Abstract
Glioblastoma (GBM) is the most prevalent and lethal adult primary central nervous system cancer. An immunosuppresive and highly heterogeneous tumor microenvironment, restricted delivery of chemotherapy or immunotherapy through the blood-brain barrier (BBB), together with the brain’s unique biochemical and anatomical features result in its universal recurrence and poor prognosis. As conventional models fail to predict therapeutic efficacy in GBM, in vitro 3D models of GBM and BBB leveraging patient- or healthy individual-derived cells and biomaterials through 3D bioprinting technologies potentially mimic essential physiological and pathological features of GBM and BBB. 3D bioprinted constructs enable investigation of cellular and cell-extracellular matrix (ECM) interactions in a species-matched, high-throughput, and reproducible manner, serving as screening or drug delivery platforms. This review provides an overview of current 3D-bioprinted GBM and BBB models and elaborates on the microenvironmental compositions of GBM and BBB, relevant biomaterials to mimic the native tissues, and bioprinting strategies to implement the model fabrication. Collectively, 3D-bioprinted GBM and BBB models are promising systems and biomimetic alternatives to traditional models for more reliable mechanistic studies and pre-clinical drug screenings that may eventually accelerate the drug development process for GBM.
Keywords: biomaterial, 3D bioprinting, 3D model, glioblastoma, blood-brain barrier
1. INTRODUCTION
Glioblastoma (GBM) is the most common and aggressive adult primary brain cancer, accounting for 14.6% of all malignant central nervous system (CNS) tumors.[1] The five-year relative survival is 6.8% for patients in the United States, ranking lowest among all primary malignant CNS tumors.[1] Despite tremendous efforts in the past decades, little advance in the outcome for patients afflicted with GBM has been achieved. Standard-of-care GBM treatment involves maximal safe surgical resection, followed by concurrent chemoradiation with the oral methylator, temozolomide (TMZ), and then adjuvant TMZ. Complete surgical removal using hemicraniectomies was previously attempted, but failed to achieve cure due to the diffuse invasion of tumor cells into the brain and the necessity to preserve essential brain function. GBM cells invade into the brain parenchyma in different modes, including as single cells, and act as reservoirs for recurrence. Extensive molecular profiling of GBM has identified distinct transcriptional subtypes that reflect heterogenous tumor genetics and epigenetics. Complex cellular and cell-matrix interactions among tumor cells, stromal cells, and the extracellular matrix (ECM) within the TME result in a dynamic and immunosuppresive GBM tumor ecosystem highly resistant to existing treatments. Universal relapse, high intratumoral and intertumoral heterogeneity, and resistance of recurrent GBM to therapies lead to poor prognoses and a dismal median survival time of patients less than 70 years old of 14.6 months.[2] Delivery of therapeutic agents to GBM tumor sites is especially challenging compared to other solid tumors due to the restricted drug and cellular transport across the unique vascular barrier of the brain, the blood-brain barrier (BBB). The BBB serves as a barrier between the circulating blood and the brain parenchyma to prevent entry of blood-borne pathogens or toxic substances into CNS and to maintain CNS homeostasis.[3] The BBB excludes over 98% of small molecule drugs and tightly regulates lymphocyte extravasation, limiting accumulation of chemotherapies and effector T-cells in the GBM tissue.[4] Regulation of the BBB or circumvention of the barriers facilitates some brain tumor therapies, suggesting that the presence of a functional BBB may be essential to accurately evaluate GBM treatments.[5–7] Growing interest in repurposing FDA-approved cancer drugs with enhanced BBB penetration for GBM treatments also demonstrates the potential role of BBB in GBM therapeutic efficacy.[7] The bottleneck in current GBM therapeutic development indicates limitations of current modeling modalities and supports development of more reliable model systems to help elucidate the pathways involved in different subtypes and provide more informative preclinical drug evaluations that will accelerate the drug development process.
GBM modeling requires the recapitulation of not only the dynamic, multi-component TME, but also the brain’s unique anatomical and biochemical features that play critical roles in GBM pathogenesis and treatment response. Traditional modeling modalities have limited capacity to reconstruct important aspects of the GBM, such as relevant tumor-stromal interactions and TME heterogeneity, or to reliably evaluate novel therapies due to the absence of BBB barriers and other features of the brain related to tumor development, drug penetration, and treatment efficacy. Patient-derived xenografts (PDXs) retain many transcriptomic and genomic signatures of the donor tumors and provide ECM-rich microenvironments conducive to cell growth.[8] However, generating PDX requires the use of immunodeficient animals, which prevents investigation of relevant immune responses, such as the interactions between GBM cells and tumor-associated microglia and macrophages (TAMs). TAMs account for about one-third of the tumor mass in recurrent GBM and modulate various cancer activities such as tumor cell migration, invasion, and drug resistance.[8,9] Development of PDXs is also time-consuming and relatively low throughput, requiring a timespan not ideal for diseases like GBM that have fast progression. In vitro models that recapitulate native tumor-stromal interactions and cell-ECM interactions of GBM in a reproducible, efficient, and high-throughput manner may serve as better alternatives to in vivo models. 2D cell cultures are the most common and accessible in vitro modeling methods, but they lack the proper dimensionality and the cell-ECM interactions critical to GBM development. 2D culture conditions also induce irreversible alterations to gene expression, cell morphologies, and cellular activities of the cultured cells, reducing their similarity to primary tumors.[8] Transwell systems have been utilized to explore the cellular interactions in tumor development, cellular dependencies, and BBB maintenance and breakdown.[10–14] However, fixed pore sizes of the transwell membrane cannot recapitulate the dynamic changes of BBB’s tight junctions, and cells cultured in transwells experience a “phenotypical shift” due to the 2D culture condition and the lack of proper interactions with ECM.[11,15,16] Organoids are 3D in vitro models with improved biomimicry compared to other in vitro culture methods. GBM organoids better maintain the cellular heterogeneity and the gene expression of primary tumors, and the tumor cells within organoids display enhanced hypoxic state and stemness compared to their counterparts 2D cultures.[17,18] Organoid fabrication protocols have been developed, but the variability among organoids and the limited control of cellular organization within organoids due to the self-assembly process limit their broader applications.[15,17,18] Traditional in vitro modeling methods are still limited in terms of recapitulating the highly heterogeneous GBM microenvironment or physiologically relevant BBB barrier properties in a reproducible and scalable fashion.
Advanced biofabrication technologies can produce customized 3D tissue models with good flexibility, reproducibility, and scalability, addressing many limitations of other modeling modalities. Biofabrication technologies can be categorized based on whether cellular components are seeded onto constructs after device fabrication or encapsulated in the biomaterials during the fabrication process. The cell-encapsulating approach enables better control of the number and the position of deposited cells and molecules than the cell-seeding approach, resulting in better reproducibility.[19,20] Cells encapsulated in hydrogels encounter ECM cues from all directions, resembling their physiologic states, while seeded cells receive ECM cues mainly from the side in contact with hydrogels. Many technologies are capable of fabricating acellular scaffolds or devices with high resolution and throughput, such as electrospinning, fused deposition modelling, and selective laser sintering, among others, but are not commonly used for cell encapsulation purposes.[21] 3D Bioprinting has emerged to advance the field of cancer and tissue modeling due to its ability to encapsulate cells in biomaterials with good viability and to precisely control tissue architecture and matrix properties.[19,22,23] 3D bioprinting enables creation of reproducible and personalized models, making it especially suitable for modeling diseases like GBM that have high intratumoral and interpatient heterogeneity.[24] Applications of bioprinting technology are not limited to living tissues, but also acellular scaffolds, microfluidic devices, and implantable constructs. Dynamic, microfluidic BBB models providing the shear stress critical to barrier functions through laminar flow have also been developed using bioprinting.[25]
Here we review recent progress in the design and fabrication of 3D-bioprinted GBM and BBB models. We first provide a detailed analysis of the microenvironment composition in GBM and BBB, focusing on cellular and ECM components and properties. We next introduce two important tools for implementing the perceived models: (1) 3D bioprinting strategies that have been utilized for biological applications, with an overview of their mechanisms, advantages, limitations, and applications, and (2) relevant biomaterials and their derivatives. We then review current studies using 3D bioprinting and biomaterials to construct GBM and/or BBB models that have demonstrated physiologically relevant properties and improved features compared to traditional models. Finally, we discuss the challenges and future perspectives of GBM drug development, current 3D-bioprinted GBM and BBB models and their future directions, and benchmarks for 3D in vitro models.
2. GBM and BBB microenvironments
Hierarchical information of native tissues is provided in this section: the building block of native tissues, i.e. the cellular and ECM components; the assembly and organization of the basic building blocks; and the collective biophysical or biochemical properties of the microenvironment from assembly.
2.1. Cellular composition and function
Cellular composition, function, and interactions with other cells in the GBM and BBB microenvironment have been extensively studied and reviewed.[26–28] Here, we provide a brief overview to introduce the essential cellular components and their roles.
2.1.1. Cellular components of GBM
The GBM TME consists of heterogeneous cell populations (Figure 1). Major non-neoplatic stromal cells within the GBM TME include TAMs, microglia, astrocytes, neurons, mesenchymal stem cells (MSCs), and perivascular cells. In the necrotic region of GBM, up to 30–50% of the tumor mass is composed of TAMs with an M2 pro-tumor phenotype.[29] The M2 phenotype is anti-inflammatory, creating an immunosuppressive TME that promotes tumor growth. Macrophages derived from circulating monocytes are recruited to the GBM site due to compromised BBBs and perturbations in brain homeostasis, whereas microglia are CNS resident immune cells that become activated in response to tumor-derived cues. These immune components promote tumor invasiveness through upregulation of matrix metalloproteases (MMPs), such as MMP-2 and MMP-9.[30] Astrocytes can be recruited and activated by tumor cells through multiple modes of communication, including extracellular vesicles and efflux transporters. Tumor-associated, reactive astrocytes promote the invasion of CD133-positive glioblastoma stem cells (GSCs) and secrete anti-inflammatory cytokines, such as TGFβ, that suppress anti-tumor immune respones, resulting in an overall immunosuppressive GBM microenvironment.[31,32] Glutamatergic synaptic communications between tumor cells and neurons promote GBM growth and invasion, and other neuronal effects on GBM have been attributed to autocrine signaling and paracrine signaling.[33] MSCs are also important stromal components in the GSC niche that can promote a mesenchymal tumor transcriptional state and mediate tumor proliferation through interlukin-6 and exosomes containing miRNA-1587.[34,35]
Figure 1.
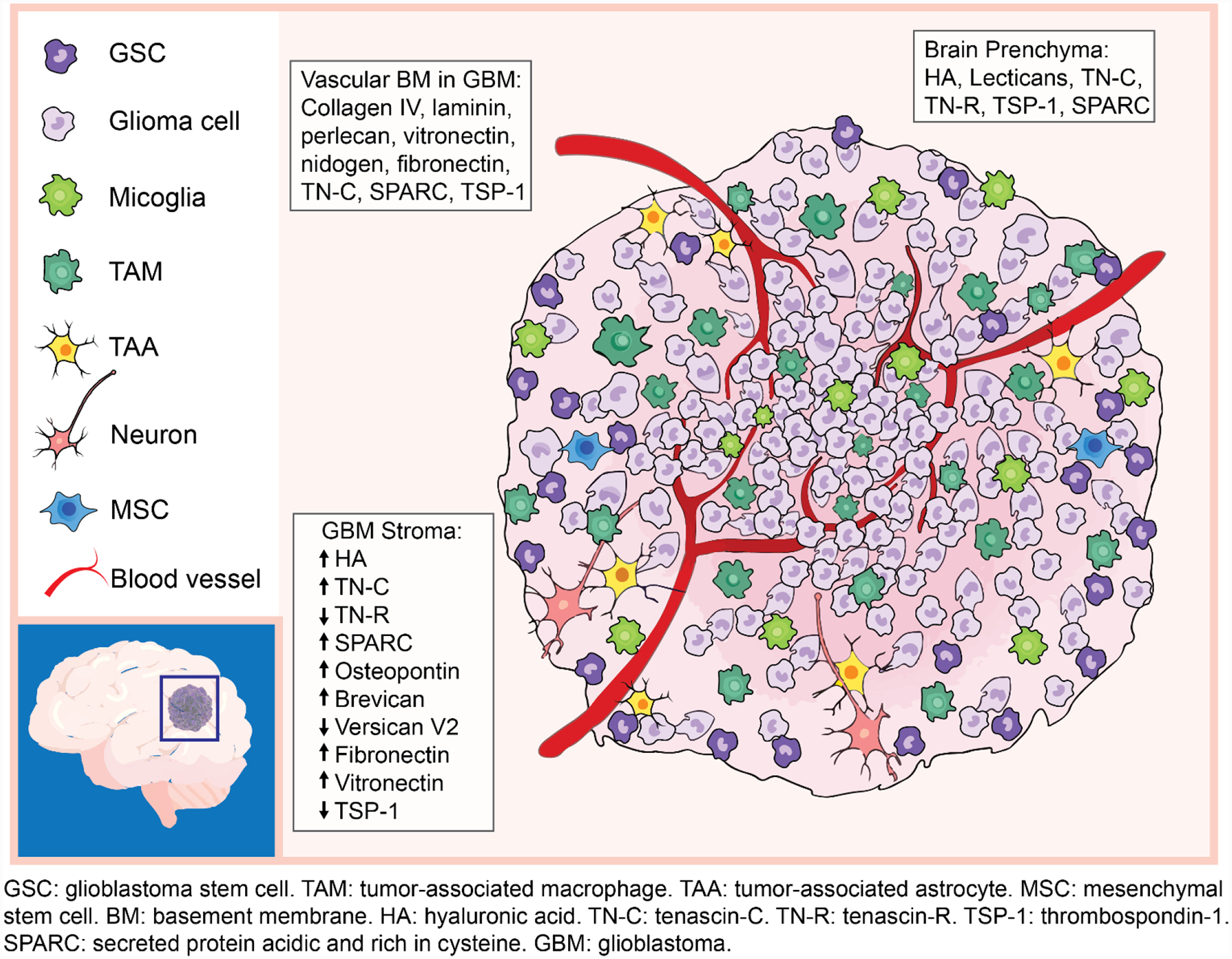
Cellular and ECM compostions of brain parenchyma and GBM microenvironments.
Neoplastic tumor cells are not homogeneous populations; single cell -omics studies confirm multiple cellular states, including stem cell-like GSCs, which promote tumor initiation, therapeutic resistance, and regrowth after therapy.[36,37] GBM cells diffusively invade into the brain parenchyma, precluding complete surgical removal. Adhesion molecules, including CD44 and receptor for hyaluronan mediated motility (RHAMM), are expressed on the cell surface of GBM cells, augmenting adhesion and migration along brain ECM rich in hyaluronic acid (HA).[38] Neoplastic cells remodel the local ECM to assist invasion through secretion of multiple proteases, such as MMPs and plasminogen activators (PAs).[39] GBM rarely metastasize outside the CNS, suggesting that neoplastic cells have adapted to the distinct CNS microenvironment.
2.1.2. Cellular components of the BBB
Cellular components of the BBB include brain microvascular endothelial cells (BMECs), pericytes, astrocytes, and neurons, which collectively form the functional neurovascular units of the CNS (Figure 2). BMECs lining the inner layer of microvessels are highly polarized endothelial cells, characterized by continuous tight junctions, adherens junctions, and limited transcytosis.[40] Junction protein complexes provide physical barriers that prevent paracellular diffusion of molecules, and the high electrical resistance of tight junctions bars entry of charged molecules.[41–43] BBB-specific influx transporters, including solute carrier proteins, and efflux transporters, including ATP-binding cassette (ABC) family members, permit uptake of nutrients into the CNS and remove substances against their concentration gradients.[44] Efflux transporters, such as ABCB1, ABCG2, and multidrug resistance-associated proteins, remove therapeutic agents from the CNS, lowering their concentrations to subtherapeutic levels.[45,46] Pericytes are mural cells embedded in the basement membrane of microvessels. Pericytes are involved in numerous functions of the BBB, regulating endothelial cell tight junction formation and astrocyte end-foot polarization.[47] Lower coverage or deficiency of pericytes increases the permeability of the BBB.[48] Astrocytes interact with microvessels through their end-feet lining along the vascular walls. Astrocytes regulate BBB diffusion barrier properties and are essential for BBB repair following injury.[42,49] BMECs, pericytes, and astrocytes synthesize the majority of BBB-specific ECM proteins. Neurons regulate the BBB permeability through neuronal activity and the release of growth factors, such as the brain-derived neurotrophic factor (BDNF).[50,51] Brain resident microglia migrate toward the brain vasculature under inflammatory conditions and play multiple roles in BBB integrity. While CCR5-dependent migration of microglia initially maintains BBB integrity, sustained inflammation leads to phagocytosis of astrocytic end-feet, which impairs BBB integrity.[52] Activated microglia secrete pro-inflammatory molecules that disrupt the BBB.[53] While the BBB is regionally defective in GBM, especially near the necrotic tumor core, the BBB remains almost intact at the proliferating and invading edges of the tumor in contact with the surrounding brain parenchyma.[4,54]
Figure 2.
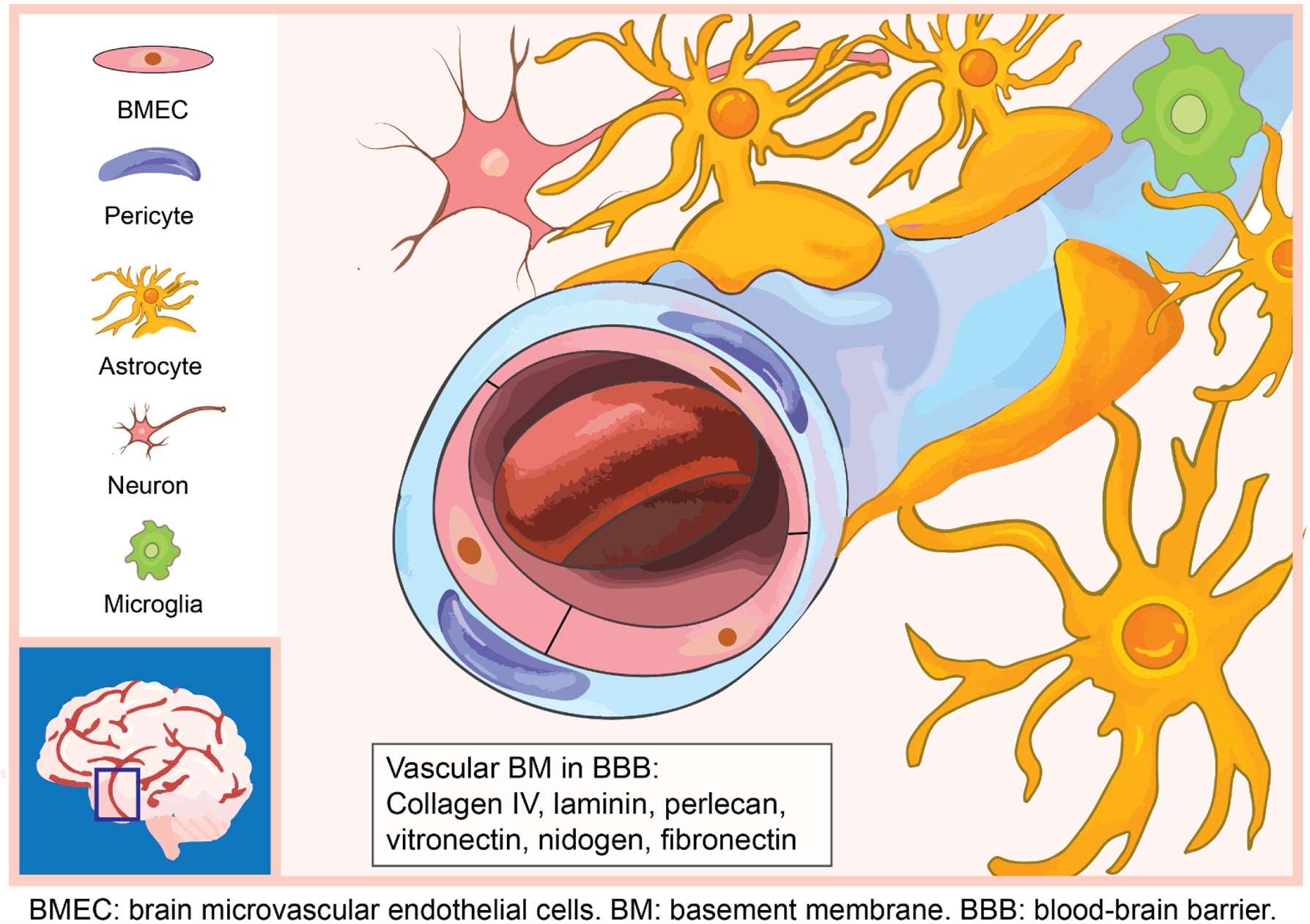
Cellular and ECM compostions of BBB microenvironment.
