Abstract
Purpose
The objective of this study is to characterize the role of microRNAs (miRNA) in the classification of head and neck squamous cell carcinoma (HNSCC).
Experimental Design
Here, we analyzed 562 HNSCC samples, 88 from a novel cohort and 474 from The Cancer Genome Atlas, using miRNA-microarray and miRNA-sequencing, respectively. Using an integrative correlations method followed by miRNA expression-based hierarchical clustering, we validated miRNA clusters across cohorts. Evaluation of clusters by logistic regression and gene ontology approaches revealed subtype-based clinical and biological characteristics.
Results
We identified two independently validated and statistically significant (p<0.01) tumor subtypes and named them ‘epithelial’ and ‘stromal’ based on associations with functional target gene ontology relating to differing stages of epithelial cell differentiation. MicroRNA-based subtypes were correlated with individual gene expression targets based on miRNA seed sequences, as well as with miRNA families and clusters including the miR-17 and miR-200 families. These correlated genes defined pathways relevant to normal squamous cell function and pathophysiology. MicroRNA clusters statistically associated with differential mutation patterns including higher proportions of TP53 mutations in the stromal class and higher NSD1 and HRAS mutation frequencies in the epithelial class. MicroRNA classes correlated with previously reported gene expression subtypes, clinical characteristics, and clinical outcomes in a multivariate Cox proportional hazards model with stromal patients demonstrating worse prognoses (HR = 1.5646, p = 0.006).
Conclusion
We report a reproducible classification of HNSCC based on miRNA that associates with known pathologically altered pathways and mutations of squamous tumors and is clinically relevant.
Keywords: microRNA, head and neck cancer, squamous, subtypes, epithelial, mesenchymal, stromal
INTRODUCTION
Squamous cell carcinoma is the most frequently occurring malignant tumor of the head and neck region, and head and neck squamous cell carcinoma (HNSCC) is the sixth leading cancer by incidence with approximately 600,000 new cases worldwide each year (1). Well-established risk factors for HNSCC include tobacco and alcohol consumption, as well as infection with human papillomavirus (HPV) (2). Given the limited means for subclassification, HNSCC follows a heterogenous clinical course for patients currently grouped into a rudimentary disease classification based on clinical stage alone. While most HNSCC patients are candidates for curative therapy, currently there are a lack of approaches for accurately selecting treatments that are tailored in intensity and mechanism to specific patient populations (3).
Outside of HPV status, there is no standardized molecular classification system for HNSCC tumors. We have previously proposed one such classification system based on distinct gene expression patterns in four mRNA-based classes, and recent data from The Cancer Genome Atlas (TCGA) study provided further insight concerning the underlying molecular biology of HNSCC (4,5). Concurrently, other groups have proposed new subtypes or refined previous molecular classifications using integrative analyses that use a variety of omic profiling methods (6,7).
MicroRNAs are small, highly conserved, non-coding RNA molecules that post-transcriptionally regulate gene expression by binding to and inhibiting target mRNAs, which can influence stem cell properties, differentiation, and tumorigenesis (8). They also form networks with other non-coding RNA molecules as competing endogenous RNA (ceRNA) that act as ‘molecular sponges’ to regulate the levels of entire miRNA families, which can influence specific biologic pathways involved in head and neck carcinogenesis (9,10). Additionally, they may be attractive biomarkers because some are potentially preserved in formalin-fixed paraffin-embedded (FFPE) tissues and some biological fluids, but quantification is limited by the stability of specific miRNAs in these routine clinical samples being influenced by many extrinsic and intrinsic factors, such as GC content (11–14). However, it should be noted that this benefit is not established in the current work which was entirely performed using assays performed on fresh frozen tissue. Increasing evidence relates the dysregulation of certain miRNAs to the development of HNSCC (15–20). However, while prior miRNA profiling efforts in HNSCC revealed differential miRNA expression between malignant and normal tissue, few have focused on miRNA differences occurring among individual tumors (21).
In this study, we identified robust miRNA clusters that can be used to subclassify HNSCC tumors as well as other squamous tumors of other anatomic locations. The analysis includes miRNA expression from two independent HNSCC cohorts, offering confidence in the validity of the results. Using expression profiling results and associated clinical variables, we investigated the relationship between the miRNA subtypes and primary tumor sites, biological and molecular signatures, and overall survival.
METHODS
Discovery cohort
An institutional discovery cohort was generated by obtaining tumor specimens acquired through protocols approved by the Institutional Review Board of the University of North Carolina (#01–1283) and written informed consent was obtained per the approved protocol. This study follows the Declaration of Helsinki. Patients included in this study were adults with histologically confirmed HNSCC of the oral cavity, oropharynx, hypopharynx, and larynx. Medical records were abstracted for clinical events and demographics, and fresh frozen tissue was obtained at the time of surgical resection. We obtained 10 normal controls, consisting of five tumor adjacent normal samples and five normal tonsil tissue samples from tonsillectomy performed for non-malignant indications.
RNA was extracted using TRIzol from Invitrogen and quantified using the NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE). RNA quality was assessed by Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA). One microgram total RNA was labeled with FlashTaq RNA labeling kit from Genisphere (Genisphere Inc. Hatfield, PA), and then poly-(A) tailing was added to RNA, before ligating a biotinylated signal molecule to the target RNA sample. Biotin-labeled RNA was hybridized to the Affymetrix GeneChip miRNA-1_0 array overnight at 48°C. After hybridization and wash, the arrays were scanned with the GeneChip Scanner 3000. Probe-level intensity files were evaluated for quality using the ‘affy’ package (22) to process and quantile normalize the raw CEL files, and the log2 RMA expression values were used for downstream analysis. A total of 88 tumor and 10 normal arrays consisting of 825 mature miRNAs remained after the removal of low-quality and non-HNSCC samples. MicroRNA array data from these samples has been deposited in the NCBI’s Gene Expression Omnibus (GEO) under accession number GSE144711 (23). We determined the HPV status of the 88 patients in the UNC cohort via in situ hybridization of tumor samples as previously reported (4).
Validation cohorts
For the purposes of independent validation, we obtained sequencing data from the TCGA HNSCC cohort including processed (level 3) mRNA-sequencing (mRNA-seq) and mature miRNA-sequencing (miRNA-seq) data from the public repository for a total of 528 tumors (474 tumors with both mRNA and miRNA) and normal controls (n=44)(24). We obtained additional miRNA expression data from related tumor cohorts from TCGA including bladder cancer, squamous lung cancer, and cervical carcinoma (24). All miRNA-seq data was obtained as normalized and log2-transformed expression in reads per million, while the mRNA-seq data included normalized and log2-transformed RSEM expression. Duplicate assays, samples with incomplete gene expression data, and unusual clinical cases were excluded as not representative of the discovery cohort (n=10).
Clinical data, including patient survival, was obtained from the public repository (24). We determined the HPV status of all 474 TCGA patients using mRNA sequencing reads aligned with all HPV reference genomes taken from the PaVE database (https://pave.niaid.nih.gov/, RRID:SCR_016599) (25), using methods that have been previously described (5). Briefly, patients were determined to have an HPV-associated tumor if the mRNA aligned counts exceeded 1,000 reads mapped to viral genes, primarily E6 and E7.
miRNA filtering, class discovery, and validation
Most miRNAs within a given cell are expressed at a relatively low baseline, and the miRNA with lower expression are less likely to be biologically active (25). Thus, we considered only the most variable miRNAs with a mean absolute deviation (MAD) > 75th percentile in our clustering analysis. In order to interrogate only those miRNAs that were well measured on both platforms (array and sequencing), we computed integrative correlation coefficients (ICC) (26). We considered a total of 50 miRNAs within the top 10% ICC to have similar expression across cohorts and used them for the remainder of the analyses. A flow chart depicting the selection of these genes is provided in Supplementary Fig. S1.
