Abstract
A wealth of biochemical and cellular data, accumulated over several years by multiple groups, has provided a great degree of insight into the molecular mechanisms of actions of GCN5 and PCAF in gene activation. Studies of these lysine acetyltransferases (KATs) in vitro, in cultured cells, have revealed general mechanisms for their recruitment by sequence-specific binding factors and their molecular functions as transcriptional co-activators. Genetic studies indicate that GCN5 and PCAF are involved in multiple developmental processes in vertebrates, yet our understanding of their molecular functions in these contexts remains somewhat rudimentary. Understanding the functions of GCN5/PCAF in developmental processes provides clues to the roles of these KATs in disease states. Here we will review what is currently known about the developmental roles of GCN5 and PCAF, as well as emerging role of these KATs in oncogenesis.
Keywords: Gcn5, Pcaf, chromatin, development, cancer
1. Introduction
1.1. GCN5 and PCAF in vertebrates
The Gcn5 locus was duplicated in vertebrates, giving rise to Gcn5 (GCN5L2, KAT2a) and Pcaf (p300/CBP Associated Factor; KAT2b). Mammalian GCN5 and PCAF exhibit high sequence similarities (75% amino acid identity), and they have similar if not identical biochemical specificities. Both acetylate multiple lysines in histones and other proteins, such as transcription factors and cytoskeletal components, and they appear to be the primary KATs for acetylation of H3K9, at least in mouse embryonic fibroblasts (MEFs) [1]. GCN5 and PCAF are both incorporated into STAGA/TFTC complexes [2, 3]; hereafter referred to as SAGA) and ATAC complexes, but in a mutually exclusive way. However, in vivo, these KATs are not interchangeable, as indicated by different phenotypes upon mutation of Gcn5 or Pcaf in mice. Loss of Gcn5 causes early embryonic lethality whereas deletion of Pcaf causes no abnormalities, although double knockouts of both KATs results in an earlier and more severe phenotype than the loss of Gcn5 alone, indicating some redundancy early during mouse embryogenesis [4, 5]. Despite numerous studies over the last 25 years, the unique and shared functions of these KATs in both gene regulation and development are still not clearly defined.
1.2. Conserved functional domains of GCN5/PCAF
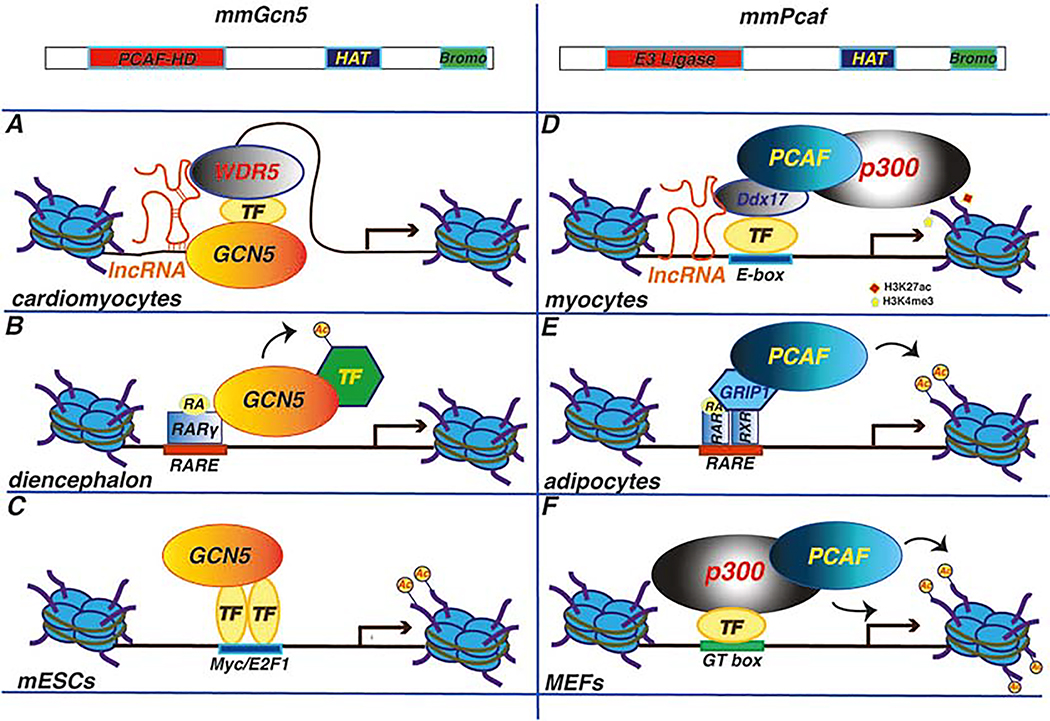
The histone acetyltransferase (HAT) domain and the bromodomain in the carboxy-terminal of both mammalian GCN5 and PCAF are highly conserved with their yeast, protozoa and fly counterparts, highlighting the critical functions of these domains throughout evolution [2, 6] (see also reviews by Brownell & Allis and Berger, Grant & Winston in this special issue). However, mammalian GCN5 and PCAF have extended an amino-terminal domain not found in lower organisms that is important for binding to nucleosomes in vitro [2]. and that houses ubiquitin ligase activity [7, 8] (Fig. 1, top panels).
Fig.1: Examples of different mechanisms of GCN5 or PCAF recruitment to promoters or enhancers in embryonic stem cells, tissues, or organs.
Highlighted are GCN5 or PCAF, transcription factors (TFs), co-activators (WDR5, p300, GRIP-1, Ddx17), DNA binding elements (E-box, GT box, RARE) and lncRNAs (linc1405, Myoparr) as well as histone modifications analyzed (H3 acetylation, H3K27ac, H3K4me3) A. GCN5 is recruited to multiple gene promoters through its direct or indirect interaction with TFs (like c-Myc and E2F1) in mouse embryonic stem cells [48]. B. GCN5-dependent acetylates TACC1 after retinoic acid (RA) induction in the developing mouse diencephalon [40]. C. Recruitment of GCN5 to the enhancer of Mesp1 in cardiomyocytes is mediated by linc1405, WDR5 and Eomes [18] D. PCAF and p300 acetylate histone H3 when recruited to Cyclin D1 promoter by KLF8 in MEFs [36]. E. PCAF-dependent histone H3 acetylation after its RA-dependent recruitment to the promoter of orphan nuclear receptor TR2 [43]. PCAF interaction with RAR/RXR in preadipocytes is mediated by GRIP1. F. Long non-coding RNA Myoparr and Ddx17 mediates the recruitment of PCAF and (possibly p300) to gene promoters with a subsequent increase in H3K4me3 and H3K27ac levels [20–22]
Both the HAT and the bromodomains of GCN5 and PCAF bridge interactions with both histones and transcription factors. For example, the PCAF HAT and bromodomains directly interact with Twist in vitro and this interaction inhibits PCAF acetyltransferase activity [9]. Despite the critical role of Twist in the inhibition of differentiation in multiple cell lineages, including muscle, bone and neuronal cells, the potential genetic and functional interactions of Twist and PCAF have not been further addressed, perhaps due to the lack of embryonic phenotypes in Pcaf null mice. In another example, the bromodomains of PCAF and GCN5 interact with two transcription factors involved in insulin response, IRF-1 and IRF-2, in vitro [10]. PCAF and GCN5 both bind acetylated tails of H3 and H4 [11] through their bromodomains, making them both “writers” and readers” of histone acetylation marks (see also review by Strahl & Briggs in this special issue).
The functional importance of the GCN5 bromodomain was first demonstrated in yeast where phenotypes of a GCN5-deleted strain were only partially complemented by a Gcn5 expression plasmid missing the bromodomain [12]. The bromodomain was later shown to facilitate nucleosome remodeling by the SWI/SNF complex [13] by anchoring the SAGA complex to nucleosomes. The bromodomain also promotes cooperative and cross-tail acetylation of H3 at K9/14/18/23 [14] (see also review by Strahl & Briggs in this special issue). However, the bromodomain of GCN5 is dispensable for HAT activity and gene activation. It remains to be seen whether the molecular functions of the bromodomain in vertebrates hold true to those in yeast. Gcn5−/− mice have a more severe phenotype than Gcn5hat/hat mice [15], indicating that GCN5 has functions outside of its HAT activity, and at least some of those functions may be mediated by the bromodomain. The GCN5/PCAF bromodomain could provide a targetable domain in disease states, following the precedent of BET domain inhibitors used to combat specific human cancers.
2. Gcn5 and Pcaf functions during development
Multiple reports have defined GCN5 and PCAF functions in vivo through genetic studies in mice and other vertebrates. Animal development offers an excellent platform to define HAT functions in gene transcription. The dynamic nature of tissue and organ formation requires the integration of multiple spatial, temporal, and quantitative inputs to progressively define and adapt the properties of cells to their final destinations and functional status. Transcriptional regulation in concert with chromatin modulations presents a critical hub for signal integration necessary for fate decisions to be made.
2.1. Gcn5 is required for mesoderm specification and survival
The analysis of mouse knockout phenotypes clearly indicates that Gcn5 plays a critical role during mouse embryogenesis, whereas PCAF does not [4, 5]. These phenotypes are consistent with expression patterns of these HATs, as Gcn5 is strongly and widely expressed early during embryogenesis, but PCAF is expressed at very low levels, indicating PCAF likely has few or limited functions at this time [4, 5].
