Abstract
The molecular basis underlying fetal programming in response to maternal nutrition remains unclear. Herein, we investigated the regulatory relationships between genes in fetal cerebrum, liver, and muscle tissues to shed light on the putative mechanisms that underlie the effects of early maternal nutrient restriction on bovine developmental programming. To this end, cerebrum, liver, and muscle gene expression were measured with RNA-Seq in 14 fetuses collected on day 50 of gestation from dams fed a diet initiated at breeding to either achieve 60% (RES, n = 7) or 100% (CON, n = 7) of energy requirements. To build a tissue-to-tissue gene network, we prioritized tissue-specific genes, transcription factors, and differentially expressed genes. Furthermore, we built condition-specific networks to identify differentially co-expressed or connected genes. Nutrient restriction led to differential tissue regulation between the treatments. Myogenic factors differentially regulated by ZBTB33 and ZNF131 may negatively affect myogenesis. Additionally, nutrient-sensing pathways, such as mTOR and PI3K/Akt, were affected by gene expression changes in response to nutrient restriction. By unveiling the network properties, we identified major regulators driving gene expression. However, further research is still needed to determine the impact of early maternal nutrition and strategic supplementation on pre- and post-natal performance.
Subject terms: Gene expression profiling, Transcriptomics
Introduction
The relevance of optimal maternal nutrient supply in the periconception period and during pregnancy has received close attention due to growing evidence showing its impact on offspring development1–3. Imbalance in the availability of uterine-derived histotroph4 as well as placental nutrient supply, either by overfeeding or underfeeding, leads the fetus to physiological and metabolic changes with long-term consequences5–7. Nutritional insults affect energy partitioning among metabolically active target tissues, such as the liver and muscle8, affecting economic traits such as lactation, reproduction, meat quality, body composition, and carcass weight5,9,10. These adverse long-term consequences could decrease the livestock industry’s profitability as the negative performance may be epigenetically inherited across generations11.
The brain, liver, and muscle tissues have different priorities in nutrient partitioning when limited resources are available, affecting their development and functionality8,12. The liver plays a key role in body energy metabolism13, whereas the brain, by regulating liver metabolism, prioritizes its own glucose needs over those of other tissues14. Thus, with restricted energy availability, more glucose is allocated for the functioning of the central nervous system14, potentially impairing muscle development and reducing muscle mass12. Reduced number of myofibers and liver sizes have been reported in nutrient restricted sheep fetuses and adults, respectively15,16. Furthermore, intrauterine growth restricted bovine fetuses showed increased brain weight as a percentage of fetal body weight in response to early maternal undernutrition6. Altogether, the nutritional regulation of fetal programming underlies a complex genomic regulatory network that modulates gene expression and leads to changes in tissue structure and function2,3,8. Differences in gene expression patterns and regulation of epigenetic mechanisms were reported as underlying the tissue-specific metabolic changes observed in nutrient restricted offspring1,15–17.
Previously, our group showed that the exposure of pregnant cows to a nutritional insult in early pregnancy changed fetal liver, cerebrum, and muscle gene expression profiles, suggesting a possible compensatory growth in response to reduced nutrient availability17. Likewise, candidate genes involved with myogenesis, muscle differentiation, and hepatic metabolism were reported as differentially expressed as a consequence of maternal nutrition1,15,18. Despite the knowledge provided by differential expression analysis, this approach does not consider the multiple interactions involved with gene expression regulation19 that are central to phenotype determination. To overcome this limitation, gene networks can be used successfully as a complementary framework to untangle the mechanisms and unravel the effects of maternal nutrition on fetal programming8. By measuring gene co-expression similarity, we can capture the relationship among genes across experimental conditions or tissue types and decipher the biologically-related gene functions19–21. Furthermore, we can assess the network ‘rewiring’ and the differences in gene–gene interactions driving changes in gene expression in response to external stimuli20,22.
Although we have a growing understanding of the molecular and physiological basis of fetal programming, it is still unclear to the extent these differences can be attributed to the coordinated function of genes and the regulation of specific pathways. Therefore, we hypothesized that maternal nutrient restriction from breeding to day 50 of gestation would negatively affect fetal cerebrum, liver, and muscle development and function through differential gene regulation. Herein, by measuring the gene–gene interactions, we uncovered the regulatory interplay between genes and showed that these fetal tissues were differentially regulated in response to maternal nutrition.
Results
We applied tissue-to-tissue and tissue condition-specific co-expression network approaches to infer the regulatory relationships between genes and to shed light on the putative mechanisms underlying the effects of early maternal nutrient restriction on bovine developmental programming (Fig. 1). Previously, Crouse et al.17 reported 373 differentially expressed genes (DEG) from fetal liver (n = 201), muscle (n = 144), and cerebrum (n = 28) by contrasting control (CON, n = 7) and nutrient restricted (RES, n = 7) groups. Herein, we report significant differentially connected and differentially co-expressed genes in 50-day fetuses. Likewise, we identified putative regulators driving the differential gene expression between the groups and the over-represented biological pathways.
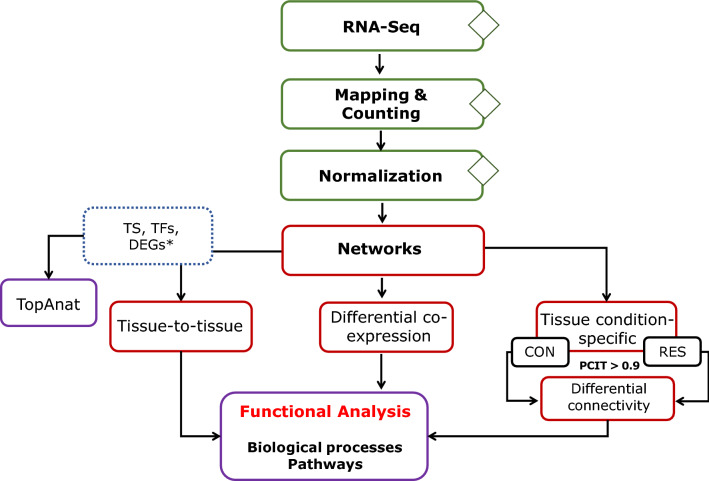
Figure 1.
Bioinformatics workflow of the multi-tissue RNA-Seq-based gene co-expression network. TS—tissue-specific genes; *DEGs—differentially expressed genes17; TFs—transcription factors; CON—control; RES—restricted.
RNA sequencing data analysis
After quality control, on average, 92.8%, 89.9%, and 93.0% of unique reads from the cerebrum, liver, and muscle tissues, respectively, were mapped to the reference genome. A summary of sequencing throughput and mapping rates per sample and tissue is reported in Tables S1, S2, and S3. Mapped reads saturation plot showed that, on average, 92% of the genes were identified, supporting an adequate read-depth per tissue in the current study (Figure S1). The multi-tissue normalization approach (see Methods) identified 17,164 genes out of 27,607 reported on the Ensembl annotation file that were clustered by tissue, as shown in Figure S2.
Tissue-specific genes
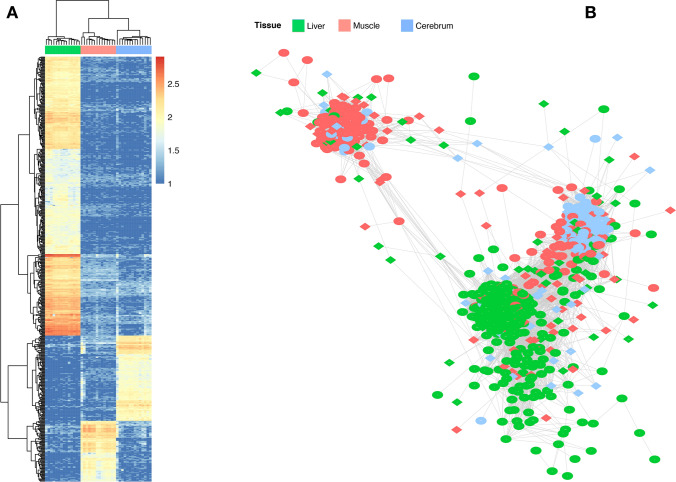
Based on the Tau index23, we identified 553 tissue-specific (TS) genes (τ ≥ 0.8). The liver (n = 363) and cerebrum (n = 111) expressed the greatest number of TS genes, followed by muscle (n = 79) (Fig. 2A). The NEUROD6, PFKFB, and MYOG genes were amongst the TS genes with the greatest τ values for the cerebrum, liver, and muscle, respectively. Furthermore, 19 and 15 TS genes were reported as differentially expressed for the liver and muscle. To support our approach, we carried out a tissue over-representation analysis. As expected, the genes were indeed over-represented for the tissues where they are preferentially expressed [prefrontal cortex, adjusted p-value (adj.pvalue) = 4.59 E−10; liver, adj.pvalue = 4.58E−05; and skeletal muscle, adj.pvalue = 2.11 E−05]. Additionally, gene ontology (GO) terms highlighted biological processes such as forebrain development (adj.pvalue = 2.26E−21), hemostasis (adj.pvalue = 1.68E−16), and muscle organ development (adj.pvalue = 1.55E−21). The over-represented Uberon ontology terms, the directed acyclic graphic, and the GO terms are reported in Tables S1, S2, and S3.
