Abstract
Birth weight (BW) represents an important clinical and toxicological measure, indicative of the overall health of the newborn as well as potential risk for later-in-life outcomes. BW can be influenced by endogenous and exogenous factors and is known to be heavily impacted in utero by the health and function of the placenta. An aspect that remains understudied is the influence of genomic and epigenomic programming within the placenta on infant BW. To address this gap, we set out to test the hypothesis that genes involved in critical placental cell signaling are associated with infant BW, and are likely regulated, in part, through epigenetic mechanisms based on microRNA (miRNA) mediation. This study leveraged a robust dataset based on 390 infants born at low gestational age (ranged 23 to 27 weeks) to evaluate genome-wide expression profiles of both mRNAs and miRNAs in placenta tissues and relate these to infant BW. A total of 254 mRNAs and 268 miRNAs were identified as associated with BW, the majority of which showed consistent associations across placentas derived from both males and females. BW-associated mRNAs were found to be enriched for important biological pathways, including glycoprotein VI (the major receptor for collagen), human growth, and hepatocyte growth factor signaling, a portion of which were predicted to be regulated by BW-associated miRNAs. These miRNA-regulated pathways highlight key mechanisms potentially linking endogenous/exogenous factors to changes in birth outcomes that may be deleterious to infant and later-in-life health.
Keywords: Birth weight, genomic signature, mechanisms, microRNAs, placenta, birth outcome
1. Introduction
Infant birth weight (BW) is an important clinical and toxicological measure that is collected shortly after birth and is strongly associated with infant mortality risk and overall health [1,2]. Alterations in BW are linked to adverse health outcomes for the child later-in-life, including type II diabetes, cardiovascular diseases, and obesity [1,3–5]. For example, low BW neonates (<2500g) have been shown to be more susceptible to developing hypertension and glucose intolerance, while high BW or macrosomic (>4000g) neonates have been shown to be more likely to develop metabolic syndrome and cancer [4,6]. Because this outcome represents a critical indicator of newborn health and is associated with health endpoints later-in-life, BW serves as a common measure evaluated in routine developmental toxicity testing in animals, further highlighting the importance of understanding factors and mechanisms that impact infant BW [7].
Changes in BW are influenced by multiple factors that can impact placental function, such as maternal nutrition, gestational age, and maternal exposures, as well as pregnancy complications association with placental insufficiency and decreased nutrient supply to the fetus [8–10]. By providing a constant supply of nutrients as well as means for waste and gas exchange, the placenta is crucial to the BW and overall health of the fetus [11]. Cells within the placenta are characterized as transcriptionally active and shown to be functionally controlled by the genome and epigenome [12]. An important type of epigenetic mechanism that can regulate gene transcription is microRNA (miRNA)-mediated regulation. Some studies have documented the importance of certain miRNAs on placental development; however the specific pathways and associated miRNA mediation involved in regulating infant BW remain elusive [8,13,14].
miRNAs are noncoding molecules, approximately 18–25 nucleotides in length, integral to the regulation of gene expression via epigenetic processes [9,11,15]. Through posttranscriptional regulation, miRNAs have been shown to control many complex biologically processes including cell cycle/cell death, cell growth and proliferation, cell differentiation, hormone signaling, and inflammation, which in turn, can influence potential disease development [16]. miRNAs can specifically regulate posttranscriptional processes by repressing mRNA translation, directing the cleavage of translated mRNA molecules, and/or increasing mRNA expression by targeting enhancer sequences [4,8,17]. The mRNAs that are regulated by miRNAs are largely dictated through complementary base pairing processes, wherein partial base pairing has been shown to halt mRNA translation, and complete base pairing has been shown to cause mRNA degradation [11,15]. miRNAs have previously been evaluated in the placenta and shown to play a role in critical biological pathways influencing placental health [18]. Since miRNAs significantly impact placental biology, it is plausible that these epigenetic regulators can affect placental processes that may influence infant health indices, including BW.
Previous studies evaluating mRNA and miRNA signatures in the placenta have provided evidence for expression alterations associated with preeclampsia, uterine growth restriction, fetal growth restriction, and spontaneous abortion [10,18–20]. There is evidence to support the role of placental miRNA expression dysregulation in affecting infant BW, where previous studies have implemented gene-specific approaches to associate select miRNA expression changes with altered BW [4,9,11,19,21–24]. For example, one study investigated the expression of placental miRNAs in small for gestational age infants that were compared to normal BW infants across a cohort of 68 neonates [8]. Select miRNAs were evaluated using quantitative real-time polymerase chain reaction (qRT-PCR), and a subset were identified as differentially expressed between the two groups. These miRNAs were noted for their roles in biological pathways necessary for cell migration and viability, inflammation, and insulin-like growth factor signaling [8]. Another study examined the relationship between in utero arsenic exposure and infant BW, finding that arsenic contributed to greater placental miRNA differences in newborns delivered earlier in gestation, in comparison to those delivered later in gestation [9]. Changes in miRNA expression associated with BW have also been identified within placentas from rats and mice, confirming the importance of placental miRNAs across species [25]. These data provide preliminary evidence for the role of placental miRNAs potentially affecting processes potentially involved in the regulation of infant BW. To our knowledge, no study has investigated genome-wide placental miRNA expression signatures in relation to BW.
The current study set out to address this research issue by testing the hypothesis that genes involved in critical placental cell signaling are associated with infant BW, and are regulated, in part, through epigenetic mechanisms based on miRNA mediation. This study leveraged a robust dataset based on 390 infants to evaluate genome-wide expression profiles of both mRNAs and miRNAs in placenta tissues and relate these to infant BW. This study is unique for the following reasons: (i) genome-wide mRNA and miRNA expression profiles were evaluated simultaneously allowing for a more comprehensive evaluation of their molecular interactions in the placenta; and (ii) data were leveraged from the previously established Extremely Low Gestational Age Newborns (ELGAN) cohort, representing a robust dataset to identify new trends in genomic and epigenomic signaling pertinent to BW. Results from this study contribute valuable information towards further understanding the role of the placenta in fetal growth and development, as well as the mechanisms impacting infant health.
2. Materials and Methods
2.1. Study cohort recruitment and BW evaluation
The study sample was derived from the ELGAN cohort, comprising of pregnancies that ended in live births prior to completing 28 weeks of gestation [26]. Methods of subject recruitment and sample collection have been described extensively elsewhere [3]. To summarize, from 2002 to 2004, pregnant women scheduled to give birth at one of 14 participating institutions were recruited for participation. Study procedures were approved by the Institutional Review Board at each of the 14 participating ELGAN sites, and consent was provided prior to hospital admission or soon after delivery [3]. In total, 1506 infants and 1249 mothers enrolled in the ELGAN study. At or prior to delivery, maternal and demographic and clinical assessments were completed. A trained research nurse measured demographic and pregnancy variables after delivery using a structured questionnaire [26]. Each infant’s anthropometric measures, including BW, were recorded throughout hospitalization and at follow-up [27]. Women participating in the ELGAN study also agreed to the collection of placenta samples. Of the original sample, placental genomic data are available for 390 individuals, representing the samples evaluated in this study.
2.2. Placenta tissue collection
Placenta samples were collected immediately after each delivery, as previously described [28,29]. In summary, placentas were deposited into a sterile exam basin and taken to the sampling room for biopsies. In acquiring the specific tissue samples, the amnion enclosing the embryonic sack was pulled away from the underlying chorion (representing fetally-derived tissue) using sterile techniques in order to expose the chorion. To determine the area, each sample was measured from the midpoint to the longest distance between the cord insertion and the edge of the placental disk. A tissue sample was trimmed from the base of the chorion after the application of traction to both the chorion and the underlying trophoblast tissue. Each sample was subsequently inserted into a sterile 2 mL cryovial that was quickly submerged in liquid nitrogen, shipped frozen to the University of North Carolina at Chapel Hill, and stored at −80°C until further processing. For the current study, a total of 390 placentas were available for subsequently used for RNA analysis.
2.3. Placental RNA extraction and sequencing analyses
The placental tissues were processed by first placing the cryotubes containing the placental biopsies on dry ice. Frozen tissue samples were sliced into approximately 0.2g segments using a sterile dermal curette and washed in 1x PBS (Fisher Scientific, Waltham, MA) to reduce any potential blood contamination. To preserve sample integrity, washed samples were then immediately snap frozen in homogenization tubes and placed back on dry ice. Tissue segments were homogenized using a sterile stainless-steel bead (Qiagen, Germantown, MD) in RLT + lysis buffer (Qiagen) with the TissueLyserII instrument (Qiagen). Samples were then clarified by spinning to collect the bead and cellular debris, and homogenates were stored at −80°C until nucleic acid extraction. RNA molecules 18 nucleotides and greater were extracted using the AllPrep DNA/RNA/miRNA Universal kit (Qiagen). RNA quantity was then measured using the NanoDrop™ 1000 Spectrophotometer (Thermo Scientific, Waltham, MA) and tested for quality based on RNA integrity scores produced by the QIAxcel system (Qiagen).
