Abstract
Nonsense-mediated mRNA decay (NMD) is a conserved translation-coupled quality control mechanism in all eukaryotes that regulates the expression of a significant fraction of both the aberrant and normal transcriptomes. In vertebrates, NMD has become an essential process owing to expansion of the diversity of NMD-regulated transcripts, particularly during various developmental processes. Surprisingly, however, some core NMD factors that are essential for NMD in simpler organisms appear to be dispensable for vertebrate NMD. At the same time, numerous NMD enhancers and suppressors have been identified in multicellular organisms including vertebrates. Collectively, the available data suggest that vertebrate NMD is a complex, branched pathway wherein individual branches regulate specific mRNA subsets to fulfill distinct physiological functions.
Introduction
Genetic pathways are governed by fidelity mechanisms that perform proofreading functions to limit errors during information flow. NMD is one such mechanism that is conserved across all eukaryotic cells, and rapidly degrades mRNAs with nonsense mutations to maintain fidelity during protein synthesis. In addition to its RNA surveillance function, NMD also serves a regulatory role by degrading a significant fraction of transcripts arising from mutation-free genes. For example, NMD regulates ~10% of the transcriptome in mammals [1,2]. Work on diverse model systems suggests that the NMD pathway has acquired an increasing importance over the course of evolution. Although Saccharomyces cerevisiae and Caenorhabditis elegans are viable without a functional NMD pathway [3,4], the pathway is essential for viability in vertebrates [5–7]. This gain of essentiality with increasing organismal complexity underscores the expansion of NMD functions from quality control to a more sophisticated regulatory mechanism. At the heart of the pathway, three conserved UPF (up-frameshift) proteins UPF1, UPF2, and UPF3 distinguish NMD substrates from non-substrates [8–10]. Surprisingly, however, evidence suggests that, unlike in yeast, UPF3 and perhaps UPF2 are not necessary for suppression of all NMD-targeted mRNAs in mammals [11–13]. Further, numerous NMD enhancer and suppressor proteins have been identified in multicellular organisms that play a crucial role in NMD, albeit only for a subset of target mRNAs (e.g., [14,15]). Based on these observations, NMD in complex organisms such as vertebrates can be conceptualized as a branched pathway in which parallel branches regulate distinct sets of transcripts. This review focuses on the evidence, possible mechanisms, and biological implications of the branched nature of the vertebrate NMD pathway.
Current State of NMD: Signals and Mechanisms
We begin with a brief summary of mRNA features, key protein factors, and molecular events that trigger NMD. Several excellent reviews provide a more detailed view of the NMD mechanism [10,16–20].
NMD Recognizes a Variety of Substrates during Translation
The first event in initiation of NMD is recognition of translation termination as an abnormal or ‘aberrant’ event. Such termination can be premature if it occurs at premature termination codons (PTCs), which can arise from mutations or from transcriptional or mRNA processing errors. In other cases, termination at normal stops can be sensed as ‘aberrant’ owing to the presence of naturally occurring features such as long 3′ untranslated regions (3′-UTRs), exon–exon junctions downstream of stop codons, and upstream open reading frames (uORFs) (see Glossary) [21] (Box 1). Mammalian NMD occurs during the early rounds of translation of transcripts that remain associated with the nuclear cap-binding complex (CBC) [22], or where the CBC has been replaced by the cytoplasmic cap-binding protein EIF4E [23,24]. An elegant investigation in human cells using single-molecule microscopy of NMD reporter RNAs further confirms this view, and even shows that each ribosome terminating translation at a PTC exhibits an equal and unexpectedly low probability of PTC recognition and NMD execution [25]. NMD-promoting features such as extended 3′-UTRs or 3′-UTR exon–exon junctions may function by increasing the likelihood that a termination event is recognized as aberrant during each round of translation.
Box 1. Gene Features That Produce NMD-Susceptible Transcripts.
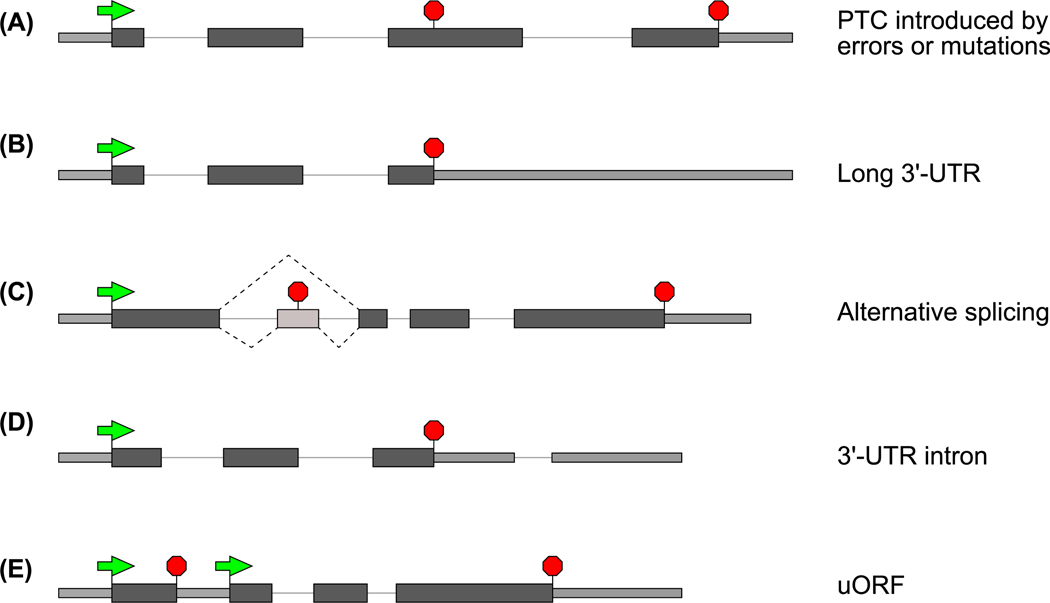
mRNAs that are subjected to NMD can arise in a variety of ways. Genes can acquire NMD-inducing premature termination codons (PTCs) via nonsense or frameshift mutations, somatic recombination (e.g., VDJ recombination), or random transcriptional errors (Figure IA) [10,16–20]. Although the introduction of a PTC shortens the ORF, this also lengthens the 3′-UTR, and this appears to be the primitive feature that is recognized by the NMD machinery [8–10]. Many endogenous transcripts with normal termination codons but longer than average 3′-UTRs are also recognized by the NMD machinery (Figure IB) [137,138,143], suggesting a regulatory role of NMD in degrading endogenous transcripts. ‘Normal’ transcripts with average-length 3′-UTRs can become NMD substrates if ribosomes undergo frameshift and encounter a PTC in another frame, thus creating a long 3′-UTR [70]. Importantly, having a long 3′-UTR does not always lead to NMD because protective mechanisms can override NMD [14,125,126]. Reinforcing the important role of NMD in regulating endogenous transcripts, PTCs can also arise as a result of alternative pre-mRNA splicing that can change the mRNA open reading frame (ORF) (Figure IC) [144]. Regulated alternative splicing events generate mRNA isoforms that are targeted for NMD to control gene expression, a mechanism that is particularly widespread among genes encoding RNA-processing machineries [144,145]. This link between splicing and NMD is particularly strong in vertebrates where NMD is strongly enhanced when a termination codon occurs upstream of an exon–exon junction (reviewed in [37–39]). Hence, in vertebrates, additional NMD targets include transcripts that contain 3′-UTR introns (Figure ID) [68]. Finally, upstream ORFs (uORFs), short reading frames in mRNA 5′-UTRs that often serve a translation regulatory function, can also induce NMD [17] (Figure IE). Translation termination at uORF stop codons perhaps presents downstream sequences as extended 3′-UTRs and/or those dotted with exon–exon junctions.
Figure I. Gene Features That Cause NMD Susceptibility.
Light-shaded thinner rectangles are untranslated regions, and dark-shaded thicker rectangles represent coding regions. Introns are shown as a black line, green arrows denote start codons, and red stop-signs indicate stop codons.
When a ribosome encounters a termination codon (either normal or premature), polypeptide release factor eRF1 enters the ribosome A-site to recognize the codon, and another release factor eRF3a (referred to here as eRF3) promotes polypeptide release to accomplish termination. Normal and aberrant translation termination events may have intrinsic differences, as suggested by a higher likelihood of translational readthrough at PTCs than at normal termination codons [10]. Earlier reports suggested that the ribosome pauses at PTCs significantly longer than it does at normal termination codons [26,27], although a recent study found no detectable difference in ribosomal stalling at premature versus normal stop codons [28]. The molecular details of the events that define aberrant (i.e., NMD-inducing) termination remain to be fully understood.
Core NMD Factors Recognize mRNAs Undergoing Premature Termination
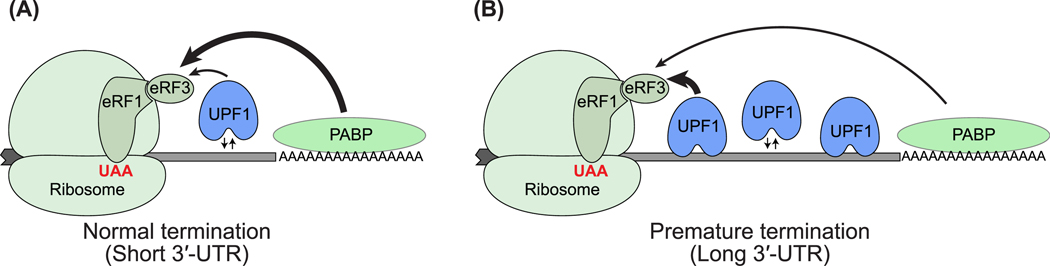
UPF1, UPF2, and UPF3 are a conserved set of core NMD factors that distinguish mRNAs undergoing aberrant termination [10]. Two mutually non-exclusive models describe how UPF1 can differentiate between transcripts undergoing aberrant versus normal termination (Figure 1). In one model, the introduction of a PTC prevents displacement of RNA-bound UPF1 from a stretch of coding sequence (CDS) during translation [29–31], causing UPF1 accumulation on RNA, more frequent engagement of UPF1 with the termination machinery, and hence abnormal termination (Figure 1A). The other model posits that an interaction between poly(A)-tail-bound poly(A)-binding protein (PABP) and ribosome-bound release factor eRF3 is crucial for efficient ribosome termination [32]. However, a longer 3′-UTR disfavors PABP–eRF3 interaction, and 3′-UTR-bound UPF1 outcompetes PABP for eRF3 interaction, thereby causing inefficient translation termination [26,27,33–36] (Figure 1B). In either model, UPF1 engaged with the terminating ribosome is then activated by UPF2 and UPF3, and NMD ensues (Figure 2A). Additional NMD enhancers such as the 3′-UTR-bound exon-junction complex (EJC), a multi-subunit complex deposited upstream of exon–exon junctions during pre-mRNA splicing [37–39], can further enhance the recruitment and activation of UPF factors on specific mRNAs (Figure 2B) (see below).
