Abstract
Nonulosonic acids (NulOs) are a diverse family of 9-carbon α-keto acid sugars that are involved in a wide range of functions across all branches of life. The family of NulOs includes the sialic acids as well as the prokaryote-specific NulOs. Select bacteria biosynthesize the sialic acid N-acetylneuraminic acid (Neu5Ac), and the ability to produce this sugar and its subsequent incorporation into cell-surface structures is implicated in a variety of bacteria-host interactions. Furthermore, scavenging of sialic acid from the environment for energy has been characterized across a diverse group of bacteria, mainly human commensals and pathogens. In addition to sialic acids, bacteria have the ability to biosynthesize prokaryote-specific NulOs, of which there are several known isomers characterized. These prokaryotic NulOs are similar in structure to Neu5Ac but little is known regarding their role in bacterial physiology. Here, we discuss the diversity in structure, the biosynthesis pathways, and the functions of bacteria-specific NulOs. These carbohydrates are phylogenetically widespread among bacteria, with numerous structurally unique modifications recognized. Despite the diversity in structure, the NulOs are also involved in functions such as motility, biofilm formation, host colonization, and immune evasion.
The Biosynthesis and Role of Sialic Acids in Human Physiology
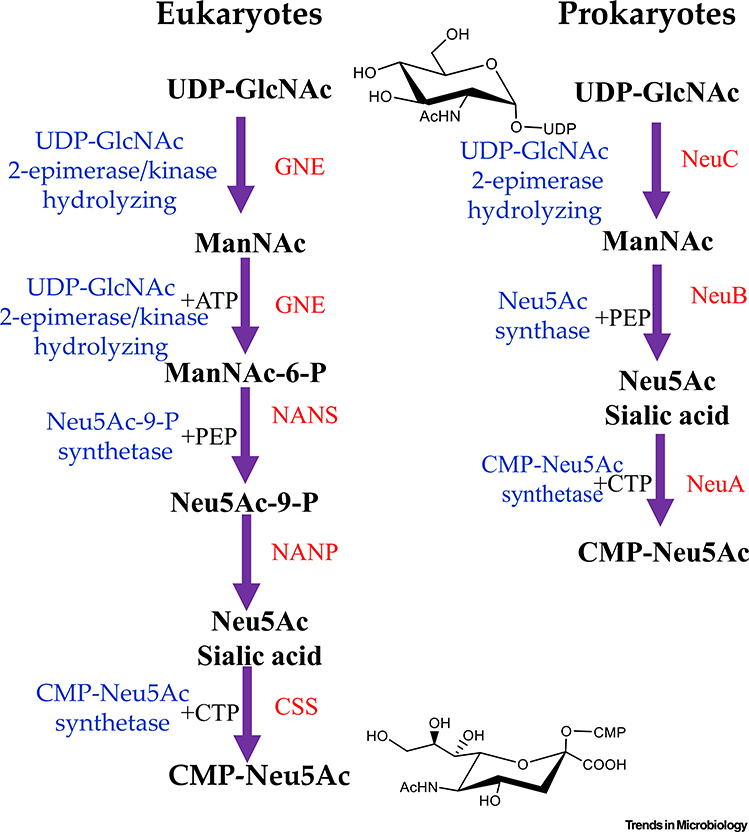
N-acetylneuraminic acid (Neu5Ac sialic acid) is a 9-carbon acidic amino sugar first identified in bovine submaxillary mucin in 1936. Neu5Ac was the first described member of a family of carbohydrates known as nonulosonic acids (NulOs). Decades of research have been dedicated to characterizing the structural diversity and biological significance of NulOs [1]. Within the sialic acids, there are over 50 different structural variations, including N-glycolylneuraminic acid (Neu5Gc), found in most nonhuman mammals, and 2-keto-3-deoxy-d-glycero-d-galacto-nononic acid (Kdn), typically found in lower ordered vertebrates, all of which perform a wide range of functions in eukaryotes [2]. The sialic acids are most commonly the terminal carbohydrate in oligosaccharide chains; thus, they are the first point of contact for many molecular interactions. Neu5Ac, the predominant NulO in humans, requires three enzymes for production. First, a UDP-N-acetylglucosamine (UDP-GlcNAc) is epimerized to form N-acetylmannosamine (ManNAc), which is phosphorylated by the same bifunctional enzyme UDP-GlcNAc epimerase (GNE), a hydrolyzing 2-epimerase, and a ManNAc kinase, forming a 6-carbon sugar intermediate ManNAc-6P [3,4] Figure 1). This 6-carbon phosphorylated intermediate is condensed with 3-carbon phosphoenolpyruvate (PEP) resulting in a 9-carbon N-acetylneuraminic acid-9P (Neu5Ac-9P) through the action of N-acetylneuraminic acid 9-phosphate synthase (NANS) [3,4]. The phosphorylated Neu5Ac-9P is dephosphorylated by N-acetylneuraminate 9-phosphate phosphohydrolase (NANP). Neu5Ac is activated by cytidine 5-monophosphate (CMP) Neu5Ac synthetase resulting in CMP-Neu5Ac, which is available for transfer to downstream targets [4] (Figure 1). For humans, Neu5Ac is present on every cell type and is involved in self-recognition and cell-cell communication. In the terminal position, sialic acids and various binding partners have evolved to play a critical role in modulating the immune response in humans and is one of the main mechanisms to distinguish self from non-self [5]. For example, the serum protein Factor H will bind to sialic acid residues on host cells and prevent the activation of the innate immunity complement cascade and cell lysis [5]. Additionally, a class of lectins, known as the sialic-acid-binding-immunoglobulin-like lectins (Siglecs) will recognize surface sialic acids in various number and linkages [6–8]. The masking effect of sialic acids is, in part, why cancerous cells will typically become hypersialylated and thus undetectable by immune cells [9,10]. In addition, sialic acids are an essential component of all mucous membranes, where the negatively charged molecules will repel one another resulting in a sliding effect as well as providing protection, particularly against bacteria [11].
Figure 1. Biosynthesis of N-acetylneuraminic (Sialic) Acid in Eukaryotes and Prokaryotes.
In eukaryotes, UDP-N-acetylglucosamine (UDP-GlcNAc) is epimerized to form N-acetylmannosamine (ManNAc), which is phosphorylated by the same enzyme, UDP-GlcNAc epimerase (GNE), a hydrolyzing 2-epimerase with a kinase domain, forming a 6-carbon sugar intermediate, ManNAc-6P [1–3]. This 6-carbon intermediate is condensed with 3-carbon phosphoenolpyruvate (PEP), resulting in a 9-carbon N-acetylneuraminic acid-9P (Neu5Ac-9P) by N-acetylneuraminic acid 9-phosphate synthase (NANS), which is dephosphorylated by N-acetylneuraminate 9-phosphate phosphohydrolase (NANP). The carbohydrate is activated by CMP-Neu5Ac synthetase (CSS). In bacteria, the biosynthesis similarly begins with UDP-GlcNAc, which is converted to ManNAc via NeuC, a hydrolyzing UDP-GlcNAc epimerase with no kinase domain. The Neu5Ac is produced via a condensation reaction with PEP catalyzed by NeuB and is activated with CMP by NeuA. CMP, cytidine 5’-monophosphate.
Bacterial Biosynthesis of N-acetylneuraminic Acid
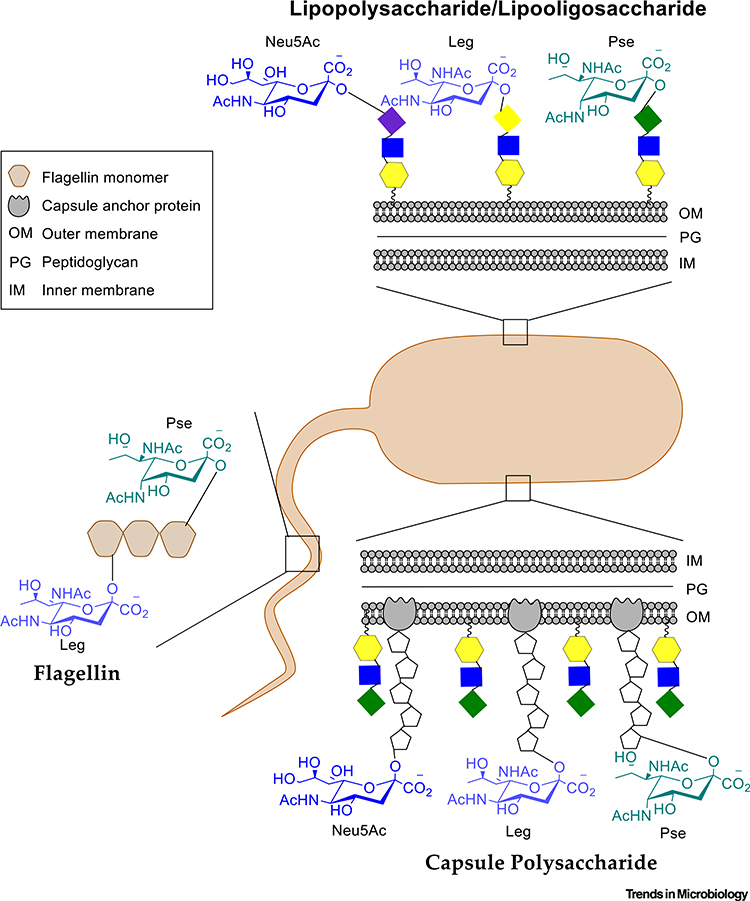
Due to the clear significance of Neu5Ac in humans, there has been considerable research to understand the role of sialic acid in microbes. Many bacteria will decorate their cell surface structures with sialic acid either by de novo biosynthesis or scavenging from the surrounding environment [12]. De novo biosynthesis of sialic acid has been demonstrated in Campylobacter jejuni, Escherichia coli, Neisseria meningitidis, and Streptococcus agalactiae or Group B streptococci (GBS) [13–21]. Scavenging of sialic acid for incorporation into surface structures was shown for Haemophilus ducreyi, Haemophilus influenzae, Pasteurella multocida, and Pseudomonas aeruginosa [22–33]. The first example of bacterial biosynthesis of sialic acid is the polysialic capsular polysaccharide of E. coli [22]. Since then, Neu5Ac has been identified in the outer-membrane lipopolysaccharide (LPS) or low-molecular-weight outer-membrane component lipooligosaccharide (LOS) and/or capsule polysaccharide (CPS) of many Gram-negative and Gram-positive bacteria that produce them (Figure 2) [13–19].
