Abstract
Despite transcriptional silencing in mature sperm and cytoplasmic expulsion of RNA during the final sperm maturation process, thousands of RNAs have been successfully identified in ejaculated sperm. Although most of RNAs’ function is still unknown, it is suggested that sperm RNAs have a vital biological role in fertilization and post-fertilization events. Nevertheless, the lack of accurate RNA isolation techniques and the resultant good quality sperm RNA has hampered the exploration of sperm RNAs function. Additionally, small non-coding RNAs are found in extracellular fluids including seminal plasma. These small RNAs may participate in cell to cell communication or intracellular and extracellular message transmission. Developing precise protocols to extract RNA from sperm and seminal plasma is critical to elucidate sperm physiology and paternal contributions to fertilization and post-fertilization events. A detailed procedure consisting of semen collection, separation of sperm and seminal plasma, extracting RNA from sperm and seminal plasma, and determining the quantity and quality of RNA for boar semen is presented here. This efficient protocol can be extrapolated to isolate RNAs from sperm and seminal plasma across mammalian species.
Keywords: Semen collection, RNA extraction, Boar sperm, Seminal plasma, Non-coding RNA, Spin-column based purification, RNA elution using nuclease free water
Background
Transcriptionally inactive mature sperm contains a complex population of RNAs including non-coding small RNAs ( Ostermeier et al., 2002 ). These RNAs can participate in fertilization and post-fertilization events including zygotic gene activation. Recently, it was suggested that sperm also can pick up small RNAs on their way during the epididymal transit and these small RNAs can influence the gene regulation in developing embryos for efficient implantation ( Kasimanickam et al., 2019b ). It is important to study the RNA profiles in immature and mature sperm harvested from various locations such as caput epididymis, cauda epididymis, ejaculated sperm and seminal plasma to know how these RNAs are shuttled between sperm cells and seminal plasma. Better understanding of coding and non-coding RNAs in sperm and seminal plasma by gene expression arrays and microarrays allows investigating the problems associated with infertility. Nevertheless, success of microarray and gene expression analyses is extremely dependent on the quality and quantity of the RNA obtained from sperm and seminal plasma. Therefore, the development of methods that isolate highly purified and intact RNA from sperm and seminal plasma is of utmost importance. Purification of sperm and seminal plasma, RNA extraction from sperm and seminal plasma and evaluation of RNA quality are the critical steps to be considered to achieve these goals. Since sperm and seminal plasma contain a very small amount of RNA, a method consisting capture of RNA by silica membrane and elution using nuclease-free water (column-based approach) is more advantageous to perform rather than a method involving precipitation of RNA. Hence, detailed protocols for semen collection, semen processing to purify sperm and seminal plasma, RNA isolation from sperm and seminal plasma adopting a column-based approach and determining the quantity and quality of RNA for boars are described here.
Materials and Reagents
Pipette tips (Eppendorf, catalog numbers: 02-717-349, 02-717-351, 02-717-352)
Disposable homogenizer (Thermo Fisher Scientific, Fisher, catalog number: 12-141-368)
Polyvinyl gloves (Thermo Fisher Scientific, Fisher Scientific, catalog number: 19-121-834B)
J-cloth (https://www.amazon.com/J-Cloth-Blue-50s/dp/B00V3UIQ3E)
Miracloth (Millipore Sigma, Millipore, catalog number: 475855)
Microscopic slides (Thermo Fisher Scientific, Thermo Scientific, catalog number: 4951PLUS4)
Microscopic cover glasses (Thermo Fisher Scientific, Thermo Scientific, catalog number: 18X18-1)
Graduated measuring cylinder
Falcon tubes, 50 ml (Thermo Fisher Scientific, Corning, catalog number: 14-432-22)
Microcentrifuge tubes (Thermo Fisher Scientific, Thermo Scientific, catalog number: 3448)
Square cuvette
-
Mature trained boars for semen collection (Landrace boars of 8 to 18 months of age were used in this protocol)
Notes:
Follow institutional guidelines for animal maintenance and use for sample collection.
Maintain clean semen collection area with appropriate human and animal safety guidelines.
Beltsville Thawing Solution (Minitube USA, catalog number: 13525)
Liquid nitrogen (Washington State University Store, WSU, catalog number: 12506)
RNeasy plus Universal Mini Kit (RNeasy mini spin column, 1.5- and 2.0-ml collection tubes, QIAzol lysis reagent, gDNA eliminator solution, RWT buffer, RPE buffer and RNase free water) (Qiagen Inc., catalog number: 73404)
Chloroform (VWR International, J.T. Baker, catalog number: 9180-1)
Ethanol (Washington State University Store, Decon, catalog number: Decon 2701)
MiRNeasy serum/plasma kit (RNeasy MinElute spin column, 1.5- and 2.0-ml collection tubes, QIAzol lysis reagent, RWT buffer, RPE buffer and RNase free water) (Qiagen Inc., catalog number: 217184)
MiRNeasy serum/plasma spike-in-controls (Qiagen Inc., Valencia, CA, USA, catalog number: 219610)
Mild disinfectant solution (Washington State University Store, Decon, catalog number: Decon 57065)
Equipment
Boar semen collection flask (IMV Technologies USA, IMV, catalog number: 017282)
Pipettes (Eppendorf, catalog numbers: 2231300034, 2231300037)
Adjustable built-in semen collection dummy
Phase-contrast microscope (Minitube, Olympus CX43, heated stage, HT 200, catalog number: 12005/0041)
Refrigerated centrifuge (Jouan refrigerated centrifuge, model: CR422)
Microcentrifuge (Eppendorf, model: 5415C)
Ultra-freezer (-80 °C)
Spectrophotometer (Thermo Fisher Scientific, model: NanoDrop 1000)
Benchtop incubator (Unico, model: Unico 30L Incubator #L-CU300)
Procedure
-
Boar semen collection
Follow the institutional guidelines to maintain boars and semen collection procedures. Use mature and trained boars for semen collection. Maintain a hygienic environment in semen collection area.