We used the ConsensusClusterPlus R package (RRID:SCR_016954) to determine a range of possible miRNA cluster assignments for each cohort by average linkage hierarchical clustering using a distance measure of one minus the Pearson’s correlation coefficient (27). The statistical significance of clusters was assessed using the SigClust R package and methodology (28). For the purpose of individual sample class assignment, we applied the centroid predictor method in which centroids were determined for each cluster in both the discovery and validation cohorts by computing the mean expression value for each miRNA over all samples in each cohort having the appropriate class label. By calculating the correlation-based distance (1 – Pearson correlation coefficient) to each centroid, we determined the class labels for each sample within a cohort and each cluster across cohorts. This miRNA-based clustering methodology was also applied to the bladder, cervical, and lung squamous carcinoma TCGA cohorts for pan-cancer validation.
Differentially expressed genes and microRNAs
To identify differentially expressed miRNAs (DEmiRs) between clusters in the TCGA cohorts we employed the likelihood ratio test method using a false discovery rate (FDR) threshold of 0.05 as implemented in the R package DEGseq (version 1.42.0) (29). Additionally, DEmiRS were determined similarly between HPV(+) and HPV(−) TCGA samples, as well as between each miRNA cluster and normal controls in the TCGA samples. Differential gene expression analysis was conducted in the TCGA cohort between the miRNA-based clusters after filtering genes with low expression variability (MAD < 75 percentile) using DEGseq with an FDR threshold of 0.05.
MicroRNA-mRNA integrative analysis
We next used the gene expression data from the TCGA cohort to quantify the effect of miRNA expression on gene expression using an approach from other miRNA studies(29). We calculated the Pearson correlation coefficients for all possible combinations of miRNAs and mRNAs, with Benjamini and Hochberg corrections (q-values) to control for multiple testing. We determined a q-value of 0.01 as the cutoff for the negative correlations between miRNAs and genes. We further annotated miRNA-gene pairs using a sequence complementarity approach derived from the seed sequence of the miRNA and predicted gene targets. Only miRNA-gene pairs with algorithmic support from at least two methods were considered for further analysis. Gene target prediction approaches included: targetScan v7.2 (RRID:SCR_010845, no percentile scores were used to filter binding sites) (30), miRwalk v3 (RRID:SCR_016509) (31), miRTarBase v7.0(32), and miRDB v6.0(33). For integrative analysis in the TCGA bladder, lung, and cervical data sets we used only the negatively correlated and predicted genes from the TCGA HNSCC data set that have an ICC value in the top 75 percentile (26).
Functional gene network analysis
Gene sets identified by either supervised or unsupervised approaches were annotated using the DAVID algorithm (https://david.ncifcrf.gov/, RRID:SCR_001881) and the parametric gene set enrichment analysis (PAGE) technique relying on 5917 gene ontology (GO) terms obtained from MSigDB (34,35). In the PAGE technique, normalized Z-scores were calculated and tested for statistical significance (FDR < 0.01) across miRNA-target specific gene sets and the resulting matrix assessed for patterns by hierarchical clustering (36). The ‘cutree’ function was then used to parse row dendrograms into clusters of GO terms representing functional categories according to the correlation similarities.
Mutations and copy number
Somatic DNA alterations for tumor samples were obtained from the public repository, including mutation annotation format files and copy number alterations as described by level 3 segmented genome regions determined by the GISTIC methodology (24). We then found the top 50 most frequently mutated genes with relevance to cancer as determined by the Cancer Gene Census and analyzed copy number profiles between the two subtypes using the GISTIC module v2.0.23 (RRID:SCR_000151) (37,38).
Statistical Analysis
All analyses were performed using R 3.6.0 (39). Categorical comparisons were performed using Fisher’s exact test, unless otherwise noted. Correlations were performed using the Pearson correlation coefficient. Survival analysis was with the R packages survival (version 3.2–3) (40) and survminer (version 0.4.8) (41). Statistical significance in survival analysis was determined by the log-rank test to compare between the two clusters, and a Cox proportional hazards model was used to estimate univariate and multivariate associations and to control for potential confounders.
RESULTS
Demographics and miRNA class discovery in HNSCC cohorts
For the purpose of discovering and validating miRNA patterns in HNSCC we developed two independent patient cohorts, one novel institutional cohort from UNC and a second cohort which was a superset of previously described TCGA samples. Patients treated at UNC were somewhat younger than in TCGA (median of 57 vs 61; P = 0.036, Mann-Whitney) and more likely to have oropharyngeal cancer (25% vs 15%; P<0.001), but were similar in overall stage distribution, HPV status, and gender. (Supplementary Table S1).
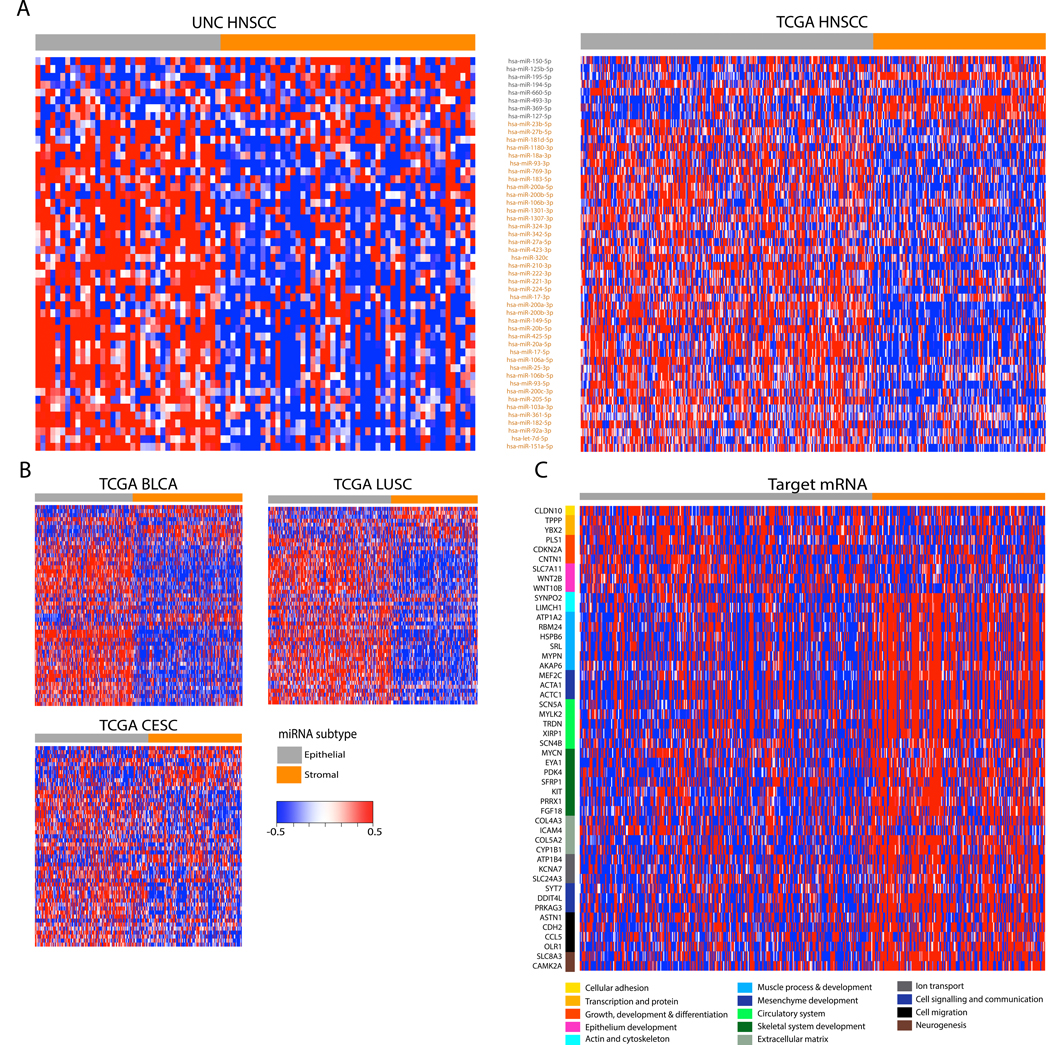
MicroRNAs were filtered to select for the most well-measured and highly-variable genes across samples and cohorts using previously described methods including the integrative correlations technique and the mean absolute deviation respectively (42,43). For the purposes of discovering reproducible and statistically significant classes based on miRNA expression we used 50 miRNAs from the filtered miRNA gene list to perform consensus clustering (unsupervised hierarchical clustering) (Supplementary Table S2A and Supplementary Fig. S1). Clusters identified by independent analysis of the UNC and TCGA cohorts were assessed for statistical significance by the SigClust method, suggesting that two clusters was the only statistically significant solution validated across these cohorts (p<0.01). Cross platform centroid-based classifiers demonstrated the mapping of class labels between the two expression platforms, with UNC clusters I and II mapping to TCGA clusters II and I respectively (Supplementary Table S2B). In other words, a predictor built in the UNC cohort sorted samples mapped to clusters independently discovered in the validation cohort and vice versa. (Fig. 1A). We named the clusters based on the miRNA associated with each of the clusters, including 8 miRNAs downregulated in UNC cluster I and TCGA cluster II (Fig. 1A in gray, labeled ‘epithelial’), and the remaining 42 miRNAs downregulated in UNC cluster II and TCGA cluster I (Fig. 1A in orange, labeled ‘stromal’). We next asked the question if similar clusters could be detected in tumors from other anatomic locations as has been done in other pan-cancer analysis.(5,44). As with gene expression data, we confirmed highly concordant miRNA-based expression patterns across multiple data sets including lung squamous cell carcinoma, cervical cancer, and bladder cancer (Fig. 1B, Supplemental Fig. S3). The reproducibility of the gene signature suggests that the biological processes underlying various carcinomas may all be influenced by a robust set of miRNAs.
