Abstract
There is currently no specific prophylaxis or vaccine against Crimean-Congo hemorrhagic fever virus (CCHFV). Crimean-Congo hemorrhagic fever (CCHF) is a severe febrile-illness transmitted by Hyalomma ticks in endemic areas, handling of infected livestock or care of infected patients. We report here the successful protection against CCHFV-mediated disease in a non-human primate disease model. Cynomolgus macaques were vaccinated with a DNA-based vaccine using in vivo electroporation-assisted delivery. The vaccine contained two plasmids encoding the glycoprotein precursor (GPC) and the nucleoprotein (NP) of CCHFV. Animals received three vaccinations and we recorded potent antibody and T-cell responses after vaccination. While all sham-vaccinated animals developed viremia, high tissue viral loads and CCHF-induced disease, the NP + GPC vaccinated animals were significantly protected. In conclusion, this is the first evidence of a vaccine that can protect against CCHFV-induced disease in a non-human primate model. This supports clinical development of the vaccine to protect groups at risk for contracting the infection.
One-sentence summary:
A DNA-based vaccine confers significant protection from CCHFV infection in Cynomolgus macaques
Introduction.
Crimean-Congo hemorrhagic fever (CCHF) is a severe febrile-illness caused by the Crimean-Congo hemorrhagic fever virus (CCHFV), a negative-sense RNA virus in the Nairoviridae family of the Bunyavirales order. Aligning with the wide geographic distribution of its Hyalomma species tick-vector and reservoir, cases of CCHF are reported throughout Eastern Europe, Africa, the Middle East and parts of Asia 1. In addition, recent cases of CCHF have been reported in Spain 2 and global climate change is likely to lead to expansion of the range of the Hyalomma tick leading to introduction of CCHFV into new areas. CCHFV-exposure typically occurs through tick bite, handling of infected livestock or during the treatment of infected patients in the health care setting. CCHF begins as a non-specific febrile illness that can rapidly progress to severe hemorrhagic manifestations 3. Case fatality rates vary among regions but typically range between 5 – 30% 3. Currently, treatment options for CCHFV are lacking. While ribavirin is recommended by the World Health Organization 4, clinical data from human cases 5 and animal models 6–8 provide conflicting evidence on the benefit of ribavirin for treatment of CCHFV and suggest better treatments are needed. The World Health Organization has put CCHFV on its blue print list to highlight the need to develop antivirals and vaccines (https://www.who.int/blueprint/).
A safe, effective vaccine is critically needed for at risk populations such as health care workers, rural inhabitants and abattoir workers. As CCHFV infection of livestock is apparently asymptomatic 9, farmers and abattoir workers may be unknowingly infected with CCHFV during the handling and care of livestock. Similarly, tick-bites by Hyalomma ticks may not be fully appreciated as a risk factor for CCHFV by rural inhabitants in endemic areas. Thus, the early clinical signs of CCHF, fever, headache and myalgia 3, may not be recognized as the early stages of CCHF. This is problematic as clinical evidence for the efficacy of ribavirin in treatment of CCHF indicates that ribavirin must be given early after symptom onset to offer clinical benefit 10,11. Consequently, by the time many patients present at health care facilities, they are exhibiting the more severe signs of CCHF, when treatment is limited to supportive care. Therefore, vaccination of at-risk populations against CCHFV likely represents the most effective therapeutic intervention to limit CCHFV-induced morbidity and mortality. While an inactivated preparation of CCHFV grown in mouse brains has been used in eastern Europe 12,13, practical and safety considerations will likely prevent this vaccine from achieving wide-spread distribution. Several vaccine platforms have been evaluated in mouse models including modified-Vaccinia Ankara-based, vesicular stomatitis virus-based, human adenovirus-based, subunit-based, DNA-based, virus-like particle-based and transgenic plant-based vaccines 14–22. We have recently developed a non-human primate model of CCHF 23,24 in which cynomolgus macaques inoculated with the clinical isolate of CCHFV strain Hoti recapitulate many aspects of human CCHF. To date no vaccine candidates have been evaluated in this immunocompetent animal disease model and evaluation of vaccine candidates in this unique model will provide important pre-clinical data on the safety, immunogenicity and protection of potential vaccine candidates for CCHFV.
DNA-based vaccines developed by our group and others have demonstrated efficacy against CCHFV in lethal mouse challenge models 16,20 and DNA-based vaccines have a favorable safety profile owing to their non-replicating nature 25. Hence, DNA-based vaccines represent a promising platform for development of CCHFV vaccines. We report here evaluation of a DNA-based vaccine consisting of plasmid-expressed CCHFV strain Hoti nucleoprotein (NP) and the CCHFV strain Hoti M-segment open-reading frame encoding the viral glycoproteins (GPC) in the cynomolgus macaque model. Plasmid DNA was delivered by intramuscular injections followed by in vivo electroporation. Vaccination appeared well-tolerated and elicited CCHFV-specific antibody and T-cell responses. Such responses in vaccinated animals correlated with absent viremia, substantially reduced viral burdens in all tissues evaluated and improved blood parameters upon CCHFV-challenge. Together our data show that this vaccine provides significant protection against CCHFV and represents the first vaccine with demonstrated efficacy in a non-human primate model of CCHF.
Results.
Vaccination of animals.
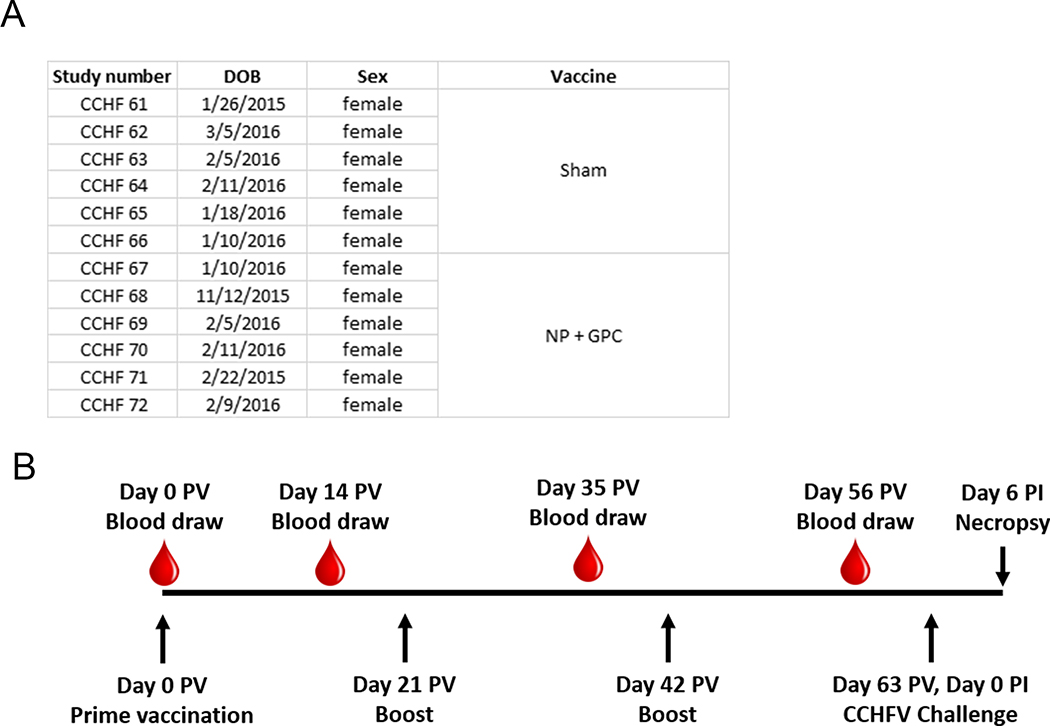
We have previously demonstrated that interferon signaling-incompetent IFNAR−/− mice immunized with DNA coding for the viral NP, Gn and Gc of CCHFV strain IbAr 10200 were protected against homologous lethal CCHFV challenge 20. The recent development of a non-human primate model for CCHF, based on CCHFV strain Hoti 23, enabled evaluation of this DNA-based vaccination approach in fully immunocompetent animals. Since our initial report on CCHFV-infection of cynomolgus macaques, we have further developed the model using combined subcutaneous and intravenous inoculation 24. To date, infection of cynomolgus macaques has demonstrated that CCHFV infection results in a spectrum of disease outcomes from mild-to-moderate-to-severe disease, similar to the spectrum of disease seen in humans. Since the model is not uniformly lethal, we designed a study with a scheduled euthanasia at day 6 post-CCHFV infection to evaluate vaccine-mediated protection, similar to the rhesus macaque model for Middle-Eastern respiratory virus and severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) 26–28 and similar to our study evaluating favipiravir in CCHFV-infected cynomolgus macaques 24. Twelve female Chinese-origin adult cynomolgus macaques, Macaca fascicularis, were used for this study (Extended Figure 1).
