Abstract
BACKGROUND:
Exposure to diabetes in utero influences future metabolic health of the offspring. MicroRNAs (miRNA) are small non-coding RNAs that may contribute mechanistically to the effects on offspring imparted by diabetes mellitus (DM) during pregnancy. We hypothesized that exposure to DM during pregnancy influences select miRNAs in fetal circulation, in human umbilical vein endothelial cells (HUVEC), and placenta.
METHODS:
miRNA abundance was quantified using real-time PCR from RNA isolated from umbilical cord serum exosomes, HUVEC and placenta exposed to diabetes or normoglycemia during pregnancy. The abundance of each of these miRNAs was determined by comparison to a known standard and the relative expression assessed using the 2−ΔΔCt method. Multivariable regression models examined the associations between exposure to diabetes during pregnancy and miRNA expression.
RESULTS:
miR-126–3p was highly abundant in fetal circulation, HUVEC and placenta. Diabetes exposure during pregnancy resulted in lower expression of miR-148a-3p and miR-29a-3p in the HUVEC. In the placenta, for miR-126–3p, there was a differential effect of DM by birthweight between DM versus control group, expression being lower at the lower birth weight, however not different at the higher birthweight.
CONCLUSION:
Exposure to DM during pregnancy alters miRNA expression in the offspring in a tissue specific manner.
INTRODUCTION
Exposure to the diabetic milieu in utero has long-term effects on the offspring, predisposing them to develop type 2 diabetes mellitus (T2DM) and obesity. Exposure to gestational diabetes mellitus (GDM) is associated with increased adiposity in the offspring during childhood, along with increased risk of metabolic syndrome by age 11 (1). Infants born to women with either GDM or T2DM have altered metabolism demonstrated by reduced resting energy expenditure and fat oxidation at 1 month of age, even though body mass and composition are unaffected (2). While the influence of the prenatal environment on postnatal growth and metabolism is well established, the mechanisms involved remain poorly understood.
Increasing evidence suggests that in utero exposures, such as maternal diabetes, influence offspring outcomes via epigenetic mechanisms, with alterations in microRNA (miRNA) expression one potential means of these epigenetic changes (3). miRNAs are small, non-coding RNAs that modify post-transcriptional target gene expression. Secreted miRNAs are packaged into microvesicles termed exosomes, protecting them from degradation by RNAases (4). Exosomes are 60–150 nm particles secreted by most cell types and are found in multiple biologic fluids including serum. Thus, exosomal miRNAs provide a means of intercellular communication both locally (i.e. within their tissue of origin) as well as systemically, similar to that of hormones (5).
Previous reports have shown the abundance of miRNAs within the circulation in adults is affected by T2DM (6) but studies during the perinatal period are limited. Therefore, the aim of the present study was to examine the effect of diabetes during pregnancy on the expression of certain miRNAs in fetal tissues and in the circulating exosomes of offspring. Since fetal endothelial cells and the trophoblast are potential sources of circulating exosomal miRNAs, HUVEC and placental trophopblast cells exposed to the diabetic milieu of pregnancy were examined. miRNA abundance was assessed simultaneously in cord blood exosomes, HUVEC, and placental tissue. We selected five miRNAs: miR-126–3p, miR-130b-3p, miR-148a-3p, miR-let-7a-5p and miR-29a-3p previously shown to be related to insulin signaling, glucose and lipid metabolism (7–10).
We hypothesized that in utero exposure to the diabetic milieu influences the miRNA expression in HUVEC and placenta as well as in exosomes isolated from fetal cord serum.
METHODS
Subjects
Pregnant women presenting for elective caesarian section at term were recruited from the OU Medical Center, Oklahoma City, OK. The diagnosis of GDM or T2DM was based on the criteria by the American Diabetes Association (11). Forty-seven women were recruited for the control group and 35 were included in the DM group. Within the DM group, 23 women had GDM and 12 had T2DM. The participants in the DM group (GDM and T2DM) did not differ with respect to their baseline characteristics such as maternal age, pre-pregnancy body mass index (BMI), gestational age and infant birth weight. Women were excluded if the infants were small for gestational age, or had a major malformation, or chromosome abnormality. They were also excluded if they had type 1 diabetes, pre-eclampsia, chronic hypertension, renal disorders or a smoking history of more than five cigarettes per day during pregnancy. This study was approved by the Institutional Review Board at OUHSC. All participants provided informed consent before participation.
Sample Collection
Cord blood, umbilical cord and placenta specimens were collected in the surgical suite within 15 minutes of delivery of the placenta. Cord blood was drained directly into collection tubes and allowed to clot at room temperature, after which the serum was separated by centrifugation and stored at −80°C until further analysis. Immediately after blood collection, the cord was separated from the placenta at the insertion site, placed into a sterile container, and stored at 4°C until processing.
For the isolation of HUVEC, umbilical cords were processed within 24h of delivery. HUVEC were isolated as previously described by Jaffe et al (12). The umbilical vein was cannulated at both ends of the cord specimen, flushed with DPBS without Ca/Mg (Gibco) and incubated with 0.1% collagenase A solution (Worthington Biochemical Corp, Lakewood, NJ) in DPBS with Ca/Mg (Gibco) at 37°C for 15 min. Isolated HUVEC were washed and immediately disrupted with Qiazol Lysis Reagent (Qiagen) according to the manufacturer’s protocol and stored at −80 °C until RNA extraction. HUVEC were identified morphologically by their cobble-stone appearance by microscopy and their vascular endothelial nature confirmed by immune-staining for von Willebrand factor (vWF; Anti-Human von Willebrand factor; Sigma-Aldrich) (13). More than 90% of the cells were vWF-positive.
