Abstract
Lysosomes are in the center of the cellular control of catabolic and anabolic processes. These membrane-surrounded acidic organelles contain around 70 hydrolases, 200 membrane proteins and numerous accessory proteins associated with the cytosolic surface of lysosomes. Accessory and transmembrane proteins assemble in signaling complexes that sense and integrate multiple signals and transmit the information to the nucleus. This communication allows cells to respond to changes in multiple environmental conditions, including nutrient levels, pathogens, energy availability and lysosomal damage, with the goal of restoring cellular homeostasis. This review summarizes our current understanding of the major molecular players and known pathways that are involved in control of metabolic and stress responses that either originate from lysosomes or regulate lysosomal functions.
Keywords: Lysosomes, nutrient sensing, mTOR, autophagy, transcription factors, TFEB
Lysosomes regulate degradation and cellular adaptation to changing environmental situations
Lysosomes constitute the main degradative organelle in eukaryotic cells, enclosing a wide repertoire of acid hydrolases capable of digesting macromolecules such as proteins, glycans, lipids, and nucleic acids. The catalytic function of lysosomes is critical for many cellular processes, including turnover of cellular components, downregulation of surface receptors, bone remodeling, inactivation of pathogenic organisms and antigen presentation.
The strategic position of lysosomes as end points of the endocytic, phagocytic and autophagic pathways, allows them to receive material from different sources, including nutrients and proteins internalized through endocytosis, viruses and bacteria trapped by phagocytosis, and cytosolic components, including entire organelles, delivered via autophagy. Importantly, lysosomes not only degrade and recycle the building blocks of this material but use it to collect information about changing environmental conditions, integrate the corresponding multiple signals, and generate a response that allow cellular adaptation and homeostatic regulation of cell physiology. This response involves interconversion of many proteins and importantly also elicits transcriptional regulation by modulating the activity of various transcription factors. Since lysosomes are directly involved in the control of autophagy, they also regulate the overall degradation of cellular macromolecules. This novel role as signaling hubs, which has also been highlighted in excellent recent review articles [1, 2], has changed our view of lysosomes, placing them in the center of the cellular response to stress. Here we provide insight into the most recent advances that have been made towards understanding of how lysosomes orchestrate different degradative, metabolic and signaling functions in health and disease.
Lysosomes function as signaling platforms
Recent evidence has shown that multiple types of signals are sensed and integrated at the lysosome-limiting membrane, allowing modulation of critical cellular processes such as nutrient sensing, energy metabolism, immune response, and adaptation to lysosomal damage. The activation of these pathways requires a sophisticated and regulated recruitment of signaling complexes to the lysosomal surface, as well as the subsequent activation of specific transcription factors that coordinate the expression of elaborate transcriptional networks.
Metabolic regulation
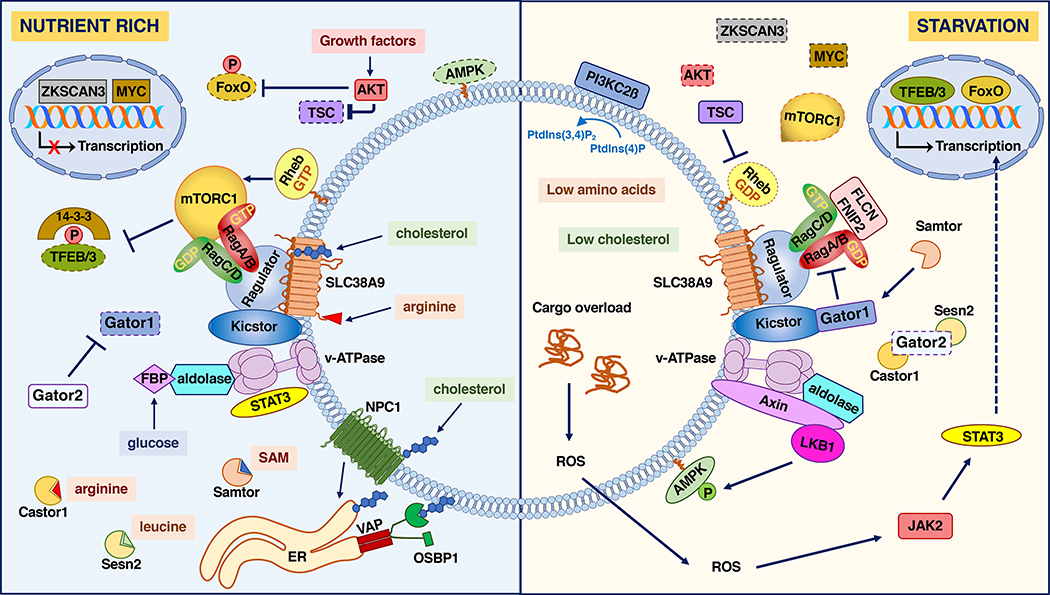
Lysosomes function as platforms for metabolic signal transduction and for detecting variations on the levels of different nutrients, including amino acids, glucose and lipids, in particular cholesterol (Figure 1).
Figure 1. Metabolic regulation in lysosomes.
Scheme of the different signaling pathways assembled on the lysosome surface in response to variations in the levels of amino acids, glucose, growth factors and cholesterol. Multiple nutrient signals, including glucose, amino acids and cholesterol converge to regulate the activity of Rag GTPases. When the levels of these nutrients are elevated (left panel), Rag GTPases activate, promoting the recruitment of the mTORC1 complex to the lysosome surface. At the same time, high levels of growth factors cause displacement of the TSC complex from the lysosomal surface, preventing its GAP activity towards Rheb. GTP-bound Rheb then promotes mTORC1 activation. A number of sensors, including Castor, Sestrin2 and Samtor, detect variations in the cytosolic pool of amino acids, whereas the lysosomal amino acid permease SLC38A9 recognizes amino acid concentration, in particular arginine, inside the lysosome lumen. Cholesterol transfer from the endoplasmic reticulum (ER) to the lysosomal membrane via OSBP1 also favors mTORC1 activation. Active mTORC1 phosphorylates members of the MiT/TFE family of transcription factors, such as TFEB and TFE3, promoting their binding to 14-3-3 and consequent cytosolic sequestration. At the same time, active AKT promotes autophagy inhibition through phosphorylation and inactivation of beclin and FoxO transcription factors. When the nutrients are scarce (right panel), GATOR1 is recruited to lysosomes inducing GDP-loading of RagA/B and inactivation of mTORC1. This allows translocation of TFEB and TFE3 to the nucleus to induce expression of multiple genes implicated in lysosomal biogenesis, autophagy, and metabolic regulation. The autophagic/lysosomal axis is also modulated by activation of FoxO and STAT3. Low glucose levels, results in recruitment of Axin-LKB1 to lysosomes and activation of AMPK, which balance energy homeostasis by promoting catabolic processed to restore ATP levels. Dashed lines indicate inactivation; P inside a red circle represents inhibitory phosphorylation, P inside a green circle represents activating phosphorylation.
Metabolic regulation by amino acids
Mechanistic target of rapamycin complex 1 (mTORC1; see Glossary) is an important regulator of cellular metabolism that is mainly activated by growth factors and amino acids. mTORC1 promotes anabolic processes, including protein synthesis, glucose metabolism and nucleotide and lipid biosynthesis through phosphorylation of numerous proteins and activation of transcription factors, such as Hypoxia-inducible factor 1-alpha (HIF1alpha) and Sterol regulatory element-binding proteins (SREBPs) [3]. At the same time, mTORC1 blocks catabolic pathways, such as autophagy via inhibition of Unc-51 like autophagy activating kinase 1 (ULK1) and the Microphthalmia family of bHLH-LZ transcription factors (MiT/TFE).
Activation of mTORC1 occurs on lysosomes (reviewed in [1]). When the levels of cellular amino acids are high, the mTORC1 complex is recruited to the cytosolic face of lysosomal membranes through interaction with active Ras-related GTP-binding protein (Rag) GTPases, which are themselves anchored to lysosomes by the pentameric protein complex, Ragulator (also called LAMTOR). Rags function as heterotetramers consisting of two heterodimers. The active complex consists of RagA/B with GTP bound to RagA and RagC/D with GDP bound to RagC. Amino acids trigger the GTP loading of RagA/B proteins, promoting binding and recruitment of mTORC1 to lysosomes [4]. Once on the lysosomal surface, mTORC1 is allosterically activated by association with the lysosomal-bound GTPase Ras homolog enriched in brain (Rheb) [5]. The association of Rheb with mTORC1 requires growth factor induced phosphorylation and dissociation of the Tuberous Sclerosis Complex (TSC1/TSC2) from the lysosomal surface. Therefore, different stimuli, i.e. growth factors and amino acids, must cooperate in order to achieve full mTORC1 activation. In the absence of amino acids, Rags turn into an inactive conformation, causing shuttling of mTORC1 to the cytosol. Inactive Rags also recruit TSC2, the GTPase activating protein (GAP) component of the TSC complex, to promote Rheb inactivation [6], indicating that Rags play an active role terminating mTORC1 signaling under starvation conditions.
It has been suggested that lysosomes act as amino acid storage sites. mTORC1, although located at the cytosolic surface of lysosomes, has the ability to respond to amino acid levels in the lysosomal lumen. According to the current model, both the vacuolar-ATPase (v-ATPase) and the membrane-spanning amino acid transporter sodium-coupled neutral amino acid transporter 9 (SLC38A9) sense intralysosomal amino acids and transmit these signals to the Ragulator complex, which in turn triggers nucleotide exchange on RagA/B to the GTP-bound state as a prerequisite for mTORC1 activation [7, 8]. A growing number of proteins have also been identified in the regulation of Rag activation in response to changes in the cytoplasmic pool of nutrients (Box 1).
