Summary:
CDK12 inactivation in prostate cancer is associated with tandem genomic duplications that may generate fusion-associated neoantigens and elicit immune responses amenable to checkpoint blockade. In the first study to comprehensively characterize the T-cell immune microenvironment of CDK12-deficient prostate cancers, subsets of immunosuppressive CD4+FOXP3- T-cells were increased compared to CDK12-proficient controls.
In this issue of Clinical Cancer Research, Rescigno and colleagues are the first to examine the immune tumor microenvironment (TME) in CDK12-mutated prostate cancers, providing key biological insights into the potential of immunotherapy in this molecular subset of tumors (1). To date, only deficiencies in DNA-damage response directly guide management of advanced prostate cancer patients. Prostate tumors with pathogenic mutations in genes essential for homologous-recombination (HR) repair (such as BRCA2) may benefit from poly(ADP-ribose) polymerase (PARP) inhibitors, leveraging a similar synthetic lethal interaction to that observed in ovarian and breast carcinoma. Given that alterations in HR occur in 20-30% of metastatic prostate cancers, this finding has generated excitement in the field. Alternatively, as seen in their colorectal and endometrial cancer counterparts, prostate cancers with mismatch repair (MMR) deficiency (2-5% of metastatic tumors) may benefit from immune checkpoint blockade due to increased tumor mutation burden with ensuing neoantigen generation and immune responsiveness.
Following these early successes, there have been intense efforts to discover additional molecular alterations that may sensitize prostate cancers to PARP or immune checkpoint inhibitors. Cyclin-dependent kinases (CDKs) comprise a family of serine/threonine kinases with two major subclasses, including the group that regulates the cell cycle, and another group of transcription-associated CDKs which includes CDK12. Transcription-associated CDKs are thought to regulate gene transcription via phosphorylation of RNA polymerase-II. Early preclinical work suggested that depletion of CDK12 resulted in decreased expression of genes involved in HR, conferring sensitivity to PARP inhibition (2). These findings were particularly interesting in light of the fact that CDK12 functions as a bona-fide tumor suppressor gene in ovarian and castration resistant prostate cancer (CRPC), where it is inactivated via somatic mutation in ~3-6% of cases. Though CDK12 mutations were initially reported as mutually exclusive with alterations in HR genes in both ovarian and prostate cancer, CDK12 inactivation was not associated with lower expression of HR genes, nor was it associated with ploidy changes or mutational signatures typical of HR deficiency in these tumor types (3). These surprising results – that CDK12 inactivation does not resemble HR deficiency at the genomic level – were further reinforced by the finding that prostate tumors with CDK12 mutations only respond modestly to PARP inhibition, although there are exceptions.
In contrast to HR-deficient tumors, CDK12-deficient ovarian and prostate tumors are associated with a unique pattern of genomic instability comprised of focal-tandem duplications (2,3). In prostate cancer, these tumors are typically associated with gain of a single duplicated DNA segment, consistent with aberrant DNA re-replication during S-phase. Intriguingly, this focal tandem duplicator phenotype leads CDK12-mutant tumors to harbor the highest gene-fusion burden of any other molecular subclass of prostate cancer, and fusion-induced neoantigens are abundant with a total predicted neoantigen burden approaching that of MMR-deficient prostate cancers. Consistent with this, CDK12-mutant tumors have gene expression profiles associated with higher levels of immune infiltration, with a greater abundance of T-cells and expanded T-cell clones as measured by T-cell receptor sequencing.
These initial molecular studies suggested the intriguing hypothesis that CDK12-deficient prostate tumors may be particularly sensitive to immune checkpoint blockade. However, two key pieces of data were missing: the clinical outcomes of CDK12-mutant tumors treated with immunotherapy, and a comprehensive investigation of the immune TME in these tumors. The former has been preliminarily addressed by recent retrospective studies confirming that a subset of CDK12-altered prostate cancers do respond favorably to PD-1 inhibitors (4), and additional clinical trials to formally address this question are underway (NCT03570619, NCT04104893). Importantly, the characteristics that distinguish tumors that respond from those that do not respond remain unknown, representing a critical area for future investigation.