Five placental samples were collected from areas evenly distributed around the circumference of the fetal side of the placenta, roughly halfway between the cord insertion site and the placental margin. Each sample was approximately 2 cm in diameter and extended approximately 1 cm below the fetal membrane. The fetal membrane was trimmed away and the remaining tissue was rinsed in cold DPBS, blotted dry, wrapped in aluminum foil and submerged in liquid nitrogen. Each of the five samples was ground to a fine powder in liquid nitrogen and then equal amounts of powder from the five samples were combined to make a representative specimen for each placenta. The representative specimen was homogenized in Qiazol Lysis Reagent (Qiagen) using a Polytron homogenizer (Kinematica, Luzern, Switzerland) then stored at −80°C until analysis.
Exosome Isolation and RNA Extraction
Exosomes were extracted from 250μl of the cord serum using Exoquick ™ Exosome Precipitation Solution (System Biosciences, Mountain View, CA) according to the manufacturer’s protocol. Total RNA was extracted from the exosomes using the SeraMir Exosome RNA Isolation kit (System Biosciences) according to the manufacturer’s instructions. Given that exosomal miRNA are hormone-like bioactive factors in circulation, we used a fixed volume of serum for RNA extraction. Our method is consistent with other studies which have also used fixed volume of serum for the RNA extraction (6). Synthetic C.elegans microRNA (cel-miR-39–3p) at a final concentration of 25 fmol was added during RNA extraction as an exogenous spike in control to account for losses during RNA extraction. Sera from total of 47 control and 35 DM group were used for exosome and RNA extraction.
Total RNA was extracted from 44 control and 27 DM HUVEC and 46 control and 33 DM placenta samples using the miRNeasy kit (Qiagen) according to the manufacturer’s instructions. Isolated total RNA was quantified by a NanoDrop ND-100 spectrophotometer (Thermo Scientific) and presence of miRNA was verified using the small RNA chip on an Agilent 2100 Bioanalyzer (Agilent Technologies). For all sample types, a pool of miRNA-specific RT primers was created according to the Applied Biosystems protocol (publication part number 4465407). RT was performed on 20μl of total RNA from the serum (14) using the TaqMan MicroRNA Reverse Transcription Kit (Life Technologies, Grand Island, NY). For HUVEC and placenta samples, 50 and 200 ng of RNA, respectively, was used going into the RT reaction.
qPCR Analysis
The miRNA sequences, Taqman assay ID along with the sequence of each of the miRNA are available in Supplemental Table S1. All quantitative PCR (qPCR) reactions were performed in triplicate using TaqMan MicroRNA Assays and TaqMan Universal Master Mix II, no UNG using the Bio-Rad (Hercules, CA) CFX96 Touch Real-Time PCR Detection System. For qPCR, 1.5 ng and 6 ng of RNA was used per reaction per well for HUVEC and placenta samples respectively. RNU48 (13) and U6 (15) were used as endogenous controls for HUVEC and placenta samples respectively. No significant difference in the expression of RNU48 and U6 between the control and DM groups was demonstrated (data not shown).
miRNA Standard Curves
Five-point standard curves were prepared from miRNA mimics (Dharmacon,Inc.,Chicago, IL) (16) in the range of 10 pM to 1 fM. Separate reverse transcription reactions were performed for each miRNA mimic and standard curve point. The curves were then used to estimate the abundance of miRNAs within the samples.
Statistical methods
Participant’s demographic and anthropometric characteristics were compared by Student’s t-test for continuous variables or Fisher’s exact test for categorical variables using IBM SPSS Statistics (SPSS Released 2009. PASW Statistics for Windows, Version 18.0. SPSS). Normality was determined using Kolmogorov-Smirnov test. Kruskal-Wallis test was conducted to examine differences in the expression of miRNAs in each tissue type given the data was not normally distributed. Relative expression by qPCR of individual miRNA species was calculated using the 2−ΔΔCt method (17), where cel-miR-39–3p was used as the exogenous control for cord sera samples and RNU48 and U6 were used as endogenous controls for HUVEC and placenta samples, respectively. 2−ΔΔCt or the fold change was log-transformed to log-base two for analysis. For interpretative clarity, results were transformed back (applying the logarithm inverse) to fold change units (presented in tables 2–4). The associations between relative expression of miRNA and exposure to maternal diabetes were assessed using the glm package of R (R Core Team (2018). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org) to estimate multiple variable regression models adjusted for the following a priori chosen covariates: maternal age, maternal pre-pregnancy BMI, gestational age, infant sex, birth weight, and ethnicity with miRNA expression as the dependent variable. Missing covariate data (ranging from 0% to 7.3%) were handled with multiple imputation using sequential predictive mean matching models of the MICE package (18) of R. Imputation models were run separately by tissue type and DM status to preserve possible interactions, and 40 imputed data sets were saved for use in later analyses. Since RNA extraction was performed in separate batches within each tissue type, all models were controlled for the possibility of batch effects. Given that this is a first study reporting miRNA abundance in fetal exosomes and currently no normative data are available for miRNA abundance in control and DM exposed groups, no outliers were excluded for interpretation of the results. Data were excluded from small batches with disproportionate DM-to-control counts to enable batch-by-group interaction tests. Mann Whitney U test was used to determine the impact of type of diabetes (GDM versus T2DM) on miRNA abundance. No effect of type of DM on the miRNA abundance was noted; therefore, they were considered as one group in all further analysis. For all tests, Rubin’s rules (19) were used to combine multiple imputation estimates and standard errors, and statistical significance was considered at p<0.05.