Figure 1. Two Models of How UPF1 Differentiates Aberrant Translation Termination.
The first model suggests that, compared with short 3′-UTRs (A), long 3′-UTRs (B) provide a greater opportunity for UPF1 binding and accumulation on RNAs, which promotes premature termination and NMD. The second model suggests that, on short 3′-UTRs (A), PABP– eRF3 is a more dominant interaction (thicker arrow) than UPF1–eRF3 (thinner arrow), leading to normal termination, whereas long 3′-UTRs (B) favor UPF1–eRF3 (thicker arrow) over PABP–eRF3 (thinner arrow) interaction, leading to premature termination and NMD. The grey shape to the left of the ribosome shows a truncated portion of mRNA coding sequence, and the grey line to the right of the ribosome is the 3′-UTR. Shapes representing protein factors are labeled. Abbreviations: NMD, nonsense-mediated decay; UTR, untranslated region.
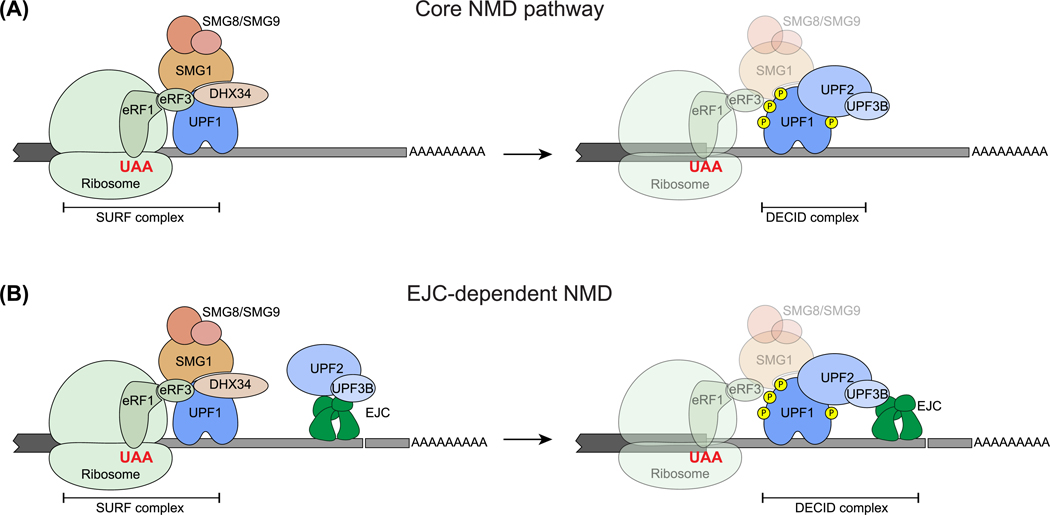
Figure 2. Mechanism of UPF1 Activation in the Two Major NMD Branches.
(A) On mRNAs without downstream exon–exon junctions, UPF1, together with eRF1 and eRF3, interacts with SMG1 to form the SURF complex [146]. The SMG1 regulatory SMG8–SMG9 heterodimer and the scaffold protein DHX34 aid in SURF complex assembly, subsequent UPF1 phosphorylation, and interaction between the SURF complex and UPF2–UPF3, leading to formation of the decay-inducing (DECID) complex and UPF1 phosphorylation (right) [147,148]. Ribosome, eRFs, SMG1 and its regulators may dissociate from the RNA at this stage, and hence are shown by more transparent shapes. (B) The exon-junction complex (EJC), a key NMD enhancer in vertebrates, when present in 3′-UTRs can recruit UPF2/UPF3B to 3′-UTR (left) and facilitate formation of the UPF complex, and hence DECID complex formation (right), leading to UPF1 phosphorylation and NMD activation. Shapes representing various protein factors are labeled. Yellow circle, phosphate. Abbreviations: NMD, nonsense-mediated decay; SURF complex, SMG1–UPF1–eRF1–eRF3; UTR, untranslated region.
UPF1 is highly conserved throughout eukaryotic evolution from fungi to humans [40], and consists of a cysteine/histidine (CH)-rich domain connected via a flexible linker to a RecA-like helicase domain. The RecA helicase domain exhibits RNA binding, ATPase, and RNA helicase activities [10,16,41]. UPF1 undergoes cycles of ATP binding and hydrolysis, and a nucleotide-induced conformational switch that converts it from a ‘loosely’ RNA-bound state to more ‘intimately’ RNA-bound form [42,43]. The intramolecular protein–protein interaction between the CH domain and the helicase domain further augments UPF1 RNA binding, and at the same time diminishes its ATPase and helicase activities, thus acting as a key regulator of the UPF1 catalytic engine [42,43]. UPF1 ATPase and helicase activities are essential for NMD [44,45]. The ATPase activity promotes UPF1 disassociation from non-NMD target RNAs, and hence mediates target discrimination [46]. This activity is also necessary to release UPF1 and other NMD factors from cleaved RNA fragments to allow completion of transcript degradation [45].
UPF2 is the next most conserved NMD factor that is maintained in almost all eukaryotes examined [40]. UPF2 interacts with UPF1 CH domain via its C-terminal intrinsically disordered region (IDR) and with UPF3 via one of its three MIF4G domains [47,48]. Simultaneous UPF2 association with both UPF1 and UPF3 mediates assembly of the UPF1–UPF2–UPF3 complex [43,49]. However, UPF2 is much more than a bridge between UPF1 and UPF3. When UPF2 engages the UPF1 CH domain, it relieves the inhibitory role of the CH domain and thus activates UPF1 ATPase and helicase activities [42,43]. Interestingly, UPF2 can also interact directly with eRF3, although the significance of this interaction for NMD remains unclear [50].
UPF3 is the third core NMD factor but is the least conserved of the three, and is even undetectable in several eukaryotes [40]. Mammalian genomes encode two UPF3 paralogs, UPF3A and UPF3B, which share a well-conserved N-terminal RNA-recognition motif-like domain that interacts with UPF2 [47,51], and a C-terminal domain for EJC interaction [52,53]. Thus, by connecting the UPF proteins with the EJC, UPF3 links translation termination with the downstream exon–exon junctions [16,17,19,20]. Compared with UPF3B, UPF3A exhibits weaker EJC binding affinity and hence weaker NMD activity [54]. UPF3A could even serve as an NMD suppressor potentially by sequestering UPF2 away from UPF3B and the NMD complex [55]. Thus, UPF3B is likely to provide the major NMD-activating function, whereas UPF3A can act as a weak NMD activator or as a suppressor [54,55]. Although the full picture of UPF3 biochemical functions remains elusive, UPF3B can enhance the ability of UPF2 to promote UPF1 ATPase and helicase activities [43]. Human UPF3B can also interact directly with eRF3 and influence different stages of translational termination reaction in vitro [56], with unknown consequences for NMD.
An essential step in NMD is UPF1 phosphorylation at its several serine/threonine-glutamine motifs [(S/T)Q], which is carried out in mammals (and most eukaryotes) by SMG1, a phosphatidylinositol 3-kinase-related protein kinase [57–60]. Because UPF1 phosphorylation licenses the mRNA for degradation, a series of events govern this crucial step (Figure 2). Phosphorylated UPF1 binds to EIF4E to inhibit new cap-dependent translation [61], and serves as a binding platform for SMG5/SMG7 and SMG6 [62,63], the effectors that initiate mRNA decay and mediate UPF1 dephosphorylation via PP2A phosphatase [61,62]. Overall, a complex series of events recruit and activate UPF1 to distinguish aberrant from normal termination events, and then recycle UPF1 and other factors such that translation termination is continuously monitored.
Multiple Parallel Routes to Activate UPF1
We turn next to observations that suggest gain of additional regulators and a surprising loss of full dependence on some core NMD factors in the vertebrate NMD pathway. An emergent view is that the NMD pathway, at least in the vertebrates, can be conceptualized as a branched network that converges at UPF1.
EJC-Enhanced NMD Is a Major Branch of the Pathway
In addition to the core NMD pathway dictated by the UPF proteins (Figure 2A), a prominent branch of the vertebrate NMD pathway is activated when exon junctions are present in 3′-UTRs (Figure 2B) [16,17,19,20]. This branch of NMD is enhanced by the EJCs, which are assembled during splicing as a trimeric core composed of EIF4A3, MAGOH, and Y14 (also known as RBM8A) (Figure 3A) [37–39], and mark the position of exon–exon junctions until they are removed in the cytoplasm by the translating ribosome. If the ribosome terminates translation (either at PTCs or at normal termination codons) >50 nt upstream of the last exon–exon junction [21], or sometimes even closer [64], 3′-UTR-bound EJC(s) can activate NMD. Although the core EJC proteins are widely conserved in eukaryotes, the EJC function in NMD has been confirmed mainly in vertebrates, where the EJC proteins are also essential for viability [64–66]. The trimeric EJC core provides a composite surface for binding UPF3B (or UPF3A) [53]. Current models of EJC-dependent NMD state that the EJC downstream of a terminated ribosome recruits UPF3B and UPF2 to the vicinity of the terminated ribosome (Figures 2B and 3A), enhancing the formation of the UPF complex and UPF1 phosphorylation. In addition, EJCs, both upstream and downstream of stop codons, can stimulate NMD indirectly by enhancing translation [25,67]. Because the vast majority of vertebrate genes contain introns, sometimes even in 3′-UTRs [64,68], and because alternative pre-mRNA splicing is widespread in these organisms [69], the EJC-dependent NMD branch regulates a large fraction of NMD substrates.