2.2. ECM composition of the brain parenchyma, GBM, and BBB
ECM modulates numerous brain functions, BBB barrier properties, and GBM initiation, progression, and invasion. ECM provides structural support to tissues, physically interacts with cells and other ECM components, and transduces signals upon binding through integrins and cell surface receptors. The brain ECM accounts for about 17% – 20% of the total brain volume,[55] and is composed of primarily HA, proteoglycans (e.g. the lectican family), and glycoproteins (e.g. tenascin proteins, secreted protein acidic and rich in cysteine (SPARC), and thrombospondin-1 (TSP-1)).[39,56–59] Brain parenchyma ECM components are present in the GBM stroma, but with distinct expression patterns. Other ECM components in the GBM stroma include vitronectin, osteopontin, and vascular ECM components due to active angiogenesis in GBM. The vascular basement membrane (BM) of the BBB displays a vastly different ECM composition from the brain parenchyma or the tumor stroma. The BBB ECM lacks HA and is mainly consisted of collagen IV, laminin, nidogen (also entactin), perlecan, fibronectin, and vitronectin.[39] Major ECM components in the brain parenchyma, GBM (Table 1), and BBB (Table 2) are dispalyed with their structural properties, crosstalk with other ECM components or cell surface receptors, expression patterns in the GBM stroma, and primary functions in regulating brain activities, GBM progression, or BBB properties. Changes in the amount or the composition of ECM occur with many CNS diseases, but the specific interactions and how they regulate the brain microenvironment on the molecular level remain an area of active investigation. Mechanical properties, such as the stiffness of the tissue, are associated with ECM composition and organization. Constructing 3D models will improve our understanding of ECMs in more realistic settings, enabling identification of novel mechanisms underlying specific interactions that drive neoplastic transformation, as variables can be precisely controlled and isolated in vitro.
Table 1.
Major ECMs in GBM and brain parenchyma (excluding BBB)
| ECM | Class | Structure | Size | Primary crosstalk | Expression in GBM stroma | Primary functions in brain and GBM | Ref. |
|---|---|---|---|---|---|---|---|
| HA | GAG | Linear polysaccharide with no protein core | >1,000 kDa in normal brain | Integrins, CD44, RHAMM, lectican, GHAP | Increased, low molecular weight forms present | GBM progression and invasion; structural and biochemical support to brain | [39,60,63,148] |
| TSP-1 | MCP | Homotrimer with three type 1 repeats | 420 kDa | Heparin, α5β1 integrin, HSPG, fibronectin, laminin | Decreased | GBM cell adhesion, migration, invasion; MMP inhibition; angiogenesis inhibition | [39] |
| TN-C | MCP | Oligomer with six monomers linked by disulfide bonds | 180–250 kDa (each monomer) | Lectican, HSPG, fibronectin, α5β1/αvβ6 integrins, CAM, phosphacan | Increased | Angiogenesis; ECM stiffness; immune suppression; EMT; GBM migration | [39,65,70,72,193] |
| TN-R | MCP | Present in monomeric, dimeric, or trimeric forms | 160 or 180 kDa (each monomer) | Lectican, fibronectin | Decreased | Brain plasticity regulation; synaptic activity stablization | [64,65,73] |
| SPARC | MCP | Trimer | 32 kDa | Collagen, vitronectin | Increased | Cell de-adhesion; tissue remodeling; angiogenesis; EMT | [39,70,194] |
| Osteopontin | MCP | Phosphorylated protein | 60kDa | CD44, integrin, heparin | Increased | Angiogenesis; GSC stemness; tumor growth and invasion | [74] |
| Lectican | CSPG | (See subclass) | 95–400 kDa | HA, TN-R, fibulin-2 | (See subclass) | (See subclass) | [56,59,64] |
| Subclass | Structure | Expression in GBM stroma | Primary functions in brain and GBM | Ref. | |||
| Aggrecan | G1, G2, and G3 domains with a center domain for CS/KS chains | - | Brain plasticity regulation | [195] | |||
| Versican | Two subdomains GAGα and GAGβ, and a central domain binds CS chains. Isoform V0 carries both; V1 with only GAGβ; V2 with only GAGα; V3 lacks both. | V0/V1 increase V2 decrease |
Cell adhesion and migration; drug resistance | [60,70] | |||
| Brevican | N- and C-terminal domains and a center domain for CS chains. | Significantly increased | GBM growth and progression | [69,196] | |||
| Neurocan | N- and C-terminal domains and a center domain binds up to 7 CS chains. | - | Inhibition of neurite outgrowth | [195] | |||
Table 2.
Major ECMs in vascular basement membrane
| ECM | Class | Structure | Size | Primary crosstalk | Expression in GBM stroma | Primary functions in BBB and GBM | Ref. |
|---|---|---|---|---|---|---|---|
| Laminin | Glycoprotein | Trimeric protein with α, β and γ chains | ~400 kDa | α1β1 and α6β1 integrin, HSPG, TSP-1, nidogen | - | BMEC differentiation; BBB integrity; GSC survival | [39,81–83] |
| Fibronectin | Insoluble glycoprotein | Dimer connected through disulfide bonds | 440 kDa | α5β1 integrin, fibrin, collagen, gelatin, TSP-1 | Increased | Cell adhesion, cohesion, and invasion | [39,70,76] |
| Perlecan | HSPG2 | Core protein with three GAG chains attached | ~500 kDa (core protein) | Nidogen, fibronectin, collagen IV heparin, heparin-binding growth factors | Increased | Tumor angiogenesis, biomechanical properties of BM | [39,81,195] |
| Type IV collagen | Protein | Trimeric with three α chains | ~180 kDa (each monomer) | Fibronectin, perlecan, nidogen | Increased (localized to BM) | Angiogenesis; structural support to vascular BM | [39,81] |
| Nidogen (entactin) | Glycoprotein | Three globular subdomains | 139 kDa | Collagen IV, perlecan, laminin, integrin | - | Collagen IV-laminin network stabilization | [81,197] |
| Vitronectin | Glycoprotein | Two polypeptides linked by disulfide bonds | 75 kDa | αvβ5 and αvβ3 integrins, PAI-1, antithrombin III | Increased | Microglia activation; tumor malignancy | [75,82,198] |
2.2.1. Brain parenchyma ECM
HA, a negatively charged glycosaminoglycan (GAG) without a protein core, is the most abundant ECM component in the brain.[39,60] Its negative charge attracts cations and leads to osmotic influx of water, which, in addition to its hydrophilicity, results in a high water retention capacity. High HA levels in the brain parenchyma and the lack of fibrillar proteins, such as type-I collagen, make the brain a very soft organ with remarkable plasticity. The normal brain parenchyma has an elastic modulus around 0.1–1 kPa.[61,62] In healthy brain, HA is usually present in its high molecular weight form, ranging from 1,000 kDa to 8,000 kDa.[63] HA binds non-covalently to other ECM components, including the lectican family proteoglycans. Proteoglycans are composed of a core protein with different GAG side chains. Lecticans are a family of chondroitin sulfate proteoglycans (CSPGs), which include versicans, aggrecans, neurocans, and brevicans. Other CNS CSPGs include phosphacan and neuroglial antigen 2.[64] Expression of neurocans and brevicans is mostly restricted to CNS, while versicans and aggrecans are more ubiquitously expressed in other parts of the body. Versicans have several isoforms; the V2 versican isoform is the predominant CSPG in the healthy adult brain. Lecticans are considered organizers of the CNS ECM because they can form ternary complexes with HA and tenascin-R (TN-R), known as the perineuronal net of the CNS. Tenascin-C (TN-C) and TN-R are two tenascin glycoproteins found in the CNS, produced by oligodendrocytes and astrocytes, respectively.[65] Tenascins belong to a family of matricellular proteins that are non-structural ECM proteins capable of modulating cell functions and cell-ECM interactions by binding to both cell surface receptors and structural ECM components. Two other important matricellular proteins in the CNS are SPARC and TSP-1. TSP-1 binds to CD36 on endothelial cell surface to inhibit angiogenesis.[66]
2.2.2. GBM ECM
The unique ECM of GBM, also predominantly composed of HA, contributes to the extensive invasion of GBMs within the CNS and constrains the very rare metastatic spread outside of the CNS.[38,39,67] HA content correlates with GBM malignancy. High and low molecular weight HA are found at elevated levels in GBM stroma, with low molecular weight HA involved in angiogenesis, tumor progression, and migration.[63] HA receptors, CD44 and RHAMM, and integrins on the tumor cell surface facilitate cell adhesion to and migration along the ECM.[39,67] Binding of tumor cells to ECM regulates the cell motility and the protease production, facilitating remodeling of the local ECM. Low molecular weight HA and HA fragments are involved in immune regulation by transducing signals through the toll-like receptors (TLR), such as TLR4, on macrophages, inducing M2-like phenotypes.[68] Expression patterns of many proteoglycans are altered in the GBM stroma. Brevicans, also known as brain-enriched hyaluronic acid binding proteins, are elevated in GBM stroma and involved in GBM growth and progression.[69,70] The V2 versican isoform is downregulated in the GBM stroma, whereas V0/V1 isoforms interact with transforming growth factor-β2 (TGF-β2) to promote tumor progression.[71] Upregulated levels of TN-C and SPARC in the pericellular ECM within the GBM stroma suggest potential roles in angiogenesis. TN-C overexpressed by tumor cells is also involved in TAM activation and correlates with GBM stiffness.[72] The expression of TN-R diminishes in higher grade gliomas, but its role remains unclear.[73] TSP-1, known to be anti-angiogenic, is downregulated in the GBM stroma, consistent with the hypervascularity in the GBM TME.[39] Osteopontin is a matricellular phosphoglycoprotein capable of promoting tumor progression and metastasis by interacting with CD44 and integrins. Overexpression of osteopontin in the GBM microenvironment induces M2 phenotypes in TAMs, maintains the stemness of GSCs, induces angiogenesis, and enhances tumor cell migration.[72,74] Fibronectin and vitronectin, which are components of the BM, are also overexpressed in GBM, reported to regulate tumor cell adhesion, cohesion, and invasion, and activate microglia.[75,76] Overall, changes in ECM composition and expression levels create positive feedback with GBM growth and invasion, resulting in fast tumor progression and poor prognosis.
The constant remodeling of ECM within the tumor stroma, the invasive edges, and the non-tumoral brain parenchyma of the GBM patient leads to detectable changes in mechanical properties of the microenvironment. Stiffness of tumor stroma ranges from 11.4 kPa to 26 kPa, and the non-tumoral brain regions of GBM patient have a stiffness of 7.3 ± 2.1 kPa, much stiffer than that of the healthy brain.[77–80]
2.2.3. BBB ECM
The primary BBB ECM network is formed by non-fibrillar network-forming collagen type IV and laminin, and stabilized by nidogen and proteoglycans. BM is usually in the form of organized ECM sheet with thickness of 50–100 nm.[81] Collagen IV, the primary structural element of the vascular BM, has a trimeric structure consisting of three α-chains, providing the structural support to the BM and maintaining the integrity of the BBB. Laminin is a trimeric protein consisting of α, β and γ chains. Different laminin isoforms are synthesized by BMECs, pericytes, and astrocytes, leading to differential distribution of laminin isoforms in the vascular BM on the two sides of embedded pericytes. The endothelial-side BM is rich in laminin-411 and laminin-511 synthesized by BMECs and pericytes, whereas the parenchyma-side ECM is rich in laminin-211 synthesized predominantly by astrocytes.[81–83] Global knockout of endothelial laminin induces embryonic lethality, whereas deficiency in α2 or β1 subunits causes BBB disruption and increased permeability. The laminin isoforms synthesized by BMECs and pericytes cannot compensate the loss of laminin-211, which may explain why severe BBB breakdown occurs when astrocytic laminin is deficient. In addition, integins bind to laminin and regulate the tight junction formation and the permeability of the BBB. Perlecan is a large heparan sulfate proteoglycan that binds to many ECM proteins including nidogen, and induces embryonic lethality if deficient. The exact role of nidogen and perlecan in BBB integrity is still under investigation.[81] Fibronectin and vitronectin levels are associated with compromised BBB and microglia activation, but their other functional roles remain unclear.[82,84,85]
3. Overview of 3D bioprinting strategies
3D bioprinting is an additive manufacturing technology capable of fabricating user-defined 3D objects based on computer-aided design (CAD) models. CAD models can be reconstructed from clinical images, such as magnetic resonance imaging (MRI) scans or computed tomography (CT) scans, or designed with CAD software to present specific geometries for individual applications. 3D models are sectioned into a series of 2D cross-sectional slices with predetermined layer thickness to be implemented by the bioprinters. The 3D bioprinting process generates well-defined structures in all three dimensions, and its high resolution, reproducibility, flexibility, and customizability, make it a powerful tool for a wide range of biological applications. For successful modeling of biological samples, these strategies must permit good cell viability and allow tissues to develop functionality after printing.[86] Biomimicry of bioprinted models requires the use of property-matching biomaterials and the incorporation of relevant cell types and other molecules. The major bioprinting methods include inkjet-based, extrusion-based, and light-assisted bioprinting processes.[19,87,88] Advantages, limitations, and important features of the bioprinting methods are summarized in Table 3. Regardless of the type of bioprinting methods and biomaterials, successful construction of cell-encapsulated tissues and disease models, biological platforms for screening or delivery of drugs, and acellular scaffolds have been realized.[19,89]
Table 3.
Brain-relevant natural and synthetic biomaterials
| Material | Type | Crosslinking mechanisms | Common modifications | Composite with other biomaterials | Brain-relevant elastic modulus | Ref. |
|---|---|---|---|---|---|---|
| HA | Natural polysaccharide | Photo-crosslinking, shear thinning | Methacrylic anhydride, glycidyl methacrylate, thiol, RGD peptide | Collagen, gelatin, GelMA, chitosan, laminin, fibrin, PEG, PU | 11 Pa to 3.5 kPa | [23,38,121–126,135,136] |
| Gelatin | Natural protein | Thermal, photo-crosslinking, enzymatic | Methacrylate | HA, PU, collagen, PEGDA, fibrin, alginate, chitosan, fibrinogen | 0.49 – 12.8 kPa | [127,129–133,199] |
| dECM | Natural mixture | Relies on composite material | - | Collagen | 78.09 ± 29.22 Pa | [142] |
| Collagen | Natural protein | Thermal, Photo-crosslinking | Methacrylate | HA, GelMA, fibrin, agarose, riboflavin | 0.9 – 3.6 kPa | [135,138,139,154] |
| Matrigel | Natural mixture | Thermal | - | PEG, gelatin, alginate, agarose | 0.4 kPa | [149–151,200] |
| Fibrin | Natural protein | Enzymatic | - | HA, collagen, laminin | 0.058 – 4 kPa | [124,152,153] |
| Silk fibroin | Natural protein | Photo-crosslinking, Thermal | Methacrylate | Collagen, gelatin | 17.1 ± 7.8 kPa | [115,159] |
| Gellan gum | Natural protein | Calcium ions, photo-crosslinking | RGD peptide, methacrylate | GelMA | 6.4 – 17.2 kPa | [160,201] |
| PNIPAAm | Synthetic polymer | Thermal | - | PEG | 1.4 – 3.8 kPa | [166,202] |
| PU | Synthetic polymer | Thermal, Photo-crosslinking | - | HA, gelatin | 0.6 – 8.1 kPa | [130,172] |
| PEG | Synthetic polymer | Photo-crosslinking, click chemistry | Methacrylate, thiol, diacrylate, RGD peptide | HA, GelMA, PNIPAAm, laminin | 1 – 26 kPa | [78] |
| SAP | Synthetic peptide | Self-assembly | - | - | 0.3 – 5.3 kPa | [174,177] |
3.1. Inkjet-based 3D bioprinting
Inkjet-based bioprinting forms 3D constructs by depositing volume-controlled droplets of bioinks from a nozzle. Inkjet bioprinting uses thermal, piezoelectric, or electrostatic mechanisms to deposit droplets onto receiving substrates (Figure 3a).[90] In thermal inkjet bioprinting, air bubbles generated by localized heating eject droplets from the nozzle. Instant heating does not substantially impact cell viability. Piezoelectric and electrostatic approaches utilize the pressure generated from a piezoelectric actuator or the deflection of a pressure plate, respectively, to eject droplets.[90] Inkjet bioprinting offers simplicity, low cost, fast printing speed, and high resolution without sacrificing cell viability.[91] However, the cell density needs to be kept below 106 cells ml−1 for this printing modality to mitigate the shear stress that may reduce cell viability during dispensing.[92] Striking a balance between target resolution, material viscosity, nozzle size, and dispensing speed is critical for this printing method. Using nozzles with a smaller diameter can lead to a better resolution, but also increases the possibility of clogging if the material viscosity is not appropriate. Biomaterials with low viscosity, below 12 mPa-s, are compatible with inkjet printing.[19] Using inkjet bioprinting, a wide range of biological applications has been demonstrated, including cancer models,[93] stem cell research,[86] tissue engineering,[94] single-cell studies,[95] cell array patterning,[96] and controlled release of molecules.[97] Inkjet bioprinting can also achieve high throughput by inclusion of multiple nozzles, making it desirable for screening applications.[98,99]
Figure 3.