FIGURE 1:
MicroRNAs identify two HNSCC subtypes related to stages of epithelial differentiation. (A) Consensus clustering of 50 microRNAs with high integrative correlations between the 88 UNC samples (left) and the 474 TCGA samples (right) with HNSCC revealed two HNSCC subtypes (also see Supplementary Fig. 1). Columns represent individual samples and are labeled by class. Rows represent mature miRNA strands and are ordered by the discovery cohort hierarchical clustering. The epithelial-stromal developmental annotations of the two miRNA clusters are shown along with the miRNAs contained within each cluster. MicroRNAs downregulated in the ‘epithelial’ cluster are shown in gray, while those downregulated in the ‘stromal’ cluster are in orange. (B) Independent validation heatmaps show similar expression patterns of the two miRNA clusters and the two miRNA-based subtypes among the TCGA bladder urothelial carcinoma (BLCA), lung squamous cell carcinoma (LUSC), and cervical squamous cell carcinoma (CESC). Rows represent mature miRNA strands and are ordered the same as Fig. 1A. (C) The expression of curated, target predicted, and negatively correlated mRNAs are displayed as a heatmap along with biological function groupings. Columns have the same samples and order as the TCGA miRNA clustering in Fig. 1A.
Pathway analyses and review of existing miRNA-based studies in squamous tumors supported the choice of the ‘epithelial’ and ‘stromal’ class labels (Supplementary Table S3A) (45,46). Half of the miRNAs downregulated in epithelial cluster (miR-150–5p, miR-125b-5p, miR-195–5p, miR-127–5p) were also downregulated in epithelial cancer tissue from colorectal tumors, suggesting that those clusters may represent a well-differentiated, epithelial-like signature across tumors. Additionally, many of the miRNAs downregulated in the stromal cluster were downregulated in stromal cancer tissue, suggesting they may represent a poorly differentiated, stromal-like pan-cancer subtype. Taken together, these patterns suggest an axis of cancer differentiation along a stromal-epithelial spectrum influenced by miRNAs and supporting a role for a miRNA-based classification system in HNSCC.
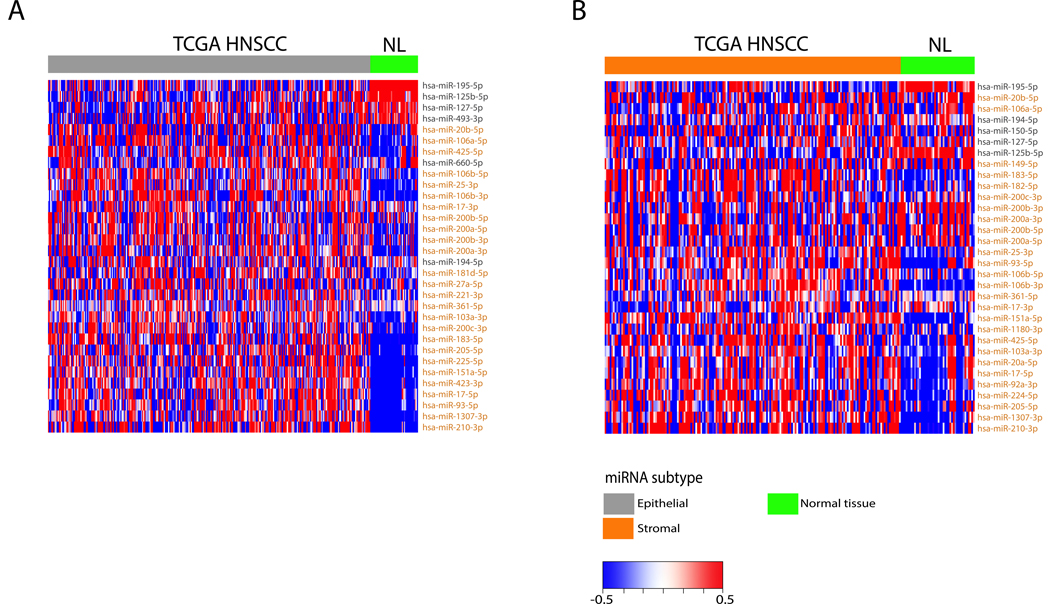
We next turned our attention to the common concern that profiling of bulk tumors might reveal results in which contamination of non-malignant adjacent epithelium may contribute to the observed patterns (Fig. 2). We document significant differentially expressed miRNAs (DEmiRS) as a function of normal versus malignant epithelium, supporting the idea that tumor subtypes are not explained by contaminating normal epithelium. This figure does not explicitly exclude the possibility of contamination by other types of non-malignant cells. In both cases, 32 significant DEmiRs (q value < 0.05) distinguished tumor from normal, with the independent process selecting largely the same genes, documenting a shared tumor-normal set of classifying genes. Many of the miRNAs that are representative of the epithelial subtype (including miRs-195, −127, and −125b) are downregulated in both tumor subtypes when compared to the normal, suggesting that they may play an overall tumor suppressive role in HNSCC. Additionally, while many of the miRNAs representative of the stromal subtype are also downregulated in the normal tissues, there is a clear upregulation of previously characterized oncogenic miRNAs (miR-17/92a cluster, miR-183–5p, miR-106b/25 cluster) in the stromal subtype when compared to the normal (47–50).
FIGURE 2:
Differential expressed miRNAs (DEmiRs) are compared between HNSCC and normal samples. Heatmaps depict miRNA expression of DEmiRs in the TCGA cohort between the epithelial subtype and normal (A) and between the stromal subtype and normal (B). MicroRNAs are color coded based on which class they represent in Figure 1A.
