Abstract
Purpose:
Long interspersed nuclear element 1 (LINE-1 or L1) is a dominant non-long terminal repeat (non-LTR) retrotransposon in the human genome that has been implicated in the overexpression of MET. Both the canonical MET and L1-MET transcripts are considered to play a role in hepatocellular carcinoma (HCC) development. The aim of this study was to assess the utility of canonical MET, L1-MET, and MET protein expressions as predictive biomarkers for chemo-sensitivity to MET-inhibitors in HCC cell lines in-vitro. Additionally, we assessed their expression in tumour tissues from Egyptian HCC patients.
Methods:
MET and L1-MET expressions were assessed by qRT-PCR in six liver cancer cell lines (SNU-387, SNU-475, SK-HEP-1, PLC/PRF/5, SNU-449 and SNU-423) and 47 HCC tumour tissues. MET protein expression was measured by western blot in cell lines and immunohistochemistry in the tumors. Cell proliferation assay was used to assess the effect of crizotinib and tivantinib on the six liver cancer cell lines in correlation with the expression of MET, L1-MET and MET.
Results:
The antitumor effect of crizotinib and tivantinib correlated with MET gene expression but not with L1-MET transcript or MET protein expressions. No significant difference was observed between HCC tumours and non-tumour samples in MET and L1-MET transcripts expression. There were no significant correlations between the 2-year overall survival rate and the MET, L1-MET transcripts and the MET protein expression.
Conclusion:
MET RNA expression could be useful biomarker for tivantinib and crizotinib targeted therapy in HCC. The value of assessment of MET protein expression is limited.
Keywords: MET gene, L1-MET transcript, MET protein, Hepatocellular carcinoma, Crizotinib, Tivantinib
1. Introduction:
Worldwide, hepatocellular carcinoma (HCC) ranks as the fifth most common cancer and represents the most lethal cancer (Ferlay et al. 2015; Siegel et al. 2016). Currently, HCC treatment outcomes are unsatisfactory. The molecular heterogeneity of HCC (Zucman-Rossi et al. 2015; Ally et al. 2017) requires a therapeutic strategy based on predictive biomarkers. Many HCC randomized phase III trials failed because they were offered to all patients rather than selected patient population based on their tumour molecular profiles to maximize the benefit of the treatment (Llovet et al. 2015). Developing molecular targets for HCC could help in improving the treatment outcomes. Several meta-analyses have demonstrated MET protein overexpression to be an adverse prognostic marker in different types of cancers (Yu et al. 2013; Liu et al. 2015; Yan et al. 2015; Pyo et al. 2016). Targeting MET activation in patients using MET inhibitors is considered the standard of care in several tumours such as advanced renal cell carcinoma.
In addition to the canonical MET transcripts, long interspersed nuclear element 1 (LINE-1 or L1) has been implicated in the overexpression of MET. L1 is a dominant non-long terminal repeat (non-LTR) retrotransposon in the human genome (Han 2010; Naufer et al. 2018). A chimeric L1-MET transcript is produced upon L1 insertion in the second intron of the MET gene (Weber et al. 2010) and it has been suggested that L1-MET plays a role in HCC development (Honda 2016). In Japanese HCC patients L1-MET transcript expression is an adverse prognostic biomarker that causes activation of the MET signalling pathway (Zhu et al. 2014).
Both tivantinib and crizotinib are small molecule MET Inhibitors. Crizotinib has been approved by U.S. Food and Drug Administration (FDA) and European Medicines Agency (EMA) for treatment of ROS1-positive advanced Non-Small Cell Lung Cancer (NSCLC) (Puccini et al. 2019). Tivantinib is currently in phase II to III clinical trials for treatment of many cancers such as HCC, small cell lung cancer, prostate cancer and others, either as monotherapy or in combination with other chemotherapies (Bouattour et al. 2018; Parikh and Ghate 2018). In HCC, it succeeded as a second line treatment in phase II trials but failed in phase III trial to meet the primary endpoint of improving the overall survival (OS) (Rimassa et al. 2018).
In that trial, MET expression was assessed by immunohistochemistry (IHC) on archival or recent biopsy samples using a score of ≥2 in ≥50% of tumour cells as a cut off point for selection of patients. Failure of that study could be due to limitation of the assay utilized and highlight the importance of development of other biomarker for patient selection (Hughes and Siemann 2018; Weekes et al. 2018).
The aim of this study was to assess the utility of canonical MET, L1-MET, and MET protein expressions as predictive biomarkers for chemo-sensitivity to MET-inhibitors in HCC cell lines in-vitro. Additionally, we assessed their expression in tumour tissues from Egyptian HCC patients.
2. Materials and Methods:
Experimental work on cell lines was carried out at Dr. Abdel-Rahman’s laboratory at The Ohio State University while studies on HCC tumour tissues were performed at the National Liver Institute-Sustainable Science Institute-Collaborative Research Centre (NLISSICRC).
2.1. Cell Lines:
Six liver cancer cell lines, SNU-387, SNU-475, SK-HEP-1, PLC/PRF/5, SNU-449 and SNU-423, were purchased from American Type Culture Collection (ATCC®). The Catalogue Of Somatic Mutations In Cancer (COSMIC) (Forbes et al. 2017) database [GRCh37· CELL_LINES v86] was used to identify variants of the MET gene (mutations, fusions, breakpoints, non-coding mutations and copy number variations [CNV] as well as MET RNA expression) in the six cell lines. Authentication of the cell lines was achieved by short tandem repeat (STR) profiling using ten highly polymorphic microsatellite STR loci and sex determination (AMEL, CSF1PO, D13S317, D16S539, D21S11, D5S818, D7S820, TH01, TPOX, and vWA) at the Ohio State University’s genomic core facility. STR profiles were compared to available profiles through Cellosaurus (Bairoch 2018).
