Abstract
The chick chorioallantoic membrane (CAM) is an extra-embryonic organ and thus well accessible for seeding and harvesting 3D cell cultures. Samples from CAM assays are suitable for protein and gene expression analysis as well as for immuno-histochemical studies. Here we present the CAM assay as a possible model to study autophagy in different types of cancer using immunohistochemistry. Compared with other 3D and xenograft models, the CAM assay displays several advantages such as lower costs, shorter experimental times, physiological environment and reproducibility. Macroautophagy hereafter simply referred to as “autophagy” is a conserved cellular catabolic process that degrades and recycles cellular components. Under basal conditions, autophagy contributes to the maintenance of cellular homeostasis whereas under cellular stress, such as starvation or hypoxia, autophagy is activated as a survival mechanism. Dysregulation of autophagy has been described in many diseases. In cancer, autophagy has been suggested to play a dual role. Whereas autophagy has been reported to play a tumor suppressive role in early stages, it seems to be rather tumor supportive in later stages. Here we provide a method to study autophagy in 3D microtumors of cancer cells grown on the CAM.
Keywords: CAM, Autophagy, p62, LC3B, OE19, Cancer, Immunohistochemistry
Background
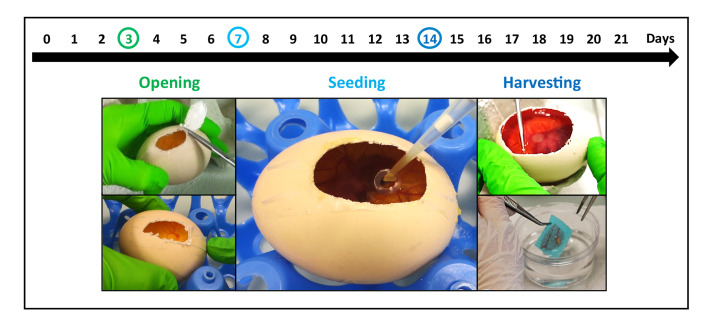
The chick chorioallantoic membrane (CAM) is a highly vascularized extra-embryonic membrane that has multiple functions during embryonic development, such as gas exchange (Romanoff, 1960). It is a well-known model to study blood vessels and angiogenesis in the context of tumor biology ( Kain et al., 2014 ; Nowak- Sliwinska et al., 2014 ). The chick development in the egg lasts 21 days (Romanoff, 1960). Three days after starting incubating the eggs, we cut a small round window into the eggshell for easy access to the CAM later in the procedure (Figure 1). At this time point the membrane is not yet formed and thus there is no risk to damage it. On Day 7 of embryo development the tumor cells are seeded on the CAM and seven days later (Day 14) the microtumors that developed on the membrane are harvested and analyzed. To this end, the tumors are excised, fixed, dehydrated, embedded and cut to apply immunohistochemical stainings for the autophagy-related markers: microtubule associated protein 1 light chain 3 beta (MAP1LC3B, short LC3B) and sequestosome 1 (SQSTM1, aka p62).
Figure 1. Overview of the workflow.
The development of the chicken in the egg takes in total 21 days. At Day 3 of incubation, when the CAM is not yet formed a window is cut into the eggshell. At Day 7 of incubation when the membrane is visible but not yet completely developed the cancer cells are seeded in a collagen scaffold and then transferred to the membrane. After another 7 days in the incubator, at Day 14 of development the tumor is harvested, fixed and embedded.
Experimentally assessing autophagy is challenging, since it is a dynamic process, and steady state expression levels of autophagy related genes do not give a complete picture of the active autophagic process ( Barth et al., 2010 ). In tissue, dot like staining of LC3B and p62 is considered a better indication of autophagic flux, defined as the degradation rate of cargo via autophagy ( Martinet et al., 2013 ). Our group previously established an automated LC3B and p62 staining and scoring protocol for tumor tissue ( Schläfli et al., 2015 ), which we apply here on microtumors generated in the CAM model. Using a 3D CAM xenograft model to broaden our knowledge on autophagy in tumor biology has several advantages. It is easy accessible for manipulation (addition of an autophagy blocking agent), for microscopy (for instance to analyze the growth and vascularization of the tumors) and it provides small tumors that are suitable for immunohistochemical analyses. Thus, the expression of different markers can be visualized in the tissue.
