SUMMARY
Lactose is an abundant dietary carbohydrate metabolized by the dental pathogen Streptococcus mutans. Lactose metabolism presents both classic diauxic behaviors and long-term memory, where the bacteria can pause for >11 h before initiating growth on lactose. Here, we explored mechanisms contributing to unusual aspects of regulation of the lac operon. The fructose phosphate metabolites, F-1-P and F-6-P, could modulate the DNA-binding activities of the lactose repressor. Recombinant LacR proteins bound upstream of lacA and Gal-6-P induced the formation of different LacR-DNA complexes. Deletion of lacR resulted in strain-specific growth phenotypes on lactose, but also on a number of mono- and di-saccharides that involve the glucose-PTS or glucokinase in their catabolism. The phenotypes were consistent with the novel findings that loss of LacR altered glucose-PTS activity and expression of the gene for glucokinase. CcpA was also shown to affect lactose metabolism in vivo and to bind to the lacA promoter region in vitro. Collectively, our study reveals complex molecular circuits controlling lactose metabolism in S. mutans, where LacR and CcpA integrate cellular and environmental cues to regulate metabolism of a variety of carbohydrates that are critical to persistence and pathogenicity of S. mutans.
Keywords: lactose repressor, fructose PTS, long-term memory, carbohydrate catabolite repression, dental caries
INTRODUCTION
Lactose is a β1,4-linked disaccharide of galactose and glucose. Due to the ubiquity of dairy products in many modern diets, lactose is one of the more common carbohydrates introduced into the oral cavity and rapidly consumed by many oral bacteria, particularly lactic acid bacteria (LAB) (de Vos & Vaughan, 1994). Although lactose can be cleaved into monosaccharides by extracellular β-galactosidases, it is often hydrolyzed intracellularly after being internalized by specialized transporters. For the major etiologic agent of human dental caries, Streptococcus mutans, lactose is transported by the phosphoenopyruvate (PEP)-dependent sugar: phosphotransferase system (PTS), yielding lactose-6-phosphate (Lac-6-P)(Vadeboncoeur & Proulx, 1984). Subsequent hydrolysis of Lac-6-P by a cytoplasmic phospho-β-galactosidase (LacG) releases a molecule of galactose-6-phosphate (Gal-6-P) and a molecule of glucose (Rosey & Stewart, 1992). It is believed that Gal-6-P serves as an allosteric effector that relieves repression by LacR of the lac operon (lacABCDFEGX), which encodes LacG, along with a lactose-specific PTS permease LacFE, and the enzymes of the tagatose pathway (LacABCD) (Hamilton & Lo, 1978, Morse et al., 1968, Zeng et al., 2010). Gal-6-P generated by cleavage of Lac-6-P, or from PTS-dependent uptake of extracellular galactose, is fermented primarily by the tagatose pathway, which bypasses the upper parts of the glycolytic pathway by converting Gal-6-P to tagatose-6-P (T-6-P), which is cleaved by a specific aldolase to 3-carbon end products (Jagusztyn-Krynicka et al., 1992). Catabolism of intracellular glucose generated from hydrolysis of Lac-6-P was thought to be carried out exclusively by an ATP-dependent glucokinase (Glk). However, much of the glucose released from Lac-6-P by S. mutans can be excreted to the extracellular environment, then internalized by the glucose-PTS (Liberman & Bleiweis, 1984, Zeng et al., 2018b).
Producing the enzymes for lactose utilization is energetically costly for bacteria, so tight regulation of expression of lac genes is common. Aside from the negative regulator LacR, the lac operon of many bacteria is subjected to carbon/carbohydrate catabolite repression (CCR), whereby the expression of the lac operon is suppressed when preferred sugars, such as glucose, are present (Deutscher, 2008, Gorke & Stulke, 2008). To trigger CCR in most Gram-positive bacteria, metabolism of glucose or certain other monosaccharides results in accumulation of glucose-6-P (G-6-P) and/or fructose-1,6-bisphosphate (F-1,6-bP), which can activate an HPr kinase/phosphatase (HPrK). HPrK phosphorylates the phospho-carrier protein HPr at a conserved Ser-46 residue, yielding HPr-Ser-P. Compared to HPr, HPr-Ser-P is a poorer substrate for the donation of a phosphoryl group at HPr-His15 by Enzyme (EI) of the PTS, thus lowering the efficiency of PTS in transporting all carbohydrates, including lactose (inducer exclusion)(Postma et al., 1993, Reizer et al., 1989). More importantly, HPr-Ser-P is a critical co-factor for catabolite control protein (CcpA), which forms a complex with HPr-Ser-P that can negatively regulate transcription of CCR-sensitive catabolic genes (Abranches et al., 2008). Previous research on CCR of lactose metabolism by S. mutans strain UA159 revealed some unique characteristics. While glucose very efficiently inhibits the expression of the lac operon, no alleviation of CCR was evident following deletion of the ccpA gene. Instead, deletion of the manL gene, encoding the EIIABMan domains of the glucose-PTS (manLMNO) (Zeng et al., 2010), led to near complete alleviation of CCR.
We recently described a long-term memory effect for the transition to lactose metabolism by S. mutans, observed in cells cultured previously on glucose, but especially pronounced in cells pre-grown on fructose (Zeng et al., 2018b). Specifically, when cells were cultured in base medium with fructose as the sole added carbohydrate source, then transferred to the same base medium with lactose, no growth occurred for 10 to 15 h; then the cells grew normally on lactose. If the population was subsequently repassaged on fructose, the memory remained; so the behavior was not associated with selection for a mutant population. The long-term memory phenotype is different from the classic diauxic growth delay when cells are cultured on a mix of, for example, glucose and lactose, where the transition to robust growth on lactose occurs in less that 1 h. Interestingly, the long lag could be eliminated by supplementing the lactose-containing medium with very low concentrations (as little as 10 μM) of a preferred carbohydrate, such as glucose. Also of note, two well-characterized laboratory strains, UA159 and GS-5, displayed significant difference in the length of time it took to initiate growth on lactose, with GS-5 showing notably longer delays (Zeng et al., 2018b). Finally, related to the effects of fructose, S. mutans UA159 expresses a fructose-phosphate phosphatase operon sppRA that is responsible for dephosphorylating metabolic intermediates of fructose, F-1-P and to a lesser degree F-6-P (Zeng & Burne, 2019). It was shown that fructose, specifically accumulation of F-1-P, alters biofilm development by S. mutans, partly by affecting bacterial autolytic activities. The latter study demonstrated the capacity of fructose metabolites, F-1-P in particular, in working as allosteric effectors for SppR function. GS-5 lacks the sppRA operon.
Utilizing both genetic and biochemical tools, we dissect here various potential mechanisms controlling the expression of the lac operon, including fructose-specific long-term memory, metabolism of glucose released from Lac-6-P, and the modulation of DNA-binding activities of LacR and CcpA. The results revealed sophisticated, but strain-specific, strategies for prioritizing carbohydrate metabolism for the purpose of maximizing bioenergetics and ecological advantages.
RESULTS AND DISCUSSION
F-1-P is required for fructose-specific memory in S. mutans GS-5.
Growth on fructose alters bacterial biofilm development and that the effects are, at least in part, attributable to accumulation of F-1-P (Zeng & Burne, 2019). As prior exposure to fructose results in an unusually long lag phase when cells are transferred onto lactose or cellobiose, we tested whether F-1-P might contribute to long-term memory. F-1-P is generated by internalization of fructose by the FruI PTS permease: one of at least 3 fructose PTS permeases in S. mutans, but the only one that generates F-1-P, as opposed to F-6-P. Interestingly, in comparison to UA159, GS-5 had significantly greater sensitivity to fructose in terms of the duration of the memory effect (Zeng et al., 2018b). A fruI mutation was therefore introduced into the S. mutans GS-5 genetic background. Cells were cultured with glucose or fructose as the sole carbohydrate, then diluted into a defined medium (FMC) with lactose or cellobiose as the sole carbohydrate. The long lag phase after growth on fructose that was typical of wild-type GS-5 was eliminated by deletion of fruI (Fig. 1A&B). The same fruI deletion was introduced into the UA159 background, but there was no significant impact of the mutation on the duration of the lag phase for transition to growth on lactose or cellobiose. As reported previously and confirmed here (Fig. 1C&D), growth on glucose or fructose resulted in similarly long lag phases for UA159 following transfer to lactose- or cellobiose-containing media.
Fig. 1.