2.2. Subjects:
The inclusion criteria of study participants were newly diagnosed HCC Egyptian patients primarily treated by surgical resection of their tumours regardless of the aetiology of their disease. Exclusion criteria were lack of tumour tissue or prior therapies. Patients were accrued from the National Liver Institute (NLI), Menoufia University from 2014 to 2016 in accordance with an institutional review board approved protocol (IRB0051/2012). Patients were followed up until December 2018 with an average follow up of 25.6 months (range=1.0-39.2 months). Matched snap-frozen tissue samples were obtained from tumour and non-tumour liver tissues of all patients. Table 1 summarizes the clinical and pathological characteristics of HCC patients included in the study.
Table 1.
Clinical and pathological characteristics of Egyptian HCC patients.
| Variable | N (%) Total=47 | |
|---|---|---|
| Age | Mean±SD | 57.02±6.92 |
| Sex | Male | 37 (21.3) |
| Female | 10 (78.7) | |
| Etiology | HCV | 42 (89.4) |
| HBV | 2 (4.3) | |
| Mixed | 1 (2.1) | |
| Negative | 2 (4.3) | |
| Site | Left | 24 (51.06) |
| Right | 22 (46.82) | |
| Unknown | 1 (2.12) | |
| Grade | II | 24 (51.06) |
| III | 23 (48.9) | |
| Margin | Free | 34 (72.3) |
| Involved | 13 (27.7) | |
| Focality | Solitary | 34 (72.3) |
| Multiple | 13 (27.7) | |
| Type | Solid | 21 (44.7) |
| Trabecular, Acinar | 25 (53.2) | |
| Trabecular, Solid | 1 (2.1) | |
| Adjacent liver | Cirrhotic | 36 (76.6) |
| Non-Cirrhotic | 10 (21.27) | |
| Unknown | 1 (2.13) | |
| Vascular Invasion | Yes | 24 (51.1) |
| No | 23 (48.9) | |
| Pathological staging | T1 | 13 (27.7) |
| T2 | 30 (63.8) | |
| T3a | 3 (6.4) | |
| T4 | 1 (2.1) | |
| Survival | Alive | 34 (72.34) |
| Dead | 13 (27.66) | |
| Recurrence | Yes | 10 (21.28) |
| No | 37 (78.72) |
2.3. DNA and RNA Extraction and cDNA synthesis:
DNA and RNA were extracted from cell lines using the AllPrep DNA/RNA/miRNA Universal Kit (Qiagen, Carlsbad, CA) according to the manufacturer’s protocol. RNA from fresh frozen HCC (tumour and non-tumour) tissue samples was isolated using TRIzol (Invitrogen, Carlsbad, CA) according to the manufacturer’s protocol. One to two μg of RNA was used for cDNA synthesis using the Superscript VILO Master Mix (Invitrogen®) according to the manufacturer’s protocol (Invitrogen, Carlsbad, CA).
2.4. Semi-quantitative analysis of conventional PCR for assessment of expression in tissue samples:
cDNA equivalent to 50ng from each patient sample (tumour and non-tumour tissues) was amplified in two sets of 15-μl reaction mixtures, one for MET and the other for L1-MET. Each reaction mixture contained 7.5 μl of HotStar Taq™ DNA Polymerase (Invitrogen®), 0.8 μl of forward primer (10 μM), 0.8 μl of reverse primer (10 μM) (Supplementary Table S1) and 2.4 μl of PCR water. PCR was run in a GSI thermal cycler according to the following protocol: HotStar Taq™ DNA Polymerase was activated by incubation for 15 minutes at 95°C, followed by 3-step cycling [denaturation for 30 s at 94°C, annealing for 45 s at 60°C, and extension for 45 s at 72°C] for 35 cycles and a final elongation step for 10 minutes at 72°C. The amplicons were visualized by gel imaging (FireReader Gel Documentation System, UVITEC, Cambridge, UK) after electrophoretic separation (100 V for 40 minutes) on a 2% agarose gel using horizontal gel electrophoresis (Cleaver Scientific, Ltd., UK). Semi-quantitative analysis was performed by digital analysis of gel images using a freely available image analysis software-ImageJ (Rueden et al. 2017). GUSB was used as a reference gene for semiquantitative assessment of expression.
2.5. Quantitative RT-PCR (qRT-PCR) for assessment of expression in cell lines:
MET and L1-MET expression in cell lines was assessed in separate reactions using qRT-PCR according to the manufacturer’s protocol (Applied Biosystems, Inc., Foster City, CA). The reactions were performed in triplicate in 15 μL, with a final dilution of 1X each of PCR universal master mix. Probes spanning exons to exclusively amplify RNA were selected (Supplementary Table S1). The expression of GUSB (Applied Biosystems, Inc., Foster City, CA) was used as an internal control. qRT-PCR was performed using an Applied Biosystems 7900HT Fast Real-Time PCR machine at 95°C for 3 minutes, followed by 45 cycles of 95°C for 15 s and 60°C for 1 minute. The relative expression levels were assessed by the comparative CT method (threshold cycle) according to our previously published protocol (Abdel-Rahman et al. 2010).
