Abstract
The therapeutic strategies of stage II/III colorectal cancer (CRC) patients after curative surgery remain controversial. In the clinical decision-making process, oncologists need to answer questions such as whether adjuvant chemotherapy is necessary or which therapeutic regimen should be given to each patient. At present, whether adjuvant chemotherapy should be applied is primarily based on histopathological features and clinical risk factors. However, only a fraction of patients can benefit from it. More rigorous stratifying biomarkers are urgently needed to help further distinguishing these populations of patients. Recent progress in next-generation sequencing and high-throughput technologies has greatly promoted biomarker discovery as well as our understanding of the underlying mechanisms in CRC. Novel genetic and epigenetic biomarkers that are associated with prognosis or therapeutic responses have emerged. In this review, we discuss the strategies of biomarker discovery and summarize the status and assess the utility of previously published biomarkers in CRC.
Keywords: colorectal cancer, biomarker, prognostic, predictive, genomic, transcriptomic, epigenetic, non-coding RNA, lncRNA, miRNA, circRNA
Graphical Abstract
Xu et al. discuss the strategies of biomarker discovery and summarize the current status and assess the clinical effectiveness of previously published genomic and transcriptomic biomarkers and signatures in stage II/III colorectal carcinoma. Novel prognostic and predictive molecular markers are highlighted and their potential applications are discussed.
Main Text
Colorectal cancer (CRC) was estimated to be the fourth most frequently diagnosed cancer and the second most common cause of cancer deaths worldwide in 2018.1 CRC has been recognized as a disease affecting mostly developed countries. However, in recent years, the incidence of CRC has kept increasing in developing countries, such as China, possibly due to changes in lifestyle and nutritional habits.2 Early-stage CRC is mostly asymptomatic, and many patients are diagnosed at advanced stages.3
The tumor-node-metastasis (TNM) stage remains the most potent prognostic factor in the clinical decision-making process. Based on current guidelines, most stage II CRC patients are treated surgically without adjuvant chemotherapy. However, almost 15% of these patients undergo tumor recurrence and death caused by disease progression.4,5 Whether a stage II CRC patient should receive adjuvant chemotherapy after surgery largely depends on the recurrence risk assessed by a group of clinical factors.6,7 Still, a considerable proportion of stage II patients who are evaluated as low-risk patients later suffer from recurrence and progression. Stage III patients routinely receive adjuvant chemotherapy after surgery.8 However, evidence showed these patients did not respond equally.9, 10, 11, 12 Therefore, the need for biomarkers to more precisely identify stage II/III CRC patients that are suitable for adjuvant chemotherapy is highlighted.
The biomarkers that doctors use to predict CRC patients’ prognosis and therapeutic response at present, such as carcinoembryonic antigen (CEA) and carbohydrate antigen 19-9 (CA19-9),13,14 have relatively low sensitivity and specificity.15,16 In this case, more meticulous biomarkers are needed. Thanks to the development of genomic high-throughput screening technologies, including next-generation sequencing and microarray analysis, a large number of molecular biomarkers and signatures with potential clinical prognostic and predictive values have been discovered via comprehensive association and bioinformatics analyses. Notably, several genomic-based biomarkers, such as the mismatch repair (MMR) or microsatellite instability (MSI) status, which plays a role as a predictive marker for adjuvant chemotherapy in patients with stage II CRC, have already entered into clinical practice and been validated.17, 18, 19, 20
In this review, we summarize novel genomic and transcriptomic biomarkers as well as signatures reported recently with prognostic and predictive potentials in stage II/III CRC. We broadly examined current studies, focusing mainly on transcriptomic markers and signatures, which include newly discovered molecules such as microRNAs (miRNAs), long noncoding RNA (lncRNAs), and circular RNAs (circRNAs). We also assessed the clinical effectiveness of those already revealed and discussed their potential applications. The purpose of this review is to highlight some of the new candidate prognostic and predictive molecular markers for stage II/III CRC and assess their clinical utility for possible future applications.
Biomarker Discovering Strategies
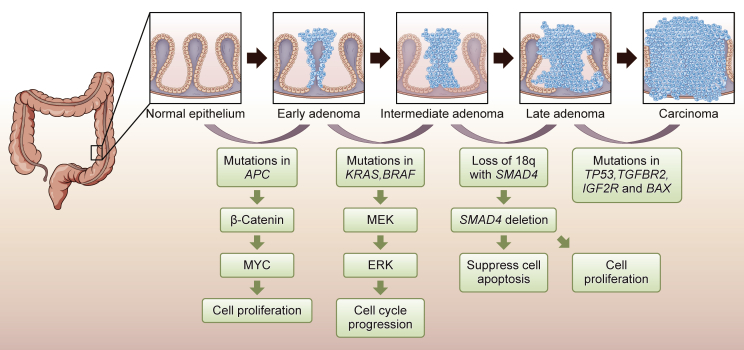
Currently, two major strategies are adopted by researchers to discover new molecular biomarkers: mechanism-based strategies and unbiased high-throughput screening. In the first scenario, a biomarker study often begins with candidate genes that play essential roles in tumorigenesis and cancer development with already defined molecular mechanisms that have been fully investigated both in vitro and in vivo. These genes are usually hallmark genes in biological processes such as the cell cycle, apoptosis, and drug metabolism.21, 22, 23 In the case of CRC, Vogelstein et al.24 proposed a model describing the sequential progression from adenoma to carcinoma process (Figure 1). Biomarkers developed by mechanism-based strategies include Kirsten rat sarcoma viral oncogene (KRAS), adenomatous polyposis coli (APC), and tumor protein p53 (TP53) that can be traced back to Vogelstein’s model.25,26 Investigators first group patients by clinical parameters (e.g., responders versus non-responders to fluorouracil-based chemotherapy), then they compare the expression levels of the candidate genes between groups and evaluate the power of these genes in distinguishing patients’ grouping status. However, only discrete genes with well-known roles, but not new molecules, can be investigated and developed as biomarkers using this hypothesis-driven, mechanism-based strategy. Furthermore, this strategy might lead to unilateral conclusions considering the heterogeneity of CRC.
Figure 1.
Sequential Progression Model in Colorectal Cancer
This is an oversimplified model describing the tumor progression sequence of CRC from adenoma to carcinoma. The development of CRC is a stepwise accumulation of genetic and epigenetic events, which align with clinical pathological changes.24,27 The loss of APC, gatekeeping gene of CRC tumorigenesis, leads to small adenoma formation. Next, mutations in KRAS promote larger adenomas and gradually early carcinomas. The loss of SMAD family member 4 (SMAD4) and mutations in TP53 as well as PIK3CA are acquired later in carcinoma. These serial alterations in genes that regulate cellular differentiation, proliferation, and apoptosis pathways give rise to CRC carcinogenesis.28
While target-based approaches can search only a small part of the genome, unbiased high-throughput profiling has opened new avenues for marker discovery by screening the whole genome. For example, whole-transcriptome expression can be profiled comprehensively by high-throughput technologies such as transcriptomic assays and RNA sequencing. Moreover, by the use of modified library construction or capture probes, as well as other bioinformatics analysis strategies, the technology can help develop novel RNA biomarkers such as miRNAs,29 lncRNAs,30 and circRNAs.31 Specifically, CRC can be better characterized by transcriptomic subtypes that encompass not only information within the tumor but also stromal and immune components.32 Genomic and bioinformatics technologies have yielded many potential biomarkers and related signatures, which may prove useful with prognostic and predictive values. Data acquired from high-throughput technologies such as microarrays are usually enormous and disordered. Researchers use hierarchical clustering analysis to examine these data.33 Hierarchical clustering analysis is considered an unsupervised clustering method that is suitable for an exploratory analysis to determine how microarray groups cluster together according to similar features without taking experimental variables into account. Patients are sorted according to similarities in their gene expression profiles to predict different clinical outcomes.
Genomic Biomarkers
Genomic instability and oncogene mutations, tumor suppressor gene mutations, and mismatch repair genes can serve as biomarkers. Here, we summarize a few clinically approved genomic biomarkers in stage II/III CRC.