Isolated RNA samples were then used to evaluate genome-wide miRNA expression profiles using the HTG EdgeSeq miRNA Whole Transcriptome Assay (HTG Molecular Diagnostics, Tucson, AZ), which utilizes next-generation sequencing technologies to analyze expression levels of 2,083 human miRNA transcripts. The counts of sequencing reads per miRNA were aligned to miRBase v20 and organized using Parser (HTG Molecular Diagnostics). Isolated RNA samples were also used to evaluate genome-wide mRNA expression profiles using the QuantSeq 3′ mRNA-Seq Library Prep Kit (Illumina) and libraries were pooled and sequenced (single-end 50 bp) on one lane of the Illumina Hiseq 2500. The counts of sequencing reads per mRNA were aligned to the GENCODE database v30 and organized using Salmon, yielding measures of 37,268 unique human RNA transcripts, including protein-coding and non-coding RNAs [30,31]. The resulting summarized count data were used in downstream data processing and statistical analyses. Sequencing data have been submitted to National Center for Biotechnology Information (NCBI) Gene Expression Omnibus repository [32].
2.4. Data processing and statistical analysis to identify miRNAs and mRNAs associated with neonatal BW
miRNA and mRNA sequencing data were processed separately using R (v3.6.2). Count data were first filtered to exclude universally lowly expressed transcripts, requiring that > 25% of the samples be expressed at signals above the overall median signal intensity, similar to our previous genome-wide mRNA and miRNA analyses [33–36]. This resulted in panels of 10,412 mRNA transcripts and 1,130 miRNA transcripts available for further evaluation. Potential sample outliers were identified through principal component analysis, in which principal components were calculated and visualized using the prcomp function within the R stats package (v3.6.2) [37]. Potential outliers were also evaluated via hierarchical clustering, in which distance metrics were calculated and visualized using the hclust function within the R stats package. The same outliers were identified using both approaches and thus excluded from the analysis. Count data were then normalized by median signal intensity using algorithms enabled within the DESeq2 package (v1.24.0), resulting in variance stabilized expression values [38]. Batch effects and potential sources of sample heterogeneity were accounted for using surrogate variable analysis (SVA) within the SVA R package (v3.32.1) [37]. Using default parameters to estimate control probes, three significant surrogate variables were calculated and included as covariates in the final statistical model [39].
To identify potential confounders, variables associated with BW and gene expression were identified via literature review. Statistical tests were performed to identify significant (p < 0.05), plausible associations between variables and BW. A directed acyclic graph (DAG) approach was then used to model causal associations between variables. To capture potential sources of bias without substantial losses in precision, a parsimonious model was constructed from covariates that were significantly associated and/or had known relationships with miRNA and/or mRNA expression levels and BW: multiple-gestation pregnancy (no/yes), gestational age (days), first or secondhand smoke exposure (no/yes), and sex of the fetus (male/female). Note that additional covariates demonstrated substantial collinearity with the included variables and were thus excluded from the analysis (e.g., maternal age). The final model evaluated BW as a continuous, log-transformed variable.
Negative binomial generalized linear models within the DESeq2 package were used to identify miRNAs and mRNAs differentially expressed in association with infant BW, while adjusting for aforementioned covariates [38]. This method, based on the Wald test, calculated shrunken logarithmic fold changes in expression, which were divided by their standard error values to produce z-statistics. Resulting z-statistics were compared against standard normal distribution curves to generate Wald test p-values. To account for multiple testing, these p-values were then adjusted using the Benjamini and Hochberg (BH) procedure [40]. All differentially expressed miRNAs and mRNAs were defined as those with false discovery rate (FDR) < 0.1, based on a BH-adjusted p-value. miRNA- and mRNA-seq data were first analyzed for statistical relationships with infant BW using all placental data from all newborns. Then, sex-stratified approaches were implemented, analyzing placenta data derived from male newborns separate from female newborns.
2.5. Predicting miRNA-mRNA interactions
The mRNAs found to be associated with BW were evaluated for potential regulation via miRNAs using an in silico approach based on experimentally observed miRNA-mRNA interactions curated from literature coupled with computational predictions. Specifically, miRNAs identified with expression levels significantly associated with BW were queried using the Ingenuity Knowledge Database (Ingenuity Systems®, Redwood City, CA) for mRNA targets predicted based on experimental observation or high predicted confidence. Experimental observations represented those gathered from TarBase, which contains approximately 670,000 unique miRNA-mRNA interactions shown through published literature [41]. Computationally predicted interactions represented those derived using algorithms generated through TargetScan Human v7.2, which identifies miRNA-mRNA interactions based on potential base pairing homologies between the 3’ untranslated mRNA regions and miRNA seed sequences [42]. The resulting potential miRNA-mRNA interactions were filtered to only include those with high predicted confidence, defined as those with cumulative weighted context scores ≤−0.4. These scores represent an aggregation of factors that influence the likelihood of miRNA-mRNA interactions, including binding site type and location, local adenine and uracil content, target site abundance, seed-pairing stability, and supplementary pairing [43]. To further evaluate the relationships between BW-associated miRNAs and their predicted mRNA targets, correlation analyses were carried out based on the Spearman Rank Correlation test (R Software). Correlations with p < 0.05 were identified as significant.
2.6. Pathway and network enrichment analysis of genomic and epigenomic profiles associated with BW
To further understand the biological implications of the BW-associated mRNAs and miRNAs, canonical pathway and network enrichment analyses were carried out, as enabled through the Ingenuity Knowledge Database. Over-represented canonical pathways were defined as those containing more BW-associated mRNAs than expected by random chance, as based on a BH-corrected p-value calculated from a right-tailed Fisher’s Exact Text [40,44]. Pathways with enrichment BH-corrected p-values <0.05 were considered significant. Similarly, networks were constructed based on known protein-protein interactions and other molecular interactions to further elucidate biological signaling that may influence infant BW. Networks were ranked based on right-tailed Fisher’s Exact test p-values, indicating the likelihood of observing a network containing at least the same number of proteins encoded by BW-associated genes by chance in comparison to random selections of other genes within the genome [44]. miRNAs that were predicted to regulate the expression of BW-associated mRNAs mapped within these networks were overlaid onto resulting network visualizations. These methods parallel previous publications [33–35,45].
3. Results
3.1. Study cohort characteristics
The overall demographic characteristics of this study cohort are largely representative of the full ELGAN cohort [26]. Subject information was included and summarized from the 390 placentas available for the current study, derived from women aged 14 to 45 years old (Table 1). Most women identified as White (61.4%), had a normal BMI ranging from 18.5 to 25.0 kg/m2 (52.8%), were not exposed to first- or second-hand smoking (76.4%), and completed between 12 and 16 years of education (48.8%). The average pregnancy ended at 26 completed weeks of gestation, and the average infant BW was 831 grams, ranging from 420 to 1418 grams. All 390 subjects were considered to have low BW as expected for children born extremely preterm. Placentas were derived from 205 (52.6%) male infants and 185 (47.4%) female infants. A total of 103 (26.4%) pregnancies represented multiple-gestations.
Table 1. Subject Characteristics.
Maternal demographic data, pregnancy characteristics, and data on birth outcomes are presented for the ELGAN subjects used in this analysis. Data are presented as the number (%) of subjects and mean [range] in the cohort.
| Overall (n = 390) | miRNA subset (n = 376) | mRNA subset (n = 378) | |
|---|---|---|---|
| Maternal age (years) | 29 [14–45] | 29 [14–45] | 29 [14–45] |
| Maternal race | |||
| White | 237 (61.4) | 228 (61.3) | 229 (61.2) |
| Non-white | 149 (38.6) | 144 (38.7) | 145 (38.8) |
| Maternal BMI (kg/m 2 ) | |||
| Underweight (< 18.5) | 27 (7.2) | 26 (7.0) | 26 (7.0) |
| Normal (18.5 – < 25.0) | 199 (52.8) | 197 (53.1) | 199 (53.4) |
| Overweight or obese (≥ 25.0) | 151 (40.1) | 148 (39.9) | 148 (39.7) |
| Smoke exposure * | |||
| No | 292 (76.4) | 287 (76.3) | 289 (76.5) |
| Yes | 90 (23.6) | 89 (23.7) | 89 (23.5) |
| Multiple-gestation pregnancy | |||
| No | 287 (73.6) | 274 (72.9) | 276 (73.0) |
| Yes | 103 (26.4) | 102 (27.1) | 102 (27.0) |
| Highest level of educational attainment | |||
| Less than 12 years | 49 (12.9) | 47 (12.6) | 47 (12.5) |
| Between 12 and 16 years | 185 (48.8) | 182 (48.8) | 184 (49.1) |
| Greater than 16 years | 145 (38.3) | 144 (38.6) | 144 (38.4) |
| Newborn sex | |||
| Male | 205 (52.6) | 197 (52.4) | 198 (52.4) |
| Female | 185 (47.4) | 179 (47.6) | 180 (47.6) |
| Newborn birth weight (grams) | 831.4 [420–1418] | 835.2 [420–1418] | 834.2 [420–1418] |
| Gestational age (weeks) | 26 [23–27] | 26 [23–27] | 26 [23–27] |
first- or second-hand smoke exposure
A total of 378 placentas were included in the final mRNA analysis and 376 in the final miRNA analysis. Subjects were removed from the mRNA and/or miRNA analyses due to the following criteria: (i) low expression values (i.e., non-detection across all genes) (n = 2 for the mRNA analysis); (ii) identified as sample outliers, based on criteria defined in the methods section (n = 2 for the mRNA, n = 2 for the miRNA analyses); and (iii) demographic data were missing for the included covariates (n = 8 for the mRNA, n = 12 for the miRNA analyses). Similar demographic distributions were apparent across all 390 evaluated placentas in comparison to the final sets of subjects included in the mRNA and miRNA analyses (Table 1).