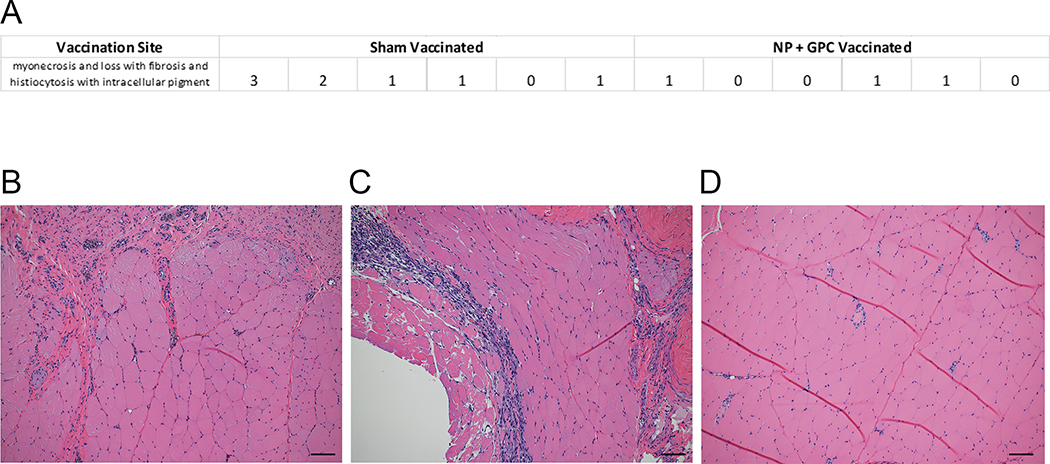
Six animals were vaccinated with two ubiquitin-antigen fusion plasmids, one encoding the CCHFV NP and the other the viral GPC, both derived from CCHFV strain Hoti. One milligram of each respective plasmid was delivered independently to opposing quadriceps. Six animals also received a sham-vaccination of an equivalent dose of plasmid encoding ubiquitin to each quadricep followed by in vivo electroporation. Animals received three identical vaccinations separated by three-week intervals (Extended Figure 1). Blood draws were taken prior to vaccination and two-weeks after each vaccination (Extended Figure 1). The electroporation delivery of the vaccine appeared well tolerated. No serious adverse events were observed, and the most common side-effect observed during clinical exams following vaccinations was firmness at the vaccination site that resolved without intervention; this induration is expected after vaccination. In one animal, a lesion developed at the vaccination site, likely due to pruritis, but this too resolved without intervention. At time of scheduled euthanasia following CCHFV challenge (27 days after last vaccination), histopathological analysis of the vaccination site indicated no pathology in 4 of the vaccinated animals. Six animals had minor, one had moderate and one had severe myonecrosis and loss with histiocytosis with intracellular pigment at the vaccination site (Extended Figure 2). These histologic lesions were limited to the immediate site of vaccination and at no time post-vaccination did any animals display clinical signs to indicate pain or discomfort associated with muscular lesions.
DNA-vaccination induces B- and T-cell responses to CCHF antigens.
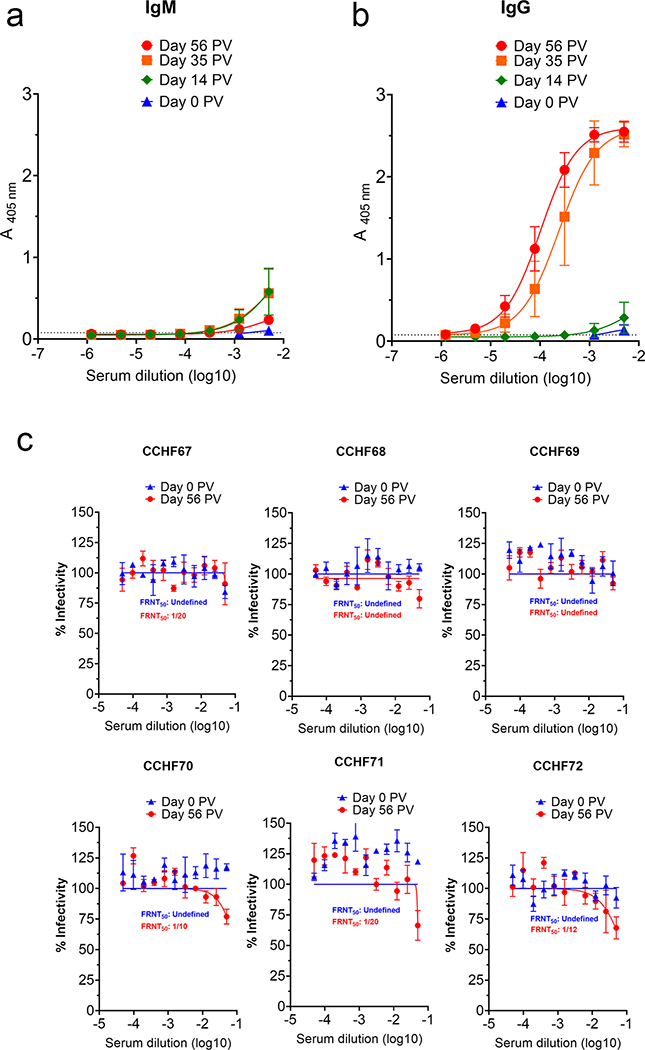
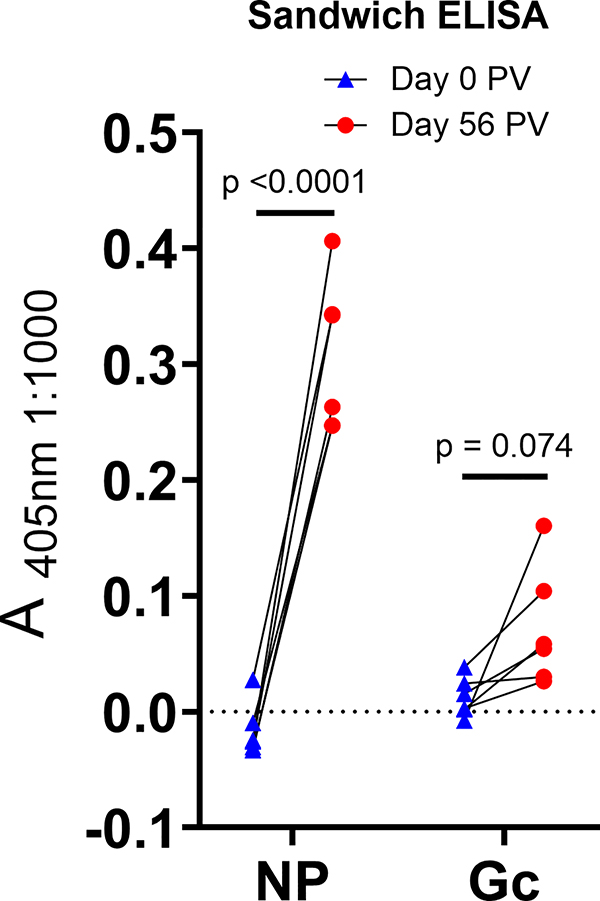
To evaluate the antibody response to the vaccine, serum was collected at time of first immunization (pre-bleed) and two weeks after each immunization thereafter (Extended Figure 1) and CCHFV-specific IgM and IgG quantified by whole-virion ELISA. Although we detected CCHFV-specific IgM two-weeks after the first immunization (day 14 post-prime-vaccination (PV)) (Figure 1A), strong CCHFV-specific IgG responses were not detected until two-weeks after the second immunization (day 35 PV) (Figure 1B). We detected slight increases in CCHFV-specific IgG titers after the third immunization (day 56 PV) (Figure 1B). As expected, sham-vaccinated animals had no change in ELISA signal between pre-bleed (day 0 PV) and day 56 PV (Extended Figure 3). We also evaluated whether the CCHFV-specific antibody response we detected in NP + GPC vaccinated animals by ELISA was neutralizing. Neutralization capacity of heat-inactivated serum collected at day 0 PV or on day 56 PV was evaluated by FRNT. Interestingly, despite high levels of antibody detected by ELISA, day 56 PV serum demonstrated poor neutralization activity against infectious CCHFV (Figure 1C). In light of the high ELISA titers but poor serum neutralizing activity, we performed a sandwich ELISA to specifically measure NP or Gc-specific responses. Consistent with the poor neutralizing activity, we did not detect significant increases in Gc-specific antibody responses between day 0 and 56 PV (Extended Figure 4). In contrast, significant increases between day 0 and 56 PV in NP-specific antibody responses were detected (Extended Figure 4), suggesting the humoral response was largely directed against NP.
