Abstract
Maternal infection and inflammation during pregnancy are associated with neurodevelopmental disorders in offspring, but little is understood about the molecular mechanisms underlying this epidemiologic phenomenon. We leveraged single-cell RNA sequencing to profile transcriptional changes in the mouse fetal brain in response to maternal immune activation (MIA) and identified perturbations in cellular pathways associated with mRNA translation, ribosome biogenesis, and stress signaling. We found that MIA activates the integrated stress response (ISR) in male, but not female, MIA offspring in an Interleukin-17a dependent manner, thereby reducing global mRNA translation and altering nascent proteome synthesis. Moreover, blockade of ISR activation prevented the behavioral abnormalities as well as an increase in cortical neural activity in MIA male offspring. Our data suggest that sex-specific activation of the ISR leads to maternal inflammation-associated neurodevelopmental disorders.
Keywords: maternal immune activation, fetal brain, neuroimmune, protein translation
INTRODUCTION
Fetal brain development is exquisitely sensitive to environmental influence. Transcriptional programs in the fetal brain regulate complex and overlapping developmental processes that ultimately shape the functional architecture of the adult brain. Perturbations in the intrauterine environment can thus have profound and long-lasting effects on neurodevelopment.1-3 Epidemiological data suggest that significant maternal infections during pregnancy are associated with an increased risk of neurodevelopmental and neuropsychiatric diseases in offspring.4-7 However, our understanding of the molecular mechanisms underpinning the detrimental effects of adverse environmental exposures on the development of the fetal brain remains limited.
This phenomenon has been studied in a rodent maternal immune activation (MIA) model in which offspring of pregnant mice injected intra-peritoneally with a mimetic of viral infection (synthetic dsRNA, polyinosinic:polycytidylic acid (poly(I:C)) exhibit pronounced neurodevelopmental consequences, including impaired social interaction and repetitive behavior.8-10 MIA-induced disruption of offspring neurodevelopment requires maternal type 17 helper T cell/interleukin 17a (TH17/IL-17a) pathways9; however, the signaling mechanisms downstream of IL-17a that give rise to MIA-associated neurodevelopmental phenotypes remain unclear.
Using a combination of single-cell transcriptomics and ribosome profiling, we systematically quantified transcriptional and translational changes in MIA offspring. We found that MIA was associated with cell type- and sex-specific transcriptional alterations in the fetal brain, and we identified a widespread disruption in the expression of genes involved in mRNA translation. These effects were associated with activation of the integrated stress response (ISR) via phosphorylation of the α subunit of eukaryotic translation initiation factor 2 (eIF2α), leading to sex-specific arrest of mRNA translation in the fetal brain. ISR dysregulation has previously been implicated in neurodegenerative conditions, including frontotemporal dementia and amyotrophic lateral sclerosis (ALS), as well as the behavioral and neurophysiological abnormalities associated with Down Syndrome.11-14 Strikingly, genetic reduction of phospho-eIF2α levels and pharmacological blockade of ISR activation in the developing fetus were found to protect against MIA-associated behavioral abnormalities. Together, our findings provide new insight into the implications of aberrant intrauterine ISR activation for MIA-associated neurodevelopmental conditions and broaden our understanding of the mechanisms by which intrauterine stress shapes neurodevelopment.
RESULTS
Single-cell RNA-seq of the fetal brain exposed to maternal inflammation
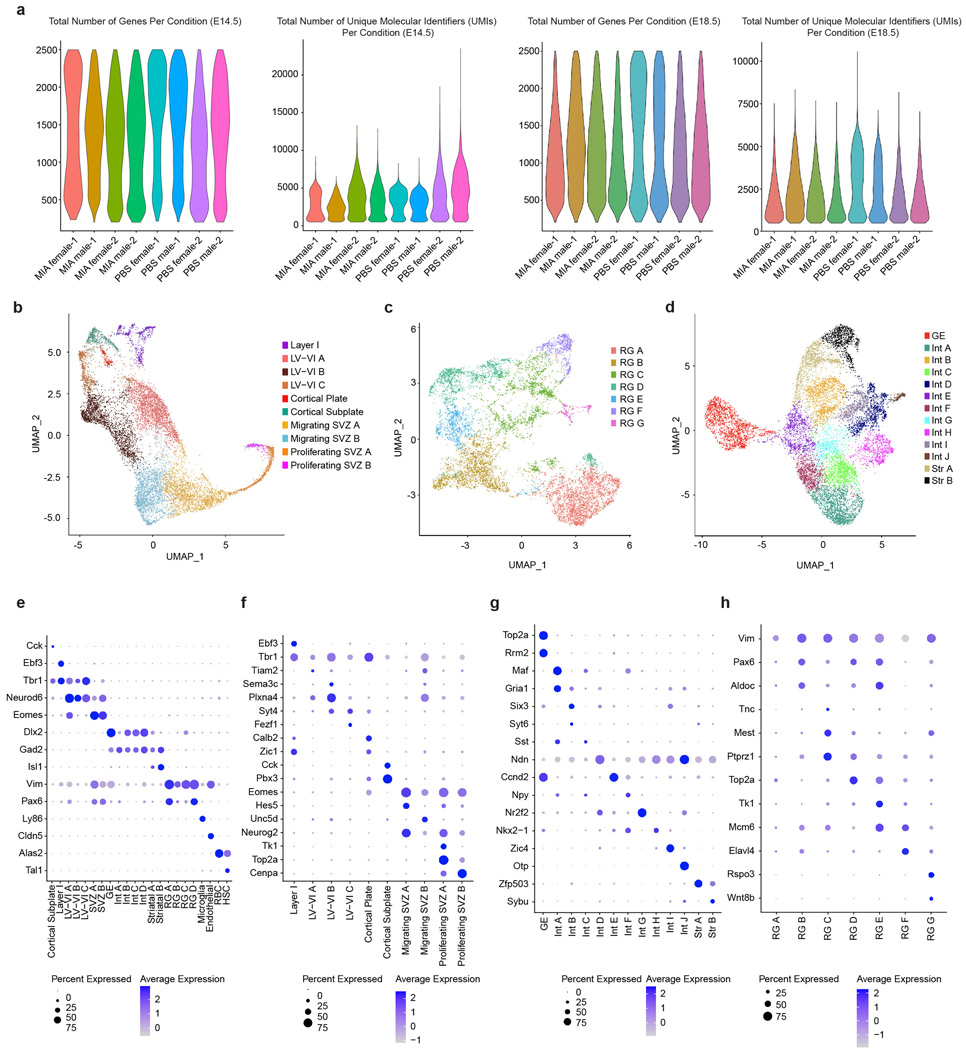
The development of the fetal cortex requires precise temporal and spatial coordination of gene expression and mRNA translation. To gain insight into the cell type-specific effects of MIA on the developing fetal brain, we employed high-throughput single-cell RNA sequencing (scRNA-seq) to profile gene expression in the fetal cortex of both male and female fetuses at embryonic days 14.5 and 18.5 (E14.5 and 18.5) in the context of MIA or a sham saline control treatment (PBS) (Fig. 1a). These time points were selected to survey critical periods in fetal brain maturation following poly(I:C) administration at E12.5. After quality and doublet filtering (quality metrics in Extended Data Fig. 1a), we analyzed the transcriptomes of 37,377 cells at E14.5 and 57,953 cells at E18.5, recovering an average of 3,529 and 2,168 unique transcripts per cell, respectively.
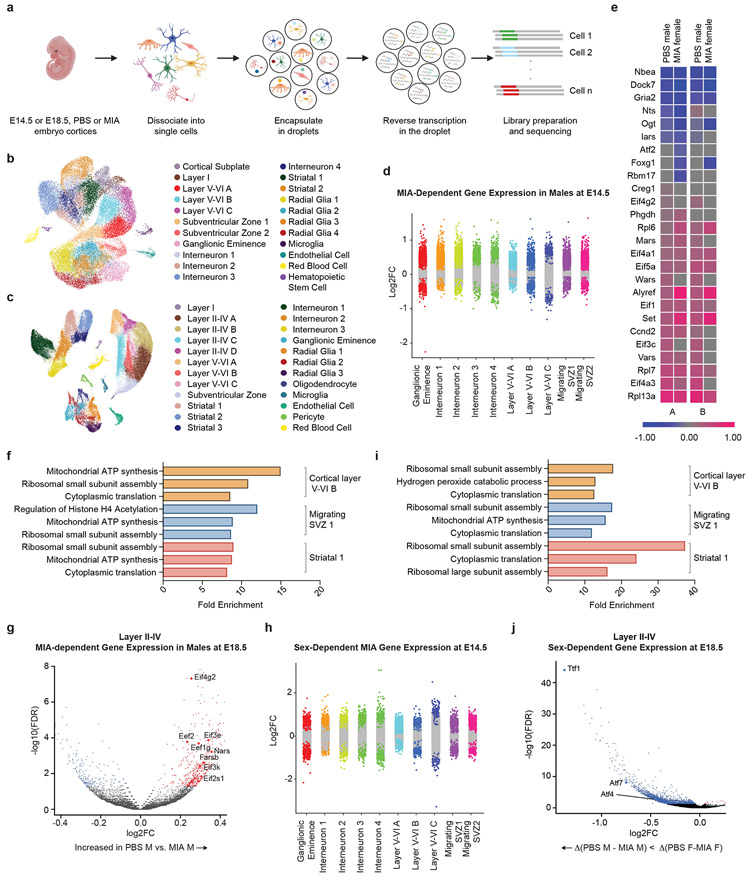
Figure 1. Single-cell sequencing of the fetal brain at E14.5 and E18.5 in the setting of Maternal Immune Activation.
a, Schematic overview of the experimental design, including dissociation of embryonic cortices, single cell encapsulation, and library preparation.
b, UMAP of all analyzed E14.5 cells, with cell types labeled by color (n = 22 clusters). Data from n = 2 mice per group.
c, UMAP of all analyzed E18.5 cells, with cell types labeled by color (n = 24 clusters). Data from n = 2 mice per group.
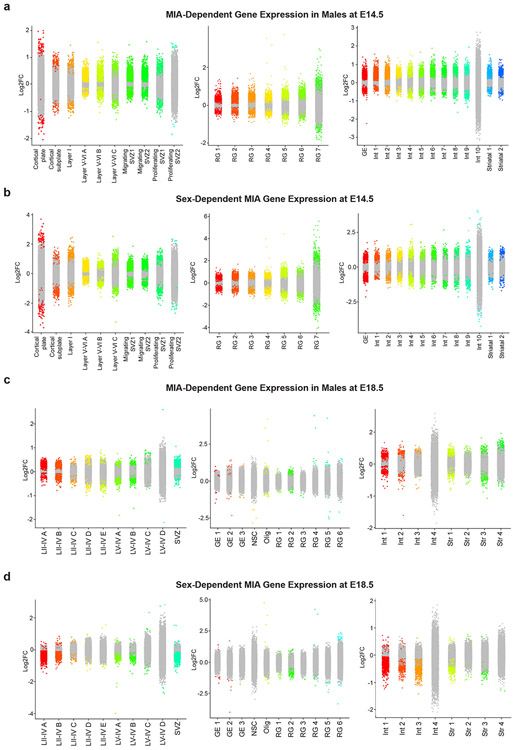
d, Strip plot displaying differential gene expression between MIA male offspring and PBS male offspring at E14.5. Colored dots represent significant genes (FDR < 0.05). X-axis displays select cortical cell types. Data from n = 2 mice per group.
e, Heat map of single cell gene expression changes of select genes in two cell types at E14.5: Neurod2-positive cortical excitatory neurons (A) and Gad2-positive inhibitory neurons (B). Comparisons labeled “PBS male” are pairwise comparisons between PBS males and MIA males. For these comparisons, blue corresponds to increased expression in MIA males, while pink corresponds to decreased expression in MIA males. Comparisons labeled “MIA female” are multi-factor comparisons measuring the difference between the changes in expression with MIA between male and female mice. For these comparisons, blue corresponds to an increase in the difference between MIA and PBS expression levels in males, while pink corresponds to a decrease in this difference. Data from n = 2 mice per group.
f, GO analysis of differentially expressed genes (FDR < 0.05) between MIA and PBS male offspring in select cell types at E14.5. X-axis displays fold enrichment relative to reference gene set. Data from n = 2 mice per group.
g, Volcano plot of MIA-dependent gene expression in Layer II-IV cluster A neurons at E18.5. Colored dots indicate statistical significance (FDR < 0.05). Positive log2FC (red dots) indicates higher gene expression in PBS males relative to MIA males, and negative log2FC (blue dots) indicates higher gene expression in MIA males relative to PBS males. Data from n = 2 mice per group.
h, Strip plot displaying sex-dependent gene expression in MIA offspring at E14.5. Colored dots represent significant genes (FDR < 0.05, log2(Fold Change) > 0.5. X-axis displays select cortical cell types. Data from n = 2 mice per group.
i, GO analysis of sex-dependent genes in MIA offspring in select cell types at E14.5 (FDR < 0.05). X-axis displays fold enrichment relative to reference gene set. Data from n = 2 mice per group.
j, Volcano plot of sex-dependent gene expression in MIA offspring in Layer II-IV cluster A neurons at E18.5. Colored dots indicate statistical significance (FDR < 0.05) by interaction term analysis to identify genes that vary by sex with MIA exposure. Negative log2FC values indicate Δ(PBS M – MIA M) < Δ (PBS F-MIA F). Data from n = 2 mice per group.