-
Use gloved hand technique to collect boar semen ( Kasimanickam et al., 2019b ). After mounting on a dummy, clean sheath and prepuce with a mild disinfectant solution. Following a couple of copulatory thrusts, grasp the corkscrew penis with a gloved hand to collect ejaculate.
Note: Use polyvinyl gloves not the latex gloves since the latex gloves are spermicidal in nature.
-
Discard the pre-sperm fraction. Collect the sperm-rich fraction into a pre-warmed container through a j-cloth filter (Thermo Fisher Scientific, Bothell, WA, USA). Maintain clean environment to prevent contamination.
Note: Pre-warm the container, filter cloths, transfer pipets, microscopic slides and coverslips at 37 °C using a benchtop incubator.
Assess the sperm motility immediately after the semen collection. Place a drop of semen on a pre-warmed (37 °C) microscopic slide and observe under a phase contrast microscope at 100x magnification. Use the semen having ≥ 80% motility for the subsequent procedures (RNA isolation from sperm and seminal plasma).
Filter the sperm-rich fraction through a double layered Miracloth (Millipore Sigma, Burlington, MA, USA) into a pre-warmed conical flask to determine the volume, concentration, motility and presence of immature sperm cells (Table 1).
Determine the semen volume using a graduated measuring cylinder.
-
Evaluate the sperm concentration using a calibrated and standardized spectrophotometer for boar semen. Pipet the diluted semen sample in 2.9% sodium citrate in a square cuvette and measure the optical density at the excitation wavelength of 550 nm. Note the sperm concentration corresponding to the OD.
Table 1. Semen volume, concentration, motility and purity in ten mature Landrace boars.
Mature boars1 Semen volume2 Sperm moncentration3 Sperm motility4 Presence of immature sperm & somatic cells5 Boar 1 120 ml 1.2 x 109/ml 90% Absence Boar 2 150 ml 0.8 x 109/ml 85% Absence Boar 3 120 ml 1.0 x 109/ml 90% Absence Boar 4 180 ml 1.5 x 109/ml 88% Absence Boar 5 130 ml 0.9 x 109/ml 90% Absence Boar 6 180 ml 1.2 x 109/ml 85% Absence Boar 7 200 ml 1.5 x 109/ml 87% Absence Boar 8 170 ml 1.0 x 109/ml 85% Absence Boar 9 150 ml 1.2 x 109/ml 90% Absence Boar 10 200 ml 0.9 x 109/ml 87% Absence Mean ± SD 160 ± 30.55 ml 1.2 x 109 ± 0.24 /ml 87.7 ± 2.21 % N/A 18 to 18 months Landrace
2Sperm-rich fraction
3Evaluated by spectrophotometer (Spectronic 20D+ spectrophotometer)
4Evaluated using phase-contrast microscope
5Evaluated using phase-contrast microscope
-
Separation of sperm and seminal plasma
-
Centrifuge undiluted semen at 1,000 × g for 20 min at 4 °C. Use pre-cold 50 ml Falcon tubes for 40 ml semen.
Note: Extended or non-extended semen should be stored at 15 °C to maintain viability to use for insemination.
-
Transfer the seminal plasma (supernatant) into a new tube. Centrifuge the seminal plasma at 16,000 × g, 4 °C to ensure removal of sperm. Use microcentrifuge tubes to use the centrifugal force of 16,000 × g at 4 °C.
Note: Any other types of centrifuges that can reach higher centrifugal force and the temperature of 4 °C can also be used to separate seminal plasma from the semen.
-
Aliquot the seminal plasma (250 μl aliquots) and store at -80 °C.
Note: The number of 250 μl aliquots that will be stored can be determined based on your experimental plan.
-
Wash the sperm pellet twice using Beltsville Thawing Solution (BTS; IMV Technologies, Maple Grove, MN, USA) and centrifuging at 1,000 × g for 20 min at 4 °C.
Note: While re-suspending the sperm pellet, dissolve the pellet in a small volume (1 ml) of BTS first, and dilute further (20 ml) with BTS for effective washings.
-
Re-suspend the final sperm pellet in BTS (20 ml; resuspend in 1 ml first, add 19 ml and mix the suspension thoroughly) at 4 °C, aliquot in micro centrifuge tubes (approximately 500 x 106 sperm). Centrifuge at 16,000 × g, 4 °C and discard the supernatant. Flash-freeze the sperm pellet in liquid nitrogen and then store at -80 °C.
