Abstract
Pathogenic RNA viruses continue to emerge owing to their rapid evolutionary rates. The family of the Flaviviridae contains enveloped, single-stranded, positive-sense RNA viruses that include mosquito borne viruses such as dengue virus and the blood-borne hepatitis C virus. Upon infection, the genomic viral RNA needs to first compete with a sea of host mRNAs for host ribosomes that synthesize the viral proteins. Then, the positive-sense template needs to be amplified and packaged into newly assembled virions. To accomplish these tasks, the virus subverts several biochemical machineries from the host. The participation of specific structures in the viral RNA mediate specific RNA-RNA and RNA-protein interactions that dictate many viral subversion strategies. In this review, we shall focus on the various mechanisms by which RNA elements in the dengue virus and hepatitis C virus untranslated regions aid the viral infectious cycle and contribute to viral fitness.
Keywords: flaviviruses, viral RNA structures, 5' to 3' end communication, translation initiation, switch from translation to replication, viral fitness
Introduction
The Flaviviridae family of enveloped, single-stranded RNA viruses are classified into three genera: Flavivirus, pestivirus and hepacivirus [1]. Dengue virus (DENV) is a flavivirus, characterized by its ability to propagate both in invertebrate and vertebrate hosts. Hepatitis C virus (HCV) belongs to the hepacivirus genus, and primarily infects humans and chimpanzees. While the virion morphology and genome organization are similar for the two viruses, they differ in their species-specificity, tissue-specificity and genomic RNA sequence/structure. RNA shapes in flaviviral genomes are important regulators of viral infection [2,3]. In this review, we discuss the similar and unique mechanisms by which RNA structures in the untranslated regions located in DENV and HCV genomes regulate the viral infectious cycle and pathogenesis.
Flaviviral untranslated regions in the regulation of translation
Most cellular mRNAs are translationally initiated by a canonical cap-dependent translation initiation mechanism. This mechanism involves the recruitment of host ribosomes and translation initiation factors to the methylated capped structure located at the 5’ end of the mRNA. In particular, the eIF4F complex, composed of eIF4E, eIF4G and eIF4A, binds preinitiation complexes (PICs), composed of 40S subunits, factors eIF3, eIF2 and initiator tRNA, via eIF4G-eIF3 protein interactions (Fig. 1A) [4]. Subsequently, the PIC scans the mRNA in a 5’ to 3’ direction until an appropriate start-site AUG codon is encountered. There, the 60S subunits join and protein synthesis commences. In most, but not all cases, the polyadenosine binding protein (PABP) binds eIF4G and facilitates the recruitment of ribosomal subunits that accumulate at the 3’ end of the mRNA to the 5’ untranslated region (UTR) (Fig. 1A).
Fig 1:
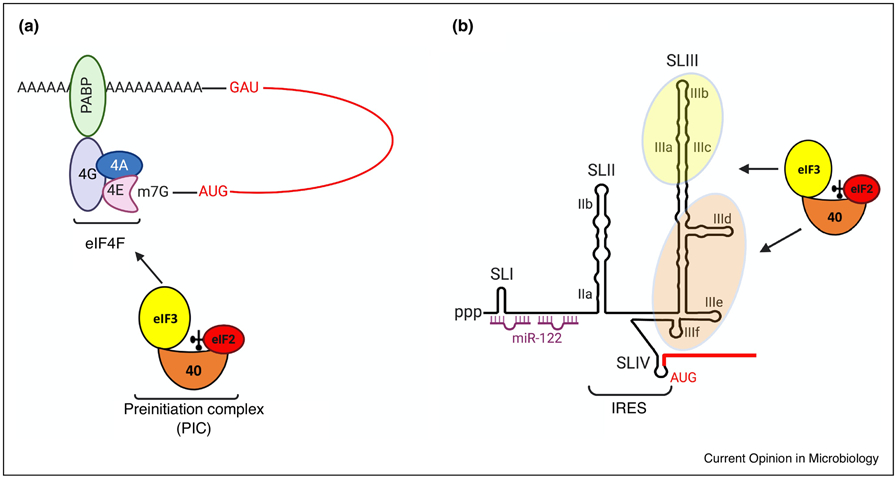
Recruitment of cellular translation machinery: (A) Translation initiation on capped, cellular mRNAs begins with the recruitment of the 43S pre-initiation complex (PIC), which contains the 40S ribosomal subunit in a complex with methionyl-initiator tRNA, GTP-bound eIF2 (depicted as a red circle bound to the 40S subunit) and eIF3 (depicted as a yellow circle bound to the 40S subunit). Capped RNAs have a 7-methyl guanosine cap at the 5’ end which interacts with the cap-binding complex (eIF4F) to recruit PICs. The eIF4F complex includes the cap binding protein eIF4E, the RNA helicase eIF4A and the scaffold protein eIF4G. eIF4G interacts with the poly A binding protein (PABP) which is usually associated with the 3’ end of polyadenylated mRNAs to promote translation initiation and ribosome recycling. The PIC scans the 5’ UTR for the start codon which is recognized through complementary base pairing with the initiator tRNA. This is followed by release of eIF2 and assembly of the 60S ribosomal subunit to form a functional 80S initiation complex that start protein synthesis. (B) HCV RNA lacks a 5’ cap structure. Instead it contains an internal ribosome entry site (IRES) that recruits PICs directly to the start codon in a sequence- and structure-dependent manner. The HCV 5’UTR is organized into four stem loop structures (SLI, SLII, SLIII and SLIV). The region spanning SLI contains two binding sites for miR-122 which promotes IRES-mediated translation. The interactions of 40S-associated eIF3 and 40S subunits with domains IIIa/b/c and IIId/e/f of the IRES, respectively, help in PIC recruitment to the start codon present in SLIV. Coding regions are indicated by red lines.