Materials and Reagents
Egg trays
Nitrile gloves (ecoSHIELD, ecoshieldTM eco nitrile PF 250, catalog number: 62512X)
Syringe 5 ml (Codan)
Needle (BC MicrolanceTM 3, 18 G)
Adhesive tape (MagicTM tape, invisible, 7.5 mm, Scotch, 8-1975R2)
Collagen sponge (AviteneTM UltrafoamTM, BARD, catalog number: 1050050)
Petri dish 10 cm (TC dish 100, standard) (Sarstedt, catalog number: 83.3902)
Tube 50 ml (Sarstedt, catalog number: 62.547.254)
Pipettes
Tissue embedding sponges (Kartell, catalog number: 02922-04)
Embedding cassette (Sakura, Tissue-Tek® Uni-Cassette®, Biopsy Grey, catalog number: 4172)
Plastic containers
Fertilized Ross 308 broiler eggs (Wüthrich Geflügel AG, Belp, Schweiz)
OE19 esophageal adenocarcinoma cells (ECACC, catalog number: 96071721)
Antibody (Dako, M7240)
Antibody (Novus Biologicals, NB600-1384)
Antibody (MBL rabbit polyclonal, LabForce, Nunningen, Switzerland, #PM0045)
RPMI-1640 (Sigma-Aldrich, Acuderm inc., catalog number: R8758)
Fetal bovine serum (Sigma-Aldrich, catalog number: F7524)
VPS34 inhibitor (VPS34-IN1, Selleckchem, catalog number: 1382716-33-3)
Accutase (StemProTM, Thermo Fisher, catalog number: A1110501)
Acetic acid (glacial, 100%) (Merck, catalog number: 100063)
Chloroform for analysis (Merck, catalog number: 102445)
Formalin (Thommen-Furler, catalog number: 612103)
Paraffin wax (Tissue-Tec®, Sakura, catalog number: 4656)
Methanol for analysis (Merck, catalog number: 106009)
Ethanol absolute (Merck, catalog number: 107017)
Xylene (Sigma, catalog number: 16446)
Mayer’s hemalum solution (Merck, catalog number: 109249)
Eosin yellowish (Gurr, catalog number: 34197) (for working solution see Recipes)
Phloxin B (Gurr, catalog number: 43064)
DAB detection kit (Venta Roche)
Polymer refine detection kit (Leica, Biosystems, catalog number: DS9800)
Carnoy’s fixative (see Recipes)
Eosin-phloxin working solution (see Recipes)
HER2 IHC (see Recipes)
Ki76 IHC (see Recipes)
LC3B IHC (see Recipes)
P62/SQSTM1 IHC (see Recipes)
Equipment
Operating scissors (14 cm, sharp, straight) (World Precision Instruments, catalog number: 501218-G)
Tweezers
Biopsy punch,8 mm (Acu-Punch, catalog number: P850)
Humidified incubator with rotating drawers (Rcom, Maru 300 MAX)
Humidified incubator stationary (Rcom, Maru hatcher & brooder 100)
Laminar flow hood (Kojair®, BioWizard Silver SL-130 Blue Series)
Water bath (GFL 1013, GFL_20007)
Stereo microscope (OLYMPUS, SZX16)
Oven (Binder, ED56)
Benchmark Ultra immunostainer (Venta Roche)
Leica Bond RX immunostainer (Leica Biosystems, Muttenz, Switzerland)
Software
Olympus CellSens Standard 1.15 (OLYMPUS CORPORATION, www.olympus-sis.com)
Procedure
-
Incubation
Clean the fertilized chicken eggs with distilled water and tissue, and incubate them in a rotating incubator (37 °C and 60% humidity).
-
Opening of the eggs (day 3 of incubation)
In the first step, clean the eggshell with ethanol on the side where the window will be made. Work under a laminar flow hood.