Gcn5 null embryos are defective in somitogenesis and neurulation. Histological analyses and characterization of lineage markers showed that although the primitive streak formed, downstream formation of paraxial mesoderm and the notochord was disrupted in the mutant embryos. Analysis of the affected mesodermal tissues revealed elevated levels of apoptosis that could be responsible for the disappearance of these lineages [4]. Subsequent studies support this idea, as elimination of p53 in Gcn5 null mice prevented apoptosis and allowed embryos to survive until mid-gestation [15].
A role for Gcn5 in the regulation of genes and pathways important for mesoderm specification is further indicated by morphological and molecular analyses of embryoid bodies derived from mouse embryonic stem cells (mESCs) devoid of Gcn5. These studies revealed deregulation of FGF signaling activity in the absence of Gcn5 [16]. FGF and downstream Erk1/2 and PI3K pathways are crucial for mouse ESC differentiation towards mesodermal and neural lineages [17]. Genome-wide analyses of Gcn5−/− embryoid bodies did not show a global change in H3K9ac levels, however, localized decreases in H3K9ac levels were observed near the promoters of multiple genes whose expression was affected by Gcn5 loss [16]. Gcn5 (but not Pcaf or p300) has also been identified as an interacting partner of linc1405, WDR5, and the transcription factor Eomes in mesoderm and cardiomyocytes in differentiating embryoid bodies. These factors are co-recruited to the well-characterized enhancer of Mesp1, which regulates a downstream gene transcriptional network responsible for cardiac lineage commitment [18]. This study offers mechanistic insights into how Gcn5 is potentially recruited to key mesodermal genes though a combinatorial action of transcription factors and long non-coding RNAs (Fig. 1A). However, increased myogenic and cardiogenic potential was observed in embryoid bodies devoid of Gcn5 [19], highlighting the need for additional studies of Gcn5 functions in these lineages in vivo. Intriguingly, Pcaf was found to interact with the lncRNA Myoparr and the transcriptional co-activator Ddx17 and regulate the transcription of myogenin in myogenic differentiation (Fig. 1D) [20]. The recruitment of PCAF to myogenin promoter probably involves interactions with MyoD and p300 [21, 22], but more experiments are required for the detailed characterization of myogenin promoter regulation.
Gcn5 involvement in the specification of mesodermal tissues is conserved in other vertebrates. Elegant experiments performed in developing Xenopus embryos to define the functions of TBP family members in actively transcribed genes discovered increased recruitment of Gcn5 to a subset of TBP-family insensitive genes upon ablation of TBP and two related factors, TLF and TBP2. These findings indicate that Gcn5 is important for non-canonical, TBP-independent transcriptional regulation of a densely connected network of developmental genes in the Xenopus mesoderm [23].
2.2. Hypomorphic mutations in SAGA components reveal functions in Hox gene regulation
While the early embryonic lethality of Gcn5 null embryos [5, 24] precludes the study of Gcn5 functions in later developmental processes, hypomorphic mutations that substantially lower Gcn5 expression or KAT function result in axial skeleton defects in lower thoracic regions, including rib fusions and homeotic transformations, along with spina bifida and exencephaly [15, 25, 26]. These rib fusions and homeotic transformations of lumbar L1 to thoracic T14 vertebrae are preceded by posterior shifts in the anterior boundary of Hoxc8 and Hoxc9 expression [25]. Interestingly, hypomorphic mutations in another SAGA component, Supt20 (Suppressor of Ty 20 Homolog), cause similar defects in rostral-caudal somite patterning [27]. Supt20 mutations affect the integrity of the SAGA complex, highlighting the importance of this Gcn5-containing complex - rather than ATAC - in somitic segmentation and Hox gene regulation. However, global Gcn5 loss of function results in a more severe phenotype in mesoderm specification than does the loss of Supt20 [28], suggesting that Gcn5 may also act outside of SAGA, either as part of a small ADA complex [29] or the ATAC complex in the developing mesoderm.
2.3. PCAF and Gcn5 interactions with p300/CBP
PCAF is so named due to its physical association in mammalian cell lines with CBP/p300 [30]. Mammalian PCAF was predicted early on to play a pivotal role in the control of cell growth and differentiation by integrating diverse signals for gene expression through direct interactions with the transcription machinery and targeted modifications of chromatin structure. Initial experiments functionally linked PCAF, together with CBP/p300, to transcriptional activators like MyoD, retinoic acid receptor, and CREB in the execution of developmental programs in vitro [31–33]. These results suggested that CBP/p300 and PCAF affect transcriptional regulation in part through histone acetylation and that PCAF is potentially recruited to a wide range of promoters via multiple protein-protein interactions (Fig. 1D, E, F). The complete lack of developmental phenotypes in Pcaf null mice suggests that Gcn5 likely provides redundancy to these functions in vivo.
The first diversification of CBP/p300 and PCAF functions was suggested by the identification of different histone acetylation patterns within nucleosomal substrates [34]. A direct comparison of the genome-wide distribution of CBP, p300, and PCAF in human T cells provided further evidence for their functional diversification, as CBP and p300 were associated both with enhancers and promoters, whereas PCAF was elevated along regions of actively transcribed genes beyond promoters [35]. Interestingly though, a high percentage of genes with PCAF bound at their promoters were also occupied by CBP and p300. PCAF and p300 (but not CBP) were found to associate with KLF8 at the Cyclin D1 promoter in MEFs. Their recruitment to the gene promoter increased the acetylation on H3 (Fig. 1F), offering an example of transcriptional activation mediated by both co-activators [36].
Gcn5 likely also interacts with p300/CBP, physically and functionally. Although mice heterozygous for either Gcn5 or p300 null alleles are viable, a significant fraction (25%) of Gcn5+/− p300+/− mice die before birth. In contrast, no abnormalities were observed in Pcaf−/− p300+/− mice, again suggesting Gcn5 is more important that Pcaf in executing early developmental programs [37]. Of course, Gcn5 and p300 also have distinct functions. For example, p300, together with BMP signaling, is important in establishing ‘pre-patterns’ of histone acetylation at liver and pancreas-specific genes in embryonic endoderm progenitors, but Gcn5 is dispensable [38].
2.4. Gcn5 KAT activity is important for neuronal development
A mouse model bearing point mutations in the catalytic acetyltransferase domain of Gcn5 provided more clues as to the importance of its KAT activity during development [15]. These Gcn5hat/hat mice exhibited defects in cranial neural tube closure with exencephaly. The neural folds normally lift and fuse at the hindbrain, the midbrain, and finally at the rostral end of the neural tube [39]. Neural folds of the mutant Gcn5hat/hat embryos remained open and dramatic exencephaly was observed from E11.5 onwards. Even though these embryos survived longer than Gcn5 null mutants, they still died at E13.5 to E16.5 [15]. A detailed study of the Gcn5hat/hat mutant phenotype revealed a significant diencephalic expansion and telencephalic compression, indicating a functional requirement for Gcn5 catalytic activity in the developing diencephalon between E9.5 and E10.5 [40]. The expansion of the diencephalic tissue was linked to the deregulation of WNT signaling repression in the Gcn5hat/hat embryos, which acts upon Gli3 to limit Shh expression. Gcn5 was found in a complex with the transcription factor TACC1 and RARα in neuroepithelial cells isolated from E9.0 mouse forebrains. Recruitment of this complex to retinoic acid-responsive elements (RARE) of Foxa1 and Rarb genes (Fig. 1B) was dependent on TACC1 acetylation mediated by Gcn5 acetyltransferase activity [40]. The Gcn5-mediated acetylation of TACC1 causes the dissociation of the transcription factor from the promoter and allows the transcription of the downstream targets. In the absence of Gcn5, the RA target genes remain repressed, the WNT signaling is not inhibited and Shh is derepressed causing an expansion of the zona limitans intrathalamica (ZLI) [40]. PCAF is also among the co-activators identified to interact with nuclear receptors and activate transcription of downstream targets [32]. Although there are implications of PCAF function in neuronal cells through its interaction with the RARβ receptor [41, 42], PCAF’s recruitment to gene promoters through RAR interactions is better characterized during the proliferation of preadipocytes (Fig.1E) [43].
Gcn5hat/hat mutant embryos also showed defects in the neural crest cell-derived craniofacial cartilage and bone. Cartilage and bone differentiation factors like sox9a, cola1, and runx2a/2b were reduced in the E12.5 mutant embryos. Interestingly the craniofacial abnormalities observed in Gcn5hat/hat mutant mouse embryos were also present in zebrafish devoid of either Gcn5 or Pcaf, with a more severe phenotype in double Gcn5/Pcaf mutants [44]. Lack of Gcn5 was also found to cause decreased osteogenic differentiation in human periodontal ligament stem cells, again linked to aberrant activation of the WNT pathway, through the transcriptional deregulation of the WNT pathway inhibitor DKK1 [45].