Biological and functional annotation of miRNA-based HNSCC subtypes
A microRNA’s primary function is the regulation of gene expression through hybridization of its seed sequence to a target RNA sequence, resulting in degradation of the resulting double-stranded RNA product. At the present time, most miRNA-mRNA pairs have not been functionally described but instead are derived based on computational approaches. In order to discover the mRNA targets of miRNAs we calculated the correlation of miRNA-seq and mRNA-seq expression in TCGA HNSCC data, detecting 150,408 significant negative correlations representing 15,654 unique mRNAs associated with one or more of the 50 miRNAs selected for inclusion in the clustering analysis (Supplementary Fig. S2A). Negative correlations only were considered as this is the expected direction of miRNA regulation of gene expression. Some miRNAs, such as miR-17–5p and miR-20a-5p, exhibited higher numbers of significant negative correlations with genes, (Supplementary Fig. S2B), suggesting they might serve as core regulators of miRNA-based phenotypes in HNSCC. Recognizing that many genes might be indirect targets of miRNA, we further filtered to a set of 3,500 genes for which there was computational support for a predicted sequence target in at least one transcript from the gene. Additionally, in order to select the genes most relevant to the subtype classification, we compared our 3,500 negatively correlated and target predicted genes to the 2,665 genes that were significantly differentially expressed (q-value < 0.05) between the two subtypes (Supplementary Fig. S2C), which left 91 and 340 mRNAs upregulated in the epithelial and stromal subtypes, respectively (Supplementary Fig. S2D).
In order to further interpret the etiology of reproducible miRNA subtypes, we applied several pathway-based techniques to the lists of genes statistically associated with miRNA-target genes that defined the classes. As predicted, DAVID analysis of the 431 differentially expressed mRNA transcripts detected functional gene ontologies supporting class labels previously defined using miRNAs (Supplementary Table S3B). Overall, miRNAs downregulated in the stromal subtype were associated with upregulation of mRNAs involved in stromal processes derived from mesenchymal cells including muscle, circulatory, skeletal, and neuronal system development (Fig. 1C). By contrast, those miRNAs downregulated in the epithelial subtype were associated with upregulation of mRNAs involved in epithelial cell growth, differentiation development, and other differentiated cell activities such as secretion and cell-to-cell adhesion. This gene expression signature is also relatively reproducible in the other three TCGA cohorts (Supplemental Fig. S4). In either case, we conclude that the strong coordination of miRNA and mRNA expression patterns was parallel biologic validation of informatically derived classes.
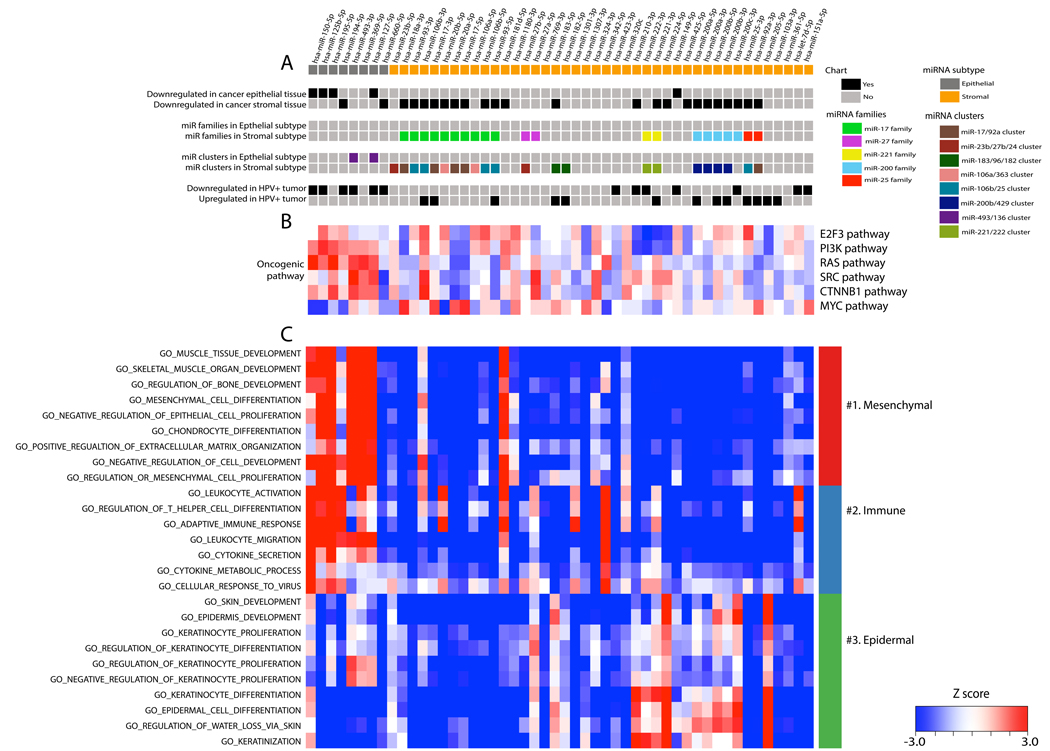
The coordination of miRNA targeting to gene expression is more complex than simple recognition of a seed sequence at the nucleic acid level. MicroRNA families have been recognized in which individual members have related seed sequences, although the miRNAs themselves may be regulated independently at the transcriptional level. By contrast, other miRNA species with non-overlapping seed sequences are regulated in a clustered manner within a single transcript, such as in the introns of a host gene or otherwise co-transcribed in physical proximity along a chromosome. Such intricate co-regulation of miRNA-gene expression suggests an even higher level of cellular pathway coordination (47,51). Additionally, many miRNA clusters and families have been associated with the development of various cancer types (52). Accordingly, we observed enrichment of numerous miRNA families and clusters associated with our newly described tumor subtypes, offering a higher order biologic interpretation to the class assignment (Fig. 3A).
FIGURE 3:
Two miRNA clusters display the distinct phenotypes of epithelial cancers. (A) The upper chart illustrates the association of 50 miRNAs with phenotypes of epithelial cancer, as well as known-miRNA clusters and families based on previously published papers (also see Supplementary Table S3B) and differentially expressed miRNAs between HPV(+) and HPV(–) tumors (also see Supplementary Table S5C). (B) The middle heatmap illustrates the extent of correlation between miRNAs in the TCGA cohort and the primary airway cell oncogenic mRNA signatures of the E2F3, PIK3CA, RAS, SRC, WNT, and MYC pathways. Red and blue represent positive and negative correlations, respectively. (C) The extent of correlation between the 50 microRNAs in the TCGA cohort and a manually curated set of GO categories was calculated and rendered as the lower heatmap. Red and blue represent positive and negative correlation between the corresponding miRNA and genes in the GO category, respectively. The GO categories are colored as groups with related functional annotations.
Whereas the DAVID analysis described above tests the hypothesis that miRNAs differentiating classes belong to differential ontologies or functions, alternative approaches have been developed to assess pathway activation status using the data embedded in expression of groups of genes across different classes. Using methods developed by Bild et. al., we used gene expression data from the cohorts to interrogate oncogenic pathways for differential activation as a function of the miRNA’s selected to define the epithelial and stromal classes (Fig. 3B) (53). In this approach, expression of target genes of each miRNA is aggregated and correlated with expression of the miRNA. In HNSCC we document that the epithelial subtype is associated with activation of the RAS, PI3K, and TNNB1 oncogenic pathways known to be relevant in epithelial tumors (54). By contrast, the miRNAs in the stromal subtype (especially those in the miR-17/92a cluster) were associated strongly with the c-MYC pathway as has been experimentally documented in numerous cases (50). We extended the approach beyond oncogenic signaling to a set of curated GO categories, detecting 4655 as associated with the miRNA network (Supplementary Table S4). A curated and representative subset is shown which supports our classification of tumors as either stromal or epithelial (Fig. 3C). GO categories representative of their clustered correlations are grouped and labeled based on enriched biological functions, specifically with mesenchymal, immune, and epidermal signatures. Interestingly, epithelial miRNAs demonstrated strong negative correlation with genes involved in keratinocyte differentiation and proliferation, while the stromal miRNAs were negatively correlated with genes responsible for mesenchymal cell differentiation and immune response. This analysis demonstrated the extended separation of the various functional roles of the miRNAs in each subtype.