Box 1. Sensing of cytosolic amino acids by Rags.
Sensing of amino acid availability by Rags is critical to modulate mTORC1 activation. Major mechanistic insight into this process was gained with the identification of the GTPase-activating protein toward Rags (GATOR) complex by protein cross-linking coupled mass spectrometry. The GATOR complex consists of two subcomplexes: the trimeric GATOR1, composed of Nitrogen permease regulator like protein (Nprl)2, Nprl3 and DEP domain containing 5 (Depdc5), which exhibits GAP activity toward RagA/B, and the pentameric GATOR2 that inhibits GATOR1 in the presence of amino acids [91]. The recruitment of GATOR1 to lysosomal membranes is mediated by KICSTOR, a tetrameric complex whose mutation results in mTORC1 hyperactivation [92]. GATOR1 and GATOR2 interact directly with cytoplasmic amino acid sensors. Under amino acid deprivation conditions, CASTOR, a sensor for arginine [93], and Sestrin2, a sensor for leucine [94], bind GATOR2, blocking its inhibitory effect on GATOR1. At the same time, the methionine sensor SAMTOR binds GATOR1, activating its GAP activity toward RagA/B and causing mTORC1 inactivation [95]. Nutrient-dependent ubiquitination of RagA by ring finger protein 152 (RNF152) and S-phase kinase-associated protein 2 (Skp2) has also been described as a mechanism to promote GATOR1 recruitment and prevent mTORC1 hyperactivation [96]. Under nutrient rich conditions the interaction of GATOR2 with Sestrin2 and CASTOR is interrupted, allowing GATOR2-mediated inhibition of GATOR1 and mTORC1 activation.
Rag activity is also regulated by the Folliculin (FLCN)/Folliculin-interacting protein (FNIP) complex, which functions as a GAP for RagC, thus promoting Rag and mTORC1 activation. FLCN/FNIP is recruited to lysosomes under starvation conditions in a GATOR1-dependent manner [97], where GDP-loaded RagA keeps the complex in an inactive conformation. Loading of RagA with GTP following nutrient accessibility, releases FLCN/FNIP to induce GDP-loading of RagC, further promoting Rag activation [98]. In addition, LeuRS, best characterized for its role in charging leucine to its cognate tRNA for protein translation, localizes to lysosomes in a leucine dependent manner and functions as a GAP for RagD [99]. Leucine has also been shown to promote glutaminolysis through allosteric binding of glutamate dehydrogenase, promoting the formation of α-ketoglutarate which induces GTP-loading of RagB through a mechanism dependent on proteins in the prolyl hydroxylase (PHD) family [100]. Overall, it has become clear that the precise control of Rag signaling is critical for cellular adaptation to constantly changing environmental demands.
Modulation of the mTORC1 response is critical, as evidenced by its well-characterized role in cancer progression. Recently, several mechanisms of mTORC1 activity attenuation have been described, including dissociation of the Rag/mTORC1 complex from lysosomes to the cytosol [9], regulation of lysosomal surface availability [10], and changes in lysosomal positioning [11] (Box 2).
Box 2. Lysosomal positioning and signaling.
Closer inspection reveals that lysosomes are preferentially localized in two pools, one which is perinuclear and the other one which is in the periphery [11]. The formation and dispersion of dynamic clusters of lysosomes regulates fusion with endosomes. This process is dependent on the active transport of lysosomes confined by the ER network [101]. Cell migration and adhesion, cancer cell invasion, microbial killing, neuronal function and immune responses are linked to a regulated intracellular distribution of lysosomes [11]. Lysosomes can move in a bidirectional manner along microtubules. They make use of retrograde-acting dynein-dynactin or anterograde-acting kinesin motors [11]. It is speculated that during intracellular transport lysosomes can physically interact with other organelles. The positioning of lysosomes is tightly linked with the available nutrient levels and also with the activation state of mTORC1 [102]. A key observation is that the cellular activation of mTORC1 by nutrients causes a scattering of lysosomes to the cell periphery. Deprivation of nutrients causes the redistribution of lysosomes to the perinuclear region characterized by an increased number of fusion events between lysosomes and autophagosomes. Retrograde dynein-mediated motility of lysosomes is occurring when amino acids or growth factors are removed. In this case mTORC1 signaling is reduced. Nuclear translocation of TFEB causes an increased expression of some dynein-dynactin-involved transport proteins. The dynein-dynactin motors can be coupled to lysosomes by a complex of the lysosomal Ca2+ channel TRPML1 and ALG2, a protein involved in Ca2+ sensing [103]. The lysosomal phosphoinoside Ptdins(3,5)P(2) activates this coupling event and triggers tubulation and reformation of lysosomes. The lysosomal membrane protein TMEM55B and the adaptor protein JIP4 can interact and contribute to the dynein-dependent transport of lysosomes [104]. Phosphorylation of TMEM55B by Erk/MAPK may regulate this transport pathway [104]. The small GTPase Rab7 couples lysosomes to dynein-dynactin via RILP. RILP can interact with folliculin and rab34 but it also interacts with ORP1L, which can cholesterol-dependently accelerate ER contact site formation. Anterograde kinesin-mediated transport of lysosomes towards the plus end of microtubules occurs when sufficient nutrients are present. It involves BLOC-1-related complex (BORC), ADP-ribosylation factor-like protein 8 (Arl8), SKI-interacting protein (SKIP) bound to kinesins [105–108]. Ragulator interacts with BORC under starvation inhibiting recruitment of kinesins and it dissociates when growth factors are present. The ER protein protrudin stimulates FYCO1 to bind to Rab7 and PtdIns3-P at the lysosomal membrane [109]. This stimulates binding to kinesin 1 and anterograde transport. A lack of amino acids reduces PtdIns3-P levels and blocks this transport pathway [24].
Metabolic regulation by glucose
AMP-activated protein kinase (AMPK) is a well-recognized energy sensor and regulator of energy metabolism [12]. Increases in Adenosine monophosphate (AMP) and Adenosine diphosphate (ADP) cellular levels, as a result of Adenosine triphosphate (ATP) consumption, trigger AMPK activation. AMP binds the AMPK-gamma subunit, inducing allosteric changes, and facilitates the phosphorylation of a threonine residue (Thr172) located in the activation loop of the AMPK-alpha subunit, inducing AMPK activation [13]. Activated AMPK phosphorylates a variety of downstream targets to promote catabolic processes (e.g. glycolysis, fatty acid-beta oxidation, autophagy) that restore ATP levels.
Lysosomes have recently emerged as a crucial place to mediate AMPK signaling [14]. A fraction of AMPK constitutively localizes to the lysosome surface, where it remains inactive under high glucose conditions. Following glucose starvation, the complex formed by AXIN and the AMPK-activating kinase, Liver kinase B1 (LKB1), is recruited to lysosomes via direct interaction with the v-ATPase and Ragulator. At the same time, binding of AMP to the AMPK-gamma subunit, induces a conformational change that favors AXIN/LKB1/AMPK interaction, resulting in AMPK activation. Interestingly, glucose starvation can induce lysosomal AMPK activation even before the cellular levels of AMP increase [15]. Fructose-1,6-biphosphate (FBP), a glycolysis intermediate that is utilized by aldolase, prevents the recruitment of AXIN/LKB1 to lysosomes. In low glucose, FBP-unoccupied aldolase facilitates the formation of a v-ATPase/Ragulator/AXIN/LKB1/AMPK super-complex that induces lysosomal AMPK phosphorylation. This suggests that lysosomes are not just platforms for AMPK activation but play a critical role in glucose sensing and in the modulation of the cellular response to glucose deprivation. Thus, AMP-independent activation of AMPK on lysosomes may be an early event following glucose starvation that is important not just to activate catalytic pathways, but to induce mTORC1 inactivation through AMPK-dependent phosphorylation of the mTORC1 subunit Raptor [16] and the TSC-1-TSC2 complex [17], allowing rapid autophagy induction. If glucose levels remain low for longer periods of time, increased AMP/ATP ratios may further contribute to the activation of AMPK not only on lysosomes but also in the cytosol and mitochondria. Therefore, compartmentalization of AMPK may permit the activation of different metabolic responses depending on the degree of energy stress.
Glucose can also regulate the lysosomal localization of mTORC1 in an AMPK-independent manner [18]. Recent evidence showed that phosphofructokinase1 (PFK1) and 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 (PFKB3), two important regulatory glycolytic enzymes, bind RagB GTPase to promote mTORC1 recruitment to lysosomes [19], further emphasizing the important role of lysosomes as sensory platforms for glucose metabolism and response to energy stress.