Note: Based on the initial concentration, the sperm can be resuspended and aliquoted. For example, when the sample of 40 ml semen contains 40 billion sperm, the final sperm pellet after washing can be resuspended in 20 ml. Therefore, an aliquot of 250 μl will contain 500 x 106 sperm. Modify your starting volume of semen based on the experimental design and the need.
-
-
Sperm RNA purification
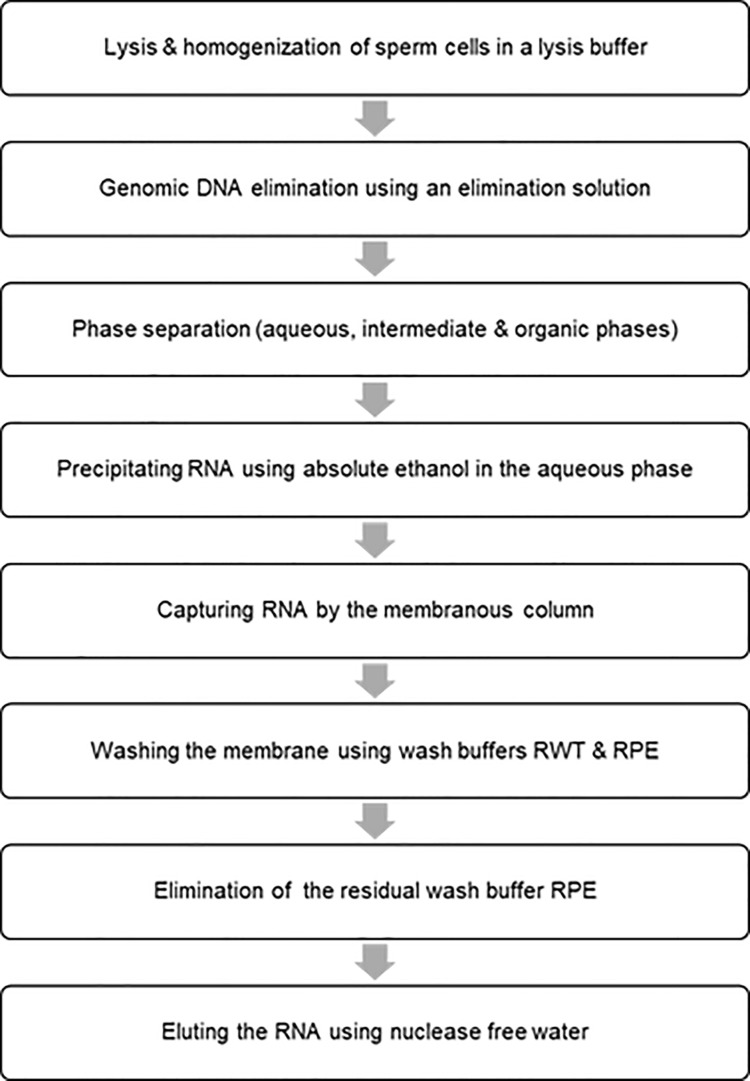
Isolate total RNA containing small RNAs including miRNAs from sperm using RNeasy plus Universal Mini Kit (Qiagen Inc., Valencia, CA, USA).
Add 900 μl QIAzol lysis reagent to the sperm pellet (approximately 500 x 106 sperm). homogenize completely using a disposable homogenizer (Thermo Fisher Scientific, San Francisco, CA, USA), and hold at room temperature for 5 min to promote dissociation of nucleoprotein complexes.
Add 100 μl genomic DNA (gDNA) eliminator solution and shake the mixture vigorously to eliminate gDNA.
Add 180 μl of chloroform. Agitate vigorously for about 1 min and incubate for 2-3 min at room temperature.
Centrifuge the mixture at 12,000 × g for 15 min at 4 °C. Harvest the upper aqueous phase (~600 μl) into a new Eppendorf tube, add 1.5 volume of 100% ethanol, and mix the mixture thoroughly by pipetting up and down.
Layer the sample (about 700 μl) on an RNeasy mini spin column (included in the RNeasy plus Universal Mini Kit), and centrifuge at ≥ 8,000 × g for 30 s at room temperature.
Discard the flow-through and repeat the centrifugation with the rest of the sample. Discard the flow-through again.
Wash the bound RNA by centrifugation (≥ 8,000 × g, 30 s, at room temperature) using buffers RWT and RPE consequently.
-
Add 700 μl buffer RWT to the RNeasy spin column. Close the lid gently, and centrifuge for 30 s at ≥ 8,000 × g to wash the membrane. Discard the flow-through.
Note: Dilute the RWT buffer by adding 2 volumes of ethanol (96-100%) before use.
-
Add 500 μl RPE buffer to the RNeasy spin column. Close the lid gently, and centrifuge for 30 s at ≥ 8,000 × g to wash the membrane. Discard the flow-through.
Note: Dilute the RPE buffer by adding 4 volumes of ethanol (96-100%) before use.