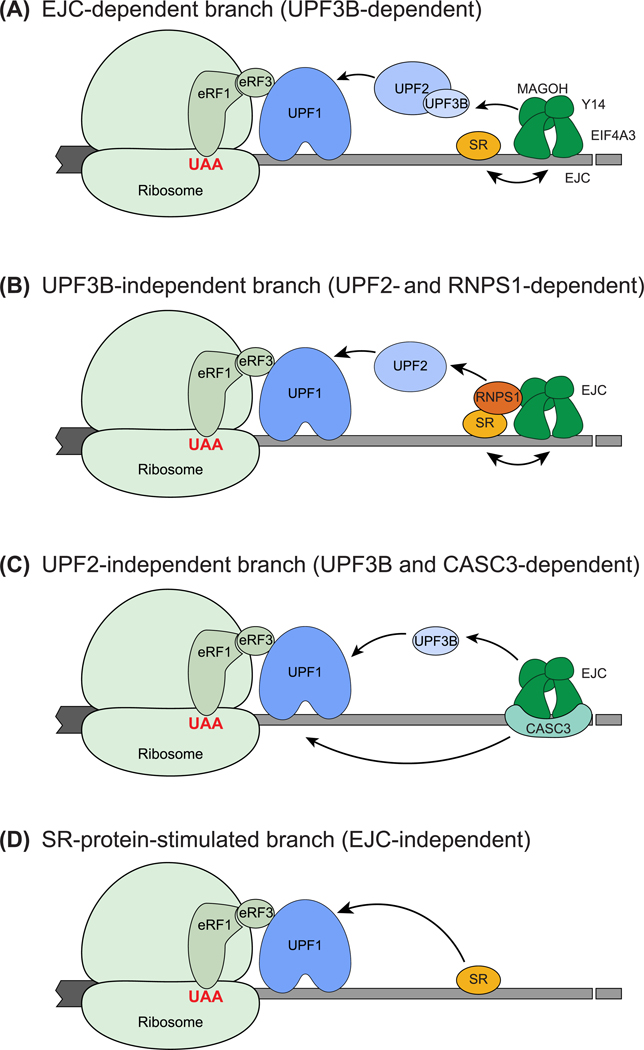
Figure 3. Loss of Complete Dependence on UPF2/3 and Gain of Enhancers of UPF Function Leads to Specific NMD Branches.
(A) The EJC-dependent NMD branch. An EJC downstream of a terminated ribosome can lead to UPF1 activation at a termination event via EJC–UPF3B–UPF2–UPF1 interaction. SR proteins can enhance this EJC-dependent NMD by boosting EJC deposition/RNA binding. (B) The UPF3B-independent NMD branch. Interaction between RNPS1-containing EJC and UPF2 can cause EJC-dependent and UPF3B-independent NMD. (C) The UPF2-independent branch. UPF3B can directly interact with UPF1 and elicit NMD. CASC3-containing EJCs may contribute to this branch. (D) SRSF1 bound to the 3′-UTR can enhance NMD activation in an EJC-independent manner by interacting directly with UPF1. Abbreviations: EJC, exon-junction complex; NMD, nonsense-mediated decay; UTR, untranslated region.
NMD Can Occur via UPF3- and UPF2-Independent Branches
An interesting development in the NMD field has been the discoveries that, although UPF2 and UPF3 are required for all NMD in S. cerevisiae [3,70], at least to some extent, mammalian NMD can occur without UPF3B, and possibly even without UPF2 (e.g., [11,13,71]). The most convincing evidence is for UPF3B dispensability. The non-essential nature of UPF3B is underscored by mutations in human UPF3B in individuals with X-linked mental retardation, some of whom almost completely lack UPF3B protein and suffer from a range of neurological disorders such as autism and intellectual disabilities [12,72–74]. Thus, UPF3B deficiency in mammals seems to be tolerable for viability but preferentially affects neuronal function [75–77]. Consistently, only a small subset of known NMD targets are upregulated in mice depleted of UPF3B via antisense oligonucleotide (ASO)-mediated knockdown [78], in UPF3B-lacking human patient cells [12,79], and in UPF3Bdepleted HeLa cells [71] (Figure 3B). Thus, it appears that UPF3B-independent NMD is likely a prominent branch in mammals, whereas UPF3B-dependent NMD may have a more specialized function.
How does the UPF3B-independent NMD pathway operate mechanistically? In a simplest scenario, UPF3A may activate NMD in the absence of UPF3B, at least on a subset of NMD targets [54,80]. Interestingly, many UPF1-sensitive NMD targets in HeLa cells can still undergo efficient NMD when both UPF3A and UPF3B are depleted [71], suggesting that some human NMD targets are likely to be independent of both UPF3A and UPF3B. Supporting this idea, in Drosophila, UPF3 is dispensable both for viability and for NMD of several mRNAs [81]. Other mechanisms of UPF3B-independent NMD may operate via NMD enhancers within the EJC. 3′-UTR tethering of RNPS1, a peripheral EJC protein, elicits strong decay of a reporter mRNA, even when UPF3B is depleted from cells (Figure 3B) [11]. In addition, the UPF3B requirement in NMD could be dictated by its function in translation termination [56]. It remains to be tested whether the UPF3B-dependence (or UPF3B-independence) of NMD is governed by distinct transcript-specific features that modulate translation.
Unlike UPF3B, UPF2 is indispensable for embryonic development in mice, and its homozygous loss leads to lethality by embryonic day 9.5 [6]. Conditional Upf2 knockouts in mice result in strong NMD inhibition and developmental defects in almost all tissues examined [6,13,82,83], suggesting an essential role for UPF2 during development. Surprisingly, conditional Upf2 knockout in mouse spermatocytes shows no global upregulation of PTC-containing transcripts, whereas transcripts containing long 3′-UTRs are preferentially upregulated [13]. Identification of partial UPF2 loss-of-function mutations in individuals with neurological deficits suggest that different cell/tissue types may have variable UPF2-dependence [84,85]. These genetic data hint at a dispensable function of UPF2 under specific conditions and/or cell types. Interestingly, in HeLa cells some NMD-targeted mRNAs are almost insensitive to UPF2 knockdown, suggesting that their NMD might be UPF2-independent or insensitive to its reduced levels [11]. Moreover, in these cells, a UPF1 mutant protein lacking UPF2-binding ability can still be phosphorylated by SMG1, and can almost completely substitute for wild-type UPF1 for efficient NMD of a β-globin reporter mRNA [36]. Consistently, there is evidence for the assembly and activation of UPF2independent NMD complexes (Figure 3C). Tethering of UPF3B or the EJC factor CASC3 (also known as MLN51 or BTZ) downstream of a stop codon triggers strong UPF1-dependent downregulation of the reporter RNA in UPF2-depleted HeLa cells [11]. Along these lines, UPF2 interaction-deficient UPF1 mutants can interact with UPF3B and CASC3 [36,86]. Further, a direct UPF3B–UPF1 interaction has been observed in vitro [56]. It remains unknown whether UPF1 ATPase and helicase activities can be activated without UPF2.
Overall, although genetic studies based on the complete loss-of-function of UPF3B [12,77] and UPF2 [13] lend strong support for their dispensability during NMD, some key evidence for this idea comes from partial protein knockdown experiments, which remains an important caveat (Box 2).
Box 2. Key Caveats of Experiments Supporting NMD Factor Independence in Specific Branches.
Thus far, many experiments investigating specific NMD factors and branches, including some of the studies that suggest UPF2 and UPF3B dispensability [11,71], have relied on RNAi-mediated knockdown of these factors. The lack of effects following NMD factor knockdown could be either because these proteins do not target the substrates examined, or because the residual amount of protein is sufficient to carry out its function. Thus, insufficient protein knockdown could lead to mischaracterization of effector-dependent substrates as effector-independent substrates. Thus, incomplete protein depletion is an important issue to be considered in the interpretation of such results, although a differential response of mRNA substrates to partial knockdown of NMD factors does suggest variable sensitivity of different substrates to key effectors. CRISPR/Cas9-mediated complete gene knockout to create cell lines and/or animal models completely lacking gene function could help to circumvent the insufficient protein depletion problem. This is exemplified by the recent analysis of NMD after CRISPR/Cas9-mediated complete SMG7 loss-of-function, which has revealed important differences in comparison to earlier knockdown studies [105,108,109] (see main text). Nonetheless, the application of complete loss-of-function approaches could be limited because many NMD factors are essential for cell viability and/or are required for early embryonic development. In future, careful investigations using conditional knockout cell/animal models will be crucial to address the complex nature of NMD branches.
Another caveat with NMD protein loss-of-function (partial or complete) studies is that many NMD factors also have functions outside the pathway. An obvious and prominent example is UPF1, which has been shown to function in a variety of mRNA degradation processes outside NMD (reviewed in [41]), and can even function as an E3 ligase that targets proteins for ubiquitination [149]. Such broad activities of UPF1 might lead to mischaracterizing of mRNAs upregulated following UPF1 knockdown as NMD substrates even though they could also be the targets of a non-NMD-related activity of UPF1. Even though functions of UPF2 and UPF3 outside NMD remain largely unknown, it is possible that both factors can carry out non-NMD-related activities. Thus, at least some of the effects on gene expression and cell function produced by inactivation of NMD factors could result from NMD-unrelated perturbations.
EJC/UPF Enhancers Further Branch the NMD Pathway
EJC Composition Splits the EJC-Dependent NMD Branch
The EJC is a dynamic complex in which its core remains the same but its complement of peripheral proteins changes during the mRNA life-cycle [37–39]. The earliest hints that EJC composition can influence NMD came from experiments noted above where mRNA tethering of the EJC proteins including RNPS1 and CASC3 suggested that there are UPF2-and UPF3B-independent routes of NMD [11]. The recent discovery that RNPS1 and CASC3 define two mutually exclusive or alternative EJC compositions in mammalian cells further explains the non-overlapping roles of these two factors in NMD [87].
Owing to the mutually exclusive nature of RNPS1- and CASC3-containing EJCs, the EJC-dependent NMD branch can now be viewed as at least two distinct branches (Figure 3A–C). The distinctive features of NMD targets that shunt them to one or the other EJC-dependent branches remain to be identified. mRNAs preferentially bound by the RNPS1-EJC as compared with the CASC3-EJC in HEK293 cells are enriched in binding sites of serine/arginine-rich (SR) proteins, which exclusively associate with RNPS1-EJC [87]. It remains to be seen whether mRNAs enriched in SR protein binding sites are preferred targets of the RNPS1-dependent NMD branch. Although no distinctive features of CASC3-bound mRNAs are known, a limited effect of CASC3 knockout on NMD targets in HeLa cells confirms that CASC3 is required for efficient NMD of only a subset of NMD-targeted mRNAs [88]. How do RNPS1 and CASC3 enhance NMD? RNPS1 interacts with the EJC indirectly via the ASAP–PSAP complex [89,90]. It is possible that it enhances NMD either indirectly by promoting deposition and/or stable binding of the EJC, or more directly via interactions with the EJC/UPF proteins. Unlike RNPS1, CASC3 directly binds to EIF4A3 [91,92] and shows a strong association with UPF3B [87], as well as a UPF2independent link to UPF1 [86]. Whether CASC3 can engage with UPF proteins independently of the EJC core factors remains unknown.