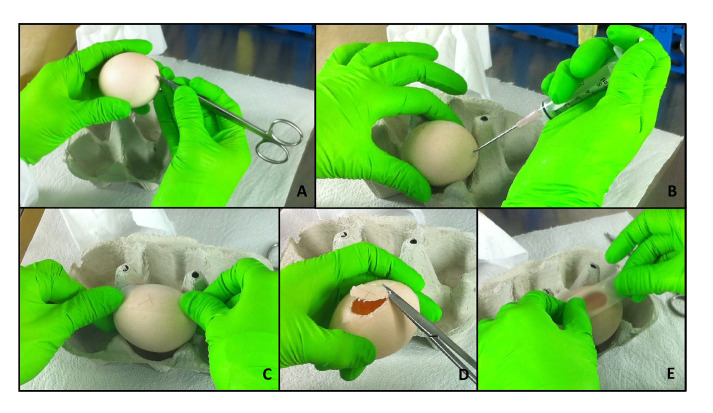
Put the egg sideways on an egg tray. Punch a hole in the bottom of the egg using a pair of sharp scissors (Figure 2A).
Now, remove approximately 3 ml of albumen with a syringe (Figure 2B). Make sure to introduce the needle in a steep angle in order to avoid injuring the egg yolk.
Discard the removed albumen and close the hole with a stripe of adhesive tape.
To avoid too much crumbling of the eggshell, put a stripe (long enough to cover the window that will be cut) of adhesive tape on the side of the egg (Figure 2C).
Again, using a sharp scissor punch a hole on one side of the scotch tape in order to insert the scissors into the egg. Now, cut out an oval window (Figure 2D).
Discard the cut out eggshell and close the window with an additional stripe of tape (Figure 2E).
From now on the egg is incubated in a stationary incubator on an egg tray (37 °C, 60% humidity).
-
Preparation of collagen scaffolds (24 h before seeding)
Work in a laminar flow hood. Punch one collagen scaffold per sample.
Use sterile tweezers and a blade to cut off an appropriate amount of the collagen sponge.
It is best to punch out the scaffolds in a sterile Petri dish. Use an 8 mm biopsy punch to get the number of scaffolds needed.
Soak the scaffolds in the medium that will be used later for cell seeding overnight or over the weekend at 4 °C in a 50 ml Falcon tube.
-
Seeding cancer cells (Day 7 of incubation)
Prepare cancer cells for seeding. To this end, the cells are detached with Accutase. Accutase is supposed to be less harsh on the cells during detachment than trypsin. We experienced better results when the cells were detached with Accutase. However, this effect could be cell line dependent and in some cases, Accutase might be replaced with Trypsin.
Cells are counted and a cell suspension of 1 x 106 cells in 20 µl (= 5 x 106 cells in 1 ml) is prepared that is needed per CAM per scaffold.
Preheat the prepared scaffolds in a water bath at 37 °C.
Prepare seeding cells on the membrane in a laminar flow hood. Prepare an egg tray, sterile tweezers, Petri dishes and pipettes.
Take the scaffolds out of the tube and drip them in a Petri dish.
Take an egg out of the incubator, put it into the hood and remove the adhesive tape that is closing the window.
Pipette 20 µl of cell suspension on a scaffold in the Petri dish and put the scaffold upside-down, so that the cells are sandwiched between the CAM and the scaffold, onto the membrane in the egg.
Close the window with a new stripe of tape and put the egg back into the incubator.
-
Harvesting of the tumors (Day 14 of incubation)
Put the eggs on ice (for at least 30 min).
During this time, prepare 200 ml (for around 30 samples) of Carnoy’s fixative (see Recipes). Work under a fume hood.
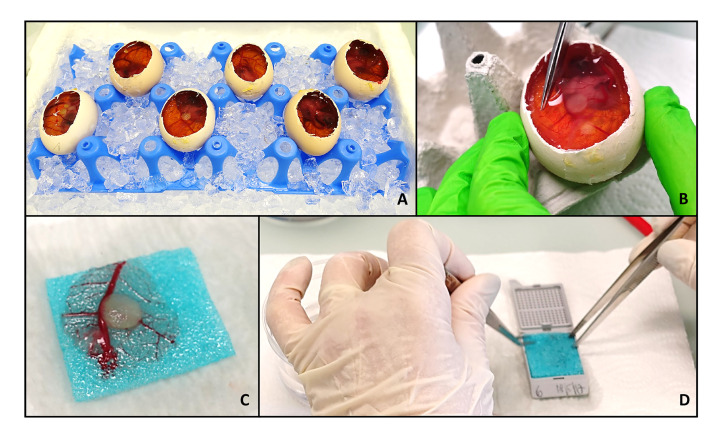
Cut the window in the eggshells wider (again use sharp scissors; the adhesive tape can be cut through), to enable easier handling (Figure 3A).