Repeat the RPE wash by centrifuging for 2 min.
Place the RNeasy spin column in a new 2 ml collection tube. Close the lid gently, and centrifuge at full speed for 1 min. Perform this step to eliminate any possible carryover of RPE buffer.
Elute the RNA using 60 μl RNase-free water by centrifuging for 1 min at ≥ 8,000 × g.
-
Measure the concentration of RNA using a Thermo Scientific NanoDrop 1000 Spectrophotometer (Table 2). Evaluate the purity of RNA by determining the ratio of absorbance at 260 and 280 nm and store the samples at -80 °C until use. Refer to the flow chart for the main steps (Figure 1).
Table 2. RNA yield from 500 x 106 sperm in ten mature Landrace boars .
Landrace Mature Boars RNA Concentration RNA Yield in 500 x 106 Sperm Boar 1 50 ng/µl 3,000 ng Boar 2 50 ng/µl 3,000 ng Boar 3 60 ng/µl 3,600 ng Boar 4 75 ng/µl 4,500 ng Boar 5 65 ng/µl 3,900 ng Boar 6 60 ng/µl 3,600 ng Boar 7 55 ng/µl 3,300 ng Boar 8 50 ng/µl 3,000 ng Boar 9 70 ng/µl 4,200 ng Boar 10 70 ng/µl 4,200 ng Mean ± SD 60.5 ± 9.26 ng/µl 3630 ± 556 ng
-
Seminal plasma RNA isolation
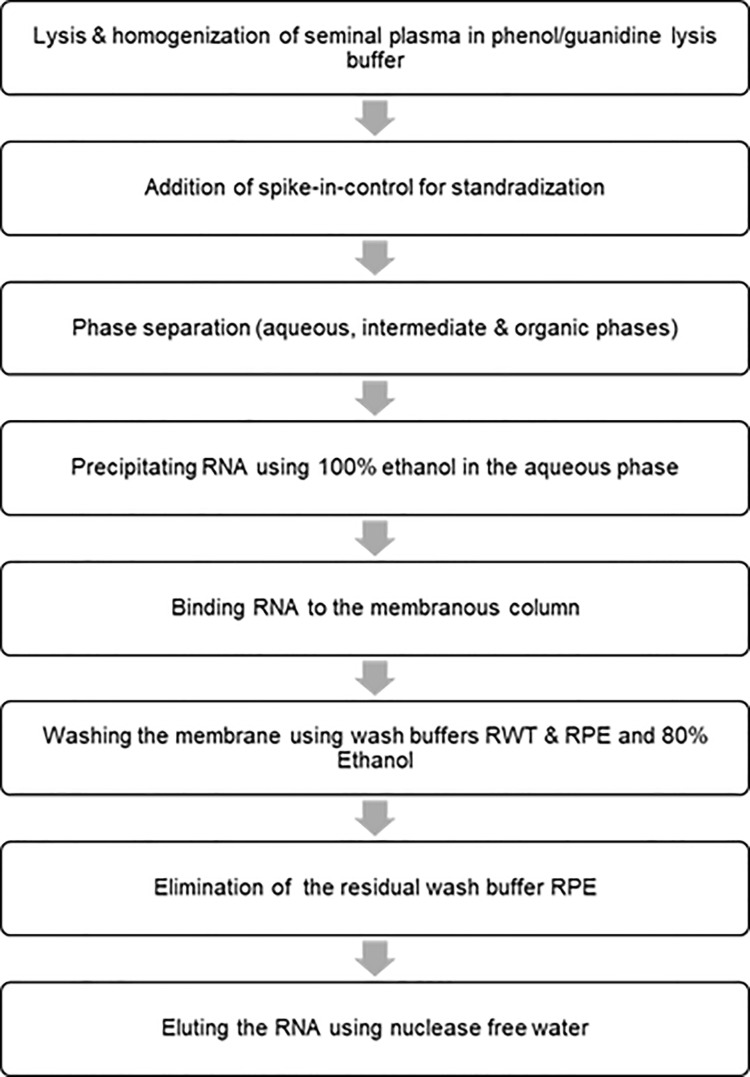
Purify total RNA from boar seminal plasma using miRNeasy serum/plasma kit (Qiagen, Valencia, CA, USA).
Thaw frozen seminal plasma samples at room temperature until it became completely liquid (~15 min).
-
Add 750 μl of QIAzol reagent (five volumes of seminal plasma) to 150 μl of seminal plasma. Mix by pipetting up and down or by vortexing briefly. Incubate for 5 min at room temperature (15-25 °C).
Note: The lysate can be stored at -80 °C for several months.
Add 3.5 μl miRNeasy serum/plasma spike-in-controls (lyophilized C. elegans miR-39 miRNA mimic; 1.6 x 108 copies/μl) and mix thoroughly.
Add 150 μl chloroform (same volume of starting material). Shake the mixture vigorously for 30 s and incubate for 2-3 min at room temperature (15-25 °C).
Centrifuge at 12,000 × g for 20 min at 4 °C. Harvest the upper aqueous phase, avoiding the interphase, into a new Eppendorf tube. Mix with ~1.5 volumes of 100% ethanol by pipetting up and down.