3.2. Differential expression of placental mRNAs associated with infant BW
Changes in genome-wide gene expression profiles were evaluated for associations with infant BW. A total of 254 genes were identified with expression levels significantly (FDR < 0.1) associated with BW (Figure 1A, Supplemental Table S1). Of these, 164 showed increased expression and 90 showed decreased expression in relation to increasing BW. Notable genes identified as associated with infant BW include Janus Kinase 1 (JAK1, β = 4.08, FDR = 0.02), Signal Transducing Adaptor Molecule 2 (STAM2, β = 6.35, FDR = 0.09), and Nuclear Cap-Binding Protein Subunit 3 (NCBP3, β = 11.00, FDR = 0.02). Here, the β estimate associated with each gene is interpreted as the change in expression associated with each one-unit increase in infant BW (log-scaled, in grams).
Figure 1. MAplots of mean (A) mRNA and (B) miRNA expression levels vs. fold change in expression associated with infant BW across human placenta tissues.
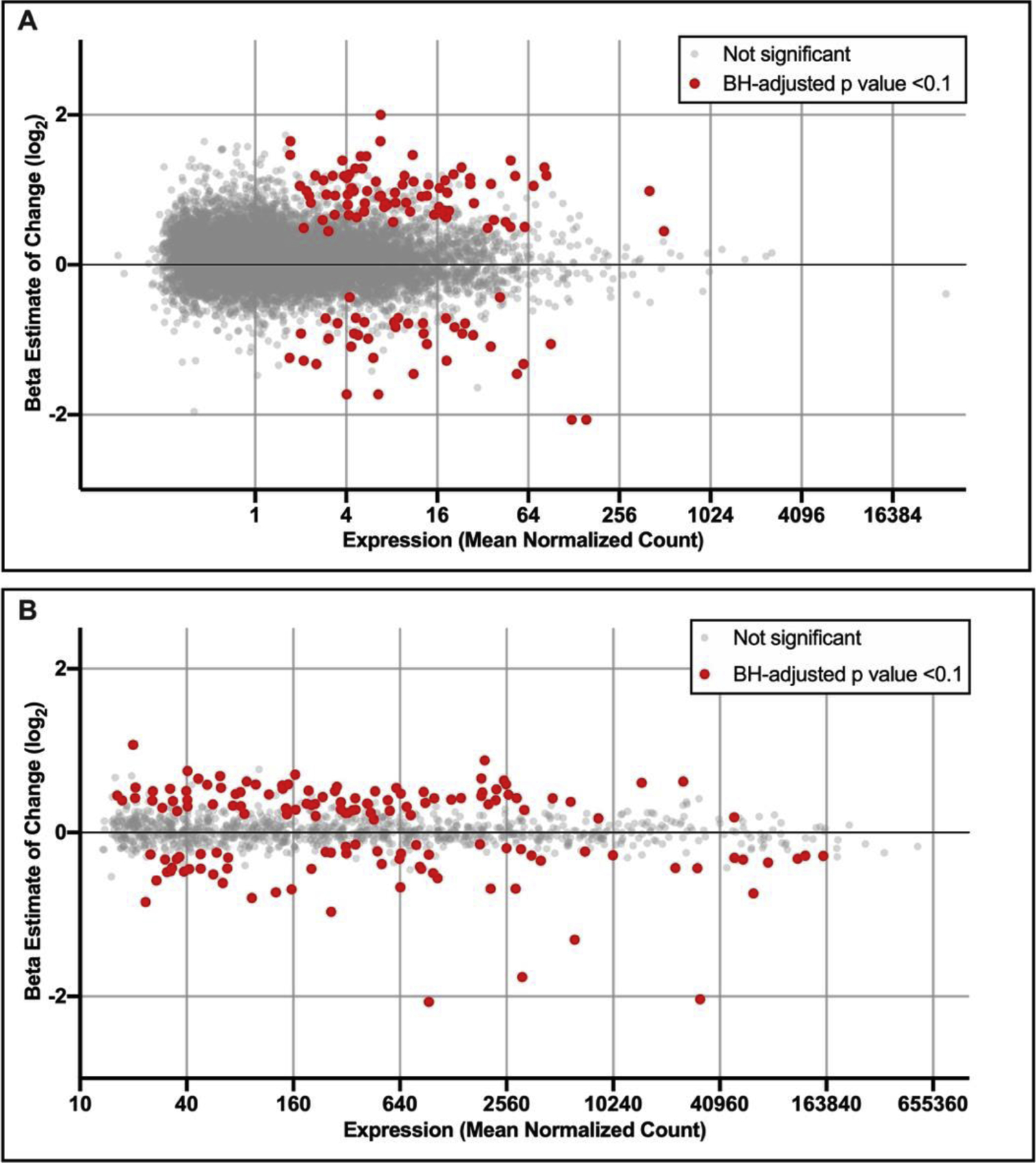
Beta estimates represent the change in expression associated with one unit increase in BW and are plotted as log2 transformed values. mRNAs and miRNAs that were identified as significantly (FDR < 0.10) associated with BW are red, and those that were not significantly associated are displayed in grey. Only mRNAs and miRNAs expressed above background are displayed.
Placentas are fetally-derived, and there is evidence for placental gene expression profiles to be highly dependent upon infant sex [22,46,47]. As such, we carried out analyses to test whether BW-associated mRNAs exhibit trends that are sex-specific (also referred to as sexually dimorphic). Stratifying data derived from male placentas separate from female placentas, we identified 42 mRNAs associated with BW in males and 31 associated with BW in females (Figure 2A, Supplemental Table S2). When compared to the global analysis, 35 (83%) of the male-derived mRNAs and 21 (68%) of the female-derived mRNAs associated with BW were also identified when analyzing all placentas collectively. Seven mRNAs overlapped between the two sex-stratified analyses, representing 17% of the male-derived mRNAs and 23% of the female-derived mRNAs associated with BW. Of these, all 7 mRNAs showed consistent directionality of change across sexes. Taken together, the results suggest that the majority of genes associated with infant BW identified through sex-stratification were also identified in the collective analysis, indicative of minimal sexual dimorphism, and thus are important to further investigate.
Figure 2.
