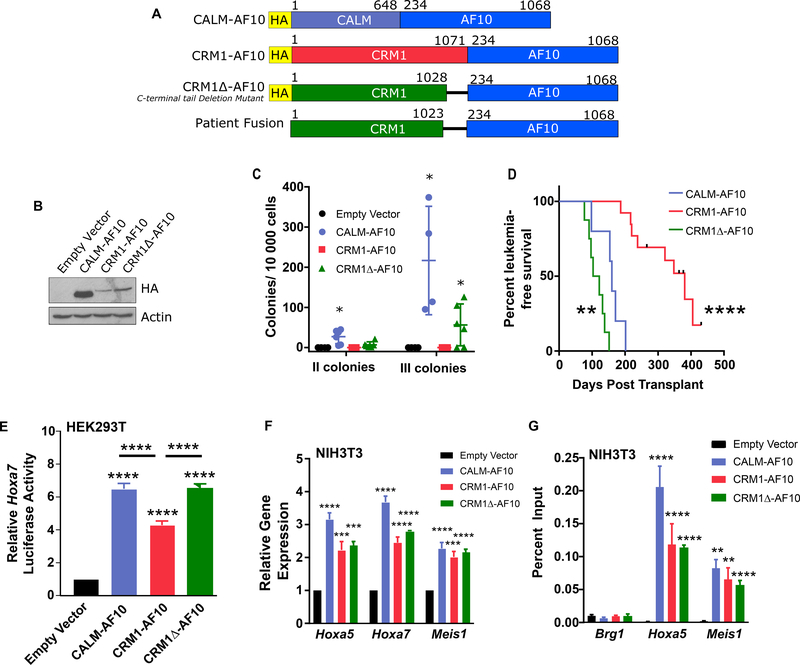
Figure 1: CRM1-AF10 and CRM1Δ-AF10 display Leukemogenic Activity.
A. Schematic representation of CALM-AF10, CRM1-AF10, CRM1Δ-AF10 and the translocation fusion found in a leukemia patient [6]. The CRM1Δ-AF10 construct that we designed is truncated at amino acid 1028, whereas the CRM1-AF10 fusion isolated in the patient is truncated at amino acid 1023. Numbers above bars indicate amino acids. B. Western Blot analysis of fusion proteins. Lysates from HEK293T cells transiently transfected with the specified proteins were probed with an anti-HA antibody. CALM-AF10 is approximately 150 kDa, CRM1-AF10 and CRM1Δ-AF10 are approximately 220 kDa. C. Clonogenic assay showing the number of secondary and tertiary colonies generated from fetal liver hematopoietic cells transduced with Empty Vector, CALM-AF10, CRM1-AF10 or CRM1Δ-AF10 following serial replating in methylcellulose. Horizontal bars indicate mean number of colonies, with error bars indicating SD. One-way ANOVA test was used to determine significance compared to Empty Vector, *p<0.05. D. Kaplan-Meier curve of mice transplanted with bone marrow progenitors transduced with CALM-AF10 (n=5), CRM1-AF10 (n=13), or CRM1Δ-AF10 (n=8). Tick marks on CRM1-AF10 survival curve indicate censored mice that died without signs of leukemia. Statistical significance was determined by log-rank test, survival of CRM1-AF10 and CRM1Δ-AF10 mice was compared to CALM-AF10 mice. E. Transcriptional activation of the Hoxa7 reporter in HEK293T cells transiently co-transfected with Empty Vector, CALM-AF10, CRM1-AF10, or CRM1Δ-AF10. Luciferase values are shown relative to empty vector. Asterisks over each fusion protein indicate significance compared to empty vector. Asterisks over horizontal bars indicate significance between noted fusion proteins. Error bars indicate SD, a minimum of 15 independent experiments (transduction replicates) were measured per sample. F. Expression levels of Hoxa5, Hoxa7, and Meis1 transcripts in NIH3T3 cells stably transduced with Empty Vector, CALM-AF10, CRM1-AF10, or CRM1Δ-AF10. Values were measured by qRT-PCR and normalized to Gapdh. Results are normalized to empty vector control. Error bars indicate S.E.M., from at least 3 biological replicates. Significance is shown in comparison with empty vector. G. Binding of CALM-AF10, CRM1-AF10, and CRM1Δ-AF10 to Hoxa5 and Meis1 genes determined by ChIP analysis. The Brg1 gene is used as a negative control. ChIP was performed using an anti-HA antibody to precipitate the HA-tagged fusion proteins expressed in stably transduced NIH3T3 cells. Error bars indicate S.E.M. from at least three biological replicates, significance is shown in comparison with empty vector. E-G: Differences determined using unpaired t-tests. **p<0.01; ***p<0.001, ****p<0.0001