Transfer approximately half of the mix (about 750 μl) into an RNeasy MinElute spin column in a 2 ml collection tube. Centrifuge at 12,000 × g for 30 s at room temperature (15-25 °C).
Discard the flow-through and repeat this step with the rest of the sample using the same column.
Wash the RNA bound to the membrane sequentially with the buffer RWT and the buffer RPE.
Add 700 μl RWT buffer to the RNeasy MinElute spin column. Close the lid gently and centrifuge for 30 s at 8,000 × g at room temperature to wash the column. Discard the flow-through.
Pipet 500 μl RPE buffer onto the RNeasy MinElute spin column. Close the lid gently and centrifuge for 30 s at 8,000 × g at room temperature to wash the column. Discard the flow-through.
Add 500 μl of 80% ethanol onto the RNeasy MinElute spin column. Close the lid gently and centrifuge for 2 min at 8,000 × g at room temperature (15-25 °C) to wash the spin column membrane. Discard the collection tube with the flow-through.
Dry the RNeasy MinElute column by centrifugation at 12,000 × g for 5 min and discard the flow-through and collection tube.
Elute RNAs by placing the membrane in 24 μl RNase-free water and centrifuging at 12,000 × g for 2 min.
-
Determine the RNA concentration (Table 3) using a spectrophotometer (Thermo Scientific NanoDrop 1000). Refer to the flow chart for the main steps for isolating RNA from the seminal plasma (Figure 2).
Table 3. RNA yield from 150 µl of seminal plasma in ten Landrace boars.
Landrace Mature Boars RNA Concentration Total RNA in 150 μl Seminal Plasma Boar 1 30 ng/μl 720 ng Boar 2 40 ng/μl 960 ng Boar 3 31 ng/μl 744 ng Boar 4 34 ng/μl 816 ng Boar 5 40 ng/μl 960 ng Boar 6 35 ng/μl 840 ng Boar 7 26 ng/μl 624 ng Boar 8 29 ng/μl 696 ng Boar 9 40 ng/μl 960 ng Boar 10 35 ng/μl 840 ng Mean ± SD 34 ± 4.99 ng/μl 816 ± 120 ng
Figure 1. Flow chart for isolating the total RNA including small RNAs from sperm cells.
Figure 2. Flow chart for isolating RNA from the seminal plasma.
Data analysis
Since this protocol only includes semen volume, sperm concentration, sperm motility, presence/absence of abnormal and immature sperm cells, RNA yield from sperm and seminal plasma from 10 Landrace boars, mean and standard deviation values of the variables are presented in Tables 1, 2 and 3.
Notes
Diploid spermatogonia differentiate into mature haploid sperm during spermatogenesis in mammals. When haploid spermatids undergo morphological modifications and transition into elongated sperm, transition proteins, protamines and histones play a unique role in condensing spermatid chromatin ( Miller et al., 2005 ). Sperm, in addition to the main contribution of paternal genome, deliver a large range of RNA molecules to the oocyte during fertilization and these RNAs may have a functional role in fertilization and early embryonic development ( Boerke et al., 2007 ; Gòdia et al., 2018 ). Non-coding RNAs also contributes to fertilization, embryogenesis and offspring health and participate in transgenerational epigenetic inheritance ( Champroux et al., 2018 ). Transcriptional activity is found to be higher in primary spermatocytes and spermatids compared to secondary spermatocytes and then the transcription is almost arrested during the nuclear elongation phase of the final maturation step in spermatogenesis ( Dadoune et al., 2004 ). Presence of coding and non-coding RNAs in sperm has been revealed by a variety of techniques, microarray (Liu, T. et al., 2012; Bansal et al., 2015 ), qRT-PCR (Kasimanickam and Kastelic, 2016; Kasimanickam et al., 2019a and 2019b) and RNA sequencing ( Du et al., 2014 ; Mao et al., 2014 ) and these RNAs may originate from germ and somatic cells and partake in a variety of functions.
Sperm transcripts can be used as candidate markers for male fertility. High fertility bulls had higher levels of sperm transcripts and their proteins were correlated to important cellular processes such as metabolism, signal transduction, translation, glycosylation, and protein degradation ( Lalancette et al., 2008 ; Kasimanickam et al., 2019a ). Five genes, RPL31, PRKCE, PAPSS2, PLP1, and R1G7 were differentially seen between fresh and frozen-thawed sperm in Holstein bulls ( Chen et al., 2015 ). Sperm-borne miRNAs were found to be needed for normal fertilization and preimplantation embryo development in mice ( Yuan et al., 2016 ). Sperm transcripts were associated with in-vitro motility and field fertility in bulls using stringent RNA isolation protocols ( Bissonnette et al., 2009 ; Kasimanickam et al., 2012 ). A total of 7721 long non-coding RNAs and 6097 mRNAs were found to be differentially expressed in sperm between diabetic and normal group of mice ( Jiang et al., 2016 ). Sperm-borne microRNA-34c was determined to be important for the first cleavage division in mice (Liu, W. M. et al., 2012). A complex population of male-derived small non-coding RNAs were seen in the zygote after fertilization ( Krawetz et al., 2011 ). Since the role of these sperm transcripts is suggested in fertilization as well as zygotic and early embryonic development, a better understanding of these RNAs is imperative. However, the study of sperm RNAs has been challenging because of the difficulty accompanying with sperm RNA isolation. Several sperm RNA isolation protocols have been shown extremely variable sperm RNA yield due not only to the different approaches used but also to the heterogeneity of RNA within an individual sperm ( Lalancette et al., 2009 ; Mao et al., 2013 , Mao et al., 2014 ; Barragan et al., 2015 ).
