Abstract
The nuclear receptor peroxisome proliferator-activated receptor γ (PPARγ) is the master transcriptional regulator in adipogenesis. PPARγ forms a heterodimer with another nuclear receptor, retinoid X receptor (RXR), to form an active transcriptional complex, and their transcriptional activity is tightly regulated by the association with either coactivators or corepressors. In this study, we identified T-cell death-associated gene 51 (TDAG51) as a novel corepressor of PPARγ-mediated transcriptional regulation. We showed that TDAG51 expression is abundantly maintained in the early stage of adipogenic differentiation. Forced expression of TDAG51 inhibited adipocyte differentiation in 3T3-L1 cells. We found that TDAG51 physically interacts with PPARγ in a ligand-independent manner. In deletion mutant analyses, large portions of the TDAG51 domains, including the pleckstrin homology-like, glutamine repeat and proline-glutamine repeat domains but not the proline-histidine repeat domain, are involved in the interaction with the region between residues 140 and 506, including the DNA binding domain, hinge, ligand binding domain and activation function-2 domain, in PPARγ. The heterodimer formation of PPARγ-RXRα was competitively inhibited in a ligand-independent manner by TDAG51 binding to PPARγ. Thus, our data suggest that TDAG51, which could determine adipogenic cell fate, acts as a novel negative regulator of PPARγ by blocking RXRα recruitment to the PPARγ-RXRα heterodimer complex in adipogenesis.
Keywords: adipocyte differentiation, adipogenesis, pleckstrin homology-like domain A family, peroxisome proliferator-activated receptor gamma, retinoid X receptor alpha, T-cell death-associated gene 51
INTRODUCTION
Adipogenesis is the process of mature adipocyte formation from adipocyte precursors, and adipocytes play a vital role in lipid metabolism, energy homeostasis and insulin sensitivity (Sarjeant and Stephens, 2012). Adipogenesis is mainly induced by the coordinated interplay among transcriptional factors, such as CCAAT/enhancer binding proteins (C/EBPs)and peroxisome proliferator-activated receptor γ (PPARγ) (Rosen et al., 2002; 2009).
C/EBPs, such as C/EBPα, C/EBPβ, and C/EBPδ, are members of the basic leucine zipper transcription factor family (Rosen et al., 2009). C/EBPβ and C/EBPδ are expressed at the early stage of adipocyte differentiation, but their expression is not sufficient for complete adipogenic differentiation (Rosen et al., 2002; 2009; Tanaka et al., 1997). C/EBPβ and C/EBPδ are required for the early stage of adipogenic differentiation by inducing C/EBPα and PPARγ expression (Rosen et al., 2002; 2009). The expression of C/EBPα is enhanced late in adipogenesis, and C/EBPα promotes adipogenesis by inducing PPARγ expression through direct binding of C/EBP recognition elements on the PPARγ promoter (Elberg et al., 2000; Wu et al., 1996). Consequently, C/EBPs are considered to be important adipogenic transcription factors together with PPARγ in adipogenesis. Interestingly, ectopic expression of PPARγ in C/EBPα-deficient fibroblasts was shown to induce adipocyte differentiation, whereas C/EBPα expression in PPARγ-deficient fibroblasts was not observed (Rosen et al., 2002; Wu et al., 1999). Thus, PPARγ, a member of the nuclear receptor gene family, acts as the master transcriptional regulator in adipogenesis (Lefterova and Lazar, 2009).
PPARγ is expressed as two major different isoforms, PPARγ1 and PPARγ2, as a result of alternative promoter usage; PPARγ2 has an extra 30 residues in its N-terminal end (Kroker and Bruning, 2015; Tontonoz et al., 1994). PPARγ2 is predominantly expressed in adipose tissue, where it regulates adipogenesis, while PPARγ1 is broadly expressed in adipose tissue, colon, retina, liver, spleen and cardiac muscles (Saladin et al., 1999). Upon PPARγ activation by PPARγ ligand binding, PPARγ forms a heterodimer with another nuclear receptor, such as retinoid X receptors (RXRs) (Chandra et al., 2008; Tontonoz et al., 1994). The formation of PPARγ-RXRα heterodimers is crucial to induce the expression of adipogenic markers, such as adipocyte fatty acid-binding protein 2 (aP2) and adiponectin, through direct binding to PPAR responsive elements (PPREs) in their promoters during adipogenesis (Juge-Aubry et al., 1997; Lefterova and Lazar, 2009). Interestingly, the transcriptional activity of PPARγ-RXRα heterodimers is tightly regulated by the association of either coactivators or corepressors in many different tissues and cell types (Lefterova and Lazar, 2009). Thus, elucidating a functional role of the novel PPARγ regulator may improve our understanding of the molecular regulatory mechanism of PPARγ in various tissues and cells and in their relevance to human metabolic diseases, such as obesity and diabetes.
T-cell death-associated gene 51 (TDAG51), also known as the pleckstrin homology-like (PHL) domain A family 1 (PHLDA1) in humans, possesses an N-terminal PHL domain and a glutamine repeat (QQ) domain (Park et al., 1996). In the C-terminus, TDAG51 contains a proline-glutamine repeat (PQ) domain and a proline-histidine repeat (PH) domain (Park et al., 1996; 2013). In eukaryotic systems, PHL domains are mainly involved in protein-protein interactions, and the QQ, PQ, and PH domains are predominantly present in transcriptional activators (Gehring et al., 1994; Scheffzek and Welti, 2012; Xiao and Jeang, 1998). Thus, TDAG51 was proposed to be a putative transcriptional regulator involved in many biological processes, such as cell proliferation, migration, apoptosis and differentiation (Chen et al., 2018; Nagai, 2016; Park et al., 1996). TDAG51 expression is modulated by various cellular stimuli, such as heat shock, oxidative stress, inflammation, tumor induction and endoplasmic reticulum stress (Hayashida et al., 2006; Hossain et al., 2013; Jiao et al., 2016; Nagai, 2016; Park et al., 2013). In addition, microarray analysis showed that TDAG51 expression is only maintained at the early stage of adipogenic lineage and is rapidly decreased during the process of adipogenesis (Burton et al., 2004). Moreover, TDAG51-deficient (TDAG51-/-) mice showed mature-onset obesity, hepatic steatosis and insulin resistance by regulating lipogenesis (Basseri et al., 2013). Thus, TDAG51 may negatively contribute to the regulation of adipogenesis. However, the exact role of TDAG51 in adipogenesis remains unclear.
In this study, we identified TDAG51 as a novel PPARγ regulator. Adipogenesis is reduced by TDAG51 expression. The heterodimer formation of PPARγ-RXRα is competitively inhibited by TDAG51 binding to PPARγ. Thus, our data suggest that TDAG51 is a crucial negative regulator of PPARγ-induced adipogenesis.
MATERIALS AND METHODS
Antibodies and cells
Specific antibodies were purchased from the following commercial sources: anti-Flag, anti-Myc, and anti-β-Actin from Sigma-Aldrich (USA); anti-Xpress (Xp) from Invitrogen (USA); and anti-GST, anti-TDAG51, and anti-PPARγ antibodies from Santa Cruz Biotechnology (USA). Murine preadipocyte 3T3-L1 and human embryonic kidney 293T and PlatE cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Welgene, Korea) supplemented with 10% fetal bovine serum (FBS; Invitrogen) and 1× antibiotic/antimycotic solution (Welgene).
Plasmids
The luciferase reporter harboring the TDAG51 promoter fragments containing nt –2,450 to +551 (PT51-3K-Luc) was described previously (Park et al., 2013). For the construction of aP2 luciferase reporter (aP2-Luc), the aP2 promoter fragments from nt –5,379 to +21 were amplified by polymerase chain reaction (PCR) from the murine genomic DNA and subcloned into the pGL3-basic vector (Promega, USA). The eukaryotic expression plasmid of Flag-tagged TDAG51 (Flag-TDAG51) was described previously (Park et al., 2013). Flag-tagged RXRα (Flag-RXRα) was kindly provided by Yong-Ho Ahn (Yonsei University College of Medicine, Korea) (Kim et al., 2009). The epitope-tagged (Flag-, Myc-, Xp-, or GST-tagged) eukaryotic expression plasmids for TDAG51, PPARγ, RXRα, and their corresponding deletion mutants were generated by PCR amplification and subcloned into pFLAG-CMV2 (Sigma-Aldrich), pcDNA3.1/myc-His/lacZ (Invitrogen), pcDNA3.1-His (Invitrogen), pEBG (Park et al., 2015), and pMXs-puro (Park et al., 2015).