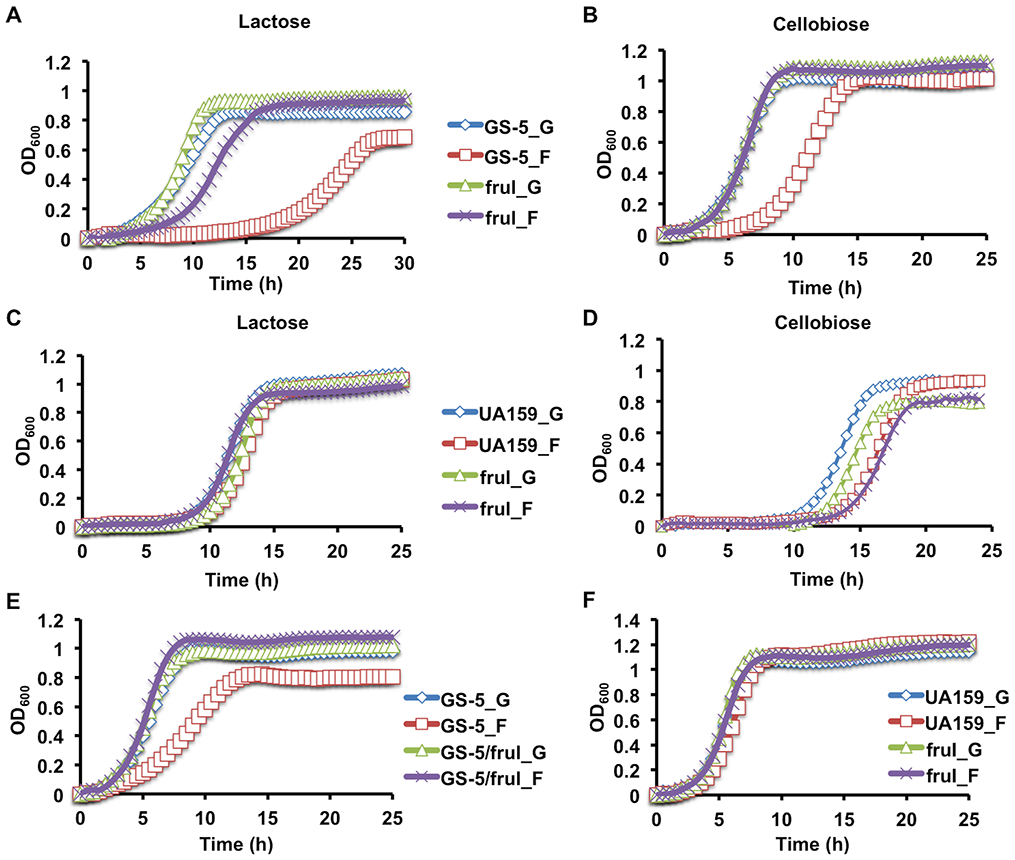
Growth curves of S. mutans fruI mutants. Wild-type strains GS-5 (A, B, E) and UA159 (C, D, F) and their respective mutants deficient in fructose-PTS permease (fruI) were first cultured to exponential phase in FMC supported with 20 mM of glucose (_G) or fructose (_F) and then diluted into fresh FMC containing 10 mM of lactose (A, C), cellobiose (B, D), or into FMC containing the same carbohydrates as that in the respective starter cultures (E, F). Optical density at 600 nm (OD600) was monitored using Bioscreen C over the period of 48 h. Results are each the average of at least three biological replicates.
We also studied the growth phenotypes of these mutants on glucose and fructose. Wild-type GS-5, but not its otherwise-isogenic fruI mutant, showed significantly slower exponential growth and lower final yields when growing on fructose, compared to glucose (Fig. 1E). No differences were seen when the fruI mutant was tested in glucose-containing medium. Similarly, UA159, as well as its fruI mutant, showed little difference in growth on glucose or fructose (Fig. 1F). These results support the hypothesis that F-1-P is required for the extended memory effect seen with strain GS-5 (Zeng et al., 2018a). The reduced yield of GS-5 growing on fructose could be due to increased autolytic activities, as was noted previously for UA159 growing in biofilms (Zeng & Burne, 2019). In fact, we previously showed that GS-5 releases more eDNA than does UA159 when grown in biofilms (Zeng & Burne, 2019). Regarding the phenotypes in the UA159 genetic background, we posit that a number of genetic differences between these two strains (Cornejo et al., 2013) could regulate metabolic activities that account for these strain-specific phenotypes. One such difference is that GS-5 lacks the fructose-phosphate phosphohydrolase operon (sppRA)(Fig. S1) that is required for dephosphorylation of F-1-P (Zeng & Burne, 2019), potentially enhancing the impact of F-1-P accumulation. The sppRA operon is present in most sequenced strains of S. mutans. It is interesting to note, though, that GS-5 was isolated prior to the inclusion of large quantities of high fructose corn syrup in the human diet (https://en.wikipedia.org/wiki/High-fructose_corn_syrup), whereas UA159 was isolated after. An exploration of the evolutionary impact of increased dietary fructose on S. mutans and other abundant members of the human oral microbiome might shed new light on the effects of the high fructose corn syrups on the composition and cariogenic potential of the oral microbiota in recent decades.
LacR coordinates lactose operon expression with glucose metabolism.
We have reported (Zeng & Burne, in press) that compared to UA159, strain GS-5 expresses the lac operon at notably lower levels, prompting us to investigate the function of LacR in the GS-5 genetic background. A lacR-deficient mutant was constructed in both UA159 and GS-5, and growth on lactose following inoculation with exponential-phase glucose-or fructose-grown cells was monitored. Consistent with our previous report (Zeng et al., 2010), the lacR mutant of UA159 initiated growth on lactose more rapidly than the wild type (Fig. 2A), reaffirming LacR as a repressor of the lac operon. Surprisingly, though, a deletion of lacR in GS-5 yielded a different behavior (Fig. 2B): Compared to the wild type, the lacR mutant initiated growth more rapidly than the parental strain when diluted into FMC-lactose from FMC-glucose, but the growth rate and the yield of the culture were both significantly reduced in the mutant. Similar outcomes were observed when FMC-fructose cultures were used as the inocula. To ensure that these phenotypes were not a result of secondary mutations, a wild-type copy of lacR was introduced into the GS-5 lacR mutant using the integration plasmid pBGE, which drives constitutive expression of inserted DNA fragments. Compared to the mutant transformed with only the pBGE vector, the lacR-complemented strain grew on lactose similarly to wild-type GS-5 (Fig. S2). Thus, in the GS-5 genetic background, LacR appears to have functions beyond repressing the lac operon.
Fig. 2.
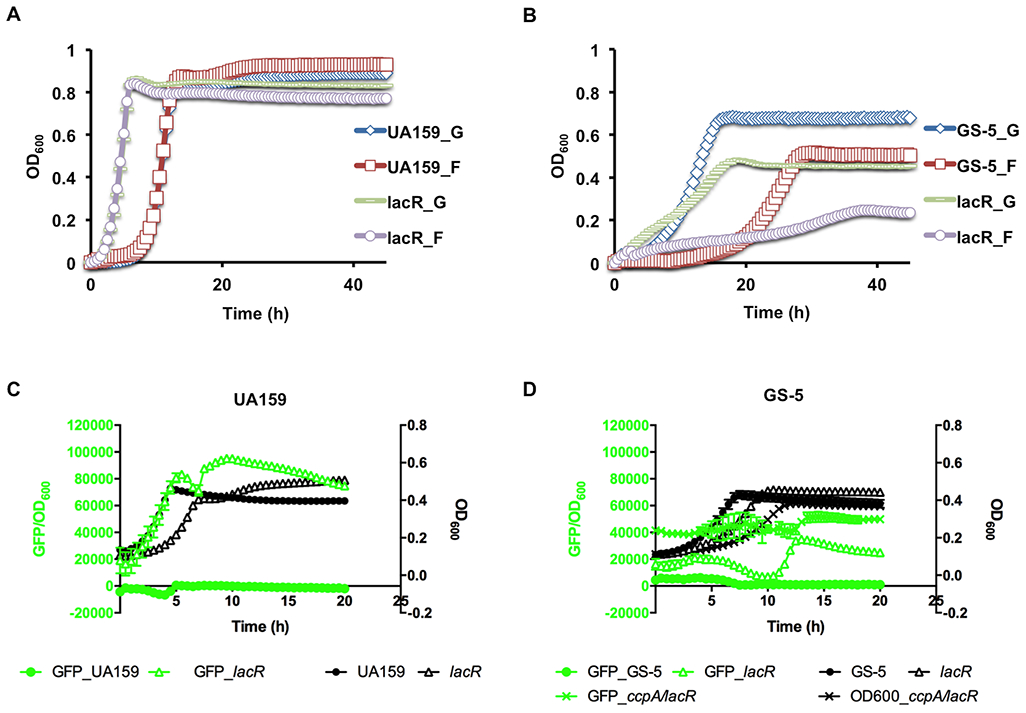
Growth phenotypes of S. mutans lacR mutants on lactose (A, B) and glucose (C, D). (A, B) Exponential phase cultures prepared in FMC constituted with glucose (_G) or fructose (_F) were diluted into fresh FMC supported with 10 mM lactose, then monitored using Bioscreen C. (C, D) Wild type strains and their mutants, each containing a PlacA::gfp fusion, were grown to exponential phase in FMC-glucose, diluted into the same medium, followed by 20-h incubation on a Synergy 2 plate reader that monitored both optical density (OD600) and green fluorescence. Results are each the average derived from three biological repeats.
