Abstract
Glucocorticoid drugs are commonly used in the treatment of several conditions, including autoimmune diseases, asthma and cancer. Despite their widespread use and knowledge of biological pathways via which they act, much remains to be learned about the cell type-specific mechanisms of glucocorticoid action and the reasons why patients respond differently to them. In recent years, human and in vitro studies have addressed these questions with genomics, transcriptomics and other omics approaches. Here, we summarize key insights derived from omics studies of glucocorticoid response, and we identify existing knowledge gaps related to mechanisms of glucocorticoid action that future studies can address.
Keywords: Epigenomics, Genomics, Glucocorticoids, Metabolomics, Proteomics, Transcriptomics
1. Introduction
Glucocorticoids, steroid hormones synthesized and released by the adrenal glands, are essential for the body to maintain homeostasis in response to internal and environmental changes, something they achieve via the regulation of metabolic, inflammatory, immune and apoptotic pathways. Glucocorticoids influence normal metabolism by promoting gluconeogenesis, increasing glycogen storage in hepatocytes, and decreasing glucose uptake and utilization in skeletal muscle and adipose tissue (Kuo, et al., 2015). Synthetic glucocorticoids, which have strong anti-inflammatory, immunosuppressive and anti-proliferative properties, have been used to treat a wide range of conditions, including autoimmune diseases (e.g., rheumatoid arthritis, psoriasis, eczema), asthma, chronic obstructive pulmonary disease (COPD) and some cancers (e.g., acute lymphoblastic leukemia (ALL)) (Hapgood, et al., 2016). Because chronic glucocorticoid use can result in detrimental side effects, including osteoporosis, hyperglycemia, diabetes mellitus, obesity, cardiovascular diseases and depression (Desmet and De Bosscher, 2017), glucocorticoids are ideally administered for short time periods and to relevant target tissues when possible. Although glucocorticoids exhibit cell- and tissue-specific responses, the tissue-targeted treatment of glucocorticoids occurs via the selection of tissue-specific routes of administration, rather than by targeting cell type-specific pathways (Hapgood, et al., 2016). While administering glucocorticoids to specific tissues can reduce systemic side effects, their chronic use may elicit resistance in the target tissues, which may lead to increased dosages and worse side effects (Barnes, 2010; Chung, et al., 2009; Schacke, et al., 2002). Thus, a better understanding of cell type-specific glucocorticoid responses would lead to more precise therapies that may overcome glucocorticoid side effects and resistance.
Inter-individual variability in response to glucocorticoids, and the development of resistance to them, have long been recognized and remain poorly understood processes (Barnes, 2013; Barnes and Adcock, 2009; Wilkinson, et al., 2018). In rare cases, some individuals have glucocorticoid insensitivity, or an inherent inability to respond to steroids, due to mutations in the glucocorticoid receptor (GR) or a fundamental response mechanism. More often, glucocorticoid resistance may indicate that higher doses of glucocorticoids are necessary in some patients, and that after prolonged glucocorticoid use, patients may require higher doses to sustain initially observed benefits. The term glucocorticoid response is used here to indicate specific effects observed after glucocorticoid exposure in the case of in vitro assays (e.g., changes in gene transcription levels), or changes in clinical symptoms after treatment with glucocorticoids in the case of the people. Although the terms insensitivity, resistance and response are sometimes used interchangeably, it is important to note the context of a specific study to understand what the term(s) used is(are) meant to represent. Some individuals are referred to as not being responsive to glucocorticoids because symptoms persist with glucocorticoid treatment, despite the fact that at a cellular level, glucocorticoids elicit expected responses. In such cases, the reason for observed glucocorticoid “resistance” or lack of “responsiveness” may be due to having a subtype of illness that involves pathways other than steroid-sensitive ones, and thus, glucocorticoids alone are not sufficient treatment.
Inhaled glucocorticoids that act directly in the lung successfully control symptoms in most patients with persistent asthma, but up to 30% of patients with severe asthma require long-term use of oral glucocorticoids to control their disease (Chung, et al., 2014). Further, approximately 10% of people with asthma remain symptomatic with high dose glucocorticoid treatment despite exhibiting negative side effects (Barnes, 2011, 2013; Barnes and Adcock, 2009; Ernst, et al., 2015). Recently developed biologics are efficacious for the treatment of some asthma subtypes (e.g., elevated IgE levels) (Holguin, et al., 2020), including some that are poorly responsive to steroids. Glucocorticoid resistance is observed in 10%−30% of untreated, and many more relapsed, ALL patients (Haarman, et al., 2003). Thus, a more thorough understanding of mechanisms underlying glucocorticoid responses would enable the development of precise approaches to recognize and treat patients who do not adequately respond to them.
Omics approaches have enabled the unbiased measure of multi-layered information in biological systems and have been applied to address a wide range of biomedical questions [Box 1], including pulmonary diseases (Kan, et al., 2017) and glucocorticoid responses: pharmacogenomics studies have sought genetic determinants of patient response to glucocorticoids; transcriptomic and epigenomics studies have measured cell type-specific transcriptional changes and related these to transcription factors that bind to DNA, and proteomics and metabolomics studies have searched for downstream biomarkers that reflect responses to glucocorticoids. Here, we review the application of omics approaches to better understand glucocorticoid responses, with a focus on applications in asthma and ALL, as these two conditions have been studied with multiple omics modalities to understand two major pathways altered by glucocorticoids: inflammation and apoptosis. Table 1 summarizes prominent findings that are described in further detail below.
Box 1. Summary of omics approaches.
Genomics
Refers to measures of DNA sequence variation. Genome-wide association studies (GWAS), initially based on genome-wide genotyping microarrays, relate frequencies of common single nucleotide polymorphisms (SNPs) (those with minor allele frequency >1–5%) to a phenotype. Next-generation sequencing (NGS) techniques, including whole-exome sequencing (WES) and whole-genome sequencing (WGS) (Goodwin, et al., 2016), are more recent techniques that enable studies of common, rare and structural variants by more comprehensively capturing genomic variation. Validation of genomics findings begins with fine mapping to identify likely causal variants by cataloging evidence from annotated resources, such as those related to linkage disequilibrium patterns, type of SNP (coding, non-coding) and expression quantitative trait loci (eQTL). Subsequent functional studies may use knock-down/knock-in techniques to determine whether a specific variant changes a functional outcome.
Transcriptomics
Refers to measures of transcript expression. Gene expression microarrays and the more recently developed RNA-Sequencing (RNA-Seq) (Z. Wang, et al., 2009) are technologies that permit genome-wide measures of transcript expression. Functional experiments to validate specific findings include RT-qPCR and immunoblots to verify transcript- and protein-level changes, respectively, as well as knock-down and/or overexpression experiments of specific transcripts to measure functional consequences of their altered levels.
Epigenomics
Refers to measures of DNA alterations beyond sequence variation. Chromatin immunoprecipitation sequencing (ChIP-Seq) measures DNA sequences bound by specific histones or transcription factors. Global run-on sequencing (GRO-Seq) measures nascent transcription by labeling transcripts engaged with RNA polymerase II (RNAP2) during the run-on process and selecting these fragments for sequencing (Core, et al., 2008). Self-transcribing active regulatory region sequencing (STARR-Seq) measures whether specific DNA fragments have enhanced ability to alter transcription by cloning a ChIP DNA library into a reporter transcript and sequencing the reporter’s RNA abundance (Arnold, et al., 2013). DNase I hypersensitive sites sequencing (DNase-Seq) assesses genome-wide open chromatin by using DNase I to cut freely accessible DNA regions and sequencing the DNA fragments (L. Song and Crawford, 2010). Hi-C assesses three-dimensional architecture of a genome where chromatin interactions occur by incorporating biotin-labeled nucleotides into formaldehyde-fixed chromatin ligation junctions, which are then selected for sequencing (Belton, et al., 2012). Although ChIP-Seq is the most commonly used technique, it is limited in that it alone cannot distinguish whether a transcription factor binds to a site directly versus while tethered to other proteins. Motif analysis helps confirm possible binding motifs, and ChIP-Seq data for co-factors helps to determine protein complexes necessary for transcription. Functional experiments to confirm binding peaks include ChIP-qPCR, and reporter assays to understand which alterations of DNA sequence influence transcription.
Proteomics
Refers to measures of protein expression. High-throughput technologies include mass spectrometry (MS)-based techniques (e.g., tandem-MS (MS/MS)) and gel-based techniques (e.g., differential in-gel electrophoresis (DIGE)) (Altelaar, et al., 2013). The newer MS-based approaches, based on liquid chromatography (LC)-MS that measure peptides via MS/MS within fractionated portions separated by LC, have advanced quantitative proteomics (Altelaar, et al., 2013). Validation of specific proteins begins with immunoblots and ELISA assays to confirm levels of a specific protein, and the subsequent design of experiments to determine whether changes in protein levels influence a functional outcome.
Metabolomics
Refers to measures of metabolite levels. The most widely used approaches are nuclear magnetic resonance (NMR) (Keun and Athersuch, 2011) and high-resolution MS (Junot, et al., 2014). NMR requires minimal sample pre-processing, but MS has higher sensitivity and resolution (Marshall and Powers, 2017). Validation of metabolomics findings includes use of pre-developed panels for specific metabolite species and design of experiments to determine whether changes in metabolite levels influence a functional outcome.
Table 1.
Summary of select findings from omics studies of glucocorticoid responses.