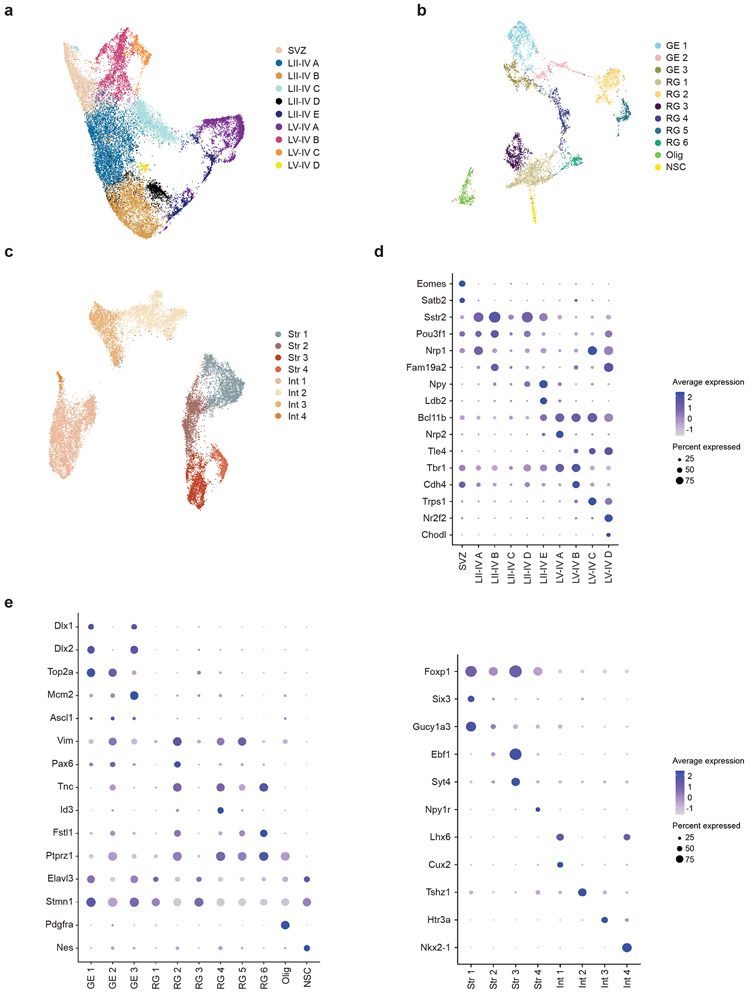
Standard graph-based clustering methods were used to classify E14.5 and E18.5 samples into 22 and 24 transcriptionally distinct cell types, respectively, which were assigned identities based on canonical marker gene expression (Fig. 1b,c). This analysis resolved developmental, anatomical, and functional cell states in the cortex, including proliferative neural precursors, migratory radial glia, as well as intermediate and mature neuronal subtypes. Moreover, further clustering of major cell classes served to reveal additional developmental and functional cellular subtype heterogeneity. At E14.5, we identified major cortical layer subtypes, interneurons, cortical plate, cortical subplate, migrating and proliferating subventricular zone (SVZ) cells, as well as multiple radial glia subtypes, all with distinct transcriptional signatures (Extended Data Fig. 1b-h). At E18.5, we identified increased diversity of cortical layer, interneuronal, and striatal subtypes as compared to E14.5 (Extended Data Fig. 2a-e). Together, the developmental hierarchy of our cell type classifications were in broad agreement with previous single-cell RNA-seq data from the embryonic mouse brain.15
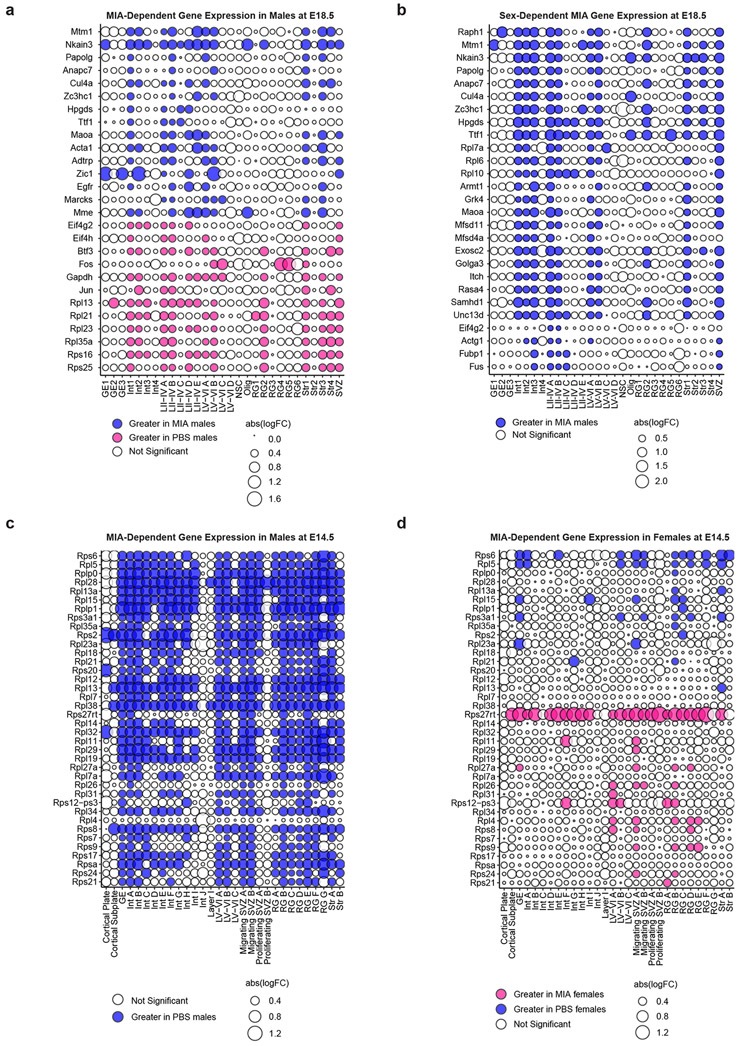
We analyzed male and female offspring independently, given previously described sex differences in the MIA phenotype.16,17 While MIA did not dramatically alter the distribution of cell types in male or female mice, differential gene expression (DGE) analysis between MIA and PBS subpopulations within defined cell types revealed profound, widespread gene expression changes in MIA samples (Supplementary Tables 1-9). We used a false discovery rate (FDR < 0.05) to define statistically significant changes in gene expression between MIA and control offspring. We were able to identify both shared and cell type-specific MIA-associated gene signatures at both E14.5 and E18.5 in neuronal and non-neuronal subtypes (Fig. 1d, e and Extended Data Fig. 3). For example, expression levels of those genes regulating translation were commonly decreased in the cortical excitatory and inhibitory neurons of MIA male offspring, compared those of PBS male or MIA female offspring (Fig. 1e). Gene ontology (GO) analysis of differentially expressed genes in select cell types suggested widespread disruption of networks associated with mRNA trafficking and cytoplasmic translation (Fig. 1f). We identified altered transcription of numerous eukaryotic translation initiation and elongation factors across several cell types comparing MIA males and PBS males. Specifically, comparing MIA males to PBS males in layer II-IV cortical neurons at E18.5, we found decreased expression of Eif3e, Eif3k, Eef2, and Eef1g, all of which are critical for protein synthesis (Fig. 1g). In this same cell cluster, we identified decreased expression of several tRNA synthetases in MIA males, including Farsb and Nars (Fig. 1g). Aminoacyl-tRNA synthetase factors help to maintain translational fidelity and are essential components of the protein biosynthesis machinery.18 Finally, we identified decreased expression of Eif2s1 in MIA males, which encodes for eIF2α – a central regulatory protein in cellular translation (Fig. 1g). Additionally, many genes encoding ribosomal subunits (e.g. Rps8, Rps10, Rpl13) were down-regulated in MIA male offspring relative to PBS male offspring at E14.5, and to a lesser extent at E18.5 (Fig. 2a, Extended Data Fig. 4a).
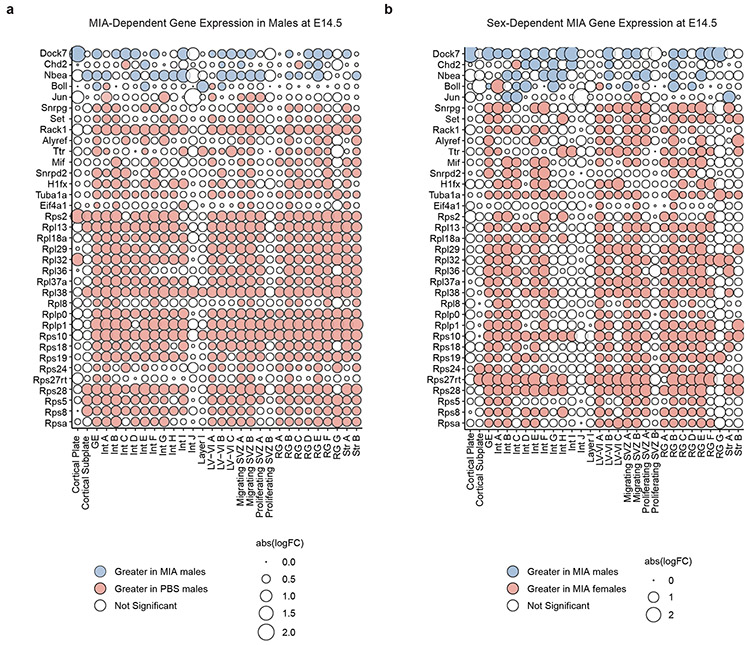
Figure 2. Single cell differential gene expression changes at E14.5.
a, Bubble plot of highly variable genes between MIA and PBS male offspring at E14.5, demonstrating a widespread decrease in expression of multiple ribosomal subunits in MIA male offspring. All significant genes FDR < 0.05. Blue indicates an increase in MIA males, and red indicates an increase in PBS males. Data from n = 2 mice per group.
b, Bubble plot of sex-dependent genes in MIA offspring at E14.5, demonstrating a widespread decrease in expression of multiple ribosomal subunits in MIA male offspring. All significant genes FDR < 0.05. Blue indicates an increase in Δ(MIA males – PBS males), and red indicates an increase in Δ(MIA females – PBS females). Data from n = 2 mice per group.
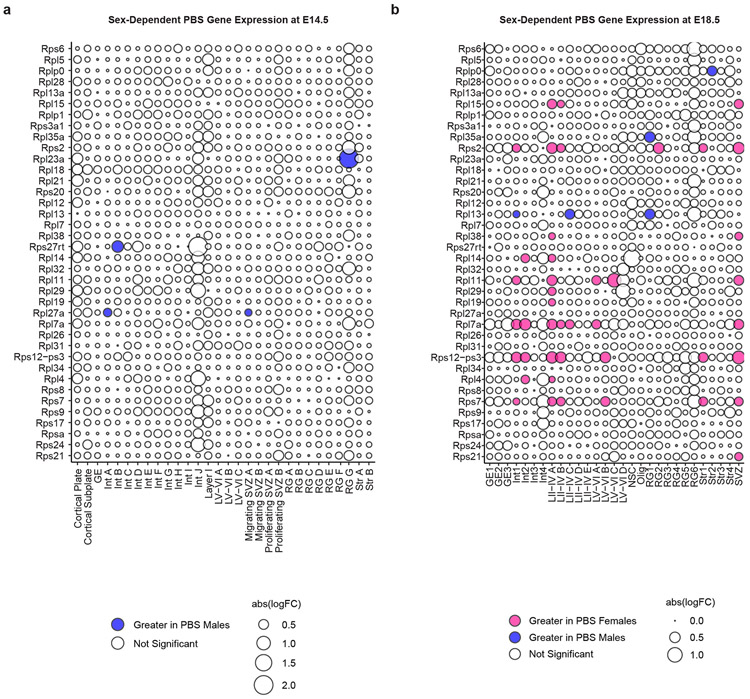
Interestingly, we found that a large portion of the transcriptional effects of MIA on male mice were sex-dependent; male and female MIA offspring demonstrate prominent differences in cell type-specific gene expression across numerous cell types (Fig. 1e,h, Extended Data Fig. 3). We applied a multi-factor statistical analysis to identify genes with altered cell type-specific expression changes with MIA exposure between males and females. Similar to comparisons between MIA and PBS male offspring, MIA male offspring exhibited profound changes in ribosomal, stress, and translation-associated genes across many cell types compared to MIA female offspring (Fig. 2b, Extended Data Fig. 4b). At E14.5 in particular, male MIA offspring demonstrated a near-global reduction in expression of ribosomal subunit genes compared to male PBS offspring, whereas female MIA offspring demonstrate few changes in ribosomal subunit genes compared to female PBS offspring (Extended Data Fig. 4c,d). Comparisons between PBS male and female offspring revealed some sex-specific expression of ribosomal subunit genes in select cell types at E14.5 and E18.5, but these changes were far less prevalent than in the MIA male to PBS male or MIA female comparisons (Extended Data Fig. 5). GO analysis of differentially expressed genes in the multi-factor analysis between MIA males and females revealed several broad and cell type-specific functional pathways, but the regulation of translation emerged as a common biologic process altered in many cell types (Fig. 1i). For example, in layer II-IV cortical neurons, we found sex-dependent expression of Ttf1, which is a nucleolar protein involved in the Pol I-mediated transcription of ribosomal genes and ribosome biogenesis (Fig. 1j).19,20 When compared within sex groups, Ttf1 was increased in MIA males (FDR 3.1x10−5) and decreased in MIA females (FDR 1.5x10−51) relative to sex-matched PBS controls. We also found sex-dependent expression of the stress-responsive factors ATF4 (decreased in MIA females relative to PBS females, FDR 0.033) and ATF7 (decreased in MIA females relative to PBS females, FDR 1.04x10−19) (Fig. 1j). ATF4 controls the expression of adaptive genes that facilitate cell survival during periods of stress, such as hypoxia or starvation, and has been implicated in neurodegeneration.21 These findings suggest that MIA may induce cell type- and sex-specific transcriptional alterations in the fetal brain.
MIA disrupts protein synthesis in the fetal brain
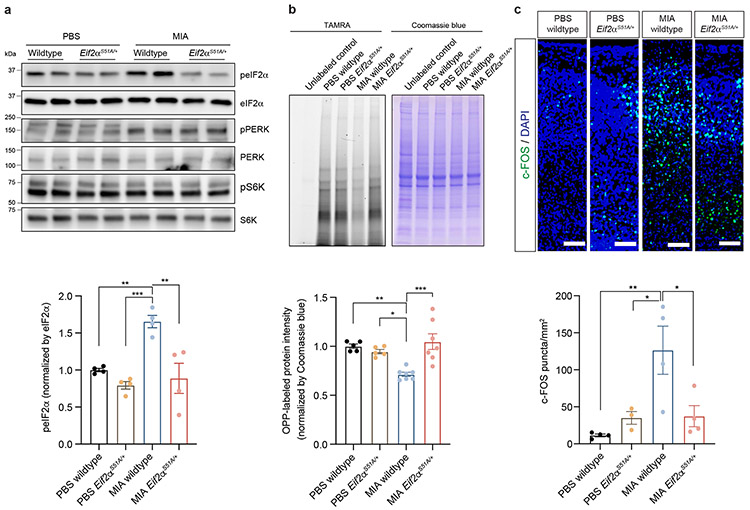
Given our finding that cytoplasmic translation was a common pathway dysregulated in male MIA offspring, we sought to assess directly the effects of MIA on global protein synthesis in the fetal mouse brain. We employed a modified version of the previously described SUnSET (Surface Sensing of Translation) method.22 In this approach, the puromycin analog O-propargyl-puromycin (OPP) serves as a structural mimic of aminoacyl tRNAs and is incorporated into nascent polypeptide chains.22 OPP labeling permits subsequent fluorescent tagging of the modified polypeptides via a click-chemistry reaction, thereby allowing for quantitative measurement of protein synthesis within tissues. Fetal cortical tissue from E18.5 controls and MIA offspring were with incubated with OPP, followed by a click chemistry reaction with TAMRA-azide and subsequent SDS-PAGE electrophoresis. Protein synthesis was quantified by TAMRA signal (new protein) over Coomassie (total protein). We found that nascent protein synthesis was decreased only in the cortex of male MIA offspring (Fig. 3a). This finding was consistent with our single-cell RNA-sequencing data that suggests deficits in ribosome subunit expression in MIA male offspring.
