Abstract
The TRIM-NHL family of proteins share a conserved domain architecture and play crucial roles in stem cell biology, fertility, and development. This review synthesizes new insights that have revolutionized our understanding of the molecular and biological functions of TRIM-NHL proteins. Multiple TRIM-NHLs have been shown to bind specific RNA sequences and structures. X-ray crystal structures of TRIM-NHL proteins in complex with RNA ligands reveal versatile modes of RNA recognition by the NHL domain. Functional and genetic analyses show that TRIM-NHL RNA-binding proteins negatively regulate the protein expression from the target mRNAs that they bind. This repressive activity plays a crucial role in controlling stem cell fate in the developing brain and differentiating germline. To highlight these paradigms, we focus on several of the most-extensively studied TRIM-NHL proteins, specifically Drosophila Brain tumor (Brat) and vertebrate TRIM71, among others. Brat is essential for development and regulates key target mRNAs to control differentiation of germline and neural stem cells. TRIM71 is also required for development and promotes stem cell proliferation while antagonizing differentiation. Moreover, TRIM71 can be utilized to help reprogram fibroblasts into induced pluripotent stem cells. Recently discovered mutations in TRIM71 cause the neurodevelopmental disease Congenital Hydrocephalus and emphasize the importance of its RNA-binding function in brain development. Further relevance of TRIM71 to disease pathogenesis comes from evidence linking it to several types of cancer, including liver and testicular cancer. Collectively, these advances demonstrate a primary role for TRIM-NHL proteins in the post-transcriptional regulation of gene expression in crucial biological processes.
Keywords: TRIM NHL, Sequence-specific RNA-binding proteins, mRNA regulation, RNA decay, translational control, stem cells, brain development, cancer, neurological disease
Graphical/Visual Abstract and Caption
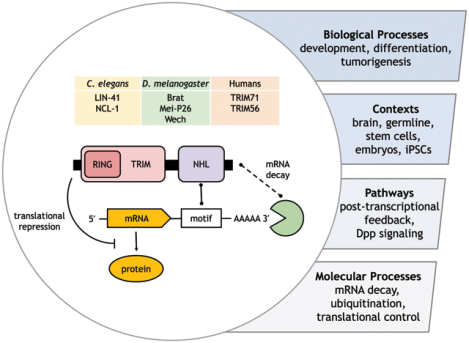
TRIM-NHL proteins were known to be involved in an array of biological processes in various contexts. This includes tuning signalling pathways and post-transcriptional gene regulation. The molecular underpinning of these activities, however, remained unknown until recent discoveries provided an answer: these proteins control gene expression by directly regulating mRNA decay and translation.
1. INTRODUCTION
TRIM-NHLs are animal-specific proteins with roles in neurogenesis, oogenesis, and embryonic development. In addition, TRIM-NHL proteins have less well-characterized roles in cell and cancer biology. In some cases, the loss of function phenotypes have been known for decades. For example, loss of Drosophila gene brain tumor (brat) causes overproliferation of larval neural stem cells (Arama et al., 2000; Betschinger et al., 2006; Kurzik-Dumke et al., 1992; Lee et al., 2006; Woodhouse et al., 1998). The molecular basis of these phenotypes, however, remained largely unknown. Recently, several TRIM-NHL proteins have been shown to bind specific RNA sequences and structures (Kumari et al., 2018; Laver, Li, et al., 2015; Loedige et al., 2013; Loedige et al., 2015; Loedige et al., 2014). This discovery has ushered in a flurry of new evidence on TRIM-NHL function and a re-evaluation of previous findings. Growing evidence indicates that several members of this protein family act as post-transcriptional repressors that reduce the stability and translation of specific mRNAs in diverse biological processes.
This review will summarize RNA-binding by TRIM-NHL proteins, and discuss the latest models of their molecular function as updated by this new information. In the first part, we focus on the molecular underpinning of NHL-RNA interactions and effects of RNA-binding on the transcriptome. The second part explores the specific biological functions of TRIM-NHL proteins that have been advanced by this discovery. Emphasis is on Drosophila Brat and human TRIM71 - and to a lesser extent Drosophila Mei-P26 and C. elegans LIN-41 - which are the most extensively characterized TRIM-NHLs. For more general overviews of the TRIM-NHL family or other RNA-binding TRIM proteins, we direct readers to recent reviews by Tocchini & Ciosk (2015) and Williams (2019). In the third part of this article, we discuss the remaining unanswered questions and future research directions.
2. GENERAL PRINCIPLES OF TRIM-NHL PROTEINS, RNA-BINDING, AND REGULATION
2.1. The TRIM-NHL family is a conserved subset of TRIM proteins
The tripartite motif (TRIM)-containing superfamily of proteins consist of an N-terminal TRIM motif appended to various C-terminal domains, by which they are routinely classified (Marin, 2012; Ozato et al., 2008; Sardiello et al., 2008; Short & Cox, 2006). The tripartite motif is defined by an ensemble of a RING-type zinc finger, one or two B-box type zinc fingers, and a coiled-coil domain (Reymond et al., 2001) (Figure 1A). RING domains often function as E3 ubiquitin ligases, while coiled-coil domains typically mediate protein-protein interactions; the function of the B-Box domains is unknown (reviewed by (Tocchini & Ciosk, 2015)). The presence and order of these domains are largely conserved throughout metazoans, suggesting the motif is a functional unit (Reymond et al., 2001).
Figure 1. Structure and phylogeny of TRIM-NHL proteins.
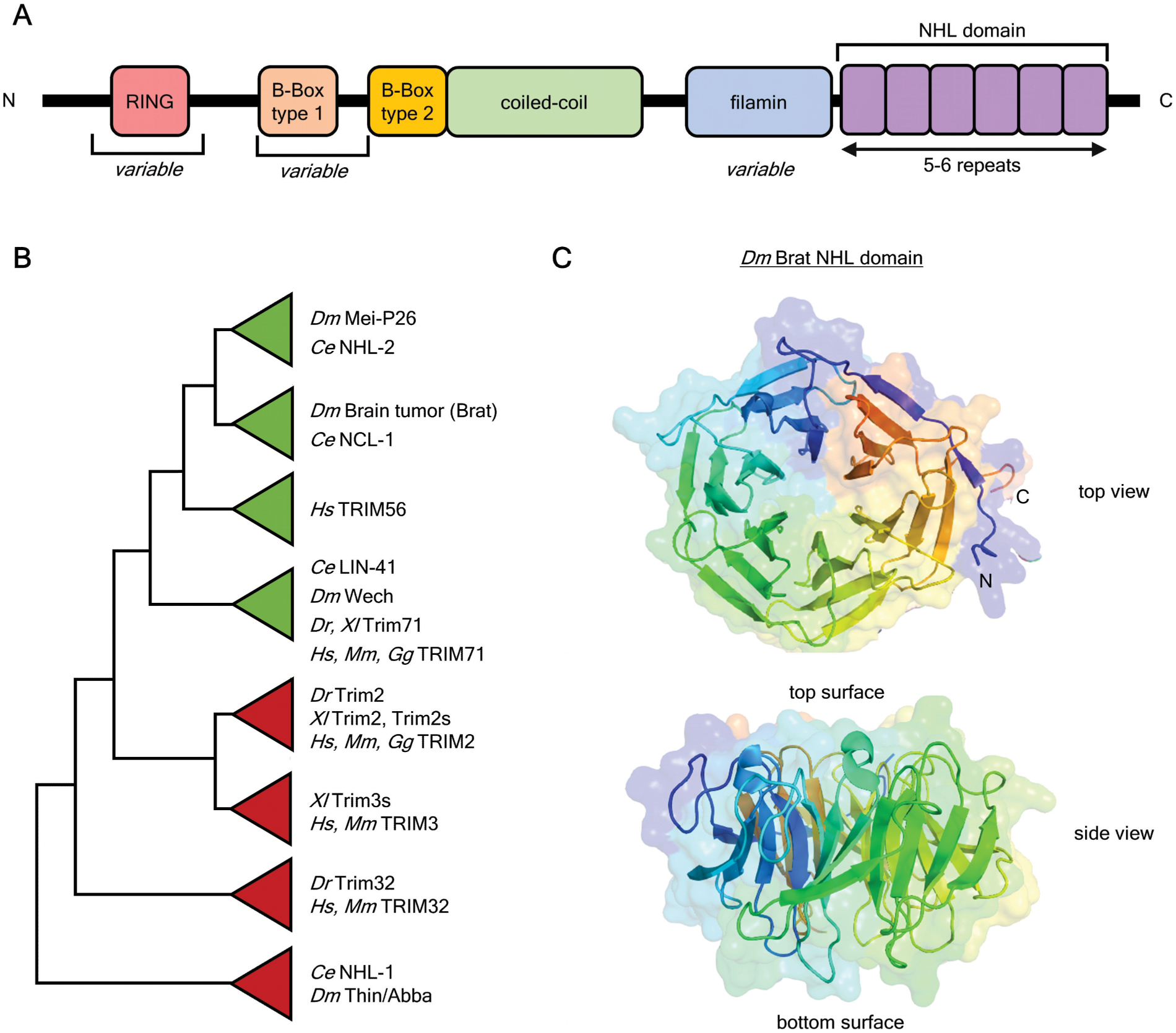
A. Architecture of a generalized TRIM-NHL protein and arrangement of conserved domains, including several that are variable within family members.
B. Cladogram of TRIM-NHL family, based on generated and published phylogenies. For generated phylogeny, all annotated full-length coding sequences of TRIM-NHL proteins were aligned using ClustalW, and Maximum Likelihood phylogeny prepared using MEGAX. Species: Drosophila melanogaster (Dm), Caenorhabditis elegans (Ce), Xenopus laevis (Xl), Danio rerio (Dr), Gallus gallus (Gg), Mus musculus (Mm), Homo sapiens (Hs)
C. X-ray crystal structure (PDB: 1Q7F) of the Brat NHL domain (Edwards et al., 2003). The coloration highlights the individual NHL repeats.
This review focuses on the subset of TRIM proteins that also contain a C-terminal NHL domain. Slack and Ruvkun (1998) defined the NHL repeats (named after founding proteins NCL-1, HT2A, LIN-41) that are appended C-terminally to TRIM domains in diverse species. Often these TRIM and NHL domains are accompanied by an intervening filamin-like domain. Furthermore, the presence of a type I B-Box domain and a functional RING domain varies between members of the TRIM-NHL family. The TRIM-NHL family can be subdivided into several clades (Figure 1B): i-ii) D. melanogaster Brat and Mei-P26 are paralogs, and their C. elegans counterparts are NCL-1 and NHL-2, respectively. These two clades are specific to invertebrates. iii) Human TRIM56, which has a poorly conserved NHL domain. iv) The LIN-41 clade consists of single homologs present in vertebrates and invertebrates (e.g. human TRIM71 and C. elegans LIN-41). v-vi) Human TRIM2 and TRIM3 clades, which have counterparts in other vertebrates. vii-viii) Human TRIM32, D. melanogaster Abba, and C. elegans NHL-1 are often grouped together, but the exact phylogenetic relationship is unclear.
Crystal structures of NHL domains, such as that of Brat (Figure 1C), show propellers of six blades, each consisting of approximately 44 residues arranged in four beta sheets (Edwards et al., 2003; Kumari et al., 2018; Loedige et al., 2015). In this regard, NHL domains are similar to other beta-propeller domains, such as WD40 repeats (Edwards et al., 2003; Slack & Ruvkun, 1998; Xu & Min, 2011). Other beta-propeller domains - such as WD40 domains - directly contact mRNAs, indicating an evolutionary precedent for RNA-binding (Loedige et al., 2015). While most propellers have six blades (e.g. Dm Brat, Dm Mei-P26, Hs TRIM71, Ce LIN-41), the number can vary - such as five (Dm Wech) or four (Hs TRIM56). Loops connecting the blades extend from the surface of the propellers, defining the “top” and “bottom” of the domain. These are the sites of protein and RNA interactions.
2.2. The NHL domain is a versatile RNA-binding platform
Early genetic analysis showed a clear role for TRIM-NHL proteins - and the NHL domain in particular - in development, differentiation, and stem cell biology. For example, the NHL domain of Brat plays a crucial role in vivo, as premature truncation codons or point mutations in this domain are lethal (Arama et al., 2000; Sonoda & Wharton, 2001). It remained unknown, however, how TRIM-NHL proteins functioned. Sonoda & Wharton (2001) showed that Brat repressed the translation of hunchback mRNA in the early embryo, as a component of an RNA-protein complex on the hunchback 3’ untranslated region (3’UTR). Subsequently, human TRIM-NHL proteins were discovered to crosslink to mRNAs (Baltz et al., 2012; Castello et al., 2012; Kwon et al., 2013). Loedige (2013) reported that human TRIM71 can reduce mRNA and protein expression levels of specific genes, and was targeted to those transcripts by the NHL domain. Shortly thereafter, the Brat NHL domain was shown to bind directly and specifically to RNA (Laver, Li, et al., 2015; Loedige et al., 2015; Loedige et al., 2014). This new paradigm of direct RNA-binding has since been expanded to include several other members of the TRIM-NHL family, though there are important differences (see below).
The Brat NHL domain forms a complex with single-stranded RNA with 1:1 stoichiometry and a dissociation constant (KD) of 100–150 nM (Loedige et al., 2014). RNA is broadly recognized by a niche across the NHL surface (Figure 2A) (Loedige et al., 2015). Specific nucleotides are discriminated by the amino acids lining the niche, causing preference for certain nucleotides at specific positions (Figure 2B). This preference defines the Brat consensus sequence (Figure 2C) (Laver, Li, et al., 2015; Loedige et al., 2015). This motif has been determined by several in vitro approaches and bioinformatics analysis of Brat-bound mRNAs (Laver, Li, et al., 2015; Loedige et al., 2015). The Brat binding site contains a core 5’-UGUU-3’ flanked with variable U/A-rich nucleotides. RNA structure appears to play little role in binding (Kumari et al., 2018), but can alter the accessibility of certain sequences (Loedige et al., 2015).
