Short summary
Peanut-specific CD8+ T cells in nonallergic individuals are not deleted, but have an expansion block that can be released by impairing regulatory T cell associated signaling pathways.
Keywords: human CD8+ T cells, food allergy, food immune tolerance
To the Editor:
It is postulated that loss of immune tolerance to food can lead to food allergies. CD8+ T cells are known to participate in pathologic immune responses to food antigen, such as seen in eosinophilic esophagitis and celiac disease.1–4 In light of this, we recently reported that peanut specific CD8+ T cells are increased in the blood of peanut-allergic individuals and express the TH2 associated chemokine receptor CCR4.1 We also identified an Ara h 1 derived peptide that triggered the activation of nearly one percent of CD8+ T cells from one HLA-A*02:01+ peanut-allergic individual, and derived Ara h 1 specific CD8+ T cell clones from the same person by in vitro expansion.1
In this follow-up study, we first wanted to better characterize the phenotype of the peanut-specific CD8+ T cell clones described above in the setting of food allergy. Five Ara h 1 specific CD8+ T cell clones were incubated for 2 days with or without phytohemagglutinin (PHA), after which their cytokine expression profile was measured by BD cytokine bead array assay. Supernatants from all five clones contained increased levels of IL-5, IL-13, and GM-CSF after stimulation (Fig E1, A). The same cytokines were increased after a 5 day incubation with the Ara h 1 peanut peptide, but not a control peptide (Fig E1, B). Thus, peanut-specific CD8+ T cells may contribute to food allergy by becoming activated and expressing TH2 cytokines after exposure to allergen. GM-CSF enables antigen cross presentation by dendritic cells, which would promote further allergen recognition by CD8+ T cells.5
The identification of a peanut epitope recognized in food allergy opened the opportunity to study the behavior of food-specific CD8+ T cells in nonallergic individuals. Doing so is challenging not only because one must first identify a food epitope that is recognized in disease, but the frequency of cells specific for a given food epitope is also low, making detection difficult. We addressed this issue by employing tetramer enrichment with HLA-A*02:01 tetramers loaded with the Ara h 1 peptide, and conjugated to phycoerythrin (PE).6 In this technique, tetramer+ cells are incubated with paramagnetic beads coated with anti-PE antibody, then enriched using a magnetized column before detection by flow cytometry. Peptide-specific cells at frequencies less than one cell per a million total CD8+ T cells can be detected by flow cytometry in this way.6
To determine if peanut-specific CD8+ T cells are present in nonallergic individuals, we performed tetramer enrichment on PBMCs from HLA-A*02:01+ blood donors from the Stanford Blood Center. The prevalence of peanut allergy is approximately 2.3% among adults, so it is unlikely that multiple donors are peanut allergic.7 We found that the frequency of Ara h 1 peptide-specific CD8+ T cells in eight different blood donors is approximately one cell in 105 to 106 total CD8+ T cells (Fig 1, A). We conclude that peanut-specific CD8+ T cells are not deleted (as a mechanism of immune tolerance) in peanut tolerant individuals and are found in peripheral blood at frequencies consistent with CD8+ T cells specific for other antigens, both foreign and self.6
FIG 1.
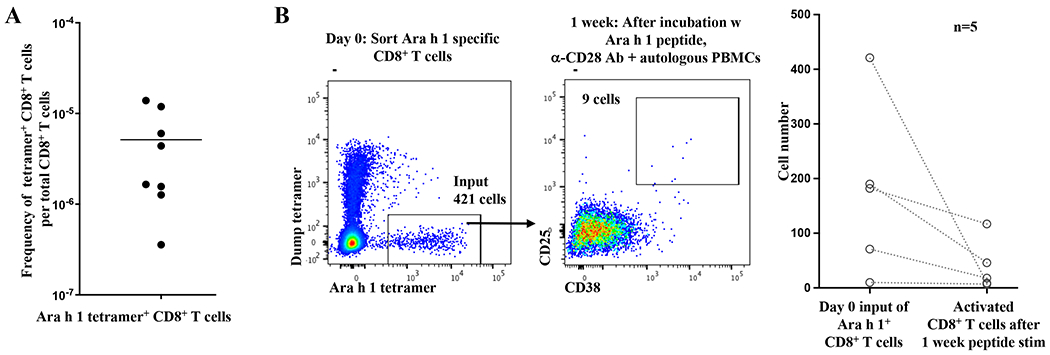
A, Ara h 1 specific CD8+ T cells are not clonally deleted in nonallergic individuals.
Tetramer enrichment using HLA-A*02:01 tetramers loaded with Ara h 1 peptide was performed on PBMCs from HLA-A*02:01+ blood bank donors, after which peanut (Ara h 1) tetramer+ CD8+ T cells were enumerated by flow cytometry. Each point represents one blood bank sample.