Table 2.
Multiple variable regression model using fold change for miRNAs in HUVEC
| miR-126-3p | miR-130b-3p | miR-148a-3p | miR-29a-3p | miR-let-7a-5p | |
|---|---|---|---|---|---|
| DM | 0.778 [0.601, 1.007] | 0.864 [0.658, 1.135] | 0.602* [0.443, 0.818] | 0.742* [0.566, 0.971] | 0.820 [0.664, 1.012] |
| Maternal age (Years) | 1.029* [1.006, 1.052] | 1.013 [0.989, 1.038] | 1.030* [1.003, 1.058] | 1.018 [0.995, 1.043] | 1.024* [1.006, 1.043] |
| Maternal BMI (kg/m2) | 1.025* [1.007, 1.044] | 1.013 [0.994, 1.033] | 1.023* [1.001, 1.045] | 1.009 [0.991, 1.029] | 1.007 [0.992, 1.022] |
| Gestational age (Weeks) | 0.997 [0.838, 1.186] | 1.037 [0.863, 1.246] | 0.979 [0.796, 1.204] | 1.110 [0.920, 1.339] | 0.997 [0.864, 1.149] |
| Infant sex (M) | 1.285 [0.978, 1.687] | 1.177 [0.883, 1.570] | 1.496* [1.082, 2.068] | 1.397* [1.051, 1.856] | 1.163 [0.931, 1.453] |
| Birth weight(kg) | 0.765* [0.606, 0.966] | 0.799 [0.625, 1.021] | 0.742* [0.562, 0.979] | 0.835 [0.653, 1.068] | 0.807* [0.667, 0.975] |
| Race/Ethnicity (Caucasian) | 0.942 [0.732, 1.213] | 1.177 [0.901, 1.538] | 1.333 [0.987, 1.800] | 1.053 [0.808, 1.371] | 1.043 [0.849, 1.283] |
Adjusted fold change [95%CI], each fold change adjusted for the above covariates
p value-<0.05, significant values are shown in bold
DM-n=27, Control-n=44
Table 4.
Multivariable regression model using fold change for individual miRNAs in exosomes
| miR-126-3p | miR-130b-3p | miR-148a-3p | miR-29a-3p | miR-let-7a-5p | |
|---|---|---|---|---|---|
| DM | 1.058 [0.787, 1.421] | 1.096 [0.693, 1.735] | 0.995 [0.689, 1.436] | 1.270 [0.077, 21.013] | 1.136 [0.794, 1.626] |
| Maternal age (years) | 1.028* [1.005, 1.051] | 1.060* [1.024, 1.098] | 1.026 [0.999, 1.053] | 1.034 [0.845, 1.266] | 1.044* [1.017, 1.073] |
| Maternal pre-pregnancy BMI (kg/m2) | 1.014 [0.993, 1.036] | 1.029 [0.997, 1.063] | 1.027* [1.002, 1.053] | 0.968 [0.802, 1.170] | 0.993 [0.969, 1.018] |
| Gestational age (weeks) | 1.001 [0.838, 1.194] | 1.010 [0.767, 1.330] | 0.872 [0.709, 1.072] | 0.913 [0.199, 4.190] | 1.145 [0.929, 1.410] |
| Infant Sex M | 1.349* [1.013, 1.796] | 1.269 [0.814, 1.980] | 1.383 [0.991, 1.930] | 1.499 [0.125, 17.996] | 1.116 [0.799, 1.558] |
| Birth weight (kg) | 0.864 [0.665, 1.122] | 0.662* [0.441, 0.994] | 0.731 [0.522, 1.024] | 0.506 [0.041, 6.187] | 0.978 [0.719, 1.330] |
| Race/Ethnicity (Caucasian) | 1.399* [1.047, 1.868] | 1.356 [0.865, 2.127] | 1.315 [0.928, 1.863] | 6.158 [0.471, 80.421] | 1.337 [0.950, 1.881] |
Adjusted fold change [95%CI], each fold change adjusted for the above covariates
p value-<0.05, significant values are shown in bold
DM-n=35, Control-n=47
RESULTS
Characteristics of Mothers and Offspring
Mothers with DM during pregnancy had an increased pre-pregnancy BMI and were older by 3 years compared to controls (Table 1). Offspring in the DM group had higher birth weights than the control group (Table 1). In addition, the proportion of female offspring was higher in both groups, but not different between groups. In general, maternal diabetes was well controlled during the pregnancy as evidenced by an average hemoglobin A1C (HbA1c) of 6.0 % in the DM group.
Table 1.
