Abstract
Airway mucus secretion is an essential innate immune response for host protection. However, overproduction and hypersecretion of mucus, mainly composed of the gel-forming MUC5AC protein, are significant risk factors for patients with asthma and chronic obstructive pulmonary disease (COPD). The transforming growth factor β (TGFβ) signaling pathway negatively regulates MUC5AC expression; however, the underlying molecular mechanism is not fully understood. Here, we showed that TGFβ significantly reduces the expression of MUC5AC mRNA and its protein in NCI-H292 cells, a human mucoepidermoid carcinoma cell line. This reduced MUC5AC expression was restored by a TGFβ receptor inhibitor (SB431542), but not by the inhibition of NF-κB (BAY11-7082 or Triptolide) or PI3K (LY294002) activities. TGFβ-activated Smad3 dose-dependently bound to MUC5AC promoter. Notably, TGFβ-activated Smad3 recruited HDAC2 and facilitated nuclear translocation of HDAC2, thereby inducing the deacetylation of NF-κB at K310, which is essential for a reduction in NF-κB transcriptional activity. Both TGFβ-induced nuclear translocation of Smad3/HDAC2 and deacetylation of NF-κB at K310 were suppressed by a Smad3 inhibitor (SIS3). These results suggest that the TGFβ-activated Smad3/HDAC2 complex is an essential negative regulator for MUC5AC expression and an epigenetic regulator for NF-κB acetylation. Therefore, these results collectively suggest that modulation of the TGFβ1/Smad3/HDAC2/NF-κB pathway axis can be a promising way to improve lung function as a treatment strategy for asthma and COPD.
Keywords: HDAC2, MUC5AC, NF-κB, Smad3, transforming growth factor β
INTRODUCTION
Asthma and chronic obstructive pulmonary disorder (COPD) commonly cause inflammation and hyperactivity of the airway. Asthma shows intermittent and reversible airway obstruction, whereas COPD results in generally progressive and irreversible lung damage, such as chronic bronchitis and emphysema (Cukic et al., 2012; Kim et al., 2019).
Overproduction and hypersecretion of mucus (sputum) are prominent pathophysiology shown by patients with asthma and COPD (Shen et al., 2018). Although mucus secretion in airway epithelial cells is an essential innate immune response for host protection against pathogens or irritants at mucosal surfaces, too much sputum contributes to increased morbidity and mortality in chronic airway diseases, including asthma and COPD (Rose and Voynow, 2006). Mucus consists of mucin proteins, which comprise heavily glycosylated proteins with high molecular weights. The major mucin protein secreted by epithelial cells in the human airway is MUC5AC. Since the level of MUC5AC expression is markedly upregulated in lung tissues of ovalbumin-induced asthmatic mice model (Bonser and Erle, 2017), as well as in the bronchiolar epithelium of COPD patients (Caramori et al., 2004), the high level of MUC5AC is considered a hallmark of chronic lung diseases. Thus, it is crucial to understand the mechanisms regulating MUC5AC expression to treat chronic airway diseases such as asthma and COPD.
Transforming growth factor β (TGFβ) is a central regulator of various cellular processes, such as cellular growth, proliferation, differentiation, migration, apoptosis, and immunity. Humans have three isoforms of TGFβ (TGFβ1, -β2, and -β3). Notably, TGFβ can either stimulate or inhibit the immune cell function, depending on its surrounding environment (Letterio and Roberts, 1998). TGFβ1 can also regulate cytokine production positively or negatively depending on the type or the differentiated states of a particular cell (Ling and Robinson, 2002; Wrzesinski et al., 2007). In asthmatic airways and epithelial cells, the level of TGFβ1 and its downstream signaling processes increase. In experimental asthma models, TGFβ1 reverses airway inflammation and hyperresponsiveness (AHR) (Branchett and Lloyd, 2019). Moreover, TGFβ1 inhibits inflammatory responses such as MUC5AC secretion in human lung adenocarcinoma cells (Alcorn et al., 2007; Sato et al., 2016). Thus, the TGFβ1-mediated signaling pathway is an essential mechanism for inhibiting inflammatory responses in the airway epithelium (Curran and Cohn, 2010).
In airway cells, TGFβ1 binds to a TGFβ receptor type 2 (TGFβR2, a serine/threonine kinase receptor) dimer, which recruits and phosphorylates another receptor, TGFβ receptor type 1 (TGFβR1) for activation (Wrana et al., 1994). TGFβR1 is also a serine/threonine kinase receptor that will, in turn, phosphorylate Smad transcription factors, such as Smad2 or Smad3. Phosphorylated Smad2/3 forms a protein complex with the co-factor Smad4 and is translocated into the cell nucleus to control gene expression (Lagna et al., 1996; Nakao et al., 1997). Smad2 is involved in the developmental process, whereas Smad3 is essential for the anti-inflammatory process (Takimoto et al., 2010), which is demonstrated by Smad2-knockout (KO) mice that are embryonic-lethal (Nomura and Li, 1998) and Smad3-KO mice that exhibit inflammatory diseases (Anthoni et al., 2007). Moreover, Smad3 activation leads to the downregulation of MUC5AC expression in the human airway epithelial cells (Jono et al., 2003). Thus, the mechanistic understanding of the TGFβ1-mediated Smad3 pathway for MUC5AC expression could be a good starting point to find out ways to relieve the symptoms of chronic airway diseases. Moreover, it is still elusive how TGFβ1 signaling regulates MUC5AC expression at the epigenetic level.
Histone deacetylases (HDACs) are enzymes that remove the acetyl group from the lysine residues of histones and transcriptional factors. Thus, they play essential roles in the epigenetic regulation of gene expression (Kuo and Allis, 1998). HDAC2, as a critical epigenetic regulator, reduces inflammatory gene expression, such as MUC5AC, by negatively modulating NF-κB activity (Mortaz et al., 2011). Thus, strategies to increase HDAC2 activity have been suggested to manage lung inflammatory diseases such as asthma and COPD (Ito et al., 2005; Thomson et al., 2004; Zwinderman et al., 2019). However, the mechanism by which HDAC2 suppresses NF-κB activity is not clearly defined.
In this study, we showed that TGFβ1 signaling recruits HDAC2 and represses MUC5AC expression by reducing NF-κB activity in NCI-H292 human airway cells. TGFβ1-activated Smad3 directly binds to the MUC5AC promoter to control MUC5AC expression. We also demonstrated that TGFβ1 promotes the physical interaction between activated Smad3 and HDAC2 and induces their nuclear translocation. Moreover, our immunofluorescence studies demonstrated for the first time that epigenetic modification on NF-κB (acetylation at K310) is considerably suppressed by TGFβ1 signaling, probably reducing full transcriptional activation of NF-κB. Collectively, our results suggest that modulating the TGFβ1/Smad3/HDAC2/NF-κB pathway axis can be a promising way to improve lung function in the treatment of asthma and COPD.
MATERIALS AND METHODS
Chemicals and reagents
Recombinant human epidermal growth factor (EGF), tumor necrosis factor-α (TNF-α), TGFβ1, and interleukin (IL)-1β were purchased from PeproTech (USA). Cigarette smoke condensate (CSC) was acquired from the Tobacco and Health Research Institute 26 (University of Kentucky, USA). Lipopolysaccharides (LPS), phorbol 12-myristate 13-acetate (PMA), Acrolein, and dimethyl sulfoxide (DMSO) were obtained from Sigma-Aldrich (USA). For the inhibition experiments, the IKK inhibitor (BAY 11-7082), PI3K inhibitor (LY294002), NF-κB inhibitor (Triptolide), and TGFβ receptor inhibitor (SB431542) were also purchased from Sigma-Aldrich. Rabbit anti-glyceraldehyde 3-phosphate dehydrogenase (GAPDH, sc-25778) was purchased from Santa Cruz Biotechnology (USA). Rabbit anti-phospho-Smad3 (#9520), mouse anti-HDAC2 (#5113), and rabbit anti-NF-κB (#8242) were obtained from Cell Signaling Technology (USA). Rabbit anti-acetyl-NF-κB at K310 (ab19870 for ChiP assay), mouse anti-MUC5AC (ab3649), and rabbit anti-Smad3 (ab28379) were acquired from Abcam (UK). Rabbit anti-acetyl-NF-κB at K310 (PA5-17264 for cell staining), Alexa Fluor 488-conjugated goat anti-rabbit IgG (A32731), and Alexa Fluor 568-conjugated goat anti-mouse (A11126) were obtained from Thermo Fisher Scientific (USA). The secondary antibodies for western analysis were acquired from GenDepot (USA).
Cell preparation and culture
NCI-H292 cells, a human pulmonary mucoepidermoid carcinoma cell line, were purchased from the American Type Culture Collection (CRL-1848; ATCC, USA). NCI-H292 cells were grown in growth medium (GM) (RPMI 1640 medium Hyclone; GE Healthcare, USA) supplemented with 10% fetal bovine serum (FBS) (Hyclone) and 100 units/ml penicillin plus 100 µg/ml streptomycin (Hyclone) at 37°C under a humidified 5% CO2 atmosphere. For the treatment with the various stimuli or inhibitors, NCI-H292 cells (1 × 104 cells/cm2 well) were seeded in GM and incubated for 16 h. Subsequently, the medium was changed to RPMI supplemented with 0.1% FBS and 100 units/ml penicillin plus 100 µg/ml streptomycin, and the cells were incubated for another 16 h.