Figure 2. Structure of Drosophila melanogaster Brat NHL domain bound to RNA.
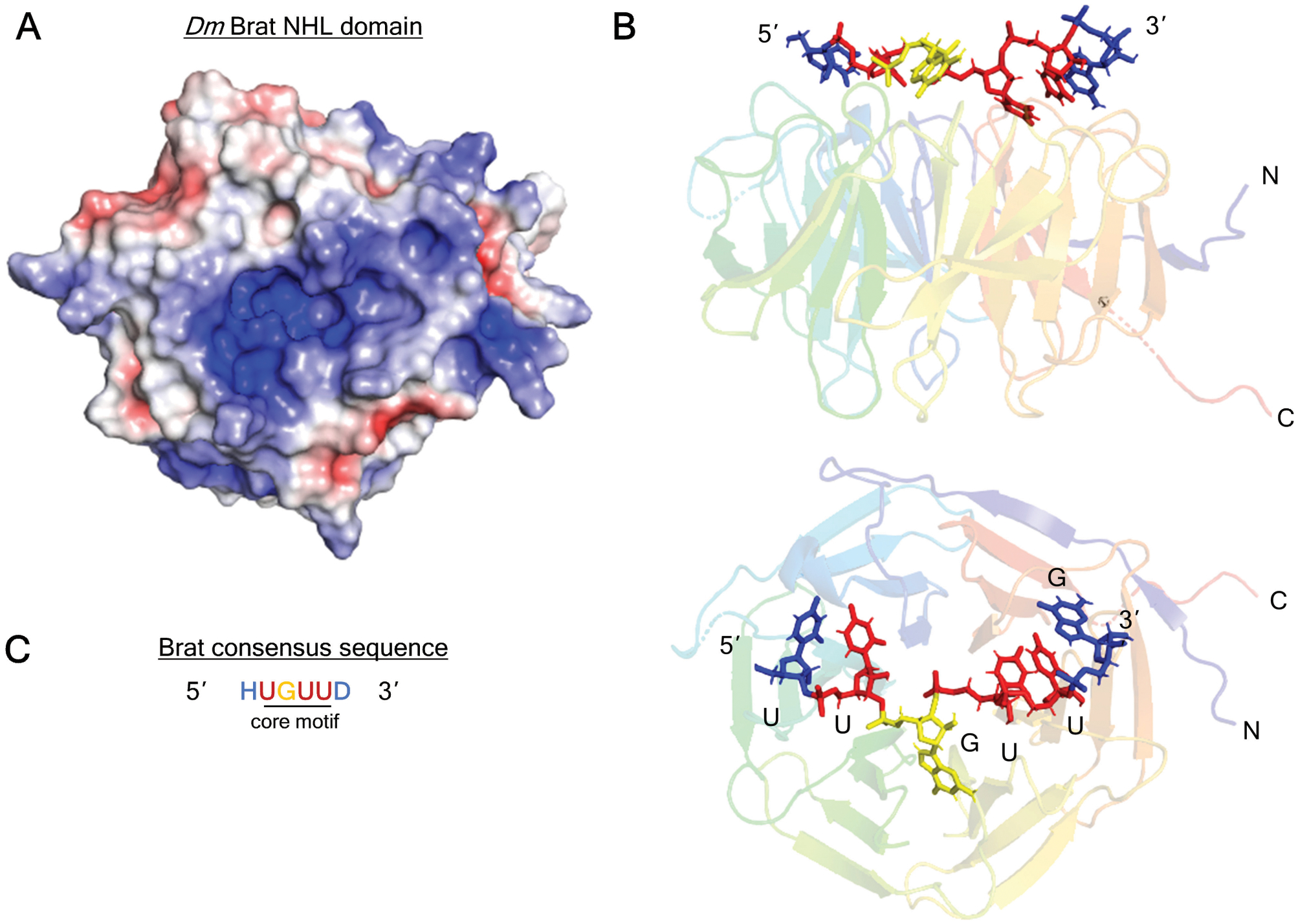
A. Positively-charged channel on the top, RNA-binding surface of Brat NHL domain (PDB: 1Q7F) (Edwards et al., 2003). The surface charge is visualized with Adaptive Poisson-Boltzmann Solver (APBS) macromolecular electrostatics calculation plugin in PyMOL. Blue denotes positive charge, red denotes negative charge.
B. Structure of the Brat NHL domain in complex with its RNA binding site from the hunchback mRNA 3’UTR (PDB: 4ZLR) (Loedige et al., 2015). The linear chain of nucleotides contact side chains of loop residues on the NHL surface.
C. Brat (and NCL-1) consensus binding motif consists of a core 5’-UGUU, with variable flanking nucleotides: H = (A/C/U), D = (A/G/U). The Brat binding motif consensus is derived from Loedige (2015) and Laver (2015).
In contrast to Brat, members of the LIN-41 clade (e.g. Human TRIM71, C. elegans LIN-41, Drosophila Wech, and Danio Trim71) recognize both RNA structure and sequence (Kumari et al., 2018; Loedige et al., 2015). The Kumari (2018) crystal structure of Danio Trim71 shows that the NHL domain contains a large, positively-charged cavity which nonspecifically interacts with the ribose and phosphates of the RNA (Figure 3A). The shape constraints imposed by this cavity ensure short RNA stem-loops are bound (Figure 3B). RNA nucleotides in and adjacent to the trinucleotide loop are recognized by amino acid side chains. While this network of protein-RNA contacts is less extensive than that of Brat, it establishes a strong preference for purines in the final loop position and a 5’-U:A first stem pair (Figure 3C). The high degree of conservation of the RNA-binding amino acid residues throughout the LIN-41 clade allows distantly related members to recognize the same RNA sequences and structures (Kumari et al., 2018; Loedige et al., 2015).
Figure 3. Structure of Danio rerio TRIM71 NHL domain bound to RNA.
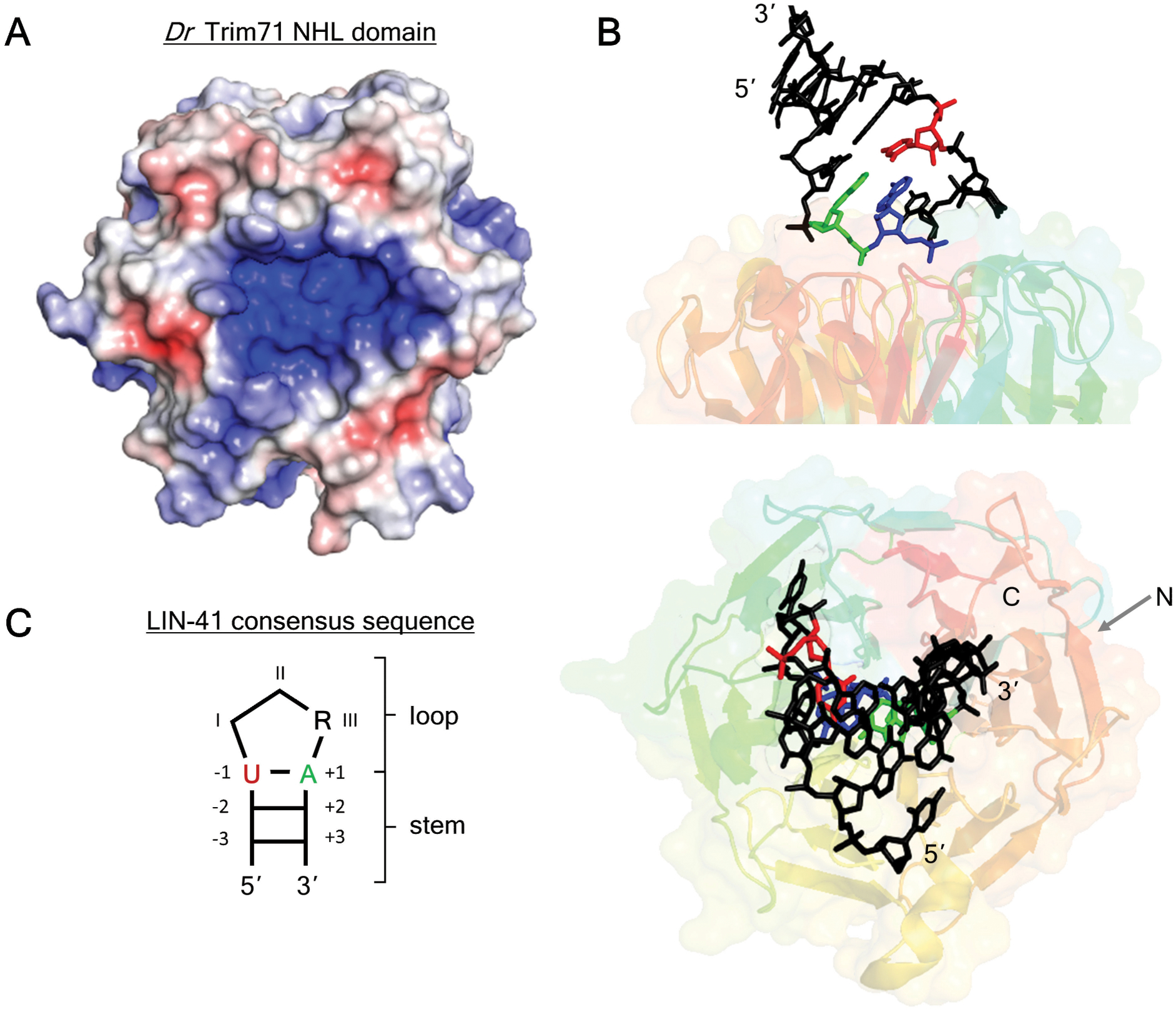
A. The positively-charged cavity on the top surface of Dr Trim71 NHL domain (PDB: 6FPT) (Loedige et al., 2015) was visualized with APBS in PyMOL. Blue denotes positive charge, red denotes negative charge.
B. X-ray crystal structure of the Dr Trim71 NHL domain in complex with the RNA-binding site from the 3’UTR of the mab-10 mRNA (PDB: 6FQL) (Kumari et al., 2018). The RNA stem loop binding site sits within an indentation on the Trim71 NHL domain. Nucleotides that are important for RNA-binding specificity are highlighted in color.
C. The LIN-41 consensus binding motif consists of a short stem with a three nucleotide loop. Specific nucleotides in the loop (position III) and stem (positions −1 &+1) are preferred by LIN-41 and TRIM71 orthologs.
The RNA-binding abilities demonstrated by TRIM71 and Brat suggest that other TRIM-NHL family members may also bind RNA. Loedige (2015) demonstrated in vitro that NCL-1 recognizes the same single-stranded RNA sequence motif as its homologue Brat. Mei-P26, on the other hand, promiscuously recognizes single-stranded U-rich motifs (Loedige et al., 2015). Further, mammalian TRIM56 is routinely identified in global screens for RNA-binding proteins (Baltz et al., 2012; Castello et al., 2012; Trendel et al., 2019), and has a preference for single-stranded RNA motifs (Kumari et al., 2018; Loedige et al., 2015). Finally, there is currently little evidence that other TRIM-NHLs, such as TRIM32, TRIM3, TRIM2, and C. elegans NHL-1 can bind RNA. While their NHL domains are structurally similar, the apparent versatility and differences in RNA-binding modes by TRIM-NHL proteins makes speculation difficult (Kumari et al., 2018).
2.3. TRIM-NHL proteins regulate a wide variety of mRNAs
Global analysis of TRIM-NHL proteins shows that they associate with and regulate many mRNAs. For example, Brat associates with transcripts from several thousand genes in fly embryos (Laver, Li, et al., 2015; Loedige et al., 2015). Hypomorphic brat mutants alter the levels of hundreds of transcripts in embryos (Laver, Li, et al., 2015) and adult brains (Juschke et al., 2013; Loop et al., 2004). Depletion of brat in neural stem cells causes a more modest dysregulation of 79 transcripts (Reichardt et al., 2018). Collectively, these transcripts are enriched in determinants of stem cell fate, embryonic development, signaling cascades, and integral membrane proteins, among others.
Similarly, TRIM71 and its homologs regulate hundreds of mRNAs in various cell types. Loss of endogenous TRIM71 in mouse embryonic stem cells (ESCs) (Loedige et al., 2013; Mitschka et al., 2015; Welte et al., 2019) and reduction of TRIM71 in human ESCs (Worringer et al., 2014) alter the levels of tens to hundreds of transcripts. A core set of twelve mRNAs are bound and regulated by TRIM71 in several cell lines (Welte et al., 2019). Additionally, ectopic TRIM71 in cultured human cells alters the levels of approximately 800 transcripts, the majority of which are decreased (Loedige et al., 2013). The gene ontologies of these transcripts indicate roles in neurogenesis, development, cell communication, and signaling (Worringer et al., 2014). The C. elegans homolog LIN-41 associates with more than a thousand mRNAs (Tsukamoto et al., 2017), but only regulates tens of genes when ectopically expressed in juvenile worms (Aeschimann et al., 2019).
With such a wide variety of effects, the challenge of proving how many of these transcripts are physiologically relevant has proven difficult to answer. The majority of affected genes are upregulated in the absence of TRIM-NHL proteins, which is consistent with repressive activity. However, it is unclear which of these are due to direct or secondary effects. The collection of mRNAs that is bound by a TRIM-NHL protein does not correlate well with the set of transcripts that change in abundance in that protein’s absence (Laver, Li, et al., 2015; Welte et al., 2019). Overall, many more transcripts appear to be bound than measurably regulated.