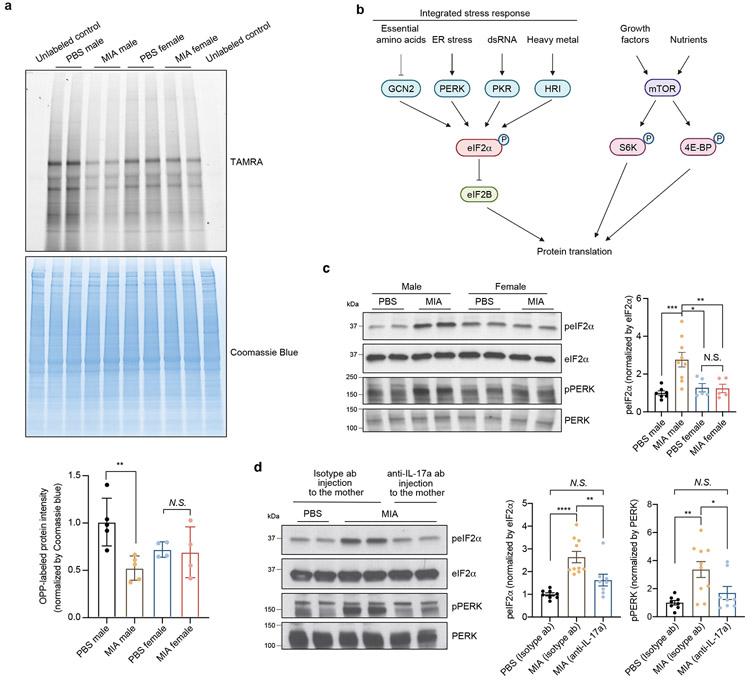
Figure 3. MIA inhibits protein translation in the fetal brain via the integrated stress response.
a, TAMRA-conjugated proteins labeled by OPP were extracted and isolated by SDS-PAGE and analyzed by fluorescence scan at 532 nm. Representative section of Coomassie-stained gel for loading comparison (Two-way ANOVA sex (male or female) × stimulus (PBS or MIA) interaction F1,14 = 5.874, P = 0.0295; effect of sex F1,14 = 0.4167, P = 0.5290; effect of stimulus F1,14 = 7.462, P = 0.0162 followed by Tukey multiple comparisons test: PBS male versus MIA male **P = 0.0082; n = 5 for PBS male and MIA male, n = 4 for PBS female and MIA female pups; 3 independent litters for PBS, 2 independent litters for MIA).
b, Schematic representation of the regulation of translation initiation through eIF2α -dependent regulation and mTOR-dependent regulation.
c, Immunoblot analysis measuring phospho-eIF2α and phospho-PERK levels and quantification for phospho-eIF2α in E18.5 PBS and MIA fetal cortices. Y-axis represents relative blot intensity to PBS male control (Two-way ANOVA sex (male or female) × stimulus (PBS or MIA) interaction F1,22 = 8.229, P = 0.0089; effect of sex F1,22 = 3.939, P = 0.0598; effect of stimulus F1,22 = 7.567, P = 0.0117 followed by Tukey multiple comparisons test: PBS male versus MIA male ***P = 0.0009, PBS female versus MIA male *P = 0.0118, MIA male versus MIA female **P = 0.0097; n = 7 for PBS male, n = 9 for MIA male, n = 5 for PBS female and MIA female pups; 3 independent litters).
d, Immunoblot analysis measuring phospho-eIF2α and phospho-PERK levels and quantification for phospho-eIF2α in isotype antibody or IL-17a-blocking antibody-treated E18.5 PBS and MIA fetal cortices. Y-axis represents relative blot intensity to the isotype antibody-treated PBS control (phospho-eIF2α: one-way ANOVA, F2,23 = 14.88, P = 7.14 x 10−5, Tukey’s multiple comparisons test: PBS isotype ab versus MIA isotype ab ****P = 5.58 x 10−5, MIA isotype ab versus MIA anti-IL-17a **P = 0.0083; phospho-PERK: one-way ANOVA, F2,23 = 7.259, P = 0.0036, Tukey’s multiple comparisons test: PBS isotype ab versus MIA isotype ab **P = 0.0036, MIA isotype ab versus MIA anti-IL-17a *P = 0.0423; n = 8 for PBS isotype ab and MIA anti-IL-17a, n = 10 for MIA isotype ab; 3 independent litters).
Data are mean ± SEM; see Supplementary Table 20 for detailed statistics. Unprocessed blots are provided as a Source Data file.
Activation of the integrated stress response in male MIA offspring
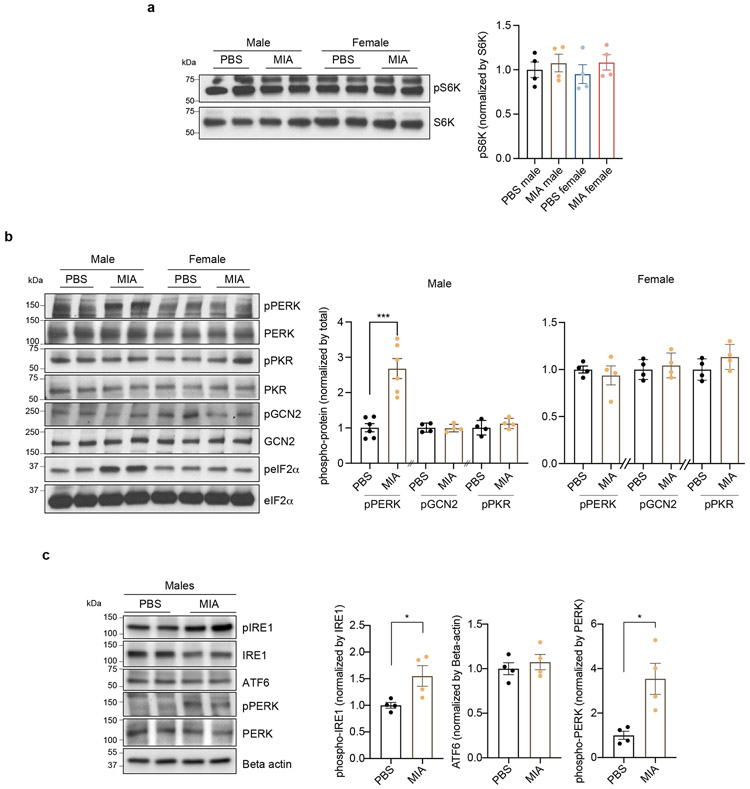
To explain the finding that there is a significant reduction in nascent protein synthesis in MIA male fetal brains, we hypothesized that alterations in fetal brain translation in response to MIA might be due to activation of the integrated stress response (ISR). The ISR is an adaptive mechanism that can be activated under physiologic or pathologic conditions to execute a global reduction in protein synthesis as a means to shift resources towards cell survival. The phosphorylation of eIF2α at serine 51 is the critical regulatory hub in the activation of the ISR (Fig. 3b).23 Probing with an eIF2α phospho-specific antibody, we found a strong induction of eIF2α phosphorylation specifically in the brain of male MIA offspring (Fig. 3c). By contrast, we observed no changes in the levels of ribosomal protein S6 kinase phosphorylation, a distinct point of translational control (Extended Data Fig. 6a).
eIF2α phosphorylation can be triggered by a diverse array of cellular stressors23 and is mediated by four cellular kinases, three of which – PKR, PERK, and GCN2 – are present in the brain.24 The activity of these eIF2α kinases can be monitored via a series of phospho-specific antibody reagents that recognize kinase auto-phosphorylation that occurs when the kinase is activated. Blotting fetal cortical lysates with these antibodies revealed that PERK (PKR-like endoplasmic reticulum kinase), but not PKR or GCN2, was specifically activated in the cortex of male MIA offspring (Fig. 3c and Extended Data Fig. 6b). PERK is triggered by ER stress and the unfolded protein response (UPR), suggesting that these processes are critical signaling mechanisms in MIA.25 To test whether MIA activates other sensors of the UPR, we performed immunoblotting for IRE-1α and ATF6 levels in the E18.5 brain. We found that phosphorylation of IRE1α was increased in the brain of male MIA offspring, while cleaved ATF6 was unchanged (Extended Data Fig. 6c).
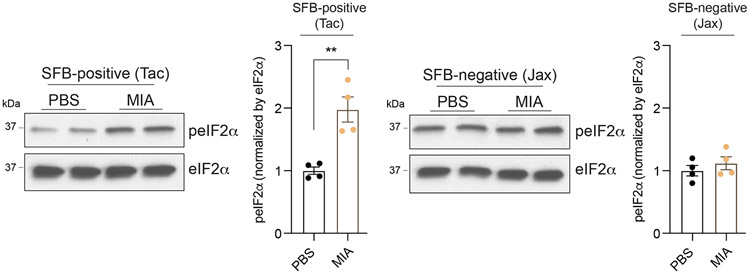
We have previously shown that behavioral abnormalities in the MIA offspring are mediated by maternal induction of the inflammatory cytokine IL-17a.9 To test whether activation of ISR in the MIA offspring is dependent on maternal IL-17a, we pre-treated pregnant dams with either a control or IL-17a-blocking antibody 5 hours before introducing poly(I:C). Administration of the IL-17a-blocking antibody, but not an isotype-matched control antibody, completely suppressed MIA-induced eIF2α and PERK phosphorylation in male fetal cortex (Fig. 3d), demonstrating that PERK-mediated ISR activation in the fetal brain is dependent on maternal IL-17a. Furthermore, previous reports demonstrated that the TH17-inducing mouse commensal bacteria segmented filamentous bacteria (SFB) is required for maternal induction of the pro-inflammatory cytokine IL-17a upon MIA.26,27 In line with this finding, we did not observe activation of the ISR in the brains of MIA offspring from SFB-negative dams (Extended Data Fig. 7). This result suggests a mechanistic link between the microbiome, maternal IL-17a, and the fetal brain stress response.
MIA alters the translational landscape of the fetal brain
Activation of the ISR restructures global mRNA translation in two stages.28 First, cellular stress induces phosphorylation of eIF2α, which transiently attenuates protein synthesis through inhibition of cap-dependent translation. After resolution of cellular stress, increased expression of GADD34 – a regulatory subunit of the phosphatase that dephosphorylates eIF2α – facilitates cap-independent translation of stress-adaptive transcripts, while also feeding back to promote dephosphorylation of eIF2α.29 Stress-adaptive transcripts necessary to promote cell survival are translated through cap-independent mechanisms, including internal ribosome entry sits (IRES) and alternative initiation mechanisms.28
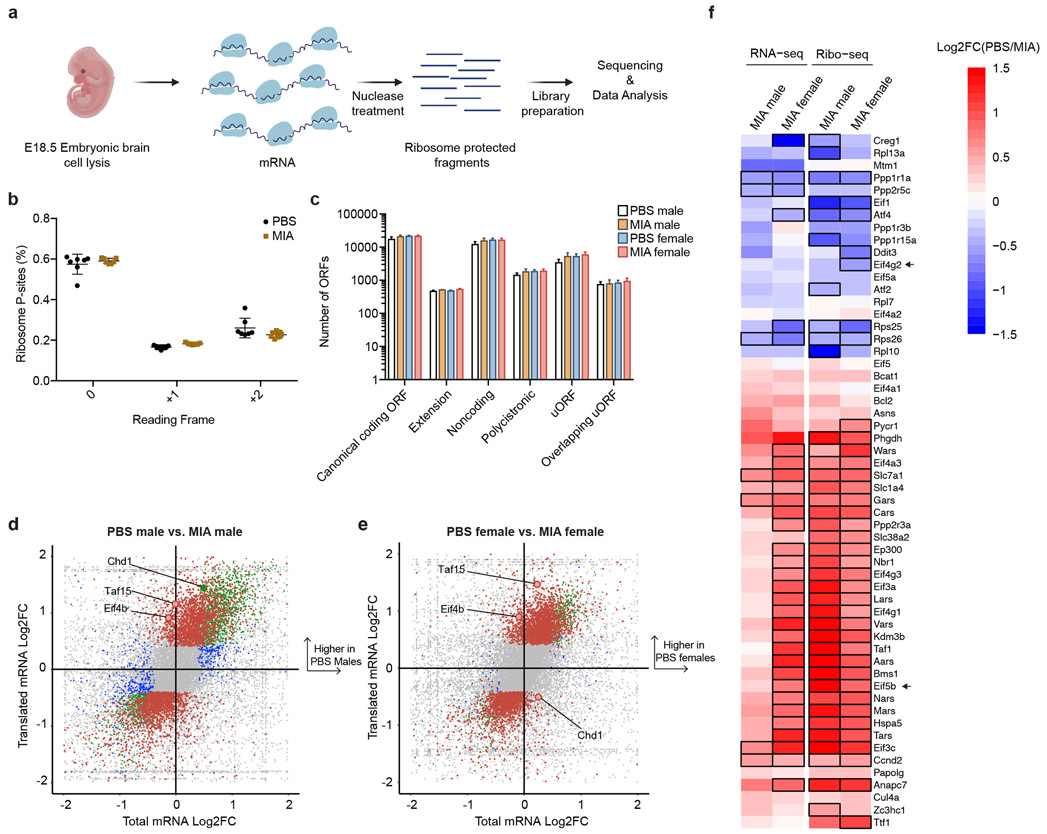
We sought to measure the changes associated with MIA in this dynamic process at codon-level resolution. Therefore, we performed deep sequencing of ribosome-protected mRNA fragments (Ribo-seq) at E18.5, in parallel with bulk RNA-seq from the same biologic sample (Fig 4a, Supplementary Tables 9-19).30 DNA libraries prepared from ribosome-protected mRNA fragments using established methods30 were sequenced to a depth of ~80 million reads per sample and showed expected periodicity indicative of high quality ribosome profiling (Fig. 4b). We characterized the translated open reading frames (ORFs) based on transcript types and ORF locations, including canonical ORFs, upstream ORFs in 5’ untranslated regions (uORFs), downstream ORFs in 3’ untranslated regions (dORFs) and annotated non-coding ORFs (ncORFs) using RiboORF (Fig. 4c).31 Although some studies have suggested that the acute ISR increases the usage of uORFs, we did not observe any statistically significant differences in the distribution of expression between experimental groups for any of the ORF classes. Such changes in uORF utilization may occur earlier than the selected E18.5 time point. We identified transcripts with significant changes in transcription, translation, or both in MIA males and females relative to sex-matched PBS controls (Fig. 4d-f). Both male and female MIA offspring showed reduced translation of Eif4b, which is an important regulator of cap-dependent translation (Fig. 4d,e).32 Additionally, both male and female MIA offspring demonstrated reduced translation of transcription initiation factor Taf15, suggesting a negative effect on global transcription (Fig. 4d,e).33 On the other hand, MIA females significantly increased translation of Chd1, which encodes a chromatin remodeler; translation of Chd1 was significantly decreased in MIA males (Fig. 4d,e). Chd1 is necessary for nucleosomal dynamics during stress, and impairment in Chd1 function may contribute to neurodegeneration.34 We also observed a number of sex-dependent translation events among ISR downstream target genes that might contribute to female resiliency to MIA. For example, female MIA offspring had significantly increased translation of Eif4g2; there was no significant change in translation in male MIA offspring (Fig. 4f). Eif4g2 encodes for the protein DAP5 and is an alternative translation initiation factor that promotes cap-independent translation.35,36 Both males and females demonstrated reduced translation of Eif5b with MIA, but this decrease was significantly greater in male MIA offspring (Fig. 4f). Eif5b is important for translation when eIF2α is phosphorylated in stress conditions.37
Figure 4. Alterations in the translational landscape of the MIA fetal brain.