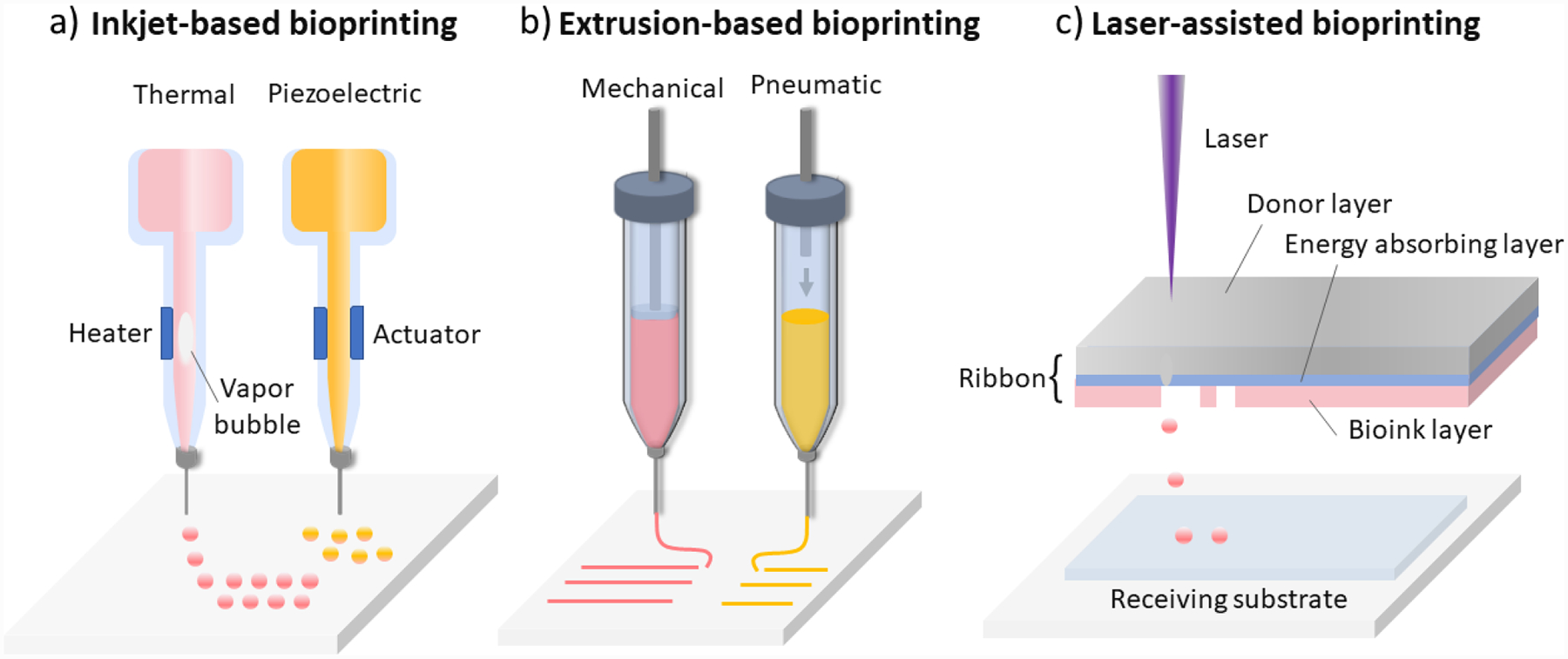
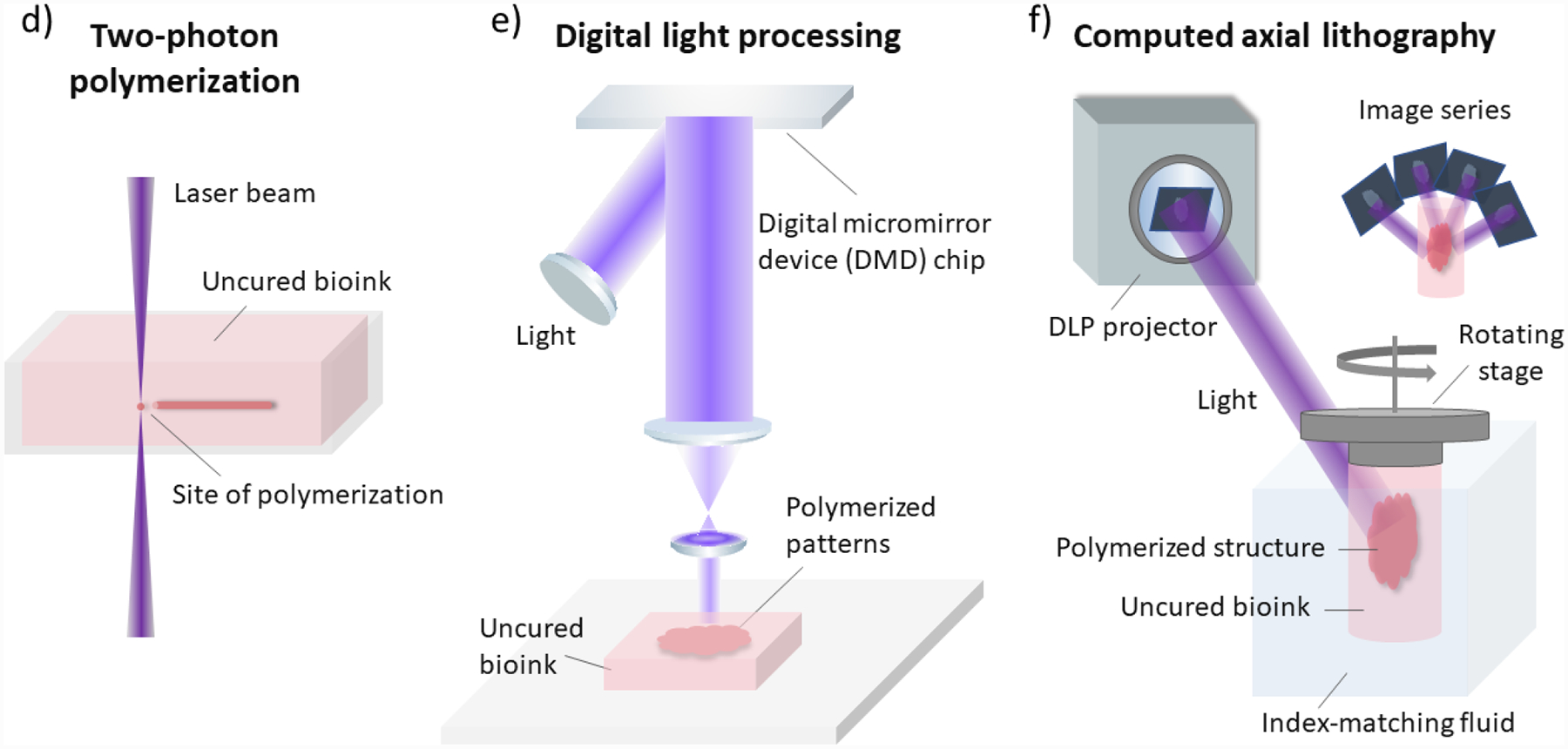
Schematic illustrations of common 3D bioprinting strategies. a) Inkjet-based bioprinting. b) Extrusion-based bioprinting. c) Laser-assisted bioprinting. d) Two-photon polymerization-based bioprinting. e) Digital light processing-based bioprinting. f) Computed axial lithography.
3.2. Extrusion-based 3D bioprinting
Extrusion-based bioprinting relies on a continuous deposition of material filaments through a nozzle. The continuous process enables it to generate constructs with an overall better interface integrity compared to inkjet bioprinting. Two main dispensing mechanisms of the extrusion-based bioprinting are pneumatic-based and mechanical-based; the latter includes piston-driven and screw-driven methods (Figure 3b).[19,92] During printing, either the stage or the bioink-filled dispensing nozzle is motorized to create 3D structures in a layer-by-layer fashion. Pneumatic dispensing is directly controlled by changes in the pressure, making it highly flexible; meanwhile, the delay in pressure change can reduce its precision in spatial control of deposited bioinks. The mechanical dispensing approach is generally better in spatial control due to the real-time impact on the material flow, while the screw-driven system is especially suitable for highly viscous materials. Versatility of extrusion-based bioprinting makes it compatible with a broad selection of biomaterials, with viscosity ranging from 30 mPa-s to 6×107 mPa-s. This printing modality also allows encapsulation of cells at a relatively high density, or even in the form of spheroids.[100,101] Despite the shear stress that occurs within the nozzle, extrusion-based bioprinting methods permit favorable cell viability in printed constructs.[19] Resolution of extrusion-based bioprinting is limited by a few factors, including nozzle diameter, gelation kinetics, and properties and composition of bioinks. While high resolution of 5 μm can be achieved for acellular scaffolds, [102] cell encapsulated samples often have compromised resolution, generally over 100 μm, [103] as a tradeoff to scale-up potential and high encapsulation capacity.[19,20] Nevertheless, extrusion-based bioprinting is the most widely used bioprinting strategy for tissue engineering applications, given its ability to generate samples with physiologically relevant dimension, mechanical properties, and cell density.[89,104]
3.3. Light-assisted 3D bioprinting
Light-assisted bioprinting uses photon energy to induce photo-polymerization of bioinks to form 3D structures. Light-assisted strategies have high resolution and precise control of the architectures in all three dimensions. Without the high sheer pressures that occur in inkjet or extrusion bioprinting, higher cell viabilities can be achieved, even for sensitive cell types, including stem cells, using light assisted bioprinting approaches. Lght-assisted bioprinting can be categorized based on fabrication processes: scanning-based and projection-based. Scanning-based strategies usually require serial movement along all three axes. First, 2D features on one layer are formed through scanning of the laser beam within the bioinks or on a donor film. The laser beam then moves along the third axis, usually the z-axis, to build up a 3D structure. Projection-based bioprinting polymerizes an entire layer at a time. Features on one plane are formed with a single projection of patterned light, so the motor movement is often only necessary along the third axis during printing. Thus, projection-based strategies generally render higher throughput and faster printing speed than scanning-based strategies.[105] Commonly used light-assisted approaches for biological applications include: (1) scanning-based strategies, such as the laser-assisted bioprinting (LAB) and the two-photon polymerization (TPP)-based bioprinting; and (2) projection-based strategies, mainly digital light processing (DLP)-based bioprinting.[105–110]
A laser-assisted bioprinter is composed of a pulsed laser source, a receiving substrate, and a ribbon consisting of a bioink layer and a metal laser-absorbing layer usually made of gold or titanium (Figure 3c).[106] During the printing process, the laser pulse induces vapor bubbles on the donor layer and, in turn, ejects droplets of bioink onto the receiving substrate parallel to the ribbon. Micron-scale structures with high cell density have been printed, and a variety of materials is compatible with this strategy.[106,107] TPP is a laser-based direct-writing strategy that uses an ultrafast laser beam (e.g. femtosecond pulse) to trace and polymerize the cross-sectional features of 3D structures layer-by-layer. TPP polymerizes bioinks by the simultaneous absorption of two photons from a near-infrared femtosecond pulsed laser (Figure 3d). The resolution of TPP is not limited by the diffraction limit of the light source, so submicron scale features can be achieved.[111] Fine features of 1 μm or smaller have been printed with Poly(ethylene glycol) diacrylate (PEGDA) using TPP.[108] The relatively high resolution makes it suitable for fine patterning of biomaterials and single cell studies, with the trade-off of a slower bioprinting speed and limitation in scalability.
DLP bioprinting is a rapid projection-based stereolithography, which can fabricate millimeter- or centimeter-scale constructs within seconds to minutes.[105] DLP printers are usually equipped with a digital micromirror device (DMD) chip, a motorized stage or bioprinting probe, a set of optical paths, and a computer to synchronize the movement of the stage or the probe to corresponding patterns (Figure 3e). The DMD chip consists of millions of micromirrors that can be independently switched on or off to display the user-defined patterns with micron-scale features. Photocurable bioinks are polymerized only at the positions where light is projected from the DMD chip, permitting a highly defined architecture with a resolution of 2 μm.[22] Functional tissue constructs integrating multiple cell types and various ECM materials have been produced with this bioprinting strategy. High cell viability has been achieved, including stem cell-derived cells.[23,112] DLP bioprinting allows precise control over material properties, such as the elastic modulus and the amount of biochemical cues, which are important aspects for biological studies.[113] Many biomaterials have been used with DLP bioprinting, including HA, gelatin, decellularized ECM, silk fibroin, poly(ethylene glycol) (PEG), PEGDA, and polyurethane (PU), whereas some require modifications to obtain photosensitivity.[22,23,114,115] Broad biological applications of DLP include controlled release of growth factors,[116] nerve regeneration,[117] high throughput drug testing,[112] and tissue and disease modeling.[23,112,113,118,119] A DLP-based volumetric 3D bioprinting strategy, named computed axial lithography (CAL), enables fabrication of an entire 3D structure through one complete rotation of bioinks with synchronized pattern projections (Figure 3f).[110] The strategy relies on the back-projection algorism of the CT reconstruction. This implementation enables improved geometric flexibility than prior attempts using field interference, allowing it to print complex non-symmetric 3D structures. Materials of high viscosity up to 90,000 mPa-s were used to avoid the necessity of supporting materials. This strategy offers many distinct advantages, such as the ability to print around an existing object and the scalability such that a centimeter-scale structure can be fabricated within a minute.
4. Relevant biomaterials for modeling GBM and BBB
Biomimetic 3D models require biomaterials with good biocompatibility and tissue-specific properties, including appropriate biophysical/biochemical properties and degradation kinetics.[19] Biomaterials form structural networks that foster cell adhesion, proliferation, and migration, and provide specific spatiotemporal cues to modulate cell behaviors. Here we discuss biomaterials in the order of their relevance to the brain microenvironment and appropriateness for 3D modeling and 3D bioprinting (Table 4). Two primary categories of biomaterials include: (1) natural materials that are constituents of the native tissue ECM, and (2) synthetic materials with good biocompatibility. Natural materials are innately biocompatible and bioactive, possessing biochemical and biophysical features that result in exceptional biomimicry, and can be remodeled or cleared through natural degradation mechanisms. Alternatively, synthetic materials have defined chemical structures and tunable properties, but lack the innate bioactivity or physiologic degradation mechanisms. However, synthetic materials can be modified to incorporate adhesive peptides or cleavable linkers to mimic the functional or structural properties of the native ECM.[72] It is common for 3D modeling to combine several biomaterials to take advantage of the collective properties of each individual component.
Table 4.
3D bioprinting strategies
| Type | Inkjet-based | Extrusion-based | Light-assisted | Light-assisted | Light-assisted |
|---|---|---|---|---|---|
| Subtype | Thermal, piezoelectric, electrostatic | Pneumatic, mechanical (piston-driven, screw-driven) | Scanning-based (LAB, TPP) | Projection-based (DLP) | Projection-based (volumetric) |
| Fabrication process | Serial: point-by-point | Serial: line-by-line | Serial: point-by-point | Parallel: layer-by-layer | Parallel: rotational |
| Advantages | Fast printing speed, high resolution, high throughput, low cost | Broad biomaterial selection, scale-up potential, high cell densities, low cost | Very high resolution, compatible with biomaterials in different phases | High resolution, very high speed, good interface integrity, broad biomaterial selection, scale-up potential | Concurrent printing of real 3D structures, scalable to large constructs |
| Limitations | Poor interface integrity, low cell densities, limited to low viscosity biomaterials | Limited interface integrity, resolution limited by nozzle diameter | High cost, limited biomaterial selection, limited scalability, low throughput | Requires photosensitive biomaterials | Limited resolution, cell density may be limited due to light scattering |
| Typical resolution | 10 μm | 100 μm (with cell), 5 μm (acellular) | 1 μm | 2 μm | mm scale |
| Bioink viscosity | Low: 3.5 to 12 mPa×s | Wide range: 30 to 6×107 mPa×s | Medium: 1 to 300 mPa×s | - | High viscosity fluids: 90,000 mPa×s, or solids |
| Cell density | Low :106 cells/ml | High | High: 108 cells/ml | High | - |
| Print speed | Fast | Medium | Medium | Fast | Fast |
| Reference | [19,88,104] | [19,88,92] | [19,88,203] | [22,105] | [110] |
For clarity, we refer to biomaterials suitable for bioprinting processes as bioinks. Developing bioinks with good printability and biomimicry is critical to 3D bioprinting applications. The printability of bioinks include various aspects, such as viscosity, thermo-sensitivity, and photosensitivity, depending on the specific bioprinting modality. Additionally, rheological properties, crosslinking mechanisms and kinetics, and post-printing mechanical properties, such as the elastic modulus and the swelling ratio, are important parameters of the bioinks.[92] To enhance biomimicry, bioinks are often integrated with cells, growth factors, cytokines, and other molecules to accommodate specific biological applications.
4.1. Natural biomaterials and their derivatives
4.1.1. HA
HA is a negatively charged, linear polysaccharide composed of alternating D-glucuronic acid and N-acetyl-D-glucosamine, synthesized at cell plasma membranes of neurons and glial cells.[120] Due to the predominance of HA in the brain and GBM stroma and its critical role in regulating diverse physiological and pathological processes through interaction with cells and other ECM components, HA-based hydrogels are the most relevant matrix materials for modeling brain tissues and brain tumors. HA hydrogels have nanoporous structures and a range of elastic modulus recapitulating the brain and the GBM stroma.[121] HA has been combined with various biomaterials, including type I collagen,[122] gelatin methacrylate (GelMA),[38] chitosan,[123] laminin, fibrin,[124] and PEG[78] to fabricate 3D GBM models. HA demonstrates size-dependent regulatory behaviors, so the range of molecular weight of HA should be considered when designing specific models. HA with over 1,000 kDa is appropriate for modeling the healthy brain tissue, while HA with lower molecular weight has been observed in GBM stroma and affects GBM progression and migration. By fixing the poroelastic properties of a series of HA-GelMA hydrogels, lower molecular weight HAs (10 kDa and 60 kDa) result in higher invasiveness compared to higher molecular weight HA (500 kDa). The molecular weights of HA did not affect the elastic modulus of HA-GelMA hydrogels; all groups were measured around 3 kPa.[38] Scaffolds made of HA, laminin, and fibrin support human neural precursor cells (NPCs) growth and vascular formation.[124]
Chemical modifications to generate HA derivatives appropriate for 3D modeling or 3D bioprinting have been previously reviewed.[121] Modifications generally target the carboxylate group on the D-glucuronic acid moiety, the N-acetyl group on the N-acetyl-D-glucosamine moiety, and the hydroxyl groups on both moieties. HA-derivatives form hydrogels through radical polymerization. For example, HA functionalized with glycidyl methacrylate or methacrylic anhydride on the C-6 hydroxyl group of the N-acetyl-D-glucosamine to form glycidyl methacrylate HA (GMHA) or methacrylated HA (MeHA) can be photopolymerized to form hydrogel in the presence of photo-initiators, such as lithium phenyl-2,4,6-trimethylbenzoylphosphinate (LAP).[121] GMHA and MeHA are suitable bioinks for light-assisted bioprinting due to their rapid photo-polymerization ability. Liver tissues and GBM models have been bioprinted using GMHA-based hydrogel mixture.[9,23] MeHA has also been functionalized with Arg-Gly-Asp (RGD) peptides to facilitate cell adhesion to the 3D matrix.[125] Another method to form HA-based hydrogel is through addition and condensation reactions. HA thiol derivatives spontaneously crosslink through disulfide bond formation in air without initiators, making it a good bioink candidate for extrusion or inkjet bioprinting.[126] Aldehyde-, dihydrazide-, and haloacetate-modified HA form biocompatible hydrogels through addition and condensation reactions.