| Omics Approach | Select Findings |
|---|---|
| Genomics | - A GLCCI1 SNP that was nominally associated with glucocorticoid response in patients treated with ICS, was in high linkage disequilibrium with a variant that was associated with decreased GLCCI1 expression in B cells (Tantisira, et al., 2011) - No genome-wide significant variants were found in the largest GWAS of glucocorticoid response in asthma published to date based on subjects in clinical trials of fluticasone furoate and fluticasone propionate (Mosteller, et al., 2017) - A CMTR1 SNP was associated with increased risk of asthma exacerbations in subjects from two Biobanks (Dahlin, et al., 2015) - A locus in the intergenic region of APOBEC3B and APOBEC3C was associated with glucocorticoid responses in Hispanic/Latino and African American children (Hernandez-Pacheco, et al., 2019) - Loci near GRIN3A and ACP1 were associated with increased risk of glucocorticoid-induced osteonecrosis in children with ALL (Karol, et al., 2015; Kawedia, et al., 2011). |
| Transcriptomics | - Many glucocorticoid-responsive genes have been identified in primary airway epithelial and ASM cells, including TSC22D3, FKBP5, PER1, KLF15, DUSP1, CRISPLD2, CEBPD (Himes, et al., 2014; Kan, et al., 2019; Leigh, et al., 2016) - Expression of the pro-apoptotic gene BIM was increased while the anti-apoptotic gene BCL2 was decreased with glucocorticoid exposure in ALL cells (Ploner, et al., 2008). - Glucocorticoid-responsive genes in ALL cells include TXNIP, ZBTB16, PFKFB2 (Carlet, et al., 2010; Schmidt, et al., 2006; Tissing, et al., 2007) |
| Epigenomics | - Some cell type-specific GR-binding sites lack a GRE and are enriched in open chromatin prior to glucocorticoid treatment (Gertz, et al., 2013). - In A549 cells, GR-binding can occur through pre-established chromatin interactions that form a DNA loop. That is, some GRs that are bound to GREs also interact with distal enhancers at so-called GR-tethered binding sites (D’Ippolito, et al., 2018; Vockley, et al., 2016). - GR and p65 cooperatively regulate transcription of anti-inflammatory genes (e.g. SERPINA1 and FOXP4) in BEAS-2B cells (Kadiyala, et al., 2016). - PLCD1 was identified as a KLF15 target gene and a novel repressor of ASM hypertrophy (Sasse, et al., 2017). |
| Proteomics | - Nasal fluid apoH was decreased in patients with allergic rhinitis after glucocorticoid treatment, especially among those who responded to glucocorticoids (H. Wang, Chavali, et al., 2011; H. Wang, Gottfries, et al., 2011) - Serum VDBP had increased expression in patients with steroid-resistant asthma and its levels were highly correlated with proportions of neutrophils and monocytes (H. Jiang, et al., 2016). - PCNA had decreased expression with glucocorticoid exposure in glucocorticoid-sensitive leukemia cell lines and was highly predictive of glucocorticoid response in children with ALL (N. Jiang, et al., 2011) |
| Metabolomics | - Plasma metabolite changes in people following administration of oral dexamethasone reflected changes in pathways related to energy, lipolysis and muscle proteolysis, endogenous steroid production (Bordag, et al., 2015) |
2. Cellular Mechanisms of Glucocorticoid Responses
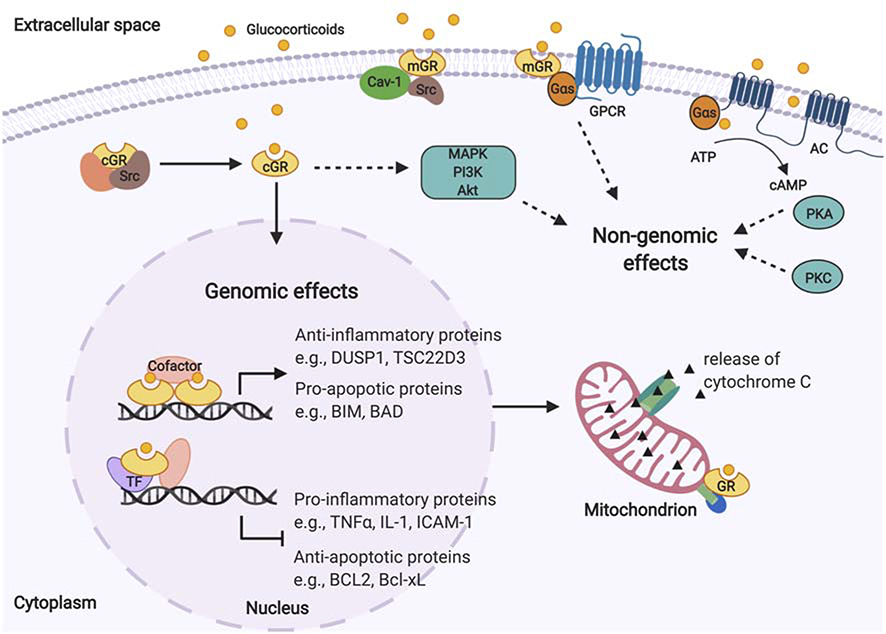
In broad terms, glucocorticoids exert their effects via genomic and non-genomic mechanisms [Figure 1]. The genomic actions involve direct modulation of DNA transcription via the GR. In the absence of glucocorticoids, GRs reside in the cytoplasm as a multi-protein complex with chaperone proteins (heat shock protein 90 [hsp90], heat shock protein 70 [hsp70], and prostaglandin E synthase 3 [p23]) and immunophilins (FK506-binding protein 51 [FKBP51], FK506-binding protein 52 [FKBP52]) (Oakley and Cidlowski, 2013). These complexes preserve GRs in a transcriptionally inactive but high-affinity ligand-binding conformation. Once a GR is bound by its glucocorticoid ligand, the GR is released from the protein complex and is translocated into a cell’s nucleus where GR-GR dimers directly bind to DNA sequences at glucocorticoid response elements (GREs), the GR-specific target motifs that activate gene transcription (Newton, 2014; Oakley and Cidlowski, 2013). In addition to binding to GREs directly, GRs interact with other transcription factors, such as activator protein 1 (AP-1), nuclear factor κB (NF-κB) and signal transducer and activator of transcription 3 (STAT3), to regulate transcription of genes (Biddie, et al., 2011; Langlais, et al., 2012; Rao, et al., 2011; Uhlenhaut, et al., 2013). A prominent effect of glucocorticoids is the reduction of inflammation, which is achieved via 1) the direct activation of anti-inflammatory proteins such as mitogen-activated protein kinase phosphatase-1 (MKP-1, aka dual specificity phosphatase 1 [DUSP1]) that dephosphorylates mitogen-activated protein kinases (MAPKs) (Moosavi, et al., 2017) and glucocorticoid-induced leucine zipper (GILZ, aka TSC22 domain family member 3 [TSC22D3]) (Bereshchenko, et al., 2019), and 2) the repression of pro-inflammatory gene transcription through interactions with transcription factors like AP-1 and/or NF-κB that lead to reduced levels of pro-inflammatory cytokines and cytokine-induced proteins such as tumor necrosis factor α (TNFα), interleukin 1 (IL-1) and intercellular adhesion molecule-1 (ICAM-1) (Newton, 2014; Newton and Holden, 2007; Oakley and Cidlowski, 2013). Glucocorticoids reduce expression of pro-inflammatory cytokines by decreasing their mRNA stability (Amano, et al., 1993; Gille, et al., 2001; Lasa, et al., 2001), and they have been postulated to bind to negative GREs (nGREs) that inhibit their transcription (Surjit, et al., 2011) although genome-wide GR-binding studies do not support this mechanism (Kadiyala, et al., 2016; Oh, et al., 2017; Rao, et al., 2011).
Figure 1. Overview of genomic and non-genomic glucocorticoid mechanisms of action.
GRs reside in the cytoplasm as a multi-protein complex. After GRs are bound by glucocorticoids, they are released and translocate into cell nuclei, where they exert genomic effects by directly binding to DNA sequences or interacting with other transcription factors, leading to anti-inflammatory and apoptotic effects. Glucocorticoids exert non-genomic effects on several signaling processes via non-specific interactions with the cell membrane, cytosolic GRs, and membrane-bound GRs. AC: adenylyl cyclase; Akt: protein kinase B; ATP: adenosine triphosphate; BCL2: BCL2 apoptosis regulator; Bcl-xL: BCL2 like 1; BIM: BCL2 like 11; BAD: BCL2 associated agonist of cell death; cAMP: cyclic adenosine monophosphate; Cav-1: caveolin-1; DUSP1: dual specificity phosphatase 1; cGR: cytosolic glucocorticoid receptor; eNOS: endothelial nitric oxide synthase; GPCR: G protein-coupled receptors; GR: glucocorticoid receptor; Gs: stimulatory G protein; ICAM-1: intercellular adhesion molecule-1; IL-1: interleukin 1; MAPK: mitogen-activated protein kinases; mGR: membrane-bound glucocorticoid receptor; PI3K: phosphoinositide 3-kinase; PKA: protein kinase A; PKC: protein kinase C; Src: proto-oncogene tyrosine-protein kinase Src; TF: transcription factor; TNFα: tumor necrosis factor α; TSC22D3: TSC22 domain family member 3.
The GR is encoded by the nuclear receptor subfamily 3 group C member 1 (NR3C1) gene, which generates two major splice variants, GRα and GRβ. Each GR mRNA variant also has a set of translation initiation sites that results in multiple GR translational isoforms (Oakley and Cidlowski, 2013). The GRα splice variant is the classic GR protein that mediates the action of glucocorticoids as described in the previous paragraph. GRβ has an incomplete ligand-binding domain and is thus unable to bind glucocorticoids. However, GRβ acts as a dominant negative inhibitor when co-expressed with GRα by competing for GREs, as GRα-GRβ heterodimers antagonize the GRα-GRα functions on glucocorticoid-responsive genes (Oakley and Cidlowski, 2013). Increased GRβ levels and a decreased GRα:GRβ ratio have been associated with glucocorticoid resistance in many cell types, suggesting that GRβ could be a biomarker of glucocorticoid response (Lewis-Tuffin and Cidlowski, 2006). The activity of the GR itself is influenced by several factors, including post-translational modifications, such as phosphorylation at certain amino acids, which alter its role in gene transcription (Oakley and Cidlowski, 2013).
Known genomic mechanisms via which glucocorticoids exert apoptotic effects include increased expression of genes encoding pro-apoptotic proteins (e.g., BCL2 like 11 [BIM], BCL2 associated agonist of cell death [BAD]) and decreased expression of genes encoding anti-apoptotic proteins (e.g., BCL2 apoptosis regulator [BCL2] and BCL2 like 1 [BCL2L, aka Bcl-xL]) that disrupt mitochondrial membrane potential and lead to caspase 9- and caspase 3-induced apoptosis (Gruver-Yates and Cidlowski, 2013). Another apoptotic mechanism is the translocation of GRs to mitochondria, where they interact with pro-apoptotic proteins of the BCL2 family in developing T cells (Prenek, et al., 2017). Apoptotic effects of glucocorticoids have been observed in B cells, T cells, macrophages and eosinophils, while anti-apoptotic effects have been observed in breast epithelium, ovarian follicular cells, and hepatocytes, as well as some cancer cells that do not respond to glucocorticoid therapy. These cell type-specific differences appear to result from differences in the balance of pro- and anti-apoptotic proteins of the BCL2 family (Gruver-Yates and Cidlowski, 2013).
Some genomic actions of glucocorticoids occur rapidly, with robust transcriptional changes observed in airway epithelial cells after 10 min of glucocorticoid exposure (Sasse, et al., 2019). Additionally, glucocorticoids elicit changes to intracellular calcium mobilization, muscle reactivity and reactive oxygen/nitrogen species via non-genomic responses within seconds to minutes that may play a role in the treatment of asthma (Panettieri, et al., 2019). These non-genomic effects have been proposed to occur via 1) non-specific interactions with the cell membrane, 2) interactions with cytosolic GRs, and 3) interactions with membrane-bound GRs (I. H. Song and Buttgereit, 2006):
Glucocorticoid molecules intercalate in cell membranes and may physicochemically interact with them to influence ion transport, as evidenced by experiments in which bronchial epithelial cells exposed to dexamethasone had reduced basal calcium (Ca2+) levels within 30 s, a process that was not prevented by the GR antagonist, RU486, or the protein synthesis inhibitor cycloheximide, suggesting the involvement of a non-genomic and GR-independent mechanism (Urbach, et al., 2002). Further, the rapid calcium decrease induced by glucocorticoids was inhibited by a Ca2+-ATPase pump inhibitor, an adenylyl cyclase (AC) inhibitor and a protein kinase A (PKA) inhibitor, suggesting that AC/PKA signaling mediated the changes in calcium flux (Urbach, et al., 2002).
In vascular endothelial cells exposed to high-dose glucocorticoids, there was rapid stimulation of phosphoinositide 3-kinase (PI3K) and protein kinase B (Akt) pathways that led to the activation of nitric oxide (NO)- and endothelial NO synthase (eNOS)-dependent vasorelaxation (Hafezi-Moghadam, et al., 2002). The increased eNOS activity that became significant after 10 min of glucocorticoid exposure was abrogated by the GR antagonist RU486, a PI3K inhibitor and an eNOS inhibitor, but not by a transcriptional inhibitor, suggesting that non-transcriptional effects of cytosolic GRs mediated the vasorelaxation (Hafezi-Moghadam, et al., 2002).
Membrane-bound GRs may activate downstream signaling on their own or through interactions with G protein-coupled receptors (GPCRs). In CCRF-CEM cells exposed to a membrane-impermeable glucocorticoid (i.e. bovine serum albumin (BSA)-conjugated cortisol), expression changes in proteins associated with cell death and apoptosis were observed at 5 min, and the presence of membrane-bound GR on the cell surface was confirmed using in situ proximity ligation assays (Vernocchi, et al., 2013). Evidence from in situ proximity ligation assays and immunoblots suggests that some GRs become membrane-bound in association with caveolin-1 (Cav-1) in A549 lung epithelial cells, U2OS osteosarcoma cells and MCF-7 breast adenocarcinoma cells, but not CCRF-CEM or Jurkat cells (Matthews, et al., 2008; Vernocchi, et al., 2013). Membrane-bound GRs that were observed in human airway smooth muscle (ASM) cells were proposed to mediate the rapid (within minutes) induction of 3’, 5’-cyclic adenosine monophosphate (cAMP) levels by glucocorticoids because both cortisol and BSA-conjugated cortisol exposure resulted in an equal rise in cAMP levels (Koziol-White, et al., 2020; Nunez, et al., 2019). Knockdown of GNAS, the gene encoding the G protein subunit alpha S (Gαs), abrogated the effect, suggesting that membrane-bound GRs interact with GPCRs to modulate cAMP levels (Nunez, et al., 2019).
At a cellular level, various mechanisms have been linked to differences in glucocorticoid responses. Increased p38 MAPK-activated GR phosphorylation in peripheral blood mononuclear cells and alveolar macrophages have been reported to contribute to decreased GR nuclear translocation in patients with severe asthma (Bhavsar, et al., 2008; Bhavsar, et al., 2010; Mercado, et al., 2012). Reduced GR translocation has also been observed in ASM cells from severe asthma patients (Chang, et al., 2015) and attributed to impaired GR phosphorylation due to high levels of the serine/threonine protein phosphatase 5 (PP5) (Bouazza, et al., 2012; Chachi, et al., 2017; Chachi, et al., 2013). Another mechanism proposed to contribute to differences in glucocorticoid responses in severe asthma is reduced nuclear histone deacetylase 2 (HDAC2) activity that may modify inflammatory gene expression (Hew, et al., 2006; L. B. Li, et al., 2010).
3. Pharmacogenomics: Genetic Variants that Influence Glucocorticoid Responses
Glucocorticoid pharmacogenomics studies are challenged by the difficulty that collecting data on sufficiently large numbers of patients with well-defined drug response phenotypes entails, selecting appropriate outcomes, ensuring that the same outcomes are used across studies to test for their generalizability, and ensuring that people are taking medications as intended while controlling for modifiers (e.g., use of other drugs). For example, glucocorticoid pharmacogenomics studies in asthma have been based on cohorts of patients taking inhaled corticosteroids (ICS) over variable time courses while using the number of asthma exacerbations and/or improved lung function over a time period as outcomes. Some of these cohorts consist of subjects in clinical trials that had wash-out periods and monitored drug use carefully (Denlinger, et al., 2007; Mosteller, et al., 2017; Strunk and Childhood Asthma Management Program Research, 2007), while other studies included subjects with varying levels of disease severity and no medication adherence data (Dahlin, et al., 2015; Hernandez-Pacheco, et al., 2019).