a, Schematic of experimental approach for ribosome profiling.
b, Percentage of ribosome profiling reads in frame 0 by experimental condition. Data are mean ± SEM. Data n = 7 for PBS and n = 7 for MIA, each from 2 independent litters.
c, Bar graph of ORF subtypes based on transcript types and ORF locations between all experimental groups. Data are mean ± SEM. For individual data underlying this plot, see Supplementary Table 21. Data n = 7 for PBS and n = 7 for MIA, each from 2 independent litters.
d, Quadrant plot of transcriptional and translational changes in MIA male offspring compared to PBS male offspring. Genes significant only in the total mRNA-seq are in blue, genes significant only in the ribo-seq are in red, and genes significant in both are in green. Significant transcripts have a FDR < 0.05. Positive log2 fold change values indicate an increase in the PBS group. Data n = 7 for PBS and n = 7 for MIA, each from 2 independent litters.
e, Quadrant plot of transcriptional and translational changes in MIA female offspring compared to PBS female offspring. Transcripts significant only in the total mRNA-seq are in blue, transcripts significant only in the ribo-seq are in red, and transcripts significant in both are in green. Significant transcripts have a FDR < 0.05. Positive log2 fold change values indicate an increase in the PBS group. Data n = 7 for PBS and n = 7 for MIA, each from 2 independent litters.
f, Heat map of log2 Fold Change of transcriptional (RNA-seq) and translational (Ribo-seq) changes in MIA males and females relative to sex-matched PBS controls. Genes are known targets of the ISR response to chronic stress49 and translational machinery. Black outlines indicate statistically significant (FDR < 0.05) changes relative to corresponding sex-matched PBS controls in a multi-factor EdgeR analysis. Positive log2 fold change values, shown in red, indicate an increase in the PBS group, while negative log2 fold change values, shown in blue, indicate an increase in the MIA group. Data n = 7 for PBS and n = 7 for MIA, each from 2 independent litters.
In summary, MIA offspring – both male and female – exhibit profound changes in mRNA translation, but female offspring exhibit distinct translational differences that may promote adaptation and resiliency to intrauterine inflammation.
Blockage of ISR activation attenuates excessive neural activity in MIA offspring
We next sought to determine whether attenuation of ISR activation could rescue MIA-associated neuro-behavioral phenotypes. To genetically dampen ISR activation, we utilized mice harboring a knock-in allele in which eIF2α serine 51 is replaced with an alanine (Eif2αS51A), rendering it refractory to ISR-induced phosphorylation and thus blocking ISR pathway activation.38 We crossed wildtype B6 females with heterozygous Eif2αS51A/+ males, then injected poly(I:C) to induce MIA at E12.5. We first examined the resulting levels of phosphorylated eIF2α in E18.5 cortices of wildtype or Eif2αS51A/+ littermate MIA offspring. Eif2αS51A/+ male MIA offspring showed similar levels of eIF2α phosphorylation to the wildtype offspring of control PBS-treated dams, confirming an attenuation of the ISR in these animals (Fig. 5a). As expected, levels of phospho-PERK remained elevated in MIA Eif2αS51A/+ offspring (Fig. 5a), confirming that PERK functions upstream of eIF2α. Accordingly, we also confirmed that decreased nascent protein synthesis in the wildtype MIA offspring was reversed in the cortex of Eif2αS51A/+ littermate MIA offspring (Fig. 5b).
Figure 5. Genetic ablation of ISR protects excessive neural activity in MIA offspring.
a, Immunoblot analysis measuring phospho-eIF2α, phospho-PERK and phospho-S6K levels and quantification for phospho-eIF2α in wildtype and eIF2αS51A/+ E18.5 male fetal cortices. Y-axis represents relative blot intensity to the wildtype PBS fetal cortices (Two-way ANOVA stimulus (PBS or MIA) x genotype (wildtype or eIF2αS51A/+) interaction F1,12 = 6.055, P = 0.03; effect of stimulus F1,12 = 10.88, P = 0.0064; effect of genotype F1,12 = 18.34, P = 0.0011 followed by Tukey multiple comparisons test: PBS wildtype versus MIA wildtype **P = 0.0073, MIA wildtype versus PBS eIF2αS51A/+ ***P = 0.0008, MIA wildtype versus MIA eIF2αS51A/+ **P = 0.0022; n = 4 pups from 2 independent litters).
b, TAMRA-conjugated proteins labeled by OPP were extracted and isolated by SDS-PAGE and analyzed by fluorescence scan at 532 nm. Representative section of Coomassie-stained gel for loading comparison (Two-way ANOVA stimulus (PBS or MIA) x genotype (wildtype or eIF2αS51A/+) interaction F1,20 = 14.35, P = 0.0012; effect of stimulus F1,20 = 3.265, P = 0.0858; effect of genotype F1,20 = 7.283, P = 0.0138 followed by Tukey multiple comparisons test: PBS wildtype versus MIA wildtype **P = 0.004, MIA wildtype versus PBS eIF2αS51A/+ *P = 0.0221, MIA wildtype versus MIA eIF2αS51A/+ ***P = 0.0004; n = 5 for PBS wildtype and PBS eIF2αS51A/+ from 3 independent litters, n = 7 for MIA wildtype and MIA eIF2αS51A/+ pups from 3 independent litters).
c, Representative images of 8-10 weeks brain tissue immunostained for c-Fos. Conditions are MIA and PBS wildtype and eIF2αS51A/+ male mice. Scale bar 100 μm. Quantification indicates c-Fos puncta/mm2 (Two-way ANOVA stimulus (PBS or MIA) x genotype (wildtype or eIF2αS51A/+) interaction F1,11 = 8.320, P = 0.0149; effect of stimulus F1,11 = 9.007, P = 0.0121; effect of genotype F1,11 = 2.798, P = 0.1226 followed by Tukey multiple comparisons test: PBS wildtype versus MIA wildtype **P = 0.0056, MIA wildtype versus PBS eIF2αS51A/+ *P = 0.0373, MIA wildtype versus MIA eIF2αS51A/+ *P = 0.0281; n = 3 for PBS eIF2αS51A/+, n = 4 for PBS wildtype, MIA wildtype and MIA eIF2αS51A/+; 2 independent litters per group.)
Data are mean ± SEM; see Supplementary Table 20 for detailed statistics. Unprocessed blots are provided as a Source Data file.
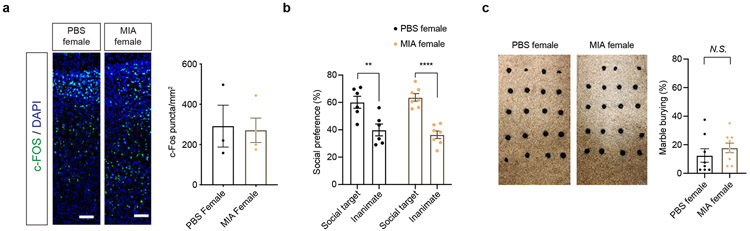
MIA offspring have previously been shown to display an abnormal increase in cortical neural activity, as measured by c-Fos, in the primary somatosensory cortex including its dysgranular zone (S1DZ).10 Optogenetic modulation of neural activity in S1DZ regulates social preference in MIA offspring.10 We tested whether ISR blockade also rescued the aberrant increase in cortical c-Fos. We confirmed that male MIA offspring demonstrate an increase in c-Fos expression in the cortex compared to male PBS offspring (Fig. 5c). By contrast, male Eif2αS51A/+ MIA offspring exhibited a statistically significant reduction in cortical c-Fos expression compared to male WT offspring (Fig. 5c). MIA female offspring did not exhibit a significant change in c-Fos expression compared to PBS offspring (Extended Data Fig. 8a). This result supports a model in which the ISR is a central regulatory hub necessary for molecular phenotypes of MIA.
Blockage of ISR activation rescues the neurobehavioral phenotype in MIA offspring
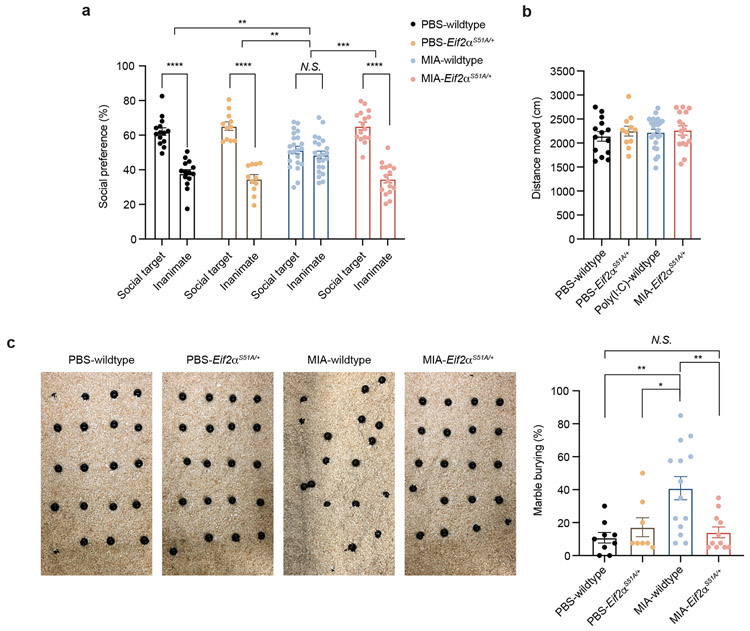
We next examined the effects of ISR attenuation on MIA-induced behavioral abnormalities. Previous studies have demonstrated that MIA male offspring have reproducible deficits in sociability and repetitive behavior (marble burying).9,10 We performed behavioral assessment of Eif2αS51A/+ and WT littermates from MIA and PBS control pregnancies. Consistent with prior results, WT MIA offspring demonstrated a loss of social preference compared to WT PBS offspring (Fig. 6a). Eif2αS51A/+ MIA offspring showed normal social preference compared to littermate wildtype MIA offspring. Total distance moved, a measurement of activity or arousal during the sociability test, was indistinguishable between Eif2αS51A/+ and wildtype (WT) littermates (Fig. 6b). In addition, WT MIA offspring exhibited increased marble burying behavior, while Eif2αS51A/+ MIA offspring exhibited similar marble burying behavior to PBS offspring (Fig. 6c). MIA-driven behavioral abnormalities in the offspring were not presented in the female offspring (Extended Data Fig. 8b,c).
Figure 6. Genetic ablation of ISR protects MIA offspring from neurobehavioral abnormalities.
a-b, Percentage of interaction (a) (Two-way ANOVA group (PBS wildtype, MIA wildtype, PBS eIF2αS51A/+ or MIA eIF2αS51A/+) x preference to the target (social target or inanimate) interaction F3,118 = 19.23, P = 3.214 x 10−10; effect of preference to the target F1,118 = 170.1, P = 1.0 x 10−15 followed by Sidak multiple comparisons test-within group: PBS wildtype **** P = 7.9 x 10−10, PBS eIF2αS51A/+ **** P = 1.19 x 10−11, MIA wildtype P = 0.7772, MIA eIF2αS51A/+ **** P = 2 x 10−15; two-way ANOVA stimulus (PBS or MIA) x genotype (wildtype or eIF2αS51A/+) interaction F1,59 = 5.275, P = 0.0252; effect of stimulus F1,59 = 5.100, P = 0.0276; effect of genotype F1,59 = 12.56, P = 0.0008 followed by Tukey multiple comparisons test: PBS wildtype versus MIA wildtype **P = 0.0054, MIA wildtype versus PBS eIF2αS51A/+ ***P = 0.0008, MIA wildtype versus MIA eIF2αS51A/+ ***P = 0.0001), and total distance moved (b) (Two-way ANOVA stimulus (PBS or MIA) x genotype (wildtype or eIF2αS51A/+) interaction F1,59 = 0.1195, P = 0.7308; effect of stimulus F1,59 = 0.2629, P = 0.6101; effect of genotype F1,59 = 0.6928, P = 0.4086) in the three-chamber sociability test of wildtype and eIF2αS51A/+ adult PBS and MIA offspring littermates (n = 14 for PBS wildtype, n = 11 for PBS eIF2αS51A/+, n = 22 for MIA wildtype, n = 16 for MIA eIF2αS51A/+ mice; 4 independent experiments).
c, Marble burying index of wildtype and eIF2αS51A/+ adult PBS and MIA offspring littermates (Two-way ANOVA stimulus (PBS or MIA) x genotype (wildtype or eIF2αS51A/+) interaction F1,38 = 8.180, P = 0.0068; effect of stimulus F1,38 = 5.410, P = 0.0255; effect of genotype F1,38 = 3.111, P = 0.0858 followed by Tukey multiple comparisons test: PBS wildtype versus MIA wildtype **P = 0.0025, MIA wildtype versus PBS eIF2αS51A/+ *P = 0.0292, MIA wildtype versus MIA eIF2αS51A/+ **P = 0.0045; n = 9 for PBS wildtype, n = 8 for PBS eIF2αS51A/+, n = 14 for MIA wildtype, n = 11 for MIA eIF2αS51A/+; 3 independent experiments).