Figure 2. Bacterial Surface Structures Decorated by Nonulosonic Acids (NulOs).
NulOs identified to date have be found in lipopolysaccharide (LPS), lipooligosaccharide (LOS), and capsule polysaccharide (CPS) or as glycans on flagellin proteins of flagellum or S-layer proteins.
These surface structures perform a variety of functions for the bacteria as well as serving as potential virulence factors. The LPS is generally a defining feature of Gram-negative bacteria and is composed of three components: the O-antigen, core oligosaccharide, and the lipid A. The O-antigen is a repeating unit oligosaccharide and is the outermost exposed feature of the LPS. Following the O-antigen is the inner core, which is a shorter oligosaccharide domain that links directly to the lipid domain known as lipid A. Some bacteria will lack the outermost O-antigen and have only the core and lipid A domains, which together constitute the LOS. The bacterial capsule is composed of organized polysaccharides secreted from the cell and is not easily dissociated.
At the biochemical and genetic level, the Neu5Ac biosynthesis pathway has been extensively studied. Biosynthesis of sialic acid in bacteria begins with the metabolite UDP-GlcNAc, a substrate of bacterial cell wall biosynthesis, which can be converted to ManNAc through the action of UDP N-acetylglucosamine hydrolyzing 2-epimerase (NeuC) (Figure 1) [4,34]. The function of NeuC is to hydrolyze the UDP moiety and isomerize the carbohydrate [35]. Where bacteria and humans differ is through the action of N-acetylneuraminate synthase (NeuB), which can directly convert ManNAc, with the addition of PEP to Neu5Ac and no phosphorylated intermediates [4,34] (Figure 1). The final step is performed by N-acetylneuraminate cytidylyltransferase (NeuA), which activates Neu5Ac by adding a CMP moiety to the hydroxyl group on C-2 of the sialic acid in a reaction homologous to the enzyme present in humans [34] (Figure 1). This step is required for the Neu5Ac to be recognized by sialyltransferases and incorporation into bacterial surface structures (Figure 1). Sialic acids can be modified, typically via NeuD within the neu gene cluster, which O-acetylates monomeric sialic acid. NeuD was first described in GBS strains but bioinformatics analysis also suggested that the O-acetyltransferase was present in E. coli, C. jejuni, and N. meningitidis [36–39]. In E. coli K1, the neuO gene, which is within a prophage, is required for O-acetylation of polymeric sialic acid [37].
Sialic Acids as Bacterial Virulence Determinants
Neu5Ac biosynthesis has been demonstrated in a growing list of bacterial species, which includes host-adapted commensals and pathogens. Most of the research has focused on the ability of this carbohydrate to act as a molecular mimic for invading bacterial human pathogens [40]. Ultimately, pathogen surface sialylation camouflages the bacteria from the host. The presence of sialylated surface structures in pathogens affects susceptibility to bactericidal antibodies and phagocytosis, as well as resulting in decreased complement activation and neutrophil adherence [18,41,42]. A model system for Neu5Ac bacterial modification is GBS, a leading cause of neonatal pneumonia, septicemia, and meningitis. GBS produce a capsular polysaccharide in which sialic acids present as terminal caps are considered essential for evading the host’s immune response and promoting survival in vivo [14]. The mechanisms include interference with the host’s complement system and engaging the host’s immune receptor Siglecs to dampen the innate immune response [41 –43]. Specifically, GBS sialylated capsular polysaccharide interacts with the inhibitory Siglec-9 to blunt neutrophil activation and bacterial activity [41]. C. jejuni is a leading cause of gastroenteritis, and many strains have a sialylated LOS that is required for invasion and serum resistance [24,44]. In addition, some C. jejuni strains expressing specific sialic linkages that are structurally similar to human gangliosides are associated with the neurological disorder Guillain— Barré syndrome [45].
Sialic acid is present in surface structures of the human pathogens N. meningitidis, Neisseria gonorrhoeae, H. influenzae, Helicobacter pylori, and Salmonella enterica amongst others [13,24,46–49]. Nontypeable H. influenzae (NTHi), a host-adapted pathogen, causes respiratory tract infections and can cause major complications in and exacerbation of chronic obstructive 115 pulmonary disease (COPD). NTHi does not biosynthesize Neu5Ac and must scavenge it from 116 its host [50]. Incorporation of Neu5Ac into the LOS in NTHi is an important mechanism for 117 protection against host adaptive and innate immune factors as well as bacterial biofilm formation 118 [51–53]. Incidentally, NTHi can acquire human dietary Neu5Gc to decorate its outer membrane, 119 and it was demonstrated to have evolved to preferentially incorporate Neu5Ac into its LOS [54]. 120 Similarly, the host-adapted pathogen Streptococcus pneumoniae also preferentially targets and 121 utilizes Neu5Ac over Neu5Gc [55]. This bias against Neu5Gc incorporation into LOS makes 122 evolutionary sense given that Neu5Gc is recognized as foreign in the human host and thus 123 would target the bacterial cell for the host’s immune response.
Beyond biosynthesis, select bacteria can catabolize sialic acid and use it as a nutrient for niche expansion [56–59]. The ability to catabolize Neu5Ac is widespread among commensal and pathogenic species, and this ability is an important phenotype for host-bacteria interactions [57–61]. This is exemplified by the pandemic pathogen Vibrio cholerae, which secretes a sialidase that cleaves sialic acids from host intestinal glycans, exposing the cholera toxin receptor to cause the diarrheal disease cholera [62]. V. cholerae catabolizes free sialic acid as a sole carbon source by uptake via a specialized transporter, and this catabolic ability is essential for optimal intestinal colonization [57,60,63].
Prokaryote-specific NulOs
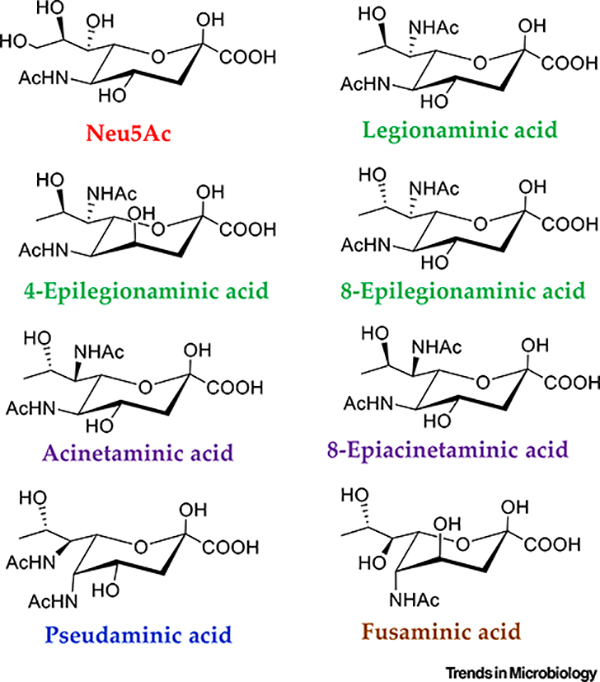
While sialic acid is present in eukaryotes and some prokaryotes, it is not the predominant NulO present on bacterial cell surfaces. This domain of life produces prokaryote-specific NulOs (Figure 3). These carbohydrates bear a strong resemblance to Neu5Ac but are derived from 5,7-diamino-3,5,7,9-tetradeoxynon-2-ulosonic acids (Figure 3). The first described prokaryotic NulO was discovered in the LPS of both Ps. aeruginosa and Shigella boydii, opportunistic and gastrointestinal pathogens respectively [64]. This NulO, named pseudaminic acid (Pse), is the L-glycero- L-manno isomer of the bacteria-specific NulOs and is present in various bacterial cell-surface structures, including the LPS and CPS, as well as glycosylated proteins of the surface layers (S-layers), flagellin, and pili (Figure 2, Table 1). The S-layer is a proteinaceous cell-envelope component, present in many bacteria, in which the proteins are frequently glycosylated [65,66]. The S-layer serves numerous functions in bacteria, including: protection, adhesion, resistance against abiotic stressors, and cell-membrane stabilization among others [65,66]. The 5,7-N-acetyl form of Pse appears to be the predominant molecule used for bacterial glycosylation of ace structures (Table 1), although several derivatives are known and utilized, which include 147 :amidino and acetylated forms, as well as a hydroxyproprionyl substituted form and a form 148 to which an N-acetylglutamine residue is attached (Table 1) [ 67,68].
Figure 3. Nonulosonic Acids (NulOs) Present in Bacteria.
The core structural isomers of NulOs commonly found in bacteria, including Neu5Ac and the prokaryotic NulOs. The variability of these structures occurs in the orientation of the core chiral carbons. Each of these carbohydrates is typically N-acetylated at position 5 for Neu5Ac or 5,7 for the prokaryotic NulOs, as depicted here. Other modifications, besides acetylation, have been identified.
Table 1.