Cell viability assay
The NCI-H292 cells were plated in 96-well plates in GM at a density of 5 × 103 cells/well and grown for 16 h. The GM was subsequently changed to a serum-reduced medium (0.1% FBS). After 16 h incubation, cells were incubated with different concentrations of TGFβ1 for 24 h. Cell viability was measured in triplicate using a Cell Counting Kit-8 (Dojindo Molecular Technologies, USA) according to the manufacturer’s protocol. The absorbance was measured using a VERSA max microplate reader (Molecular Devices, USA), and the measured absorbance was converted to the percentage (%) of the control value.
MUC5AC protein enzyme-linked immunosorbent assay (ELISA)
MUC5AC protein in the cell culture supernatant or cell lysate was measured using a method described previously with slight modifications (Sikder et al., 2014). In brief, the culture supernatants (100 µl) were incubated and dried at 50°C in a 96-well plate (Costar, USA). The plate was washed three times with wash buffer (0.05% Tween20 in phosphate-buffered saline [PBS]) and was subsequently blocked with blocking buffer (1% bovine serum albumin in PBS) for 1 h at room temperature. The plate was washed three times with wash buffer and incubated with 100 µl of a mouse monoclonal MUC5AC antibody (1:500 in blocking buffer; Abcam) in each well. After 2 h, the plate was washed three times with wash buffer, and 100 µl of horseradish peroxidase-conjugated goat anti-mouse IgG (1:2,000 in blocking buffer) was added to each well. After 1 h, the plate was washed three times with wash buffer. The color reaction was generated with 3,3’,5,5’-tetramethylbenzidine peroxide solution, and stopped with 2N H2SO4. The absorbance was measured at 450 nm using a VERSA max microplate reader. The measured absorbance was converted to the percentage (%) of the control value.
Western blot analysis
NCI-H292 cells (5 × 105 cells/well) were seeded in 6-well plates. The cells were incubated for 12 h in GM, and the medium was subsequently changed to serum-reduced GM medium (0.1% FBS). After 16 h, the cells were treated with the respective concentration of TGFβ1 for 30 min. Proteins were prepared and loaded as described elsewhere (Lee et al., 2014). At least 30 µg of protein from the whole cell lysate was used per sample for western blot analysis. The band intensity was visualized using a LAS-4000 luminescent image analyzer (Fujifilm, Japan) and quantified by densitometry (Fuji Multi Gauge software ver. 3.0).
Evaluation of the mRNA expression level
Total RNA was isolated with TRIzol reagent (Invitrogen, USA) according to the manufacturer’s protocol. The total RNA concentration and purity were calculated using the absorbance at 260 nm and 280 nm using a NanoDrop (Thermo Fisher Scientific). The first cDNA strand was synthesized with 2 µg of total RNA and 1 µM of Oligo-dT18 primer using Omniscript Reverse Transcriptase (Qiagen, Germany). SYBR green-based quantitative real-time polymerase chain reaction (qRT-PCR) amplification was performed using an S1000 thermal cycler real-time PCR system and iQ SYBR Green supermix (Bio-Rad, USA) in the presence of 1:25 diluted first-strand cDNA and 20 pmol of primers according to the manufacturer’s protocols. The following primers were used to amplify human MUC5AC-specific products: (forward) 5'-TGA TCA TCC AGC AGG GCT-3' and (reverse) 5'-CCG AGC TCA GAG GAC ATA TGG G-3'. The primers for human GAPDH were used as quantitative controls: (forward) 5'-CAA AAG GGT CAT CTC TG-3' and (reverse) 5'-CCT GCT TCA CCA CCT TCT TG-3'. The PCR conditions consisted of three segments (Lee et al., 2018). All reactions were run in triplicate, and data were analyzed by the 2-ΔΔCT method (Livak and Schmittgen, 2001).
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation (ChIP) using antibodies against Smad3 (ab28379), NF-κB (#8242), Ac-NF-κB (ab19870), or HDAC2 (sc-7899) was performed using a ChIP Assay Kit according to the manufacturer’s protocol (Cell Signaling Technology). In brief, a total of 3 × 107 NCI-H292 cells were fixed in 1% formaldehyde, lysed, and sonicated five times for 2 s in ice-cold lysis buffer using a sonicator (550 Ultrasonic Dismembrator; Thermo Fisher Scientific). Following centrifugation of the extract for 10 min at 10,000g at 4°C, the extract was incubated with control rabbit IgG (#2729), Smad3 (ab28379), NF-κB (#8242), Ac-NF-κB (ab19870), or HDAC2 (sc-7899) antibody for 16 h at 4°C. The samples were incubated with ChIP-Grade Protein G Magnetic Beads for 2 h at 4°C. The immunoprecipitated complexes were eluted with elution buffer for 30 min at 65°C. The eluates were combined, and the DNA was reverse cross-linked by incubation for 2 h at 65°C after the addition of proteinase K and NaCl. Finally, the immunoprecipitated DNA was extracted using spin columns. The following oligonucleotide PCR primers were designed to include the Smad3 binding region (–234 to –134) and NF-κB binding region (–349 to –145) of the MUC5AC promoter (NCBI reference sequence: NM_001304359) in the PCR product: Smad3 binding region, (forward) 5'-TGG GCA CCA GGA ACT CAC-3' and (reverse) 5'-CGG GCT GGC CAG CGG CCG-3'; NF-κB binding region, (forward) 5'-ACT TCT GGG CAC CAG GAA CTC ACA-3' and (reverse) 5'-ACC CAA GTA AAC AGT GGG TGC TCA-3'. The PCR conditions consisted of three stages: the first stage (95°C for 5 min) activated the polymerase; the second stage included three-step cycling (34 cycles) at 95°C for 30 s (denaturation), 62°C for 30 s (annealing), and 72°C for 30 s (extension); and the third stage was a final extension step at 72°C for 5 min. Non-immunoprecipitated chromatin was used as an “input” control. The amplified PCR products were separated using 1.2% agarose gel electrophoresis and visualized by RedSafe (iNtRON Biotechnology, Korea).
Co-immunoprecipitation assay
NCI-H292 cells were extracted in ice-cold Nonidet P40 (NP40) extraction buffer (20 mM HEPES, pH 8.0, 1 mM DTT, 5% glycerol, 0.5 mM EDTA, 100 mM KCl, 0.2% NP40) (Hong and Choi, 2016). For immunoprecipitation (IP), protein extracts (1 mg) were incubated with 2 μg of anti-Smad3 antibody at 4°C for 3 h. Protein A/G-agarose beads (30 μl) were added and mixed at 4°C for 3 h. The immune complexes were washed three times with the extraction buffer and boiled in protein sample buffer for 5 min. After SDS-PAGE, proteins were transferred to a polyvinylidene difluoride membrane (Millipore, USA). The membrane was incubated with anti-HDAC2 or anti-Smad3 antibodies for western blotting assay. The protein bands were visualized using a LAS-4000 luminescent image analyzer (Fujifilm).
Immunofluorescence staining
For immunostaining, NCI-H292 cells were cultured on coverslips and fixed 3.7% paraformaldehyde (10 min, room temperature) in PBS (pH 7.4). After washing three times with PBS, the cells were permeabilized with 0.1% Triton-X100 (Sigma-Aldrich) for 10 min at room temperature. After three washes with PBS for 5 min, the cells were incubated with a blocking solution (3% BSA in PBS) for 30 min. For primary antibody, rabbit anti-Ac-NF-κB p65 at K310 (PA5-17264), rabbit anti-Smad3 (ab28379), or mouse anti-HDAC2 (#5113) antibody was used. The primary antibodies were incubated overnight at 4°C. The next day, the cells were washed 5 times for 5 min each time with PBS and then incubated with the secondary antibody such as Alexa Fluor 488-conjugated goat anti-rabbit IgG or Alexa Fluor 568-conjugated goat anti-mouse (Thermo Fisher Scientific) for 1 h. The cells were washed three times with PBS for 5 min each time. Then, nuclei were stained with 10 μg/ml DAPI for 30 min at room temperature. After three washes with PBS, the coverslips were mounted on slides using Fluoro-GEL (Electron Microscopy Sciences, USA). The image was obtained using a confocal fluorescence microscope (LSM800; Carl Zeiss, Germany). Higher-resolution airyscan processed images were acquired using an LSM 880 with Airyscan (Carl Zeiss) system with GaAsP detectors and a module for airyscan imaging. In Airyscan modes, a 63× Plan Apochromat (1.4 NA) oil objective was used. Confocal imaging was sequential for different fluorophore channels to obtain a series of axial images. Images were adjusted for contrast and brightness using the Zen software (Carl Zeiss).
Statistical analysis
Data are presented as mean ± SD. Student’s t-test using Prism 5 software (GraphPad Software, USA) were used for statistical analyses. Differences were considered significant at P < 0.05 (*), P < 0.01 (**), and P < 0.001 (***).