Figure 1: NP + GPC vaccination induces robust antibody responses to CCHFV.
(A & B) At indicated time-points, CCHFV-specific IgM and IgG antibody responses in the serum were measured by whole-virion ELISA. Data is shown as the mean plus the standard deviation of duplicate measurements. Connecting lines were derived from unconstrained non-linear regression. Dashed line indicates average absorbance of background wells receiving no serum. (C) FRNT was performed to measure neutralizing activity of day −7 serum from NP + GPC vaccinated animals. Individual neutralization curves for each NP + GPC vaccinated animals are shown. Non-linear regression with a top constraint of 100 and bottom constraint of 0 was calculated and the FRNT50 determined. Data shown as mean plus standard error of measurement from triplicate technical replicates. N = 6 animals per group.
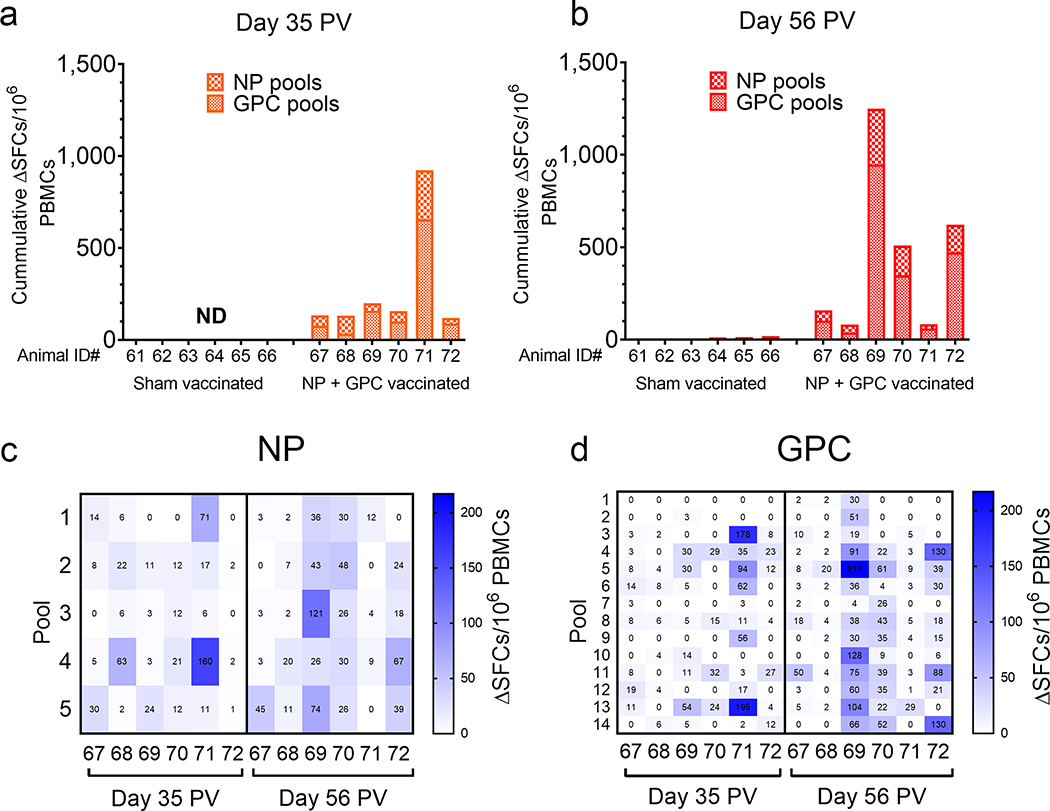
To evaluate T-cell responses, we performed an IFNγ ELISpot on cryo-preserved PBMCs collected two-weeks after the second (day 35 PV) and third vaccinations (day 56 PV) (Figure 2). Compared to sham-vaccinated animals, all NP + GPC vaccinated animals had detectable in vitro recall responses measured as IFNγ-producing SFCs to CCHFV-peptides, indicating the priming of CCHFV-specific T-cell responses by the vaccination (Figure 2A & B). Recall responses against both NP and GPC-derived peptide pools were detected in all animals. Responses were equally distributed across NP (Figure 2C). Across the GPC, we did not identify an immunodominant peptide pool although minimal responses were directed against N-terminal pools #1 or 2 or pool #7 (Figure 2D). Together, our ELISpot data demonstrate that NP + GPC vaccination induced CCHFV-specific IFNγ recall responses against both vaccine antigens.
Figure 2: NP + GPC vaccination induces CCHFV-specific IFNγ T-cell responses.
Cryopreserved PBMCs from day −28 (A) or day −7 (B) were stimulated with pooled overlapping peptides derived from the CCHFV GPC or NP at 1μg/mL per peptide. Background was ascertained by wells stimulated with DMSO-vehicle alone. After 20 hours, IFNγ producing cells were detected by ELISpot. Spot forming cells (SFCs) were counted, background count subtracted from peptide-stimulated wells (ΔSFCs) and normalized to 1×106 PBMCs. All measurements were performed in duplicate for each animal. (A & B) Data is shown as the sum of SFCs counted among all NP and GPC peptide pools. (C & D) For NP + GPC vaccinated animals the number of SFCs against each peptide pool for NP (C) and GPC (D) is shown. ND = not done.
NP + GPC DNA-vaccination protects against CCHFV induced disease.
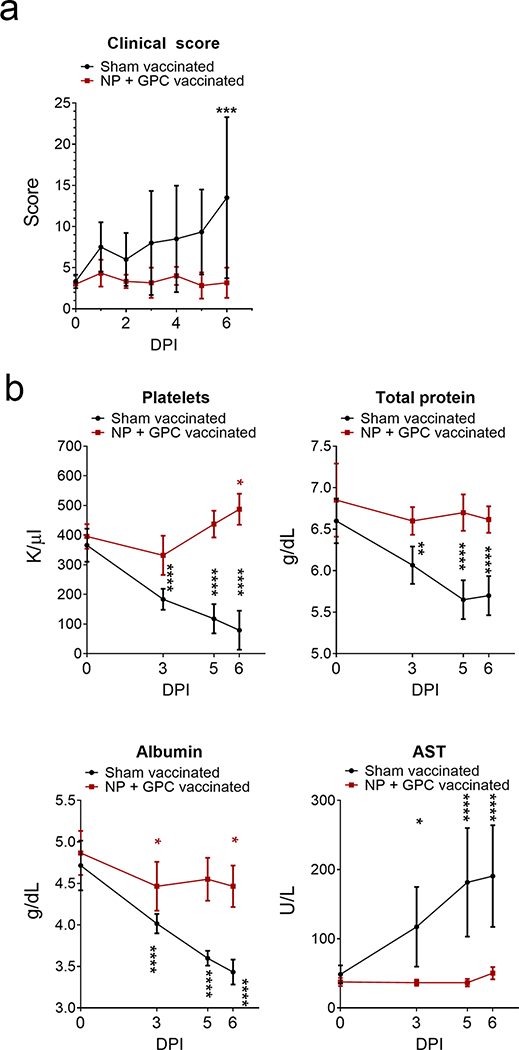
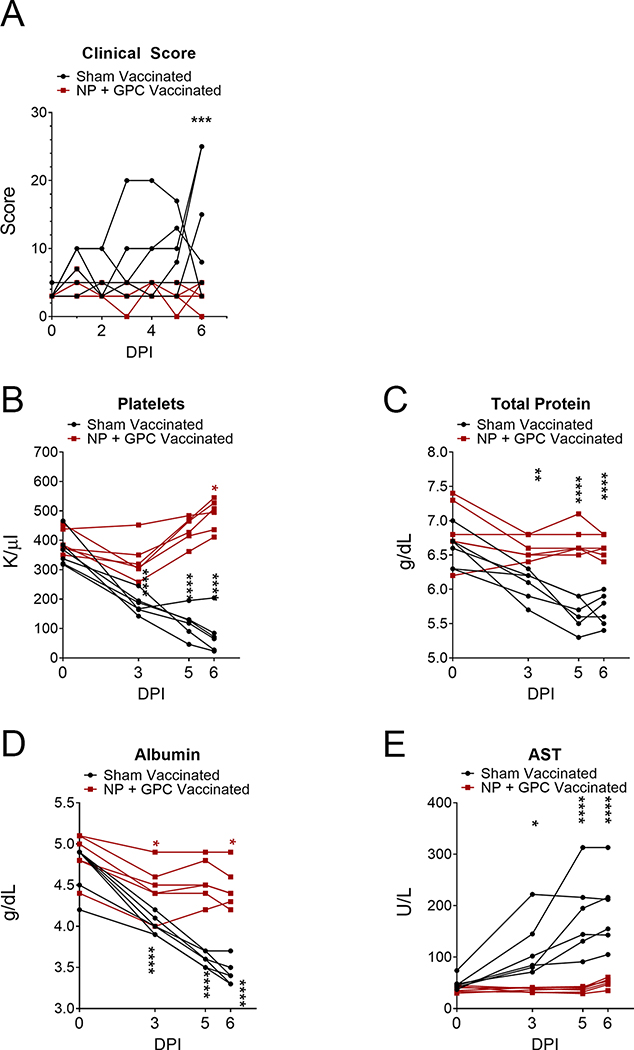
To evaluate the protection afforded by NP + GPC vaccination, on day 0 relative to CCHFV challenge (post-infection (PI)) and day 63 PV, sham-vaccinated and NP + GPC-vaccinated animals were challenged with 100,000 TCID50 of CCHFV strain Hoti via simultaneous subcutaneous and intravenous injections 23. Although this model is not uniformly lethal, inoculation of naïve animals via these routes results in consistent development of viremia, blood hematology and chemistry perturbations and significant viral burdens in a variety of tissues following CCHFV-infection 23. Animals were comprehensively scored daily and NP + GPC vaccinated animals had significantly reduced clinical signs of disease on day +6 (Figure 3A and Extended figure 5A). Clinical exams on days 0, 3, 5 and 6 PI were performed and blood collected for evaluation of blood chemistry and hematology changes between groups. When compared to baseline values obtained on day 0, we found that sham-vaccinated animals had significantly reduced platelets, total protein and albumin levels along with significantly elevated AST values at days 3, 5 and 6 PI (Figure 3B – E and Extended figure 5B – E). In contrast, these values in NP + GPC vaccinated animals remained largely unchanged from day 0 with slight decreases in albumin on days 3 and 6 PI and even significantly elevated platelets on day 6 (Figure 5B–E and Extended figure 5B–E), although these values remained within normal ranges for NP + GPC vaccinated animals. These data demonstrate that NP + GPC vaccination prevents changes in blood chemistry that are often associated with poor outcome in human CCHF cases 3,29. Other blood chemistry and hematology parameters were largely similar between groups. Complete blood chemistry and hematology profiles are provided in supplemental files.