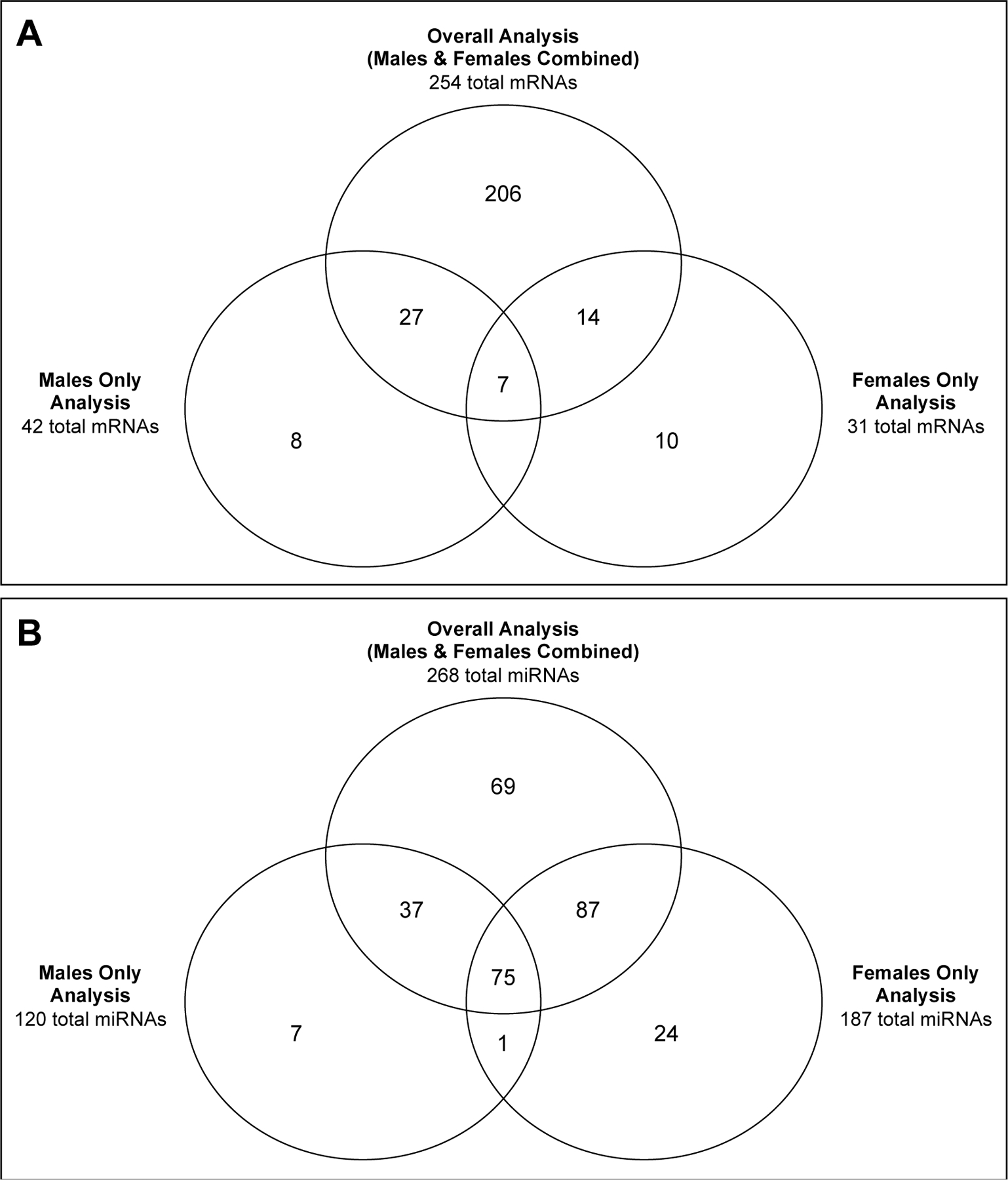
Overlap between (A) mRNAs and (B) miRNAs identified as associated with BW within the overall, collective analyses, in comparison to the sex-stratified analyses.
3.3. Differential expression of placental miRNAs associated with infant BW
Changes in genome-wide miRNA expression profiles were evaluated for associations with infant BW. A total of 268 miRNAs were identified with expression levels significantly (FDR < 0.10) associated with BW (Figure 1B, Supplemental Table S3). Of these, 163 showed increased expression and 105 showed decreased expression in relation to increasing BW. Notable miRNAs identified as associated with infant BW include miR-6773–5p (β = 35, FDR = 0.005), miR-6081 (β = 134, FDR = 0.01), and miR-519c-3p (β = 26,739, FDR = 0.04). Here, the β estimate associated with each miRNA is interpreted as the change in expression associated with each one-unit increase in infant BW (log-scaled, in grams).
Similar to the gene expression analysis, we carried out separate statistical analyses to test whether BW-associated miRNAs exhibit trends that are sexually dimorphic. Stratifying data derived from male placentas separate from female placentas, we identified 120 miRNAs associated with BW in males and 187 associated with BW in females (Figure 2B, Supplemental Table S4). When compared to the global analysis, 113 (94%) of the male-derived miRNAs and 163 (87%) of the female-derived miRNAs associated with BW were also identified when analyzing all placentas collectively. Many of these miRNAs overlapped between the two sexes. Specifically, 76 miRNAs overlapped between the two sex-stratified analyses, representing 63% of the male-derived miRNAs and 41% of the female-derived miRNAs. Of these, 34 (45%) miRNAs showed consistent directionality of change across sexes. Similar to the mRNA analysis, these results suggest that the majority of miRNAs associated with infant BW show largely consistent trends, independent of sex, and thus represent consistent epigenetic changes that might influence BW regardless of infant sex.
3.4. Comparing placental mRNAs and miRNAs associated with BW to previous cohorts
There are a limited number of studies that have previously evaluated relationships between mRNA and miRNA expression profiles in the placenta and infant BW in human cohorts [4,8,9,11,19,20,48]. A recent study by Cox et al. (2019) carried out a transcriptome-wide analysis of 183 placental samples collected as part of the ENVIRONAGE birth cohort study in Belgium [20]. This cohort included infants that had an average gestational age of 38.9 weeks, though 24 of the infants were born at preterm (gestational age < 37 weeks). Investigators reported 85 mRNAs associated with BW that also overlapped with those associated with pre-pregnancy BMI. Though this investigation’s cohort and analysis focus differed from the current study, it is important to highlight that four mRNAs were identified as associated with BW across studies. Specifically, EGF like domain multiple 6 (EGFL6), nuclear receptor subfamily 2 group F member 1 (NR2F1), protein tyrosine phosphatase receptor type D (PTPRD), and spondin 1 (SPON1) all showed increased expression associated with increased BW across cohorts.
Select studies have also employed gene-specific approaches to identify miRNAs in the placenta that are associated with infant BW [4,8,9,11,19]. Some of the miRNAs identified by these studies were also identified in the current investigation as associated with BW, including miR-105–5p, miR-1290, miR-27a, miR-193b-3p, miR-195, miR-335–3p, miR-324–5p, miR-328, and miR-517a [8,9,19]. Of these, miR-105–5p, miR-195, and miR-335–3p showed consistent directional changes in expression, all showing increased expression associated with increasing BW across studies.
3.5. Placental miRNAs predicted to regulate mRNAs associated with BW
To evaluate which of the BW-associated mRNAs were predicted to be regulated by miRNAs, miRNA-mRNA interactions were identified based on known, published interactions as well as computationally predicted interactions, largely based on base pairing homologies. Analyses focused on the lists of mRNAs/miRNAs identified as associated with BW using all subject data, as opposed to sex-stratified results, since sex-specific responses were limited. From this global analysis, a total of 172 BW-associated miRNAs were predicted to regulate the expression levels of 161 BW-associated mRNAs that have either been experimentally observed and/or predicted with high levels of confidence (Supplemental Table S5). As a result, 63% of the mRNAs associated with BW were predicted as possibly under miRNA mediation.
A total of 405 miRNA-mRNA interactions were predicted from this analysis, representing 160 interactions with the expected inversely correlated relationships. To elaborate, 160 miRNA-mRNA predicted interactions represented either miRNAs with increased expression predicted to regulate mRNAs with decreased expression associated with BW, or miRNAs with decreased expression predicted to regulate mRNAs with increased expression associated with BW. These directional trends are important to note, as miRNAs are classically understood to regulate mRNA expression through silencing interactions [8,17]; although there have been instances when miRNAs have shown to induce/increase mRNA expression [4,8]. Of the 160 inversely related interactions that were predicted, 104 showed significant (p<0.05) correlation between expression profiles (Figure 3, Supplemental Table S5). Of the 245 positively related interactions that were predicted, 176 showed significant (p<0.05) correlation between expression profiles (Figure 3, Supplemental Table S5).
Figure 3. Example BW-associated mRNAs predicted to be regulated by miRNAs.