B, Poor activation of Ara h 1 specific CD8+ T cells in nonallergic individuals.
Left: Ara h 1 specific CD8+ T cells were isolated from a HLA-A*02:01+ blood bank donor by tetramer enrichment followed by FACS. Ara h 1 specific CD8+ T cells were incubated one week with Ara h 1 peptide (1.5ug/ml), anti-CD28 antibody (5ug/ml), and autologous PBMCs as feeder cells before analysis. Flow cytometry panels are gated on CD8+ T cells.
Right: Cumulative results from 5 blood bank samples. Each pair of points connected by a line represents one sample. In some cases, FACS was not performed and instead 1/11th of the tetramer enriched cells was analyzed by flow cytometry to calculate the number of Ara h 1 specific CD8+ T cells, and the remainder used for tissue culture. The p value for a 2 tailed Wilcoxon signed rank test was 0.0625, which is the minimum value possible for n=5.
To measure the activation of Ara h 1 specific CD8+ T cells in nonallergic individuals, we incubated tetramer enriched cells with autologous feeder cells, anti-CD28 antibody, and peanut peptide – the same conditions which previously activated nearly one percent of CD8+ T cells from a HLA-A*02:01+ peanut allergic individual.1 After one week, we measured the number of activated CD8+ T cells by the expression of CD25 and CD38. In five blood bank donor samples, the number of activated CD8+ T cells after stimulation was less than the original input of Ara h 1 specific CD8+ T cells (Fig 1, B). This poor activation is similar to what we have previously observed in CD8+ T cells specific for self antigen,6 and is consistent with the need for immune tolerance to both food antigens and endogenous antigens. We conclude that Ara h 1 specific CD8+ T cells are poorly activated by stimulation through the TCR and CD28 costimulatory receptor in nonallergic individuals, consistent with a mechanism of tolerance to food.
We hypothesized that CD4+ regulatory T cells (Tregs) might contribute to the suppression of Ara h 1 specific CD8+ T cells in nonallergic individuals. To test this, we mixed Ara h 1 specific CD8+ T cells with either whole autologous PBMCs (as before) or CD25-depleted, autologous PBMCs before stimulating with peanut peptide and anti-CD28 antibody (Fig 2, A). Because FOXP3+ Tregs are CD25hiCD127lo, removing CD25+ cells by bead depletion should lift suppression due to this cell type. An analysis of 8 blood donor samples showed two outcomes: in the case of 4 samples there was a 3.4 to 10.5-fold increase in the number of Ara h 1 specific CD8+ T cells when CD25+ cells were depleted; in the case of the other samples there was little change (Fig 2, A, middle and right). We conclude that in a proportion of individuals, CD25+ cells suppress the activation of Ara h 1 specific CD8+ T cells.
FIG 2.
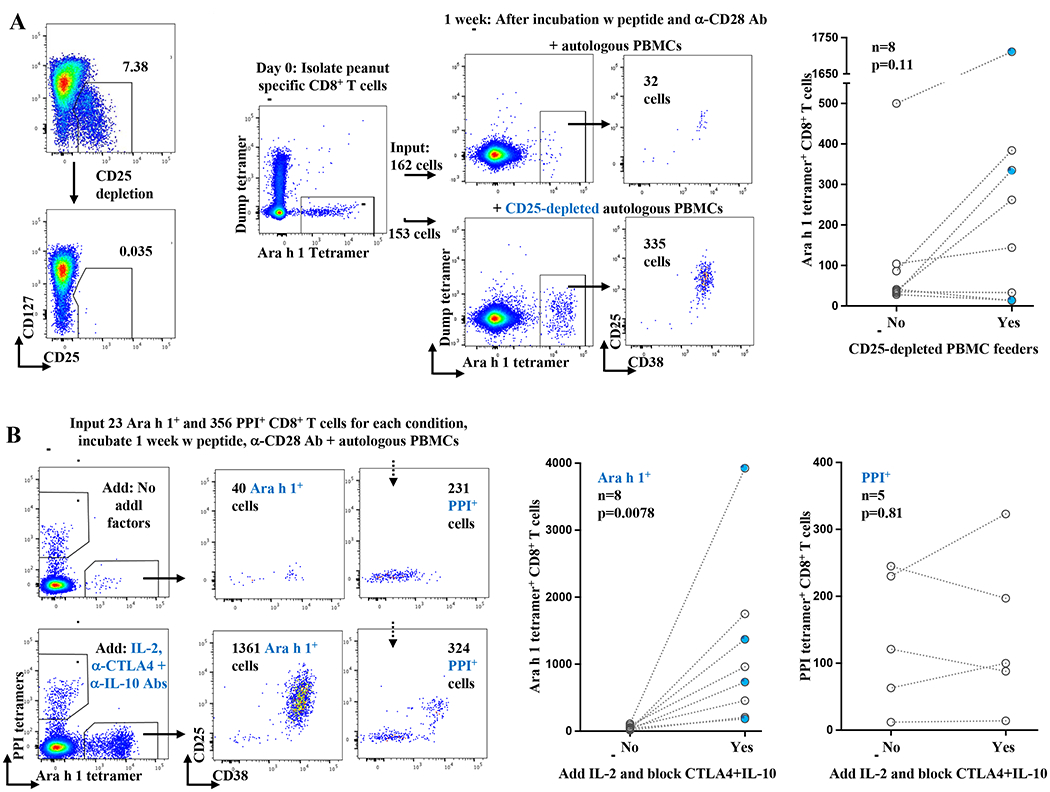
A, Enhanced expansion of CD8+ T cells specific for Ara h 1 after the depletion of CD25+ cells from PBMC feeders in the case of some nonallergic individuals, but not others.