One possible interpretation is that only a biologically-relevant subset of bound transcripts is targeted for destabilization. Indeed, LIN-41 mediates the juvenile-to-adult transition in C. elegans by repressing only four transcripts (Aeschimann et al., 2019). The lethality associated with loss of brat in fly neural stem cells can also be rescued by reducing the level of several different Brat-associated transcripts (Reichardt et al., 2018; Song & Lu, 2011). These specific developmental processes may involve unique mRNA targets.
The challenge of identifying regulated, functionally significant mRNA targets can be interrogated by combining several types of evidence. First, there should be reproducible evidence of an effect of the TRIM-NHL protein on either mRNA abundance and/or translation (see below), often assessed by knockout, knockdown, or mutation of the TRIM-NHL. Such analysis has been conducted for brat using hypomorphic mutants (Laver, Li, et al., 2015) or tissue specific RNAi (Reichardt et al., 2018). Second, direct targets should have one or more binding consensus sequences which confer responsiveness to the TRIM-NHL. For example, the 3’UTRs of relevant target transcripts contain functional Brat consensus binding sites that confer regulation in fly neural stem cells (Komori et al., 2018; Reichardt et al., 2018) and embryos (reviewed in (Arvola et al., 2017)). Third, there should be experimental evidence demonstrating RNA-binding to the target transcript. Fourth, there should be a rescuable loss-of-function phenotype in which the levels of potential targets can be experimentally manipulated. An elegant example was employed by Reichardt (2018). Viability of adult Drosophila was measured following simultaneous RNAi-induced silencing of both the repressor (Brat) and various up-regulated targets, demonstrating that downregulation of these targets are crucial for the brat mutant phenotype.
2.4. Control of mRNA level or translation?
TRIM-NHL RNA-binding proteins regulate gene expression by reducing translation and causing degradation of their target mRNAs. Brat and TRIM71 are prime examples. Both are predominantly cytoplasmic, where they likely regulate target mRNAs (Chang et al., 2012; Loedige et al., 2013; Reichardt et al., 2018; Rybak et al., 2008). Decreasing mRNA levels will necessarily decrease expression of the encoded protein. Alternatively, translation can be prevented without altering transcript levels during certain developmental stages (e.g. early embryos). It is important to acknowledge that mRNA decay and translation are intimately linked, and therefore reducing the efficiency of translation can make mRNAs susceptible to more rapid mRNA decay. Careful analysis of TRIM-NHL regulatory activity is necessary to discriminate these potential mechanisms. Thus far, transcriptome-wide measurements of mRNA half-life in the absence of any TRIM-NHL have not been reported; however, multiple studies have now examined their effects on steady state mRNA levels.
In the most extensively studied case, Brat is reported to both repress translation and increase degradation of mRNAs. The ability of Brat to repress translation is well documented in the early embryo and cultured cells (Cho et al., 2006; Harris et al., 2011; Laver, Li, et al., 2015; Loedige et al., 2015; Loedige et al., 2014; Muraro et al., 2008; Sonoda & Wharton, 2001). Transcripts associated with Brat in embryos have lower translation efficiency, compared to all expressed mRNAs, as determined by polysome profiling (Laver, Li, et al., 2015). Further, Brat-bound mRNAs are unstable and prone to degradation (Laver, Li, et al., 2015; Reichardt et al., 2018). Additionally, Brat mediated mRNA degradation has been demonstrated in cultured cells (Laver, Li, et al., 2015).
TRIM71 also reduces both the protein and mRNA levels of reporter genes (Chang et al., 2012; Kumari et al., 2018; Loedige et al., 2013; Mitschka et al., 2015; Torres-Fernandez et al., 2019). Increased Trim71 during mouse ESC differentiation coincides with reduction in target mRNA levels (Welte et al., 2019). Consistent with these effects, ectopic TRIM71 decreased the half-life of several target mRNAs (Loedige et al., 2013). The broad impact of TRIM71 repressive activity on the transcriptome has been determined in cultured cell lines, embryonic stem cells, and induced pluripotent cells (Loedige et al., 2013; Mitschka et al., 2015; Welte et al., 2019; Worringer et al., 2014). Further, comparison of global measurements of mRNA level (by RNA-seq) and translation efficiency (by ribosome profiling) indicates that TRIM71 primarily controls abundance of mRNAs, whereas there is no evidence that TRIM71 alters ribosome occupancy of these transcripts (Welte et al., 2019). In contrast, C. elegans LIN-41 is capable of exercising both RNA decay and translational control, depending on the transcript (discussed below) (Aeschimann et al., 2017). It is likely that nature utilizes different mechanisms to alter protein levels - reducing RNA stability or reducing translation - by different TRIM-NHLs and in different biological contexts.
2.5. Regulating translation and degradation through the 3’UTR
The majority of transcripts bound by TRIM-NHL proteins have 3’UTRs that are enriched with consensus binding motifs. This is true for both Brat and TRIM71 (Laver, Li, et al., 2015; Loedige et al., 2015; Welte et al., 2019). For C. elegans LIN-41, motifs are enriched in the coding sequence (Kumari et al., 2018), but motifs in the 3’UTR are functionally relevant (Aeschimann et al., 2017). Sites in the 3’UTR are sufficient to confer responsiveness to TRIM-NHL proteins, as demonstrated by reporter genes that bear the 3’UTR of specific target mRNAs. Reporters containing the 3’UTRs of Brat-bound transcripts (e.g. hunchback, mad, myc, shn, etc.) are destabilized in the presence of ectopic Brat. This activity depends on the NHL domain binding to the consensus motifs (Komori et al., 2018; Loedige et al., 2013; Loedige et al., 2015). The same observation has been made with TRIM71 in cultured cells (Chang et al., 2012; Loedige et al., 2013; Torres-Fernandez et al., 2019) and endogenous LIN-41 in worms (Kumari et al., 2018). Artificial 3’UTRs with only the consensus motifs are similarly repressed (Komori et al., 2018; Kumari et al., 2018; Laver, Li, et al., 2015; Welte et al., 2019), indicating that these sites are necessary and sufficient for repression. This was demonstrated by Welte (2019) in a particularly elegant experiment: Trim71 was found to destabilize Mbnl1 transcript in mouse ESCs, and this regulation can be disrupted by mutating either the RNA-binding domain of Trim71 or its binding sites in the Mbnl1 3’UTR via CRISPR/Cas9.
Surprisingly, the majority of transcripts that are bound by TRIM-NHLs are not increased when the TRIM-NHL is absent (Laver, Li, et al., 2015; Welte et al., 2019). Instead, only a subset of transcripts appear to be regulated. One explanation may be that while a single site can be bound by the TRIM-NHL protein, multiple copies of motifs may be required for regulation. Indeed, multiple binding sites are necessary for effective repression by Trim71 (Kumari et al., 2018), and the level of repression scales proportionally with the number of motifs (Welte et al., 2019). This has been proposed to compensate for the relatively low (Kd ~1 μM) affinity of the TRIM71 NHL domain for a single motif (Kumari et al., 2018). Certain LIN-41 binding motifs are regulated, but others are not, depending on the transcript (Aeschimann et al., 2017). This is true for other RNA-binding proteins, and the reasons remain unclear. The extent of mRNA destabilization appears to vary across transcripts that are bound equally well by Trim71 (Welte et al., 2019), indicating that differences in occupancy of a transcript cannot fully account for the variation. Together these observations indicate that additional factors mediate regulation, which remain to be discovered.
2.6. TRIM-NHL proteins can act through the 5’ UTR
Recently, C. elegans LIN-41 was found to repress translation, without altering mRNA levels, upon being recruited to the 5’UTR (Aeschimann et al., 2017). LIN-41 binds to specific motifs in the lin-29A 5’UTR, and transplanting this 5’UTR into an unregulated 3’UTR destabilizes the transcript. Conversely, transplanting fragments of the target mab-10 3’UTR into the 5’UTR of reporters is sufficient to inhibit translation. These elegant experiments show the position of binding motifs in the transcript can tip the balance between RNA degradation and translational inhibition. This is particularly relevant for TRIM-NHL proteins, as Brat and Trim71 have also shown enrichment of binding sites in the 5’UTR, albeit to lower levels than the 3’UTR (Arvola et al., 2017; Laver, Marsolais, et al., 2015; Loedige et al., 2015; Welte et al., 2019). In light of this observation, position-dependent regulation of mRNAs by TRIM-NHLs remains an exciting avenue of research.
2.7. Several domains of TRIM-NHL proteins confer mRNA repression
Functional analysis of the domains of TRIM-NHL proteins indicates specific roles for individual modules. The NHL domain is necessary and sufficient to bind target mRNAs, and is therefore necessary for mRNA repression (Kumari et al., 2018; Loedige et al., 2013; Loedige et al., 2015; Loedige et al., 2014; Welte et al., 2019). Several individual domains are sufficient for repression when artificially tethered to reporter mRNAs. Such reporter assays in effect bypass the TRIM-NHL protein’s RNA-binding domains (Coller & Wickens, 2007). When tethered, Brat and TRIM71 readily destabilize transcripts even when the NHL domain is deleted (Chang et al., 2012; Loedige et al., 2013; Loedige et al., 2015; Loedige et al., 2014). Instead, the TRIM domain - particularly the coiled-coil domain - and NHL domain autonomously repress mRNAs (Chang et al., 2012; Loedige et al., 2013; Torres-Fernandez et al., 2019). Likewise, the N-terminus of TRIM32 has repressive activity (Loedige et al., 2013). The RING domain (discussed further below) and B-Box zinc fingers of TRIM71 are largely dispensable for RNA regulation (Chang et al., 2012; Loedige et al., 2013). It will be interesting to compare whether these autonomous repressive domains are necessary for mRNA repression in vivo. Finally, the TRIM71 coiled-coil, NHL, and B-box zinc fingers contribute to TRIM71 localization in cytoplasmic RNA-protein granules, known as Processing bodies (P-bodies); though it is unclear if this is necessary for mRNA repression (Chang et al., 2012; Rybak et al., 2008; Torres-Fernandez et al., 2019).
3. THIS KNOWLEDGE HAS SHAPED OUR UNDERSTANDING OF TRIM-NHL FUNCTIONS
3.1. Brat regulates maternal mRNAs during embryogenesis
The initial connection between Brat and mRNA control came from work of Sonoda & Wharton (2001) in their studies of fly embryogenesis. Embryos of all animals undergo an initial period of transcriptional inactivity immediately following fertilization (reviewed in (Tadros & Lipshitz, 2009; Vastenhouw et al., 2019)). During this period, all cellular processes are governed by proteins and mRNAs deposited into the oocyte during oogenesis. In the absence of mRNA transcription and widespread RNA decay, changes in gene expression solely depend on altering mRNA translation (Tadros & Lipshitz, 2009). In this context, maternally provided Brat reduces translation of target mRNAs.
For some transcripts, Brat is assisted in translational control by other RNA-binding proteins. In the posterior of the embryo, RNA-binding repressor proteins Nanos (Nos) and Pumilio (Pum) assist Brat in preventing translation of the transcription factor Hunchback (Figure 4A) (Arvola et al., 2017; Laver, Li, et al., 2015; Sonoda & Wharton, 2001). Ectopic Hunchback causes a lethal loss of abdominal segments in the resulting larva (Sonoda & Wharton, 2001). In this unique molecular environment, hunchback mRNA is kept stable and translationally silenced by a two-pronged mechanism (Figure 4B). First, the poly-adenosine tail of the hunchback mRNA is enzymatically shortened to reduce protein expression (Wreden et al., 1997), as length correlates with translation efficiency in early embryos (Eichorn et al., 2016; Subtelny et al., 2014). Second, Brat is assisted by the 5’ cap-binding protein eIF4E homologous protein (4EHP) to reduce translation of Hunchback (Cho et al., 2006). In addition to hunchback, Brat and Pum co-regulate a subset of bound mRNAs which are enriched in both binding motifs (Laver, Li, et al., 2015). Further, Brat and Pum independently regulate distinct sets of mRNAs which are enriched with their individual, respective binding motifs (Laver, Li, et al., 2015).
Figure 4. Brat regulates translation of hunchback mRNA during early embryogenesis.