Adipogenic differentiation and analysis
Differentiation of adipocytes was performed as described previously (Basseri et al., 2013). Briefly, 3T3-L1 preadipocytes (1 × 105 cells/ml) were cultured in 10% FBS/DMEM supplemented with differential cocktail (1 µM dexamethasone, 0.5 mM 3-isobutyl-1-methylxanthine, 100 µM indomethacin, and 10 µg/ml insulin [Sigma-Aldrich]) for 6 days. For the ectopic expression of TDAG51, retroviral supernatants harboring the Flag-tagged TDAG51 transgene were generated from PlatE cells as described previously (Yu et al., 2016). 3T3-L1 cells (1 × 105 cells/ml) were infected by retroviral supernatants with 8 µg/ml polybrene for 6 h. At 24 h postinfection, infected 3T3-L1 cells were selected by puromycin (2 µg/ml) for 2 days. After puromycin selection, 3T3-L1 cells were seeded at a density of 1 × 105 cells/ml in adipocyte differentiation media and differentiated into mature adipocytes for 6 days. Finally, adipocyte differentiation was analyzed by oil red O staining and solution assays as described previously (Basseri et al., 2013; Hossain et al., 2013).
Real-time PCR analysis
Real-time PCR was performed as described previously (Park et al., 2015). Briefly, differentiated 3T3-L1 cells were harvested, and total RNA was isolated using TRI reagent (MRC, USA) according to the manufacturers’ instructions. Reverse transcription was performed with 1 µg of total RNA and M-MLV reverse transcriptase (USB, USA) for 1 h at 42°C. The synthesized cDNAs were then subjected to real-time PCR analysis using a CFX Connect Real-time PCR Detection System (Bio-Rad, USA) with the appropriate primers: PPARγ, 5'-ACC AGG GAG TTC CTC AAA AG-3' and 5'-TTG TCT TGG ATG TCC TCG AT-3'; adiponectin, 5'-ACT CCT GGA GAG AAG GGA GA-3' and 5'-GAA TGG GTA CAT TGG GAA CA-3'; and β-actin, 5'-ATG AAG ATC CTC CTG ACC GAG CG-3' and 5'-TAC TTG CGC TGA GGA GC-3'. β-Actin was used as an internal normalization control.
Luciferase reporter assay
A luciferase reporter assay was performed as described previously (Kim et al., 2019; Yu et al., 2016). Briefly, 293T (3.5 × 105 cells/ml) or 3T3-L1 (1 × 105 cells/ml) cells cultured in 24-well plates were cotransfected with reporters and an appropriate combination of epitope-tagged eukaryotic expression plasmids for TDAG51, PPARγ or RXRα in triplicate using TurboFect reagent (Fermentas, USA) according to the manufacturers’ instructions. Plasmids PT51-3K-Luc (0.1 µg), PPARγ-Luc (0.1 µg), aP2-Luc (0.1 µg), and pcDNA3.1/His/LacZ (0.1 µg, Invitrogen) were used as reporters. The reporters were cotransfected with the epitope-tagged expression plasmids (0.1-0.5 µg). The β-galactosidase activity derived from the expression of pcDNA3.1/His/LacZ was used as an internal normalization control for transfection. For the reporter assay in 3T3-L1 cells, PT51-3K-Luc-transfected 3T3-L1 cells were differentiated with adipocyte differentiation media for 2 days. For the treatment of rosiglitazone, the transfected cells were treated with 5 µM rosiglitazone for 36 h, as described previously (Dowell et al., 2003). At 24 h posttransfection, reporter activities were measured using a luciferase assay kit (Promega) and β-galactosidase assay kit (Applied Biosystems, USA) according to the manufacturers’ instructions.
Protein interaction and immunoblotting
Protein-protein interaction and immunoprecipitation (IP) assays were performed as described previously (Yu et al., 2016). Briefly, 293T cells seeded at 3.5 × 105 cells/ml in 6-well plates were cotransfected with an appropriate combination of epitope-tagged eukaryotic expression plasmids for TDAG51 (0.2-2.5 µg), PPARγ (1.5 µg), or RXRα (0.25-1.0 µg). For the treatment of rosiglitazone, the transfected cells were treated with 5 µM rosiglitazone for 36 h as described previously (Dowell et al., 2003). At 24 h posttransfection, the cells were lysed in cold lysis buffer (250 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1 mM EDTA, 1 mM NaF, 1 mM sodium orthovanadate, 1 mM phenylmethylsulfonyl fluoride, 5% glycerol, and 0.5% Triton X-100). The lysates were pulled down (PD) by glutathione S-transferase (GST) beads or immunoprecipitated by anti-Flag beads. The bead-bound proteins were washed three times with lysis buffer and finally analyzed by immunoblotting (IB) with antibodies against epitopes, as described previously (Park et al., 2015; Son et al., 2020). For the endogenous protein-protein interaction assay, 3T3-L1 cells seeded at 1 × 105 cells/ml were cultured with adipocyte differentiation media for 2 days. The differentiated cells were lysed in cold lysis buffer, and the lysates were incubated with anti-TDAG51 or anti-PPARγ antibody overnight at 4°C with rocking. Then, the lysates were precipitated with protein G beads (50% slurry) for 2 h at 4°C. The beads were washed three times with lysis buffer, and the interacting proteins were analyzed by IB with antibodies against anti-TDAG51 or anti-PPARγ antibody.
Statistical analysis
Student’s t-test was used to determine the significance of differences between experimental samples (n = 3 per group) using IBM SPSS Statistics (ver. 24.0; IBM, USA). The data represent the mean ± SD. P values < 0.05 were considered statistically significant.
RESULTS
TDAG51 is negatively involved in the regulation of PPARγ- induced adipocyte formation
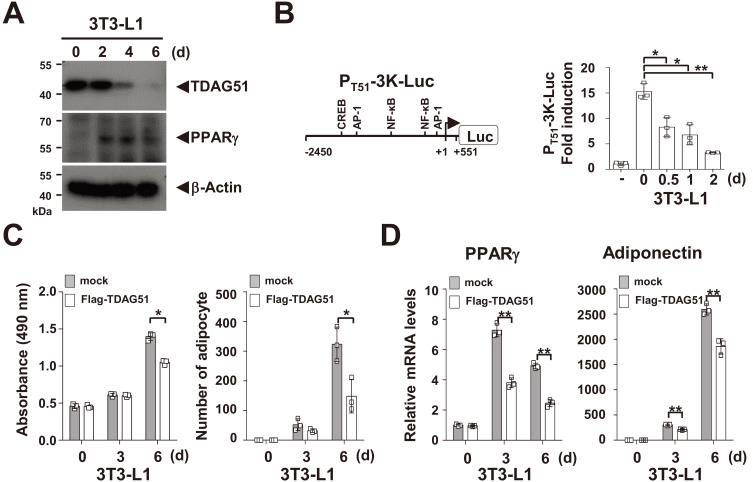
Because TDAG51 expression is decreased in adipogenesis, as revealed by a microarray study (Burton et al., 2004), we determined whether the expression of TDAG51 is downregulated in adipogenesis using 3T3-L1 preadipocytes. We observed that TDAG51 expression was abundantly maintained at day 0 and was then rapidly reduced after day 2 of adipogenic differentiation, while PPARγ expression was induced after day 2 (Fig. 1A). In a TDAG51 reporter assay, TDAG51 promoter activities were reduced (41.6%-79.3% reduction, P < 0.05 or P < 0.01) in a differentiation-dependent manner in 3T3-L1 cells (Fig. 1B). In addition, adipocyte formation was significantly inhibited (25.0% or 45.9% reduction at day 6, P < 0.05) by the forced expression of TDAG51 in 3T3-L1 preadipocytes (Fig. 1C). Consistent with these results, we observed that the mRNA levels of the adipogenic markers PPARγ and adiponectin were inversely correlated with the levels of TDAG51 expression during adipogenic differentiation in 3T3-L1 preadipocytes (Fig. 1D). These results indicate that the downregulation of TDAG51 expression during adipogenesis is crucial for adipocyte formation and adipogenic marker expression.