RESULTS
TGFβ1 negatively regulates MUC5AC expression in NCI-H292 cells
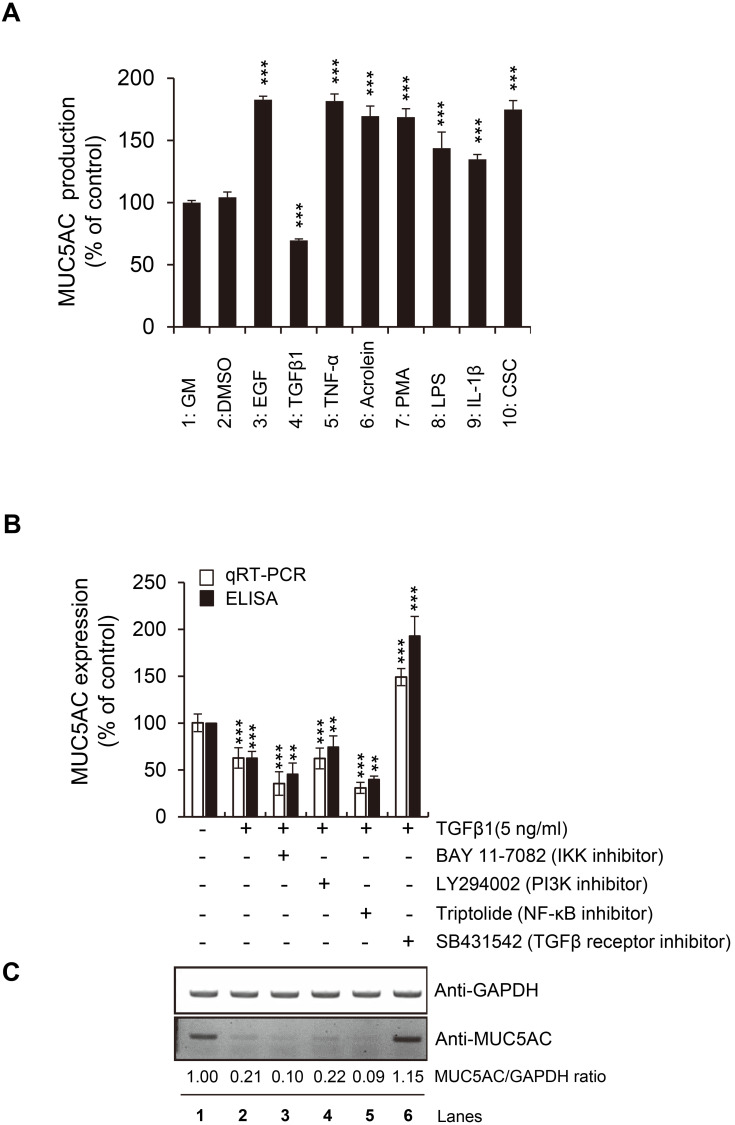
Various ligands and environmental stimuli regulate inflammatory responses in the human airway (Bonser and Erle, 2017). Several factors, such as EGF, TNF-α, Acrolein, PMA, LPS, IL-1β, CSC, or TGFβ are reported to modulate mucin production and secretion in NCI-H292 mucoepidermoid carcinoma cells derived from human lungs (Hewson et al., 2004; Kanai et al., 2015; Lee et al., 2018; Voynow and Rubin, 2009). However, the molecular mechanisms regulating mucin production and secretion in the airway cells are not fully understood. Thus, we evaluated how these factors regulate MUC5AC expression in NCI-H292 cells to understand further the molecular mechanism for MUC5AC expression. When these factors were incubated with NCI-H292 cells, seven of them (EGF, TNF-α, Acrolein, PMA, LPS, IL-1β, and CSC) enhanced the MUC5AC production (secretion into growth media) significantly by about 2-fold (Fig. 1A). This result was consistent with previous reports, including ours (Lee et al., 2018; 2019; Samsuzzaman et al., 2019), demonstrating that these factors increase the nuclear translocation of NF-κB, thereby overproducing MUC5AC in the airway cells.
Fig. 1. TGFβ1-induced reduction of MUC5AC expression.
(A) For 24 h, NCI-H292 cells were incubated with various ligands or stimuli, including EGF (100 ng/ml, bin 3), TGFβ1 (10 ng/ml, bin 4), TNF-α (20 ng/ml, bin 5), Acrolein (30 nM, bin 6), PMA (100 nM, bin 7), LPS (1 µg/ml, bin 8), IL-1β (20 ng/ml, bin 9), and CSC (10 µg/ml, bin 10). Bar graphs represent the mean ± SD of three independent experiments (***P < 0.001 compared with the controls, such as GM with blank or GM with 0.05% DMSO). The secretion of MUC5AC was assayed using ELISA. (B and C) TGFβ1-induced reduction of MUC5AC was restored by adding a TGFβ receptor inhibitor (SB431542). In contrast, other inhibitors for IKK (BAY11-7082), PI3K (LY294002), or NF-κB (Triptolide) showed no such effect. The mRNA or protein expression level of MUC5AC was evaluated using qRT-PCR, ELISA, or western blot analysis.
In the above assay, interestingly, only TGFβ1 ligand showed a reduction in MUC5AC production or secretion (Fig. 1A, bin 4). This reduction by TGFβ1 was confirmed by qRT-PCR, western blot analysis, and ELISA on whole cell lysates (Figs. 1B and 1C). TGFβ1 treatment on NCI-H292 cells dramatically reduced the mRNA and protein levels of MUC5AC (Figs. 1B and 1C, lanes 1 and 2). In contrast, additional treatment with SB431542, an inhibitor for the TGFβ receptor, markedly blocked the suppressive effect of TGFβ1 on MUC5AC production (Figs. 1B and 1C, lane 6), indicating that TGFβ1 signaling negatively regulates MUC5AC expression in the airway cells. Another inhibitor for PI3K (LY294002) did not show any additional effect on TGFβ1 treatment (Figs. 1B and 1C, lane 4), suggesting that TGFβ1-induced MUC5AC reduction is not non-specific.
Reduction of NF-κB activity by BAY 11-7082 or Triptolide marginally enhanced the inhibition of MUC5AC expression by TGFβ1 (Figs. 1B and 1C, lanes 3 and 5), indicating that TGFβ1 signaling might interact with NF-κB signaling to control MUC5AC expression.
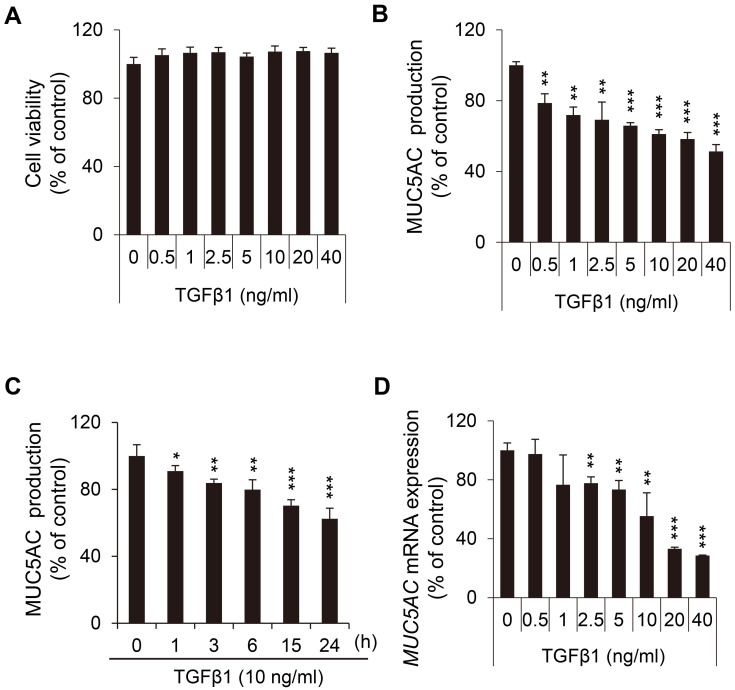
Because the negative effect of TGFβ1 on MUC5AC production was clearly distinguishable from the positive effect of the other factors, we focused on the role of TGFβ1 for further study. Before scrutinizing the suppressive role of TGFβ1 on MUC5AC production, we evaluated the viability of NCI-H292 cells after TGFβ1 treatment (Fig. 2A). Because TGFβ1 showed no cytotoxicity at concentrations of less than 40 ng/ml, we applied TGFβ1 at this concentration range in the subsequent experiments. In NCI-H292 airway cells, the inhibitory effect of TGFβ1 on MUC5AC production was both concentration and time-dependent, as demonstrated by ELISA data. Responding to various concentrations of TGFβ1, MUC5AC secretion was significantly reduced in a concentration-dependent manner (Fig. 2B). Besides, TGFβ1 at 10 ng/ml decreased MUC5AC production in a time-dependent manner (Fig. 2C). Interestingly, the level of MUC5AC transcripts was more strongly affected by TGFβ1, compared with that of MUC5AC secretion (Fig. 2D). After TGFβ1 addition, MUC5AC mRNA expression decreased to about 30% of the control. In contrast, the reduced level of MUC5AC production was about 50% of control (Figs. 2B and 2D). Altogether, these results suggested that TGFβ1 inhibits MUC5AC expression at the transcriptional level in NCI-H292 airway cells.
Fig. 2. TGFβ1 negatively affects MUC5AC expression at the transcriptional level.