2.5. Gcn5/PCAF functions in stem cell pluripotency and differentiation
mESCs have the capacity to self-renew indefinitely and the potential to give rise to all the somatic lineages and germ cells of a multicellular organism [46]. Both their pluripotency and differentiation potential in vitro is supported by a network of signaling, transcriptional, and epigenetic regulatory interactions [47]. Gcn5 has been identified as a critical component of a feedforward loop that controls Myc-dependent circuits in stem cells. Chromatin immunoprecipitation for the analysis of Gcn5 occupancy in undifferentiated ES cells revealed preferred recruitment to Myc and E2F1 binding motifs, and genes affected by Gcn5 loss were enriched in Myc and E2F1 targets (Fig.1C). However, Gcn5 loss of function in mouse embryos did not hinder the development of the blastocyst, and embryonic stem cells that lack Gcn5 can be derived and cultured in vitro [19, 48], perhaps reflecting redundancies with Pcaf in ESCs. Connections with Myc functions are also indicated by overlapping gene expression programs observed in neurospheres devoid of Gcn5 or N-myc in vitro. Many of the genes affected by both N-Myc and Gcn5 loss encoded components of signaling cascades, including the Ras and Wnt pathways. Downregulation of these genes in the absence of Gcn5 or N-myc was associated with reduced levels of H3K9ac at their promoters, further highlighting a key role for Gcn5 in the regulation of these genes [49].
Myc is one of the four canonical “Yamanaka” factors that drive reprogramming of differentiated cells towards pluripotency. c-Myc was later found to be dispensable for inducing pluripotency in somatic cells [50], but it likely enhances reprogramming by augmenting transcription of the other pluripotency inducing factors, Sox2, Oct4, or KLF4, via direct promoter binding. A siRNA screen identified Gcn5 as required for reprogramming of fibroblasts to induced pluripotent stem cells (iPSCs) upon induction of expression of the Yamanaka factors [48]. Overexpression of GCN5 alone did not induce reprogramming in the absence of MYC, consistent with a need for MYC-dependent recruitment of GCN5 to downstream target genes. Interestingly, multiple other SAGA components were also identified as essential for somatic cell reprogramming in this screen (e.g. Taf12, Sgf29, and Trrap), indicating the importance of SAGA in iPSC formation. These findings are consistent with previous studies in vitro that indicated the TRRAP subunit of SAGA interacts directly with Myc [51, 52], although these interactions have not yet been confirmed in a developmental context. Interestingly, genome-wide analysis of TRRAP occupancy in mESCs shows significant overlap with Oct4 binding sites, supporting a functional role of TRRAP in the maintenance of self-renewal [53] that may reflect TRRAP functions in the Tip60 HAT complex [54].
3. Functional connections with other modifiers during development
Epigenetic writers and readers often work combinatorially to achieve specific developmental outcomes [55], but at present, we do not have a clear understanding of how GCN5 and PCAF might cooperate with other histone modifiers during embryogenesis. GCN5 and PCAF were identified as key writers for H3K9 and K14 acetylation [1] in mouse fibroblasts. H3K9/14ac and H3K4me3 are enriched along with RNA Pol II at the promoters of most active genes in human embryonic stem cells, suggesting GCN5 and PCAF might cooperate with MLL family members in this context [56]. Precedence for MLL-KAT interactions comes from observation of synergetic H3K4 methylation and histone acetylation at active genes, facilitated by physical interaction between trithorax and CBP, in drosophila [57]. Interactions between MLL1 and CBP also occur in human cells [58]. Besides, MLL1 physically interacts with another HAT, MOF, and joint recruitment of these factors regulates expression of Hoxa9 in MEFs as well as the closely correlated distribution of H3K4me3 and H4K16ac at other active genes [59].
Global deletion of the Moz KAT in mice showed a profound homeotic transformation in the skeleton and defects in the nervous system [60], somewhat similar to the phenotype of Gcn5hat/hat mutants. In the absence of Moz, H3K9ac levels were decreased at Hox gene promoters, even though Moz is generally thought of as an H4-specific HAT. MLL recruitment was also decreased at the Hox genes upon Moz deletion, suggesting functional interplay between MOZ and Mll1. Mll1-mediated methylation is stimulated by the presence of histone acetylation (H3K9ac/H3K14ac) on peptides [61]. Interestingly, despite the obvious deregulation of Hox genes patterning in Moz mutant embryos, the role of MOF acetyltransferase in the regulation of Hox genes remains elusive, as MOF gene deletion causes early embryonic lethality at implantation [62, 63]. How or if Gcn5/PCAF mediated acetylation of H3K9/K14 is connected to Moz functions has not been directly addressed, but the similarities in the homeotic phenotypes caused by loss or diminution of MLL1, MOZ and GCN5 are consistent with overlapping or synergistic functions. One major obstacle in dissecting the genome-wide functions of GCN5 and PCAF in different developmental contexts remains the low reproducibility of the available antibodies against Gcn5 and Pcaf in chromatin immunoprecipitation experiments in mammals. Genome-wide studies of GCN5 and PCAF, in conjunction with H3K9ac and H3K4me3 or H3K4me1 and H3K27ac in different developmental contexts, will shed light on their critical functions in promoters and enhancers and will help define developmental gene regulatory networks and/or their global impact on different steps of transcriptional progression.
4. Gcn5 and Pcaf functions in adult tissues and disease
In addition to their functions in developing embryos, both Gcn5 and Pcaf have important functions in adult cells. The use of conditional alleles has revealed functions for Gcn5 in specific tissues, such as immune cells, and molecular analyses have uncovered additional, non-histone substrates for Gcn5 and Pcaf. The importance of these KATs is also reflected by emerging studies implicating their importance in cancer.
4.1. Gcn5/Pcaf functions in hematopoietic cell development and immune function
Both Gcn5 and Pcaf are implicated in the development and function of immune cells. Deletion of Pcaf in Foxp3+ inducible T regulatory cells (iTregs) reduces the production of CD4+ Foxp3+ Tregs [64]. Gcn5 deletion in iTregs does not appear to reduce the production of absolute numbers of Treg in vivo, however, loss of GCN5 and/or PCAF does affect their function as evidenced by allograft rejection and loss of IL-2 production [64, 65]. Gcn5 deletion also impairs T cell development at other distinct stages. While mice with Gcn5 conditional knockout in CD4 CD8 double-negative (DN) T cells do not show significant alteration to development or survival of this population [66], there is a decrease in single-positive CD4+ and CD8+ cells as well as a reduction of Treg cells in the spleen [65]. There is also an accumulation of DN3 stage cells and a reduction of DN4 stage cells suggesting that GCN5 regulates this transition. GCN5 is also required for the maturation of these cells into invariant natural killer T (iNKT) cells via acetylation of early response growth protein 2 (EGR2), a key transcription factor in the development of iNKT cells [66]. GCN5 enhances the interaction between steroid receptor coactivator 1 (SRC1) and retinoic acid receptor-related orphan receptor gamma (RORγt) in T helper 17 (TH17) cells [67]. Loss of Gcn5 in TH17 cells impairs differentiation by downregulating genes essential to TH17 cells. In most of these studies, GCN5/PCAF appears to facilitate the function of T cells subsets by regulating the transcription of key factors such as interleukin- 2 (IL-2) and interferon-gamma (IFN-γ). Double knockout of Gcn5 and Pcaf in iTregs leads to smaller body size and thymic atrophy, as well as autoimmunity and premature death in mice. These effects seem to be due to the loss of peripheral Tregs and activation of T effector cells [64]. These findings illustrate that proper regulation of T cell development and function is dependent on expression of Gcn5/Pcaf.
Gcn5 is also associated with the function of other myeloid cells. Acetylation of the transcription factor CCAAT Enhancer Binding Protein Alpha (C/EBPα) by GCN5 inhibits its ability to bind to DNA and transcribe target genes leading to impairment of granulocyte differentiation [68]. GCN5 interacts and colocalizes with transcription factor Zinc Finger DNA-Binding Protein 89 (Zbp-89) in human erythroid cells at genes that are necessary for globin regulation and terminal maturation of erythroid cells [69]. Gcn5 expression also increases during the “differentiation phase” of an erythroid differentiation time course of human CD34 cells further indicating a potential role for GCN5 in erythroid maturation [69]. The SAGA complex protein SUPT20 (also known as P38-Interacting protein (P38IP)) regulates monocyte and macrophage differentiation in part by regulating the stability of GCN5 in the SAGA complex [70]. GCN5/PCAF regulates innate immune cell signaling by targeting Tank-binding kinase 1 (TBK1), a kinase that regulates the production of the cytokine IFN-β, in MEFs [71]. Regulation of interferons are tightly controlled as overproduction may lead to the development of autoimmune diseases such as lupus erythematosus and multiple sclerosis. These findings highlight that Gcn5/Pcaf plays significant roles in different lineages of hematopoietic cells and that early deletion of these KATs has a significant impact on the development of many types of blood cells.
Taken together, these studies indicate that Gcn5 and Pcaf play important roles in both the innate and adaptive immune systems. In most studies, these KATs appear to play an essential role in the differentiation of hematopoietic cells into mature cells. The contrasting effects of deletion of Gcn5 in the different subsets of T cells reveal the dynamic manner by which expression of and gene regulation by GCN5 can affect immune cell development in vertebrates. The results of these studies also highlight the differences in the functions of Gcn5 and Pcaf in development and how loss of one KAT may compensate for the other in some contexts. In summary, Gcn5 and Pcaf regulate the development of immune cells and function through the transcription of genes that are essential for immune system homeostasis. Furthermore, the studies implicate both Gcn5 and Pcaf in the development of autoimmune diseases and suggest that they may be attractive therapeutic targets in the treatment of these maladies.