Metabolic regulation by lipids
Lysosomes play a central role in the regulation of cholesterol homeostasis. Several lysosomal membrane proteins, including Niemann Pick Disease, type C1 (NPC1), Lysosomal associated membrane proteins (LAMPs) and Lysosomal integral membrane protein type 2 (LIMP2/SCARB2) are involved in the transport of internalized cholesterol from the lysosomal lumen to its limiting membrane, where a complex machinery implicated in lysosomal positioning, cholesterol binding, and contact with other organelles, contributes to the transfer of cholesterol to different cellular membranes [20]. Importantly, the levels of lysosomal cholesterol modulate mTORC1 activity. Increased cholesterol levels inside lysosomes are sensed by SLC38A9 via its CARC and CRAC cholesterol-binding motifs, leading to Rag GTPase activation and increased mTORC1 recruitment [21]. At the same time, and in concert with its ER membrane anchors VAMP-associated protein (VAP)A and (VAP)B, Oxysterol-binding protein (OSBP) delivers cholesterol from the ER to lysosomes in exchange for Phosphatidylinositol 4-phosphate [PI(4)P]. Cholesterol deposited on lysosomes interacts with the mTORC1 scaffolding machinery to promote Rag GTPase-mediated mTORC1 activation, suggesting that ER-lysosome contacts play an important role in cholesterol sensing by mTORC1 [22]. In contrast, NPC1 inhibits cholesterol-mediated mTORC1 activation by limiting levels of cholesterol in the lysosomal limiting membrane [22].
Changes in levels of lysosomal phosphoinositides (PIPs) have also an important impact on lysosome function, affecting processes such as lysosome-autophagosome fusion, lysosome positioning and lysosomal reformation (reviewed in [23]). While PIPs generally function as allosteric activators of proteins or by promoting recruitment of specific factors to cellular membranes, recent evidence suggests that they also directly modulate nutrient signaling. For example, full activation of mTORC1 requires generation of PI(3)P via Vacuolar protein sorting 34 (VPS34), a process that is thought to facility ER-lysosome contact sites via FYCO-protrudin interactions [24]. Conversely, recruitment of PI3KC2ß to lysosomal membranes under growth factors deprived conditions, increases PI(3,4)P2 levels, resulting in binding of 14-3-3 to raptor and mTORC1 inactivation [25]. When growth factors are abundant, mTORC2-dependent activation of protein kinase N2 (PKN2) results in PI3KC2 ß phosphorylation, leading to its binding to 14-3-3 and cytoplasmic sequestration [26]. At the same time, mTORC2 represses activation of chaperone-mediated autophagy (CMA) in a particular set of lysosomes by activating AKT and negatively regulating the assembly of LAMP2A into the CMA translocation complex [27].
A mechanism of regulating lipid metabolism via lysosome-to-nucleus signaling has recently been described in C. elegans [28]. Under starvation conditions, activation of the transcription factors FoxO/DAF-16 and TFEB/HLH-30 leads to increased levels of the lysosomal lipase LIPA/LIPL-4, which generates the lipid oleoylethanolamide (OEA). Binding of OEA to the fatty acid-lipid binding protein LBP-8 induces translocation of the OEA/lipid binding protein-8 (LBP-8) complex from the lysosome to the nucleus. In the nucleus, OEA binds its nuclear hormone receptor (NHR)-80, which cooperates with NHR-49 to induce the expression of multiple genes implicated in mitochondrial beta-oxidation and response to oxidative stress. These metabolic adjustments improve lipid catabolism and resistance to stress, resulting in increased longevity [29].
Immune responses
The contribution of lysosomes to different types of immune responses is well established. For example, tubulation of lysosomes in dendritic cells is critical for delivery of Major Histocompatibility Complex (MHC) class II-peptide complexes to the plasma membrane, favoring antigen presentation [30], while certain lysosomal membrane proteins, such as NPC1, LAMP1 and LIMP-2 regulate viral entry, infection and pathogenesis [31–33]. The MHCII invariant chain (CD74) is also proteolyzed within the membrane of lysosomes by the signal peptide peptidase type 2A (SPPL2A), thereby promoting B-cell development and function [34]. Lysosomes are also the site of degradation for many pathogens. Products of microbial degradation are then transported to the cytosol where they are detected by specific sensors. Degradation of microbial membranes produces lipopeptides that are delivered to the cytosol by SLC15A3 and SLC15A4 to activate Nucleotide-binding oligomerization domain-containing protein 2 (NOD2) [35]. Likewise, double stranded (ds)RNAs are transported via SIDT2 to activate the cytosolic sensors Retinoic acid inducible gene 1 (RIG-1) and melanoma differentiation-associated protein 5 (MDA5) [36].
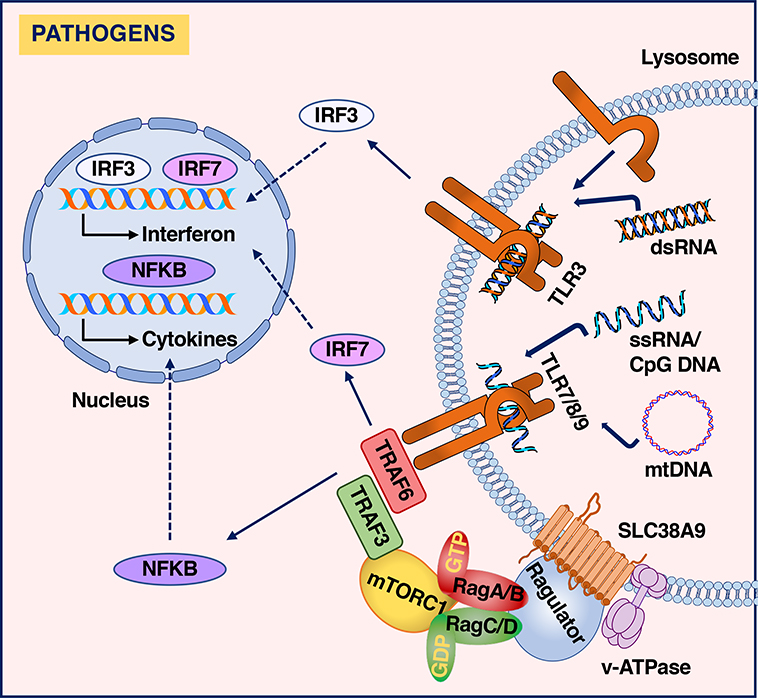
Microbial nucleic acids also activate Toll-like receptors (TLRs). Five members of the TLR family, TLR3, TLR7, TLR8, TLR9, and TLR13, recognize different nucleic acid features in the lumen of endolysosomes. Upon binding to their specific ligands, TLRs cluster, leading to the dimerization and oligomerization of their cytoplasmic domains. This promotes the recruitment of multiple cytosolic signaling adaptors and ultimately results in the expression of inflammatory cytokines and interferons [37]. Therefore, lysosomes act as signaling platforms that facilitate the efficient and localized activation of the TLR-mediated immune responses (Figure 2).
Figure 2. Activation of the immune response in lysosomes.
Multiple cellular stress responses are initiated on lysosome membranes. Sensing of nucleic acids in the lysosome is critical for detection of microbial infections and activation of immune responses. Binding of double-stranded RNA (dsRNA) to TLR3, single-stranded RNA (ssRNA) to TLR7 and TLR8, and single-stranded unmethylated DNA containing the cytosine-phosphate-guanine (CpG) to TLR9 in the lumen of endolysosomes induces TLR dimerization and oligomerization of their cytoplasmic motifs, resulting in recruitment of signaling adaptor proteins and activation of signaling cascades that trigger transcription of interferons and inflammatory cytokines. TLR9 also senses mitochondrial DNA (mtDNA) as a readout of the autophagy-mediated delivery of mitochondria to lysosomes.
Emerging evidence suggests an important cross-regulation between immune and metabolic sensors. Anabolic processes supported by mTORC1, such as glucose uptake, mitochondrial activity or protein and lipid synthesis, are essential to promote immune responses [38]. Furthermore, the innate immune kinase TANK binding kinase 1 (TBK1) interacts and phosphorylates mTORC1 to increase its catalytic activity, placing mTORC1 as a direct downstream effector of TBK1 in the regulation of innate immunity [39]. mTORC1 associates with several interferon signaling molecules, including TNF receptor-associated factor (TRAF)3 and IKKalpha, bringing them to lysosomal membranes. After ligand stimulation, recruitment of TRAF6 by TLR7 and TLR3, allows TRAF6/TRAF3 interaction, resulting in efficient interferon production [40]. In agreement with this model, interferon production in response to TLR3, TLR7 and TLR9 activation is severely impaired by mTORC1 inhibition [39, 40]. Another example of communication between immunity and metabolism in lysosomes is the recent discovery that TLR9 binds mitochondrial DNA, which is delivered to the lysosomal lumen via autophagy. TLR9 activation by mitochondrial DNA initiates a cargo response that results in remodeling of lysosomal membranes and modulation of autophagosome/lysosome fusion [41]. When lysosomal degradation is impaired, persistent TLR9 activation may lead to nuclear factor-κB (NF-κB) nuclear translocation, resulting in inflammation. Thus, TLR9 functions as a sensor of autophagic delivery to lysosomes and orchestrates adaptation of cellular degradative capabilities.
Response to lysosomal damage
Intracellular pathogens, aggregates and crystals, may rupture lysosomal membranes, leading to the release of lysosomal luminal components, such as cathepsins, to the cytosol and resulting in cell death [42]. Therefore, cells have evolved mechanisms to recognize, repair and eliminate damaged lysosomes (Figure 3).
Figure 3. Detection and elimination of damaged lysosomes.
Cell must be able to detect, repair and eliminate damaged lysosomes. Minor damage of the lysosomal membrane results in the calcium-dependent recruitment of ALIX and ESCRT-I, followed by the recruitment of ESCRT-III and VPS4. This results in the sealing or repair of the membrane by a mechanism yet to be characterized, although it has been suggested that may implicate the formation of luminal vesicles. Lysosomal damage that cannot be repaired by the ESCRT complex triggers the recruitment of galectins, which recruit adaptors to promote lysosome degradation via autophagy (lysophagy). In addition, galectin 8 and galectin 9 inactivate mTORC1 and activate AMPK, respectively, to further activate autophagy and modulate metabolism and removal of damaged lysosomes.