Figure 3: NP + GPC vaccination improves clinical scores and blood chemistry following CCHFV challenge.
(A) Animals were comprehensively scored for evaluation of overt clinical disease and cumulative clinical scores are shown. Data shown as mean plus standard deviation. Statistical comparison was performed by two-way ANOVA with Sidak’s multiple comparison test at each time point. Day 6 PI P value = 0.0003. (B) At indicated time points platelets were enumerated in EDTA-treated whole-blood (normal range 300 – 500k/uL) and serum was evaluated for total protein (normal range 7.5 g/dL ±0.57), albumin (4.2 g/dL ±0.28) and aspartate aminotransferase (AST) (48.2IU/L ±36). Data is shown as mean plus standard deviation. Statistical comparison was performed between baseline values on day 0 and indicated time-point. N = 6 animals per group. Platelets, **** P < 0.0001, * P = 0.0129. Total Protein, **** P < 0.0001, ** P = 0.0027. Albumin, **** P < 0.0001, * P = 0.0131. AST, **** P < 0.0001, * P = 0.0280. P-values calculated with a two-tailed two-way ANOVA with Sidak’s multiple comparison test.
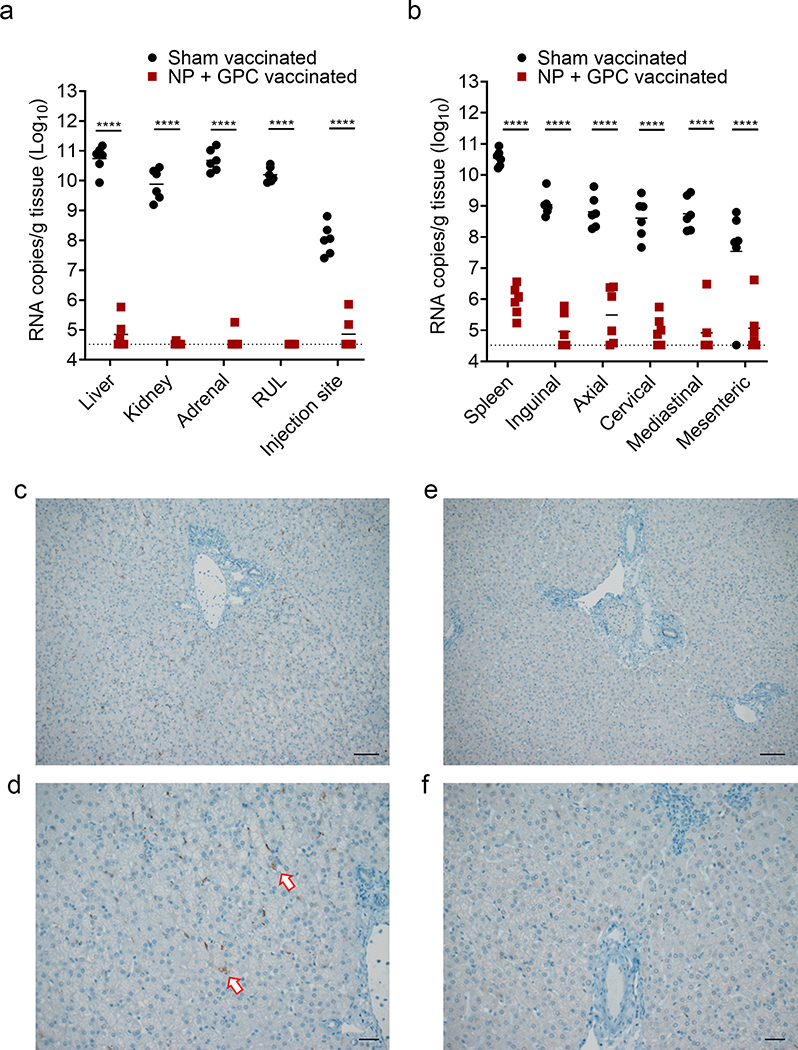
Figure 5: NP + GPC vaccination reduces viral burden in multiple tissues.
(A & B) At day +6 PI a scheduled necropsy was performed, and viral genomes quantified in the indicated tissues by qRT-PCR. RUL = right upper lung. Individual values are shown, bar indicates mean and dashed line indicates limit of detection. **** p < 0.0001. P-values calculated with a two-tailed two-way ANOVA with Sidak’s multiple comparison test. (C – F) Immunohistochemistry to detect CCHFV antigen in the liver was performed and representative images of sham vaccinated (C & D) and NP + GPC vaccinated (E & F) animals are shown at 100x (C & E) or 200x (D & F) magnification. Arrows indicate antigen positive cells. Six sections from six animals per group were stained for CCHFV antigen and representative images from one animal per group shown. Scale bars indicate 50μm (C & E) or 100μm (D & F).
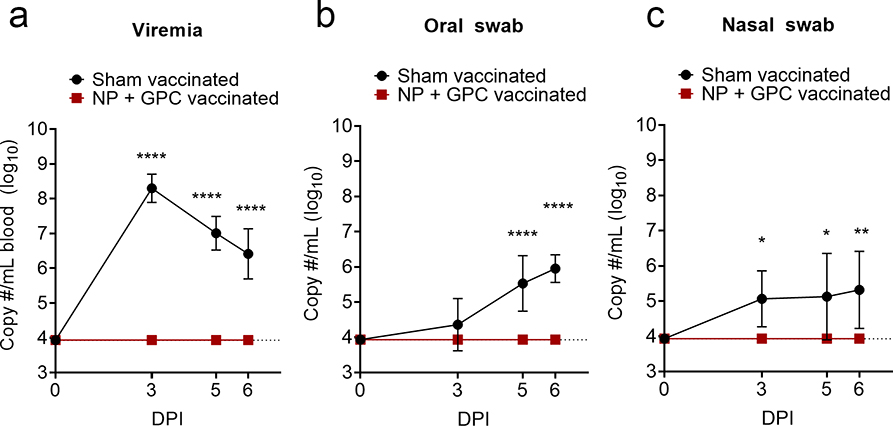
DNA-vaccination prevents viremia, viral shedding and reduces viral RNA burdens in multiple tissues.