2.6. Western Blot Assay:
Total protein was extracted by incubating cell lines in 1x ice-cold cell lysis buffer (Cell Signalling Technology, Boston, MA), spiked with 1 mM PMSF and 1x phosphatase inhibitor cocktail 2 (Sigma, St. Louis, MO) immediately before use according to a previously published protocol (Abdel-Rahman et al. 2010). Transfer efficiency was assessed by Ponceau S staining (0.1 % (w/v) in 5% acetic acid. After blocking the membranes were incubated separately overnight at 4°C with two monoclonal anti-MET antibodies (1:200, clone D-4: sc-514148, Santa Cruz Biotechnology, US), (1:250, MAB3729: clone-4AT44, Millipore, US) in addition to one polyclonal anti-MET antibody (1:100, C-28: sc-161, Santa Cruz Biotechnology, US). The IRDye 800CW conjugated goat anti-mouse secondary antibody was used at a 1:2000 dilution (LI-COR, US) for the two monoclonal antibodies. The IRDye®) 680RD goat anti-Rabbit IgG secondary antibody was used at a 1:1000 dilution (LI-COR, US) for the polyclonal antibody. The equality of loading was assessed by a rabbit monoclonal antibody for GAPDH (clone 14C10, Cell Signalling Technology). Signals were captured using the Odyssey CLx system along with Image studio™ 5.2 software (LI-COR Biosciences).
2.7. Cell Proliferation Assay:
The cell proliferation assay was performed in triplicate with a cell density of 4 × 103 cells/well in 96 well plates and eight 3X serial dilutions of the drug, ranging from 100 to 0.046 μM. Two MET tyrosine kinase inhibitors, crizotinib and tivantinib, were purchased from MedChemExpress (NJ, USA). Serial dilutions of Curcumin (Sigma-Aldrich, US) ranging from 50 to 0.781 μg/ml were used as standards to ensure the consistency of the experiments across replicates. Cells were incubated at 37°C for 72 hours in complete media and cell viability was assessed using the CellTiter Aqueous Non-Radioactive Cell Proliferation Assay, (Promega, Madison, WI, USA), according to the manufacturer’s protocol. The absorbance at a wavelength of 490 nm was measured using the Epoch Microplate Spectrophotometer (BioTek Instruments, Inc., US). The half-maximum inhibitory concentration (IC50) was assessed using Gen5 software, version 1.11.5. This program calculates the IC50 using the dose-response equation [Y = {(A – D)/[l + (X/C) B ]} + D], where X is the drug concentration, Y is the absorbance at 490 nm, A is the upper asymptote, B is the slope, C is the IC50, and D is the lower asymptote.
2.8. Immunohistochemistry (IHC) Analysis:
A rabbit polyclonal MET antibody optimized from immunohistochemistry (C-28, Santa Cruz Biotechnology, Inc.) was used to evaluate the total MET protein expression in the HCC tumour and non-tumour tissues. IHC was performed according to our previously published protocol (Abdel-Rahman et al. 2010) using the Dako HRP kit (Glostrup, Denmark). MET staining was evaluated by two independent pathologists (MHA and DM). The IHC staining intensity was scored according to a four-tier system: 0, no staining; 1+, weak; 2+, moderate; and 3+, strong. In brief, the H-score of each sample was calculated as the sum of each intensity (0-3) multiplied by the percentage of positive cells (0-100%) determined by IHC. The score ranged from 0-300. The median value of H-score was calculated. We used the score method defined in the MetMAb phase III trial (NCT01456325). MET overexpression was defined as a MET IHC staining score ≥ 2+ and/or ≥ 50% of tumour cells positive for membranous or cytoplasmic MET and it is called MetMAb assessment (Spigel et al. 2012).
2.9. Statistical Analysis:
Much of the data for assays described herein were largely descriptive in nature with a focus on validation of the presence or absence of a finding in human tissue that we observed in cell cultures. Qualitative data are summarized as proportions and percentages. Differences between the proportions of the tumour and non-tumour samples for the same patient were analysed by McNemar’s test (Hazra and Gogtay 2016a). Comparisons among unrelated categorical variables were performed using the chi-square test or Fisher’s exact test. Overall survival (OS) was defined as time from date of diagnosis till date of death or lost follow up, whichever comes first. Univariate analysis using Cox proportional hazards regression was performed to analyse several clinicopathological characteristics and the MET gene and L1-MET transcript expression and H-score of MET by IHC as independent prognostic factor of survival. The hazard ratio (HR) and 95%confidence interval (CI) were calculated. Correlation was done using Karl Pearson’s correlation coefficient method (Hazra and Gogtay 2016b). All tests were two-sided, and the level of statistical significance was P<0.05. R Foundation for Statistical Computing, Vienna, Austria version 3.6.0 and SPSS software version 20 (IBM corporation) were used for the statistical analysis. R packages used in the analysis were ‘survminer’ version 0.4.4 for survival analysis. SPSS was used for the remaining analyses.
3. Results:
3.1. MET, L1-MET Transcripts and MET protein Expression in cell lines:
Using the COSMIC database, we didn’t identify any variant in the six liver cancer cell lines.-Comparison of the STR genotyping results to the DNA typing data on Cellosaurus verified the authentication of the six cell lines used.
Qualitative PCR identified the expression of both MET and L1-MET transcripts in all cell lines at the expected product sizes of 117 and 109 base pairs, respectively.
Quantitative assessment of the expression of MET and L1-MET transcripts in cell lines showed marked variation with SNU-387 showing the highest expression of L1-MET and SNU-449 showing the highest expression MET (Table 2).
Table 2.