As per current evidence, the RNPS1-containing EJC likely represents a predominantly nuclear and early cytoplasmic stage of the EJC that switches to a CASC3-containing EJC either before or during translation [87]. Therefore, the dependence of NMD targets on RNPS1-versus CASC3-EJC will potentially be influenced by the rate of the EJC compositional switch on individual mRNAs, and the rate of mRNA entry into the translation pool. mRNAs that enter translation soon after their export may depend on RNPS1, whereas those that spend an extended time as pre-translation mRNAcontaining ribonucleoproteins (mRNPs) may depend on CASC3. Whether this compositional switch also affects UPF2/UPF3-dependence remains to be investigated.
SR Proteins Can Enhance EJC-Dependent and -Independent NMD Branches
Since the first report that overexpression of canonical SR proteins enhances NMD of reporter transcripts [15], several lines of evidence have emerged that support their role in NMD. Both nucleocytoplasmic shuttling SR proteins (e.g., SRSF1) [93] and the nucleus-retained SRSF2 [94] can enhance NMD. Thus, SR proteins likely promote NMD via multiple mechanisms. First, these proteins can act in concert with the EJC to enhance NMD. With the exception of SRSF2, all canonical SR proteins interact with the EJC [95,96], specifically the RNPS1-EJC [87]. SR proteins and EJC could stabilize each other’s binding to spliced RNAs to thereby enhance NMD (Figure 3A,B). Such a model is supported by the enrichment of SR protein binding sites at noncanonical EJC binding positions close to the EJC deposition site [87,95,96], and by the enhancement of EJC deposition by nucleus-retained SRSF2 [94]. Second, SRSF1 can interact with UPF1 in both the nucleus and cytoplasm, and a direct interaction has been reported between the proteins in vitro [93]. Thus, SRSF1 can enhance NMD independently of the EJC, and even independently of UPF2 and UPF3B (Figure 3D). SRSF1 may also potentially enhance UPF1 activity by recruiting phosphatase PP2A [93]. Lastly, overexpression of SRSF1 can alter the location of NMD and the pioneer round of translation from cytoplasm to nucleus-associated [97], suggesting another potential mechanism of NMD regulation by enhancing translation. Although it is clear that SR proteins act as NMD enhancers, much remains to be learned about their function in NMD.
Multiple Routes to mRNA Degradation
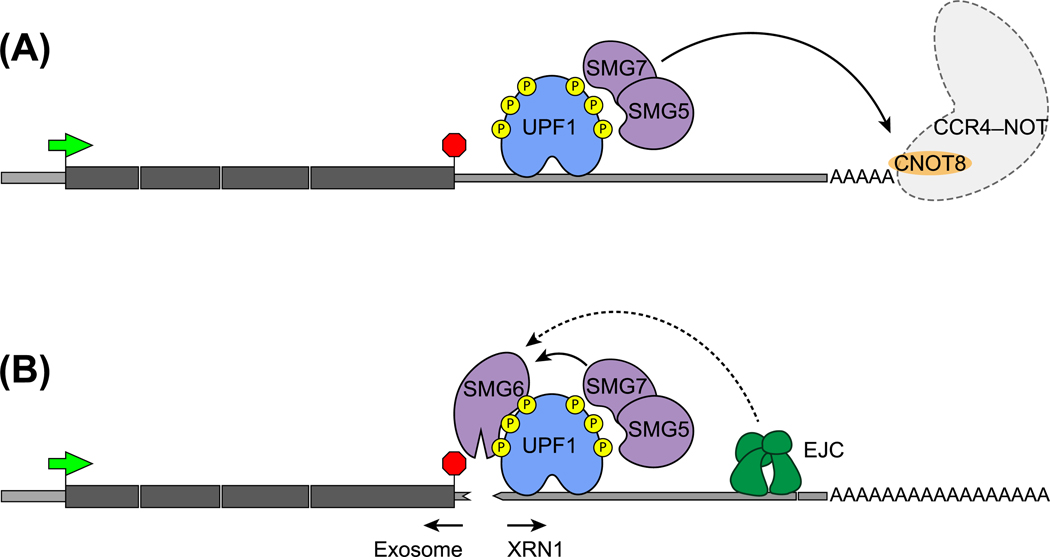
Following UPF1 phosphorylation, the NMD pathway enters the mRNA degradation phase, which can also proceed via multiple routes. The key functions in this phase are carried out by the three SMG proteins – SMG5, SMG6, and SMG7 (Figure 4). Each of these proteins can bind to phosphorylated UPF1 via their 14–3-3 domain but can also interact with UPF1 in a phosphorylation-independent manner [98,99]. SMG6 cleaves the mRNA into two fragments [100], at or immediately downstream of the PTC [101,102], via the endonuclease activity of its PIN domain. SMG5/SMG7 are recruited to UPF1 as a heterodimer, and a direct interaction between SMG7 and CNOT8 (also known as POP2) can connect the target mRNA to the CCR4/NOT deadenylation complex (Figure 4A) [103,104]. Recruitment of the CCR4/NOT complex to NMD-targeted mRNAs can initiate rapid deadenylation, followed by mRNA decapping and degradation by XRN1, the 5′–3′ exonuclease (Figure 4A).
Figure 4. Two Main Routes for mRNA Degradation after UPF1 Activation.
(A) SMG5–SMG7 heterodimer recruited to phosphorylated UPF1 can interact with CNOT8 protein (also known as POP2) in the CCR4–NOT complex. The CCR4–NOT complex initiates mRNA degradation by deadenylating the poly(A) tails. (B) The SMG6 endonuclease recruited to phosphorylated UPF1 acts in SMG5/SMG7-dependent manner to cleave mRNA in the vicinity of PTCs. The action of SMG6 could further be enhanced by the presence of downstream NMD enhancer EJC. After cleavage of mRNAs, 5′ and 3′ fragments are degraded by exosomes and exonuclease XRN1 respectively. Green arrow, initiation codon; red stop-sign, stop codon. Abbreviations: EJC, exon-junction complex; NMD, nonsense-mediated decay; PTC, premature termination codon; UTR, untranslated region.
The relative flux of NMD through mRNA degradation routes and their target specificity is not yet completely understood. Earlier reports based on SMG6 and SMG7 knockdown suggested that the SMG5/SMG7 and SMG6 routes act essentially on the same set of transcripts and thus may work in a partially redundant and independent manner [105]. This view is also supported by an increase in decapped mRNA, and hence the increased flux through SMG5/SMG7 route, under SMG6 limiting conditions [102,106]. Consistently, Smg6 is essential for mouse embryonic stem cell (ESC) differentiation but Smg6 knockout does not affect somatic cell viability [107], suggesting redundant roles of SMG6 versus SMG5/SMG7, at least in some cell types. Interestingly, mRNAs targeted for NMD because they contain PTCs appear to be more sensitive to SMG6 levels [108], and the addition of EJCs to a long 3′-UTR leads to much more efficient SMG6-mediated cleavage of mRNAs [101]. Thus, the presence of downstream EJCs may confer increased sensitivity to decay via the SMG6 route.
A recent study has challenged the complete independence of SMG5/SMG7 and SMG6 functions within the NMD pathway [109]. Under SMG7 loss-of-function conditions, SMG6-mediated cleavage was found to be inactivated, suggesting that SMG6 function depends on SMG5/SMG7. The essential nature of the SMG5–SMG7 interaction for the NMD pathway further underscores this dependence, whereas the SMG7–CNOT8 interaction is dispensable for NMD. Interestingly, in Smg7 deletion cells, UPF1 phosphorylation is altered and its release from non-target mRNAs is also impaired, possibly because of disruption of PP2A-mediated UPF1 dephosphorylation. Thus, it appears that, even though SMG5/SMG7 and SMG6 are capable of defining independent mRNA decay routes, their functions within the NMD pathway are more intertwined such that SMG5/SMG7 perform additional functions to authenticate and allow SMG6-mediated decay to proceed.
Phosphorylated UPF1 can also be linked to mRNA decay proteins via a third possible route in which UPF1 interacts directly with the decapping complex components DCP2 and PNRC2 [103,104]. However, this route may play only a minor role because NMD reporter RNAs are unaffected by PNRC2 knockdown [104].
Implications of the Branched Nature of the NMD Pathway
The alternative routes to initiate and execute the NMD pathway (Figure 5) provide opportunities for regulating specific mRNAs in particular cells/tissues, cellular states, and subcellular locations, and can also boost the robustness of the pathway in the face of genetic variation imposed by intrinsic and extrinsic factors. The branched nature of the pathway also provides an opportunity to specifically target a particular NMD branch for developing therapeutic interventions (Box 3).
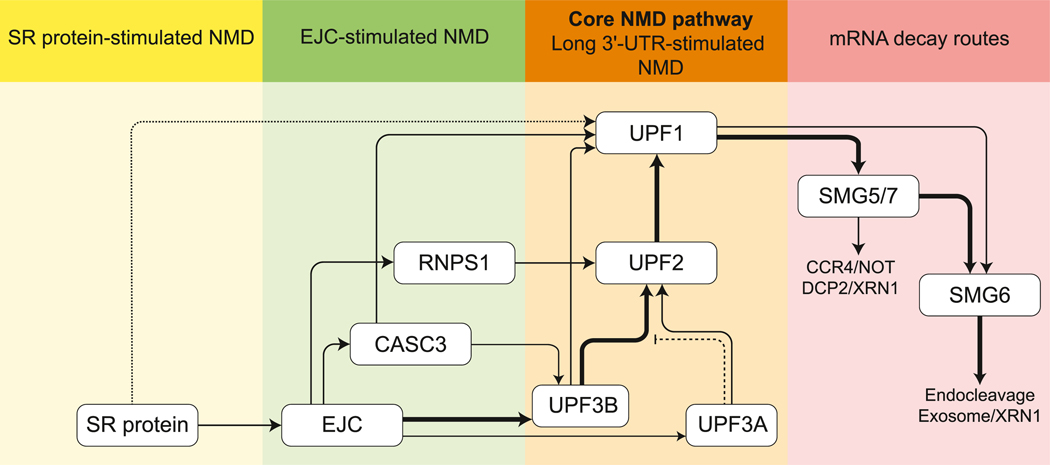
Figure 5. A Schematic Summarizing the Flux of NMD Targets through Possible NMD Branches.
Individual branches/routes are labeled above each shaded region. The branch requiring only the UPF proteins may constitute the ‘core’ NMD branch that is maintained in most eukaryotes and remains active in vertebrates. Thick arrows indicate major NMD routes in mammals. UPF3A can serve as an NMD activator (thin arrow) or as a repressor (inhibitory line) of UPF3B function. SRSF1 may activate UPF1 independently of the EJC, but this constitutes only an optional nucleation point for the NMD pathway (indicated by dotted lines). Abbreviations: EJC, exon-junction complex; NMD, nonsense-mediated decay; UTR, untranslated region.
Box 3. Inhibition of Individual NMD Branches for Therapeutics.