To determine shedding of virus following CCHFV challenge, we evaluated viral RNA burdens in blood and oral and nasal swabs on days 0, 3, 5 and 6 PI (Figure 4). We found that NP + GPC vaccinated animals had no detectable viral RNA in the blood at any time point evaluated (Figure 4A). In contrast, sham-vaccinated animals had significant levels of viral RNA at 3, 5- and 6-days PI (Figure 4A). Similarly, NP + GPC vaccinated animals had no detectable viral RNA in the oral or nasal cavities whereas most sham-vaccinated animals had detectable viral RNA in these samples (Figure 4B and C). Cumulatively, NP + GPC vaccinated animals had no detectable viral RNA in the blood, oral cavity or nasal cavity at any time point evaluated suggesting NP + GPC vaccinated animals exhibited robust control of the CCHFV challenge.
Figure 4: NP + GPC vaccination prevents viremia and viral shedding.
(A – C) At indicated time-points viral genomes were quantified by qRT-PCR. Data is shown as mean plus standard deviation. Dashed line indicates limit of detection. Viremia and Oral swab, **** p < 0.0001. Nasal swab, * P = 0.0169 (3 DPI) or 0.0109 (5 DPI), ** P = 0.0025. P-values calculated with a two-tailed two-way ANOVA with Sidak’s multiple comparison test. N = 6 animals per group.
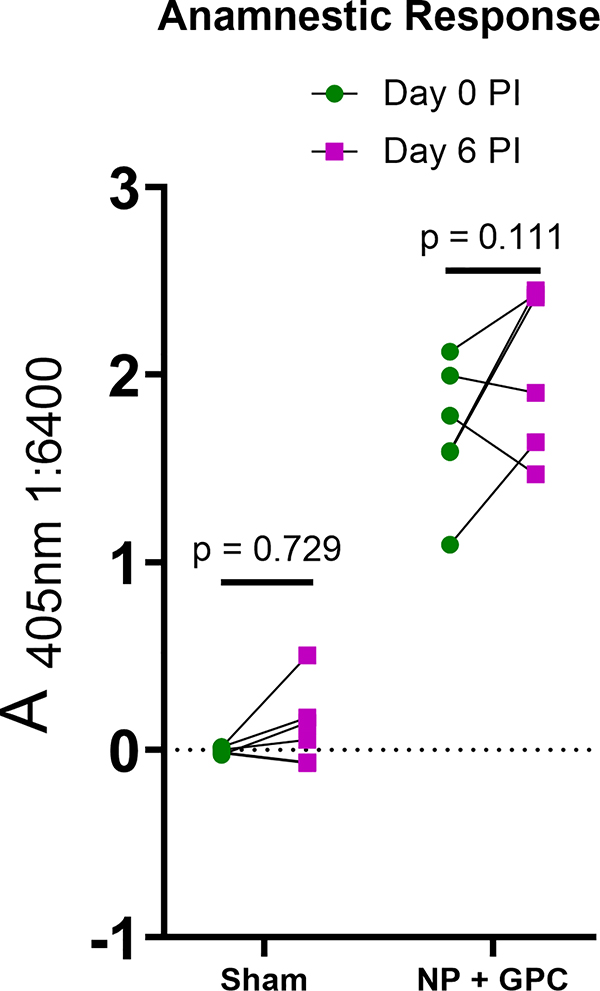
Since our study was not powered to detect differences in survival between sham- and NP + GPC-vaccinated groups, a scheduled necropsy was performed on day 6 PI to evaluate viral burdens and histopathology in a variety of tissues. We found that NP + GPC vaccination significantly reduced viral RNA burdens in all tissues evaluated (Figure 5). In key organs such as the liver, kidney, lung and adrenal gland, NP + GPC vaccination reduced viral RNA to undetectable levels in the majority of vaccinated animals (Figure 5A). Remarkably, NP + GPC vaccinated animals also had significantly reduced viral burdens compared to sham-vaccinated animals at the injection site of the subcutaneous CCHFV inoculation (Figure 5A). Four of 6 NP + GPC vaccinated animals had no detectable CCHFV RNA at the injection site suggesting these animals effectively cleared CCHFV RNA from the site of inoculation within 6 DPI. We also collected the spleen and lymph nodes to evaluate CCHFV viral burdens in lymphoid tissues. In these tissues, NP + GPC vaccination significantly reduced CCHFV RNA burdens compared to sham-vaccinated animals, with a greater than 100-fold reduction in viral burden in all tissues (Figure 5B). To further evaluate CCHFV burdens in the liver, we performed immunohistochemistry to detect CCHFV NP antigen. All 6 sham-vaccinated animals but no NP + GPC vaccinated animals had detectable CCHFV antigen in the liver (Figure 5C – F), consistent with our qRT-PCR data. Lastly, when we evaluated serum antibody responses after challenge on day 6 PI, we found no significant increases in CCHFV-specific titers (Extended Figure 6) indicating no anamnestic antibody response, at least through day 6 PI. These data are consistent with the significantly reduced viral loads in multiple tissues and substantial vaccine-mediated protection against CCHFV challenge. Cumulatively, our data demonstrated that NP + GPC vaccination significantly reduced CCHFV viremia, viral shedding and viral burdens in a multitude of tissues.
Discussion
There is a critical need for vaccines against CCHFV particularly for populations at risk of exposure to CCHFV. We report here a DNA-based vaccine that elicits humoral and cellular immunity against CCHFV. These responses provided significant protection against CCHFV-challenge. Although the cynomolgus macaque model of CCHF is not uniformly lethal and we were unable to power the study to demonstrate improvements in survival, NP + GPC vaccination improved several key parameters that are associated with poor outcome in human CCHF cases including viremia, platelet count and liver enzymes 30,31. High viral loads are often associated with poor outcome in human CCHF cases 30–32 and NP + GPC vaccinated animals had no detectable viremia at any time point evaluated, even when using a sensitive qRT-PCR assay. In addition to absent CCHFV viremia, NP + GPC vaccinated animals had significantly reduced CCHFV loads in a multitude of tissues.
Thrombocytopenia is a common clinical feature of CCHF and low platelet counts typically correlate with prolonged clotting times and poor outcome 29,33,34. Further, decreases in total protein and albumin have also been noted in severe CCHFV cases 29. While sham-vaccinated animals showed significant declines in platelet counts, total protein and albumin following CCHFV-infection, NP + GPC vaccinated animals exhibited no such decline. Although we did not measure clotting times in our study, platelet counts of NP + GPC vaccinated animals remained within normal ranges suggesting vaccination protected against dysregulated clotting function. Similarly, AST levels, often elevated in severe and fatal human cases of CCHF 32,34, were significantly elevated in sham-vaccinated but not NP + GPC vaccinated animals. These data indicate that the immune responses induced by NP + GPC vaccination were highly effective in controlling the CCHFV challenge, reducing viral loads in multiple tissues and preventing perturbations to blood chemistry and hematology that are often associated with poor outcome in humans.
It has been previously shown that both B and T-cell responses contribute to protection conferred by a modified vaccinia virus vaccine for CCHFV 14,35. Our data demonstrate that NP + GPC vaccination induced a robust but non-neutralizing antibody response, indicating neutralizing antibody responses, in the context of this vaccine, are dispensable for vaccine-mediated protection. Non-neutralizing antibodies can contribute to protection against CCHFV as non-neutralizing antibodies against Gn or GP38 were protective when administered therapeutically in a lethal mouse model 36,37. Further, a subunit vaccine induced neutralizing responses but failed to protect lethally infected mice 18 indicating that responses beyond neutralizing antibodies may be required for vaccine mediated protection from CCHFV. The role of T-cells in vaccine-mediated protection from CCHFV is less clear. In studies evaluating a modified vaccinia Ankara virus expressing CCHFV antigens, both humoral and cellular immunity was required for protection 14,35. Cumulatively, our data indicate that all NP + GPC vaccinated animals developed CCHFV-specific antibody and IFNγ recall responses.
We chose to vaccinate animals with plasmids expressing NP and GPC. Although GPC contains the glycoproteins Gn and Gc, which are the target of protective and neutralizing antibodies 36,38, NP based vaccines can confer significant protection on their own 15. However, this may depend on the vaccine platform 39. Furthermore, the GPC of CCHFV exhibits the lowest sequence conservation among the viral segments with some strains differing by up to 15% in Gn and Gc at the amino acid level 1,40. This sequence variation can lead to impairment of antibody-mediated neutralization 38 and could lead to incomplete protection when the vaccine antigens are mismatched to the infecting virus strain. Thus, the greater sequence conservation of NP and potentially protective immune responses directed against this antigen may favor the inclusion of NP in CCHFV-vaccine preparations. Interestingly, DNA-immunization of mice with full length GPC only provided partial protection in lethally infected mice 16 whereas mice immunized with the glycoproteins and NP were completely protected 20. While we cannot exclude the possibility that differences in vaccine delivery, mouse strain or challenge dose may contribute to these distinct outcomes, it suggests inclusion of NP may improve vaccine-mediated protection.