4.1.2. Gelatin
Gelatin is a partial hydrolysis product of collagen. Gelatin and its derivatives are widely used in 3D tissue modeling due to their inherent bioactive features including integrin binding RGD sequences and MMP digestion sites. Coculture of perivascular niche (PVN) cells and GBM cells in a 3D gelatin matrix demonstrated elevated levels of angiogenesis and ECM remodeling compared to tumor cells or PVN cells cultured alone.[127] Due to good rheological properties and thermally responsive characteristics, gelatin-based materials are popular bioinks used in extrusion-based bioprinting.[128] Encapsulation of hepatocytes has been achieved with gelatin hydrogel, and the 3D-printed tissue remains viable and functional over two months of culture.[129] Gelatin can also be combined with synthetic materials, such as PU, to improve its printability in terms of longer bioprinting window and higher resolution. A gelatin-PU matrix allowed high viability and proliferation of MSCs.[130] GelMA is a versatile derivative of gelatin also popular for 3D bioprinting. GelMA is developed by modifying the lysine and hydroxyl groups with methacrylamide and methacrylate side groups, rendering the prepolymer GelMA bioink photo-polymerizable in the presence of photo-initiators under UV exposure.[131] GelMA preserves the biological features of gelatin and enables tunable mechanical properties of 3D matrices. GelMA can serve as the base matrix material to facilitate investigation of other functional ECM such as HA in brain-related studies. The effects of biochemical cues from HA on tumor growth have been investigated by mixing gelatin-based matrix with different amounts of soluble or immobilized HA.[132] The expression of angiogenic markers and hypoxia markers demonstrate biphasic peaks when HA concentration falls between 0.3% and 0.5%.[133] GelMA-based hydrogels can generate gradients of HA, crosslinking density, and GBM cell density.[133] Spatially gradated matrix reveals that tumor cell proliferation and pro-angiogenic expressions correlate with the local crosslinking density and tumor cell density, whereas the local MMP2 expression inversely correlated with the cell density. GelMA has also been combined with PEGDA to generate cardiac patch for the treatment of myocardial infarction.[134]
4.1.3. Collagen
Collagen is a ubiquitous ECM component in most body tissues. Although the brain is virtually absent of the fibrillar collagen type I, the vascular basement membrane is abundant with collagen type IV and some collagen type V. Thus, collagen-derived biomaterials are appropriate for modeling the BBB. Nonetheless, various GBM studies have exploited collagen biomaterials due to their well-studied gelation mechanism, including pH-based and temperature-based, abundance of cell binding sites, and tunable mechanical properties to match tissue-specific requirements. GBM cells adopt different morphologies in 3D matrices by collagen types: round in type IV and spindle-like in type I/III.[135] Collagen is commonly combined with other biomaterials, including HA, agarose, and synthetic materials, for tissue modeling. In a hybrid matrix with HA, only collagen type IV, not type III, supports GBM cell proliferation.[136] Pure collagen solutions have relatively slow gelation process and low viscosity.[137] Increasing the concentration of collagen or including riboflavin in the pre-polymer solution improves bioprinting accuracy.[138,139] Inclusion of riboflavin increases the storage modulus of collagen bioinks, improving printability. Gelation of collagen-based bioinks is usually thermally controlled or pH-driven, and collagen-based bioprinting has been used in tissue engineering applications, including heart regeneration and liver modeling.[137,140,141] The hydrogel elastic modulus can be tailored between 0.9 kPa and 3.6 kPa, which is suitable for brain tissues.[139]
4.1.4. Decellularized ECM (dECM)
DECM is obtained by removing all cellular components of a tissue while preserving most of the tissue-specific and patient/host-specific ECM structures and components, retaining native ECM cues conducive to cell growth.[113] The analysis of GBM patient brain tissue-derived dECM has demonstrated that GAGs, HA, collagen IV, laminin, and fibronectin are not significantly disturbed after processing, thus appropriate as an in vitro modeling biomaterial.[142] Patient brain dECM has been mixed with collagen to achieve better gelation through extrusion-based biopriting. Compared to cells in the collagen control, disseminated single cells have heterogeneous and rounded morphologies in the patient dECM-based matrix. Moreover, GBM cells in dECM-based matrix express increased level of matrix remodeling protein MMP9 and HA-related genes, including Hyal1, Hyal2, HAS2, and CD44.[142] While the slow gelation kinetics of dECM-based hydrogels often necessitate the integration of dECM bioinks with other biomaterial to improve printability, recent studies have induced thermal gelation of the dECM bioinks alone.[143–145] Bioinks based on dECM have been developed for various tissues, such as cartilage, heart, adipose, liver, and tumors, and demonstrate good printability on extrusion-based and DLP-based bioprinters.[113,144–147] However, dECM is usually derived from an individual’s tissue and contains a variety of natural proteins and polysaccharides, so variation is inevitable and control over specific variables is challenging. Despite the limitations, dECM with its potential in GBM modeling for individual patients, remains an exceptional choice of biomaterial for precision medicine applications.
4.1.5. Matrigel
Matrigel is a thermally curable mixture of ECM components derived from murine Engelbreth-Holm-Swarm sarcoma, composed of about 60% laminin, 30% collagen type IV, 8% nidogen, and other growth factors and proteoglycans. Its similarity to the vascular ECM composition makes it especially suitable for BBB modeling.[148] As a result, Matrigel is broadly used for vascular formation and related studies in vitro. GBM organoids have also been developed in Matrigel, with cells within the organoid displaying hypoxic gradients and heterogeneity in stemness and proliferation.[17] However, the majority of proteins in Matrigel are present in low amounts in the brain (excluding the BBB) or GBM, making it a suboptimal choice for GBM modeling, even though many studies have demonstrated good GBM cell viability and proliferation in the matrix. Limitations of Matrigel include its animal origin, batch variation that reduce experimental reproducibility, and limited control over the physiochemical properties of the formed 3D matrix. Matrigel also has limited printability due to its relatively poor mechanical properties and lack of photo-sensitivity, and thus it is often combined with other biomaterials, such as agarose, alginate, and gelatin, to fabricate scaffolds or tissue models using 3D bioprinting technologies.[149–151]
4.1.6. Fibrin
Fibrin is formed by crosslinking of fibrinogen and thrombin. Mechanical properties of fibrin hydrogels depend mainly on the concentration of fibrinogen and to the lesser extent on the thrombin. Stiffness ranging from 0.058 kPa to 4 kPa, a relevant range for brain applications, can be achieved with a fibrin matrix.[152] Co-culture models of GBM spheroids and endothelial cells in a fibrin matrix have been used to test anti-angiogenic compounds.[153] Vascular endothelial growth factor (VEGF)-loaded fibrin hydrogels support neural stem cell growth and migration compared to fibrin matrix with no VEGF or VEGF-loaded collagen hydrogel, demonstrating beneficial properties of fibrin matrices to embed growth factors for extended culture time.[154] Similarly, improved cell proliferation and prolonged persistence have been observed for cytotoxic human MSCs cultured in fibrin matrices, enabling MSC-based GBM therapy to suppress post-surgical recurrence.[155] Fibrin-based bioinks are popular with extrusion-based bioprinting. Fibrin bioinks have been mixed with gelatin, alginate, or HA to improve its mechanical and biochemical properties, and have generated various tissue models including GBM models, cardiac tissues, and dentin-pulp complex.[156–158]
4.1.7. Others
Other natural biomaterials that are not native in the brain but with good biocompatibility and printability have also been explored for CNS studies. Silk fibroin (SF) have been used for neural network formation and gellan gum for multilayer neural circuit formation.[159,160] The range of stiffness of the SF hydrogels and GG hydrogels is appropriate for modeling the GBM stroma. A human GBM cell line exhibited distinct responses in two types of SF hydrogels – enhanced viability and proliferation in the random coil type and induced apoptosis in the crystalline type.[161] SF hydrogels with tunable mechanical properties and post-printing degradation rates can also be adapted to different bioprinting applications.[162] Other non-network-forming ECM components present in the native tumor stroma or the BBB may be incorporated into 3D matrices with the above-mentioned hydrogel-forming biomaterials to improve the material biomimicry in future studies.
4.2. Synthetic biomaterials
Despite a non-biological origin, synthetic biomaterials can be readily modified to have mechanically and biochemically robust properties and degradation kinetics for biological modeling. By functionalizing with cell adhesion peptides and MMP-cleavable sequences, or mixing with other natural biomaterials, synthetic biomaterials-based hydrogels can create microenvironments with comparable properties to native ones. Models based on synthetic biomaterials generally have good scalability and reproducibility due to their synthetic nature. In addition to cell-encapsulating models, synthetic materials are suitable for fabricating cell culture scaffolds, microfluidic devices, or implantable devices. GBM cells cultured on polystyrene scaffolds have generated more clinically relevant drug efficacy predictions for TMZ, erlotinib, and bevacizumab than traditional 2D cultures.[163]
4.2.1. Synthetic polymers
Poly(N-isopropylacrylamide) (PNIPAAm), PEG, and PU are synthetic biomaterials that have been used for GBM studies. PNIPAAm and its composite materials are thermo-responsive hydrogels and demonstrate good printability on extrusion-based bioprinters.[164] PNIPAAm embedded with gold nanorods can be printed with multiphoton lithography to achieve a nano-scale resolution and post-printing dynamic modulations.[165] Primary GSCs cultured in a PNIPAAm-PEG matrix retain stemness over long-term culture and can be easily retrieved and re-encapsulated by adjusting the temperature of the hydrogel.[166] The hydrogel can expand GSCs into large numbers necessary for screening purposes. PEG is a popular biomaterial for 3D tissue modeling due to its good biocompatibility, inert biochemical properties, and tunable mechanical properties.[114] PEG and its deriviatives can be readily modified with bioactive components to enhance its biomimicry and printbility as bioinks.[167–169] PEG hydrogels mixed with fixed concentration of HA and functionalized by RGD peptides and MMP degradation crosslinkers, have been used to investigate the stiffness impacts on GBM progression.[78] GBM cells cultured in a stiff PEG hydrogel (26 kPa) form denser tumor spheroids compared to the cells in a softer structure (1 kPa). PEGDA is a derivative of PEG that have demonstrated broad applications in 3D bioprinting due to its biocompatibility and photo-polymerizability.[117] PEGDA has been used to form microwells for the in vitro culture of glioblastoma cells or co-culture of glioblastoma cells with endothelial cells for high throughput drug screening.[170,171] PU hydrogels are thermo-responsive and biodegradable. Neural stem cells embedded in a water-based PU hydrogesl through 3D bioprinting have demonstrated excellent growth and differentiation potential.[172]
4.2.2. Self-assembled peptides
Self-assembled peptide (SAP)-based hydrogels are crosslinked by physical or chemical bonding of the peptides, forming organized nanofibrous β-sheets resembling the native ECM structures.[173] Peptides are chains of amino acids that possess innate biological properties. Fibrous SAP hydrogels have tunable mechanical properties and controllable stimuli-responsive gelation processes (e.g. enzymatic triggering), making them promising bioinks for extrusion-based bioprinting.[174,175] Proof-of-concept extrusion-based printing of fluorecent SAP hydrogels demonstrate good mechanical stability and low erosion rate in solutions.[176] The injectability of SAP and its ability to adapt to irregular shapes also makes it a good candidate for CNS regenerations, such as BBB repair or brain tissue repair after GBM surgery. A peptide RADA16-SVVYGLR-forming hydrogel with a stiffness between 0.326 kPa and 5.336 kPa injected into the brain of a zebrafish brain injury model induces both angiogenesis and neurogenesis.[177]
5. 3D bioprinting for GBM and BBB
5.1. 3D bioprinting for GBM modeling
3D bioprinting has emerged as a promising tool for modeling and developing treatments for various cancer types, such as breast cancer, [178,179] pancreatic cancer, [180] liver cancer, [113] ovarian cancer,[98] and metastatic models.[179,181] The ability of 3D bioprinting to fabricate complex 3D architectures with living cells and biomaterials makes it especially suitable for recapitulating the heterogeneous GBM TME. 3D-bioprinted GBM models have been developed with engineered biomaterials and cell types, appropriate for different applications, such as mechanistic studies, cell-matrix interactions, cellular crosstalk within specific niches, treatment evaluations, or as screening platforms. While the selection of engineered materials to mimic the natural ECM environment might have been relatively limited for early studies, various cell types have been well-studie and available for use. The rationales for the selection of cell types in each study have been justified, and thus the cellular compositions are used to classify these models in our review. In this section, we comprehensively review the current 3D-bioprinted GBM models (Table 5) based on their cellular complexity and provide perspectives on the type of biological questions they may address.
Table 5.
3D-bioprinted GBM models
| Cellular components | Species | Biomaterials | Printing method | Features of 3D-printed models | Applications | Ref. |
|---|---|---|---|---|---|---|
| GSCs (GSC23) | Human | Gelatin, alginate, fibrinogen | Extrusion-based | Increased angiogenic potential, enhanced stemness, morphological changes | Mechanistic studies | [182] |
| GSCs (SU3) or GBM cell line (U87) | Human | Gelatin, alginate, fibrinogen | Extrusion-based | Increased angiogenic expression, higher drug resistance, enhanced tumorigenicity | Mechanistic studies, drug responses | [183] |
| GBM cell line (U118) | Human | Gelatin, alginate, fibrinogen | Extrusion-based | Enhanced stemness, increased EMT markers, higher drug resistance and tumorigenicity | GSC enrichment by 3D ECM, mechanistic studies | [184] |
| GBM cells (GL261), macrophages | Mouse | GelMA, gelatin | Extrusion-based | Macrophage recruitment, higher matrix remodelling and EMT activities | Tumor-macrophage interactions, drug responses | [185] |
| GBM cell line (U87), HUVECs | Human | Brain dECM, collagen | Extrusion-based | Hypoxic gradient, proliferation gradient, higher pro-angiogenic markers | Angiogenesis events, cellular responses to dECM, drug responses | [186] |
| GSCs (GSC23), MSCs | Human | Alginate, gelatin, fibrinogen | Extrusion-based | Spontaneous tumor fiber formation, enhanced stemness and EMT | Tumor-MSC interactions, cell-ECM interactions | [187] |
| GSCs (GSC23), GBM cell line (U118) | Human | Alginate | Extrusion-based | Elevated expression of matrix remodeling and angiogenic markers, enhanced drug resistance | GSC impact on regular tumor cells, drug responses | [158] |
| GSCs (CW468, GSC23, 2907, 3264), macrophages, astrocytes, NPCs | Human | GM-HA, GelMA | DLP-based | Enhanced invasion and drug resistance, macrophage polarization to M2 | Tumor-stromal interactions, drug response, clinical predictions | [9] |
| GSCs (G144, G166, G7), GASCs, microglia | Human | RGD-alginate, collagen, HA | Extrusion-based | Higher drug resistance | Tumor-stromal interactions, drug responses | [188] |
5.1.1. Mono-culture 3D-bioprinted GBM models
Mono-culture 3D GBM models are good for investigating tumor-ECM interactions and mechanistic studies. GSCs have been encapsulated in a gelatin-alginate-fibrinogen (GAF) hydrogel by a multi-nozzle extrusion-based bioprinter (Figure 4a).[182] Gelatin, alginate, and fibrinogen were crosslinked sequentially by transglutaminase, calcium chloride, and thrombin, respectively, to achieve high post-printing cell viability and proliferation. Elevated levels of angiogenic regulators and stemness markers, CD31, VEGFR2, HIF-1a, and CD133, were detected in 3D-cultured GSCs, compared to suspension cultured controls (Figure 4b). Morphological changes were observed in 3D cultured GSCs, including higher contents of endoplasmic reticulum and mitochondrion and increased amount of microvilli (Figure 4c), which played an important role tumor cell survival. Patient-derived GSCs and glioma cell lines were bioprinted using the same extrusion-based method and GAF hydrogel.[183,184] GSCs expressed elevated levels of VEGF released by tumor cells in vivo to trigger angiogenesis and exhibited higher resistance to TMZ treatment compared to 2D cultured controls. 3D hydrogel-enriched stem cell populations expressed stemness markers CD133 and Nestin (Figure 4d). Enhanced epithelial-mesenchymal transition (EMT), associated with the transition of non-GSCs towards GSCs, occured in 3D models with increased levels of Twist and Snail (Figure 4e).[184] GBM cells derived from 3D models exhibited enhanced in vivo tumorigenicity compared to 2D cultured cells (Figure 4f).
Figure 4.
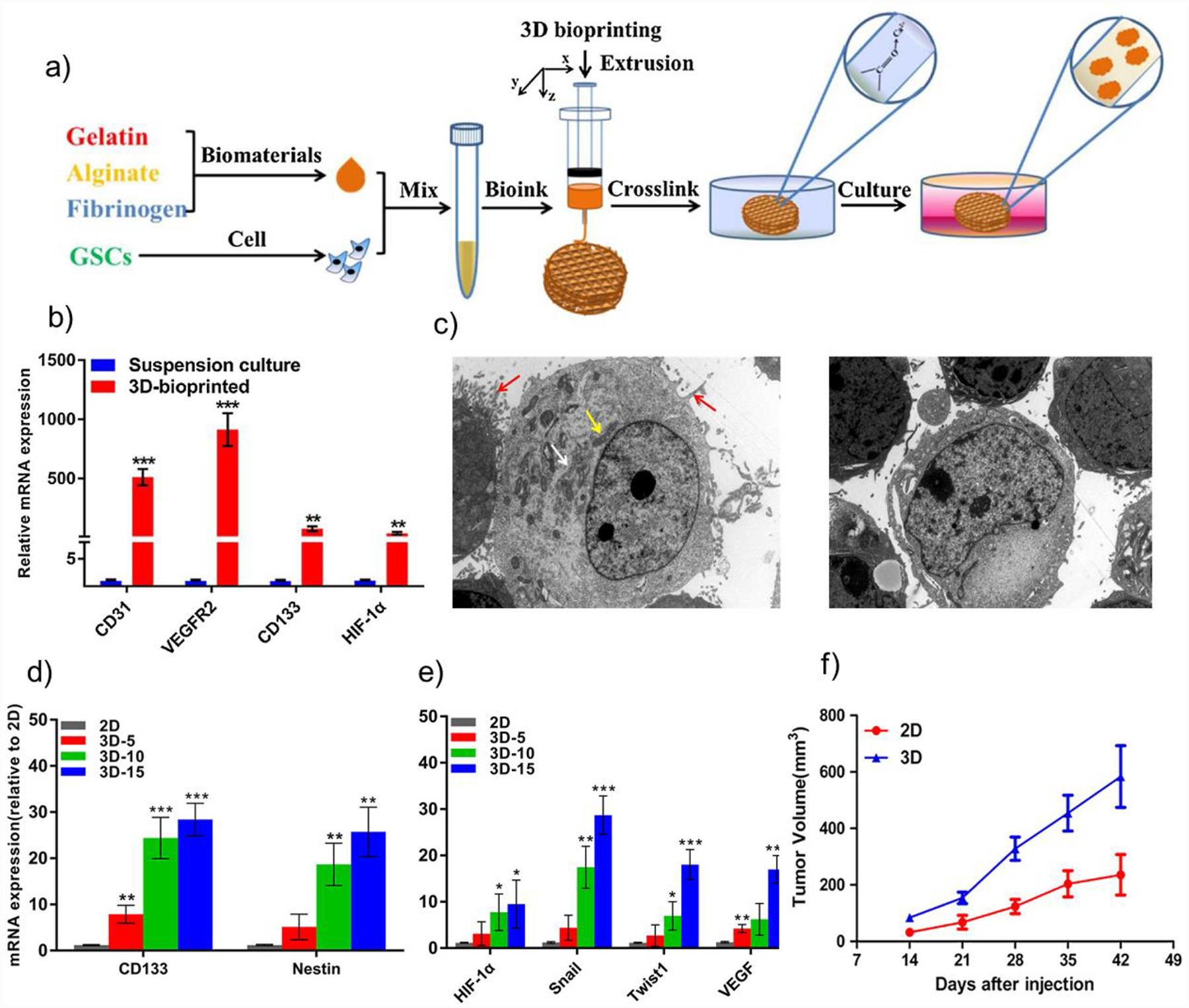
Schematic representation of the bioprinting process and functional evaluations of a mono-culture GBM model. a) GSCs were encapsulated in a gelatin-alginate-fibrinogen hydrogel by an extrusion bioprinter and crosslinked post-printing. b) Expression of CD31, VEGFR2, HIF-1a, and CD133 were significantly elevated in 3D-bioprinted GSCs compared to suspension cultures. c) Increased endoplasmic reticulum, mitochondrion, and microvilli were observed in 3D-bioprinted cells (left) compared to suspension culture (right). Reproduced with permission.[182] 2018, Elsevier. d) 3D hydrogel enriched stem cell population in the U118 cells with increasing expression of CD133 and Nestin at day 5, 10, and 15. e) 3D culture enhanced expression of EMT markers (Twist, Snail), hypoxia marker (HIF-1a), and angiogenesis marker (VEGF). f) U118 cells derived from 3D models exhibited enhanced in vivo tumorigenicity compared to 2D cultured cells. Copyright Wiley, 2018.[184] Reproduced with permission.