Small disruptions in lysosomal membrane integrity trigger a lysosomal repair mechanism that allows membrane sealing through recruitment of the endosomal sorting complex required for transport (ESCRT) complex [43]. Alternatively, if the harm is extensive, exposed luminal glyco-conjugates are recognized by galectins, which bind and recruit autophagy receptors (e.g. Nuclear dot protein 52 kDa (NDP52) and Tripartite Motif Containing 16 (TRIM16) in the case of galectin-8 and galectin-3, respectively) to assure elimination of damaged lysosomes by lysophagy [44]. Importantly, galectins do not just play a passive role in recruiting the autophagy machinery. Binding of galectin-8 to luminal lysosomal glycans includes glycosylated residues in SLC38A9, which affects the activation status of Rag GTPases and causes mTORC1 inactivation. At the same time, galectin-9 associates with TAK1, an upstream activating kinase of AMPK, inducing AMPK activation [45]. In this way, galectins transmit lysosome damage signals directly to mTORC1 and AMPK, a process that results in autophagy induction and consequent removal of damaged lysosomes, but likely extends to a broader modulation of cellular metabolism. In addition, intracellular galectins play an important role in the innate response against pathogens, mainly by promoting autophagic clearance of bacteria-containing vesicles [46]. However, they also modulate non-canonical inflammasome activation [47], as well as TLR7/TLR9 mTOR-dependent endosomal signaling [48]. This suggests that galectins play a dual role coordinating lysosomal membrane disruption and inflammation.
Recent evidence indicates that regulated leakage of cathepsins from lysosomes may have important physiological functions. For example, release of cathepsin B from a set of lysosomes adjacent to chromatin during prometaphase seems critical for efficient chromosome segregation and maintenance of genomic integrity in mammalian cells and tissues [49]. In addition, several lysosomal cathepsins have been implicated in NLRP3 inflammasome activation [50], further suggesting that controlled lysosomal membrane permeability may be a critical contributor to both, physiological and pathological processes. How such a selective cathepsin escape is mediated is still unknown.
Transcription factors modulate lysosome function
Lysosomes not only sense and integrate a wide variety of environmental cues, they must also transmit this information to allow cellular response and adaptation. Part of this response controlled by signaling pathways initiated on lysosomes results in changes in the distribution and activity of different transcription factors, some of which directly regulate lysosome numbers, lysosomal protein expression levels and lysosomal activity as a way to further enhance or attenuate lysosomal signaling.
MiT/TFE transcription factor family
The MiT/TFE family of basic helix-loop-helix leucine-zipper transcription factors, which includes Microphthalmia-associated transcription factor (MITF), Transcription factor (TF)EB, Transcription factor binding to IGHM enhancer 3 (TFE3), and TFEC, is present in most metazoan organisms. Under conditions of stress, the MiT/TFE members are activated and coordinate the initiation of multiple stress response pathways, promoting elimination of damaged macromolecules and organelles, preservation of cellular functions and, ultimately, restoration of cellular homeostasis (reviewed in [51]). TFEB and TFE3 bind directly to the promoter of numerous autophagic and lysosomal genes [52–54] causing autophagy induction, lysosomal biogenesis and modulation of lysosomal activity (e.g. by regulating lysosomal acidification, exocytosis, degradative capability, Ca2+ homeostasis, and positioning).
The main regulatory mechanism for the transcription factors TFEB and TFE3 is the control of their translocation from the cytosol to the nucleus, although changes in their stability, dimerization, degradation and nuclear export are also important contributors (reviewed in [2]). Retention of TFEB and TFE3 in the cytosol is achieved by phosphorylation of specific residues that mediate binding to the cytosolic chaperone 14-3-3. Under basal (non-stressed) conditions, TFEB and TFE3 are recruited to the surface of lysosomes through their interaction with active Rag GTPases [55], allowing mTORC1-dependent phosphorylation and binding to 14-3-3 [54, 56–58]. In addition to mTORC1, other kinases likely contribute to TFEB retention in the cytosol [59]. Dephosphorylation of TFEB and TFE3 under stress conditions, for example by inactivation of mTORC1 and/or activation of specific phosphatases, such as calcineurin [60] or protein phosphatase 2A (PP2A) [61], causes a rapid translocation of these transcription factors to the nucleus. A wide variety of stressors induce TFEB and TFE3 activation, including nutrient deprivation [53, 54, 56–58], inflammation [62], pathogens [63–66], oxidative stress [61, 67], accumulation of unfolded proteins [68], physical exercise [69], DNA damage [70, 71], increased cytosolic Ca2+ [60], and mitochondrial damage [72], suggesting that the modulation of the autophagic/lysosomal pathway is a critical component of the cellular stress response.
Signal transducer and activator of transcription-3 (STAT3)
STAT3 was originally identified as a latent cytosolic transcription factor that translocates to the nucleus in response to different cytokines and interferons to regulate inflammation and immunity. Later on, a more pleiotropic role for STAT3 was established, with a particular relevance in cancer cells, where it functions as an oncogene by activating genes involved in proliferation, differentiation, metastasis, apoptosis, angiogenesis and metabolism [73].
Recently, it has been suggested that STAT3 may also act as an important regulator of lysosomal function. Accumulation of undegraded cargo inside the lysosome lumen or defective lysosomal protease activity is sufficient to induce accumulation of STAT3 in the nucleus, resulting in STAT3-dependent/TFEB-independent expression of multiple lysosomal hydrolases. The mechanisms of STAT3 stimulation by cargo overload requires the generation of oxidative stress inside lysosomes and the consequent activation of the JAK2-STAT3 pathway [74]. Interestingly, while TFEB and TFE3 simultaneously coordinate the autophagic and lysosomal pathways, activation of STAT3 does not result in autophagy induction. This could be important to prevent further cargo accumulation inside lysosomes in conditions in which lysosomal proteolysis is impaired.
STAT3 plays an important role regulating lysosomal-mediated cell death during post-lactational regression of the mammary gland. During involution, activation of STAT3 in the epithelial cells of the mammary gland results in a dramatic upregulation of the lysosomal proteases cathepsin B and cathepsin L, as well as downregulation of their serine protease inhibitor (Spi)2A [75]. STAT3 also promotes uptake of milk-fat globules that are delivered to lysosomes for degradation [76] and overall stimulation of endolysosomal trafficking [77]. Hyperactivation of cathepsins, combined with high concentration of free fatty acids, may increase lysosomal membrane permeability (LMP), leading to lysosomal leakiness and caspase-independent cell death. Based on these studies, it has been suggested that the development of small molecules with the ability to induce STAT3 transcriptional activity might be a good approach to induce lysosome-mediated cell death in cancer cells [78].
STAT3 also regulates lysosomal homeostasis in a transcription-independent manner. Cytosolic STAT3 directly associates with the lysosomal v-ATPase, stimulating its ATPase activity and contributing to the acidification of the lysosomal lumen [79]. This regulation seems to be particularly relevant in cancer cells, where acidification of the cytosol due to increased glycolysis promotes rapid recruitment of STAT3 to lysosomal membranes, thus allowing increased proton pumping activity of the v-ATPase and neutralization of the acidified cytosol. Therefore, STAT3 may contribute to the reverse pH gradient (alkaline cytosol and acidic extracellular medium) known to be critical for malignant transformation and cancer progression.
The forkhead box O (FoxO) family
The FoxO family of transcriptions factors includes four members in mammals (FoxO1, FoxO3, FoxO4, and FoxO6) and has orthologs in worm, fly, rodent, and zebrafish [80]. While the important role of FoxO proteins in regulation of oxidative stress, proliferation, cell differentiation, survival, metabolic homeostasis, and autophagy is well-established, recent evidence suggests that they may also modulate lysosome function.
FoxO-dependent autophagy regulation is crucial for tissue homeostasis. In many organs, such as liver, neurons and kidney, FoxO-induced autophagy has a protective role; however, it also contributes to atrophy of cardiac and skeletal muscle. The FoxO proteins regulate autophagy by three different mechanism, including direct binding to the promoter of autophagy genes causing increased transcription, induction of epigenetic changes, and direct interaction with autophagy proteins (reviewed in [81]). Several post-translational modifications assure retention of FoxO proteins in the cytosol under normal conditions. This includes phosphorylation of key residues by AKT, as well as acetylation via p300 and binding to X-box binding protein 1 (XBP1). Following stress, phosphorylation of FoxO by AMPK and JNK, together with methylation and Poly ADP-ribosylation (PARylation), result in increased FoxO nuclear accumulation and transcriptional activity. FoxO proteins transactivate multiple genes implicated in different stages of the autophagic process by binding to their promoters. Moreover FoxO also increases the expression of Ras-related in brain (Rab)7, a late endosome/lysosome protein important for different trafficking processes, as well as the expression of TFEB, thus directly modulating lysosomal function [82]. FoxO also increases expression of Sestrin3, which activates AMPK and inactivates mTORC1, potentially inducing TFEB/TFE3 activation [83].