Examples of Bacteria Experimentally Shown to Contain Pseudaminic Acid and Its Derivatives
| Pseudaminic acid | Structure | Functional groups | Refs |
|---|---|---|---|
| Vibrio vulnificus 27562 | LPSa | 5,7-N-acetyl | [95] |
| Escherichia coli O136 | LPS | 5,7-N-acetyl | [96] |
| Proteus vulgaris O39 | LPS | 5,7-N-acetyl | [97] |
| Piscirickettsia salmonis 70 | LPS | 5,7-N-acetyl | [98] |
| Pseudoalteromonas atlantica IAM 14165 | LPS | 5,7-N-acetyl | [99] |
| Enterobacter cloacae C5529 | LPS | 5,7-N-acetyl | [100] |
| Cellulophaga fucicola 68 | LPS | 5,7-N-acetyl | [101] |
| Campylobacter jejuni 11168 | Flagellin | 5,7-N-acetyl | [102] |
| Campylobacter coli VC167 | Flagellin | 5,7-N-acetyl | [103] |
| Campylobacter jejuni 81-176 | Flagellin | 5,7-N-acetyl | [82] |
| Helicobacter pylori 106 | Flagellin | 5,7-N-acetyl | [83] |
| Aeromonas caviae UU51 | Flagellin | 5,7-N-acetyl | [104] |
| Bacillus thuringiensis ATCC 35646 | Flagellin | 5,7-N-acetyl | [105] |
| Rhizobium sp. NGR234 | CPSb | 5,7-N-acetyl | [106] |
| Acinetobacter baumannii A74 | CPS | 5,7-N-acetyl | [107] |
| Kribbella spp. VKM | Cell wall | 7-3-hydroxybutyrate | [108] |
| Actinoplanes utahensis | Cell wall | 7-3-hydroxybutyrate | [109] |
| Pseudomonas aeruginosa O10a | LPS | 7-3-hydroxybutyrate | [64] |
| Shigella boydii type 7 | LPS | 7-3-hydroxybutyrate | [64] |
| Pseudomonas aeruginosa O9a, O9b | LPS | 7-3-hydroxybutyrate | [110] |
| Plesiomonas shigelloides O36 | LPS | 7-3-hydroxybutyrate | [111] |
| Pseudomonas chlororaphis UCM B-106 | LPS | 7-3-hydroxybutyrate | [112] |
| Escherichia coli O165 | LPS | 7-3-hydroxybutyrate | [113] |
| Acinetobacter baumannii B11911 | CPS | 7-3-hydroxybutyrate | [114] |
| Pseudomonas aeruginosa O7a, 7b, 7d, 7d | LPS | 7-formamido | [115] |
| Pseudoalteromonas distincta KMM 638 | LPS | 7-formamido | [116] |
| Vibrio cholerae O:2 | LPS | 5-acetamidino | [117] |
| Campylobacter jejuni 81-176 | Flagellin | 5-acetamidino | [82] |
| Campylobacter coli VC167 | Flagellin | 5-acetamidino | [103] |
| Sinorhizobium fredii HH103 | LPS | 5-acetamido-7-3-hydroxybutyramido | [118] |
| Vibrio vulnificus 27562 | LPS | 7-glycerate | [95] |
| Tannerella forsythia ATCC 43037 | S-layer | 5-Am-7-glycerate | [76] |
| Campylobacter jejuni 11168 | Flagellin | O-methylglyceric acid | [102] |
| Campylobacter coli VC167 | Flagellin | 5-7-2N-2,3-dihydroxypropionyl | [103] |
| Campylobacter jejuni 81-176 | Flagellin | 5-7-2N-2,3-dihydroxypropionyl | [82] |
| Campylobacter jejuni 81-176 | Flagellin | 5-Am-7Ac-8-N-acetylglutamine | [104] |
| Treponema denticola | Flagellin | 7-(2-methoxy-4,5,6-trihydroxy-hexanoyl) | [86] |
LPS, lipopolysaccharide.
CPS, capsule polysaccharide.
The d-glycero-d-galacto NulO isomer named legionaminic acid (Leg) was first identified in Legionella pneumophila, the causative agent of the respiratory infection Legionnaires’ disease (Figure 3) [69]. Similar to Pse, Leg is present in the LPS of various bacteria as well as in CPS and the flagellin protein of the flagellum (Figure 2, Table 2). Furthermore, the first example of a legionaminic biosynthesis pathway in Archaea was the recent demonstration of N-linked glycosylation of the S-layer from Halorubrum sp. PV6 [67]. Pse biosynthesis previously had been demonstrated in the archaean Methanobrevibacter smithii [16]. Several epimers of Leg have been characterized - 4-epi legionaminic acid and 8-epi legionaminic acid - which are much more limited in their distribution (Figure 3, Table 2). More recently, a novel isomer named acinetaminic acid was identified in the CPS of the multidrug-resistant pathogen Acinetobacter baumannii. ThisNulO is the 7,8-epimer of Leg taking on the l-glycero-l-altro confirmation (Figure 3, Table 3) [70]. In another A. baumannii strain, an 8-epiacinetaminic acid, the 7-epimer of Leg, was present 161 in the CPS (Figure 3, Table 3) [ 71]. A novel NulO, described as fusaminic acid, is present in the LPS of Fusobacterium nucleatum strain 25586, a bacterium that is associated with periodontitis. Fusaminic acid was proposed to have the l-glycero-l-gluco confirmation, but lacked the amino group of C-7; rather, this position is occupied with a hydroxyl group (Figure 3, Table 3) [ 72]. This was the first description of a 9-deoxynonulosonic acid with a hydroxyl group at C-7 [72].
Table 2.
Examples of Bacteria Experimentally Shown to Contain Legionaminic Acid and Its Derivatives
| Legionaminic acid | Structure | Functional groups | Refs |
|---|---|---|---|
| Acinetobacter baumannii O24 | CPSa | 5,7-N-acetyl | [119] |
| Campylobacter jejuni 11168 | Flagellin | 5,7-N-acetyl | [102] |
| Vibrio alginolyticus 945–80 | LPSb | 5,7-N-acetyl | [119] |
| Vibrio parahaemolyticus | O2 | LPS 5,7-N-acetyl | [120] |
| Enterobacter cloacae C6285 | LPS | 5,7-N-acetyl | [121] |
| Enterococcus faecium | CPS | 5,7-N-acetyl | [122] |
| Tannerella forsythia FDC92A2 | S-layer | 5,7-N-acetyl | [76] |
| Halorubrum sp. PV6 | S-layer | N-formylated | [67] |
| Campylobacter coli VC167 | Flagellin | 5-acetamidino | [123] |
| Campylobacter jejuni 11168 | Flagellin | 5-acetamidino | [102] |
| Legionella pneumophila serogroup 1 | LPS | 5-acetamidino | [69] |
| Pseudomonas fluorescens ATCC 49271 | LPS | 5-acetamidino | [119] |
| Vibrio salmonicida NCMB 2262 | LPS | 5-acetamidino | [119] |
| Acinetobacter baumannii O24 | CPS | 5-(3-hydroxybutyrate) | [119] |
| Campylobacter coli VC167 | Flagellin | 5-N-methylacetamidino | [123] |
| Campylobacter coli 11168 | Flagellin | 5-N-methylacetamidino | [102] |
| Clostridium botulinum | Flagellin | 5-N-methyl-glutam-4-yl | [124] |
| Vibrio parahaemolyticus KX-V212 | LPS | 7-N-acetyl-D-alanyl | [120] |
| Escherichia coli O161 | LPS | 7-D-alanyl | [125] |
| Vibrio vulnificus CMCP6 | LPS | 7-N-acetyl-D-alanyl | [80] |
| 4-epi-legionaminic acid | |||
| Legionella pneumophila serogroups 1, 3–12, 14 | LPS | 5,7-N-acetyl | [126,127] |
| Shewanella japonica KMM 3601 | LPS | 5,7-N-acetyl | [128] |
| Legionella pneumophila serogroup 2 | LPS | Acetamidino | [127] |
| 8-epi-legionaminic acid | |||
| Pseudomonas aeruginosa O12 | LPS | 5,7-N-acetyl | [110] |
| Providencia stuartii O20 | LPS | 5,7-N-acetyl | [129] |
| Escherichia coli O108 | LPS | 5,7-N-acetyl | [130] |
| Vibrio fischeri ES114 | LPS | 5,7-N-acetyl | [29] |
| Acinetobacter baumannii LAC-4 | LPS | 5,7-N-acetyl | [131] |
| Morganella morganii KF 1676 | LPS | 5-Acetamidino | [132] |
| Shewanella putrefaciens A6 | LPS | 7-Acetamidino | [133] |
| Salmonella arizonae O61 | LPS | 5-(3-hydroxybutyrate) | [134] |
| Yersinia ruckeri O1 | LPS | 5-(4-hydroxybutyramido) | [135] |
CPS, capsule polysaccharide.
LPS, lipopolysaccharide.
Table 3.
Examples of Bacteria Experimentally Shown to Contain Acinetaminic, 8-Epiacinetaminic, or Fusaminic Acid
| Acinetaminic acid | Structure | Functional groups | Refs |
|---|---|---|---|
| Acinetobacter baumannii D36 | CPSa | 5,7-N-acetyl | [70] |
| 8-epi-acinetaminic acid | |||
| Acinetobacter baumannii SGH 0703 | CPS | 5,7-N-acetyl | [71] |
| Fusaminic acid | |||
| Fusobacterium nucleatum ATCC25586 | LPSb | 5,7-N-acetyl | [72] |
CPS, capsule polysaccharide.
LPS, lipopolysaccharide.