Maternal and Infant demographics and clinical variables
| DM (n=35)a | Control (n=47) | ||
|---|---|---|---|
| Maternal age, years, Mean (SD) | 32.1 (± 5.5) * | 29.0 ±5.8 | |
| Maternal Race/Ethnicity | Caucasian, n (%) | 11 (31%) | 20 (42%) |
| Hispanic, n (%) | 14 (40%) | 15 (32%) | |
| African American, n (%) | 6 (17%) | 7 (15%) | |
| Native American, n (%) | 4 (11%) | 2 (4.3%) | |
| Asian, n (%) | 0 | 3 (6.4%) | |
| Maternal pre-pregnancy weightb, kg, Mean (SD) | 93.9 ± 23.5* | 81.5 ± 23.8 | |
| Maternal pre-pregnancy BMI, kg/m2, Mean (SD) | 35.7 ± 7.8 * | 30.9 ± 6.0 | |
| Sex of infant F/M (%F), Mean (SD) | 20/15 (57%) | 32/15 (68%) | |
| Birth weight, kg, Mean (SD) | 3.70 ± 0.55 * | 3.47 ± 0.48 | |
| Gestational age, weeks, Mean (SD) | 38.9 ± 0.9 | 39.0 ± 0.7 | |
| Maternal HbAlC, %c, Mean (SD) | 6.0 ± 1.4 * | 5.1 ± 0.3 | |
p value < 0.05, significant values are shown in bold
GDM n=23, T2DM n=12
By maternal report
DM n=29, Control n=23
Relative abundance of miRNA species differs among the tissue sources and circulation
All five miRNAs (miR-126–3p, miR-130b-3p, miR-148a-3p, miR-let-7a-5p, and miR-29a-3p) were detectable in HUVEC, placenta, and exosomes isolated from cord sera (average PCR cycle threshold below 32 for all; Figure 1). miR-126–3p was the most abundant miRNA across all tissue types and in cord serum exosomes. Though detectable, miR-130b-3p had the lowest measured abundance compared to the other miRNAs measured across all three tissue types.
Figure 1.
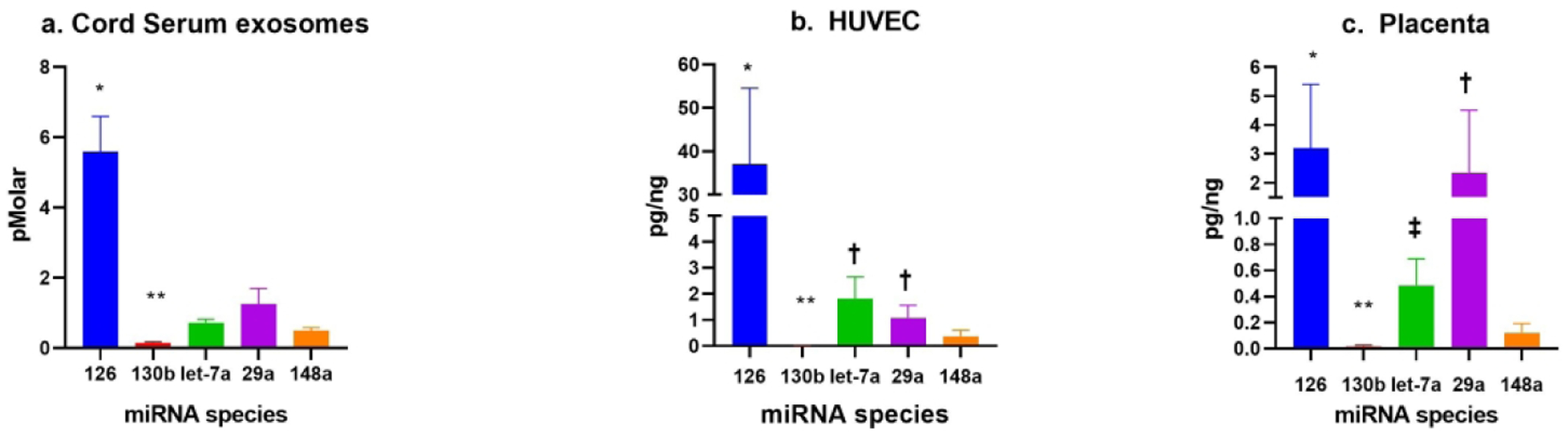
Abundance of miRNA within fetal serum exosomes, HUVEC and Placenta respectively (a-c).
a. Mean ± SEM. * = p<0.05: Abundance of miR-126 compared to all other miRNAs. ** =p<0.05: Abundance of miR-130b compared to all other miRNAs. Actual concentration of miR-130b in fetal exosomes was 0.15 pMolar.
b. Mean ± SEM. * = p<0.05: Abundance of miR-126 compared to all other miRNAs. † = p<0.05: Abundance of miR-let-7a-5p and miR-29a-3p compared to miR-148a-3p. ** = p<0.05: Abundance of miR-130b compared to all other miRNAs. Actual concentration of miR-130b in HUVEC was 0.014 pg/ng.
c. Mean ± SEM. * = p<0.05: Abundance of miR-126–3p as compared to miR-let-7a-5p, miR-148a-3p and miR-130b-3p. † = p<0.05: Abundance of miR-29a-3p as compared to miR-let-7a-5p and miR-148a-3p. ‡ = p<0.05: Abundance of miR-let-7a 5p as compared to miR-148a-3p. **= p<0.05: Abundance of miR-130b compared to all other miRNAs. Actual concentration of miR-130b in placenta was 0.015 pg/ng.
Exposure to diabetes alters miRNA in HUVEC and placenta
The expression of miR-148a-3p and miR-29a-3p were lower by 40% and 26% respectively in HUVEC from infants exposed to diabetes compared to the control group (Table 2) after adjusting for covariates. In the placenta, for miR-126–3p, there was a moderated effect of DM by birth weight, with larger expression differences appearing among smaller infants. For instance, at one standard deviation below the birth weight mean, a simple effect estimate indicated a 62% decline in expression among DM infants relative to controls (Table 3).