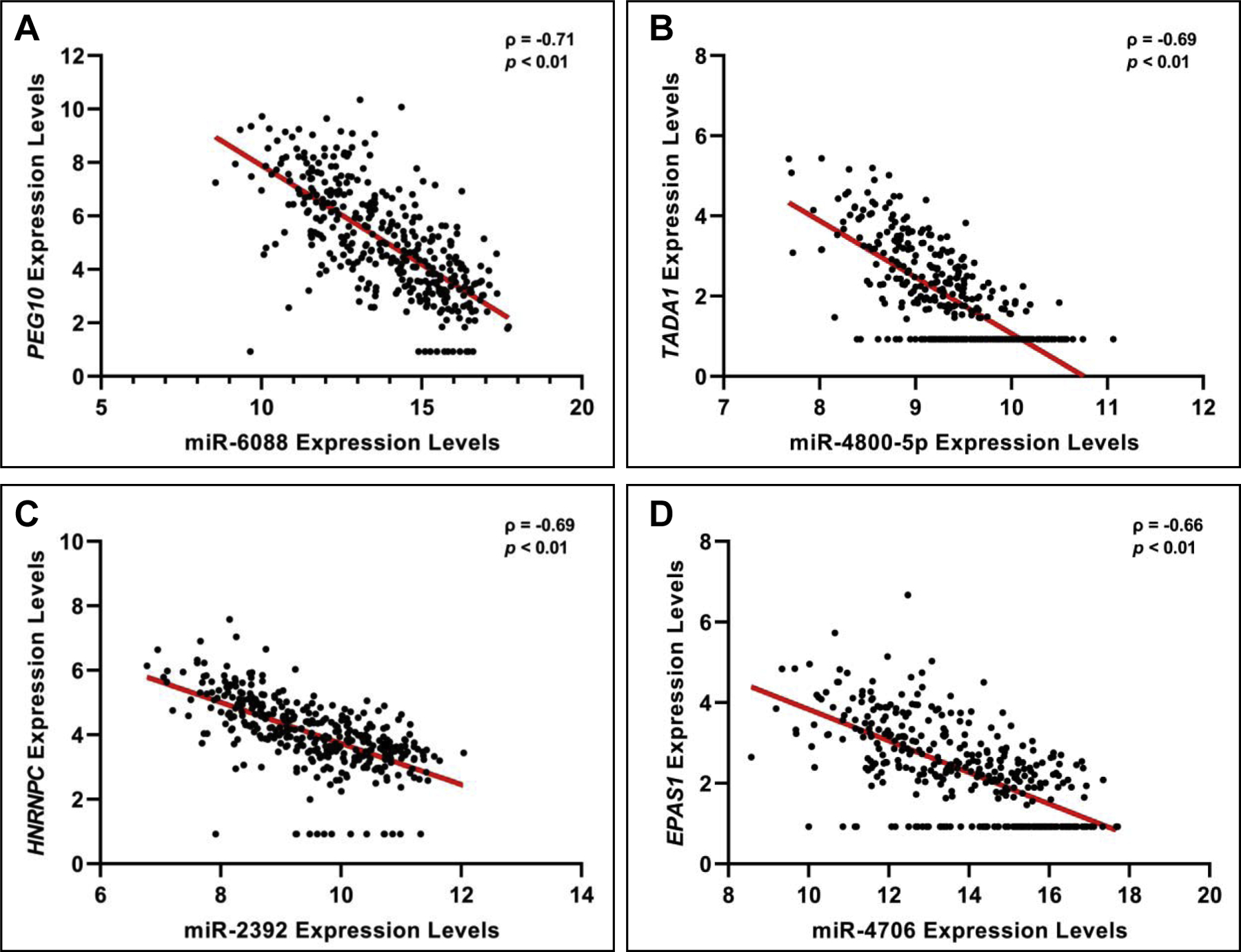
The four most significantly correlated expression level pairings are shown, specifically including (A) paternally expressed 10 (PEG10) and miR-6088; (B) transcriptional adaptor 1 (TADA1) and miR-4800–5p; (C) heterogeneous nuclear ribonucleoprotein C (HNRNPC) and miR-2392; and (D) COMM domain containing 2 (COMMD2) and miR-6088. These graphs were made with variance stabilized normalized counts.
3.6. Canonical pathways and networks enriched within genomic profiles associated with BW
To further understand the functional implications of placental genomic alterations associated with BW, canonical pathway and network analyses were carried out on the BW-associated mRNAs. Several canonical pathways were identified as significantly associated with the BW-associated genes (Supplemental Table S6), with the glycoprotein VI (GP6) pathway as being one of the pathways identified with the most significant enrichment (BH p-value = 0.002), alongside nine other pathways. GP6 is a glycoprotein receptor for collagen [49], and the GP6 pathway enriched here included the following BW-associated genes: Calmodulin 2 (CALM1), Collagen Type VI Alpha 3 (COL6A3), Laminin Subunit Beta 1 (LAMB1), Laminin Subunit Gamma 1 (LAMC1), Phosphatidylinositol-4,5-Bisphosphate 3-Kinase Catalytic Subunit Beta (PIK3CB), Protein Kinase C Alpha (PRKCA), Protein Kinase C Eta (PRKCH), and Protein Kinase D3 (PRKD3). These genes are notably involved in the collagen complex, laminin family, calmodulin complex, phosphoinositide 3-kinases (PI3K) complex, and the protein kinase C (PKC) complex (Figure 4).
Figure 4. Proteins encoded by BW-associated genes involved in GP6 signaling in the placenta.
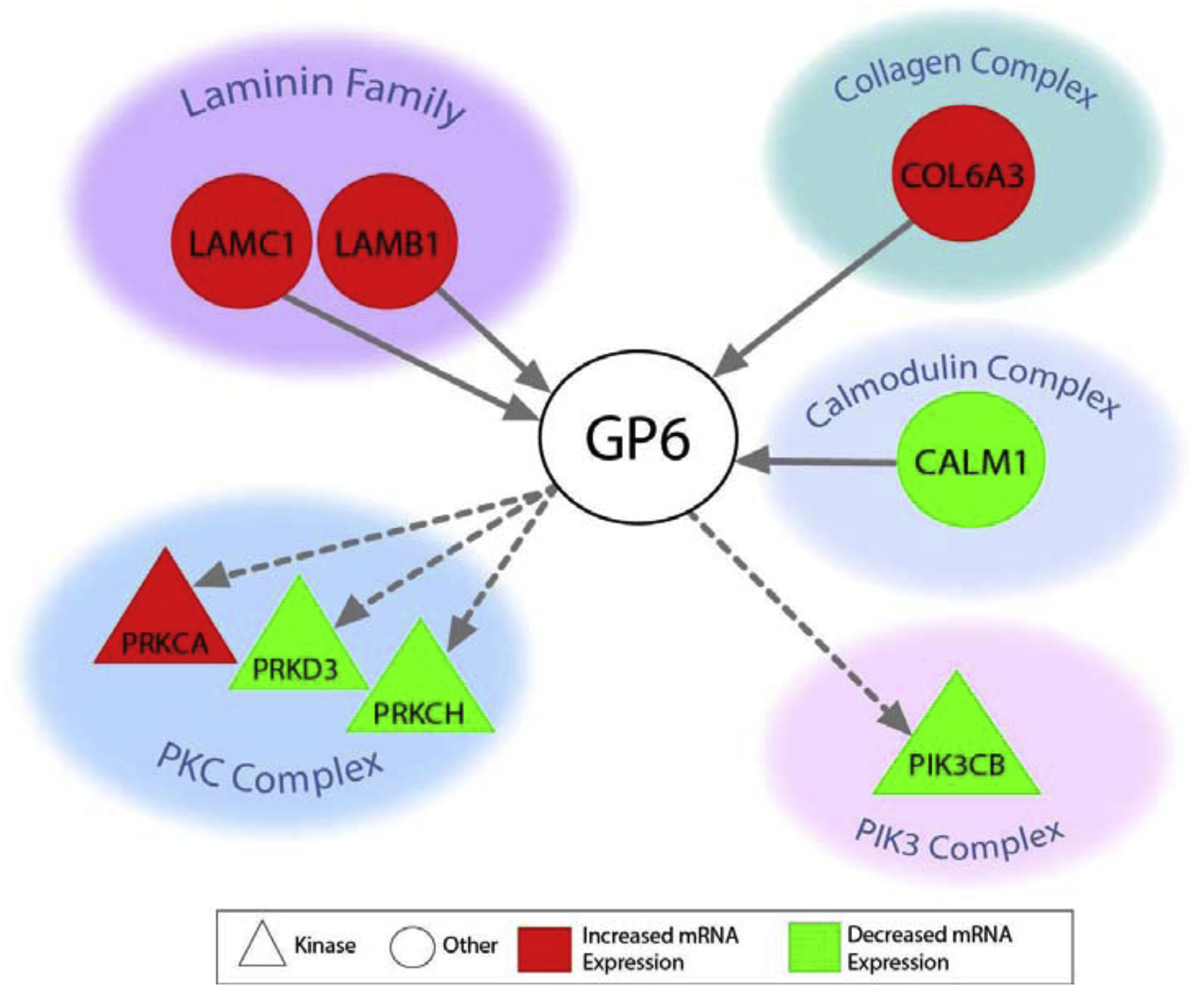
Direct interactions are shown in solid lines and indirect in dashed lines.
Other notable pathways related to GP6 signaling included those relevant to growth signaling, including Hepatocyte Growth Factor Signaling (HGF) (BH p-value = 0.005) and Growth Hormone (GH) Signaling (BH p-value = 0.01). Protein-protein interaction networks were also constructed to illustrate known and predicted molecular interactions associated with the BW-associated genes and related pathways. A network was identified to contain a portion of the GP6-relevant signaling, as well as other signaling relevant to growth signaling. A portion of the genes encoding proteins in the network were also predicted to be regulated by the BW-associated miRNAs (Figure 5). These data therefore highlight the potential novel role of GP6, HGF, and GH signaling involved in placental-mediated influences on infant BW.
Figure 5. Network showing biological interactions between proteins encoded by mRNAs associated with infant BW, a portion of which are predicted to be regulated by BW-associated miRNAs in human placenta tissues.