Biosynthesis of Prokaryotic NulOs
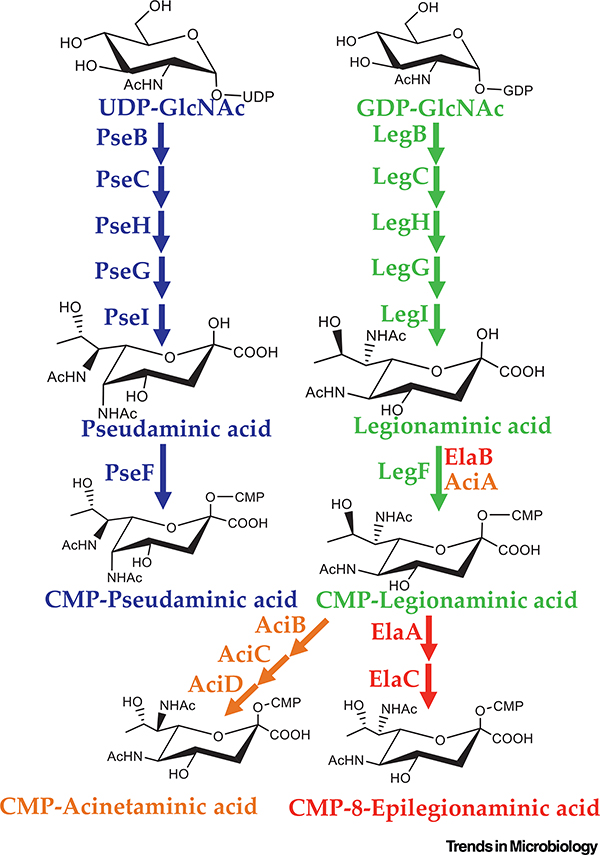
The biosynthetic pathways for both Leg and Pse acid are characterized fully in a number of bacterial species [73–76]. The complete pathway for Pse was first demonstrated in its entirety via chemienzymatic synthesis from H. pylori (Figure 4) [75]. The pathway initiates with a UDP-GlcNAc and requires six enzymes: PseB (a dehydratase/epimerase), PseC (an aminotransferase), PseH (an N-acetyltransferase), PseG (an NDP-sugar hydrolase removing UDP), Psel (which condenses the product of PseG with pyruvate to generate pseudaminic acid), and finally PseF (which activates Pse with a CMP moiety allowing for incorporation of the carbohydrate in downstream targets) (Figure 4) [68,77]. Not long after the characterization of the pseudaminic acid pathway, the total biosynthesis of legionaminic acid was described in its namesake bacterium L. pneumophila and in C. jejuni [73,74]. Interestingly, the biosynthesis of Leg in C. jejuni proceeds via a GDP-nucleotide linkage opposed to the more typical UDP carrier [74]. The authors speculated that utilizing multiple nucleotide carriers is a mechanism to separate similar biosynthetic pathways within a given cell. In addition to the difference in nucleotide carrier, there are multiple alternative epimerization reactions, accounting for the hallmark stereoisomers of Leg and Pse (Figure 4) [74].
Figure 4. Biosynthesis Pathways of Pseudaminic Acid (Pse) and Legionaminic Acid (Leg), and Predicted Pathways for 8-epilegionaminic Acid and Acinetaminic Acid.
The biosynthesis pathway for both Pse (blue) and Leg (green) acid requires six enzymes to form activated CMP-NulO. The generation of CMP-8-epilegionaminic acid (red) and CMP-acinetaminic acid (orange) was first predicted in Acinetobacter baumannii and has not yet been validated. CMP, cytidine 5’-monophosphate.
The more recent discovery of acinetaminic acid and 8-epiacinetaminic acid in A. baumannii has provided further insight into the alternative biosynthetic pathways of these unique carbohydrates [70,71]. Based on carbohydrate content and genetic analysis, pathways for both 8-epilegionaminic acid and acinetaminic acid were proposed (Figure 4) [70]. In both cases it is expected that biosynthesis proceeds through Leg prior to a series of dehydration/reduction reactions to change the stereochemistry (Figure 4). Specifically, ElaA is proposed to act as a C-8 dehydrogenase forming the keto group on CMP-legionaminic acid prior to the reduction proposed to be catalyzed by ElaC resulting in CMP-8-epilegionaminic acid (Figure 4). A biosynthetic route for 4-epilegionaminic acid has yet to be proposed; however, it is likely dependent on dehydrogenase/reductase activity, and the gene candidates are likely clustered with the other NulO biosynthesis genes. Similar to 8-epilegionaminic acid, the generation of CMP-acinetaminic acid is predicted to depend on the dehydrogenase/reductase activity of AciB, dehydrogenase activity of AciC, and reductase activity of AciD (Figure 4). It will be of interest to characterize these enzymes to fully assign their functions and role in the biosynthesis of 8-epilegionaminic acid and acinetaminic acid.
Following the identification of Aci, a second novel isomer, 8-epiAci, was characterized from the CPS from an additional strain of A. baumannii [71]. The authors characterized the NulO biosynthetic genomic region between the strains and found that a single gene was variable between 220 two [71]. Specifically, the gene aciB, annotated as a dehydrogenase and predicted to be involved in the formation of Aci, was replaced with aciE, a gene predicted to encode a nonorthologous dehydrogenase. It can be surmised that the formation 8-epiAci is via a similar pathway as predicted for Aci proceeding first through Leg and the epimerization is due to the action of AciE rather than AciB. Within A. baumannii, a wide array of NulOs have been identified, including Pse, Leg, 8-epiLeg, as well as the aforementioned Aci and 8-epiAci, which to date have only been identified within this species [68,70,71]. The array and prevalence of NulOs within the species suggests that this class of carbohydrates is an important feature for this clinical nosocomial pathogen.
Genomic and metabolic profiling of NulOs from members of the family Vibrionaceae showed that these carbohydrates were phylogenetically widespread, highly diverse, and that many species produced di-N-acetylated NulOs [78,79]. In addition, in the marine genera Vibrio, Aliivibrio, and Photobacterium, genes with homology to neuD are present within the NulO gene clusters, suggesting that O-acetylation is likely to be an important modification of these molecules. Within one species, Vibrio vulnificus, a deadly opportunistic pathogen of humans, several divergent NulO gene clusters were described [78,80]. These data suggested that NulOs have diverse roles in environmental persistence and/or virulence of these species. It was demonstrated that, in V. vulnificus, CMCP6, a novel modified Leg with an N-acetyl-d-alanyl modification at the C-7 position named 5-N-acetyl-7-N-acetyl-d-alanyl-legionaminic acid (Leg5Ac7AcAla), is produced [80]. Bioinformatics, genetic and functional analyses suggest that the Leg5Ac7AcAla biosynthesis pathway differs from the canonical Leg pathway (Figure 4). Many of the steps in Leg5Ac7AcAla biosynthesis are predicted to be carried out by homologs (LegB, LegC, LegG, LegI, and LegF) of the Leg biosynthesis proteins from previously described Leg pathways (Figure 4). However, several novel genes are present within the operon, named nab4, nab5, and nab6, suggesting key differences with the Leg pathway. The nab4 gene is predicted to encode an alanyl-transferase that adds an alanyl-moiety to the carbohydrate prior to epimerization by LegG. It is speculated that the sugar carrier UDP is hydrolyzed by the product of nab6, which, based on HHpred, contains a nucleotide-binding domain as well as a nucleotidyl-87 transferase domain [80]. The alanine-containing intermediates continue through the canonical Leg pathway, resulting in CMP-Leg5Ac7Ala [80]. In typical NulO biosynthesis pathways, activation by CMP is the final step prior to incorporation into downstream glycans. In V. vulnificus CMCP6, acetylation of the d-alanine group via Nab5, which contains an acetyltransferase (GNAT) domain and has been shown to be required for NulO biosynthesis in this strain, occurs following CMP activation [80]. Incidentally, it was also shown that V. vulnificus CMCP6 can catabolize modified Leg, and this is one of the few examples demonstrating bacterial catabolism of bacteria-specific NulOs [80]. Bioinformatics analysis demonstrated that the CMCP6 Leg5Ac7AcAla biosynthesis pathway is present in several Vibrio species as well as in other Gamma Proteobacteria [80]. Recent genomic and biochemical analyses within the families Vibrionaceae and Moritellaceae, which contain major fish pathogens, has identified novel NulO biosynthesis pathways, presenting the potential for additional structures to be uncovered. In addition, this same study identified a NulO pathway in Aliivibrio wodanis homologous to V. vulnificus CMCP6, and this strain is predicted to produce Leg5Ac7AlaAc [79].
Beyond the seven currently described isomers of bacterial NulOs, many modifications of these carbohydrates are identified in various glycan structures further adding to the diversity of these unique sugars (Tables 1 and 2). Much is to be learnt regarding these modifications and the enzymes required in the biosynthesis pathways. Nevertheless, as more NulOs are identified and paired with genomic sequence data, insights into these modifications are being revealed. As exemplified by the pathway described above for V. vulnificus, the modifying enzymes are often associated within the same genomic context as canonical NulO biosynthesis genes. Again, this is also the case for the proposed pathways for Aci and 8-epilegionaminic acid in A. baumannii isolates where the enzymes involved in the epimerization reactions are clustered with the canonical Leg genes [70].