Table 3.
Multiple variable regression model using fold change for individual miRNAs in Placenta
| miR-126-3p | miR-130b-3p | miR-148a-3p | miR-29a-3p | miR-let-7a-5p | |
|---|---|---|---|---|---|
| DM | a | 1.156 [0.936, 1.429] | 1.266 [0.961, 1.667] | 1.188 [0.954, 1.479] | 1.066 [0.815, 1.394] |
| Maternal age (Years) | 0.998 [0.964, 1.033] | 0.991 [0.974, 1.008] | 0.985 [0.964, 1.007] | 1.001 [0.983, 1.018] | 1.001 [0.980, 1.022] |
| Maternal BMI (kg/m2) | 1.008 [0.979, 1.038] | 1.010 [0.995, 1.024] | 1.007 [0.989, 1.026] | 0.997 [0.982, 1.012] | 1.007 [0.988, 1.026] |
| Gestational age (Weeks) | 0.897 [0.687, 1.171] | 1.015 [0.892, 1.156] | 0.959 [0.813, 1.132] | 1.010 [0.879, 1.162] | 0.943 [0.800, 1.110] |
| Infant sex (M) | 0.929 [0.616, 1.402] | 1.160 [0.946, 1.422] | 1.170 [0.897, 1.527] | 0.824 [0.666, 1.018] | 0.935 [0.725, 1.206] |
| Birth Weight (kg) | 0.648 [0.377, 1.112] | 0.807* [0.671, 0.971] | 0.824 [0.652, 1.042] | 0.789* [0.651, 0.958] | 0.663* [0.526, 0.835] |
| Race/Ethnicity (Caucasian) | 0.921 [0.615, 1.380] | 0.951 [0.778, 1.164] | 0.971 [0.747, 1.262] | 0.962 [0.776, 1.193] | 1.155 [0.894, 1.492] |
| Interactions: | |||||
| DM: birth weight (kg) | 3.388*a [1.571, 7.305] | NA | NA | NA | NA |
Adjusted fold change [95%CI], each fold change adjusted for the above covariates
p value-<0.05, significant values are shown in bold
Simple effect of birth weight on miR-126 in the DM group
DM-n=33, Control-n=46
miRNA expression in fetal exosomes, HUVEC and placenta is affected by other maternal and infant factors independent of DM exposure
In the multivariable regression models maternal age, maternal BMI, offspring gender, birth weight and ethnicity were significant predictors of miRNA abundance (Table 2, 3 and 4).
Maternal Age:
With every one-year increase in maternal age, the relative abundance of miR-126–3p, miR-130b-3p and let-7a-5p increased by 2.8%, 6% and 4.4% respectively in circulating exosomes. Similarly, in the HUVEC, with increase in maternal age, there was an increase in abundance of miR-126–3p, 148a-3p and let-7a-5p by 2.9%, 3% and 2.4% respectively. There was no effect of maternal age on expression of miRNAs in placenta.
Maternal BMI:
With every 1 kg/m2 increase in maternal pre-pregnancy BMI, expression of miR-148a-3p increased by 2.7% in cord serum exosomes. In the HUVEC, with every 1 kg/m2 increase in maternal pre-pregnancy BMI, expression of miR-126–3p and miR-148a-3p increased by 2.5% and 2.3% respectively. There was no effect of maternal pre-pregnancy BMI on miRNA expression in placenta.
Infant Birth weight:
Regarding offspring birth weight, with every 1 kg increase in infant weight, expression of miR-130b-3p decreased by 34% in circulating exosomes. miR-126–3p, miR-148a-3p and miR-let-7a-5p decreased by 24%, 26% and 20% in HUVEC. miR-130b-3p, miR-29a-3p and miR-let-7a-5p decreased by 20%, 21% and 34% in placenta respectively.
Infant Sex:
Males had higher expression of miR-126–3p by 35% in the exosomes. In the HUVEC, males had higher expression of miR-148a-3p (50%) and miR-29a-3p (40%) as compared to females when controlled for other covariates. There was no effect of infant sex on expression of the miRNAs in placenta.
Race/Ethnicity:
In the cord serum exosomes, race/ethnicity was also a significant predictor of miR-126–3p as it was higher by 40% respectively in Caucasians than other ethnic and racial groups. There was no effect of race/ethnicity on miRNA expression in the HUVEC or placenta.
DISCUSSION
For patients with diabetes, mounting evidence indicates that miRNAs respond to metabolic status and are involved in the pathogenesis of diabetic complications. However, the vast majority of such studies are conducted in adults, with only limited investigation during the prenatal and immediate postnatal period. Our principal findings are that the relative abundance of the miRNAs is dependent on their source and the abundance of certain metabolically relevant miRNAs is altered by in utero diabetes exposure as well as other variables independent of diabetes.
Although we did not study the impact of miRNA changes on placental function or infant metabolic state, their potential significance is intriguing because a single miRNA may target multiple elements in a single pathway or may partner with another miRNA impacting a single target. Indeed several of the miRNAs that were affected by maternal DM (i.e. miR-148a-3p, miR-130b-3p, miR-29a-3p and miR-126–3p) have the potential to impact insulin and insulin like growth factor-1 (IGF1) signaling (9). Glucose metabolism per se may also be affected as miR-29 inhibits GLUT4 and hexokinase 2, and miR-130b targets glucokinase which, in aggregate, could reduce glucose metabolism (9). Thus, these changes in miRNAs could be interpreted as a defense against caloric excess involving attenuation of insulin action and glucose metabolism.