Fig. 1. TDAG51 inhibits adipogenesis.
(A) Downregulation of TDAG51 expression in 3T3-L1 preadipocyte differentiation. The 3T3-L1 preadipocytes were cultured in adipocyte differentiation media for 6 days (d). The expressed proteins were analyzed by immunoblotting against anti-PPARγ or anti-TDAG51 antibody indicated by an arrow. β-Actin was used as the loading control. (B) Downregulation of TDAG51 transcription in 3T3-L1 preadipocytes. A schematic diagram of the luciferase reporter of the TDAG51 promoter (PT51-3K-Luc) is shown in the left panel. The putative transcription factor binding sites are illustrated, and the nucleotide positions were numbered based on the transcriptional start site. NF-κB, nuclear factor kappa B; CREB, cAMP response element-binding protein; AP-1, activator protein 1. TDAG51 promoter activities during 3T3-L1 preadipocyte differentiation were analyzed in PT51-3K-Luc-transfected 3T3-L1 preadipocytes using reporter assay kits (right panel). (C) Inhibition of 3T3-L1 preadipocyte differentiation by TDAG51 expression. The Flag-tagged TDAG51-transduced 3T3-L1 preadipocytes were cultured in adipocyte differentiation media for 6 days. The differentiated cells were stained with oil red O. The oil red O levels in the differentiated cells were monitored at 490 nm (left panel). The number of stained cells was counted (right panel). Mock, gray box; Flag-TDAG51 expression, open box. (D) Downregulation of adipogenic markers in 3T3-L1 preadipocyte differentiation by TDAG51 expression. The expression levels of the adipogenic markers PPARγ and adiponectin were analyzed by real-time PCR. *P < 0.05; **P < 0.01.
As shown in Fig. 1, we observed that the mRNA levels of PPARγ were downregulated by forced TDAG51 expression. Furthermore, Basseri et al. (2013) demonstrated that TDAG51-/- mice exhibited greater lipogenic potential and mature-onset obesity. Thus, we postulated that TDAG51 is negatively involved in the regulation of the master transcription factor PPARγ in adipogenesis. To address this possibility, we constructed a luciferase reporter of the PPARγ2 promoter (PPARγ-Luc) harboring two putative PPREs in the promoter fragment from nt –2,983 to +68, and the functionality of these putative PPREs was confirmed by site-directed mutagenesis and reporter assays (Supplementary Fig. S1). We next analyzed PPARγ-induced PPARγ-Luc activation by forced TDAG51 expression using reporter assays. Our results showed that PPARγ-induced PPARγ-Luc activation was dose-dependently inhibited (13.6%-29.0% reduction, P < 0.01 or P < 0.001) by forced TDAG51 expression (Supplementary Fig. S2A). Contrary to the TDAG51 inhibitory effect on PPARγ promoter activity, however, TDAG51 promoter activities were not inhibited by the forced expression of PPARγ (Supplementary Fig. S2B). Taken together, these results indicate that TDAG51 expression is negatively involved in the regulation of PPARγ-induced transcriptional activation.
TDAG51 interacts with PPARγ
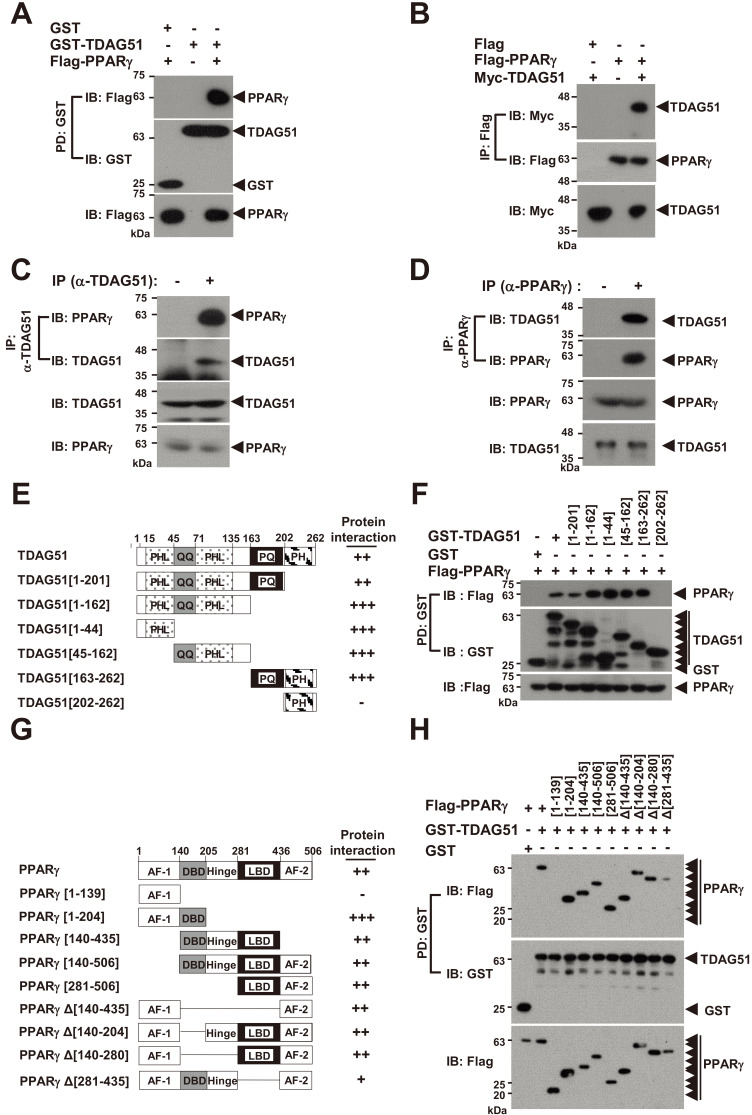
We next analyzed whether TDAG51 physically interacts with PPARγ. In the GST bead pulldown (PD) assay with GST-tagged TDAG51 (GST-TDAG51), we observed an interaction between TDAG51 and PPARγ (Fig. 2A). Consistent with these results, the anti-Flag IP assay with Flag-tagged PPARγ (Flag-PPARγ) revealed that Flag-PPARγ interacted with Myc-TDAG51 (Fig. 2B). To further confirm this observation, we examined their endogenous interaction using an IP assay in 3T3-L1 cells. Endogenous TDAG51 was immunoprecipitated by anti-TDAG51 antibodies, and the bound proteins were analyzed by IB with anti-PPARγ antibodies. Our results showed that endogenous TDAG51 was associated with endogenous PPARγ in 3T3-L1 cells (Fig. 2C). In an IP assay using anti-PPARγ antibodies, we also observed that endogenous PPARγ was associated with endogenous TDAG51 in 3T3-L1 cells (Fig. 2D). We next constructed deletion mutants of TDAG51, as shown in Fig. 2E, and analyzed particular domains involved in interacting with PPARγg using a GST-PD assay. All TDAG51 deletion mutants, except for TDAG51 [202-262] containing the PH domain only, could interact with PPARγ, indicating that the PHL, QQ, and PQ domains in TDAG51 are involved in the PPARγ interaction (Fig. 2F). To determine the precise domain of PPARγ that interacts with TDAG51, we constructed expression plasmids for PPARγ deletion mutants lacking the N-terminal, C-terminal, and internal regions (Fig. 2G). GST-PD assays for the N- and C-terminal deletion mutants of PPARγ revealed that only the PPARγ [1-139] mutant was not bound to TDAG51, indicating that the activation function-1 (AF-1) domain in PPARγ is not involved in the TDAG51 interaction (Fig. 2H). In the GST-PD assay with the internal deletion mutants, all PPARγ internal deletion mutants could interact with TDAG51, although the PPARγ Δ[281-435] mutant lacking the ligand binding domain (LBD) showed a markedly reduced interaction with TDAG51 (Fig. 2H). Collectively, we observed that the region between residues 140 and 506, including the DNA binding domain (DBD), hinge, LBD, and AF-2 domain, in PPARγ is required for the TDAG51 interaction (Fig. 2H). Taken together, these results indicate that TDAG51 is a binding partner of PPARγ.
Fig. 2. TDAG51 interacts with PPARγ.