To further the understanding of the effects of lacR deletion, the lacR mutation was introduced into strains of UA159 and GS-5 that each contained their cognate lacA promoters fused to a gfp gene (PlacA::gfp) (Zeng et al., 2018b). This allowed us to use a fluorescence-capable plate reader to monitor optical density and promoter activities of lacA over time. When tested in the UA159 genetic background growing on FMC-glucose (Fig. 2C), deletion of lacR resulted in significantly enhanced, constitutive expression of the promoter fusion, as lactose (or galactose) was no longer required for induction of the promoter. Conversely, and consistent with the growth phenotype (Fig. 2B) for GS-5, deletion of lacR resulted in only a minor increase in the activities of the lac promoter (Fig. 2D).
Surprisingly, the deficiency of LacR also resulted in a notable reduction in the growth rate in FMC-glucose for GS-5 and UA159 (Fig. 2C&D). We hypothesized that the slower growth on glucose could be associated with reduced efficiency in glucose transport. To test this possibility, phosphoenolpyruvate (PEP)-dependent glucose-phosphorylation assays were performed using permeablized, glucose-grown cells of the lacR mutants derived from GS-5 or UA159. For GS-5, the results (Fig. 3A) clearly indicated a significant reduction (~75%, p<0.01) in glucose-PTS activity associated with the loss of LacR. For UA159, a reduction in glucose PTS activity in the lacR mutant was consistently noted, but was not statistically significant. When we compared the glucose-PTS activities in these strains growing on lactose, however, glucose PTS activity was too low to allow meaningful comparisons (data not shown). Thus, LacR-dependent control of glucose uptake may be relevant under conditions that were not evaluated here; e.g. when lactose is present at steady-state levels in concentrations 10- to 100-fold lower than those used here, which would not elicit CCR via EIIMan. To further understand the underlying mechanism for how LacR may impact glucose PTS activity, we examined the mRNA and protein levels of the primary glucose-PTS (EIIMan) in the lacR mutants of UA159 and GS-5. After measuring mRNA levels of all 4 genes encoding EIIMan (manLMNO) using RT-qPCR, no difference was found for the lacR mutants in either background in cells cultured on glucose (Fig. S3A). Similarly, Western blots of lysates from cells growing on glucose showed no significant differences in ManL protein levels between the lacR mutants and their respective wild-type parents, as measured by densitometry (Fig. S3B). It therefore seems likely that LacR or a gene(s) under the control of LacR affects PTS-dependent transport of glucose at the post-translation level.
Fig. 3.
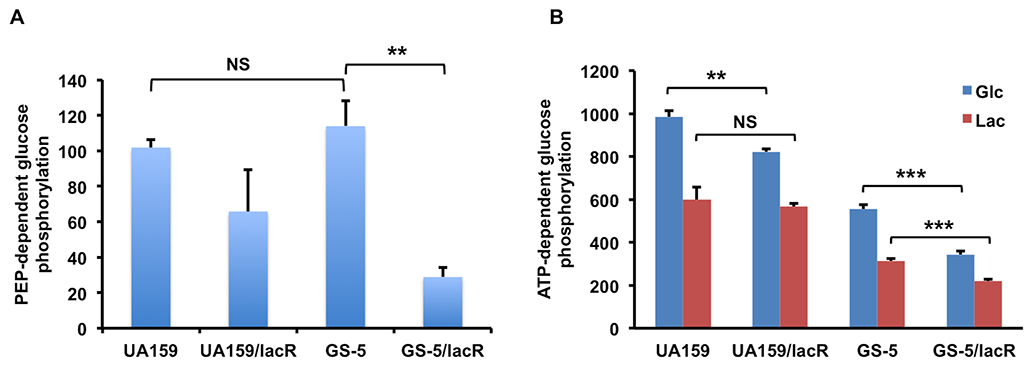
Glucose phosphorylation activities dependent on PEP (A) or ATP (B). Bacteria were cultivated to mid-exponential phase in TV medium supplemented with 0.5% of glucose (A, B), or lactose(B), before being harvested for assays. For PEP-dependent phosphorylation of glucose via PTS, cell suspensions were permeabilized with toluene: acetone (1:9); for ATP-dependent phosphorylation of glucose via glucokinase, cell lysates were prepared by bead-beating. In both cases, protein concentrations measured in the samples were used to normalize the activities. The results are each the average of at least three biological repeats, with statistical significance derived from Student’s t-test (* P<0.05, ** P<0.01, *** P<0.001).
Another potential mechanism by which LacR affects glucose metabolism is through glucokinase. While we have reported elsewhere (Zeng & Burne, in press) that Glk activities affect the expression of the lac operon, here we investigated if LacR could regulate glucose metabolism, particularly since Glk is responsible for phosphorylating the free glucose that is released from hydrolysis of lactose-6-P. It is worth noting that S. mutans can internalize unphosphorylated glucose via other PEP-independent pathways (Cvitkovitch et al., 1995), albeit not as efficiently as for the PTS, so Glk would also be needed to phosphorylate any glucose that entered via such transporters. When ATP-dependent glucose phosphorylation was measured in glucose-grown cells, lacR mutants of both UA159 and GS-5 produced lower Glk activities than their respective parents (Fig. 3B). When lactose-grown cells were used in the same assays however, only the lacR mutant of GS-5 showed significantly reduced Glk activities, compared to the parental strain. Transcriptional analysis via RT-qPCR also confirmed the difference in glK mRNA levels under glucose and lactose conditions (data not shown). Taken together with the PTS data, it appears that LacR exerts positive regulation on the metabolism of glucose, allowing the cells to coordinate the regulation of the expression of the lactose operon with glucose-PTS and glucokinase activities in a way that optimizes cell growth. It is notable that growth on lactose tends to lower these glucose-PTS and Glk activities compared to growth on glucose, indicating additional regulatory circuits may participate, e.g. the ability of glucose to induce expression of the glucose-PTS and glucokinase.
Deletion of lacR broadly affects carbohydrate metabolism by streptococci.
To better understand the roles of LacR in carbohydrate metabolism, we expanded our study to include a number of carbohydrates that influence the cariogenicity of S. mutans. Both wild-type strains and their lacR mutants were grown in BHI, then diluted into FMC media containing 10 mM glucose, fructose, sucrose, mannose, galactose, lactose, cellobiose, glucosamine (GlcN), N-acetylglucosamine (GlcNAc), maltose, or trehalose. When growth curves were recorded using a Bioscreen C system, significant changes in the length of lag phases, growth rates, and final optical densities were observed on a number of carbohydrate sources in the lacR mutants, as compared to parental strains growing on the same carbohydrate (Table 1 and Fig. S4). Specifically, the lacR mutants of UA159 and GS-5 each showed significant growth defects on glucose, mannose, GlcNAc, or cellobiose. The lacR mutants of both strains showed enhanced growth on galactose (Fig. 2). Specific for GS-5, various growth defects were also noted on fructose, GlcN, sucrose, maltose and trehalose. This experiment was repeated with two additional S. mutans strains, V403 and 11SSST2, and lacR deletion mutants thereof. The lacR mutant of V403 had reduced growth on glucose, mannose and GlcN, as well as minor defects on GlcNAc, maltose, trehalose and cellobiose. In contrast, the lacR mutant of 11SSST2 only showed growth defects on mannose, and a moderate reduction in yield on cellobiose. GS-5 was the only S. mutans strain in which the lacR mutation caused reduced growth on lactose, whereas the other three lacR mutant strains all had improved growth on lactose (Table 1). Taken together, these results indicated a pleiotropic effect on carbohydrate metabolism by S. mutans associated with the loss of LacR, apparently by affecting growth on carbohydrates that can be transported by the glucose-PTS. For example, glucose, mannose, GlcNAc, and GlcN all utilize EIIMan as the primary transporter in S. mutans (Moye et al., 2014b). As catabolism of lactose, maltose, trehalose, and cellobiose each involves generation of glucose, substantial glucose efflux and re-internalization by the PTS of the excreted glucose [(Zeng et al., 2018) and data not shown], the various growth phenotypes seen with the lacR mutants on these carbohydrates could, at least in part, be attributable to effects of loss of LacR on glucokinase and glucose-PTS activities. Alternatively, as deletion of lacR in these bacteria likely resulted in increased expression of the tagatose pathway and lactose-PTS to varying degrees, some of these phenotypes could also be due to the nonspecific activities of certain Lac enzymes.
Table 1.