Left: Example depletion of CD25hiCD127lo CD4+ regulatory T cells from autologous PBMC feeder cells. Flow cytometry plots gated on CD4+ T cells.
Middle: CD8+ T cells specific for Ara h 1 were isolated by tetramer enrichment from a HLA-A*02:01+ blood bank donor as in Figure 1B. An equal number of Ara h 1 specific CD8+ T cells was then incubated for one week with cognate peptide, anti-CD28 antibody, and either whole autologous PBMCs or autologous PBMCs that had been depleted of CD25+ cells. Flow cytometry plots gated on CD8+ T cells.
Right: Cumulative results from 8 blood bank donors. Each pair of points connected by a line represents one sample. Blue points represent samples for which donors were confirmed peanut IgE negative. P value calculated by Wilcoxon signed rank test, 2 tailed.
B, CTLA-4 and IL-10 blockade combined with the addition of IL-2 promote the expansion of Ara h 1 (i.e., food specific) CD8+ T cells, but not self antigen specific CD8+ T cells.
Left: CD8+ T cells specific for Ara h 1 or human preproinsulin peptide(s) (PPI 2:10 and 15:24) were isolated by tetramer enrichment from a HLA-A*02:01+ blood bank donor. An equal number of enriched cells was then incubated with cognate peptides, anti-CD28 antibody, and autologous PBMCs (without CD25 depletion) +/− IL-2 with blocking antibodies to both CTLA-4 and IL-10. Flow cytometric plots gated on CD8+ T cells show analysis after one week.
Middle and Right: Cumulative results of CD8+ T cells specific for either Ara h 1 (middle) or PPI (right). Each pair of points connected by a line represents one blood bank sample. IgE negative. Blue points represent samples for which donors were confirmed peanut P value calculated by Wilcoxon signed rank test, 2 tailed.
We sought to determine which intercellular signals could relieve the activation block in Ara h 1 specific CD8+ T cells. Tregs use multiple mechanisms to suppress other cell types including IL-2 sequestration, the inhibitory coreceptor CTLA-4, and IL-10 secretion.8 To test these mechanisms in food specific CD8+ T cells, we added IL-2 (50 to 100U/ml) as well as blocking antibodies against CTLA-4 (20ug/ml) and IL10 (40ug/ml) to the standard in vitro assay of Ara h 1 CD8+ T cells, peanut peptide, anti-CD28 antibody, and autologous PBMCs (including CD25+ Tregs). Blocking IL-10 and CTLA-4 signaling while adding IL-2 resulted in robust proliferation of Ara h 1 specific CD8+ T cells from 8 blood bank donor samples, again consistent with a model where Tregs contribute to the suppression of peanut specific CD8+ T cells (Fig 2, B, left and middle, p = 0.0078).
Because previously we had shown that self-specific CD8+ T cells fail to expand after stimulation with peptide and anti-CD28 antibody,6 we tested whether self-specific CD8+ T cells would proliferate (as peanut specific CD8+ T cells do) after the addition of IL-2 combined with blocking antibodies to CTLA-4 and IL-10. We used tetramer enrichment to isolate CD8+ T cells specific for endogenous peptides derived from human preproinsulin (PPI). An analysis of 5 blood bank samples showed minimal expansion of self-specific CD8+ T cells even with the addition of IL-2 combined with blocking antibodies to CTLA-4 and IL-10 (Fig 2, B, left and right). We conclude that despite the need to develop CD8+ T cell tolerance to two parallel classes of antigens - food or endogenous – different mechanisms are used to achieve this goal.