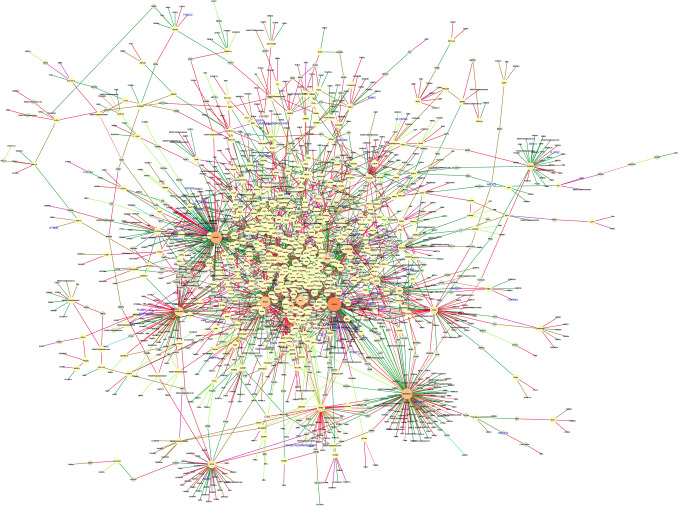
Figure 2.
Tissue-to-tissue co-expression network of the bovine fetal organ transcriptome. (A) Heat map of 553 tissue-specific genes from the liver, muscle, and cerebrum tissues. (B) Co-expression network of 992 significantly co-expressed genes. Only nodes with a correlation greater than |0.9| are shown. Overlapped genes between analysis were colored based on the tissue with its maximum expression. Transcription factors are represented by a diamond shape. Nodes with few connections not linked to the main network are not showed. Heat map was constructed using pheatmap v.1.010.93 on R79, whereas gene network was created on Cytoscape v.3.789.
Key regulatory genes (transcription factors, TFs)
Regulatory impact factors, RIF1 and RIF224, were used to identify TFs potentially modulating the expression of DEG and TS genes. Based on those metrics, we identified 129, 119, and 90 TFs for liver, muscle, and cerebrum tissues, respectively, grouped into 52 families (p ≤ 0.01). Shared TFs were identified between cerebrum and muscle (n = 10), muscle and liver (n = 8), and liver and cerebrum (n = 7). The HOXA4 (z-score = − 6.88), ZBTB33 (z-score = − 4.37), and GATA1 (z-score = − 3.85) genes showed the most extreme value for RIF1 in liver, muscle, and cerebrum, respectively. A list of all RIF1 and RIF2 significant TFs is presented in Tables S1, S2, and S3. Herein, those genes encoding TFs and identified as key regulators will be called TFs throughout the text. The over-represented BP and pathways in which the TFs are involved with DEGs and TS genes are reported in the Tab8 of the Supplementary Tables S1 (muscle), S2 (cerebrum), and S3 (liver).
Tissue-to-tissue co-expression network
To build a network across tissues based on the PCIT framework, we prioritized 1,160 unique informative genes considering the following criteria: (1) DEG between RES and CON groups; (2) tissue-specific; and (3) key TFs based on RIF1 or RIF2. Based on that, a significant network with 992 nodes and 105,449 interactions (r >|0.9|) was retrieved (Table S4, Fig. S3). The network had a high clustering coefficient (0.8) and was clustered by tissue (Fig. 2B), mainly driven by TS genes, followed by DEGs and TFs. Genes from the liver showed higher average connectivity (284.5) compared to the cerebrum (144.7) or muscle (100.06). The GATA1, HSF2, and PLEK TFs showed the greatest connectivity within liver, muscle, and cerebrum tissues, respectively. Furthermore, these genes shared all the first neighbors of one another, creating a sub-network with 427 nodes and 81,308 connections (Fig. S4).
Differential gene co-expression network analysis
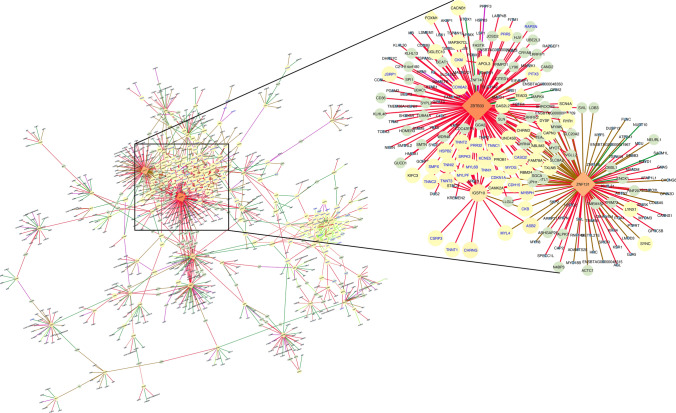
To uncover the functional relationship between gene pairs across conditions, we applied a differential gene co-expression analysis for each tissue separately. From muscle, we identified 17,282 differentially co-expressed (DC) gene pairs (corresponding to 7,479 unique genes out of 11,439 tested; q ≤ 0.05; Table S1). Among them, 101 and 91 genes were identified as DEGs and TFs, respectively. Figure 3 shows the network of DC genes filtered by those gene pairs assigned either DEG or TF for muscle. We used the connectivity as a measure of centrality to select the genes topologically important in the network (“hubs”). Accordingly, 225 DC genes were identified as hubs (p < 0.05). The top five hub genes included IGSF10, ZBTB33, CD2AP, PLEKHG1, and ZBTB1, and were associated with more than 120 genes in muscle. The DEGs were mainly differentially co-expressed with the genes ZBTB33 (n = 24), IGSF10 (n = 24), HACD3 (n = 16), and ZNF131 (n = 8). Interestingly, the co-expression of ZBTB33 and ZNF131 with the DEGs were classified as inversely correlated between the maternal diet groups based on the correlation classes proposed by the R-package DGCA22 (Figs. 3, S5).
Figure 3.
Muscle regulatory network of differentially co-expressed genes from the bovine transcriptome. Nodes are genes with significant changes in the correlation between control and nutrient restricted fetus (q ≤ 0.05). The node size and color (from light to dark) are proportional to the number of connections for each gene. Nodes with few connections not linked to the main network are not shown. Transcription factors are represented by a diamond shape. Differentially expressed genes are labeled in blue. Edges are colored based on the differential correlation class (+/− , red; +/0, salmon; − /+ , green; − /0, green-yellow; 0/+ , magenta; 0/− , brown). Gene network was created on Cytoscape v.3.789.
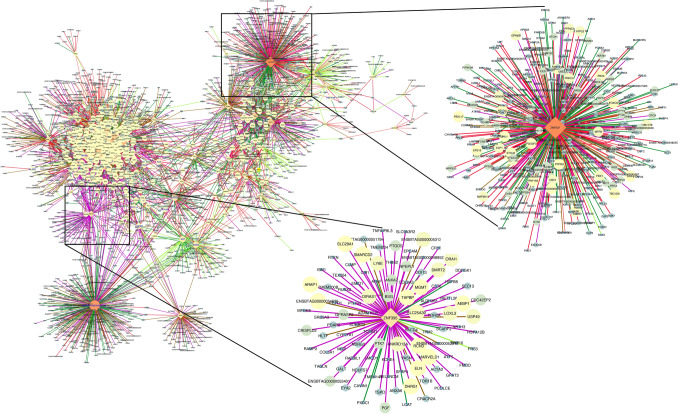
We found 284,738 DC gene pairs by applying the same approach to the cerebrum. To reduce the data dimensionality, these pairs were then filtered by DEG, TS, or TF genes. Based on that, 14,423 DC pairs (4676 unique genes) (q ≤ 0.05) remained for further analysis (Table S2). The TS genes ELAV4, PAK5, SLC17A6, GPR12, and CLVS2, were the most differentially co-expressed ones (hubs). Among the 61 genes classified as hubs (p < 0.05), 21 were TFs identified as putative regulators of brain gene expression. From these TFs, ZNF207, ETS2, and the novel gene ENSBTAG00000003447 were highlighted as targeting 412, 250, and 248 genes (Fig. 4). Unlike the muscle, no clear pattern of correlation was observed for TFs and DEGs differentially co-expressed between conditions.
Figure 4.
Cerebrum regulatory network of differentially co-expressed genes from the bovine transcriptome. Nodes are genes with significant changes in the correlation between control and nutrient restricted fetus (q ≤ 0.05). The node size and color (from light to dark) are proportional to the number of connections for each gene. Nodes with few connections not linked to the main network are not showed. Only those gene pairs assigned as DEG, TS, or TF are shown. Transcription factors are represented by a diamond shape. Differentially expressed genes are labeled in blue. Edges are colored based on the differential correlation class (+/− , red; + /0, salmon; − /+ , green; − /− , deep sky blue; − /0, green-yellow; 0/+ , magenta; 0/− , brown). Gene network was created on Cytoscape v.3.789.