Early genetics studies of glucocorticoid responses focused on single nucleotide polymorphisms (SNPs) of the NR3C1 gene due to its direct role in glucocorticoid action. NR3C1 polymorphisms reported to be associated with glucocorticoid responses include the functional variants N363S (rs56149945) and ER22/23EK (rs6189 and rs6190), and the intronic variant rs41423247 (aka BclI polymorphism) (Oakley and Cidlowski, 2013). N363S and ER22/23EK were first identified by polymerase chain reaction/single-strand conformation analysis of NR3C1 in a genetic association study of 20 healthy subjects who had reduced response according to a dexamethasone suppression test versus 20 control subjects (Koper, et al., 1997). Carriers of the BclI polymorphism, originally identified by the digestion of DNA restriction enzyme BclI (Murray, et al., 1987), and carriers of N363S were found to have increased sensitivity to glucocorticoids in candidate gene studies (Huizenga, et al., 1998; van Rossum, et al., 2003), while healthy individuals who were carriers of ER22/23EK had decreased sensitivity (van Rossum, et al., 2002). Beyond results of candidate gene studies of dexamethasone suppression tests, the only NR3C1 SNP that has been reported as significantly associated with therapeutic response to glucocorticoids is the BclI polymorphism among ALL patients (Pavlovic, et al., 2019). None of these candidate gene associations have been observed in subsequent genome-wide association studies (GWAS).
Several loci associated with glucocorticoid responses in asthma patients have been identified via GWAS (Farzan, et al., 2017; Keskin, et al., 2019). Tantisira and colleagues performed a GWAS of ICS responsiveness in 181 white asthma-proband trios from the Childhood Asthma Management Program (CAMP) and four independent clinical trial cohorts for attempted replication. In a discovery stage, the quantitative outcome selected was residuals of the difference between the forced expiratory volume in 1 second (FEV1) following months of budesonide treatment after adjustment for age, sex, and height. Of the 13 top variants from this stage that were selected for replication, the rs37972 SNP near glucocorticoid induced 1 (GLCCI1), was the only nominally associated one (p <0.05). Functional analysis showed that SNP rs37973, which was in high linkage disequilibrium with rs37972, was associated with decreased GLCCI1 expression in B cells, suggesting that rs37973 leads to decreased glucocorticoid responsiveness via changes in GLCCI1 expression (Tantisira, et al., 2011). The GLCCI1 associations have not been replicated in some independent studies (Hosking, et al., 2014; Vijverberg, et al., 2014). A GWAS of ICS response defined as the change in percent FEV1 after four weeks of ICS treatment performed in 189 Korean subjects with asthma found that the top SNPs (p-value <10−6) mapped to allantoicase (ALLC) (T. J. Park, et al., 2014). A GWAS of this same cohort with changes in asthma symptom scores after ICS treatment as the outcome identified three SNPs near rhabdomyosarcoma 2 associated transcript (RMST), long intergenic non-protein coding RNA 2140 (LINC02140), and F-box and leucine rich repeat protein 7 (FBXL7) as nominally replicated in children but not adults (H. W. Park, et al., 2014). A subsequent GWAS that considered ICS dose was performed in 120 randomized asthma patients who received varying doses of ICS by modeling association with dose-dependent response to ICS (measured as percent change in FEV1) (Y. Wang, et al., 2015). The authors identified five loci that mapped to metabolic genes related to lung function and asthma risk, three of which reached genome-wide significance (p-value <5 × 10−8), indicating that the pharmacological model had increased statistical power to detect potential associations (Y. Wang, et al., 2015). To date, the largest published GWAS of ICS response, defined as change in FEV1 after 8–12 weeks, conducted using data from 2,675 asthma subjects who were part of GlaxoSmithKline clinical trials of fluticasone furoate and fluticasone propionate, found that no genetic variants met pre-specified genome-wide significant statistical criteria (Mosteller, et al., 2017). Although this study found that GLCCI1 variants were nominally associated with change in FEV1, its main results suggest that common genetic variants are unlikely to serve as biomarkers of steroid responsiveness in asthma.
In several population studies and biobanks in which glucocorticoid responses GWAS could be performed, longitudinal pulmonary function data is not available. Such studies often use number of asthma exacerbations in a defined time period as an outcome. For example, a GWAS based on two biobanks (i.e., BioVU at Vanderbilt University Medical Center in Tennessee (369 patients), and the Personalized Medicine Research Project at the Marshfield Clinic in Wisconsin (437 patients)) identified a SNP in cap methyltransferase 1 (CMTR1) as associated with increased risk of exacerbations in both populations (Dahlin, et al., 2015). The first and largest GWAS study of ICS response in Hispanics/Latinos and African Americans was carried out in 1,347 children treated with ICS; using meta-analysis of GWAS conducted in each racial/ethnic group that examined whether asthma exacerbations occurred during the 12 months preceding study enrollment, a novel locus located in the intergenic region of apolipoprotein B mRNA editing enzyme catalytic subunit 3B (APOBEC3B) and apolipoprotein B mRNA editing enzyme catalytic subunit 3C (APOBEC3C) was significant (Hernandez-Pacheco, et al., 2019).
GWAS relating side effects of glucocorticoids to genetic variants have also been conducted. To address the fact that long-term, intermittent oral corticosteroid use in children with asthma leads to significant decrements in bone mineral accretion (BMA), a GWAS was conducted to relate multiple oral corticosteroid bursts with bone mineral density in 489 children from the CAMP trial (Park, et al., 2015). Two SNPs that mapped to tubulin folding cofactor D (TBCD) and tubulin gamma 1 (TUBG1) were identified as associated with BMA, and decreased BMA was more strongly correlated with increased dose of prednisone as the number of risk alleles from these two SNPs increased from zero to three (Park, et al., 2015). A recent study sought to identify variants involved in ICS-induced adrenal suppression by conducting a discovery GWAS in 499 children with asthma and attempting to replicate results in 81 children with asthma and 78 adults with COPD (cases were 7% children and 22% adults who developed adrenal suppression) (Hawcutt, et al., 2018). A single locus near platelet derived growth factor D (PDGFD) was significantly associated with adrenal suppression in both the discovery (p-value =5.8 × 10−8) and replication cohorts (p <0.05) (Hawcutt, et al., 2018).
In ALL patients, GWAS have been performed to identify variants associated with side effects following glucocorticoid treatment (Pavlovic, et al., 2019). Osteonecrosis is a major side effect of glucocorticoids in the remission-induction therapy of childhood ALL within the first 2 years of treatment. GWAS of glucocorticoid-induced osteonecrosis found that variants in the genes glutamate [NMDA] receptor subunit 3A (GRIN3A) and acid phosphatase-1 (ACP1) were associated with increased risk of osteonecrosis (Karol, et al., 2015; Kawedia, et al., 2011). Because dexamethasone has become the preferred glucocorticoid in ALL treatment due to its greater anti-leukemic efficacy, studies have sought to identify genetic variants that contribute to the development of dexamethasone-induced side effects to guide clinical practice. A recent GWAS that fully considered relationships among 14 pleiotropic phenotypes in 391 patients with ALL identified the F2R-like trypsin receptor (F2RL1) locus as having pleiotropic effects on osteonecrosis and thrombosis, adverse effects of dexamethasone treatment (Ramsey, et al., 2017). Further validation of these associations may lead to biomarker tests of glucocorticoid sensitivity and likelihood of developing side effects induced by glucocorticoids in the treatment of ALL.
Relating genetic variants identified via GWAS to mechanisms of glucocorticoid responses is challenging. Such variants, as well as those associated with asthma and/or ALL, may influence expression of the GR or its primary or secondary targets. For example, the asthma-associated gene tumor necrosis factor α-induced protein 3 (TNFAIP3) (X. Li, et al., 2012) is a primary GR target gene and key regulator of GR signaling whose glucocorticoid-induced expression is further enhanced by the addition of a long-acting β2-agonist, a combination that is used in asthma therapy (Altonsy, et al., 2017). The association of TNFAIP3 with asthma may thus reflect an influence on pathways related to glucocorticoid responses.
4. Transcriptomics and Epigenomics: Cell Type-Specific Gene Transcription and its Regulation by Glucocorticoid Receptors
Measures of the transcriptome, or RNA expressed in a system, can be captured by technologies such as gene expression microarrays and RNA sequencing (RNA-Seq) (Kan, et al., 2017). Many transcriptomic studies of glucocorticoid responses in humans and in vitro models have been conducted for various cell and tissue types, including macrophages and primary cells derived from ASM and airway epithelium, as well as transformed cell lines such as ALL, lymphoblastoid cells, the human mammary epithelial cell line MCF10A-Myc, and osteosarcoma U2OS cells (Kan, et al., 2019). Chromatin immunoprecipitation sequencing (ChIP-Seq) is an epigenomic technique used to measure transcription factor binding sites on a genome-wide scale. Most GR ChIP-Seq studies integrate transcriptomic and ChIP-Seq data from cells under the same treatment conditions because 1) transcriptomic data helps to identify gene targets of GR-binding events that are actually differentially expressed, and 2) ChIP-Seq results can be used to distinguish genes that are primary versus downstream GR targets. Alternatively, ChIP-Seq for RNA polymerase II (RNAP2) can be captured along with that of the GR to identify which transcription factor binding sites are more likely to be active, as RNAP2 binding to DNA provides some evidence of dynamic transcription (Kadiyala, et al., 2016; Sasse, et al., 2017). More recent techniques such as global run-on sequencing (GRO-Seq) can directly measure nascent transcription and offer more direct evidence that a site has ongoing mRNA transcription (Core, et al., 2008). Other approaches that have been applied to study glucocorticoid responses include 1) STARR-Seq, which measures enhancer ability of identified GR-binding sites by cloning ChIP fragments into a transcript reporter (Arnold, et al., 2013), 2) DNase I hypersensitive sites sequencing (DNase-Seq), which measures open chromatin structures (L. Song and Crawford, 2010), and 3) Hi-C, which measures topological structures of chromatin interactions (Belton, et al., 2012).
Consistent with the fact that genomic actions of glucocorticoids include direct modulation of gene transcription, transcriptomic studies have readily identified gene expression changes induced by glucocorticoids. Dexamethasone is the most commonly used synthetic glucocorticoid in transcriptomic studies, although some have involved the clinically used formulations prednisolone and budesonide. While different glucocorticoids may have distinct actions, transcriptomic studies have found that cellular responses to exposure to various glucocorticoid drugs are similar (Bindreither, et al., 2014; Leigh, et al., 2016). Asthma-related transcriptomic studies have largely focused on airway structural cells (i.e., ASM and airway epithelium). An early microarray study investigated the effects of dexamethasone on ASM cells and identified Krüppel like factor 15 (KLF15), a gene encoding a transcription factor, as a novel modulator of airway hyperresponsiveness (Masuno, et al., 2011). Functional studies related to KLF15 have been conducted since its discovery, showing that it is a primary GR target with GR-binding sites, and that the physical interaction between GR and KLF15 contributes to combinatorial transcriptional regulation of genes such as those encoding amino acid-metabolizing enzymes (Sasse, et al., 2017; Sasse, et al., 2013).
RNA-Seq studies comparing transcriptomic changes in ASM cells derived from donors without any chronic disease that were treated with budesonide versus vehicle control have found thousands of differentially expressed genes, including cysteine rich secretory protein LCCL domain containing 2 (CRISPLD2) and CCAAT enhancer binding protein delta (CEBPD) (Himes, et al., 2014; Kan, et al., 2019). ASM cells derived from fatal asthma donors retain differences in proliferative and contractile outcomes in vitro, suggesting that their responses to glucocorticoids differ from that of donors without asthma (Burgess, et al., 2008; Goncharova, et al., 2006; Joubert, et al., 2005; Oliver, et al., 2006; Panettieri, et al., 1989). Our recent study comparing transcriptomic changes in response to budesonide exposure between fatal asthma and non-asthma donor-derived ASM cells, however, found that gene expression profiles in response to glucocorticoid exposure did not differ according to asthma status (Kan, et al., 2019). While transcriptomic changes were consistent across ASM cells, an integrative analysis of seven cell types found that only two well-known glucocorticoid-responsive genes (FK506 binding protein 5 [FKBP5] and TSC22D3) had increased levels across ASM cells, childhood ALL cells, macrophages, lymphoblastoid cell lines and U2OS; other glucocorticoid-responsive genes such as period circadian clock 1 (PER1), KLF15 and CRISPLD2 had increased levels across some cell types (Kan, et al., 2019). These results demonstrate that some of the cell type-specific responses to glucocorticoids occur at the level of gene expression. Given the large number of differentially expressed genes identified with glucocorticoid exposure, it is difficult to prioritize genes for further study based on transcriptomic data alone.