Growth phenotypes of lacR mutants in comparison to their respective wild-type parents (see Fig. S4 for representative growth curves). Each strain was prepared as BHI cultures at exponential phase, and diluted into FMC media constituted with 10 mM of specified carbohydrates for growth monitoring. Results presented are the scored from at least three biological replicates.
| UA159 | GS-5 | V403 | 11SSST2 | DL1 | SK36 | |
|---|---|---|---|---|---|---|
| Glucose | − | -- | -- | +/− | +/−* | -- |
| Fructose | +/− | -- | +/− | − | +/−* | +/− |
| Mannose | --- | -- | --- | ---- | +/−* | +/−* |
| Galactose | +++ | ++ | ++ | − (yield only) | +/−* | − |
| GlcNAc | − | -- | − | +/− | +/−* | ---- |
| GlcN | +/− | -- | -- | +/− | +/− | ---- |
| Lactose | ++ | -- | + | + | +/− | − |
| Sucrose | +/− | -- | +* | +* | +/− | − |
| Maltose | +/− | − | − | +/− | +/−* | − |
| Trehalose | +/− | − | − | +/− | +/− | − |
| Cellobiose | -- | − | − | -- (yield only) | +/− | -- |
+/−: no change; −, --, ---, ----: minor, intermediate, major defect, or near flat line
increased yield; +, ++, +++: minor, intermediate, or major enhancement
Lastly, we investigated the effect of deletion of lacR in two well-studied commensal streptococci, Streptococcus gordonii DL1 and Streptococcus sanguinis SK36. DL1 possesses a 17-gene, tandemly-arranged gene cluster encoding two systems dedicated to metabolism of lactose and galactose (Zeng et al., 2012). The SK36 genome harbors a lac operon similar in size to that of S. mutans. The LacR sequences in these three organisms are highly similar, with sequence identity ranging from 59% to 85%. In addition to a LacR homologue, DL1 and SK36 each harbors a PRD (PTS-responsive domain)-containing transcriptional anti-terminator LacT (Zeng et al., 2012) that is absent in S. mutans. When lacR was deleted in DL1, the mutant showed no defect in growth on any of the carbohydrates tested here (Table 1). Instead, the mutation increased final optical density for a number of the carbohydrates, perhaps associated with the fact that the mutant tended to have increased chain length. The lacR mutant of SK36, however, presented significant growth defects in all but fructose and mannose, and had most severe defects when growing on the amino sugars GlcN and GlcNAc (Table 1 and Fig. S4). Collectively, these findings point to a broad role for LacR in carbohydrate metabolism by oral streptococci that merits further analysis and comparisons, and perhaps a careful assessment of the effects of lactose on the oral microbiome is warranted.
Fructose-phosphate intermediates affect DNA binding by LacR in vitro.
LacR belongs to a family of DeoR-type transcriptional regulators that have their DNA-binding activities modified by binding to a variety of sugar-phosphates (Skerlova et al., 2014). To study the potential capacity of F-1-P and other metabolic intermediates to affect the ability of LacR to bind its cognate targets, a recombinant His-tagged LacR protein was engineered by cloning the structural gene from strain GS-5 into plasmid pQE30 and overexpressing it in E. coli (Fig. S5A). A biotin-labeled DNA fragment containing the intergenic region (IGR) upstream of lacA from GS-5 was used in an electrophoretic mobility shift assay (EMSA) with the His-LacR protein. As shown in Fig. 4A, addition of His-LacR protein reduced the migration of the lacA probe in a concentration-dependent manner, yielding one band of higher molecular mass (complex I) with 0.2 μM His-LacR, and two higher bands (complexes I & II) when 0.4 μM protein was used. As is common with EMSAs, complexes with slower rates of migration generally represent binding by one or more molecules of protein. Since DeoR-family regulators are known to form dimers or even oligomers, it is possible that complex II results from protein-protein interactions.
Fig. 4.
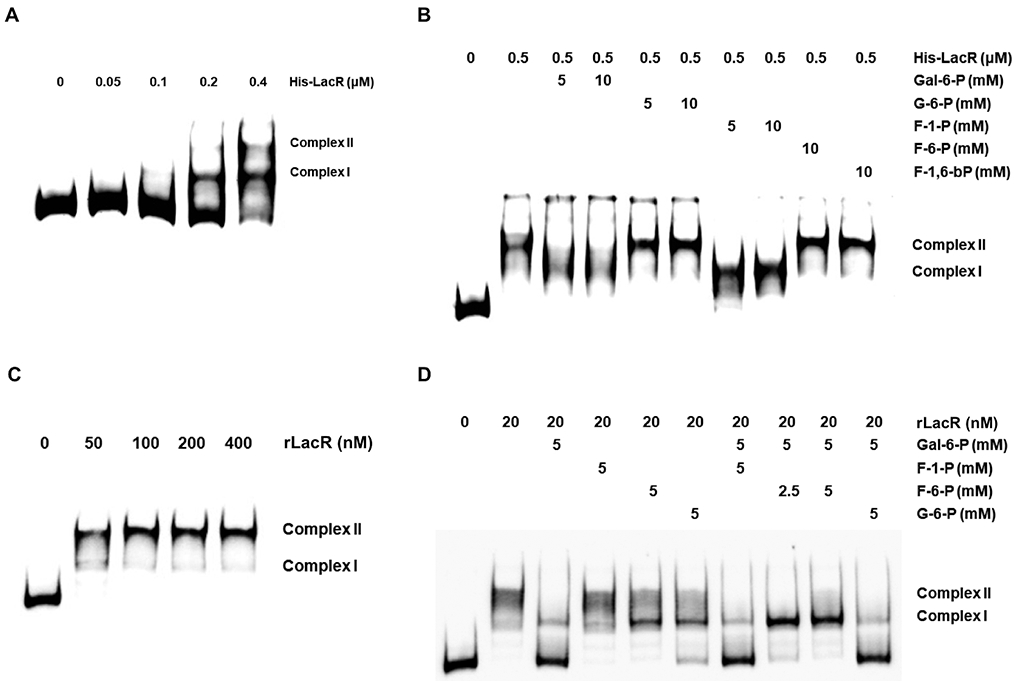
Electrophoretic mobility shift assay (EMSA) performed with recombinant LacR proteins and IGR of lacA, both derived from GS-5 background. Reactions were performed using varying amounts of His-LacR protein (A, B) or rLacR released from MBP-LacR fusion protein (C, D), in the presence (B, D) or absence (A, C) of various metabolic intermediates.
The binding of the His-LacR protein to the lacA probe was then investigated in the presence of the following metabolic intermediates: Gal-6-P, G-6-P, F-1-P, F-6-P or F-1,6-bP. Addition of 5 or 10 mM Gal-6-P (Fig. 4B), which is an allosteric regulator of LacR in related bacteria (Morse et al., 1968), reduced or eliminated the formation of complex II, without affecting the formation of complex I. In contrast, G-6-P, F-6-P and F-1,6-bP showed no effect on His-LacR DNA-binding activity. Interestingly, 5 or 10 mM F-1-P showed similar effects to that of Gal-6-P, allowing the formation of only complex I.
Recognizing that the His-tag could affect the DNA-binding or effector-binding activity of LacR, we constructed an MBP-LacR fusion protein derived from GS-5, from which the MBP portion was subsequently removed using Factor Xa (Fig. S5B&C). The resultant LacR protein had only 6 additional amino acids (ISEFGS) at the N-terminus of the sequence, and the EMSA was repeated using this new recombinant LacR protein (henceforth named rLacR). Compared to the His-tagged version, this rLacR protein bound to the lacA probe with apparently higher affinity than the His-LacR, forming both complexes (I & II) with as little as 50 nM rLacR (Fig. 4C). When tested for response to the same metabolic intermediates, however, only Gal-6-P was able to inhibit the binding of rLacR to the lacA probe, while none of the other compounds showed significant effects when tested individually (Fig. 4D). Interestingly, when used together with Gal-6-P, each at 5 mM, F-6-P abolished the ability of Gal-6-P to inhibit the binding of rLacR to lacA probe. None of the other compounds, including F-1-P, showed any such effects.
For comparison, another set of recombinant LacR proteins derived from S. mutans UA159 were engineered with His or MBP tags and similarly purified. A DNA probe containing the lacA IGR from UA159 was also prepared. LacR proteins from UA159 and GS-5 have only 1 amino acid residue difference (2nd residue: Arg in UA159, Lys in GS-5). The lacA probe from GS-5 is 8-bp longer than that from UA159, due to the presence of two contiguous (T)7G motifs in GS-5, but only one (T)7G in UA159 (Zeng et al., 2018a); otherwise the probes were identical in sequence. Similar results in EMSAs were obtained using materials from UA159 and GS-5 (Fig. S6), except that the MBP-derived rLacR of GS-5 showed slightly higher apparent affinity for the lacA probe from GS-5, compared to the UA159 interactions.