For miR-126–3p, we observed a differential effect of DM on the placental expression in infants with lower birth weight, suggesting that lowering of miR-126–3p in placenta in infants with lower birth weight might be protective against growth promoting effect of DM and maternal over nutrition. We found miR-126–3p to be most abundant across all the tissue types and in circulating exosomes. The findings in HUVEC are consistent with a prior report showing that miR-126–3p is highly enriched in endothelial cells and plays a key role in maintaining endothelial homeostasis and vascular integrity (20). The idea that high levels in circulation could be indicative of an endothelial cell source is intriguing. miR-126–3p targets insulin receptor substrate 1 (IRS1) and phosphatidylinositol 3-kinase (PI3K) which are involved in insulin signaling (9). miR-148a-3p expression was lower in HUVEC exposed to DM during pregnancy. Although miR-148a-3p is elevated in circulation of adults with frank T2DM (21), it is decreased in adults with early stages of DM with impaired fasting glucose (22). The lower miR-148a-3p in the HUVEC exposed to the diabetic milieu in our study, is perhaps analogous to that observed in in early stages of dysglycemia, rather than overt DM. miR-148a-3p targets AMP-activated protein kinase (AMPKα1) which is a major regulator of energy metabolism (e.g. stimulating glucose uptake and fatty acid oxidation) as well as insulin-like growth factor-1 receptor (IGF1R), insulin receptor substrate 1/2 (IRS1/2), peroxisome proliferator-activated receptors-γ (PPARγ) involved in insulin signaling and adipogenesis pathways (13,23).
The miR-29 family (including 29a and 29b) is known to regulate insulin-stimulated glucose metabolism and targets insulin induced gene 1 and PI3K, downstream elements involved in insulin signaling (8,9). miR-29 is upregulated by glucose in skeletal muscle, liver, and white adipose tissue and overexpression leads to insulin resistance in rodents (8). In humans, circulating levels and skeletal muscle expression are increased in T2DM (24). We found lower expression of miR-29a-3p in the HUVEC exposed to DM. This may also be a potential mechanism by which the fetus is protected against exposure to maternal hyperglycemia very early on in life. Further studies will be required to understand biological significance.
Prior studies suggest a role for miR-130b-3p in the pathogenesis of obesity and T2DM as it is increased in circulation in adults with T2DM and obese children (25). miR-130b-3p is known to target AMPKα1 as well (7,13). Jiang et al have shown that exposure to high glucose increases exosomal secretion of miR-130b-3p from placental trophoblast (BeWo) cells and that miR-130b-3p reduces expression of proteins involved in energy metabolism, such as AMPKα1 and peroxisome proliferator activated receptor-g coactivator-1a (PGC-1α) (26,27). In the current study, we did not observe differential expression of miR-130b in DM exposed group. This could be explained by lower abundance of miR-130b in each tissue type as well in circulating exosomes.
The present study did not show an effect of DM on miR-let-7a expression. This could be due to the fact that let-7 is a family of genes which also encodes other miRNAs such as let-7b, let-7f which are metabolically relevant and abundantly expressed in islet cells (10). We did not measure expression of other let-7 family of miRNAs. The let-7 family of genes have been shown to regulate glucose metabolism and insulin signaling and targets insulin receptor, IRS1, IRS2, forkhead box protein 1 (FOXO1) and peroxisome proliferator-activated receptors (PPARs) (9,10,13).
During the course of our analysis, we also to examined the influence of maternal age and adiposity, infant birthweight and sex, as well as race/ethnicity on miRNA abundance. These results should be considered exploratory, as they were not part of our a priori hypotheses. Nevertheless, the findings are of potential interest.
Maternal BMI was associated with increases in miR-148a-3p in fetal exosomes and miR-126–3p and miR-148a in the HUVEC, independent of the effect of diabetes. Effect of maternal pre-pregnancy BMI on placental expression of miRNAs was measured by Tsammou et al who observed inverse association between maternal pre-pregnancy BMI and placental expression of select miRNAs (miR-20a, miR-34a, miR-146a, miR-210 and miR-222) involved in obesity, inflammation, oxidative stress and angiogenesis in a sex-specific pattern specifically in females (28). While different species of miRNA were measured by Tsammou, both studies have demonstrated the impact of maternal BMI on miRNA expression suggesting that isolated maternal obesity alters miRNA expression in the offspring.
Male sex was associated with increases in miR-126–3p in circulating exosomes and miR-148a-3p and miR-29a-3p in HUVEC, independent of other factors. Recent research has indicated that miRNA patterns are different based on the sex of the individual. For instance, sex-dependent differences in free circulating miRNAs have been identified in children (29) and healthy adults (30). This would indicate that epigenetic regulation can be sex specific.