Transcriptomic studies of glucocorticoid responses in people have also been performed. One study investigated the effects of a 2-week course of oral prednisolone on patients with mild asthma by applying RNA-Seq to ASM extracted via laser capture microdissection from bronchoscopy samples (Yick, et al., 2013). Comparing samples from 6 patients assigned to glucocorticoid treatment versus 6 patients assigned to placebo, this study found that 15 genes were significantly differentially expressed between groups, and two of them, family with sequence similarity 12 (FAM129A) and synaptopodin 2 (SYNPO2), were also associated with airway hyperresponsiveness. The relatively small number of genes identified by this study relative to in vitro studies was likely due to the difficulty of obtaining ASM via laser capture microdissection and low sequencing depth. Comparison of airway epithelial transcriptomes in patients with asthma receiving a 10-week course of inhaled fluticasone versus placebo found that chloride channel accessory 1 (CLCA1), periostin (POSTN), and serine family B member 2 (SERPINB2) had decreased expression in those who took fluticasone (Woodruff, et al., 2007). Another study that compared transcriptomic changes in airway epithelial cells of healthy men receiving inhaled budesonide versus placebo identified several of the same differentially expressed genes (e.g., TSC22D3, PER1, KLF15, DUSP1, FKBP5, CEBPD, CRISPLD2) (Leigh, et al., 2016) that have been observed in in vitro ASM studies (Himes, et al., 2014; Kan, et al., 2019).
Transcriptomic studies in ALL cells have identified glucocorticoid-responsive genes that are largely distinct from those observed in airway structural cells. An early transcriptomic study that compared expression changes in prednisolone-resistant vs. prednisolone-sensitive ALL cells derived from 173 children with ALL found increased expression of the anti-apoptotic gene MCL1 apoptosis regulator (MCL1) of the BCL2 family and decreased expression of several transcription-associated genes (e.g. CCCTC-binding factor [CTCF]) that are thought to affect apoptosis in prednisolone-resistant cells (Holleman, et al., 2004). Comparison of transcriptomic data from 13 children with ALL prior to versus after glucocorticoid treatment found that genes from the BCL2 family related to apoptotic pathways were differentially expressed: the pro-apoptotic gene BIM had increased expression, while the anti-apoptotic gene BCL2 had decreased expression with glucocorticoid treatment (Ploner, et al., 2008). Other glucocorticoid-responsive genes in ALL included the tumor suppressor genes thioredoxin interacting protein (TXNIP), zinc finger and BTB domain containing 16 (ZBTB16), and 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2 (PFKFB2) (Carlet, et al., 2010; Schmidt, et al., 2006; Tissing, et al., 2007). A subsequent study of these genes found that PFKFB2 was not an essential regulator of the anti-leukemic effects of glucocorticoids, as its overexpression in a human T-cell lymphoblastic leukemia cell line (i.e., CCRF-CEM) had little effect on glucocorticoid-induced apoptosis (Carlet, et al., 2010). Via integration of transcriptomic data with a genome-wide RNA-interference screen, a recent study of B-cell precursor ALL cells confirmed that glucocorticoids altered expression of apoptosis-related genes such as BCL2, BCL2 like 11 (BCL2L11), TXNIP and ZBTB16, and B-cell development genes (e.g. phosphatidylinositol 3-kinase δ [PI3Kδ]) (Kruth, et al., 2017). The ALL-specific transcriptional signatures related to apoptosis are consistent across these studies, but further work is needed to determine whether any specific genes can serve as biomarkers of glucocorticoid responses or as novel drug targets.
Several GR ChIP-Seq studies have used the lung epithelial carcinoma A549 cell line, including an early study that used both ChIP-Seq and RNA-Seq to identify genes that were direct GR targets (Reddy, et al., 2009). Major findings of this study were that 1) there were many more GR-binding sites than genes differentially expressed in response to dexamethasone exposure (i.e. 4392 vs. 234); 2) 70% of the 234 differentially expressed genes had GR-binding sites that were further than 10 kb of gene a transcription start site (TSS); and 3) the GR-binding sites of genes with decreased expression had a median distance of 100 kb to the TSS, which was much greater than the distance of genes with increased expression (Reddy, et al., 2009). A subsequent study that used ChIP-Seq and DNase-Seq data to identify shared and cell type-specific GR-binding sites between A549 and human endometrium-derived ECC-1 cells found that only a small proportion of binding sites (i.e., 7.7% of 20,190) were shared (Gertz, et al., 2013). Most cell type-specific GR-binding sites lacked GREs and were in open chromatin regions prior to glucocorticoid treatment, while the shared GR-binding sites were more likely to have GREs (Gertz, et al., 2013). According to a STARR-Seq study of A549 cells, only 13% of GR-binding sites had dexamethasone-induced activity, and 80% of these sites had a GRE (Vockley, et al., 2016). Introduction of a GRE to non-responsive GR-binding sites greatly increased their enhancer activity, leading authors to propose a model whereby non-responsive GR-binding sites modulate gene transcription via distal interactions with GREs (Vockley, et al., 2016). Recent Hi-C studies have found that GR-binding occurs through pre-established chromatin interactions rather than ones established after glucocorticoid exposure, supporting a model in which looped GR-binding sites regulate gene expression via existing chromatin structures (D’Ippolito, et al., 2018; McDowell, et al., 2018).
Efforts to identify primary GR gene targets via which glucocorticoids exert asthma-related therapeutic effects have been undertaken with ChIP-Seq in airway structural cells. To investigate glucocorticoid-induced interactions between GR and NF-κB, one study performed ChIP-Seq for GR, p65 (a subunit of NF-κB) and RNAP2 in human epithelial Beas-2B cells treated with dexamethasone, TNFα or both (Kadiyala, et al., 2016). The authors found that GR recruited p65 to dimeric GR binding sites across the genome, and this GR-p65 cooperation increased expression of genes such as the well-known anti-inflammatory and injury response genes serpin family A member 1 (SERPINA1) and forkhead box P4 (FOXP4). Consistent with previous reports, the authors also identified GR-p65 cooperative binding sites in two genes encoding negative regulators of NF-κB, interleukin-1 receptor-associated kinase 3 (IRAK3) (Miyata, et al., 2015) and TNFAIP3 (Sasse, et al., 2016). For genes whose expression was repressed by glucocorticoids, however, the study by Kadiyala et al, as well as a ChIP-Seq study in mouse bone-marrow-derived macrophages (Oh, et al., 2017), support mechanisms whereby glucocorticoids activate transcription of negative regulators of NF-κB that inhibit its function directly, rather than via GR-p65 tethering to suppress expression of pro-inflammatory genes. To further understand how GR and NF-κB interact to repress cytokines, Sasse and colleagues used GRO-Seq to measure nascent transcription in BEAS-2B cells after 10 min and 30 min of glucocorticoid and/or TNFα exposure, along with high-resolution micrococcal nuclease (MNase) accessibility assays to measure the chromatin accessibility at identified enhancers (Sasse, et al., 2019). Glucocorticoids repressed activity of TNFα-induced enhancers after 10 min. Among the enhancers overlapping with GR-binding peaks, GR-repressed enhancers had increased chromatin accessibility but not significant GR occupancy at 10 min, suggesting that early repression by GR occurred via direct internucleosomal accessibility rather than local GR occupancy (Sasse, et al., 2019). Negative regulators of NF-κB such as TNFAIP3 had increased transcription, but not protein expression, after 30 min of glucocorticoid exposure, suggesting that a secondary wave of repression was due to later effects of genes induced by the transcriptional cooperation of GR and NF-κB (Sasse, et al., 2019). ChIP-Seq studies have also been undertaken to investigate asthma-related candidate genes, including one study that measured the effect of KLF15 on ASM glucocorticoid responses by obtaining transcriptomic and ChIP-Seq data for GR and RNAP2, with and without dexamethasone exposure and KLF15 overexpression (Sasse, et al., 2017). It found that phospholipase C delta 1 (PLCD1) was a KLF15 target and novel repressor of ASM hypertrophy.
5. Proteomics and Metabolomics: Identifying Downstream Biomarkers of Glucocorticoid Responses
Proteomics and metabolomics approaches provide snapshots of many proteins and metabolites in a cell or tissue, which may reflect current physiological status more closely than the genome, transcriptome or epigenome (Kan, et al., 2017). Proteomics and metabolomics are not fully unbiased, in that the identification of specific species of proteins and metabolites relies on having reference databases to link measured spectra to them, and it is not currently possible to measure all proteins or metabolites in a system simultaneously (Kan, et al., 2017). Nonetheless, proteomics and metabolomics have tremendous potential to aid in biomarker discovery, disease phenotyping, and identification of targets for mechanistic studies of disease and drug responses.
Several proteomics studies based on patients with asthma who used glucocorticoids have sought to identify response biomarkers. By comparing levels of proteins in endobronchial biopsies from asthma patients taking budesonide versus placebo, one study identified a protein signature representing the acute phase response pathway (O’Neil, et al., 2011). This pathway was also found in two studies that compared nasal fluid proteomic differences 1) in patients with intermittent allergic rhinitis before and after fluticasone treatment (H. Wang, Chavali, et al., 2011), and 2) in patients with seasonal allergic rhinitis who showed high versus low response to fluticasone (H. Wang, Gottfries, et al., 2011). Apolipoprotein H (apoH) was a particularly promising candidate biomarker corresponding to the acute phase response pathway that had decreased levels in patients after fluticasone treatment, especially among those who had the greatest clinical response (H. Wang, Chavali, et al., 2011; H. Wang, Gottfries, et al., 2011). Seven differentially expressed proteins were identified by comparing serum proteomes from 6 patients with “steroid-resistant asthma” versus 6 with “steroid-sensitive asthma” using differential in-gel electrophoresis (DIGE) and MS approaches (H. Jiang, et al., 2016). Of these proteins, vitamin D-binding protein (VDBP) had the greatest increase in the “steroid-resistant” asthma group, and serum VDBP levels were highly correlated with proportions of neutrophils and monocytes, suggesting that VDBP may distinguish asthma patients who respond clinically to glucocorticoids (H. Jiang, et al., 2016). An in vitro proteomics study sought to characterize changes in protein expression due to dexamethasone by using high-resolution MS in healthy donor-derived peripheral blood mononuclear cells exposed to: 1) vehicle control, 2) inflammatory agents (lipopolysaccharide and phytohemagglutinin) for 1 hour, and 3) inflammatory agents for 1 hour with subsequent addition of dexamethasone for 3 hours (Bileck, et al., 2014). The authors found that known pro-inflammatory proteins such as interleukin 1β (IL-1β), interleukin 6 (IL-6), C-X-C motif chemokine ligand 2 (CXCL2), C-X-C motif chemokine ligand 1 (GROα), pentraxin 3 (PTX3) and TNF alpha induced protein 6 (TSG6) had increased levels after exposure to the inflammatory agents. After exposure to dexamethasone, some proteins (i.e. IL-1β, IL-6, CXCL2, GROα) had decreased levels, while levels of others (i.e. PTX3, TSG6) remained the same (Bileck, et al., 2014). These proteomics studies offer hypotheses regarding drug response biomarkers that could be tested prospectively or serve as candidates for mechanistic studies.
Proteomics studies in ALL cell lines that are resistant versus sensitive to glucocorticoids have yielded other candidate biomarkers. A study that compared proteomic changes in four clinically relevant leukemia cell lines (i.e., the glucocorticoid-resistant cell line REH; the glucocorticoid-sensitive cell lines PreB 697, Sup-B15 and RS4;11) before and after exposure to prednisolone identified 77 significantly differentially expressed proteins in the glucocorticoid-sensitive cell lines and 17 in the glucocorticoid-resistant cell line (N. Jiang, et al., 2011). Among those proteins, proliferating cell nuclear antigen (PCNA) showed decreased expression in the glucocorticoid-sensitive cell lines and was validated in 43 paired bone marrow samples from children with newly diagnosed ALL versus 7 days after prednisolone treatment. The difference in PCNA expression was highly predictive of prednisolone response in patients independent of their molecular subtypes, suggesting that it could be a universal prognostic marker for treatment outcome (N. Jiang, et al., 2011). Another study that applied an MS-based approach to compare proteomes of the glucocorticoid-sensitive cell line PreB 697 versus its glucocorticoid-resistant sub-clone R3F9 when exposed to dexamethasone versus vehicle control found that paired box 5 (PAX5), a transcription factor critical to B-cell development, had significantly decreased expression in the glucocorticoid-resistant cell line, as did its transcription target CD19 (Nicholson, et al., 2015).
Comparison of proteins in cytosolic versus nuclear fractions of CCRF-CEM cells stimulated with BSA-conjugated cortisol or vehicle control for 5, 15 and 90 minutes revealed that proteins activated by the membrane-bound GR were specific to cell death and apoptosis pathways (Vernocchi, et al., 2013). These protein expression changes were confirmed to be independent of cytosolic GR activity: expression of the well-known cytosolic GR-target gene GILZ was induced as expected by cortisol but remained unchanged after stimulation with (membrane-impermeable) BSA-conjugated cortisol. Because non-genomic effects of glucocorticoids are also modulated by protein post-translational modifications, future phosphoproteomics studies that focus on protein modifications may yield insights into additional mechanisms that link non-genomic priming events to genomic glucocorticoid responses. Further, such studies may reveal differences in glucocorticoid responses among those with and without asthma.