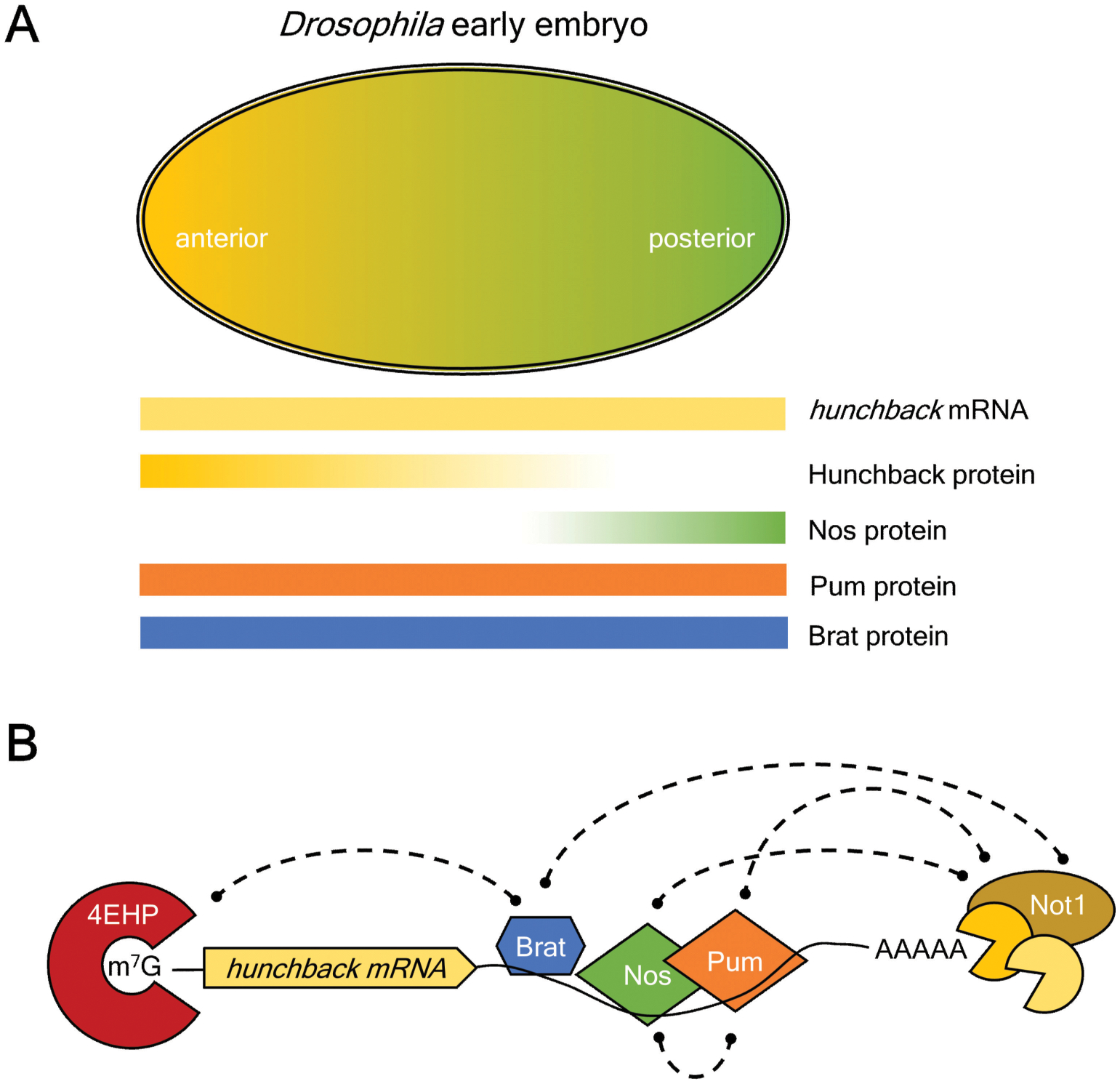
A. Gradients of key maternal mRNAs and proteins in the early Drosophila embryo control polarity of the body plan. Hunchback mRNA is ubiquitous, but Hunchback protein is restricted to the anterior half, mediated by translational repression by the combination of Brat, Pum, and Nanos. Pum and Brat protein are distributed throughout the embryo, whereas Nanos protein forms a gradient emanating from the posterior.
B. Model of translational repression mechanism by Brat, Pum, and Nanos. The poly(A)-tail of hunchback mRNA is shortened the CNOT deadenylase, which can be recruited by the three RNA-binding proteins bound to the 3’UTR. Brat can also utilize the 5’ 7-methyl guanosine (m7G) cap binding protein, 4EHP, to repress translation initiation.
Following this period of genome silence, zygotic transcription and RNA decay are initiated, causing a massive clearing of maternal mRNA and proteins (Vastenhouw et al., 2019). During this maternal-to-zygotic transition (MZT), Brat-associated transcripts are destabilized (Laver, Li, et al., 2015). Specifically, Brat binds to distinct sets of transcripts which are targeted by either maternal or zygotic decay machinery, implying Brat can associate with components of both (Laver, Li, et al., 2015). Compared to normal embryos, a statistically significant fraction of Brat-associated mRNAs are upregulated in a maternal brat mutant. However, this observation is complicated by the enigmatic nature of the particular brat mutation - bratfs1- that was used for this analysis out of necessity (Edwards et al., 2003; Lee et al., 2006; Loedige et al., 2015; Loedige et al., 2014; Reichardt et al., 2018; Sonoda & Wharton, 2001). The basis of Brat’s mRNA decay activity during MZT remains an area of active research.
3.2. Brat represses self-renewal factors during larval neurogenesis
The namesake phenotype of brat loss-of-function mutants is an over-proliferation of larval neural stem cells (neuroblasts), at the expense of neurons (Bello et al., 2006; Betschinger et al., 2006; Komori et al., 2014; Lee et al., 2006; Xiao et al., 2012). As a result, these tumor-filled larval brains rapidly grow in size, becoming metastatic and ultimately lethal (Arama et al., 2000; Beaucher et al., 2007; Caussinus & Gonzalez, 2005; Kurzik-Dumke et al., 1992; Mukherjee et al., 2016; Spradling et al., 1999; Stathakis et al., 1995; Woodhouse et al., 1998; Wright et al., 1981; Wright et al., 1976). Although this phenotype was identified decades ago, the function of Brat on the molecular level remained a mystery. The discovery of Brat’s RNA-binding ability has dramatically increased our understanding of its function, while also raising new questions.
Brat ensures larval neuroblasts complete differentiation into the next cell type. Larval neuroblasts divide asymmetrically, so that the two daughter cells have different developmental fates (Figure 5A). One daughter cell remains a neuroblast; while the other further differentiates and eventually forms the neurons that make up the adult nervous system. For reviews of this process, Homem & Knoblich (2012) and Jannsens & Lee (2014) are suggested. Brat is dispensable in Type I neuroblasts, which divide asymmetrically to form a Ganglion Mother Cell (GMC) that subsequently divides to produce neurons. In contrast, Brat is necessary for the appropriate differentiation of type II neuroblasts (Bowman et al., 2008). These neuroblasts divide asymmetrically to produce secondary neuroblasts called Intermediate Neural Progenitors (INPs) (Bowman et al., 2008). While type II neuroblasts are far fewer in number than type I, these additional transit-amplifying INPs allow increased proliferative capacity.
Figure 5. Brat regulates neurogenesis.
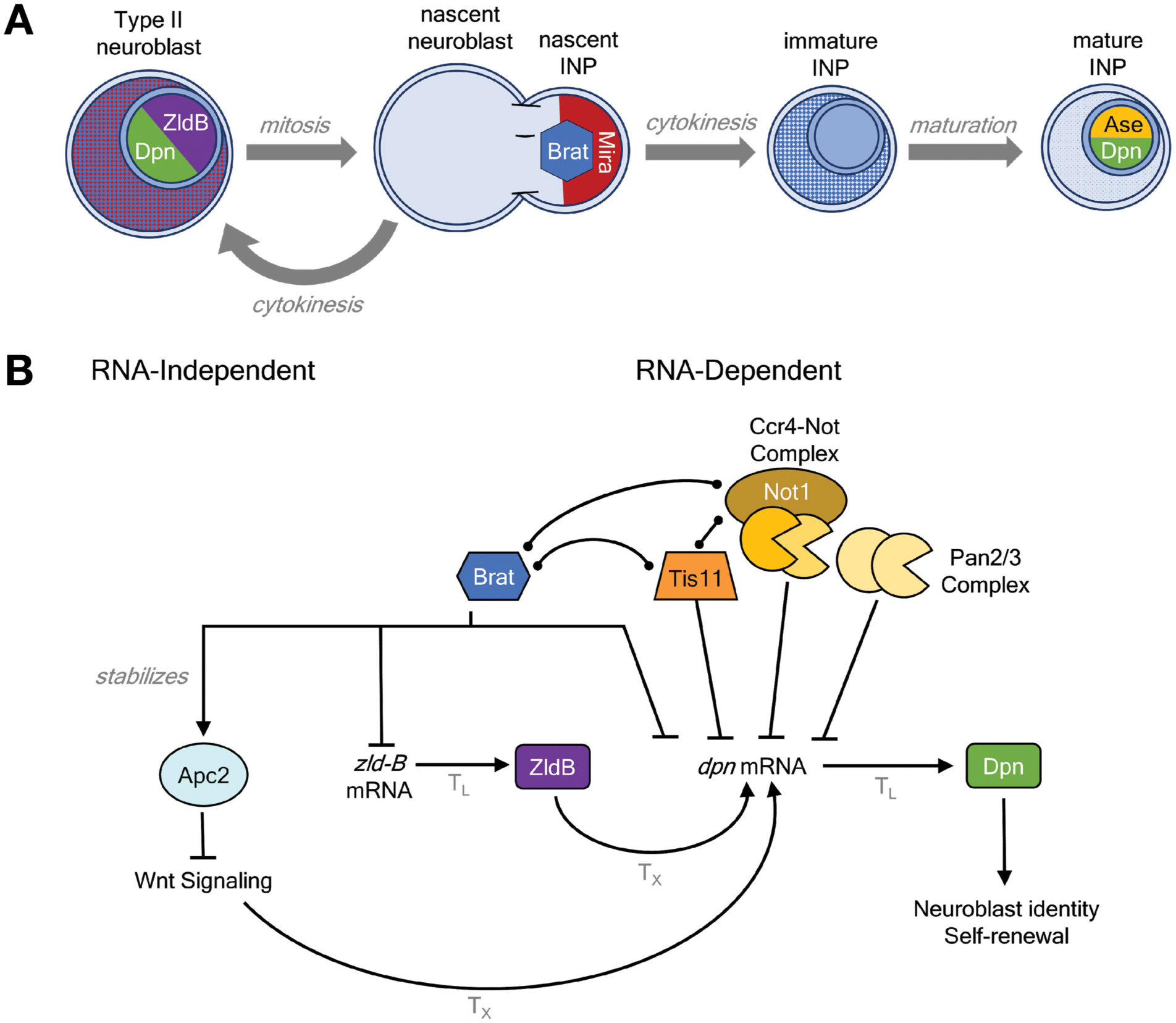
A. Brat controls early differentiation of Drosophila Type II neuroblasts in the larval brain. During asymmetric cell division, Miranda (Mira) localizes and concentrates Brat in the nascent Intermediate Neural Progenitor (INP). Following cytokinesis, Mira is degraded, releasing Brat. During INP maturation, Brat is reduced and transcription factors Asense and Deadpan are expressed. These transcription factors promote further asymmetric divisions of INPs, forming glial cells and neurons.
B. Proposed RNA-dependent and -independent regulatory roles of Brat in INP maturation. Activation is indicated by pointed arrows whereas repression is marked with blunted arrows. Observed physical associations of Brat with the RNA-binding protein Tis11 and CNOT deadenylase complex are marked with bulb-end lines. Translational control (TL) and transcriptional control (TX) are indicated on the respective arrows.
During asymmetric neuroblast division, Brat localizes to one side of the dividing neuroblast through the adapter protein Miranda (Mira). This has been observed in embryonic and larval neuroblasts (Betschinger et al., 2006; Lee et al., 2006), and is proposed to enrich Brat in immature INPs (iINPs) following cytokinesis (Reichardt et al., 2018). INPs mature over a period of hours, during which the cytoplasm is cleared of factors that previously maintained neuroblast identity. In the absence of brat, type II neuroblasts divide normally. But brat mutant INPs fail to down-regulate these factors, and revert to neuroblast-like cells with an abnormally high mitotic index (Bello et al., 2006; Betschinger et al., 2006; Komori et al., 2014; Lee et al., 2006; Xiao et al., 2012).. These ‘ectopic neuroblasts’ are indistinguishable from normal neuroblasts by cell markers (Bello et al., 2006) but over-proliferate and form tumors (Betschinger et al., 2006). In effect, for a nascent INP, Brat closes the door on a neuroblast fate.
What is Brat doing in INPs? Brat was long assumed to be a translational repressor, though the exact function was not understood (Arama et al., 2000; Sonoda & Wharton, 2001). After Brat’s RNA-binding ability was discovered, evidence for direct post-transcriptional regulation began to emerge: Reichardt (2018) showed that Brat downregulates Deadpan and Zelda isoform B in neuroblasts. Brat was previously shown to associate with deadpan (Loedige et al., 2015) and zelda (Laver, Li, et al., 2015) mRNAs. This regulation depends on Brat binding to sites in the 3’UTRs of deadpan and zelda-B transcripts (Komori et al., 2018; Loedige et al., 2015; Reichardt et al., 2018). These transcription factors are markers of neuroblast identity, and promote neuroblast proliferation and asymmetric cell divisions (Homem & Knoblich, 2012). Simultaneous reduction of deadpan or zelda-B with brat rescues adult survival (Reichardt et al., 2018), showing Brat represses key neuroblast determinants to promote the maturation of INPs.
As ectopic Deadpan is not sufficient to cause brain tumors (Komori et al., 2018; Reichardt et al., 2018), other Brat targets must also exist. Brat represses the transcription factor myc in type II neuroblasts (Reichardt et al., 2018), and reduction of myc partially reduces the ectopic neuroblasts of brat mutants (Neumuller et al., 2013; Reichardt et al., 2018; Song & Lu, 2011). Among its many functions, Myc upregulates transcription of the translation factor eIF4E; reduction of which also rescues brat mutants (Song & Lu, 2011). This is consistent with the unique dependency of ectopic neuroblasts on eIF4E and Myc (Song & Lu, 2011), potentially due to the demands of increased growth and proliferation (Betschinger et al., 2006; Song & Lu, 2011).
Parallel, but nonredundant, pathways also exist to ensure maturation of iINPs. In GMCs and iINPs, Notch signalling is repressed by the Notch inhibitor Numb (Haenfler et al., 2012; Wang et al., 2006), which is asymmetrically segregated similarly to Brat (Haenfler et al., 2012). Overactivation of Notch signalling in type II neuroblasts phenocopies brat mutants (Bowman et al., 2008; Neumuller et al., 2011). Loss of numb enhances the brat mutant phenotype – and vice versa (Komori et al., 2018). Furthermore, the vacuolar ATPase Vha68–2 enhances Notch signalling in type I neuroblasts, and reduction of vha68–2 suppresses brat tumors (Wissel et al., 2018). Notch signalling also upregulates myc, eIF4E, and dpn (San-Juan & Baonza, 2011; Song & Lu, 2011). Despite the overlap, Brat and Numb play non-redundant roles during INP maturation (Komori et al., 2014), indicating Brat independently regulates some yet-unknown targets.