NMD can be a double-edged sword. Although it limits the production of truncated and potentially harmful proteins from PTC-containing transcripts, in some cases truncated polypeptides may still be functional. However, suppression of PTC-containing mRNAs by NMD can limit the amount of protein produced in such cases. For example, in cancer cells, nonsense mutations are disproportionately enriched in tumor-suppressor genes [150], presumably inhibiting their expression and activity via NMD. A potential therapeutic approach is to combine inhibition of NMD of PTC-containing mRNAs with translational readthrough to promote full-length protein expression. One problem with such an approach however is that NMD is essential in mammals, and therefore broad NMD inhibition would cause undesirable changes in gene expression. Alternatively, an individual NMD branch can be inhibited to target a more limited set of genes. In one such approach, application of NMDi-14, an NMD inhibitor that disrupts SMG7–UPF1 association [151], together with a readthrough-promoting drug G418, increases full-length protein expression from a PTC-containing TP53 allele in N417 cell lines [151]. This restores sufficient p53 activity, resulting in p53-mediated programmed cell death. Despite the promising outcome, NMDi-14 treatment upregulates nearly 1000 mRNAs in human U2OS cell lines [151], suggesting that SMG7 targets a relatively broad range of NMD targets. It remains desirable to inhibit a more specific branch to block NMD of an even smaller set of transcripts. Recently, Huang et al. reported that ASO-mediated inhibition of UPF3B significantly alters the expression of only ~250 genes in mouse liver [78]. UPF3B inhibition using ASOs in hemophilia B mice model, which expresses a human PTC-containing FIX transgene, increases FIX mRNA levels by threeto fourfold. Combined with translational readthrough treatments (G418 and eRF3-inhibiting ASO), FIX protein production can be increased to up to 4% of the wild-type level, compared with <1% in untreated mice [78]. This small increase in full-length protein production leads to a slight but significant improvement in blood coagulation. These results underscore the promise of inhibition of specific NMD branches for developing new therapeutics for cancers and genetic diseases. Greater understanding of the molecular mechanisms and target mRNAs of specific NMD branches will provide more such opportunities.
Regulation via the UPF3B-Dependent NMD Pathway
The UPF3B-dependent branch is the best-understood for its regulatory nature and for the mechanisms by which its activity can be controlled. As noted above, the UPF3B-dependent branch plays an essential role in neuronal tissues [12,75–77], although the underlying basis remains to be understood. It is possible that some neuron-specific transcripts that are important for the identity/function of these cells have specific features, so far unknown, that confer UPF3B dependence. Alternatively, such cells could be more reliant on a ubiquitous UPF3B function (e.g., regulation of unfolded protein response (UPR) [110]).
Various mechanisms can limit UPF3B function to select cells/tissues. A simple mode may be to regulate UPF3B expression levels to tune the NMD flux through this branch. Compared with pluripotent cells, UPF3B mRNA levels are ~twofold lower in non-pluripotent cells [111], suggesting that UPF3B-dependent NMD is dampened as cells exit from the pluripotent state to differentiate. Intriguingly, when C2C12 cells undergo myogenesis, Staufen-mediated mRNA decay competes with NMD for UPF1, and perhaps UPF2 [112,113], and slows down NMD [112]. Under these conditions, UPF3B becomes upregulated, with consequent downregulation of UPF3B-dependent targets. The UPF3B-dependent NMD branch can also be regulated by the antagonistic activity of UPF3A to repress UPF3B function [55]. During mouse spermatocyte maturation, the UPF3B-to-UPF3A ratio decreases dramatically, leading to NMD suppression which is crucial for male germ cell development [55]. Interestingly, the UPF3A:UPF3B ratio is also regulated by negative feedback regulation of UPF3A by UPF3B [80], which may fine-tune UPF3-dependent NMD. Another trans-acting factor that can stimulate the UPF3B-dependent NMD branch is ICE1, which interacts with the EJC via its putative MIF4G domain [114]. Although ICE1 does not stably associate with any NMD factors, normal levels of ICE1 are necessary for efficient association of UPF2/UPF3B with the EJC and for maintaining sufficient cytoplasmic UPF3B levels [114]. It will be interesting to test whether ICE1 function in NMD shows any variation in a transcript/cell/tissue-specific manner.
Regulation via the EJC-Dependent NMD Pathway
Several mechanisms are emerging whereby the EJC-dependent NMD branch could be regulated. One such mechanism acts via phosphorylation of EIF4A3 at threonine 163 by CDK1 and CDK2 [115], thereby inhibiting EIF4A3 RNA binding and hence its assembly into the EJC. Moreover, the level of EIF4A3 phosphorylation changes during the cell cycle and negatively correlates with NMD efficiency. Several other EJC factors are post-translationally modified (e.g., Y14 [116,117] and RNPS1 [118]), although the significance of these modifications on EJC function during NMD remains unknown. Modulation of the two alternative EJC factors RNPS1 and CASC3 can also influence the efficiency of EJC-dependent NMD. In different HeLa cell strains, RNPS1 levels are directly proportional to the NMD efficiency of an EJC-dependent NMD reporter RNA [119]. Notably, like UPF3B, RNPS1 mRNA levels are downregulated ~twofold in non-pluripotent cells compared with pluripotent cells [111]. Further, Casc3 mRNA levels are much lower than those of the other core EJC factors in several murine tissues [120], and CACS3 overexpression modestly slows down NMD in HeLa cells [87]. Because the RNPS1- and CASC3-dependent phases of NMD are non-overlapping, the RNPS1-to-CACS3 ratio could also tune NMD efficiency. Regulation of CASC3 mRNA by miR-128 in neural cell types [121], and assembly of CASC3 protein into stress/cytoplasmic granules upon stress [122,123], exemplify other regulatory modes that may affect NMD. Finally, given the extensive EJC–SR protein nexus within mRNPs [95], it is important to understand whether the EJC-dependent branch could be regulated by signaling pathways that converge on the SR proteins [124].
Transcript- or Location-Specific Control of NMD Branches
Cis-acting mRNA features can also modulate NMD, some of which may preferentially affect one of the NMD branches. Some mRNAs with long 3′-UTRs evade NMD via specific 3′-UTR sequence elements, thus exemplifying how the core NMD pathway can be regulated in a transcript-specific manner [14,125,126]. Some of these NMD-inhibitory elements occur in the vicinity of stop codons and are binding sites for HNRNP factors such as PTBP1 and HNRNPL [14,126]. PTBP1 binding close to stop codons promotes UPF1 dissociation from substrate mRNAs to inhibit NMD [127]. Notably, similar 3′-UTR binding of HNRNPL can override NMD induction even by downstream EJCs [126]. The increased density of binding sites of other HNRNP factors in the 3′-UTRs of mRNAs that show reduced UPF1 binding suggests that this NMD-protective mechanism is probably a more widespread function of HNRNP proteins [126]. Notably, HNRNPs are well-known regulators of cell- and tissue-specific mRNA processing and translation [128], and hence may also regulate NMD in a context-dependent manner. In addition to 3′-UTR-based elements, mRNA features such as translation reinitiation downstream of PTCs can cause escape from NMD [129–131]. Systematic analysis of large-scale omic datasets has confirmed these previous observations, and has also uncovered many other potential NMD evasion strategies [21,132,133], which remain to be further dissected to understand whether they operate in a branch-specific manner.
Evidence suggests that the NMD pathway and its different branches can also regulate gene expression in a spatial manner at subcellular locations. A noteworthy example is the regulation of endoplasmic reticulum (ER) homeostasis and UPR by NMD [134,135] where the UPF3dependent NMD branch has a prominent role in regulating the mRNAs encoding three transmembrane UPR sensors ATF6, PERK, and IRE-α [110]. UPF3B and SMG6 have been constitutively observed at the ER [134], and NBAS, a protein involved in ER-to-Golgi trafficking, can directly recruit UPF1 and other SURF (SMG1–UPF1–eRF1–eRF3) complex components to termination events at the ER [136]. Indeed, NBAS regulates a fivefold higher number of membrane-associated mRNAs than cytosolic mRNAs, whereas UPF2 appears to be more specific for cytosolic mRNAs, supporting the spatial organization of different NMD branches.
Branched NMD Pathways and Genetic Robustness
In multicellular organisms, transcripts encoding several NMD factors, including the three UPF proteins and the SMG proteins, are themselves under negative feedback regulation by the pathway [137–139]. The evolution of such a feedback mechanism has been suggested to tune the magnitude of NMD in situations where genetic or environmental factors dampen the pathway [20]. Interestingly, the feedback regulation of NMD factor genes is controlled by different branches, and this also varies according to cell and tissue type [137]. For example, the EJC-dependent branch regulates UPF1 and SMG1 mRNAs, whereas the other NMD factor mRNAs are regulated by the EJC-independent branch. Among all feedback-regulated NMD factor mRNAs, only UPF1 and SMG7 mRNAs are degraded by the UPF3B-dependent branch. As the ES cells differentiate, feedback regulation of some NMD factor mRNAs switches from the UPF3B-independent to the UPF3B-dependent pathway. Such autoregulatory controls can buffer the pathway for phenotypic advantage, as suggested by the amplification of NMD factor genes and NMD activity during tumor evolution to suppress novel immunogenic epitopes [140]. It is reasonable to predict that the branched organization of the pathway can further extend the robustness of these controls over evolutionary timescales. Moreover, NMD branches may also contribute to robustness by offering at least partial functional redundancy, which can buffer developmental reactions and stabilize information flow networks during evolution [141,142].
Concluding Remarks
The field has made tremendous progress in understanding the roles of the NMD pathway in mRNA quality control and in gene regulation. In this review we have discussed how, with the acquisition of new enhancers and suppressors, and the loss of full dependence on some core factors, NMD has evolved from a simpler pathway into a complex intricate regulatory network (Figure 5). Further, as NMD became enmeshed within developmental pathways, this may have brought about the evolution of autoregulatory feedback loops and its parallel branches. We (here) and others [20] have conceptualized the parallel mechanisms as a branched network, but it can also be viewed as an ensemble of mRNP assembly and modification states that determine whether a translation termination event will be labeled as aberrant [18]. Nonetheless, many fundamental questions remain about NMD mechanism and function (see Outstanding Questions), perhaps the most profound being how NMD and its various branches contribute to biological and developmental outcomes.
Outstanding Questions.
When and how do UPF2 and UPF3 recognize and activate UPF1 following SURF complex assembly on NMD substrates?
When and how are UPF2 and UPF3 recruited to NMD substrates during EJC-independent NMD?
How can UPF1 activation occur in the absence of UPF2 or UPF3? Are there additional factors that similarly activate UPF1 in UPF2- and/or UPF3independent NMD?
How is mRNA substrate specificity established within different NMD branches? What are the contributions of trans-acting factors and RNA ciselements to such substrate specificity?