When controlling for other covariates, an increase in birth weight (BW) was associated with a decline in miR-130b-3p expression in fetal exosomes. For the HUVEC, expression of miR-126–3p, miR-148a-3p and miR-let-7a-5p decreased with increasing BW. Likewise, in placenta miR-130b-3p, miR-29a-3p and miR-let7a-5p expression decreased as BW increased. BW is also an early predictor of metabolic diseases and recent studies have demonstrated that BW impacts expression of miRNAs associated to metabolic dysfunction (31). Several other studies have reported association between infant BW and placental expression of another subset of miRNAs (32); however, this is the first study to demonstrate a relationship of BW with expression of miRNA in fetal exosomes and HUVEC. Multiple studies have shown that increased birth weight is associated with increased long term risk of developing obesity and T2DM (33). Given the potential targets of these miRNAs, a decrease in the miRNA expression with increasing birth weight might lead to increased expression of target genes, which might be protective against the nutrient excess of the in utero environment.
Some results of the present study differ from those of our prior study where we observed increased expression of miR-126–3p and miR-148a-3p in HUVEC isolated from offspring born to mothers with DM (13). The current study involves miRNA analysis from a newly recruited patient population with a much larger sample size. The previous study involved miRNA analysis only in the HUVEC whereas; the current study includes more extensive analysis involving different tissues such as placenta and fetal exosomes in addition to HUVEC. In addition to the larger sample size in the current report, the studies differ in at least two fundamental ways. In the previous study, the majority of samples were collected after an uncomplicated vaginal delivery, whereas samples were obtained following elective caesarian section in the absence of labor for this study. A prior report has shown that labor affects placenta-specific miRNA levels in maternal plasma (34). Therefore, the stress of labor may also have affected HUVEC miRNA expression and influenced the previous results. In addition, the previous cohort had predominately-male offspring in the DM group. In the present analysis, male sex is associated with increased abundance of miR-148a-3p within the HUVEC. Thus, the preponderance of males may also have affected results of the previous study.
Our approach has several strengths. First, we measured miRNA profile in relatively large number of subjects exposed to the diabetic milieu. Second, all the babies were born by elective caesarian section, thereby eliminating potential confounding effects of labor. Third, we have simultaneously measured expression of these miRNAs across various fetal tissues exposed to DM and estimated the miRNA abundance in tissues and exosomes.
However, limitations to the present study are also acknowledged. First, some maternal factors that might influence miRNA expression, such as weight gain during pregnancy (28), physical activity, and diet, were not captured. Second, though most mothers in the diabetic group were well controlled based on their HbA1C, this may not the best measure of glycemic control during pregnancy (35). Further, we did not measure other markers of metabolic dysfunction such as cholesterol and triglyceride levels in mothers, which could potentially influence metabolic function in the offspring. In addition, treatment of diabetes varied greatly in the cohort including diet, metformin, glyburide and/or insulin. Due to small sample size within each treatment group, subgroup analysis was not performed to determine the specific impact of therapy on miRNA. Future studies with larger samples size will be performed to measure impact of type of DM (GDM versus T2DM), different therapies such as diet, metformin and/or insulin on miRNA expression. Finally, though BW showed associations with miRNA abundance, other infant outcome measures such as body composition were not obtained nor do we have long-term follow up of the infants.
CONCLUSION
This study has identified several miRNAs that are differentially expressed by exposure to in utero diabetes in HUVEC, placenta and cord serum exosomes. Many of the genes targeted by these miRNAs influence metabolic pathways and could potentially impact infant growth and metabolism by targeting downstream targets. We also identified how several maternal and infant factors can impact expression of these miRNAs when controlled for disease status. The miRNA changes we have observed are presumably adaptive responses to the energy surfeit experienced during fetal life and may be involved in the development of metabolic diseases in later life.
Supplementary Material
Impact.
miRNAs are differentially expressed in fetal tissues from offspring exposed to in utero diabetes mellitus compared to those who were not exposed.
miRNA expression differs among tissue types (Human Umbilical Vein Endothelial cells, Placenta and Circulation Exosomes) and response to diabetes exposure varies according to tissue of origin.
miRNA expression is also affected by maternal and infant characteristics such as infant birth weight, infant sex, maternal age and maternal BMI.
miRNAs might be one of the potential mechanisms by which offspring’s future metabolic status may be influenced by maternal diabetes mellitus.
ACKKNOWLEDGEMENTS
We thank our study participants.
Statement of financial support: Funding was received from NIH 1K23DK106533-01A1, Endocrine Fellow Foundation Grant, Children Hospital Foundation Fellow Research Award
Footnotes
Disclosure statement: Authors do not have any conflict of interest.