Several non-canonical mechanisms for translation initiation are observed for a few cellular and viral mRNAs that are translated by a cap-independent translation initiation mechanism. In these instances, internal ribosome entry sites (IRES), first discovered in viral mRNAs, located in the 5’UTRs of the mRNAs can recruit either PICs or empty 40S subunits, without the aid of the cap binding complex eIF4F (Fig. 1B) [4].
Recruitment of host translation machinery
Curiously, the mechanisms by which RNA structures in the genome of Flaviviridae regulate translation initiation involve both cap-dependent and cap-independent mechanims. DENV genomic RNA has a methylated cap structure at the 5’ terminus (Fig. 2) which was expected to be translated by the canonical cap-dependent translation. However, the DENV RNA does not require the eIF4E cap binding protein component of eIF4F (Fig. 1A) for translation [5].
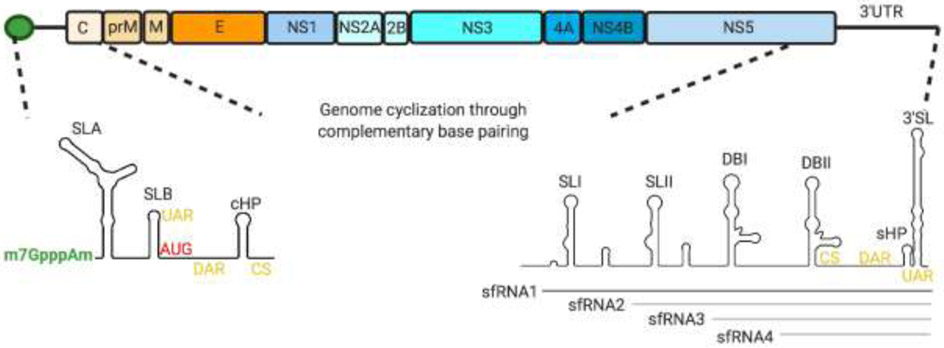
Fig 2:
DENV genome organization. The DENV genomic RNA contains an open reading frame that codes for three structural (C (capsid), prM/M (membrane) and E (envelope)) proteins and seven non structural (NS1, NS2A, NS2B NS3, NS4A, NS4B, NS5) proteins flanked by 5’ and 3’ untranslated regions. The RNA is capped at the 5’ end but lacks a 3’ poly A tail. The 5’UTR is organized into two defined stem loops, SLA which recruits NS5 and SLB. The capsid-coding region hairpin (cHP) downstream of the start codon stalls the PIC at the AUG triplet for translation initiation. The 3’ UTR is organized into stem loops SLI, SLII, dumbbell structures DBI, DBII, a small hairpin (sHP) and a terminal 3’ stem loop (3’SL). There are at least three conserved RNA sequences in the UTRs that mediate genome cyclization through complementary base pairing: 5’-3’ sequence upstream of the AUG region (5’-3’UAR), 5’-3’ sequence downstream of the AUG region (5’-3, DAR) and 5’-3’ cyclization sequence (5’-3’CS) (shown in yellow). The 3’UTR folds to form nuclease resistant structures that cannot be degraded by the host nuclease XrnI, resulting in accumulation of subgenomic flaviviral RNAs (sfRNAs). Four different sfRNA isoforms (sfRNA1-4) can be generated by XrnI stalling at different stem loops on the 3’UTR. sfRNA1 (shown in dark grey) is predominantly formed in human cells while sfRNAs 2-4 (shown in light grey) accumulate in mosquito-adapted variants.