Monoculture 3D GBM models are not optimal due to lack of cellular interactions between stromal cells and tumor cells, and biomaterials being used may not be the major native ECM components of the GBM stroma. However, monoculture 3D models enable tumor cell interactions with natural ECM components within a matrix of the native dimensionality. The matrix- or dimensionality-induced changes in gene expression and cell morphology result in elevated expression of angiogenesis and stemness markers, enhanced EMT, and higher drug resistance of 3D cultured cells compared to their traditional cultured controls.
5.1.2. Co-culture 3D-bioprinted GBM models
Co-culture 3D-bioprinted GBM models have been used to study specific cellular interactions between neoplastic cells and stromal cells, such as macrophages, MSCs, and ECs, in biomimetic 3D context. Miniaturized brains (mini-brains) fabricated with the extrusion-based bioprinting of GelMA-gelatin bioink were used to investigate mouse GBM-macrophage interactions (Figure 5a).[185] For investigation of the crosstalk between macrophages and GBM cells, the brain portion was bioprinted with mouse macrophages, and the cavity was filled with mouse GBM cells (Figure 5b). Macrophage recruitment towards GBM cells and the polarization of macrophages were observed (Figure 5c). Cells expressed elevated expression of matrix remodeling markers, TAM-specific markers, and GBM-specific markers in 3D co-culture models compared to their 2D controls (Figure 5d–5f). Paracrine signaling was also investigated by co-culturing mini-brains fabricated with individual cell types. GBM cells cultured in the mini-brain system demonstrated enhanced mesenchymal features with expression of vimentin (Vim) and decreased expression level of E-cadherin (Cdh1). Several immunomodulatory and chemotherapeutic compounds that target either the proliferating GBM cells or the macrophages were evaluated with the mini-brain and demonstrated clinical relevance (Figure 5g).
Figure 5.
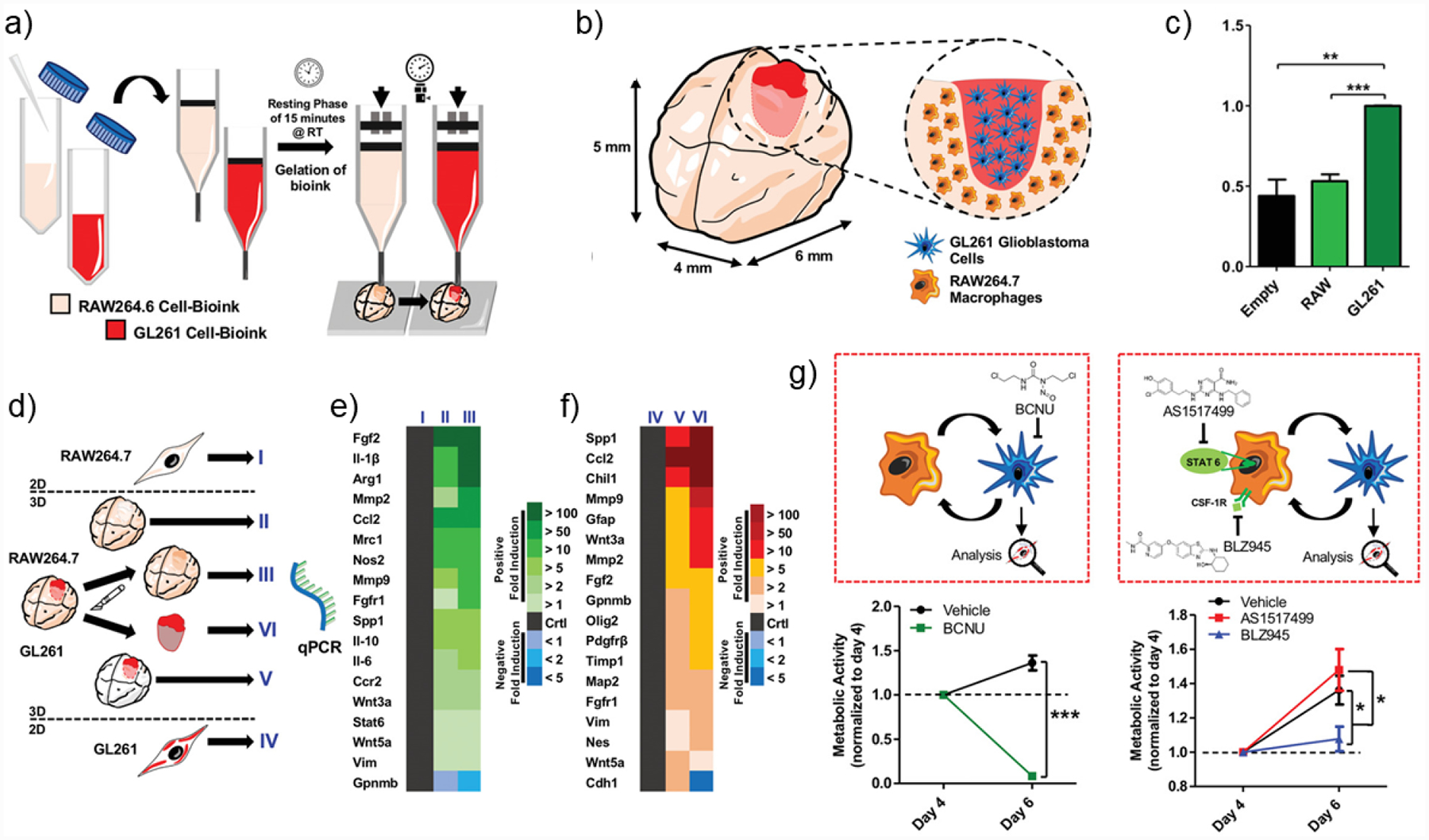
Schematic representation of the bioprinting process and features of a co-culture GBM model. a) Preparation of the GelMA-gelatin two bioinks encapsulated with RAW264.7 mouse macrophages GL261 mouse GBM cells, respectively. Co-culture model was fabricated by a two-step bioprinting process. b) Co-culture model and its cross-sectional view. The brain was bioprinted with RAW264.7, and the cavity was filled with GL261. c) Quantification for macrophage migration toward empty control, RAW (macrophages), and GL261 (GBM cells). **p < 0.01, ***p < 0.001. d) Schematic of the experimental groups for the co-culture model. e,f) Gene expression of RAW264.7 in (I) 2D culture, (II) 3D mono-culture, and (III) 3D co-culture, and of GL261 in (IV) 2D culture, (V) 3D mono-culture, and (VI) 3D co-culture model. g) Drug evaluation using the co-culture model. Schematic illustration of BCNU, AS1517499, or BLZ945 treatment to the co-culture GBM-macrophage model and measured metabolic activities of GL261 after 3D co-culture and treatment, respectively. Copyright Wiley, 2019.[185] Reproduced with permission.
To predict patient-specific treatment response, a GBM-on-a-chip model was developed by extrusion-based bioprinting using pig brain dECM (BdECM), patient-derived GBM cells, human umbilical vein endothelial cells (HUVECs), and silicone bioink.[186] A circular tumor core was encompassed by a ring of HUVECs, encapsulated in the BdECM hydrogel. To create a radial oxygen gradient, a layer of gas-permeable silicone ink was printed around the cellular parts, and a gas-impermeable glass covered the entire printed structures on the top so that oxygen can only reach the tumor core after passing through the silicone layer and the HUVEC layer. The BdECM preserved the majority of biochemical cues of the HA-rich brain ECM microenvironment and demonstrated superiority in enhancing cellular behaviors. GBM cells expressed higher levels of pro-angiogenic markers and increased proliferation and invasion. HUVECs expressed higher angiogenesis markers in the BdECM compared to their counterparts in the collagen hydrogel. A hypoxia gradient and an inversely correlated proliferation of tumor cells were observed in the co-culture model. Differential treatment resistance was predicted using the patient-specific 3D GBM-on-a-chip.
A co-axial extrusion bioprinting approach was used to produce a core-shell tube with alginate-gelatin bioinks as the shell and a fibrinogen core encapsulating GSCs and MSCs.[187] GSCs and MSCs spontaneously formed tumor fibers in a co-axial model, and the tumor fibers expressed high levels of Nestin, CD44, and Vim. In the control group where cells were directly mixed with the alginate hydrogel without a core-shell structure, GSCs and MSCs did not interact with each other spontaneously, indicating the role of ECM in modulating tumor-stromal interactions. The same co-axial printing method was used to produce a core-shell structure of GSCs and non-GSC tumor cells.[158] The shell was composed of alginate hydrogel with or without GSCs and the core with an established GBM cell line. The expression patterns of makers related to GBM invasion, including MMP2, MM9, VEGF2, were elevated in the GBM cell line co-cultured with GSCs. Drug resistance genes, including a regulator of TMZ resistance, O6-methylguanine-DNA methyltransferase (MGMT), were also enhanced in the cocultured GBM cells, indicating that the presence of GSCs could enhance the invasiveness and drug resistance of non stem-like tumor cells.
Co-culture 3D models are simplified versions of the native GBM microenvironment. Using brain dECM demonstrated that recent advances in biomaterials enable modeling of GBM not only with relevant cell types, but also with relevant ECM components, leading to improved biomimcry. The limitation of co-culture models is that cell types do not interact one-to-one in the native TME. However, these models offer the opportunity to interrogate specific niche interactions, such as the perivascular niche, immune niche, or stem cell niche, in isolation from other interactions. They can potentially uncover pathways underlying specific cellular interactions and be used to screen therapeutic compounds that target the stromal components of GBM in a biomimetic 3D context.
5.1.3. Multilineage 3D-bioprinted GBM models
Multilineage bioprinted 3D GBM models are GBM models composed of tumor cells and multiple stromal cell types with a defined 3D structural organization of proper biomaterials. Multilineage models are in vitro models that capture a high level of heterogeneity of GBM. Human multicellular GBM models recapitulating complex immune interactions and functional dependencies were developed using DLP-based bioprinting.[9] In addition to GSCs, stromal cells, including monocyte-/iPSC-derived macrophages or primary macrophages, astrocytes, and NPCs, were printed with spatial separation. A GMHA-GelMA matrix encapsulating tumor and stromal cells was designed to mimic the native tissue stiffness and HA content (Figure 6a). GSCs with macrophages (tetra-culture) or without macrophages (tri-culture) formed the tumor core and were encompassed by NPCs and astrocytes mimicking the brain parenchyma. GSCs in the tetra-culture recapitulated transcriptional profiles of clinical GBM tumor tissues better than the traditional sphere culture (Figure 6b). Correlating the gene expression profiles of GSCs in the tetra-culture with drug sensitivity data from the Cancer Therapeutic Response Platform (CTRP) and patient survival data in either The Cancer Genome Atlas (TCGA) or the Chinese Glioma Genome Atlas (CGGA) enabled predictions of drug sensitivity and patient prognosis. Macrophages promoted invasiveness, drug resistance, and hypoxic expression of the GSCs in the tetra-culture (Figure 6c and 6d). Undifferentiated monocytes, when bioprinted in the tetra-culture model, spontaneously polarized toward an M2 macrophage phenotype without extra external stimulations, indicating that stromal cells respond to the multicellular TME (Figure 6e). The tetra-culture model was used as a whole genome CRISPR-Cas9 screening platform to uncover novel functional dependencies and pathways. Individual gene knockout of several candidates indicated by the tetraculture model were validated in GSC sphere cultures, 3D-bioprinted models, and xenografts. Specifically, reduced cell viability in vitro and prolonged survival in xenografts were observed with knowout of the PAG1 gene (Figure 6f and 6g).
Figure 6.
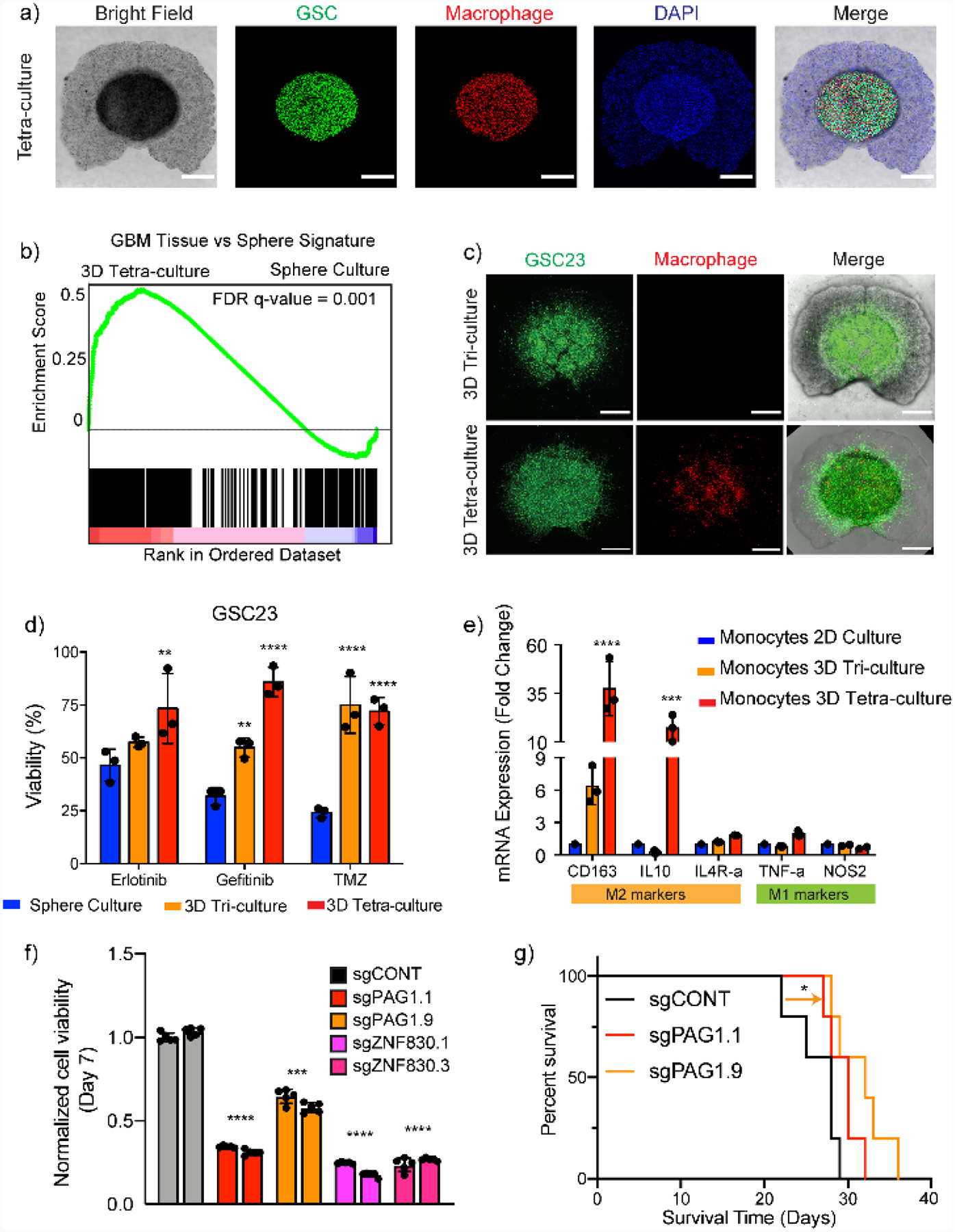
Schematic representation of the bioprinting process and features of a multi-cellular GBM model. a) GSCs, macrophages, astrocytes, and NPCs were bioprinted in an HA-GelMA hydrogel with spatial separations. b) GSCs in 3D-bioprinted model recapitulated transcriptional profiles of tumor tissue. c,d) 3D TME with macrophages promoted invasiveness and drug resistance of the GSCs. e) 3D TME differentially polarized monocytes to M2 macrophage phenotype. f,g) Novel functional dependencies indicated by 3D-bioprinted models were validated both in vitro and in animal models. Reproduced with permission. Copyright Springer Nature, 2020.[9] Reproduced with permission.
Another multi-nozzle extrusion-based bioprinting method was used to produce a 3D model with GBM cell lines or GSCs, patient-derived GBM-associated stromal cells (GASCs), and microglia in an alginate-based hydrogel.[188] The alginate was functionalized with RGDs, and in some groups with HA and collagen. The 3D multicellular models showed moderately enhanced resistance to TMZ and enhanced resistance to cisplatin, a compound that failed many clinical trials but showed promising results in 2D cultures, indicating potential application of this system for more reliable preclinical drug efficacy evaluations.
As more cellular components and biomaterials are integrated in the 3D-bioprinted models, isolating factors that drive tumor phenotypes becomes more complicated, but biomimicry improves with the increased heterogeneity. By enabling multiple cell-cell and cell-matrix interactions in the 3D models, it is possible to reproduce physiologically relevant tumor growth and invasion patterns, recapitulate native tumor transcriptomic profiles, suggest personalized treatment plans, and predict prognosis that parallel clinical outcomes. These highly biomimetic and heterogeneous models are promising in vitro platforms for reproducible, reliable, and high throughput drug screening and CRISPR screenings that interrogate functional dependencies in more clinically relevant settings.
5.2. 3D bioprinting for BBB modeling
A functional 3D BBB model should recapitulate critical BBB properties comparable to their physiological levels. Primary features of this brain vascular barrier include the tightness, the integrity, the selective permeability, and the transport mechanisms. Various assays developed for traditional in vitro BBB models can be deployed to evaluate the properties of 3D models.[189] Transepithelial/transendothelial electrical resistance (TEER) quantitatively measures the integrity of tight junctions in real time. Tightness and transport mechanisms can be measured by the expression of BBB-specific junction and transporter proteins, including: (1) tight junction proteins, such as claudins, occludins, zonula occludens-1 (ZO-1), ZO-2, and ZO-3; and (2) transporters, such as GLUT1, P-gp, BCRP, and MRP. Permeability can be assessed with fluorescent molecules with defined size such as dextrans and sodium fluoresceins.
A biohybrid microfluidic device fabricated with the two-photon lithography approach was seeded with mouse brain endothelial cells and GBM cells (Figure 7a).[190] Several tubular structures with 10 μm diameter, comparable to the brain microcapillaries, and 1 μm pores on the channel wall, were fabricated in parallel between the inlet and the outlet (Figure 7b and 7c). Numerical simulations demonstrated uniform and physiologically relevant flow rates in the microcapillaries (Figure 7d). Endothelial cells in the biohybrid system formed tight junctions and exhibited barrier properties, verified by the ZO-1 expression and the dextran diffusion (Figure 7e). TEER also increased in the presence of endothelial cells compared to the acellular device. Porous areas along the microvessels were significantly reduced after cell seeding.