If wanted, take pictures of the tumor in the CAM using a stereomicroscope. If the tumor cells express green or red fluorescent protein (GFP or RFP) and the respective channels are available on your stereomicroscope, the fluorescence can be helpful to localize the microtumor.
Cut out the area of the CAM containing the tumor using sharp scissors and tweezers (Figure 3B).
Put the removed CAM with the tumor in formalin and place it on an embedding sponge (Figure 3C). Formalin is used to presoak the embedding sponge and to spread out the CAM on the sponge. Carnoy’s fixative used later is quite aggressive and changes the rigidity of the sponges, and is therefore not recommended to use for spreading out the membrane.
Take the sample out and drip dry. Put the sponge into a labeled embedding mold and cover the sample with an additional sponge, so that the tumor is sandwiched between two sponges (Figure 3D).
Close the mold and put the sample in Carnoy’s fixative for 12 h.
Melt paraffin pellets of about 260 g in two suitable plastic containers (final volume about 400 ml) at 56-60 °C, overnight.
-
Dehydration and embedding (12 h after harvesting)
After 12 h of fixation start the dehydration process.
Put the fixed tissue 2 times in methanol for 30 min each.
Afterwards, put the samples 2 times in ethanol for 20 min each.
This is followed by 2 Xylene baths for 15 min each.
Now, drip-dry cassettes on paper tissue, to remove as much Xylene as possible.
Incubate the samples twice each for 30 min at 56 °C in warm paraffin to allow soaking of the tissue.
-
Cutting and staining
For further processing (embedding, cutting and staining), the samples were given to a routine histology pathology laboratory.
Figure 2. Opening of the eggs.
At day 3 of incubation, the eggs are opened and further kept in a stationary incubator. In a first step, a small hole is punched into the eggshell at the “bottom” of the egg (A). Then, around 3 ml of albumen are removed using a syringe and the hole is closed with adhesive tape (B). Another stripe of tape is glued on the side of the egg (C). A window is cut in the area of the scotch to avoid eggshell crumbs to fall on the membrane, using sharp scissors (D). Finally, the window is closed with an additional stripe of tape (E).
Figure 3. Harvesting of the tumor.
To harvest the tumors, the eggs are put on ice for at least 30 min. During this time, the windows are enlarged to enable a more precise handling (A). If desired, pictures of the tumors in the egg can now be taken. Labeling of the tumor cells with a fluorophore may help to identify the area of tumor if the scaffold is barely visible. Then the part of the CAM around the tumor is excised (B), sandwiched between two embedding sponges (C) and placed in an embedding mold (D). The samples are now ready to be fixed, plunged in paraffin and embedded.
Data analysis
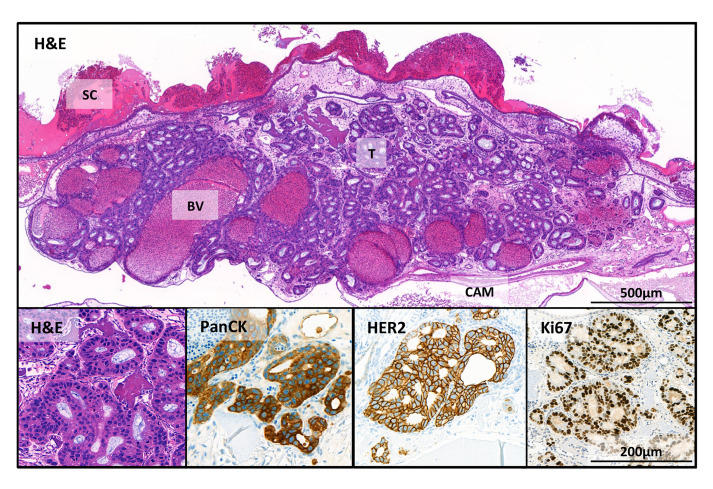
The tumor cells can be well visualized after hematoxylin-eosin staining (Figure 4). In the overview picture, glandular structures newly formed by the OE19 esophageal adenocarcinoma tumor cells (T) can be observed. The micro tumors that developed in the CAM assay are comparable to tumors formed from OE19 cells in mouse xenograft models ( Wang et al., 2014 ). Some tumor OE19 cells show invasive growth into the stroma. In addition, parts of the collagen scaffold (SC), blood vessels (BV) in the lower part of the micro tumor and the epithelial layers of the CAM can be seen. Tumor cells are made visible by immunohistochemistry (IHC) using pan-Cytokeratin staining. A high proliferation activity was demonstrated by Ki67 staining (human specific). Overexpression of Her2 in tumor cells was also demonstrated by IHC staining (for antibody information see “Recipes”).