It is clear that some inconsistency exits between the behaviors of His-LacR and the rLacR released from MBP-LacR fusion protein, which were modified by the N-terminal addition of a positively charged 6-histidine tag and a small peptide (ISEFGS), respectively. Despite this limitation, these EMSA analyses support that metabolites derived from fructose metabolism, F-1-P and/or F-6-P, can modulate DNA-binding activities of LacR, either directly or by interfering with the effects of the presumptive effector molecule, Gal-6-P. The fact that Gal-6-P does not always abolish the binding of His-LacR to the lacA probe is consistent with the notion that LacR could be required for expression of the lac operon even in the presence of lactose, at least in the GS-5 genetic background.
More details of the LacR-DNA interactions were obtained using DNase I footprinting. Fluorescently-labeled DNA probes containing the same IGR of lacA were allowed to interact with rLacR that was released from MBP-LacR under conditions similar to that used in EMSAs, followed by treatment with DNase I and capillary electrophoresis (DFACE). As shown in Fig. 5, rLacR reduced access of DNase I to a 50-bp, A/T-rich region of the DNA probe (site A, ATA ATA CCA AAA ATG TTT AAA AAA TAA ACA AAA ATG TGT TGA TTT TTT AA) encompassing the region located between 78 and 127 bp upstream of the start codon of lacA in UA159, although the bound region was not uniformly protected from DNase I digestion. About 40 bp further upstream, an approximately 20-bp region (site B, 147~167bp) was also protected by rLacR. A putative −35 motif (TTGATT) overlaps site A (Zeng et al., 2018b). Although Gal-6-P clearly reduced binding of LacR to the same DNA fragment in EMSA (Fig. 4), addition of 5 mM Gal-6-P together with rLacR did not significantly change the DNAase I footprints on the lacA probe at these two sites. Notably, though, addition of Gal-6-P resulted in the appearance of a DNase I hyper-sensitive site about 20-nucleotides upstream of site B (Fig. 5, site C), indicative of a shift in the accessibility of the DNA. No effect was observed when F-1-P, F-6-P, or G-6-P was used individually with rLacR. Interestingly, when both F-6-P and Gal-6-P were added to the reaction, there was a substantial reduction in the height of the peaks in hyper-sensitive site C, compared to Gal-6-P alone. When the GS-5-derived rLacR was used in the DFACE assay, the same two binding sites upstream of lacA in GS-5 were protected and the effects from Gal-6-P and F-6-P were the same (Fig. S7). Given the impact of F-1-P on the interaction of His-LacR with the lacA probe (Fig. 4B), we also conducted a DFACE assay using His-LacR in the presence of F-1-P. His-LacR protected site A and F-1-P had no effect when added to the reaction (data not shown).
Fig. 5.
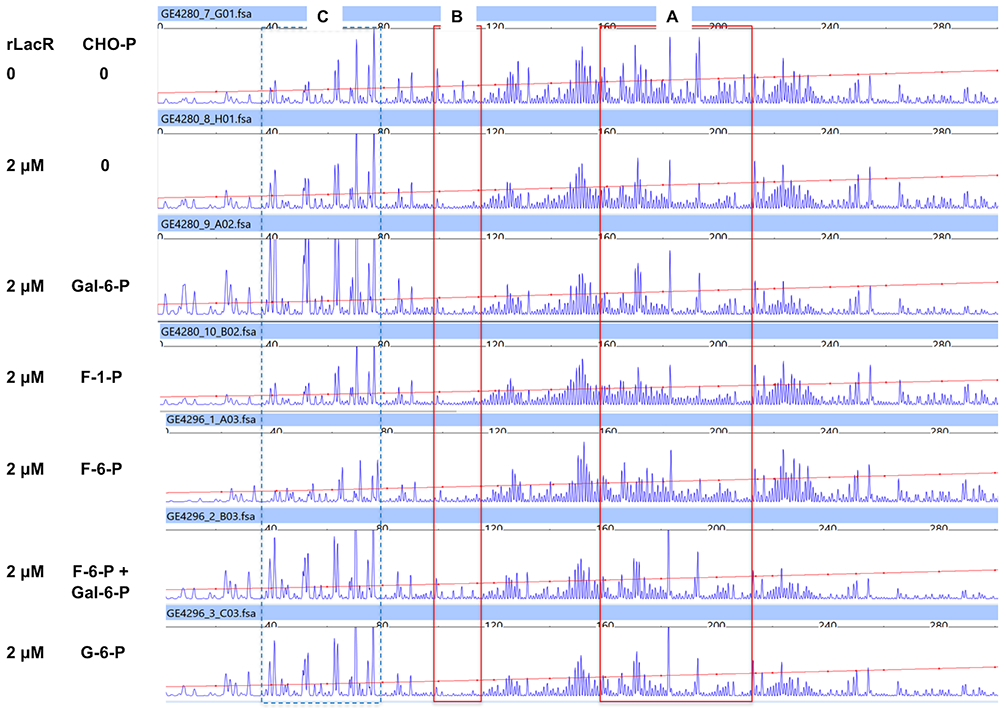
DNase I footprints of rLacR protein mapped to the IGR of lacA in the sense strand. Results were obtained using rLacR protein released from MBP-LacR and a fluorescently-labeled lacA probe, each derived from UA159 background. Each lane of the graph represents a reaction that contained specified amounts of rLacR protein, and 0 or 5 mM each of metabolites (CHO-P). Three regions of the probe are highlighted by boxes, denoting two sites (A, B) that are protected by rLacR, and one (C) showing hypersensitivity to DNase I in the presence of Gal-6-P.
Therefore, while it is likely that Gal-6-P and F-1-P bind to recombinant LacR and decrease its binding affinity in vitro, the interactions may not result in complete loss of binding by LacR to its cognate targets, nor do the sugar phosphates alter the site that is bound by LacR. Instead, Gal-6-P seems to affect the interaction in a way that changes the configuration of the DNA-protein complex, an effect that could be alleviated by the presence of F-6-P. As Gal-6-P serves as an inducer for the expression of the lac operon, it is probably safe to assume that the outcome of such reconfiguration would lead to enhanced promoter activity. The effect of F-1-P, on the other hand, could be more subtle than that of F-6-P and not detectable under our current assay conditions, or involve other transcriptional regulators or co-factors in vivo that are absent in our in vitro assays.
Interaction of CcpA with the 5’ region of lacA.
Although deletion of ccpA in UA159 failed to alleviate glucose-dependent CCR of the lac operon (Zeng et al., 2010), changes in glucokinase activities do affect lac operon expression and CcpA does affect the duration of memory in GS-5 (Zeng & Burne, in press). One possible scenario is that both LacR and CcpA exert direct control over lac transcription, but to various extents in different strains. Specifically, the expression pattern of the lac operon in GS-5 is distinct from that of UA159, with relatively low levels of induction by lactose or when lacR is deleted. The lower level of responsiveness to lactose can, at least in part, be explained by CCR. In particular, the lacR mutant of GS-5 grows more efficiently than the wild type on galactose (Fig. S8); a sugar that induces the lac operon without triggering CCR (Zeng et al., 2010). However, it is only when both ccpA and lacR are deleted from GS-5 that there is significant constitutive expression of the lacA promoter (Fig. 2D), indicating a strong influence by CcpA that is not apparent in UA159.
To obtain more information regarding how CcpA and LacR may function together, we performed EMSA and DFACE assays using the aforementioned lacA probes, and a His-tagged CcpA protein utilized in a previous study (Abranches et al., 2008). The amino acid sequences of the CcpA proteins of UA159 and GS-5 are identical. Migration of the biotin-labeled probes was impeded by the addition of increasing amounts of CcpA protein, with both the GS-5 and UA159 fragments showing similar interactions with as little as 50 nM CcpA (Fig. 6A). A notable characteristic of these EMSA results was the ladder of bands formed when CcpA protein was present in the range of 0.05 and 0.2 μM. DFACE assays were performed under similar conditions. The digestion patterns of both DNA probes were comparable overall, with the footprint of CcpA covering a 66-bp span of DNA (254-188 bp) upstream of lacA in UA159, including the (T)7G motif near the 3’ end of the probe. For GS-5, the footprint was about the same size as that for UA159, and both (T)7G motifs were protected by CcpA, in effect shifting the footprint to the right by about 8 bp (Fig. 6B). This result, together with the “laddering” effect in EMSAs, could indicate that binding of CcpA initiated near the (T)7G motif, followed by protein oligomerization. Another interesting difference was the appearance of two DNaseI hyper-sensitive regions in GS-5 that were absent from the UA159 samples; indicative of potential differences in the conformation of the DNA. Closer examination also indicated that the last 13 nucleotides of the footprints, TTT TTT TGC GTA A, constitute an imperfect consensus cre element (Hueck et al., 1994). The fact that these (T)7G motifs are part of the region bound by CcpA echoes our previous finding that glucose-PTS-dependent regulation of the lac operon in GS-5 requires the duplicate (T)7G element (Zeng et al., 2018b). We recognize the usual limitations of using a recombinant protein in vitro, however it seems likely that metabolism of glucose released from lactose hydrolysis, funneled into glycolysis by either Glk or the glucose-PTS, has the potential to trigger CCR. In particular, increased levels of the metabolic intermediates G-6-P and F-1,6-bP generated from glucose metabolism would increase the proportion of cellular HPr that is phosphorylated on Ser46 by HprK. HPr-Ser-P and these metabolic intermediates can enhance binding of CcpA to its cognate targets (Deutscher, 2008).
