Abstract
Background and Aims:
High-resolution microendoscopy (HRME) is an optical biopsy technology that provides subcellular imaging of esophageal mucosa but requires expert interpretation of these histopathology-like images. We compared endoscopists with an automated software algorithm in esophageal squamous cell neoplasia (ESCN) detection and evaluated the endoscopists’ accuracy with and without input from the software algorithm.
Methods:
Thirteen endoscopists (6 experts, 7 novices) were trained and tested on 218 post-hoc HRME images from 130 consecutive patients undergoing ESCN screening/surveillance. The automated software algorithm interpreted all images as neoplastic (high-grade dysplasia, ESCN) or non-neoplastic. All endoscopists provided their interpretation (neoplastic or non-neoplastic) and confidence level (high or low) without and with knowledge of the software overlay highlighting abnormal nuclei and software interpretation. The criterion standard was histopathology consensus diagnosis by 2 pathologists.
Results:
The endoscopists had a higher mean sensitivity (84.3%, standard deviation [SD] 8.0% vs 76.3%, p=0.004), lower specificity (75.0%, SD 5.2% vs 85.3%, p<0.001) but no significant difference in accuracy (81.1%, SD 5.2% vs 79.4%, p=0.26) in ESCN detection compared with the automated software algorithm. With knowledge of the software algorithm, the endoscopists significantly increased their specificity (75.0% to 80.1%, p=0.002) but not sensitivity (84.3% to 84.8%, p=0.75) or accuracy (81.1% to 83.1%, p=0.13). The increase in specificity was among novices (p=0.008) but not experts (p=0.11).
Conclusions:
The software algorithm had lower sensitivity but higher specificity for ESCN detection than endoscopists. Using computer-assisted diagnosis, the endoscopists maintained high sensitivity while increasing their specificity and accuracy compared with their initial diagnosis. Automated HRME interpretation would facilitate widespread usage in resource-poor areas where this portable, low-cost technology is needed.
Keywords: artificial intelligence, computer-assisted diagnosis, high-resolution microendoscopy, esophageal squamous cell neoplasia, esophageal cancer
Introduction:
Esophageal squamous cell neoplasia (ESCN) is the sixth leading cause of cancer mortality worldwide [1]. Despite improvements in screening and treatment, most patients present with advanced disease, and the 5-year survival rates remain <15% [2]. Although the disease carries a significant burden in the United States, there is a markedly greater burden in the developing world. The cancer belt, which includes parts of Northern China and Iran, has an annual incidence of up to 183 per 100,000 [3], whereas the United States has an incidence rate of 2.1 per 100,000 [4]. The current criterion standard for ESCN screening is Lugol iodine chromoendoscopy (LCE) where abnormal areas appear unstained with application of Lugol iodine to the esophageal mucosa. Although LCE is the most sensitive method for ESCN detection (92%-98%), the specificity is quite low (37%-63%) [5–7]. Areas of benign inflammation also appear unstained, indistinguishable from cancerous lesions, leading to unnecessary biopsies, costs, and high false-positive rates (82%) [8].
High-resolution microendoscopy (HRME) is a low-cost (<$1500), portable, tablet-based advanced imaging technology that provides subcellular images of the esophageal mucosa, thus, providing an “optical biopsy” in real-time [9–11]. The addition of HRME to LCE increased the specificity up to 88% in clinical trials when used by experienced endoscopists for ESCN detection [8]. However, a significant limitation of widespread dissemination of HRME to low-resource settings is the availability of expert microendoscopists who can interpret these histopathology-like images in real-time.
Automated software algorithms have been used to assist physicians in interpretation of radiographic and endoscopic images. Computer-assisted algorithms have been used in still radiographic images and endoscopic videos for diagnosis of colon polyps, gastric cancer, and breast cancer [12–14]. In particular, for low-cost, portable technologies, designed for under-resourced or community-based settings, such as HRME, a computer-assisted diagnosis would help overcome issues of end user expertise, assuming the automated diagnosis was noninferior to expert endoscopists. We developed an automated HRME image analysis algorithm to discriminate between neoplastic and non-neoplastic tissue using nuclear metrics (ie, abnormal nuclear density) [15]. An automated software algorithm that interprets microendoscopic images in real-time would provide an immediate computer-assisted diagnosis without waiting for biopsy results, thus resulting in more accurate and selective biopsies, and facilitating immediate, minimally invasive endoscopic therapy (ie, resection, ablation).
The purpose of this study was to compare the accuracy of endoscopists (ie, experts and novices) with the automated software algorithm in ESCN detection using post-hoc HRME images. Additionally, we evaluated the accuracy and confidence level of endoscopists without and with input from the automated software algorithm to determine the effect of the software on endoscopist performance and confidence in real-time decision making (eg, decision to forego biopsy or decision for endoscopic therapy) using the computer-assisted diagnosis provided by the automated software.
Methods
HRME Imaging and Pathologic Correlation
Images were obtained sequentially from patients enrolled in an ongoing, clinical trial at three sites: First Hospital of Jilin University (Changchun, China), the Cancer Institute at The Chinese Academy of Medical Sciences (Beijing, China), and Baylor College of Medicine (Houston, Texas) from December 2014 to November 2016. All subjects underwent LCE followed by HRME of Lugol unstained areas. HRME captures high-resolution microscopic imaging of tissue at a subcellular level, described previously in detail [8]. We used still HRME images due to the comparable performance of still images and video clip in diagnostic performance [16]. The operator pauses the video feed using a foot pedal, visually assesses the captured still image for quality before saving the image. The high-resolution images are transmitted to a Galaxy tablet which displays the optical biopsy image (Figure 1). Each imaged site was biopsied and sent for histopathology. All biopsies were interpreted by 2 expert gastrointestinal pathologists. For disagreements, a consensus interpretation was reached. All pathologists were blinded to results of LCE and HRME. All HRME images and biopsy histopathology was interpreted using a binary classification of neoplastic versus non-neoplastic, consistent with previous studies [17, 18]. High-grade dysplasia and ESCN were classified as neoplastic; normal squamous epithelium, esophagitis and low-grade dysplasia were classified as non-neoplastic. The study was approved by the Institutional Review Boards at all 3 institutions, and all patients provided informed consent.
Figure 1.