Lastly, observation of immunized animals together with our immunology and challenge data indicate in vivo electroporation of plasmid DNA was well tolerated and immunogenic. Noserious adverse events were observed in any of the vaccinated animals. Vaccination site reactions were mild and histopathological analysis of the vaccination sites at time of necropsy indicated absent-to-mild pathology in most animals. These data are consistent with other preclinical data and human clinical trials demonstrating the safety and immunogenicity of in vivo electroporation to deliver DNA-based vaccine platforms 41–43.
There are several important limitations of our study that will require further investigation. First, in many CCHFV endemic regions, the three-vaccination regimen evaluated here may be difficult to achieve in at-risk populations due to remoteness and lack of infrastructure. Vaccinations providing protection after one or two immunizations would certainly be preferred for these communities. Some CCHFV experimental vaccines have shown efficacy in type I IFN-deficient mice after a single-dose 14,21,22. However, our immunogenicity data certainly suggest that the third vaccination may not be needed and the use of immunodeficient mice may not strictly predict efficacy in immunocompetent NHPs and humans 44,45. Second, our study design of performing a timed necropsy on day 6 PI was chosen to maximize readout parameters but it prevented us from evaluating the possibility of delayed disease development in sham and NP + GPC vaccinated animals. However, prior to euthanasia no NP + GPC vaccinated animals exhibited any indications of worsening clinical disease or declining immunity to suggest viral escape from the host response and even naïve animals begin to recover around day 7 PI 23. Lastly, we did not determine the immune correlates of protection in our study. NP + GPC vaccinated animals developed both humoral and cellular immunity against CCHFV and it is possible that either or both mediated the protection we observed in this study. Similarly, it is unclear if both NP and GPC vaccine-expressed antigens are needed for the observed protection. A vaccine containing just one antigen would simplify production and clinical evaluation and further studies are planned to evaluate vaccination with just NP or GPC alone.
In summary, our data demonstrate that a DNA-based vaccine delivered by in vivo electroporation against CCHFV is well-tolerated, immunogenic and protects against disease in a pre-clinical non-human primate model of CCHF. Vaccinated animals showed CCHFV-specific antibody and T cell responses and upon CCHFV-challenge showed improved clinical parameters, significantly reduced viremia, viral shedding and viral burdens in several key tissues including the liver. Further studies are needed to evaluate prime/boost versus prime-only vaccination strategies, durability of immune responses and to determine the correlates of protection in this model. However, our data indicate that this NP + GPC DNA-driven vaccine platform may be a safe and effective vaccine to prevent the significant morbidity and mortality caused by CCHFV. The current study supports that this vaccine can be advanced into human clinical trials.
Materials and Methods.
Animals, Biosafety and Ethics.
All infectious work with CCHFV and sample inactivation was performed in the maximum containment laboratory in accordance with standard operating procedures approved by the Rocky Mountain Laboratories Institutional Biosafety Committee, Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health (Hamilton, MT, USA). All animal work was performed in strict accordance with the recommendations described in the Guide for the Care and Use of Laboratory Animals of the Office of Animal Welfare, National Institutes of Health and the Animal Welfare Act of the US Department of Agriculture, in an Association for Assessment and Accreditation of Laboratory Animal Care International (AAALAC)-accredited facility. Twelve female cynomolgus macaques of Chinese origin, three to four years of age were used in this study. Animals were housed in adjoining individual primate cages that enabled social interaction, under controlled conditions of humidity, temperature and light (12-h light/12-h dark cycles). Food and water were available ad libitum. Animals were monitored at least twice daily (pre- and post-infection) and fed commercial monkey chow, treats and fruit twice a day by trained personnel. Environmental enrichment consisted of manipulanda, visual enrichment and audio enrichment.
Plasmids.
To create the pVAX1-Ub universal fusion vector, the ubiquitin sequence was subcloned from plasmid pCMV_Ub_F1F2 46 into pVAX1 (Invitrogen), using NheI restriction enzyme. A Kozak sequence was added in 5’, and a linker with a KpnI recognition site was added before the ubiquitin stop codon to allow subsequent cloning of antigen sequence. Protein expression from this construct will result in 5’ -ubiquitin-antigen fusion 47 (Extended Figure 6). The ubiquitin protein has the mutation G76A to prevent its cleavage from the fusion protein, and all pVAX1 plasmids are Kanamycin resistant. cDNA sequences from CCHFV strain Hoti S and M segments (Genbank Accession MH483984, MH483985) were codon-optimized (CO) for expression in human cells and cloned in a shuttle vector (BioCat GmbH, Germany). All inserts were fully sequenced. To create the plasmids used for immunization, the pVAX1-Ub vector was linearized by using restriction enzymes KpnI and NotI, which also remove the stop codon from the ubiquitin. Then the cDNA sequences of the antigens were amplified by PCR using the high fidelity Phusion HotStartII polymerase (FinnZymes, Thermo Fisher Scientific), subcloned in frame on the 3’ end of the ubiquitin sequence, using the same enzymes and a Rapid DNA ligation kit (Fermentas). All DNA vaccine plasmids were transformed into competent DH10b bacteria (Thermo Fisher Scientific) and Gigapreps were prepared using EndoFree Plasmid Giga Kit (Qiagen AB) according to the manufacturer’s protocol. Each plasmid batch was fully sequenced. Sequences of primers used for cloning and sequences of plasmids are available upon request.
Vaccination.
At time of vaccination, animals were anesthetized with ketamine or telazol. In some animals, sedation was maintained via inhalational isoflurane as necessary. Vaccination was performed by disinfection of the injection site followed by an injection of 0.5 to 1mL of plasmid DNA diluted in saline to the quadriceps muscle. Immediately following instillation, the injection site was subject to electroporation by delivery of 0.6ms 600V/cm pulse followed by a 400ms 60V/cm pulse using a four-electrode array at a depth of 1 cm. Electric pulses were delivered by the Genedrive device (IGEA, Carpi, Italy). Animals were boosted at three-week intervals for a total of three vaccinations. Vaccinations were performed by trained personnel under supervision of veterinary staff. Animals were routinely monitored by veterinary staff, and detailed clinical exams performed weekly after vaccinations.
Virus.
Animals were challenged with 1×105 TCID50 of CCHFV strain Hoti divided between subcutaneous injections to the cranial dorsum and intravenously through the saphenous vein as previously described 23. Our challenge stock of CCHFV Hoti was propagated, titered and sequenced as previously described 6,23.
Interferon gamma ELISpot.
Peripheral blood mononuclear cells (PBMCs) were isolated from EDTA-treated whole-blood spun over a Histopaque 1077 gradient (Sigma). Red blood cells were lysed with ACK lysis buffer (Gibco) and PBMCs frozen in fetal bovine serum supplemented with 10% dimethyl sulfoxide (Hybrimax grade, Sigma) in liquid nitrogen vapor phase. Cryopreserved PBMCs were evaluated for interferon gamma (IFNγ) production in response to CCHFV-peptides by commercial ELISpot (Cellular Technologies Limited). PBMCs were thawed and plated at 100,000 – 400,000 cells per well in CTL-Test media. 15-mer peptides overlapping by 11 amino acids derived from the CCHFV NP or GPC were synthesized (Genscript), resuspended in DMSO (Hybrimax-grade, Sigma) and pooled at 19 – 31 peptides per pool. Cells were stimulated with peptide pools at a final concentration of 1μg/mL each peptide. As positive control, cells were stimulated with concanavalin A (Life Technologies) or DMSO-vehicle alone. Cells were incubated for 20 hours at 37°C in 5% CO2 before plates were developed according to manufacturer’s protocol. Spots were counted using an S6 Universal analyzer (CTL) and data normalized per 1×106 cells. Upper limit of detection was set at 750 SFCs per well. Background was defined as number of spots in DMSO-vehicle alone stimulated wells and this value subtracted from counts measured in peptide- and concanavalin A-stimulated wells. Each measurement was performed in duplicate.
ELISA.