Metabolomics studies have been performed to predict cellular responses and side effects of glucocorticoids. One such study compared plasma metabolomes from 20 healthy male volunteers before and after receiving a single dose of 4 mg oral dexamethasone. Samples were drawn at three pre-defined points (i.e. morning, midday, and evening) per day from day 1 to day 7, and dexamethasone was administered on day 3. In total, seven untreated (during days 1–3) and four treated (during days 3–4) samples were obtained per volunteer and underwent MS (Bordag, et al., 2015). Using a mixed-effects analysis of variance (ANOVA) model, the authors found that 150 of 214 measured metabolites significantly changed at least one time point after treatment. They confirmed that dexamethasone had an impact on circadian rhythm: the levels of cortisol and corticosterone fluctuated between mornings and evenings before treatment with dexamethasone. After treatment, however, levels of cortisol and corticosterone were substantially decreased at all-time points (Bordag, et al., 2015). In addition to this suppression of endogenous steroids, other metabolite changes reflected altered energy pathways, lipolysis, and muscle proteolysis that may be related to glucocorticoid side effects (Bordag, et al., 2015). Another metabolomics study that used rats injected with dexamethasone (2.5 mg/kg twice a week) and control saline for 14 weeks found distinctive metabolomics changes in dexamethasone-treated rats that were associated with observed phenotypic changes (Malkawi, et al., 2018). For example, plasma amino acid changes (reduction in phenylalanine, lysine, and arginine levels and increased tyrosine and hydroxyproline levels) were correlated with weight loss and reduced bone formation, while increased sorbitol levels were correlated with hyperglycemia (Malkawi, et al., 2018). These studies support various metabolites as potential biomarkers of glucocorticoid-induced side effects.
To understand the role of metabolic reprogramming in ALL cell death induced by glucocorticoids, a recent study examined metabolomics profiles of human ALL RS4;11 cells exposed to dexamethasone using MS-based proteomics, metabolomics and isotope-tracing experiments (Dyczynski, et al., 2018). Metabolite changes resulting from dexamethasone exposure that were associated with growth arrest, autophagy and catabolism prior to onset of apoptosis included: decreased de novo purine synthesis and polyamine synthesis, decreased fatty acid and sterol synthesis, and increased fatty-acid oxidation. Glucocorticoids also suppressed entry of glucose and glutamine into the citric acid cycle (TCA) cycle. In contrast, the expression of glutamine-ammonia ligase (GLUL) and cellular glutamine content was increased by dexamethasone, suggesting that glutamine synthesis plays a role in autophagy and onset of apoptosis in response to glucocorticoids (Dyczynski, et al., 2018).
Few metabolomics studies have focused on glucocorticoid responses among asthma patients, and most published reports are based on small sample sizes and lack thorough validation. Interesting findings from these studies point to their potential, however, including the identification of metabolites related to lipid and amino acid metabolism that are associated with asthma severity and inhaled corticosteroid dose (Kelly, et al., 2017; Reinke, et al., 2017), which is consistent with gene expression patterns observed in transcriptomic studies (Kan, et al., 2019; Sasse, et al., 2013). Specific findings across omics studies that may reflect major metabolic effects of glucocorticoids include expression changes in some solute carrier (SLC) transporter family genes (e.g. SLC6A6, and SLC2A5) (Kan, et al., 2019) whose products are transporters of amino acid and glucose, thus, may influence metabolism (Bordag, et al., 2015; Malkawi, et al., 2018). GLUL, an enzyme involved in the synthesis of glutamine and amino acid metabolism, has been observed to have both increased gene expression (Himes, et al., 2014; Kan, et al., 2019) and metabolite levels (Dyczynski, et al., 2018) in omics studies.
6. Conclusion and Future Perspectives
Glucocorticoid responses in people and in vitro systems have been studied via several omics approaches, most often to characterize single layers of biological information, but increasingly, data obtained via complementary assays are being integrated. Our ability to generate omics data far exceeds our ability to validate findings that are biologically actionable. However, carefully designed omics experiments can accelerate the discovery of novel mechanisms by focusing experiments on top-ranked genes/proteins that would not otherwise be studied.
Promising findings of genomics studies of glucocorticoid responses include loci associated with changes in lung function after ICS treatment in asthma (e.g. GLCCI1) and side effects in ALL (e.g. GRIN3A). Further replication and experimental validation is necessary for these loci to be translated into clinically impactful findings. Transcriptomic and epigenomic studies have identified reproducible cell type-specific glucocorticoid-responsive genes, including TSC22D3, FKBP5, PER1, DUSP1, KLF15, CRISPLD2 and CEBPD that are the focus of ongoing mechanistic studies. Integration of epigenomic and transcriptomic data has identified novel patterns of GR regulation and helped to distinguish primary from secondary targets of the GR. Thus far, genome-wide transcription factor-binding and histone posttranslational modification studies have not identified asthma-specific signatures. Future studies that do so may be fruitful, as suggested by the fact that epigenetic studies of candidate genes such as CXCL8 and VEGF have found that changes in histone H3 acylation or methylation in their promoter regions leads to enhanced secretion of IL-8 and VEGF in ASM cells from donors with asthma (Clifford, et al., 2012; Clifford, et al., 2015; Kaczmarek, et al., 2019).
Relatively few proteomics and metabolomics glucocorticoid studies have been published. The identification of protein signatures that arise after treatment with glucocorticoids, as well as those that distinguish glucocorticoid-responsive patients, may lead to the identification of novel mechanisms of glucocorticoid action and candidate biomarkers that are more efficacious than those arising from other omics approaches. Metabolomics approaches are also promising for the development of non-invasive biomarkers in exhaled breath condensate, which may have advantages over the use of sputum eosinophils or exhaled nitric oxide (Maniscalco and Motta, 2017; van der Schee, et al., 2013).
Genomic and non-genomic mechanisms of glucocorticoid responses are complementary but have not been connected in detail. Non-genomic mechanisms involve rapid changes in some protein and metabolite levels, and thus, ongoing studies that apply proteomics and metabolomics approaches to measure short-term changes in responses to glucocorticoid exposure are underway. Time course experiments that link these short-term non-genomic changes to short- and long-term genomic processes could address knowledge gaps regarding initial versus sustained glucocorticoid responses and lead to more precise treatments that activate specific glucocorticoid-related pathways. An additional area that has not been adequately explored with omics data is the overlap of glucocorticoid and β2-agonist mechanisms of action to better understand the synergistic effects of these two drugs in the treatment of asthma (Giembycz and Newton, 2015; Newton, et al., 2010; Rider, et al., 2018).
As the volume of omics data continues to increase, its integration offers a cost-effective and convenient means to develop and test new hypotheses. Meta-analyses of single-modality data, which can reduce noise and identify more robust signatures than single datasets, are a first step toward multi-layered integration of data (Diwadkar, et al., 2019; Kan, et al., 2018; Shumyatcher, et al., 2017). The meaningful integration of multi-omics datasets that follows is challenging though, particularly when based on datasets in the public domain that are not fully described. Ideally, multi-omics studies would involve various layers of data collected from the same subjects, so that the fact that some samples arose from the same individual could be taken into account explicitly during analyses, thereby reducing noise due to inter-individual variability that is not due to the condition or treatment in question. Although efforts to collect such comprehensive datasets are underway, most currently available omics studies have been conducted with cells/tissues derived from different people. Raw data from transcriptomic and epigenomic studies is maintained in public repositories such as the Gene Expression Omnibus (GEO) (Clough and Barrett, 2016) and Sequence Read Archive (SRA) (Leinonen, et al., 2011). Due to its much larger size and concerns regarding potential subject re-identification, genomic data is not as readily accessible. However, summary statistics for most published studies are available in resources such as the GWAS Catalog (Buniello, et al., 2019; MacArthur, et al., 2017). Proteomics and metabolomics data are not widely distributed, partly because there is less consensus regarding their format and places where data should be deposited. ProteomeXchange has arisen as a viable repository for proteomics data (Vizcaino, et al., 2014), and the National Metabolomics Data Repository now contains a lot of metabolomics data (Sud, et al., 2016). With ongoing and future multi-omics studies covering a greater number of more diverse patients, integrative analyses including machine learning-based strategies (Y. Li, et al., 2018) will become critical for translating omics data into key insights that improve our understanding of glucocorticoid responses.
Acknowledgments
This work was supported by National Institutes of Health grants R01 HL133433, R01 HL141992 and P30 ES013508.
Abbreviations
- ALL
acute lymphoblastic leukemia
- ASM
airway smooth muscle
- ChIP-Seq
chromatin immunoprecipitation sequencing
- FEV1
forced expiratory volume in 1 second
- GWAS
genome-wide association study
- GR
glucocorticoid receptor
- GRE
glucocorticoid response element
- MS
mass spectrometry
- ICS
inhaled corticosteroids
- RNA-Seq
RNA sequencing
- RNAP2
RNA polymerase II
- SNP
single nucleotide polymorphism
Footnotes
Conflicts of Interest Statement
The authors declare that there are no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Altelaar AF, Munoz J, & Heck AJ (2013). Next-generation proteomics: towards an integrative view of proteome dynamics. Nat Rev Genet, 14, 35–48. [DOI] [PubMed] [Google Scholar]
- Altonsy MO, Mostafa MM, Gerber AN, & Newton R (2017). Long-acting beta2-agonists promote glucocorticoid-mediated repression of NF-kappaB by enhancing expression of the feedback regulator TNFAIP3. Am J Physiol Lung Cell Mol Physiol, 312, L358–L370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amano Y, Lee SW, & Allison AC (1993). Inhibition by glucocorticoids of the formation of interleukin-1 alpha, interleukin-1 beta, and interleukin-6: mediation by decreased mRNA stability. Mol Pharmacol, 43, 176–182. [PubMed] [Google Scholar]
- Arnold CD, Gerlach D, Stelzer C, Boryn LM, Rath M, & Stark A (2013). Genome-wide quantitative enhancer activity maps identified by STARR-seq. Science, 339, 1074–1077. [DOI] [PubMed] [Google Scholar]
- Barnes PJ (2010). Inhaled Corticosteroids. Pharmaceuticals (Basel), 3, 514–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes PJ (2011). Glucocorticosteroids: current and future directions. Br J Pharmacol, 163, 29–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes PJ (2013). Corticosteroid resistance in patients with asthma and chronic obstructive pulmonary disease. J Allergy Clin Immunol, 131, 636–645. [DOI] [PubMed] [Google Scholar]
- Barnes PJ, & Adcock IM (2009). Glucocorticoid resistance in inflammatory diseases. Lancet, 373, 1905–1917. [DOI] [PubMed] [Google Scholar]
- Belton JM, McCord RP, Gibcus JH, Naumova N, Zhan Y, & Dekker J (2012). Hi-C: a comprehensive technique to capture the conformation of genomes. Methods, 58, 268–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bereshchenko O, Migliorati G, Bruscoli S, & Riccardi C (2019). Glucocorticoid-Induced Leucine Zipper: A Novel Anti-inflammatory Molecule. Front Pharmacol, 10, 308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhavsar P, Hew M, Khorasani N, Torrego A, Barnes PJ, Adcock I, & Chung KF (2008). Relative corticosteroid insensitivity of alveolar macrophages in severe asthma compared with non-severe asthma. Thorax, 63, 784–790. [DOI] [PubMed] [Google Scholar]