It is known that several cellular mRNAs are efficiently translated by a cap-dependent mechanism when eIF4E is inactivated, suggesting that additional cap-binding proteins exist in cells. In a search for additional cap-binding proteins, Lee et al [6] discovered that the eIF3d subunit of the eIF3 complex has affinity for the 5’ cap structure and can direct the assembly of translation complexes on certain mRNAs, such as cell proliferation regulator c-Jun mRNA. It is likely that, like c-Jun mRNA, DENV mRNA harbors structures that directly recruit eIF3d as a component of an alternate cap-binding protein complex. The eIF4E-independent translation of DENV mRNA is dependent on the presence of both the 5’ and 3’ UTRs of the viral RNA [5] indicating that RNA-RNA or RNA-protein interactions at the UTRs are important regulators of viral mRNA translation. Furthermore, a conserved stem loop structure located downstream of the start-site AUG codon (cHP, Fig.2) has been shown to direct efficient translation initiation by stalling ribosomes at the DENV AUG start codon, which is in suboptimal context for translation initiation [7]. Thus, DENV mRNA translation has several features that are common with cap-dependent translation mechanism. However, lack of 3’ polyadenosine sequences in the viral RNA, and eIF4E-independent translation suggest that RNA-RNA and RNA-protein interactions dictate this unusual translation initiation mechanism (see below).
In an interesting twist, Song and co-workers reported recently that the DENV 5’UTR contains IRES activity that is not enhanced by the viral 3’UTR [8]. The different outcome to what was reported by Edgil et al. [5] could be explained that the former study employed chimeric mRNAs containing the UTRs of DENV. DENV IRES activity was not observed in insect cells [8], suggesting that the viral mRNA can be translated by two distinct translation initiation mechanisms that is dependent on the host. The Zika virus 5’UTR also mediates translation initiation via an IRES element [8]. The chosen mechanism is likely controlled by the formation of higher order RNA structures and the availability of canonical and non-canonical translation initiation factors.
Hepatitis C viral RNA is not capped and has evolved an IRES element that can autonomously recruit ribosomes (Fig. 1B, Fig. 3). This allows the viral RNA to be translated even when host cap-dependent translation is inhibited during virus infection and stress of the endoplasmic reticulum (ER) that accomplishes it. Employing an IRES for translation initiation also reduces competition with the host cell for canonical translation initiation factors required for cap-dependent translation. The HCV IRES can recruit either free 40S ribosomal subunits or PICs, depending on the physiological state of the infected cell to assemble 40S subunits at the AUG start codon [9]. The three-way junction domain IIIa/b/c in the HCV 5’ UTR recruits eIF3, and domain IIId/e/f directly binds the 40S subunit (Fig. 1B)[9]. When PICs are in high abundance, the HCV IRES recruits PICs. However, the HCV IRES also functions efficiently when PIC abundance is low, such as during ER stress and innate immune responses when the eIF2α subunit of eIF2 is phosphorylated, and initiator-tRNA-eIF2 complexes are in low abundance [4].
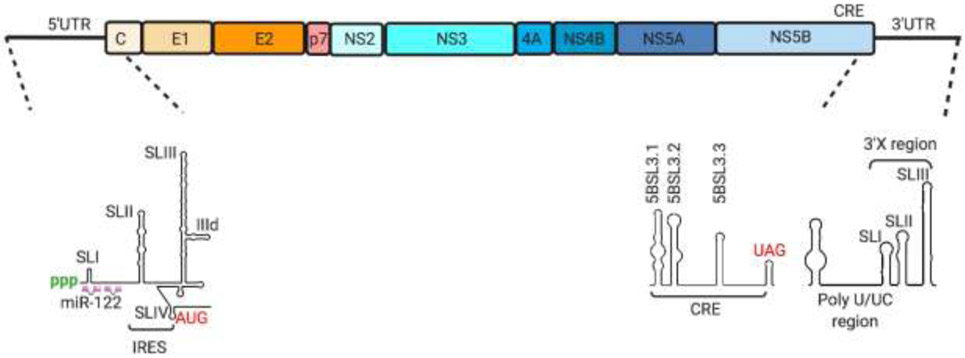
Fig 3.