Chromosomal Instability (CIN) and MSI
CIN and MSI are two types of genomic instabilities. CIN refers to the increased rate of chromosomal gain and loss that leads to numerical or structural chromosome aberrations. MSI is defined as tumors having a defective DNA mismatch repair system due to the inactivation of either one of the following genes as the dominant genomic feature: mutL homolog 1 (MLH1), MLH3, mutS homolog 2 (MSH2), MSH3, MSH6, or PMS1 homolog 2 (PMS2).26,34,35 CIN is more commonly presented in CRC than MSI.36 The prognostic value of both CIN and MSI has been validated in a large meta-analysis.37,38 Patients with CIN disease have a worse prognosis than those without CIN. In addition, MSI-high (MSI-H) patients have a better prognosis than microsatellite stable (MSS) patients in stage II CRCs39 and maintain their survival advantages in the presence of 5-fluorouracil (5-FU) treatment;40 however, MSI has no prognostic value in stage III CRCs.41
CpG Island Methylation Phenotype (CIMP)
Aberrant DNA methylation is the most broadly studied epigenetic alteration in cancer. The CIMP was first described in CRC in 1999 by Toyota et al.42 It was characterized as a cluster of exceptionally hypermethylated CpG dinucleotides. Weisenberger et al.43 later defined CIMP in CRC based on the methylation status of five genes: calcium channel voltage dependent alpha 1G (CACNA1G), insulin-like growth factor 2 (IGF2), neurogenin 1 (NEUROG1), RUNX family transcription factor 3 (RUNX3), and suppressor of cytokine signaling (SOCS). Multiple, extensive clinical studies have demonstrated that specific methylated DNA signatures could be developed as prognostic and predictive biomarkers in CRC. Several independent studies reported that CIMP-positive cancers were correlated with an unfavorable prognosis, including a cohort of more than 600 MSS CRC patients by Lee et al.44 and a cohort of 206 stage III CRC patients by van Rijnsoever et al.45 This conclusion was validated later in two other cohorts of stage II/III patients.46,47 However, the opinions differ: some studies have suggested that the prognosis of CIMP-positive CRC patients depends on the MSI status of the tumor,48,49 while other studies have suggested that the poor prognosis of CIMP-positive CRCs was from coexisting B-Raf proto-oncogene (BRAF) V600E mutations.44,50, 51, 52 These contrary findings may be due to the heterogeneity of patient cohorts or the different CIMP criteria used in different studies. However, the CIMP status is still promising and can be further investigated as a prognostic factor.
KRAS and BRAF
The KRAS proto-oncogene encodes a small GTPase that affects cell proliferation and differentiation.53 KRAS mutations in codons 12, 13, and 61 impair the GTPase activity of the encoded protein, which leads to the consistent activation of RAS/RAF signaling.54 KRAS mutations occur at the early stage of CRC, accounting for one-third of all cases, and a large majority of the mutations are located in codon 12 (approximately 80%), followed by codon 13.55 Many studies have evaluated KRAS as a prognostic biomarker based on its association with CRC outcomes.50,55, 56, 57, 58 The RASCAL study, a large meta-analysis, found that the glycine to valine substitution in codon 12 of KRAS was associated with a poor prognosis in stage III CRC patients.56 However, several other large studies found no association.50,57,58 In stage II CRC, no prognostic significance was found of the KRAS mutation type.59 BRAF is the downstream target of KRAS. Mutations in KRAS and BRAF seem to be mutually exclusive.60,61 BRAF mutation was considered a prognosticator for poor survival in stage II/III CRC patients receiving adjuvant chemotherapy.62 The most famous BRAF V600E mutation was also reported to confer a worse prognosis in stage II/III CRC patients.63
Transcriptomic Biomarkers
More than 90% of the human genome is actively transcribed, but only less than 2% encodes protein-coding genes that produce translational messenger RNA (mRNA) transcripts, while most of these transcripts are noncoding RNAs (ncRNAs).64 As protein-coding mRNAs have a short half-life and their expression changes enormously according to the physiological status, they are not an ideal prognosticator. Only a few studies have applied mRNA levels as biomarkers. For example, Takahashi et al.65 demonstrated that high never in mitosis A (NIMA)-related kinase 2 (NEK2) mRNA levels were associated with a poor prognosis in 180 CRC patients.
ncRNAs have more advantages than translational mRNAs and thus can be developed into biomarkers based on their various types and remarkable stability. The characteristics of different types of RNA molecules and their potential as biomarkers are summarized in Table 1. In addition to the well-established types, such as transfer RNAs (tRNAs) and ribosomal RNAs (rRNAs), ncRNAs also comprise many recently identified novel transcripts, including miRNAs, circRNAs, small nucleolar RNAs (snoRNAs), PIWI interaction RNAs (piRNAs), and lncRNAs. Many studies have suggested that ncRNAs play important roles in various biological processes, such as the cell cycle, proliferation, migration, and apoptosis.66 Several investigators have observed altered ncRNA expression patterns in human diseases, including CRC. ncRNA alterations can act as biomarkers to predict patient outcomes. Their potential is enormous. Compared to genomic markers, transcriptomic biomarkers are mostly quantitative markers rather than markers of a discrete gene status. Thus, the disease status can be better characterized by a continuous change than by a positive-negative index. This discovery is attributed to the rapid development of unbiased high-throughput screening methods. Quantitative real-time PCR is generally considered the “gold standard” method used to measure target RNA expression. It facilitates the possible clinical application of transcriptomic biomarkers. Here, we summarize the studies that identified ncRNA biomarkers with clinical prognostic and predictive values using various patient cohorts with different clinical characteristics.
Table 1.
Characteristics of Different RNA Molecules as CRC Biomarkers
| Molecule | mRNA | miRNA | lncRNA | circRNA |
|---|---|---|---|---|
| Abundance | high | Low | low | low |
| Stability | + | ++ | + | +++ |
| Detection method | qPCR sequencing | microarray (array-CGH), sequencing, qPCR | microarray (array-CGH), sequencing, qPCR | qPCR |
| Supporting evidence | weak | strong | intermediate | weak |
CGH, comparative genomic hybridization.
miRNAs
miRNAs mediate mRNA degradation and the inhibition of mRNA translation.67, 68, 69 Croce and colleagues70 first described the role of miRNAs in cancer in 2002. They found that the expression of miR-15 and miR-16 was decreased in patients with chronic lymphocytic leukemia. Since then, hundreds and thousands of miRNAs have been reported to be dysregulated in various human malignant diseases, including CRC, with a regulatory role in the expression of important oncogenes and tumor suppressor genes.71 miRNAs are involved in multiple pathways that are important in CRC development, including the TP53, Wnt/β-catenin, RAS/mitogen-activated protein kinase (MAPK), and phosphatidylinositol 3-kinase (PI3K)/AKT signaling pathways.72, 73, 74, 75 They also play important roles in regulating drug resistance.76, 77, 78 However, most of these results are based on in vitro studies and remain to be assessed in clinical sample sets.
miRNA biomarker studies in human cancer have emerged in large numbers during the past decade. The underlying reasons for this increasing interest are based on some of their unique characteristics. First, miRNAs are tolerant to RNase-mediated degradation because of their short length and hairpin-loop structure,79 enabling extraction from a number of clinical specimens, including formalin-fixed, paraffin embedded (FFPE) tissues and various kinds of body fluids, including blood, saliva, urine, and feces. Second, miRNA stability is remarkable under a variety of laboratory conditions. Cell-free miRNAs are often contained in high-density lipoprotein particles, apoptotic bodies, microvesicles, and exosomes80, 81, 82, 83 and are thus protected from degradation. In addition, their binding to argonaute-2 (Ago-2) also increases their stability.80 Third, cancer cells can secrete miRNAs into the blood and digestive tract.84,85 Therefore, the stability of miRNAs and their extensive presence in the body (e.g., blood, feces, cancer cells, cells near the cancer) have made miRNAs promising candidates for prognostic and predictive biomarkers; thus, miRNAs have been widely investigated.