(A) TDAG51 binds to PPARγ. 293T cells were transfected with GST-TDAG51 (1.5 µg) with or without Flag-PPARγ (2.5 µg). GST-TDAG51 was pulled down (PD) using GST beads. The bound proteins were visualized via anti-Flag (top) or anti-GST (middle) immunoblotting (IB). The level of PPARγ expression in whole cell lysates was detected via anti-Flag IB (bottom). GST alone (mock) was used as a control. Protein expression is indicated by an arrow. (B) PPARγ binds to TDAG51. 293T cells were transfected with Flag-PPARγ (1.5 µg) with or without Myc-TDAG51 (2.5 µg). The bound proteins by anti-Flag IP were visualized via anti-Myc (top) or anti-Flag IB (middle). The level of TDAG51 expression in whole cell lysates was detected via anti-Myc IB (bottom). Flag alone (mock) was used as a control. (C) Identification of the endogenous TDAG51-PPARγ interaction. Endogenous TDAG51 was immunoprecipitated (IP) with an anti-(α-)TDAG51 antibody and visualized via anti-TDAG51 IB (second). Proteins bound to TDAG51 were visualized via anti-PPARγ IB (top). The levels of endogenous TDAG51 and PPARγ in whole cell lysates were detected via anti-TDAG51 (third) and anti-PPARγ IB (bottom), respectively. (D) Identification of the endogenous PPARγ-TDAG51 interaction. Endogenous PPARγ was immunoprecipitated with an anti-(α-)PPARγ antibody and visualized via anti-PPARγ IB (second). Proteins bound to PPARγ were visualized via anti-TDAG51 IB (top). The levels of endogenous PPARγ and TDAG51 in whole cell lysates were detected via anti-PPARγ (third) and anti-TDAG51 IB (bottom), respectively. (E) Schematic diagram of TDAG51 mutants. PHL, pleckstrin homology-like domain; QQ, glutamine repeat; PQ, proline-glutamine repeat; PH, proline-histidine repeat. The number of amino acid residues is shown. The interactions of deletion mutants are summarized at the right panel. (F) Analysis of TDAG51 deletion mutants interacting with PPARγ. The bound proteins by GST-PD were visualized via anti-Flag (top) or anti-GST IB (middle). The level of PPARγ expression in whole cell lysates was detected via anti-Flag IB (bottom). (G) Schematic diagram of PPARγ mutants. AF-1, activation function-1 domain; DBD, DNA binding domain; Hinge, hinge domain; AF-2, activation function-2 domain; LBD, ligand binding domain. The interactions of deletion mutants are summarized at the right panel. (H) Analysis of PPARγ deletion mutants interacting with TDAG51. The bound proteins by GST-PD were visualized via anti-Flag (top) or anti-GST IB (middle). The PPARγ expression level in whole cell lysates was detected via anti-Flag IB (bottom).
The interaction of TDAG51 and PPARγ inhibits PPARγ-induced aP2 promoter activity
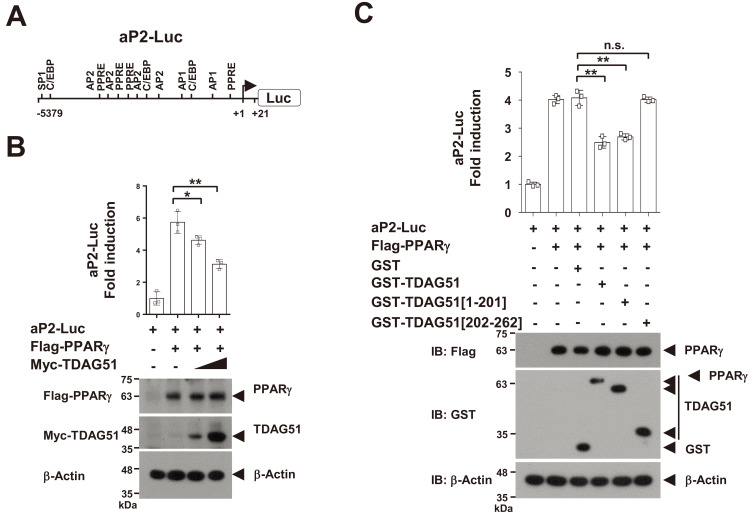
PPARγ is a crucial transcriptional regulator of the induction of adipogenic markers, such as aP2 and adiponectin (Juge-Aubry et al., 1997; Lefterova and Lazar, 2009). Thus, we investigated whether the interaction of TDAG51-PPARγ could regulate PPARγ-induced aP2 transcriptional activation using a luciferase reporter of the aP2 promoter (aP2-Luc) harboring transcriptional regulatory elements, such as PPREs and C/EBPs, in the promoter fragment from nt –5,379 to +21 (Fig. 3A). In a PPARγ-induced aP2-Luc reporter assay, PPARγ-induced aP2-Luc activities were inhibited (19.5%-45.5% reduction, P < 0.05 or P < 0.01) in a dose-dependent manner by forced TDAG51 expression (Fig. 3B). Consistent with the results shown in Fig. 2F, the inhibitory effect of TDAG51 in the PPARγ-induced aP2-Luc reporter assay was not observed by the PPARγ-nonbinding mutant of TDAG51 (GST-TDAG51[202-262]), while the wild-type TDAG51 (GST-TDAG51) or its PPARγ-binding mutant (GST-TDAG51[1-201]) showed inhibitory effects (39.0% or 34.1% reduction, P < 0.01) (Fig. 3C). Taken together, these results indicate that the binding of TDAG51 to PPARγ is negatively involved in the regulation of PPARγ-induced adipogenic marker expression.
Fig. 3. TDAG51 inhibits PPARγ-induced aP2 promoter activity.
(A) Schematic diagram of the aP2 promoter (aP2-Luc). The putative transcription factor binding sites are illustrated, and the nucleotide positions are numbered based on the transcriptional start site. PPRE, PPARγ responsive element; C/EBP, CCAAT/enhancer binding protein; SP1, specificity protein 1; AP-2, activator protein 2. (B) The inhibitory effect of TDAG51 on PPARγ-induced aP2-Luc reporter activity. Reporter plasmids (aP2-Luc [0.1 µg] and pcDNA3.1/His/LacZ [0.1 µg]) were cotransfected with epitope-tagged expression plasmids (Flag-PPARγ [0.3 µg] and/or Myc-TDAG51 [0.2 and 0.5 µg]) into 293T cells. The expression of transfected plasmids was analyzed by immunoblotting against anti-epitope antibodies. β-Actin was used as the loading control. Protein expression is indicated by an arrow. (C) The inhibitory effects of TDAG51 deletion mutants on PPARγ-induced aP2-Luc reporter activity. Reporter plasmids were cotransfected with epitope-tagged expression plasmids (Flag-PPARγ [0.3 µg] and/or GST-TDAG51 [0.5 µg]) into 293T cells. GST alone (mock) was used as a control. *P < 0.05; **P < 0.01; n.s., not significant.
TDAG51 binding to PPARγ inhibits the formation of PPARγ-RXRα heterodimers
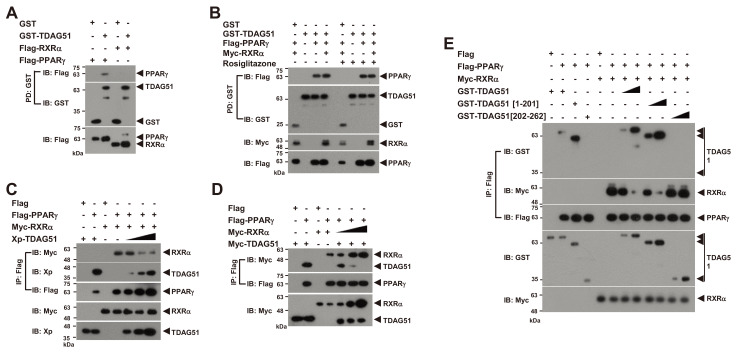
The formation of PPARγ-RXRα heterodimers by PPARγ ligand stimulation is crucial to induce the expression of adipogenic markers through binding to PPREs in their promotors in adipogenesis (Chandra et al., 2008; Lefterova and Lazar, 2009; Tontonoz et al., 1994). Thus, we examined whether TDAG51 acts as a corepressor of the PPARγ-RXRα heterodimer. In a GST-PD assay, we observed a direct interaction of TDAG51 with PPARγ but not with RXRα (Fig. 4A). Moreover, the interaction of TDAG51 and PPARγ was not blocked by treatment with the PPARγ ligand rosiglitazone (Fig. 4B). Thus, TDAG51 binding to PPARγ per se may negatively affect the formation of PPARγ-RXRα heterodimers. To address this possibility, we performed competitive IP assays with Flag-PPARγ by dose-dependent expression of TDAG51 or RXRα (Figs. 4C-4E). Our results showed that the formation of PPARγ-RXRα heterodimers was inhibited in a dose-dependent manner by forced TDAG51 expression (Fig. 4C), whereas the interaction between PPARγ and TDAG51 was dose-dependently reduced by forced RXRα expression (Fig. 4D). Consistent with the results shown in Figs. 2F and 3C, in IP assays with TDAG51 deletion mutants, the formation of the PPARγ-RXRα heterodimer was not inhibited by the forced expression of the PPARγ- non-binding mutant (GST-TDAG51[202-262]), while wild-type TDAG51 (GST-TDAG51) or its PPARγ-binding mutant (GST-TDAG51[1-201]) showed competitive inhibitory effects on the formation of the PPARγ-RXRα heterodimer (Fig. 4E). Taken together, these results indicate that TDAG51 inhibits the formation of PPARγ-RXRα heterodimers through direct interaction with PPARγ.