Surprisingly, ectopic neuroblasts in brat mutants can be reduced by expressing a brat transgene lacking the NHL domain (Komori et al., 2014). This observation brings up several questions: First, is Brat localization required to control INP fate? The NHL domain is required to concentrate Brat in iINPs (Betschinger et al., 2006; Lee et al., 2006). This may be obviated by transgenic overexpression. Similar alterations in localization may be the basis of several brat point mutations (Arama et al., 2000; Reichardt et al., 2018). Second, can Brat post-transcriptionally repress deadpan without its NHL domain? Komori (2018) reported that Brat partners with the RNA-binding protein Tis11 to repress deadpan, and hypothesized that this collaboration enables ectopic Brat to bypass the requirement for its NHL domain. Brat interacts with Tis11 through a region of its TRIM domain (Komori et al., 2018), and this region is essential for rescuing brat mutants (Komori et al., 2014). While Tis11 represses Deadpan in INPs, Tis11 binding sites are largely unnecessary for repression of the deadpan 3’UTR reporters (Komori et al., 2018). This indicates Tis11 may serve as a back-up system in the absence of functional Brat. Third, could Brat regulate cell fate independent of its RNA-regulatory function? Brat’s TRIM domain also stabilizes Apc2 to the interface of dividing neuroblasts and embryonic eagle neurons, downregulating Armadillo signalling in INPs (Arbeille & Bashaw, 2018; Komori et al., 2014). In summary, Brat function in neuroblasts may involve collaborations with other RNA-binding proteins or additional RNA-independent roles (Figure 5B).
3.3. TRIM71 maintains the identity of embryonic stem cells
Mammalian TRIM71, like Drosophila Brat, regulates differentiation in stem cells during neurogenesis. However, their roles are opposite: Brat promotes differentiation of stem cells, whereas TRIM71 prevents differentiation. TRIM71 is highly expressed in embryonic stem cells (ESCs), where it promotes proliferation and antagonizes differentiation (Chang et al., 2012; Worringer et al., 2014). Loss of TRIM71 in mouse ESCs increases expression of neurogenesis factors, thereby promoting differentiation into neuroectoderm cells (Mitschka et al., 2015). The ability of TRIM71 to antagonize differentiation is further exemplified by its ability to promote reprogramming of fibroblasts into induced pluripotent stem cells (iPCSs) when overexpressed along with pluripotency factors Oct4, Sox2, and Klf4 (Worringer et al., 2014).
TRIM71, like all LIN-41 homologs, is repressed by the let-7 microRNA (Maller Schulman et al., 2008; Rybak et al., 2008; Schulman et al., 2005). This conserved regulation – reviewed by Ecesdi & Groβhans (2015) is crucial for tuning the levels of LIN-41 homologs in various biological processes. For example, LIN-41 expression in C. elegans is regulated by let-7 through a specific seed site in the lin-41 3’UTR (Aeschimann et al., 2017). In the context of vulval development, lin-41 mRNA is the only functional target of let-7 (Ecsedi et al., 2015). Similarly, in mammalian ESCs, let-7 and TRIM71 form an opposing axis. The high level of TRIM71 in ESCs is repressed during development and differentiation by the increased expression of let-7 (Worringer et al., 2014). These two types of post-transcriptional regulators - a TRIM-NHL protein and a microRNA - form an opposing axis that regulates stem cell fate.
The precise role of TRIM-NHL RNA-binding activity in ESCs has recently been examined. Both the NHL and RING domains are required to induce human iPCS (Worringer et al., 2014). Depletion of TRIM71 in human ESCs resulted in altered levels of more than one thousand mRNAs, which are enriched for functions in development and differentiation. In mouse ESCs, a single mutation abolishing RNA-binding mimics the transcriptome changes of complete TRIM71 knock-out (Welte et al., 2019). Together, these results highlight the important regulatory role of TRIM71 in mammalian stem cells.
3.4. Brat & Mei-P26 promote differentiation of the Drosophila germline
The importance of TRIM-NHLs is evident in Drosophila oogenesis, wherein paralogs Brat and Mei-P26 ensure that germline stem cells (GSCs) differentiate properly. Like neuroblasts, GSCs divide asymmetrically to produce two nearly identical daughter cells with different developmental fates. For reviews of this process, Bastock & St. Johnston (2008), Harris & Ashe (2011), and Slaidina & Lehmann (2014) are recommended. External and internal cues determine the fate of these daughter cells (Figure 6A), with Brat and Mei-P26 playing different roles in different stages.
Figure 6. Brat and Mei-P26 regulate Drosophila oogenesis.
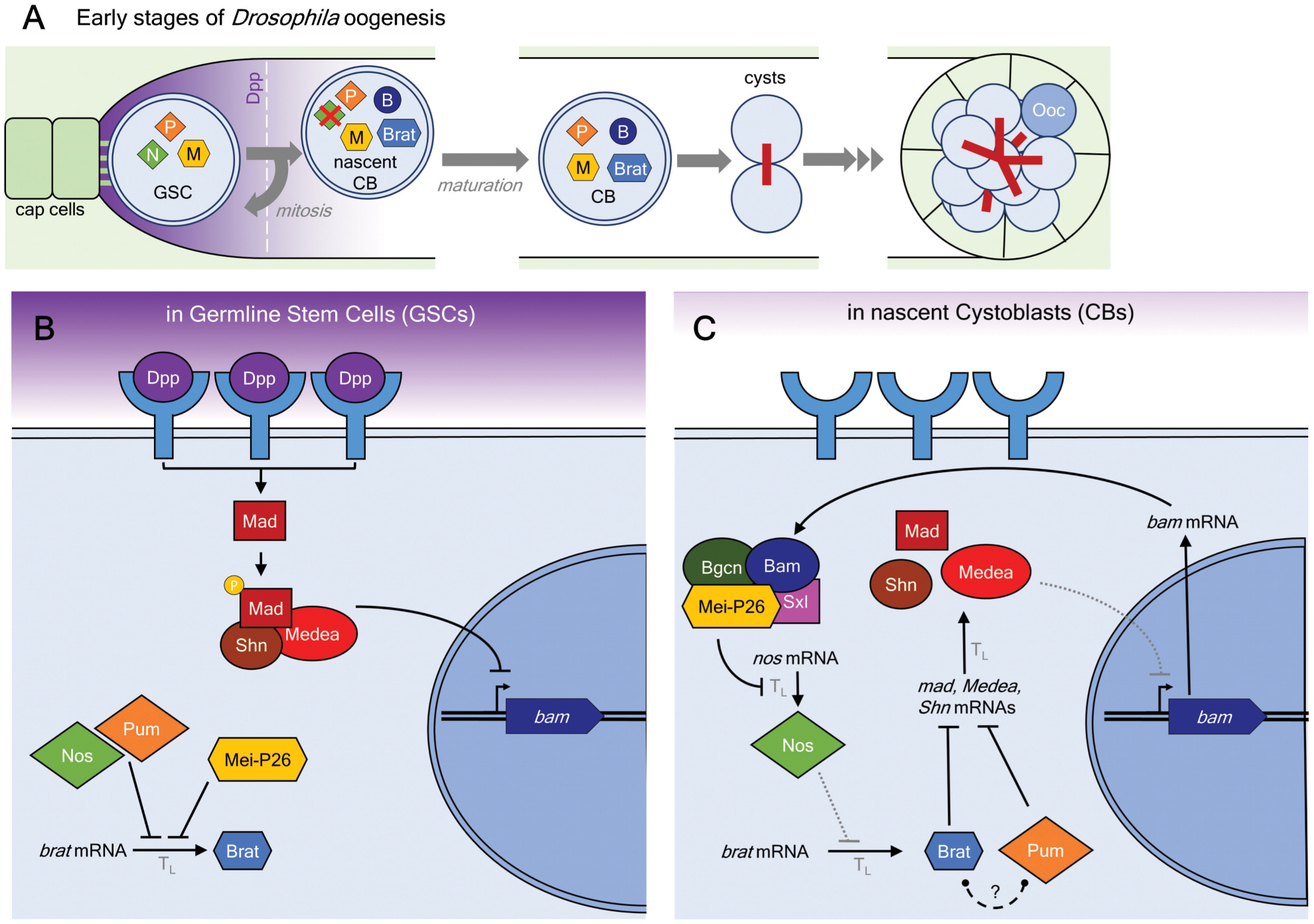
A. Early oogenesis in adult Drosophila. Germline Stem Cells (GSCs), in contact with somatic cap cells, receive Decapentaplegic (Dpp) signal that is necessary for their maintenance. The resulting signaling cascade ensures expression of Nanos (N), Pumilio (P), and Mei-P26 (M). Asymmetric cell division pushes one daughter cell below the signal threshold (dashed line), causing expression of Bam (B) and Brat. The nascent cystoblast (CB) further divides incompletely into cysts, one of which is designated to become the oocyte (Ooc).
B. Molecular pathways maintaining GSC identity. Dpp signaling promotes Mothers against Dpp (Mad)-mediated transcriptional (Tx) regulation of Bag of marbles (bam). At the same time, Nanos (Nos), Pum, and Mei-P26 repress translation (TL) of brat mRNA.
C. Molecular pathways promoting differentiation of nascent cystoblasts. Loss of Dpp signal in the cystoblast allows expression of Bam. In turn, Bam collaborates with Mei-P26, Sex-lethal (Sxl), and benign gonial cell neoplasm (Bgcn) proteins to repress translation of Nanos mRNA. As a result, translational repression of Brat is alleviated. Brat protein collaborates with Pum to repress expression of mad, medea, and schnurri (shn) transcription factors. Dotted lines represent reduced strength of interactions.
The identity of GSCs is maintained in a niche created by Decapentaplegic (Dpp) signaling from nearby somatic cells (Figure 6B) (Slaidina & Lehmann, 2014). This signaling activates transcription factors Mad, Medea, and Schnurri (Shn), which prevent transcription of the fate determinant bam. Bam levels are tightly controlled in GSCs, since Bam is necessary and sufficient to prompt GSC differentiation (Li et al., 2012; Li et al., 2009; Ohlstein & McKearin, 1997). Brat antagonizes this signal cascade (Harris et al., 2011; Newton et al., 2015), so translation of brat mRNA is post-transcriptionally inhibited by the RNA-binding repressors Pum, Nanos, and Mei-P26 (Harris et al., 2011; Li et al., 2012). Mei-P26 further enhances Dpp signaling; while this depends on its NHL domain, the relevant target transcripts remain unknown (Li et al., 2012). Overall, a moderate Mei-P26/low Brat environment maintains GSC identity (Figure 6B).
When a GSC divides, one daughter cell moves away from the Dpp source and below the signal threshold (Figure 6C). In response, Bam is expressed. Rearrangements in RNA-protein complexes form a two-step positive feedback loop to ensure this cell differentiates into a cystoblast and not another GSC (Slaidina & Lehmann, 2014). Conceptually, this process is similar to that documented in iINPs immediately after cell division. First, a Mei-P26-containing complex lowers Nanos levels. Mei-P26 and another RNA-binding protein, Sex lethal (Sxl), bind to the 3’UTR of nanos mRNA (Chau et al., 2009; Li et al., 2012; Li et al., 2009; Li et al., 2013). These proteins, in turn, recruit Bam, Bgcn, and Brat to repress nanos expression (Li et al., 2013; Malik et al., 2017; Neumuller et al., 2008). Bam interacts with the CNOT deadenylase complex (Sgromo et al., 2018; Sgromo et al., 2017), which removes the poly(A) tail of mRNAs (Temme et al., 2004; Temme et al., 2010). This directly connects the complex to the cellular RNA decay machinery. Second, the resulting decrease in Nanos allows Brat to be translated (Harris et al., 2011). Brat dampens BMP signaling by post-transcriptionally repressing mad, medea, and schnurri (Harris et al., 2011; Newton et al., 2015). The 3’UTRs of these transcripts are sufficient to confer sensitivity to Brat, implying these are direct targets. Pum also represses these mRNAs, though it is unclear if Brat and Pum function together or in parallel. In a proposed model, Brat and Pum recruit the CNOT deadenylase complex to reduce the levels of these transcripts. The reduction in Dpp signaling allows further Bam transcription, forming an all-or-nothing switch for differentiation into a cystoblast (Slaidina & Lehmann, 2014).
Mei-P26 is also crucial for other events in germline differentiation. For example, cystoblasts further undergo several cell divisions with incomplete cytokinesis, forming cysts. In 16-cell cysts, one cell is specified as the oocyte and the remaining 15 support the developing egg as nurse cells. This coincides with peak Mei-P26 levels (Neumuller et al., 2008). In addition to reduced GSCs, mei-P26 mutant germaria are filled with cysts, but lack oocytes and nurse cells (Li et al., 2012; Neumuller et al., 2008; Page et al., 2000). The RNA-binding protein Orb is expressed in the one cell per cyst that is destined to become oocyte (Barr et al., 2019). Mei-P26 appears to ensure specification of a single oocyte by repressing orb translation (Li et al., 2012; Neumuller et al., 2008). Additionally, the namesake phenotype of mei-P26 mutants derives from defects in meiotic recombination, which occur later during oogenesis (Page et al., 2000; Sekelsky et al., 1999). Finally, Mei-P26 has also been reported to function in the male germline (Insco et al., 2012). Strong loss of function mei-P26 mutants are male sterile (Page et al., 2000). In transit-amplifying spermatogonia, Mei-P26 facilitates the accumulation of Bam, prompting differentiation of these cells (D. Chen et al., 2014; Insco et al., 2012). These activities are still under investigation.