In summary, these initial results show that Ara h 1 specific CD8+ T cells from nonallergic individuals are not deleted, but expand poorly after stimulation through the TCR and CD28 coreceptor. Their expansion is augmented in some individuals by the deletion of CD25+ cells, and more generally by the addition of IL-2 combined with a block on CTLA-4 and IL-10. These interventions are consistent with a role for Treg cells. Interestingly, in one mouse food allergy model, blocking CTLA-4 during sensitization with peanut protein and cholera toxin enhanced the IgE response against Ara h 1, while CTLA-4 blockade combined with peanut exposure resulted in increased TH2 cytokines.9 We propose a model where food antigen specific CD8+ T cells are susceptible to suppression by Tregs in an antigen independent manner via a combination of IL-10, CTLA-4 and IL-2 signaling. A breakdown of this tolerance mechanism would enable CD8+ T cells to augment an allergic response to food via the secretion of TH2 cytokines and the cross presentation of food allergen. The expansion of this study to a larger population will provide additional confirmation of these mechanisms.
Supplementary Material
Acknowledgments
Sources of funding: We thank the NIH (grants R01AI140134 and U19AI104209) and the Sean N. Parker Center for allergy and Asthma at Stanford University for funding the work.
Conflict of interest disclosure statement: Dr. Nadeau reports grants from National Institute of Allergy and Infectious Diseases (NIAID), Food Allergy Research & Education (FARE), End Allergies Together (EAT), Allergenis, and Ukko Pharma; Grant awardee at NIAID, National Institute of Environmental Health Sciences (NIEHS), National Heart, Lung, and Blood Institute (NHLBI), and the Environmental Protection Agency (EPA); is involved in Clinical trials with Regeneron, Genentech, AImmune Therapeutics, DBV Technologies, AnaptysBio, Adare Pharmaceuticals, and Stallergenes-Greer; Research Sponsorship by Novartis, Sanofi, Astellas, Nestle; Data and Safety Monitoring Board member at Novartis and NHLBI; Cofounded Before Brands, Alladapt, ForTra, and Iggenix; Chief Intellectual Office at FARE, Director of the World Health Organization (WAO) Center of Excellence at Stanford, Personal fees from Regeneron, Astrazeneca, ImmuneWorks, and Cour Pharmaceuticals; Consultant and Advisory Board Member at Ukko, Before Brands, Alladapt, IgGenix, Probio, Vedanta, Centecor, Seed, Novartis, NHBLI, EPA, National Scientific Committee of ITN and NIH Programs; US patents (patent numbers 62/647,389; 62/119,014; 12/610,940, 12/686,121, 10/064,936, 62/767,444; application numbers S10-392). All other authors declare no conflict of interest.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Yu W, Zhou X, Dunham D, Lyu SC, Manohar M, Zhang W, et al. Allergen-specific CD8(+) T cells in peanut-allergic individuals. J Allergy Clin Immunol 2019;143:1948–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mazzarella G, Stefanile R, Camarca A, Giliberti P, Cosentini E, Marano C, et al. Gliadin activates HLA class I-restricted CD8+ T cells in celiac disease intestinal mucosa and induces the enterocyte apoptosis. Gastroenterology 2008;134:1017–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Han A, Newell EW, Glanville J, Fernandez-Becker N, Khosla C, Chien YH, et al. Dietary gluten triggers concomitant activation of CD4+ and CD8+ alphabeta T cells and gammadelta T cells in celiac disease. Proc Natl Acad Sci U S A 2013;110:13073–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wen T, Aronow BJ, Rochman Y, Rochman M, Kc K, Dexheimer PJ, et al. Single-cell RNA sequencing identifies inflammatory tissue T cells in eosinophilic esophagitis. J Clin Invest 2019;129:2014–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dresch C, Leverrier Y, Marvel J, Shortman K. Development of antigen cross-presentation capacity in dendritic cells. Trends Immunol 2012;33:381–8. [DOI] [PubMed] [Google Scholar]
- 6.Yu W, Jiang N, Ebert PJ, Kidd BA, Muller S, Lund PJ, et al. Clonal Deletion Prunes but Does Not Eliminate Self-Specific alpha beta CD8(+) T Lymphocytes. Immunity 2015;42:929–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gupta RS, Warren CM, Smith BM, Jiang J, Blumenstock JA, Davis MM, et al. Prevalence and Severity of Food Allergies Among US Adults. JAMA Netw Open 2019;2:e185630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sakaguchi S, Mikami N, Wong JB, Tanaka A, Ichiyama K, Ohkura N. Regulatory T cells and Human Disease. Ann Rev Immunol 2020;38:541–66. [DOI] [PubMed] [Google Scholar]
- 9.van Wijk F, Hoeks S, Nierkens S, Koppelman SJ, van Kooten P, Boon L, et al. CTLA-4 signaling regulates the intensity of hypersensitivity responses to food antigens, but is not decisive in the induction of sensitization. J Immunol 2005;174:174–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