Schetter et al.86 evaluated the expression levels of 389 miRNAs in 84 CRC patients and matched normal tissues by a microarray-based approach and validated the results by quantitative PCR. They found that high miR-21 expression was associated with poor survival in CRC patients. They also discovered that miR-21 overexpression was associated with a poor response to 5-FU-based adjuvant chemotherapy. Several other studies confirmed their conclusions independently. miR-21 has been proposed as both a prognostic and predictive marker for CRC.87 Furthermore, Hur et al.88 and Toiyama et al.89 found that miR-200c was elevated in CRC, and they conducted a three-phase study using 446 colorectal specimens from both the serum and primary tumor from stage II and III CRC patients. They found that high serum miR-200c expression levels indicated lymph node metastasis and a poor prognosis. The let-7 family has also been broadly studied in the tumorigenesis of CRC. Members of this family were identified as tumor-suppressive miRNAs and downregulated in CRC tumor tissues.90,91 Nakajima et al.92 proposed that let-7g and miR-181b could act as indicators for the response to 5-FU-based adjuvant chemotherapy in CRC patients. In addition, many dysregulated miRNAs have been proposed as prognostic and predictive CRC markers by different researchers, as shown in Table S1.
lncRNAs
A lncRNA is another type of single-stranded ncRNA transcript that consists of more than 200 nt.93 Only a few lncRNAs, such as H19 and Xist, were characterized in the pregenomic era. However, since the early 2000s, the elucidation of the human genome and transcriptome has led to an increasing number of studies on lncRNAs. Thousands of lncRNAs have been identified in the human genome, and this number continues to grow.94,95 lncRNAs are the second most commonly studied ncRNA, following miRNAs. Many studies have suggested that lncRNAs are involved in multiple processes of cancer biology.96 These studies have shown that lncRNAs function through a variety of regulatory mechanisms, including translational activation or inhibition, mRNA degradation, acting as RNA decoys or miRNA sponges, recruiting chromatin modifiers as scaffolds, and in the regulation of protein activity or stability.97
A growing body of literature indicates that the dysregulated expression of lncRNAs may be functional in human malignancies, including CRC, with clinical implications. For instance, HOX antisense intergenic RNA (HOTAIR) is reported to be an oncogenic lncRNA. HOTAIR cooperates with polycomb repressive complex 2 (PRC2) to reprogram chromatin organization and promote metastasis in breast cancer.98 Kogo et al.99 first demonstrated in CRC that high HOTAIR expression was associated with distant metastasis and a poor prognosis. Their finding was later validated by two other groups.100,101 Li et al.102 also discovered that a high expression level of HOTAIR was associated with a poor response to 5-FU-based treatment and predicted poor overall survival (OS) and recurrence-free survival (RFS) in CRC patients who received 5-FU-based chemotherapy. HOTAIR can be established as a both prognostic and predictive marker in CRC.
Another lncRNA, colorectal neoplasia differentially expressed (CRNDE), is overexpressed in a variety of cancers, including CRC.103 Several independent studies conducted by different groups of researchers reached the same conclusion: high CRNDE expression is associated with a poor prognosis in CRC patients.104,105 Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) was originally identified as a metastasis predictor in non-small cell lung cancer patients.106 Later, Zheng et al.107 found MALAT1 to be prognostic in CRC patients. In addition, several other lncRNAs were found to be prognostic in CRC, as shown in Table S2. lncRNAs have been detected in extracellular vesicles,108,109 and some of these studies successfully detected and measured certain lncRNA levels in the serum or plasma,30,110 indicating the potential of lncRNAs to serve as minimally invasive biomarkers in CRC.
circRNAs
circRNAs, with lengths of hundreds to thousands of nucleotides, are a group of naturally occurring endogenous ncRNAs.111 circRNAs were first discovered in 1976 in viroids of RNA viruses as single-stranded covalently closed circRNA molecules.112 Since then, circRNAs have been broadly found in different species, including humans.113 The development of next-generation sequencing techniques and bioinformatics has allowed researchers to further investigate the functions and mechanisms of circRNAs.
Currently, circRNAs are much less studied than miRNAs and lncRNAs, with their functions remaining mostly unknown. A few circRNAs have been reported to be functional in CRC and may act as potential prognostic biomarkers. For example, the circRNA ciRS-7, one of the most studied circRNAs, is a miR-7 sponge that results in reduced miR-7 activity and increased levels of miR-7 targets.114,115 miR-7 is considered a tumor suppressor in CRC, and it negatively controls the expression of several oncogenes.115 Impairing miR-7 activity would have an important impact on the cell phenotype. Weng et al.116 reported that ciRS-7 was overexpressed in CRC cancerous tissue and that high ciRS-7 expression indicated a poor patient prognosis. They performed a multivariate survival analysis in both training and validation cohorts and found that ciRS-7 expression was an independent risk factor for OS. A similar case was reported by Zeng et al.,117 which indicated that circHIPK3 also acts as a miR-7 sponge, and that a high expression level of circHIPK3 is an independent prognostic factor of poor OS in CRC patients. The most reported mechanism of circRNAs is acting as miRNA sponges. They may also encode proteins with biological functions.118 It has been reported that circRNAs can be transferred to exosomes and secreted into the circulatory system as well as other body fluids, indicating the possibility of circRNAs acting as noninvasive biomarkers.119 However, investigations on circRNAs are mostly in vitro studies and lack validation in clinical specimens and patient cohorts, and the potential for prognostic biomarker discovery in circRNAs has yet to be exploited.
Transcriptomic Signatures
In addition to individual markers, gene panels comprised of ncRNAs have been developed to identify high-risk CRC patients. Specific expression patterns of several genes may yield higher power in distinguishing patients with different prognosis than a single gene. Signatures have higher sensitivity and specificity than do single gene biomarkers. CRC is not a one-gene show, as hundreds if not thousands of genes have been effective during the genesis and progression of the disease. It is too complicated to be simply represented by the status of one single gene. Using signatures as biomarkers, we generate scores that could more accurately represent the disease status, which allows clinicians to stratify patients more meticulously into groups, instead of just two groups depending on the positive/negative status of one gene.
Zhang et al.29 examined the expression of 1,849 miRNAs in 40 paired stage II colon tumors and adjacent normal mucosa tissues using miRNA microarrays and found 35 differentially expressed miRNAs. They subsequently built a miRNA-based classifier comprising six miRNAs (miR-20a-5p, miR-21-5p, miR-103a-3p, miR-106a-5p, miR-143-5p, and miR-215) in a testing cohort of 138 patients and an independent validation cohort of 460 patients. The six-miRNA signature was able to identify stage II CRC patients with a high risk of recurrence. We recently developed a four-circRNA-based risk score (cirScore) (i.e., hsa_circ_0122319, hsa_circ_0087391, hsa_circ_0079480, and hsa_circ_0008039) to predict postoperative recurrence in stage II/III colon cancer patients using a cohort of 667 patients. Patients with high cirScores had shorter DFS and OS than did patients with low cirScores in the training cohort of 249 patients. The prognostic capacity of the classifier was validated in internal and external cohorts of 122 and 180 patients, respectively.31
Several other studies also developed different classifiers for patient stratification, including miRNA signatures, lncRNA signatures, and signatures comprised of both miRNAs and lncRNAs. However, there were more or less inherent differences in the patient characteristics of the cohorts; details are shown in Table S3.
Conclusions
Currently, the TNM stage and histopathological and clinical risk factors remain the most commonly applied risk factors in the clinical decision-making process. However, the recurrent risk assessment system of patients with stage II CRC needs further improvement to optimize the treatment strategy. Also, all stage III patients are unreasonably given adjuvant chemotherapy after curative surgery indistinguishably. With more accurate biomarkers, clinicians could better identify stage II patients who are more likely to go through recurrence and also spare those stage III patients who would not benefit from chemotherapy from the toxicities. Novel types of biomarkers are arising. Molecular-based biomarkers, such as genetic markers, epigenetic markers, and their signatures, enable physicians to more precisely stratify patients for personalized treatment. In addition, new methods of biomarker detection are also developing rapidly, such as liquid biopsy, which is minimally invasive and characterized by the isolating cancer-derived components from peripheral blood or other body fluids.120 However, there are still issues remaining to be addressed before broad clinical application, such as the low amounts of circulating tumor cells or circulating tumor DNA or other molecules derived from the tumor.120 However, it is reasonable to think that the liquid biopsy could play an important role in predicting relapses and monitoring metastases and treatment responses in CRC in the foreseeable future. Additionally, liquid biopsy could provide real-time genetic information of the tumor, which can help physicians better tailor the treatment to each patient.121 Yet, with the discovery of novel biomarkers and ongoing development of detection methods, the application of biomarker-guided treatment for patients with CRC remains limited. CRC therapies are becoming increasingly target specific. In the era of personalized treatment, biomarkers will inevitably develop in tandem to play greater roles in predicting the prognosis and treatment response of CRC patients.
