of Republic of Korea.
Footnotes
AUTHOR CONTRIBUTIONS
H.S.L. and M.W.K. performed and analyzed experiments and wrote the manuscript. K.S.J. performed and analyzed experiments. H.C.S. and W.K.K. provided expertise and feedback. S.C.L. provided expertise and feedback and secured funding. S.J.K. provided supervision. E.W.L. conceived and analyzed experiments, secured funding, and provided supervision. B.K. conceived and analyzed experiments, wrote the manuscript, secured funding, and provided supervision.
CONFLICT OF INTEREST
The authors have no potential conflicts of interest to disclose.
REFERENCES
- Avvakumov N., Torchia J., Mymryk J.S. Interaction of the HPV E7 proteins with the pCAF acetyltransferase. Oncogene. 2003;22:3833–3841. doi: 10.1038/sj.onc.1206562. [DOI] [PubMed] [Google Scholar]
- Berezutskaya E., Bagchi S. The human papillomavirus E7 oncoprotein functionally interacts with the S4 subunit of the 26 S proteasome. J. Biol. Chem. 1997;272:30135–30140. doi: 10.1074/jbc.272.48.30135. [DOI] [PubMed] [Google Scholar]
- Bodily J.M., Mehta K.P., Laimins L.A. Human papillomavirus E7 enhances hypoxia-inducible factor 1-mediated transcription by inhibiting binding of histone deacetylases. Cancer Res. 2011;71:1187–1195. doi: 10.1158/0008-5472.CAN-10-2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brehm A., Nielsen S.J., Miska E.A., McCance D.J., Reid J.L., Bannister A.J., Kouzarides T. The E7 oncoprotein associates with Mi2 and histone deacetylase activity to promote cell growth. EMBO J. 1999;18:2449–2458. doi: 10.1093/emboj/18.9.2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlucci A., Gedressi C., Lignitto L., Nezi L., Villa-Moruzzi E., Avvedimento E.V., Gottesman M., Garbi C., Feliciello A. Protein-tyrosine phosphatase PTPD1 regulates focal adhesion kinase autophosphorylation and cell migration. J. Biol. Chem. 2008;283:10919–10929. doi: 10.1074/jbc.M707248200. [DOI] [PubMed] [Google Scholar]
- Carlucci A., Porpora M., Garbi C., Galgani M., Santoriello M., Mascolo M., di Lorenzo D., Altieri V., Quarto M., Terracciano L., et al. PTPD1 supports receptor stability and mitogenic signaling in bladder cancer cells. J. Biol. Chem. 2010;285:39260–39270. doi: 10.1074/jbc.M110.174706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carra G., Lingua M.F., Maffeo B., Taulli R., Morotti A. P53 vs NF-κB: the role of nuclear factor-kappa B in the regulation of p53 activity and vice versa. Cell. Mol. Life Sci. 2020;77:4449–4458. doi: 10.1007/s00018-020-03524-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho Y.C., Kim B.R., Cho S. Protein tyrosine phosphatase PTPN21 acts as a negative regulator of ICAM-1 by dephosphorylating IKKβ in TNF-α-stimulated human keratinocytes. BMB Rep. 2017;50:584–589. doi: 10.5483/bmbrep.2017.50.11.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatterschide J., Bohidar A.E., Grace M., Nulton T.J., Kim H.W., Windle B., Morgan I.M., Munger K., White E.A. PTPN14 degradation by high-risk human papillomavirus E7 limits keratinocyte differentiation and contributes to HPV-mediated oncogenesis. Proc. Natl. Acad. Sci. U. S. A. 2019;116:7033–7042. doi: 10.1073/pnas.1819534116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J.M., Nagatomo I., Suzuki E., Mizuno T., Kumagai T., Berezov A., Zhang H., Karlan B., Greene M.I., Wang Q. YAP modifies cancer cell sensitivity to EGFR and survivin inhibitors and is negatively regulated by the non-receptor type protein tyrosine phosphatase 14. Oncogene. 2013;32:2220–2229. doi: 10.1038/onc.2012.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh K., Zhou X., Hayakawa H., Cho J.Y., Libermann T.A., Jin J., Harper J.W., Munger K. Human papillomavirus type 16 E7 oncoprotein associates with the cullin 2 ubiquitin ligase complex, which contributes to degradation of the retinoblastoma tumor suppressor. J. Virol. 2007;81:9737–9747. doi: 10.1128/JVI.00881-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang H., Park S., Kim J., Kim J.H., Kim S.Y., Cho S., Park S.G., Park B.C., Kim S., Kim J.H. The tumor suppressor, p53, negatively regulates non-canonical NF-κB signaling through miRNA-induced silencing of NF-κB-inducing kinase. Mol. Cells. 2020;43:23–33. doi: 10.14348/molcells.2019.0239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K.W., Kim J., Yun Y.D., Ahn H., Min B., Kim N.H., Rah S., Kim H.Y., Lee C.S., Seo I.D., et al. Small-angle X-ray scattering beamline BL4C SAXS at Pohang Light Source II. Biodesign. 2017;5:24–29. [Google Scholar]