- Bhavsar P, Khorasani N, Hew M, Johnson M, & Chung KF (2010). Effect of p38 MAPK inhibition on corticosteroid suppression of cytokine release in severe asthma. Eur Respir J, 35, 750–756. [DOI] [PubMed] [Google Scholar]
- Biddie SC, John S, Sabo PJ, Thurman RE, Johnson TA, Schiltz RL, Miranda TB, Sung MH, Trump S, Lightman SL, Vinson C, Stamatoyannopoulos JA, & Hager GL (2011). Transcription factor AP1 potentiates chromatin accessibility and glucocorticoid receptor binding. Mol Cell, 43, 145–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bileck A, Kreutz D, Muqaku B, Slany A, & Gerner C (2014). Comprehensive assessment of proteins regulated by dexamethasone reveals novel effects in primary human peripheral blood mononuclear cells. J Proteome Res, 13, 5989–6000. [DOI] [PubMed] [Google Scholar]
- Bindreither D, Ecker S, Gschirr B, Kofler A, Kofler R, & Rainer J (2014). The synthetic glucocorticoids prednisolone and dexamethasone regulate the same genes in acute lymphoblastic leukemia cells. BMC Genomics, 15, 662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordag N, Klie S, Jurchott K, Vierheller J, Schiewe H, Albrecht V, Tonn JC, Schwartz C, Schichor C, & Selbig J (2015). Glucocorticoid (dexamethasone)-induced metabolome changes in healthy males suggest prediction of response and side effects. Sci Rep, 5, 15954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouazza B, Krytska K, Debba-Pavard M, Amrani Y, Honkanen RE, Tran J, & Tliba O (2012). Cytokines alter glucocorticoid receptor phosphorylation in airway cells: role of phosphatases. Am J Respir Cell Mol Biol, 47, 464–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buniello A, MacArthur JAL, Cerezo M, Harris LW, Hayhurst J, Malangone C, McMahon A, Morales J, Mountjoy E, Sollis E, Suveges D, Vrousgou O, Whetzel PL, Amode R, Guillen JA, Riat HS, Trevanion SJ, Hall P, Junkins H, Flicek P, Burdett T, Hindorff LA, Cunningham F, & Parkinson H (2019). The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res, 47, D1005–D1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess JK, Lee JH, Ge Q, Ramsay EE, Poniris MH, Parmentier J, Roth M, Johnson PR, Hunt NH, Black JL, & Ammit AJ (2008). Dual ERK and phosphatidylinositol 3-kinase pathways control airway smooth muscle proliferation: differences in asthma. J Cell Physiol, 216, 673–679. [DOI] [PubMed] [Google Scholar]
- Carlet M, Janjetovic K, Rainer J, Schmidt S, Panzer-Grumayer R, Mann G, Prelog M, Meister B, Ploner C, & Kofler R (2010). Expression, regulation and function of phosphofructo-kinase/fructose-biphosphatases (PFKFBs) in glucocorticoid-induced apoptosis of acute lymphoblastic leukemia cells. BMC Cancer, 10, 638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chachi L, Abbasian M, Gavrila A, Alzahrani A, Tliba O, Bradding P, Wardlaw AJ, Brightling C, & Amrani Y (2017). Protein phosphatase 5 mediates corticosteroid insensitivity in airway smooth muscle in patients with severe asthma. Allergy, 72, 126–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chachi L, Shikotra A, Duffy SM, Tliba O, Brightling C, Bradding P, & Amrani Y (2013). Functional KCa3.1 channels regulate steroid insensitivity in bronchial smooth muscle cells. J Immunol, 191, 2624–2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang PJ, Michaeloudes C, Zhu J, Shaikh N, Baker J, Chung KF, & Bhavsar PK (2015). Impaired nuclear translocation of the glucocorticoid receptor in corticosteroid-insensitive airway smooth muscle in severe asthma. Am J Respir Crit Care Med, 191, 54–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung KF, Caramori G, & Adcock IM (2009). Inhaled corticosteroids as combination therapy with beta-adrenergic agonists in airways disease: present and future. Eur J Clin Pharmacol, 65, 853–871. [DOI] [PubMed] [Google Scholar]
- Chung KF, Wenzel SE, Brozek JL, Bush A, Castro M, Sterk PJ, Adcock IM, Bateman ED, Bel EH, Bleecker ER, Boulet LP, Brightling C, Chanez P, Dahlen SE, Djukanovic R, Frey U, Gaga M, Gibson P, Hamid Q, Jajour NN, Mauad T, Sorkness RL, & Teague WG (2014). International ERS/ATS guidelines on definition, evaluation and treatment of severe asthma. Eur Respir J, 43, 343–373. [DOI] [PubMed] [Google Scholar]
- Clifford RL, John AE, Brightling CE, & Knox AJ (2012). Abnormal histone methylation is responsible for increased vascular endothelial growth factor 165a secretion from airway smooth muscle cells in asthma. J Immunol, 189, 819–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifford RL, Patel JK, John AE, Tatler AL, Mazengarb L, Brightling CE, & Knox AJ (2015). CXCL8 histone H3 acetylation is dysfunctional in airway smooth muscle in asthma: regulation by BET. Am J Physiol Lung Cell Mol Physiol, 308, L962–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough E, & Barrett T (2016). The Gene Expression Omnibus Database. Methods Mol Biol, 1418, 93–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, & Lis JT (2008). Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science, 322, 1845–1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Ippolito AM, McDowell IC, Barrera A, Hong LK, Leichter SM, Bartelt LC, Vockley CM, Majoros WH, Safi A, Song L, Gersbach CA, Crawford GE, & Reddy TE (2018). Pre-established Chromatin Interactions Mediate the Genomic Response to Glucocorticoids. Cell Syst, 7, 146–160 e147. [DOI] [PubMed] [Google Scholar]
- Dahlin A, Denny J, Roden DM, Brilliant MH, Ingram C, Kitchner TE, Linneman JG, Shaffer CM, Weeke P, Xu H, Kubo M, Tamari M, Clemmer GL, Ziniti J, McGeachie MJ, Tantisira KG, Weiss ST, & Wu AC (2015). CMTR1 is associated with increased asthma exacerbations in patients taking inhaled corticosteroids. Immun Inflamm Dis, 3, 350–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denlinger LC, Sorkness CA, Chinchilli VM, & Lemanske RF Jr. (2007). Guideline-defining asthma clinical trials of the National Heart, Lung, and Blood Institute’s Asthma Clinical Research Network and Childhood Asthma Research and Education Network. J Allergy Clin Immunol, 119, 3–11; quiz 12–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmet SJ, & De Bosscher K (2017). Glucocorticoid receptors: finding the middle ground. J Clin Invest, 127, 1136–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diwadkar AR, Kan M, & Himes BE (2019). Facilitating Analysis of Publicly Available ChIP-Seq Data for Integrative Studies. AMIA Annu Symp Proc, 2019, 371–379. [PMC free article] [PubMed] [Google Scholar]
- Dyczynski M, Vesterlund M, Bjorklund AC, Zachariadis V, Janssen J, Gallart-Ayala H, Daskalaki E, Wheelock CE, Lehtio J, Grander D, Tamm KP, & Nilsson R (2018). Metabolic reprogramming of acute lymphoblastic leukemia cells in response to glucocorticoid treatment. Cell Death Dis, 9, 846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst P, Saad N, & Suissa S (2015). Inhaled corticosteroids in COPD: the clinical evidence. Eur Respir J, 45, 525–537. [DOI] [PubMed] [Google Scholar]
- Farzan N, Vijverberg SJ, Arets HG, Raaijmakers JA, & Maitland-van der Zee AH (2017). Pharmacogenomics of inhaled corticosteroids and leukotriene modifiers: a systematic review. Clin Exp Allergy, 47, 271–293. [DOI] [PubMed] [Google Scholar]
- Gertz J, Savic D, Varley KE, Partridge EC, Safi A, Jain P, Cooper GM, Reddy TE, Crawford GE, & Myers RM (2013). Distinct properties of cell-type-specific and shared transcription factor binding sites. Mol Cell, 52, 25–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giembycz MA, & Newton R (2015). Potential mechanisms to explain how LABAs and PDE4 inhibitors enhance the clinical efficacy of glucocorticoids in inflammatory lung diseases. F1000Prime Rep, 7, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gille J, Reisinger K, Westphal-Varghese B, & Kaufmann R (2001). Decreased mRNA stability as a mechanism of glucocorticoid-mediated inhibition of vascular endothelial growth factor gene expression by cultured keratinocytes. J Invest Dermatol, 117, 1581–1587. [DOI] [PubMed] [Google Scholar]
- Goncharova EA, Lim P, Goncharov DA, Eszterhas A, Panettieri RA Jr., & Krymskaya VP (2006). Assays for in vitro monitoring of proliferation of human airway smooth muscle (ASM) and human pulmonary arterial vascular smooth muscle (VSM) cells. Nat Protoc, 1, 2905–2908. [DOI] [PubMed] [Google Scholar]
- Goodwin S, McPherson JD, & McCombie WR (2016). Coming of age: ten years of next-generation sequencing technologies. Nat Rev Genet, 17, 333–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruver-Yates AL, & Cidlowski JA (2013). Tissue-specific actions of glucocorticoids on apoptosis: a double-edged sword. Cells, 2, 202–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haarman EG, Kaspers GJ, & Veerman AJ (2003). Glucocorticoid resistance in childhood leukaemia: mechanisms and modulation. Br J Haematol, 120, 919–929. [DOI] [PubMed] [Google Scholar]
- Hafezi-Moghadam A, Simoncini T, Yang Z, Limbourg FP, Plumier JC, Rebsamen MC, Hsieh CM, Chui DS, Thomas KL, Prorock AJ, Laubach VE, Moskowitz MA, French BA, Ley K, & Liao JK (2002). Acute cardiovascular protective effects of corticosteroids are mediated by non-transcriptional activation of endothelial nitric oxide synthase. Nat Med, 8, 473–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hapgood JP, Avenant C, & Moliki JM (2016). Glucocorticoid-independent modulation of GR activity: Implications for immunotherapy. Pharmacol Ther, 165, 93–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawcutt DB, Francis B, Carr DF, Jorgensen AL, Yin P, Wallin N, O’Hara N, Zhang EJ, Bloch KM, Ganguli A, Thompson B, McEvoy L, Peak M, Crawford AA, Walker BR, Blair JC, Couriel J, Smyth RL, & Pirmohamed M (2018). Susceptibility to corticosteroid-induced adrenal suppression: a genome-wide association study. Lancet Respir Med, 6, 442–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Pacheco N, Farzan N, Francis B, Karimi L, Repnik K, Vijverberg SJ, Soares P, Schieck M, Gorenjak M, Forno E, Eng C, Oh SS, Perez-Mendez L, Berce V, Tavendale R, Samedy LA, Hunstman S, Hu D, Meade K, Farber HJ, Avila PC, Serebrisky D, Thyne SM, Brigino-Buenaventura E, Rodriguez-Cintron W, Sen S, Kumar R, Lenoir M, Rodriguez-Santana JR, Celedon JC, Mukhopadhyay S, Potocnik U, Pirmohamed M, Verhamme KM, Kabesch M, Palmer CNA, Hawcutt DB, Flores C, Maitland-van der Zee AH, Burchard EG, & Pino-Yanes M (2019). Genome-wide association study of inhaled corticosteroid response in admixed children with asthma. Clin Exp Allergy, 49, 789–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hew M, Bhavsar P, Torrego A, Meah S, Khorasani N, Barnes PJ, Adcock I, & Chung KF (2006). Relative corticosteroid insensitivity of peripheral blood mononuclear cells in severe asthma. Am J Respir Crit Care Med, 174, 134–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himes BE, Jiang X, Wagner P, Hu R, Wang Q, Klanderman B, Whitaker RM, Duan Q, Lasky-Su J, Nikolos C, Jester W, Johnson M, Panettieri RA Jr., Tantisira KG, Weiss ST, & Lu Q (2014). RNA-Seq transcriptome profiling identifies CRISPLD2 as a glucocorticoid responsive gene that modulates cytokine function in airway smooth muscle cells. PLoS One, 9, e99625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holguin F, Cardet JC, Chung KF, Diver S, Ferreira DS, Fitzpatrick A, Gaga M, Kellermeyer L, Khurana S, Knight S, McDonald VM, Morgan RL, Ortega VE, Rigau D, Subbarao P, Tonia T, Adcock IM, Bleecker ER, Brightling C, Boulet LP, Cabana M, Castro M, Chanez P, Custovic A, Djukanovic R, Frey U, Frankemolle B, Gibson P, Hamerlijnck D, Jarjour N, Konno S, Shen H, Vitary C, & Bush A (2020). Management of severe asthma: a European Respiratory Society/American Thoracic Society guideline. Eur Respir J, 55. [DOI] [PubMed] [Google Scholar]
- Holleman A, Cheok MH, den Boer ML, Yang W, Veerman AJ, Kazemier KM, Pei D, Cheng C, Pui CH, Relling MV, Janka-Schaub GE, Pieters R, & Evans WE (2004). Gene-expression patterns in drug-resistant acute lymphoblastic leukemia cells and response to treatment. N Engl J Med, 351, 533–542. [DOI] [PubMed] [Google Scholar]