Whole-CCHFV-virion-specific IgM and IgG responses in serum were quantified by an in-house ELISA as previously described 23. To measure NP or Gc specific responses we performed a sandwich ELISA. Nunc Maxisorp plates were coated with 200ng/well of mouse monoclonal antibodies against NP (clone 9D5, BEI Resources) or Gc (11E7, BEI resources). Plates were then blocked with 5% skim milk in PBS 0.05% Tween and then semi-purified whole virus lysate 23 applied. Plates were washed and a 1:1000 dilution of serum applied. Bound antibodies were detected with goat horseradish peroxidase conjugated anti-monkey IgG (Seracare, catalog KPL 074–11-021) at 1:2000 and plates developed with ABTS solution (Seracare). Development was stopped with 5% sodium dodecyl sulfate in water and absorbance at 405nm read on a Synergy HTX (Biotek). For data analysis, absorbance values of wells coated with anti-NP or anti-Gc but receiving no CCHFV lysate were subtracted from wells receiving lysate. All measurements were performed in duplicate.
Serum neutralization titers.
Focus reduction neutralization tests (FRNT) were performed as previously described 23,48.
qRT-PCR.
Viral RNA in indicated tissues was quantified by qRT-PCR as previously described 6.
Blood chemistry and hematology.
Hematology was evaluated on EDTA-treated whole-blood using a Procyte DX (IDEXX Laboratories) and serum chemistries were completed on a Vetscan 2 (Abaxis) using Preventive care profile disks (Abaxis). Indicated normal ranges for blood parameters were obtained from published resources 49,50.
Immunohistochemistry.
Immunohistochemistry (IHC) to detect viral antigen was performed as described previously 48.
Statistics and Reproducibility.
Indicated statistical tests were performed using Prism 8 (GraphPad). All data is derived from one experiment.
Data availability.
The data that support the findings of this study are available from the corresponding author upon reasonable request and source data for figures 1, 3, 4 and 5 are provided.
Extended Data
Extended Data Fig. 1. Animal information and vaccination schedule.
Animal information and vaccination schedule. (A) Animal number, date of birth (DOB), sex and vaccination. Animals were randomly assigned to either group prior to start of study. (B) Vaccination, blood draw and CCHFV challenge schedule. PV = post prime-vaccination, PI = post-CCHFV infection
Extended Data Fig. 2. Vaccination Site Histology.
Vaccination Site Histology. At Day +6 CCHFV challenge, 27 days after last vaccination, tissue from the vaccination site was fixed in 10% formalin and stained with hematoxylin and eosin. Slides were scored by a pathologist (A). 0 = absent, 1 = mild, 2 = moderate, 3 = severe. (B-D) Representative images of the one severe (B), one moderate (C) and one of six mild animals (D) are shown at 100x magnification.
Extended Data Fig. 3. CCHFV ELISA on serum from sham-vaccinated animals.
CCHFV ELISA on serum from sham-vaccinated animals. At indicated time points, serum was collected from sham-vaccinated animals and CCHFV-specific IgG measured by ELISA on day 0 or 56 post-prime vaccination. N = 6 animals per timepoint. Data shown as mean plus standard deviation.
Extended Data Fig. 4. Sandwich ELISA.
Sandwich ELISA. A sandwich ELISA was performed on NP + GPC vaccinated animals to measure NP or Gc specific responses at day 0 post-prime vaccination (PV) and day 56 PV. N = 6 animals per timepoint. Statistical comparison performed using a two-way ANOVA with Sidak’s multiple comparison test.
Extended Data Fig. 5. Individual data for clinical scores and blood chemistry.
Individual data for clinical scores and blood chemistry. The data from figure 3 is shown again but with individual data points shown.
Extended Data Fig. 6. Anamnestic response to CCHFV challenge.
Anamnestic response to CCHFV challenge. A whole-virion ELISA was used to measure antibody responses in animals after CCHFV challenge on day 6 post-infection (PI). CCHFV-specific IgG was quantified in a 1:6400 dilution of serum. Statistical tests performed using a two-way ANOVA with Sidak’s multiple comparison test.
Extended Data Fig. 7.
General plasmid schematics of plasmids used in this study.
Supplementary Material
Acknowledgements
The authors thank the staff of the Rocky Mountain Veterinary Branch for their help with animal care and veterinary clinical and pathology work.
Funding: Our work was supported by the CCHVaccine consortium that has received funding from the European Union’s Horizon 2020 research and innovation program under grant agreement no. 732732 (AM, MS and FW), by the Swedish Research Council (Vetenskapsrådet; VR, MS) section Research Environment, Infection Biology (contract number 2018-05766) (AM and FW), the Swedish Cancer Foundation (MS), the Stockholm County Council ALF and CIMED grants (MS), Vinnova CAMP grant (MS), and by the Intramural Research Program of the NIAID, NIH. The funders had no role in study design, data interpretation or decision to publish.
Footnotes
Competing Interests
MS is a founder and chairman of the board of Svenska Vaccinfabriken Produktion AB.
References
- 1.Bente DA et al. Crimean-Congo hemorrhagic fever: history, epidemiology, pathogenesis, clinical syndrome and genetic diversity. Antiviral Res 100, 159–189, doi: 10.1016/j.antiviral.2013.07.006 (2013). [DOI] [PubMed] [Google Scholar]
- 2.Negredo A et al. Autochthonous Crimean–Congo Hemorrhagic Fever in Spain. New England Journal of Medicine 377, 154–161, doi: 10.1056/NEJMoa1615162 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Ergonul O Crimean-Congo haemorrhagic fever. Lancet Infect Dis 6, doi: 10.1016/s1473-3099(06)70435-2 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stuart MC, Kouimtzi M & Hill SR WHO Model Formulary 2008. (World Health Organization, 2008). [Google Scholar]
- 5.Johnson S et al. Ribavirin for treating Crimean Congo haemorrhagic fever. The Cochrane database of systematic reviews 6, CD012713–CD012713, doi: 10.1002/14651858.CD012713.pub2 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hawman DW et al. Favipiravir (T-705) but not ribavirin is effective against two distinct strains of Crimean-Congo hemorrhagic fever virus in mice. Antiviral Res 157, 18–26, doi: 10.1016/j.antiviral.2018.06.013 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oestereich L et al. Evaluation of Antiviral Efficacy of Ribavirin, Arbidol, and T-705 (Favipiravir) in a Mouse Model for Crimean-Congo Hemorrhagic Fever. PLOS Neglected Tropical Diseases 8, e2804, doi: 10.1371/journal.pntd.0002804 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bente DA et al. Pathogenesis and immune response of Crimean-Congo hemorrhagic fever virus in a STAT-1 knockout mouse model. Journal of virology 84, 11089–11100, doi: 10.1128/jvi.01383-10 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spengler JR, Bergeron É & Rollin PE Seroepidemiological Studies of Crimean-Congo Hemorrhagic Fever Virus in Domestic and Wild Animals. PLOS Neglected Tropical Diseases 10, e0004210, doi: 10.1371/journal.pntd.0004210 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ergonul O et al. Systematic Review and Meta-analysis of Postexposure Prophylaxis for Crimean-Congo Hemorrhagic Fever Virus among Healthcare Workers. Emerg Infect Dis 24, 1642–1648, doi: 10.3201/eid2409.171709 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson S et al. Ribavirin for treating Crimean Congo haemorrhagic fever. The Cochrane database of systematic reviews 6, Cd012713, doi: 10.1002/14651858.CD012713.pub2 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mousavi-Jazi M, Karlberg H, Papa A, Christova I & Mirazimi A Healthy individuals’ immune response to the Bulgarian Crimean-Congo hemorrhagic fever virus vaccine. Vaccine 30, 6225–6229, doi: 10.1016/j.vaccine.2012.08.003 (2012). [DOI] [PubMed] [Google Scholar]
- 13.Papa A, Papadimitriou E & Christova I The Bulgarian vaccine Crimean-Congo haemorrhagic fever virus strain. Scandinavian journal of infectious diseases 43, 225–229, doi: 10.3109/00365548.2010.540036 (2011). [DOI] [PubMed] [Google Scholar]