- Kobayashi K., Hisamatsu K., Suzui N., Hara A., Tomita H., Miyazaki T. A review of HPV-related head and neck cancer. J. Clin. Med. 2018;7:241. doi: 10.3390/jcm7090241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozin M.B., Svergun D.I. Automated matching of high-and low-resolution structural models. J. Appl. Crystallogr. 2001;34:33–41. [Google Scholar]
- Lee J.O., Russo A.A., Pavletich N.P. Structure of the retinoblastoma tumour-suppressor pocket domain bound to a peptide from HPV E7. Nature. 1998;391:859–865. doi: 10.1038/36038. [DOI] [PubMed] [Google Scholar]
- Liu X., Clements A., Zhao K., Marmorstein R. Structure of the human Papillomavirus E7 oncoprotein and its mechanism for inactivation of the retinoblastoma tumor suppressor. J. Biol. Chem. 2006;281:578–586. doi: 10.1074/jbc.M508455200. [DOI] [PubMed] [Google Scholar]
- Liu X., Yang N., Figel S.A., Wilson K.E., Morrison C.D., Gelman I.H., Zhang J. PTPN14 interacts with and negatively regulates the oncogenic function of YAP. Oncogene. 2013;32:1266–1273. doi: 10.1038/onc.2012.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Zapien D., Ruiz F.X., Poirson J., Mitschler A., Ramirez J., Forster A., Cousido-Siah A., Masson M., Vande Pol S., Podjarny A., et al. Structure of the E6/E6AP/p53 complex required for HPV-mediated degradation of p53. Nature. 2016;529:541–545. doi: 10.1038/nature16481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello S.S., Valente L.J., Raj N., Seoane J.A., Flowers B.M., McClendon J., Bieging-Rolett K.T., Lee J., Ivanochko D., Kozak M.M., et al. A p53 super-tumor suppressor reveals a tumor suppressive p53-Ptpn14-Yap axis in pancreatic cancer. Cancer Cell. 2017;32:460–473. doi: 10.1016/j.ccell.2017.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaloglou C., Lehmann W., Martin T., Delaunay C., Hueber A., Barys L., Niu H., Billy E., Wartmann M., Ito M., et al. The tyrosine phosphatase PTPN14 is a negative regulator of YAP activity. PLoS One. 2013;8:e61916. doi: 10.1371/journal.pone.0061916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittal S., Banks L. Molecular mechanisms underlying human papillomavirus E6 and E7 oncoprotein-induced cell transformation. Mutat. Res. Rev. Mutat. Res. 2017;772:23–35. doi: 10.1016/j.mrrev.2016.08.001. [DOI] [PubMed] [Google Scholar]
- Munger K., Werness B.A., Dyson N., Phelps W.C., Harlow E., Howley P.M. Complex formation of human papillomavirus E7 proteins with the retinoblastoma tumor suppressor gene product. EMBO J. 1989;8:4099–4105. doi: 10.1002/j.1460-2075.1989.tb08594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz N., Bosch F.X., de Sanjose S., Herrero R., Castellsague X., Shah K.V., Snijders P.J., Meijer C.J. International Agency for Research on Cancer Multicenter Cervical Cancer Study Group, author. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N. Engl. J. Med. 2003;348:518–527. doi: 10.1056/NEJMoa021641. [DOI] [PubMed] [Google Scholar]
- Ohlenschlager O., Seiboth T., Zengerling H., Briese L., Marchanka A., Ramachandran R., Baum M., Korbas M., Meyer-Klaucke W., Durst M., et al. Solution structure of the partially folded high-risk human papilloma virus 45 oncoprotein E7. Oncogene. 2006;25:5953–5959. doi: 10.1038/sj.onc.1209584. [DOI] [PubMed] [Google Scholar]
- Pal S., Bhattacharjee A., Ali A., Mandal N.C., Mandal S.C., Pal M. Chronic inflammation and cancer: potential chemoprevention through nuclear factor kappa B and p53 mutual antagonism. J. Inflamm. 2014;11:23. doi: 10.1186/1476-9255-11-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plani-Lam J.H., Chow T.C., Fan Y.H., Garcia-Bloj B., Cheng L., Jin D.Y., Hancock W., Fanayan S., Ingley E., Song Y.Q. High expression of PTPN21 in B-cell non-Hodgkin's gastric lymphoma, a positive mediator of STAT5 activity. Blood Cancer J. 2016;6:e388. doi: 10.1038/bcj.2015.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirson J., Biquand E., Straub M.L., Cassonnet P., Nomine Y., Jones L., van der Werf S., Trave G., Zanier K., Jacob Y., et al. Mapping the interactome of HPV E6 and E7 oncoproteins with the ubiquitin-proteasome system. FEBS J. 2017;284:3171–3201. doi: 10.1111/febs.14193. [DOI] [PubMed] [Google Scholar]