4.2. Gcn5/Pcaf acetylate non-histone proteins
Although GCN5 and PCAF are primarily known as histone acetyltransferases, both proteins also acetylate non-histone proteins (see also review by Michael Downey in this special issue). Fournier and colleagues identified 398 proteins that are acetylated by GCN5 and PCAF using shotgun proteomic analysis comparing control versus Gcn5 and Pcaf knockdown cells [72]. The targets identified are involved in multiple cellular processes beyond transcription including mitotic cell cycle, DNA replication and repair, and cell death. Their study revealed that acetylation of PLK4 by GCN5/PCAF prevents centrosome amplification. GCN5 also acetylates α-tubulin, a reaction that is enhanced by its interaction with MYC, increasing the stability of the protein and subsequently the microtubules formed that are necessary for spindle formation and cell cycle progression [73, 74]. PCAF acetylates PTEN on lysines 125 and 128, which inhibits its ability to regulate AKT activation and to regulate G1 arrest [75]. GCN5 acetylates c-MYC, stabilizing this oncogenic transcription factor involved in cell division, growth, and survival [76].
Transcriptional regulation by GCN5/PCAF is also facilitated by acetylation of transcription factors. Acetylation of β-Catenin by PCAF improves its stability by preventing its ubiquitination and degradation through the proteasomal pathway and regulating its transcriptional activity [77]. Autoacetylation by PCAF enables its translocation to the nucleus where it facilitates transcription of target genes [78]. PCAF acetylates E2F1 in its DNA-binding domain enabling its ability to interact with the DNA and subsequently increases its transcriptional function [79]. GCN5/PCAF acetylate C/EBPβ in 3T3 L1 preadipocyte cells leading to transcription of C/EBPα, another GCN5 target [80, 81]. This modification was necessary for the differentiation of the cells into mature adipocytes when stimulated. Acetylation of the SAGA HAT module structural protein Alteration in activation 3 (ADA3) by GCN5, PCAF, and P300 is necessary to maintain global histone acetylation levels thus impacting transcription and cell proliferation [82]. Gcn5/Pcaf expression is necessary for the proper regulation of cell cycle progression and cell division via both the transcription of essential genes and acetylation and stabilization of proteins necessary for these processes.
4.3. Emerging Roles of Gcn5 and PCAF in Cancer
Unsurprisingly, many of the genes regulated by GCN5/PCAF in development have roles in cancer. Knockdown of PCAF in glioma reduced proliferation through transcriptional regulation of Hedgehog-Gli dependent genes [83]. GCN5 regulates the expression of E2F1, Cyclin D1, and Cyclin E1 in lung cancer [84]. GCN5 also regulates the expression of genes related to epithelial to mesenchymal transition (MMP-9 and E2F1) induced by TGFβ signaling in breast cancer cells [85]. Interestingly, ADA3 is overexpressed in breast cancer cells which also correlates to c-MYC overexpression [86]. Most genes regulated by GCN5 in cancer are associated with cell cycle progression, which MYC is a master regulator of. MYC is known to be the most overexpressed gene in cancer and GCN5 acting as a transcriptional coactivator as well as mediating acetylation of MYC most certainly enhances the role of MYC in cancer. The partnership between MYC and GCN5 in cancer has been shown so far in colon cancer [87], Burkitt lymphoma [88], and lung cancer [84, 89–91]. The association of GCN5 with c-MYC provides a direct link to the transcription of genes associated with cancer development and progression.
Consistent with its role in the development of blood cell types, GCN5 functions are critical in leukemias and lymphomas. Inhibition of GCN5 in Burkitt lymphoma reduced viability and induced apoptosis through regulation of the PI3K pathway [88]. A CRISPR dropout screen identified GCN5 as an essential gene in the survival of acute myeloid leukemia (AML) cells [92]. GCN5 expression is necessary for the expression of genes related to leukemogenesis ensuring proliferation and cell survival. Kahl and colleagues also showed that GCN5 is overexpressed in AML cells from patients [93]. Several studies showed that inhibition of Gcn5 caused differentiation and eventual cell death of AML cells [89, 92, 93]. Chemical inhibition of GCN5 combined with all-trans retinoic acid (ATRA) induces apoptosis in AML cells. Interestingly, acute promyelocytic leukemia (APL) cells treated with ATRA induced PCAF expression and differentiation of the APL cells into granulocytic cells. Knockdown of PCAF prevented this ATRA induced differentiation. Also knockdown of GCN5, but not PCAF, in some cancer cell types, reduces proliferation of the cells [84, 94]. This further demonstrates the differences in the roles of GCN5 and PCAF in developmental as well as cancer phenotypes.
PCAF has been identified as a potential tumor suppressor in some settings. PCAF is downregulated in gastric cancer (GC) tissues and its loss is associated with poor survival [95]. PCAF overexpression in GC cells promotes expression of the tumor suppressor p16 as well as impairing the interaction between CDK4 and Cyclin D1 inhibiting proliferation and colony formation. It will be important to determine in which particular cancers GCN5 and PCAF play roles as tumor suppressors versus oncogenes and whether those roles are dependent on their inclusion in either the SAGA or ATAC complexes.
Bondey-Chorney and colleagues also profiled the non-histone protein targets of GCN5/PCAF that are implicated in the progression of cancer, thus we will highlight a few key interactions [96]. PCAF acetylates EZH2 which prevents it from regulating its target genes leading to the progression of lung adenocarcinoma [97]. PCAF-mediated acetylation of EZH2 was also associated with poor patient survival. PCAF acetylates both p53 and hypoxia inducible-1α (HIF-1α) influencing transcriptional activity of both and regulating proapoptotic pathways under hypoxic conditions [98, 99]. E2F1 acetylation at lysines 117, 120, and 125 is partially mediated by PCAF as well as by CBP/p300 and TIP60 stabilizes the protein and is necessary for efficient binding to DNA [79, 100]. In MEFs these acetylation sites act as docking points for the bromodomains of p300 and CBP at double-strand breaks, however, are not transcriptionally linked to its role in DNA damage response but in the transcription of genes involved in nervous system development and differentiation [101]. The acetylation of E2F1 by PCAF, in addition to the co-transcriptional role played by GCN5 and PCAF, certainly plays a role in its oncogenic function. It would be interesting to ascertain what the transcriptional effects would be with mutations of these residues in a cancer cell context. Gcn5 also acetylates c-MYC stabilizing the protein which contributes to the transcriptional role in promoting cancer progression [76]. Targeting GCN5 and PCAF in cancer will have multiple repercussions in addition to transcription affects and these processes continue to be an active area of study.
GCN5/PCAF acetyltransferase activity is being increasingly linked with survival of various human cancer cell types. Currently, there are no clinically useful inhibitors of GCN5/PCAF. Inhibitors used in in vitro studies such as CPTH6 and MB-3 have IC50 values that are too high to be used in either animal models or humans. The implications of these KATs in many disease models underscore the need to develop compounds that could be used to treat humans for autoimmune disorders or cancer.
5. Future directions
Because Gcn5 and Pcaf are part of multiple, multisubunit complexes, the phenotypes observed upon mutation of these KATs reflect a concurrent loss of these activities. In vitro assays that monitor changes in cell proliferation, survival, or differentiation upon loss of Gcn5 or Pcaf must also be interpreted with the complexities of SAGA and ATAC in mind (see also review by Helmlinger et al in this special issue). A key question for the future is the definition of genes that depend specifically on each of these complexes for activation, and the definition of specific domains, partner proteins, and transcription factors that direct them to those genes.
Another unanswered question is how the various activities within SAGA work together to activate gene expression through histone acetylation, deubiquitination of H2B, and delivery of TBP (see also reviews by Goswami et al and Marc Timmers in this special issue). The elegant integration of these functional modules is highlighted by recent high-resolution cryo-EM structures of two different yeast SAGA complexes [102, 103]. The increased complexity of SAGA composition in vertebrates raises the possibility of even more specialized functions for these modules (see also review by Nuno-Cabanes and Rodriguez-Navarro in this special issue). Post-translational modifications of SAGA components in mammalian systems are also understudied at present, yet such modifications might affect both the structure and function of SAGA and its interactions with other factors. Meshing our increased understanding of pathways that require SAGA (Fig. 2), as defined by genetic studies, with an increased understanding of the structural plasticity of the complex, as highlighted by the recent structures, will perhaps allow us to finally determine how SAGA assimilates inputs from multiple developmental signals to deliver properly calibrated transcriptional outputs, and in turn, how co-opting of those functions by oncogenes such as Myc facilitates cancer and other disease states.
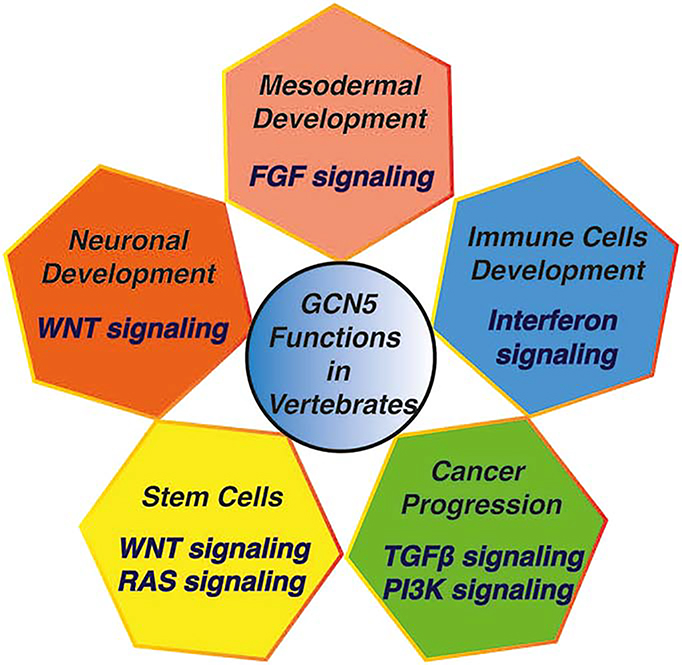
Fig.2: Signaling Pathways Regulated by Gcn5.