Data are mean ± SEM; see Supplementary Table 20 for detailed statistics.
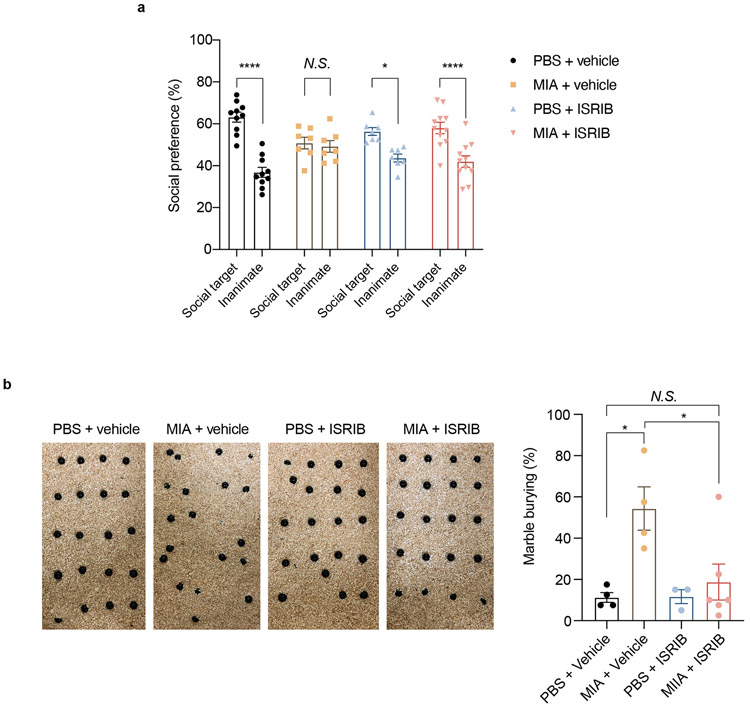
To further corroborate the importance of the ISR to the behavioral phenotype in MIA male offspring, we treated pregnant dams with the integrated stress response inhibitor ISRIB every other day after PBS or poly(I:C) injection. ISRIB rescues translation in the setting of eIF2α phosphorylation by promoting assembly of eIF2B.39-41 The male MIA offspring from ISRIB-treated mothers exhibit normalization of social interaction and marble burying behavior (Extended Data Fig. 9a,b). These data suggest that partial genetic and pharmacological blockade of the ISR are able to prevent MIA-associated behavioral abnormalities.
DISCUSSION
In this study, we explore the mechanisms by which intrauterine inflammation shape early neurodevelopment. We profile the transcriptional and translational signatures of MIA and identify key pathways perturbed by intrauterine inflammation. Dysregulation of protein homeostasis (proteostasis) has previously been implicated in a wide variety of cognitive and neurodevelopmental disorders,42 and the coordination between protein synthesis and breakdown plays a critical role in the regulation of neural circuit development and synaptic plasticity.43,44 Here, we show that MIA exerts an intrauterine, sexually dimorphic effect on cell type-specific transcription, with a disruption in genes associated with translation. These results have notable similarities to cell type-specific transcriptional changes observed in the ageing brain, including changes in ribosome biogenesis-associated genes.45 Our results also support the earlier findings by Lombardo et al., who identified altered expression of translation initiation factors in the fetal brain in response to maternal LPS exposure.46 Disruption of proteostasis may therefore be a common pathway in both neurodevelopmental and neurodegenerative disease.
In the present study, we identify activation of the ISR in male MIA offspring, and find that ISR activation is maternal IL-17a (and gut bacteria promoting Th17 cells)-dependent, which sheds new mechanistic insight into the role of IL-17a in mediating MIA-associated neurodevelopmental disorders. We also demonstrate that activation of the ISR in MIA offspring is necessary for the core behavioral phenotype, as genetic attenuation of the ISR rescues social and repetitive behavioral dysfunction in MIA male offspring. We observe activation of PERK, an upstream eIF2α kinase, in male MIA offspring, which suggests ER stress is a fundamental component of the early cellular response to MIA. This is in contrast to recent studies in Down Syndrome mice, which identified PKR as the primary ISR-associated kinase.11 Based on our results, we propose a model by which maternal-derived IL-17a triggers ER stress in the fetal cortex, thereby activating the ISR, arresting protein synthesis, and promoting chronic, unresolved stress at this critical stage of neurodevelopment. Moreover, our data suggest that there is a sex-specific vulnerability to ISR activation in the fetal brain. While male offspring have a more prominent disruption of translation machinery than females, both male and female offspring exhibit transcriptional changes in response to MIA. Female offspring may possess resiliency factors that attenuate chronic stress, which warrants further study. The sexually dimorphic response to MIA has been previously described16,17,47, and our results are consistent with prior reports suggesting sex-specific effects of intrauterine stress.48 The present study adds molecular insight to this phenomena and expands our knowledge of sex-specific transcription in the developing brain at single cell resolution. Additional studies are necessary to elucidate the cellular mechanisms by which the male fetal brain is especially susceptible to IL-17a, including possible roles of intrauterine neuroendocrine signaling.
EXPERIMENTAL PROCEDURES
Animals
All animals were housed in specific pathogen free (SPF) facility with 20-22°C, 40-55% humidity and 12-hour light/12-hour dark cycle. All experiments were conducted in accordance with procedures approved by the Institutional Animal Care and Use Committees of Harvard University. All wild-type C57BL/6 mice were obtained from Taconic (USA) except for those used in Extended Data Fig. 7 (ordered from Jackson laboratory (USA)). The generation and validation of Eif2αS51A/+ mice has been previously described.38
Maternal immune activation
Mice were mated overnight with 8 to 12-week-old C57BL/6 females carrying a mouse commensal, segmented filamentous bacteria (SFB).26,27 For Extended Data Fig. 7, both C57BL/6 males and females were directly purchased from Jackson laboratory, mated overnight and maintained as SFB-free. To generate wildtype and Eif2αS51A/+ littermates, we mated wildtype C57BL/6 Taconic females with SFB-positive Eif2αS51A/+ males. On E12.5, pregnant female mice were weighed and injected with a single dose (20 mg/kg, intraperitoneal injection) of poly(I:C) (P9582 or P1530, Sigma Aldrich) or PBS. Each dam was returned to its cage and left undisturbed until the birth of its litter. All pups remained with the mother until weaning on postnatal day 21–28, at which time mice were group-housed at a maximum of 5 per cage with same-sex littermates. For collecting embryonic tissues, pregnant dams were sacrificed either on E14.5 or E18.5. Embryonic cortices were freshly dissected and processed. For the IL-17 cytokine blockade experiment, monoclonal IL-17a blocking antibody (clone 50104; R&D) or isotype control antibody (IgG2a, clone 54447; R&D) were administered 5 h before maternal immune activation via i.p. route (500 μg/animal). For ISRIB treatment, pregnant female mice were injected with 2.5 mg/kg ISRIB or vehicle followed by PBS or poly(I:C) injection every two days from E12.5 to E18.5 as previously described.11 Sex of the embryos were determined by SRY gene by PCR (Forward: 5’- ACAAGTTGGCCCAGCAGAAT-3’, Reverse: 5’- GGGATATCAACAGGCTGCCA-3’).
Single cell RNA-sequencing
Samples were processed for single cell sequencing as described previously.50 Embryonic cortices at E14.5 and E18.5 were transferred to dissociation media: HBSS (Life Technologies), 10 mM HEPES (Sigma), 172 mg/L kynurenic acid (Sigma), 0.86 g/L MgCl2·6H2O (Sigma), and 6.3 g/L D-glucose (Sigma), 1uM TTX, 100 uM AP-V (Thermo Fisher Scientific), 5 ug/ml Actinomycin D (Sigma), 10 uM Triptolide (Sigma) and 10 ug/ml Anicomycin (Sigma). This solution was saturated with 95% oxygen and 5% CO2 and was pH-adjusted to 7.35 before use. Papain (20 U/mL; Worthington), Pronase from Streptomyces griseus (1 mg/mL; Sigma), proteinase XXIII from Aspergillus melleus (3 mg/mL; Sigma), and DNase (2 mg/mL; Worthington) were added to the dissociation media. Dissociation was carried out at 37 °C for 1 h. The samples were then triturated, filtered, and spun at 300 × g for 5 min. The pellet was resuspended in trypsin inhibitor solution (dissociation media with 1% BSA and 1% ovomucoid). Following gradient centrifugation (70 × g for 10 min), the cells were washed in dissociation media containing 0.04% BSA and resuspended in dissociation media containing 0.04% BSA and 15% OptiPrep (Sigma). Individual cells were captured and barcoded using the inDrops platform as previously described.51 Briefly, single-cell suspensions were fed into a microfluidic device that packaged the cells with barcoded hydrogel microspheres and reverse transcriptase/lysis reagents. After cell encapsulation, primers were photo-released by UV exposure. Indexed libraries were pooled and sequenced on a NextSeq 500 (Illumina).
Single cell RNA-sequencing data analysis
Sequencing data were aligned to the genome and processed according to a previously published pipeline (https://github.com/indrops/indrops). Briefly, this pipeline was used to build a custom transcriptome from Ensembl GRCm38 genome and GRCm38.84 annotation using Bowtie 1.1.1, after filtering the annotation gtf file (gencode.v17.annotation.gtf filtered for feature_type=”gene”, gene_type="protein_coding" and gene_status="KNOWN"). Unique molecular identifiers (UMIs) were used to link sequence reads back to individual captured molecules. All steps of the pipeline were run using default parameters unless explicitly stated.
All cells were combined into a single dataset. All mitochondrial genes were filtered from the dataset. Cells with fewer than 250 or more than 2500 unique genes were excluded. Cells were then clustered using the Seurat R package. The data were log normalized and scaled to 10,000 transcripts per cell. Variable genes were identified using the following parameters: x.low.cutoff = 0.0125, x.high.cutoff = 3, y.cutoff = 0.5. We limited the analysis to the top 30 principal components (PCs). Clustering resolution was set to 1. Clusters containing fewer than 100 cells were discarded, as were clusters for which one sample accounted for greater than 30 percent of the cells. Doublets were removed using Scrublet, and the remaining cells were re-clustered.52 The expression of known marker genes was used to assign each cluster to one of the main cell types. Clusters from each cell type were combined and the Seurat-based clustering repeated to characterize subtype diversity. The Seurat FindMarkers function was used to identify genetic markers of cellular subtypes.
Identification of differentially expressed genes in single cell sequencing data
To identify differentially expressed genes by cell type, we performed a differential gene expression analysis using edgeR (3.28.1).53,54 The analysis was conducted on each cell type and also certain unions of cell types with common traits. To detect sex-dependent effects of maternal immune activation on gene expression, the difference between the change in gene expression due to MIA in males and the same change in females was used. Next, pairwise comparisons were performed to measure the effect of MIA on gene expression in each sex, and also the baseline differences between sexes without treatment. The data were modeled and normalized using a negative binomial distribution, and counts data was normalized for gene length and read depth. Genes whose false discovery rate (FDR) was less than 5% were considered statistically significant. GO analysis was performed using the PANTHER classification system (FDR < 0.05 for significant GO terms).55
Three-chamber social approach assay
8~12-week-old male or female mice were tested for social behavior using a three-chamber social approach paradigm. A day before the test, experimental mice were introduced into a three-chamber arena (50 cm x 35 cm x 30 cm) with only empty object containment cages (circular metallic cages, Stoelting Neuroscience) for a 10 min acclimation phase. The following day, the mice underwent 5 min exploration period with empty object containment cages. Immediately after, the mice were confined to the center chamber, while a social object (unfamiliar C57BL/6 male mouse) in one chamber and an inanimate object (rubber object of a similar size as the social object) in the other chamber were placed. Barriers to the adjacent chambers were removed, and the experimental mice were given 10 min to explore both chambers and measured for approach behavior as interaction time (i.e. sniffing, approach) with targets in each chamber (within 2 cm). Sessions were video-recorded and object exploration time and total distance moved were analyzed using the Noldus tracking system. A social preference was calculated as the percentage of time spent investigating the social target out of the total exploration time of both objects.56 Arenas and contents were thoroughly cleaned between testing sessions. Multiple social targets from different home cages were used for testing to prevent potential odorant confounds from target home cages.
Marble burying test
8~12-week-old male or female mice were placed in a testing arena (arena size: 40 × 20 cm2, bedding depth: 3 cm) containing 20 glass marbles, which were laid out in five rows of four marbles equidistant from one another. At the end of a 15 min exploration period, mice were gently removed from the testing cages and the number of marbles buried was recorded. A marble burying index was scored as 1 for marbles covered >50% by bedding, 0.5 for around 50% covered, or 0 for anything less. Percentage of buried marbles is plotted on the y-axis.
Immunoblotting
For immunoblot analyses, embryonic corticies were lysed in lysis buffer consisting of 1% Triton X-100, 150 mM NaCl, 50 mM Tris-HCl (pH7.4), 1 mM EDTA, 0.25% sodium deoxycholate, 0.1% SDS, phosphatase inhibitor cocktail (Thermo Fisher Scientific), and protease inhibitor cocktail (Roche). Protein concentrations were determined by the bicinchoninic acid assay (Thermo Fisher Scientific). Proteins were denatured by the addition of SDS containing Laemmli sample buffer (Boston Bioproduct) and boiled for 5 mins at 95°C. Antibodies against the following proteins were obtained from the indicated sources: phospho-S6K (9205), S6K (9202), phospho–eIF2α (9721), eIF2α (9722), phospho-PERK (3179), PERK (5683), GCN2 (3302), IRE1α (3294), β-Actin (4970), Horseradish peroxidase (HRB)–conjugated anti-rabbit secondary antibodies (7074) (Cell Signaling); phospho-GCN2 (ab75836), phospho-PKR (ab32036) (Abcam); pIRE1α (NB100-2323), ATF6 (NBP1-40256) (Novus Biologicals); and PKR (18244-1-AP; Proteintech). All primary antibodies were used in 1:1000 dilution in 10% blocking buffer (Thermo Fisher Scientific) in TBS-T (Tris-buffered saline, 0.1% Tween-20). The HRP-conjugated secondary antibody was used in 1:2000 dilution. The density of the resulting bands was quantified using ImageJ version 2.0.0.