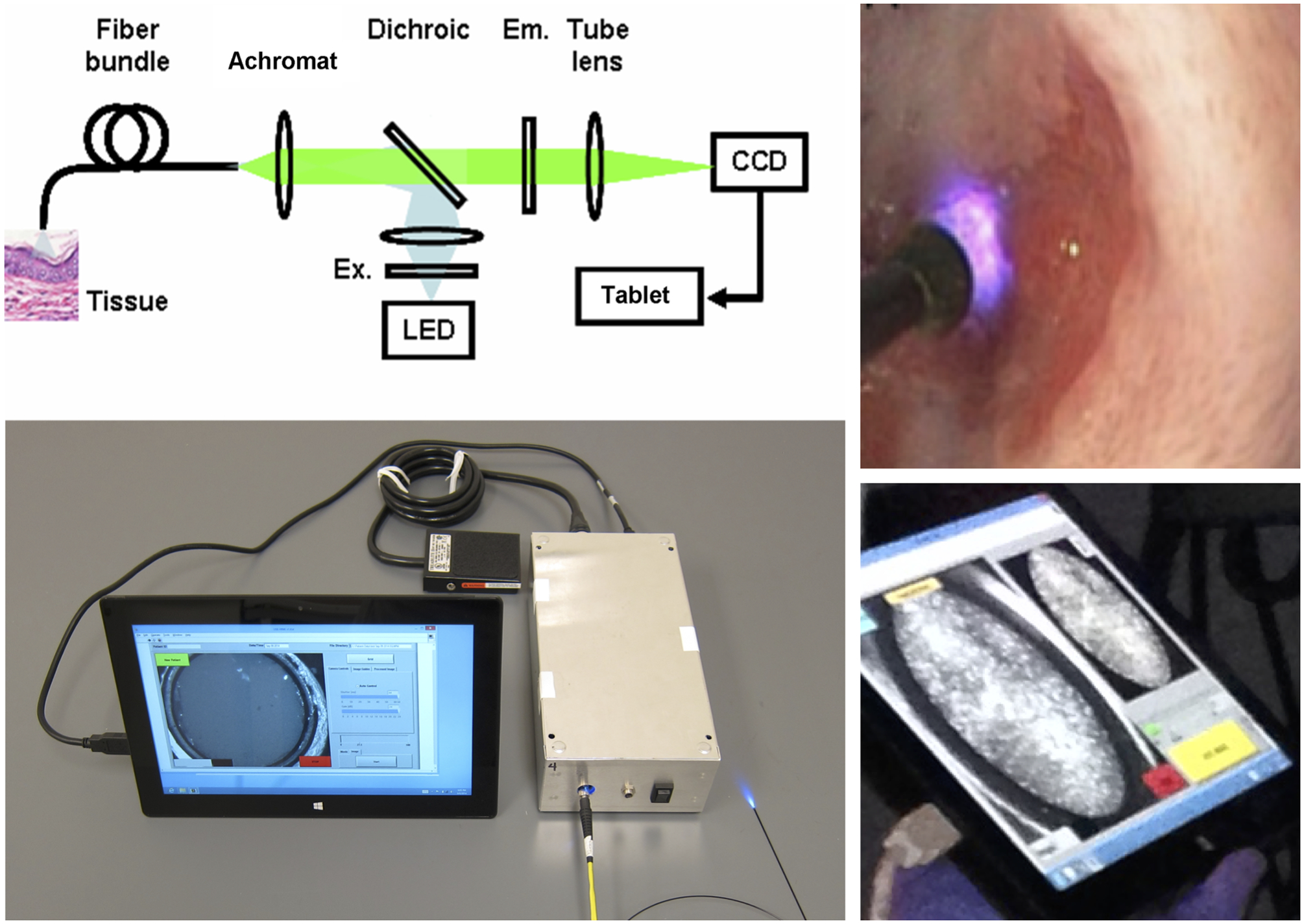
High-resolution microendoscopy (HRME) consists of a 1mm coherent fiber bundle inserted through the endoscope biopsy channel and placed in contact with esophageal tissue to acquire high-resolution images when used with a topical fluorescent agent. High-resolution images are transmitted to a Galaxy tablet, which displays the optical biopsy image.
Quality Control and Image Selection
HRME images obtained for each biopsy site were reviewed by a panel of expert microendoscopists and software development engineers who were blinded to both histopathology and automated software interpretation. All images underwent quality review and were only selected if >50% of the nuclei were visible and >50% of the image was clearly visible. The intent was to remove images with significant artifact (eg, blurring, mucus) making the image uninterpretable by both the software algorithm and the endoscopists. A final 218 high quality HRME images from 150 biopsy sites from 130 consecutively enrolled subjects were included.
Automated Software Algorithm for HRME Image Interpretation
An inbuilt automated software algorithm was developed to classify HRME images as neoplastic or non-neoplastic. Development of this image analysis algorithm has been described in previous work [15]. Briefly, the algorithm identifies a region of interest within the HRME field of view that excludes debris. All nuclei are then segmented from the background in the entire region of interest. We calculate morphometric criteria for all nuclei in order to classify normal and abnormal nuclei to discriminate between normal and neoplastic tissue [15, 19]. Previous iterations of the algorithm were semiautomated [20], and the first version of the fully automated algorithm classified images as normal or abnormal by calculating the percentage of abnormal nuclei in the field of view. In that study, abnormal nuclei were defined as nuclei with a nuclear area and eccentricity above set thresholds [15]. This initial algorithm was retrospectively evaluated with a training (104 biopsy sites), test (104 biopsy sites), and validation (167 biopsy sites) dataset obtained from 177 patients using the metric “% abnormal nuclei” [15]. We further refined the algorithm using abnormal nuclear density (ie, #abnormal nuclei/mm2), which uses the abnormal nuclei scaled relative to the physical area and better characterizes images with low total number of nuclei. We found an abnormal nuclear density cutoff of 218 corresponded to a sensitivity of 73% and specificity of 80% in predicting ESCN. We chose an abnormal nuclear density cutoff that prioritized specificity as HRME functions as a high-resolution optical technology used to image abnormal areas on LCE, which is highly sensitive but not specific. All 218 high-quality HRME images were analyzed post-hoc using the automated software algorithm which provided an image overlay highlighting abnormal nuclei and the raw abnormal nuclear density value. The images were classified binarily as neoplastic or non-neoplastic based on an abnormal nuclear density cutoff of 218.
Training of Endoscopists in HRME Interpretation
A total of 13 endoscopists (ie, 6 experts, 7 novices) underwent standardized training in HRME image interpretation. Expert endoscopists were defined as having previously performed >50 HRME cases, whereas novices were new to the technology. All endoscopists viewed a training slideshow that demonstrated the features of neoplastic and non-neoplastic classification of HRME images, including nuclear size, crowding, and pleomorphism. In addition, several examples of neoplastic (high-grade dysplasia, ESCN) and non-neoplastic HRME images (normal, inflammation, low-grade dysplasia) were shown in addition to potential confounders (eg, differentiating esophagitis from fragmented ESCN nuclei) and difficult images. Endoscopists were also familiarized with the testing procedure including how to interpret the automated software overlay and abnormal nuclear density value.