Gcn5 influences the development of different cell types and tissues by regulating essential signaling pathways. Regulation of these signaling pathways may also affect the development of diseases in a cell-type specific manner.
Table 1:
Representative examples of mouse models with global or conditional deletions of Gcn5/Kat2a allele and global deletion of Pcaf/Kat2b in different tissues and organs and their observed phenotypes.
| Year | Allele Name | Allele Type | Cre | Phenotype | REF |
|---|---|---|---|---|---|
| 2000 | Gcn5Δ | loss of function | NA | embryonic lethal E11.0 | [4] |
| 2000 | PcafΔ | loss of function | NA | viable and fertile | [4] |
| 2000 | Gcn5Δ and PcafΔ | loss of function (double knockout) | NA | embryonic lethal E8.0 | [4] |
| 2000 | PcafΔ | loss of function | NA | viable and fertile | [5] |
| 2000 | Gcn5ΔHAT&Bromo | loss of function | NA | embryonic lethal E11.5 | [5] |
| 2007 | Gcn5HAT | catalytic inactive | NA | neural tube closure defects | [15] |
| 2008 | Gcn5flox(neo) | hypomorphic | NA | Embryonic lethal E18.5 | [26] |
| 2008 | Gcn5flox | conditional | NA | NA | [26] |
| 2008 | Gcn5Δ3-19 | loss of function | NA | embryonic lethal E11.0 | [26] |
| 2008 | Gcn5 flox(neo) | hypomorphic | NA | abnormal patterning of mouse skeleton | [26] |
| 2008 | Gcn5 flox(neo)/Gcn5Δ3-19 | hypomorphic | NA | abnormal patterning of mouse skeleton | [26] |
| 2011 | Gcn5flox and PcafΔ | loss of function (double knockout) | retroviral Cre | no effect on morphology or growth of MEFs | [1] |
| 2012 | Gcn5flox | neural progenitor cell specific loss of function | Nestin-Cre | impaired brain growth and microcephaly | [49] |
| 2012 | Gcn5flox | Purkinje-cell specific loss of function | Pcp2-Cre | impaired brain growth and microcephaly | [104] |
| 2014 | Gcn5flox and PcafΔ | brown fat and skeletal muscle specific loss of function (double knockout) | Myf5-Cre | prevention of adipocyte differentiation | [105] |
| 2014 | Gcn5flox | adult forebrain specific loss of function | CamKIIα-cre | impaired hippocampal synaptic plasticity and long-term memory consolidation | [106] |
| 2017 | Gcn5flox | muscle-specific loss of function | MCK-Cre | no effect on metabolic remodeling in mouse skeletal muscle | [107] |
| 2017 | Gcn5flox | T-Cell-specific loss of function | Lck-Cre (CD4 CD8 DN T cells) | impaired T cell activation | [65] |
| 2017 | Gcn5flox | tamoxifen-induced T cell specific loss of function | UB/ESR-Cre (CD4+ T cells) | impaired T cell differentiation in vitro | [65] |
| 2018 | Gcn5HAT | catalytic inactive | NA | craniofacial cartilage and bone defects | [44] |
| 2019 | Gcn5flox | T-Cell-specific loss of function | Foxp3-YFP Cre | inhibitory effects on Treg function | [64] |
| 2019 | Gcn5flox | T-Cell-specific loss of function | CD4-Cre | impaired proliferation | [64] |
| 2019 | PcafΔ | loss of function | NA | impaired Treg peripheral conversion in vitro | [64] |
| 2019 | Gcn5flox and PcafΔ | T-Cell-specific loss of function (double knockout) | Foxp3-YFP Cre | lethal autoimmunity | [64] |
| 2019 | Gcn5flox | sperm-specific | Stra8-Cre | abnormal sperm formation and male infertility | [108] |
| 2020 | Gcn5flox | tamoxifen-induced muscle specific loss of function | iHSA-Cre (human a-skeletal actin) | no effect on metabolic remodeling in mouse skeletal muscle with combined overexpression of SIRT1 | [109] |
| 2020 | Gcn5flox | neural crest cell specific | Wnt1-Cre | severe craniofacial defects and neural tube closure defects | [110] |
| 2020 | Gcn5flox/Gcn5HAT | neural crest cell specific | Wnt1-Cre | defects in the mandibular portion of the craniofacial skeleton | [110] |
Highlights.
Gcn5 null embryos are defective in somitogenesis and neurulation, whereas Pcaf null mice show a complete lack of developmental phenotypes.
Hypomorphic mutations that substantially lower Gcn5 expression or abrogate KAT function result in axial skeleton defects, including rib fusions and homeotic transformations, along with spina bifida and exencephaly.
Chromatin immunoprecipitation of GCN5 in undifferentiated ES cells reveals preferred recruitment to Myc and E2F1 binding motifs, and genes affected by Gcn5 loss were enriched in Myc and E2F1 targets.
Gcn5 and Pcaf play important roles in both the innate and adaptive immune systems.
GCN5/PCAF KAT activity is being increasingly linked with the survival of various human cancer cell types, however, no clinically useful inhibitors of GCN5/PCAF exist to date.
Acknowledgements
We dedicate this manuscript to the memory of Dr. Susan Abmayr for her seminal contributions to TFIID and SAGA research. We apologize to the many colleagues whose important findings could not be cited due to space limitations. We thank Drs Michael J. McAndrew and Li Wang for useful comments and discussions on the manuscript. This study was supported by the National Institutes of Health grant R35GM131678 to S.Y.R.D.
Footnotes
Conflict of interest statement
The authors declare they have no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Jin Q, Yu LR, Wang L, Zhang Z, Kasper LH, Lee JE, Wang C, Brindle PK, Dent SY, Ge K, Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation, EMBO J 30(2) (2011) 249–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Xu W, Edmondson DG, Roth SY, Mammalian GCN5 and P/CAF acetyltransferases have homologous amino-terminal domains important for recognition of nucleosomal substrates, Mol Cell Biol 18(10) (1998) 5659–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Ogryzko VV, Kotani T, Zhang X, Schiltz RL, Howard T, Yang XJ, Howard BH, Qin J, Nakatani Y, Histone-like TAFs within the PCAF histone acetylase complex, Cell 94(1) (1998) 35–44. [DOI] [PubMed] [Google Scholar]
- [4].Xu W, Edmondson DG, Evrard YA, Wakamiya M, Behringer RR, Roth SY, Loss of Gcn5l2 leads to increased apoptosis and mesodermal defects during mouse development, Nature Genetics 26(2) (2000) 229–232. [DOI] [PubMed] [Google Scholar]
- [5].Yamauchi T, Yamauchi J, Kuwata T, Tamura T, Yamashita T, Bae N, Westphal H, Ozato K, Nakatani Y, Distinct but overlapping roles of histone acetylase PCAF and of the closely related PCAF-B/GCN5 in mouse embryogenesis, Proc Natl Acad Sci U S A 97(21) (2000) 11303–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Koutelou E, Hirsch CL, Dent SY, Multiple faces of the SAGA complex, Curr Opin Cell Biol 22(3) (2010) 374–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Linares LK, Kiernan R, Triboulet R, Chable-Bessia C, Latreille D, Cuvier O, Lacroix M, Le Cam L, Coux O, Benkirane M, Intrinsic ubiquitination activity of PCAF controls the stability of the oncoprotein Hdm2, Nat Cell Biol 9(3) (2007) 331–8. [DOI] [PubMed] [Google Scholar]
- [8].Mao X, Gluck N, Li D, Maine GN, Li H, Zaidi IW, Repaka A, Mayo MW, Burstein E, GCN5 is a required cofactor for a ubiquitin ligase that targets NF-kappaB/RelA, Genes Dev 23(7) (2009) 849–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Hamamori Y, Sartorelli V, Ogryzko V, Puri PL, Wu HY, Wang JY, Nakatani Y, Kedes L, Regulation of histone acetyltransferases p300 and PCAF by the bHLH protein twist and adenoviral oncoprotein E1A, Cell 96(3) (1999) 405–13. [DOI] [PubMed] [Google Scholar]
- [10].Masumi A, Wang IM, Lefebvre B, Yang XJ, Nakatani Y, Ozato K, The histone acetylase PCAF is a phorbol-ester-inducible coactivator of the IRF family that confers enhanced interferon responsiveness, Mol Cell Biol 19(3) (1999) 1810–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Dhalluin C, Carlson JE, Zeng L, He C, Aggarwal AK, Zhou MM, Structure and ligand of a histone acetyltransferase bromodomain, Nature 399(6735) (1999) 491–6. [DOI] [PubMed] [Google Scholar]
- [12].Marcus GA, Silverman N, Berger SL, Horiuchi J, Guarente L, Functional similarity and physical association between GCN5 and ADA2: putative transcriptional adaptors, EMBO J 13(20) (1994) 4807–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Syntichaki P, Topalidou I, Thireos G, The Gcn5 bromodomain co-ordinates nucleosome remodelling, Nature 404(6776) (2000) 414–7. [DOI] [PubMed] [Google Scholar]