(A) No adverse effect of TGFβ1 on NCI-H292 cell viability at concentration ranges of 0 to 40 ng/ml. (B and C) The effects of TGFβ1 on MUC5AC secretion were analyzed by ELISA. In NCI-H292 airway cells, TGFβ1 decreased MUC5AC secretion in a concentration- (B) and time- (C) dependent manner. NCI-H292 cells were incubated with an increasing or a fixed concentration of TGFβ1 for 24 h. (D) The effects of TGFβ1 on MUC5AC mRNA expression were evaluated using qRT-PCR. NCI-H292 cells were pretreated with various concentrations of TGFβ1 for 12 h. The expression of MUC5AC transcripts was more prominently reduced by TGFβ1 addition, compared with the secretion of MUC5AC proteins in Fig. 2B. Data represent mean ± SD of at least three individual experiments (*P < 0.05, **P < 0.01, and ***P < 0.001 compared with the control, without TGFβ1).
TGFβ1-activated Smad3 binds to the MUC5AC promoter
There was a report that activated TGFβ receptor-Smad3 signaling reduces MUC5AC expression in human epithelial cells exposed to a human bacterial pathogen (non-typeable Haemophilus influenza, or NTHi) (Jono et al., 2003). This report showed that NTHi-induced TLR2 receptor phosphorylates p38 MAPK to positively regulate MUC5AC expression and showed that additive TGFβ receptor-Smad3 signaling in this condition results in a reduction in MUC5AC expression by inducing the expression of MKP-1 phosphatase, an inhibitor for p38. However, the overexpression effect of MKP-1 on MUC5AC reduction was relatively mild compared with MUC5AC reduction by TGFβ or Smad3 overexpression. Thus, we inferred that there could be another player to regulate MUC5AC reduction by TGFβ signaling.
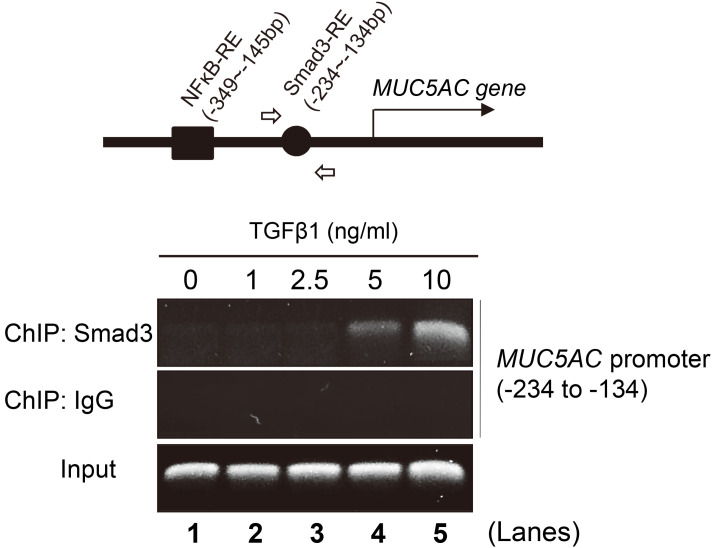
The data depicted in Fig. 2D suggest that Smad3 could suppress MUC5AC transcription by directly binding to the promoter region of the MUC5AC gene. Therefore, we tested whether TGFβ1-activated Smad3 binds to the MUC5AC promoter in NCI-H292 cells using a ChIP assay with a Smad3 antibody. When the MUC5AC promoter regions between –1384 and +31 bp were analyzed using the TFBIND (http://tfbind.hgc.jp), we found distinct binding sites for two transcription factors: a Smad3-responsive element (RE) between –234 and –134 bp and an NF-κB-RE between –349 and –145 bp. PCR primers for detecting the Smad3-RE were designed, and TGFβ1-induced Smad3 binding to the MUC5AC promoter was assayed (Fig. 3, upper diagram, open arrows). Notably, consistent with our prediction, Smad3 bound to the MUC5AC promoter after TGFβ1 addition. This interaction was positively dependent on TGFβ1 concentration (Fig. 3, lower panels). Combined with Fig. 2D, our result indicated that in human airway cells, the signaling pathway connecting TGFβ1, TGFβ1 receptor, and Smad3 directly and negatively regulates MUC5AC promoter activity.
Fig. 3. TGFβ1-activated Smad3 binds to the MUC5AC promoter.
ChIP assay using the anti-Smad3 antibody on the MUC5AC promoter region. The binding of Smad3 to the MUC5AC promoter was increased by TGFβ1 addition in NCI-H292 cells. Upper diagram: The MUC5AC promoter between -1384 and +31 bp contains two responsive elements (RE) for transcription factors such as Smad3 and NF-κB, predicted by the TFBIND. Lower panels: NCI-H292 cells were treated with each indicated concentration of TGFβ1 for 30 min. PCR primers were designed to identify the Smad3 RE within the MUC5AC promoter region between –234 to –134 bp (arrows in the upper diagram). Rabbit IgG was used as a negative control. Input samples (unprecipitated chromatin) were used as positive controls for PCR amplification.
TGFβ1 negatively regulates NF-κB acetylation at K310 by Smad3/HDAC2 complex formation
To elucidate the mechanism by which TGFβ1-Smad3 signaling reduces MUC5AC expression in NCI-H292 cells, we inferred three assumptions. TGFβ1-activated Smads combine with additional factors to control the expression of TGFβ1-responsive target genes (Hill, 2016). Among these factors, HDACs form a complex with Smads and serve as a negative regulator for the target genes (Bai and Xi, 2018). Thus, we firstly reasoned that Smad3 binding to the MUC5AC promoter could recruit HDAC proteins to reduce MUC5AC production in human airway cells. Secondly, our results with both inhibitors for NF-κB activity (Figs. 1B and 1C) and promoter prediction (Fig. 3) suggested that Smad3 could functionally interact with NF-κB to inhibit MUC5AC expression. NF-κB is a positive regulator for MUC5AC production in human airway cells (Lee et al., 2016; 2018; Samsuzzaman et al., 2019). Lastly, there were also reports that HDAC2 suppresses NF-κB activity to reduce the expression of pro-inflammatory genes, although the precise epigenetic mechanism is not fully elucidated (Ashburner et al., 2001; Ito et al., 2000; 2001; Moodie et al., 2004; Mortaz et al., 2011; Yang et al., 2006). Thus, we hypothesize that TGFβ1-activated Smad3 may form a protein complex with HDAC2 to suppress NF-κB activity.
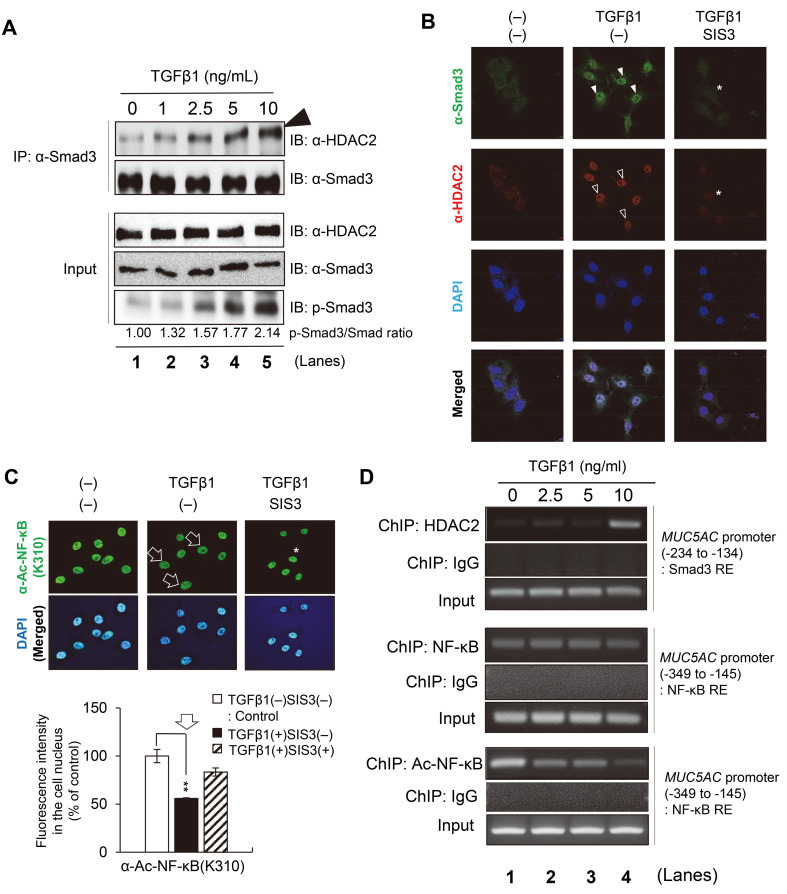
To attest this hypothesis, we first checked the physical binding between Smad3 and HDAC2 proteins with a co-immunoprecipitation assay using NCI-H292 cell extracts. TGFβ1 addition to cell culture media increased Smad3 phosphorylation in a concentration-dependent manner in NCI-H292 cells (Fig. 4A, Input). In this condition, a co-immunoprecipitation assay was performed using the Smad3 antibody. Notably, HDAC2 binding to Smad3 was prominently increased by TGFβ1 addition in a concentration-dependent manner (Fig. 4A, arrowhead). Consistent with this, the physical interaction between Smad3 and HDAC2 caused nuclear translocation of this protein complex. Co-immunostaining of NCI-H292 cells using Smad3 and HDAC2 antibodies showed that both Smad3 and HDAC2 proteins co-localize in the cell nucleus in the presence of TGFβ1 compared with a non-stimulated control (Fig. 4B, closed and open arrowheads). These data suggest that the activated Smad3 recruits HDAC2 to make a protein complex in the airway cells, and that nuclear translocation of the Smad3–HDAC2 complex may suppress the effect of NF-κB on MUC5AC expression by epigenetic regulation.