Testing of Endoscopists in HRME Interpretation
Of the 218 high-quality HRME images, 19 images were used in the training set and 199 images in the test set. All endoscopists were asked their initial read (ie, pre-software) of the HRME image as neoplastic or non-neoplastic along with their confidence level. The pre-software read was based on the HRME image alone without the software overlay or the algorithm diagnosis (Figure 2A). Confidence level (ie, high or low) was determined by the endoscopist’s confidence to act based on the HRME optical diagnosis: to not biopsy a non-neoplastic diagnosis and to ablate or resect immediately for a neoplastic diagnosis.
Figure 2.
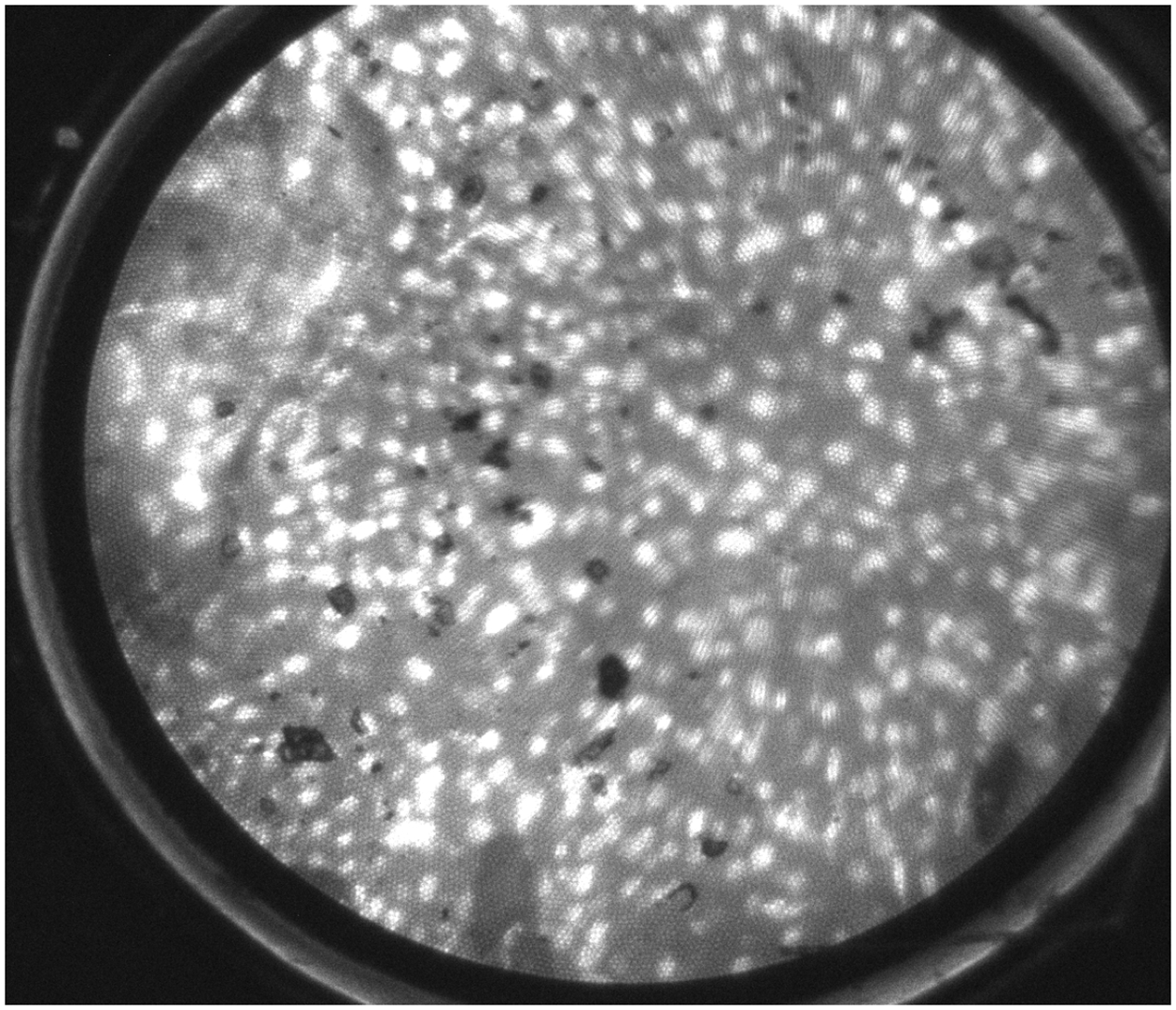
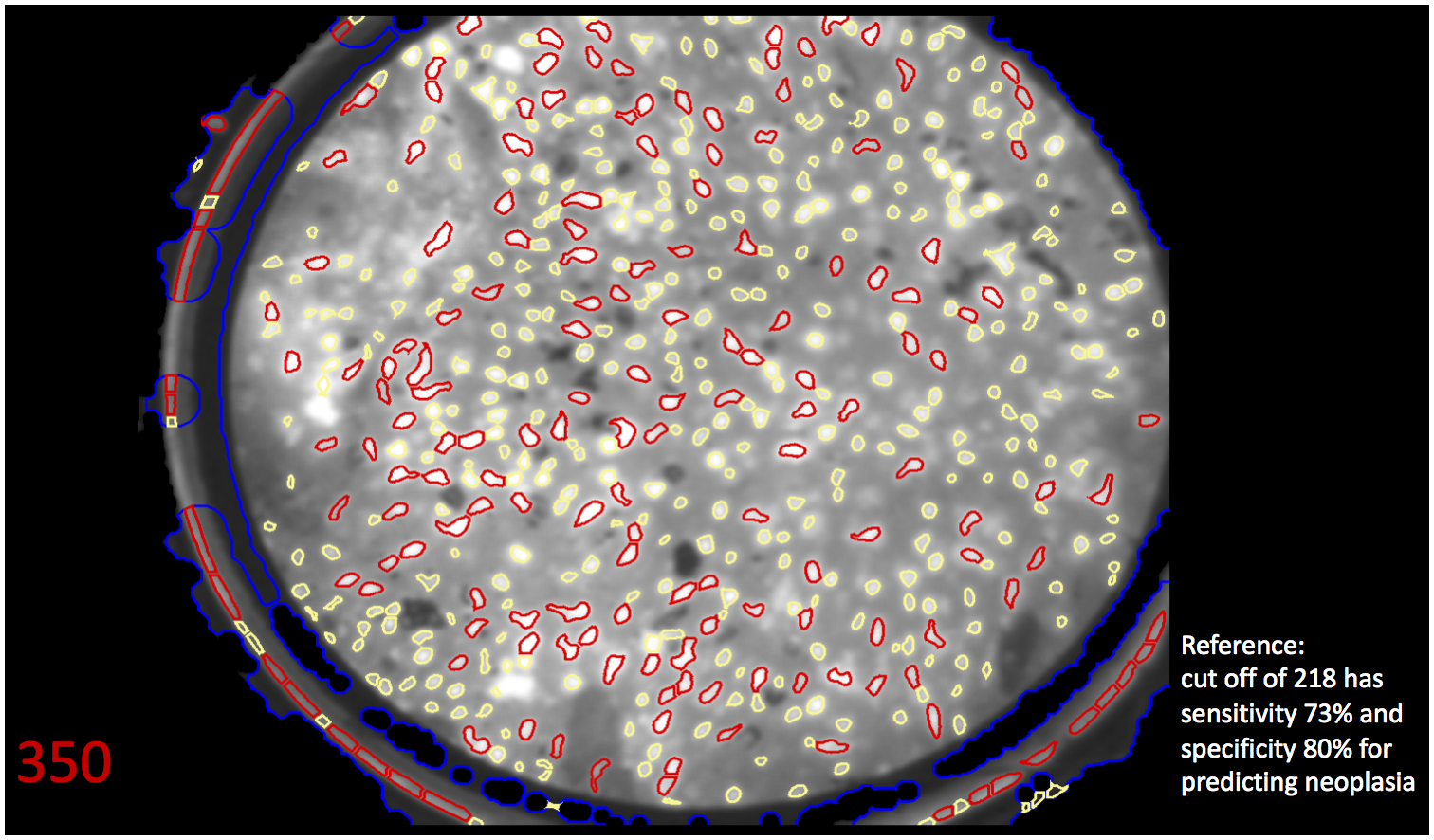
A, A high-resolution image obtained using high-resolution microendoscopy (HRME) of neoplastic esophageal mucosa, which demonstrates large, crowded, pleomorphic nuclei. Endoscopists were shown HRME images and asked their initial (pre-software) read. B, The same HRME image with the software overlay highlighting abnormal nuclei in red and abnormal nuclear density of 350. Based on abnormal nuclear density cutoff of 218, the software algorithm interpreted this image as neoplastic (red font). Endoscopists were shown this image along with reference algorithm performance and asked their second (post-software) read.
They were then shown the same image but with the automated software algorithm overlay outlining abnormal nuclei in red, abnormal nuclear density number, and the software algorithm’s interpretation as neoplastic (shown in red font) or non-neoplastic (shown in green font) (Figure 2B). Additionally, the sensitivity and specificity of the automated software algorithm for abnormal nuclear density cutoff of 218 (ie, 73% and 80%) were shown with every image as a reference. The endoscopist was provided both the image overlay to independently assess for neoplasia (subjective interpretation) and the algorithm diagnosis (objective interpretation) to make their post-software read.
At the end of testing, all endoscopists additionally answered a questionnaire to assess their attitudes toward the automated software algorithm. Responses were graded on a 5-point Likert scale ranging from Highly Agree (5) to Highly Disagree (1).
Statistical Analysis
Assuming 80% power, alpha=0.05, 2-sided test to establish equivalence between endoscopist and automated algorithm read with equivalence limit of 0.10, a sample size of 172 images was calculated using sample-based variance estimates without continuity correction for paired binary data [21].