- [14].Li S, Shogren-Knaak MA, The Gcn5 bromodomain of the SAGA complex facilitates cooperative and cross-tail acetylation of nucleosomes, J Biol Chem 284(14) (2009) 9411–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Bu P, Evrard YA, Lozano G, Dent SY, Loss of Gcn5 acetyltransferase activity leads to neural tube closure defects and exencephaly in mouse embryos, Mol Cell Biol 27(9) (2007) 3405–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Wang L, Koutelou E, Hirsch C, McCarthy R, Schibler A, Lin K, Lu Y, Jeter C, Shen J, Barton MC, Dent SYR, GCN5 Regulates FGF Signaling and Activates Selective MYC Target Genes during Early Embryoid Body Differentiation, Stem Cell Reports 10(1) (2018) 287–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Chen Y, Li X, Eswarakumar VP, Seger R, Lonai P, Fibroblast growth factor (FGF) signaling through PI 3-kinase and Akt/PKB is required for embryoid body differentiation, Oncogene 19(33) (2000) 3750–6. [DOI] [PubMed] [Google Scholar]
- [18].Guo X, Xu Y, Wang Z, Wu Y, Chen J, Wang G, Lu C, Jia W, Xi J, Zhu S, Jiapaer Z, Wan X, Liu Z, Gao S, Kang J, A Linc1405/Eomes Complex Promotes Cardiac Mesoderm Specification and Cardiogenesis, Cell Stem Cell 22(6) (2018) 893–908 e6. [DOI] [PubMed] [Google Scholar]
- [19].Lin W, Srajer G, Evrard YA, Phan HM, Furuta Y, Dent SY, Developmental potential of Gcn5(−/−) embryonic stem cells in vivo and in vitro, Dev Dyn 236(6) (2007) 1547–57. [DOI] [PubMed] [Google Scholar]
- [20].Hitachi K, Nakatani M, Takasaki A, Ouchi Y, Uezumi A, Ageta H, Inagaki H, Kurahashi H, Tsuchida K, Myogenin promoter-associated lncRNA Myoparr is essential for myogenic differentiation, EMBO Rep 20(3) (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Sartorelli V, Puri PL, Hamamori Y, Ogryzko V, Chung G, Nakatani Y, Wang JY, Kedes L, Acetylation of MyoD directed by PCAF is necessary for the execution of the muscle program, Mol Cell 4(5) (1999) 725–34. [DOI] [PubMed] [Google Scholar]
- [22].Dilworth FJ, Seaver KJ, Fishburn AL, Htet SL, Tapscott SJ, In vitro transcription system delineates the distinct roles of the coactivators pCAF and p300 during MyoD/E47-dependent transactivation, Proc Natl Acad Sci U S A 101(32) (2004) 11593–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Gazdag E, Jacobi UG, van Kruijsbergen I, Weeks DL, Veenstra GJ, Activation of a T-box-Otx2-Gsc gene network independent of TBP and TBP-related factors, Development 143(8) (2016) 1340–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Xu W, Edmondson DG, Evrard YA, Wakamiya M, Behringer RR, Roth SY, Loss of Gcn5l2 leads to increased apoptosis and mesodermal defects during mouse development, Nat Genet 26(2) (2000) 229–32. [DOI] [PubMed] [Google Scholar]
- [25].Lin W, Zhang Z, Chen CH, Behringer RR, Dent SY, Proper Gcn5 histone acetyltransferase expression is required for normal anteroposterior patterning of the mouse skeleton, Dev Growth Differ 50(5) (2008) 321–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Lin W, Zhang Z, Srajer G, Chen YC, Huang M, Phan HM, Dent SY, Proper expression of the Gcn5 histone acetyltransferase is required for neural tube closure in mouse embryos, Dev Dyn 237(4) (2008) 928–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Warrier S, Nuwayhid S, Sabatino JA, Sugrue KF, Zohn IE, Supt20 is required for development of the axial skeleton, Dev Biol 421(2) (2017) 245–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Zohn IE, Li Y, Skolnik EY, Anderson KV, Han J, Niswander L, p38 and a p38-interacting protein are critical for downregulation of E-cadherin during mouse gastrulation, Cell 125(5) (2006) 957–69. [DOI] [PubMed] [Google Scholar]
- [29].Soffers JHM, Li X, Saraf A, Seidel CW, Florens L, Washburn MP, Abmayr SM, Workman JL, Characterization of a metazoan ADA acetyltransferase complex, Nucleic Acids Res 47(7) (2019) 3383–3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Yang X-J, Ogryzko VV, Nishikawa J-I, Howard BH, Nakatani Y, A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A, Nature 382(6589) (1996) 319–324. [DOI] [PubMed] [Google Scholar]
- [31].Puri PL, Sartorelli V, Yang XJ, Hamamori Y, Ogryzko VV, Howard BH, Kedes L, Wang JY, Graessmann A, Nakatani Y, Levrero M, Differential roles of p300 and PCAF acetyltransferases in muscle differentiation, Mol Cell 1(1) (1997) 35–45. [DOI] [PubMed] [Google Scholar]
- [32].Blanco JCG, Minucci S, Lu J, Yang XJ, Walker KK, Chen H, Evans RM, Nakatani Y, Ozato K, The histone acetylase PCAF is a nuclear receptor coactivator, Genes & Development 12(11) (1998) 1638–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Korzus E, Torchia J, Rose DW, Xu L, Kurokawa R, McInerney EM, Mullen T-M, Glass CK, Rosenfeld MG, Transcription Factor-Specific Requirements for Coactivators and Their Acetyltransferase Functions, Science 279(5351) (1998) 703–707. [DOI] [PubMed] [Google Scholar]
- [34].Schiltz RL, Mizzen CA, Vassilev A, Cook RG, Allis CD, Nakatani Y, Overlapping but distinct patterns of histone acetylation by the human coactivators p300 and PCAF within nucleosomal substrates, J Biol Chem 274(3) (1999) 1189–92. [DOI] [PubMed] [Google Scholar]
- [35].Wang F, Marshall CB, Li GY, Yamamoto K, Mak TW, Ikura M, Synergistic interplay between promoter recognition and CBP/p300 coactivator recruitment by FOXO3a, ACS Chem Biol 4(12) (2009) 1017–27. [DOI] [PubMed] [Google Scholar]
- [36].Urvalek AM, Wang X, Lu H, Zhao J, KLF8 recruits the p300 and PCAF co-activators to its amino terminal activation domain to activate transcription, Cell Cycle 9(3) (2010) 601–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Phan HM, Xu AW, Coco C, Srajer G, Wyszomierski S, Evrard YA, Eckner R, Dent SY, GCN5 and p300 share essential functions during early embryogenesis, Dev Dyn 233(4) (2005) 1337–47. [DOI] [PubMed] [Google Scholar]
- [38].Xu CR, Cole PA, Meyers DJ, Kormish J, Dent S, Zaret KS, Chromatin “prepattern” and histone modifiers in a fate choice for liver and pancreas, Science 332(6032) (2011) 963–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Copp AJ, Greene ND, Murdoch JN, Dishevelled: linking convergent extension with neural tube closure, Trends Neurosci 26(9) (2003) 453–5. [DOI] [PubMed] [Google Scholar]
- [40].Wilde JJ, Siegenthaler JA, Dent SY, Niswander LA, Diencephalic Size Is Restricted by a Novel Interplay Between GCN5 Acetyltransferase Activity and Retinoic Acid Signaling, J Neurosci 37(10) (2017) 2565–2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Puttagunta R, Di Giovanni S, Retinoic acid signaling in axonal regeneration, Front Mol Neurosci 4 (2011) 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Puttagunta R, Schmandke A, Floriddia E, Gaub P, Fomin N, Ghyselinck NB, Di Giovanni S, RA-RAR-beta counteracts myelin-dependent inhibition of neurite outgrowth via Lingo-1 repression, J Cell Biol 193(7) (2011) 1147–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Gupta P, Park SW, Farooqui M, Wei LN, Orphan nuclear receptor TR2, a mediator of preadipocyte proliferation, is differentially regulated by RA through exchange of coactivator PCAF with corepressor RIP140 on a platform molecule GRIP1, Nucleic Acids Res 35(7) (2007) 2269–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Sen R, Pezoa SA, Carpio Shull L, Hernandez-Lagunas L, Niswander LA, Artinger KB, Kat2a and Kat2b Acetyltransferase Activity Regulates Craniofacial Cartilage and Bone Differentiation in Zebrafish and Mice, J Dev Biol 6(4) (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Li B, Sun J, Dong Z, Xue P, He X, Liao L, Yuan L, Jin Y, GCN5 modulates osteogenic differentiation of periodontal ligament stem cells through DKK1 acetylation in inflammatory microenvironment, Sci Rep 6 (2016) 26542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Dunn SJ, Martello G, Yordanov B, Emmott S, Smith AG, Defining an essential transcription factor program for naive pluripotency, Science 344(6188) (2014) 1156–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Morey L, Santanach A, Di Croce L, Pluripotency and Epigenetic Factors in Mouse Embryonic Stem Cell Fate Regulation, Mol Cell Biol 35(16) (2015) 2716–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Hirsch CL, Coban Akdemir Z, Wang L, Jayakumaran G, Trcka D, Weiss A, Hernandez JJ, Pan Q, Han H, Xu X, Xia Z, Salinger AP, Wilson M, Vizeacoumar F, Datti A, Li W, Cooney AJ, Barton MC, Blencowe BJ, Wrana JL, Dent SY, Myc and SAGA rewire an alternative splicing network during early somatic cell reprogramming, Genes Dev 29(8) (2015) 803–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Martinez-Cerdeno V, Lemen JM, Chan V, Wey A, Lin W, Dent SR, Knoepfler PS, N-Myc and GCN5 regulate significantly overlapping transcriptional programs in neural stem cells, PLoS One 7(6) (2012) e39456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Nakagawa M, Koyanagi M, Tanabe K, Takahashi K, Ichisaka T, Aoi T, Okita K, Mochiduki Y, Takizawa N, Yamanaka S, Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts, Nat Biotechnol 26(1) (2008) 101–6. [DOI] [PubMed] [Google Scholar]