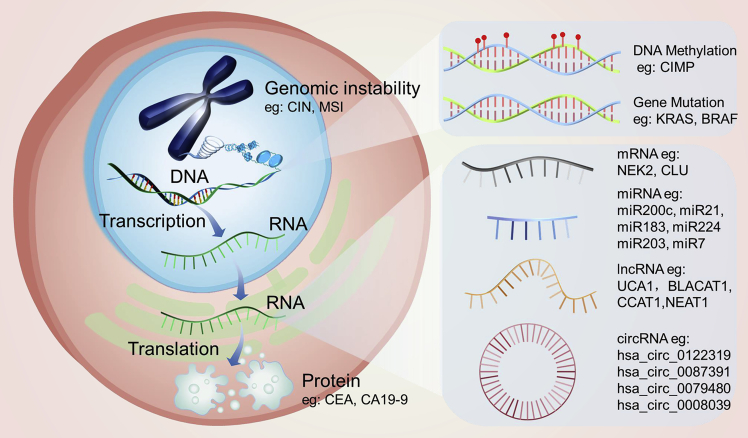
Advances in high-throughput technology and bioinformatics have opened a gate for understanding the genetic and epigenetic alterations in CRC. In the last two decades, epigenetic alterations have become a burgeoning research hotspot for biomarker discovery. Epigenetic biomarkers, including methylated DNAs, miRNAs, lncRNAs, and circRNAs, for CRC prognosis and treatment response have emerged in large numbers (Figure 2). However, few markers have been validated and integrated into clinical practice. Most established biomarkers have a low clinical prevalence. Currently, the only marker with sufficient evidence to justify its use in routine clinical applications is KRAS mutation and the selection for anti-epidermal growth factor receptor (EGFR) therapy.122 Different sample types, small cohorts, algorithms with poor efficiency, as well as high heterogeneity of the individuals pose challenges for biomarker discovery and development. In this setting, large-scale comprehensive molecular profiling from translational studies combined with artificial intelligence may offer an intriguing opportunity to develop improved molecular biomarkers and prediction algorithms and efficacious therapeutic targets. The transfer of these new biomarkers and targets from bench to bedside also necessitates larger-scale and multicenter trials to confirm their advantages. The discovery of potential biomarkers is opening the way to a more individualized practice of CRC, although the practical problems are many and difficult. The next frontier for novel molecular biomarkers is perhaps to answer the question of whether they might act as predictive and prognostic biomarkers for immunotherapy in CRC.
Figure 2.
Different Categories of Prognostic and Predictive CRC Biomarkers in Bimolecular Level
Different categories in the bimolecular level of CRC biomarkers with potential prognostic and predictive values, which include genomic and transcriptomic aspects mainly, are summarized. Also, representative candidate genes that are currently under research subordinate to each category are enumerated.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (81930065 and 81871951), the Natural Science Foundation of Guangdong Province (2014A030312015), the Science and Technology Program of Guangdong (2019B020227002), and by the Science and Technology Program of Guangzhou (201904020046, 201803040019, and 201704020228).
Declaration of Interests
The authors declare no competing interests.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.ymthe.2020.12.017.
Supplemental Information
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Chen W., Zheng R., Zeng H., Zhang S., He J. Annual report on status of cancer in China, 2011. Chin. J. Cancer Res. 2015;27:2–12. doi: 10.3978/j.issn.1000-9604.2015.01.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benitez-Majano S., Fowler H., Maringe C., Di Girolamo C., Rachet B. Deriving stage at diagnosis from multiple population-based sources: colorectal and lung cancer in England. Br. J. Cancer. 2016;115:391–400. doi: 10.1038/bjc.2016.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benson A.B., 3rd, Schrag D., Somerfield M.R., Cohen A.M., Figueredo A.T., Flynn P.J., Krzyzanowska M.K., Maroun J., McAllister P., Van Cutsem E. American Society of Clinical Oncology recommendations on adjuvant chemotherapy for stage II colon cancer. J. Clin. Oncol. 2004;22:3408–3419. doi: 10.1200/JCO.2004.05.063. [DOI] [PubMed] [Google Scholar]
- 5.Figueredo A., Coombes M.E., Mukherjee S. Adjuvant therapy for completely resected stage II colon cancer. Cochrane Database Syst. Rev. 2008;(3):CD005390. doi: 10.1002/14651858.CD005390.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oñate-Ocaña L.F., Montesdeoca R., López-Graniel C.M., Aiello-Crocifoglio V., Mondragón-Sánchez R., Cortina-Borja M., Herrera-Goepfert R., Oros-Ovalle C., Gallardo-Rincón D. Identification of patients with high-risk lymph node-negative colorectal cancer and potential benefit from adjuvant chemotherapy. Jpn. J. Clin. Oncol. 2004;34:323–328. doi: 10.1093/jjco/hyh054. [DOI] [PubMed] [Google Scholar]
- 7.O’Connor E.S., Greenblatt D.Y., LoConte N.K., Gangnon R.E., Liou J.I., Heise C.P., Smith M.A. Adjuvant chemotherapy for stage II colon cancer with poor prognostic features. J. Clin. Oncol. 2011;29:3381–3388. doi: 10.1200/JCO.2010.34.3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jonker D.J., Spithoff K., Maroun J., Gastrointestinal Cancer Disease Site Group of Cancer Care Ontario’s Program in Evidence-based Care Adjuvant systemic chemotherapy for stage II and III colon cancer after complete resection: an updated practice guideline. Clin. Oncol. (R. Coll. Radiol.) 2011;23:314–322. doi: 10.1016/j.clon.2011.02.010. [DOI] [PubMed] [Google Scholar]
- 9.Gray R., Barnwell J., McConkey C., Hills R.K., Williams N.S., Kerr D.J., Quasar Collaborative Group Adjuvant chemotherapy versus observation in patients with colorectal cancer: a randomised study. Lancet. 2007;370:2020–2029. doi: 10.1016/S0140-6736(07)61866-2. [DOI] [PubMed] [Google Scholar]
- 10.Moertel C.G., Fleming T.R., Macdonald J.S., Haller D.G., Laurie J.A., Tangen C.M., Ungerleider J.S., Emerson W.A., Tormey D.C., Glick J.H. Intergroup study of fluorouracil plus levamisole as adjuvant therapy for stage II/Dukes’ B2 colon cancer. J. Clin. Oncol. 1995;13:2936–2943. doi: 10.1200/JCO.1995.13.12.2936. [DOI] [PubMed] [Google Scholar]
- 11.Schippinger W., Samonigg H., Schaberl-Moser R., Greil R., Thödtmann R., Tschmelitsch J., Jagoditsch M., Steger G.G., Jakesz R., Herbst F., Austrian Breast and Colorectal Cancer Study Group A prospective randomised phase III trial of adjuvant chemotherapy with 5-fluorouracil and leucovorin in patients with stage II colon cancer. Br. J. Cancer. 2007;97:1021–1027. doi: 10.1038/sj.bjc.6604011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shiovitz S., Bertagnolli M.M., Renfro L.A., Nam E., Foster N.R., Dzieciatkowski S., Luo Y., Lao V.V., Monnat R.J., Jr., Emond M.J., Alliance for Clinical Trials in Oncology CpG island methylator phenotype is associated with response to adjuvant irinotecan-based therapy for stage III colon cancer. Gastroenterology. 2014;147:637–645. doi: 10.1053/j.gastro.2014.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sener S.F., Imperato J.P., Chmiel J., Fremgen A., Sylvester J. The use of cancer registry data to study preoperative carcinoembryonic antigen level as an indicator of survival in colorectal cancer. CA Cancer J. Clin. 1989;39:50–57. doi: 10.3322/canjclin.39.1.50. [DOI] [PubMed] [Google Scholar]
- 14.Zhong W., Yu Z., Zhan J., Yu T., Lin Y., Xia Z.S., Yuan Y.H., Chen Q.K. Association of serum levels of CEA, CA199, CA125, CYFRA21-1 and CA72-4 and disease characteristics in colorectal cancer. Pathol. Oncol. Res. 2015;21:83–95. doi: 10.1007/s12253-014-9791-9. [DOI] [PubMed] [Google Scholar]