Overall, circuits of multiple RNA-binding proteins (i.e. Brat, Mei-P26, Pum, Nos, & Sxl) and transcription factors specify GSC identity. Dynamic changes in expression of these factors control the differentiation of a GSC to an oocyte.
3.5. Brat & Mei-P26 converge on nucleoli
Brat and Mei-P26 regulate the transcription factor Myc and the size of nucleoli, the site of ribosome biogenesis (Figure 7A). Myc is a transcription factor that stimulates ribosome biogenesis (reviewed in Oskarsson & Trumpp (2005) and Campbell & White (2014)), which in turn causes enlarged nucleoli. Regulation of Myc and/or nucleolar size by Brat and Mei-P26 has been observed in various Drosophila tissues, such as female GSCs (Harris et al., 2011), female cystoblasts and cysts (Neumuller et al., 2008), male germline stem cells (Insco et al., 2012), neuroblasts (Betschinger et al., 2006; Neumuller et al., 2013; Reichardt et al., 2018), and wing imaginal discs (Abidi & Smith-Bolton, 2019; Ferreira et al., 2014; Frank et al., 2002). Crucially, Brat binds myc mRNA (Loedige et al., 2015) and can repress the myc 3’UTR in cultured fly cells (Harris et al., 2011; Loedige et al., 2015), indicating myc is a direct target of Brat. Furthermore, suppression of brat-derived ectopic neuroblasts can be achieved by reducing myc, eIF4E, or nucleolar components (Neumuller et al., 2013; Song & Lu, 2011). Similar results for myc and eIF4E have been obtained for mei-P26-derived ectopic GSCs (Song & Lu, 2011). As wild-type neuroblasts and GSCs remain unaffected by reduction of myc or eIF4E, it has been proposed that the cancer stem cells formed in the absence of Brat and Mei-P26 depend on higher rates of ribosome biogenesis (Song & Lu, 2011).
Figure 7. TRIM-NHL proteins control nucleolar size in Drosophila and C. elegans.
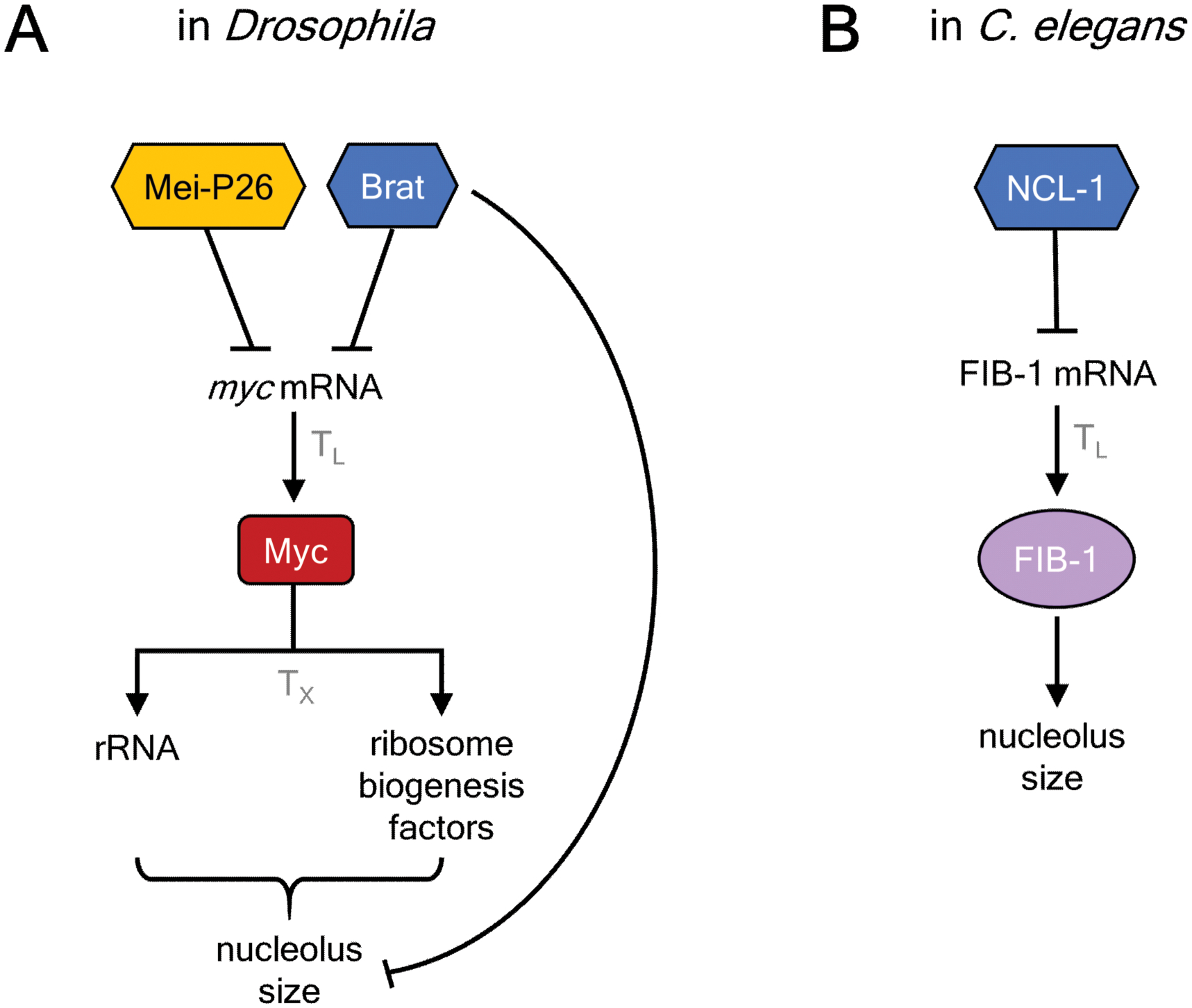
A. Drosophila Brat and Mei-P26 repress myc expression, which controls expression of rRNA and ribosome biogenesis factors to affect nucleolar size, particularly wing imaginal discs.
B. C. elegans NCL-1 represses the FIB-1 mRNA to control nucleolar size.
The connection between nucleolar size and Myc regulation in normal cells, however, is not always apparent. For example, loss of brat in wing imaginal disc cells causes enlarged nucleoli (Frank et al., 2002), but Brat does not normally regulate Myc levels in these cells (Abidi & Smith-Bolton, 2019). Brat can repress Myc when Mei-P26 is absent (Ferreira et al., 2014), indicating Brat and Mei-P26 function redundantly in this case. Recently, Abidi & Smith-Bolton (2019) showed injuries induce myc transcription to allow regrowth, and Brat prevents overgrowth by tuning Myc levels.
The role of Brat in repressing nucleolar size and ribosomal rRNA transcription is evolutionarily conserved (Figure 7B). The C. elegans homolog of brat is ncl-1. Loss of function ncl-1 mutants are named for enlarged nucleoli, and have higher rates of ribosomal RNA transcription (Frank & Roth, 1998). Brat transgenes expressed in C. elegans can even rescue the ncl-1 phenotype, emphasizing their orthologous relationship (Frank et al., 2002). NCL-1 represses translation of the fibrillarin homolog fib-1, though binding sites in the 3’UTR of the transcript (Y. H. Yi et al., 2015).
3.6. TRIM71 is required for brain development
TRIM71 expression is dynamic, and temporally controlled during embryonic development. In mice, TRIM71 is abundant in limb and tail buds, dorsal root ganglia, branchial arches, eye spots, and the developing brain, including the neuroepithelium (J. Chen et al., 2012; Maller Schulman et al., 2008; Mitschka et al., 2015). Expression is particularly high in undifferentiated stem cell populations, and declines upon differentiation and throughout embryogenesis (Chang et al., 2012; J. Chen et al., 2012; Rybak et al., 2008). Overall, TRIM71 levels decrease throughout development in many tissues (Cardoso-Moreira et al., 2019).
The crucial role of TRIM71 in embryonic development was revealed by genetic analysis in mice. Homozygous loss of TRIM71 causes defects in neural tube closure (J. Chen et al., 2012; Cuevas et al., 2015). As a result, the mouse embryos develop exencephaly, a condition where the brain is outside of the skull, and perish prenatally. Their brains also exhibit altered morphology and reduced growth of neural tissue (J. Chen et al., 2012). These abnormalities are associated with decreased neural progenitor cell proliferation and increased differentiation.
The importance of TRIM71’s RNA-binding activity is evident in human brain development. Children born with the disease congenital hydrocephalus (CH) exhibit enlarged heads with altered brain morphology, marked by enlarged cerebrospinal fluid-filled spaces within the brain (ventricles). With an incidence of 1 in 1000 live births, approximately 40% of CH cases are attributed to genetic causes (Duy et al., 2019). Furey et al. (2018) identified heterozygosity for several de novo TRIM71 mutations that account for 10% of total CH cases. Among these, R608H and R796H are located on the RNA-binding interface of the NHL domain (Furey et al., 2018; Kumari et al., 2018). While the effect of these TRIM71 mutations on RNA-binding have not been measured directly, they do impair the ability of TRIM71 to repress target mRNAs (Welte et al., 2019). The specific target mRNAs that TRIM71 regulates during brain development, however, remain to be identified. It is worth noting that, unlike in mouse models, only one copy of these TRIM71 mutants is sufficient to produce the CH phenotype (Furey et al., 2018). Thus, CH pathogenesis could be a result of reduced functional TRIM71 level or perhaps a dominant negative effect of the mutant protein.
4. FUTURE DIRECTIONS AND REMAINING QUESTIONS
4.1. Combinatorial control of mRNAs
The untranslated regions of mRNAs act as landing pads for different RNA-binding regulatory factors, including TRIM-NHL proteins and RNA-Induced Silencing Complexes (RISCs) (for reviews, Bartel (2018) and Mitchell & Parker (2014) are recommended). These factors can co-occupy a single transcript and alter the activities of each other. Alternatively, RNA-binding proteins can act in parallel on mRNAs encoding the same protein. This principle of combinatorial control is relevant to TRIM-NHL proteins, supported by growing evidence linking them to other RNA-binding factors.
Combinatorial control has been observed between Brat and several RNA-binding proteins. For example, interactions between Brat, Pum, and Nanos have recently been reviewed in Arvola (2017). In Drosophila embryos, a subset of approximately 200 mRNAs are bound by both Brat and Pum (Laver, Li, et al., 2015). For hunchback mRNA specifically, pre-bound Pum or Brat enhances the other’s affinity by modulating RNA structure (Loedige et al., 2015; Loedige et al., 2014). Brat and Pum also repress several of the same targets in the fly germline (Harris et al., 2011; Newton et al., 2015). It is noteworthy that while Brat and Pum can collaborate, the vast majority of Pum and Brat targets are independently regulated by one or the other (Laver, Li, et al., 2015). Brat can additionally repress hunchback and mad reporters independently of Pum (Loedige et al., 2014). In larval neural stem cells, Brat collaborates with Tis11 to repress deadpan expression (Komori et al., 2018). Tis11 is a well-characterized AU-rich RNA-binding protein that causes mRNA degradation (Fu & Blackshear, 2017). This relationship is intriguing, as the C. elegans LIN-41 and orthologs of Tis11, OMA-1 and OMA-2, are also functionally and physically connected in regulating mRNAs in the germline [(Tsukamoto et al., 2017).
Additional protein partners can regulate Brat activity. Mira binds the RNA-binding face of the NHL domain, preventing repression of mRNAs by Brat (Betschinger et al., 2006; Lee et al., 2006; Loedige et al., 2015; Reichardt et al., 2018). Mira is expressed in neuroblasts and is rapidly degraded in iINPs. This may be the basis by which Brat activity is restricted to iINP maturation.
Combinatorial control is also relevant to human TRIM71, which has been shown to associate with multiple RNA-binding factors involved in RNA metabolism. These include human homologs of Pum, PUM1 and PUM2, and Argonaute proteins, which are central components of RISC (Loedige et al., 2013; Torres-Fernandez et al., 2019). These interactions appear to be linked by RNA, and TRIM71 does not require PUM1/2 or Argonuates to repress reporter mRNAs (Chang et al., 2012; Loedige et al., 2013; Torres-Fernandez et al., 2019). Because these partners can inhibit translation or cause mRNA decay on their own, it is plausible that they function in parallel with TRIM71 to suppress specific targets. For example, TRIM71 collaborates with the Argonaute-containing RISC to promote embryonic stem cell proliferation (Chang et al., 2012). In this context, TRIM71 and microRNA miR285/302 repress the CDKN1A mRNA, which encodes the tumor suppressor protein p21, a cyclin dependent kinase inhibitor, thereby promoting proliferation (Chang et al., 2012; Torres-Fernandez et al., 2019).
Recent evidence indicates that TRIM71 also collaborates with components of the nonsense codon mediated mRNA decay (NMD) pathway to repress select mRNAs, in particular CDKN1A (Torres-Fernandez et al., 2019). Normally, NMD causes degradation of mutant or aberrant transcripts that contain a premature termination codon (Kurosaki et al., 2019). As in the case of CDKN1A mRNA, NMD factors can also act to degrade certain mRNAs that do not have a premature termination codon. TRIM71 interacts with NMD factors in a RNA-dependent manner, indicating that they likely co-occupy target mRNAs. The presence of TRIM71 binding sites is crucial for regulation of CDKN1A mRNA; however, the determinants that recruit NMD factors in this instance are not known (Torres-Fernandez et al., 2019). Further, only a subset of TRIM71 repressed target mRNAs appear to be affected by NMD factors. Thus, TRIM71 and NMD factors can collaborate to repress certain mRNAs, but not all of them.