Regarding the liver, we identified 2,989 DC pairs (2319 unique genes) that included 56 DEGs and 23 TFs (q ≤ 0.05) (Table S3). The top five hub genes among the 27 identified included: PGAP3, B3GNT4, MARVELD1, BCAM, and NIT1. The PGAP3 gene stood out as the main DC gene with 11 DEGs. These pairs were classified as positively correlated in the RES group and negative or not significant in the CON group (−/+ or 0/+) (Fig. 5). Additionally, the ZNF175, HSF1, and HSF2 TFs were among the hubs.
Figure 5.
Liver regulatory network of differentially co-expressed genes from the bovine transcriptome. Nodes are genes with significant changes in the correlation between control and nutrient restricted fetus (q ≤ 0.05). The node size and color (from light to dark) are proportional to the number of connections for each gene. Nodes with few connections not linked to the main network are not showed. Transcription factors are represented by a diamond shape. Differentially expressed genes are labeled in blue. Edges are colored based on the differential correlation class (+/− , red; + /0, salmon; − /+ , green; − /0, green-yellow; 0/+ , magenta; 0/− , brown). Gene network was created on Cytoscape v.3.789.
Differential connectivity analysis
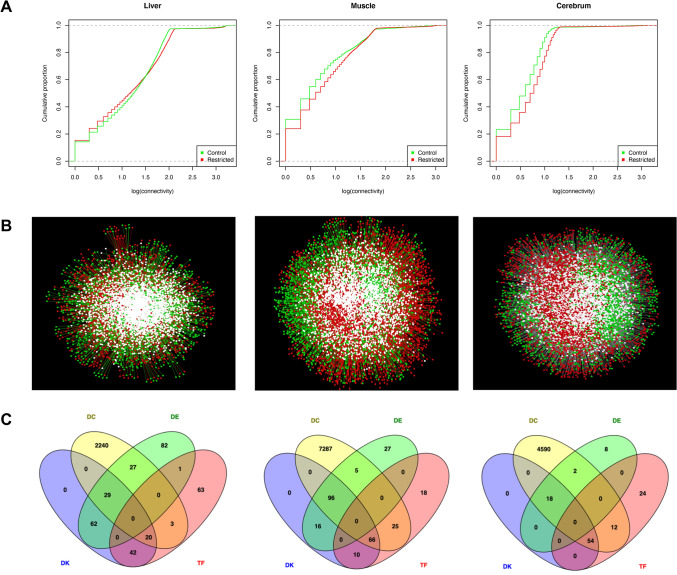
To identify condition-specific connections, we inferred networks in the CON and RES groups separately for each tissue. Based on the concept of differential connectivity25, we identified the most rewired genes between the groups (Tables S1, S2, S3). Despite the similar topological behavior, overall, we found a gain of connectivity in networks from RES (Fig. 6A,B). These networks followed a scale-free model with R-squared ranging from 0.67 to 0.89.
Figure 6.
Network topology of co-expressed genes between control and nutrient restricted fetuses from the liver, muscle, and cerebrum tissues in bovine. (A) Cumulative distribution functions of the gene connectivity. (B) Central reference union networks. Only those gene pairs assigned as DEG or TF are shown. Unique nodes are shown in green (control) or red (restricted). Shared nodes are shown in white; (C) Overlapping genes among the different analyses. TF—transcription factors; DE – differentially expressed gene; DC—differentially co-expressed; and DK—differentially connected. Cumulative distribution was created on R v.3.5.179; Gene network was created on Cytoscape v.3.789; and Venn diagram was created using Venny v.2.1.94 (https://rb.gy/jxxufy).
From the liver, the networks filtered for either DEG or TF genes resulted in 8,350 and 8,632 nodes and 261,099 and 231,270 edges for RES and CON groups, respectively. By contrasting the connectivity between the groups, we found 153 differentially connected (DK) genes (p < 0.05). Similarly, we identified 188 DK genes in muscle by contrasting 6,925 nodes and 63,105 edges with 8515 nodes and 94,186 edges from CON and RES groups, respectively. Regarding the cerebrum, the CON network was composed of 6,390 nodes and 29,531 edges, whereas the RES one had 7,555 nodes and 48,662 edges. Accordingly, we identified 72 DK genes (p < 0.05). Several DEGs from the liver and muscle tissues were also identified as DK (Fig. 6C).
Functional over-representation analysis
Functional analysis revealed a total of 374, 212, and 143 significant biological process (BP) terms over-represented in the liver, muscle, and cerebrum gene lists, respectively (adj.pvalue ≤ 0.05). In the liver, common BP across DE, TFs, and TS genes included categories such as blood coagulation, immune response (defense and inflammatory response), and energy metabolism (fatty acid metabolic process and cellular glucose homeostasis). Tissue specific genes highlighted BP related to metabolism, transport, and blood coagulation, whereas TFs underlined regulation of RNA biosynthetic process and hormone metabolic process (Table S3). The pathway analysis included complement and coagulation cascade (adj.pvalue = 2.9E−13), metabolic pathways (adj.pvalue = 2.64E−02), and maturity onset diabetes of the young (adj.pvalue = 2.64E−02) (Table S3). Moreover, MAPK (adj.pvalue = 3.32E−02), non-alcoholic fatty liver disease (NAFLD) (adj.pvalue = 3.68E−02), and oxidative phosphorylation (adj.pvalue = 3.32E−02) pathways were retrieved from DC genes.
Regarding the muscle, the identified GO terms pointed out the muscle structure development and myotube and myoblast differentiation as common BP among the gene lists. Similarly, TS and TF genes included skeletal system morphogenesis and regulation of transcription from RNA polymerase II promoter (Table S1). Furthermore, FoxO (adj.pvalue = 8.32E−06), MAPK (adj.pvalue = 3.69E−09), and PI3K/Akt (adj.pvalue = 3.94E−07) signaling pathways were among the over-represented pathways from the DC genes.
From the cerebrum, central nervous system development (adj.pvalue = 1.68E−13) and brain development (adj.pvalue = 4.60E−08) were among the over-represented BP underlying the TS genes. The TF gene list retrieved BP such as cellular response to steroid hormone stimulus (adj.pvalue = 6.47E−07) and negative regulation of DNA binding transcription factor activity (adj.pvalue = 6.47E−07) (Table S2). Moreover, signaling pathways from the DC genes highlighted, for example, ribosome (adj.pvalue = 8.86E−21), PI3K-Akt (adj.pvalue = 3.19E−06), insulin (adj.pvalue = 3.11E−03), mTOR (adj.pvalue = 2.41E−04), and HIF-1 signaling pathways (adj.pvalue = 2.17E−03). Tissue-specific and overlapping pathways across tissues are showed in Figure S6.
Discussion
Dietary restriction has a negative effect on the offspring’s development, metabolism and genome function with long-term consequences2,3,5,9. Previously, we reported a significant impact on genome-wide expression profiles underpinning tissue metabolism, accretion, and function in fetal muscle, liver, and cerebrum from dietary intake-restricted heifers17. Herein, based on the co-expression network, we uncovered the regulatory interplay between genes and shed light on the putative mechanisms underlying the effects of early maternal nutrient restriction on bovine developmental programming on day 50 of gestation. We found that these tissues were affected by dietary intake restriction leading to differences in transcriptional regulation between CON and RES groups. Furthermore, we pointed out TFs potentially modulating the expression of DEGs and TS genes.
Tissue specificity results from a tight regulation of gene expression26 driven by different regulatory mechanisms that include TFs and cross-tissue communication20. The TFs play a significant role in determining differential expression in response to nutrient availability7, although they are not themselves usually DEG26. We successfully mapped 338 TFs as putative regulators of TS and DEG. It is worth mentioning that these are not the only TFs regulating the tissue development and gene transcription; however, they were consistently most differentially co-expressed (RIF1) or could predict (RIF2) the abundance of the TS and DEG24. The regulatory role of these TFs on day 50 of gestation was perturbed due to maternal nutrient restriction, as evidenced by their identification either as DC or DK. The liver showed the greatest number of TS genes, followed by the cerebrum and muscle. Moreover, the greatest number of DEG and TF genes were found in the liver, emphasizing its wide range of essential metabolic functions.
To provide an overview of the gene expression relationship across the fetal tissues studied, we prioritized informative genes from TS, TF, and DEG analyses. This integrative framework allowed us to dissect the gene–gene interactions within and across tissues, uncovering new insights into how these relationships may be rewired due to maternal nutrition8,27,28. Our tissue-to-tissue network exhibited a small-world behavior29, mainly driven by the TS genes that showed a strong clustering. Based on this guilty-by-association assumption, we pinpointed tissue-specific functions related to tissue development, structure, and metabolism.