Surface Sensing of Translation (SUnSET) and click conjugation with TAMRA-azide
Protein synthesis was measured using modified-SUnSET method, a non-radioactive labeling method to monitor protein synthesis, as previously described.22 Briefly, the fetal cortex was microdissected and incubated for 1 hour at room temperature in oxygenated (95% O2, 5% CO2) ACSF followed by incubation at 32°C for 1 hour in oxygenated (95% O2, 5% CO2) ACSF. O-propargyl-puromycin (OP-Puro, 25 μM) was bath applied to the slices for 20 min followed by a wash with untreated oxygenated ACSF. The slices were then snap-frozen on dry ice and stored at −80 C until use. Frozen slices were lysed in homogenization buffer (1% Triton X-100, 150 mM NaCl, 50 mM Tris-HCl (pH7.4), and protease inhibitor cocktail). OP-Puro incorporation was detected by click chemistry (Invitrogen Cat. C10276) according to the manufacturer’s recommendations. After click conjugation with TAMRA-azide, 20 μg of protein samples was treated with 6x Laemmli sample buffer and resolved by SDS-PAGE gels. The gel was visualized by Bio-Rad ChemiDoc imaging system using excitation wavelength 647 nm, then stained with Coomassie blue reagent. The density of the resulting bands was quantified using ImageJ version 2.0.0.
Ribosome Profiling and RNA-seq
We employed a modified version of the ribosome profiling protocol described by McGlincy et al.30 Flash-frozen embryonic brain tissue was thawed in 400 μl lysis buffer (150 mM NaCl, 20 mM Tris, pH 7.4, 5mM MgCl2, 1mM DTT, 100 μg/ml cyclohexamide, 1% v/v Triton X-100, and 25 U/ml Turbo DNase I (ThermoFisher Scientific). Tissue was pipetted ten times in lysis buffer and incubated for 10 minutes on ice. Tissue was then triturated ten times through a 26-gauge needle. Lysate were clarified by centrifugation for 10 min at 20000 × g at 4°C and supernatant was recovered. RNA concentration was quantified using Qubit RNA assay (ThermoFisher Scientific). 5% input was reserved, and total RNA purified using the Direct-zol RNA MiniPrep kit (Zymo) with DNaseI treatment. RNA-seq libraries were prepared from 10 ng total RNA using the SMARTer® Stranded Total RNA-Seq Kit v2 (Takara) and sequenced on a Next-seq 500 using paired-end 2 x 37nt sequencing runs.
For ribosome profiling, 1.5μl RNase I (10U/μl, ThermoFisher Scientific) was add to lysate containing 30 μg RNA and incubated for 45 minutes at room temperature with gentle agitation. 10 μl SUPERaseIn RNase inhibitor (20U/μl, ThermoFisher Scientific) was added after 45 minutes and samples transferred to ice to stop nuclease digestion. Lysate was transferred to a polycarbonate ultracentrifuge tube and underlayed with 0.9 ml sucrose cushion (1 M sucrose, 20 U/ml SUPERaseIn in Polysome buffer, 150 mM NaCl, 20 mM Tris, pH 7.4, 5mM MgCl2, 1mM DTT, 100 μg/ml cycohexamide). Ribosome-protected RNAs were pelleted by centrifugation at 52,000 rpm, 4°C for 2 h. Supernatant was removed, and pellet resuspended in 300 μl TRIzol reagent. RNA fragments were purified using a Direct-zol RNA MiniPrep kit (Zymo Research) and eluted in 100 μl RNase-free water. RNA was precipitated with one volume of isopropanol, 30 μg GlycoBlue, and 100 mM NaOAc pH 5.5 (300 μl total volume), and chilled at −20°C for at least 2 hours, or overnight. RNA was pelleted by centrifugation for 30 min at 20000 × g, 4°C, supernatant was removed, and pellets were air-dried for 10 min. Purified RNA was resuspended in 5 μl 10 mM Tris pH 8, and RNA 17-34 nucleotides was size selected using a 15% TBE-Urea gel (Life Technologies). RNA was extracted from gel sections in 400 ul RNA extraction buffer (300 mM NaOAc, pH 5.5, 1mM EDTA, 0.25% v/v SDS), then precipitated with 30 μg GlycoBlue and one volume of isopropanol. Precipitations were left at −20°C overnight. Size-selected RNA was resuspended in 4 μl 10 mM Tris pH 8 and transferred to a clean non-stick RNase-free microfuge tube. Size-selected RNA was dephosphorylated with T4 PNK (NEB) at 37°C for 1 hour. The reaction was brought up to 50 μl with nuclease-free water and purified using 1.8 volumes of Agencourt RNAClean XP beads (Beckman Coulter) and 5.4 volumes of isopropanol. Beads were captured on a magnetic rack and washed twice with 85% EtOH. Beads were air-dried and resuspended in 7 μl nuclease-free water. Supernatant containing purified RNA was removed from beads and quantified by Qubit RNA assay (ThermoFisher Scientific). Sequencing libraries prepared using the SMARTer® smRNA-Seq Kit for Illumina® (Takara). Libraries were sequenced on a Novaseq 6000 using single-end 1 x 50 nt sequencing runs.
RNA-seq and ribosome profiling libraries were analyzed in parallel. Reads were aligned to the mm10 genome using STAR Version 2.5.4a.57 Data was then analyzed by RibORF Version 1.0, which confirmed periodicity of ribo-seq data.58 The RibORF script was run using SAM files, read lengths 22, 28, 29, and 30 nucleotides, and an offset value of 12 nucleotides. Htseq Version 0.9.159 was used with default parameters to generate raw counts files from bam files for both RNA-seq and Ribo-seq. Counts were filtered to remove mitochondrial and ribosomal RNAs and differential gene expression analysis was performed using edgeR with the same scheme as the single cell analysis. For ribosome profiling reads, ribosomal RNA was filtered using Bowtie260, followed by alignment of the non-ribosomal reads to the mm10 genome using STAR Version 2.5.4a.57
Immunohistochemistry
Mice were transcardially perfused with ice-cold PBS first, and then paraformaldehyde (PFA) (4% in PBS). Brains were kept in PFA overnight at 4°C and moved to PBS until sectioned. Brains were submerged in PBS with 3% agarose, and then sliced on a vibratome (Leica VT1000S) into 50 μm coronal sections. Coronal sections were washed three times with PBS containing 0.3% TritonX-100 (PBST) and blocked for 1 hr at room temperature with PBST containing 5% donkey serum. Sections were incubated overnight at 4°C with rabbit anti-Fos antibody 1:2000 (Cedarlane # 226003(SY)), washed again three times with PBST, and incubated for 1 hr at room temperature with 1:500 donkey anti-rabbit 647 secondary antibody (Life Technologies # A31573). After washing in PBST and PBS, samples were mounted onto SuperFrost Plus glass slides (VWR) using DAPI Fluoromount-G.
Images of brain slices were taken using a 10X magnification lens on Olympus VS-120 Slide Scanner using a 50 ms exposure time for DAPI signal and 3 s exposure time for CY5 signal corresponding to Alexa 647 secondary antibody. Image analysis was performed by a blinded investigator. CY5 signal images were loaded into ImageJ and reduced to include only fluorescent puncta using the native “adjust threshold” function. Brains with technical/quality issues were excluded. One cortical region of interest (ROI) from each side of the brain slice was drawn using the freehand ROI maker tool in ImageJ. Area within each drawn ROI was measured and recorded. Fluorescent puncta were automatically counted using the “analyze particles” function in ImageJ with 0.7 circularity threshold and 3 square micron size threshold to exclusively count biologically relevant fluorescent puncta. Of note, analysis of male brain tissue (Figure 5c) was performed independently from analysis of female brain tissue (Extended Figure 8a). Therefore, these results cannot be directly compared.
Statistics and reproducibility
No statistical methods were used to predetermine sample size, but the sample sizes were determined based on similar, previously conducted studies.10,11 Data distribution was assumed to be normal, but this was not formally tested. Animals were randomized into different groups with approximately comparable numbers of animals in each group whenever possible. Analysis of c-Fos immunohistochemistry was performed by a blinded investigator. For the behavioral assays, experiments were run in a semi-blinded way, which means mice were numbered earlier at the weaning and their genotype information was matched after completion of the behavioral experiment. In the case of all other experiments, investigators were not blinded during group allocation and data analysis due to the nature of the experiments. The statistical analyses were performed using GraphPad Prism software V8, using two-way ANOVA or one-way ANOVA followed by least significance difference post-hoc tests, or using Student’s t-test, both two-sided. Details of specific statistical comparisons are listed in the relevant figure legends.
Reporting summary
Further information on research design is available in the Life Sciences Reporting Summary linked to this article.
Code availability
All custom code is available at https://github.com/bkalishneuro/Maternal-Immune-Activation-Project/blob/main/README.md.
Extended Data
Extended Data Fig. 1. Quality Control and E14.5 Sub-clustering.
a, Total number of genes per condition and total number of unique molecular identifiers (UMIs) per condition in the final E14.5 and E18.5 single cell sequencing datasets. Data from n = 2 mice per group.
b, UMAP of sub-clustering of mature and immature (SVZ, cortical plate, cortical subplate) neurons at E14.5. Data from n = 2 mice per group.
c, UMAP of sub-clustering of radial glia (RG) at E14.5. Data from n = 2 mice per group.
d, UMAP of sub-clustering of interneurons (Int), ganglionic eminence (GE), and striatal neurons at E14.5. Data from n = 2 mice per group.
e, Dot plot of marker genes associated with all cells at E14.5. Data from n = 2 mice per group.
f, Dot plot of marker genes associated with the cells in (b): mature and immature (SVZ, cortical plate, cortical subplate) neurons at E14.5. Data from n = 2 mice per group.
g, Dot plot of marker genes associated with the cells in (d): interneurons (Int), ganglionic eminence (GE), and striatal neurons at E14.5. Data from n = 2 mice per group.
h, Dot plot of marker genes associated with the cells in (c): radial glia (RG) at E14.5. Data from n = 2 mice per group.
Extended Data Fig. 2. E18.5 sub-clustering.
a, UMAP of sub-clustering of mature and immature (SVZ) neurons at E18.5. Data from n = 2 mice per group.
b, UMAP of sub-clustering of radial glia (RG), ganglionic eminence (GE), oligodendrocytes (Olig), and neural stem cells (NSC) at E18.5. Data from n = 2 mice per group.
c, UMAP of sub-clustering of striatal neurons (Str) and interneurons (Int) at E18.5. Data from n = 2 mice per group.
d, Dot plot of marker genes associated with cells in (a): mature and immature (SVZ) neurons at E18.5. Data from n = 2 mice per group.
e, (left) Dot plot of marker genes associated with cells in (b): radial glia (RG), ganglionic eminence (GE), oligodendrocytes (Olig), and neural stem cells (NSC) at E18.5; and (right) Dot plot of marker genes associated with cells in (c): striatal neurons (Str) and interneurons (Int) at E18.5. Data from n = 2 mice per group.
Extended Data Fig. 3. Differential gene expression strip plots.
a, Strip plot displaying differential gene expression between MIA male offspring and PBS male offspring at E14.5. Colored dots represent significant genes (FDR < 0.05). X-axis displays select cortical cell types. Cell groups (left to right): (1) mature and immature (SVZ, cortical plate, cortical subplate) neurons, (2) radial glia (RG), and (3) interneurons (Int), ganglionic eminence (GE), and striatal neurons. Data from n = 2 mice per group.
b, Strip plot displaying sex-dependent gene expression in MIA offspring at E14.5. Colored dots represent significant genes (FDR < 0.05). X-axis displays select cortical cell types. Cell groups (left to right): (1) mature and immature (SVZ, cortical plate, cortical subplate) neurons, (2) radial glia (RG), and (3) interneurons (Int), ganglionic eminence (GE), and striatal neurons. Data from n = 2 mice per group.
c, Strip plot displaying differential gene expression between MIA male offspring and PBS male offspring at E18.5. Colored dots represent significant genes (FDR < 0.05). X-axis displays select cortical cell types. Cell groups (left to right): (1) mature and immature (SVZ) neurons, (2) radial glia (RG), ganglionic eminence (GE), oligodendrocytes (Olig), and neural stem cells (NSC), and (3) striatal neurons (Str) and interneurons (Int). Data from n = 2 mice per group.
d, Strip plot displaying sex-dependent gene expression in MIA offspring at E18.5. Colored dots represent significant genes (FDR < 0.05). X-axis displays select cortical cell types. Cell groups (left to right): (1) mature and immature (SVZ) neurons, (2) radial glia (RG), ganglionic eminence (GE), oligodendrocytes (Olig), and neural stem cells (NSC), and (3) striatal neurons (Str) and interneurons (Int). Data from n = 2 mice per group.
Extended Data Fig. 4. Single cell differential gene expression changes.
a, Bubble plot of highly variable genes between MIA and PBS male offspring at E18.5. All significant genes FDR < 0.05. Blue indicates an increase in MIA males relative to PBS males. Data from n = 2 mice per group.
b, Bubble plot of sex-dependent genes in MIA offspring at E18.5. All significant genes FDR < 0.05. Blue indicates an increase in Δ(MIA males – PBS males) relative to females. Data from n = 2 mice per group.
c, Bubble plot of ribosome subunit genes between MIA male and PBS male offspring at E14.5, demonstrating a widespread decrease in expression of multiple ribosomal subunits in MIA male offspring. All significant genes FDR < 0.05. Blue indicates an increase in PBS males relative to MIA males. Data from n = 2 mice per group.
d, Bubble plot of ribosome subunit genes between MIA female and PBS female offspring at E14.5, demonstrating an increase in expression of multiple ribosomal subunits in MIA female offspring. All significant genes FDR < 0.05. Blue indicates an increase in PBS females relative to MIA females. Data from n = 2 mice per group.
Extended Data Fig. 5. Sex-specific comparisons in control (PBS) conditions.
a, Bubble plot of ribosome subunit genes between PBS male and PBS female offspring at E14.5. All significant genes FDR < 0.05. Data from n = 2 mice per group.
b, Bubble plot of ribosome subunit genes between PBS male and PBS female offspring at E18.5. All significant genes FDR < 0.05. Data from n = 2 mice per group.