HCV genome organization. The HCV genomic RNA contains an open reading frame that codes for three structural proteins, core (C) and envelope (E1, E2) and seven non-structural proteins (p7, NS2, NS3, NS4A, NS4B, NS5A, NS5B) flanked by 5’ and 3’ untranslated regions. The 5’UTR is organized into stem loops I-IV and harbors the internal ribosome entry site as well as binding sites for miR-122. The 3’UTR is organized into the variable region, the poly U/UC region and the highly conserved 3’X tail, composed of stem loops I-III. The NS5B coding region contains a cis-acting replication element (CRE) at its 3’ end just prior to the 3’UTR. The CRE is organized into three stem loop structures (5BSL3.1, 3.2 and 3.3) which are involved in dynamic interactions with the 5’UTR, 3’UTR and other upstream sequences in the NS5B coding region to regulate translation and replication of the HCV RNA.
How is the initiator-tRNA positioned into the ribosomal P-site when initiator-tRNA-eIF2 complexes are low in abundance? Recently, Jafaar et al. showed that initiation factor eIF1A, likely in concert with factor eIF5B, stabilizes initiator-tRNA molecules and facilitates codon-anticodon interactions in the ribosomal P-site in the absence of eIF2 [10]. Thus, the HCV IRES has subverted eIF1A, which, in addition to stabilizing initiator tRNA, also enhances ribosome dissociation into subunits, to allow viral mRNA translation when innate immune responses inhibit cap-dependent translation.
Flaviviral untranslated regions in the regulation of the switch from RNA translation to RNA replication
Directing 5’ to 3’ end communications
There are several reasons why 5’ to 3’ interactions in viral RNAs might be important. End-to-end interactions a) confer stability to the viral RNA, b) are convenient in the recycling of ribosomes, c) provide mechanisms to prevent RNA replication before translation products have accumulated, d) block translation initiation once negative-strand RNA synthesis has commenced, and e) provide mechanisms to create dedicated complexes for either translation or RNA replication, but not both. However, the dynamics of 5’ and 3’ end communications in even host cell mRNAs that modulate RNA decay and mRNA translation are not well known [11]. These dynamics are even more complicated in viral RNAs which assemble replication machineries and a balance between translating and replicating genomes needs to be established.
Viral mRNAs have developed alternative strategies for 5' to 3' end communications that include recruitment of host proteins to the UTRs to bridge the DENV and HCV RNA ends. For example, the non-polyadenylated DENV 3’ UTR binds PABP which could enhance viral translation possibly by bridging the 5’ and 3’ ends [12]. Genetic and biochemical analyses have shown that long-range RNA interactions in DENV are mediated by at least three highly conserved 5’ to 3’ RNA interactions, i.e. sequences upstream of the AUG region (UAR), sequences downstream of the AUG region (DAR) and cyclization sequences (CS), that are involved in long-distance base pairing with sequences located at the 3’ end of the viral genome (highlighted in yellow in Fig. 2) [13]. Abolishing these interactions affects replication of DENV RNA to different degrees in the mosquito and mammalian host. These 5’ to 3’ end interactions are predicted to change local structures, suggesting that the viral RNA assumes different shapes in infected cells that dictate their biological roles. Indeed, cell-free reconstitution experiments have shown that linear viral RNA conformations favor translation while 5’ to 3’ conformations inhibit translation [14].
In case of HCV, 5’ to 3’ communication is mediated by both RNA-protein and long-range RNA interactions between the IRES and cis-acting RNA elements both in the 3’ UTR (3’ X region) and internal to the NS5B protein coding region (CRE) (Fig. 3) [15]. Similar to DENV, the HCV 3’UTR also stimulates HCV-IRES mediated translation by up to 25-fold. A specific interaction between the 5BSL3.2 domain in the CRE and the 3’ X tail has been shown to be important for translation stimulation possibly by recruiting components of the translation machinery [16]. In particular, the HCV 3’UTR can directly bind eIF3 and 40S ribosomal subunits. It is hypothesized that this could be a rapid transfer mechanism to facilitate subsequent rounds of translation [17].