Fig. 4. The interaction of TDAG51 and PPARγ inhibits the formation of the PPARγ-RXRα heterodimer.
(A) No interaction between TDAG51 and RXRα. First, 293T cells were cotransfected with GST-TDAG51 (1.5 µg) with or without Flag-PPARγ (1.5 µg) or Flag-RXRα (0.1 µg). The bound proteins by GST-PD were visualized via anti-Flag (top) or anti-GST IB (middle). The PPARγ and RXRα expression levels in whole cell lysates were detected via anti-Flag IB (bottom). GST alone (mock) was used as a control. Protein expression is indicated by an arrow. (B) No effect of rosiglitazone treatment on the TDAG51-PPARγ interaction. The 293T cells cotransfected with epitope-tagged expression plasmids were treated with 5 μM rosiglitazone for 36 h. The protein interaction was monitored by GST-PD and IB analysis. (C) The inhibitory effect of TDAG51 on the formation of PPARγ-RXRα heterodimers. The Xp-tagged TDAG51 plasmid (Xp-TDAG51 [0.2, 0.5, and 2.0 µg]) was dose-dependently cotransfected with Flag-PPARγ (1.5 µg) and Myc-RXRα (0.1 µg) expression plasmids into 293T cells. The protein interactions were monitored by anti-Flag IP and IB analysis (top three panels). The TDAG51 and RXRα expression levels in whole cell lysates were detected via anti-Xp and anti-Myc IB, respectively (bottom two panels). (D) The inhibitory effect of RXRα on the interaction of PPARγ and TDAG51. The Myc-tagged RXRα plasmid (Myc-RXRα [0.25, 0.5, and 1.0 µg]) was dose-dependently cotransfected with Flag-PPARγ (1.5 µg) and Myc-TDAG51 (1.5 µg) expression plasmids into 293T cells. (E) The inhibitory effects of TDAG51 deletion mutants on the formation of PPARγ-RXRα heterodimers. The GST-tagged TDAG51 deletion mutant plasmids (0.5 and 2.0 µg) were dose-dependently cotransfected with Flag-PPARγ (1.5 µg) and Myc-RXRα (0.1 µg) expression plasmids into 293T cells.
TDAG51 inhibits aP2 promoter activity induced by the PPARγ-RXRα heterodimer
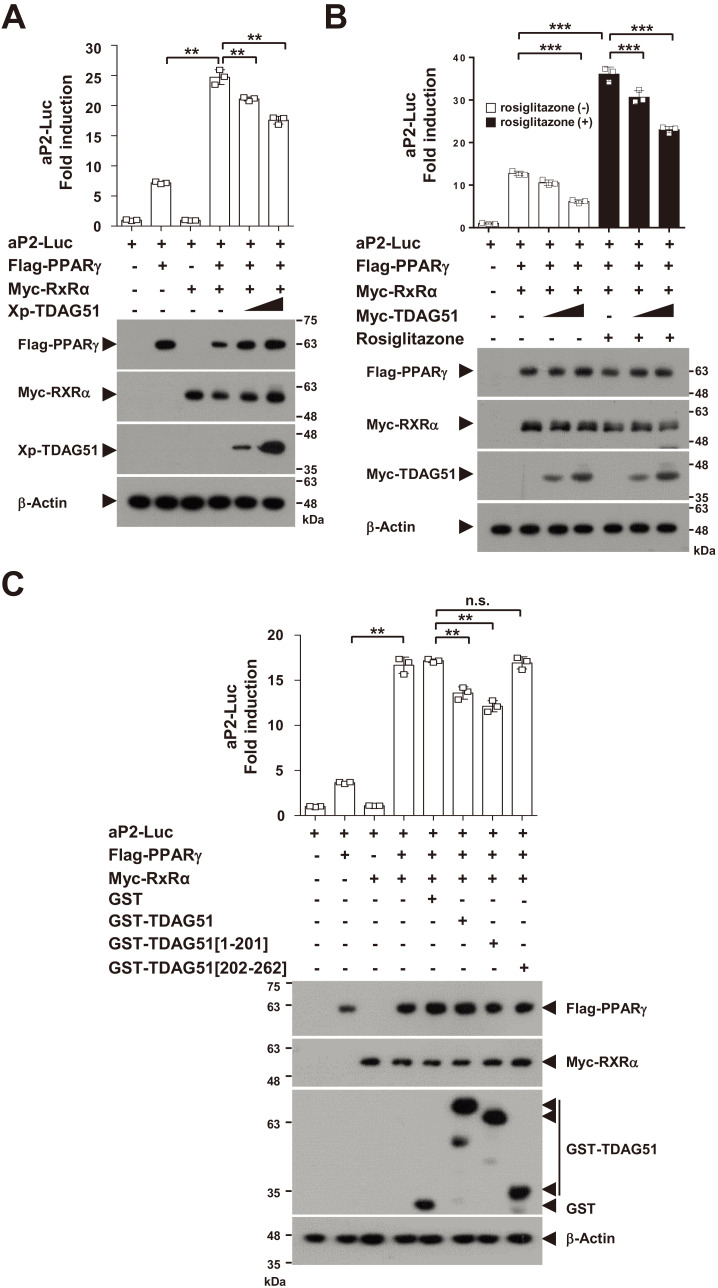
We next examined whether TDAG51 could inhibit aP2 promoter activity induced by the PPARγ-RXRα heterodimer using an aP2-Luc reporter assay (Fig. 5A). In the aP2-Luc reporter assay, we observed enhanced promoter activities of PPARγ-induced aP2-Luc (3.4-fold induction, P< 0.01) by coexpressing RXRα compared to PPARγ alone (Fig. 5A). These enhanced aP2 promoter activities induced by the PPARγ-RXRα heterodimer were dose-dependently inhibited (14.6%-28.9% reduction, P < 0.01) by forced TDAG51 expression (Fig. 5A). Consistent with the results shown in Fig. 4B, treatment with rosiglitazone did not affect the inhibitory effect of TDAG51 on the activation of PPARγ-induced aP2 promoter activity (Fig. 5B). Finally, we analyzed the inhibitory effects of TDAG51 using TDAG51 deletion mutants. Similar to the results shown in Figs. 2F, 3C, and 4E, the inhibitory effects of TDAG51 on the activation of aP2 promoter activity induced by the PPARγ-RXRα heterodimer were observed (20.8% or 29.4% reduction, P < 0.01) with wild type or PPARγ-binding mutant (GST-TDAG51[1-201]) expression but not with the PPARγ-nonbinding mutant (GST-TDAG51[202-262]) (Fig. 5C). Taken together, these results indicate that TDAG51 functions as a corepressor of PPARγ in adipogenic marker expression by inhibiting the transcriptional activity of the PPARγ-RXRα heterodimer.
Fig. 5. TDAG51 inhibits aP2 promoter activity induced by the PPARγ-RXRα heterodimer.