Clinical and prognostic correlations in miRNA-based HNSCC subtypes
Having defined reproducible miRNA subtypes of HNSCC, we investigated their associations with a variety of previously described molecular markers and clinical phenotypes (Fig. 4A, Supplemental Fig. S5, Table 1). There were statistical differences in miRNA subtype as a function of anatomic tumor site, with a higher fraction of oral cavity cancers found in the stromal subtype but more oropharyngeal, hypopharyngeal, and larynx tumors in the epithelial subtype (p <0.001). Similarly, we observed an increased percentage of HPV(+) tumors among the epithelial subtype (p<0.001) (55). Additionally, we observed a significant difference in histological grade, with more well differentiated tumors in the epithelial subtype, and more poorly differentiated in the stromal subtype (p = 0.003); however, the difference in advanced T-staging between the two subtypes was marginal (p = 0.10). There was no significant difference in lymph node status among the subtypes (p=0.20), but among those tumors with TP53 mutations, there were a higher proportion with LN metastasis in the stromal subtype when compared to the epithelial (58.2% vs 48.9%; p = 0.033).
FIGURE 4:
Distinct genetic alterations and clinical parameters characterize HNSCC subtypes in the TCGA cohort. (A) The distribution of clinical parameters and somatic mutations is shown. (B) Kaplan–Meier survival analysis with log-rank p-values for HNSCC patients in each subtype is shown. The survival (left) of patients with epithelial subtype (grey) versus stromal subtype (orange) is distinguished. The stratification of HPV infection status was then applied to the survival analysis with HPV(+) patients (right).
TABLE 1:
Patient demographics and tumor characteristics by cluster assignment in TCGA cohort.
| Characteristics | TCGA (n = 474) | ||
|---|---|---|---|
| Epithelial subtype (n=298) | Stromal subtype (n=176) | P-value | |
| Age, median (range), year | 60 (24–87) | 61 (24–88) | 0.300 |
| Gender | 0.300 | ||
| Male | 224 (75.2) | 124 (70.5) | |
| Female | 74 (24.8) | 52 (29.5) | |
| Smoking status | 0.100 | ||
| Never | 69 (23.2) | 41 (23.3) | |
| Smoker ( < 10 pack-years) | 59 (19.8) | 50 (28.4) | |
| Smoker ( >= 10 pack-years) | 163 (54.7) | 80 (45.5) | |
| Primary site | <0.001 | ||
| Oral cavity | 162 (54.4) | 128 (72.7) | |
| Oropharynx | 58 (19.5) | 13 (7.4) | |
| Larynx | 71 (23.8) | 33 (18.8) | |
| Hypopharynx | 7 (2.3) | 2 (1.1) | |
| Pathologic T stage | 0.010 | ||
| T0 - T2 | 97 (34.9) | 72 (41.4) | |
| T3 – T4 | 151 (54.3) | 96 (55.2) | |
| Tx | 30 (10.8) | 6 (3.4) | |
| Pathologic N stage | 0.200 | ||
| N negative | 106 (38.4) | 52 (29.9) | |
| N positive | 130 (47.1) | 93 (53.4) | |
| Nx | 40 (14.5) | 29 (16.7) | |
| TNM stage | 0.100 | ||
| I -II | 50 (20.6) | 46 (27.7) | |
| III - IV | 193 (79.4) | 120 (72.3) | |
| Histological grade | 0.003 | ||
| Well | 46 (15.6) | 13 (7.4) | |
| Moderately | 167 (56.8) | 105 (59.7) | |
| Poorly | 62 (21.1) | 54 (30.7) | |
| Undifferentiated | 7 (2.4) | 0 | |
| GX | 12 (4.1) | 4 (2.3) | |
| HPV status | <0.001 | ||
| Negative | 245 (82.2) | 165 (93.8) | |
| Positive | 53 (17.8) | 11 (6.2) | |
We then considered the clinical impact of tumor subtype assignment and observed that overall patients in the stromal subtype had statistically worse outcomes in the univariate analyses (Fig. 4B, log-rank), as did patients with higher T staging and classical gene expression subtype as expected (Supplementary Table S5A, all p <0.05). Patients with oropharynx anatomic site and HPV(+) had improved outcomes (p< 0.05). We further explored the prognostic value of the miRNA-based subtypes above previously reported gene expression subtypes (Supplementary Fig. S6) and found survival differences between the epithelial and stromal subtypes in both the atypical (p = 0.021, log-rank) and basal (p = 0.041, log-rank) mRNA classes. We considered the impact of miRNA subtype on HPV(+) patients in a subset analysis, and demonstrated that HPV(+) stromal patients had far inferior outcomes with median survival of just over 3 years, compared to nearly 90% at that same time point in epithelial patients who were HPV(+), although the difference was not significant due to small numbers with sufficient follow up (Fig. 4B). Quite unexpectedly, in the multivariate Cox proportional hazards model, only miRNA class and T stage remained statistically significant predictors of outcome, as epithelial subtype assumed the favorable prognosis of both anatomic site and HPV status (Supplementary Table S5A) (HR = 1.5646, 95% CI = 1.14–2.15, p = 0.006). For completeness, we also investigated the impact of individual miRNAs on survival (Supplementary Table S5B), and four miRNAs (miRs-493, −369, −127, −103a) were significantly associated with poorer prognosis, while five miRNAs (miRs-150, −125b, −27b, −200a, −342) had a significant positive influence on survival. The UNC cohort is considerably smaller and more heterogeneous than TCGA, including recurrent and metastatic tumors, as well as more oropharynx tumors. In univariate analysis there was no effect of miRNA subtype although the confidence interval was wide and included the effect seen in the TCGA dataset (HR = 1.001, 95% CI = 0.35–2.81). The small cohort size did not support multivariate analysis.
Correlation between miRNA-based subtypes and HPV infection
We observed a strong statistical association between epithelial subtype and both oropharyngeal anatomic site, and HPV(+) status in tumors. To measure the extent to which these factors included shared miRNA signatures, we first determined those miRNAs associated with HPV status using a supervised approach. At a significance level of q-values < 0.01, 26 miRNAs were differentially expressed among our 50 miRNAs used in the study (Supplementary Table S5C). Thirteen miRNAs were upregulated and downregulated, respectively, in the HPV(+) tumors compared to HPV(−), and many of them have been reported as differentially expressed in HPV(+) cancers, documenting our results agree with previous findings (15,17,18). Six of the eight miRNAs (miRs-150, −194, −660, −127, −125b, and −493) downregulated in the epithelial subtype were also downregulated in the HPV(+) tumors. Interestingly, these same miRNAs were expressed at low levels in HPV(−) patients of the epithelial subtype compared to the stromal. Likewise, HPV(+) patients in the stromal class did not appear to have the same low levels of these miRNA. This finding suggests a molecular signature independent of HPV status that portends a favorable prognosis.
Genomic alterations in miRNA-based HNSCC subtypes
Using the expression data described above (Fig. 2), we documented that the expression patterns in miRNA subtypes differ from normal controls such that normal contamination alone is unlikely to be the etiology of the expression subtypes. Whole tumor analysis, as opposed to single cell approaches, does not exclude the possibility that other infiltrating cell types other than normal epithelium might explain some or all of the signatures as opposed to those transcripts inherent to the tumor. In fact, a recent report suggests that after computational removal of gene expression signatures associated with inflammatory cells rather than normal epithelium, the gene expression signature of the basal and mesenchymal subtypes of HNSCC were indistinguishable (7). We therefore considered the possibility that the stromal subtype of miRNA, which is highly correlated with the mesenchymal gene expression subtype, might be explained by contamination of normal cells other than epithelium.