Importance of Prokaryotic NulO Biosynthesis
The role of prokaryotic NulO biosynthesis has been elucidated in a number of species, predomi-nately human pathogens. In bacteria, NulOs decorate several different cell-surface structuresdepending on the species and/or strain examined, and they play a role in survival and colonization of host systems. The gastroenteritis-causing bacteria Aeromonas hydrophila, C. jejuni, and H. pylori, and the oral pathogen Treponema denticola heavily glycosylate their flagellin proteins 277 either, and in some cases both, Pse and Leg residues [81–84]. Deletion mutants in NulO biosynthetic pathways of these pathogens are nonmotile and avirulent [24,81,85]. Specifically, the absence of Pse5Am7Ac from C. jejuni 81–176 resulted in a mutant that was deficient in adherence and invasion of intestinal epithelial cells as well as attenuated in a diarrheal disease model [81]. Similar findings were demonstrated in the spirochete T. denticola, in which the inability to produce 2 unique Pse residue resulted in amotility and no detectable flagellin proteins [86]. In Aeromonas species, marine bacteria and human pathogens, the flagellum is also glycosylated with Pse 284 [84, Mutant Aeromonas strains that were unable to produce Pse were nonmotile; however, 285 ir caviae, a flagella glycosyltransferase mutant was hypermotile [84,87]. It was speculated that 286 glycosylation of flagellin with NulOs is part of a regulatory pathway to control motility [83,87]. 287 necessity of NulOs in the motility of these bacteria is critical because motility is often a required virulence factor in numerous bacterial pathogens [88].
There are limited studies that have determined whether the structurally similar Leg and Pse variants or other bacterial NulOs have the same ability to interact with host immune cells and cause immunosuppressive effects as sialic acid. In L. pneumophila serogroup 1, where Leg was first identified in the LPS as a homopolymer, the repeating Leg units aid in adherence to the alveolar macrophages in the lung and serve a sa virulence factor for this serogroup [89]. A recent study in the periodontitis-causing bacterium Tannerella forsythia demonstrated that there are strain-specific differences within the species in which some strains glycosylate the S-layer with a modified Leg and others with a modified Pse [76] (Tables 1 and 2). In T. forsythia, the S-layer was shown to aid in survival in the natural environment as well as serving as a virulence factor in infections by downregulating immune responses [90]. Researchers demonstrated that the absence of Leg or Pse residues in the glycosylated S-layer result in differential release of several cytokines and chemokines compared to the parental strains [91]. In particular, a Leg-deficient strain resulted in elevated release of MIP-1a and RANTES, suggesting a possible role in dampening the immune detection for T. forsythia strains 302 [91]. Interestingly, in T. forsythia strains which glycosylate their S-layers with either Leg or Pse 303 there were variable levels of biofilm formation [76]. It appears that NulO biosynthesis is an important 304 virulence determinant for T. forsythia, and all pathogenic strains have a NulO pathway whereas non-305 pathogenic strains do not [92]. A study in C. jejuni also demonstrated that Pse residues on the 306 flagella can interact with Siglec-10 on bone-marrow-derived dendritic cells, resulting in an anti-307 inflammatory response through the increased production of the cytokine IL-10 [93]. This is one of 308 the few demonstrations of a prokaryotic NulO interacting with a Siglec, suggesting the potential 309 for many more interactions that may be occurring during infections.
NulO biosynthesis gene clusters are widespread within species belonging to the family Vibrionaceae, and studies have demonstrated that a variety of di-N-acetylated NulOs are produced by multiple strains of Vibrio parahaemolyticus, Aliivibrio fischeri, and V. vulnificus among others [79,80,94]. The functional and physiological significance of NulO biosynthesis was inves-tigated in two clinical V. vulnificus strains, CMCP6 and YJ016, both predicted to contain Leg butwith very different NulO biosynthesis gene clusters [80,94]. BothCMCP6 and YJ016 have NulO in the LPS, but CMCP6 produces 100-fold more NulO than does YJ016 [94]. In strain CMCP6, the NulO operon consists of 11 genes, and in YJ016 it consists of 14 genes. The first two genes, legB and legC, in the operon in both strains are conserved and are involved in the first two steps of the biosynthesis pathway [80]. However, the rest of the NulO operons share less than 40% sequence homology and have different gene complements [80]. The legI gene, named nab2 in these strains, encodes a legionaminic acid synthase required for legionaminic acid synthesis through condensation of its precursor with PEP. In both V. vulnificus strains, when legI was deleted, these mutants did not produce NulO [94]. The mutants showed increased sensitivity to the antimicrobial peptide polymyxin B and were defective in biofilm formation and motility [80,94]. Utilizing a mouse bloodstream model of infection, the NulO-deficient strains were attenuated and were outcompeted in vivo compared to the wild-type parental strains [94]. To date, the biological significance of the more recently discovered NulOs (i.e., Aci, 8-epiAci, or Fus) (Table 3) has not been elucidated. It will be interesting to determine if the loss of these carbohydrates results in physiological and pathogenic defects, as is described for Leg, Pse, and Neu5Ac derivatives.
Concluding Remarks and Future Perspectives
Since their discovery over three decades ago, tremendous work has been accomplished to better understand and characterize prokaryotic NulOs. It is clear that the biosynthesis of these carbohydrates is widespread within the bacterial kingdom across highly diverse families, suggesting the potential for a broad significance of these carbohydrates in nature. Furthermore, there has been a wide diversity of prokaryote-specific NulOs identified to date. As of now, there are seven isomers within the family with countless further modifications. Because of the uniqueness and physiological significance of these carbohydrates, the biosynthetic pathways for canonical Leg and Pse were fully elucidated. The identification of key enzymes in the synthesis of prokaryotic NulOs allowed genetic manipulation, resulting in a better understanding of the physiological role of the NulOs. These carbohydrates are frequently identified as glycosylation motifs of bacterial flagellin, and S-layer proteins and modifications of LPS and CPS, where their presence is required for motility, biofilm formation, antimicrobial resistance, and in many cases, virulence.
The investigations involving prokaryotic NulO biosynthesis have uncovered many key findings; however, there is much left to be characterized regarding the significance of prokaryotic NulOs. While these carbohydrates were identified in diverse bacterial species along with numerous modifications, the biosynthesis pathways remain largely unknown outside of the canonical pathways of Leg and Pse. Bioinformatics analyses can reveal homologs of the essential genes of the pathways; however, there is little information regarding how the various modifications fit into currently known routes of biosynthesis. A concerted effort is needed to fully elucidate the biosynthetic pathways of these unique carbohydrates; fortunately, the addition of synthetic approaches holds promise in aiding these efforts [77].
Moreover, compared to Neu5Ac, the biological significance of prokaryotic NulOs is understood much less. It is clear that, when strains and species of bacteria lose the ability to produce prokaryotic NulOs, there are dramatic physiological defects. Despite these observed phenotypes, the exact mechanism of how the prokaryotic NulOs are involved has yet to be characterized. In addition, more work is needed to determine whether known sialic acid-recognizing proteins, such as Siglecs or components of the innate immune system, will also interact with prokaryotic NulOs. Beyond this, the predominance of modifications will alter the types of interaction that can occur with regard to recognition motifs. Ultimately, understanding the biosynthetic routes and characterizing the interactions between prokaryotic NulOs and the environment are integral steps in developing therapeutics that can target these key carbohydrates in numerous human and animal pathogens.
Highlights.
Nonulosonic acids (NulOs) are a diverse family of 9-carbon a-keto acid sugars that encompass the sialic acids (i.e., Neu5Ac) and prokaryote-specific NulOs.
In this review, we discuss bacterial biosynthesis of Neu5Ac, and its incorporation into cell-surface structures, as well as the scavenging of this carbohydrate. We review the importance of sialic acids in surface structures for host-pathogen interactions.
We discuss how bacteria have the ability to biosynthesize prokaryote-specific NulOs, describe the known isomers, and review the diversity of modification in structures. Wereviewthebiosynthesis pathways and examine the functions of bacteria-specific NulOs, such as their role in motility, biofilm formation, and host immune avoidance.
Outstanding Questions.
Many of the NulO gene clusters are embedded within CPS clusters. How are these diverse gene sequences acquired?
How does gene and protein sequence diversity translate into diversity of structure and modification?
Prokaryotic NulOs are very much understudied, and the role of NulOs in the natural environment, as well as in the host environment, requires more attention.
The distinct functions of NulO-modifying enzymes, and the significant role of NulOs in pathogens, makes the NulO biosynthetic proteins ideal targets for novel antimicrobial therapies.
Acknowledgments
This research was supported in part by a National Science Foundation grant (award IOS-1656688) to E.F.B. N.D.M. was funded by a University of Delaware graduate fellowship award and a Chemistry-Biology Interface predoctoral training pro-3 gram grant: 5T32GM008550. We thank four anonymous reviewers for their expert feedback and suggestions, it was greatly 3 appreciated.