- [51].McMahon SB, Wood MA, Cole MD, The essential cofactor TRRAP recruits the histone acetyltransferase hGCN5 to c-Myc, Mol Cell Biol 20(2) (2000) 556–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Zhang N, Ichikawa W, Faiola F, Lo SY, Liu X, Martinez E, MYC interacts with the human STAGA coactivator complex via multivalent contacts with the GCN5 and TRRAP subunits, Biochim Biophys Acta 1839(5) (2014) 395–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Sawan C, Hernandez-Vargas H, Murr R, Lopez F, Vaissiere T, Ghantous AY, Cuenin C, Imbert J, Wang ZQ, Ren B, Herceg Z, Histone acetyltransferase cofactor Trrap maintains self-renewal and restricts differentiation of embryonic stem cells, Stem Cells 31(5) (2013) 979–91. [DOI] [PubMed] [Google Scholar]
- [54].Ravens S, Yu C, Ye T, Stierle M, Tora L, Tip60 complex binds to active Pol II promoters and a subset of enhancers and co-regulates the c-Myc network in mouse embryonic stem cells, Epigenetics Chromatin 8 (2015) 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Qiu Y, Liu L, Zhao C, Han C, Li F, Zhang J, Wang Y, Li G, Mei Y, Wu M, Wu J, Shi Y, Combinatorial readout of unmodified H3R2 and acetylated H3K14 by the tandem PHD finger of MOZ reveals a regulatory mechanism for HOXA9 transcription, Genes Dev 26(12) (2012) 1376–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA, A chromatin landmark and transcription initiation at most promoters in human cells, Cell 130(1) (2007) 77–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Petruk S, Sedkov Y, Smith S, Tillib S, Kraevski V, Nakamura T, Canaani E, Croce CM, Mazo A, Trithorax and dCBP acting in a complex to maintain expression of a homeotic gene, Science 294(5545) (2001) 1331–4. [DOI] [PubMed] [Google Scholar]
- [58].Ernst P, Wang J, Huang M, Goodman RH, Korsmeyer SJ, MLL and CREB bind cooperatively to the nuclear coactivator CREB-binding protein, Mol Cell Biol 21(7) (2001) 2249–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Dou Y, Milne TA, Tackett AJ, Smith ER, Fukuda A, Wysocka J, Allis CD, Chait BT, Hess JL, Roeder RG, Physical association and coordinate function of the H3 K4 methyltransferase MLL1 and the H4 K16 acetyltransferase MOF, Cell 121(6) (2005) 873–85. [DOI] [PubMed] [Google Scholar]
- [60].Voss AK, Collin C, Dixon MP, Thomas T, Moz and retinoic acid coordinately regulate H3K9 acetylation, Hox gene expression, and segment identity, Dev Cell 17(5) (2009) 674–86. [DOI] [PubMed] [Google Scholar]
- [61].Milne TA, Briggs SD, Brock HW, Martin ME, Gibbs D, Allis CD, Hess JL, MLL targets SET domain methyltransferase activity to Hox gene promoters, Mol Cell 10(5) (2002) 1107–17. [DOI] [PubMed] [Google Scholar]
- [62].Thomas T, Dixon MP, Kueh AJ, Voss AK, Mof (MYST1 or KAT8) is essential for progression of embryonic development past the blastocyst stage and required for normal chromatin architecture, Mol Cell Biol 28(16) (2008) 5093–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Gupta A, Guerin-Peyrou TG, Sharma GG, Park C, Agarwal M, Ganju RK, Pandita S, Choi K, Sukumar S, Pandita RK, Ludwig T, Pandita TK, The mammalian ortholog of Drosophila MOF that acetylates histone H4 lysine 16 is essential for embryogenesis and oncogenesis, Mol Cell Biol 28(1) (2008) 397–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Liu Y, Bao C, Wang L, Han R, Beier UH, Akimova T, Cole PA, Dent SYR, Hancock WW, Complementary Roles of GCN5 and PCAF in Foxp3+ T-Regulatory Cells, Cancers (Basel) 11(4) (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Gao B, Kong Q, Zhang Y, Yun C, Dent SYR, Song J, Zhang DD, Wang Y, Li X, Fang D, The Histone Acetyltransferase Gcn5 Positively Regulates T Cell Activation, J Immunol 198(10) (2017) 3927–3938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Wang Y, Yun C, Gao B, Xu Y, Zhang Y, Wang Y, Kong Q, Zhao F, Wang CR, Dent SYR, Wang J, Xu X, Li HB, Fang D, The Lysine Acetyltransferase GCN5 Is Required for iNKT Cell Development through EGR2 Acetylation, Cell Rep 20(3) (2017) 600–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].He Z, Zhang J, Huang Z, Du Q, Li N, Zhang Q, Chen Y, Sun Z, Sumoylation of RORgammat regulates TH17 differentiation and thymocyte development, Nat Commun 9(1) (2018) 4870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Bararia D, Kwok HS, Welner RS, Numata A, Sarosi MB, Yang H, Wee S, Tschuri S, Ray D, Weigert O, Levantini E, Ebralidze AK, Gunaratne J, Tenen DG, Acetylation of C/EBPalpha inhibits its granulopoietic function, Nat Commun 7 (2016) 10968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Woo AJ, Kim J, Xu J, Huang H, Cantor AB, Role of ZBP-89 in human globin gene regulation and erythroid differentiation, Blood 118(13) (2011) 3684–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Yu X, Wang QL, Li YF, Wang XD, Xu A, Li Y, A novel miR-200b-3p/p38IP pair regulates monocyte/macrophage differentiation, Cell Discov 2 (2016) 15043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Jin Q, Zhuang L, Lai B, Wang C, Li W, Dolan B, Lu Y, Wang Z, Zhao K, Peng W, Dent SY, Ge K, Gcn5 and PCAF negatively regulate interferon-beta production through HAT-independent inhibition of TBK1, EMBO Rep 15(11) (2014) 1192–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Fournier M, Orpinell M, Grauffel C, Scheer E, Garnier JM, Ye T, Chavant V, Joint M, Esashi F, Dejaegere A, Gonczy P, Tora L, KAT2A/KAT2B-targeted acetylome reveals a role for PLK4 acetylation in preventing centrosome amplification, Nat Commun 7 (2016) 13227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Conacci-Sorrell M, Ngouenet C, Eisenman RN, Myc-nick: a cytoplasmic cleavage product of Myc that promotes alpha-tubulin acetylation and cell differentiation, Cell 142(3) (2010) 480–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Liu X, Xiao W, Wang XD, Li YF, Han J, Li Y, The p38-interacting protein (p38IP) regulates G2/M progression by promoting alpha-tubulin acetylation via inhibiting ubiquitination-induced degradation of the acetyltransferase GCN5, J Biol Chem 288(51) (2013) 36648–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Okumura K, Mendoza M, Bachoo RM, DePinho RA, Cavenee WK, Furnari FB, PCAF modulates PTEN activity, J Biol Chem 281(36) (2006) 26562–8. [DOI] [PubMed] [Google Scholar]
- [76].Patel JH, Du Y, Ard PG, Phillips C, Carella B, Chen CJ, Rakowski C, Chatterjee C, Lieberman PM, Lane WS, Blobel GA, McMahon SB, The c-MYC oncoprotein is a substrate of the acetyltransferases hGCN5/PCAF and TIP60, Mol Cell Biol 24(24) (2004) 10826–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Ge X, Jin Q, Zhang F, Yan T, Zhai Q, PCAF acetylates {beta}-catenin and improves its stability, Mol Biol Cell 20(1) (2009) 419–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Blanco-Garcia N, Asensio-Juan E, de la Cruz X, Martinez-Balbas MA, Autoacetylation regulates P/CAF nuclear localization, J Biol Chem 284(3) (2009) 1343–52. [DOI] [PubMed] [Google Scholar]