Fig. 4. Smad3/HDAC2 complex inhibits NF-κB activation.
(A) Co-immunoprecipitation (IP) using the Smad3 antibody. NCI-H292 cells were treated with the indicated concentrations of TGFβ1 for 30 min and subsequently lysed for IP assays. The immunoprecipitants collected by the Smad3 antibody were immunoblotted (IB) with an antibody against HDAC2 or Smad3. The input panels represent 4% of the cell extracts used for IP, immunoblotted using antibodies against phosphorylated HDAC2, Smad3, and phosphorylated Smad3. TGFβ-induced Smad3 phosphorylation increased in a concentration-dependent manner. Smad3 antibody was used as the loading control. The numbers at the bottom of each lane represent the relative band intensity normalized to the control (Smad3 without phosphorylation). (B) TGFβ1-induced colocalization of SMAD3 and HDAC2 proteins in the cell nucleus (closed and open arrowheads, respectively). NCI-H292 cells were co-labeled using antibodies against Smad3 (green) and HDAC2 (red). The cell nucleus was stained with DAPI (blue). The merged confocal images were shown at the bottom. SIS3 (1 µM), a specific Smad3 inhibitor, was treated to suppress the nuclear colocalization of Smad3 or HDAC2 (asterisks). (C) The acetylated level at lysine 310 of NF-κB (Ac-NF-κB (K310)) was reduced by TGFβ1 (arrows) and restored by an additional Smad3 inhibitor, SIS3 (asterisk). NCI-H292 cells were stained with anti-Ac-NF-κB (K310) antibody (green). Cell nuclei were stained with DAPI (blue). Lower histogram: Quantification of the confocal images. The signal intensity for Ac-NF-κB (K310) in the cell nucleus was normalized to the control (without TGFβ1 treatment). The bar graph represents the mean ± SD of three independent experiments (**P < 0.01). (D) ChIP assay using anti-HDAC2, anti-NF-κB, or anti-Ac-NF-κB antibody on the MUC5AC promoter region. NCI-H292 cells were treated with each indicated concentration of TGFβ1 for 30 min. PCR primers were designed to identify the Smad3 RE (–234 to –134 bp) or NF-κB RE (–349 to –145 bp) within the MUC5AC promoter region. Upper panels: The recruitment of HDAC2 to the Smad3 RE on the MUC5AC promoter was increased by TGFβ1 addition in NCI-H292 cells. Middle panels: The binding of NF-κB to the MUC5AC promoter seemed not to be affected by TGFβ1 addition. Lower panels: Contrary to NF-κB binding to the MUC5AC promoter, epigenetic modification on NF-κB (Ac-NF-κB at K310) was prominently reduced by TGFβ1. Rabbit IgG was used as a negative control. Input samples (unprecipitated chromatin) were used as positive controls for PCR amplification.
Next, we checked whether the Smad3–HDAC2 complex suppresses NF-κB activity epigenetically. Acetylation on NF-κB itself or histones near chromatin-bound NF-κB is critical for the transcriptional activity of NF-κB. For example, acetylation of NF-κB p65 subunit at lysine 310 (Ac-NF-κB at K310) is essential for the transcriptional activity of the NF-κB complex (Ashburner et al., 2001; Chen et al., 2002; Ito et al., 2000). Thus, we immunostained NCI-H292 cells to examine the NF-κB activity using a specific antibody for Ac-NF-κB at K310. In control NCI-H292 cells, NF-κB acetylation at K310 was highly detected in the cell nucleus, whereas TGFβ1 addition reduced the acetylation level to about a half of the control (Fig. 4C and a histogram, arrows), suggesting that TGFβ1 suppresses the transcriptional activity of NF-κB by inducing HDAC2-mediated deacetylation.
Both TGFβ1-induced nuclear translocation of HDAC2 and subsequent deacetylation of NF-κB at K310 were evidently dependent on TGFβ1–Smad3 signaling. When TGFβ1-treated NCI-H292 cells were combined with a selective Smad3 inhibitor (SIS3), they showed considerable restoration of both nuclear translocations of HDAC2–Smad3 complex and the acetylation level of NF-κB (Figs. 4B and 4C, asterisks).
Lastly, we checked whether TGFβ1 regulates the recruitment of HDAC2, NF-κB, and Ac-NF-κB at K310 on the MUC5AC promoter region using a ChIP assay (Fig. 4D). In this assay, we found that HDAC2 binding to the Smad3-RE on the MUC5AC promoter was increased by TGFβ1 addition in a concentration-dependent manner (Fig. 4D, upper panels), suggesting that Smad3 can recruit HDAC2 to the MUC5AC promoter. This result was consistent with the data above in Figs. 3 and 4A. Interestingly, the recruitment of NF-κB to the MUC5AC promoter was not severely affected by TGFβ1 addition (Fig. 4D, middle panels). Instead, K310 acetylation for the transcriptional activity of NF-κB was considerably reduced by TGFβ1 on the MUC5AC promoter (Fig. 4D, lower panels), suggesting that TGFβ1 epigenetically suppresses NF-κB activity to inhibit MUC5AC expression.
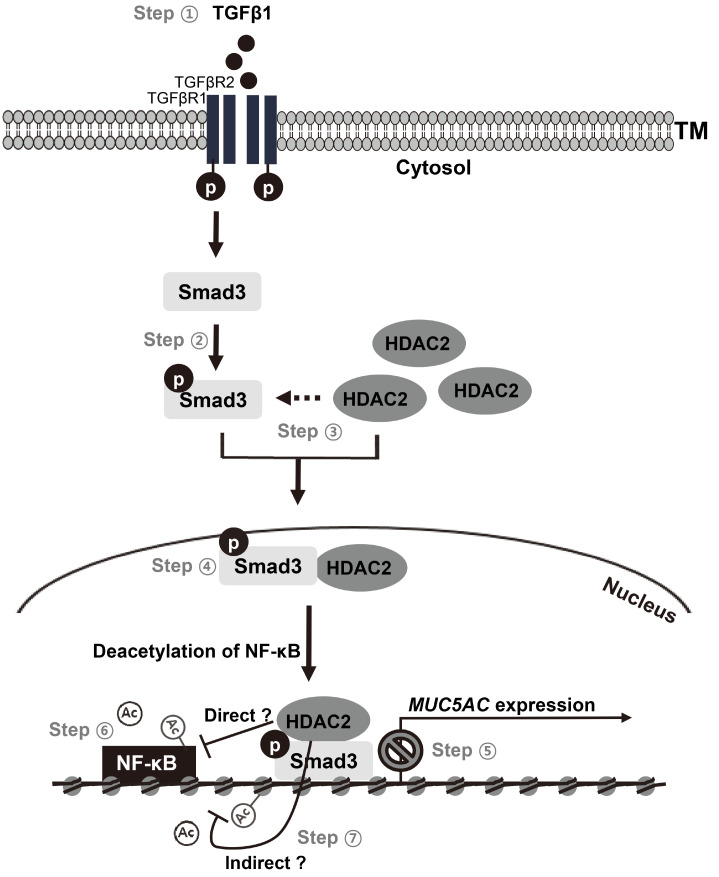
Altogether, these results suggest that TGFβ1-activated Smad3 recruits HDAC2, thereby making a Smad3–HDAC2 protein complex that translocates to the cell nucleus to inhibit both NF-κB activity and expression of its downstream target genes, including MUC5AC (Fig. 5).
Fig. 5. Smad3-recruited HDAC2 induces NF-κB deacetylation at K310.
TGFβ1 signaling decreases MUC5AC expression transcriptionally in NCI-H292 cells. Steps ① and ②: TGFβ1-binding to TGFβ receptors activates Smad3 by phosphorylation. Steps ③ and ④: Activated Smad3 binds to HDAC2 and makes a protein complex (Smad3/HDAC2 complex), which translocates to the cell nucleus. Step ⑤: Activated Smad3 in the cell nucleus binds to the promoter region of MUC5AC and reduces its expression. Step ⑥: Smad3/HDAC2 complex induces the deacetylation of Ac-NF-κB at K310, thereby reducing the transcriptional activity of NF-κB. Deacetylation of Ac-NF-κB at K310 is considerably restored by adding a Smad3 inhibitor (SIS3). Step ⑦: This study is undefined whether the deacetylation of NF-κB at K310 is mediated by HDAC2 directly or indirectly.
DISCUSSION
Airway mucus hypersecretion is a visible marker of lung inflammatory diseases such as asthma and COPD (Kesimer et al., 2017). In particular, MUC5AC is the predominant mucin protein that is increased in asthmatic and COPD patients (Mata et al., 2005). Thus, understanding the exact molecular mechanism for regulating MUC5AC expression is important for the development of effective treatments for asthma and COPD.