Induction of changes in chromatin structure constitute an efficient way to modulate gene expression. In response to glucose starvation, AMPK-mediated phosphorylation of FoxO3 causes its translocation to the nucleus to induce downregulation of S-phase kinase associated protein 2 (SKP2), a component of the SCF E3 ligase complex that mediates degradation of coactivator-associated arginine methyltransferase (CARM1). As a consequence, the levels of CARM1 are upregulated, causing demethylation of histone H3 to foster gene expression. CARM1 also functions as a transcriptional co-activator of TFEB, resulting in upregulation of autophagic and lysosomal genes [84].
The cooperation between FoxO and TFEB for regulating gene expression is likely more complex than currently recognized. In C. elegans, DAF-16 and HLH-30, the orthologs of FoxO and TFEB, respectively, are activated in response to a variety of distress signals. In some cases, they target and modulate expression of genes involved in response to stress and development independently. However, regulation of other genes, particularly those implicated in longevity and response to oxidative stress, requires interaction and combinatorial recruitment of DAF-16 and HLH-30 to promoters, suggesting a tight coordination of the activation of these transcription factors [85].
MYC and Zinc finger protein with KRAB and SCAN domains 1 (ZKSCAN3)
MYC is a member of the basic-helix-loop-helix leucine-zipper class of transcription factors implicated in controlling the expression of many genes important for proliferation, biomass accumulation, energy metabolism and stem-cell self-renewal. Recently, a novel role for MYC as an antagonist of MiT/TFE transcription factors has been reported. MYC binds E-boxes in the promoter sites of lysosomal and autophagy genes, preventing occupancy of the same sites by the MiT/TFE members. In addition, MYC recruits chromatin-modifying complexes, such as histone deacetylases, to further stabilize its binding to promoters. This MYC-dependent repression of autophagy and lysosomal biogenesis seems to be essential to prevent differentiation [86]. Under stress, inactivation of mTORC1 by amino acid deprivation or AMPK activation, causes not only activation of MiT/TFE members but also nuclear translocation of GSK3beta, which phosphorylates MYC in Thr58 to induce its ubiquitin-dependent proteasome degradation [87].
The zinc-finger with KRAB and SCAN domains 3 transcription factor, ZKSCAN3, is a negative regulator of autophagic and lysosomal gene expression. Silencing of ZKSCAN3 in cancer cells induces autophagy, increased lysosomal biogenesis, and senescence [88]. Furthermore, induction of lysosomal genes in TFEB-overexpressing cells is further augmented by ZKSCAN3 depletion, suggesting that both transcription factors work in conjunction. Mechanistically, it was described that stress-induced activation of the protein kinase C/c-Jun N-terminal kinase/p28 (PKC/JNK/p28) axis results in phosphorylation of the nuclear export sequence of ZKSCAN, causing its translocation from the nucleus to the cytosol. At the same time PKC inactivates glycogen synthase kinase 3 beta (GSK3beta) leading to TFEB activation [89]. It is important to note that some questions remain with regard to the role of ZKSCAN3 in vivo, since a recently generated ZKSCAN3 knockout mouse did not show alterations in the expression of autophagy or lysosomal genes [90].
Concluding remarks
Having regarded lysosomes as mere degradative endpoint for many decades, it is becoming increasingly clear that these organelles are at the center of sensing and controlling cellular stress events to allow for a rapid cellular response to adapt to changing extracellular conditions. Evolution has developed an intriguing machinery to employ the cytosolic surface of lysosomes to act as a signaling hub to integrate the information about levels of nutrients and to translate it into signal transduction events and transcription factor shuttling to the nucleus. Open questions (see related Box with Outstanding questions) in the field need to clarify which metabolic, nutritional and stress modulators are acting at the lysosomal surface under which specific condition. This leads to the question of which transcription factors signal transduction pathways and corresponding gene networks are activated by which of these stressors. It will also be interesting to more exactly determine the role of lysosomal signaling in LSDs (Box 3) and also in common diseases (Box 4) where lysosome dysfunction has been observed. A relevant albeit still hypothetical question is if a modulation of single or combined lysosomal signaling events can also be beneficial for treating patients suffering from LSDs, cancer, neurodegeneration and metabolic disorders.
Outstanding questions.
What additional metabolites, nutrients or stress conditions are sensed on the lysosomal surface? Could those vary depending on the cell type?
How does contact of lysosomes with other organelles, such as endoplasmic reticulum, mitochondria, nucleus or lipid droplets modulate metabolic pathways?
Can the nutrient-dependent regulation of mTORC1 and MiT/TFE transcription factors be overridden depending on specific cellular needs? For example, do cancer cells count with additional mechanisms to prevent mTORC1 inactivation by starvation to facilitate proliferation and survival?
Which physiological processes require controlled release of lysosomal hydrolases and how is this release regulated?
How the different transcription factors regulating lysosomal activity coordinate their activity in response to different stress conditions?
Given that MiT/TFE, FoxO, and STAT3 transcription factors regulate expression of genes, not only implicated in the autophagic/lysosomal pathways, but also in cell cycle, survival, homeostasis and inflammation, what is the specific contribution of lysosomes to immunity, longevity, proliferation, and cell differentiation?
How aberrant lysosomal signaling contribute to LSDs? Is correction of metabolic pathways, either alone or in combination with enzyme replacement therapy, a valid therapeutic strategy?
How does lysosomal signaling contribute to disease progression in cancer, neurodegeneration and autoimmunity?
Box 3. Lysosomal signaling and lysosomal storage disease.
It is not so much of a surprise that signaling from lysosomes and to lysosomes is affected in dysfunctional lysosomes. The prototype of such a dysfunction is storage lysosomes as observed in lysosomal storage disorders (LSDs) where lysosomal hydrolases, accessory proteins, membrane transporters or trafficking proteins can be mutated [110]. An intriguing discovery was that TFEB controls Ca2+-mediated lysosomal exocytosis, a fusion of lysosomes with the plasma membrane. This led to a rescue of the lysosomal storage in different types of LSD models [111]. In Pompe disease a TSC knockdown or arginine supply reversed the pathological autophagic buildup [112]. Similarly, an induced expression of TFEB reduced lysosomal storage, increased exocytosis of lysosomes and accelerated autophagic clearance [113]. It is of special note that the forced expression of TFEB is linked with cancer development and control of cell growth, differentiation and survival [114]. In sialidosis, an exacerbation of lysosomal exocytosis may affect the integrity of the plasma membrane and extracellular matrix remodeling and degradation which possibly promotes precancerous conditions [115, 116]. In human iPSC-neuronal Gaucher Disease cells mTORC1 seems hyperactive most likely caused by the glycosphingolipid (GSL) storage. This could be reversed by inhibition of mTORC1 which increased autophagic clearance [117]. GSL accumulation is also evident in Krabbe disease models leading to an inhibition of the IGF-1-PI3K-Akt-mTORC2 signaling and increased neuronal vulnerability [118]. In ceroid lipofuscinosis (CLN)7 disease the reactivation of mTORC1 after starvation was negatively affected [119]. Sonic hedgehog (Shh) signaling and associated developmental processes are affected in MPSII and NPC diseases due to an accumulation of glycosaminoglycans (GAGs) and cholesterol, respectively. Whereas GAGs may interfere extracellularly with the Shh ligand binding to patched cholesterol accumulation in lysosomes reduces the activation of Smoothened and Shh. Both pathways seem connected to a disturbed autophagic flow [120]. In the MPS disorders, MPSI and MPSVII defects in bone development are explained by dysregulated hedgehog signaling [120]. In Gaucher Disease (GD) models developmental defects due to a downregulation of the Wnt/β-catenin pathway were found. Pharmacological Wnt/β-catenin activation rescued osteoblast abnormalities [121]. An enhanced lysosomal exocytosis in sialidosis myofibroblasts causes an increase in the release of exosomes that contain profibrotic signaling molecules, including TGF-beta. This may explain the pathological expansion of connective tissue and generalized fibrosis [122]. Finally, calcium signaling is also altered in LSDs [123]. Reduced lysosomal calcium levels and release from lysosomes were reported for NPC1. Calcium release from lysosomes and more indirectly from the ER may regulate endocytosis, membrane fusion events and autophagy.
Box 4. Lysosomal signaling in neurodegeneration, cancer and autoimmunity.