Results of RT-qPCR, proliferation assay for six Liver Cancer cell lines used in the study.
| Variables | Liver Cancer cell lines | ||||||
|---|---|---|---|---|---|---|---|
| SK-HEP-1 | PLC-PRF-5 | SNU-387 | SNU-423 | SNU-449 | SNU-475 | ||
| RT-qPCR (Relative gene expression, ±SD). | RNA-MET | 5.16±0.29 | 0.47±0.04 | 1.66±0.04 | 1.46±0.15 | 11.79±2.71 | 1.38±0.16 |
| RNA, L1-MET | 0.68±0.10 | 1.35±0.11 | 8.13±0.83 | 0.298±0.02 | 2.49±0.42 | 4.96±0.82 | |
| Western Blot- Relative Band Intensity: Clone D-4, Santa Cruz Biotechnology. | MET Protein (132KD) | 0.064 | 0.017 | 0.065 | 0.238 | 0.005 | 0.027 |
| Western Blot -Relative Band Intensity: MAB3729, Millipore. | MET Protein (100KD) | 1.06 | 0.014 | 0.62 | 0.35 | 0.5 | 0.21 |
| Western Blot -Relative Band Intensity: C-28: sc-161, Santa Cruz Biotechnology. | MET Protein (140KD) | 2.40 | 2.0 | 8.0 | 4.5 | 3.33 | 2.75 |
| Proliferation Assay “Anti-Met Drugs” “IC50,μM, ±SD”. | Crizotinib | 8.32±1.44 | 8.45±0.33 | 7.98±0.65 | 4.59±1.09 | 3.48±0.66 | 3.74±0.71 |
| Tivantinib | 5.65±1.20 | 8.24±0.09 | 5.12±10 | 1.31±0.56 | 0.37±0.21 | 1.42±0.04 | |
Three different antibodies were used for assessment of MET expression (Figure. 1). Significant variation was observed between the three antibodies.
Fig. 1.
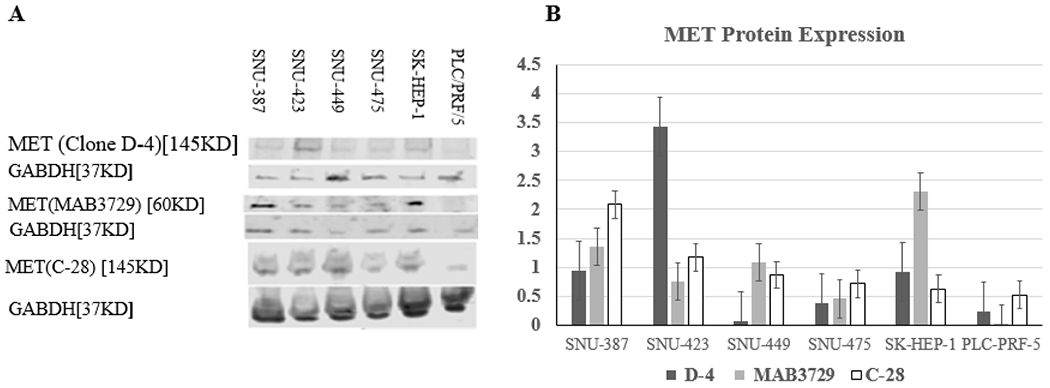
Relative protein expression of MET and GAPDH gene by Western blot in six liver cancer cell lines using three antibodies showing marked variation in the expression: clone D-4, Santa Cruz Biotechnology, MAB3729, Millipore and C-28: sc-161, Santa Cruz Biotechnology. Note that clone D-4 and C-28 detects full-length protein while MAB3729 detects only a truncating protein at 60 KDs.
Quantitative assessment of the MET transcript and the relative protein expression of MET showed was no significant correlation by the MAB3729, the D-4 clone and C-28 antibodies (P=0.43, P=0.51, P=0.76; respectively) [r= 0.40, r= −0.34 and r=−0.162, respectively].
3.2. Cell Proliferation Assay:
Table 2 shows the IC50 results of crizotinib and tivantinib in the six liver cancer cell lines. The IC50 of each cell line represents the average of at least two independent experiments, each performed in triplicate. SNU-449 had the lowest IC50 for both drugs while PLC/PRF/5 had the highest IC50 for both drugs (Table 2). There is a weak correlation between MET protein expression and the IC50 of crizotinib (r= 0.28 for MAB3729, r= −0.125 for clone D-4, r=−0.121 for C-28) and IC50 of tivantinib (r= 0.013 for MAB3729, r= −0.258 for clone D-4, r=−0.077 for C-28). While RNA expression of MET showed a stronger correlation with IC50 of both crizotinib (r=−0.398) and tivantinib (r=−0.463) but it didn’t reach statistical significance (P=0.43 and P=0.36. respectively) due to the small sample size used (n=6).
3.3. MET gene, L1-MET transcript and MET protein expression in HCC patients:
We assessed the expression of MET and L1-MET transcripts as well as MET protein expression in each tumour compared to its matching non-tumour tissue. When compare to matching non-tumour tissues, tumours had statistically significant higher expression of MET (28, 59.6%), Li-MET (31, 66%) and MET protein (22, 46.8%) expressions, Table 3. This suggests that MET overexpression is important in HCC tumorigenesis. However, variation in the expression in the non-tumour tissues was observed with a subset of non-tumour liver showing higher expression of MET, L1i-MET and MET compared the average of the non-tumour controls. This suggests that MET activation occurs also in premalignant liver tissues. MET protein expression was mostly cytoplasmic (Figure. 3), with weak nuclear staining detected in six (12.8%) HCC samples. The median H-score values for the tumour and non-tumour samples were 130 and 100, respectively. According to the defined criterion for the MetMAb assessment, 25 HCC cases (53.19%) had low MET expression, while 22 cases (46.80%) had high MET expression. There was no significant correlation between MET RNA transcript and MET protein expression of tumour tissues (r=0.011). Association of MET RNA and protein expression with both survival and recurrence:
Table 3.