In summary, these observations emphasize that TRIM-NHL proteins function in the context of a multitude of cis-acting elements and RBPs to regulate the fate of target mRNAs. Future research should focus on analyzing the biological impacts of these overlapping post-transcriptional regulatory networks.
4.2. Mechanism of mRNA repression
Since the discovery of their RNA-binding function, abundant research has shown TRIM-NHL proteins can alter mRNA translation and stability (discussed above). Yet comparatively little is known about how this repression occurs. Still, important clues are emerging in the literature. Generally, cellular mRNA levels are controlled by one or more mRNA decay pathways, such as the predominant deadenylation-dependent decay pathway (Garneau et al., 2007). Frequently mRNA decay is initiated by deadenylase enzymes that shorten the 3’ poly-adenosine tail of mRNAs (Goldstrohm & Wickens, 2008). This reduces translation and destabilizes the transcripts, leading to digestion by exoribonucleases. The CNOT deadenylase complex is a major determinant of poly(A) tail length (Goldstrohm & Wickens, 2008; Webster et al., 2018; H. Yi et al., 2018). In Drosophila, this complex consists of deadenylases Ccr4 and Pop2, and accessory proteins - all bound by the Not1 scaffold protein (Temme et al., 2014). Additionally, flies express the Pan2/Pan3 deadenylase complex, which contributes to shortening of long poly(A) tails (Goldstrohm & Wickens, 2008).
A growing paradigm in post-transcriptional regulation is that sequence specific RNA-binding proteins repress mRNAs by recruiting mRNA degradation enzymes (Goldstrohm & Wickens, 2008). Indeed, Brat was shown to physically associate with the Not1 component of the CNOT deadenylase complex (Temme et al., 2010). Functional evidence also supports the role of mRNA decay factors. In larval neuroblasts, reduction of subunits of the major deadenylase complexes not1, pop2, pan2, pan3, or RNA helicase mei31B enhances the brat mutant phenotype (Komori et al., 2018). Conversely, overexpression of catalytically-active Pan2 deadenylase partially rescues the brat phenotype. In the female germline, reduction of Pop2 or Ccr4 deadenylases phenocopies loss of brat (Newton et al., 2015). These observations provide physical and genetic evidence that Brat utilizes factors which deadenylate target transcripts, ultimately leading to mRNA decay.
Evidence for other repressive mechanisms also exists. For example, repression of hunchback translation by Brat in the early embryo involves 4EHP, a cap-binding translational repressor (Cho et al., 2006). In cultured cells; however, mutations that prevent the 4EHP interaction do not affect Brat-mediated repression of hunchback reporter mRNA (Loedige et al., 2014). Brat’s function in neural stem cells is also independent of 4EHP (Reichardt et al., 2018). Thus, the utilization of 4EHP may be cell-type specific. The molecular environment, which changes during development, may therefore determine which repressive mechanism is utilized.
TRIM71 likely recruits mRNA degradation factors to its target mRNAs. This is bolstered by the observation that TRIM71 in mouse ESCs reduces mRNA levels but not ribosome occupancy (Welte et al., 2019). At this time, no functionally relevant cofactors have been verified. Leodige (2013) reported that the 5’ exoribonuclease, XRN1, associates with TRIM71 but did not test its role in TRIM71 mediated repression. Several publications have reported that TRIM71 localizes to P-bodies, which are enriched with mRNA decay enzymes (Chang et al., 2012; Rybak et al., 2008; Torres-Fernandez et al., 2019). This localization suggests that TRIM71 may direct or sequester target mRNAs in P-bodies.
It is noteworthy that an early proposed function for TRIM71 in ESCs was to inhibit the RNAi interference pathway. This model was based on the observation that TRIM71 ubiquitinated Argonaute proteins AGO2 and AGO4 (Rybak et al., 2008). As this causes proteasomal degradation of AGO2, TRIM71 was therefore thought to reduce let-7 activity. Such a feedback loop would create a bistable switch between ESC proliferation and differentiation. While an attractive model, multiple followup studies were unable to corroborate TRIM71-mediated turnover of Argonaute (Chang et al., 2012; J. Chen et al., 2012; Loedige et al., 2013; Mitschka et al., 2015; Torres-Fernandez et al., 2019; Worringer et al., 2014). Multiple studies have confirmed the RNA-dependent association of TRIM71 with Argonaute; therefore, an alternative explanation is that both TRIM71 and Argonautes simply co-occupy transcripts (Chang et al., 2012; Loedige et al., 2013).
Both Brat and Mei-P26 have been found to associate with Ago1 through the NHL domain (Neumuller et al., 2008). For Mei-P26, this interaction is RNA-independent (Li et al., 2012). The consequences of these interactions, however, have not been determined. For example, Mei-P26 has been proposed to work in combination with Ago1 to repress orb (Li et al., 2012). On the other hand, mei-P26 overexpression was found to alleviate repression by the bantam miRNA (Neumuller et al., 2008).
4.3. A role for the RING domain?
The RING domain is a defining feature of the TRIM motif (Marin, 2012; Ozato et al., 2008; Reymond et al., 2001; Sardiello et al., 2008; Short & Cox, 2006). Several family members - TRIM71, TRIM2, TRIM3, and TRIM32 - have now been shown to be active ubiquitin ligases (Deshaies & Joazeiro, 2009; Loedige et al., 2013; Rybak et al., 2008). Recently, other RNA-binding E3 ubiquitin ligases have also been identified (Williams et al., 2019). It is therefore surprising that the RING domain is unnecessary for RNA-binding and mRNA repression in the few cases where it has been interrogated. Deletion of the TRIM71 RING domain does not alter its ability to repress mRNAs (Loedige et al., 2013). Transcriptome changes caused by TRIM71 loss in mouse ESCs are largely copied by introducing a single point mutation that prevents RNA-binding, whereas mutation of the RING domain has little effect (Welte et al., 2019). Additionally, several TRIM-NHL family members (Drosophila Brat and Wech, C. elegans LIN-41) lack functional RING domains altogether (Arama et al., 2000; Loer & Hoch, 2008; Tocchini & Ciosk, 2015).
While the function of the TRIM71 RING domain in gene regulation remains largely a mystery, it has been shown to ubiquitinate specific proteins. A recent search of proteins ubiquitinated by TRIM71 identified components of the p53 tumor suppressor pathway, among others (Nguyen et al., 2017). TRIM71 binds and ubiquitinates p53 on multiple sites, leading to proteasomal degradation. This is germane to TRIM71 and p53 having opposing effects in neural stem cells: TRIM71 promotes self-renewal whereas p53 limits it. TRIM71 also promotes neural stem cell proliferation by enhancing Fibroblast Growth Factor (FGF) signalling (J. Chen et al., 2012). Specifically, TRIM71 stabilizes SHCBP1, which enhances FGF signalling. This is dependent on the TRIM71 RING domain and SHCBP1 ubiquitination, though the molecular basis remains to be discovered.
In certain contexts, the function of the RING domain may even be independent of ubiquitin ligation. Worringer et al. (2014) showed the RING domain is important for TRIM71 to induce iPSCs. While deletion of the RING domain abrogated TRIM71-mediated reprogramming activity, mutating key active site cysteines did not.
In summary, TRIM71 may have two separate activities: one post-transcriptional and one post-translational. Or, perhaps these activities collaborate, suggested by the observation that RNA decay and translation factors were among the TRIM71-ubiquitinated proteins (Nguyen et al., 2017). To dissect the contributions of each function, genetic models should interrogate the loss of RNA-binding and ubiquitin ligase activities of TRIM71 relative to the null phenotypes. While this has been explored in cell culture, it is nontrivial to assess in model organisms.
4.4. Potential links between TRIM71 and cancer
The ability of TRIM71 to regulate cancer genes and pathways, including tumor suppressors CDKN1A/p21 and TP53/p53, suggests potential connections to cancer (Chang et al., 2012; Torres-Fernandez et al., 2019). Several reports suggest that TRIM71 may enhance the severity of hepatocellular carcinoma (Y. L. Chen et al., 2013; Torres-Fernandez et al., 2019). Further, data from the Cancer Genome Atlas (TCGA) indicates that TRIM71 is overexpressed in testicular cancer. This observation is intriguing because TRIM71 is abundant in the male germline - specifically in germline stem cells and to a lesser extent in other cell types (Rybak et al., 2008; The Human Protein Atlas). Further research is necessary to establish a role in the male germline.
5. Conclusions
Future research on TRIM-NHL proteins should focus on exploring the impact of RNA-binding activities in biologically relevant contexts, such as dissecting the contributions of RNA-binding, mRNA repression, mRNA localization, and protein ubiquitination. This includes characterizing the regulatory networks of each TRIM-NHL and identifying the mechanism(s) of gene regulation and crucial co-repressors. Genetic engineering of model organisms including Drosophila, C. elegans, and mice will be crucial in these endeavours, as these organisms offer extensive toolkits for tissue specific knockouts and rationally-designed mutations. This information will be necessary to understand how TRIM-NHLs contribute to normal development, and how dysfunction contributes to diseases like CH and cancer.
Acknowledgments
We thank members of the Goldstrohm lab for discussions of this topic. We also thank Dr. Christopher Tilmann for advice on mammalian germline and Drs. Katherine McKenney and Zachary Campbell for comments and critique of the work prior to submission. We sincerely apologize to colleagues whose work we were unable to include due to space limitations.
Funding Information
This work was supported by grant R01GM105707 from the National Institute of General Medical Sciences, National Institutes of Health (A.C.G.).
References
- Abidi SNF, & Smith-Bolton RK (2019). The gene brain tumor constrains growth to ensure proper patterning during regeneration in Drosophila imaginal discs. bioRxiv, 615948. doi: 10.1101/615948 [DOI] [Google Scholar]
- Aeschimann F, Kumari P, Bartake H, Gaidatzis D, Xu L, Ciosk R, & Grosshans H (2017). LIN41 Post-transcriptionally Silences mRNAs by Two Distinct and Position-Dependent Mechanisms. Mol Cell, 65(3), 476–489 e474. doi: 10.1016/j.molcel.2016.12.010 [DOI] [PubMed] [Google Scholar]
- Aeschimann F, Neagu A, Rausch M, & Grosshans H (2019). let-7 coordinates the transition to adulthood through a single primary and four secondary targets. Life Sci Alliance, 2(2). doi: 10.26508/lsa.201900335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arama E, Dickman D, Kimchie Z, Shearn A, & Lev Z (2000). Mutations in the beta-propeller domain of the Drosophila brain tumor (brat) protein induce neoplasm in the larval brain. Oncogene, 19(33), 3706–3716. doi: 10.1038/sj.onc.1203706 [DOI] [PubMed] [Google Scholar]
- Arbeille E, & Bashaw GJ (2018). Brain Tumor promotes axon growth across the midline through interactions with the microtubule stabilizing protein Apc2. PLoS Genet, 14(4), e1007314. doi: 10.1371/journal.pgen.1007314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arvola RM, Weidmann CA, Tanaka Hall TM, & Goldstrohm AC (2017). Combinatorial control of messenger RNAs by Pumilio, Nanos and Brain Tumor Proteins. RNA Biol, 1–12. doi: 10.1080/15476286.2017.1306168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltz AG, Munschauer M, Schwanhausser B, Vasile A, Murakawa Y, Schueler M, … Landthaler M (2012). The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol Cell, 46(5), 674–690. doi: 10.1016/j.molcel.2012.05.021 [DOI] [PubMed] [Google Scholar]
- Barr J, Gilmutdinov R, Wang L, Shidlovskii Y, & Schedl P (2019). The Drosophila CPEB Protein Orb Specifies Oocyte Fate by a 3’UTR-Dependent Autoregulatory Loop. Genetics, 213(4), 1431–1446. doi: 10.1534/genetics.119.302687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP (2018). Metazoan MicroRNAs. Cell, 173(1), 20–51. doi: 10.1016/j.cell.2018.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastock R, & St Johnston D (2008). Drosophila oogenesis. Curr Biol, 18(23), R1082–1087. doi: 10.1016/j.cub.2008.09.011 [DOI] [PubMed] [Google Scholar]
- Beaucher M, Goodliffe J, Hersperger E, Trunova S, Frydman H, & Shearn A (2007). Drosophila brain tumor metastases express both neuronal and glial cell type markers. Dev Biol, 301(1), 287–297. doi: 10.1016/j.ydbio.2006.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bello B, Reichert H, & Hirth F (2006). The brain tumor gene negatively regulates neural progenitor cell proliferation in the larval central brain of Drosophila. Development, 133(14), 2639–2648. doi: 10.1242/dev.02429 [DOI] [PubMed] [Google Scholar]
- Betschinger J, Mechtler K, & Knoblich JA (2006). Asymmetric segregation of the tumor suppressor brat regulates self-renewal in Drosophila neural stem cells. Cell, 124(6), 1241–1253. doi: 10.1016/j.cell.2006.01.038 [DOI] [PubMed] [Google Scholar]