Fig. 6.
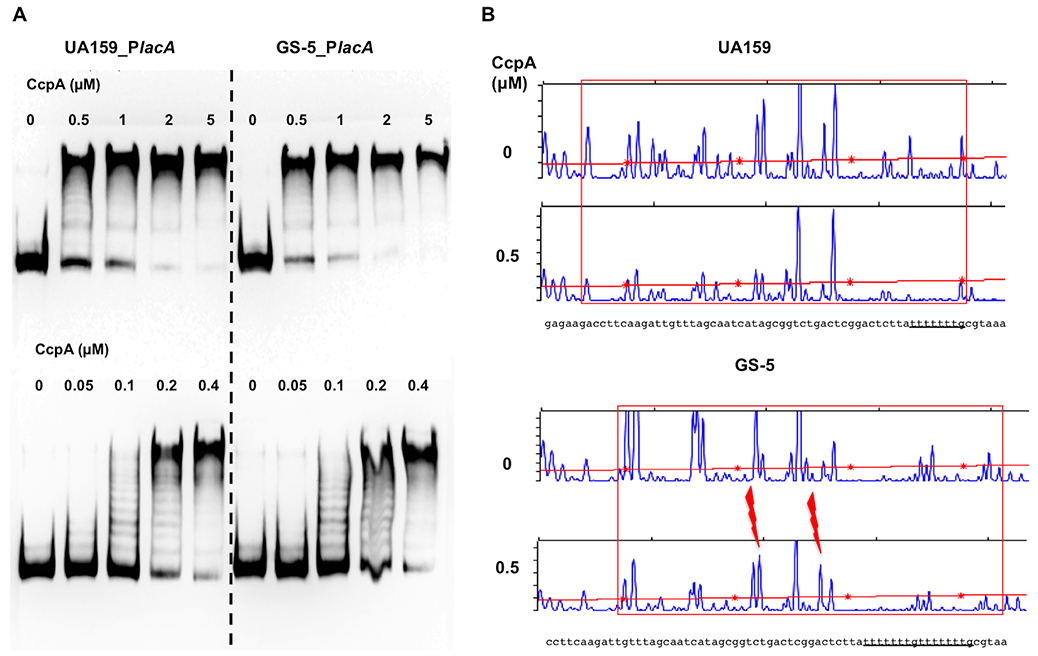
EMSA (A) and DFACE (B) performed with CcpA and IGR of lacA. Various concentrations of recombinant protein His-CcpA was used to interact with biotin-labeled probes (A) or fluorescent probes (B) that encompass the IGR of lacA, followed by electrophoresis (A) or DNase I treatment and fragment analysis (B). Red boxes denote the region protected by CcpA against DNase I digestion, and two red bolts represent hyper-sensitive sites.
It is important to note that the binding sites for LacR and CcpA identified in these studies do not appear to overlap, which could allow these regulators to act concomitantly to control lacA transcription (Fig. 7A). The fact that two (T)7G motifs are present in the IGR of lacA in GS-5, but just one is in UA159, suggests that the occupancy by and orientation of CcpA protein at this location could differ between the strains. Together with LacR and its allosteric effectors, CcpA could differentially affect how accessible the promoter becomes to RNA polymerase when growing on certain carbohydrates. We developed a working model to explain the mechanisms that allow metabolic intermediates from glucose, fructose and lactose to modulate the functions of LacR and CcpA (Fig. 7B). This model does not necessarily exclude additional mechanisms that allow the glucose-PTS to influence the regulation of the lac operon independent of CcpA. We also performed additional EMSAs using both rLacR and CcpA proteins in the same reaction, together with biotin-labeled lacA probes from GS-5 or UA159. The results (Fig. S9) gave some support for simultaneous binding by these regulators, however binding by rLacR appeared more prominent, especially in the GS-5 background. We recognize that without HPr-Ser-P as a co-factor, the affinity of CcpA to the putative cre could be reduced. Considering the fact that more than one copy of these proteins binds a single DNA molecule, methods with greater resolution and sensitivities will be needed in the future to refine our working model.
Fig. 7.
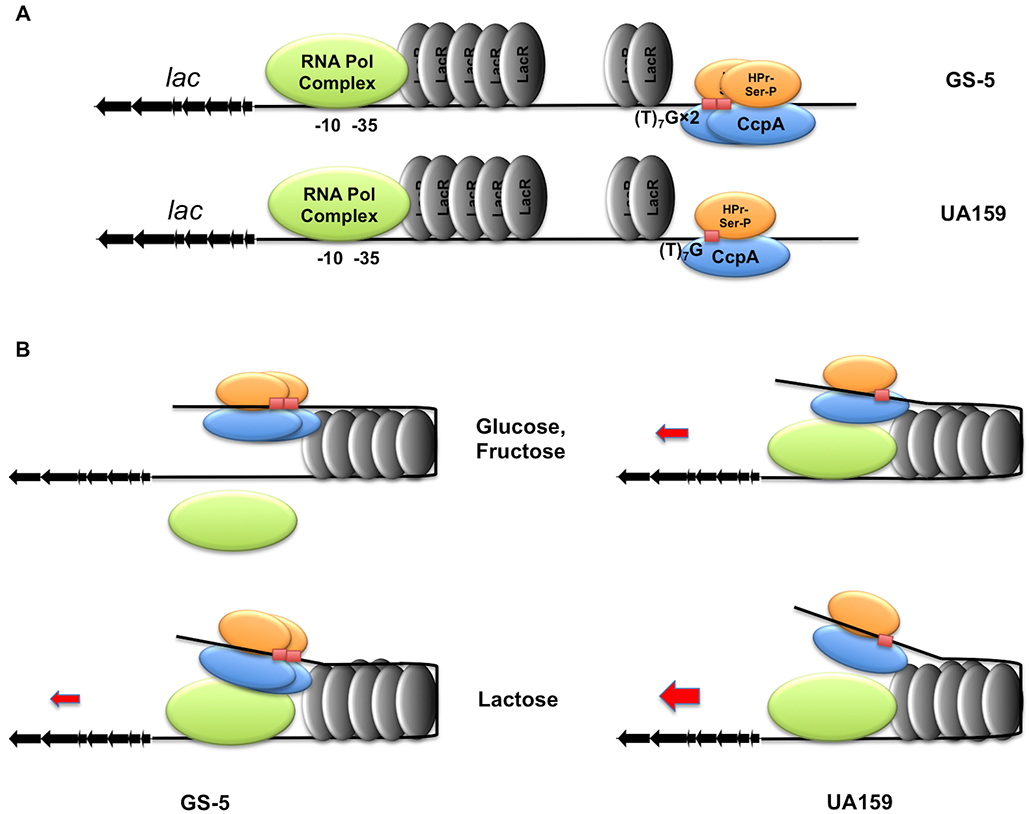
A schematic model depicting mechanisms controlling expression of the lac operon in two S. mutans strains. (A) Locations of LacR and CcpA upstream of lacA as deduced from EMSA and DFACE assays. (B) In the presence of glucose or fructose, metabolites G-6-P, F-1-P or F-6-P allows a closed configuration of the DNA-protein complex. This results in loss of lac expression in GS-5 and suppressed expression in UA159. Metabolism of lactose or presence of Gal-6-P reconfigures the complex, which allows transcription to proceed in UA159, and allows partial expression in GS-5 due to the influence of CCR. This model does not rule out the influence of the glucose-PTS.
Concluding remarks.
After intracellular hydrolysis, Lac-6-P metabolism by S. mutans proceeds with degradation of Gal-6-P via the tagatose pathway, and phosphorylation and glycolysis of intracellular glucose. It is clear from this study that the major transcriptional regulator LacR controls both pathways, and that glucose released from hydrolysis of Lac-6-P contributes to CCR of the lac operon. By investigating the fate of excreted glucose, we have (Zeng & Burne, in press) identified a stochastic phenotype in lac gene expression and a subpopulation behavior in lactose metabolism that leads to cheating, a strategy favored by GS-5. Here, we uncovered the molecular mechanisms behind some of these phenotypes. Compared to UA159, GS-5 shows a greater tendency to recognize and respond to metabolic signals derived from the preferred carbohydrates, glucose and fructose, both in terms of modulating LacR activity, and triggering memory and CCR. This, in turn, results in suppressed expression of the lac operon in GS-5, thus favoring the branch that catabolizes glucose. In addition, we showed the ability of LacR to regulate metabolism of an array of carbohydrates important to the persistence of both the caries pathogen and certain commensals, supporting its potential as a therapeutic target for oral and other infectious diseases. Together our research has illuminated the complex nature of lactose metabolism by oral bacteria, both in terms of molecular mechanisms, and at population and inter-bacterial levels. And since dental caries is considered a multi-microbial, microbiome-based infectious disease that is often driven by carbohydrate-rich diets, continued effort in understanding how common carbohydrates are metabolized by major constituent bacteria, as well as its ecological impact, could help us develop better strategy in managing or combatting this important disease.