- Hosking L, Bleecker E, Ghosh S, Yeo A, Jacques L, Mosteller M, & Meyers D (2014). GLCCI1 rs37973 does not influence treatment response to inhaled corticosteroids in white subjects with asthma. J Allergy Clin Immunol, 133, 587–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huizenga NA, Koper JW, De Lange P, Pols HA, Stolk RP, Burger H, Grobbee DE, Brinkmann AO, De Jong FH, & Lamberts SW (1998). A polymorphism in the glucocorticoid receptor gene may be associated with and increased sensitivity to glucocorticoids in vivo. J Clin Endocrinol Metab, 83, 144–151. [DOI] [PubMed] [Google Scholar]
- Jiang H, Chi X, Zhang X, & Wang J (2016). Increased serum VDBP as a risk predictor for steroid resistance in asthma patients. Respir Med, 114, 111–116. [DOI] [PubMed] [Google Scholar]
- Jiang N, Kham SK, Koh GS, Suang Lim JY, Ariffin H, Chew FT, & Yeoh AE (2011). Identification of prognostic protein biomarkers in childhood acute lymphoblastic leukemia (ALL). J Proteomics, 74, 843–857. [DOI] [PubMed] [Google Scholar]
- Joubert P, Lajoie-Kadoch S, Labonte I, Gounni AS, Maghni K, Wellemans V, Chakir J, Laviolette M, Hamid Q, & Lamkhioued B (2005). CCR3 expression and function in asthmatic airway smooth muscle cells. J Immunol, 175, 2702–2708. [DOI] [PubMed] [Google Scholar]
- Junot C, Fenaille F, Colsch B, & Becher F (2014). High resolution mass spectrometry based techniques at the crossroads of metabolic pathways. Mass Spectrom Rev, 33, 471–500. [DOI] [PubMed] [Google Scholar]
- Kaczmarek KA, Clifford RL, & Knox AJ (2019). Epigenetic Changes in Airway Smooth Muscle as a Driver of Airway Inflammation and Remodeling in Asthma. Chest, 155, 816–824. [DOI] [PubMed] [Google Scholar]
- Kadiyala V, Sasse SK, Altonsy MO, Berman R, Chu HW, Phang TL, & Gerber AN (2016). Cistrome-based Cooperation between Airway Epithelial Glucocorticoid Receptor and NF-kappaB Orchestrates Anti-inflammatory Effects. J Biol Chem, 291, 12673–12687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan M, Koziol-White C, Shumyatcher M, Johnson M, Jester W, Panettieri RA Jr., & Himes BE (2019). Airway Smooth Muscle-Specific Transcriptomic Signatures of Glucocorticoid Exposure. Am J Respir Cell Mol Biol, 61, 110–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan M, Shumyatcher M, Diwadkar A, Soliman G, & Himes BE (2018). Integration of Transcriptomic Data Identifies Global and Cell-Specific Asthma-Related Gene Expression Signatures. AMIA Annu Symp Proc, 2018, 1338–1347. [PMC free article] [PubMed] [Google Scholar]
- Kan M, Shumyatcher M, & Himes BE (2017). Using omics approaches to understand pulmonary diseases. Respir Res, 18, 149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karol SE, Yang W, Van Driest SL, Chang TY, Kaste S, Bowton E, Basford M, Bastarache L, Roden DM, Denny JC, Larsen E, Winick N, Carroll WL, Cheng C, Pei D, Fernandez CA, Liu C, Smith C, Loh ML, Raetz EA, Hunger SP, Scheet P, Jeha S, Pui CH, Evans WE, Devidas M, Mattano LA Jr., & Relling MV (2015). Genetics of glucocorticoid-associated osteonecrosis in children with acute lymphoblastic leukemia. Blood, 126, 1770–1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawedia JD, Kaste SC, Pei D, Panetta JC, Cai X, Cheng C, Neale G, Howard SC, Evans WE, Pui CH, & Relling MV (2011). Pharmacokinetic, pharmacodynamic, and pharmacogenetic determinants of osteonecrosis in children with acute lymphoblastic leukemia. Blood, 117, 2340–2347; quiz 2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly RS, Dahlin A, McGeachie MJ, Qiu W, Sordillo J, Wan ES, Wu AC, & Lasky-Su J (2017). Asthma Metabolomics and the Potential for Integrative Omics in Research and the Clinic. Chest, 151, 262–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keskin O, Farzan N, Birben E, Akel H, Karaaslan C, Maitland-van der Zee AH, Wechsler ME, Vijverberg SJ, & Kalayci O (2019). Genetic associations of the response to inhaled corticosteroids in asthma: a systematic review. Clin Transl Allergy, 9, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keun HC, & Athersuch TJ (2011). Nuclear magnetic resonance (NMR)-based metabolomics. Methods Mol Biol, 708, 321–334. [DOI] [PubMed] [Google Scholar]
- Koper JW, Stolk RP, de Lange P, Huizenga NA, Molijn GJ, Pols HA, Grobbee DE, Karl M, de Jong FH, Brinkmann AO, & Lamberts SW (1997). Lack of association between five polymorphisms in the human glucocorticoid receptor gene and glucocorticoid resistance. Hum Genet, 99, 663–668. [DOI] [PubMed] [Google Scholar]
- Koziol-White C, Johnstone TB, Corpuz ML, Cao G, Orfanos S, Parikh V, Deeney B, Tliba O, Ostrom RS, Dainty I, & Panettieri RA Jr. (2020). Budesonide enhances agonist-induced bronchodilation in human small airways by increasing cAMP production in airway smooth muscle. Am J Physiol Lung Cell Mol Physiol, 318, L345–L355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruth KA, Fang M, Shelton DN, Abu-Halawa O, Mahling R, Yang H, Weissman JS, Loh ML, Muschen M, Tasian SK, Bassik MC, Kampmann M, & Pufall MA (2017). Suppression of B-cell development genes is key to glucocorticoid efficacy in treatment of acute lymphoblastic leukemia. Blood, 129, 3000–3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo T, McQueen A, Chen TC, & Wang JC (2015). Regulation of Glucose Homeostasis by Glucocorticoids. Adv Exp Med Biol, 872, 99–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langlais D, Couture C, Balsalobre A, & Drouin J (2012). The Stat3/GR interaction code: predictive value of direct/indirect DNA recruitment for transcription outcome. Mol Cell, 47, 38–49. [DOI] [PubMed] [Google Scholar]
- Lasa M, Brook M, Saklatvala J, & Clark AR (2001). Dexamethasone destabilizes cyclooxygenase 2 mRNA by inhibiting mitogen-activated protein kinase p38. Mol Cell Biol, 21, 771–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leigh R, Mostafa MM, King EM, Rider CF, Shah S, Dumonceaux C, Traves SL, McWhae A, Kolisnik T, Kooi C, Slater DM, Kelly MM, Bieda M, Miller-Larsson A, & Newton R (2016). An inhaled dose of budesonide induces genes involved in transcription and signaling in the human airways: enhancement of anti- and proinflammatory effector genes. Pharmacol Res Perspect, 4, e00243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leinonen R, Sugawara H, Shumway M, & International Nucleotide Sequence Database, C. (2011). The sequence read archive. Nucleic Acids Res, 39, D19–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis-Tuffin LJ, & Cidlowski JA (2006). The physiology of human glucocorticoid receptor beta (hGRbeta) and glucocorticoid resistance. Ann N Y Acad Sci, 1069, 1–9. [DOI] [PubMed] [Google Scholar]
- Li LB, Leung DY, Martin RJ, & Goleva E (2010). Inhibition of histone deacetylase 2 expression by elevated glucocorticoid receptor beta in steroid-resistant asthma. Am J Respir Crit Care Med, 182, 877–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Ampleford EJ, Howard TD, Moore WC, Torgerson DG, Li H, Busse WW, Castro M, Erzurum SC, Israel E, Nicolae DL, Ober C, Wenzel SE, Hawkins GA, Bleecker ER, & Meyers DA (2012). Genome-wide association studies of asthma indicate opposite immunopathogenesis direction from autoimmune diseases. J Allergy Clin Immunol, 130, 861–868 e867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Wu FX, & Ngom A (2018). A review on machine learning principles for multi-view biological data integration. Brief Bioinform, 19, 325–340. [DOI] [PubMed] [Google Scholar]
- MacArthur J, Bowler E, Cerezo M, Gil L, Hall P, Hastings E, Junkins H, McMahon A, Milano A, Morales J, Pendlington ZM, Welter D, Burdett T, Hindorff L, Flicek P, Cunningham F, & Parkinson H (2017). The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Res, 45, D896–D901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkawi AK, Alzoubi KH, Jacob M, Matic G, Ali A, Al Faraj A, Almuhanna F, Dasouki M, & Abdel Rahman AM (2018). Metabolomics Based Profiling of Dexamethasone Side Effects in Rats. Front Pharmacol, 9, 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniscalco M, & Motta A (2017). Metabolomics of exhaled breath condensate: a means for phenotyping respiratory diseases? Biomark Med, 11, 405–407. [DOI] [PubMed] [Google Scholar]
- Marshall DD, & Powers R (2017). Beyond the paradigm: Combining mass spectrometry and nuclear magnetic resonance for metabolomics. Prog Nucl Magn Reson Spectrosc, 100, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuno K, Haldar SM, Jeyaraj D, Mailloux CM, Huang X, Panettieri RA Jr., Jain MK, & Gerber AN (2011). Expression profiling identifies Klf15 as a glucocorticoid target that regulates airway hyperresponsiveness. Am J Respir Cell Mol Biol, 45, 642–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews L, Berry A, Ohanian V, Ohanian J, Garside H, & Ray D (2008). Caveolin mediates rapid glucocorticoid effects and couples glucocorticoid action to the antiproliferative program. Mol Endocrinol, 22, 1320–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell IC, Barrera A, D’Ippolito AM, Vockley CM, Hong LK, Leichter SM, Bartelt LC, Majoros WH, Song L, Safi A, Kocak DD, Gersbach CA, Hartemink AJ, Crawford GE, Engelhardt BE, & Reddy TE (2018). Glucocorticoid receptor recruits to enhancers and drives activation by motif-directed binding. Genome Res, 28, 1272–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercado N, Hakim A, Kobayashi Y, Meah S, Usmani OS, Chung KF, Barnes PJ, & Ito K (2012). Restoration of corticosteroid sensitivity by p38 mitogen activated protein kinase inhibition in peripheral blood mononuclear cells from severe asthma. PLoS One, 7, e41582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata M, Lee JY, Susuki-Miyata S, Wang WY, Xu H, Kai H, Kobayashi KS, Flavell RA, & Li JD (2015). Glucocorticoids suppress inflammation via the upregulation of negative regulator IRAK-M. Nat Commun, 6, 6062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moosavi SM, Prabhala P, & Ammit AJ (2017). Role and regulation of MKP-1 in airway inflammation. Respir Res, 18, 154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosteller M, Hosking L, Murphy K, Shen J, Song K, Nelson M, & Ghosh S (2017). No evidence of large genetic effects on steroid response in asthma patients. J Allergy Clin Immunol, 139, 797–803 e797. [DOI] [PubMed] [Google Scholar]
- Murray JC, Smith RF, Ardinger HA, & Weinberger C (1987). RFLP for the glucocorticoid receptor (GRL) located at 5q11–5q13. Nucleic Acids Res, 15, 6765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton R (2014). Anti-inflammatory glucocorticoids: changing concepts. Eur J Pharmacol, 724, 231–236. [DOI] [PubMed] [Google Scholar]
- Newton R, & Holden NS (2007). Separating transrepression and transactivation: a distressing divorce for the glucocorticoid receptor? Mol Pharmacol, 72, 799–809. [DOI] [PubMed] [Google Scholar]
- Newton R, Leigh R, & Giembycz MA (2010). Pharmacological strategies for improving the efficacy and therapeutic ratio of glucocorticoids in inflammatory lung diseases. Pharmacol Ther, 125, 286–327. [DOI] [PubMed] [Google Scholar]
- Nicholson L, Evans CA, Matheson E, Minto L, Keilty C, Sanichar M, Case M, Schwab C, Williamson D, Rainer J, Harrison CJ, Kofler R, Hall AG, Redfern CP, Whetton AD, & Irving JA (2015). Quantitative proteomic analysis reveals maturation as a mechanism underlying glucocorticoid resistance in B lineage ALL and re-sensitization by JNK inhibition. Br J Haematol, 171, 595–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunez FJ, Johnstone TB, Corpuz ML, Kazarian AG, Mohajer NN, Tliba O, Panettieri RA Jr., Koziol-White C, Roosan MR, & Ostrom RS (2019). Glucocorticoids rapidly activate cAMP production via Galphas to initiate non-genomic signaling that contributes to one-third of their canonical genomic effects. FASEB J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neil SE, Sitkauskiene B, Babusyte A, Krisiukeniene A, Stravinskaite-Bieksiene K, Sakalauskas R, Sihlbom C, Ekerljung L, Carlsohn E, & Lotvall J (2011). Network analysis of quantitative proteomics on asthmatic bronchi: effects of inhaled glucocorticoid treatment. Respir Res, 12, 124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley RH, & Cidlowski JA (2013). The biology of the glucocorticoid receptor: new signaling mechanisms in health and disease. J Allergy Clin Immunol, 132, 1033–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh KS, Patel H, Gottschalk RA, Lee WS, Baek S, Fraser IDC, Hager GL, & Sung MH (2017). Anti-Inflammatory Chromatinscape Suggests Alternative Mechanisms of Glucocorticoid Receptor Action. Immunity, 47, 298–309 e295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver BG, Johnston SL, Baraket M, Burgess JK, King NJ, Roth M, Lim S, & Black JL (2006). Increased proinflammatory responses from asthmatic human airway smooth muscle cells in response to rhinovirus infection. Respir Res, 7, 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panettieri RA, Murray RK, DePalo LR, Yadvish PA, & Kotlikoff MI (1989). A human airway smooth muscle cell line that retains physiological responsiveness. Am J Physiol, 256, C329–335. [DOI] [PubMed] [Google Scholar]