(A) The inhibitory effects of TDAG51 on the promoter activity of aP2-Luc induced by PPARγ-RXRα coexpression. Reporter plasmids (aP2-Luc [0.1 µg] and pcDNA3.1/His/LacZ [0.1 µg]) were cotransfected with epitope-tagged expression plasmids (Flag-PPARγ [0.25 µg], Myc-RXRα [0.1 µg], and/or Xp-TDAG51 [0.25-0.5 µg]) into 293T cells. The expression of transfected plasmids was analyzed by immunoblotting against anti-epitope antibodies. β-Actin was used as the loading control. (B) The inhibitory effects of TDAG51 on the promoter activity of aP2-Luc induced by PPARγ expression with rosiglitazone treatment. (C) The inhibitory effects of TDAG51 deletion mutants on the promoter activity of aP2-Luc induced by PPARγ-RXRα coexpression. Reporter plasmids were cotransfected with epitope-tagged expression plasmids (Flag-PPARγ [0.25 µg], Myc-RXRα [0.1 µg], and/or GST-TDAG51 [0.5 µg]) into 293T cells. GST alone (mock) was used as a control. **P < 0.01; ***P < 0.001; n.s., not significant.
DISCUSSION
PPARγ plays a central role in regulating the expression of a wide range of genes involved in adipogenesis, lipid metabolism, insulin sensitivity, energy storage, inflammation and differentiation (Kroker and Bruning, 2015; Lefterova and Lazar, 2009). In particular, the crucial role of PPARγ in adipogenesis has been extensively studied by many researchers (Lefterova and Lazar, 2009). Although PPARγ functions as the master transcriptional regulator of adipocyte differentiation, few genes have been validated as direct PPARγ regulators or targets (Lefterova and Lazar, 2009). Thus, the identification of novel regulators of PPARγ is important for understanding the transcriptional regulatory mechanism of PPARγ in various cell types or tissues. PPARγ contains AF-1 and DBD in the N-terminal region, the flexible hinge domain in the middle, and LBD and AF-2 in the C-terminal region (Kroker and Bruning, 2015). Upon the binding of the PPARγ ligand, PPARγ forms a heterodimer with RXRα through their LBD domain in the C-terminal region (Chandra et al., 2008). In this study, we identified TDAG51 as a novel negative regulator of PPARγ that blocks PPARγ-RXRα heterodimer formation in adipogenesis. We showed that the region between residues 140 and 506, including the DBD, hinge, LBD and AF-2 domain, of the PPARγ protein is involved in direct interactions with the PHL, QQ, and PQ domains, except for the PH domain, in TDAG51 (Fig. 2). Furthermore, we observed that the TDAG51-PPARγ interaction is not inhibited by the binding of synthetic PPARγ ligand and that TDAG51 is more strongly associated with PPARγ alone than the PPARγ-RXRα heterodimer in protein-protein interaction assays (Fig. 4). To date, no DNA binding motif in TDAG51 has been found. TDAG51, which has evolutionarily conserved PHL, PQ, and PH domains, is considered a putative transcriptional regulator (Nagai, 2016). Thus, based on our results and previous studies, TDAG51 can function as a ligand-independent competitive inhibitor of RXRα recruitment toward assembly as a PPARγ-RXRα heterodimer complex in adipogenesis. Interestingly, we showed that TDAG51 expression is downregulated in a differentiation-dependent manner during adipogenic differentiation and that this downregulation is not directly mediated by the adipogenic transcription factor PPARγ (Fig. 1, Supplementary Fig. S2). Hence, further exploration of the transcriptional regulatory pathways involved in the downregulation of TDAG51 at the early stage of adipogenesis would be of interest.
PPARγ activity is modulated by several posttranslational modifications, including phosphorylation, sumoylation and ubiquitination (Ahmadian et al., 2013). Interestingly, the phosphorylation of PPARγ can repress or enhance its activity depending on the phosphorylation site or kinase activation, while PPARγ sumoylation has been shown to repress PPARγ activity by the recruitment of corepressors with histone deacetylases (Hu et al., 1996; Iankova et al., 2006; Pascual et al., 2005). Notably, TDAG51 was reported to be involved in regulating several kinase pathways, such as aurora A kinase, ErbB2 tyrosine kinase, Akt and receptor tyrosine kinase, in cancer cells (Fearon et al., 2018; Johnson et al., 2011; Li et al., 2014). Hence, based on previous studies, the inhibitory effect of TDAG51 on PPARγ activity is indirectly mediated by possible recruitment of another corepressor or kinase. Thus, further studies are needed to elucidate whether any other regulators or kinases are involved in indirect regulation of the TDAG51-mediated negative effects on PPARγ-induced adipogenesis. Ubiquitination and proteasome-dependent degradation are also linked to the regulation of PPARγ activity by ligand binding, IFNγ stimulation or recruitment of mediators, including Wnt1-inducible signaling pathway protein-1 (Hauser et al., 2000; Waite et al., 2001). However, we assumed that TDAG51 function is not linked to the proteasomal degradation of PPARγ because the levels of PPARγ were not altered by the TDAG51 interaction (Figs. 2 and 4).
Several transcription factors have been shown to regulate PPARγ expression and activity in adipogenesis (Miard and Fajas, 2005). Of the six GATA family transcription factors, GATA-2 and GATA-3 were shown to downregulate PPARγ expression by directly interacting with the PPARγ2 promoter and C/EBP family, thereby inhibiting adipogenic differentiation (Tong et al., 2000). The expression of GATA-2/3 is abundant in preadipocytes and progressively decreases during adipocyte differentiation (Tong et al., 2000). Thus, GATA-2/3 have a potential role in commitment to the adipogenic lineage (Kamata et al., 2014). Similar to the pattern of GATA-2/3 expression shown in the work of Tong et al. (2000), the most abundant expression of TDAG51 was observed only in the early stage of adipogenic differentiation (Fig. 1). Although we have not yet experimentally determined the preadipocyte restriction by TDAG51 expression in the early stage of adipogenic differentiation, we propose that TDAG51 may also have a potential role in commitment to the adipogenic lineage. FoxO1, a member of the forkhead transcription factor family, is known to be a negative regulator of adipogenesis (Nakae et al., 2003). This molecule represses both PPARγ expression through direct binding to the FoxO1 binding consensus sequences on the PPARγ2 promoter and PPARγ activity by inhibiting the DNA binding activity of the PPARγ-RXRα heterodimer through direct interaction with PPARγ (Armoni et al., 2006; Dowell et al., 2003; Fan et al., 2009). Furthermore, the physical interaction among FoxO1, C/EBPα and small heterodimer partner (SHP) has been reported (Park et al., 2007; Qiao and Shao, 2006; Yamagata et al., 2004). Interestingly, SHP, an orphan nuclear receptor that lacks a DBD, is also known to repress PPARγ activity by directly interacting with RXRα, but not with PPARγ, which blocks RXRα recruitment to the PPARγ-RXRα heterodimer, although the precise role of SHP in PPARγ activity modulation remains controversial (Kim et al., 2009; Nishizawa et al., 2002). Consequently, the mechanism of FoxO1 or SHP that represses PPARγ activity is quite distinct from the inhibitory mechanism of TDAG51 that blocks the recruitment of RXRα to the PPARγ-RXRα heterodimer complex by directly interacting with PPARγ.
In conclusion, we demonstrated that TDAG51 is a novel negative regulator of PPARγ in adipogenesis. In this study, we showed that TDAG51 expressed abundantly in the early stage of adipocyte diferentiation inhibits adipogenesis. TDAG51 interacts with PPARγ in a ligand-independent manner, thereby inhibiting PPARγ activity by blocking the formation of the PPARγ-RXRα heterodimer complex. Thus, these results indicate that TDAG51, which could determine adipogenic cell fate, is a ligand-independent competitive inhibitor of PPARγ in adipogenesis, expanding the importance of TDAG51 in the regulation in metabolic diseases, including obesity and diabetes.
Supplemental Materials
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
ACKNOWLEDGMENTS
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) and was funded by the Ministry of Education (NRF-2017R1A2B4007327, 2019R1A2C1084311, 2019M3F6A1109486) and by the research fund of Chungnam National University.
Footnotes
AUTHOR CONTRIBUTIONS
S.K. and N.L. performed experiments and wrote the manuscript. E.S.P., H.Y., T.U.H., H.J., J.Y., S.C., B.S., and J.Y. performed experiments or analyzed the data. S.D.R. and Y.C. contributed essential reagents or tools and designed the research study. J.R. supervised the research and edited the manuscript.