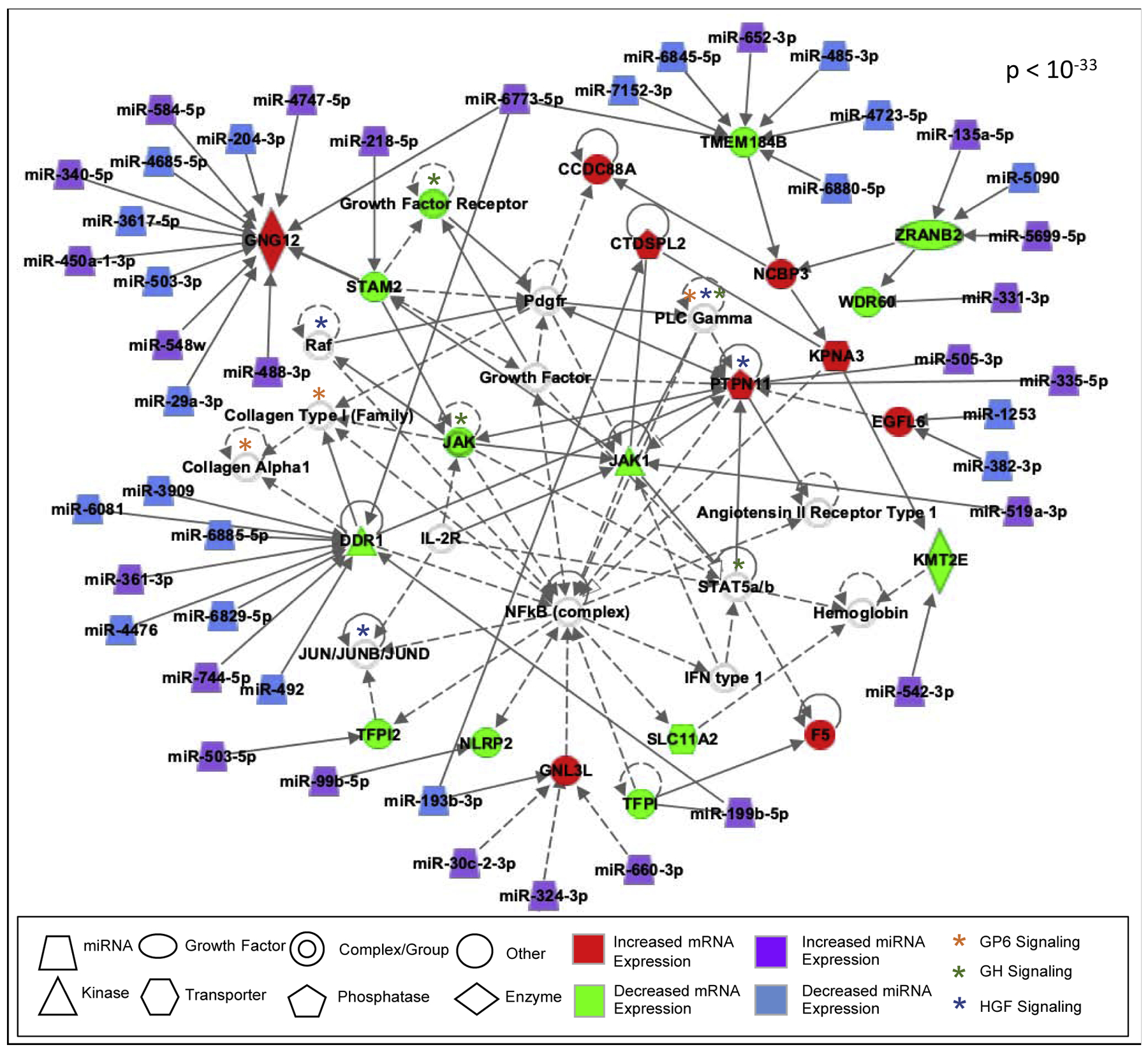
Molecules with BW-associated changes are colored, and molecules with associated signaling are white. Direct interactions are shown in solid lines and indirect in dashed lines.
4. Discussion
This study set out to evaluate molecular signatures associated with infant BW, focusing on transcriptomic (mRNA) and epigenomic (miRNA) signatures within placentas derived from extremely premature infants. We hypothesized that genes involved in critical placental cell signaling are associated with infant BW, and are regulated, in part, through epigenetic mechanisms based on miRNA mediation. Reduced fetal growth resulting in low BW is specifically linked to placental insufficiency, and is associated with multiple chronic conditions in adulthood [50–52]. Therefore, identifying mechanisms that regulate placental health and relate to changes in BW is of vital importance. Our study provides novel evidence that the placental genome and epigenome is highly linked to infant BW. Specifically, we identified 254 mRNAs and 268 miRNAs associated with infant BW, the majority of which showed consistent associations across placentas derived from both males and females. Gene expression patterns associated with BW were identified as enriched for critical pathways, including GP6, GH, and HGF signaling. A portion of these signaling alterations were predicted to be regulated by miRNAs that were also identified as associated with infant BW; specifically, 161 of the BW-associated mRNAs were predicted as regulated by 172 miRNAs. Together, these data highlight important mechanisms potentially regulating infant BW through the placenta.
In this study, we found that the majority of mRNA and miRNA signatures associated with BW were largely consistent across infant sexes. These data suggest that, unlike other infant outcomes, BW may be regulated by placental transcriptomic mechanisms that are largely consistent across infant sex. Furthermore, these genomic and epigenomic alterations were enriched for other biological processes besides oxidative stress; a notable finding given that sexually dimorphic phenotypes have been attributable to oxidative stress [53,54]. For example, males have been shown to have fewer antioxidant defense mechanisms contributing to poorer health outcomes in preterm infants [55–57]. The current study highlighted other mechanisms besides oxidative stress as being sex-independent and associated with infant BW.
Our results yielded several canonical pathways significantly associated with mRNA expression profiles related to BW, including GP6 signaling. Specific genes involved in GP6 signaling that were identified with expression levels associated with BW included those within the collagen complex (e.g., COL6A3), laminin family (e.g., LAMB1 and LAMC1), and PKC complex (e.g., PRKCA, PRKCH and PRKD3), among others. GP6 plays a role in inducing platelet activation and thrombus formation and is the major signaling receptor for collagen [49]. In the placenta, collagen is essential for the construction of its underlying structure and is primarily supported by collagen providing tensile strength [58]. A decrease in placental collagen content has been found to contribute to remodeling of the extra cellular matrix, resulting in cervical softening and fetal membrane activation [58]. Collagen-degrading enzymes have specifically been identified as active during labor, promoting the separation of the placenta from the uterus during delivery [59]. Though the role of collagen is known in terms of influencing placental structure, what remains understudied is the potential role of collagen and GP6 signaling on fetal outcomes, such as BW.
Growth signaling pathways were also identified as enriched within the BW-associated genes, including GH and HGF signaling. GH is secreted by the placenta during gestation and is well recognized for its role in regulating fetal development [60,61]. For example, a clinical study identified lower levels of placental GH levels in infants born at lower BW; and levels of placental GH, insulin-like growth factor I, and insulin-like growth factor-binding protein-3 accounted for a total of 40% of the variance in infant BW [61]. Interestingly, these proteins were also linked to maternal glycemic status, indicating potential cross-talk between maternal metabolic processes and fetal BW via placental GH signaling [61]. Animal studies have also shown relationships between circulating GH levels during pregnancy and altered BW in offspring (e.g., mouse and bovine models) [60]. Additionally, HGF is strongly expressed in the placenta, particularly in the amnionic epithelium, extravillous trophoblast, and villous syncytium, and is known to act in concert with its tyrosine kinase receptor, Met, to regulate cell proliferation, migration, and morphogenesis [62,63]. A study in nonhuman primates found that blocking HGF signaling through Onartuzumab treatment during pregnancy resulted in decreased BW in offspring [64]. Our study highlights an enrichment for BW-associated genes involved in GP6, GH, and HGF pathways, a portion of which were predicted as regulated by miRNAs.
Though this study has several advantages, including a robust dataset of transcriptomic and epigenomic human placental signatures derived from 390 infants, this study is not without limitations. When interpreting findings from this analysis, it is important to consider that study participants included infants from the ELGAN cohort, representing infants born at extremely low gestational age. Future studies should evaluate whether these findings are apparent within placentas from infants born at full-term. However, comparing our findings against previous studies that have evaluated mRNA and miRNA expression alterations through gene-specific approaches within placentas of full-term newborns identified a portion of expression changes associated with BW consistent across cohorts. In addition, analyses from human cohorts have inherent limitations surrounding potentially unmeasured confounding. For example, select socioeconomic factors, maternal nutrition, and level of prenatal healthcare may have influenced lower BW [65–67], representing factors that may not be fully captured in the covariates that were collected and tested. Still, this study contributes valuable information towards advancing our understanding of placental mechanisms associated with the critical outcome, infant BW. Future studies could further evaluate the impact of these placental genomic and epigenomic alterations using controlled in vitro and/or animal models [7] and test potential strategies for therapeutic intervention to enhance the health of newborns at risk for low BW.