To have tissue genome-scale information, we identified specific co-expression differences between CON and RES groups based on DGCA and DK analyses. We identified hub genes with important biological roles. Furthermore, we found a gain of connectivity in the networks from fetuses from the RES dams. The rewiring of major regulators likely modulates the expression of gene targets as an adaptive response to nutrient availability, impacting tissue development. These findings are supported by the DGCA results that showed TFs differentially coordinating gene transcription between the groups. For example, we found transcriptional repressors in muscle, such as ZBTB33, REST, and RCOR3, more connected in the RES network. Likewise, we identified ARID2, whose protein is involved with chromatin remodeling that is important for metabolic programming2. Herein, we will mainly focus our discussion on the TFs considering their topological importance in the network for each tissue, the over-represented pathways, and their regulatory role in fetal tissue programming.
The liver plays a pivotal role in energy metabolism13. Energy imbalances affect liver function and trigger a cascade of events in insulin sensitive tissues, such as the brain and muscle7,18. As a response to a moderate maternal nutrient restriction, the fetuses from the RES group tended to have a reduced level of glucose in allantoic fluid30. In goats, Zhou et al.18 found several glucogenic amino acids (AA) differentially abundant in the liver of nutrient-restricted fetuses. Supporting these findings, we identified pathways related to glucogenic AA, such as arginine biosynthesis and tryptophan metabolism. Additionally, the liver is the main hematopoietic organ during pre-natal development18. We found GATA1 as the most connected TF, and this gene is important for early hematopoiesis31. Furthermore, the liver is the primary site of complement component synthesis32. The complement system is essential for host defense, and its failure has been associated with adverse pregnancy and fetal growth restriction in humans32. The complement and coagulation cascade pathway, as well as several hematopoietic genes were pinpointed in our findings.
We identified master regulators of energy homeostasis, such as FOXA3, PPARA, and SREBF2. Among the pathways important for energy homeostasis, we found MAPK, which acts as a PPAR regulator33. The PPAR protein is suggested to have an adaptive role during tissue development in response to nutrient availability7. The transcriptional complex formed by PPARs and nuclear receptors, like RXRG, is the key activator of target genes involved with fatty acid oxidation, insulin secretion, and inflammation7,13. The relationship of PPAR-RXRG with energy imbalance underlies metabolic abnormalities, such as diabetes and chronic inflammation34. Moreover, FOXA3 participates on the pathway maturity onset diabetes as well as carbohydrate and glucose homeostasis. In addition, SREBF2 plays a role in energy metabolism and also lipid storage, regulation of sterol and cholesterol transport, lipid and cholesterol homeostasis. The protein coded by this gene regulates the cholesterol biosynthesis7. Cholesterol is the precursor of all steroid hormones and it plays a role in early fetal development by moderating nuclear receptors35.
Besides the imbalances in energy homeostasis, maternal nutrient restriction has been associated with an inflammatory response in the dam and the offspring36. We identified TGFB1 and HDAC8 differentially co-expressed and positively correlated in RES fetuses. The TGFB1 gene encodes a multifunctional cytokine with immunoregulatory and anti-inflammatory properties37. It is also important in pregnancy maintenance37, hepatic stellate cell activation, and hepatic fibrosis38. An increased expression of MEF2D in activated hepatic stellate cells in liver and the fibrotic response was reported previously38. In the current study, we identified the MEF2D gene as a key TF, more connected in RES, and positively correlated with TGFB1 and HDAC8. The TGFB1 and MEFD2 proteins have been associated with liver fibrosis activation38,39. Likewise, HDAC8 protein inhibition suppresses TGFB1-induced genes in pulmonary fibrosis40.
In response to cellular stress, HSF1 and HSF2 TFs activate genes encoding heat shock proteins (HSPs) to stabilize nascent proteins41. These TFs in liver showed high connectivity in RES fetuses. Since misfolded proteins are eliminated either by ubiquitination or autophagy41, the over-representation of the ubiquitin-mediated proteolysis pathway found here is likely acting in concert with the HSPs. Thus, these evidences suggest that RES fetuses may have shown hepatic cellular stress in response to lipid metabolism imbalances.
Maternal nutritional insults may have more dramatic effects on muscle development as it has low priority in nutrient partitioning, resulting in greater concern due to the economic importance of muscle tissue12. Offspring of nutrient-restricted dams showed a reduced number of muscle fibers and muscle mass, as well as altered muscle function and gene regulation1,12,17,42. Among the TS genes, well known upstream regulators of myogenesis, such as MYOG, MYOD1, PAX1, and MYF543, were found by our approach. Interestingly, some of these TS regulators along with DEGs were negatively correlated with the ZBTB33 and ZNF131 TFs in the RES group. The ZBTB33 gene encodes a methyl-CpG-binding protein (Kaiso) that recognizes both unmethylated and methylated CpGs, acting as a transcriptional repressor44. The role of Kaiso in regulating myogenic genes is still to be examined; however, Ruzov et al.45 showed that Kaiso is necessary during early Xenopus laevis development. Moreover, CIBZ, a Kaiso-like family member, represses MYOG in a methylation-dependent manner44. Although we do not have the methylation profile for these animals, Crouse et al.30 reported reduced methionine and increased homocysteine concentrations in amniotic fluid and serum, respectively, in RES heifers. The imbalance in one-carbon metabolism would likely affect the gene transcription by altering chromatin remodeling and DNA methylation46.
Although ZNF131 acts as a transcriptional activator, Kaiso, a transcriptional regulator, was implicated in its repression, and together they fine-tune the transcription of Kaiso targets47. Muscle structure development and skeletal muscle tissue growth were among the BP over-represented by the 34 co-regulated genes between ZNF131 and ZBTB33. The myogenic regulatory factors MYOD1, MYF5, and MYOG were negatively and differentially correlated with ZBTB33 and/or ZNF131 in RES fetuses. The CDKN1A gene was negatively correlated with these TFs in the RES group and also DEG17. The protein encoded by the ZNF131 gene has been associated with skeletal muscle atrophy48. Moreover, CDKN1A gene expression is synergistically activated by SMAD449 that was identified here as DK and negatively co-expressed with MYOG and MYOD1. The protein encoded by SMAD4 has an inhibitory effect on protein synthesis and muscle growth50. Genes encoding proteins involved in muscle contraction, such as those from the troponin and myosin families, were also negatively regulated in the RES group. We collected the fetal tissues on day 50 of pregnancy, which coincides with the beginning of primary myogenesis12. Therefore, our findings show the potential mechanism by which maternal nutrient restriction negatively modulates myogenic developmental programming in cattle.
We found the DC genes from the fetal muscle over-represented in the nutrient-sensing signaling pathways PI3K-Akt, MAPK, and FoxO. These pathways along with mTOR, play a major role in muscle development, energy metabolism, gene expression, and metabolic epigenetic programming50,51. The Akt-mTOR pathway acts as positive regulator of protein synthesis, whereas FoxO signaling is required for muscle atrophy50. We found the ELK3 TF more connected and negatively and differentially correlated with AKT1 in fetuses from RES dams. The ELK3 protein is a transcriptional repressor whose knockdown induced the activation of the PI3k/Akt/mTOR pathway in breast cancer cell line52. In addition, intrauterine growth restriction in rats has been associated with reduced activity of this pathway53. We found FoxO6 significantly more connected in the RES group. FoxO induction leads to the transcription of atrophy-related genes50. Additionally, SMAD3/SMAD4 heterodimers may repress mTOR50. We identified the SMAD4 gene negatively co-expressed with EIF4EBP1 that acts as protein synthesis inhibitor50. Altogether, it seems that the TFs were acting redundantly and coordinately to repress muscle formation and differentiation in response to nutrient restriction.
Like the muscle, our findings evidenced a severe impact of a moderate maternal nutrient restriction in the day 50 fetal brain genes involved with development and metabolism. First, we found a large number of DC genes between CON and RES, and several DK TFs. Second, pathways such as PI3K/AKT and mTOR seem to be differentially regulated between the CON and RES groups. The brain plays a central role in the body’s energy homeostasis by integrating metabolic signals from other tissues and coordinating adaptive changes under nutrient deficiency54. These signals are integrated by the mTOR pathway that together with the PI3K/Akt signaling pathway governs neuronal fate by regulating protein synthesis and energy homeostasis55,56. Furthermore, insulin signaling plays a role in neuronal differentiation by activating mTOR55. Nutrient restriction has been associated with reduced transcription rate modulated by mTOR57. We found the ribosome pathway over-represented by the DC genes encoding ribosomal proteins, and intriguingly these genes were mainly negatively correlated in the RES group. Impaired ribosome biogenesis has been associated with neurodevelopmental disorders and reduced protein synthesis57,58. The PAK5, ELAVL4, and ZNF207 genes were DC with the ribosomal genes identified in the current study. The ELAVL4 gene encodes an RNA-binding protein (HuD) important for neuronal development and plasticity, acting on mRNA stability, translation, and alternative polyadenylation of target genes59. The PAK5 gene encodes a brain-specific protein involved with neurite development60. Furthermore, the PAK family members are regulators of PI3K and via the PI3K/Akt pathway modulate HuD abundance post-transcriptionally59. The ZNF207 TF plays a role in cell pluripotency by regulating neuronal TFs61.