When the autophagic flux is inhibited in the central nervous system degeneration of neurons is a common hallmark [124, 125]. Therefore, the continuous autophagic/lysosomal removal and clearance of protein aggregates and damaged organelles in postmitotic neurons is an essential process [1]. In common human neurodegenerative disorders such as Parkinson Disease (PD), Alzheimer Disease (AD), Amytrophic Lateral Sclerosis (ALS), Frontotemporal Dementia (FTD) and Huntington Disease (HD) dysfunctional lysosomes and an impaired autophagic pathway have been reported. Mice with neuronal targeted TFEB are characterized by reduced levels of amyloid beta peptides and amyloid plaques [126]. In PD degeneration in dopaminergic neurons is linked to reduced lysosome functions leading to cytoplasmic retention of TFEB [127]. Overexpression of TFEB led to neuroprotection and cleared pathological α-synuclein aggregates. Huntingtin (Htt) and poly-Q-expanded Htt which are linked to HD pathology promote the signaling by mTORC1 [128]. In human knockout cells of C9orf72, a gene mutated in familial FTD and/or ALS, enlarged lysosomes and an impaired response to mTORC1 signaling were observed [129]. The recruitment of C9orf72 to lysosomes is mediated by an interaction with the lysosomal cationic amino acid transporter PQLC2 and the availability of the amino acids arginine, lysine and histidine [130]. Interestingly, TMEM106b, another risk factor for FTD, is a lysosomal protein and its deficiency causes an impaired sorting of lysosomes and axonal transport in motor neurons [131]. A transcriptional activation of autophagy by MiT/TFEB is also observed in pancreatic cancer where this degradative pathway is required to supply metabolites for cell growth and survival. The transcription factors are pushed towards nuclear import, enhancing lysosomal biogenesis, degradation and sufficient amino acid supply. MiT/TFE can also directly promote tumorigenesis and amplifications of MITF are frequently found in melanomas [132]. Mutations in the gene encoding for Folliculin (FLCN) cause a rare tumor form (Birt-Hogg-Dubé syndrome). FLCN interacts with RagA/B GTPases and is involved in the amino-acid-dependent activation of mTORC1 [133]. Similarly, a wide spectrum of mutations in the TSC genes, which act as tumor suppressors can lead to brain, kidney, heart, lung and skin tumors. Also, mutations in the genes encoding for members of the GATOR complex may cause cancer. Inactivating mutations in GATOR1 led to an hyperactivation of mTORC1, insensitivity to amino acid starvation and hypersensitivity to the mTORC1 inhibitor rapamycin [134]. An upregulation of the activity of lysosome enzymes is also one of the hallmarks of autoimmune diseases such as lupus, rheumatoid arthritis and multiple sclerosis [135]. Lysosomes may affect both anti-inflammatory (phospholipase A2, cyclooxygenase 2) and pro-inflammatory (IL1-β-caspase 1) pathways [136].
Highlights.
Lysosomes are key sensors and mediators to quickly adapt cellular metabolism to environmental changes. They collect information about changing environmental conditions, translating it into cellular adaptations.
mTORC1 is activated on lysosomes by growth factors and amino acids, promoting many anabolic processes and inhibiting autophagy. The lysosomal membrane proteins v-ATPase and SLC38A9 sense intraluminal amino acids and transmit signals to the Ragulator complex.
Lysosomes act as platforms for AMPK activation, glucose sensing and regulation of the cellular response after glucose deprivation and energy stress. AMPK activation promotes catabolic processes.
Cholesterol in lysosomal membranes interacts with the mTORC1 machinery and promotes Rag-GTPase-mediated mTORC1 activation. Also, phosphoinositides impact lysosome function and modulate nutrient signaling.
Some Toll-like receptors sense intraluminal nucleic acids in lysosomes and regulate immune-responses. They are also involved in cross-regulation between immune and metabolic sensors controlling mTORC1.
Damaged lysosomes are repaired or removed by mechanisms involving the ESCRT complex machinery or recognition by galectins leading to autophagy activation, respectively.
The MiT/TFE transcription factor family members can be activated by changes in environmental conditions, thereby regulating stress response pathways and contributing to the restoration of cellular homeostasis.
Oxidative stress inside lysosomes, as observed in lysosomal-mediated cell death and cancer, activates STAT3, triggering the expression of multiple lysosomal hydrolases.
The FoxO family of transcriptions factors controls autophagy by causing increased transcription of autophagy genes after promoter binding, induction of epigenetic changes, and interaction with autophagy proteins.
MYC- and ZKSCAN3 transcription factors antagonize MiT/TFE transcription factors and repress autophagy and lysosomal biogenesis.
Acknowledgements
We thank Volkmar Gieselmann and Volker Haucke for critical reading and commenting our manuscript. This work was supported by the Deutsche Forschungsgemeinschaft (DFG; FOR2625) (PS) and the Intramural Research Program of the NHLBI of the National Institutes of Health (RP).
Glossary
- Mammalian target of rapamycin complex (mTORC1)
is a huge intracellular kinase protein complex acting as a central regulator of cell metabolism, growth, proliferation and survival. In general, mTORC1 activation switches metabolism towards an anabolic state by phosphorylation of multiple targets. Its activation depends on a complex protein machinery located on the cytosolic surface of lysosomes
- Microphthalmia family of bHLH-LZ transcription factors (MiT/TFE)
is a family of transcription factors composed of four members (MITF, TFEB, TFE3 and TFEC) containing DNA binding domains. They play important roles in the regulation of cellular processes such as lysosomal homeostasis and autophagy induction
- vacuolar-type H+-ATPase (v-ATPase)
is an ATP-dependent proton pump which drives organellar acidification. It is composed of at least 13 protein subunits in a membrane and cytosolic subcomplexes. It participates in processes of membrane trafficking, protein degradation, small molecule transport, autophagy and cell signaling
- AMP-activated protein kinase (AMPK)
is a heterotrimeric enzyme complex reacting on low energy situations. Accordingly, it switches the metabolism towards a catabolic state by phosphorylating multiple targets. Thus, AMPK is an mTORC1 antagonist. A subset of AMPK is also activated on the lysosomal cytosolic surface
- Toll-like receptors (TLRs)
are central players for innate immunity. They recognize pathogen-associated molecular patterns and can lead to activation of transcription factors which control immune reactions
- Lysosomal membrane permeability (LMP)
is a process where different types of stress disrupt the lysosomal membrane and lead to release of intraluminal proteins. There are specific responses leading to rapid repair or elimination of such damaged organelles
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lawrence RE and Zoncu R (2019) The lysosome as a cellular centre for signalling, metabolism and quality control. Nat Cell Biol 21 (2), 133–142. [DOI] [PubMed] [Google Scholar]
- 2.Ballabio A and Bonifacino JS (2020) Lysosomes as dynamic regulators of cell and organismal homeostasis. Nat Rev Mol Cell Biol 21 (2), 101–118. [DOI] [PubMed] [Google Scholar]
- 3.Duvel K et al. (2010) Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol Cell 39 (2), 171–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rogala KB et al. (2019) Structural basis for the docking of mTORC1 on the lysosomal surface. Science 366 (6464), 468–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang H et al. (2017) Mechanisms of mTORC1 activation by RHEB and inhibition by PRAS40. Nature 552 (7685), 368–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Demetriades C et al. (2014) Regulation of TORC1 in response to amino acid starvation via lysosomal recruitment of TSC2. Cell 156 (4), 786–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zoncu R et al. (2011) mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science 334 (6056), 678–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rebsamen M et al. (2015) SLC38A9 is a component of the lysosomal amino acid sensing machinery that controls mTORC1. Nature 519 (7544), 477–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lawrence RE et al. (2018) A nutrient-induced affinity switch controls mTORC1 activation by its Rag GTPase-Ragulator lysosomal scaffold. Nat Cell Biol 20 (9), 1052–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mutvei AP et al. (2020) Rap1-GTPases control mTORC1 activity by coordinating lysosome organization with amino acid availability. Nat Commun 11 (1), 1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cabukusta B and Neefjes J (2018) Mechanisms of lysosomal positioning and movement. Traffic 19 (10), 761–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herzig S and Shaw RJ (2018) AMPK: guardian of metabolism and mitochondrial homeostasis. Nat Rev Mol Cell Biol 19 (2), 121–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hawley SA et al. (1995) 5’-AMP activates the AMP-activated protein kinase cascade, and Ca2+/calmodulin activates the calmodulin-dependent protein kinase I cascade, via three independent mechanisms. J Biol Chem 270 (45), 27186–91. [DOI] [PubMed] [Google Scholar]
- 14.Zhang CS et al. (2014) The lysosomal v-ATPase-Ragulator complex is a common activator for AMPK and mTORC1, acting as a switch between catabolism and anabolism. Cell Metab 20 (3), 526–40. [DOI] [PubMed] [Google Scholar]
- 15.Zhang CS et al. (2017) Fructose-1,6-bisphosphate and aldolase mediate glucose sensing by AMPK. Nature 548 (7665), 112–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gwinn DM et al. (2008) AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell 30 (2), 214–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Inoki K et al. (2003) TSC2 mediates cellular energy response to control cell growth and survival. Cell 115 (5), 577–90. [DOI] [PubMed] [Google Scholar]
- 18.Efeyan A et al. (2013) Regulation of mTORC1 by the Rag GTPases is necessary for neonatal autophagy and survival. Nature 493 (7434), 679–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Almacellas E et al. (2019) Phosphofructokinases Axis Controls Glucose-Dependent mTORC1 Activation Driven by E2F1. iScience 20, 434–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meng Y et al. (2020) Cholesterol Handling in Lysosomes and Beyond. Trends Cell Biol 30 (6), 452–466. [DOI] [PubMed] [Google Scholar]