The sensitivity, specificity, and accuracy of HRME image interpretation of individual endoscopists and the automated software algorithm was compared with histopathology as the criterion standard. Comparison of mean sensitivity, specificity, and accuracy along with standard deviation (SD) of all endoscopists and the automated software algorithm was determined using one-sample t-test. Differences in individual endoscopist’s pre-software and post-software sensitivity, specificity, and accuracy was compared using paired t-test. Diagnostic performance of endoscopists was stratified based on confidence level (ie, images reported with high confidence and low confidence). Inter-rater agreement between endoscopists was calculated using Cohen’s kappa (k) coefficient.
Statistical analyses were performed using Stata version 14 (StataCorp, College Station, Tex, USA). Statistical significance was determined at α = 0.05, and all tests for statistical significance were 2-sided.
Results:
Patient and Image Characteristics
We obtained 218 high quality HRME images from 130 consecutive patients consisting of 101 men (78.3%) and with mean age 59.6 years (SD 8.5 years). Of the 199 HRME images included in the testing set, 131 images (68.5%) were neoplastic (68 high-grade dysplasia, 63 ESCN) and 68 images (35.6%) were non-neoplastic (55 normal, 3 esophagitis, 10 low-grade dysplasia).
Endoscopists vs. Software HRME Image Interpretation
Overall, the 13 endoscopists had a mean pre-software sensitivity of 84.3% (SD 8.0%), specificity of 75.0% (SD 5.2%), and accuracy of 81.1% (SD 5.2%) for ESCN detection. The software had lower sensitivity (76.3%, p=0.004), higher specificity (85.3%, p<0.001), but no significant difference in accuracy (79.4%, p=0.26) compared with all endoscopists. The software algorithm had lower sensitivity than both experts (83.3%, p=0.16) and novices (85.1%, p=0.01) and higher specificity than both experts (76.7%, p=0.01) and novices (73.5%, p=0.001). However, the software algorithm was not significantly different in overall accuracy compared with experts (81.1%, p=0.54) and novices (81.1%, p=0.36) (Table 1). There was substantial congruence between endoscopists and software (k=0.63) and among the endoscopists (k=0.64).
Table 1.
Diagnostic performance of the software algorithm and endoscopists in diagnosing esophageal squamous cell neoplasia using high-resolution microendoscopy (HRME) images. Endoscopists provided an initial read (pre-software) and a second read (post-software) before and after knowledge of the software algorithm image overlay and diagnosis.
| Sensitivity | Specificity | Accuracy | |
|---|---|---|---|
| Software algorithm | 76.3% | 85.3% | 79.4% |
| All endoscopists pre-software | 84.3% (8.0%)^ | 75.0% (5.2%)^* | 81.1% (5.2%) |
| Experts pre-software | 83.3% (10.5%) | 76.7% (5.0%)^ | 81.1% (6.2%) |
| Novices pre-software | 85.1% (5.8%)^ | 73.5% (5.3%)^* | 81.1% (4.6%) |
| All endoscopists post-software | 84.8% (4.5%)^ | 80.1% (4.1 %)^* | 83.1% (2.1%)^ |
| Experts post-software | 84.4% (6.2%)^ | 79.7% (5.0%)^ | 82.7% (2.7%)^ |
| Novices post-software | 85.2% (2.9%)^ | 80.5% (3.5%)^* | 83.5% (1 .5%)^ |
p<0.05 for comparison of endoscopists and software.
p<0.05 for comparison of pre-software and post-software read among endoscopists.