Metabolic stress resulting from energy restriction can activate the transcription of inflammatory genes57. The TGF-β family is pivotal for organogenesis, and it was shown to negatively affect brain development by inhibiting neurite elongation62. We identified TGFB1 and TGFB3 DC and negatively correlated with NKX2-1, ELAVL4, and SLC17A6 in the RES group. The SLC17A6 gene encodes a glutamate transporter protein63, while NKX2-1 encodes a TF that regulates target genes involved with the development of interneurons64. In response to inflammatory processes, hypoxia-inducible factors (HIFs) modulate metabolic homeostasis by activating hypoxia-induced genes65. Additionally, these genes underlie cell differentiation and vascular development in the fetal brain65. We identified the HIF pathway over-represented from the DC genes. Among them, the ZNF395 TF was positively correlated with most of the gene pairs DC in the RES fetuses. ZNF395 is targeted by the HIF-1α TF activating hypoxia-induced pro-inflammatory genes66. On the other hand, the ETS2 and NFKB2 TFs that were both DC and DK showed mainly negative correlations in the RES group. The ETS2 protein has been associated with neuronal apoptosis and neurodegeneration in neurons of Down syndrome67. However, in macrophages, it had a negative effect on the inflammatory response67. Regarding the NFKB2 protein, it has a pleiotropic effect and acts in processes such as synaptic plasticity under normal physiology; however, in disease, it is involved with neuroinflammation as well as neuronal cell death68. Considering the findings, the moderate maternal dietary restriction induced changes in gene wiring and expression in the brain may have altered BP related to neuronal development and energy homeostasis. Although the effects of these changes in animal health and performance are still unknown, they could affect animal reactivity, stress responses, and feeding behavior17.
Our findings reinforce that fetal metabolic programming is affected by maternal nutritional imbalances even in early pregnancy. Energy metabolism-related pathways were over-represented in the cerebrum, liver, and muscle, suggesting a multi-tissue coordinated adaptive response mediated by differential gene expression. Likewise, pathways related to myogenesis seems to be negatively regulated in nutrient restricted fetuses. Factors affecting fetal muscle development can reduce animal performance, muscle growth, and consequently, meat production and quality12. Furthermore, fetal adaptive response by affecting insulin metabolism may adversely affect animal feed efficiency. It’s worth mentioning that due to the similarities underlying embryonic development in humans and cattle and the conserved genetic architecture of complex traits in both species69, our findings can help understanding the effects of maternal nutrition on human fetal development. Obesity, molecular disorders, and cardiovascular diseases later in life have been associated with pre-natal maternal stressors8,70. Pathways related to human diseases, such as maturity onset diabetes of the young and non-alcoholic fatty liver disease, were over-represented among the differentially regulated genes in our study. These findings reinforce the potential adverse effects of early maternal nutrient restriction on offspring health. Additionally, it suggests that pathways affected by maternal nutrition in cows are likely to be regulated similarly in humans.
Altogether, our data suggest that nutrient restricted animals will later in life be metabolically compromised, leading to long-term consequences probably across generations. Although the genetic basis of fetal programming is still under investigation, our results point to the key role of epigenetic mechanisms driving the genome regulation. We hypothesize that RES fetuses have impaired one-carbon metabolism, as demonstrated by the reduced availability of methionine in allantoic fluid30. Further research is warranted to investigate the role of non-coding RNAs and histone modifications in addition to DNA methylation as the mechanisms underlying fetal programming and the transmission of intergenerational effects.
Conclusions
Our approach identified the complex gene network on day 50 of gestation that underlies cross-tissue communication in response to maternal nutritional restriction. We identified major regulators driving gene expression and found differences in the regulatory mechanisms involved with developmental programming between fetuses gestated in nutrient restricted and control dams. In particular, moderate dam nutrient restriction led to differential regulation of myogenic factors by the ZBTB33 and ZNF131 TFs that may negatively affect myogenesis. In addition to tissue-specific pathways, the over-representation of nutrient-sensing signaling pathways across tissues, such as mTOR, PI3K/Akt, and insulin, shed light on their role in fetal programming. Further research is still needed to determine the role of epigenetics and chromatin remodeling in developmental programming, their interplay with nutrition, and their impact on animal health and performance.
Methods
Animals, tissue collection, library preparation, and sequencing
All experiments and methods were performed in accordance with relevant guidelines and regulations. The experimental design, animal management, and tissue collection were approved by the North Dakota State University Institutional Animal Care and Use Committee and previously reported by Crouse et al.17. In brief, 14 Angus-cross heifers with average initial body weight = 313 ± 24.9 kg were randomly assigned at breeding to receive dietary intake of either 100% of energy requirement71 for 0.45 kg/day of body weight gain (control; CON, n = 7) or were fed a diet to delivery 60% of CON intake (restricted; RES, n = 7). The diet was delivered via total mixed ration (TMR), and consisted of grass hay, corn silage, alfalfa haylage, grain, and mineral mix. Dried distillers grains with solubles (53.4% NDF, 31.3% CP) were supplemented in addition to the TMR and fed to achieve the target nutrient content of the CON and RES diets17. The two nutritional levels supplied to the heifers were chosen to represent two nutritional states (weight gain versus maintenance) as well as conditions applicable to beef production systems.
On day 50 of gestation, cerebrum (n = 7/group), liver (n = 7/group), and muscle (n = 7/group) were collected from fetuses through ovariohysterectomy72. Tissue collection was performed at this time because fetal organogenesis have already finished and it is in the peak primary myogenesis12. The tissues were snap-frozen in liquid nitrogen, stored at − 80 °C, and later on, RNA isolation and sequencing were performed by the University of Minnesota Genomics Center (Minneapolis-St. Paul, MN), as reported elsewhere17. After RNA quality control, strand-specific RNA libraries were prepared, and sequencing was carried out on the Illumina HiSeq 2500 platform. Sequencing was pair-end (50-bp reads) at a depth of 2 × 10.4 M reads/sample in both forward and reverse directions17. Fetal sex was determined by PCR based on the hind limb DNA amplification of the DDX3X and DDX3Y genes located on the X and Y chromosomes. The PCR product was analyzed on agarose gel electrophoresis, where males showed two bands, and females resulted in only one. This analysis was based on the primers, and PCR parameters reported elsewhere73. Thus, were evaluated 6 males and 1 female in the CON group and 5 males and 2 females in the RES group.
RNA-Seq data analyses and differentially expressed genes
Data quality control was performed with FastQC v.0.11.874 (https://rb.gy/lxqcwa) and MultiQC v.1.875 (https://multiqc.info/) software. Filtered reads were mapped to the bovine reference genome (ARS-UCD 1.2)76 and gene annotation file (release 100) from the Ensembl database. The STAR aligner version 2.7.3a77 (https://rb.gy/dlgdva) using the –quantMode GeneCounts flag was used to obtain the raw counts per gene.
The RStudio78 v. 1.1.442 environment for R79 v.3.5.1. was used for data analyses as described below. Post-mapping quality control was performed using MultiQC, NOISeq v.2.26.080 (10.18129/B9.bioc.NOISeq), and edgeR v.3.24.081 (10.18129/B9.bioc.edgeR). Read counts were transformed to counts per million (CPM), and lowly expressed genes (CPM < 1 in 80% of the samples) were filtered out. Gene expression normalization was performed under two approaches: (1) across all samples and the three tissues together and (2) across all samples but individually for each tissue. Both procedures were performed using the VST function from DESeq2 v.1.22.182 (10.18129/B9.bioc.DESeq2). To control gene expression for sex effect, fetal sex was included in the DESeq2 model as a factor. Hierarchical clustering and Principal Component Analysis on normalized data were performed by using NOISeq v.2.26.080 and DESeq2 v.1.22.182. Normalized gene expression (n = 17,164) from the first approach was used to predict TS genes, to predict the key regulators (TFs), and to build the tissue-to-tissue network. On the second approach, besides removing genes with low expression based on the CPM criterion, genes with low dispersion after normalization were filtered out when the quantile dispersion measure was < 0.2. Further, the normalized data were used for tissue condition-specific networks construction, differential connectivity, and differential gene co-expression analyses. A summary of the workflow is shown in Fig. 1.
Differential expression analysis for the tissues and animals used here were reported elsewhere17. In brief, by using the Tuxedo Suite pipeline, the authors identified 373 DEGs between the CON and RES groups for fetal liver (n = 201), muscle (n = 144), and cerebrum (n = 28). Herein, these DEGs were used for further analyses.
Identification of tissue-specific genes (TS)
The normalized expression was used to calculate a mean value for each gene from each tissue separately. To summarize the gene specificity among tissues, the Tau index (τ) was computed using the R-package tispec v.0.99.083 (https://rb.gy/ch0h4w) considering the following Eq. 23:
in which n is the number of tissues and is the gene expression in tissue i. Tau values range from zero (genes ubiquitously expressed) to one (TS genes)84. Accordingly, we defined the TS genes with τ ≥ 0.884.