- Panettieri RA, Schaafsma D, Amrani Y, Koziol-White C, Ostrom R, & Tliba O (2019). Non-genomic Effects of Glucocorticoids: An Updated View. Trends Pharmacol Sci, 40, 38–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HW, Dahlin A, Tse S, Duan QL, Schuemann B, Martinez FD, Peters SP, Szefler SJ, Lima JJ, Kubo M, Tamari M, & Tantisira KG (2014). Genetic predictors associated with improvement of asthma symptoms in response to inhaled corticosteroids. J Allergy Clin Immunol, 133, 664–669 e665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HW, Ge B, Tse S, Grundberg E, Pastinen T, Kelly HW, & Tantisira KG (2015). Genetic risk factors for decreased bone mineral accretion in children with asthma receiving multiple oral corticosteroid bursts. J Allergy Clin Immunol, 136, 1240–1246 e1241–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park TJ, Park JS, Cheong HS, Park BL, Kim LH, Heo JS, Kim YK, Kim KU, Uh ST, Lee HS, Na JO, Seo KH, Choi JS, Kim YH, Kim MS, Park CS, & Shin HD (2014). Genome-wide association study identifies ALLC polymorphisms correlated with FEV(1) change by corticosteroid. Clin Chim Acta, 436, 20–26. [DOI] [PubMed] [Google Scholar]
- Pavlovic S, Kotur N, Stankovic B, Zukic B, Gasic V, & Dokmanovic L (2019). Pharmacogenomic and Pharmacotranscriptomic Profiling of Childhood Acute Lymphoblastic Leukemia: Paving the Way to Personalized Treatment. Genes (Basel), 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ploner C, Rainer J, Niederegger H, Eduardoff M, Villunger A, Geley S, & Kofler R (2008). The BCL2 rheostat in glucocorticoid-induced apoptosis of acute lymphoblastic leukemia. Leukemia, 22, 370–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prenek L, Boldizsar F, Kugyelka R, Ugor E, Berta G, Nemeth P, & Berki T (2017). The regulation of the mitochondrial apoptotic pathway by glucocorticoid receptor in collaboration with Bcl-2 family proteins in developing T cells. Apoptosis, 22, 239–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey LB, Pounds S, Cheng C, Cao X, Yang W, Smith C, Karol SE, Liu C, Panetta JC, Inaba H, Rubnitz JE, Metzger ML, Ribeiro RC, Sandlund JT, Jeha S, Pui CH, Evans WE, & Relling MV (2017). Genetics of pleiotropic effects of dexamethasone. Pharmacogenet Genomics, 27, 294–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao NA, McCalman MT, Moulos P, Francoijs KJ, Chatziioannou A, Kolisis FN, Alexis MN, Mitsiou DJ, & Stunnenberg HG (2011). Coactivation of GR and NFKB alters the repertoire of their binding sites and target genes. Genome Res, 21, 1404–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy TE, Pauli F, Sprouse RO, Neff NF, Newberry KM, Garabedian MJ, & Myers RM (2009). Genomic determination of the glucocorticoid response reveals unexpected mechanisms of gene regulation. Genome Res, 19, 2163–2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinke SN, Gallart-Ayala H, Gomez C, Checa A, Fauland A, Naz S, Kamleh MA, Djukanovic R, Hinks TS, & Wheelock CE (2017). Metabolomics analysis identifies different metabotypes of asthma severity. Eur Respir J, 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rider CF, Altonsy MO, Mostafa MM, Shah SV, Sasse S, Manson ML, Yan D, Karrman-Mardh C, Miller-Larsson A, Gerber AN, Giembycz MA, & Newton R (2018). Long-Acting beta2-Adrenoceptor Agonists Enhance Glucocorticoid Receptor (GR)-Mediated Transcription by Gene-Specific Mechanisms Rather Than Generic Effects via GR. Mol Pharmacol, 94, 1031–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasse SK, Altonsy MO, Kadiyala V, Cao G, Panettieri RA Jr., & Gerber AN (2016). Glucocorticoid and TNF signaling converge at A20 (TNFAIP3) to repress airway smooth muscle cytokine expression. Am J Physiol Lung Cell Mol Physiol, 311, L421–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasse SK, Gruca M, Allen MA, Kadiyala V, Song T, Gally F, Gupta A, Pufall MA, Dowell RD, & Gerber AN (2019). Nascent transcript analysis of glucocorticoid crosstalk with TNF defines primary and cooperative inflammatory repression. Genome Res, 29, 1753–1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasse SK, Kadiyala V, Danhorn T, Panettieri RA Jr., Phang TL, & Gerber AN (2017). Glucocorticoid Receptor ChIP-Seq Identifies PLCD1 as a KLF15 Target that Represses Airway Smooth Muscle Hypertrophy. Am J Respir Cell Mol Biol, 57, 226–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasse SK, Mailloux CM, Barczak AJ, Wang Q, Altonsy MO, Jain MK, Haldar SM, & Gerber AN (2013). The glucocorticoid receptor and KLF15 regulate gene expression dynamics and integrate signals through feed-forward circuitry. Mol Cell Biol, 33, 2104–2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schacke H, Docke WD, & Asadullah K (2002). Mechanisms involved in the side effects of glucocorticoids. Pharmacol Ther, 96, 23–43. [DOI] [PubMed] [Google Scholar]
- Schmidt S, Rainer J, Riml S, Ploner C, Jesacher S, Achmuller C, Presul E, Skvortsov S, Crazzolara R, Fiegl M, Raivio T, Janne OA, Geley S, Meister B, & Kofler R (2006). Identification of glucocorticoid-response genes in children with acute lymphoblastic leukemia. Blood, 107, 2061–2069. [DOI] [PubMed] [Google Scholar]
- Shumyatcher M, Hong R, Levin J, & Himes BE (2017). Disease-Specific Integration of Omics Data to Guide Functional Validation of Genetic Associations. AMIA Annu Symp Proc, 2017, 1589–1596. [PMC free article] [PubMed] [Google Scholar]
- Song IH, & Buttgereit F (2006). Non-genomic glucocorticoid effects to provide the basis for new drug developments. Mol Cell Endocrinol, 246, 142–146. [DOI] [PubMed] [Google Scholar]
- Song L, & Crawford GE (2010). DNase-seq: a high-resolution technique for mapping active gene regulatory elements across the genome from mammalian cells. Cold Spring Harb Protoc, 2010, pdb prot5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strunk RC, & Childhood Asthma Management Program Research, G. (2007). Childhood Asthma Management Program: lessons learned. J Allergy Clin Immunol, 119, 36–42. [DOI] [PubMed] [Google Scholar]
- Sud M, Fahy E, Cotter D, Azam K, Vadivelu I, Burant C, Edison A, Fiehn O, Higashi R, Nair KS, Sumner S, & Subramaniam S (2016). Metabolomics Workbench: An international repository for metabolomics data and metadata, metabolite standards, protocols, tutorials and training, and analysis tools. Nucleic Acids Res, 44, D463–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surjit M, Ganti KP, Mukherji A, Ye T, Hua G, Metzger D, Li M, & Chambon P (2011). Widespread negative response elements mediate direct repression by agonist-liganded glucocorticoid receptor. Cell, 145, 224–241. [DOI] [PubMed] [Google Scholar]
- Tantisira KG, Lasky-Su J, Harada M, Murphy A, Litonjua AA, Himes BE, Lange C, Lazarus R, Sylvia J, Klanderman B, Duan QL, Qiu W, Hirota T, Martinez FD, Mauger D, Sorkness C, Szefler S, Lazarus SC, Lemanske RF Jr., Peters SP, Lima JJ, Nakamura Y, Tamari M, & Weiss ST (2011). Genomewide association between GLCCI1 and response to glucocorticoid therapy in asthma. N Engl J Med, 365, 1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissing WJ, den Boer ML, Meijerink JP, Menezes RX, Swagemakers S, van der Spek PJ, Sallan SE, Armstrong SA, & Pieters R (2007). Genomewide identification of prednisolone-responsive genes in acute lymphoblastic leukemia cells. Blood, 109, 3929–3935. [DOI] [PubMed] [Google Scholar]
- Uhlenhaut NH, Barish GD, Yu RT, Downes M, Karunasiri M, Liddle C, Schwalie P, Hubner N, & Evans RM (2013). Insights into negative regulation by the glucocorticoid receptor from genome-wide profiling of inflammatory cistromes. Mol Cell, 49, 158–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbach V, Walsh DE, Mainprice B, Bousquet J, & Harvey BJ (2002). Rapid non-genomic inhibition of ATP-induced Cl- secretion by dexamethasone in human bronchial epithelium. J Physiol, 545, 869–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Schee MP, Palmay R, Cowan JO, & Taylor DR (2013). Predicting steroid responsiveness in patients with asthma using exhaled breath profiling. Clin Exp Allergy, 43, 1217–1225. [DOI] [PubMed] [Google Scholar]
- van Rossum EF, Koper JW, Huizenga NA, Uitterlinden AG, Janssen JA, Brinkmann AO, Grobbee DE, de Jong FH, van Duyn CM, Pols HA, & Lamberts SW (2002). A polymorphism in the glucocorticoid receptor gene, which decreases sensitivity to glucocorticoids in vivo, is associated with low insulin and cholesterol levels. Diabetes, 51, 3128–3134. [DOI] [PubMed] [Google Scholar]
- van Rossum EF, Koper JW, van den Beld AW, Uitterlinden AG, Arp P, Ester W, Janssen JA, Brinkmann AO, de Jong FH, Grobbee DE, Pols HA, & Lamberts SW (2003). Identification of the BclI polymorphism in the glucocorticoid receptor gene: association with sensitivity to glucocorticoids in vivo and body mass index. Clin Endocrinol (Oxf), 59, 585–592. [DOI] [PubMed] [Google Scholar]
- Vernocchi S, Battello N, Schmitz S, Revets D, Billing AM, Turner JD, & Muller CP (2013). Membrane glucocorticoid receptor activation induces proteomic changes aligning with classical glucocorticoid effects. Mol Cell Proteomics, 12, 1764–1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijverberg SJ, Tavendale R, Leusink M, Koenderman L, Raaijmakers JA, Postma DS, Koppelman GH, Turner SW, Mukhopadhyay S, Palmer CN, & Maitland-van der Zee AH (2014). Pharmacogenetic analysis of GLCCI1 in three north European pediatric asthma populations with a reported use of inhaled corticosteroids. Pharmacogenomics, 15, 799–806. [DOI] [PubMed] [Google Scholar]
- Vizcaino JA, Deutsch EW, Wang R, Csordas A, Reisinger F, Rios D, Dianes JA, Sun Z, Farrah T, Bandeira N, Binz PA, Xenarios I, Eisenacher M, Mayer G, Gatto L, Campos A, Chalkley RJ, Kraus HJ, Albar JP, Martinez-Bartolome S, Apweiler R, Omenn GS, Martens L, Jones AR, & Hermjakob H (2014). ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat Biotechnol, 32, 223–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vockley CM, D’Ippolito AM, McDowell IC, Majoros WH, Safi A, Song L, Crawford GE, & Reddy TE (2016). Direct GR Binding Sites Potentiate Clusters of TF Binding across the Human Genome. Cell, 166, 1269–1281 e1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Chavali S, Mobini R, Muraro A, Barbon F, Boldrin D, Aberg N, & Benson M (2011). A pathway-based approach to find novel markers of local glucocorticoid treatment in intermittent allergic rhinitis. Allergy, 66, 132–140. [DOI] [PubMed] [Google Scholar]
- Wang H, Gottfries J, Barrenas F, & Benson M (2011). Identification of novel biomarkers in seasonal allergic rhinitis by combining proteomic, multivariate and pathway analysis. PLoS One, 6, e23563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Tong C, Wang Z, Wang Z, Mauger D, Tantisira KG, Israel E, Szefler SJ, Chinchilli VM, Boushey HA, Lazarus SC, Lemanske RF, & Wu R (2015). Pharmacodynamic genome-wide association study identifies new responsive loci for glucocorticoid intervention in asthma. Pharmacogenomics J, 15, 422–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, & Snyder M (2009). RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet, 10, 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson L, Verhoog NJD, & Louw A (2018). Disease and treatment associated acquired glucocorticoid resistance. Endocr Connect. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff PG, Boushey HA, Dolganov GM, Barker CS, Yang YH, Donnelly S, Ellwanger A, Sidhu SS, Dao-Pick TP, Pantoja C, Erle DJ, Yamamoto KR, & Fahy JV (2007). Genome-wide profiling identifies epithelial cell genes associated with asthma and with treatment response to corticosteroids. Proc Natl Acad Sci U S A, 104, 15858–15863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yick CY, Zwinderman AH, Kunst PW, Grunberg K, Mauad T, Fluiter K, Bel EH, Lutter R, Baas F, & Sterk PJ (2013). Glucocorticoid-induced changes in gene expression of airway smooth muscle in patients with asthma. Am J Respir Crit Care Med, 187, 1076–1084. [DOI] [PubMed] [Google Scholar]