- 15.Sanders D.S., Kerr M.A. Lewis blood group and CEA related antigens; coexpressed cell-cell adhesion molecules with roles in the biological progression and dissemination of tumours. Mol. Pathol. 1999;52:174–178. doi: 10.1136/mp.52.4.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ritts R.E., Jr., Del Villano B.C., Go V.L., Herberman R.B., Klug T.L., Zurawski V.R., Jr. Initial clinical evaluation of an immunoradiometric assay for CA 19-9 using the NCI serum bank. Int. J. Cancer. 1984;33:339–345. doi: 10.1002/ijc.2910330310. [DOI] [PubMed] [Google Scholar]
- 17.Hewish M., Lord C.J., Martin S.A., Cunningham D., Ashworth A. Mismatch repair deficient colorectal cancer in the era of personalized treatment. Nat. Rev. Clin. Oncol. 2010;7:197–208. doi: 10.1038/nrclinonc.2010.18. [DOI] [PubMed] [Google Scholar]
- 18.Sinicrope F.A. DNA mismatch repair and adjuvant chemotherapy in sporadic colon cancer. Nat. Rev. Clin. Oncol. 2010;7:174–177. doi: 10.1038/nrclinonc.2009.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zaanan A., Shi Q., Taieb J., Alberts S.R., Meyers J.P., Smyrk T.C., Julie C., Zawadi A., Tabernero J., Mini E. Role of deficient DNA mismatch repair status in patients with stage III colon cancer treated with FOLFOX adjuvant chemotherapy: a pooled analysis from 2 randomized clinical trials. JAMA Oncol. 2018;4:379–383. doi: 10.1001/jamaoncol.2017.2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vilar E., Gruber S.B. Microsatellite instability in colorectal cancer-the stable evidence. Nat. Rev. Clin. Oncol. 2010;7:153–162. doi: 10.1038/nrclinonc.2009.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Singh A., Sweeney M.F., Yu M., Burger A., Greninger P., Benes C., Haber D.A., Settleman J. TAK1 inhibition promotes apoptosis in KRAS-dependent colon cancers. Cell. 2012;148:639–650. doi: 10.1016/j.cell.2011.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Whitfield M.L., George L.K., Grant G.D., Perou C.M. Common markers of proliferation. Nat. Rev. Cancer. 2006;6:99–106. doi: 10.1038/nrc1802. [DOI] [PubMed] [Google Scholar]
- 23.Popat S., Matakidou A., Houlston R.S. Thymidylate synthase expression and prognosis in colorectal cancer: a systematic review and meta-analysis. J. Clin. Oncol. 2004;22:529–536. doi: 10.1200/JCO.2004.05.064. [DOI] [PubMed] [Google Scholar]
- 24.Vogelstein B., Fearon E.R., Hamilton S.R., Kern S.E., Preisinger A.C., Leppert M., Nakamura Y., White R., Smits A.M., Bos J.L. Genetic alterations during colorectal-tumor development. N. Engl. J. Med. 1988;319:525–532. doi: 10.1056/NEJM198809013190901. [DOI] [PubMed] [Google Scholar]
- 25.Wang M.T., Holderfield M., Galeas J., Delrosario R., To M.D., Balmain A., McCormick F. K-Ras promotes tumorigenicity through suppression of non-canonical Wnt signaling. Cell. 2015;163:1237–1251. doi: 10.1016/j.cell.2015.10.041. [DOI] [PubMed] [Google Scholar]
- 26.Cancer Genome Atlas Network Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fearon E.R., Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 28.Vogelstein B., Papadopoulos N., Velculescu V.E., Zhou S., Diaz L.A., Jr., Kinzler K.W. Cancer genome landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang J.X., Song W., Chen Z.H., Wei J.H., Liao Y.J., Lei J., Hu M., Chen G.Z., Liao B., Lu J. Prognostic and predictive value of a microRNA signature in stage II colon cancer: a microRNA expression analysis. Lancet Oncol. 2013;14:1295–1306. doi: 10.1016/S1470-2045(13)70491-1. [DOI] [PubMed] [Google Scholar]
- 30.Wang R., Du L., Yang X., Jiang X., Duan W., Yan S., Xie Y., Zhu Y., Wang Q., Wang L. Identification of long noncoding RNAs as potential novel diagnosis and prognosis biomarkers in colorectal cancer. J. Cancer Res. Clin. Oncol. 2016;142:2291–2301. doi: 10.1007/s00432-016-2238-9. [DOI] [PubMed] [Google Scholar]
- 31.Ju H.Q., Zhao Q., Wang F., Lan P., Wang Z., Zuo Z.X., Wu Q.N., Fan X.J., Mo H.Y., Chen L. A circRNA signature predicts postoperative recurrence in stage II/III colon cancer. EMBO Mol. Med. 2019;11:e10168. doi: 10.15252/emmm.201810168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dienstmann R., Vermeulen L., Guinney J., Kopetz S., Tejpar S., Tabernero J. Consensus molecular subtypes and the evolution of precision medicine in colorectal cancer. Nat. Rev. Cancer. 2017;17:79–92. doi: 10.1038/nrc.2016.126. [DOI] [PubMed] [Google Scholar]
- 33.Oh S.C., Park Y.Y., Park E.S., Lim J.Y., Kim S.M., Kim S.B., Kim J., Kim S.C., Chu I.S., Smith J.J. Prognostic gene expression signature associated with two molecularly distinct subtypes of colorectal cancer. Gut. 2012;61:1291–1298. doi: 10.1136/gutjnl-2011-300812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grady W.M., Carethers J.M. Genomic and epigenetic instability in colorectal cancer pathogenesis. Gastroenterology. 2008;135:1079–1099. doi: 10.1053/j.gastro.2008.07.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boland C.R., Goel A. Microsatellite instability in colorectal cancer. Gastroenterology. 2010;138:2073–2087.e3. doi: 10.1053/j.gastro.2009.12.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boland C.R., Thibodeau S.N., Hamilton S.R., Sidransky D., Eshleman J.R., Burt R.W., Meltzer S.J., Rodriguez-Bigas M.A., Fodde R., Ranzani G.N., Srivastava S. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998;58:5248–5257. [PubMed] [Google Scholar]
- 37.Walther A., Houlston R., Tomlinson I. Association between chromosomal instability and prognosis in colorectal cancer: a meta-analysis. Gut. 2008;57:941–950. doi: 10.1136/gut.2007.135004. [DOI] [PubMed] [Google Scholar]
- 38.Popat S., Hubner R., Houlston R.S. Systematic review of microsatellite instability and colorectal cancer prognosis. J. Clin. Oncol. 2005;23:609–618. doi: 10.1200/JCO.2005.01.086. [DOI] [PubMed] [Google Scholar]
- 39.Petrelli F., Ghidini M., Cabiddu M., Pezzica E., Corti D., Turati L., Costanzo A., Varricchio A., Ghidini A., Barni S., Tomasello G. Microsatellite instability and survival in stage II colorectal cancer: a systematic review and meta-analysis. Anticancer Res. 2019;39:6431–6441. doi: 10.21873/anticanres.13857. [DOI] [PubMed] [Google Scholar]
- 40.Klingbiel D., Saridaki Z., Roth A.D., Bosman F.T., Delorenzi M., Tejpar S. Prognosis of stage II and III colon cancer treated with adjuvant 5-fluorouracil or FOLFIRI in relation to microsatellite status: results of the PETACC-3 trial. Ann. Oncol. 2015;26:126–132. doi: 10.1093/annonc/mdu499. [DOI] [PubMed] [Google Scholar]
- 41.Wang B., Li F., Zhou X., Ma Y., Fu W. Is microsatellite instability-high really a favorable prognostic factor for advanced colorectal cancer? A meta-analysis. World J. Surg. Oncol. 2019;17:169. doi: 10.1186/s12957-019-1706-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Toyota M., Ahuja N., Ohe-Toyota M., Herman J.G., Baylin S.B., Issa J.P. CpG island methylator phenotype in colorectal cancer. Proc. Natl. Acad. Sci. USA. 1999;96:8681–8686. doi: 10.1073/pnas.96.15.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weisenberger D.J., Siegmund K.D., Campan M., Young J., Long T.I., Faasse M.A., Kang G.H., Widschwendter M., Weener D., Buchanan D. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat. Genet. 2006;38:787–793. doi: 10.1038/ng1834. [DOI] [PubMed] [Google Scholar]