Extended Data Fig. 6. Upstream regulators of phospho-eIF2α and UPR sensors in E18.5 PBS and MIA fetal cortices.
a, Immunoblot analysis measuring phospho-S6K and quantification in E18 PBS and MIA fetal cortices. Y-axis represents relative blot intensity to PBS male control (Two-way ANOVA sex (male or female) × stimulus (PBS or MIA) interaction F1,12 = 0.08413, P = 0.7767; effect of sex F1,12 = 1.188, P = 0.2971; effect of stimulus F1,12 = 0.04729, P = 0.8315 followed by Tukey multiple comparisons test; n = 4 pups from 2 litters).
b, Immunoblot analysis measuring upstream regulators of phospho-eIF2α and quantification in E18.5 PBS and MIA fetal cortices. Y-axis represents relative blot intensity to PBS control for each sex (Two-tailed unpaired t-test male: pPERK t = 5.485, df = 10, ***P = 0.0003, n = 6 from 3 litters; pGCN2 t = 0.1503, df = 6, n = 4 pups from 2 litters; pPKR t = 0.9501, df = 6, n = 4 pups from 2 litters; Two-tailed unpaired t-test female: pPERK t = 0.5761, df = 6; pGCN2 t = 0.5204, df = 6; pPKR t = 1.545, df = 6; n = 4 pups from 2 litters).
c, Immunoblot analysis measuring UPR sensors (IRE1α, ATF6 and PERK) and quantification in E18.5 PBS and MIA male fetal cortices. Y-axis represents relative blot intensity to PBS control (Two-tailed unpaired t-test: pIRE t = 2.745, df = 6, *P = 0.0335; ATF6 t = 0.6822, df = 6; pPERK t = 3.516, df = 6, * P = 0.0126; n = 4 pups from 2 litters).
Data are mean ± SEM; see Supplementary Table 20 for detailed statistics. Unprocessed blots are provided as a Source Data file.
Extended Data Fig. 7. ISR activation of E18.5 PBS and MIA male fetus in SFB-negative dams.
Immunoblot analysis measuring phospho-eIF2α and quantification in E18.5 PBS and MIA male fetal cortices, either from SFB-positive or SFB-negative B6 dams. SFB-positive dams were obtained from Taconic Biosciences (Tac) whereas SFB-negative dams from Jackson laboratory (Jax) and maintained in SFB-positive and negative conditions, respectively. Y-axis represents relative blot intensity to each PBS control (Two-tailed unpaired t-test: Tac t = 4.659, df = 6, **P = 0.0035; Jax t = 0.9013, df = 6; n = 4 pups from 2 litters). Data are mean ± SEM; see Supplementary Table 20 for detailed statistics. Unprocessed blots are provided as a Source Data file.
Extended Data Fig. 8. Female MIA offspring neither show ISR activation nor neurodevelopmental abnormalities.
a, Representative images of 8-10 weeks old MIA and PBS female offspring brain tissue immunostained for c-Fos. Scale bar 100 μm. Quantification indicates c-Fos puncta/mm2 (Two-tailed unpaired t-test PBS female versus MIA female: t = 0.1829, df = 5, P = 0.8621; n= 3 for PBS female, n = 4 for MIA female mice).
b, Percentage of interaction in the three-chamber sociability test of adult PBS and MIA female offspring littermates (Two-way ANOVA group (PBS female or MIA female) x preference to the target (social target or inanimate) interaction F1,22= 0.986, P = 0.3315; effect of preference to the target F1,22 = 45.24 , P = 9.2454 x 10−7 followed by Sidak multiple comparisons test-within group: PBS female ** P = 0.0015, MIA female **** P = 2.06 x 10−5; Two-tailed unpaired t-test social score between PBS female versus MIA female: t = 0.7021, df = 11, P = 0.4972; n = 6 for PBS female, n = 7 for MIA female mice; 2 independent experiments).
c, Marble burying index of adult PBS and MIA female offspring littermates (Two-tailed unpaired t-test PBS female versus MIA female: t = 0.9278, df = 15, P = 0.3682; n = 8 for PBS female, n = 9 for MIA female mice; 2 independent experiments).
Data are mean ± SEM; see Supplementary Table 20 for detailed statistics.
Extended Data Fig. 9. Pharmacological inhibition of ISR protects MIA offspring from neurobehavioral abnormalities.
a, Percentage of interaction in the three-chamber sociability test of vehicle and ISRIB-treated adult PBS and MIA offspring littermates (Two-way ANOVA group (PBS vehicle, MIA vehicle, PBS ISRIB or MIA ISRIB) x preference to the target (social target or inanimate) interaction F3,62 = 7.401, P = 0.003; effect of preference to the target F1,62 = 57.62 , P = 2 x 10−10 followed by Bonferroni multiple comparisons test-within group: PBS vehicle **** P = 4.9 x 10−10, PBS ISRIB * P = 0.0119, MIA vehicle P > 0.9999, MIA ISRIB **** P = 2.76 x 10−5; two-way ANOVA stimulus (PBS or MIA) x treatment (vehicle or ISRIB) interaction F1,31 = 7.111, P = 0.0121; effect of stimulus F1,31 = 4.109, P = 0.0513; effect of treatment F1,31 = 0.003618, P = 0.9524 followed by Dunnett multiple comparisons test: PBS vehicle versus PBS ISRIB P = 0.19, MIA vehicle versus PBS vehicle **P = 0.0071, PBS vehicle versus MIA ISRIB P = 0.3031; n = 10 for PBS vehicle, n = 7 for MIA vehicle, n = 7 for PBS ISRIB, n = 11 for MIA ISRIB; 2 independent experiments).
b, Marble burying index of vehicle and ISRIB-treated adult PBS and MIA offspring littermates (Two-way ANOVA stimulus (PBS or MIA) x treatment (vehicle or ISRIB) interaction F1,13 = 4.549, P = 0.0526; effect of stimulus F1,13 = 8.829, P = 0.0108; effect of treatment F1,13 = 4.341, P = 0.0575 followed by Tukey multiple comparisons test: PBS vehicle versus MIA vehicle *P = 0.0147, PBS ISRIB versus MIA vehicle *P = 0.0255, MIA vehicle vs. MIA ISRIB *P = 0.0275; n = 4 for PBS vehicle, n = 4 for MIA vehicle, n = 3 for PBS ISRIB, n = 6 for MIA ISRIB mice; 2 independent experiments).
Data are mean ± SEM; see Supplementary Table 20 for detailed statistics.
Supplementary Material
Supplementary Table 1 Differential gene expression between MIA male and PBS male offspring in E14 single cell RNA-seq. Data analyzed using EdgeR with Quasi-likelihood F test. Data includes Log2 fold change, logCPM (counts per million), F statistic, p value (unadjusted), and false discovery rate (FDR) adjusted for multiple comparisons.
Supplementary Table 2 Differential gene expression between MIA female and PBS female offspring in E14 single cell RNA-seq. Data analyzed using EdgeR with Quasi-likelihood F test. Data includes Log2 fold change, logCPM (counts per million), F statistic, p value (unadjusted), and false discovery rate (FDR) adjusted for multiple comparisons.
Supplementary Table 3 Differential gene expression between PBS male and PBS female offspring in E14 single cell RNA-seq. Data analyzed using EdgeR with Quasi-likelihood F test. Data includes Log2 fold change, logCPM (counts per million), F statistic, p value (unadjusted), and false discovery rate (FDR) adjusted for multiple comparisons.
Supplementary Table 4 Differential gene expression between MIA male and MIA female offspring in E14 single cell RNA-seq using interaction term analysis. Data analyzed using EdgeR with Quasi-likelihood F test. Data includes Log2 fold change, logCPM (counts per million), F statistic, p value (unadjusted), and false discovery rate (FDR) adjusted for multiple comparisons.
Supplementary Table 5 Differential gene expression between MIA male and PBS male offspring in E18 single cell RNA-seq. Data analyzed using EdgeR with Quasi-likelihood F test. Data includes Log2 fold change, logCPM (counts per million), F statistic, p value (unadjusted), and false discovery rate (FDR) adjusted for multiple comparisons.
Supplementary Table 6 Differential gene expression between MIA female and PBS female offspring in E18 single cell RNA-seq. Data analyzed using EdgeR with Quasi-likelihood F test. Data includes Log2 fold change, logCPM (counts per million), F statistic, p value (unadjusted), and false discovery rate (FDR) adjusted for multiple comparisons.
Supplementary Table 7 Differential gene expression between PBS male and PBS female offspring in E18 single cell RNA-seq. Data analyzed using EdgeR with Quasi-likelihood F test. Data includes Log2 fold change, logCPM (counts per million), F statistic, p value (unadjusted), and false discovery rate (FDR) adjusted for multiple comparisons.
Supplementary Table 8 Differential gene expression between MIA male and MIA female offspring in E18 single cell RNA-seq using interaction term analysis. Data analyzed using EdgeR with Quasi-likelihood F test. Data includes Log2 fold change, logCPM (counts per million), F statistic, p value (unadjusted), and false discovery rate (FDR) adjusted for multiple comparisons.
Supplementary Table 9 Quality control metrics for ribosome profiling data
Supplementary Table 10 Differential gene expression between MIA male and PBS male offspring in E18 bulk RNA-seq. Data analyzed using EdgeR with Quasi-likelihood F test. Data includes Log2 fold change, logCPM (counts per million), F statistic, p value (unadjusted), and false discovery rate (FDR) adjusted for multiple comparisons.
Supplementary Table 11 Differential gene expression between MIA female and PBS female offspring in E18 bulk RNA-seq. Data analyzed using EdgeR with Quasi-likelihood F test. Data includes Log2 fold change, logCPM (counts per million), F statistic, p value (unadjusted), and false discovery rate (FDR) adjusted for multiple comparisons.
Supplementary Table 12 Differential gene expression between PBS male and PBS female offspring in E18 bulk RNA-seq. Data analyzed using EdgeR with Quasi-likelihood F test. Data includes Log2 fold change, logCPM (counts per million), F statistic, p value (unadjusted), and false discovery rate (FDR) adjusted for multiple comparisons.
Supplementary Table 13 Differential gene expression between MIA male and MIA female offspring in E18 bulk RNA-seq using interaction term analysis. Data analyzed using EdgeR with Quasi-likelihood F test. Data includes Log2 fold change, logCPM (counts per million), F statistic, p value (unadjusted), and false discovery rate (FDR) adjusted for multiple comparisons.
Supplementary Table 14 Differential gene expression between MIA male and MIA female offspring in E18 bulk RNA-seq using a pairwise comparison. Data analyzed using EdgeR with Quasi-likelihood F test. Data includes Log2 fold change, logCPM (counts per million), F statistic, p value (unadjusted), and false discovery rate (FDR) adjusted for multiple comparisons.
Supplementary Table 15 Differential translation analysis between MIA male and PBS male offspring in E18 ribosome profiling. Data analyzed using EdgeR with Quasi-likelihood F test. Data includes Log2 fold change, logCPM (counts per million), F statistic, p value (unadjusted), and false discovery rate (FDR) adjusted for multiple comparisons.
Supplementary Table 16 Differential translation analysis between MIA female and PBS female offspring in E18 ribosome profiling. Data analyzed using EdgeR with Quasi-likelihood F test. Data includes Log2 fold change, logCPM (counts per million), F statistic, p value (unadjusted), and false discovery rate (FDR) adjusted for multiple comparisons.
Supplementary Table 17 Differential translation analysis between PBS male and PBS female offspring in E18 ribosome profiling. Data analyzed using EdgeR with Quasi-likelihood F test. Data includes Log2 fold change, logCPM (counts per million), F statistic, p value (unadjusted), and false discovery rate (FDR) adjusted for multiple comparisons.
Supplementary Table 18 Differential translation analysis between MIA male and MIA female offspring in E18 ribosome profiling using interaction term analysis. Data analyzed using EdgeR with Quasi-likelihood F test. Data includes Log2 fold change, logCPM (counts per million), F statistic, p value (unadjusted), and false discovery rate (FDR) adjusted for multiple comparisons.
Supplementary Table 19 Differential translation analysis between MIA male and MIA female offspring in E18 ribosome profiling using a pairwise comparison. Data analyzed using EdgeR with Quasi-likelihood F test. Data includes Log2 fold change, logCPM (counts per million), F statistic, p value (unadjusted), and false discovery rate (FDR) adjusted for multiple comparisons.
Supplementary Table 21 Source data for Fig. 4C
ACKNOWLEDGEMENTS
BTK was supported by the Pediatric Scientist Development Program and the March of Dimes. EK was supported by the Basic science research program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2018R1A6A3A03010693). MEG and BTK were supported by R01 NS115965 from the National Institute of Neurological Disorders and Stroke. RJK was supported by NIH grants R01CA198103-04, R01DK113171-03 and R01AG062190-02. EED was supported by the Damon Runyon Foundation. GBC and JRH were supported by the Jeongho Kim Neurodevelopmental Research Fund, the Simons Foundation Autism Research Initiative and the National Institute of Mental Health grants (R01MH115037 and R01MH119459, respectively). JRH was also supported by the PEW Scholars Program, by N of One: Autism Research Foundation and by the Burroughs Wellcome Fund. Figures 1a, 3b, 4a were created using Biorender.com.
Footnotes
Accession Codes
All sequencing data has been deposited in the Gene Expression Omnibus under accession GSE148237
COMPETING INTERESTS STATEMENT
The authors do not report any conflicts of interest related to the content of this work.
Data Availability
Sequence data that support the findings of this study have been deposited in the Gene Expression Omnibus with the accession code GSE148237. Publicly available datasets from PANTHER (http://pantherdb.org/) were used for gene ontology analysis.