What fraction of the NMD-regulated transcriptome is directly influenced by various NMD enhancers (e.g., SR proteins) and suppressors (e.g., HNRNP proteins)? What are the contributions of these factors to the EJC-dependent versus EJC-independent NMD branches?
Do NMD suppressors and enhancers undergo dynamic regulation such that their activities can be rapidly reversed under particular conditions to regulate NMD?
What is the relative flux through various NMD activation and execution branches? Do these branches act in a redundant fashion?
What biological functions are governed by the individual NMD branches?
Highlights.
Core factors such as UPF3 and even UPF2 that are necessary for yeast NMD can be dispensable for vertebrate NMD.
Vertebrate NMD is a branched pathway that converges at UPF1, wherein individual branches target different transcript subsets.
Several NMD enhancers and suppressors have been identified that function in a branch-, transcript-, and/or location-specific manner to provide an additional layer of NMD regulation.
The heterogeneous composition of the exon-junction complex, a key NMD enhancer in vertebrates, can further influence specific NMD branches.
The mRNA degradation phase of NMD proceeds via multiple parallel routes that may function in transcript-, branch-, and/or cell/tissue-specific manner.
Inhibition of specific NMD branches can provide therapeutic avenues to treat diseases caused by nonsense mutations by reducing toxicity caused by total inhibition of the pathway.
Acknowledgments
This work was supported by a National Institutes of Health grant to G.S. (R01GM120209). Z.Y. is supported by fellowships from the Ohio State University Center for RNA Biology and the Pelotonia program.
Glossary
- Alternative pre-mRNA splicing:
the process of creating multiple mRNA isoforms from a single pre-mRNA via different exon combinations.
- Antisense oligonucleotide (ASO):
short RNA-like oligomer that binds to its complementary sequence in the target RNA to block RNA–protein or RNA– RNA interactions.
- ATPase:
enzymes that catalyze the decomposition of ATP into ADP and inorganic phosphate, often using the energy from ATP to alter protein conformation to perform functions.
- Deadenylation:
shortening of the poly (A) tails at mRNA 3′-ends.
- Decapping:
removal of the m7GpppN cap at mRNA 5′-ends, which exposes the 5′-end for decay.
- Endonuclease:
an enzyme that cleaves the phosphodiester bond within a polynucleotide chain (DNA or RNA).
- Exonuclease:
an enzyme that cleaves nucleotides one by one from the ends (5′ or 3′) of a polynucleotide chain.
- Frameshift mutations:
mutations caused by insertions or deletions of nucleotides that are not a multiple of three. Owing to the triplet nature of the genetic code, this results in the ribosome decoding the mRNA in a different frame and completely changes the resulting protein sequence.
- Intrinsically disordered region (IDR):
a stretch of amino acids within a protein that does not form a defined structure, but instead generates an ensemble of disordered conformations.
- mRNA tethering:
an experimental tool wherein a protein of interest can be attached to a desired location in an mRNA of interest. It can be accomplished by fusing a protein of interest with a viral capsid protein, and embedding the capsid protein RNA recognition elements in an RNA of interest.
- mRNA-containing ribonucleoproteins (mRNPs):
the ribonucleoprotein (RNP) complexes formed by the assemblage of mRNA and its bound proteins.
- Nuclear cap-binding complex (CBC):
a protein complex consisting of CBP80 and CBP20 that binds to the mRNA m7GpppN cap in the nucleus.
- Open reading frame (ORF):
the part of an mRNA sequence that is flanked by a start codon and a stop codon and that is translated into protein.
- Ribosome A-site:
the site within the ribosome where charged tRNAs bind and pair with the respective codons on the mRNA during translation.
- RNA helicase:
an enzyme that uses ATP to unwind or remodel RNA:RNA interactions or RNA:protein complexes; such an enzyme is therefore also an ATPase.
- Translational readthrough:
the event of a ribosome translating through and beyond a stop codon.
- VDJ recombination:
the process of genetic recombination in immune cells that randomly joins gene segments (dubbed variable, V; diversity, D; and junction, J) to create novel sequences of immunoglobulin genes in individual B or T cells to ultimately generate a diverse repertoire of antibodies and T cell receptors.
References
- 1.Mendell JT et al. (2004) Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat. Genet 36, 1073–1078 [DOI] [PubMed] [Google Scholar]
- 2.Wittmann J et al. (2006) hUPF2 silencing identifies physiologic substrates of mammalian nonsense-mediated mRNA decay. Mol. Cell. Biol 26, 1272–1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.He F et al. (1997) Upf1p, Nmd2p, and Upf3p are interacting components of the yeast nonsense-mediated mRNA decay pathway. Mol. Cell. Biol 17, 1580–1594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pulak R and Anderson P. (1993) mRNA surveillance by the Caenorhabditis elegans smg genes. Genes Dev. 7, 1885–1897 [DOI] [PubMed] [Google Scholar]
- 5.Medghalchi SM et al. (2001) Rent1, a trans-effector of nonsense-mediated mRNA decay, is essential for mammalian embryonic viability. Hum. Mol. Genet 10, 99–105 [DOI] [PubMed] [Google Scholar]
- 6.Weischenfeldt J et al. (2008) NMD is essential for hematopoietic stem and progenitor cells and for eliminating by-products of programmed DNA rearrangements. Genes Dev. 22, 1381–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wittkopp N et al. (2009) Nonsense-mediated mRNA decay effectors are essential for zebrafish embryonic development and survival. Mol. Cell. Biol 29, 3517–3528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conti E and Izaurralde E. (2005) Nonsense-mediated mRNA decay: molecular insights and mechanistic variations across species. Curr. Opin. Cell Biol. 17, 316–325 [DOI] [PubMed] [Google Scholar]
- 9.Mühlemann O. (2008) Recognition of nonsense mRNA: towards a unified model. Biochem. Soc. Trans 36, 497–501 [DOI] [PubMed] [Google Scholar]
- 10.He F and Jacobson A. (2015) Nonsense-mediated mRNA decay: degradation of defective transcripts is only part of the story. Annu. Rev. Genet 49, 339–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gehring NH et al. (2005) Exon-junction complex components specify distinct routes of nonsense-mediated mRNA decay with differential cofactor requirements. Mol. Cell 20, 65–75 [DOI] [PubMed] [Google Scholar]
- 12.Tarpey PS et al. (2007) Mutations in UPF3B, a member of the nonsense-mediated mRNA decay complex, cause syndromic and nonsyndromic mental retardation. Nat. Genet 39, 1127–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bao J et al. (2016) UPF2-dependent nonsense-mediated mRNA decay pathway is essential for spermatogenesis by selectively eliminating longer 3′UTR transcripts. PLoS Genet. 12, e1005863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ge Z et al. (2016) Polypyrimidine tract binding protein 1 protects mRNAs from recognition by the nonsense-mediated mRNA decay pathway. Elife 5, e11155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Z and Krainer AR (2004) Involvement of SR proteins in mRNA surveillance. Mol. Cell 16, 597–607 [DOI] [PubMed] [Google Scholar]
- 16.Karousis ED et al. (2016) Nonsense-mediated mRNA decay: novel mechanistic insights and biological impact. Wiley Interdiscip. Rev. RNA 7, 661–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lykke-Andersen S and Jensen TH (2015) Nonsensemediated mRNA decay: an intricate machinery that shapes transcriptomes. Nat. Rev. Mol. Cell Biol. 16, 665–677 [DOI] [PubMed] [Google Scholar]
- 18.Kishor A et al. (2019) Nonsense‐mediated mRNA decay: the challenge of telling right from wrong in a complex transcriptome. Wiley Interdiscip. Rev. RNA 10, 997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kurosaki T et al. (2019) Quality and quantity control of gene expression by nonsense-mediated mRNA decay. Nat. Rev. Mol. Cell Biol. 20, 406–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang L and Wilkinson MF (2012) Regulation of nonsensemediated mRNA decay. Wiley Interdiscip. Rev. RNA 3, 807–828 [DOI] [PubMed] [Google Scholar]
- 21.Lindeboom RGH et al. (2016) The rules and impact of nonsense-mediated mRNA decay in human cancers. Nat. Genet 48, 1112–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ishigaki Y et al. (2001) Evidence for a pioneer round of mRNA translation: mRNAs subject to nonsense-mediated decay in mammalian cells are bound by CBP80 and CBP20. Cell 106, 607–617 [DOI] [PubMed] [Google Scholar]
- 23.Durand S et al. (2016) Hyperphosphorylation amplifies UPF1 activity to resolve stalls in nonsense-mediated mRNA decay. Nat. Commun 7, 12434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rufener SC and Mühlemann O. (2013) eIF4E-bound mRNPs are substrates for nonsense-mediated mRNA decay in mammalian cells. Nat. Struct. Mol. Biol 20, 710–717 [DOI] [PubMed] [Google Scholar]
- 25.Hoek TA et al. (2019) Single-molecule imaging uncovers rules governing nonsense-mediated mRNA decay. Mol. Cell 75, 324–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Amrani N et al. (2004) A faux 3′-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature 432, 112–118 [DOI] [PubMed] [Google Scholar]
- 27.Peixeiro I et al. (2012) Interaction of PABPC1 with the translation initiation complex is critical to the NMD resistance of AUG-proximal nonsense mutations. Nucleic Acids Res. 40, 1160–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karousis ED et al. (2020) Human NMD ensues independently of stable ribosome stalling. Nat. Commun 11, 4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hogg JR and Goff SP et al. (2010) Upf1 senses 3′UTR length to potentiate mRNA decay. Cell 143, 379–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zünd D et al. (2013) Translation-dependent displacement of UPF1 from coding sequences causes its enrichment in 3′ UTRs. Nat. Struct. Mol. Biol 20, 936–943 [DOI] [PubMed] [Google Scholar]
- 31.Kurosaki T and Maquat LE (2013) Rules that govern UPF1 binding to mRNA 3′ UTRs. Proc. Natl. Acad. Sci. U. S. A 110, 3357–3362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ivanov A et al. (2016) PABP enhances release factor recruitment and stop codon recognition during translation termination. Nucleic Acids Res. 44, 7766–7776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Behm-Ansmant I et al. (2007) A conserved role for cytoplasmic poly(A)-binding protein 1 (PABPC1) in nonsense-mediated mRNA decay. EMBO J. 26, 1591–1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eberle AB et al. (2008) Posttranscriptional gene regulation by spatial rearrangement of the 3′ untranslated region. PLoS Biol. 6, e92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Singh G et al. (2008) A competition between stimulators and antagonists of Upf complex recruitment governs human nonsense-mediated mRNA decay. PLoS Biol. 6, e111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ivanov PV et al. (2008) Interactions between UPF1, eRFs, PABP and the exon junction complex suggest an integrated model for mammalian NMD pathways. EMBO J. 27, 736–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boehm V and Gehring NH (2016) Exon junction complexes: supervising the gene expression assembly line. Trends Genet. 32, 724–735 [DOI] [PubMed] [Google Scholar]