Purification of sperm, RNA extraction and evaluation of RNA quality are the critical steps that need to be focused to obtain quality RNA. Besides sperm, an ejaculate also contains somatic cells, including immature germ cells, leucocytes and epithelial cells. Consequently, somatic cell removal is performed to avoid contamination of somatic cell RNAs and to derive only sperm transcripts. However, superior quality semen does not contain either immature germ cells or somatic cells (leucocytes or epithelial cells). Several methods such as swim-up procedure, density gradient centrifugation and lysis protocol are used to remove somatic cells from the ejaculate. Since seminal plasma also contains RNA, complete removal of seminal plasma is furthermore imperative.
To extract RNA from purified sperm, there are two methods primarily being used. One method is based on phase separation and precipitation whereas the other method is dependent on phase separation and column capture of RNAs. Since sperm contains a very small amount of RNA, capturing RNAs by a spin column and then eluting in a small volume of nuclease-free water yield reproducible results in specific to RNA’s quantity and quality. Sperm should be devoid of contaminant of somatic cells such as leukocytes and epithelial cells. Semen collected from mature and healthy domestic animals does not generally contain somatic cells. Presence of somatic cells can be evaluated by microscopic examination. RNA’s quantity and quality can be determined using a NanoDrop spectrophotometer (Fisher Thermo Scientific, Bothell, WA, USA) or an Agilent BioAnalyzer (Agilent Technologies, Santa Clara, CA, USA). Sperm does not contain ribosomal RNAs, 18S and 28S ribosomal subunits ( Goodrich et al., 2013 ) in contrast to other types of cells. The absence of these ribosomal RNAs in the elute can validate the purity of sperm RNAs.
Messenger RNAs have been found in mature sperm in different animal species. Transcriptome analysis of ejaculated sperm has been performed in pigs, many mRNAs have been identified in sperm, and these mRNAs have been associated with biological processes, molecular functions and cellular components of spermatogenesis and other testicular functions ( Yang et al., 2009 ). Various sperm transcripts at different expression levels have been noticed between capacitated and non-capacitated sperm and variation in mRNAs level in sperm caused different cleavage rate in pigs ( Hwang et al., 2013 ). Sperm containing abundant levels of MYC, CYP19, ADAM2, PRM1 and PRM2 instigated the increased cleavage rate. Often physical stress, environmental stress and infections affect the boar semen quality. In addition to the conventional semen parameters, sperm transcriptomics could serve as a reliable biomarker to evaluate sperm quality and fertility. Sperm delivers not only DNA to the oocyte but also bring thousands of RNAs to the oocyte, and these coding and non-coding RNA species greatly influence gene regulation in developing embryos for efficient implantation and placental development. Although small quantity of RNA is detected in seminal plasma when compared to sperm RNA in each semen sample ( Kasimanickam et al., 2019b ), comparative analysis of expression levels will be crucial to elucidate their functional significance and mechanistic interpretation of cell to cell communication and shuttling between endogenous and exogenous environment.
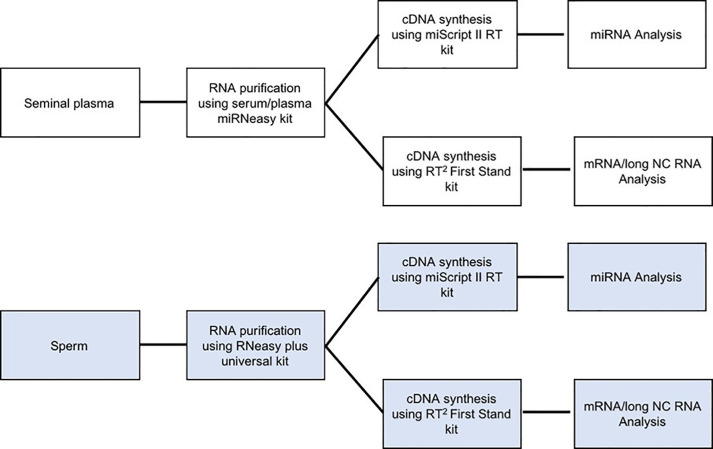
Therefore, efficient methods of sperm purification, RNA extraction from sperm and characterizing RNA’s quality are essential to elucidate their clinical significance. In this protocol, details procedures for the separation of pure sperm population and seminal plasma from high-quality semen via washing steps, extraction of measurable quantities of sperm and seminal plasma RNAs using membrane column capturing and determining the quantity and quality are presented. The outcome of these procedures appears to be reproducible for different boars. In summary, these techniques can be repeated for superior quality semen to obtain a detectable amount of good quality sperm and seminal plasma RNAs to utilize for downstream applications (Figure 3).