Influence of Software Algorithm on the Endoscopists’ HRME Image Interpretation
After exposure to the image overlay and automated software interpretation, experts changed their read 9.2% of the time and novices 9.4%. On the post-software read, the endoscopists maintained their high sensitivity (84.8%, SD 4.5%; p<0.001 compared with the software algorithm) and overall accuracy (83.1%, SD 2.1%; p<0.001) but had lower specificity (80.1%, SD 4.1%; p=0.001) compared with the software algorithm. Compared with their pre-software read, the endoscopists significantly increased their specificity (change from pre- to post-software read p=0.002) but did not significantly change their sensitivity (p=0.75) or overall accuracy (p=0.13) (Figure 3). Although no significant difference in pre- and post-software read was seen among experts (sensitivity 83.3% to 84.4%, p=0.65; specificity 76.7% to 79.7%, p=0.11; accuracy 81.1% to 82.7%, p=0.36), a significant increase in specificity was seen among novices (sensitivity 85.1% to 85.2%, p=0.97; specificity 73.5% to 80.5%, p= 0.008; accuracy 81.1% to 83.5%, p=0.27) after knowledge of the automated software algorithm (Figure 4). Examining individual endoscopists, those with initial sensitivity better than the software algorithm tended to drop their sensitivity after knowledge of the software interpretation; whereas, those with sensitivity lower than the software algorithm tended to raise their sensitivity after knowledge of the software interpretation. Of 13 endoscopists, 12 improved their specificity after exposure to the automated software algorithm interpretation (Supplementary Figure 1).
Figure 3.
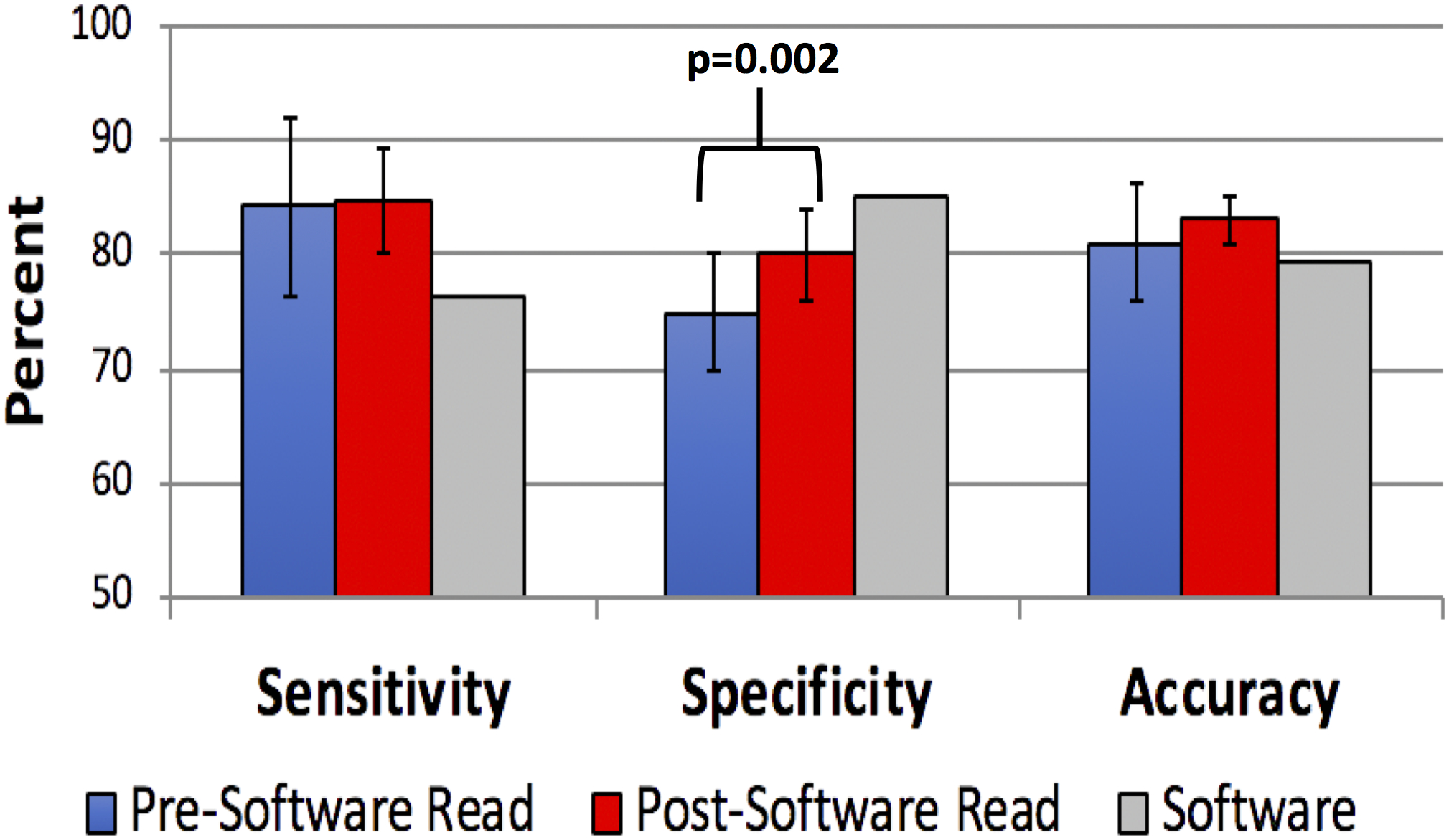
Accuracy of endoscopists’ high-resolution microendoscopy (HRME) image diagnosis before and after input from the software algorithm. Gray bars represent the software algorithm’s performance in relation to the endoscopists’.
Figure 4.