- 38.Woodward LA et al. (2017) The exon junction complex: a lifelong guardian of mRNA fate. Wiley Interdiscip. Rev. RNA 8, e1411. [DOI] [PubMed] [Google Scholar]
- 39.Le Hir H et al. (2016) The exon junction complex as a node of post-transcriptional networks. Nat. Rev. Mol. Cell Biol 17, 41–54 [DOI] [PubMed] [Google Scholar]
- 40.Causier B et al. (2017) Conservation of nonsense-mediated mRNA decay complex components throughout eukaryotic evolution. Sci. Rep 7, 16692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim YK and Maquat LE (2019) UPFront and center in RNA decay: UPF1 in nonsense-mediated mRNA decay and beyond. RNA 25, 407–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chakrabarti S et al. (2011) Molecular mechanisms for the RNA-dependent ATPase activity of Upf1 and its regulation by Upf2. Mol. Cell 41, 693–703 [DOI] [PubMed] [Google Scholar]
- 43.Chamieh H et al. (2008) NMD factors UPF2 and UPF3 bridge UPF1 to the exon junction complex and stimulate its RNA helicase activity. Nat. Struct. Mol. Biol 15, 85–93 [DOI] [PubMed] [Google Scholar]
- 44.Weng Y et al. (1996) Genetic and biochemical characterization of mutations in the ATPase and helicase regions of the Upf1 protein. Mol. Cell. Biol 16, 5477–5490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Franks TM et al. (2010) Upf1 ATPase-dependent mRNP disassembly is required for completion of nonsense-mediated mRNA decay. Cell 143, 938–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee SR et al. (2015) Target discrimination in nonsensemediated mRNA decay requires Upf1 ATPase activity. Mol. Cell 59, 413–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kadlec J et al. (2004) The structural basis for the interaction between nonsense-mediated mRNA decay factors UPF2 and UPF3. Nat. Struct. Mol. Biol 11, 330–337 [DOI] [PubMed] [Google Scholar]
- 48.Clerici M et al. (2014) Structural and functional analysis of the three MIF4G domains of nonsense-mediated decay factor UPF2. Nucleic Acids Res. 42, 2673–2686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Singh G et al. (2007) Communication with the exon-junction complex and activation of nonsense-mediated decay by human Upf proteins occur in the cytoplasm. Mol. Cell 27, 780–792 [DOI] [PubMed] [Google Scholar]
- 50.López-Perrote A et al. (2016) Human nonsensemediated mRNA decay factor UPF2 interacts directly with eRF3 and the SURF complex. Nucleic Acids Res. 44, 1909–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Serin G et al. (2001) Identification and characterization of human orthologues to Saccharomyces cerevisiae Upf2 protein and Upf3 protein (Caenorhabditis elegans SMG-4). Mol. Cell. Biol 21, 209–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gehring NH et al. (2003) Y14 and hUpf3b form an NMDactivating complex. Mol. Cell 11, 939–949 [DOI] [PubMed] [Google Scholar]
- 53.Buchwald G et al. (2010) Insights into the recruitment of the NMD machinery from the crystal structure of a core EJC–UPF3b complex. Proc. Natl. Acad. Sci. U. S. A 107, 10050–10055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kunz JB et al. (2006) Functions of hUpf3a and hUpf3b in nonsense-mediated mRNA decay and translation. RNA 12, 1015–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shum EY et al. (2016) The antagonistic gene paralogs Upf3a and Upf3b govern nonsense-mediated RNA decay. Cell 165, 382–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Neu-Yilik G et al. (2017) Dual function of UPF3B in early and late translation termination. EMBO J. 36, 2968–2986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yamashita A et al. (2001) Human SMG-1, a novel phosphatidylinositol 3-kinase-related protein kinase, associates with components of the mRNA surveillance complex and is involved in the regulation of nonsense-mediated mRNA decay. Genes Dev. 15, 2215–2228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Denning G et al. (2001) Cloning of a novel phosphatidylinositol kinase-related kinase: characterization of the human SMG-1 RNA surveillance protein. J. Biol. Chem 276, 22709–22714 [DOI] [PubMed] [Google Scholar]
- 59.Grimson A et al. (2004) SMG-1 is a phosphatidylinositol kinase-related protein kinase required for nonsense-mediated mRNA decay in Caenorhabditis elegans. Mol. Cell. Biol 24, 7483–7490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lloyd JPB (2018) The evolution and diversity of the nonsensemediated mRNA decay pathway. F1000Res. 7, 1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Isken O et al. (2008) Upf1 phosphorylation triggers translational repression during nonsense-mediated mRNA decay. Cell 133, 314–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ohnishi T et al. (2003) Phosphorylation of hUPF1 induces formation of mRNA surveillance complexes containing hSMG-5 and hSMG-7. Mol. Cell 12, 1187–1200 [DOI] [PubMed] [Google Scholar]
- 63.Okada-Katsuhata Y et al. (2012) N- and C-terminal Upf1 phosphorylations create binding platforms for SMG-6 and SMG-5:SMG-7 during NMD. Nucleic Acids Res. 40, 1251–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gangras P et al. (2020) Zebrafish rbm8a and magoh mutants reveal EJC developmental functions and new 3′UTR introncontaining NMD targets. PLoS Genet. 16, e1008830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Silver DL et al. (2010) The exon junction complex component Magoh controls brain size by regulating neural stem cell division. Nat. Neurosci 13, 551–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McMahon JJ et al. (2016) The exon junction complex in neural development and neurodevelopmental disease. Int. J. Dev. Neurosci 55, 117–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nott A et al. (2004) Splicing enhances translation in mammalian cells: an additional function of the exon junction complex. Genes Dev. 18, 210–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bicknell AA et al. (2012) Introns in UTRs: why we should stop ignoring them. Bioessays 34, 1025–1034 [DOI] [PubMed] [Google Scholar]
- 69.Baralle FE and Giudice J. (2017) Alternative splicing as a regulator of development and tissue identity. Nat. Rev. Mol. Cell Biol. 18, 437–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Celik A et al. (2017) High-resolution profiling of NMD targets in yeast reveals translational fidelity as a basis for substrate selection. RNA 23, 735–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chan W-K. et al. (2007) An alternative branch of the nonsense-mediated decay pathway. EMBO J. 26, 1820–1830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Laumonnier F et al. (2010) Mutations of the UPF3B gene, which encodes a protein widely expressed in neurons, are associated with nonspecific mental retardation with or without autism. Mol. Psychiatry 15, 767–776 [DOI] [PubMed] [Google Scholar]
- 73.Lynch SA et al. (2012) Broadening the phenotype associated with mutations in UPF3B: two further cases with renal dysplasia and variable developmental delay. Eur. J. Med. Genet 55, 476–479 [DOI] [PubMed] [Google Scholar]
- 74.Xu X et al. (2013) Exome sequencing identifies UPF3B as the causative gene for a Chinese non-syndrome mental retardation pedigree. Clin. Genet 83, 560–564 [DOI] [PubMed] [Google Scholar]
- 75.Jolly LA et al. (2013) The UPF3B gene, implicated in intellectual disability, autism, ADHD and childhood onset schizophrenia regulates neural progenitor cell behaviour and neuronal outgrowth. Hum. Mol. Genet 22, 4673–4687 [DOI] [PubMed] [Google Scholar]
- 76.Alrahbeni T et al. (2015) Full UPF3B function is critical for neuronal differentiation of neural stem cells. Mol. Brain 8, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Huang L et al. (2018) A Upf3b-mutant mouse model with behavioral and neurogenesis defects. Mol. Psychiatry 23, 1773–1786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Huang L et al. (2018) Antisense suppression of the nonsense mediated decay factor Upf3b as a potential treatment for diseases caused by nonsense mutations. Genome Biol. 19, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nguyen LS et al. (2012) Transcriptome profiling of UPF3B/ NMD-deficient lymphoblastoid cells from patients with various forms of intellectual disability. Mol. Psychiatry 17, 1103–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chan W-K. et al. (2009) A UPF3-mediated regulatory switch that maintains RNA surveillance. Nat. Struct. Mol. Biol 16, 747–753 [DOI] [PubMed] [Google Scholar]
- 81.Avery P et al. (2011) Drosophila Upf1 and Upf2 loss of function inhibits cell growth and causes animal death in a Upf3-independent manner. RNA 17, 624–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Thoren LA et al. (2010) UPF2 is a critical regulator of liver development, function and regeneration. PLoS One 5, e11650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Weischenfeldt J et al. (2012) Mammalian tissues defective in nonsense-mediated mRNA decay display highly aberrant splicing patterns. Genome Biol. 13, R35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nguyen LS et al. (2013) Contribution of copy number variants involving nonsense-mediated mRNA decay pathway genes to neuro-developmental disorders. Hum. Mol. Genet 22, 1816–1825 [DOI] [PubMed] [Google Scholar]
- 85.Johnson JL et al. (2019) Inhibition of Upf2-dependent nonsense-mediated decay leads to behavioral and neurophysiological abnormalities by activating the immune response. Neuron 104, 665–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gehring NH et al. (2009) The hierarchy of exon-junction complex assembly by the spliceosome explains key features of mammalian nonsense-mediated mRNA decay. PLoS Biol. 7, e1000120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mabin JW et al. (2018) The exon junction complex undergoes a compositional switch that alters mRNP structure and nonsense-mediated mRNA decay activity. Cell Rep. 25, 2431–2446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gerbracht JV et al. (2020) CASC3 promotes transcriptomewide activation of nonsense-mediated decay by the exon junction complex. Nucleic Acids Res. 48, 8626–8644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Boehm V et al. (2018) Exon junction complexes suppress spurious splice sites to safeguard transcriptome integrity. Mol. Cell 72, 482–495 [DOI] [PubMed] [Google Scholar]
- 90.Wang Z et al. (2018) Exon junction complexes can have distinct functional flavours to regulate specific splicing events. Sci. Rep 8, 9509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bono F et al. (2006) The crystal structure of the exon junction complex reveals how it maintains a stable grip on mRNA. Cell 126, 713–725 [DOI] [PubMed] [Google Scholar]
- 92.Andersen CBF et al. (2006) Structure of the exon junction core complex with a trapped DEAD-box ATPase bound to RNA. Science 313, 1968–1972 [DOI] [PubMed] [Google Scholar]