CONFLICT OF INTEREST
The authors have no potential conflicts of interest to disclose.
REFERENCES
- Ahmadian M., Suh J.M., Hah N., Liddle C., Atkins A.R., Downes M., Evans R.M. PPARgamma signaling and metabolism: the good, the bad and the future. Nat. Med. 2013;19:557–566. doi: 10.1038/nm.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armoni M., Harel C., Karni S., Chen H., Bar-Yoseph F., Ver M.R., Quon M.J., Karnieli E. FOXO1 represses peroxisome proliferator-activated receptor-gamma1 and -gamma2 gene promoters in primary adipocytes. A novel paradigm to increase insulin sensitivity. J. Biol. Chem. 2006;281:19881–19891. doi: 10.1074/jbc.M600320200. [DOI] [PubMed] [Google Scholar]
- Basseri S., Lhoták Š., Fullerton M.D., Palanivel R., Jiang H., Lynn E.G., Ford R.J., Maclean K.N., Steinberg G.R., Austin R.C. Loss of TDAG51 results in mature-onset obesity, hepatic steatosis, and insulin resistance by regulating lipogenesis. Diabetes. 2013;62:158–169. doi: 10.2337/db12-0256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton G.R., Nagarajan R., Peterson C.A., McGehee R.E., Jr. Microarray analysis of differentiation-specific gene expression during 3T3-L1 adipogenesis. Gene. 2004;329:167–185. doi: 10.1016/j.gene.2003.12.012. [DOI] [PubMed] [Google Scholar]
- Chandra V., Huang P., Hamuro Y., Raghuram S., Wang Y., Burris T.P., Rastinejad F. Structure of the intact PPAR-gamma-RXR- nuclear receptor complex on DNA. Nature. 2008;456:350–356. doi: 10.1038/nature07413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Takikawa M., Tsutsumi S., Yamaguchi Y., Okabe A., Shimada M., Kawase T., Sada A., Ezawa I., Takano Y., et al. PHLDA1, another PHLDA family protein that inhibits Akt. Cancer Sci. 2018;109:3532–3542. doi: 10.1111/cas.13796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowell P., Otto T.C., Adi S., Lane M.D. Convergence of peroxisome proliferator-activated receptor gamma and Foxo1 signaling pathways. J. Biol. Chem. 2003;278:45485–45491. doi: 10.1074/jbc.M309069200. [DOI] [PubMed] [Google Scholar]
- Elberg G., Gimble J.M., Tsai S.Y. Modulation of the murine peroxisome proliferator-activated receptor γ2 promoter activity by CCAAT/enhancer-binding proteins. J. Biol. Chem. 2000;275:27815–27822. doi: 10.1074/jbc.M003593200. [DOI] [PubMed] [Google Scholar]
- Fan W., Imamura T., Sonoda N., Sears D.D., Patsouris D., Kim J.J., Olefsky J.M. FOXO1 transrepresses peroxisome proliferator-activated receptor gamma transactivation, coordinating an insulin-induced feed-forward response in adipocytes. J. Biol. Chem. 2009;284:12188–12197. doi: 10.1074/jbc.M808915200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fearon A.E., Carter E.P., Clayton N.S., Wilkes E.H., Baker A.M., Kapitonova E., Bakhouche B.A., Tanner Y., Wang J., Gadaleta E., et al. PHLDA1 mediates drug resistance in receptor tyrosine kinase-driven cancer. Cell Rep. 2018;22:2469–2481. doi: 10.1016/j.celrep.2018.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring W.J., Affolter M., Burglin T. Homeodomain proteins. Annu. Rev. Biochem. 1994;63:487–526. doi: 10.1146/annurev.bi.63.070194.002415. [DOI] [PubMed] [Google Scholar]
- Hauser S., Adelmant G., Sarraf P., Wright H.M., Mueller E., Spiegelman B.M. Degradation of the peroxisome proliferator-activated receptor gamma is linked to ligand-dependent activation. J. Biol. Chem. 2000;275:18527–18533. doi: 10.1074/jbc.M001297200. [DOI] [PubMed] [Google Scholar]
- Hayashida N., Inouye S., Fujimoto M., Tanaka Y., Izu H., Takaki E., Ichikawa H., Rho J., Nakai A. A novel HSF1-mediated death pathway that is suppressed by heat shock proteins. EMBO J. 2006;25:4773–4783. doi: 10.1038/sj.emboj.7601370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain G.S., Lynn E.G., Maclean K.N., Zhou J., Dickhout J.G., Lhotak S., Trigatti B., Capone J., Rho J., Tang D., et al. Deficiency of TDAG51 protects against atherosclerosis by modulating apoptosis, cholesterol efflux, and peroxiredoxin-1 expression. J. Am. Heart Assoc. 2013;2:e000134. doi: 10.1161/JAHA.113.000134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu E., Kim J.B., Sarraf P., Spiegelman B.M. Inhibition of adipogenesis through MAP kinase-mediated phosphorylation of PPARgamma. Science. 1996;274:2100–2103. doi: 10.1126/science.274.5295.2100. [DOI] [PubMed] [Google Scholar]
- Iankova I., Petersen R.K., Annicotte J.S., Chavey C., Hansen J.B., Kratchmarova I., Sarruf D., Benkirane M., Kristiansen K., Fajas L. Peroxisome proliferator-activated receptor gamma recruits the positive transcription elongation factor b complex to activate transcription and promote adipogenesis. Mol. Endocrinol. 2006;20:1494–1505. doi: 10.1210/me.2005-0222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao H.W., Jia X.X., Zhao T.J., Rong H., Zhang J.N., Cheng Y., Zhu H.P., Xu K.L., Guo S.Y., Shi Q.Y., et al. Up-regulation of TDAG51 is a dependent factor of LPS-induced RAW264.7 macrophages proliferation and cell cycle progression. Immunopharmacol. Immunotoxicol. 2016;38:124–130. doi: 10.3109/08923973.2016.1138968. [DOI] [PubMed] [Google Scholar]
- Johnson E.O., Chang K.H., de Pablo Y., Ghosh S., Mehta R., Badve S., Shah K. PHLDA1 is a crucial negative regulator and effector of Aurora A kinase in breast cancer. J. Cell Sci. 2011;124:2711–2722. doi: 10.1242/jcs.084970. [DOI] [PubMed] [Google Scholar]
- Juge-Aubry C., Pernin A., Favez T., Burger A.G., Wahli W., Meier C.A., Desvergne B. DNA binding properties of peroxisome proliferator-activated receptor subtypes on various natural peroxisome proliferator response elements. Importance of the 5'-flanking region. J. Biol. Chem. 1997;272:25252–25259. doi: 10.1074/jbc.272.40.25252. [DOI] [PubMed] [Google Scholar]
- Kamata M., Okitsu Y., Fujiwara T., Kanehira M., Nakajima S., Takahashi T., Inoue A., Fukuhara N., Onishi Y., Ishizawa K., et al. GATA2 regulates differentiation of bone marrow-derived mesenchymal stem cells. Haematologica. 2014;99:1686–1696. doi: 10.3324/haematol.2014.105692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.I., Kaufman R.J., Back S.H., Moon J.Y. Development of a reporter system monitoring regulated intramembrane proteolysis of the transmembrane bZIP transcription factor ATF6alpha. Mol. Cells. 2019;42:783–793. doi: 10.14348/molcells.2019.0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.H., Kim H., Park J.M., Im S.S., Bae J.S., Kim M.Y., Yoon H.G., Cha J.Y., Kim K.S., Ahn Y.H. Interrelationship between liver X receptor alpha, sterol regulatory element-binding protein-1c, peroxisome proliferator-activated receptor gamma, and small heterodimer partner in the transcriptional regulation of glucokinase gene expression in liver. J. Biol. Chem. 2009;284:15071–15083. doi: 10.1074/jbc.M109.006742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroker A.J., Bruning J.B. Review of the structural and dynamic mechanisms of PPARgamma partial agonism. PPAR Res. 2015;2015:816856. doi: 10.1155/2015/816856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefterova M.I., Lazar M.A. New developments in adipogenesis. Trends Endocrinol. Metab. 2009;20:107–114. doi: 10.1016/j.tem.2008.11.005. [DOI] [PubMed] [Google Scholar]