After the viral mRNAs are translated, they need to function as templates for negative-strand synthesis in RNA replication. The switch from translating to replicating RNA is crucial for positive-strand RNA viruses, because it is known that 5’ to 3’ translating ribosomes inhibit replication, as was first shown for poliovirus [18]. Given the importance of avoiding viral RNA polymerase collusions with host ribosomes, the mechanism of the switch may be distinct for each class of virus. In DENV, the interactions of the viral polymerase with the UTRs can act as a switch by directly competing with ribosomes for binding to the 5’UTR while initiating replication at the 3’UTR [19,20]. RNA cyclization and other conformational changes in the UTRs triggered by 5’-3’ end communications could regulate sequential interactions of NS5 with either of the UTRs [20,21]. In HCV, the interaction between the CRE 5BSL3.2 and the 3’ X region (Fig. 3) that favors translation can be disrupted by a CRE-IRES domain IIId interaction [22]. The temporal control of these dynamic interactions involving the CRE, the 3’ X region and the HCV IRES are likely important for the switch from translation to replication. In addition to long-range RNA interactions, the CRE also sequesters 40S ribosomal subunits and stalls polysome elongation at the HCV 3’ end through interactions with ribosomal proteins RPSA and RPS29, thus downregulating translation [23]. Thus, both changes in long-range RNA interactions and sequestration of ribosomal subunits accompany the switch from translation to replication in flavivirus-infected cells.
Interaction with host cellular miRNAs
Flaviviral RNAs bind cellular microRNAs (miRs) to regulate viral RNA stability, RNA translation, RNA replication, or in combination thereof. In the HCV genome, the 5’ sequences in the 5’ UTR have conserved, tandem binding sites for the highly abundant, liver-specific miR-122 [24]. The binding of miR-122 protects the uncapped 5’ end of the viral RNA from degradation by cellular pyrophosphatases DUSP11 and DOM3Z [25] and 5’-3’ exonucleases XrnI and Xrn2 [26]. However, miR-122 also stimulates IRES-mediated translation independently of its effect on stability [27]. Mechanistically, miR-122 assists in folding a dynamic HCV RNA structure that enhances IRES activity in one conformation and modulates replication in an alternate structure [28]. Thus, it is likely that a structural re-arrangement of the viral 5’UTR by a small microRNA aids in the switch from translation to RNA replication in HCV.
Contribution of DENV 3’ UTR-derived RNAs to viral fitness and adaptation
Curiously, all invertebrate flaviviruses 3’UTRs form three-way junctions and pseudoknot structures that block degradation of the viral RNA by 5’ to 3’ exonuclease XRN1. This results in accumulation of 3’UTR-derived, noncoding subgenomic RNA fragments (sfRNA) that are 300-500 nucleotides in length [29]. Using genetic analyses, it was shown that DENV sfRNAs disrupt host immune response pathways and are important determinants of viral fitness and adaptation. In particular, the abundance of each sfRNA isoform (Fig. 2) is an important regulator of viral infection. In the mosquito host, a high sfRNA:genomic RNA ratio correlates with better virus transmission efficiency and higher epidemiological fitness of DENV [30]. Specific adaptive mutations in a stem loop structure in the viral 3’UTR lead to generation of the shorter sfRNA3 and sfRNA4 (Fig. 2) that are under positive selection in the mosquito host [31]. In the human host, mutations that reduce the accumulation of sfRNAs make viral RNA amplification more susceptible to interferon responses [32]. Infection of human cells with mosquito adapted DENV, which produces sfRNA3 and sfRNA4, grows poorly, triggers the interferon pathway leading to lower viral fitness, and is quickly outcompeted by DENV that expresses the long sfRNA1 [31]. These findings suggest that the accumulation of different isoforms of sfRNAs correlate with the pathogenic signatures of the virus in mosquito and human hosts.
What are the mechanisms by which sfRNAs contribute to species-specific growth of DENV? In the mosquito host, DENV sfRNAs interfere with the antiviral responses by at least two mechanisms. First, sfRNAs sequester Dicer and Ago2 [33] thereby inhibiting the antiviral RNAi pathway. Secondly, sfRNAs derived from Zika virus and West Nile virus have been shown to sequester the mosquito DDX6 ortholog, ME31B [34]. ME31B is located in processing bodies (PB) where it is associated with RNA decapping enzymes. Because PBs are dispersed during flaviviral infection, ME31B and associated RNA degradation enzymes have been shown to bind to the viral genomic RNA, resulting in the generation of sfRNAs. The sfRNAs sequester ME31B protein complexes, allowing efficient amplification of the genomic RNA. Several mechanisms have been shown for sfRNAs to aid in viral RNA amplification in human cells. In contrast to ME31B, DDX6 has a proviral role in the DENV life cycle through interactions with the dumbbell structures DBI and DBII located in the DENV 3’UTR and, likely, in sfRNA1-3 (Fig. 2) [35]. In addition, the variable sequences in the 3’UTR interact with stress granule (SG) proteins G3BP1, G3BP2, Caprin 1 and USP10 [36]. Immunofluorescence studies have shown that these proteins colocalize with the viral replication complex. Thus, DENV 3' UTR is a site for assembly of PB and SG proteins that modulate viral gene expression in a species-specific manner. Finally, using epidemiological and molecular studies showed that DENV sfRNAs sequester E3 ligase tripartite motif 25 (TRIM25) [37]. TRIM25, upon its deubiquitylation by ubiquitin-specific peptidase 15, polyubiquitylates RIG-I which activates type I interferon responses. However, sfRNA-bound TRIM25 cannot be deubiquitylated, resulting in enhanced fitness of the virus.