To support the gene tissue specificity analysis, we performed a TS over-representation analysis using TopAnat (https://rb.gy/6qvwla). This tool allowed us to identify where genes from a set are preferentially expressed85, considering the anatomical structures and the Uberon ontology annotations86. Thus, for each TS gene list, we used TopAnat based on bovine as the target species and the Begee database for B. taurus as background85.
Identification of key transcription factors (TFs)
To identify TFs differentially regulating gene expression in CON and RES groups for each tissue, we used the regulatory impact factor (RIF) algorithms, RIF1 and RIF224. These metrics assume that master regulators in a network change their behavior in different biological conditions contributing to differential gene expression19,26. RIF algorithm was implemented in FORTRAN 90 based on the source code available from the RIF’s main author24. The RIF1 score highlights regulators differentially connected to the targets, whereas RIF2 measures those TFs with the potential to act as predictors of target abundance24,26. To identify the regulators, we first downloaded 1396 TFs from the Animal Transcription Factor Database (Animal TFDB v3.0)87. After filtering out those TFs not expressed in our dataset, 1164 TFs were contrasted to a unique list of DEG and TS genes. The RIF comparisons were performed separately for each tissue, comparing CON versus RES groups.
Gene networks
To determine the co-expression profile of gene pairs across tissues and experimental groups, we used the partial correlation and information theory (PCIT) algorithm28. FORTRAN 90 source code to perform the PCIT algorithm is available as Supplementary File 1 at https://rb.gy/vyfqrv. This approach compares all possible triplets of genes by exploring the concepts of partial correlation and mutual information reporting the significantly correlated pairs after accounting for all the other genes in the network19,28,88. For the tissue-to-tissue network inference, all the genes (n = 1,160) selected as DEG, TS genes, or TFs from the previous analyses were used considering all animals and tissues together. Significant co-expressed pairs were determined based on partial correlation values greater than |0.9| (p < 0.01).
To shed light on the coordinated gene expression and identify the conditional regulatory relationships underlying the differences between RES and CON groups, a differential gene co-expression network analysis was performed by using the R-package DGCA v.1.0.222 (https://rb.gy/qadgby). To this end, all the genes kept after filtering and normalization (cerebrum, n = 11,432; liver, n = 10,146; and muscle, n = 11,439) were used. Significantly correlated gene pairs were determined based on the adjusted p-values (q ≤ 0.05). The DC gene pairs were grouped in one of the eight differential correlation classes proposed by McKenzie et al.22. These classes (+/+ ; +/− ; +/0; − /− ; −/+ ; −/0; 0/+ ; 0/−) show the correlation signal [positive, negative, or not significant (0)] for each gene and condition when contrasting the groups (CON versus RES).
To identify changes in the gene network topology between conditions, we used PCIT to build CON and RES networks separately for each tissue from the same dataset used for DGCA analysis. Significantly correlated pairs were selected when DEGs or TFs were present and showed a partial correlation greater than |0.9| (p < 0.01). To explore the DK genes between the maternal dietary intake conditions, we first standardized the connectivity (K) measures for each network by dividing each gene connectivity by the maximum connectivity25. Next, the DK measure was defined as . To assign a significance level, each score was transformed into a Z-score, and values located ± 1.96 from the standard deviation (SD) were considered significant (p < 0.05). The DyNet27 Cytoscape plugin v.1.0 (https://rb.gy/9vp9js) was used to visualize the most rewiring nodes between the conditions considering the Dn-score. This score ranks the variability of connections and highlights the most rewired nodes based on a central reference network27 constructed from the union of RES and CON networks. Networks were visualized in Cytoscape (v.3.7)89 (https://cytoscape.org/). Highly connected genes or “hubs” were defined considering the degree measure (2 SD, p < 0.05) retrieved from the Cytoscape Network Analyzer tool v.2.790 (https://rb.gy/twossl).
Functional over-representation analyses
We carried out functional over-representation analyses to identify the biological process and pathways underpinning relevant functions related to fetal programming. The gene lists from DEG, TS, and TF analyses were queried by using ClueGO v. 2.5.7 (https://rb.gy/x5cb7q), considering a cluster analysis framework91. This approach allowed us to underline specific and common biological functions within and among gene lists for each tissue. To reduce the redundancy of the GO terms, we applied the fusion of similar biological processes. Likewise, the terms were grouped based on the kappa score = 0.4. Significant results were taken considering the group p-value after Bonferroni step down correction (adj.pvalue ≤ 0.05). Pathway analysis was performed for DC genes by using the ShinyGO v0.6192 (https://rb.gy/o97axc). This analysis was performed separately for each tissue considering the B. taurus annotation as background and adj.pvalue ≤ 0.05 as significant.
Supplementary Information
Acknowledgements
This project was supported by Agriculture and Food Research Initiative Competitive Grant No. 2016-67016-24946 from the USDA National Institute of Food and Agriculture as well as the USDA-NIFA-AFRI (2018-67011-31708). Mention of a trade name, proprietary product, or specific agreement does not constitute a guarantee or warranty by the USDA and does not imply approval to the inclusion of other products that may be suitable. USDA is an equal opportunity provider and employer. This work used resources of the Center for Computationally Assisted Science and Technology (CCAST) at North Dakota State University.
Author contributions
W.J.S.D. designed and implemented the bioinformatic analyses and drafted the manuscript. M.S.C. collected tissues and performed bioinformatic analysis to identify differentially expressed genes. R.A.C. performed bioinformatic analysis to identify differentially expressed genes. J.S.C., K.J.M., C.R.D., L.P.R., A.K.W. designed and implemented the treatment structure and collection of tissues. A.K.W. supervised W.J.S.D. All authors collaborated with the interpretation of results and discussion. All authors have read, reviewed, and approved the final manuscript.
Data availability
All relevant data are within the paper and its Supplementary Information files. All sequencing data is publicly available on NCBI’s Gene Expression Omnibus through GEO Series accession number GSE154299 (Bioproject PRJNA645822). All additional datasets generated and analyzed during this study are available from the corresponding author.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-82156-w.
References
- 1.Paradis F, et al. Maternal nutrient restriction in mid-to-late gestation influences fetal mRNA expression in muscle tissues in beef cattle. BMC Genom. 2017;18:1–14. doi: 10.1186/s12864-017-4051-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reynolds, L. P., Ward, A. K. & Caton, J. S. Epigenetics and developmental programming in ruminants: Long-term iimpacts on growth and development. in Biology of Domestic Animals (eds. Scanes, C. G. & Hill, R. A.) 85–120 (CRC Press, Boca Raton, 2017). 10.1201/9781315152080
- 3.Caton JS, et al. Maternal nutrition and programming of offspring energy requirements. Transl. Anim. Sci. 2019;3:976–990. doi: 10.1093/tas/txy127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bazer FW, Wang X, Johnson GA, Wu G. Select nutrients and their effects on conceptus development in mammals. Anim. Nutr. 2015;1:85–95. doi: 10.1016/j.aninu.2015.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chavatte-Palmer P, Tarrade A, Kiefer H, Duranthon V, Jammes H. Breeding animals for quality products: Not only genetics. Reprod. Fertil. Dev. 2016;28:94–111. doi: 10.1071/RD15353. [DOI] [PubMed] [Google Scholar]
- 6.Long NM, Vonnahme KA, Hess BW, Nathanielsz PW, Ford SP. Effects of early gestational undernutrition on fetal growth, organ development, and placentomal composition in the bovine. J. Anim. Sci. 2009;87:1950–1959. doi: 10.2527/jas.2008-1672. [DOI] [PubMed] [Google Scholar]
- 7.Maloney CA, Rees WD. Gene-nutrient interactions during fetal development. Reproduction. 2005;130:401–410. doi: 10.1530/rep.1.00523. [DOI] [PubMed] [Google Scholar]
- 8.Sookoian S, Gianotti TF, Burgueño AL, Pirola CJ. Fetal metabolic programming and epigenetic modifications: A systems biology approach. Pediatr. Res. 2013;73:531–542. doi: 10.1038/pr.2013.2. [DOI] [PubMed] [Google Scholar]
- 9.Greenwood, P. L., Hearnshwaw, H., Cafe, L. M., Hennessy, D. W. & Harper. Nutrition in utero and pre-weaning has long term consequences for growth and size of Piedmontese and Wagyu-sired steers. J. Anim. Sci.82 (Suppl. 1), 408 (2004).