Semi quantitative expression of MET, L1-MET transcript and MET protein in HCC tumors relative to non-tumor tissues from the same patient.
| Variable (Total number of patients =47) | Less than NT | Equal to NT | Higher than NT | P Value |
|---|---|---|---|---|
| Canonical MET | 11 (23.4%) | 8 (17.0%) | 28 (59.6%) | 0.006 |
| L1-MET | 8 (17%) | 8 (17%) | 31 (66%) | 0.0001 |
| MET protein | 8 (17%) | 17 (36. 2%) | 22 (46.8%) | 0.011 |
NT: matching non-tumour tissue
Fig. 3.
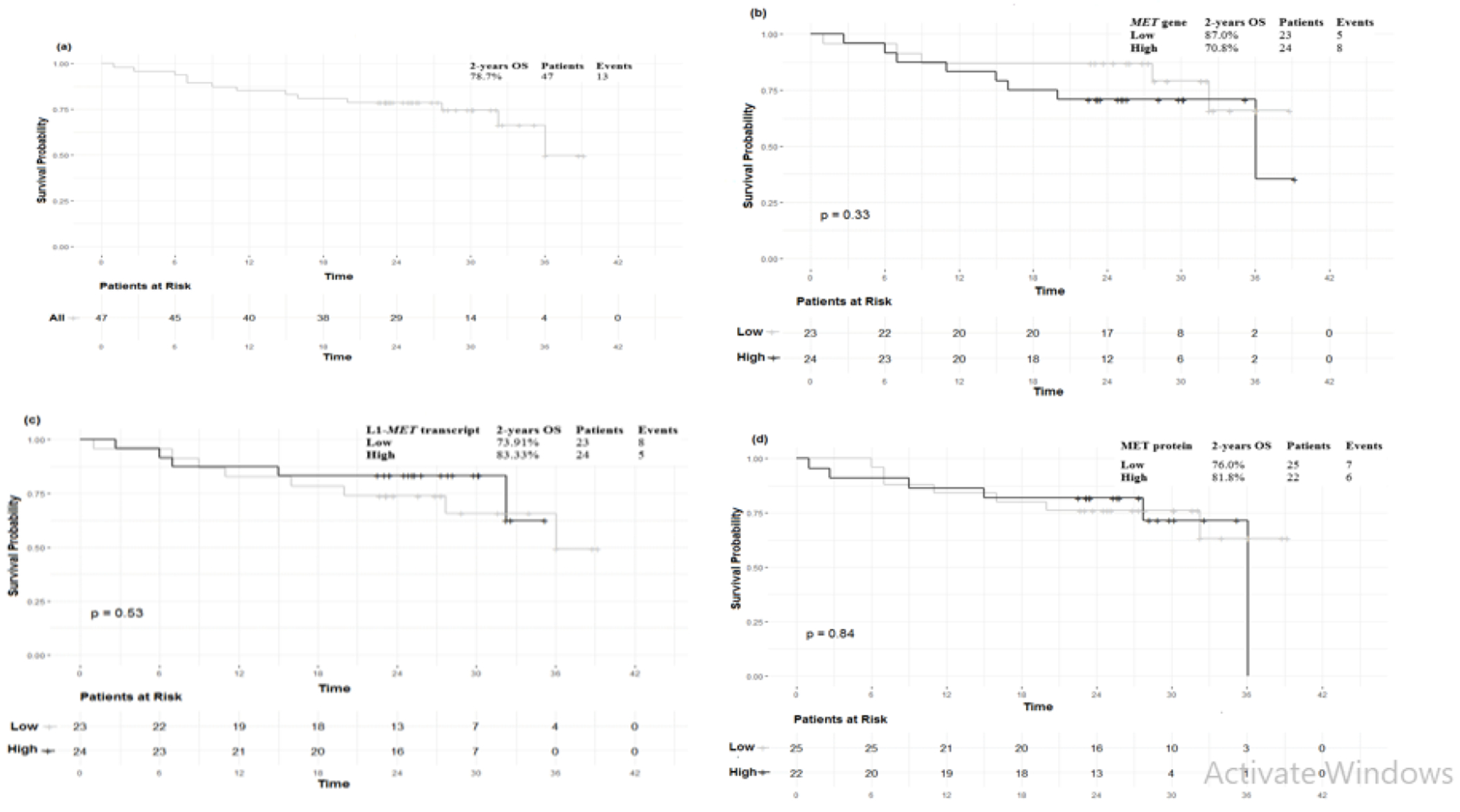
Kaplan-Meier analysis of two-years overall survival (OS). (a). two-year OS (78.7%), (b). low vs high tumour canonical MET gene expression (80.0%vs 78.1%, P=0.88), (c). low vs high tumour L1-MET transcript expression (75.9% vs 83.3%, P=0.72), (d). low vs high tumour MET protein expression (76.0% vs 81.8%, P=0.84). No significant association was observed between MET and L1-MET transcripts, as well as, MET protein expressions and OS of HCC patients
The two-year OS of HCC patients was 78.7%%, and the standard error (SE) was 1.87 (Figure. 3a). Using univariate Cox proportional hazards regression analysis, no significant statistical association were detected between the 2-year OS and the expression of MET and L1-MET transcript or the MET protein (P=0.86, 0.82, and 0.99, respectively) (Figure. 3b, 3c and 3d).
Ten patients showed recurrence of their tumours in the follow up period. No significant statistical association was detected between tumour recurrence and MET, L1-MET transcripts or MET protein expression (P=0.48, 1.0, and 1.0, respectively).
4. Discussion:
The most significant finding in our study is that the response to the tyrosine kinase inhibitors in HCC cell lines correlated with MET canonical transcript expression but not L1-MET or MET protein expressions. Given the reported variation in MET protein expression between different antibodies (Pozner-Moulis et al. 2007), we utilized two monoclonal antibodies and one polyclonal for detection of MET protein expression. Significant variations in the expression of the three antibodies were detected, Figure. 1. Similar to the previous report (Pozner-Moulis et al. 2007), clone MAB3729 detected only a truncated protein and not the full length transcript. This clone was suggested to be used for IHC assessment of MET expression based on the low lot to lot variability compared to other MET antibodies (Pozner-Moulis et al. 2007).