- 44.Lee S., Cho N.Y., Choi M., Yoo E.J., Kim J.H., Kang G.H. Clinicopathological features of CpG island methylator phenotype-positive colorectal cancer and its adverse prognosis in relation to KRAS/BRAF mutation. Pathol. Int. 2008;58:104–113. doi: 10.1111/j.1440-1827.2007.02197.x. [DOI] [PubMed] [Google Scholar]
- 45.Van Rijnsoever M., Elsaleh H., Joseph D., McCaul K., Iacopetta B. CpG island methylator phenotype is an independent predictor of survival benefit from 5-fluorouracil in stage III colorectal cancer. Clin. Cancer Res. 2003;9:2898–2903. [PubMed] [Google Scholar]
- 46.Tan I.B., Ivanova T., Lim K.H., Ong C.W., Deng N., Lee J., Tan S.H., Wu J., Lee M.H., Ooi C.H. Intrinsic subtypes of gastric cancer, based on gene expression pattern, predict survival and respond differently to chemotherapy. Gastroenterology. 2011;141:476–485. doi: 10.1053/j.gastro.2011.04.042. 485.e1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Min B.H., Bae J.M., Lee E.J., Yu H.S., Kim Y.H., Chang D.K., Kim H.C., Park C.K., Lee S.H., Kim K.M., Kang G.H. The CpG island methylator phenotype may confer a survival benefit in patients with stage II or III colorectal carcinomas receiving fluoropyrimidine-based adjuvant chemotherapy. BMC Cancer. 2011;11:344. doi: 10.1186/1471-2407-11-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Suzuki H., Yamamoto E., Maruyama R., Niinuma T., Kai M. Biological significance of the CpG island methylator phenotype. Biochem. Biophys. Res. Commun. 2014;455:35–42. doi: 10.1016/j.bbrc.2014.07.007. [DOI] [PubMed] [Google Scholar]
- 49.Juo Y.Y., Johnston F.M., Zhang D.Y., Juo H.H., Wang H., Pappou E.P., Yu T., Easwaran H., Baylin S., van Engeland M., Ahuja N. Prognostic value of CpG island methylator phenotype among colorectal cancer patients: a systematic review and meta-analysis. Ann. Oncol. 2014;25:2314–2327. doi: 10.1093/annonc/mdu149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ogino S., Nosho K., Kirkner G.J., Kawasaki T., Meyerhardt J.A., Loda M., Giovannucci E.L., Fuchs C.S. CpG island methylator phenotype, microsatellite instability, BRAF mutation and clinical outcome in colon cancer. Gut. 2009;58:90–96. doi: 10.1136/gut.2008.155473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pai R.K., Jayachandran P., Koong A.C., Chang D.T., Kwok S., Ma L., Arber D.A., Balise R.R., Tubbs R.R., Shadrach B., Pai R.K. BRAF-mutated, microsatellite-stable adenocarcinoma of the proximal colon: an aggressive adenocarcinoma with poor survival, mucinous differentiation, and adverse morphologic features. Am. J. Surg. Pathol. 2012;36:744–752. doi: 10.1097/PAS.0b013e31824430d7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lochhead P., Kuchiba A., Imamura Y., Liao X., Yamauchi M., Nishihara R., Qian Z.R., Morikawa T., Shen J., Meyerhardt J.A. Microsatellite instability and BRAF mutation testing in colorectal cancer prognostication. J. Natl. Cancer Inst. 2013;105:1151–1156. doi: 10.1093/jnci/djt173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Worthley D.L., Whitehall V.L., Spring K.J., Leggett B.A. Colorectal carcinogenesis: road maps to cancer. World J. Gastroenterol. 2007;13:3784–3791. doi: 10.3748/wjg.v13.i28.3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Markowitz S.D., Bertagnolli M.M. Molecular origins of cancer: molecular basis of colorectal cancer. N. Engl. J. Med. 2009;361:2449–2460. doi: 10.1056/NEJMra0804588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Andreyev H.J., Norman A.R., Cunningham D., Oates J.R., Clarke P.A. Kirsten ras mutations in patients with colorectal cancer: the multicenter “RASCAL” study. J. Natl. Cancer Inst. 1998;90:675–684. doi: 10.1093/jnci/90.9.675. [DOI] [PubMed] [Google Scholar]
- 56.Andreyev H.J., Norman A.R., Cunningham D., Oates J., Dix B.R., Iacopetta B.J., Young J., Walsh T., Ward R., Hawkins N. Kirsten ras mutations in patients with colorectal cancer: the “RASCAL II” study. Br. J. Cancer. 2001;85:692–696. doi: 10.1054/bjoc.2001.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Samowitz W.S., Curtin K., Schaffer D., Robertson M., Leppert M., Slattery M.L. Relationship of Ki-ras mutations in colon cancers to tumor location, stage, and survival: a population-based study. Cancer Epidemiol. Biomarkers Prev. 2000;9:1193–1197. [PubMed] [Google Scholar]
- 58.Ince W.L., Jubb A.M., Holden S.N., Holmgren E.B., Tobin P., Sridhar M., Hurwitz H.I., Kabbinavar F., Novotny W.F., Hillan K.J., Koeppen H. Association of k-ras, b-raf, and p53 status with the treatment effect of bevacizumab. J. Natl. Cancer Inst. 2005;97:981–989. doi: 10.1093/jnci/dji174. [DOI] [PubMed] [Google Scholar]
- 59.Wang C., van Rijnsoever M., Grieu F., Bydder S., Elsaleh H., Joseph D., Harvey J., Iacopetta B. Prognostic significance of microsatellite instability and Ki-ras mutation type in stage II colorectal cancer. Oncology. 2003;64:259–265. doi: 10.1159/000069311. [DOI] [PubMed] [Google Scholar]
- 60.Rajagopalan H., Bardelli A., Lengauer C., Kinzler K.W., Vogelstein B., Velculescu V.E. Tumorigenesis: RAF/RAS oncogenes and mismatch-repair status. Nature. 2002;418:934. doi: 10.1038/418934a. [DOI] [PubMed] [Google Scholar]
- 61.Di Nicolantonio F., Martini M., Molinari F., Sartore-Bianchi A., Arena S., Saletti P., De Dosso S., Mazzucchelli L., Frattini M., Siena S., Bardelli A. Wild-type BRAF is required for response to panitumumab or cetuximab in metastatic colorectal cancer. J. Clin. Oncol. 2008;26:5705–5712. doi: 10.1200/JCO.2008.18.0786. [DOI] [PubMed] [Google Scholar]
- 62.Zhu L., Dong C., Cao Y., Fang X., Zhong C., Li D., Yuan Y. Prognostic role of BRAF mutation in stage II/III colorectal cancer receiving curative resection and adjuvant chemotherapy: a meta-analysis based on randomized clinical trials. PLoS ONE. 2016;11:e0154795. doi: 10.1371/journal.pone.0154795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fariña-Sarasqueta A., van Lijnschoten G., Moerland E., Creemers G.J., Lemmens V.E.P.P., Rutten H.J.T., van den Brule A.J.C. The BRAF V600E mutation is an independent prognostic factor for survival in stage II and stage III colon cancer patients. Ann. Oncol. 2010;21:2396–2402. doi: 10.1093/annonc/mdq258. [DOI] [PubMed] [Google Scholar]
- 64.International Human Genome Sequencing Consortium Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 65.Takahashi Y., Iwaya T., Sawada G., Kurashige J., Matsumura T., Uchi R., Ueo H., Takano Y., Eguchi H., Sudo T. Up-regulation of NEK2 by microRNA-128 methylation is associated with poor prognosis in colorectal cancer. Ann. Surg. Oncol. 2014;21:205–212. doi: 10.1245/s10434-013-3264-3. [DOI] [PubMed] [Google Scholar]
- 66.Costa F.F. Non-coding RNAs: new players in eukaryotic biology. Gene. 2005;357:83–94. doi: 10.1016/j.gene.2005.06.019. [DOI] [PubMed] [Google Scholar]