To shed light on this question, Rescigno et al. performed a detailed investigation of the baseline immune TME in CDK12-mutated prostate cancers, correlating these findings with clinical and genomic analysis. In a retrospective study examining 913 patients with metastatic CRPC who underwent targeted DNA sequencing, the authors identified 43 patients (4.7%) with pathogenic alterations in CDK12, of which 31 were biallelic mutations expected to be associated with the tandem-duplicator phenotype. Clinically, biallelic CDK12-deficient tumors had worse survival compared to a control group of CDK12-proficient CRPC patients with available clinical and genomic data. However, this may have been driven by their higher-risk pathologic features, including higher Gleason scores, as CDK12 status was not significantly associated with clinical outcomes in multivariable analyses. In contrast to previous reports in prostate and ovarian cancers, the authors found that prostate tumors with biallelic CDK12 alterations frequently harbored other pathogenic mutations associated with HR or other DNA-repair pathways, including PALB2, BRCA2 and, most strikingly, ATM, which is lost by immunohistochemistry in ~25% of cases. This raises the question of whether CDK12 deficiency might synergize with other alterations to promote defective DNA repair in some contexts, and that a subset of these patients might be sensitive to PARP inhibitors.
Using a combination of immunohistochemistry and multispectral immunofluorescence, Rescigno et al found that while the intratumoral CD3+ T-cell density was numerically higher in biallelic CDK12-deficient cases compared to controls, this difference was not statistically significant, and there was no apparent overall enrichment in CD8+ cytotoxic T-cells. Unexpectedly, when the authors examined regulatory T-cell subsets, they found that CDK12-aberrant tumors were significantly enriched for potentially immunosuppressive CD4+FOXP3- T-cells, with a higher CD4/CD8 ratio than seen in controls. Higher CD4+FOXP3- T-cell densities were associated with worse survival in the overall cohort. These data are surprising in light of the fact that the high neoantigen burden observed in CDK12-deficient tumors is only slightly lower than that of MMR-deficient prostate cancers, and the latter have significantly increased tumor-associated CD8+ T-cells (5) and also generally respond favorably to anti–PD-1 therapies. However other conditions – such as androgen deprivation – may increase effector T-cells in prostate cancer also lead to proportional increases in immunosuppressive T-cell subsets, potentially contributing to immune evasion. Notably, Rescigno et al. report significant variability in T-cell densities across both CDK12-deficient and control cohorts, the source of which remains unclear. Indeed, there was no apparent correlation of TIL density with measures of genome fragmentation on whole-exome sequencing (i.e., copy-number breaks) which are associated with tandem duplications and which the authors found can vary among CDK12-altered tumors.
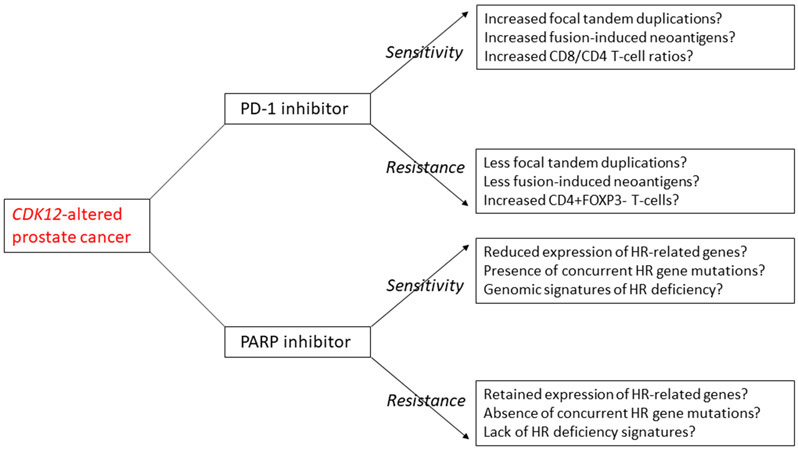