- [79].Martinez-Balbas MA, Bauer UM, Nielsen SJ, Brehm A, Kouzarides T, Regulation of E2F1 activity by acetylation, EMBO J 19(4) (2000) 662–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Wiper-Bergeron N, Salem HA, Tomlinson JJ, Wu D, Hache RJ, Glucocorticoid-stimulated preadipocyte differentiation is mediated through acetylation of C/EBPbeta by GCN5, Proc Natl Acad Sci U S A 104(8) (2007) 2703–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Abdou HS, Atlas E, Hache RJ, Liver-enriched inhibitory protein (LIP) actively inhibits preadipocyte differentiation through histone deacetylase 1 (HDAC1), J Biol Chem 286(24) (2011) 21488–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Mohibi S, Srivastava S, Bele A, Mirza S, Band H, Band V, Acetylation of Mammalian ADA3 Is Required for Its Functional Roles in Histone Acetylation and Cell Proliferation, Mol Cell Biol 36(19) (2016) 2487–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Malatesta M, Steinhauer C, Mohammad F, Pandey DP, Squatrito M, Helin K, Histone acetyltransferase PCAF is required for Hedgehog-Gli-dependent transcription and cancer cell proliferation, Cancer Res 73(20) (2013) 6323–33. [DOI] [PubMed] [Google Scholar]
- [84].Chen L, Wei T, Si X, Wang Q, Li Y, Leng Y, Deng A, Chen J, Wang G, Zhu S, Kang J, Lysine acetyltransferase GCN5 potentiates the growth of non-small cell lung cancer via promotion of E2F1, cyclin D1, and cyclin E1 expression, J Biol Chem 288(20) (2013) 14510–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Zhao L, Pang A, Li Y, Function of GCN5 in the TGF-beta1-induced epithelial-to-mesenchymal transition in breast cancer, Oncol Lett 16(3) (2018) 3955–3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Griffin NI, Sharma G, Zhao X, Mirza S, Srivastava S, Dave BJ, Aleskandarany M, Rakha E, Mohibi S, Band H, Band V, ADA3 regulates normal and tumor mammary epithelial cell proliferation through c-MYC, Breast Cancer Res 18(1) (2016) 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Yin YW, Jin HJ, Zhao W, Gao B, Fang J, Wei J, Zhang DD, Zhang J, Fang D, The Histone Acetyltransferase GCN5 Expression Is Elevated and Regulated by c-Myc and E2F1 Transcription Factors in Human Colon Cancer, Gene Expr 16(4) (2015) 187–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Farria AT, Mustachio LM, Akdemir ZHC, Dent SYR, GCN5 HAT inhibition reduces human Burkitt lymphoma cell survival through reduction of MYC target gene expression and impeding BCR signaling pathways, Oncotarget 10(56) (2019) 5847–5858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Trisciuoglio D, Ragazzoni Y, Pelosi A, Desideri M, Carradori S, Gabellini C, Maresca G, Nescatelli R, Secci D, Bolasco A, Bizzarri B, Cavaliere C, D’Agnano I, Filetici P, Ricci-Vitiani L, Rizzo MG, Del Bufalo D, CPTH6, a thiazole derivative, induces histone hypoacetylation and apoptosis in human leukemia cells, Clin Cancer Res 18(2) (2012) 475–86. [DOI] [PubMed] [Google Scholar]
- [90].Mustachio LM, Roszik J, Farria AT, Guerra K, Dent SY, Repression of GCN5 expression or activity attenuates c-MYC expression in non-small cell lung cancer, Am J Cancer Res 9(8) (2019) 1830–1845. [PMC free article] [PubMed] [Google Scholar]
- [91].Kumari R, Deshmukh RS, Das S, Caspase-10 inhibits ATP-citrate lyase-mediated metabolic and epigenetic reprogramming to suppress tumorigenesis, Nat Commun 10(1) (2019) 4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Tzelepis K, Koike-Yusa H, De Braekeleer E, Li Y, Metzakopian E, Dovey OM, Mupo A, Grinkevich V, Li M, Mazan M, Gozdecka M, Ohnishi S, Cooper J, Patel M, McKerrell T, Chen B, Domingues AF, Gallipoli P, Teichmann S, Ponstingl H, McDermott U, Saez-Rodriguez J, Huntly BJP, Iorio F, Pina C, Vassiliou GS, Yusa K, A CRISPR Dropout Screen Identifies Genetic Vulnerabilities and Therapeutic Targets in Acute Myeloid Leukemia, Cell Rep 17(4) (2016) 1193–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Kahl M, Brioli A, Bens M, Perner F, Kresinsky A, Schnetzke U, Hinze A, Sbirkov Y, Stengel S, Simonetti G, Martinelli G, Petrie K, Zelent A, Bohmer FD, Groth M, Ernst T, Heidel FH, Scholl S, Hochhaus A, Schenk T, The acetyltransferase GCN5 maintains ATRA-resistance in non-APL AML, Leukemia 33(11) (2019) 2628–2639. [DOI] [PubMed] [Google Scholar]
- [94].Koutsogiannouli EA, Wagner N, Hader C, Pinkerneil M, Hoffmann MJ, Schulz WA, Differential Effects of Histone Acetyltransferase GCN5 or PCAF Knockdown on Urothelial Carcinoma Cells, Int J Mol Sci 18(7) (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Fei HJ, Zu LD, Wu J, Jiang XS, Wang JL, Chin YE, Fu GH, PCAF acts as a gastric cancer suppressor through a novel PCAF-p16-CDK4 axis, Am J Cancer Res 6(12) (2016) 2772–2786. [PMC free article] [PubMed] [Google Scholar]
- [96].Bondy-Chorney E, Denoncourt A, Sai Y, Downey M, Nonhistone targets of KAT2A and KAT2B implicated in cancer biology (1), Biochem Cell Biol 97(1) (2019) 30–45. [DOI] [PubMed] [Google Scholar]
- [97].Wan J, Zhan J, Li S, Ma J, Xu W, Liu C, Xue X, Xie Y, Fang W, Chin YE, Zhang H, PCAF-primed EZH2 acetylation regulates its stability and promotes lung adenocarcinoma progression, Nucleic Acids Res 43(7) (2015) 3591–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Liu L, Scolnick DM, Trievel RC, Zhang HB, Marmorstein R, Halazonetis TD, Berger SL, p53 sites acetylated in vitro by PCAF and p300 are acetylated in vivo in response to DNA damage, Mol Cell Biol 19(2) (1999) 1202–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Xenaki G, Ontikatze T, Rajendran R, Stratford IJ, Dive C, Krstic-Demonacos M, Demonacos C, PCAF is an HIF-1alpha cofactor that regulates p53 transcriptional activity in hypoxia, Oncogene 27(44) (2008) 5785–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Ianari A, Gallo R, Palma M, Alesse E, Gulino A, Specific role for p300/CREB-binding protein-associated factor activity in E2F1 stabilization in response to DNA damage, J Biol Chem 279(29) (2004) 30830–5. [DOI] [PubMed] [Google Scholar]
- [101].Manickavinayaham S, Velez-Cruz R, Biswas AK, Bedford E, Klein BJ, Kutateladze TG, Liu B, Bedford MT, Johnson DG, E2F1 acetylation directs p300/CBP-mediated histone acetylation at DNA double-strand breaks to facilitate repair, Nat Commun 10(1) (2019) 4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Papai G, Frechard A, Kolesnikova O, Crucifix C, Schultz P, Ben-Shem A, Structure of SAGA and mechanism of TBP deposition on gene promoters, Nature 577(7792) (2020) 711–716. [DOI] [PubMed] [Google Scholar]
- [103].Wang H, Dienemann C, Stutzer A, Urlaub H, Cheung ACM, Cramer P, Structure of the transcription coactivator SAGA, Nature 577(7792) (2020) 717–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Chen YC, Gatchel JR, Lewis RW, Mao CA, Grant PA, Zoghbi HY, Dent SY, Gcn5 loss-of-function accelerates cerebellar and retinal degeneration in a SCA7 mouse model, Hum Mol Genet 21(2) (2012) 394–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Jin Q, Wang C, Kuang X, Feng X, Sartorelli V, Ying H, Ge K, Dent SY, Gcn5 and PCAF regulate PPARgamma and Prdm16 expression to facilitate brown adipogenesis, Mol Cell Biol 34(19) (2014) 3746–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Stilling RM, Ronicke R, Benito E, Urbanke H, Capece V, Burkhardt S, Bahari-Javan S, Barth J, Sananbenesi F, Schutz AL, Dyczkowski J, Martinez-Hernandez A, Kerimoglu C, Dent SY, Bonn S, Reymann KG, Fischer A, K-Lysine acetyltransferase 2a regulates a hippocampal gene expression network linked to memory formation, EMBO J 33(17) (2014) 1912–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Dent JR, Martins VF, Svensson K, LaBarge SA, Schlenk NC, Esparza MC, Buckner EH, Meyer GA, Hamilton DL, Schenk S, Philp A, Muscle-specific knockout of general control of amino acid synthesis 5 (GCN5) does not enhance basal or endurance exercise-induced mitochondrial adaptation, Mol Metab 6(12) (2017) 1574–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Luense LJ, Donahue G, Lin-Shiao E, Rangel R, Weller AH, Bartolomei MS, Berger SL, Gcn5-Mediated Histone Acetylation Governs Nucleosome Dynamics in Spermiogenesis, Dev Cell 51(6) (2019) 745–758 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Svensson K, Tahvilian S, Martins VF, Dent JR, Lemanek A, Barooni N, Greyslak K, McCurdy CE, Schenk S, Combined overexpression of SIRT1 and knockout of GCN5 in adult skeletal muscle does not affect glucose homeostasis or exercise performance in mice, Am J Physiol Endocrinol Metab 318(2) (2020) E145–E151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Pezoa SA, Artinger KB, Niswander LA, GCN5 acetylation is required for craniofacial chondrocyte maturation, Dev Biol (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]