- 67.He L., Hannon G.J. MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 68.Mendell J.T. MicroRNAs: critical regulators of development, cellular physiology and malignancy. Cell Cycle. 2005;4:1179–1184. doi: 10.4161/cc.4.9.2032. [DOI] [PubMed] [Google Scholar]
- 69.Vasudevan S., Tong Y., Steitz J.A. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 70.Calin G.A., Dumitru C.D., Shimizu M., Bichi R., Zupo S., Noch E., Aldler H., Rattan S., Keating M., Rai K. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Slaby O., Svoboda M., Michalek J., Vyzula R. MicroRNAs in colorectal cancer: translation of molecular biology into clinical application. Mol. Cancer. 2009;8:102. doi: 10.1186/1476-4598-8-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Slattery M.L., Mullany L.E., Sakoda L.C., Wolff R.K., Stevens J.R., Samowitz W.S., Herrick J.S. The PI3K/AKT signaling pathway: Associations of miRNAs with dysregulated gene expression in colorectal cancer. Mol. Carcinog. 2018;57:243–261. doi: 10.1002/mc.22752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Slattery M.L., Mullany L.E., Sakoda L.C., Samowitz W.S., Wolff R.K., Stevens J.R., Herrick J.S. Expression of Wnt-signaling pathway genes and their associations with miRNAs in colorectal cancer. Oncotarget. 2017;9:6075–6085. doi: 10.18632/oncotarget.23636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cha S.T., Tan C.T., Chang C.C., Chu C.Y., Lee W.J., Lin B.Z., Lin M.T., Kuo M.L. Retracted. Nat. Cell Biol. 2016;19:76. doi: 10.1038/ncb3455. [DOI] [PubMed] [Google Scholar]
- 75.Ji S., Ye G., Zhang J., Wang L., Wang T., Wang Z., Zhang T., Wang G., Guo Z., Luo Y. miR-574-5p negatively regulates Qki6/7 to impact β-catenin/Wnt signalling and the development of colorectal cancer. Gut. 2013;62:716–726. doi: 10.1136/gutjnl-2011-301083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rasmussen M.H., Lyskjær I., Jersie-Christensen R.R., Tarpgaard L.S., Primdal-Bengtson B., Nielsen M.M., Pedersen J.S., Hansen T.P., Hansen F., Olsen J.V. miR-625-3p regulates oxaliplatin resistance by targeting MAP2K6-p38 signalling in human colorectal adenocarcinoma cells. Nat. Commun. 2016;7:12436. doi: 10.1038/ncomms12436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ye L., Jiang T., Shao H., Zhong L., Wang Z., Liu Y., Tang H., Qin B., Zhang X., Fan J. miR-1290 is a biomarker in DNA-mismatch-repair-deficient colon cancer and promotes resistance to 5-fluorouracil by directly targeting hMSH2. Mol. Ther. Nucleic Acids. 2017;7:453–464. doi: 10.1016/j.omtn.2017.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Boussios S., Ozturk M.A., Moschetta M., Karathanasi A., Zakynthinakis-Kyriakou N., Katsanos K.H., Christodoulou D.K., Pavlidis N. The developing story of predictive biomarkers in colorectal cancer. J. Pers. Med. 2019;9:12. doi: 10.3390/jpm9010012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Creemers E.E., Tijsen A.J., Pinto Y.M. Circulating microRNAs: novel biomarkers and extracellular communicators in cardiovascular disease? Circ. Res. 2012;110:483–495. doi: 10.1161/CIRCRESAHA.111.247452. [DOI] [PubMed] [Google Scholar]
- 80.Arroyo J.D., Chevillet J.R., Kroh E.M., Ruf I.K., Pritchard C.C., Gibson D.F., Mitchell P.S., Bennett C.F., Pogosova-Agadjanyan E.L., Stirewalt D.L. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl. Acad. Sci. USA. 2011;108:5003–5008. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Turchinovich A., Weiz L., Langheinz A., Burwinkel B. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 2011;39:7223–7233. doi: 10.1093/nar/gkr254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Vickers K.C., Palmisano B.T., Shoucri B.M., Shamburek R.D., Remaley A.T. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat. Cell Biol. 2011;13:423–433. doi: 10.1038/ncb2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Valadi H., Ekström K., Bossios A., Sjöstrand M., Lee J.J., Lötvall J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 84.Toiyama Y., Takahashi M., Hur K., Nagasaka T., Tanaka K., Inoue Y., Kusunoki M., Boland C.R., Goel A. Serum miR-21 as a diagnostic and prognostic biomarker in colorectal cancer. J. Natl. Cancer Inst. 2013;105:849–859. doi: 10.1093/jnci/djt101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ogata-Kawata H., Izumiya M., Kurioka D., Honma Y., Yamada Y., Furuta K., Gunji T., Ohta H., Okamoto H., Sonoda H. Circulating exosomal microRNAs as biomarkers of colon cancer. PLoS ONE. 2014;9:e92921. doi: 10.1371/journal.pone.0092921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Schetter A.J., Leung S.Y., Sohn J.J., Zanetti K.A., Bowman E.D., Yanaihara N., Yuen S.T., Chan T.L., Kwong D.L., Au G.K. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299:425–436. doi: 10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mima K., Nishihara R., Yang J., Dou R., Masugi Y., Shi Y., da Silva A., Cao Y., Song M., Nowak J. MicroRNA MIR21 (miR-21) and PTGS2 expression in colorectal cancer and patient survival. Clin. Cancer Res. 2016;22:3841–3848. doi: 10.1158/1078-0432.CCR-15-2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hur K., Toiyama Y., Takahashi M., Balaguer F., Nagasaka T., Koike J., Hemmi H., Koi M., Boland C.R., Goel A. MicroRNA-200c modulates epithelial-to-mesenchymal transition (EMT) in human colorectal cancer metastasis. Gut. 2013;62:1315–1326. doi: 10.1136/gutjnl-2011-301846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Toiyama Y., Hur K., Tanaka K., Inoue Y., Kusunoki M., Boland C.R., Goel A. Serum miR-200c is a novel prognostic and metastasis-predictive biomarker in patients with colorectal cancer. Ann. Surg. 2014;259:735–743. doi: 10.1097/SLA.0b013e3182a6909d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Takahashi M., Sung B., Shen Y., Hur K., Link A., Boland C.R., Aggarwal B.B., Goel A. Boswellic acid exerts antitumor effects in colorectal cancer cells by modulating expression of the let-7 and miR-200 microRNA family. Carcinogenesis. 2012;33:2441–2449. doi: 10.1093/carcin/bgs286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Li Z., Pan W., Shen Y., Chen Z., Zhang L., Zhang Y., Luo Q., Ying X. IGF1/IGF1R and microRNA let-7e down-regulate each other and modulate proliferation and migration of colorectal cancer cells. Cell Cycle. 2018;17:1212–1219. doi: 10.1080/15384101.2018.1469873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nakajima G., Hayashi K., Xi Y., Kudo K., Uchida K., Takasaki K., Yamamoto M., Ju J. Non-coding microRNAs hsa-let-7g and hsa-miR-181b are associated with chemoresponse to S-1 in colon cancer. Cancer Genomics Proteomics. 2006;3:317–324. [PMC free article] [PubMed] [Google Scholar]
- 93.Pauli A., Rinn J.L., Schier A.F. Non-coding RNAs as regulators of embryogenesis. Nat. Rev. Genet. 2011;12:136–149. doi: 10.1038/nrg2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Derrien T., Johnson R., Bussotti G., Tanzer A., Djebali S., Tilgner H., Guernec G., Martin D., Merkel A., Knowles D.G. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Esteller M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 97.Akhade V.S., Pal D., Kanduri C. Long noncoding RNA: genome organization and mechanism of action. Adv. Exp. Med. Biol. 2017;1008:47–74. doi: 10.1007/978-981-10-5203-3_2. [DOI] [PubMed] [Google Scholar]