Figure 4. Immunohistochemical stains of the tumor in the CAM.
OE19 esophageal adenocarcinoma cells (1 x 106 inoculated on CAM for 7 days). SC = scaffold; BV = blood vessels; T = tumor and CAM = chorioallantoic membrane. Shown here are stains with hematoxylin and eosin (H&E); and human specific IHC stains: pan-cytokeratin (PanCK), an epithelial marker; HER2, a receptor tyrosine kinase, which is amplified in OE19 cells and Ki-67 that is associated with proliferation.
Before seeding OE19 cells, one group was treated with an early stage autophagy inhibitor VPS34-IN1, which is considered rather specific for macroautophagy ( Bago et al., 2014 ). The inhibitor was applied at a concentration of 5 µM to the cells before seeding them on the CAM.
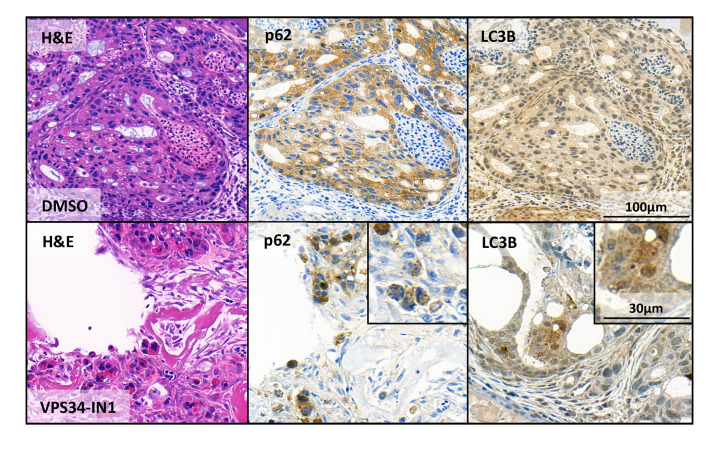
These are the same concentrations we used before for in vitro experiments ( Janser et al., 2018 ). After excision and staining with the autophagy markers LC3B and p62 untreated and pretreated tumors were compared (Figure 5). Where in the untreated sample the stainings for p62 and LC3 are continuous, the staining pattern of the VPS34-IN1 treated sample shows clearly dot formation. We already showed that VPS34-IN1 inhibits macroautophagy in vitro in OE19 cells ( Janser et al., 2018 ), thus this is a clear indication that we can capture blocked autophagy in microtumors grown in the CAM model. As the CAM represents a relatively cheap 3D tumor model allowing to assess several parameters (IHC, microscopy), this could be the method of choice to complement in vitro data and gain more insight into the role of autophagy in cancer.
Figure 5. Assessment of macroautophagy using the CAM assay.
OE19 esophageal adenocarcinoma control cells (upper panel “DMSO”) and treated with the macroautophagy inhibitor VPS34-IN1 (lower panel “VPS34-IN1”). Shown are stains with hematoxylin and eosin (H&E) and the two macroautophagy markers p62 and LC3B. In the magnifications, the dot-like structures of the autophagy markers indicative for ongoing macroautophagy are clearly visible.