We also observed no significant correlation between MET transcript and MET protein expression for the three antibodies. The strong correlation between MET transcript expression and in vitro antitumor effect of tivantinib was also noted by Gao et al., on HCC cell lines with higher mRNA expression level of MET (Gao et al. 2019).
Based on our in-vitro studies, we suggest that canonical MET transcript, but not L1-MET, could predict chemo-sensitivity to tivantinib and crizotinib therapies in HCC patients, however, further in-vivo studies are required to support this observation. RNA based clinical testing is currently available for prognostication of several tumours such as breast cancer (Harris et al. 2016; Vieira and Schmitt 2018) and BCR-ABL-positive leukaemias (Ozemri Sag et al. 2015). RNA expression is objective and reproducible and can be carried out using relatively small sample size (Xi et al. 2017).
In a phase II clinical trial, tivantinib resulted in improved OS in HCC patients with high MET protein expression (Santoro et al. 2013). However, in a randomized, double-blind phase III (METIV-HCC) clinical trial tivantinib failed to improve the OS of in advanced-stage HCC patients with high MET expression (Rimassa et al. 2018). Although both studies used the same antibody (The Ventana CONFIRM anti total MET (SP44) and the same scoring system for MET protein (Spigel et al. 2012) but concerns about the optimum cut-off for identifying HCC patients sensitive to MET inhibitors has been raised (Weekes et al. 2018). Our results on the superiority of RNA expression as biomarker as well as reported variability in the quantitative detection of MET protein (Pozner-Moulis et al. 2007), that we also observed, support such concern. Of note, 22 (46.80%) of the tumours in our cohort had high MET expression based on currently used criteria for assessment of MET protein overexpression by immunohistochemistry. However, when we used the average expression in the non-tumour tissues, we identified only 6 tumours (12.8%) with higher expression (SD+1) for canonical MET transcript. This could suggest that a much smaller number of patients may benefit from MET inhibitor targeted therapies than initially thought.
Our study showed no significant correlation between canonical MET and L1-MET chimeric transcripts (r=−0.171) in HCC patients. Also, L1-MET chimeric transcript didn’t show significant association with OS and recurrence (P=0.53 and 1.0, respectively). Similar to Zhu et al. we observed statistical significant difference between L1-MET transcript expression in HCC tumours compared to non-tumour tissues (P<0.001) (Zhu et al. 2014). However, contrary to their findings, we didn’t observe a significant positive correlation between MET gene expression and L1-MET transcript expression in HCC tumour samples. Thiscould be attributed to the etiological difference between HCC in our cohort, which was mostly from HCV patients, and the HBV associated tumours in Zhu et al.’s cohort (Zhu et al. 2014).
We detected high MET protein expression in 46.8% of the HCC patient tumour samples, which is within the range reported by a recently published meta-analysis (Kim et al. 2017). However, contrary to the conclusion of that study, we didn’t observe prognostic value for either MET protein or RNA expression.
Our study has certain limitations, including the following: (1) The sample size of HCC patients was small, (2) We didn’t test MET inhibitors on patients, (3) Data regarding the exact date of disease recurrence were unavailable.
Conclusion
Our in-vitro studies suggest that canonical MET transcript could predict chemo-sensitivity to tivantinib and crizotinib in HCC. Further in-vivo assessments are required for validation. The expression of the canonical MET, L1-MET transcripts and MET protein are not prognostic biomarkers in Egyptian HCC patients.
Supplementary Material
Fig. 2.
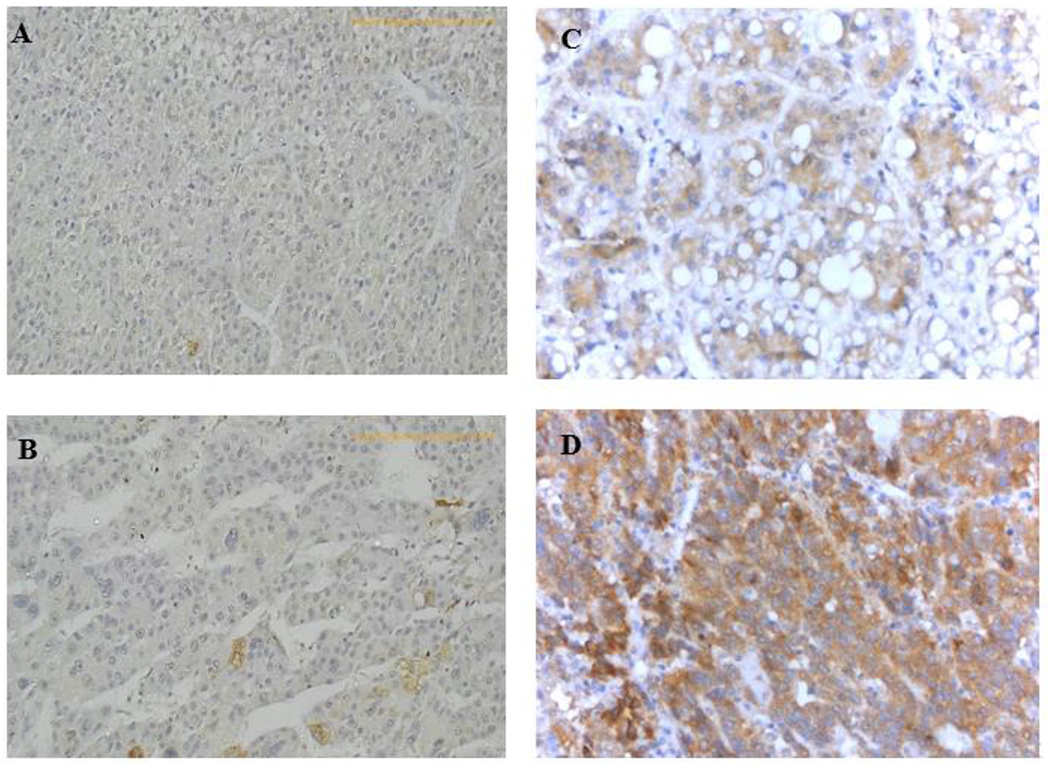
Representative examples of MET protein expression in HCC tissues assessed by immunohistochemistry. (a) Negative with no expression of MET detected (b) Mild/focal cytoplasmic and occasional nuclear expression, (c) Moderate cytoplasmic and nuclear expression, (d) Strong cytoplasmic with occasional nuclear expression. (200x, IMP)
Acknowledgements
The authors would like to thank Asmaa Mosbeh, PhD, from the SSI Team at the National Liver Institute [NLI], Menoufia University, for her help in sample preparation and practical work. Also, the authors would like to thank Getachew Boru, PhD and J. Brandon Massengill (Department of Ophthalmology and Visual Science, Havener Eye Institute, Ohio State University) for help with experimental setup and critical reading and English editing of the manuscript.