BIBLIOGRAPHY
- 1.Boney CM, Verma A, Tucker R, Vohr BR. Metabolic syndrome in childhood: association with birth weight, maternal obesity, and gestational diabetes mellitus. Pediatrics 2005;115:e290–6. [DOI] [PubMed] [Google Scholar]
- 2.Short KR, et al. Lower resting energy expenditure and fat oxidation in Native American and Hispanic infants born to mothers with diabetes. J Pediatr 2015;166:884–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kappil M, Chen J. Environmental exposures in utero and microRNA. Curr Opin Pediatr 2014;26:243–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen X, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell research 2008;18:997–1006. [DOI] [PubMed] [Google Scholar]
- 5.Rottiers V, Naar AM. MicroRNAs in metabolism and metabolic disorders. Nature reviews Molecular cell biology 2012;13:239–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zampetaki A, et al. Plasma microRNA profiling reveals loss of endothelial miR-126 and other microRNAs in type 2 diabetes. Circ Res 2010;107:810–7. [DOI] [PubMed] [Google Scholar]
- 7.Wagschal A, et al. Genome-wide identification of microRNAs regulating cholesterol and triglyceride homeostasis. Nat Med 2015;21:1290–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He A, et al. Overexpression of micro ribonucleic acid 29, highly up-regulated in diabetic rats, leads to insulin resistance in 3T3-L1 adipocytes. Mol Endocrinol 2007;21:2785–94. [DOI] [PubMed] [Google Scholar]
- 9.Mirra P, et al. The Destiny of Glucose from a MicroRNA Perspective. Front Endocrinol (Lausanne) 2018;9:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu H, et al. The Lin28/let-7 axis regulates glucose metabolism. Cell 2011;147:81–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.American Diabetes A Gestational diabetes mellitus. Diabetes care 2003;26 Suppl 1:S103–5. [DOI] [PubMed] [Google Scholar]
- 12.Jaffe EA, Nachman RL, Becker CG, Minick CR. Culture of human endothelial cells derived from umbilical veins. Identification by morphologic and immunologic criteria. J Clin Invest 1973;52:2745–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tryggestad JB, et al. Influence of gestational diabetes mellitus on human umbilical vein endothelial cell miRNA. Clin Sci (Lond) 2016;130:1955–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kroh EM, Parkin RK, Mitchell PS, Tewari M. Analysis of circulating microRNA biomarkers in plasma and serum using quantitative reverse transcription-PCR (qRT-PCR). Methods 2010;50:298–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duan ZY, et al. U6 can be used as a housekeeping gene for urinary sediment miRNA studies of IgA nephropathy. Sci Rep 2018;8:10875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bissels U, et al. Absolute quantification of microRNAs by using a universal reference. RNA 2009;15:2375–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001;25:402–8. [DOI] [PubMed] [Google Scholar]
- 18.van Buuren S, Groothuis-Oudshoorn K. mice: Multivariate Imputation by Chained Equations in R. J Stat Softw 2011;45:1–67. [Google Scholar]
- 19.Mehrotra DV, Li X, Liu J, Lu K. Analysis of longitudinal clinical trials with missing data using multiple imputation in conjunction with robust regression. Biometrics 2012;68:1250–9. [DOI] [PubMed] [Google Scholar]
- 20.Wang S, et al. The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev Cell 2008;15:261–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Candia P, et al. A unique plasma microRNA profile defines type 2 diabetes progression. PLoS One 2017;12:e0188980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mononen N, et al. Whole blood microRNA levels associate with glycemic status and correlate with target mRNAs in pathways important to type 2 diabetes. Scientific reports 2019;9:8887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Novikova DS, et al. AMP-activated protein kinase: structure, function, and role in pathological processes. Biochemistry (Mosc) 2015;80:127–44. [DOI] [PubMed] [Google Scholar]
- 24.Kong L, et al. Significance of serum microRNAs in pre-diabetes and newly diagnosed type 2 diabetes: a clinical study. Acta Diabetol 2011;48:61–9. [DOI] [PubMed] [Google Scholar]
- 25.Prabu P, et al. Circulating MiRNAs of ‘Asian Indian Phenotype’ Identified in Subjects with Impaired Glucose Tolerance and Patients with Type 2 Diabetes. PLoS One 2015;10:e0128372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang S, et al. Effects of maternal diabetes and fetal sex on human placenta mitochondrial biogenesis. Placenta 2017;57:26–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang S, Teague AM, Tryggestad JB, Chernausek SD. Role of microRNA-130b in placental PGC-1alpha/TFAM mitochondrial biogenesis pathway. Biochem Biophys Res Commun 2017;487:607–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsamou M, et al. Mother’s Pre-pregnancy BMI and Placental Candidate miRNAs: Findings from the ENVIRONAGE Birth Cohort. Sci Rep 2017;7:5548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lizarraga D, et al. miRNAs differentially expressed by next-generation sequencing in cord blood buffy coat samples of boys and girls. Epigenomics 2016;8:1619–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ameling S, et al. Associations of circulating plasma microRNAs with age, body mass index and sex in a population-based study. BMC Med Genomics 2015;8:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rodil-Garcia P, Arellanes-Licea EDC, Montoya-Contreras A, Salazar-Olivo LA. Analysis of MicroRNA Expression in Newborns with Differential Birth Weight Using Newborn Screening Cards. Int J Mol Sci 2017;18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li J, et al. The role, mechanism and potentially novel biomarker of microRNA-17–92 cluster in macrosomia. Sci Rep 2015;5:17212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weng SF, et al. Systematic review and meta-analyses of risk factors for childhood overweight identifiable during infancy. Arch Dis Child 2012;97:1019–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morisaki S, et al. Effect of labor on plasma concentrations and postpartum clearance of cell-free, pregnancy-associated, placenta-specific microRNAs. Prenat Diagn 2015;35:44–50. [DOI] [PubMed] [Google Scholar]
- 35.Hashimoto K, Koga M. Indicators of glycemic control in patients with gestational diabetes mellitus and pregnant women with diabetes mellitus. World J Diabetes 2015;6:1045–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu Y, Gao G, Yang C, et al. The role of circulating microRNA-126 (miR-126): a novel biomarker for screening prediabetes and newly diagnosed type 2 diabetes mellitus. Int J Mol Sci 2014;15:10567–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang T, et al. Circulating miR-126 is a potential biomarker to predict the onset of type 2 diabetes mellitus in susceptible individuals. Biochem Biophys Res Commun 2015;463:60–3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