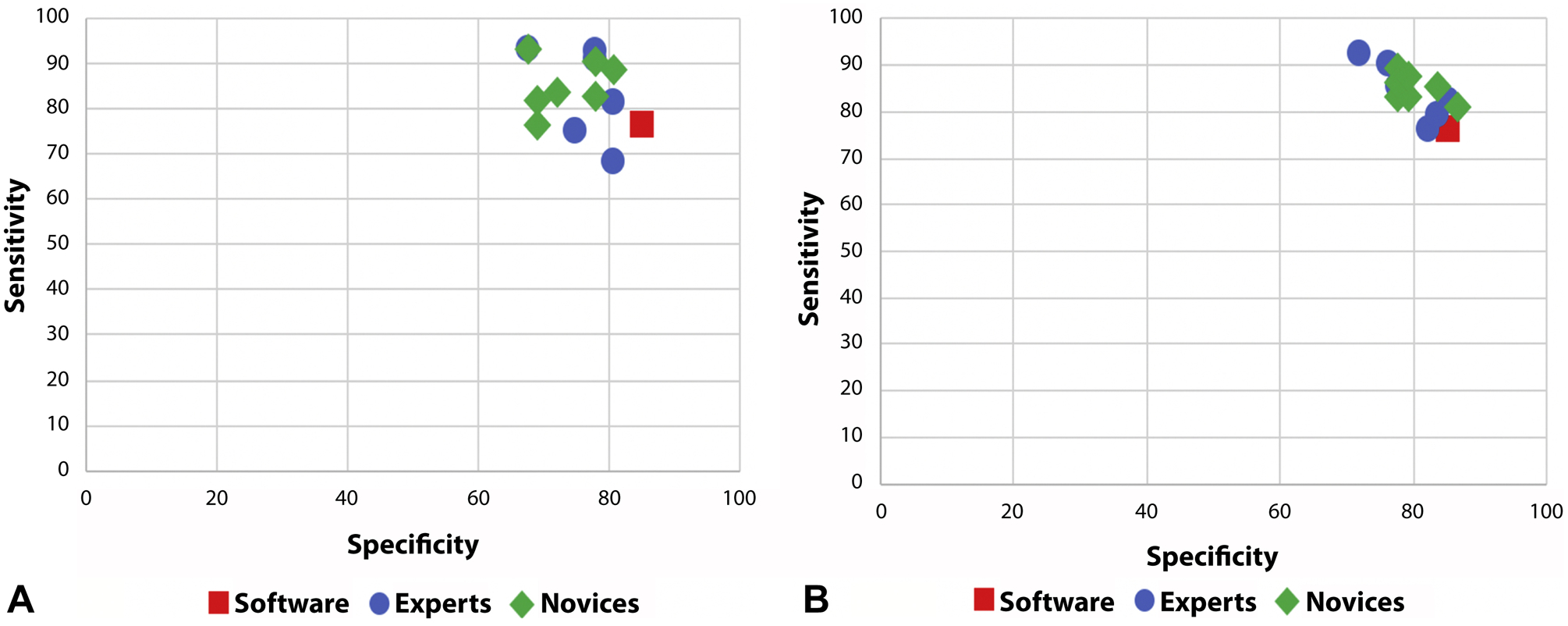
Sensitivity and specificity of high-resolution microendoscopy (HRME) image diagnosis among individual expert and novice endoscopists before (A) and after (B) input from the automated software algorithm.
Endoscopist Confidence in HRME Interpretation
Overall, experts had high confidence in mean 81.2% (SD 10.3%) of HRME images without assistance of the software algorithm, whereas novices had high confidence in 50.3% (SD 20.8%) of images. Individual endoscopist performance among high and low confidence images is shown in Supplementary Figure 2. Among high confidence images, expert endoscopists did not significantly change their sensitivity (86.3% to 86.7%, p=0.79) or specificity (82.7% to 82.7%, p=0.96) with input from the software algorithm. Conversely, novices did not significantly change their sensitivity (89.4% to 90.1%, p=0.77) but increased their specificity (81.1% to 83.4%, p=0.09) with input from the software algorithm.
For low confidence images, experts did not significantly change their sensitivity (75.9% to 70.6%; p=0.66) but trended toward an increase in specificity (49.0% to 64.4%; p=0.08) with assistance from the software algorithm. Similarly, among the novices there was not a difference in sensitivity (77.1% to 77.6%; p=0.90) but a significant increase in specificity (65.7% to 76.7%; p=0.03) with input from the software algorithm.
Endoscopist Acceptance of Software Image Interpretation
Overall, most endoscopists agreed that they trusted the automated software interpretation (84.6%), but 66.7% of experts tended to trust themselves more than the software, whereas novices showed more variation. The confidence level of the endoscopists increased when the software had the same read, whereas confidence level of the endoscopists decreased when the software had a different read. In general, endoscopists did not change their diagnosis if the software disagreed with their diagnosis in high confidence images but did change their diagnosis in low confidence images. All experts and 57.1% of novices stated they would use the software algorithm to aid in diagnosis when screening for ESCN (Supplementary Figure 3).
Conclusion:
The dissemination of any diagnostic technology into rural, underserved or community-based settings requires overcoming the need for expert interpretation. The application of computer-assisted diagnoses to portable diagnostic technologies offers the potential to increase access, reduce cost and infrastructure, and facilitate real-time decision making. In this post-hoc study, we found that a software algorithm, based on nuclear density and designed for microendoscopic evaluation of ESCN, was noninferior in overall accuracy to endoscopists, improved the specificity of novices, and increased the endoscopists’ confidence in their diagnosis. The software algorithm had lower sensitivity (76.3% vs 84.3%) but higher specificity (85.3% vs 75.0%) than the endoscopists, and when endoscopists were provided the software algorithm diagnosis, expert endoscopists did not significantly change their sensitivity (83.3% to 84.4%) or specificity (76.7% to 79.7%). However, novices tended to rely on the software algorithm and significantly increased their specificity (73.5% to 80.5%) while maintaining high sensitivity (85.1% to 85.2%). Additionally, for difficult (ie, low confidence) images, both experts and novices relied on the software algorithm, resulting in increased specificity.
Similar to our findings, computer-assisted diagnosis for cancer screening has shown to be either superior or equivalent to human interpretation alone. In a post-hoc study of colon polyps imaged using confocal laser endomicroscopy, an automated software classification had equivalent sensitivity and specificity as 2 expert endoscopists in neoplasia detection [18]. In other post-hoc studies of endoscopic and pathologic imaging technologies, deep learning algorithms outperformed endoscopists in predicting depth of gastric neoplasia invasion [13] and detecting micrometastases of breast cancers in digitized slides [14], respectively. When computer-assisted image analysis using convolutional neural networks were applied to colonoscopy videos, the convolutional neural networks identified an additional 20% of polyps that were not detected by expert endoscopists [12].
Furthermore, the greatest benefit of computer-assisted diagnosis may be assisting or training more junior or novice diagnosticians. Our study found that novice endoscopists could increase their accuracy to the level of experts with the assistance of the software algorithm. Similarly, computer-aided detection of breast cancers on mammography outperformed junior radiologists in sensitivity but did not significantly improve the accuracy of senior radiologists [22]. A computer-aided diagnosis system to evaluate endocytoscopy (ie, ultra-magnified) images of colon polyps had higher diagnostic accuracy than trainees but comparable accuracy with experts [17]. This trend persisted even for high-confidence images.