- 10.Reynolds LP, Vonnahme KA. TRIENNIAL REPRODUCTION SYMPOSIUM: Developmental programming of fertility. J. Anim. Sci. 2016;94:2699–2704. doi: 10.2527/jas.2015-0131. [DOI] [PubMed] [Google Scholar]
- 11.Thompson RP, Nilsson E, Skinner MK. Environmental epigenetics and epigenetic inheritance in domestic farm animals. Anim. Reprod. Sci. 2020 doi: 10.1016/j.anireprosci.2020.106316. [DOI] [PubMed] [Google Scholar]
- 12.Du M, et al. Fetal programming of skeletal muscle development in ruminant animals. J. Anim. Sci. 2009 doi: 10.2527/jas.2009-2311. [DOI] [PubMed] [Google Scholar]
- 13.Rui L. Energy metabolism in the liver. Compr. Physiol. 2014;176:177–197. doi: 10.1002/cphy.c130024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Longman D, Stock JT, Wells JCK. A trade-off between cognitive and physical performance, with relative preservation of brain function. Sci. Rep. 2017;7:13709. doi: 10.1038/s41598-017-14186-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hyatt MA, et al. Maternal nutrient restriction in early pregnancy programs hepatic mRNA expression of growth-related genes and liver size in adult male sheep. J. Endocrinol. 2007;192:87–97. doi: 10.1677/joe.1.06801. [DOI] [PubMed] [Google Scholar]
- 16.Zhu MJ, et al. Maternal nutrient restriction affects properties of skeletal muscle in offspring. J. Physiol. 2006;575:241–250. doi: 10.1113/jphysiol.2006.112110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crouse MS, et al. Moderate nutrient restriction of beef heifers alters expression of genes associated with tissue metabolism, accretion, and function in fetal liver, muscle, and cerebrum by day 50 of gestation. Transl. Anim. Sci. 2019;3:855–866. doi: 10.1093/tas/txz026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou X, et al. Evidence for liver energy metabolism programming in offspring subjected to intrauterine undernutrition during midgestation. Nutr. Metab. 2019;16:1–14. doi: 10.1186/s12986-018-0330-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hudson NJ, Dalrymple BP, Reverter A. Beyond differential expression: The quest for causal mutations and effector molecules. BMC Genomics. 2012;13:356. doi: 10.1186/1471-2164-13-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gaiteri C, Ding Y, French B, Tseng GC, Sibille E. Beyond modules and hubs: The potential of gene coexpression networks for investigating molecular mechanisms of complex brain disorders. Genes Brain Behav. 2014;13:13–24. doi: 10.1111/gbb.12106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Dam, S., Võsa, U., van der Graaf, A., Franke, L. & de Magalhães, J. P. Gene co-expression analysis for functional classification and gene–disease predictions. Brief. Bioinform. bbw139 (2017). 10.1093/bib/bbw139 [DOI] [PMC free article] [PubMed]
- 22.McKenzie AT, Katsyv I, Song W-M, Wang M, Zhang B. DGCA: A comprehensive R package for differential gene correlation analysis. BMC Syst. Biol. 2016;10:106. doi: 10.1186/s12918-016-0349-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yanai I, et al. Genome-wide midrange transcription profiles reveal expression level relationships in human tissue specification. Bioinformatics. 2005;21:650–659. doi: 10.1093/bioinformatics/bti042. [DOI] [PubMed] [Google Scholar]
- 24.Reverter A, Hudson NJ, Nagaraj SH, Pérez-Enciso M, Dalrymple BP. Regulatory impact factors: Unraveling the transcriptional regulation of complex traits from expression data. Bioinformatics. 2010;26:896–904. doi: 10.1093/bioinformatics/btq051. [DOI] [PubMed] [Google Scholar]
- 25.Fuller TF, et al. Weighted gene coexpression network analysis strategies applied to mouse weight. Mamm. Genome. 2007;18:463–472. doi: 10.1007/s00335-007-9043-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pérez-Montarelo D, et al. Porcine tissue-specific regulatory networks derived from meta-analysis of the transcriptome. PLoS ONE. 2012;7:e46159. doi: 10.1371/journal.pone.0046159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goenawan IH, Bryan K, Lynn DJ. DyNet: Visualization and analysis of dynamic molecular interaction networks. Bioinformatics. 2016;32:2713–2715. doi: 10.1093/bioinformatics/btw187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reverter A, Chan EKF. Combining partial correlation and an information theory approach to the reversed engineering of gene co-expression networks. Bioinformatics. 2008;24:2491–2497. doi: 10.1093/bioinformatics/btn482. [DOI] [PubMed] [Google Scholar]
- 29.Aarstad J, Ness H, Haugland SA. In what ways are small-world and scale-free networks interrelated? Proc. IEEE Int. Conf. Ind. Technol. 2013 doi: 10.1109/ICIT.2013.6505891. [DOI] [Google Scholar]
- 30.Crouse MS, et al. Maternal nutrition and stage of early pregnancy in beef heifers: Impacts on hexose and AA concentrations in maternal and fetal fluids. J. Anim. Sci. 2019;97:1296–1316. doi: 10.1093/jas/skz013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Choi M, et al. Overexpression of human GATA-1 and GATA-2 interferes with spine formation and produces depressive behavior in rats. PLoS ONE. 2014;9:e109253. doi: 10.1371/journal.pone.0109253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Regal JF, Gilbert JS, Burwick RM. The complement system and adverse pregnancy outcomes. Mol. Immunol. 2015;67:56–70. doi: 10.1016/j.molimm.2015.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lawan A, et al. Hepatic mitogen-activated protein kinase phosphatase 1 selectively regulates glucose metabolism and energy homeostasis. Mol. Cell. Biol. 2015;35:26–40. doi: 10.1128/MCB.00503-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Plutzky J. The PPAR-RXR transcriptional complex in the vasculature. Circ. Res. 2011;108:1002–1016. doi: 10.1161/CIRCRESAHA.110.226860. [DOI] [PubMed] [Google Scholar]
- 35.Baardman ME, et al. The role of maternal-fetal cholesterol transport in early fetal life: Current insights. Biol. Reprod. 2013;88:1–9. doi: 10.1095/biolreprod.112.102442. [DOI] [PubMed] [Google Scholar]
- 36.Jones AK, et al. Gestational restricted- and over-feeding promote maternal and offspring inflammatory responses that are distinct and dependent on diet in sheep. Biol. Reprod. 2018;98:184–196. doi: 10.1093/biolre/iox174. [DOI] [PubMed] [Google Scholar]
- 37.McLennan IS, Koishi K. Fetal and maternal transforming growth factor-β1 may combine to maintain pregnancy in Mice. Biol. Reprod. 2004;70:1614–1618. doi: 10.1095/biolreprod.103.026179. [DOI] [PubMed] [Google Scholar]
- 38.Wang X, et al. Regulation of hepatic stellate cell activation and growth by transcription factor myocyte enhancer factor 2. Gastroenterology. 2004;127:1174–1188. doi: 10.1053/j.gastro.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 39.Kaimori A, et al. Histone deacetylase inhibition suppresses the transforming growth factor β1-induced epithelial-to-mesenchymal transition in hepatocytes. Hepatology. 2010;52:1033–1045. doi: 10.1002/hep.23765. [DOI] [PubMed] [Google Scholar]
- 40.Saito S, et al. HDAC8 inhibition ameliorates pulmonary fibrosis. Am. J. Physiol. Cell. Mol. Physiol. 2019;316:L175–L186. doi: 10.1152/ajplung.00551.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gomez-Pastor R, Burchfiel ET, Thiele DJ. Regulation of heat shock transcription factors and their roles in physiology and disease. Nat. Rev. Mol. Cell Biol. 2017 doi: 10.1038/nrm.2017.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gauvin MC, et al. Poor maternal nutrition during gestation in sheep alters prenatal muscle growth and development in offspring. J. Anim. Sci. 2020;98:1–15. doi: 10.1093/jas/skz388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Buckingham M, Rigby PWJ. Gene regulatory networks and transcriptional mechanisms that control myogenesis. Dev. Cell. 2014;28:225–238. doi: 10.1016/j.devcel.2013.12.020. [DOI] [PubMed] [Google Scholar]
- 44.Oikawa Y, et al. The methyl-CpG-binding protein CIBZ suppresses myogenic differentiation by directly inhibiting myogenin expression. Cell Res. 2011;21:1578–1590. doi: 10.1038/cr.2011.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ruzov A, et al. The non-methylated DNA-binding function of Kaiso is not required in early Xenopus laevis development. Development. 2009;136:729–738. doi: 10.1242/dev.025569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mentch SJ, Locasale JW. One-carbon metabolism and epigenetics: understanding the specificity. Ann. N. Y. Acad. Sci. 2016;1363:91–98. doi: 10.1111/nyas.12956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Robinson SC, et al. The POZ-ZF transcription factor Znf131 is implicated as a regulator of Kaiso-mediated biological processes. Biochem. Biophys. Res. Commun. 2017;493:416–421. doi: 10.1016/j.bbrc.2017.09.007. [DOI] [PubMed] [Google Scholar]
- 48.Ebert SM, Al-Zougbi A, Bodine SC, Adams CM. Skeletal muscle atrophy: Discovery of mechanisms and potential therapies. Physiology. 2019;34:232–239. doi: 10.1152/physiol.00003.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chi X-Z, et al. RUNX3 suppresses gastric epithelial cell growth by inducing p21WAF1/Cip1 expression in cooperation with transforming growth factor β-activated SMAD. Mol. Cell. Biol. 2005;25:8097–8107. doi: 10.1128/MCB.25.18.8097-8107.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schiaffino S, Dyar KA, Ciciliot S, Blaauw B, Sandri M. Mechanisms regulating skeletal muscle growth and atrophy. FEBS J. 2013;280:4294–4314. doi: 10.1111/febs.12253. [DOI] [PubMed] [Google Scholar]