What are the effects of Xrn1 sequestration by flaviviral RNAs on the host? As predicted, depletion of XRN1 leads to accumulation of uncapped cellular mRNAs and affects mRNA turnover [38]. However, putative roles for uncapped, cytoplasmic mRNAs are not known. Curiously, depletion of XRN1 leads to upregulation of the endoplasmic reticulum-associated oligosaccharyltransferase (OST) complex which is essential for efficient flaviviral RNA replication [39]. Unlike arthropod-borne flaviviruses, HCV doesn’t generate sfRNAs; instead, it has evolved structures in the 5’UTR that can stall and inhibit XrnI activity. Because HCV RNAs sequesters XRN1, specific unstable host mRNAs that are involved in oncogenesis accumulate in infected cells, likely contributing to HCV-induced liver cancer [40]. Thus, sequestration of Xrn1 activity seems to be an important and conserved aspect of cellular pathogenesis in the Flaviviridae family.
Conclusions
Since the physiological and immunological processes in humans and mosquitos are fundamentally different, arthropod-borne flaviviruses face different selective pressures in the two hosts. The interaction of flaviviral UTRs with host proteins is essential for subverting biochemical machineries and evading the immune responses in different hosts. In the process, host proteins are sequestered away from their normal cellular function and the consequent deregulation of cellular pathways are signatures of viral pathogenesis. Flaviviral UTRs and UTR-derived subgenomic RNA fragments, and microRNAs are important regulators of viral fitness and host pathogenesis and provide novel targets for antiviral interventions.
Highlights.
Dengue viral and hepatitis C viral RNAs have evolved to recruit ribosomal subunits by non-canonical translation initiation mechanisms
5’ to 3’ end communications in viral genomes are dynamic and modulate the switch from translation to replication of the viral RNAs
Hepatitis C virus RNA binds two molecules of microRNAs miR-122 at the 5’ end to protect the RNA from degradation and to fold the viral RNA into a translation-competent structure
Specific subgenomic RNAs in the Dengue viral genome confer selective fitness in the invertebrate and vertebrate hosts
Acknowledgements
We are grateful to Drs. Karla Kirkegaard and Andrea Gamarnik for critical comments on the manuscript. Work in the authors’ laboratory was supported by grants from the NIH (R01 AI069000, R37 AI47365).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Simmonds P, Becher P, Bukh J, Gould EA, Meyers G, Monath T, Muerhoff S, Pletnev A, Rico-Hesse R, Smith DB, et al. : ICTV Virus Taxonomy Profile: Flaviviridae. J Gen Virol 2017, 98:2–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ng WC, Soto-Acosta R, Bradrick SS, Garcia-Blanco MA, Ooi EE: The 5' and 3' Untranslated Regions of the Flaviviral Genome. Viruses 2017, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu ZY, Qin CF: Structure and function of cis-acting RNA elements of flavivirus. Rev Med Virol 2020, 30:e2092. [DOI] [PubMed] [Google Scholar]
- 4.Hinnebusch AG, Ivanov IP, Sonenberg N: Translational control by 5'-untranslated regions of eukaryotic mRNAs. Science 2016, 352:1413–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Edgil D, Diamond MS, Holden KL, Paranjape SM, Harris E: Translation efficiency determines differences in cellular infection among dengue virus type 2 strains. Virology 2003, 317:275–290. [DOI] [PubMed] [Google Scholar]
- 6.Lee AS, Kranzusch PJ, Doudna JA, Cate JH: eIF3d is an mRNA cap-binding protein that is required for specialized translation initiation. Nature 2016, 536:96–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clyde K, Harris E: RNA secondary structure in the coding region of dengue virus type 2 directs translation start codon selection and is required for viral replication. J Virol 2006, 80:2170–2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •8.Song Y, Mugavero J, Stauft CB, Wimmer E: Dengue and Zika Virus 5' Untranslated Regions Harbor Internal Ribosomal Entry Site Functions. mBio 2019, 10.The authors use monocistronic and bicistronic reporter RNA constructs to demonstrate cap-independent translation initiation at the DENV and ZIKV 5'UTRs in mammalian cells, possibly mediated by an internal ribosome entry site. Surprisingly, IRES activity couldn't be detected in mosquito cells.