Currently, there are no diagnostic thresholds to direct advanced imaging technologies for ESCN detection. The American Society for Gastrointestinal Endoscopy has established Preservation and Incorporation of Valuable Endoscopic Innovations (PIVI) criteria for endoscopic technologies in Barrett’s esophagus imaging [23]. The PIVI thresholds of sensitivity 90% and specificity 80% must be met for an imaging technology to replace the current standard of care. If similar thresholds are applied in advanced imaging of ESCN, the addition of software diagnostic algorithms may bridge the gap to increase the diagnostic accuracy of clinicians in interpreting HRME images. Reliable diagnosis from HRME optical biopsy images in real-time would allow for more selective biopsies and immediate endoscopic treatment (ie, ablation or resection), resulting in less procedures, patients lost to follow-up, and cost.
For endoscopists to integrate computer-assisted diagnostic algorithms into routine practice, they need to trust and to understand limitations of the technology. Our study found that although endoscopists overall trusted the software algorithm, experts tended to trust their own diagnosis more than the software’s diagnosis. Thus, computer-assisted diagnosis in advanced imaging technologies is likely best used in 2 scenarios: (1) assisting expert endoscopists when interpreting difficult HRME images (ie, low confidence images); and (2) in training and assisting novice endoscopists in HRME interpretation. Furthermore, in low-resource countries without expert microendoscopists and with high rates of ESCN, an automated diagnosis provided by the software algorithm will provide real-time feedback and training in HRME until the novices can reach an expert level. This is the ultimate goal of the HRME technology whose cost (<$1500) and portability were designed for low-resource settings with less experienced clinicians.
Strengths of this study include HRME image acquisition from a clinical setting using consecutive patients undergoing ESCN screening and surveillance. We conducted standardized testing of the endoscopists (1) compared with the software algorithm, and (2) a second time with input from the software algorithm, allowing each endoscopist to serve as their own control. The endoscopists and the software algorithm were blinded to the criterion standard histopathology. All HRME images underwent quality control procedure, eliminating those with insufficient image quality. Additionally, we interviewed endoscopists about acceptance of computer-assisted diagnosis, which has not been widely reported.
Limitations of this post-hoc study include use of still post-hoc HRME images to test endoscopists and the software algorithm, which may not be representative of real-time HRME interpretation; all HRME images were obtained in real-time but our testing was done in a controlled setting without the obstacles encountered during real-time endoscopy. Each endoscopist rated the software overlay image immediately after the corresponding nonenhanced image, which may have introduced bias. However, the automated software algorithm is meant to be used in real-time by the endoscopist who would interpret the nonenhanced image followed immediately by the software overlay image to make the computer-assisted diagnosis. We included both the image overlay highlighting abnormal nuclei and the binary computer diagnosis (ie, software algorithm) in order to allow the endoscopist to interpret for themselves the degree and location of abnormal nuclei. Including the image overlay also allows the endoscopist to assess the quality of the algorithm’s diagnosis, especially in indeterminate or difficult images. We recognize that experts are more likely to use the overlay given their experience with HRME images and that novices are more likely to rely on the computer diagnosis. Additionally, our study was enriched with neoplastic images and was not representative of ESCN prevalence in the population. Because disease prevalence determines the pre-test probability of a clinician to have suspicion for a disease, our results may be best generalized to a population with high ESCN prevalence (eg, China, Iran, east Africa). We used linear discriminant analysis to choose an abnormal nuclear density cutoff that prioritized specificity as HRME is meant to be used as a high-resolution optical technology to image red-flag areas seen using LCE. More intensive deep learning approaches are currently being evaluated but was not done during the timeframe of this study.
The intent of the quality control image selection process was to only include interpretable HRME images without motion artifact or blurring, but this ‘image selection’ process may have introduced a bias favoring the software algorithm. However, previous studies evaluating computer-assisted diagnosis using advanced imaging technologies used similar methods for image selection (eg, narrow band imaging [24], optical coherence tomography [25], volumetric laser endomicroscopy [26]). Additionally, in any microendoscopic evaluation, a subjective diagnosis by a clinician would not be rendered using an image with significant artifact (eg, mucus, blurring). Thus, the bias presented by the image selection process was likely nondifferential between the software algorithm and endoscopists as poor image quality would have similarly affected interpretations by both. To address this potential source of bias, we are evaluating HRME interpretation prospectively in larger cohorts using real-time image quality assessments.
Our study found that the automated software algorithm’s performance was noninferior to expert microendoscopists but superior to novices in interpreting HRME images. Additionally, assistance from the software algorithm increased the specificity of both experts and novices when interpreting difficult images (ie, low confidence images). Computer-assisted diagnosis can be an asset in cancer detection and a validated software algorithm may obviate the need for an expert’s interpretation. Automated diagnostic ability would allow for wider dissemination of HRME to resource-poor areas where this portable, low-cost technology is most needed.
Supplementary Material
Supplementary Figure 1. Change in sensitivity (A) and specificity (B) of high-resolution microendoscopy (HRME) image diagnosis among individual expert (solid line) and novice (dotted line) endoscopists before and after input from the automated software algorithm. Sensitivity and specificity that increased and decreased after input from the software algorithm are shown in green and red, respectively.
Supplementary Figure 2. Sensitivity and specificity of experts and novices in high-confidence images before (A) and after (B) input from the automated software algorithm and in low-confidence images before (C) and after (D) input from the automated software algorithm.
Supplementary Figure 3. Survey results demonstrate endoscopist acceptance of the automated software algorithm in interpreting high-resolution microendoscopy (HRME) images.
Acknowledgments:
We would like to thank Tariq Hammad, Anam Khan, Sunina Nathoo, Rehman Sheikh, Hyunseok Kim, Jessica Bernica, Niharika Mallepally, and pathologists Sanford M. Dawsey and Daniel G. Rosen for their contribution to this work.
Funding Source: The research reported here was supported by grant NIH R01 CA181275-01 (SA).
Acronyms
- CI
confidence interval
- ESCN
esophageal squamous cell neoplasia
- HRME
high-resolution microendoscopy
- LCE
Lugol’s iodine chromoendoscopy
- PIVI
Preservation and Incorporation of Valuable Endoscopic Innovations
- SD
standard deviation
- U.S.
United States
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures: All authors have no disclosures relevant to this manuscript.