We considered several measures of purity other than the gene expression signatures discussed above. Primarily, mutation assessment can be instructive both for assessment of tumor purity and pathophysiology. First, it is important to note that canonical TP53 mutation was found more often in stromal tumors than epithelial tumors, 77.8% versus 68.5% respective (p = 0.04), a fact which is inconsistent with stomal tumors being more infiltrated with non-malignant cells. Infiltrations of normal or inflammatory cells would make mutation detection more difficult and one would see a lower, not higher rate of detection of canonical mutations. By extension we considered the allele frequency of TP53 mutations in stromal versus epithelial subtypes, observing 32.4% and 31.9% respectively, suggesting overall similar tumor purity as has been previously described (56). We further considered whether, in addition to TP53 frequency, there were other mutational differences across the subtypes, and we observed that the miRNA-based HNSCC subtype demonstrated a distinct pattern of somatic mutations (Fig. 5A and Supplementary Table S6). In addition to TP53 (p = 0.041), NSD1 (p < 0.001), HRAS (p = 0.018) and ZNF521 (p = 0.003) were significantly differentially mutated between the subtypes. The epithelial subtype harbored a majority of the NSD1 mutations (82.5% of the 57 total mutations), and it contained a higher proportion of tumors with NSD1 mutations compared to the stromal subtype (16.4% vs 5.8%). Conversely, the stromal subtype displayed lower rate of both HRAS (2.9% vs 8.5%) and ZNF521 (0.5% vs 6.3%) mutations.
FIGURE 5:
Distinct genetic alterations characterize HNSCC subtypes in the TCGA cohort. (A) The distribution of somatic mutations is shown for 13 frequently mutated genes across two HNSCC subtypes (blue bars). Each column contains data from a single tumor (n = 457). (B) Genome-wide copy number alterations for HNSCC across 2 subtypes are illustrated in a heatmap using the GISTIC algorithm. Each column contains data from a single tumor (n = 470), and red and blue represent copy number gains and losses, respectively.
In parallel to mutational analysis, we considered a qualitative estimate of purity as defined by assessment of copy number alterations in the genome (Fig. 5B). Both stromal and epithelial tumors demonstrated similar intensities and distribution of chromosomal gains (3q, 5p, 8q) and losses (3p and 8p) previously characterized in HNSCC (Fig. 5B) (5). We conclude that miRNA subtypes are unlikely to be primarily defined by either normal stromal contamination or inflammatory infiltrates, but more likely to be the result of intrinsic tumor factors such as mutations, stage of differentiation, and other features directing heterogenous states of gene and miRNA expression.
DISCUSSION
The current manuscript validates two statistically significant, reproducible, and clinically relevant miRNA subtypes of HNSCC based on a previously unreported dataset of miRNA expression paired with an extended characterization of TCGA miRNA sequencing data. We document that two subtypes is the largest number that can be reproducibly validated using methods that our group has previously executed in similar unsupervised reports (4,42,43). A growing body of evidence reveals that miRNAs are aberrantly expressed in many human cancers where they critically contribute to initiation, growth, metastasis, and prognosis(15–20,47–50,57). Previous studies of miRNA profiling in HNSCC have focused on comparing normal tissues to tumor samples. Here, we focus on miRNAs that define the heterogeneity of cancer rather than the differences between tumor and normal. Our results indicate the miRNA-based profiles can distinguish between epithelial and stromal subtypes in tumors, suggesting that the classification in cancer may mirror processes relevant in normal physiology although coopted by malignant transformation. While we acknowledge that this observation likely represents only a small component of the molecular heterogeneity of this class of tumors, we nonetheless recognize the importance of previously unrecognized reproducible patterns in cancer biology. Such patterns may offer new clues into the origins of cancer or potential treatments.
The patterns detected in HNSCC are broadly reproducible across a spectrum of related epithelial cancers and reflect an axis spanning two fundamental processes in both normal and malignant pathophysiology: epithelial cell differentiation and stromal infiltration. In selecting TCGA cohorts to interrogate for similar miRNA expression profiles, we considered both tumors with a known squamous morphology and/or evidence of HPV as an etiology. Interestingly, the evidence for miRNA subtypes was strongest in the tobacco-associated malignancies (BLCA and LUSC) and weaker in the HPV-associated cohort (CESC) (Fig. 1B, Supplemental Fig. S3). Additionally, the miRNA subtypes are highly correlated with gene expression subtypes, as might be expected given that gene expression subtypes define samples with highly coordinated gene expression patterns (Supplementary Fig. S4). We document through gene network analysis that predicted and validated miRNA-RNA pairs are co-regulated at the individual gene level, as well as in programs of miRNA families and clusters to regulate broad categories of coordinated gene expression. For example, we find that the miR-200 family, represented by miRs-200a-5p/−3p, −200b-5p/−3p, and −200c-3p in our data, along with miR-205–5p and miR-27b-5p, serves as a potent regulator of epithelial versus stromal plasticity in HNSCC. Other studies have shown that the miR-200 family and miR-205 regulate epithelium differentiation, and reduction or loss of these miRNAs, which downregulate E-cadherin specifically targeting ZEB1 and ZEB2, results in a more mesenchymal-like, highly mobile and aggressive phenotype of cancer cells, often described as an epithelial-mesenchymal transition (EMT) (57).
In addition, the epithelial subtype appears to subsume at least part of the favorable prognosis associated with HPV infection seen in clinical outcomes. The epithelial miRNA signature associated with favorable outcomes in HNSCC is highly concordant with the signature associated with HPV infection, suggesting these subtype-specific miRNAs regulate similar biological processes involved in the pathogenesis and prognosis of HPV-infected tumors of epithelial cells. HPV infection has been previously associated with expression of certain miRNAs in HNSCC and cervical cancers, and miRNAs can modulate the expression of HPV genes and vice versa (58–60). The difference in prognosis may be explained by multiple mechanisms, such as increased EMT, cell motility/proliferation, and angiogenesis in the stromal subtype. Our observation was strongly consistent to that of the study by Jung et al (61), in which a poor prognosis subtype of HNSCC was characterized by alterations of pathways involved in cell-cell adhesion, extracellular matrix (ECM), EMT, immune response, and apoptosis. While the current study is a retrospective tumor classification study that produced a list of miRNAs associated with clinical outcomes, future clinical studies could validate the centroid predictor of this list as a biomarker and further elucidate the role of miRNAs in HNSCC prognosis (Supplementary Table S7).
We have documented in the current analysis that miRNA subtypes are statistically significant in multiple independent cohorts, reproducible in terms of a defined signature, and associated with interesting phenotypes such as patient survival or mutation patterns. These three criteria are the minimum for suggesting that such patterns might warrant further investigation in terms of their implications for tumorigenesis, tumor progression, and possibly therapeutic intervention. Our work suggests in Figure 2 that contaminating normal epithelium is an unlikely explanation for the reproducible signatures seen. Additionally, the finding of similar rates of TP53 mutant allele fraction suggests that other reproducible, yet potentially biologically uninteresting phenomena such as tumor necrosis are unlikely explanations for the signatures seen. It is possible that tumor subtypes in bulk tumor analysis such as we have done could be explained by differing proportions of non-malignant infiltrating cells such as inflammatory or stromal cells. However, such an explanation is not supported by the finding of differential proportions of known driver mutations such as NSD1 and HRAS, not known to be present in such stromal cells. Thus, we conclude that the most likely interpretation is differences in underlying tumor pathophysiology, although other options have not been fully excluded.