Figure 7.
a) Microfluidic BBB system consisted of porous microcapillaries fabricated with TPP-based bioprinting. b) Scanning electron microscopy image of the microcapillaries with pores. c) Design of the microfluidic system with inlet, outlet, and multiple microcapillaries aligned in parallel. d) Numerical simulation of the fluid rate in the microfluidic device (half domain shown). e) Immunofluorescence staining against ZO-1 and f-actin of the microcapillaries. Copyright Wiley, 2018.[190] Reproduced with permission.
Another BBB device was assembled with three layers of 3D-printed chambers and a cell insert with iPSC-derived BMECs and astrocytes cultured on the two sides of a porous membrane to mimic the native architecture of BBB (Figure 8a and 8b).[25] A peek TEER of 4000 Ω·cm2 was measured on day 3 of coculturing, which was among the highest reported TEER values in in vitro models and within the range of in vivo values (Figure 8c). The permeability tested with dextrans of different molecular weights and small drug compounds correlated with clinical data (Figure 8d).
Figure 8.
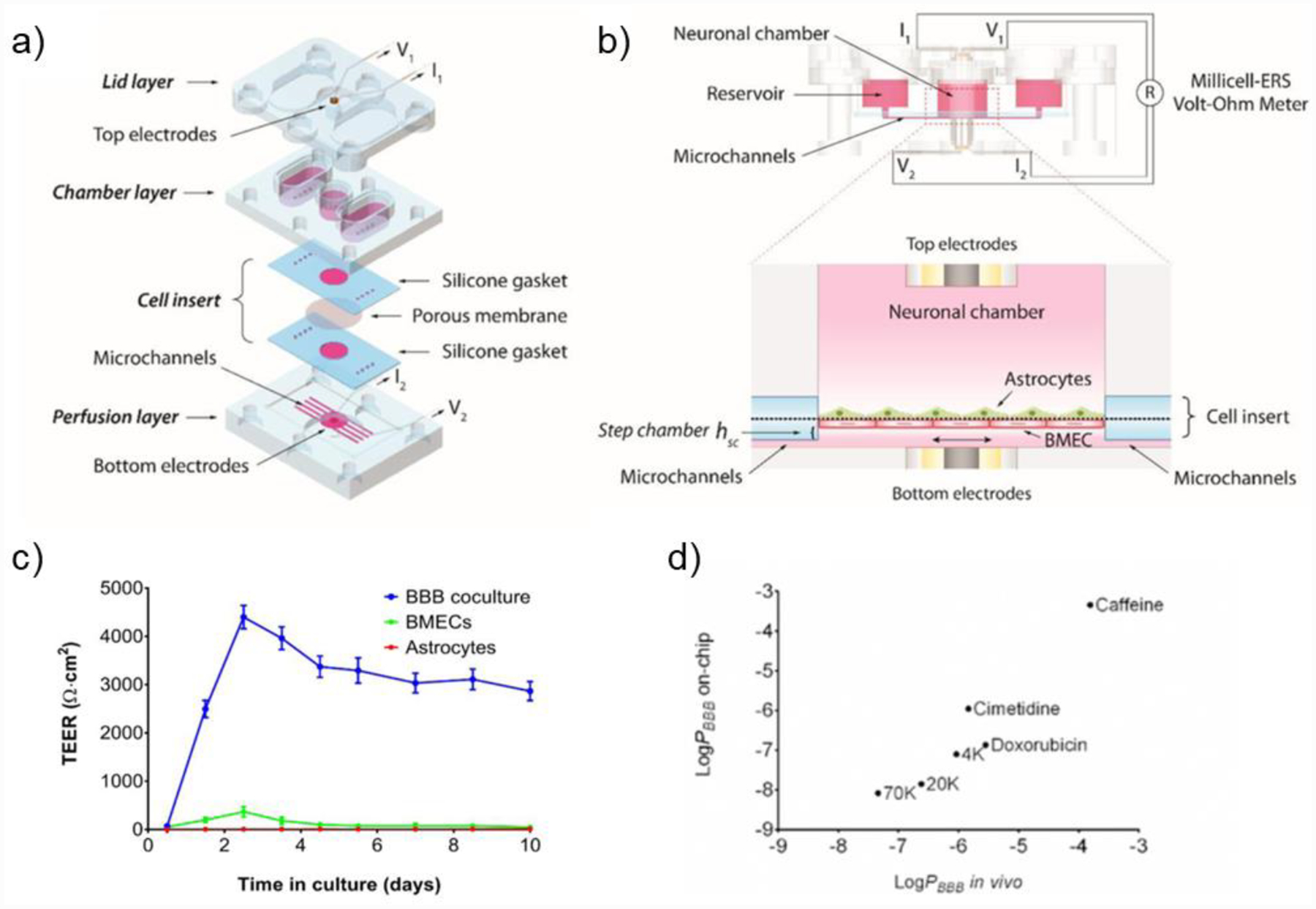
a) Schematic design of the microfluidic BBB-on-a-chip consisted of a set of cell insert and three 3D-printed parts: lid, chamber, and perfusion layer. b) Side view (top) and enlarged cross-sectional view (bottom) of the BBB system to show the arrangement of electrodes, cell layers, and fluid pathways. c) 3D microfluidic BBB demonstrated significantly higher TEER values by day 3 compared to BMEC or astrocyte monocultures, and the TEER remained high up to 10 days. d) BBB-on-a-chip demonstrated permeability to dextrans and drug molecules at various sizes consistent with in vivo data. Copyright Wiley, 2017.[25] Reproduced with permission.
A BBB model that combined ECs and microarrays made of collagen type I within an extrusion-printed frame demonstrated BBB barrier functions (Figure 9a–d).[191] The expression of tight junction protein ZO-1 increased by 2 weeks of culture (Figure 9e), and the transendothelial permeability was verified with no leakage of 40 kDa dextran from the vessels starting from one week of culture (Figure 9f). This model allowed a time-dependent observation of BBB maturation, indicated by tight junction formation and BBB disruption/recovery. System residence time was based on the actual blood residence time in the brain, enabling clinically relevant evaluation of the compound permeability if integrated with a GBM model. Current 3D bioprinting attempts for BBB mainly utilize the technology to microfluidic devices that more closely resembles the native anatomy. The advantage of 3D-bioprinted microfluidics over traditional microfluidic technology is that more complex geometries can be precisely and reproducibly produced by 3D printing, with reduced operating time and cost.[189]
Figure 9.
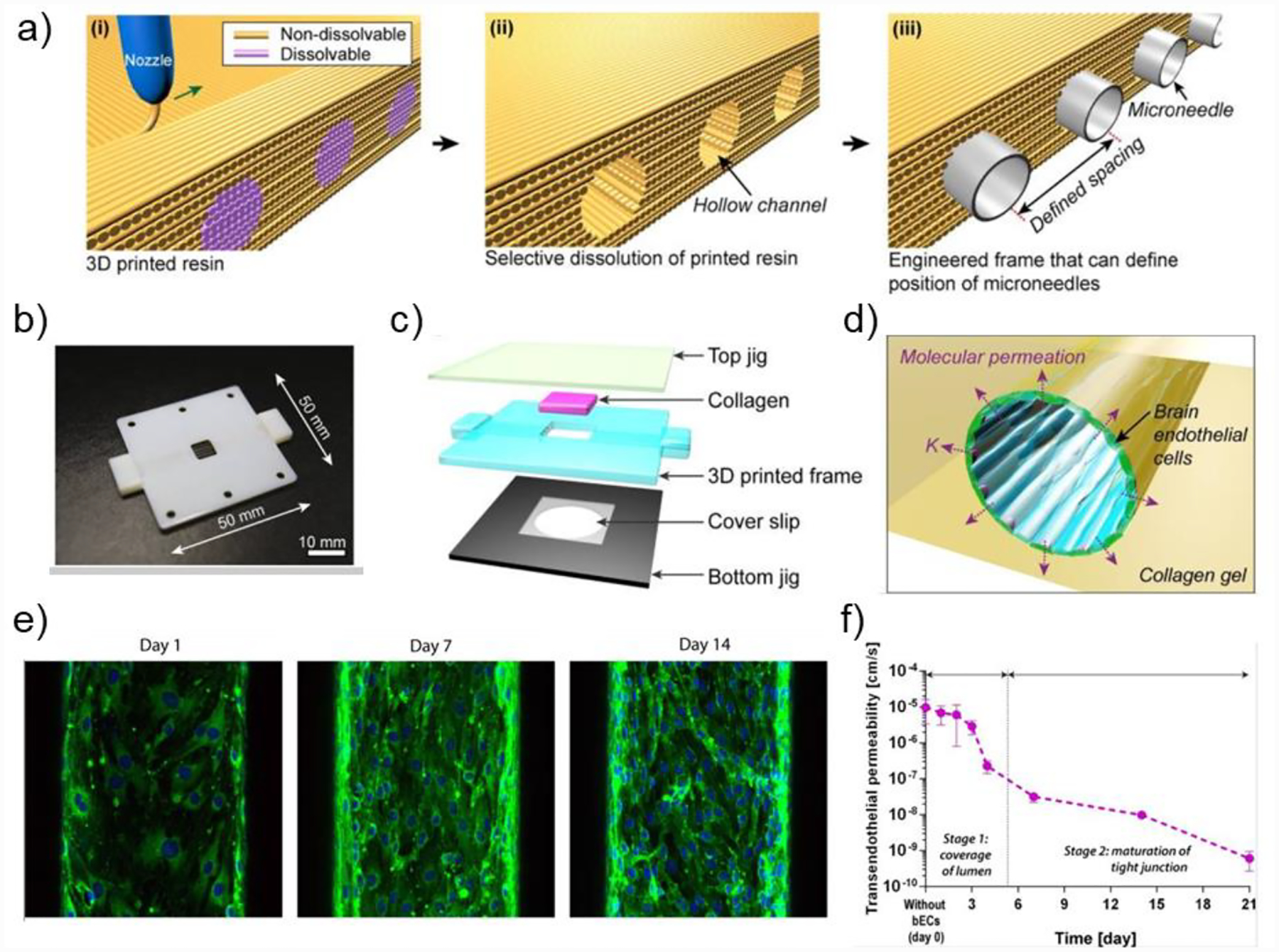
a) Schematic illustration of the 3D printing process to fabricate the main frame for BBB system. (i) Printing the frame with non-dissolvable resin and channels with dissolvable resin. (ii) Removal of the dissolvable resin. (iii) Microneedles positioned in the channels. b) A photograph of the printed frame with dimensions labled. c) Schematic llustration of the assembly of the BBB system. d) Conceptual illustration of the microvasculature features. e) Immunofluorescence stainining against ZO-1 on days 1, 7, and 14. f) The BBB system displayed consistent decrease of transendothelial permeability over 21 days after EC seeding. Copyrith AIP Publishing, 2015.[191] Reproduced with permission.
6. Conclusions
Treatment failure in GBM result from numerous factors, including high genetic heterogeneity of GBM microenvironment, fast progression and inherent drug resistance of GBM, and insufficient delivery of therapeutic agents to the GBM sites due to the barrier properties of the BBB. The currently stagnant drug development process for GBM could be improved by reducing the attrition rate of novel compounds during clinical trials and developing drugs or treatment plans specific to different GBM subtypes. The latter requires more profound understanding of the molecular mechanisms of the GBM subtypes. High attrition rate of drugs indicate that the current preclinical models are insufficient to provide clinically relevant evaluations. For in vivo GBM models, lack of species-matched cellular interactions reduces their validity in predicting therapeutic outcomes in clinical trials. For in vitro 3D GBM models, functional BBBs have not been reproducibly incorporated yet, and thus limiting their capacity to evaluate the penetration efficiency of compounds, which also impacts therapeutic outcomes. Advances in 3D bioprinting technologies and engineered biomaterials offer clinically relevant modeling capacity to develop integrative, biomimetic, and human-based model systems. These model systems potentially recapitulate species-matched and tissue-specific features, such as dimensionality, organization, cell-cell interactions, and cell-matrix interactions of their physiologic counterparts. 3D-bioprinted GBM models customized to recapitulate cellular and ECM microenvironments of patient tumors will help elucidate pathways involved in the GBM subtypes. Integrated GBM-BBB systems can potentially eliminate compounds that will fail the clinical trials but demonstrate success in static 2D cultures, stand-alone in vitro models, or animal models. Models incorporating the BBB and other stromal components of GBM into ECM-derived biomaterials will enable simultaneous evaluation of therapeutic efficacy of drugs to tumor cells, the efficiency of drug penetration across the BBB, as well as the drug toxicity to stromal cells within the tumor microenvironment. Moreover, an integrative model could recreate the non-homogeneous barrier properties of the BBB within and around the tumor tissue to mimic the native physiologic features, including compromised vessels near the necrotic tumor core and intact BBB near the invasive boundaries. The integrity of the BBB along the proliferative boundaries protects the highly invasive and stem-like GSCs from effective drug delivery. Characterization not applicable to current in vitro models but can potentially be considered with integrated models include: the tissue-to-blood ratio (TBR) that shows the delivery of a compound that reaches the tumor compared to the amount in blood, the brain efflux index (BEI) that shows how likely the drugs will be pumped back into the blood, and the penetration of compounds into different regions of the tumor after passing through the BBB.[192] Assessments based on an integrated GBM-BBB system will also empower optimization strategies to bypass the BBB and enhance delivery and efficacy of novel compounds. In conclusion, 3D-bioprinted models have great potentials to facilitate mechanistic studies and clinical applications to eventually accelerate GBM therapeutic advances.
Here, we provided a comprehensive review of current 3D-bioprinted GBM models and BBB models, covering biomaterials, biofabrication technologies, cell types, model features, and appropriate applications of each. Monoculture GBM models enable mechanistic studies and investigations of cell responses to the ECM and the dimensionality. Co-culture models allow the investigation of specific cellular interactions between tumor cells and certain stromal components and are good tools for evaluating therapies that regulate these stromal components or their related interactions. Multicellular GBM models capture the highest level of heterogeneity and biomimicry among the in vitro 3D-bioprinted models, thus possessing greater potential as drug screening platforms or for the interrogation of cellular dependencies that have enhanced clinical relevance. In addition to summarizing the recent progresses of 3D bioprinted GBM/BBB models, this review provided essential information for future design and implementation of in vitro 3D GBM and BBB models. The information includes the cellular and ECM compositions of the two native microenvironments, bioprinting methods that have demonstrated success in organotypic modeling, and relevant biomaterials for in vitro modeling of the brain, the GBM, and the BBB. Researchers in relevant fields may refer to this report to develop the most cost-efficient strategy that addresses their specific biological questions.
However, further advancements in bioink development and printing technologies are necessary to enable broader applications of bioprinting, despite its various advantages including versatility, precise control, biocompatibility, reproducibility, and high throughputness. Many 3D-bioprinted GBM models have been developed with alginate, gelatin, and GelMA hydrogels due to their good printibility, despite HA being the most abundant ECM component in the GBM microenvironment. It remains challenging for extrusion-based or inkjet-based bioprinting to print HA constructs with high resolution or structural integrity due to poor mechanical properties of HA. Encouragingly, DLP-based bioprinting has recently demonstrated success in fabricating an HA-rich multicellular GBM model, and many studies have showed that chemical modifications can improve the rheological properties of HA-based bioinks for printing. Development of novel bioinks or modification methods to existing biomaterials to improve their printability, including but not limited to the viscosity and the crosslinking mechanism, to accommodate bioprinting modalities will expand the material diversity for bioprinting and eventually enhance the material biomimicry of 3D models. For BBB modeling, 3D bioprinting improves the customizability and throughput of traditional microfluidic systems, and 3D-bioprinted BBB exhibits improved barrier properties. To date, the technology has mainly been used to facilitate device fabrication with cells seeded afterwards. While micron-scale structures, perfusable structures, and cell alignments have been achieved separately using bioprinting, the consolidation of these features are necessary for a successful cell-encapsulation printing of the BBB. In addition, proper molecular interventions with growth factors or small-molecule inhibitors that are often utilized in organoid development may also be introduced to post-printing cellular constructs to facilitate desired cellular activites, such as BBB tight junction formation.
Lastly, we believe that a benchmark, including the standardized data analysis and the evaluation of model properties, should be established for 3D-bioprinted models to ensure their clinical relevance and provide guidance for future model designs. 3D models are perceived as promising alternatives to traditional 2D models and animal models, with advantages including well-defined structures and compositions, shorter time frame of production, and species-matched modeling which provides more reliable pre-clinical data. Theoretically, to include as many components as possible and assemble them in a way comparable to the native physiology can generate a structurally similar construct to the original tissue. However, whether the structural resemblance gives rise to functional resemblance requires more strict functional evaluations. Both qualitative and quantitative standards, such as percentage of matching and correlation to the clinical data, should be established for functional parameters used to evaluate the validity and the extent of clinical relevance of individual model. Examples of functional parameters are genomic and transcriptional profiles, drug responses, and specific features of each individual tissue, such as barrier properties of the BBB and invasiveness or tumorigenesis capcity of GBM. With the collective data, it may be possible and beneficial for the research community to determine the minimal components and aspects that can reliably approximate the physiological environments, thus reducing the cost and time for building a highly complex in vitro model.
Acknowledgements
We thank our funding sources: CA253615 and EB021857 (to S.C.), CA197718, CA154130, CA169117, CA171652, NS087913, NS089272, and NS103434 (to J.N.R.). The authors thank Jiayi Wang for assistance on the digital illustration of tissue microenvironments.
Biographies
Author biographies and photographs

Dr. Shaochen Chen is a Professor and Chairman of the NanoEngineering Department and the founding director of the Biomaterials and Tissue Engineering Center at the University of California San Deigo (UCSD). Before joining UCSD, he was a professor and Pearlie D. Henderson Centennial Endowed Faculty Fellow in Engineering at the University of Texas at Austin. From 2008 to 2010, Dr. Chen served as the Program Director for the Nanomanufacturing Program in the US National Science Foundation. His research interests include 3D printing and bioprinting, biomaterials and nanomaterials, stem cells and tissue engineering.

Dr. Jeremy N. Rich, MD, MHS, MBA, is the director of Neuro-Oncology and Brain Tumor Institute, and a professor of the Department of Medicine at UCSD. Dr. Rich was the chair of the Department of Stem Cell Biology and Regenerative Medicine at Cleveland Clinic and co-director of the National Center for Regenerative Medicine at Case Western Reserve University. Dr. Rich seeks to identify novel therapies for malignant brain tumors, through the investigation of cancer stem cells and core signal transduction pathways amenable to pharmacologic targeting.

Min Tang is a graduate student in Dr. Shaochen Chen’s lab at UCSD. She obtained B.S. in Biomedical Engineering from the Washington University in St. Louis, with research in photoacoustic imaging with Dr. Lihong Wang and stem cell niche with Dr. Ting Xie. Min obtained M.S. in biomedical engineering, with research in tissue engineering with Dr. Kam Leong at the Columbia University. Her current research focuses on disease modeling and tissue engineering using 3D bioprinting.