- 14.Buttigieg KR et al. A Novel Vaccine against Crimean-Congo Haemorrhagic Fever Protects 100% of Animals against Lethal Challenge in a Mouse Model. PloS one 9, e91516, doi: 10.1371/journal.pone.0091516 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zivcec M, Safronetz D, Scott DP, Robertson S & Feldmann H Nucleocapsid protein-based vaccine provides protection in mice against lethal Crimean-Congo hemorrhagic fever virus challenge. PLOS Neglected Tropical Diseases 12, e0006628, doi: 10.1371/journal.pntd.0006628 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garrison AR et al. A DNA vaccine for Crimean-Congo hemorrhagic fever protects against disease and death in two lethal mouse models. PLOS Neglected Tropical Diseases 11, e0005908, doi: 10.1371/journal.pntd.0005908 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ghiasi SM, Salmanian AH, Chinikar S & Zakeri S Mice orally immunized with a transgenic plant expressing the glycoprotein of Crimean-Congo hemorrhagic fever virus. Clinical and vaccine immunology : CVI 18, 2031–2037, doi: 10.1128/cvi.05352-11 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kortekaas J et al. Crimean-Congo Hemorrhagic Fever Virus Subunit Vaccines Induce High Levels of Neutralizing Antibodies But No Protection in STAT1 Knockout Mice. Vector borne and zoonotic diseases (Larchmont, N.Y.) 15, 759–764, doi: 10.1089/vbz.2015.1855 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Canakoglu N et al. Immunization of Knock-Out α/β Interferon Receptor Mice against High Lethal Dose of Crimean-Congo Hemorrhagic Fever Virus with a Cell Culture Based Vaccine. PLOS Neglected Tropical Diseases 9, e0003579, doi: 10.1371/journal.pntd.0003579 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hinkula J et al. Immunization with DNA Plasmids Coding for Crimean-Congo Hemorrhagic Fever Virus Capsid and Envelope Proteins and/or Virus-Like Particles Induces Protection and Survival in Challenged Mice. Journal of virology 91, e02076–02016, doi: 10.1128/JVI.02076-16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scholte FEM et al. Single-dose replicon particle vaccine provides complete protection against Crimean-Congo hemorrhagic fever virus in mice. Emerg Microbes Infect 8, 575–578, doi: 10.1080/22221751.2019.1601030 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rodriguez SE et al. Vesicular Stomatitis Virus-Based Vaccine Protects Mice against Crimean-Congo Hemorrhagic Fever. Scientific Reports 9, 7755, doi: 10.1038/s41598-019-44210-6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haddock E et al. A cynomolgus macaque model for Crimean-Congo haemorrhagic fever. Nature microbiology 3, 556–562, doi: 10.1038/s41564-018-0141-7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hawman DW et al. Efficacy of favipiravir (T-705) against Crimean-Congo hemorrhagic fever virus infection in cynomolgus macaques. Antiviral Research, 104858, doi: 10.1016/j.antiviral.2020.104858 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kutzler MA & Weiner DB DNA vaccines: ready for prime time? Nature reviews. Genetics 9, 776–788, doi: 10.1038/nrg2432 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Falzarano D et al. Treatment with interferon-alpha2b and ribavirin improves outcome in MERS-CoV-infected rhesus macaques. Nat Med 19, 1313–1317, doi: 10.1038/nm.3362 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Wit E et al. Prophylactic and therapeutic remdesivir (GS-5734) treatment in the rhesus macaque model of MERS-CoV infection. Proc Natl Acad Sci U S A 117, 6771–6776, doi: 10.1073/pnas.1922083117 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Williamson BN et al. Clinical benefit of remdesivir in rhesus macaques infected with SARS-CoV-2. 2020.2004.2015.043166, doi: 10.1101/2020.04.15.043166 %J bioRxiv (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Swanepoel R et al. The clinical pathology of Crimean-Congo hemorrhagic fever. Reviews of infectious diseases 11 Suppl 4, S794–800 (1989). [DOI] [PubMed] [Google Scholar]
- 30.Duh D et al. Viral Load as Predictor of Crimean-Congo Hemorrhagic Fever Outcome. Emerging Infectious Diseases 13, 1769–1772, doi: 10.3201/eid1311.070222 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hasanoglu I et al. Crucial parameter of the outcome in Crimean Congo hemorrhagic fever: Viral load. Journal of clinical virology : the official publication of the Pan American Society for Clinical Virology 75, 42–46, doi: 10.1016/j.jcv.2015.12.006 (2016). [DOI] [PubMed] [Google Scholar]
- 32.Ergonul O, Celikbas A, Baykam N, Eren S & Dokuzoguz B Analysis of risk-factors among patients with Crimean-Congo haemorrhagic fever virus infection: severity criteria revisited. Clinical microbiology and infection : the official publication of the European Society of Clinical Microbiology and Infectious Diseases 12, 551–554, doi: 10.1111/j.1469-0691.2006.01445.x (2006). [DOI] [PubMed] [Google Scholar]
- 33.Aksoy F, Yilmaz G, Kaya S, Karahan SC & Koksal I The prognostic importance of platelet indices in patients with Crimean-Congo Hemorrhagic Fever. Open Forum Infectious Diseases 4, S352–S353, doi: 10.1093/ofid/ofx163.850 (2017). [DOI] [Google Scholar]
- 34.Çevik MA et al. Clinical and laboratory features of Crimean-Congo hemorrhagic fever: predictors of fatality. International Journal of Infectious Diseases 12, 374–379, doi: 10.1016/j.ijid.2007.09.010 (2008). [DOI] [PubMed] [Google Scholar]
- 35.Dowall SD et al. Protective effects of a Modified Vaccinia Ankara-based vaccine candidate against Crimean-Congo Haemorrhagic Fever virus require both cellular and humoral responses. PloS one 11, e0156637, doi: 10.1371/journal.pone.0156637 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bertolotti-Ciarlet A et al. Cellular localization and antigenic characterization of crimean-congo hemorrhagic fever virus glycoproteins. Journal of virology 79, 6152–6161, doi: 10.1128/jvi.79.10.6152-6161.2005 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Golden JW et al. GP38-targeting monoclonal antibodies protect adult mice against lethal Crimean-Congo hemorrhagic fever virus infection. Science Advances 5, eaaw9535, doi: 10.1126/sciadv.aaw9535 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zivcec M et al. Identification of broadly neutralizing monoclonal antibodies against Crimean-Congo hemorrhagic fever virus. Antiviral Res 146, 112–120, doi: 10.1016/j.antiviral.2017.08.014 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dowall SD et al. A Crimean-Congo hemorrhagic fever (CCHF) viral vaccine expressing nucleoprotein is immunogenic but fails to confer protection against lethal disease. Human Vaccines & Immunotherapeutics 12, 519–527, doi: 10.1080/21645515.2015.1078045 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Deyde VM, Khristova ML, Rollin PE, Ksiazek TG & Nichol ST Crimean-Congo hemorrhagic fever virus genomics and global diversity. Journal of virology 80, doi: 10.1128/jvi.00752-06 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.André F & Mir LM DNA electrotransfer: its principles and an updated review of its therapeutic applications. Gene Therapy 11, S33–S42, doi: 10.1038/sj.gt.3302367 (2004). [DOI] [PubMed] [Google Scholar]
- 42.Weiland O et al. Therapeutic DNA Vaccination Using In Vivo Electroporation Followed by Standard of Care Therapy in Patients With Genotype 1 Chronic Hepatitis C. Molecular Therapy 21, 1796–1805, doi: 10.1038/mt.2013.119 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sardesai NY & Weiner DB Electroporation delivery of DNA vaccines: prospects for success. Current Opinion in Immunology 23, 421–429, doi: 10.1016/j.coi.2011.03.008 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zivcec M, Spiropoulou CF & Spengler JR The use of mice lacking type I or both type I and type II interferon responses in research on hemorrhagic fever viruses. Part 2: Vaccine efficacy studies. Antiviral Research 174, 104702, doi: 10.1016/j.antiviral.2019.104702 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Palacio N et al. Early type I IFN blockade improves the efficacy of viral vaccines. Journal of Experimental Medicine 217, doi: 10.1084/jem.20191220 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boshra H, Lorenzo G, Rodriguez F & Brun A A DNA vaccine encoding ubiquitinated Rift Valley fever virus nucleoprotein provides consistent immunity and protects IFNAR(−/−) mice upon lethal virus challenge. Vaccine 29, 4469–4475, doi: 10.1016/j.vaccine.2011.04.043 (2011). [DOI] [PubMed] [Google Scholar]
- 47.Rodriguez F, Zhang J & Whitton JL DNA immunization: ubiquitination of a viral protein enhances cytotoxic T-lymphocyte induction and antiviral protection but abrogates antibody induction. Journal of virology 71, 8497 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hawman DW et al. A Crimean-Congo Hemorrhagic Fever Mouse Model Recapitulating Human Convalescence. JVI.00554–00519, doi: 10.1128/JVI.00554-19 %J Journal of Virology (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sasseville VG, Hotchkiss CE, Levesque PC & Mankowski JL in Nonhuman Primates in Biomedical Research (Second Edition) (eds Abee Christian R., Mansfield Keith, Tardif Suzette, & Morris Timothy) 357–384 (Academic Press, 2012). [Google Scholar]
- 50.Choi K et al. Reference values of hematology, biochemistry, and blood type in cynomolgus monkeys from cambodia origin. Laboratory Animal Research 32, 46–55, doi: 10.5625/lar.2016.32.1.46 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request and source data for figures 1, 3, 4 and 5 are provided.