EXPERIMENTAL PROCEDURES
Bacterial strains and culture conditions.
Brain heart infusion (BHI) medium (Difco Laboratories, Detroit, MI) was used to maintain S. mutans and other streptococcal strains (Table 2). Antibiotics were used at the following concentrations: for BHI agar plates: kanamycin (Km) 1 mg/mL, erythromycin (Em) 10 μg/mL, and spectinomycin (Sp) 1 mg/mL; only half of the concentrations were used in liquid BHI. For cultures requiring specific carbohydrates, a semi-defined Tryptone-Vitamin (TV) medium (Burne et al., 1999) or a fully synthetic medium FMC (Terleckyj et al., 1975) was utilized, each constituted with specified carbohydrates. All cultures were maintained at 37°C in an ambient atmosphere that was supplemented with 5% CO2. L-broth and L-agar were used for cultivating Escherichia coli, with antibiotics added for plasmid maintenance: ampicillin 100 μg/mL, and Km 50 μg/mL.
Table 2.
Bacterial strains and plasmids used in this study.
| Strains or plasmids | Relevant genotypesa | Source or reference |
|---|---|---|
| Strains | ||
| UA159 | Wild type S. mutans, serotype c | (Ajdic et al., 2002) |
| GS-5 | Wild type S. mutans, serotype c | (Biswas & Biswas, 2012) |
| V403 | Wild type S. mutans, serotype c | (Macrina et al., 1991) |
| 11SSST2 | Wild type S. mutans, serotype c | (Cornejo et al., 2013) |
| SK36 | Wild type Streptococcus sanguinis | (Kilian et al., 1989) |
| DL1 | Wild type Streptococcus gordonii | (LeBlanc & Hassell, 1976) |
| MMZ1285 | UA159 PlacA::gfp | (Zeng et al., 2018a) |
| MMZ1480 | GS-5 PlacA::gfp | This study |
| MMZ1654 | UA159 fruI::Kmr | This study |
| MMZ1685 | GS-5 fruI::Kmr | This study |
| MMZ1728 | UA159 lacR::Emr | This study |
| MMZ1761 | GS-5 lacR::Kmr | This study |
| MMZ1767 | GS-5 lacR::Emr PlacA::gfp | This study |
| MMZ1768 | UA159 lacR::Emr PlacA::gfp | From MMZ1285 |
| MMZ1769 | GS-5 lacR::Kmr ccpA::Emr | From MMZ1761 |
| MMZ1770 | GS-5 lacR::Emr ccpA::Spr PlacA::gfp | From MMZ1767 |
| MMZ1771 | GS-5 lacR::Kmr pBGE | From MMZ1761 |
| MMZ1772 | GS-5 lacR::Kmr pBGE-lacR | From MMZ1761 |
| MMZ1774 | SK36 lacR::Kmr | This study |
| MMZ1624 | DL1 lacR::Kmr | (Zeng et al., 2012) |
| MMZ1781 | V403 lacR::Kmr | This study |
| MMZ1782 | 11SSST2 lacR::Kmr | This study |
| Plasmids | ||
| pQE30 | E. coli vector for His-tag protein | Qiagen |
| pMAL-p2X | E. coli vector for MBP-fusion | NEB |
| pBGE | S. mutans integration vector, Emr | (Zeng & Burne, 2009) |
Kmr kanamycin resistance; Spr, spectinomycin resistance; Emr, erythromycin resistance.
For growth curves, bacterial cultures were covered with mineral oil and maintained at 37°C in a Bioscreen C lab system (Labsystems Oy, Helsinki, Finland) that monitored and recorded the optical density at 600 mM (OD600) every 30 min. To simultaneously monitor the growth of the bacterium and the expression levels of the lac operon over time using a PlacA::gfp reporter fusion (Zeng et al., 2018), bacterial cultures from the exponential phase were diluted (at 1:20) into FMC containing specified amounts of carbohydrate(s). For 20 h, optical density (OD600) and the relative fluorescence units (RFU) of the cultures were recorded using a fluorescence-capable Synergy 2 Multi-Mode reader from BioTek (Winooski, VT). The RFU of each sample was subtracted by the RFU of a control strain that lacked the reporter fusion, and the results were normalized using the OD600, following a procedure detailed elsewhere (Zeng et al., 2018a).
Construction of genetically modified strains.
Standard molecular cloning protocols (Sambrook & Russell, 2001) were followed during genetic manipulation of E. coli and streptococcal bacteria. To engineer allelic exchange mutants in S. mutans, a set of primers (Table S1) were synthesized for PCR amplification of two DNA fragments flanking the coding sequence of the target gene. Usually a 30~50 bp region from either end of the gene was excluded from deletion. These primers were designed so that the two PCR products shared sufficient overlap (~30 bp) with the 5’ and 3’ ends of an antibiotic resistance cassette to ensure successful ligation. After a Gibson assembly reaction (New England BioLabs, Beverly, MA), the ligation product could be further amplified by PCR, or used directly for transformation of S. mutans. A similar procedure was followed for the creation of genetic mutants in S. gordonii and S. sanguinis, except that longer flanking DNA fragments and higher concentrations of DNA were used to ensure successful transformation. The competent state of S. mutans was induced with either a synthetic XIP (comX/sigX-inducing peptide) in bacteria cultivated in a chemically defined medium CDM (Chang et al., 2011), or using a competence-stimulating peptide (CSP) in BHI cultures. For transformation of S. gordonii and S. sanguinis, bacteria were cultured in BHI supplemented with 10% heat-inactivated horse serum to induce competence. DNA Sanger sequencing was carried out to validate each mutant, and to ensure that no unintended mutations were created in regions flanking the target gene.
To complement the genetic deficiency in a GS-5 lacR mutant, a pair of primers, lacR-5’GA and lacR-3’GA, were designed for amplification of the structural gene and ribosome-binding site of lacR. At the same time, plasmid pBGE, which allows for double crossover recombination to integrate DNA into the gtfA gene (Zeng & Burne, 2009), was digested with restriction enzymes XbaI, BsrGI and NdeI, yielding two DNA fragments, 1.5 Kbp and 3 Kbp in size that share ~30-bp overlaps with both ends of the PCR product. Subsequently, a Gibson assembly reaction was carried out using these two DNA fragments and the PCR product of lacR, followed by a further PCR amplification using primers pBGE-GA5’ and pBGE-GA3’. The resultant recombinant DNA containing the intact copy of lacR, an erythromycin cassette, and two flanking DNA fragments homologous to gtfA, was used to transform GS-5 lacR. The empty vector pBGE was linearized by NdeI digestion and used as a control to transform GS-5 lacR. The resultant strains were sequenced in the gtfA region to validate the insert.
Engineering and preparation of recombinant proteins.
To construct a recombinant His-tagged LacR protein for overexpression in E. coli, a pair of primers, lacR-5’Bm and lacR-3’PstI, were designed to amplify lacR from the genomes of UA159 and GS-5. Each PCR product was then treated with restriction enzymes BamHI and PstI, followed by ligation with a pQE30 plasmid that was digested with the same enzymes. The ligated product was used to transform an E. coli host M15 pREP4. After validation of the clones via Sanger sequencing, each strain was cultivated to exponential phase with L-broth and then treated with 0.05 mM of IPTG (Isopropyl-β-D-thiogalactoside) for 4 h, for induction of recombinant His-LacR protein. A similar procedure was followed to clone LacR into the vector pMAL-p2X for overexpression of MBP-LacR fusion protein.
Purification of the recombinant proteins His-LacR and MBP-LacR was carried out using a nickel-nitrilotriacetic (Ni-NTA) agarose (Qiagen) and an amylose resin (NEB), respectively, according to protocols provided by the providers. MBP-LacR was subsequently cleaved using a protease Factor Xa (NEB), frequently forming aggregates of released rLacR protein. This allowed for separation of rLacR from the soluble MBP protein by centrifugation at 5000 rpm using a tabletop centrifuge. The purified proteins were analyzed by polyacrylamide gel electrophoresis (Fig. S5) to confirm their size and purity. The recombinant CcpA protein was constructed previously (Abranches et al., 2008) and purified by following the same protocol.
EMSA and DNA footprint analysis by automated capillary electrophoresis (DFACE).
The interactions between the recombinant LacR, CcpA proteins and the intergenic region (IGR) upstream of lacA were studied using EMSA (Zeng & Burne, 2015) and DFACE (Zianni et al., 2006), following protocols detailed elsewhere. A biotin-labeled DNA probe containing the IGR of lacA was created using a pair of primers, lacAEMSA-1 and lacAEMSA-2bio. Similarly, another pair of primers, lacADFACE-5 and lacADFACE-3, allowed for amplification and differential labeling of the sense (6-FAM) and antisense (VIC) strand of the same DNA.