In conclusion, our findings indicate that specific miRNAs influence HNSCC subtypes through their ability to regulate developmental growth and differentiation programs in epithelial cells. These differential patterns of miRNA regulation associate with clinical factors and canon molecular signatures previously described in HNSCC and can influence patient prognosis. Ultimately, these results delineate developmental miRNA expression signatures that characterize and yield the phenotypic heterogeneity of cancer subtypes, thus providing an expanded framework for understanding the pathogenesis and treatment of these malignancies. Discerning miRNA-based subtypes clinically will allow for the stratification of patients beyond anatomy and cytopathology, thus opening the door for personalized treatments to improve patient outcomes.
Supplementary Material
Translational relevance.
Highly coordinated, dysregulated expression of microRNAs (miRNA) has been implicated in the development of various cancers, including head and neck squamous cell carcinoma (HNSCC). Outside of HPV infection, molecular classification of HNSCC is lacking, and patient stratification by anatomical location and stage does not account for the observed heterogeneity in clinical outcomes. Here, we analyzed miRNA expression in two independent HNSCC patient cohorts using two confirmatory wet lab platforms, miRNA microarray and miRNA sequencing, respectively. Unsupervised class discovery identified two distinct miRNA-based phenotypes, termed ‘epithelial’ and ‘stromal’, that provide an expanded framework regarding the pathogenesis and heterogeneity of these tumors and help to better classify them in the clinical setting. Further analysis of these subtypes revealed distinct biological and clinical features of HNSCC tumors based on the miRNA expression profiles, which suggests they may help as biomarkers to aid decisions regarding therapeutic strategies and improve patient outcomes.
ACKNOWLEDGEMENTS
Research reported in this publication was supported by the National Cancer Institute under award number U10CA181009, CA211939, CA210988, UG1CA233333.
Footnotes
The authors declare no potential conflicts of interest.
REFERENCES
- 1.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer 2010;127(12):2893–917 doi 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 2.Suh Y, Amelio I, Guerrero Urbano T, Tavassoli M. Clinical update on cancer: molecular oncology of head and neck cancer. Cell Death Dis 2014;5:e1018 doi 10.1038/cddis.2013.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ang KK, Chen A, Curran WJ Jr., Garden AS, Harari PM, Murphy BA, et al. Head and neck carcinoma in the United States: first comprehensive report of the Longitudinal Oncology Registry of Head and Neck Carcinoma (LORHAN). Cancer 2012;118(23):5783–92 doi 10.1002/cncr.27609. [DOI] [PubMed] [Google Scholar]
- 4.Walter V, Yin X, Wilkerson MD, Cabanski CR, Zhao N, Du Y, et al. Molecular subtypes in head and neck cancer exhibit distinct patterns of chromosomal gain and loss of canonical cancer genes. PLoS One 2013;8(2):e56823 doi 10.1371/journal.pone.0056823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Network TCGA. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 2015;517(7536):576–82 doi 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Keck MK, Zuo Z, Khattri A, Stricker TP, Brown CD, Imanguli M, et al. Integrative analysis of head and neck cancer identifies two biologically distinct HPV and three non-HPV subtypes. Clin Cancer Res 2015;21(4):870–81 doi 10.1158/1078-0432.Ccr-14-2481. [DOI] [PubMed] [Google Scholar]
- 7.Puram SV, Tirosh I, Parikh AS, Patel AP, Yizhak K, Gillespie S, et al. Single-Cell Transcriptomic Analysis of Primary and Metastatic Tumor Ecosystems in Head and Neck Cancer. Cell 2017;171(7):1611–24.e24 doi 10.1016/j.cell.2017.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004;116(2):281–97 doi 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 9.Fang XN, Yin M, Li H, Liang C, Xu C, Yang GW, et al. Comprehensive analysis of competitive endogenous RNAs network associated with head and neck squamous cell carcinoma. Sci Rep 2018;8(1):10544 doi 10.1038/s41598-018-28957-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qi X, Zhang DH, Wu N, Xiao JH, Wang X, Ma W. ceRNA in cancer: possible functions and clinical implications. J Med Genet 2015;52(10):710–8 doi 10.1136/jmedgenet-2015-103334. [DOI] [PubMed] [Google Scholar]
- 11.Azzalini E, De Martino E, Fattorini P, Canzonieri V, Stanta G, Bonin S. Reliability of miRNA Analysis from Fixed and Paraffin-Embedded Tissues. Int J Mol Sci 2019;20(19) doi 10.3390/ijms20194819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haile S, Pandoh P, McDonald H, Corbett RD, Tsao P, Kirk H, et al. Automated high throughput nucleic acid purification from formalin-fixed paraffin-embedded tissue samples for next generation sequence analysis. PLoS One 2017;12(6):e0178706 doi 10.1371/journal.pone.0178706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kakimoto Y, Tanaka M, Kamiguchi H, Ochiai E, Osawa M. MicroRNA Stability in FFPE Tissue Samples: Dependence on GC Content. PLoS One 2016;11(9):e0163125 doi 10.1371/journal.pone.0163125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.El-Mogy M, Lam B, Haj-Ahmad TA, McGowan S, Yu D, Nosal L, et al. Diversity and signature of small RNA in different bodily fluids using next generation sequencing. BMC Genomics 2018;19(1):408 doi 10.1186/s12864-018-4785-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cao P, Zhou L, Zhang J, Zheng F, Wang H, Ma D, et al. Comprehensive expression profiling of microRNAs in laryngeal squamous cell carcinoma. Head Neck 2013;35(5):720–8 doi 10.1002/hed.23011. [DOI] [PubMed] [Google Scholar]
- 16.Hui AB, Lenarduzzi M, Krushel T, Waldron L, Pintilie M, Shi W, et al. Comprehensive MicroRNA profiling for head and neck squamous cell carcinomas. Clin Cancer Res 2010;16(4):1129–39 doi 10.1158/1078-0432.Ccr-09-2166. [DOI] [PubMed] [Google Scholar]
- 17.Lajer CB, Garnaes E, Friis-Hansen L, Norrild B, Therkildsen MH, Glud M, et al. The role of miRNAs in human papilloma virus (HPV)-associated cancers: bridging between HPV-related head and neck cancer and cervical cancer. Br J Cancer 2012;106(9):1526–34 doi 10.1038/bjc.2012.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lajer CB, Nielsen FC, Friis-Hansen L, Norrild B, Borup R, Garnaes E, et al. Different miRNA signatures of oral and pharyngeal squamous cell carcinomas: a prospective translational study. Br J Cancer 2011;104(5):830–40 doi 10.1038/bjc.2011.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nurul-Syakima AM, Yoke-Kqueen C, Sabariah AR, Shiran MS, Singh A, Learn-Han L. Differential microRNA expression and identification of putative miRNA targets and pathways in head and neck cancers. Int J Mol Med 2011;28(3):327–36 doi 10.3892/ijmm.2011.714. [DOI] [PubMed] [Google Scholar]
- 20.Ramdas L, Giri U, Ashorn CL, Coombes KR, El-Naggar A, Ang KK, et al. miRNA expression profiles in head and neck squamous cell carcinoma and adjacent normal tissue. Head Neck 2009;31(5):642–54 doi 10.1002/hed.21017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koshizuka K, Hanazawa T, Fukumoto I, Kikkawa N, Okamoto Y, Seki N. The microRNA signatures: aberrantly expressed microRNAs in head and neck squamous cell carcinoma. J Hum Genet 2017;62(1):3–13 doi 10.1038/jhg.2016.105. [DOI] [PubMed] [Google Scholar]
- 22.Gautier L, Cope L, Bolstad BM, Irizarry RA. affy—analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 2004;20(3):307–15 doi 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 23.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 2002;30(1):207–10 doi 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Broad-Institute-TCGA-Genome-Data-Analysis-Center. Analysis-ready standardized TCGA data from Broad GDAC Firehose 2016_01_28 run. Broad Institute of MIT and Harvard; 2016. doi 10.7908/C11G0KM9. [DOI] [Google Scholar]