The level of MUC5AC is positively regulated by various stimuli associated with lung inflammatory diseases (Voynow and Rubin, 2009). Lung inflammatory stimuli, such as EGF (Perrais et al., 2002), TNF-α (Lee et al., 2016), Acrolein (Borchers et al., 1998), PMA (Hewson et al., 2004), LPS (Zen et al., 2002), IL-1β (Chen et al., 2014), and CSC (Kanai et al., 2015), promote MUC5AC expression and secretion. We also obtained similar results in human airway epithelial cells, NCI-H292. In MUC5AC expression, NF-κB signaling has a central role. Receptor-bound EGF activates ERK kinase-SP1 or ERK kinase-AP1 transcription factor axis to enhance NF-κB-mediated transcription of the MUC5AC gene. Cytokines such as TNF-α and IL-1β also positively regulate NF-κB activity for MUC5AC transcription via activating IKK (NF-κB activator) or ERK kinase. Smokes from fossil fuels or cigarettes (Acrolein or CSC) stimulate the EGF receptor and result in NF-κB activation for MUC5AC expression (Choi et al., 2011; 2018; Lee et al., 2019). PMA-induced reactive oxygen species or bacterial endotoxins (LPS) also stimulate JNK or P38 kinase to enhance NF-κB-mediated MUC5AC expression (Krishn et al., 2018).
However, the TGFβ1 stimulus showed a unique feature compared with the other stimuli: TGFβ1 decreased the expression of MUC5AC, at both the mRNA and protein levels, in a time- and dose-dependent manner. Consistently, the TGFβ receptor inhibitor (SB431542) restored TGFβ1-induced MUC5AC reduction to control levels.
The molecular mechanisms explaining the effect of TGFβ signaling in MUC5AC expression were, to date, not clearly defined (Tong and Gu, 2020). Several studies described that TGFβ could suppress or promote MUC5AC expression. For instance, the addition of TGFβ increased MUC5AC endogenous expression in murine rectal cancer cells via Smad4 and SP1 pathway and human bronchial epithelial BEAS-2B cells via JNK pathway (Jonckheere et al., 2004; Park et al., 2015). In contrast, in human colon and lung epithelial cells, TGFβ/Smad signaling reduced p38-mediated MUC5AC expression by inducing p38 inhibitor, MKP-1 protein (Jono et al., 2003). As mentioned right above, NF-κB signaling is critical for MUC5AC expression. However, it is not clearly defined whether TGFβ1 signaling suppresses the transcriptional activity of NF-NF-κB for MUC5AC expression and how Smad mediators for TGFβ signaling regulate this NF-κB activity (Krishn et al., 2018; Luo, 2017). This ambiguous role of TGFβ signaling in MUC5AC expression led us to scrutinize the molecular mechanism by which TGFβ down-regulated MUC5AC expression in human lung epithelial cells (NCI-H292).
Our findings demonstrate that TGFβ1 signaling decreases MUC5AC expression transcriptionally in a dose-dependent manner in NCI-H292 cells via epigenetic regulation on NF-κB activity: TGFβ1 binding to TGFβ receptors (R1/R2) activates Smad3 via phosphorylation (Fig. 5, Steps ① and ②), which induce the physical interaction between Smad3 and HDAC2, forming a Smad3/HDAC2 complex (Fig. 5, Step ③ and Fig. 4A). To the best of our knowledge, this Smad3/HDAC2 complex formation in human airway cells has not been previously reported. Subsequently, the Smad3/HDAC2 complex translocates to the cell nucleus (Fig. 5, Step ④), where Smad3 will directly binding to the Smad3-responsive element in the MUC5AC promoter region (Fig. 5, Step ⑤). In NCI-H292 cells, NF-κB directly regulates MUC5AC expression transcriptionally (Fig. 5, Step ⑥). It is well known that epigenetic modifications on NF-κB subunit p65 (acetylation at the multiple positions) are critical for NF-κB function (Buerki et al., 2008; Chen et al., 2002). Among them, acetylation at lysine (K) 310 of the p65 subunit for NF-κB (Ac-NF-κB at K310) is essential for the transcriptional activity of NF-κB. Interestingly, we found for the first time that NF-κB acetylation at K310 is considerably reduced in NCI-H292 cells (Fig. 5, Step ⑥), which could be the result of the Smad3/HDAC2 complex activity by inducing the deacetylation of Ac-NF-κB at K310. Ultimately, this sequence of events will result in a reduction of MUC5AC expression, although we did not test whether Ac-NF-κB at K310 is a direct substrate for HDAC2. We did not also exclude that HDAC2 might affect NF-κB acetylation indirectly through regulating histone acetylation nearby Smad3 (Fig. 5, Step ⑦). Nevertheless, collectively, these data suggest that TGFβ1/Smad3 negatively regulates MUC5AC in airway epithelial cells.
HDACs function as negative regulators of acetylation for histones and other transcription factors, thereby downregulates the expression of various genes, including TGFβ responsive genes (Bai and Xi, 2018). In particular, HDAC2 is implicated in several epigenetic silencing complexes that are closely associated with lung inflammatory diseases such as asthma and COPD (Barnes, 2009). Currently, the most effective way of anti‐inflammatory therapy for asthma is glucocorticoids, such as dexamethasone (Dex). Dex represses mucin concentrations in lung epithelial cells via the activation of the glucocorticoid receptor (GR) (Barnes, 2011). In turn, Dex-activated GR interacts with HDAC2 to attenuate NF-κB activity, thereby effectively suppressing inflammatory gene expression such as MUC5AC (Chen et al., 2012). Thus, understanding the molecular mechanism regulating HDAC2 activity is also crucial to develop new strategies to treat lung inflammatory diseases (Mortaz et al., 2011).
In chronic and progressive lung diseases such as asthma or COPD, TGF-β’s role in MUC5AC expression is somewhat controversial (Saito et al., 2018). Some groups reported that the upregulated TGFβ level in asthmatic patients causes an increase in proliferation and extracellular matrix deposition in the human airway smooth muscle cells, suggesting that ectopic TGFβ results in stiffness and irreversible structural alteration or damage in lung tissues (Ojiaku et al., 2018; Vignola et al., 1997). On the other hand, human patients with reduced TGFβ signaling showed the opposite results. Patients with loss of function mutations in TGFβ receptors (Loeys-Dietz syndrome) frequently develop allergic diseases such as asthma, suggesting TGFβ’s protective roles for lung diseases (Frischmeyer-Guerrerio et al., 2013).
Collectively, our results suggest that the Smad3/HDAC2 complex is a promising candidate for improving lung function in the treatment of asthma and COPD by reducing the expression of NF-κB-targeted genes, including MUC5AC.
ACKNOWLEDGMENTS
This work was supported by the KRIBB Research Initiative Program funded by the Ministry of Science ICT (MSIT), the R&D Convergence Program of the National Research Council of Science and Technology (CAP-18-02KRIBB), and the National Research Foundation (NRF-2020R1A2C2006664 and NRF2020R1C1C1011146) of Republic of Korea.
Footnotes
AUTHOR CONTRIBUTIONS
S.U.L., M.O.K., M.J.K., and E.S.O. performed the experiments and analyzed the data. H.R. and S.Y.L. interpreted the results. R.W.L., Y.N.S., S.J., J.W.L., and T.B. provided technical support and performed the experiments. S.U.L. and S.T.H. conceived the study and wrote the manuscript. S.T.H. and T.D.K. supervised the project and analyzed the data. All authors were involved in writing and critical review of the paper, and approved the final version of the manuscript.
CONFLICT OF INTEREST
The authors have no potential conflicts of interest to disclose.