Glucokinase and PTS assays.
In vitro phosphorylation of glucose via glucokinase (Sato et al., 2015) or the PTS (Moye et al., 2014a) was performed according to established protocols. Bacterial strains were cultivated in TV base medium supplemented with the specified carbohydrates, and harvested at exponential phase (OD600 ≈ 0.5) before conducting the assays.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by DE12236 from the National Institute of Dental and Craniofacial Research.
Footnotes
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- Abranches J, Nascimento MM, Zeng L, Browngardt CM, Wen ZT, Rivera MF, and Burne RA (2008) CcpA regulates central metabolism and virulence gene expression in Streptococcus mutans. J Bacteriol 190: 2340–2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ajdic D, McShan WM, McLaughlin RE, Savic G, Chang J, Carson MB, Primeaux C, Tian R, Kenton S, Jia H, Lin S, Qian Y, Li S, Zhu H, Najar F, Lai H, White J, Roe BA, and Ferretti JJ (2002) Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc Natl Acad Sci U S A 99: 14434–14439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas S, and Biswas I (2012) Complete genome sequence of Streptococcus mutans GS-5, a serotype c strain. J Bacteriol 194: 4787–4788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burne RA, Wen ZT, Chen YY, and Penders JE (1999) Regulation of expression of the fructan hydrolase gene of Streptococcus mutans GS-5 by induction and carbon catabolite repression. J Bacteriol 181: 2863–2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JC, LaSarre B, Jimenez JC, Aggarwal C, and Federle MJ (2011) Two group A streptococcal peptide pheromones act through opposing Rgg regulators to control biofilm development. PLoS Pathog 7: e1002190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornejo OE, Lefebure T, Pavinski Bitar PD, Lang P, Richards VP, Eilertson K, Do T, Beighton D, Zeng L, Ahn SJ, Burne RA, Siepel A, Bustamante CD, and Stanhope MJ (2013) Evolutionary and population genomics of the cavity causing bacteria Streptococcus mutans. Mol Biol Evol 30: 881–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvitkovitch DG, Boyd DA, Thevenot T, and Hamilton IR (1995) Glucose transport by a mutant of Streptococcus mutans unable to accumulate sugars via the phosphoenolpyruvate phosphotransferase system. J Bacteriol 177: 2251–2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vos WM, and Vaughan EE (1994) Genetics of lactose utilization in lactic acid bacteria. FEMS Microbiol Rev 15: 217–237. [DOI] [PubMed] [Google Scholar]
- Deutscher J (2008) The mechanisms of carbon catabolite repression in bacteria. Curr Opin Microbiol 11: 87–93. [DOI] [PubMed] [Google Scholar]
- Gorke B, and Stulke J (2008) Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat Rev Microbiol 6: 613–624. [DOI] [PubMed] [Google Scholar]
- Hamilton IR, and Lo GC (1978) Co-induction of Beta-galactosidase and the lactose-P-enolpyruvate phosphotransferase system in Streptococcus salivarius and Streptococcus mutans. J Bacteriol 136: 900–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hueck CJ, Hillen W, and Saier MH Jr. (1994) Analysis of a cis-active sequence mediating catabolite repression in Gram-positive bacteria. Res Microbiol 145: 503–518. [DOI] [PubMed] [Google Scholar]
- Jagusztyn-Krynicka EK, Hansen JB, Crow VL, Thomas TD, Honeyman AL, and Curtiss R 3rd (1992) Streptococcus mutans serotype c tagatose 6-phosphate pathway gene cluster. J Bacteriol 174: 6152–6158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilian M, Mikkelsen L, and Henrichen J (1989) Taxonomic study of viridans streptococci: description of Streptococcus gordonii sp. nov. and emended descriptions of Streptococcus sanguis (White and Niven 1946), Streptococcus oralis (Bridge and Sneath 1982), and Streptococcus mitis (Andrewes and Horder 1906). Int J Syst Evol Microbiol 39: 471–484. [Google Scholar]
- LeBlanc DJ, and Hassell FP (1976) Transformation of Streptococcus sanguis Challis by plasmid deoxyribonucleic acid from Streptococcus faecalis. J Bacteriol 128: 347–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberman ES, and Bleiweis AS (1984) Role of the phosphoenolpyruvate-dependent glucose phosphotransferase system of Streptococcus mutans GS5 in the regulation of lactose uptake. Infect Immun 43: 536–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macrina FL, Jones KR, Alpert CA, Chassy BM, and Michalek SM (1991) Repeated DNA sequence involved in mutations affecting transport of sucrose into Streptococcus mutans V403 via the phosphoenolpyruvate phosphotransferase system. Infect Immun 59: 1535–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse ML, Hill KL, Egan JB, and Hengstenberg W (1968) Metabolism of lactose by Staphylococcus aureus and its genetic basis. J Bacteriol 95: 2270–2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moye ZD, Burne RA, and Zeng L (2014a) Uptake and metabolism of N-acetylglucosamine and glucosamine by Streptococcus mutans. Appl Environ Microbiol 80: 5053–5067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moye ZD, Zeng L, and Burne RA (2014b) Fueling the caries process: carbohydrate metabolism and gene regulation by Streptococcus mutans. J Oral Microbio 6: 24878–24892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postma PW, Lengeler JW, and Jacobson GR (1993) Phosphoenolpyruvate:carbohydrate phosphotransferase systems of bacteria. Microbio Rev 57: 543–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reizer J, Sutrina SL, Saier MH, Stewart GC, Peterkofsky A, and Reddy P (1989) Mechanistic and physiological consequences of HPr(Ser) phosphorylation on the activities of the phosphoenolpyruvate:sugar phosphotransferase system in Gram-positive bacteria: studies with site-specific mutants of HPr. Embo J 8: 2111–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosey EL, and Stewart GC (1992) Nucleotide and deduced amino acid sequences of the lacR, lacABCD, and lacFE genes encoding the repressor, tagatose 6-phosphate gene cluster, and sugar-specific phosphotransferase system components of the lactose operon of Streptococcus mutans. J Bacteriol 174: 6159–6170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, and Russell DW, (2001) Molecular cloning: a laboratory manual, p. 3 v. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [Google Scholar]
- Sato Y, Okamoto-Shibayama K, and Azuma T (2015) Glucose-PTS involvement in maltose metabolism by Streptococcus mutans. Bull Tokyo Dent Coll 56: 93–103. [DOI] [PubMed] [Google Scholar]
- Skerlova J, Fabry M, Hubalek M, Otwinowski Z, and Rezacova P (2014) Structure of the effector-binding domain of deoxyribonucleoside regulator DeoR from Bacillus subtilis. Febs J 281: 4280–4292. [DOI] [PubMed] [Google Scholar]
- Terleckyj B, Willett NP, and Shockman GD (1975) Growth of several cariogenic strains of oral streptococci in a chemically defined medium. Infect Immun 11: 649–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vadeboncoeur C, and Proulx M (1984) Lactose transport in Streptococcus mutans: isolation and characterization of factor IIIlac, a specific protein component of the phosphoenolpyruvate-lactose phosphotransferase system. Infect Immun 46: 213–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L, and Burne RA (2009) Transcriptional regulation of the cellobiose operon of Streptococcus mutans. J Bacteriol 191: 2153–2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L, and Burne RA (2015) NagR differentially regulates the expression of the glmS and nagAB genes required for amino sugar metabolism by Streptococcus mutans. J Bacteriol 197: 3533–3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L, and Burne RA (2019) Essential roles of the sppRA fructose-phosphate phosphohydrolase operon in carbohydrate metabolism and virulence expression by Streptococcus mutans. J Bacteriol 201: e00586–00518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L, Chen L, and Burne RA (2018a) Preferred hexoses influence long-term memory and induction of lactose catabolism by Streptococcus mutans. Appl Environ Microbiol 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L, Chen L, and Burne RA (2018b) Preferred hexoses influence long-term memory in and induction of lactose catabolism by Streptococcus mutans. Appl Environ Microbiol 84: e00864–00818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L, Das S, and Burne RA (2010) Utilization of lactose and galactose by Streptococcus mutans: transport, toxicity, and carbon catabolite repression. J Bacteriol 192: 2434–2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L, Martino NC, and Burne RA (2012) Two gene clusters coordinate galactose and lactose metabolism in Streptococcus gordonii. Appl Environ Microbiol 78: 5597–5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L, and Burne RA (2020) Subpopulation behaviors in lactose metabolism by Streptococcus mutans. Mol Microbiol (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zianni M, Tessanne K, Merighi M, Laguna R, and Tabita FR (2006) Identification of the DNA bases of a DNase I footprint by the use of dye primer sequencing on an automated capillary DNA analysis instrument. Journal of biomolecular techniques : JBT 17: 103–113. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