References
- 1.Angata T and Varki A (2002) Chemical diversity in the sialic acids and related alpha-keto acids: an evolutionary perspective. Chem. Rev 102, 439–469 [DOI] [PubMed] [Google Scholar]
- 2.Varki A et al. (2017) Sialic acids and other nonulosonic acidsIn Essentials of Glycobiology (3rd edn) (Varki A et al. , eds), pp. 179–195, Cold Spring Harbor Laboratory Press [Google Scholar]
- 3.Varki A and Schauer R (2009) Sialic acids In Essentials of Glycobiology, Cold Spring Harbor Laboratory Press; [PubMed] [Google Scholar]
- 4.Tanner ME (2005) The enzymes of sialic acid biosynthesis. Bioorgan. Chem 33, 216–228 [DOI] [PubMed] [Google Scholar]
- 5.Blaum BS et al. (2015) Structural basis for sialic acid-mediated self-recognition by complement factor H. Nat. Chem. Biol 11, 77–82 [DOI] [PubMed] [Google Scholar]
- 6.Zhou JY et al. (2018) The glycoscience of immunity. Trends Immunol. 39, 523–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Laubli H and Varki A (2020) Sialic acid-binding immunoglobulin-like lectins (Siglecs) detect self-associated molecular patterns to regulate immune responses. Cell. Mol. Life Sci 77, 593–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crocker PR et al. (2007) Siglecs and their roles in the immune system. Nat. Rev. Immunol 7, 255–266 [DOI] [PubMed] [Google Scholar]
- 9.Pearce OM and Laubli H (2016) Sialic acids in cancer biology and immunity. Glycobiology 26, 111–128 [DOI] [PubMed] [Google Scholar]
- 10.Boligan KF et al. (2015) Cancer intelligence acquired (CIA): tumor glycosylation and sialylation codes dismantling antitumor defense. Cell. Mol. Life Sci 72, 1231–1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lewis AL and Lewis WG (2012) Host sialoglycans and bacterial sialidases: a mucosal perspective. Cell. Microbiol 14, 1174–1182 [DOI] [PubMed] [Google Scholar]
- 12.Severi E et al. (2007) Sialic acid utilization by bacterial pathogens. Microbiology 153, 2817–2822 [DOI] [PubMed] [Google Scholar]
- 13.Parsons NJ et al. (1988) Cytidine 5’-monophospho-N-acetyl neuraminic acid and a low molecular weight factor from human blood cells induce lipopolysaccharide alteration in gonococci when conferring resistance to killing by human serum. Microb. Pathog 5, 303–309 [DOI] [PubMed] [Google Scholar]
- 14.Wessels MR et al. (1987) Structure and immunochemistry of an oligosaccharide repeating unit of the capsular polysaccharide of type III group B Streptococcus. A revised structure for the type III group B streptococcal polysaccharide antigen. J. Biol. Chem 262, 8262–8267 [PubMed] [Google Scholar]
- 15.Lewis AL et al. (2016) Discovery and characterization of de novo sialic acid biosynthesis in the phylum Fusobacterium. Glycobiology 26, 1107–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lewis AL et al. (2009) Innovations in host and microbial sialic acid biosynthesis revealed by phylogenomic prediction of nonulosonic acid structure. Proc. Natl. Acad. Sci. U. S. A 106, 13552–13557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Inzana TJ et al. (2002) Incorporation of N-acetylneuraminic acid into Haemophilus somnus lipooligosaccharide (LOS): enhancement of resistance to serum and reduction of LOS antibody binding. Infect. Immun 70, 4870–4879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Inzana TJ et al. (2012) Decoration of Histophilus somni lipooligosaccharide with N-acetyl-5-neuraminic acid enhances bacterial binding of complement factor H and resistance to killing by serum and polymorphonuclear leukocytes. Vet. Microbiol 161,113–121 [DOI] [PubMed] [Google Scholar]
- 19.Kondadi PK et al. (2012) Identification and characterization of a lipopolysaccharide alpha,2,3-sialyltransferase from the human pathogen Helicobacter bizzozeronii. J. Bacteriol 194, 2540–2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murkin AS et al. (2004) Identification and mechanism of a bacterial hydrolyzing UDP-N-acetylglucosamine 2-epimerase. Biochemistry 43, 14290–14298 [DOI] [PubMed] [Google Scholar]
- 21.Gunawan J et al. (2005) Structural and mechanistic analysis of sialic acid synthase NeuB from Neisseria meningitidis in complex with Mn2+, phosphoenolpyruvate, and N-acetylmannosaminitol. J. Biol. Chem. 280, 3555–3563 [DOI] [PubMed] [Google Scholar]
- 22.Barry GT and Goebel WF (1957) Colominic acid, a substance of bacterial origin related to sialic acid. Nature 179, 206. [DOI] [PubMed] [Google Scholar]
- 23.Vimr E et al. (1995) Biosynthesis of the polysialic acid capsule in Escherichia coii K1. J. Ind. Microbiol 15, 352–360 [DOI] [PubMed] [Google Scholar]
- 24.Guerry P et al. (2000) Sialylation of lipooligosaccharide cores affects immunogenicity and serum resistance of Campylobacter jejuni. Infect. Immun 68, 6656–6662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mandrell RE et al. (1991) Endogenous sialylation of the lipooligosaccharides of Neisseria meningitidis. J. Bacteriol 173, 2823–2832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Linton D et al. (2000) Multiple N-acetyl neuraminic acid synthetase (neuB) genes in Campylobacter jejuni: identification and characterization of the gene involved in sialylation of lipo-oligosaccharide. Mol. Microbiol 35, 1120–1134 [DOI] [PubMed] [Google Scholar]
- 27.Knirel YA et al. (2006) Conserved and variable structural features in the lipopolysaccharide of Pseudomonas aeruginosa. J. Endotoxin Res 12, 324–336 [DOI] [PubMed] [Google Scholar]
- 28.Khatua B et al. (2010) Sialic acids acquired by Pseudomonasaeruginosa are involved in reduced complement deposition and siglec mediated host-cell recognition. FEBS Lett. 584, 555–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Post DM et al. (2012) O-antigen and core carbohydrate of Vibrio fischeri lipopolysaccharide: composition and analysis of their role in Euprymna scolopes light organ colonization. J. Biol. Chem 287, 8515–8530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Post DM et al. (2005) Identification of a novel sialic acid transporter in Haemophilus ducreyi. Infect. Immun 73, 6727–6735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Steenbergen SM et al. (2005) Sialic acid metabolism and systemic pasteurellosis. Infect. Immun 73, 1284–1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mandrell RE et al. (1992) Lipooligosaccharides (LOS) of some Haemophilus species mimic human glycosphingolipids, and some LOS are sialylated. Infect. Immun 60, 1322–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bouchet V et al. (2003) Host-derived sialic acid is incorporated into Haemophilus influenzae lipopolysaccharide and is a major virulence factor in experimental otitis media. Proc. Natl. Acad. Sci. U. S. A 100, 8898–8903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vann WF et al. (1987) Purification, properties, and genetic location of Escherichia coli cytidine 5’-monophosphate N-acetylneuraminic acid synthetase. J. Biol. Chem 262, 17556–17562 [PubMed] [Google Scholar]
- 35.Vann WF et al. (2004) The NeuC protein of Escherichia coli K1 is a UDP N-acetylglucosamine 2-epimerase. J. Bacteriol 186, 706–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Claus H et al. (2004) Genetics of capsule O-acetylation in serogroup C, W-135 and Y meningococci. Mol. Microbiol 51,227–239 [DOI] [PubMed] [Google Scholar]
- 37.Steenbergen SM et al. (2006) Separate pathways for O acetylation of polymeric and monomeric sialic acids and identification of sialyl O-acetyl esterase in Escherichia coli K1. J. Bacteriol 188, 6195–6206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deszo EL et al. (2005) Escherichia coli K1 polysialic acid O-acetyltransferase gene, neuO, and the mechanism of capsule form variation involving a mobile contingency locus. Proc. Natl. Acad. Sci. U. S. A 102, 5564–5569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.King MR et al. (2007) Escherichia coli K1-specific bacteriophage CUS-3 distribution and function in phase-variable capsular polysialic acid O acetylation. J. Bacteriol 189, 6447–6456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vimr ER et al. (2004) Diversity of microbial sialic acid metabolism. Microbiol. Mol. Biol. Rev 68, 132–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chang YC et al. (2014) Group B Streptococcus engages an inhibitory Siglec through sialic acid mimicry to blunt innate immune and inflammatory responses in vivo. PLoS Pathog. 10, e1003846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carlin AF et al. (2009) Molecular mimicry of host sialylated glycans allows a bacterial pathogen to engage neutrophil Siglec-9 and dampen the innate immune response. Blood 113,3333–3336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carlin AF et al. (2007) Group B streptococcal capsular sialic acids interact with siglecs (immunoglobulin-like lectins) on human leukocytes. J. Bacteriol 189,1231–1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Louwen R et al. (2008) The sialylated lipooligosaccharide outer core in Campylobacter jejuni is an important determinant for epithelial cell invasion. Infect. Immun 76, 4431–4438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Heikema AP et al. (2010) Characterization of the specific interaction between sialoadhesin and sialylated Campylobacter jejuni lipooligosaccharides. Infect. Immun 78, 3237–3246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vogel U et al. (1997) Complement factor C3 deposition and serum resistance in isogenic capsule and lipooligosaccharide sialic acid mutants of serogroup B Neisseria meningitidis. Infect. Immun 65, 4022–4029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jenkins GA et al. (2010) Sialic acid mediated transcriptional modulation of a highly conserved sialometabolism gene cluster in Haemophilus influenzae and its effect on virulence. BMC Microbiol. 10, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pawlak A et al. (2017) Salmonella O48 serum resistance is connected with the elongation of the lipopolysaccharide O-antigen containing sialic acid. Int. J. Mol. Sci 18, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Colicchio R et al. (2019) Virulence traits of a serogroup C meningococcus and isogenic cssA mutant, defective in surface-exposed sialic acid, in a murine model of meningitis. Infect. Immun 87, e00688–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Greiner LL et al. (2004) Nontypeable Haemophilus influenzae strain 2019 produces a biofilm containing N-acetylneuraminic acid that may mimic sialylated O-linked glycans. Infect. Immun 72, 4249–4260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tangvoranuntakul P et al. (2003) Human uptake and incorporation of an immunogenic nonhuman dietary sialic acid. Proc. Natl. Acad. Sci. U. S. A 100, 12045–12050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bergfeld AK et al. (2012) Metabolism of vertebrate amino sugars with N-glycolyl groups: elucidating the intracellular fate of the non-human sialic acid N-glycolylneuraminic acid. J. Biol. Chem 287, 28865–28881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Apicella MA (2012) Nontypeable Haemophilus influenzae: the role of N-acetyl-5-neuraminic acid in biology. Front. Cell. Infect. Microbiol 2, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ng PSK et al. (2019) Nontypeable Haemophilus influenzae has evolved preferential use of N-acetylneuraminic acid as a host adaptation. mBio 10, e00422–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hentrich K et al. (2016) Streptococcus pneumoniae senses a human-like sialic acid profile via the response regulator CiaR. Cell Host Microbe 20, 307–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Almagro-Moreno S and Boyd EF (2009) Sialic acid catabolism confers a competitive advantage to pathogenic Vibrio cholerae in the mouse intestine. Infect. Immun 77, 3807–3816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chowdhury N et al. (2012) The VC1777-VC1779 proteins are members of a sialic acid-specific subfamily of TRAP transporters (SiaPQM) and constitute the sole route of sialic acid uptake in the human pathogen Vibrio cholerae. Microbiology 158, 2158–2167 [DOI] [PubMed] [Google Scholar]
- 58.Lubin JB et al. (2012) Sialic acid catabolism and transport gene clusters are lineage specific in Vibrio vulnificus. Appl. Environ. Microbiol 78, 3407–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Haines-Menges BL et al. (2015) Host sialic acids: a delicacy for the pathogen with discerning taste. Microbiol. Spectr Published online July 2, 2015. 10.1128/microbiolspec.MBP-0005-2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McDonald ND et al. (2016) Host-derived sialic acids are an important nutrient source required for optimal bacterial fitness in vivo. mBio 7, e02237–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thomas GH (2016) Sialic acid acquisition in bacteria - one substrate, many transporters. Biochem. Soc. Trans 44, 760–765 [DOI] [PubMed] [Google Scholar]
- 62.Galen JE et al. (1992) Role of Vibrio cholerae neuraminidase in the function of cholera toxin. Infect. Immun 60, 406–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Almagro-Moreno S and Boyd EF (2009) Insights into the evolution of sialic acid catabolism among bacteria. BMC Evol. Biol 9, 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Knirel YA et al. (1984) Sialic acids of a new type from the lipo-polysaccharides of Pseudomonas aeruginosa and Shigella boydii. Carbohydr. Res 133, C5–C8 [DOI] [PubMed] [Google Scholar]
- 65.Fagan RP and Fairweather NF (2014) Biogenesis and functions of bacterial S-layers. Nat. Rev. Microbiol 12, 211–222 [DOI] [PubMed] [Google Scholar]
- 66.Messner P et al. (2008) S-layer nanoglycobiology of bacteria.Carbohydr. Res 343, 1934–1951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zaretsky M et al. (2018) Sialic acid-like sugars in archaea: legionaminic acid biosynthesis in the halophile Halorubrum sp. PV6. Front. Microbiol 9, 2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chidwick HS and Fascione MA (2020) Mechanistic and structural studies into the biosynthesis of the bacterial sugar pseudaminic acid (Pse5Ac7Ac). Organ. Biomolec. Chem 18, 799–809 [DOI] [PubMed] [Google Scholar]
- 69.Knirel YA et al. (1994) The structure of the O-specific chain of Legionella pneumophila serogroup 1 lipopolysaccharide. Eur. J. Biochem 221,239–245 [DOI] [PubMed] [Google Scholar]
- 70.Kenyon JJ et al. (2015) 5,7-di-N-acetyl-acinetaminic acid: A novel non-2-ulosonic acid found in the capsule of an Acinetobacter baumannii isolate. Glycobiology 25, 644–654 [DOI] [PubMed] [Google Scholar]
- 71.Kenyon JJ et al. (2017) 5,7-Di-N-acetyl-8-epiacinetaminic acid: A new non-2-ulosonic acid found in the K73 capsule produced by an Acinetobacter baumannii isolate from Singapore. Sci. Rep 7, 11357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vinogradov E et al. (2017) The structure of the LPS O-chain of Fusobacterium nucleatum strain 25586 containing two novel monosaccharides, 2-acetamido-2,6-dideoxy-l-altrose and a 5-acetimidoylamino-3,5,9-trideoxy-gluco-non-2-ulosonic acid. Carbohydr. Res 440–441,10–15 [DOI] [PubMed] [Google Scholar]
- 73.Glaze PA et al. (2008) Biosynthesis of CMP-N,N’-diacetyllegionaminic acid from UDP-N,N’-diacetylbacillosamine in Legionella pneumophila. Biochemistry 47, 3272–3282 [DOI] [PubMed] [Google Scholar]
- 74.Schoenhofen IC et al. (2009) The CMP-legionaminic acid pathway in Campylobacter: biosynthesis involving novel GDP-linked precursors. Glycobiology 19, 715–725 [DOI] [PubMed] [Google Scholar]
- 75.Schoenhofen IC et al. (2006) Elucidation of the CMP-pseudaminic acid pathway in Helicobacter pylori: synthesis from UDP-N-acetylglucosamine by a single enzymatic reaction. Glycobiology 16, 8c–14c [DOI] [PubMed] [Google Scholar]
- 76.Friedrich V et al. (2017) Tannerella forsythia strains display different cell-surface nonulosonic acids: biosynthetic pathway characterization and first insight into biological implications. Glycobiology 27, 342–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Flack EK et al. (2020) Synthetic approaches for accessing pseudaminic acid (Pse) bacterial glycans. ChemBioChem 21, 1397–1407 [DOI] [PubMed] [Google Scholar]
- 78.Lewis AL et al. (2011) Genomic and metabolic profiling of nonulosonic acids in Vibrionaceae reveal biochemical phenotypes of allelic divergence in Vibrio vulnificus. Appl. Environ. Microbiol 77, 5782–5793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Halsor M-JH et al. (2020) Sequence analysis of nonulosonic acid biosynthetic gene clusters in Vibrionaceae and Moritella viscosa. Sci. Rep 10, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.McDonald ND et al. (2018) Structural and functional characterization of a modified legionaminic acid involved in glycosylation of a bacterial lipopolysaccharide. J. Biol. Chem 293, 19113–19126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Guerry P et al. (2006) Changes in flagellin glycosylation affect Campylobacter autoagglutination and virulence. Moi. Microbioi 60, 299–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Thibault P et al. (2001) Identification of the carbohydrate moieties and glycosylation motifs in Campylobacter jejuni flagellin. J. Biol. Chem 276, 34862–34870 [DOI] [PubMed] [Google Scholar]
- 83.Schirm M et al. (2003) Structural, genetic and functional characterization of the flagellin glycosylation process in Helicobacter pylori. Mol. Microbiol 48, 1579–1592 [DOI] [PubMed] [Google Scholar]
- 84.Wilhelms M et al. (2012) Differential glycosylation of polar and lateral flagellins in Aeromonas hydrophiia AH-3. J. Biol. Chem 287, 27851–27862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Naito M et al. (2010) Effects of sequential Campylobacter jejuni 81–176 lipooligosaccharide core truncations on biofilm formation, stress survival, and pathogenesis. J. Bacteriol 192, 2182–2192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kurniyati K et al. (2017) A novel glycan modifies the flagellar filament proteins of the oral bacterium Treponema denticoia. Mol. Microbiol 103, 67–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lowry RC et al. (2015) The Aeromonas caviae AHA0618 gene modulates cell length and influences swimming and swarming motility. Microbioiogyopen 4, 220–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Josenhans C and Suerbaum S (2002) The role of motility as a virulence factor in bacteria. Int. J. Med. Microbioi 291, 605–614 [DOI] [PubMed] [Google Scholar]
- 89.Zahringer U et al. (1995) The lipopolysaccharide of Legionella pneumophila serogroup 1 (strain Philadelphia 1): chemical structure and biological significance. Prog. Clin. Biol. Res 392, 113–139 [PubMed] [Google Scholar]
- 90.Sekot G et al. (2011) Potential of the Tannerella forsythia S-layer to delay the immune response. J. Dent. Res 90,109–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bloch S et al. (2018) Immune response profiling of primary monocytes and oral keratinocytes to different Tannerella forsythia strains and their cell surface mutants. Mol Oral Microbiol 33, 155–167 [DOI] [PubMed] [Google Scholar]