- 93.Aznarez I et al. (2018) Mechanism of nonsense-mediated mRNA decay stimulation by splicing factor SRSF1. Cell Rep. 23, 2186–2198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rahman MA et al. (2020) Recurrent SRSF2 mutations in MDS affect both splicing and NMD. Genes Dev. 34, 413–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Singh G et al. (2012) The cellular EJC interactome reveals higher-order mRNP structure and an EJC-SR protein nexus. Cell 151, 750–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Saulière J et al. (2012) CLIP-seq of eIF4AIII reveals transcriptome-wide mapping of the human exon junction complex. Nat. Struct. Mol. Biol 19, 1124–1131 [DOI] [PubMed] [Google Scholar]
- 97.Sato H et al. (2008) Efficiency of the pioneer round of translation affects the cellular site of nonsense-mediated mRNA decay. Mol. Cell 29, 255–262 [DOI] [PubMed] [Google Scholar]
- 98.Chakrabarti S et al. (2014) Phospho-dependent and phospho-independent interactions of the helicase UPF1 with the NMD factors SMG5-SMG7 and SMG6. Nucleic Acids Res. 42, 9447–9460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Nicholson P et al. (2014) A novel phosphorylation-independent interaction between SMG6 and UPF1 is essential for human NMD. Nucleic Acids Res. 42, 9217–9235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Eberle AB et al. (2009) SMG6 promotes endonucleolytic cleavage of nonsense mRNA in human cells. Nat. Struct. Mol. Biol 16, 49–55 [DOI] [PubMed] [Google Scholar]
- 101.Boehm V et al. (2014) 3′ UTR length and messenger ribonucleoprotein composition determine endocleavage efficiencies at termination codons. Cell Rep. 9, 555–568 [DOI] [PubMed] [Google Scholar]
- 102.Lykke-Andersen S et al. (2014) Human nonsense-mediated RNA decay initiates widely by endonucleolysis and targets snoRNA host genes. Genes Dev. 28, 2498–2517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Loh B et al. (2013) The SMG5–SMG7 heterodimer directly recruits the CCR4–NOT deadenylase complex to mRNAs containing nonsense codons via interaction with POP2. Genes Dev. 27, 2125–2138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Nicholson P et al. (2018) Dissecting the functions of SMG5, SMG7, and PNRC2 in nonsense-mediated mRNA decay of human cells. RNA 24, 557–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Colombo M et al. (2016) Transcriptome-wide identification of NMD-targeted human mRNAs reveals extensive redundancy between SMG6- and SMG7-mediated degradation pathways. RNA 23, 189–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Schmidt SA et al. (2015) Identification of SMG6 cleavage sites and a preferred RNA cleavage motif by global analysis of endogenous NMD targets in human cells. Nucleic Acids Res. 43, 309–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Li T et al. (2015) Smg6/Est1 licenses embryonic stem cell differentiation via nonsense-mediated mRNA decay. EMBO J. 34, 1630–1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ottens F et al. (2017) Transcript-specific characteristics determine the contribution of endo- and exonucleolytic decay pathways during the degradation of nonsense-mediated decay substrates. RNA 23, 1224–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Boehm V et al. (2020) Nonsense-mediated mRNA decay relies on ‘two-factor authentication’ by SMG5–SMG7. bioRxiv Published online July 7, 2020. 10.1101/2020.07.07.191437 [DOI] [Google Scholar]
- 110.Karam R et al. (2015) The unfolded protein response is shaped by the NMD pathway. EMBO Rep. 16, 599–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lou C-H. et al. (2016) Nonsense-mediated RNA decay influences human embryonic stem cell fate. Stem Cell Rep. 6, 844–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Gong C et al. (2009) SMD and NMD are competitive pathways that contribute to myogenesis: effects on PAX3 and myogenin mRNAs. Genes Dev. 23, 54–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Gowravaram M et al. (2019) Insights into the assembly and architecture of a Staufen-mediated mRNA decay (SMD)-competent mRNP. Nat. Commun 10, 5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Baird TD et al. (2018) ICE1 promotes the link between splicing and nonsense-mediated mRNA decay. Elife 7, e33178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ryu I et al. (2019) eIF4A3 phosphorylation by CDKs affects NMD during the cell cycle. Cell Rep. 26, 2126–2139 [DOI] [PubMed] [Google Scholar]
- 116.Hsu I-W. et al. (2005) Phosphorylation of Y14 modulates its interaction with proteins involved in mRNA metabolism and influences its methylation. J. Biol. Chem 280, 34507–34512 [DOI] [PubMed] [Google Scholar]
- 117.Tatsuno T and Ishigaki Y. (2018) C-terminal short arginine/serine repeat sequence-dependent regulation of Y14 (RBM8A) localization. Sci. Rep 8, 612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Trembley JH et al. (2005) Activation of pre-mRNA splicing by human RNPS1 is regulated by CK2 phosphorylation. Mol. Cell. Biol 25, 1446–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Viegas MH et al. (2007) The abundance of RNPS1, a protein component of the exon junction complex, can determine the variability in efficiency of the nonsense mediated decay pathway. Nucleic Acids Res. 35, 4542–4551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zetoune AB et al. (2008) Comparison of nonsense-mediated mRNA decay efficiency in various murine tissues. BMC Genet. 9, 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bruno IG et al. (2011) Identification of a microRNA that activates gene expression by repressing nonsense-mediated RNA decay. Mol. Cell 42, 500–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Baguet A et al. (2007) The exon-junction-complex-component metastatic lymph node 51 functions in stress-granule assembly. J. Cell Sci. 120, 2774–2784 [DOI] [PubMed] [Google Scholar]
- 123.Cougot N et al. (2014) Overexpression of MLN51 triggers P-body disassembly and formation of a new type of RNA granules. J. Cell Sci. 127, 4692–4701 [DOI] [PubMed] [Google Scholar]
- 124.Zhou Z and Fu X-D. (2013) Regulation of splicing by SR proteins and SR protein-specific kinases. Chromosoma 122, 191–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Toma KG et al. (2015) Identification of elements in human long 3′ UTRs that inhibit nonsense-mediated decay. RNA 21, 887–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kishor A et al. (2019) hnRNP L-dependent protection of normal mRNAs from NMD subverts quality control in B cell lymphoma. EMBO J. 38, e99128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Fritz SE et al. (2020) The RNA-binding protein PTBP1 promotes ATPase-dependent dissociation of the RNA helicase UPF1 to protect transcripts from nonsense-mediated mRNA decay. J. Biol. Chem Published online June 22, 2020. 10.1074/jbc.RA120.013824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Geuens T et al. (2016) The hnRNP family: insights into their role in health and disease. Hum. Genet 135, 851–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zhang J and Maquat LE (1997) Evidence that translation reinitiation abrogates nonsense-mediated mRNA decay in mammalian cells. EMBO J. 16, 826–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Inácio A et al. (2004) Nonsense mutations in close proximity to the initiation codon fail to trigger full nonsense-mediated mRNA decay. J. Biol. Chem 279, 32170–32180 [DOI] [PubMed] [Google Scholar]
- 131.Neu-Yilik G et al. (2011) Mechanism of escape from nonsensemediated mRNA decay of human beta-globin transcripts with nonsense mutations in the first exon. RNA 17, 843–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Jagannathan S and Bradley RK (2016) Translational plasticity facilitates the accumulation of nonsense genetic variants in the human population. Genome Res. 26, 1639–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Dyle MC et al. (2020) How to get away with nonsense: mechanisms and consequences of escape from nonsense‐mediated RNA decay. Wiley Interdiscip. Rev. RNA 11, 5884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Sakaki K et al. (2012) RNA surveillance is required for endoplasmic reticulum homeostasis. Proc. Natl. Acad. Sci. U. S. A 109, 8079–8084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Sieber J et al. (2016) Proteomic analysis reveals branchspecific regulation of the unfolded protein response by nonsense-mediated mRNA decay. Mol. Cell. Proteomics 15, 1584–1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Longman D et al. (2020) Identification of a nonsense-mediated decay pathway at the endoplasmic reticulum. Genes Dev. 34, 1075–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Huang L et al. (2011) RNA homeostasis governed by cell type-specific and branched feedback loops acting on NMD. Mol. Cell 43, 950–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Yepiskoposyan H et al. (2011) Autoregulation of the nonsense-mediated mRNA decay pathway in human cells. RNA 17, 2108–2118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Longman D et al. (2013) DHX34 and NBAS form part of an autoregulatory NMD circuit that regulates endogenous RNA targets in human cells, zebrafish and Caenorhabditis elegans. Nucleic Acids Res. 41, 8319–8331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Zhao B and Pritchard JR (2019) Evolution of the nonsensemediated decay pathway is associated with decreased cytolytic immune infiltration. PLoS Comput. Biol. 15, e1007467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kafri R et al. (2009) Genetic redundancy: new tricks for old genes. Cell 136, 389–392 [DOI] [PubMed] [Google Scholar]
- 142.Albergante L et al. (2014) Buffered qualitative stability explains the robustness and evolvability of transcriptional networks. Elife 3, e02863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Kebaara BW and Atkin AL (2009) Long 3′-UTRs target wild-type mRNAs for nonsense-mediated mRNA decay in Saccharomyces cerevisiae. Nucleic Acids Res. 37, 2771–2778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.McGlincy NJ and Smith CWJ (2008) Alternative splicing resulting in nonsense-mediated mRNA decay: what is the meaning of nonsense? Trends Biochem. Sci. 33, 385–393 [DOI] [PubMed] [Google Scholar]
- 145.Saltzman AL et al. (2008) Regulation of multiple core spliceosomal proteins by alternative splicing-coupled nonsensemediated mRNA decay. Mol. Cell. Biol 28, 4320–4330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Kashima I et al. (2006) Binding of a novel SMG-1–Upf1– eRF1–eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes Dev. 20, 355–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Melero R et al. (2016) The RNA helicase DHX34 functions as a scaffold for SMG1-mediated UPF1 phosphorylation. Nat. Commun 7, 10585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Hug N and Cáceres JF (2014) The RNA helicase DHX34 activates NMD by promoting a transition from the surveillance to the decay-inducing complex. Cell Rep. 8, 1845–1856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Feng Q et al. (2017) The RNA surveillance factor UPF1 represses myogenesis via its E3 ubiquitin ligase activity. Mol. Cell 67, 239–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Mort M et al. (2008) A meta-analysis of nonsense mutations causing human genetic disease. Hum. Mutat 29, 1037–1047 [DOI] [PubMed] [Google Scholar]
- 151.Martin L et al. (2014) Identification and characterization of small molecules that inhibit nonsense-mediated RNA decay and suppress nonsense p53 mutations. Cancer Res. 74, 3104–3113 [DOI] [PMC free article] [PubMed] [Google Scholar]