What are the clinical implications of this study? First, these data might explain why responses to anti–PD-1 therapies are generally modest in CDK12-altered mCRPC patients despite high neoantigen load (although responses can be profound in some), suggesting that alternative immunotherapeutic strategies will need to better address the immunosuppressive milieu in these cancers. Second, it seems plausible that the relative abundance of cytolytic versus suppressive TILs (e.g. CD8/CD4 ratio) will be important in predicting efficacy of immune checkpoint inhibitors, and for determining in whom treatment with anti–PD-1 monotherapy might need to be augmented. Third, future therapeutic strategies in CDK12-altered patients should aim to quantify fusion-associated neoantigens, functional HR deficiency, and TIL subpopulations in order to determine which subsets will respond best to PD-1 inhibitors versus PARP inhibitors versus the combination of both approaches (Figure). To date, tissue-based predictive biomarkers of immunotherapy responsiveness have remained elusive, consistent with the underlying complexity of the immune TME. Multiplex assays will likely be required, not only to determine the absolute density of the immune constituents (including both myeloid and lymphoid compartments), but also to discern the spatial relationships between cells in the TME. Finally, it is almost certain that the metastatic site and prior therapies are associated with significant changes in the immune TME, requiring bone lesions to be studied separately from prostatic primary or lymph-node metastases in future studies. Nevertheless, the current study provides an important proof-of-principle that molecular changes in a traditionally immunologically “cold” tumor such as prostate cancer may influence the immune response in ways that are not always predictable, but are certainly clinically relevant.
Figure.
Putative mechanisms of sensitivity or resistance to PD-1 inhibitor therapy or PARP inhibitor therapy among CDK12-deficient prostate cancer patients. Understanding the potential genomic, transcriptomic and immune microenvironment differences in CDK12-altered prostate cancers may help to guide treatment decisions for this subset of patients.
Acknowledgements:
This work was supported in part by the Patrick Walsh Prostate Cancer Research Fund (ESA, TLL), and the NCI Cancer Center Support Grant 5P30CA006973-52 (ESA, TLL).
Footnotes
Disclosure/Conflict of Interest: TLL has received research support from Roche/Ventana Medical Systems and DeepBio for other studies. ESA has served as a paid consultant/advisor for Janssen, Pfizer, Sanofi, Dendreon, Merck, Bristol-Myers Squibb, AstraZeneca, Clovis, Eli Lilly and Amgen; has received research funding to his institution from Janssen, Johnson & Johnson, Sanofi, Dendreon, Genentech, Novartis, Merck, Bristol-Myers Squibb, AstraZeneca and Constellation; and is a co-inventor of an AR-V7 biomarker technology that has been licensed to Qiagen.
References:
- 1.Rescigno P, Gurel B, Pereira R, Crespo M, Rekowski J, Rediti M, et al. Characterizing CDK12-mutated prostate cancers. Clin Cancer Res. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bajrami I, Frankum JR, Konde A, et al. Genome-wide profiling of genetic synthetic lethality identifies CDK12 as a novel determinant of PARP1/2 inhibitor sensitivity. Cancer Res. 2014;74:287–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Quigley DA, Dang HX, Zhao SG, Lloyd P, Aggarwal R, Alumkal JJ, et al. Genomic hallmarks and structural variation in metastatic prostate cancer. Cell 2018;174:758–69 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Antonarakis ES, Isaacsson Velho P, Fu W, Wang H, Agarwal N, Sacristan Santos V, et al. CDK12-altered prostate cancer: Clinical features and therapeutic outcomes to standard systemic therapies, poly (ADP-ribose) polymerase inhibitors, and PD-1 inhibitors. JCO Precis Oncol. 2020;4:370–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guedes LB, Antonarakis ES, Schweizer MT, Mirkheshti N, Almutairi F, Park JC, et al. MSH2 loss in primary prostate cancer. Clin Cancer Res. 2017;23:6863–74. [DOI] [PMC free article] [PubMed] [Google Scholar]