- 51.Yu JSL, Cui W. Proliferation, survival and metabolism: the role of PI3K/AKT/mTOR signalling in pluripotency and cell fate determination. Development. 2016;143:3050–3060. doi: 10.1242/dev.137075. [DOI] [PubMed] [Google Scholar]
- 52.Park JH, Kim KP, Ko JJ, Park KS. PI3K/Akt/mTOR activation by suppression of ELK3 mediates chemosensitivity of MDA-MB-231 cells to doxorubicin by inhibiting autophagy. Biochem. Biophys. Res. Commun. 2016;477:277–282. doi: 10.1016/j.bbrc.2016.06.057. [DOI] [PubMed] [Google Scholar]
- 53.Xing Y, et al. Reduction of the PI3K/Akt related signaling activities in skeletal muscle tissues involves insulin resistance in intrauterine growth restriction rats with catch-up growth. PLoS ONE. 2019;14:e0216665. doi: 10.1371/journal.pone.0216665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Roh E, Song DK, Kim MS. Emerging role of the brain in the homeostatic regulation of energy and glucose metabolism. Exp. Mol. Med. 2016;48:e216–e216. doi: 10.1038/emm.2016.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Licausi F, Hartman NW. Role of mTOR complexes in neurogenesis. Int. J. Mol. Sci. 2018;19:1544. doi: 10.3390/ijms19051544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Takei N, Nawa H. mTOR signaling and its roles in normal and abnormal brain development. Front. Mol. Neurosci. 2014;7:28. doi: 10.3389/fnmol.2014.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gameiro PA, Struhl K. Nutrient deprivation elicits a transcriptional and translational inflammatory response coupled to decreased protein synthesis. Cell Rep. 2018;24:1415–1424. doi: 10.1016/j.celrep.2018.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Slomnicki LP, et al. Requirement of neuronal ribosome synthesis for growth and maintenance of the dendritic tree. J. Biol. Chem. 2016;291:5721–5739. doi: 10.1074/jbc.M115.682161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bronicki LM, Jasmin BJ. Emerging complexity of the HuD/ELAVl4 gene; implications for neuronal development, function, and dysfunction. RNA. 2013;19:1019–1037. doi: 10.1261/rna.039164.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Civiero L, Greggio E. PAKs in the brain: Function and dysfunction. Biochim. Biophys. Acta Mol. Basis Dis. 2018;1864:444–453. doi: 10.1016/j.bbadis.2017.11.005. [DOI] [PubMed] [Google Scholar]
- 61.Fang F, et al. A distinct isoform of ZNF207 controls self-renewal and pluripotency of human embryonic stem cells. Nat. Commun. 2018;9:1–14. doi: 10.1038/s41467-017-02088-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nakashima H, et al. Canonical TGF-β signaling negatively regulates reuronal morphogenesis through TGIF/Smad complex-mediated CRMP2 suppression. J. Neurosci. 2018;38:4791–4810. doi: 10.1523/JNEUROSCI.2423-17.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wallen-Mackenzie Å, Wootz H, Englund H. Genetic inactivation of the vesicular glutamate transporter 2 (VGLUT2) in the mouse: What have we learnt about functional glutamatergic neurotransmission? Upsala J. Med. Sci. 2010;115:11–20. doi: 10.3109/03009730903572073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Du T, Xu Q, Ocbina PJ, Anderson SA. NKX2.1 specifies cortical interneuron fate by activating Lhx6. Development. 2008;135:1559–1567. doi: 10.1242/dev.015123. [DOI] [PubMed] [Google Scholar]
- 65.Trollmann R, Gassmann M. The role of hypoxia-inducible transcription factors in the hypoxic neonatal brain. Brain Dev. 2009;31:503–509. doi: 10.1016/j.braindev.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 66.Herwartz C, Castillo-Juárez P, Schröder L, Barron BL, Steger G. The transcription factor ZNF395 is required for the maximal hypoxic induction of proinflammatory cytokines in U87-MG cells. Mediat. Inflamm. 2015;2015:1–9. doi: 10.1155/2015/804264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ma X, et al. Ets2 suppresses inflammatory cytokines through MAPK/NF-kB signaling and directly binds to the IL-6 promoter in macrophages. Aging (Albany. NY). 2019;11:10610–10625. doi: 10.18632/aging.102480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dresselhaus EC, Meffert MK. Cellular specificity of NF-κB function in the nervous system. Front. Immunol. 2019;10:1043. doi: 10.3389/fimmu.2019.01043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu S, et al. Epigenomics and genotype-phenotype association analyses reveal conserved genetic architecture of complex traits in cattle and human. BMC Biol. 2020;18:1–16. doi: 10.1186/s12915-019-0728-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Caton, J. S. et al. Maternal periconceptual nutrition, early pregnancy, and developmental outcomes in beef cattle. J. Anim. Sci. 10.1093/jas/skaa358/5962128. [DOI] [PMC free article] [PubMed]
- 71.National Academies of Sciences, Engineering, and M. Nutrient Requirements of Beef Cattle, 8th Revised Edition. (National Academies Press, 2016). 10.17226/19014.
- 72.McLean KJ, et al. Technical note: A new surgical technique for ovariohysterectomy during early pregnancy in beef heifers. J. Anim. Sci. 2016;94:5089–5096. doi: 10.2527/jas.2016-0761. [DOI] [PubMed] [Google Scholar]
- 73.Gokulakrishnan P, Kumar RR, Sharma BD, Mendiratta SK, Sharma D. Sex determination of cattle meat by polymerase chain reaction amplification of the DEAD box protein (DDX3X/DDX3Y) gene. Asian-Austr. J. Anim. Sci. 2012;25:733–737. doi: 10.5713/ajas.2012.12003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Andrews, S. FASTQC. A quality control tool for high throughput sequence data. (2010). Available at: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- 75.Ewels P, Magnusson M, Lundin S, Käller M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics. 2016;32:3047–3048. doi: 10.1093/bioinformatics/btw354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rosen BD, et al. De novo assembly of the cattle reference genome with single-molecule sequencing. Gigascience. 2020;9:1–9. doi: 10.1093/gigascience/giaa021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dobin A, et al. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.RStudio Team. RStudio: Integrated Development Environment for R. (2020).
- 79.R Core Team. R: A Language and Environment for Statistical Computing. (2018).
- 80.Tarazona S, et al. Data quality aware analysis of differential expression in RNA-seq with NOISeq R/Bioc package. Nucl. Acids Res. 2015 doi: 10.1093/nar/gkv711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Robinson MD, McCarthy DJ, Smyth GK. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Condon, K. Tispec. (2019). Available at: https://github.com/roonysgalbi/tispec/blob/master/vignettes/UserGuide.Rmd.
- 84.Kryuchkova-Mostacci N, Robinson-Rechavi M. A benchmark of gene expression tissue-specificity metrics. Brief. Bioinform. 2017;18:205–214. doi: 10.1093/bib/bbw008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bastian, F. et al. Bgee: Integrating and comparing heterogeneous transcriptome data among species. Data Integration in the Life Sciences 124–131 (2008).
- 86.Haendel MA, et al. Unification of multi-species vertebrate anatomy ontologies for comparative biology in Uberon. J. Biomed. Semantics. 2014;5:21. doi: 10.1186/2041-1480-5-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hu H, et al. AnimalTFDB 3.0: A comprehensive resource for annotation and prediction of animal transcription factors. Nucl. Acids Res. 2018;47:33–38. doi: 10.1093/nar/gky822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Alexandre, P. A. et al. Systems biology reveals NR2F6 and TGFB1 as key regulators of feed efficiency in beef cattle. Front. Genet.10, 230 (2019). [DOI] [PMC free article] [PubMed]
- 89.Shannon P. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Assenov Y, Ramírez F, Schelhorn S-E, Lengauer T, Albrecht M. Computing topological parameters of biological networks. Bioinforma. Appl. 2008;24:282–284. doi: 10.1093/bioinformatics/btm554. [DOI] [PubMed] [Google Scholar]
- 91.Bindea G, et al. ClueGO: A cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25:1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ge SX, Jung D, Yao R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics. 2020;36:2628–2629. doi: 10.1093/bioinformatics/btz931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kolde, R. pheatmap: Pretty Heatmaps. (2018).
- 94.Oliveros, J. C. Venny. An interactive tool for comparing lists with Venn’s diagrams.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data are within the paper and its Supplementary Information files. All sequencing data is publicly available on NCBI’s Gene Expression Omnibus through GEO Series accession number GSE154299 (Bioproject PRJNA645822). All additional datasets generated and analyzed during this study are available from the corresponding author.