REFERENCES
- Alcorn J.F., Rinaldi L.M., Jaffe E.F., van Loon M., Bates J.H.T., Janssen-Heininger Y.M.W., Irvin C.G. Transforming growth factor-beta1 suppresses airway hyperresponsiveness in allergic airway disease. Am. J. Respir. Crit. Care Med. 2007;176:974–982. doi: 10.1164/rccm.200702-334OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthoni M., Wang G., Leino M.S., Lauerma A.I., Alenius H.T., Wolff H.J. Smad3 -signalling and Th2 cytokines in normal mouse airways and in a mouse model of asthma. Int. J. Biol. Sci. 2007;3:477–485. doi: 10.7150/ijbs.3.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner B.P., Westerheide S.D., Baldwin A.S., Jr. The p65 (RelA) subunit of NF-kappaB interacts with the histone deacetylase (HDAC) corepressors HDAC1 and HDAC2 to negatively regulate gene expression. Mol. Cell. Biol. 2001;21:7065–7077. doi: 10.1128/MCB.21.20.7065-7077.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai J., Xi Q. Crosstalk between TGF-beta signaling and epigenome. Acta Biochim. Biophys. Sin. (Shanghai) 2018;50:60–67. doi: 10.1093/abbs/gmx122. [DOI] [PubMed] [Google Scholar]
- Barnes P.J. Histone deacetylase-2 and airway disease. Ther. Adv. Respir. Dis. 2009;3:235–243. doi: 10.1177/1753465809348648. [DOI] [PubMed] [Google Scholar]
- Barnes P.J. Glucocorticosteroids: current and future directions. Br. J. Pharmacol. 2011;163:29–43. doi: 10.1111/j.1476-5381.2010.01199.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonser L.R., Erle D.J. Airway mucus and asthma: the role of MUC5AC and MUC5B. J. Clin. Med. 2017;6:112. doi: 10.3390/jcm6120112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borchers M.T., Wert S.E., Leikauf G.D. Acrolein-induced MUC5ac expression in rat airways. Am. J. Physiol. 1998;274:L573–L581. doi: 10.1152/ajplung.1998.274.4.L573. [DOI] [PubMed] [Google Scholar]
- Branchett W.J., Lloyd C.M. Regulatory cytokine function in the respiratory tract. Mucosal Immunol. 2019;12:589–600. doi: 10.1038/s41385-019-0158-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buerki C., Rothgiesser K.M., Valovka T., Owen H.R., Rehrauer H., Fey M., Lane W.S., Hottiger M.O. Functional relevance of novel p300-mediated lysine 314 and 315 acetylation of RelA/p65. Nucleic Acids Res. 2008;36:1665–1680. doi: 10.1093/nar/gkn003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caramori G., Gregorio C., Carlstedt I., Casolari P., Guzzinati I., Adcock I., Barnes P., Ciaccia A., Cavallesco G., Chung K.F., et al. Mucin expression in peripheral airways of patients with chronic obstructive pulmonary disease. Histopathology. 2004;45:477–484. doi: 10.1111/j.1365-2559.2004.01952.x. [DOI] [PubMed] [Google Scholar]
- Chen L.F., Mu Y., Greene W.C. Acetylation of RelA at discrete sites regulates distinct nuclear functions of NF-kappaB. EMBO J. 2002;21:6539–6548. doi: 10.1093/emboj/cdf660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Garvin L.M., Nickola T.J., Watson A.M., Colberg-Poley A.M., Rose M.C. IL-1β induction of MUC5AC gene expression is mediated by CREB and NF-κB and repressed by dexamethasone. Am. J. Physiol. Lung Cell. Mol. Physiol. 2014;306:L797–L807. doi: 10.1152/ajplung.00347.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Watson A.M., Williamson C.D., Rahimi M., Liang C., Colberg-Poley A.M., Rose M.C. Glucocorticoid receptor and histone deacetylase-2 mediate dexamethasone-induced repression of MUC5AC gene expression. Am. J. Respir. Cell Mol. Biol. 2012;47:637–644. doi: 10.1165/rcmb.2012-0009OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi B.S., Kim Y.J., Yoon Y.P., Lee H.J., Lee C.J. Tussilagone suppressed the production and gene expression of MUC5AC mucin via regulating nuclear factor-kappa B signaling pathway in airway epithelial cells. Korean J. Physiol. Pharmacol. 2018;22:671–677. doi: 10.4196/kjpp.2018.22.6.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J.H., Hwang Y.P., Han E.H., Kim H.G., Park B.H., Lee H.S., Park B.K., Lee Y.C., Chung Y.C., Jeong H.G. Inhibition of acrolein-stimulated MUC5AC expression by Platycodon grandiflorum root-derived saponin in A549 cells. Food Chem. Toxicol. 2011;49:2157–2166. doi: 10.1016/j.fct.2011.05.030. [DOI] [PubMed] [Google Scholar]
- Cukic V., Lovre V., Dragisic D., Ustamujic A. Asthma and chronic obstructive pulmonary disease (COPD) - differences and similarities. Mater. Sociomed. 2012;24:100–105. doi: 10.5455/msm.2012.24.100-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran D.R., Cohn L. Advances in mucous cell metaplasia: a plug for mucus as a therapeutic focus in chronic airway disease. Am. J. Respir. Cell Mol. Biol. 2010;42:268–275. doi: 10.1165/rcmb.2009-0151TR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frischmeyer-Guerrerio P.A., Guerrerio A.L., Oswald G., Chichester K., Myers L., Halushka M.K., Oliva-Hemker M., Wood R.A., Dietz H.C. TGFbeta receptor mutations impose a strong predisposition for human allergic disease. Sci. Transl. Med. 2013;5:195ra194. doi: 10.1126/scitranslmed.3006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewson C.A., Edbrooke M.R., Johnston S.L. PMA induces the MUC5AC respiratory mucin in human bronchial epithelial cells, via PKC, EGF/TGF-α, Ras/Raf, MEK, ERK and Sp1-dependent mechanisms. J. Mol. Biol. 2004;344:683–695. doi: 10.1016/j.jmb.2004.09.059. [DOI] [PubMed] [Google Scholar]
- Hill C.S. Transcriptional control by the SMADs. Cold Spring Harb. Perspect. Biol. 2016;8:a022079. doi: 10.1101/cshperspect.a022079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S.T., Choi K.W. Antagonistic roles of Drosophila Tctp and Brahma in chromatin remodelling and stabilizing repeated sequences. Nat. Commun. 2016;7:12988. doi: 10.1038/ncomms12988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K., Barnes P.J., Adcock I.M. Glucocorticoid receptor recruitment of histone deacetylase 2 inhibits interleukin-1beta-induced histone H4 acetylation on lysines 8 and 12. Mol. Cell. Biol. 2000;20:6891–6903. doi: 10.1128/mcb.20.18.6891-6903.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K., Ito M., Elliott W.M., Cosio B., Caramori G., Kon O.M., Barczyk A., Hayashi S., Adcock I.M., Hogg J.C., et al. Decreased histone deacetylase activity in chronic obstructive pulmonary disease. N. Engl. J. Med. 2005;352:1967–1976. doi: 10.1056/NEJMoa041892. [DOI] [PubMed] [Google Scholar]
- Ito K., Lim S., Caramori G., Chung K.F., Barnes P.J., Adcock I.M. Cigarette smoking reduces histone deacetylase 2 expression, enhances cytokine expression, and inhibits glucocorticoid actions in alveolar macrophages. FASEB J. 2001;15:1110–1112. [PubMed] [Google Scholar]
- Jonckheere N., Van Der Sluis M., Velghe A., Buisine M.P., Sutmuller M., Ducourouble M.P., Pigny P., Buller H.A., Aubert J.P., Einerhand A.W., et al. Transcriptional activation of the murine Muc5ac mucin gene in epithelial cancer cells by TGF-beta/Smad4 signalling pathway is potentiated by Sp1. Biochem. J. 2004;377:797–808. doi: 10.1042/BJ20030948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jono H., Xu H., Kai H., Lim D.J., Kim Y.S., Feng X.H., Li J.D. Transforming growth factor-beta-Smad signaling pathway negatively regulates nontypeable Haemophilus influenzae-induced MUC5AC mucin transcription via mitogen-activated protein kinase (MAPK) phosphatase-1-dependent inhibition of p38 MAPK. J. Biol. Chem. 2003;278:27811–27819. doi: 10.1074/jbc.M301773200. [DOI] [PubMed] [Google Scholar]
- Kanai K., Koarai A., Shishikura Y., Sugiura H., Ichikawa T., Kikuchi T., Akamatsu K., Hirano T., Nakanishi M., Matsunaga K., et al. Cigarette smoke augments MUC5AC production via the TLR3-EGFR pathway in airway epithelial cells. Respir Investig. 2015;53:137–148. doi: 10.1016/j.resinv.2015.01.007. [DOI] [PubMed] [Google Scholar]
- Kesimer M., Ford A.A., Ceppe A., Radicioni G., Cao R., Davis C.W., Doerschuk C.M., Alexis N.E., Anderson W.H., Henderson A.G., et al. Airway mucin concentration as a marker of chronic bronchitis. N. Engl. J. Med. 2017;377:911–922. doi: 10.1056/NEJMoa1701632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.Y., Kim W.J., Kim J.H., Hong S.H., Choi S.S. Identification of putative regulatory alterations leading to changes in gene expression in chronic obstructive pulmonary disease. Mol. Cells. 2019;42:333–344. doi: 10.14348/molcells.2019.2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishn S.R., Ganguly K., Kaur S., Batra S.K. Ramifications of secreted mucin MUC5AC in malignant journey: a holistic view. Carcinogenesis. 2018;39:633–651. doi: 10.1093/carcin/bgy019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo M.H., Allis C.D. Roles of histone acetyltransferases and deacetylases in gene regulation. Bioessays. 1998;20:615–626. doi: 10.1002/(SICI)1521-1878(199808)20:8<615::AID-BIES4>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Lagna G., Hata A., Hemmati-Brivanlou A., Massague J. Partnership between DPC4 and SMAD proteins in TGF-beta signalling pathways. Nature. 1996;383:832–836. doi: 10.1038/383832a0. [DOI] [PubMed] [Google Scholar]