REFERENCES
- 1.Buffington SA et al. Microbial Reconstitution Reverses Maternal Diet-Induced Social and Synaptic Deficits in Offspring. Cell 165, 1762–1775 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hashimoto-Torii K et al. Roles of heat shock factor 1 in neuronal response to fetal environmental risks and its relevance to brain disorders. Neuron 82, 560–572 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gladwyn-Ng I et al. Stress-induced unfolded protein response contributes to Zika virus-associated microcephaly. Nat. Neurosci 21, 63–71 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Al-Haddad BJS et al. Long-term Risk of Neuropsychiatric Disease After Exposure to Infection In Utero. JAMA Psychiatry 76, 594–602 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Atladóttir HO et al. Maternal infection requiring hospitalization during pregnancy and autism spectrum disorders. J. Autism Dev. Disord 40, 1423–1430 (2010). [DOI] [PubMed] [Google Scholar]
- 6.Patterson PH Immune involvement in schizophrenia and autism: etiology, pathology and animal models. Behav. Brain Res 204, 313–321 (2009). [DOI] [PubMed] [Google Scholar]
- 7.Lee BK et al. Maternal hospitalization with infection during pregnancy and risk of autism spectrum disorders. Brain. Behav. Immun 44, 100–105 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith SEP, Li J, Garbett K, Mirnics K & Patterson PH Maternal immune activation alters fetal brain development through interleukin-6. J. Neurosci. Off. J. Soc. Neurosci 27, 10695–10702 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choi GB et al. The maternal interleukin-17a pathway in mice promotes autism-like phenotypes in offspring. Science 351, 933–939 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shin Yim Y et al. Reversing behavioural abnormalities in mice exposed to maternal inflammation. Nature 549, 482–487 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu PJ et al. Activation of the ISR mediates the behavioral and neurophysiological abnormalities in Down syndrome. Science 366, 843–849 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Larhammar M et al. Dual leucine zipper kinase-dependent PERK activation contributes to neuronal degeneration following insult. eLife 6, e20725 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Green KM et al. RAN translation at C9orf72-associated repeat expansions is selectively enhanced by the integrated stress response. Nat. Commun 8, 2005 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheng W et al. C9ORF72 GGGGCC repeat-associated non-AUG translation is upregulated by stress through eIF2α phosphorylation. Nat. Commun 9, 51 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Loo L et al. Single-cell transcriptomic analysis of mouse neocortical development. Nat. Commun 10, 134 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carlezon WA et al. Maternal and early postnatal immune activation produce sex-specific effects on autism-like behaviors and neuroimmune function in mice. Sci. Rep 9, 16928 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gogos A, Sbisa A, Witkamp D & van den Buuse M Sex differences in the effect of maternal immune activation on cognitive and psychosis-like behaviour in Long Evans rats. Eur. J. Neurosci 52, 2614–2626 (2020) [DOI] [PubMed] [Google Scholar]
- 18.Rubio Gomez MA & Ibba M Aminoacyl-tRNA synthetases. RNA N. Y. N 26, 910–936 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Evers R, Smid A, Rudloff U, Lottspeich F & Grummt I Different domains of the murine RNA polymerase I-specific termination factor mTTF-I serve distinct functions in transcription termination. EMBO J. 14, 1248–1256 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lessard F, Stefanovsky V, Tremblay MG & Moss T The cellular abundance of the essential transcription termination factor TTF-I regulates ribosome biogenesis and is determined by MDM2 ubiquitinylation. Nucleic Acids Res. 40, 5357–5367 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pitale PM, Gorbatyuk O & Gorbatyuk M Neurodegeneration: Keeping ATF4 on a Tight Leash. Front. Cell. Neurosci 11, 410 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmidt EK, Clavarino G, Ceppi M & Pierre P SUnSET, a nonradioactive method to monitor protein synthesis. Nat. Methods 6, 275–277 (2009). [DOI] [PubMed] [Google Scholar]
- 23.Pakos-Zebrucka K et al. The integrated stress response. EMBO Rep. 17, 1374–1395 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ounallah-Saad H, Sharma V, Edry E & Rosenblum K Genetic or pharmacological reduction of PERK enhances cortical-dependent taste learning. J. Neurosci. Off. J. Soc. Neurosci 34, 14624–14632 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hetz C & Papa FR The Unfolded Protein Response and Cell Fate Control. Mol. Cell 69, 169–181 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Kim S et al. Maternal gut bacteria promote neurodevelopmental abnormalities in mouse offspring. Nature 549, 528–532 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lammert CR et al. Cutting Edge: Critical Roles for Microbiota-Mediated Regulation of the Immune System in a Prenatal Immune Activation Model of Autism. J. Immunol 201, 845–850 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ryoo HD & Vasudevan D Two distinct nodes of translational inhibition in the Integrated Stress Response. BMB Rep. 50, 539–545 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rojas M, Vasconcelos G & Dever TE An eIF2α-binding motif in protein phosphatase 1 subunit GADD34 and its viral orthologs is required to promote dephosphorylation of eIF2α. Proc. Natl. Acad. Sci. U. S. A 112, E3466–3475 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McGlincy NJ & Ingolia NT Transcriptome-wide measurement of translation by ribosome profiling. Methods San Diego Calif 126, 112–129 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Calviello L et al. Detecting actively translated open reading frames in ribosome profiling data. Nat. Methods 13, 165–170 (2016). [DOI] [PubMed] [Google Scholar]
- 32.Merrick WC Eukaryotic protein synthesis: an in vitro analysis. Biochimie 76, 822–830 (1994). [DOI] [PubMed] [Google Scholar]
- 33.Hoffmann A & Roeder RG Cloning and characterization of human TAF20/15. Multiple interactions suggest a central role in TFIID complex formation. J. Biol. Chem 271, 18194–18202 (1996). [DOI] [PubMed] [Google Scholar]
- 34.Berson A et al. TDP-43 Promotes Neurodegeneration by Impairing Chromatin Remodeling. Curr. Biol. CB 27, 3579–3590.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marash L et al. DAP5 promotes cap-independent translation of Bcl-2 and CDK1 to facilitate cell survival during mitosis. Mol. Cell 30, 447–459 (2008). [DOI] [PubMed] [Google Scholar]
- 36.Henis-Korenblit S et al. The caspase-cleaved DAP5 protein supports internal ribosome entry site-mediated translation of death proteins. Proc. Natl. Acad. Sci. U. S. A 99, 5400–5405 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Terenin IM, Dmitriev SE, Andreev DE & Shatsky IN Eukaryotic translation initiation machinery can operate in a bacterial-like mode without eIF2. Nat. Struct. Mol. Biol 15, 836–841 (2008). [DOI] [PubMed] [Google Scholar]
- 38.Scheuner D et al. Translational control is required for the unfolded protein response and in vivo glucose homeostasis. Mol. Cell 7, 1165–1176 (2001). [DOI] [PubMed] [Google Scholar]
- 39.Sidrauski C et al. Pharmacological brake-release of mRNA translation enhances cognitive memory. eLife 2, e00498 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sidrauski C et al. Pharmacological dimerization and activation of the exchange factor eIF2B antagonizes the integrated stress response. eLife 4, e07314 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sekine Y et al. Stress responses. Mutations in a translation initiation factor identify the target of a memory-enhancing compound. Science 348, 1027–1030 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen Y-C, Chang Y-W & Huang Y-S Dysregulated Translation in Neurodevelopmental Disorders: An Overview of Autism-Risk Genes Involved in Translation. Dev. Neurobiol 79, 60–74 (2019). [DOI] [PubMed] [Google Scholar]
- 43.Alvarez-Castelao B & Schuman EM The Regulation of Synaptic Protein Turnover. J. Biol. Chem 290, 28623–28630 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Costa-Mattioli M, Sossin WS, Klann E & Sonenberg N Translational Control of Long-Lasting Synaptic Plasticity and Memory. Neuron 61, 10–26 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ximerakis M et al. Single-cell transcriptomic profiling of the aging mouse brain. Nat. Neurosci 22, 1696–1708 (2019). [DOI] [PubMed] [Google Scholar]
- 46.Lombardo MV et al. Maternal immune activation dysregulation of the fetal brain transcriptome and relevance to the pathophysiology of autism spectrum disorder. Mol. Psychiatry 23, 1001–1013 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Braun AE et al. ‘Females Are Not Just “Protected” Males’: Sex-Specific Vulnerabilities in Placenta and Brain after Prenatal Immune Disruption. eNeuro 6, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sutherland S & Brunwasser SM Sex Differences in Vulnerability to Prenatal Stress: a Review of the Recent Literature. Curr. Psychiatry Rep 20, 102 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guan B-J et al. A Unique ISR Program Determines Cellular Responses to Chronic Stress. Mol. Cell 68, 885–900.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Method-only References
- 50.Hrvatin S et al. Single-cell analysis of experience-dependent transcriptomic states in the mouse visual cortex. Nat. Neurosci 21, 120–129 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Klein AM et al. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell 161, 1187–1201 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wolock SL, Lopez R & Klein AM Scrublet: Computational Identification of Cell Doublets in Single-Cell Transcriptomic Data. Cell Syst 8, 281–291.e9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Robinson MD, McCarthy DJ & Smyth GK edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McCarthy DJ, Chen Y & Smyth GK Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 40, 4288–4297 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thomas PD et al. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 13, 2129–2141 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nygaard KR, Maloney SE & Dougherty JD Erroneous inference based on a lack of preference within one group: Autism, mice, and the social approach task. Autism Res 12, 1171–1183 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dobin A et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ji Z RibORF: Identifying Genome-Wide Translated Open Reading Frames Using Ribosome Profiling. Curr Protoc Mol Biol 124, e67 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Anders S, Pyl PT & Huber W HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Langmead B & Salzberg SL Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1 Differential gene expression between MIA male and PBS male offspring in E14 single cell RNA-seq. Data analyzed using EdgeR with Quasi-likelihood F test. Data includes Log2 fold change, logCPM (counts per million), F statistic, p value (unadjusted), and false discovery rate (FDR) adjusted for multiple comparisons.
Supplementary Table 2 Differential gene expression between MIA female and PBS female offspring in E14 single cell RNA-seq. Data analyzed using EdgeR with Quasi-likelihood F test. Data includes Log2 fold change, logCPM (counts per million), F statistic, p value (unadjusted), and false discovery rate (FDR) adjusted for multiple comparisons.
Supplementary Table 3 Differential gene expression between PBS male and PBS female offspring in E14 single cell RNA-seq. Data analyzed using EdgeR with Quasi-likelihood F test. Data includes Log2 fold change, logCPM (counts per million), F statistic, p value (unadjusted), and false discovery rate (FDR) adjusted for multiple comparisons.
Supplementary Table 4 Differential gene expression between MIA male and MIA female offspring in E14 single cell RNA-seq using interaction term analysis. Data analyzed using EdgeR with Quasi-likelihood F test. Data includes Log2 fold change, logCPM (counts per million), F statistic, p value (unadjusted), and false discovery rate (FDR) adjusted for multiple comparisons.
Supplementary Table 5 Differential gene expression between MIA male and PBS male offspring in E18 single cell RNA-seq. Data analyzed using EdgeR with Quasi-likelihood F test. Data includes Log2 fold change, logCPM (counts per million), F statistic, p value (unadjusted), and false discovery rate (FDR) adjusted for multiple comparisons.
Supplementary Table 6 Differential gene expression between MIA female and PBS female offspring in E18 single cell RNA-seq. Data analyzed using EdgeR with Quasi-likelihood F test. Data includes Log2 fold change, logCPM (counts per million), F statistic, p value (unadjusted), and false discovery rate (FDR) adjusted for multiple comparisons.
Supplementary Table 7 Differential gene expression between PBS male and PBS female offspring in E18 single cell RNA-seq. Data analyzed using EdgeR with Quasi-likelihood F test. Data includes Log2 fold change, logCPM (counts per million), F statistic, p value (unadjusted), and false discovery rate (FDR) adjusted for multiple comparisons.
Supplementary Table 8 Differential gene expression between MIA male and MIA female offspring in E18 single cell RNA-seq using interaction term analysis. Data analyzed using EdgeR with Quasi-likelihood F test. Data includes Log2 fold change, logCPM (counts per million), F statistic, p value (unadjusted), and false discovery rate (FDR) adjusted for multiple comparisons.
Supplementary Table 9 Quality control metrics for ribosome profiling data
Supplementary Table 10 Differential gene expression between MIA male and PBS male offspring in E18 bulk RNA-seq. Data analyzed using EdgeR with Quasi-likelihood F test. Data includes Log2 fold change, logCPM (counts per million), F statistic, p value (unadjusted), and false discovery rate (FDR) adjusted for multiple comparisons.
Supplementary Table 11 Differential gene expression between MIA female and PBS female offspring in E18 bulk RNA-seq. Data analyzed using EdgeR with Quasi-likelihood F test. Data includes Log2 fold change, logCPM (counts per million), F statistic, p value (unadjusted), and false discovery rate (FDR) adjusted for multiple comparisons.
Supplementary Table 12 Differential gene expression between PBS male and PBS female offspring in E18 bulk RNA-seq. Data analyzed using EdgeR with Quasi-likelihood F test. Data includes Log2 fold change, logCPM (counts per million), F statistic, p value (unadjusted), and false discovery rate (FDR) adjusted for multiple comparisons.
Supplementary Table 13 Differential gene expression between MIA male and MIA female offspring in E18 bulk RNA-seq using interaction term analysis. Data analyzed using EdgeR with Quasi-likelihood F test. Data includes Log2 fold change, logCPM (counts per million), F statistic, p value (unadjusted), and false discovery rate (FDR) adjusted for multiple comparisons.
Supplementary Table 14 Differential gene expression between MIA male and MIA female offspring in E18 bulk RNA-seq using a pairwise comparison. Data analyzed using EdgeR with Quasi-likelihood F test. Data includes Log2 fold change, logCPM (counts per million), F statistic, p value (unadjusted), and false discovery rate (FDR) adjusted for multiple comparisons.
Supplementary Table 15 Differential translation analysis between MIA male and PBS male offspring in E18 ribosome profiling. Data analyzed using EdgeR with Quasi-likelihood F test. Data includes Log2 fold change, logCPM (counts per million), F statistic, p value (unadjusted), and false discovery rate (FDR) adjusted for multiple comparisons.
Supplementary Table 16 Differential translation analysis between MIA female and PBS female offspring in E18 ribosome profiling. Data analyzed using EdgeR with Quasi-likelihood F test. Data includes Log2 fold change, logCPM (counts per million), F statistic, p value (unadjusted), and false discovery rate (FDR) adjusted for multiple comparisons.
Supplementary Table 17 Differential translation analysis between PBS male and PBS female offspring in E18 ribosome profiling. Data analyzed using EdgeR with Quasi-likelihood F test. Data includes Log2 fold change, logCPM (counts per million), F statistic, p value (unadjusted), and false discovery rate (FDR) adjusted for multiple comparisons.
Supplementary Table 18 Differential translation analysis between MIA male and MIA female offspring in E18 ribosome profiling using interaction term analysis. Data analyzed using EdgeR with Quasi-likelihood F test. Data includes Log2 fold change, logCPM (counts per million), F statistic, p value (unadjusted), and false discovery rate (FDR) adjusted for multiple comparisons.
Supplementary Table 19 Differential translation analysis between MIA male and MIA female offspring in E18 ribosome profiling using a pairwise comparison. Data analyzed using EdgeR with Quasi-likelihood F test. Data includes Log2 fold change, logCPM (counts per million), F statistic, p value (unadjusted), and false discovery rate (FDR) adjusted for multiple comparisons.
Supplementary Table 21 Source data for Fig. 4C
Data Availability Statement
Sequence data that support the findings of this study have been deposited in the Gene Expression Omnibus with the accession code GSE148237. Publicly available datasets from PANTHER (http://pantherdb.org/) were used for gene ontology analysis.