Funding: This work was supported by the National Liver Institute Sustainable Science Institute (NLI-SSI) of Menoufia University and by a UICC Technical Fellowship (UICC-TF/18/575818) awarded to WMR.
Footnotes
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
Ethical Statement: All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards. The study was approved by the Institutional Ethical Review Board-approved protocols of the NLI, Menoufia University (IRB0051/2012).
Consent to participate: Informed consent was obtained from all individual participants included in the study.
Consent to publish: Patients signed informed consent regarding publishing their data.
Conflict of Interest: None.
References:
- Abdel-Rahman MH, Boru G, Massengill J, et al. (2010) MET oncogene inhibition as a potential target of therapy for uveal melanomas. Investigative ophthalmology & visual science 51:3333–9. doi: 10.1167/iovs.09-4801 [DOI] [PubMed] [Google Scholar]
- Ally A, Balasundaram M, Carlsen R, et al. (2017) Comprehensive and Integrative Genomic Characterization of Hepatocellular Carcinoma. Cell 169:1327–1341 e23. doi: 10.1016/j.cell.2017.05.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bairoch A (2018) The Cellosaurus, a Cell-Line Knowledge Resource. Journal of biomolecular techniques : JBT 29:25–38. doi: 10.7171/jbt.18-2902-002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouattour M, Raymond E, Qin S, et al. (2018) Recent developments of c-Met as a therapeutic target in hepatocellular carcinoma. Hepatology (Baltimore, Md) 67:1132–1149. doi: 10.1002/hep.29496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferlay J, Soerjomataram I, Dikshit R, et al. (2015) Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. International Journal of Cancer 136:E359–E386. doi: 10.1002/ijc.29210 [DOI] [PubMed] [Google Scholar]
- Forbes SA, Beare D, Boutselakis H, et al. (2017) COSMIC: somatic cancer genetics at high-resolution. Nucleic acids research 45:D777–D783. doi: 10.1093/nar/gkw1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Chen H, Huang X, et al. (2019) ARQ-197 enhances the antitumor effect of sorafenib in hepatocellular carcinoma cells via decelerating its intracellular clearance. OncoTargets and therapy 12:1629–1640. doi: 10.2147/OTT.S196713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han JS (2010) Non-long terminal repeat (non-LTR) retrotransposons: mechanisms, recent developments, and unanswered questions. Mobile DNA 1:15. doi: 10.1186/1759-8753-1-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris LN, Ismaila N, McShane LM, et al. (2016) Use of Biomarkers to Guide Decisions on Adjuvant Systemic Therapy for Women With Early-Stage Invasive Breast Cancer: American Society of Clinical Oncology Clinical Practice Guideline. Journal of Clinical Oncology 34:1134–1150. doi: 10.1200/JCO.2015.65.2289 [DOI] [PubMed] [Google Scholar]
- Hazra A, Gogtay N (2016a) Biostatistics Series Module 4: Comparing Groups - Categorical Variables. Indian journal of dermatology 61:385–92. doi: 10.4103/0019-5154.185700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazra A, Gogtay N (2016b) Biostatistics series module 6: Correlation and linear regression. Indian Journal of Dermatology 61:593. doi: 10.4103/0019-5154.193662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda T (2016) Links between Human LINE-1 Retrotransposons and Hepatitis Virus-Related Hepatocellular Carcinoma. Frontiers in chemistry 4:21. doi: 10.3389/fchem.2016.00021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes VS, Siemann DW (2018) Have Clinical Trials Properly Assessed c-Met Inhibitors? Trends in Cancer 4:94–97. doi: 10.1016/j.trecan.2017.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Kim HS, Kim BJ, et al. (2017) Prognostic value of c-Met overexpression in hepatocellular carcinoma: a meta-analysis and review. Oncotarget 8:90351–90357. doi: 10.18632/oncotarget.20087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Yu X-F, Zou J, Luo Z-H (2015) Prognostic value of c-Met in colorectal cancer: a meta-analysis. World journal of gastroenterology 21:3706–10. doi: 10.3748/wjg.v21.i12.3706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llovet JM, Villanueva A, Lachenmayer A, Finn RS (2015) Advances in targeted therapies for hepatocellular carcinoma in the genomic era. Nature Reviews Clinical Oncology 12:408–424. doi: 10.1038/nrclinonc.2015.103 [DOI] [PubMed] [Google Scholar]