Appendix. Glossary
| Abbreviation | Complete Term |
|---|---|
| 2D | Two-dimensional |
| 3D | Three-dimensional |
| ABC | ATP-binding cassette |
| BBB | Blood-brain barrier` |
| BdECM | Brain dECM |
| BM | Basement membrane |
| BMEC | Brain microvascular endothelial cell |
| CAD | Computer-aided design |
| CAL | Computed axial lithography |
| CGGA | Chinese Glioma Genome Atlas |
| CNS | Central nervous system |
| CSPG | Chondroitin sulfate proteoglycans |
| CT | Computed tomography |
| CTRP | Cancer Therapeutic Response Platform |
| DLP | Digital light processing |
| DMD | Digital micromirror device |
| ECM | Extracellular matrix |
| EMT | Epithelial-mesenchymal transition |
| GAF | Gelatin-alginate-fibrinogen |
| GBM | Glioblastoma |
| GelMA | Gelatin methacrylate |
| GSC | Glioblastoma stem cell |
| HA | Hyaluronic acid |
| HUVEC | Human umbilical vein endothelial cells |
| LAP | Lithium phenyl-2,4,6-trimethylbenzoylphosphinate |
| MGMT | O6-methylguanine-DNA methyltransferase |
| MMP | Matrix metalloprotease |
| MSC | Mesenchymal stem cell |
| NPC | Neural precursor cell |
| PA | Plasminogen activator |
| PDX | Patient-derived xenograft |
| PEG | Poly(ethylene glycol) |
| PEGDA | Poly(ethylene glycol) diacrylate |
| PLGA | Poly(lactic-co-glycolic acid) |
| PNIPAAm | Poly(N-isopropylacrylamide) |
| PU | Polyurethane |
| PVN | Perivascular niche |
| RGD | Arg-Gly-Asp |
| RHAMM | Receptor for hyaluronan mediated motility |
| SAP | Self-assembled peptide |
| SF | Silk fibroin |
| SPARC | Secreted protein acidic and rich in cysteine |
| TAM | Tumor-associated macrophage |
| TCGA | The cancer genome atlas |
| TME | Tumor microenvironment |
| TMZ | Temozolomide |
| TN-C | Tenascin-C |
| TN-R | Tenascin-R |
| TPP | Two-photon polymerization |
| TSP-1 | Thrombospondin-1 |
| VEGF | Vascular endothelial growth factor |
Footnotes
Conflict of Interests
The authors declare no conflict of interest.
Contributor Information
Min Tang, Department of NanoEngineering, University of California San Diego, La Jolla, CA, 92093, USA.
Jeremy N. Rich, Division of Regenerative Medicine, Department of Medicine, Department of Neurosciences, University of California San Diego, La Jolla, CA, 92093, USA Sanford Consortium for Regenerative Medicine, La Jolla, CA, 92093, USA.
Shaochen Chen, Department of NanoEngineering, University of California San Diego, La Jolla, CA, 92093, USA; Department of Bioengineering, Materials Science and Engineering Program, Chemical Engineering Program, University of California San Diego, La Jolla, CA, 92093, USA.
References
- [1].Ostrom QT, Cioffi G, Gittleman H, Patil N, Waite K, Kruchko C, Barnholtz-Sloan JS, Neuro-oncology 2019, 21, v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Koshy M, Villano JL, Dolecek TA, Howard A, Mahmood U, Chmura SJ, Weichselbaum RR, McCarthy BJ, J Neurooncol 2012, 107, 207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Sarkaria JN, Hu LS, Parney IF, Pafundi DH, Brinkmann DH, Laack NN, Giannini C, Burns TC, Kizilbash SH, Laramy JK, Swanson KR, Kaufmann TJ, Brown PD, Agar NYR, Galanis E, Buckner JC, Elmquist WF, Neuro-Oncology 2018, 20, 184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Bhowmik A, Khan R, Ghosh MK, “Blood Brain Barrier: A Challenge for Effectual Therapy of Brain Tumors,” DOI https://doi.org/10.1155/2015/320941 can be found under https://doi.org/10.1155/2015/320941https://www.hindawi.com/journals/bmri/2015/320941/ can be found under https://www.hindawi.com/journals/bmri/2015/320941/, 2015. [DOI] [PMC free article] [PubMed]
- [5].Zhao X, Chen R, Liu M, Feng J, Chen J, Hu K, Acta Pharm Sin B 2017, 7, 541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].van Tellingen O, Yetkin-Arik B, de Gooijer MC, Wesseling P, Wurdinger T, de Vries HE, Drug Resistance Updates 2015, 19, 1. [DOI] [PubMed] [Google Scholar]
- [7].Harder BG, Blomquist MR, Wang J, Kim AJ, Woodworth GF, Winkles JA, Loftus JC, Tran NL, Front Oncol 2018, 8, DOI 10.3389/fonc.2018.00462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Hidalgo M, Amant F, Biankin AV, Budinska E, Byrne AT, Caldas C, Clarke RB, de Jong S, Jonkers J, Maelandsmo GM, Roman-Roman S, Seoane J, Trusolino L, Villanueva A, for the EurOPDX Consortium, Cancer Discovery 2014, 4, 998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Tang M, Xie Q, Gimple RC, Zhong Z, Tam T, Tian J, Kidwell RL, Wu Q, Prager BC, Qiu Z, Yu A, Zhu Z, Mesci P, Jing H, Schimelman J, Wang P, Lee D, Lorenzini MH, Dixit D, Zhao L, Bhargava S, Miller TE, Wan X, Tang J, Sun B, Cravatt BF, Muotri AR, Chen S, Rich JN, Cell Res 2020, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Curet MA, Watters JJ, J. Neurooncol 2018, 137, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Stone NL, England TJ, O’Sullivan SE, Front. Cell. Neurosci 2019, 13, DOI 10.3389/fncel.2019.00230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Prolo LM, Li A, Owen SF, Parker JJ, Foshay K, Nitta RT, Morgens DW, Bolin S, Wilson CM, Vega LJCM, Luo EJ, Nwagbo G, Waziri A, Li G, Reimer RJ, Bassik MC, Grant GA, Sci Rep 2019, 9, 14020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Merolle M, Mongiardi MP, Piras M, Levi A, Falchetti ML, Int J Mol Sci 2020, 21, DOI 10.3390/ijms21041490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Farrell K, Mahajan G, Srinivasan P, Lee M-Y, Kothapalli CR, Exp. Cell Res 2018, 362, 159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Ruck T, Bittner S, Meuth SG, Neural Regen Res 2015, 10, 889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Wang C, Li J, Sinha S, Peterson A, Grant GA, Yang F, Biomaterials 2019, 202, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Hubert CG, Rivera M, Spangler LC, Wu Q, Mack SC, Prager BC, Couce M, McLendon RE, Sloan AE, Rich JN, Cancer Res. 2016, 76, 2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Jacob F, Salinas RD, Zhang DY, Nguyen PTT, Schnoll JG, Wong SZH, Thokala R, Sheikh S, Saxena D, Prokop S, Liu D, Qian X, Petrov D, Lucas T, Chen HI, Dorsey JF, Christian KM, Binder ZA, Nasrallah M, Brem S, O’Rourke DM, Ming G, Song H, Cell 2020, 180, 188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Murphy SV, Atala A, Nature Biotechnology 2014, 32, 773. [DOI] [PubMed] [Google Scholar]
- [20].Malda J, Visser J, Melchels FP, Jüngst T, Hennink WE, Dhert WJA, Groll J, Hutmacher DW, Advanced Materials 2013, 25, 5011. [DOI] [PubMed] [Google Scholar]
- [21].Gebinoga M, Katzmann J, Fernekorn U, Hampl J, Weise F, Klett M, Läffert A, Klar TA, Schober A, Engineering in Life Sciences 2013, 13, 368. [Google Scholar]
- [22].Zhu W, Qu X, Zhu J, Ma X, Patel S, Liu J, Wang P, Lai CSE, Gou M, Xu Y, Zhang K, Chen S, Biomaterials 2017, 124, 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Ma X, Qu X, Zhu W, Li Y-S, Yuan S, Zhang H, Liu J, Wang P, Lai CSE, Zanella F, Feng G-S, Sheikh F, Chien S, Chen S, Proc. Natl. Acad. Sci. U.S.A 2016, 113, 2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Hosseinzadeh R, Mirani B, Pagan E, Mirzaaghaei S, Nasimian A, Kawalec P, da S. Rosa SC, Hamdi D, Fernandez NP, Toyota BD, Gordon JW, Ghavami S, Akbari M, Advanced Therapeutics 2019, 2, 1900113. [Google Scholar]
- [25].Wang YI, Abaci HE, Shuler ML, Biotechnol Bioeng 2017, 114, 184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Quail DF, Joyce JA, Cancer Cell 2017, 31, 326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Zhao Z, Nelson AR, Betsholtz C, Zlokovic BV, Cell 2015, 163, 1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Charles NA, Holland EC, Gilbertson R, Glass R, Kettenmann H, Glia 2011, 59, 1169. [DOI] [PubMed] [Google Scholar]
- [29].Matias D, Balça-Silva J, da Graça GC, Wanjiru CM, Macharia LW, Nascimento CP, Roque NR, Coelho-Aguiar JM, Pereira CM, Dos Santos MF, Pessoa LS, Lima FRS, Schanaider A, Ferrer VP, de S. e Spohr TCL, Moura-Neto V, Front. Cell. Neurosci 2018, 12, DOI 10.3389/fncel.2018.00235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Brandao M, Simon T, Critchley G, Giamas G, Glia 2019, 67, 779. [DOI] [PubMed] [Google Scholar]
- [31].Rath BH, Fair JM, Jamal M, Camphausen K, Tofilon PJ, PLOS ONE 2013, 8, e54752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Henrik Heiland D, Ravi VM, Behringer SP, Frenking JH, Wurm J, Joseph K, Garrelfs NWC, Strähle J, Heynckes S, Grauvogel J, Franco P, Mader I, Schneider M, Potthoff A-L, Delev D, Hofmann UG, Fung C, Beck J, Sankowski R, Prinz M, Schnell O, Nat Commun 2019, 10, 2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Venkataramani V, Tanev DI, Strahle C, Studier-Fischer A, Fankhauser L, Kessler T, Körber C, Kardorff M, Ratliff M, Xie R, Horstmann H, Messer M, Paik SP, Knabbe J, Sahm F, Kurz FT, Acikgöz AA, Herrmannsdörfer F, Agarwal A, Bergles DE, Chalmers A, Miletic H, Turcan S, Mawrin C, Hänggi D, Liu H-K, Wick W, Winkler F, Kuner T, Nature 2019, 573, 532. [DOI] [PubMed] [Google Scholar]
- [34].Hossain A, Gumin J, Gao F, Figueroa J, Shinojima N, Takezaki T, Priebe W, Villarreal D, Kang S-G, Joyce C, Sulman E, Wang Q, Marini FC, Andreeff M, Colman H, Lang FF, Stem Cells 2015, 33, 2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Figueroa J, Phillips LM, Shahar T, Hossain A, Gumin J, Kim H, Bean AJ, Calin GA, Fueyo J, Walters ET, Kalluri R, Verhaak RG, Lang FF, Cancer Res. 2017, 77, 5808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, Dewhirst MW, Bigner DD, Rich JN, Nature 2006, 444, 756. [DOI] [PubMed] [Google Scholar]
- [37].Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkelman RM, Cusimano MD, Dirks PB, Nature 2004, 432, 396. [DOI] [PubMed] [Google Scholar]
- [38].Chen J-WE, Pedron S, Shyu P, Hu Y, Sarkaria JN, Harley BAC, Front. Mater 2018, 5, DOI 10.3389/fmats.2018.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Bellail AC, Hunter SB, Brat DJ, Tan C, Van Meir EG, Int. J. Biochem. Cell Biol 2004, 36, 1046. [DOI] [PubMed] [Google Scholar]
- [40].Daneman R, Zhou L, Kebede AA, Barres BA, Nature 2010, 468, 562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Rubin LL, Staddon JM, Annu. Rev. Neurosci 1999, 22, 11. [DOI] [PubMed] [Google Scholar]
- [42].Bush TG, Puvanachandra N, Horner CH, Polito A, Ostenfeld T, Svendsen CN, Mucke L, Johnson MH, Sofroniew MV, Neuron 1999, 23, 297. [DOI] [PubMed] [Google Scholar]
- [43].Price L, Wilson C, Grant G, in Translational Research in Traumatic Brain Injury (Eds.: Laskowitz D, Grant G), CRC Press/Taylor And Francis Group, Boca Raton (FL), 2016. [PubMed] [Google Scholar]
- [44].Chow BW, Gu C, Trends Neurosci 2015, 38, 598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Gottesman MM, Fojo T, Bates SE, Nature Reviews Cancer 2002, 2, 48. [DOI] [PubMed] [Google Scholar]
- [46].Dallas S, Miller DS, Bendayan R, Pharmacol. Rev 2006, 58, 140. [DOI] [PubMed] [Google Scholar]
- [47].Sweeney MD, Ayyadurai S, Zlokovic BV, Nat Neurosci 2016, 19, 771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Winkler EA, Bell RD, Zlokovic BV, Nat. Neurosci 2011, 14, 1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Nuriya M, Shinotsuka T, Yasui M, Cereb Cortex 2013, 23, 2118. [DOI] [PubMed] [Google Scholar]
- [50].Nishijima T, Piriz J, Duflot S, Fernandez AM, Gaitan G, Gomez-Pinedo U, Verdugo JMG, Leroy F, Soya H, Nuñez A, Torres-Aleman I, Neuron 2010, 67, 834. [DOI] [PubMed] [Google Scholar]
- [51].Villabona-Rueda A, Erice C, Pardo CA, Stins MF, Front Cell Neurosci 2019, 13, DOI 10.3389/fncel.2019.00405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Haruwaka K, Ikegami A, Tachibana Y, Ohno N, Konishi H, Hashimoto A, Matsumoto M, Kato D, Ono R, Kiyama H, Moorhouse AJ, Nabekura J, Wake H, Nature Communications 2019, 10, 5816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Shigemoto-Mogami Y, Hoshikawa K, Sato K, Front Cell Neurosci 2018, 12, DOI 10.3389/fncel.2018.00494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Groothuis DR, Vriesendorp FJ, Kupfer B, Warnke PC, Lapin GD, Kuruvilla A, Vick NA, Mikhael MA, Patlak CS, Ann. Neurol 1991, 30, 581. [DOI] [PubMed] [Google Scholar]
- [55].Cragg B, Neuroscience Letters 1979, 15, 301. [DOI] [PubMed] [Google Scholar]
- [56].Yamaguchi Y, CMLS, Cell. Mol. Life Sci 2000, 57, 276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Bonneh-Barkay D, Wiley CA, Brain Pathology 2009, 19, 573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Eroglu C, Cell Commun J. Signal. 2009, 3, 167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Jones CL, Liu J, Xu D, in Comprehensive Natural Products II, Elsevier, 2010, pp. 407–427. [Google Scholar]
- [60].Bignami A, Hosley M, Dahl D, Anat Embryol 1993, 188, DOI 10.1007/BF00190136. [DOI] [PubMed] [Google Scholar]
- [61].Engler AJ, Sen S, Sweeney HL, Discher DE, Cell 2006, 126, 677. [DOI] [PubMed] [Google Scholar]
- [62].Wang T-W, Spector M, Acta Biomaterialia 2009, 5, 2371. [DOI] [PubMed] [Google Scholar]
- [63].Cowman MK, Lee H-G, Schwertfeger KL, McCarthy JB, Turley EA, Front Immunol 2015, 6, DOI 10.3389/fimmu.2015.00261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Lau LW, Cua R, Keough MB, Haylock-Jacobs S, Yong VW, Nature Reviews Neuroscience 2013, 14, 722. [DOI] [PubMed] [Google Scholar]
- [65].Faissner A, Cell Tissue Res 1997, 290, 331. [DOI] [PubMed] [Google Scholar]
- [66].Dawson DW, Pearce SFA, Zhong R, Silverstein RL, Frazier WA, Bouck NP, J Cell Biol 1997, 138, 707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Gilg AG, Tye SL, Tolliver LB, Wheeler WG, Visconti RP, Duncan JD, Kostova FV, Bolds LN, Toole BP, Maria BL, Clin. Cancer Res 2008, 14, 1804. [DOI] [PubMed] [Google Scholar]
- [68].Zhang B, Du Y, He Y, Liu Y, Zhang G, Yang C, Gao F, Cancer Immunol. Immunother 2019, 68, 189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Dwyer CA, Bi WL, Viapiano MS, Matthews RT, J Neurooncol 2014, 120, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Theocharis AD, Skandalis SS, Gialeli C, Karamanos NK, Advanced Drug Delivery Reviews 2016, 97, 4. [DOI] [PubMed] [Google Scholar]
- [71].Arslan F, Bosserhoff A-K, Nickl-Jockschat T, Doerfelt A, Bogdahn U, Hau P, Br J Cancer 2007, 96, 1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Wolf KJ, Chen J, Coombes JD, Aghi MK, Kumar S, Nat Rev Mater 2019, 4, 651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Brösicke N, Faissner A, Cell Adhesion & Migration 2015, 9, 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Shevde LA, Samant RS, Matrix Biology 2014, 37, 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Gladson CL, Cheresh DA, J. Clin. Invest 1991, 88, 1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Serres E, Debarbieux F, Stanchi F, Maggiorella L, Grall D, Turchi L, Burel-Vandenbos F, Figarella-Branger D, Virolle T, Rougon G, Van Obberghen-Schilling E, Oncogene 2014, 33, 3451. [DOI] [PubMed] [Google Scholar]
- [77].Polacheck WJ, Zervantonakis IK, Kamm RD, Cell. Mol. Life Sci 2013, 70, 1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Wang C, Tong X, Yang F, Mol. Pharmaceutics 2014, 11, 2115. [DOI] [PubMed] [Google Scholar]
- [79].Chauvet D, Imbault M, Capelle L, Demene C, Mossad M, Karachi C, Boch A-L, Gennisson J-L, Tanter M, Ultraschall in Med 2015, 37, 584. [DOI] [PubMed] [Google Scholar]
- [80].Netti PA, Baxter LT, Boucher Y, Skalak R, Jain RK, Cancer Res. 1995, 55, 5451. [PubMed] [Google Scholar]
- [81].Xu L, Nirwane A, Yao Y, Stroke Vasc Neurol 2019, 4, 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Willis CM, Crocker SJ, Composition and Function of the Extracellular Matrix in the Human Body 2016, DOI 10.5772/62706. [DOI] [Google Scholar]
- [83].Lathia JD, Li M, Hall PE, Gallagher J, Hale JS, Wu Q, Venere M, Levy E, Rani MRS, Huang P, Bae E, Selfridge J, Cheng L, Guvenc H, McLendon RE, Nakano I, Sloan AE, Phillips HS, Lai A, Gladson CL, Bredel M, Bao S, Hjelmeland AB, Rich JN, Annals of Neurology 2012, 72, 766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Rosenberg GA, Clinical Science 2017, 131, 425. [DOI] [PubMed] [Google Scholar]
- [85].Milner R, Crocker SJ, Hung S, Wang X, Frausto RF, del Zoppo GJ, J. Immunol 2007, 178, 8158. [DOI] [PubMed] [Google Scholar]
- [86].Tasoglu S, Demirci U, Trends in Biotechnology 2013, 31, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Boland T, Xu T, Damon B, Cui X, Biotechnology Journal 2006, 1, 910. [DOI] [PubMed] [Google Scholar]