- 92.Bloch S et al. (2019) Nonulosonic acids contribute to the pathogenicity of the oral bacterium Tannerella forsythia. Interface Focus 9, 20180064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Stephenson HN et al. (2014) Pseudaminic acid on Campyiobacter jejuni flagella modulates dendritic cell IL-10 expression via Siglec-10 receptor: a novel flagellin-host interaction. J. Infect. Dis 210, 1487–1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lubin JB et al. (2015) Host-like carbohydrates promote bloodstream survival of Vibrio vulnificus in vivo. Infect. Immun 83, 3126–3136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Vinogradov E et al. (2009) Structure of the lipopolysaccharide core of Vibrio vulnificus type strain 27562. Carbohydr. Res 344, 484–490 [DOI] [PubMed] [Google Scholar]
- 96.Staaf M et al. (1999) Structure determination of the O-antigenic polysaccharide from the enteroinvasive Escherichia coii O136. Eur. J. Biochem 263, 656–661 [DOI] [PubMed] [Google Scholar]
- 97.Kondakova AN et al. (2001) Structure of the acidic O-specific polysaccharide from Proteus vulgaris O39 containing 5,7-diacetamido-3,5,7,9-tetradeoxy-L-glycero-L-manno-non-2-ulosonic acid. Carbohydr. Res 333, 241–249 [DOI] [PubMed] [Google Scholar]
- 98.Vinogradov E et al. (2013) Chemical structure of the carbohydrate backbone of the lipopolysaccharide from Piscirickettsia salmonis. Carbohydr. Res 378, 108–113 [DOI] [PubMed] [Google Scholar]
- 99.Perepelov AV et al. (2005) Structure of an acidic polysaccharide from the agar-decomposing marine bacterium Pseudoalteromonas atlantica strain IAM 14165 containing 5,7-diacetamido-3,5,7,9-tetradeoxy-L-glycero-L-manno-non-2-ulosonic acid. Carbohydr. Res 340, 69–74 [DOI] [PubMed] [Google Scholar]
- 100.Han R et al. (2017) Structural and genetic characterization of the O-antigen of Enterobacter cloacae C5529 related to the O-antigen of E. cloacae G3054. Carbohydr. Res 443–444, 49–52 [DOI] [PubMed] [Google Scholar]
- 101.Perepelov AV et al. (2007) A pseudoaminic acid-containing O-specific polysaccharide from a marine bacterium Cellulophaga fucicola. Carbohydr. Res 342, 1378–1381 [DOI] [PubMed] [Google Scholar]
- 102.Logan SM et al. (2009) Identification of novel carbohydrate modifications on Campylobacter jejuni 11168 flagellin using metabolomics-based approaches. FEBS J. 276, 1014–1023 [DOI] [PubMed] [Google Scholar]
- 103.Logan SM et al. (2002) Structural heterogeneity of carbohydrate modifications affects serospecificity of Campylobacter flagellins. Mol. Microbiol 46, 587–597 [DOI] [PubMed] [Google Scholar]
- 104.Schirm M et al. (2005) Identification of unusual bacterial glycosylation by tandem mass spectrometry analyses of intact proteins. Anal. Chem 77, 7774–7782 [DOI] [PubMed] [Google Scholar]
- 105.Li Z et al. (2015) Pen and Pal are nucleotide-sugar dehydratases that convert UDP-GlcNAc to UDP-6-deoxy-D-GlcNAc-5,6-ene and then to UDP-4-keto-6-deoxy-L-AltNAc for CMP-pseudaminic acid synthesis in Bacillus thuringiensis. J. Biol. Chem 290, 691–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Le Quere AJ et al. (2006) Structural characterization of a Kantigen capsular polysaccharide essential for normal symbiotic infection in Rhizobium sp. NGR234: deletion of the rkpMNO locus prevents synthesis of 5,7-diacetamido-3,5,7,9-tetradeoxy-non-2-ulosonic acid. J. Biol. Chem 281, 28981–28992 [DOI] [PubMed] [Google Scholar]
- 107.Kenyon JJ et al. (2014) Structure of the K2 capsule associated with the KL2 gene cluster of Acinetobacter baumannii. Glycobiology 24, 554–563 [DOI] [PubMed] [Google Scholar]
- 108.Shashkov AS et al. (2009) New cell wall glycopolymers of the representatives of the genus Kribbella. Carbohydr. Res 344, 2255–2262 [DOI] [PubMed] [Google Scholar]
- 109.Shashkov AS et al. (2012) Teichulosonic acid, an anionic polymer of a new class from the cell wall of Actinoplanes utahensis VKM Ac-674iJ). Biochemistry (Mosc) 77, 511–517 [DOI] [PubMed] [Google Scholar]
- 110.Knirel YA et al. (1986) Somatic antigens of Pseudomonas aeruginosa. The structure of O-specific polysaccharide chains of P. aeruginosa O10 (Lanyi) lipopolysaccharides. Eur. J. Biochem 157,129–138 [DOI] [PubMed] [Google Scholar]
- 111.Kaszowska M et al. (2016) The O-antigen of Plesiomonas shigelloides serotype O36 containing pseudaminic acid. Carbohydr. Res 434, 1–5 [DOI] [PubMed] [Google Scholar]
- 112.Zdorovenko EL et al. (2016) Structure of the O-specific polysaccharides of Pseudomonas chlororaphis subsp. chlororaphis UCM B-106. Carbohydr. Res. 433,1–4 [DOI] [PubMed] [Google Scholar]
- 113.Senchenkova SN et al. (2015) Structure and gene cluster of the O-antigen of Escherichia coii O165 containing 5-N-acetyl-7-N-[(R)-3-hydroxybutanoyl]pseudaminic acid. Glycobiology 26, 335–342 [DOI] [PubMed] [Google Scholar]
- 114.Kasimova AA et al. (2017) Structure and gene cluster of the K93 capsular polysaccharide of Acinetobacter baumannii B11911 containing 5-N-Acetyl-7-N-[(R)-3-hydroxybutanoyl] pseudaminic acid. Biochemistry (Mosc) 82, 483–489 [DOI] [PubMed] [Google Scholar]
- 115.Knirel’ Iu A et al. (1987) Antigenic polysaccharides of bacteria. 21. The structure of O-specific polysaccharide chains and serological specificity of lipopolysaccharides from 7 Pseudomonas aeruginosa immunotypes. Bioorg. Khim 13, 88–96 [PubMed] [Google Scholar]
- 116.Muldoon J et al. (2001) Structure of an acidic polysaccharide from a marine bacterium Pseudoalteromonas distincta KMM 638 containing 5-acetamido-3,5,7,9-tetradeoxy-7-formamido-L-glycero-L-manno-nonulosonic acid. Carbohydr. Res 330, 231–239 [DOI] [PubMed] [Google Scholar]
- 117.Kenne L et al. (1988) Structural studies of the O-antigen from Vibrio cholerae O:2. Carbohydr. Res 180, 285–294 [DOI] [PubMed] [Google Scholar]
- 118.Di Lorenzo F et al. (2020) Structure of the unusual Sinorhizobium fredii HH103 lipopolysaccharide and its role in symbiosis. J. Biol. Chem 295, 10969–10987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Tsvetkov YE et al. (2001) Synthesis and identification in bacterial lipopolysaccharides of 5,7-diacetamido-3,5,7,9-tetradeoxy-D-glycero-D-galacto- and -D-glycero-D-talo-non-2-ulosonic acids. Carbohydr. Res 331, 233–237 [DOI] [PubMed] [Google Scholar]
- 120.Hashii N et al. (2003) Structural characterization of the carbohydrate backbone of the lipopolysaccharide of Vibrio parahaemoiyticus O-untypeable strain KX-V212 isolated from a patient. Carbohydr. Res 338 2711–279 [DOI] [PubMed] [Google Scholar]
- 121.Filatov AV et al. (2014) Structure and genetics of the O-antigen of Enterobacter cloacae C6285 containing di-N-acetyllegionaminic acid. Carbohydr. Res 392, 21–24 [DOI] [PubMed] [Google Scholar]
- 122.Kodali S et al. (2015) A vaccine approach for the prevention of infections by multidrug-resistant Enterococcus faecium. J. Biol. Chem 290, 19512–19526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.McNally DJ et al. (2007) Targeted metabolomics analysis of Campylobacter coli VC167 reveals legionaminic acid derivatives as novel flagellar glycans. J. Biol. Chem 282,14463–14475 [DOI] [PubMed] [Google Scholar]
- 124.Twine SM et al. (2008) Flagellar glycosylation in Clostridium botulinum. FEBS J. 275, 4428–4444 [DOI] [PubMed] [Google Scholar]
- 125.Li X et al. (2010) Structural and genetic characterization of the O-antigen of Escherichia coli O161 containing a derivative of a higher acidic diamino sugar, legionaminic acid. Carbohydr. Res 345,1581–1587 [DOI] [PubMed] [Google Scholar]
- 126.Knirel YA et al. (1996) Structure of a decasaccharide isolated by mild acid degradation and dephosphorylation of the lipopolysaccharide of Pseudomonas fluorescens strain ATCC 49271. Carbohydr. Res 283,129–139 [DOI] [PubMed] [Google Scholar]
- 127.Knirel YA et al. (2001) Identification of a homopolymer of 5-acetamidino-7-acetamido-3,5,7,9-tetradeoxy-D-glycero-D-talo-nonulosonic acid in the lipopolysaccharides of Legionella pneumophila Non-1 serogroups. Biochemistry (Mosc) 66, 1035–1041 [DOI] [PubMed] [Google Scholar]
- 128.Nazarenko EL et al. (2011) Structure of the O-Specific polysaccharide from Shewanella japonica KMM 3601 containing 5.7-diacetamido-3,5,7,9-tetradeoxy-D-glycero-D-talo-non-2-ulosonic acid. Biochemistry (Mosc) 76, 791–796 [DOI] [PubMed] [Google Scholar]
- 129.Shashkov AS et al. (2007) Structure of the O-antigen of Providencia stuartii O20, a new polysaccharide containing 5.7-diacetamido-3,5,7,9-tetradeoxy-l-glycero-d-galacto-non-2-ulosonic acid. Carbohydr. Res 342, 653–658 [DOI] [PubMed] [Google Scholar]
- 130.Perepelov AV et al. (2010) Structure of the O-antigen and characterization of the O-antigen gene cluster of Escherichia coli O108 containing 5,7-diacetamido-3,5,7,9-tetradeoxy-L-glycero-D-galacto-non-2-ulosonic (8-epilegionaminic) acid. Biochemistry (Mosc) 75, 19–24 [DOI] [PubMed] [Google Scholar]
- 131.Vinogradov E et al. (2014) The structure of the polysaccharide isolated from Acinetobacter baumannii strain LAC-4. Carbohydr. Res 390, 42–45 [DOI] [PubMed] [Google Scholar]
- 132.Kilcoyne M et al. (2002) Structural investigation of the O-specific polysaccharides of Morganella morganii consisting of two higher sugars. Carbohydr. Res 337, 1697–1702 [DOI] [PubMed] [Google Scholar]
- 133.Shashkov AS et al. (2002) Structure of the phenol-soluble polysaccharide from Shewanella putrefaciens strain A6. Carbohydr. Res 337, 1119–1127 [DOI] [PubMed] [Google Scholar]
- 134.Vinogradov EV et al. (1992) The structure of the O-specific polysaccharide chain of the lipopolysaccharide of Salmonella arizonae O61. Carbohydr. Res 231,1–11 [DOI] [PubMed] [Google Scholar]
- 135.Beynon LM et al. (1994) The structure of the lipopolysaccharide O antigen from Yersinia ruckeri serotype 01. Carbohydr.Res 256, 303–317 [DOI] [PubMed] [Google Scholar]