- 21.Castellano BM et al. (2017) Lysosomal cholesterol activates mTORC1 via an SLC38A9-Niemann-Pick C1 signaling complex. Science 355 (6331), 1306–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lim CY et al. (2019) ER-lysosome contacts enable cholesterol sensing by mTORC1 and drive aberrant growth signalling in Niemann-Pick type C. Nat Cell Biol 21 (10), 1206–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ebner M et al. (2019) Phosphoinositides in the control of lysosome function and homeostasis. Biochem Soc Trans 47 (4), 1173–1185. [DOI] [PubMed] [Google Scholar]
- 24.Hong Z et al. (2017) PtdIns3P controls mTORC1 signaling through lysosomal positioning. J Cell Biol 216 (12), 4217–4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marat AL et al. (2017) mTORC1 activity repression by late endosomal phosphatidylinositol 3,4-bisphosphate. Science 356 (6341), 968–972. [DOI] [PubMed] [Google Scholar]
- 26.Wallroth A et al. (2019) Protein kinase N controls a lysosomal lipid switch to facilitate nutrient signalling via mTORC1. Nat Cell Biol 21 (9), 1093–1101. [DOI] [PubMed] [Google Scholar]
- 27.Arias E et al. (2015) Lysosomal mTORC2/PHLPP1/Akt Regulate Chaperone-Mediated Autophagy. Mol Cell 59 (2), 270–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Folick A et al. (2015) Aging. Lysosomal signaling molecules regulate longevity in Caenorhabditis elegans. Science 347 (6217), 83–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramachandran PV et al. (2019) Lysosomal Signaling Promotes Longevity by Adjusting Mitochondrial Activity. Dev Cell 48 (5), 685–696 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vyas JM et al. (2007) Tubulation of class II MHC compartments is microtubule dependent and involves multiple endolysosomal membrane proteins in primary dendritic cells. J Immunol 178 (11), 7199–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carette JE et al. (2011) Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature 477 (7364), 340–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jae LT et al. (2014) Virus entry. Lassa virus entry requires a trigger-induced receptor switch. Science 344 (6191), 1506–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamayoshi S et al. (2009) Scavenger receptor B2 is a cellular receptor for enterovirus 71. Nat Med 15 (7), 798–801. [DOI] [PubMed] [Google Scholar]
- 34.Schneppenheim J et al. (2013) The intramembrane protease SPPL2a promotes B cell development and controls endosomal traffic by cleavage of the invariant chain. J Exp Med 210 (1), 41–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakamura N et al. (2014) Endosomes are specialized platforms for bacterial sensing and NOD2 signalling. Nature 509 (7499), 240–4. [DOI] [PubMed] [Google Scholar]
- 36.Nguyen TA et al. (2017) SIDT2 Transports Extracellular dsRNA into the Cytoplasm for Innate Immune Recognition. Immunity 47 (3), 498–509 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Majer O et al. (2017) Nucleic acid-sensing TLRs: trafficking and regulation. Curr Opin Immunol 44, 26–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Linke M et al. (2017) mTORC1 and mTORC2 as regulators of cell metabolism in immunity. FEBS Lett 591 (19), 3089–3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bodur C et al. (2018) The IKK-related kinase TBK1 activates mTORC1 directly in response to growth factors and innate immune agonists. EMBO J 37 (1), 19–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saitoh SI et al. (2017) TLR7 mediated viral recognition results in focal type I interferon secretion by dendritic cells. Nat Commun 8 (1), 1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.De Leo MG et al. (2016) Autophagosome-lysosome fusion triggers a lysosomal response mediated by TLR9 and controlled by OCRL. Nat Cell Biol 18 (8), 839–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang F et al. (2018) Lysosomal membrane permeabilization and cell death. Traffic 19 (12), 918–931. [DOI] [PubMed] [Google Scholar]
- 43.Skowyra ML et al. (2018) Triggered recruitment of ESCRT machinery promotes endolysosomal repair. Science 360 (6384). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chauhan S et al. (2016) TRIMs and Galectins Globally Cooperate and TRIM16 and Galectin-3 Co-direct Autophagy in Endomembrane Damage Homeostasis. Dev Cell 39 (1), 13–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jia J et al. (2020) AMPK, a Regulator of Metabolism and Autophagy, Is Activated by Lysosomal Damage via a Novel Galectin-Directed Ubiquitin Signal Transduction System. Mol Cell 77 (5), 951–969 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li FY et al. (2020) Galectins in Host Defense Against Microbial Infections. Adv Exp Med Biol 1204, 141–167. [DOI] [PubMed] [Google Scholar]
- 47.Meunier E et al. (2014) Caspase-11 activation requires lysis of pathogen-containing vacuoles by IFN-induced GTPases. Nature 509 (7500), 366–70. [DOI] [PubMed] [Google Scholar]
- 48.Panda SK et al. (2018) Galectin-9 inhibits TLR7-mediated autoimmunity in murine lupus models. J Clin Invest 128 (5), 1873–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hamalisto S et al. (2020) Spatially and temporally defined lysosomal leakage facilitates mitotic chromosome segregation. Nat Commun 11 (1), 229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Campden RI and Zhang Y (2019) The role of lysosomal cysteine cathepsins in NLRP3 inflammasome activation. Arch Biochem Biophys 670, 32–42. [DOI] [PubMed] [Google Scholar]
- 51.Raben N and Puertollano R (2016) TFEB and TFE3: Linking Lysosomes to Cellular Adaptation to Stress. Annu Rev Cell Dev Biol 32, 255–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sardiello M et al. (2009) A gene network regulating lysosomal biogenesis and function. Science 325 (5939), 473–7. [DOI] [PubMed] [Google Scholar]
- 53.Settembre C et al. (2011) TFEB links autophagy to lysosomal biogenesis. Science 332 (6036), 1429–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Martina JA et al. (2014) The nutrient-responsive transcription factor TFE3 promotes autophagy, lysosomal biogenesis, and clearance of cellular debris. Sci Signal 7 (309), ra9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Martina JA and Puertollano R (2013) Rag GTPases mediate amino acid-dependent recruitment of TFEB and MITF to lysosomes. J Cell Biol 200 (4), 475–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Martina JA et al. (2012) MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy 8 (6), 903–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Roczniak-Ferguson A et al. (2012) The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci Signal 5 (228), ra42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Settembre C et al. (2012) A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J 31 (5), 1095–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Puertollano R et al. (2018) The complex relationship between TFEB transcription factor phosphorylation and subcellular localization. EMBO J 37 (11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Medina DL et al. (2015) Lysosomal calcium signalling regulates autophagy through calcineurin and TFEB. Nat Cell Biol 17 (3), 288–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Martina JA and Puertollano R (2018) Protein phosphatase 2A stimulates activation of TFEB and TFE3 transcription factors in response to oxidative stress. J Biol Chem 293 (32), 12525–12534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pastore N et al. (2016) TFEB and TFE3 cooperate in the regulation of the innate immune response in activated macrophages. Autophagy 12 (8), 1240–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gray MA et al. (2016) Phagocytosis Enhances Lysosomal and Bactericidal Properties by Activating the Transcription Factor TFEB. Curr Biol 26 (15), 1955–1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Visvikis O et al. (2014) Innate host defense requires TFEB-mediated transcription of cytoprotective and antimicrobial genes. Immunity 40 (6), 896–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Campbell GR et al. (2015) Human Immunodeficiency Virus Type 1 Nef Inhibits Autophagy through Transcription Factor EB Sequestration. PLoS Pathog 11 (6), e1005018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.El-Houjeiri L et al. (2019) The Transcription Factors TFEB and TFE3 Link the FLCN-AMPK Signaling Axis to Innate Immune Response and Pathogen Resistance. Cell Rep 26 (13), 3613–3628 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang X et al. (2016) Lysosome calcium in ROS regulation of autophagy. Autophagy 12 (10), 1954–1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Martina JA et al. (2016) TFEB and TFE3 are novel components of the integrated stress response. EMBO J 35 (5), 479–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mansueto G et al. (2017) Transcription Factor EB Controls Metabolic Flexibility during Exercise. Cell Metab 25 (1), 182–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Brady OA et al. (2018) The transcription factors TFE3 and TFEB amplify p53 dependent transcriptional programs in response to DNA damage. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Slade L et al. (2020) A lysosome independent role for TFEB in activating DNA repair and inhibiting apoptosis in breast cancer cells. Biochem J 477 (1), 137–160. [DOI] [PubMed] [Google Scholar]
- 72.Nezich CL et al. (2015) MiT/TFE transcription factors are activated during mitophagy downstream of Parkin and Atg5. J Cell Biol 210 (3), 435–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yu H et al. (2014) Revisiting STAT3 signalling in cancer: new and unexpected biological functions. Nat Rev Cancer 14 (11), 736–46. [DOI] [PubMed] [Google Scholar]
- 74.Martinez-Fabregas J et al. (2018) Lysosomal protease deficiency or substrate overload induces an oxidative-stress mediated STAT3-dependent pathway of lysosomal homeostasis. Nat Commun 9 (1), 5343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kreuzaler PA et al. (2011) Stat3 controls lysosomal-mediated cell death in vivo. Nat Cell Biol 13 (3), 303–9. [DOI] [PubMed] [Google Scholar]
- 76.Sargeant TJ et al. (2014) Stat3 controls cell death during mammary gland involution by regulating uptake of milk fat globules and lysosomal membrane permeabilization. Nat Cell Biol 16 (11), 1057–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lloyd-Lewis B et al. (2018) Stat3-mediated alterations in lysosomal membrane protein composition. J Biol Chem 293 (12), 4244–4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li L et al. (2018) STAT3 contributes to lysosomal-mediated cell death in a novel derivative of riccardin D-treated breast cancer cells in association with TFEB. Biochem Pharmacol 150, 267–279. [DOI] [PubMed] [Google Scholar]