- Roda-Navarro P., Bastiaens P.I. Dynamic recruitment of protein tyrosine phosphatase PTPD1 to EGF stimulation sites potentiates EGFR activation. PLoS One. 2014;9:e103203. doi: 10.1371/journal.pone.0103203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozenblatt-Rosen O., Deo R.C., Padi M., Adelmant G., Calderwood M.A., Rolland T., Grace M., Dricot A., Askenazi M., Tavares M., et al. Interpreting cancer genomes using systematic host network perturbations by tumour virus proteins. Nature. 2012;487:491–495. doi: 10.1038/nature11288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider G., Henrich A., Greiner G., Wolf V., Lovas A., Wieczorek M., Wagner T., Reichardt S., von Werder A., Schmid R.M., et al. Cross talk between stimulated NF-κB and the tumor suppressor p53. Oncogene. 2010;29:2795–2806. doi: 10.1038/onc.2010.46. [DOI] [PubMed] [Google Scholar]
- Semenyuk A.V., Svergun D.I. GNOM - a program package for small-angle scattering data processing. J. Appl. Crystallogr. 1991;24:537–540. doi: 10.1107/S0021889812007662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svergun D., Barberato C., Koch M.H.J. CRYSOL - a program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Crystallogr. 1995;28:768–773. [Google Scholar]
- Svergun D.I., Petoukhov M.V., Koch M.H. Determination of domain structure of proteins from X-ray solution scattering. Biophys. J. 2001;80:2946–2953. doi: 10.1016/S0006-3495(01)76260-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szalmas A., Tomaic V., Basukala O., Massimi P., Mittal S., Konya J., Banks L. The PTPN14 tumor suppressor is a degradation target of human papillomavirus E7. J. Virol. 2017;91:e00057–17. doi: 10.1128/JVI.00057-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todorovic B., Hung K., Massimi P., Avvakumov N., Dick F.A., Shaw G.S., Banks L., Mymryk J.S. Conserved region 3 of human papillomavirus 16 E7 contributes to deregulation of the retinoblastoma tumor suppressor. J. Virol. 2012;86:13313–13323. doi: 10.1128/JVI.01637-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tommasino M. The human papillomavirus family and its role in carcinogenesis. Semin. Cancer Biol. 2014;26:13–21. doi: 10.1016/j.semcancer.2013.11.002. [DOI] [PubMed] [Google Scholar]
- Tumban E. A current update on human papillomavirus-associated head and neck cancers. Viruses. 2019;11:922. doi: 10.3390/v11100922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Huang X., Zhang Y. Involvement of human papillomaviruses in cervical cancer. Front. Microbiol. 2018;9:2896. doi: 10.3389/fmicb.2018.02896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werness B.A., Levine A.J., Howley P.M. Association of human papillomavirus types 16 and 18 E6 proteins with p53. Science. 1990;248:76–79. doi: 10.1126/science.2157286. [DOI] [PubMed] [Google Scholar]
- White E.A., Munger K., Howley P.M. High-risk human papillomavirus E7 proteins target PTPN14 for degradation. mBio. 2016;7:e01530–16. doi: 10.1128/mBio.01530-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White E.A., Sowa M.E., Tan M.J., Jeudy S., Hayes S.D., Santha S., Munger K., Harper J.W., Howley P.M. Systematic identification of interactions between host cell proteins and E7 oncoproteins from diverse human papillomaviruses. Proc. Natl. Acad. Sci. U. S. A. 2012;109:E260–E267. doi: 10.1073/pnas.1116776109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson K.E., Li Y.W., Yang N., Shen H., Orillion A.R., Zhang J. PTPN14 forms a complex with Kibra and LATS1 proteins and negatively regulates the YAP oncogenic function. J. Biol. Chem. 2014;289:23693–23700. doi: 10.1074/jbc.M113.534701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z.Z., Lu H.P., Chao C.C. Identification and functional analysis of genes which confer resistance to cisplatin in tumor cells. Biochem. Pharmacol. 2010;80:262–276. doi: 10.1016/j.bcp.2010.03.029. [DOI] [PubMed] [Google Scholar]
- Yun H.Y., Kim M.W., Lee H.S., Kim W., Shin J.H., Kim H., Shin H.C., Park H., Oh B.H., Kim W.K., et al. Structural basis for recognition of the tumor suppressor protein PTPN14 by the oncoprotein E7 of human papillomavirus. PLoS Biol. 2019;17:e3000367. doi: 10.1371/journal.pbio.3000367. [DOI] [PMC free article] [PubMed] [Google Scholar]