Notes
The protocol was established with OE19 esophageal adenocarcinoma cells. Additionally, we tested a panel of breast cancer cell lines (SKBR3, T47D and MCF7) and two colon cancer cell lines (LS174T, CaCo2) with which we observed good results as well.
The collagen scaffold is easier to handle than Matrigel® also resulting in better grafting. Since the collagen sponge absorbs the cell suspensions, fewer cells were lost in the seeding step compared to our experiments with Matrigel® scaffolds.
During manipulation outside the incubator the eggs should not get cold. Thus, there is a much higher success rate in terms of survival of embryos if only one egg is handled at a time and every working step is well prepared.
Depending on which cell line will be used tumor cells will grow better on the CAM when seeded in serum free medium.
The cell number seeded onto the collagen scaffold on the CAM can vary from cell line to cell line. Usually, between 1 x 106 and 2 x 106 cells are seeded.
This CAM protocol has been optimized to assess autophagy in a 3D-model. Generally, this improved CAM protocol allows to determine other aspects of tumor biology as well, e.g., invasion of genetically modified or treated cancer cells into the CAM, and cell death by immunohistochemical staining for cleaved caspase-3. Using a collagen sponge as a scaffold could improve CAM assays for therapy response studies of cancer cells in 3D.
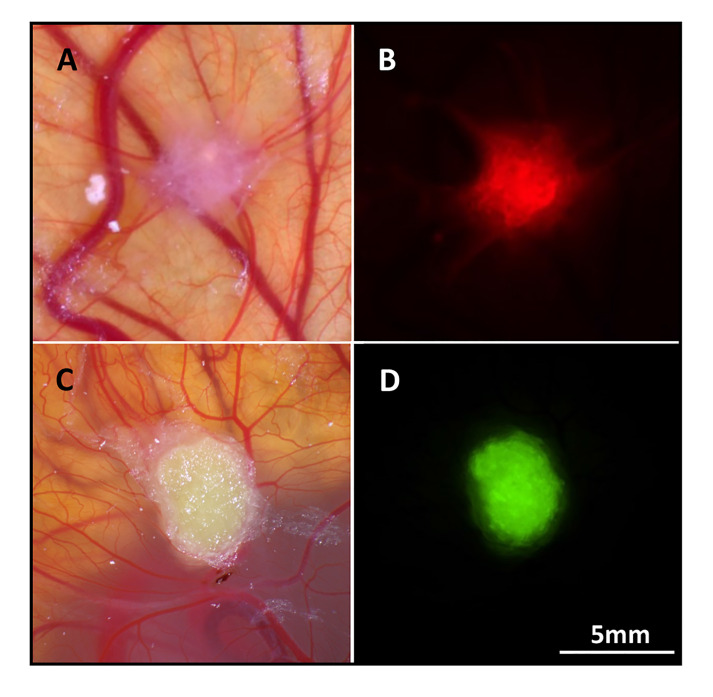
The tumor cells can be labeled with EGFP, mCherry or other fluorescent proteins (Figure 6). This will facilitate localization of the microtumors on the CAM for excision (sometimes the tumors are transparent and difficult to find). Another advantage of labeling the cells is that there is an indication whether a microtumor formed and how big it is. If there is a new tumor visible, there is no need to continue with the procedure. Moreover, the intensity of the fluorescence can be used as a viability read out as well.
Figure 6. mCherry and EGFP labeled tumor cells in the CAM.
mCherry labeled MCF7 breast cancer cells. Bright-field (A) and red fluorescence (B) picture. EGFP labeled OE19 esophageal adenocarcinoma cells, bright field (C) and green fluorescence (D) picture.
Recipes
-
Carnoy’s fixative
Mix 60% methanol absolute, 30% chloroform and 10% acetic acid glacial.
-
Eosin-phloxin working solution
Prepare a 1% stock solution with 10 g of the eosin powder and 1,000 ml distilled water.
Prepare a 1% stock solution with 10 g of the phloxin powder and 1,000ml distilled water.
Prepare the working solution with 10% of the eosin stock solution, 1% of the phloxin stock solution, 88.5% ethanol and 0.5% acetic acid glacial.