- 79.Liu B et al. (2018) STAT3 associates with vacuolar H(+)-ATPase and regulates cytosolic and lysosomal pH. Cell Res 28 (10), 996–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Webb AE and Brunet A (2014) FOXO transcription factors: key regulators of cellular quality control. Trends Biochem Sci 39 (4), 159–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cheng Z (2019) The FoxO-Autophagy Axis in Health and Disease. Trends Endocrinol Metab 30 (9), 658–671. [DOI] [PubMed] [Google Scholar]
- 82.Liu L et al. (2016) FoxO1 interacts with transcription factor EB and differentially regulates mitochondrial uncoupling proteins via autophagy in adipocytes. Cell Death Discov 2, 16066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Miki Y et al. (2018) Autophagy mediators (FOXO1, SESN3 and TSC2) in Lewy body disease and aging. Neurosci Lett 684, 35–41. [DOI] [PubMed] [Google Scholar]
- 84.Shin HJ et al. (2016) AMPK-SKP2-CARM1 signalling cascade in transcriptional regulation of autophagy. Nature 534 (7608), 553–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lin XX et al. (2018) DAF-16/FOXO and HLH-30/TFEB function as combinatorial transcription factors to promote stress resistance and longevity. Nat Commun 9 (1), 4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Annunziata I et al. (2019) MYC competes with MiT/TFE in regulating lysosomal biogenesis and autophagy through an epigenetic rheostat. Nat Commun 10 (1), 3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bautista SJ et al. (2018) mTOR complex 1 controls the nuclear localization and function of glycogen synthase kinase 3beta. J Biol Chem 293 (38), 14723–14739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chauhan S et al. (2013) ZKSCAN3 is a master transcriptional repressor of autophagy. Mol Cell 50 (1), 16–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Li Y et al. (2016) Protein kinase C controls lysosome biogenesis independently of mTORC1. Nat Cell Biol 18 (10), 1065–77. [DOI] [PubMed] [Google Scholar]
- 90.Pan H et al. (2017) The role of ZKSCAN3 in the transcriptional regulation of autophagy. Autophagy 13 (7), 1235–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Shen K et al. (2018) Architecture of the human GATOR1 and GATOR1-Rag GTPases complexes. Nature 556 (7699), 64–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wolfson RL et al. (2017) KICSTOR recruits GATOR1 to the lysosome and is necessary for nutrients to regulate mTORC1. Nature 543 (7645), 438–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Saxton RA et al. (2016) Mechanism of arginine sensing by CASTOR1 upstream of mTORC1. Nature 536 (7615), 229–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wolfson RL et al. (2016) Sestrin2 is a leucine sensor for the mTORC1 pathway. Science 351 (6268), 43–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gu X et al. (2017) SAMTOR is an S-adenosylmethionine sensor for the mTORC1 pathway. Science 358 (6364), 813–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Deng L et al. (2015) The ubiquitination of rag A GTPase by RNF152 negatively regulates mTORC1 activation. Mol Cell 58 (5), 804–18. [DOI] [PubMed] [Google Scholar]
- 97.Meng J and Ferguson SM (2018) GATOR1-dependent recruitment of FLCN-FNIP to lysosomes coordinates Rag GTPase heterodimer nucleotide status in response to amino acids. J Cell Biol 217 (8), 2765–2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lawrence RE et al. (2019) Structural mechanism of a Rag GTPase activation checkpoint by the lysosomal folliculin complex. Science 366 (6468), 971–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Han JM et al. (2012) Leucyl-tRNA synthetase is an intracellular leucine sensor for the mTORC1-signaling pathway. Cell 149 (2), 410–24. [DOI] [PubMed] [Google Scholar]
- 100.Duran RV et al. (2012) Glutaminolysis activates Rag-mTORC1 signaling. Mol Cell 47 (3), 349–58. [DOI] [PubMed] [Google Scholar]
- 101.Ba Q et al. (2018) Whole-Cell Scale Dynamic Organization of Lysosomes Revealed by Spatial Statistical Analysis. Cell Rep 23 (12), 3591–3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Korolchuk VI et al. (2011) Lysosomal positioning coordinates cellular nutrient responses. Nat Cell Biol 13 (4), 453–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Li X et al. (2016) A molecular mechanism to regulate lysosome motility for lysosome positioning and tubulation. Nat Cell Biol 18 (4), 404–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Willett R et al. (2017) TFEB regulates lysosomal positioning by modulating TMEM55B expression and JIP4 recruitment to lysosomes. Nat Commun 8 (1), 1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Guardia CM et al. (2016) BORC Functions Upstream of Kinesins 1 and 3 to Coordinate Regional Movement of Lysosomes along Different Microtubule Tracks. Cell Rep 17 (8), 195–01961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Pu J et al. (2015) BORC, a multisubunit complex that regulates lysosome positioning. Dev Cell 33 (2), 176–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Filipek PA et al. (2017) LAMTOR/Ragulator is a negative regulator of Arl8b- and BORC-dependent late endosomal positioning. J Cell Biol 216 (12), 4199–4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Pu J et al. (2017) A Ragulator-BORC interaction controls lysosome positioning in response to amino acid availability. J Cell Biol 216 (12), 4183–4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Raiborg C et al. (2015) Repeated ER-endosome contacts promote endosome translocation and neurite outgrowth. Nature 520 (7546), 234–8. [DOI] [PubMed] [Google Scholar]
- 110.Marques ARA and Saftig P (2019) Lysosomal storage disorders - challenges, concepts and avenues for therapy: beyond rare diseases. J Cell Sci 132 (2). [DOI] [PubMed] [Google Scholar]
- 111.Medina DL et al. (2011) Transcriptional activation of lysosomal exocytosis promotes cellular clearance. Dev Cell 21 (3), 421–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lim JA et al. (2017) Modulation of mTOR signaling as a strategy for the treatment of Pompe disease. EMBO Mol Med 9 (3), 353–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Spampanato C et al. (2013) Transcription factor EB (TFEB) is a new therapeutic target for Pompe disease. EMBO Mol Med 5 (5), 691–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Di Malta C and Ballabio A (2017) TFEB-mTORC1 feedback loop in metabolism and cancer. Cell Stress 1 (1), 7–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Yogalingam G et al. (2008) Neuraminidase 1 is a negative regulator of lysosomal exocytosis. Dev Cell 15 (1), 74–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Machado E et al. (2015) Regulated lysosomal exocytosis mediates cancer progression. Sci Adv 1 (11), e1500603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Brown RA et al. (2019) mTOR hyperactivity mediates lysosomal dysfunction in Gaucher’s disease iPSC-neuronal cells. Dis Model Mech 12 (10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sural-Fehr T et al. (2019) Inhibition of the IGF-1-PI3K-Akt-mTORC2 pathway in lipid rafts increases neuronal vulnerability in a genetic lysosomal glycosphingolipidosis. Dis Model Mech 12 (5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Danyukova T et al. (2018) Loss of CLN7 results in depletion of soluble lysosomal proteins and impaired mTOR reactivation. Hum Mol Genet 27 (10), 1711–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Fiorenza MT et al. (2018) The pathogenesis of lysosomal storage disorders: beyond the engorgement of lysosomes to abnormal development and neuroinflammation. Hum Mol Genet 27 (R2), R119–R129. [DOI] [PubMed] [Google Scholar]
- 121.Panicker LM et al. (2018) Gaucher disease iPSC-derived osteoblasts have developmental and lysosomal defects that impair bone matrix deposition. Hum Mol Genet 27 (5), 811–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.van de Vlekkert D et al. (2019) Excessive exosome release is the pathogenic pathway linking a lysosomal deficiency to generalized fibrosis. Sci Adv 5 (7), eaav3270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lloyd-Evans E and Waller-Evans H (2019) Lysosomal Ca(2+) Homeostasis and Signaling in Health and Disease. Cold Spring Harb Perspect Biol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hara T et al. (2006) Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 441 (7095), 885–9. [DOI] [PubMed] [Google Scholar]
- 125.Komatsu M et al. (2006) Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature 441 (7095), 880–4. [DOI] [PubMed] [Google Scholar]
- 126.Xiao Q et al. (2015) Neuronal-Targeted TFEB Accelerates Lysosomal Degradation of APP, Reducing Abeta Generation and Amyloid Plaque Pathogenesis. J Neurosci 35 (35), 12137–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Decressac M et al. (2013) TFEB-mediated autophagy rescues midbrain dopamine neurons from alpha-synuclein toxicity. Proc Natl Acad Sci U S A 110 (19), E1817–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Pryor WM et al. (2014) Huntingtin promotes mTORC1 signaling in the pathogenesis of Huntington’s disease. Sci Signal 7 (349), ra103. [DOI] [PubMed] [Google Scholar]
- 129.Amick J et al. (2016) C9orf72 binds SMCR8, localizes to lysosomes, and regulates mTORC1 signaling. Mol Biol Cell 27 (20), 3040–3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Amick J et al. (2020) PQLC2 recruits the C9orf72 complex to lysosomes in response to cationic amino acid starvation. J Cell Biol 219 (1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Luningschror P et al. (2020) The FTLD Risk Factor TMEM106B Regulates the Transport of Lysosomes at the Axon Initial Segment of Motoneurons. Cell Rep 30 (10), 3506–3519 e6. [DOI] [PubMed] [Google Scholar]
- 132.Levy C et al. (2006) MITF: master regulator of melanocyte development and melanoma oncogene. Trends Mol Med 12 (9), 406–14. [DOI] [PubMed] [Google Scholar]
- 133.Petit CS et al. (2013) Recruitment of folliculin to lysosomes supports the amino acid-dependent activation of Rag GTPases. J Cell Biol 202 (7), 1107–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Bar-Peled L et al. (2013) A Tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science 340 (6136), 1100–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Ge W et al. (2015) The Roles of Lysosomes in Inflammation and Autoimmune Diseases. Int Rev Immunol 34 (5), 415–31. [DOI] [PubMed] [Google Scholar]
- 136.Bonam SR et al. (2019) Lysosomes as a therapeutic target. Nat Rev Drug Discov 18 (12), 923–948. [DOI] [PMC free article] [PubMed] [Google Scholar]