- 98.Gupta R.A., Shah N., Wang K.C., Kim J., Horlings H.M., Wong D.J., Tsai M.C., Hung T., Argani P., Rinn J.L. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kogo R., Shimamura T., Mimori K., Kawahara K., Imoto S., Sudo T., Tanaka F., Shibata K., Suzuki A., Komune S. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 2011;71:6320–6326. doi: 10.1158/0008-5472.CAN-11-1021. [DOI] [PubMed] [Google Scholar]
- 100.Svoboda M., Slyskova J., Schneiderova M., Makovicky P., Bielik L., Levy M., Lipska L., Hemmelova B., Kala Z., Protivankova M. HOTAIR long non-coding RNA is a negative prognostic factor not only in primary tumors, but also in the blood of colorectal cancer patients. Carcinogenesis. 2014;35:1510–1515. doi: 10.1093/carcin/bgu055. [DOI] [PubMed] [Google Scholar]
- 101.Wu Z.H., Wang X.L., Tang H.M., Jiang T., Chen J., Lu S., Qiu G.Q., Peng Z.H., Yan D.W. Long non-coding RNA HOTAIR is a powerful predictor of metastasis and poor prognosis and is associated with epithelial-mesenchymal transition in colon cancer. Oncol. Rep. 2014;32:395–402. doi: 10.3892/or.2014.3186. [DOI] [PubMed] [Google Scholar]
- 102.Li P., Zhang X., Wang L., Du L., Yang Y., Liu T., Li C., Wang C. lncRNA HOTAIR contributes to 5FU resistance through suppressing miR-218 and activating NF-κB/TS signaling in colorectal cancer. Mol. Ther. Nucleic Acids. 2017;8:356–369. doi: 10.1016/j.omtn.2017.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhang J., Yin M., Peng G., Zhao Y. CRNDE: an important oncogenic long non-coding RNA in human cancers. Cell Prolif. 2018;51:e12440. doi: 10.1111/cpr.12440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Han P., Li J.W., Zhang B.M., Lv J.C., Li Y.M., Gu X.Y., Yu Z.W., Jia Y.H., Bai X.F., Li L. The lncRNA CRNDE promotes colorectal cancer cell proliferation and chemoresistance via miR-181a-5p-mediated regulation of Wnt/β-catenin signaling. Mol. Cancer. 2017;16:9. doi: 10.1186/s12943-017-0583-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Jiang H., Wang Y., Ai M., Wang H., Duan Z., Wang H., Zhao L., Yu J., Ding Y., Wang S. Long noncoding RNA CRNDE stabilized by hnRNPUL2 accelerates cell proliferation and migration in colorectal carcinoma via activating Ras/MAPK signaling pathways. Cell Death Dis. 2017;8:e2862. doi: 10.1038/cddis.2017.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ji P., Diederichs S., Wang W., Böing S., Metzger R., Schneider P.M., Tidow N., Brandt B., Buerger H., Bulk E. MALAT-1, a novel noncoding RNA, and thymosin β4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031–8041. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- 107.Zheng H.T., Shi D.B., Wang Y.W., Li X.X., Xu Y., Tripathi P., Gu W.L., Cai G.X., Cai S.J. High expression of lncRNA MALAT1 suggests a biomarker of poor prognosis in colorectal cancer. Int. J. Clin. Exp. Pathol. 2014;7:3174–3181. [PMC free article] [PubMed] [Google Scholar]
- 108.Takahashi K., Yan I.K., Haga H., Patel T. Modulation of hypoxia-signaling pathways by extracellular linc-RoR. J. Cell Sci. 2014;127:1585–1594. doi: 10.1242/jcs.141069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kogure T., Yan I.K., Lin W.L., Patel T. Extracellular vesicle-mediated transfer of a novel long noncoding RNA TUC339: a mechanism of intercellular signaling in human hepatocellular cancer. Genes Cancer. 2013;4:261–272. doi: 10.1177/1947601913499020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Liu T., Zhang X., Gao S., Jing F., Yang Y., Du L., Zheng G., Li P., Li C., Wang C. Exosomal long noncoding RNA CRNDE-h as a novel serum-based biomarker for diagnosis and prognosis of colorectal cancer. Oncotarget. 2016;7:85551–85563. doi: 10.18632/oncotarget.13465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lei B., Tian Z., Fan W., Ni B. Circular RNA: a novel biomarker and therapeutic target for human cancers. Int. J. Med. Sci. 2019;16:292–301. doi: 10.7150/ijms.28047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sanger H.L., Klotz G., Riesner D., Gross H.J., Kleinschmidt A.K. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc. Natl. Acad. Sci. USA. 1976;73:3852–3856. doi: 10.1073/pnas.73.11.3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zaphiropoulos P.G. Exon skipping and circular RNA formation in transcripts of the human cytochrome P-450 2C18 gene in epidermis and of the rat androgen binding protein gene in testis. Mol. Cell. Biol. 1997;17:2985–2993. doi: 10.1128/mcb.17.6.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hansen T.B., Jensen T.I., Clausen B.H., Bramsen J.B., Finsen B., Damgaard C.K., Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 115.Hansen T.B., Kjems J., Damgaard C.K. Circular RNA and miR-7 in cancer. Cancer Res. 2013;73:5609–5612. doi: 10.1158/0008-5472.CAN-13-1568. [DOI] [PubMed] [Google Scholar]
- 116.Weng W., Wei Q., Toden S., Yoshida K., Nagasaka T., Fujiwara T., Cai S., Qin H., Ma Y., Goel A. Circular RNA ciRS-7-A promising prognostic biomarker and a potential therapeutic target in colorectal cancer. Clin. Cancer Res. 2017;23:3918–3928. doi: 10.1158/1078-0432.CCR-16-2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zeng K., Chen X., Xu M., Liu X., Hu X., Xu T., Sun H., Pan Y., He B., Wang S. circHIPK3 promotes colorectal cancer growth and metastasis by sponging miR-7. Cell Death Dis. 2018;9:417. doi: 10.1038/s41419-018-0454-8. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 118.Zheng X., Chen L., Zhou Y., Wang Q., Zheng Z., Xu B., Wu C., Zhou Q., Hu W., Wu C., Jiang J. A novel protein encoded by a circular RNA circPPP1R12A promotes tumor pathogenesis and metastasis of colon cancer via Hippo-YAP signaling. Mol. Cancer. 2019;18:47. doi: 10.1186/s12943-019-1010-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lasda E., Parker R. Circular RNAs: diversity of form and function. RNA. 2014;20:1829–1842. doi: 10.1261/rna.047126.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Vacante M., Ciuni R., Basile F., Biondi A. The liquid biopsy in the management of colorectal cancer: an overview. Biomedicines. 2020;8:308. doi: 10.3390/biomedicines8090308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Osumi H., Shinozaki E., Yamaguchi K. Circulating tumor DNA as a novel biomarker optimizing chemotherapy for colorectal cancer. Cancers (Basel) 2020;12:1566. doi: 10.3390/cancers12061566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Van Cutsem E., Köhne C.H., Hitre E., Zaluski J., Chang Chien C.R., Makhson A., D’Haens G., Pintér T., Lim R., Bodoky G. Cetuximab and chemotherapy as initial treatment for metastatic colorectal cancer. N. Engl. J. Med. 2009;360:1408–1417. doi: 10.1056/NEJMoa0805019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.