5. Conclusion
Together, this study represents the largest investigation of genome-wide placental mRNA and miRNA expression signatures related to infant birth weight, to date. Findings demonstrated that BW is associated with changes in the placental transcriptome and epigenome, with many changes known and/or predicted to interact and regulate placental cell health via critical pathways. These pathways included GP6, HG, and HGF signaling, which are known to influence cellular growth and tissue development. A portion of the genes involved in these critical pathways were predicted to be regulated by miRNAs, which also showed expression levels associated with BW. These miRNA-regulated molecular patterns are of high importance and should be evaluated in future studies for the potential to be impacted by internal risk factors and/or external environmental insults that may induce harm to the growing fetus and impact later-in-life health consequences.
Supplementary Material
Highlights.
Placental mRNAs and miRNAs showed birth weight-related differential expression
Genomic and epigenomic changes were largely consistent across infant sex
Associated pathways included glycoprotein VI and growth factor signaling
Birth weight-associated changes were predicted to be regulated by miRNAs
Acknowledgements
Funding
This study was supported by grants from the National Institutes of Health (NIH) the Office of the NIH Director [UG3OD023348, UH3OD023348], the National Institute of Child Health and Human Development [R01HD092374], the National Institute of Nursing Research [K23NR017898], and the National Institute of Environmental Health Sciences [P42ES031007].
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest
The authors declare no competing interests.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
REFERENCES
- 1.Horikoshi M, Beaumont RN, Day FR, Warrington NM, Kooijman MN, Fernandez-Tajes J, Feenstra B, van Zuydam NR, Gaulton KJ, et al. Genome-wide associations for birth weight and correlations with adult disease. Nature. 2016;538(7624):248–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Premru-Srsen T, Verdenik I, Ponikvar BM, Steblovnik L, Gersak K, Cerar LK. Infant mortality and causes of death by birth weight for gestational age in non-malformed singleton infants: a 2002–2012 population-based study. J Perinat Med. 2018;46(5):547–53. [DOI] [PubMed] [Google Scholar]
- 3.Beaumont RN, Horikoshi M, McCarthy MI, Freathy RM. How Can Genetic Studies Help Us to Understand Links Between Birth Weight and Type 2 Diabetes? Curr Diab Rep. 2017;17(4):22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rodil-Garcia P, Arellanes-Licea EDC, Montoya-Contreras A, Salazar-Olivo LA. Analysis of MicroRNA Expression in Newborns with Differential Birth Weight Using Newborn Screening Cards. Int J Mol Sci. 2017;18(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reynolds LJ, Pollack RI, Charnigo RJ, Rashid CS, Stromberg AJ, Shen S, O’Brien JM, Pearson KJ. Increased birth weight is associated with altered gene expression in neonatal foreskin. J Dev Orig Health Dis. 2017;8(5):575–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ramadhani MK, Grobbee DE, Bots ML, Castro Cabezas M, Vos LE, Oren A, Uiterwaal CS. Lower birth weight predicts metabolic syndrome in young adults: the Atherosclerosis Risk in Young Adults (ARYA)-study. Atherosclerosis. 2006;184(1):21–7. [DOI] [PubMed] [Google Scholar]
- 7.Fry RC, Bangma J, Szilagyi J, Rager JE. Developing novel in vitro methods for the risk assessment of developmental and placental toxicants in the environment. Toxicol Appl Pharmacol. 2019;378:114635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ostling H, Kruse R, Helenius G, Lodefalk M. Placental expression of microRNAs in infants born small for gestational age. Placenta. 2019;81:46–53. [DOI] [PubMed] [Google Scholar]
- 9.Rahman ML, Liang L, Valeri L, Su L, Zhu Z, Gao S, Mostofa G, Qamruzzaman Q, Hauser R, et al. Regulation of birthweight by placenta-derived miRNAs: evidence from an arsenic-exposed birth cohort in Bangladesh. Epigenetics. 2018;13(6):573–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cai M, Kolluru GK, Ahmed A. Small Molecule, Big Prospects: MicroRNA in Pregnancy and Its Complications. J Pregnancy. 2017;2017:6972732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang D, Na Q, Song WW, Song GY. Altered Expression of miR-518b and miR-519a in the placenta is associated with low fetal birth weight. Am J Perinatol. 2014;31(9):729–34. [DOI] [PubMed] [Google Scholar]
- 12.Peng S, Deyssenroth MA, Di Narzo AF, Cheng H, Zhang Z, Lambertini L, Ruusalepp A, Kovacic JC, Bjorkegren JLM, et al. Genetic regulation of the placental transcriptome underlies birth weight and risk of childhood obesity. PLoS Genet. 2018;14(12):e1007799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vaiman D. Genes, epigenetics and miRNA regulation in the placenta. Placenta. 2017;52:127–33. [DOI] [PubMed] [Google Scholar]
- 14.Lycoudi A, Mavreli D, Mavrou A, Papantoniou N, Kolialexi A. miRNAs in pregnancy-related complications. Expert Rev Mol Diagn. 2015;15(8):999–1010. [DOI] [PubMed] [Google Scholar]
- 15.Nakamura M, Ferreira SH. A peripheral sympathetic component in inflammatory hyperalgesia. Eur J Pharmacol. 1987;135(2):145–53. [DOI] [PubMed] [Google Scholar]
- 16.Tsamou M, Vrijens K, Madhloum N, Lefebvre W, Vanpoucke C, Nawrot TS. Air pollution-induced placental epigenetic alterations in early life: a candidate miRNA approach. Epigenetics. 2018;13(2):135–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu B, Li J, Cairns MJ. Identifying miRNAs, targets and functions. Brief Bioinform. 2014;15(1):1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brooks SA, Martin E, Smeester L, Grace MR, Boggess K, Fry RC. miRNAs as common regulators of the transforming growth factor (TGF)-beta pathway in the preeclamptic placenta and cadmium-treated trophoblasts: Links between the environment, the epigenome and preeclampsia. Food Chem Toxicol. 2016;98(Pt A):50–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barchitta M, Maugeri A, Quattrocchi A, Agrifoglio O, Agodi A. The Role of miRNAs as Biomarkers for Pregnancy Outcomes: A Comprehensive Review. Int J Genomics. 2017;2017:8067972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deyssenroth MA, Peng S, Hao K, Lambertini L, Marsit CJ, Chen J. Whole-transcriptome analysis delineates the human placenta gene network and its associations with fetal growth. BMC Genomics. 2017;18(1):520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones JM 2nd. Advances in the treatment of non-Hodgkin’s lymphoma. J Med Assoc Ga. 1989;78(2):105–8. [PubMed] [Google Scholar]
- 22.Gonzalez TL, Sun T, Koeppel AF, Lee B, Wang ET, Farber CR, Rich SS, Sundheimer LW, Buttle RA, et al. Sex differences in the late first trimester human placenta transcriptome. Biol Sex Differ. 2018;9(1):4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meher AP, Wadhwani N, Randhir K, Mehendale S, Wagh G, Joshi SR. Placental DHA and mRNA levels of PPARgamma and LXRalpha and their relationship to birth weight. J Clin Lipidol. 2016;10(4):767–74. [DOI] [PubMed] [Google Scholar]
- 24.Vrijens K, Tsamou M, Madhloum N, Gyselaers W, Nawrot TS. Placental hypoxia-regulating network in relation to birth weight and ponderal index: the ENVIRONAGE Birth Cohort Study. J Transl Med. 2018;16(1):2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ilekis JV, Tsilou E, Fisher S, Abrahams VM, Soares MJ, Cross JC, Zamudio S, Illsley NP, Myatt L, et al. Placental origins of adverse pregnancy outcomes: potential molecular targets: an Executive Workshop Summary of the Eunice Kennedy Shriver National Institute of Child Health and Human Development. Am J Obstet Gynecol. 2016;215(1 Suppl):S1–S46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McElrath TF, Hecht JL, Dammann O, Boggess K, Onderdonk A, Markenson G, Harper M, Delpapa E, Allred EN, et al. Pregnancy disorders that lead to delivery before the 28th week of gestation: an epidemiologic approach to classification. Am J Epidemiol. 2008;168(9):980–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Streimish IG, Ehrenkranz RA, Allred EN, O’Shea TM, Kuban KC, Paneth N, Leviton A, Investigators ES. Birth weight- and fetal weight-growth restriction: impact on neurodevelopment. Early Hum Dev. 2012;88(9):765–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Addo KA, Bulka C, Dhingra R, Santos HP Jr., Smeester L, O’Shea TM, Fry RC. Acetaminophen use during pregnancy and DNA methylation in the placenta of the extremely low gestational age newborn (ELGAN) cohort. Environ Epigenet. 2019;5(2):dvz010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Onderdonk AB, Delaney ML, DuBois AM, Allred EN, Leviton A, Extremely Low Gestational Age Newborns Study I. Detection of bacteria in placental tissues obtained from extremely low gestational age neonates. Am J Obstet Gynecol. 2008;198(1):110 e1–7. [DOI] [PubMed] [Google Scholar]
- 30.Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa A, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22(9):1760–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods. 2017;14(4):417–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.NCBI. Entrez Gene 2019. [cited 2019 Dec 1]. Available from: https://www.ncbi.nlm.nih.gov/gene.