- Lee J.W., Ryu H.W., Lee S.U., Kim M.G., Kwon O.K., Kim M.O., Oh T.K., Lee J.K., Kim T.Y., Lee S.W., et al. Pistacia weinmannifolia ameliorates cigarette smoke and lipopolysaccharideinduced pulmonary inflammation by inhibiting interleukin8 production and NFkappaB activation. Int. J. Mol. Med. 2019;44:949–959. doi: 10.3892/ijmm.2019.4247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.U., Ahn K.S., Sung M.H., Park J.W., Ryu H.W., Lee H.J., Hong S.T., Oh S.R. Indacaterol inhibits tumor cell invasiveness and MMP-9 expression by suppressing IKK/NF-kappaB activation. Mol. Cells. 2014;37:585–591. doi: 10.14348/molcells.2014.0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.U., Lee S., Ro H., Choi J.H., Ryu H.W., Kim M.O., Yuk H.J., Lee J., Hong S.T., Oh S.R. Piscroside C inhibits TNF-alpha/NF-kappaB pathway by the suppression of PKCdelta activity for TNF-RSC formation in human airway epithelial cells. Phytomedicine. 2018;40:148–157. doi: 10.1016/j.phymed.2018.01.012. [DOI] [PubMed] [Google Scholar]
- Lee S.U., Sung M.H., Ryu H.W., Lee J., Kim H.S., In H.J., Ahn K.S., Lee H.J., Lee H.K., Shin D.H., et al. Verproside inhibits TNF-alpha-induced MUC5AC expression through suppression of the TNF-alpha/NF-kappaB pathway in human airway epithelial cells. Cytokine. 2016;77:168–175. doi: 10.1016/j.cyto.2015.08.262. [DOI] [PubMed] [Google Scholar]
- Letterio J.J., Roberts A.B. Regulation of immune responses by TGF-beta. Annu. Rev. Immunol. 1998;16:137–161. doi: 10.1146/annurev.immunol.16.1.137. [DOI] [PubMed] [Google Scholar]
- Ling E., Robinson D.S. Transforming growth factor-beta1: its anti-inflammatory and pro-fibrotic effects. Clin. Exp. Allergy. 2002;32:175–178. doi: 10.1046/j.1365-2222.2002.01287.x. [DOI] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Luo K. Signaling cross talk between TGF-beta/Smad and other signaling pathways. Cold Spring Harb. Perspect. Biol. 2017;9:a022137. doi: 10.1101/cshperspect.a022137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mata M., Sarriá B., Buenestado A., Cortijo J., Cerdá M., Morcillo E.J. Phosphodiesterase 4 inhibition decreases MUC5AC expression induced by epidermal growth factor in human airway epithelial cells. Thorax. 2005;60:144–152. doi: 10.1136/thx.2004.025692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moodie F.M., Marwick J.A., Anderson C.S., Szulakowski P., Biswas S.K., Bauter M.R., Kilty I., Rahman I. Oxidative stress and cigarette smoke alter chromatin remodeling but differentially regulate NF-kappaB activation and proinflammatory cytokine release in alveolar epithelial cells. FASEB J. 2004;18:1897–1899. doi: 10.1096/fj.04-1506fje. [DOI] [PubMed] [Google Scholar]
- Mortaz E., Masjedi M.R., Barnes P.J., Adcock I.M. Epigenetics and chromatin remodeling play a role in lung disease. Tanaffos. 2011;10:7–16. [PMC free article] [PubMed] [Google Scholar]
- Nakao A., Imamura T., Souchelnytskyi S., Kawabata M., Ishisaki A., Oeda E., Tamaki K., Hanai J., Heldin C.H., Miyazono K., et al. TGF-beta receptor-mediated signalling through Smad2, Smad3 and Smad4. EMBO J. 1997;16:5353–5362. doi: 10.1093/emboj/16.17.5353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura M., Li E. Smad2 role in mesoderm formation, left-right patterning and craniofacial development. Nature. 1998;393:786–790. doi: 10.1038/31693. [DOI] [PubMed] [Google Scholar]
- Ojiaku C.A., Cao G., Zhu W., Yoo E.J., Shumyatcher M., Himes B.E., An S.S., Panettieri R.A., Jr. TGF-beta1 evokes human airway smooth muscle cell shortening and hyperresponsiveness via Smad3. Am. J. Respir. Cell Mol. Biol. 2018;58:575–584. doi: 10.1165/rcmb.2017-0247OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.H., Gong J.H., Choi Y.J., Kang M.K., Kim Y.H., Kang Y.H. Kaempferol inhibits endoplasmic reticulum stress-associated mucus hypersecretion in airway epithelial cells and ovalbumin-sensitized mice. PLoS One. 2015;10:e0143526. doi: 10.1371/journal.pone.0143526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrais M., Pigny P., Copin M.C., Aubert J.P., Van Seuningen I. Induction of MUC2 and MUC5AC mucins by factors of the epidermal growth factor (EGF) family is mediated by EGF receptor/Ras/Raf/extracellular signal-regulated kinase cascade and Sp1. J. Biol. Chem. 2002;277:32258–32267. doi: 10.1074/jbc.M204862200. [DOI] [PubMed] [Google Scholar]
- Rose M.C., Voynow J.A. Respiratory tract mucin genes and mucin glycoproteins in health and disease. Physiol. Rev. 2006;86:245–278. doi: 10.1152/physrev.00010.2005. [DOI] [PubMed] [Google Scholar]
- Saito A., Horie M., Nagase T. TGF-beta signaling in lung health and disease. Int. J. Mol. Sci. 2018;19:2460. doi: 10.3390/ijms19082460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samsuzzaman M., Uddin M.S., Shah M., Mathew B. Natural inhibitors on airway mucin: molecular insight into the therapeutic potential targeting MUC5AC expression and production. Life Sci. 2019;231:1–14. doi: 10.1016/j.lfs.2019.05.041. [DOI] [PubMed] [Google Scholar]
- Sato R., Semba T., Kohrogi H., Saya H., Arima Y. Abstract 4192: The association between gastric Mucin 5AC (MUC5AC) expressions, transforming growth factor-β (TGF-β) signal, and histopathological phenotypes of lung cancer in xenograft models. Cancer Res. 2016;76:4192. [Google Scholar]
- Shen Y., Huang S., Kang J., Lin J., Lai K., Sun Y., Xiao W., Yang L., Yao W., Cai S., et al. Management of airway mucus hypersecretion in chronic airway inflammatory disease: Chinese expert consensus (English edition) Int. J. Chron. Obstruct. Pulmon. Dis. 2018;13:399–407. doi: 10.2147/COPD.S144312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikder M.A., Lee H.J., Mia M.Z., Park S.H., Ryu J., Kim J.H., Min S.Y., Hong J.H., Seok J.H., Lee C.J. Inhibition of TNF-α-induced MUC5AC mucin gene expression and production by wogonin through the inactivation of NF-κB signaling in airway epithelial cells. Phytother. Res. 2014;28:62–68. doi: 10.1002/ptr.4954. [DOI] [PubMed] [Google Scholar]
- Takimoto T., Wakabayashi Y., Sekiya T., Inoue N., Morita R., Ichiyama K., Takahashi R., Asakawa M., Muto G., Mori T., et al. Smad2 and Smad3 are redundantly essential for the TGF-beta-mediated regulation of regulatory T plasticity and Th1 development. J. Immunol. 2010;185:842–855. doi: 10.4049/jimmunol.0904100. [DOI] [PubMed] [Google Scholar]
- Thomson N.C., Chaudhuri R., Livingston E. Asthma and cigarette smoking. Eur. Respir. J. 2004;24:822–833. doi: 10.1183/09031936.04.00039004. [DOI] [PubMed] [Google Scholar]
- Tong J., Gu Q. Expression and clinical significance of mucin gene in chronic rhinosinusitis. Curr. Allergy Asthma Rep. 2020;20:63. doi: 10.1007/s11882-020-00958-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignola A.M., Chanez P., Chiappara G., Merendino A., Pace E., Rizzo A., la Rocca A.M., Bellia V., Bonsignore G., Bousquet J. Transforming growth factor-beta expression in mucosal biopsies in asthma and chronic bronchitis. Am. J. Respir. Crit. Care Med. 1997;156:591–599. doi: 10.1164/ajrccm.156.2.9609066. [DOI] [PubMed] [Google Scholar]
- Voynow J.A., Rubin B.K. Mucins, mucus, and sputum. Chest. 2009;135:505–512. doi: 10.1378/chest.08-0412. [DOI] [PubMed] [Google Scholar]
- Wrana J.L., Attisano L., Wieser R., Ventura F., Massagué J. Mechanism of activation of the TGF-β receptor. Nature. 1994;370:341–347. doi: 10.1038/370341a0. [DOI] [PubMed] [Google Scholar]
- Wrzesinski S.H., Wan Y.Y., Flavell R.A. Transforming growth factor-beta and the immune response: implications for anticancer therapy. Clin. Cancer Res. 2007;13:5262–5270. doi: 10.1158/1078-0432.CCR-07-1157. [DOI] [PubMed] [Google Scholar]
- Yang S.R., Chida A.S., Bauter M.R., Shafiq N., Seweryniak K., Maggirwar S.B., Kilty I., Rahman I. Cigarette smoke induces proinflammatory cytokine release by activation of NF-kappaB and posttranslational modifications of histone deacetylase in macrophages. Am. J. Physiol. Lung Cell. Mol. Physiol. 2006;291:L46–L57. doi: 10.1152/ajplung.00241.2005. [DOI] [PubMed] [Google Scholar]
- Zen Y., Harada K., Sasaki M., Tsuneyama K., Katayanagi K., Yamamoto Y., Nakanuma Y. Lipopolysaccharide induces overexpression of MUC2 and MUC5AC in cultured biliary epithelial cells: possible key phenomenon of hepatolithiasis. Am. J. Pathol. 2002;161:1475–1484. doi: 10.1016/S0002-9440(10)64423-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwinderman M.R.H., de Weerd S., Dekker F.J. Targeting HDAC complexes in asthma and COPD. Epigenomes. 2019;3:19. doi: 10.3390/epigenomes3030019. [DOI] [PMC free article] [PubMed] [Google Scholar]