- •9.Yokoyama T, Machida K, Iwasaki W, Shigeta T, Nishimoto M, Takahashi M, Sakamoto A, Yonemochi M, Harada Y, Shigematsu H, et al. : HCV IRES Captures an Actively Translating 80S Ribosome. Mol Cell 2019, 74:1205–1214 e1208.The authors use cryoEM and single molecule fluorescence studies to demonstrate that the HCV IRES not only interacts with free 40S ribosomal subunits but also with 40S subunits that are actively involved in translation of cellular or viral mRNAs as a part of the 80S initiation complex.
- 10.Jaafar ZA, Oguro A, Nakamura Y, Kieft JS: Translation initiation by the hepatitis C virus IRES requires eIF1A and ribosomal complex remodeling. Elife 2016, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vicens Q, Kieft JS, Rissland OS: Revisiting the Closed-Loop Model and the Nature of mRNA 5'-3' Communication. Mol Cell 2018, 72:805–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Polacek C, Friebe P, Harris E: Poly(A)-binding protein binds to the non-polyadenylated 3' untranslated region of dengue virus and modulates translation efficiency. J Gen Virol 2009, 90:687–692. [DOI] [PubMed] [Google Scholar]
- 13.de Borba L, Villordo SM, Iglesias NG, Filomatori CV, Gebhard LG, Gamarnik AV: Overlapping local and long-range RNA-RNA interactions modulate dengue virus genome cyclization and replication. J Virol 2015, 89:3430–3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sanford TJ, Mears HV, Fajardo T, Locker N, Sweeney TR: Circularization of flavivirus genomic RNA inhibits de novo translation initiation. Nucleic Acids Res 2019, 47:9789–9802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Romero-Lopez C, Berzal-Herranz A: The Role of the RNA-RNA Interactome in the Hepatitis C Virus Life Cycle. Int J Mol Sci 2020, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tuplin A, Struthers M, Cook J, Bentley K, Evans DJ: Inhibition of HCV translation by disrupting the structure and interactions of the viral CRE and 3' X-tail. Nucleic Acids Res 2015, 43:2914–2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bai Y, Zhou K, Doudna JA: Hepatitis C virus 3'UTR regulates viral translation through direct interactions with the host translation machinery. Nucleic Acids Res 2013, 41:7861–7874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gamarnik AV, Andino R: Switch from translation to RNA replication in a positive-stranded RNA virus. Genes Dev 1998, 12:2293–2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •19.Fajardo T, Sanford TJ, Mears HV, Jasper A, Storrie S, Mansur DS, Sweeney TR: The flavivirus polymerase NS5 regulates translation of viral genomic RNA. Nucleic Acids Res 2020, 48:5081–5093.NS5 is recruited to promoter elements in the flaviviral 5'UTR and subsequently translocated to the 3'UTR to initiate viral RNA replication. In this study, the authors demonstrate that NS5 recruitment to the 5'UTR can inhibit translation initation by blocking 48S ribosome complex assembly and aid the viral genome to switch from translation to replication.
- 20.Liu ZY, Li XF, Jiang T, Deng YQ, Ye Q, Zhao H, Yu JY, Qin CF: Viral RNA switch mediates the dynamic control of flavivirus replicase recruitment by genome cyclization. Elife 2016, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Filomatori CV, Lodeiro MF, Alvarez DE, Samsa MM, Pietrasanta L, Gamarnik AV: A 5' RNA element promotes dengue virus RNA synthesis on a circular genome. Genes Dev 2006, 20:2238–2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Romero-Lopez C, Berzal-Herranz A: A long-range RNA-RNA interaction between the 5' and 3' ends of the HCV genome. RNA 2009, 15:1740–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Romero-Lopez C, Rios-Marco P, Berzal-Herranz B, Berzal-Herranz A: The HCV genome domains 5BSL3.1 and 5BSL3.3 act as managers of translation. Sci Rep 2018, 8:16101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P: Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science 2005, 309:1577–1581. [DOI] [PubMed] [Google Scholar]