Figure 3. Flow charts showing the use of sperm and seminal plasma RNA in downstream experiments.
Critical parameter and troubleshooting:
Although high quality ejaculated semen from superior genetics mature boars are devoid of somatic cells such as leukocytes and epithelial cells, many semen samples including samples with the history of infertility have a significant number of immature germ cells, somatic cells and bacteria. Removal of somatic and immature cells is critical to prevent the contamination of RNA from these immature, epithelial, inflammatory and bacterial cells. In those circumstances, density gradient separation technique, somatic cell lysis strategy, and swim-up protocol can be performed to purify mature sperm. For instance, discontinuous Percoll (90% and 45% Percoll layers) gradient separation of pure sperm can be employed and selection of these procedures is based on the subsequent application of the study. Swim-up protocol is better for the in-vitro fertilization and the follow-up transcriptome analysis whereas Percoll gradient separation is appropriate for a transcriptome study to determine the sperm quality. Since sperm contains very scanty amount of RNA, total sperm RNA can be captured by a membranous column and subsequently the RNA can be eluted in a small volume of nuclease-free water. When the small volume elution is difficult, a larger volume of nuclease-free water can be used to elute the RNA and subsequently the diluted samples can be concentrated using an RNA concentration protocol. RNAs dissociate into the aqueous phase and hence phase separation is critical. The phase separation should be carefully performed not touching the interphase. When the quantity and purity of the RNA are evaluated using NanoDrop spectrophotometer, the ratio of absorbance at 260 and 280 nm wavelength should be close to 2.0. A small change in pH can overestimate/underestimate the ratio. If the results are not reproducible, another type of spectrophotometer can be used to evaluate the quality. The absorbance spectral graph should be ideal and should indicate the perfect 260/280 ratio.
Superior genetics mature boars produce 100-250 ml of sperm-rich fraction. Semen can be collected thrice weekly for an optimal quantity and quality. Concentration can range from 0.7 to 2 billion sperm/ml based on the secretions of accessory glands. Sperm motility varies from 80% to 90%. A total of 500 million sperm yield RNA ranging from 3,000 ng to 4,500 ng. The ratio of absorbance at 260/280 nm wavelength ranges from 1.92 to 2.08. RNA yield from 150 μl of seminal plasma varies from 720 ng to 960 ng whereas the purity ranges from 1.92 to 2.08.
Semen collection, separation and purification of sperm and seminal plasma, total RNA extraction and characterization can be performed in two weeks for ten boars, provided trained mature boars for semen collection, other supplies, equipment for laboratory work and personnel are available.
Acknowledgments
This detailed protocol presented here is expanded from our publications, Kasimanickam and Kastelic (2016) and Kasimanickam et al. (2019b).
Competing interests
The authors declare no competing interests.
Ethics
This study protocol strictly followed the standard ethics and use of animal cells for research. The protocol was approved by the institutional animal care and use committee of Washington State University (Protocol Number: 04070-001).
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1. Bansal S. K., Gupta N., Sankhwar S. N. and Rajender S.(2015). Differential genes expression between fertile and infertile spermatozoa revealed by transcriptome analysis. PLoS One 10(5): e0127007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Barragan M., Martinez A., Llonch S., Pujol A., Vernaeve V. and Vassena R.(2015). Effect of ribonucleic acid(RNA) isolation methods on putative reference genes messenger RNA abundance in human spermatozoa. Andrology 3(4): 797-804. [DOI] [PubMed] [Google Scholar]
- 3. Bissonnette N., Levesque-Sergerie J. P., Thibault C. and Boissonneault G.(2009). Spermatozoal transcriptome profiling for bull sperm motility: a potential tool to evaluate semen quality. Reproduction 138(1): 65-80. [DOI] [PubMed] [Google Scholar]
- 4. Boerke A., Dieleman S. J. and Gadella B. M.(2007). A possible role for sperm RNA in early embryo development. Theriogenology 68 Suppl 1: S147-155. [DOI] [PubMed] [Google Scholar]
- 5. Champroux A., Cocquet J., Henry-Berger J., Drevet J. R. and Kocer A.(2018). A decade of exploring the mammalian sperm epigenome: paternal epigenetic and transgenerational inheritance. Front Cell Dev Biol 6: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen X., Wang Y., Zhu H., Hao H., Zhao X., Qin T. and Wang D.(2015). Comparative transcript profiling of gene expression of fresh and frozen-thawed bull sperm. Theriogenology 83(4): 504-511. [DOI] [PubMed] [Google Scholar]