- 25.Van Doorslaer K, Li Z, Xirasagar S, Maes P, Kaminsky D, Liou D, et al. The Papillomavirus Episteme: a major update to the papillomavirus sequence database. Nucleic Acids Research 2016;45(D1):D499–D506 doi 10.1093/nar/gkw879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parmigiani G, Garrett-Mayer ES, Anbazhagan R, Gabrielson E. A cross-study comparison of gene expression studies for the molecular classification of lung cancer. Clin Cancer Res 2004;10(9):2922–7 doi 10.1158/1078-0432.ccr-03-0490. [DOI] [PubMed] [Google Scholar]
- 27.Wilkerson MD, Hayes DN. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics 2010;26(12):1572–3 doi 10.1093/bioinformatics/btq170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kimes PK, Cabanski CR, Wilkerson MD, Zhao N, Johnson AR, Perou CM, et al. SigFuge: single gene clustering of RNA-seq reveals differential isoform usage among cancer samples. Nucleic Acids Res 2014;42(14):e113 doi 10.1093/nar/gku521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fu J, Tang W, Du P, Wang G, Chen W, Li J, et al. Identifying microRNA-mRNA regulatory network in colorectal cancer by a combination of expression profile and bioinformatics analysis. BMC Syst Biol 2012;6:68 doi 10.1186/1752-0509-6-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife 2015;4 doi 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sticht C, De La Torre C, Parveen A, Gretz N. miRWalk: An online resource for prediction of microRNA binding sites. PLoS One 2018;13(10):e0206239 doi 10.1371/journal.pone.0206239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chou CH, Shrestha S, Yang CD, Chang NW, Lin YL, Liao KW, et al. miRTarBase update 2018: a resource for experimentally validated microRNA-target interactions. Nucleic Acids Res 2018;46(D1):D296–d302 doi 10.1093/nar/gkx1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen Y, Wang X. miRDB: an online database for prediction of functional microRNA targets. Nucleic Acids Res 2020;48(D1):D127–d31 doi 10.1093/nar/gkz757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences 2005;102(43):15545–50 doi 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liberzon A, Subramanian A, Pinchback R, Thorvaldsdóttir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics 2011;27(12):1739–40 doi 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim TM, Huang W, Park R, Park PJ, Johnson MD. A developmental taxonomy of glioblastoma defined and maintained by MicroRNAs. Cancer Res 2011;71(9):3387–99 doi 10.1158/0008-5472.Can-10-4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tate JG, Bamford S, Jubb HC, Sondka Z, Beare DM, Bindal N, et al. COSMIC: the Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Research 2018;47(D1):D941–D7 doi 10.1093/nar/gky1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol 2011;12(4):R41 doi 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.R-Core-Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: URL https://wwwR-projectorg/ 2019. [Google Scholar]
- 40.Therneau TM. A Package for Survival Analysis in S. version 2.38, https://CRAN.R-project.org/package=survival. 2015.
- 41.Kassambara A, Kosinski M, Biecek P. survminer: Drawing Survival Curves using ‘ggplot2’. R package version 0.4.6. https://CRAN.R-project.org/package=survminer. 2019. [Google Scholar]
- 42.Hayes DN, Monti S, Parmigiani G, Gilks CB, Naoki K, Bhattacharjee A, et al. Gene expression profiling reveals reproducible human lung adenocarcinoma subtypes in multiple independent patient cohorts. J Clin Oncol 2006;24(31):5079–90 doi 10.1200/jco.2005.05.1748. [DOI] [PubMed] [Google Scholar]
- 43.Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 2010;17(1):98–110 doi 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Network TCGA. Comprehensive genomic characterization of squamous cell lung cancers. Nature 2012;489(7417):519–25 doi 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nishida N, Nagahara M, Sato T, Mimori K, Sudo T, Tanaka F, et al. Microarray analysis of colorectal cancer stromal tissue reveals upregulation of two oncogenic miRNA clusters. Clin Cancer Res 2012;18(11):3054–70 doi 10.1158/1078-0432.Ccr-11-1078. [DOI] [PubMed] [Google Scholar]
- 46.Della Vittoria Scarpati G, Calura E, Di Marino M, Romualdi C, Beltrame L, Malapelle U, et al. Analysis of differential miRNA expression in primary tumor and stroma of colorectal cancer patients. Biomed Res Int 2014;2014:840921 doi 10.1155/2014/840921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guo L, Yang S, Zhao Y, Zhang H, Wu Q, Chen F. Global analysis of miRNA gene clusters and gene families reveals dynamic and coordinated expression. BioMed research international 2014;2014:782490 doi 10.1155/2014/782490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ma Y, Liang AJ, Fan Y-P, Huang Y-R, Zhao X-M, Sun Y, et al. Dysregulation and functional roles of miR-183–96-182 cluster in cancer cell proliferation, invasion and metastasis. Oncotarget 2016;7(27):42805–25 doi 10.18632/oncotarget.8715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mogilyansky E, Rigoutsos I. The miR-17/92 cluster: a comprehensive update on its genomics, genetics, functions and increasingly important and numerous roles in health and disease. Cell Death And Differentiation 2013;20:1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van Haaften G, Agami R. Tumorigenicity of the miR-17–92 cluster distilled. Genes Dev 2010;24(1):1–4 doi 10.1101/gad.1887110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu J, Wong C. A computational screen for mouse signaling pathways targeted by microRNA clusters. RNA 2008;14(7):1276–83 doi 10.1261/rna.997708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tan W, Liu B, Qu S, Liang G, Luo W, Gong C. MicroRNAs and cancer: Key paradigms in molecular therapy. Oncol Lett 2018;15(3):2735–42 doi 10.3892/ol.2017.7638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bild AH, Yao G, Chang JT, Wang Q, Potti A, Chasse D, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature 2006;439(7074):353–7 doi 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- 54.Castellano E, Downward J. RAS Interaction with PI3K: More Than Just Another Effector Pathway. Genes Cancer 2011;2(3):261–74 doi 10.1177/1947601911408079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Elrefaey S, Massaro MA, Chiocca S, Chiesa F, Ansarin M. HPV in oropharyngeal cancer: the basics to know in clinical practice. Acta Otorhinolaryngol Ital 2014;34(5):299–309. [PMC free article] [PubMed] [Google Scholar]
- 56.Patel NM, Jo H, Eberhard DA, Yin X, Hayward MC, Stein MK, et al. Improved Tumor Purity Metrics in Next-generation Sequencing for Clinical Practice: The Integrated Interpretation of Neoplastic Cellularity and Sequencing Results (IINCaSe) Approach. Appl Immunohistochem Mol Morphol 2019;27(10):764–72 doi 10.1097/pai.0000000000000684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol 2008;10(5):593–601 doi 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 58.Greco D, Kivi N, Qian K, Leivonen SK, Auvinen P, Auvinen E. Human papillomavirus 16 E5 modulates the expression of host microRNAs. PLoS One 2011;6(7):e21646 doi 10.1371/journal.pone.0021646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Honegger A, Schilling D, Bastian S, Sponagel J, Kuryshev V, Sultmann H, et al. Dependence of intracellular and exosomal microRNAs on viral E6/E7 oncogene expression in HPV-positive tumor cells. PLoS Pathog 2015;11(3):e1004712 doi 10.1371/journal.ppat.1004712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jimenez-Wences H, Peralta-Zaragoza O, Fernandez-Tilapa G. Human papilloma virus, DNA methylation and microRNA expression in cervical cancer (Review). Oncol Rep 2014;31(6):2467–76 doi 10.3892/or.2014.3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jung AC, Job S, Ledrappier S, Macabre C, Abecassis J, de Reynies A, et al. A poor prognosis subtype of HNSCC is consistently observed across methylome, transcriptome, and miRNome analysis. Clin Cancer Res 2013;19(15):4174–84 doi 10.1158/1078-0432.Ccr-12-3690. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.