- [88].Derakhshanfar S, Mbeleck R, Xu K, Zhang X, Zhong W, Xing M, Bioact Mater 2018, 3, 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Ma X, Liu J, Zhu W, Tang M, Lawrence N, Yu C, Gou M, Chen S, Advanced Drug Delivery Reviews 2018, 132, 235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Gudapati H, Dey M, Ozbolat I, Biomaterials 2016, 102, 20. [DOI] [PubMed] [Google Scholar]
- [91].Xu T, Jin J, Gregory C, Hickman JJ, Boland T, Biomaterials 2005, 26, 93. [DOI] [PubMed] [Google Scholar]
- [92].Hölzl K, Lin S, Tytgat L, Vlierberghe SV, Gu L, Ovsianikov A, Biofabrication 2016, 8, 032002. [DOI] [PubMed] [Google Scholar]
- [93].Park T-M, Kang D, Jang I, Yun W-S, Shim J-H, Jeong YH, Kwak J-Y, Yoon S, Jin S, Int J Mol Sci 2017, 18, DOI 10.3390/ijms18112348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Xu T, Zhao W, Zhu J-M, Albanna MZ, Yoo JJ, Atala A, Biomaterials 2013, 34, 130. [DOI] [PubMed] [Google Scholar]
- [95].Sun Y, Song W, Sun X, Zhang S, ACS Appl. Mater. Interfaces 2018, 10, 31054. [DOI] [PubMed] [Google Scholar]
- [96].Roth EA, Xu T, Das M, Gregory C, Hickman JJ, Boland T, Biomaterials 2004, 25, 3707. [DOI] [PubMed] [Google Scholar]
- [97].Cooper GM, Miller ED, DeCesare GE, Usas A, Lensie EL, Bykowski MR, Huard J, Weiss LE, Losee JE, Campbell PG, Tissue Engineering Part A 2010, 16, 1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Xu F, Celli J, Rizvi I, Moon S, Hasan T, Demirci U, Biotechnology Journal 2011, 6, 204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Suntivich R, Drachuk I, Calabrese R, Kaplan DL, Tsukruk VV, Biomacromolecules 2014, 15, 1428. [DOI] [PubMed] [Google Scholar]
- [100].Mironov V, Visconti RP, Kasyanov V, Forgacs G, Drake CJ, Markwald RR, Biomaterials 2009, 30, 2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Chang CC, Boland ED, Williams SK, Hoying JB, J. Biomed. Mater. Res 2011, 98B, 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].De Coppi P, Bartsch G, Siddiqui MM, Xu T, Santos CC, Perin L, Mostoslavsky G, Serre AC, Snyder EY, Yoo JJ, Furth ME, Soker S, Atala A, Nat. Biotechnol 2007, 25, 100. [DOI] [PubMed] [Google Scholar]
- [103].Ozbolat IT, Hospodiuk M, Biomaterials 2016, 76, 321. [DOI] [PubMed] [Google Scholar]
- [104].Pereira RF, Bártolo PJ, Journal of Applied Polymer Science 2015, 132, DOI 10.1002/app.42458. [DOI] [Google Scholar]
- [105].Raman R, Bashir R, in Essentials of 3D Biofabrication and Translation (Eds.: Atala A, Yoo JJ), Academic Press, Boston, 2015, pp. 89–121. [Google Scholar]
- [106].Guillotin B, Souquet A, Catros S, Duocastella M, Pippenger B, Bellance S, Bareille R, Rémy M, Bordenave L, Amédée J, Guillemot F, Biomaterials 2010, 31, 7250. [DOI] [PubMed] [Google Scholar]
- [107].Barron JA, Ringeisen BR, Kim H, Spargo BJ, Chrisey DB, Thin Solid Films 2004, 453–454, 383. [Google Scholar]
- [108].Hribar KC, Soman P, Warner J, Chung P, Chen S, Lab Chip 2013, 14, 268. [DOI] [PubMed] [Google Scholar]
- [109].Knowlton S, Onal S, Yu CH, Zhao JJ, Tasoglu S, Trends in Biotechnology 2015, 33, 504. [DOI] [PubMed] [Google Scholar]
- [110].Kelly BE, Bhattacharya I, Heidari H, Shusteff M, Spadaccini CM, Taylor HK, Science 2019, 363, 1075. [DOI] [PubMed] [Google Scholar]
- [111].Zhang W, Chen S, MRS Bulletin 2011, 36, 1028. [Google Scholar]
- [112].Ma X, Dewan S, Liu J, Tang M, Miller KL, Yu C, Lawrence N, McCulloch AD, Chen S, Acta Biomater 2019, 95, 319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Ma X, Yu C, Wang P, Xu W, Wan X, Lai CSE, Liu J, Koroleva-Maharajh A, Chen S, Biomaterials 2018, 185, 310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Soman P, Kelber JA, Lee JW, Wright TN, Vecchio KS, Klemke RL, Chen S, Biomaterials 2012, 33, 7064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Kim SH, Yeon YK, Lee JM, Chao JR, Lee YJ, Seo YB, Sultan MT, Lee OJ, Lee JS, Yoon S-I, Hong I-S, Khang G, Lee SJ, Yoo JJ, Park CH, Nat Commun 2018, 9, 1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Wang P, Berry D, Moran A, He F, Tam T, Chen L, Chen S, Adv. Healthcare Mater 2019, 1900977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Koffler J, Zhu W, Qu X, Platoshyn O, Dulin JN, Brock J, Graham L, Lu P, Sakamoto J, Marsala M, Chen S, Tuszynski MH, Nat Med 2019, 25, 263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Qu X, Zhu W, Huang S, Li JY-S, Chien S, Zhang K, Chen S, Biomaterials 2013, 34, 9812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Huang TQ, Qu X, Liu J, Chen S, Biomed Microdevices 2014, 16, 127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Perkins KL, Arranz AM, Yamaguchi Y, Hrabetova S, Rev Neurosci 2017, 28, 869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Burdick JA, Prestwich GD, Advanced Materials 2011, 23, H41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Herrera-Perez RM, Voytik-Harbin SL, Sarkaria JN, Pollok KE, Fishel ML, Rickus JL, PLoS ONE 2018, 13, e0194183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Florczyk SJ, Wang K, Jana S, Wood DL, Sytsma SK, Sham J, Kievit FM, Zhang M, Biomaterials 2013, 34, 10143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Arulmoli J, Wright HJ, Phan DTT, Sheth U, Que RA, Botten GA, Keating M, Botvinick EL, Pathak MM, Zarembinski TI, Yanni DS, Razorenova OV, Hughes CCW, Flanagan LA, Acta Biomaterialia 2016, 43, 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Ananthanarayanan B, Kim Y, Kumar S, Biomaterials 2011, 32, 7913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Shu XZ, Liu Y, Palumbo F, Prestwich GD, Biomaterials 2003, 24, 3825. [DOI] [PubMed] [Google Scholar]
- [127].Ngo MT, Harley BAC, Biomaterials 2019, 198, 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Wang X, Ao Q, Tian X, Fan J, Tong H, Hou W, Bai S, Polymers 2017, 9, 401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Wang X, Yan Y, Pan Y, Xiong Z, Liu H, Cheng J, Liu F, Lin F, Wu R, Zhang R, Lu Q, Tissue Eng. 2006, 12, 83. [DOI] [PubMed] [Google Scholar]
- [130].Hsieh C-T, Hsu S-H, ACS Appl Mater Interfaces 2019, 11, 32746. [DOI] [PubMed] [Google Scholar]
- [131].Van Den Bulcke AI, Bogdanov B, De Rooze N, Schacht EH, Cornelissen M, Berghmans H, Biomacromolecules 2000, 1, 31. [DOI] [PubMed] [Google Scholar]
- [132].Chen J-WE, Pedron S, Harley BAC, Macromol Biosci 2017, 17, DOI 10.1002/mabi.201700018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Pedron S, Becka E, Harley BA, Advanced Materials 2015, 27, 1567. [DOI] [PubMed] [Google Scholar]
- [134].Cui H, Liu C, Esworthy T, Huang Y, Yu Z, Zhou X, San H, Lee S, Hann SY, Boehm M, Mohiuddin M, Fisher JP, Zhang LG, Science Advances 2020, 6, eabb5067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Rao SS, DeJesus J, Short AR, Otero JJ, Sarkar A, Winter JO, ACS Appl Mater Interfaces 2013, 5, 9276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Bonnesœur S, Morin-Grognet S, Thoumire O, Le Cerf D, Boyer O, Vannier J-P, Labat B, J Biomed Mater Res A 2020, DOI 10.1002/jbm.a.36899. [DOI] [PubMed] [Google Scholar]
- [137].Mazzocchi A, Devarasetty M, Huntwork R, Soker S, Skardal A, Biofabrication 2018, 11, 015003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Diamantides N, Wang L, Pruiksma T, Siemiatkoski J, Dugopolski C, Shortkroff S, Kennedy S, Bonassar LJ, Biofabrication 2017, 9, 034102. [DOI] [PubMed] [Google Scholar]
- [139].Tirella A, Liberto T, Ahluwalia A, Materials Letters 2012, 74, 58. [Google Scholar]
- [140].Lee A, Hudson AR, Shiwarski DJ, Tashman JW, Hinton TJ, Yerneni S, Bliley JM, Campbell PG, Feinberg AW, Science 2019, 365, 482. [DOI] [PubMed] [Google Scholar]
- [141].Moncal KK, Ozbolat V, Datta P, Heo DN, Ozbolat IT, J Mater Sci Mater Med 2019, 30, 55. [DOI] [PubMed] [Google Scholar]
- [142].Koh I, Cha J, Park J, Choi J, Kang S-G, Kim P, Sci Rep 2018, 8, 4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Yu C, Ma X, Zhu W, Wang P, Miller KL, Stupin J, Koroleva-Maharajh A, Hairabedian A, Chen S, Biomaterials 2019, 194, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Toprakhisar B, Nadernezhad A, Bakirci E, Khani N, Skvortsov GA, Koc B, Macromol Biosci 2018, 18, e1800024. [DOI] [PubMed] [Google Scholar]
- [145].Kim BS, Kim H, Gao G, Jang J, Cho D-W, Biofabrication 2017, 9, 034104. [DOI] [PubMed] [Google Scholar]
- [146].Nam SY, Park S-H, Adv. Exp. Med. Biol 2018, 1064, 335. [DOI] [PubMed] [Google Scholar]
- [147].Lee H, Han W, Kim H, Ha D-H, Jang J, Kim BS, Cho D-W, Biomacromolecules 2017, 18, 1229. [DOI] [PubMed] [Google Scholar]
- [148].Xiao W, Sohrabi A, Seidlits SK, Future Science OA 2017, 3, FSO189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Fan R, Piou M, Darling E, Cormier D, Sun J, Wan J, J Biomater Appl 2016, 31, 684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Pascoal JF, Fernandes TG, Nierode GJ, Diogo MM, Dordick JS, Cabral JMS, Methods Mol. Biol 2018, 1771, 69. [DOI] [PubMed] [Google Scholar]
- [151].Berg J, Hiller T, Kissner MS, Qazi TH, Duda GN, Hocke AC, Hippenstiel S, Elomaa L, Weinhart M, Fahrenson C, Kurreck J, Sci Rep 2018, 8, 13877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Duong H, Wu B, Tawil B, Tissue Engineering Part A 2009, 15, 1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Bayat N, Izadpanah R, Ebrahimi-Barough S, Javidan AN, Ai A, Ardakan MMM, Saberi H, Ai J, Asian Pac J Cancer Prev 2018, 19, 2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Lee Y-B, Polio S, Lee W, Dai G, Menon L, Carroll RS, Yoo S-S, Experimental Neurology 2010, 223, 645. [DOI] [PubMed] [Google Scholar]
- [155].Bagó JR, Pegna GJ, Okolie O, Hingtgen SD, Biomaterials 2016, 84, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Han J, Kim DS, Jang H, Kim H-R, Kang H-W, J Tissue Eng 2019, 10, 204173141984584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Wang Z, Lee SJ, Cheng H-J, Yoo JJ, Atala A, Acta Biomater 2018, 70, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Wang X, Li X, Dai X, Zhang X, Zhang J, Xu T, Lan Q, Colloids and Surfaces B: Biointerfaces 2018, 171, 291. [DOI] [PubMed] [Google Scholar]
- [159].Tang-Schomer MD, White JD, Tien LW, Schmitt LI, Valentin TM, Graziano DJ, Hopkins AM, Omenetto FG, Haydon PG, Kaplan DL, Proceedings of the National Academy of Sciences 2014, 111, 13811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Lozano R, Stevens L, Thompson BC, Gilmore KJ, Gorkin R, Stewart EM, in het Panhuis M, Romero-Ortega M, Wallace GG, Biomaterials 2015, 67, 264. [DOI] [PubMed] [Google Scholar]
- [161].Ribeiro VP, Silva-Correia J, Gonçalves C, Pina S, Radhouani H, Montonen T, Hyttinen J, Roy A, Oliveira AL, Reis RL, Oliveira JM, PLoS ONE 2018, 13, e0194441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Wang Q, Han G, Yan S, Zhang Q, Materials (Basel) 2019, 12, DOI 10.3390/ma12030504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Gomez-Roman N, Stevenson K, Gilmour L, Hamilton G, Chalmers AJ, Neuro-Oncology 2017, 19, 229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Lanzalaco S, Armelin E, Gels 2017, 3, DOI 10.3390/gels3040036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Nishiguchi A, Zhang H, Schweizerhof S, Schulte MF, Mourran A, Möller M, ACS Appl Mater Interfaces 2020, 12, 12176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Li Q, Lin H, Wang O, Qiu X, Kidambi S, Deleyrolle LP, Reynolds BA, Lei Y, Sci Rep 2016, 6, DOI 10.1038/srep31915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Soman P, Fozdar DY, Lee JW, Phadke A, Varghese S, Chen S, Soft Matter 2012, 8, 4946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Soman P, Lee JW, Phadke A, Varghese S, Chen S, Acta Biomater 2012, 8, 2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Soman P, Tobe BTD, Lee JW, Winquist AM, Singec I, Vecchio KS, Snyder EY, Chen S, Biomed Microdevices 2012, 14, 829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Avci NG, Fan Y, Dragomir A, Akay YM, Akay M*, IEEE Transactions on NanoBioscience 2015, 14, 790. [DOI] [PubMed] [Google Scholar]
- [171].Fan Y, Nguyen DT, Akay Y, Xu F, Akay M, Sci Rep 2016, 6, DOI 10.1038/srep25062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Hsieh F-Y, Lin H-H, Hsu S, Biomaterials 2015, 71, 48. [DOI] [PubMed] [Google Scholar]
- [173].Li J, Xing R, Bai S, Yan X, Soft Matter 2019, 15, 1704. [DOI] [PubMed] [Google Scholar]
- [174].Raphael B, Khalil T, Workman VL, Smith A, Brown CP, Streuli C, Saiani A, Domingos M, Materials Letters 2017, 190, 103. [Google Scholar]
- [175].Loo Y, Hauser CAE, Biomed. Mater 2015, 11, 014103. [DOI] [PubMed] [Google Scholar]
- [176].Xia Y, Xue B, Qin M, Cao Y, Li Y, Wang W, Sci Rep 2017, 7, DOI 10.1038/s41598-017-10162-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Wang T-W, Chang K-C, Chen L-H, Liao S-Y, Yeh C-W, Chuang Y-J, Nanoscale 2017, 9, 16281. [DOI] [PubMed] [Google Scholar]
- [178].Hribar KC, Finlay D, Ma X, Qu X, Ondeck MG, Chung PH, Zanella F, Engler AJ, Sheikh F, Vuori K, Chen SC, Lab Chip 2015, 15, 2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Zhu W, Holmes B, Glazer RI, Zhang LG, Nanomedicine 2016, 12, 69. [DOI] [PubMed] [Google Scholar]
- [180].Yi H-G, Choi Y-J, Kang KS, Hong JM, Pati RG, Park MN, Shim IK, Lee CM, Kim SC, Cho D-W, J Control Release 2016, 238, 231. [DOI] [PubMed] [Google Scholar]
- [181].Meng F, Meyer CM, Joung D, Vallera DA, McAlpine MC, Panoskaltsis-Mortari A, Adv. Mater. Weinheim 2019, 31, e1806899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [182].Wang X, Li X, Dai X, Zhang X, Zhang J, Xu T, Lan Q, Colloids and Surfaces B: Biointerfaces 2018, 171, 629. [DOI] [PubMed] [Google Scholar]
- [183].Dai X, Ma C, Lan Q, Xu T, Biofabrication 2016, 8, 045005. [DOI] [PubMed] [Google Scholar]
- [184].Wang X, Dai X, Zhang X, Ma C, Li X, Xu T, Lan Q, Journal of Biomedical Materials Research Part A 2019, 107, 383. [DOI] [PubMed] [Google Scholar]
- [185].Heinrich MA, Bansal R, Lammers T, Zhang YS, Michel Schiffelers R, Prakash J, Adv. Mater 2019, 31, 1806590. [DOI] [PubMed] [Google Scholar]
- [186].Yi H-G, Jeong YH, Kim Y, Choi Y-J, Moon HE, Park SH, Kang KS, Bae M, Jang J, Youn H, Paek SH, Cho D-W, Nat Biomed Eng 2019, 3, 509. [DOI] [PubMed] [Google Scholar]
- [187].Dai X, Liu L, Ouyang J, Li X, Zhang X, Lan Q, Xu T, Scientific Reports 2017, 7, DOI 10.1038/s41598-017-01581-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [188].Hermida MA, Kumar JD, Schwarz D, Laverty KG, Di Bartolo A, Ardron M, Bogomolnijs M, Clavreul A, Brennan PM, Wiegand UK, Melchels FPW, Shu W, Leslie NR, Advances in Biological Regulation 2019, 100658. [DOI] [PubMed] [Google Scholar]
- [189].Sivandzade F, Cucullo L, Journal of Cerebral Blood Flow & Metabolism 2018, 38, 1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [190].Marino A, Tricinci O, Battaglini M, Filippeschi C, Mattoli V, Sinibaldi E, Ciofani G, Small 2018, 14, DOI 10.1002/smll.201702959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [191].Kim JA, Kim HN, Im S-K, Chung S, Kang JY, Choi N, Biomicrofluidics 2015, 9, DOI 10.1063/1.4917508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [192].Kakee A, Terasaki T, Sugiyama Y, J Pharmacol Exp Ther 1996, 277, 1550. [PubMed] [Google Scholar]
- [193].Pas J, Wyszko E, Rolle K, Rychlewski L, Nowak S, Żukiel R, Barciszewski J, The International Journal of Biochemistry & Cell Biology 2006, 38, 1594. [DOI] [PubMed] [Google Scholar]
- [194].Brekken RA, Sage EH, Matrix Biol. 2001, 19, 816. [DOI] [PubMed] [Google Scholar]
- [195].Iozzo RV, Schaefer L, Matrix Biology 2015, 42, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [196].Jaworski DM, Kelly GM, Piepmeier JM, Hockfield S, Cancer Res. 1996, 56, 2293. [PubMed] [Google Scholar]
- [197].Patel TR, Bernards C, Meier M, McEleney K, Winzor DJ, Koch M, Stetefeld J, Matrix Biology 2014, 33, 60. [DOI] [PubMed] [Google Scholar]
- [198].Shin K, Lechtenberg BC, Fujimoto LM, Yao Y, Bartra SS, Plano GV, Marassi FM, Science Advances 2019, 5, eaax5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [199].Tan Z, Parisi C, Di Silvio L, Dini D, Forte AE, Sci Rep 2017, 7, 16293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [200].Grundy TJ, De Leon E, Griffin KR, Stringer BW, Day BW, Fabry B, Cooper-White J, O’Neill GM, Scientific Reports 2016, 6, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [201].Xu Z, Li Z, Jiang S, Bratlie KM, ACS Omega 2018, 3, 6998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [202].Comolli N, Neuhuber B, Fischer I, Lowman A, Acta Biomaterialia 2009, 5, 1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [203].Miri AK, Mostafavi E, Khorsandi D, Hu S-K, Malpica M, Khademhosseini A, Biofabrication 2019, 11, 042002. [DOI] [PMC free article] [PubMed] [Google Scholar]