- 25.Amador-Canizares Y, Bernier A, Wilson JA, Sagan SM: miR-122 does not impact recognition of the HCV genome by innate sensors of RNA but rather protects the 5' end from the cellular pyrophosphatases, DOM3Z and DUSP11. Nucleic Acids Res 2018, 46:5139–5158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kincaid RP, Lam VL, Chirayil RP, Randall G, Sullivan CS: RNA triphosphatase DUSP11 enables exonuclease XRN-mediated restriction of hepatitis C virus. Proc Natl Acad Sci U S A 2018, 115:8197–8202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sarnow P, Sagan SM: Unraveling the Mysterious Interactions Between Hepatitis C Virus RNA and Liver-Specific MicroRNA-122. Annu Rev Virol 2016, 3:309–332. [DOI] [PubMed] [Google Scholar]
- 28.Schult P, Roth H, Adams RL, Mas C, Imbert L, Orlik C, Ruggieri A, Pyle AM, Lohmann V: microRNA-122 amplifies hepatitis C virus translation by shaping the structure of the internal ribosomal entry site. Nat Commun 2018, 9:2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chapman EG, Moon SL, Wilusz J, Kieft JS: RNA structures that resist degradation by Xrn1 produce a pathogenic Dengue virus RNA. Elife 2014, 3:e01892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pompon J, Manuel M, Ng GK, Wong B, Shan C, Manokaran G, Soto-Acosta R, Bradrick SS, Ooi EE, Misse D, et al. : Dengue subgenomic flaviviral RNA disrupts immunity in mosquito salivary glands to increase virus transmission. PLoS Pathog 2017, 13:e1006535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Filomatori CV, Carballeda JM, Villordo SM, Aguirre S, Pallares HM, Maestre AM, Sanchez-Vargas I, Blair CD, Fabri C, Morales MA, et al. : Dengue virus genomic variation associated with mosquito adaptation defines the pattern of viral non-coding RNAs and fitness in human cells. PLoS Pathog 2017, 13:e1006265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bustos-Arriaga J, Gromowski GD, Tsetsarkin KA, Firestone CY, Castro-Jimenez T, Pletnev AG, Cedillo-Barron L, Whitehead SS: Decreased accumulation of subgenomic RNA in human cells infected with vaccine candidate DEN4Delta30 increases viral susceptibility to type I interferon. Vaccine 2018, 36:3460–3467. [DOI] [PubMed] [Google Scholar]
- 33.Moon SL, Dodd BJ, Brackney DE, Wilusz CJ, Ebel GD, Wilusz J: Flavivirus sfRNA suppresses antiviral RNA interference in cultured cells and mosquitoes and directly interacts with the RNAi machinery. Virology 2015, 485:322–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goertz GP, van Bree JWM, Hiralal A, Fernhout BM, Steffens C, Boeren S, Visser TM, Vogels CBF, Abbo SR, Fros JJ, et al. : Subgenomic flavivirus RNA binds the mosquito DEAD/H-box helicase ME31B and determines Zika virus transmission by Aedes aegypti. Proc Natl Acad Sci U S A 2019, 116:19136–19144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ward AM, Bidet K, Yinglin A, Ler SG, Hogue K, Blackstock W, Gunaratne J, Garcia-Blanco MA: Quantitative mass spectrometry of DENV-2 RNA-interacting proteins reveals that the DEAD-box RNA helicase DDX6 binds the DB1 and DB2 3' UTR structures. RNA Biol 2011, 8:1173–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bidet K, Dadlani D, Garcia-Blanco MA: G3BP1, G3BP2 and CAPRIN1 are required for translation of interferon stimulated mRNAs and are targeted by a dengue virus noncoding RNA. PLoS Pathog 2014, 10:e1004242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Manokaran G, Finol E, Wang C, Gunaratne J, Bahl J, Ong EZ, Tan HC, Sessions OM, Ward AM, Gubler DJ, et al. : Dengue subgenomic RNA binds TRIM25 to inhibit interferon expression for epidemiological fitness. Science 2015, 350:217–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gilbertson S, Federspiel JD, Hartenian E, Cristea IM, Glaunsinger B: Changes in mRNA abundance drive shuttling of RNA binding proteins, linking cytoplasmic RNA degradation to transcription. Elife 2018, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marceau CD, Puschnik AS, Majzoub K, Ooi YS, Brewer SM, Fuchs G, Swaminathan K, Mata MA, Elias JE, Sarnow P, et al. : Genetic dissection of Flaviviridae host factors through genome-scale CRISPR screens. Nature 2016, 535:159–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moon SL, Blackinton JG, Anderson JR, Dozier MK, Dodd BJ, Keene JD, Wilusz CJ, Bradrick SS, Wilusz J: XRN1 stalling in the 5' UTR of Hepatitis C virus and Bovine Viral Diarrhea virus is associated with dysregulated host mRNA stability. PLoS Pathog 2015, 11:e1004708. [DOI] [PMC free article] [PubMed] [Google Scholar]