- 33.Rager JE, Auerbach SS, Chappell GA, Martin E, Thompson CM, Fry RC. Benchmark Dose Modeling Estimates of the Concentrations of Inorganic Arsenic That Induce Changes to the Neonatal Transcriptome, Proteome, and Epigenome in a Pregnancy Cohort. Chem Res Toxicol. 2017;30(10):1911–20. [DOI] [PubMed] [Google Scholar]
- 34.Rager JE, Bailey KA, Smeester L, Miller SK, Parker JS, Laine JE, Drobna Z, Currier J, Douillet C, et al. Prenatal arsenic exposure and the epigenome: altered microRNAs associated with innate and adaptive immune signaling in newborn cord blood. Environ Mol Mutagen. 2014;55(3):196–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klaren WD, Ring C, Harris MA, Thompson CM, Borghoff S, Sipes NS, Hsieh JH, Auerbach SS, Rager JE. Identifying Attributes That Influence In Vitro-to-In Vivo Concordance by Comparing In Vitro Tox21 Bioactivity Versus In Vivo DrugMatrix Transcriptomic Responses Across 130 Chemicals. Toxicol Sci. 2019;167(1):157–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rager JE, Moeller BC, Miller SK, Kracko D, Doyle-Eisele M, Swenberg JA, Fry RC. Formaldehyde-associated changes in microRNAs: tissue and temporal specificity in the rat nose, white blood cells, and bone marrow. Toxicol Sci. 2014;138(1):36–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stats: RDocumentation.org; [cited 2020 2020, April 9]. Available from: https://www.rdocumentation.org/packages/stats/versions/3.6.2.
- 38.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leek JT. svaseq: removing batch effects and other unwanted noise from sequencing data. Nucleic Acids Res. 2014;42(21). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B. 1995;57. [Google Scholar]
- 41.Karagkouni D, Paraskevopoulou MD, Chatzopoulos S, Vlachos IS, Tastsoglou S, Kanellos I, Papadimitriou D, Kavakiotis I, Maniou S, et al. DIANA-TarBase v8: a decade-long collection of experimentally supported miRNA-gene interactions. Nucleic Acids Res. 2018;46(D1):D239–D45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Agarwal V, Bell GW, Nam J-W, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4:e05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.WIBR. TargetScanHuman: Prediction of microRNA targets. Release 7.2: March 2018: Whitehead Institute for Biomedical Research; 2019. [cited 2019 Dec 1]. Available from: http://www.targetscan.org/vert_72/. [Google Scholar]
- 44.IPA. Understand complex ‘omics data with Ingenuity Pathway Analysis (IPA) 2019. [cited 2019 Dec 1]. Available from: https://www.qiagenbioinformatics.com/products/ingenuity-pathway-analysis/.
- 45.Rager JE, Suh M, Chappell GA, Thompson CM, Proctor DM. Review of transcriptomic responses to hexavalent chromium exposure in lung cells supports a role of epigenetic mediators in carcinogenesis. Toxicol Lett. 2019;305:40–50. [DOI] [PubMed] [Google Scholar]
- 46.Cvitic S, Strutz J, Appel HM, Weiss E, Brandl WT, Thuringer A, Bernhart EM, Lassance L, Wadsack C, et al. Sexual dimorphism of miRNA signatures in feto-placental endothelial cells is associated with altered barrier function and actin organization. Clin Sci (Lond). 2020;134(1):39–51. [DOI] [PubMed] [Google Scholar]
- 47.Joshi A, Azuma R, Akumuo R, Goetzl L, Pinney SE. Gestational diabetes and maternal obesity are associated with sex-specific changes in miRNA and target gene expression in the fetus. Int J Obes (Lond). 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cox B, Tsamou M, Vrijens K, Neven KY, Winckelmans E, de Kok TM, Plusquin M, Nawrot TS. A Co-expression Analysis of the Placental Transcriptome in Association With Maternal Pre-pregnancy BMI and Newborn Birth Weight. Front Genet. 2019;10:354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nurden AT. Clinical significance of altered collagen-receptor functioning in platelets with emphasis on glycoprotein VI. Blood Rev. 2019;38:100592. [DOI] [PubMed] [Google Scholar]
- 50.Magalhaes E, Meio M, Moreira MEL. Hormonal Biomarkers for Evaluating the Impact of Fetal Growth Restriction on the Development of Chronic Adult Disease. Rev Bras Ginecol Obstet. 2019;41(4):256–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Calkins K, Devaskar SU. Fetal origins of adult disease. Curr Probl Pediatr Adolesc Health Care. 2011;41(6):158–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.de Boo HA, Harding JE. The developmental origins of adult disease (Barker) hypothesis. Aust N Z J Obstet Gynaecol. 2006;46(1):4–14. [DOI] [PubMed] [Google Scholar]
- 53.Evans L, Myatt L. Sexual dimorphism in the effect of maternal obesity on antioxidant defense mechanisms in the human placenta. Placenta. 2017;51:64–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Myatt L, Thornburg KL. Effects of Prenatal Nutrition and the Role of the Placenta in Health and Disease. Methods Mol Biol. 2018;1735:19–46. [DOI] [PubMed] [Google Scholar]
- 55.Peacock JL, Marston L, Marlow N, Calvert SA, Greenough A. Neonatal and infant outcome in boys and girls born very prematurely. Pediatr Res. 2012;71(3):305–10. [DOI] [PubMed] [Google Scholar]
- 56.Binet ME, Bujold E, Lefebvre F, Tremblay Y, Piedboeuf B, Canadian Neonatal N. Role of gender in morbidity and mortality of extremely premature neonates. Am J Perinatol. 2012;29(3):159–66. [DOI] [PubMed] [Google Scholar]
- 57.Matheson H, Veerbeek JH, Charnock-Jones DS, Burton GJ, Yung HW. Morphological and molecular changes in the murine placenta exposed to normobaric hypoxia throughout pregnancy. J Physiol. 2016;594(5):1371–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hirayama H Biochemical studies on collagen in human placenta--relation of collagen to the construction and function of human placenta. Nihon Sanka Fujinka Gakkai Zasshi. 1983;35(12):2395–403. [PubMed] [Google Scholar]
- 59.Weiss A, Goldman S, Shalev E. The matrix metalloproteinases (MMPS) in the decidua and fetal membranes. Front Biosci. 2007;12:649–59. [DOI] [PubMed] [Google Scholar]
- 60.Oberbauer AM. Developmental programming: the role of growth hormone. J Anim Sci Biotechnol. 2015;6(1):8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McIntyre HD, Serek R, Crane DI, Veveris-Lowe T, Parry A, Johnson S, Leung KC, Ho KK, Bougoussa M, et al. Placental growth hormone (GH), GH-binding protein, and insulin-like growth factor axis in normal, growth-retarded, and diabetic pregnancies: correlations with fetal growth. J Clin Endocrinol Metab. 2000;85(3):1143–50. [DOI] [PubMed] [Google Scholar]
- 62.Kauma S, Hayes N, Weatherford S. The differential expression of hepatocyte growth factor and met in human placenta. J Clin Endocrinol Metab. 1997;82(3):949–54. [DOI] [PubMed] [Google Scholar]
- 63.Wolf HK, Zarnegar R, Oliver L, Michalopoulos GK. Hepatocyte growth factor in human placenta and trophoblastic disease. Am J Pathol. 1991;138(4):1035–43. [PMC free article] [PubMed] [Google Scholar]
- 64.Prell RA, Dybdal N, Arima A, Chihaya Y, Nijem I, Halpern W. Placental and Fetal Effects of Onartuzumab, a Met/HGF Signaling Antagonist, When Administered to Pregnant Cynomolgus Monkeys. Toxicol Sci. 2018;165(1):186–97. [DOI] [PubMed] [Google Scholar]
- 65.Chumnijarakij T, Nuchprayoon T, Chitinand S, Onthuam Y, Quamkul N, Dusitsin N, Viputsiri OA, Chotiwan P, Limpongsanurak S, et al. Maternal risk factors for low birth weight newborn in Thailand. J Med Assoc Thai. 1992;75(8):445–52. [PubMed] [Google Scholar]
- 66.Hirve SS, Ganatra BR. Determinants of low birth weight: a community based prospective cohort study. Indian Pediatr. 1994;31(10):1221–5. [PubMed] [Google Scholar]
- 67.Habibov NN, Fan L. Does prenatal healthcare improve child birthweight outcomes in Azerbaijan? Results of the national Demographic and Health Survey. Econ Hum Biol. 2011;9(1):56–65. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