Writing assistance: No writing assistance was used in the preparation of this manuscript
References
- 1.Wang Q-L, Xie S-H, Wahlin K, et al. , Global time trends in the incidence of esophageal squamous cell carcinoma. Clin Epidemiol 2018:0:717–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Khushalani NI, Cancer of the Esophagus and Stomach. Mayo Clinic Proceedings, 2008. 83: p. 712–722. [PubMed] [Google Scholar]
- 3.Lambert R and Hainaut P, Esophageal cancer: cases and causes (part I). Endoscopy, 2007. 39: p. 550–5. [DOI] [PubMed] [Google Scholar]
- 4.Holmes RS and Vaughan TL, Epidemiology and pathogenesis of esophageal cancer. Semin Radiat Oncol, 2007. 17: p. 2–9. [DOI] [PubMed] [Google Scholar]
- 5.Inoue H, Rey J, and Lightdale CJE, Lugol chromoendoscopy for esophageal squamous cell cancer. Endoscopy, 2001. 33: p. 75–79. [PubMed] [Google Scholar]
- 6.Yoshimura N, Yoshida Y, Goda K, et al. , Assessment of novel endoscopic techniques for visualizing superficial esophageal squamous cell carcinoma: autofluorescence and narrow-band imaging. Diseases of the Esophagus, 2009. 22: p. 439–446. [DOI] [PubMed] [Google Scholar]
- 7.Morita FH, Bernardo WM, Ide E, et al. , Narrow band imaging versus lugol chromoendoscopy to diagnose squamous cell carcinoma of the esophagus: a systematic review and meta-analysis. BMC Cancer, 2017. 17: p. 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Protano M-A, Xu H, Wang G, et al. , Low-Cost High-Resolution Microendoscopy for the Detection of Esophageal Squamous Cell Neoplasia: An International Trial. Gastroenterology, 2015. 149: p. 321–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Louie JS, Richards-Kortum R, and Anandasabapathy S, Applications and advancements in the use of high-resolution microendoscopy for detection of gastrointestinal neoplasia. Clin Gastroenterol Hepatol, 2014. 12: p. 1789–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Quinn MK, Bubi TC, Pierce MC, et al. , High-resolution microendoscopy for the detection of cervical neoplasia in low-resource settings. PLoS One, 2012. 7: p. e44924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vila PM, Kingsley MJ, Polydorides AD, et al. , Accuracy and interrater reliability for the diagnosis of Barrett’s neoplasia among users of a novel, portable high-resolution microendoscope. Dis Esophagus, 2014. 27: p. 55–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Urban G, Tripathi P, Alkayali T, et al. , Deep Learning Localizes and Identifies Polyps in Real Time With 96% Accuracy in Screening Colonoscopy. Gastroenterology, 2018. 155: p. 1069–1078.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu Y, Wang Q-C, Xu M-D, et al. , Application of convolutional neural network in the diagnosis of the invasion depth of gastric cancer based on conventional endoscopy. Gastrointestinal Endoscopy, 2019. 89: p. 806–815.e1. [DOI] [PubMed] [Google Scholar]
- 14.Steiner DF, MacDonald R, Liu Y, et al. , Impact of Deep Learning Assistance on the Histopathologic Review of Lymph Nodes for Metastatic Breast Cancer. 2018. 42: p. 1636–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Quang T, Schwarz RA, Dawsey SM, et al. , A tablet-interfaced high-resolution microendoscope with automated image interpretation for real-time evaluation of esophageal squamous cell neoplasia. Gastrointestinal endoscopy, 2016. 84: p. 834–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ishijima A, Schwarz RA, Shin D, et al. , Automated frame selection process for high-resolution microendoscopy. J Biomed Opt, 2015. 20: p. 46014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Misawa M, Kudo S-e, Mori Y, et al. , Accuracy of computer-aided diagnosis based on narrow-band imaging endocytoscopy for diagnosing colorectal lesions: comparison with experts. International journal of computer assisted radiology and surgery, 2017. 12: p. 757–766. [DOI] [PubMed] [Google Scholar]
- 18.Andre B, Vercauteren T, Buchner AM, et al. , Software for automated classification of probe-based confocal laser endomicroscopy videos of colorectal polyps. World J Gastroenterol, 2012. 18: p. 5560–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grant BD, Fregnani JH, Possati Resende JC, et al. , High-resolution microendoscopy: a point-of-care diagnostic for cervical dysplasia in low-resource settings. Eur J Cancer Prev, 2017. 26: p. 63–70. [DOI] [PubMed] [Google Scholar]
- 20.Shin D, Protano MA, Polydorides AD, et al. , Quantitative analysis of high-resolution microendoscopic images for diagnosis of esophageal squamous cell carcinoma. Clin Gastroenterol Hepatol, 2015. 13: p. 272–279.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu JP, Hsueh HM, Hsieh E, et al. , Tests for equivalence or non-inferiority for paired binary data. Stat Med, 2002. 21: p. 231–45. [DOI] [PubMed] [Google Scholar]
- 22.Balleyguier C, Kinkel K, Fermanian J, et al. , Computer-aided detection (CAD) in mammography: does it help the junior or the senior radiologist? Eur J Radiol, 2005. 54: p. 90–6. [DOI] [PubMed] [Google Scholar]
- 23.Sharma P, Savides TJ, Canto MI, et al. , The American Society for Gastrointestinal Endoscopy PIVI (Preservation and Incorporation of Valuable Endoscopic Innovations) on imaging in Barrett’s Esophagus. Gastrointest Endosc, 2012. 76: p. 252–4. [DOI] [PubMed] [Google Scholar]
- 24.Gross S, Trautwein C, Behrens A, et al. , Computer-based classification of small colorectal polyps by using narrow-band imaging with optical magnification. Gastrointest Endosc, 2011. 74: p. 1354–9. [DOI] [PubMed] [Google Scholar]
- 25.Qi X, Sivak MV, Isenberg G, et al. , Computer-aided diagnosis of dysplasia in Barrett’s esophagus using endoscopic optical coherence tomography. J Biomed Opt, 2006. 11: p. 044010. [DOI] [PubMed] [Google Scholar]
- 26.Swager AF, van der Sommen F, Klomp SR, et al. , Computer-aided detection of early Barrett’s neoplasia using volumetric laser endomicroscopy. Gastrointest Endosc, 2017. 86: p. 839–846. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Change in sensitivity (A) and specificity (B) of high-resolution microendoscopy (HRME) image diagnosis among individual expert (solid line) and novice (dotted line) endoscopists before and after input from the automated software algorithm. Sensitivity and specificity that increased and decreased after input from the software algorithm are shown in green and red, respectively.
Supplementary Figure 2. Sensitivity and specificity of experts and novices in high-confidence images before (A) and after (B) input from the automated software algorithm and in low-confidence images before (C) and after (D) input from the automated software algorithm.
Supplementary Figure 3. Survey results demonstrate endoscopist acceptance of the automated software algorithm in interpreting high-resolution microendoscopy (HRME) images.