-
HER2 IHC
Performed with a Benchmark Ultra immunostainer (Venta Roche). The antibody (clone 4B5, Venta Roche, Rotkreuz, Switzerland) was applied undiluted, with EDTA antigen retrieval for 35 min
-
Ki76 IHC
Performed with a Leica Bond RX immunostainer (Leica Biosystems, Muttenz, Switzerland). The antibody (M7240, Dako) was diluted 1:200, with Tris antigen retrieval for 30 min at 95 °C
-
LC3B IHC
As well done with the Leica Bond RX immunostainer. The antibody (#NB600-1384, Novus Biologicals) was diluted 1:4,000 and incubated at 95 °C for 30 min
-
P62/SQSTM1 IHC
Performed with the Leica Bond RX immunostainer. The antibody (#PM0045, MBL rabbit polyclonal, LabForce, Nunningen, Switzerland) was diluted 1:9,000 and incubated at 95 °C for 30 min
Note: For visualization, the Bond Polymer Refine Detection kit (Leica, Biosystems, DS9800) and the Ultra view DAB detection kit (Venta Roche) were used for visualization according to the instructions of the manufacturer.
Acknowledgments
The authors thank Deborah Krauer and Silvia Rhis for their excellent technical assistance. The authors acknowledge the Translational Research Unit (TRU) of the Institute of Pathology, University of Bern, particularly Dr. José Galván Hernándes and Carmen Cardozo for the excellent technical support on this project. The establishment of this method was supported by a grant from the Swiss Cancer League (KFS 3700-08-2015).
Competing interests
The authors declare no conflict of interest. The funders had no role in the design of the study, the development of the method, in the writing of the manuscript, and in the decision to publish the results.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1. Bago R., Malik N., Munson M. J., Prescott A. R., Davies P., Sommer E., Shpiro N., Ward R., Cross D., Ganley I. G. and Alessi D. R.(2014). Characterization of VPS34-IN1, a selective inhibitor of Vps34, reveals that the phosphatidylinositol 3-phosphate-binding SGK3 protein kinase is a downstream target of class III phosphoinositide 3-kinase. Biochem J 463(3): 413-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Barth S., Glick D. and Macleod K. F.(2010). Autophagy: assays and artifacts. J Pathol 221(2): 117-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Janser F. A., Adams O., Bütler V., Schläfli A. M., Dislich B., Seiler C. A., Kröll D., Langer R. and Tschan M. P.(2018). Her2-targeted therapy induces autophagy in esophageal adenocarcinoma cells. Int J Mol Sci 19(10): e3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kain K. H., Miller J. W., Jones-Paris C. R., Thomason R. T., Lewis J. D., Bader D. M., Barnett J. V. and Zijlstra A.(2014). The chick embryo as an expanding experimental model for cancer and cardiovascular research. Dev Dyn 243(2): 216-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Martinet W., Schrijvers D. M., Timmermans J. P., Bult H. and De Meyer G. R.(2013). Immunohistochemical analysis of macroautophagy: recommendations and limitations. Autophagy 9(3): 386-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nowak-Sliwinska P., Segura T. and Iruela-Arispe M. L.(2014). The chicken chorioallantoic membrane model in biology, medicine and bioengineering. Angiogenesis 17(4): 779-804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Romanoff A. L.(1960). The avian embryo: structural and functional development. The Macmillan Co. New York [Google Scholar]
- 8. Schläfli A. M., Berezowska S., Adams O., Langer R. and Tschan M. P.(2015). Reliable LC3 and p62 autophagy marker detection in formalin fixed paraffin embedded human tissue by immunohistochemistry. Eur J Histochem 59(2): 2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang Z., Da Silva T. G., Jin K., Han X., Ranganathan P., Zhu X., Sanchez-Mejias A., Bai F., Li B., Fei D. L., Weaver K., Carpio R. V., Moscowitz A. E., Koshenkov V. P., Sanchez L., Sparling L., Pei X. H., Franceschi D., Ribeiro A., Robbins D. J., Livingstone A. S. and Capobianco A. J.(2014). Notch signaling drives stemness and tumorigenicity of esophageal adenocarcinoma. Cancer Res 74(21): 6364-6374. [DOI] [PMC free article] [PubMed] [Google Scholar]