- Naufer MN, Furano AV, Williams MC (2018) Protein-nucleic acid interactions of LINE-1 ORF1p. Seminars in Cell & Developmental Biology. doi: 10.1016/j.semcdb.2018.03.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozemri Sag S, Yakut T, Gorukmez O, et al. (2015) Qualitative and Quantitative Evaluation of the BCR-ABL Fusion Gene in Chronic Myelogenous Leukemia by Flourescence In Situ Hybridization and Molecular Genetic Methods. Genetic Testing and Molecular Biomarkers 19:584–588. doi: 10.1089/gtmb.2015.0056 [DOI] [PubMed] [Google Scholar]
- Parikh PK, Ghate MD (2018) Recent advances in the discovery of small molecule c-Met Kinase inhibitors. European Journal of Medicinal Chemistry 143:1103–1138. doi: 10.1016/j.ejmech.2017.08.044 [DOI] [PubMed] [Google Scholar]
- Pozner-Moulis S, Cregger M, Camp RL, Rimm DL (2007) Antibody validation by quantitative analysis of protein expression using expression of Met in breast cancer as a model. Laboratory Investigation 87:251–260. doi: 10.1038/labinvest.3700515 [DOI] [PubMed] [Google Scholar]
- Puccini A, Marín-Ramos NI, Bergamo F, et al. (2019) Safety and Tolerability of c-MET Inhibitors in Cancer. Drug Safety, doi: 10.1007/s40264-018-0780-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyo J-S, Kang G, Cho WJ, Choi SB (2016) Clinicopathological significance and concordance analysis of c-MET immunohistochemistry in non-small cell lung cancers: A meta-analysis. Pathology - Research and Practice 212:710–716. doi: 10.1016/j.prp.2016.05.006 [DOI] [PubMed] [Google Scholar]
- Rimassa L, Assenat E, Peck-Radosavljevic M, et al. (2018) Tivantinib for second-line treatment of MET-high, advanced hepatocellular carcinoma (METIV-HCC): a final analysis of a phase 3, randomised, placebo-controlled study. The Lancet Oncology 19:682–693. doi: 10.1016/S1470-2045(18)30146-3 [DOI] [PubMed] [Google Scholar]
- Rueden CT, Schindelin J, Hiner MC, et al. (2017) ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinformatics 18:529. doi: 10.1186/s12859-017-1934-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro A, Rimassa L, Borbath I, et al. (2013) Tivantinib for second-line treatment of advanced hepatocellular carcinoma: a randomised, placebo-controlled phase 2 study. The Lancet Oncology 14:55–63. doi: 10.1016/S1470-2045(12)70490-4 [DOI] [PubMed] [Google Scholar]
- Siegel RL, Miller KD, Jemal A (2016) Cancer statistics, 2016. CA: A Cancer Journal for Clinicians 66:7–30. doi: 10.3322/caac.21332 [DOI] [PubMed] [Google Scholar]
- Spigel DR, Edelman MJ, Mok T, et al. (2012) Treatment Rationale Study Design for the MetLung Trial: A Randomized, Double-Blind Phase III Study of Onartuzumab (MetMAb) in Combination With Erlotinib Versus Erlotinib Alone in Patients Who Have Received Standard Chemotherapy for Stage IIIB or IV Met-Positive Non-Small-Cell Lung Cancer. Clinical Lung Cancer 13:500–504. doi: 10.1016/j.cllc.2012.05.009 [DOI] [PubMed] [Google Scholar]
- Vieira AF, Schmitt F (2018) An Update on Breast Cancer Multigene Prognostic Tests-Emergent Clinical Biomarkers. Frontiers in medicine 5:248. doi: 10.3389/fmed.2018.00248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber B, Kimhi S, Howard G, et al. (2010) Demethylation of a LINE-1 antisense promoter in the cMet locus impairs Met signalling through induction of illegitimate transcription. Oncogene 29:5775–5784. doi: 10.1038/onc.2010.227 [DOI] [PubMed] [Google Scholar]
- Weekes CD, Clark JW, Zhu AX (2018) Tivantinib for advanced hepatocellular carcinoma: is MET still a viable target? The Lancet Oncology 19:591–592. doi: 10.1016/S1470-2045(18)30249-3 [DOI] [PubMed] [Google Scholar]
- Xi X, Li T, Huang Y, et al. (2017) RNA Biomarkers: Frontier of Precision Medicine for Cancer. Non-Coding RNA 3:9. doi: 10.3390/ncrna3010009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan S, Jiao X, Zou H, Li K (2015) Prognostic significance of c-Met in breast cancer: a meta-analysis of 6010 cases. Diagnostic Pathology 10:62. doi: 10.1186/s13000-015-0296-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S, Yu Y, Zhao N, et al. (2013) c-Met as a Prognostic Marker in Gastric Cancer: A Systematic Review and Meta-Analysis. PLoS ONE 8:e79137. doi: 10.1371/journal.pone.0079137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C, Utsunomiya T, Ikemoto T, et al. (2014) Hypomethylation of Long Interspersed Nuclear Element-1 (LINE-1) is Associated with Poor Prognosis via Activation of c-MET in Hepatocellular Carcinoma. Annals of Surgical Oncology 21:729–735. doi: 10.1245/s10434-014-3874-4 [DOI] [PubMed] [Google Scholar]
- Zucman-Rossi J, Villanueva A, Nault J-C, Llovet JM (2015) Genetic Landscape and Biomarkers of Hepatocellular Carcinoma. Gastroenterology 149:1226–1239.e4. doi: 10.1053/j.gastro.2015.05.061 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