- 7. Dadoune J. P., Siffroi J. P. and Alfonsi M. F.(2004). Transcription in haploid male germ cells. Int Rev Cytol 237: 1-56. [DOI] [PubMed] [Google Scholar]
- 8. Du Y., Wang X., Wang B., Chen W., He R., Zhang L., Xing X., Su J., Wang Y. and Zhang Y.(2014). Deep sequencing analysis of microRNAs in bovine sperm. Mol Reprod Dev 81(11): 1042-1052. [DOI] [PubMed] [Google Scholar]
- 9. Gòdia M., Swanson G. and Krawetz S. A.(2018). A history of why fathers' RNA matters. Biol Reprod 99(1): 147-159. [DOI] [PubMed] [Google Scholar]
- 10. Goodrich R. J., Anton E. and Krawetz S. A.(2013). Isolating mRNA and small noncoding RNAs from human sperm. Methods Mol Biol 927: 385-396. [DOI] [PubMed] [Google Scholar]
- 11. Hwang J. Y., Mulligan B. P., Kim H. M., Yang B. C. and Lee C. K.(2013). Quantitative analysis of sperm mRNA in the pig: relationship with early embryo development and capacitation. Reprod Fertil Dev 25(5): 807-817. [DOI] [PubMed] [Google Scholar]
- 12. Jiang G. J., Zhang T., An T., Zhao D. D., Yang X. Y., Zhang D. W., Zhang Y., Mu Q. Q., Yu N., Ma X. S. and Gao S. H.(2016). Differential expression of long noncoding RNAs between sperm samples from diabetic and non-diabetic mice. PLoS One 11(4): e0154028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kasimanickam R. K., Kasimanickam V. R., Arangasamy A. and Kastelic J. P.(2019). Sperm and seminal plasma proteomics of high- versus low-fertility Holstein bulls. Theriogenology 126: 41-48. [DOI] [PubMed] [Google Scholar]
- 14. Kasimanickam V. and Kastelic J.(2016). MicroRNA in sperm from Duroc, Landrace and Yorkshire boars. Sci Rep 6: 32954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kasimanickam V., Buhr M. and Kasimanickam R.(2019). Patterns of expression of sperm and seminal plasma microRNAs in boar semen. Theriogenology 125: 87-92. [DOI] [PubMed] [Google Scholar]
- 16. Kasimanickam V., Kasimanickam R., Arangasamy A., Saberivand A., Stevenson J. S. and Kastelic J. P.(2012). Association between mRNA abundance of functional sperm function proteins and fertility of Holstein bulls. Theriogenology 78(9): 2007-2019 e2002. [DOI] [PubMed] [Google Scholar]
- 17. Krawetz S. A., Kruger A., Lalancette C., Tagett R., Anton E., Draghici S. and Diamond M. P.(2011). A survey of small RNAs in human sperm. Hum Reprod 26(12): 3401-3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lalancette C., Platts A. E., Johnson G. D., Emery B. R., Carrell D. T. and Krawetz S. A.(2009). Identification of human sperm transcripts as candidate markers of male fertility. J Mol Med(Berl) 87(7): 735-748. [DOI] [PubMed] [Google Scholar]
- 19. Lalancette C., Thibault C., Bachand I., Caron N. and Bissonnette N.(2008). Transcriptome analysis of bull semen with extreme nonreturn rate: use of suppression-subtractive hybridization to identify functional markers for fertility. Biol Reprod 78(4): 618-635. [DOI] [PubMed] [Google Scholar]
- 20. Liu T., Cheng W., Gao Y., Wang H. and Liu Z.(2012). Microarray analysis of microRNA expression patterns in the semen of infertile men with semen abnormalities. Mol Med Rep 6(3): 535-542. [DOI] [PubMed] [Google Scholar]
- 21. Liu W. M., Pang R. T., Chiu P. C., Wong B. P., Lao K., Lee K. F. and Yeung W. S.(2012). Sperm-borne microRNA-34c is required for the first cleavage division in mouse. Proc Natl Acad Sci U S A 109(2): 490-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mao S., Goodrich R. J., Hauser R., Schrader S. M., Chen Z. and Krawetz S. A.(2013). Evaluation of the effectiveness of semen storage and sperm purification methods for spermatozoa transcript profiling. Syst Biol Reprod Med 59(5): 287-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mao S., Sendler E., Goodrich R. J., Hauser R. and Krawetz S. A.(2014). A comparison of sperm RNA-seq methods. Syst Biol Reprod Med 60(5): 308-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Miller D., Ostermeier G. C. and Krawetz S. A.(2005). The controversy, potential and roles of spermatozoal RNA. Trends Mol Med 11(4): 156-163. [DOI] [PubMed] [Google Scholar]
- 25. Ostermeier G. C., Dix D. J., Miller D., Khatri P. and Krawetz S. A.(2002). Spermatozoal RNA profiles of normal fertile men. Lancet 360(9335): 772-777. [DOI] [PubMed] [Google Scholar]
- 26. Yang C. C., Lin Y. S., Hsu C. C., Wu S. C., Lin E. C. and Cheng W. T.(2009). Identification and sequencing of remnant messenger RNAs found in domestic swine(Sus scrofa) fresh ejaculated spermatozoa. Anim Reprod Sci 113(1-4): 143-155. [DOI] [PubMed] [Google Scholar]
- 27. Yuan S., Schuster A., Tang C., Yu T., Ortogero N., Bao J., Zheng H. and Yan W.(2016). Sperm-borne miRNAs and endo-siRNAs are important for fertilization and preimplantation embryonic development. Development 143(4): 635-647. [DOI] [PMC free article] [PubMed] [Google Scholar]