- Li G., Wang X., Hibshoosh H., Jin C., Halmos B. Modulation of ErbB2 blockade in ErbB2-positive cancers: the role of ErbB2 Mutations and PHLDA1. PLoS One. 2014;9:e106349. doi: 10.1371/journal.pone.0106349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miard S., Fajas L. Atypical transcriptional regulators and cofactors of PPARgamma. Int. J. Obes. (Lond.) 2005;29(Suppl 1):S10–S12. doi: 10.1038/sj.ijo.0802906. [DOI] [PubMed] [Google Scholar]
- Nagai M.A. Pleckstrin homology-like domain, family A, member 1 (PHLDA1) and cancer. Biomed. Rep. 2016;4:275–281. doi: 10.3892/br.2016.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakae J., Kitamura T., Kitamura Y., Biggs W.H., 3rd, Arden K.C., 3rd, Accili D., 3rd The forkhead transcription factor Foxo1 regulates adipocyte differentiation. Dev. Cell. 2003;4:119–129. doi: 10.1016/s1534-5807(02)00401-x. [DOI] [PubMed] [Google Scholar]
- Nishizawa H., Yamagata K., Shimomura I., Takahashi M., Kuriyama H., Kishida K., Hotta K., Nagaretani H., Maeda N., Matsuda M., et al. Small heterodimer partner, an orphan nuclear receptor, augments peroxisome proliferator-activated receptor gamma transactivation. J. Biol. Chem. 2002;277:1586–1592. doi: 10.1074/jbc.M104301200. [DOI] [PubMed] [Google Scholar]
- Park C.G., Lee S.Y., Kandala G., Lee S.Y., Choi Y. A novel gene product that couples TCR signaling to Fas(CD95) expression in activation-induced cell death. Immunity. 1996;4:583–591. doi: 10.1016/s1074-7613(00)80484-7. [DOI] [PubMed] [Google Scholar]
- Park E.S., Choi S., Shin B., Yu J., Hwang J.M., Yun H., Chung Y.H., Choi J.S., Choi Y., Rho J. Tumor necrosis factor (TNF) receptor-associated factor (TRAF)-interacting protein (TRIP) negatively regulates the TRAF2 ubiquitin-dependent pathway by suppressing the TRAF2-sphingosine 1-phosphate (S1P) interaction. J. Biol. Chem. 2015;290:9660–9673. doi: 10.1074/jbc.M114.609685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park E.S., Kim J., Ha T.U., Choi J.S., Soo Hong K., Rho J. TDAG51 deficiency promotes oxidative stress-induced apoptosis through the generation of reactive oxygen species in mouse embryonic fibroblasts. Exp. Mol. Med. 2013;45:e35. doi: 10.1038/emm.2013.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park M.J., Kong H.J., Kim H.Y., Kim H.H., Kim J.H., Cheong J.H. Transcriptional repression of the gluconeogenic gene PEPCK by the orphan nuclear receptor SHP through inhibitory interaction with C/EBPalpha. Biochem. J. 2007;402:567–574. doi: 10.1042/BJ20061549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual G., Fong A.L., Ogawa S., Gamliel A., Li A.C., Perissi V., Rose D.W., Willson T.M., Rosenfeld M.G., Glass C.K. A SUMOylation-dependent pathway mediates transrepression of inflammatory response genes by PPAR-gamma. Nature. 2005;437:759–763. doi: 10.1038/nature03988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao L., Shao J. SIRT1 regulates adiponectin gene expression through Foxo1-C/enhancer-binding protein alpha transcriptional complex. J. Biol. Chem. 2006;281:39915–39924. doi: 10.1074/jbc.M607215200. [DOI] [PubMed] [Google Scholar]
- Rosen E., Eguchi J., Xu Z. Transcriptional targets in adipocyte biology. Expert Opin. Ther. Targets. 2009;13:975–986. doi: 10.1517/14728220903039706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen E.D., Hsu C.H., Wang X., Sakai S., Freeman M.W., Gonzalez F.J., Spiegelman B.M. C/EBPalpha induces adipogenesis through PPARgamma: a unified pathway. Genes Dev. 2002;16:22–26. doi: 10.1101/gad.948702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saladin R., Fajas L., Dana S., Halvorsen Y.D., Auwerx J., Briggs M. Differential regulation of peroxisome proliferator activated receptor gamma1 (PPARgamma1) and PPARgamma2 messenger RNA expression in the early stages of adipogenesis. Cell Growth Differ. 1999;10:43–48. [PubMed] [Google Scholar]
- Sarjeant K., Stephens J.M. Adipogenesis. Cold Spring Harb. Perspect. Biol. 2012;4:a008417. doi: 10.1101/cshperspect.a008417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffzek K., Welti S. Pleckstrin homology (PH) like domains - versatile modules in protein-protein interaction platforms. FEBS Lett. 2012;586:2662–2673. doi: 10.1016/j.febslet.2012.06.006. [DOI] [PubMed] [Google Scholar]
- Son H.E., Min H.Y., Kim E.J., Jang W.G. Fat mass and obesity-associated (FTO) stimulates osteogenic differentiation of C3H10T1/2 cells by inducing mild endoplasmic reticulum stress via a positive feedback loop with p-AMPK. Mol. Cells. 2020;43:58–65. doi: 10.14348/molcells.2019.0136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T., Yoshida N., Kishimoto T., Akira S. Defective adipocyte differentiation in mice lacking the C/EBPbeta and/or C/EBPdelta gene. EMBO J. 1997;16:7432–7443. doi: 10.1093/emboj/16.24.7432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong Q., Dalgin G., Xu H., Ting C.N., Leiden J.M., Hotamisligil G.S. Function of GATA transcription factors in preadipocyte-adipocyte transition. Science. 2000;290:134–138. doi: 10.1126/science.290.5489.134. [DOI] [PubMed] [Google Scholar]
- Tontonoz P., Hu E., Graves R.A., Budavari A.I., Spiegelman B.M. mPPAR gamma 2: tissue-specific regulator of an adipocyte enhancer. Genes Dev. 1994;8:1224–1234. doi: 10.1101/gad.8.10.1224. [DOI] [PubMed] [Google Scholar]
- Waite K.J., Floyd Z.E., Arbour-Reily P., Stephens J.M. Interferon-gamma-induced regulation of peroxisome proliferator-activated receptor gamma and STATs in adipocytes. J. Biol. Chem. 2001;276:7062–7068. doi: 10.1074/jbc.M007894200. [DOI] [PubMed] [Google Scholar]
- Wu Z., Bucher N.L., Farmer S.R. Induction of peroxisome proliferator-activated receptor gamma during the conversion of 3T3 fibroblasts into adipocytes is mediated by C/EBPbeta, C/EBPdelta, and glucocorticoids. Mol. Cell. Biol. 1996;16:4128–4136. doi: 10.1128/mcb.16.8.4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z., Rosen E.D., Brun R., Hauser S., Adelmant G., Troy A.E., McKeon C., Darlington G.J., Spiegelman B.M. Cross-regulation of C/EBP alpha and PPAR gamma controls the transcriptional pathway of adipogenesis and insulin sensitivity. Mol. Cell. 1999;3:151–158. doi: 10.1016/s1097-2765(00)80306-8. [DOI] [PubMed] [Google Scholar]
- Xiao H., Jeang K.T. Glutamine-rich domains activate transcription in yeast Saccharomyces cerevisiae. J. Biol. Chem. 1998;273:22873–22876. doi: 10.1074/jbc.273.36.22873. [DOI] [PubMed] [Google Scholar]
- Yamagata K., Daitoku H., Shimamoto Y., Matsuzaki H., Hirota K., Ishida J., Fukamizu A. Bile acids regulate gluconeogenic gene expression via small heterodimer partner-mediated repression of hepatocyte nuclear factor 4 and Foxo1. J. Biol. Chem. 2004;279:23158–23165. doi: 10.1074/jbc.M314322200. [DOI] [PubMed] [Google Scholar]
- Yu J., Yun H., Shin B., Kim Y., Park E.S., Choi S., Yu J., Amarasekara D.S., Kim S., Inoue J., et al. Interaction of tumor necrosis factor receptor-associated factor 6 (TRAF6) and Vav3 in the receptor activator of nuclear factor kappaB (RANK) signaling complex enhances osteoclastogenesis. J. Biol. Chem. 2016;291:20643–20660. doi: 10.1074/jbc.M116.728303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.