- Bowman SK, Rolland V, Betschinger J, Kinsey KA, Emery G, & Knoblich JA (2008). The tumor suppressors Brat and Numb regulate transit-amplifying neuroblast lineages in Drosophila. Dev Cell, 14(4), 535–546. doi: 10.1016/j.devcel.2008.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell KJ, & White RJ (2014). MYC regulation of cell growth through control of transcription by RNA polymerases I and III. Cold Spring Harb Perspect Med, 4(5). doi: 10.1101/cshperspect.a018408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoso-Moreira M, Halbert J, Valloton D, Velten B, Chen C, Shao Y, … Kaessmann H (2019). Gene expression across mammalian organ development. Nature, 571(7766), 505–509. doi: 10.1038/s41586-019-1338-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castello A, Fischer B, Eichelbaum K, Horos R, Beckmann BM, Strein C, … Hentze MW (2012). Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell, 149(6), 1393–1406. doi: 10.1016/j.cell.2012.04.031 [DOI] [PubMed] [Google Scholar]
- Caussinus E, & Gonzalez C (2005). Induction of tumor growth by altered stem-cell asymmetric division in Drosophila melanogaster. Nat Genet, 37(10), 1125–1129. doi: 10.1038/ng1632 [DOI] [PubMed] [Google Scholar]
- Chang HM, Martinez NJ, Thornton JE, Hagan JP, Nguyen KD, & Gregory RI (2012). Trim71 cooperates with microRNAs to repress Cdkn1a expression and promote embryonic stem cell proliferation. Nat Commun, 3, 923. doi: 10.1038/ncomms1909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chau J, Kulnane LS, & Salz HK (2009). Sex-lethal facilitates the transition from germline stem cell to committed daughter cell in the Drosophila ovary. Genetics, 182(1), 121–132. doi: 10.1534/genetics.109.100693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Wu C, Zhao S, Geng Q, Gao Y, Li X, … Wang Z (2014). Three RNA binding proteins form a complex to promote differentiation of germline stem cell lineage in Drosophila. PLoS Genet, 10(11), e1004797. doi: 10.1371/journal.pgen.1004797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Lai F, & Niswander L (2012). The ubiquitin ligase mLin41 temporally promotes neural progenitor cell maintenance through FGF signaling. Genes Dev, 26(8), 803–815. doi: 10.1101/gad.187641.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YL, Yuan RH, Yang WC, Hsu HC, & Jeng YM (2013). The stem cell E3-ligase Lin-41 promotes liver cancer progression through inhibition of microRNA-mediated gene silencing. J Pathol, 229(3), 486–496. doi: 10.1002/path.4130 [DOI] [PubMed] [Google Scholar]
- Cho PF, Gamberi C, Cho-Park YA, Cho-Park IB, Lasko P, & Sonenberg N (2006). Cap-dependent translational inhibition establishes two opposing morphogen gradients in Drosophila embryos. Curr Biol, 16(20), 2035–2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller J, & Wickens M (2007). Tethered function assays: an adaptable approach to study RNA regulatory proteins. Methods Enzymol, 429, 299–321. doi: 10.1016/S0076-6879(07)29014-7 [DOI] [PubMed] [Google Scholar]
- Cuevas E, Rybak-Wolf A, Rohde AM, Nguyen DT, & Wulczyn FG (2015). Lin41/Trim71 is essential for mouse development and specifically expressed in postnatal ependymal cells of the brain. Front Cell Dev Biol, 3, 20. doi: 10.3389/fcell.2015.00020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies RJ, & Joazeiro CA (2009). RING domain E3 ubiquitin ligases. Annu Rev Biochem, 78, 399–434. doi: 10.1146/annurev.biochem.78.101807.093809 [DOI] [PubMed] [Google Scholar]
- Duy PQ, Furey CG, & Kahle KT (2019). Trim71/lin-41 Links an Ancient miRNA Pathway to Human Congenital Hydrocephalus. Trends Mol Med, 25(6), 467–469. doi: 10.1016/j.molmed.2019.03.004 [DOI] [PubMed] [Google Scholar]
- Ecsedi M, Rausch M, & Grosshans H (2015). The let-7 microRNA directs vulval development through a single target. Dev Cell, 32(3), 335–344. doi: 10.1016/j.devcel.2014.12.018 [DOI] [PubMed] [Google Scholar]
- Edwards TA, Wilkinson BD, Wharton RP, & Aggarwal AK (2003). Model of the brain tumor-Pumilio translation repressor complex. Genes Dev, 17(20), 2508–2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichorn N, Marton K, Schwartz RG, Melara RD, & Pirutinsky S (2016). Does Working Memory Enhance or Interfere With Speech Fluency in Adults Who Do and Do Not Stutter? Evidence From a Dual-Task Paradigm. J Speech Lang Hear Res, 59(3), 415–429. doi: 10.1044/2015_JSLHR-S-15-0249 [DOI] [PubMed] [Google Scholar]
- Ferreira A, Boulan L, Perez L, & Milan M (2014). Mei-P26 mediates tissue-specific responses to the Brat tumor suppressor and the dMyc proto-oncogene in Drosophila. Genetics, 198(1), 249–258. doi: 10.1534/genetics.114.167502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank DJ, Edgar BA, & Roth MB (2002). The Drosophila melanogaster gene brain tumor negatively regulates cell growth and ribosomal RNA synthesis. Development, 129(2), 399–407. [DOI] [PubMed] [Google Scholar]
- Frank DJ, & Roth MB (1998). ncl-1 is required for the regulation of cell size and ribosomal RNA synthesis in Caenorhabditis elegans. J Cell Biol, 140(6), 1321–1329. doi: 10.1083/jcb.140.6.1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu M, & Blackshear PJ (2017). RNA-binding proteins in immune regulation: a focus on CCCH zinc finger proteins. Nat Rev Immunol, 17(2), 130–143. doi: 10.1038/nri.2016.129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furey CG, Choi J, Jin SC, Zeng X, Timberlake AT, Nelson-Williams C, … Kahle KT (2018). De Novo Mutation in Genes Regulating Neural Stem Cell Fate in Human Congenital Hydrocephalus. Neuron, 99(2), 302–314 e304. doi: 10.1016/j.neuron.2018.06.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garneau NL, Wilusz J, & Wilusz CJ (2007). The highways and byways of mRNA decay. Nat Rev Mol Cell Biol, 8(2), 113–126. [DOI] [PubMed] [Google Scholar]
- Goldstrohm AC, & Wickens M (2008). Multifunctional deadenylase complexes diversify mRNA control. Nat Rev Mol Cell Biol, 9(4), 337–344. doi:nrm2370 [pii] 10.1038/nrm2370 [DOI] [PubMed] [Google Scholar]
- Haenfler JM, Kuang C, & Lee CY (2012). Cortical aPKC kinase activity distinguishes neural stem cells from progenitor cells by ensuring asymmetric segregation of Numb. Dev Biol, 365(1), 219–228. doi: 10.1016/j.ydbio.2012.02.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris RE, & Ashe HL (2011). Cease and desist: modulating short-range Dpp signalling in the stem-cell niche. EMBO Rep, 12(6), 519–526. doi: 10.1038/embor.2011.80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris RE, Pargett M, Sutcliffe C, Umulis D, & Ashe HL (2011). Brat promotes stem cell differentiation via control of a bistable switch that restricts BMP signaling. Dev Cell, 20(1), 72–83. doi: 10.1016/j.devcel.2010.11.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homem CC, & Knoblich JA (2012). Drosophila neuroblasts: a model for stem cell biology. Development, 139(23), 4297–4310. doi: 10.1242/dev.080515 [DOI] [PubMed] [Google Scholar]
- Insco ML, Bailey AS, Kim J, Olivares GH, Wapinski OL, Tam CH, & Fuller MT (2012). A self-limiting switch based on translational control regulates the transition from proliferation to differentiation in an adult stem cell lineage. Cell Stem Cell, 11(5), 689–700. doi: 10.1016/j.stem.2012.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens DH, & Lee CY (2014). It takes two to tango, a dance between the cells of origin and cancer stem cells in the Drosophila larval brain. Semin Cell Dev Biol, 28, 63–69. doi: 10.1016/j.semcdb.2014.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juschke C, Dohnal I, Pichler P, Harzer H, Swart R, Ammerer G, … Knoblich JA (2013). Transcriptome and proteome quantification of a tumor model provides novel insights into post-transcriptional gene regulation. Genome Biol, 14(11), r133. doi: 10.1186/gb-2013-14-11-r133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komori H, Golden KL, Kobayashi T, Kageyama R, & Lee CY (2018). Multilayered gene control drives timely exit from the stem cell state in uncommitted progenitors during Drosophila asymmetric neural stem cell division. Genes Dev, 32(23–24), 1550–1561. doi: 10.1101/gad.320333.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komori H, Xiao Q, McCartney BM, & Lee CY (2014). Brain tumor specifies intermediate progenitor cell identity by attenuating beta-catenin/Armadillo activity. Development, 141(1), 51–62. doi: 10.1242/dev.099382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari P, Aeschimann F, Gaidatzis D, Keusch JJ, Ghosh P, Neagu A, … Ciosk R (2018). Evolutionary plasticity of the NHL domain underlies distinct solutions to RNA recognition. Nat Commun, 9(1), 1549. doi: 10.1038/s41467-018-03920-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurosaki T, Popp MW, & Maquat LE (2019). Quality and quantity control of gene expression by nonsense-mediated mRNA decay. Nat Rev Mol Cell Biol, 20(7), 406–420. doi: 10.1038/s41580-019-0126-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurzik-Dumke U, Phannavong B, Gundacker D, & Gateff E (1992). Genetic, cytogenetic and developmental analysis of the Drosophila melanogaster tumor suppressor gene lethal(2)tumorous imaginal discs (1(2)tid). Differentiation, 51(2), 91–104. doi: 10.1111/j.1432-0436.1992.tb00685.x [DOI] [PubMed] [Google Scholar]
- Kwon SC, Yi H, Eichelbaum K, Fohr S, Fischer B, You KT, … Kim VN (2013). The RNA-binding protein repertoire of embryonic stem cells. Nat Struct Mol Biol, 20(9), 1122–1130. doi: 10.1038/nsmb.2638 [DOI] [PubMed] [Google Scholar]
- Laver JD, Li X, Ray D, Cook KB, Hahn NA, Nabeel-Shah S, … Smibert CA (2015). Brain tumor is a sequence-specific RNA-binding protein that directs maternal mRNA clearance during the Drosophila maternal-to-zygotic transition. Genome Biol, 16, 94. doi: 10.1186/s13059-015-0659-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laver JD, Marsolais AJ, Smibert CA, & Lipshitz HD (2015). Regulation and Function of Maternal Gene Products During the Maternal-to-Zygotic Transition in Drosophila. Curr Top Dev Biol, 113, 43–84. doi: 10.1016/bs.ctdb.2015.06.007 [DOI] [PubMed] [Google Scholar]
- Lee CY, Wilkinson BD, Siegrist SE, Wharton RP, & Doe CQ (2006). Brat is a Miranda cargo protein that promotes neuronal differentiation and inhibits neuroblast self-renewal. Dev Cell, 10(4), 441–449. doi: 10.1016/j.devcel.2006.01.017 [DOI] [PubMed] [Google Scholar]
- Li Y, Maines JZ, Tastan OY, McKearin DM, & Buszczak M (2012). Mei-P26 regulates the maintenance of ovarian germline stem cells by promoting BMP signaling. Development, 139(9), 1547–1556. doi: 10.1242/dev.077412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Minor NT, Park JK, McKearin DM, & Maines JZ (2009). Bam and Bgcn antagonize Nanos-dependent germ-line stem cell maintenance. Proc Natl Acad Sci U S A, 106(23), 9304–9309. doi: 10.1073/pnas.0901452106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zhang Q, Carreira-Rosario A, Maines JZ, McKearin DM, & Buszczak M (2013). Mei-p26 cooperates with Bam, Bgcn and Sxl to promote early germline development in the Drosophila ovary. PLoS ONE, 8(3), e58301. doi: 10.1371/journal.pone.0058301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loedige I, Gaidatzis D, Sack R, Meister G, & Filipowicz W (2013). The mammalian TRIM-NHL protein TRIM71/LIN-41 is a repressor of mRNA function. Nucleic Acids Res, 41(1), 518–532. doi: 10.1093/nar/gks1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loedige I, Jakob L, Treiber T, Ray D, Stotz M, Treiber N, … Meister G (2015). The Crystal Structure of the NHL Domain in Complex with RNA Reveals the Molecular Basis of Drosophila Brain-Tumor-Mediated Gene Regulation. Cell Rep, 13(6), 1206–1220. doi: 10.1016/j.celrep.2015.09.068 [DOI] [PubMed] [Google Scholar]
- Loedige I, Stotz M, Qamar S, Kramer K, Hennig J, Schubert T, … Meister G (2014). The NHL domain of BRAT is an RNA-binding domain that directly contacts the hunchback mRNA for regulation. Genes Dev, 28(7), 749–764. doi: 10.1101/gad.236513.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loer B, & Hoch M (2008). Wech proteins: roles in integrin functions and beyond. Cell Adh Migr, 2(3), 177–179. doi: 10.4161/cam.2.3.6579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loop T, Leemans R, Stiefel U, Hermida L, Egger B, Xie F, … Hirth F (2004). Transcriptional signature of an adult brain tumor in Drosophila. BMC Genomics, 5(1), 24. doi: 10.1186/1471-2164-5-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik S, Jang W, & Kim C (2017). Protein Interaction Mapping of Translational Regulators Affecting Expression of the Critical Stem Cell Factor Nos. Dev Reprod, 21(4), 449–456. doi: 10.12717/DR.2017.21.4.449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maller Schulman BR, Liang X, Stahlhut C, DelConte C, Stefani G, & Slack FJ (2008). The let-7 microRNA target gene, Mlin41/Trim71 is required for mouse embryonic survival and neural tube closure. Cell Cycle, 7(24), 3935–3942. doi: 10.4161/cc.7.24.7397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin I (2012). Origin and diversification of TRIM ubiquitin ligases. PLoS ONE, 7(11), e50030. doi: 10.1371/journal.pone.0050030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell SF, & Parker R (2014). Principles and properties of eukaryotic mRNPs. Mol Cell, 54(4), 547–558. doi: 10.1016/j.molcel.2014.04.033 [DOI] [PubMed] [Google Scholar]
- Mitschka S, Ulas T, Goller T, Schneider K, Egert A, Mertens J, … Kolanus W (2015). Co-existence of intact stemness and priming of neural differentiation programs in mES cells lacking Trim71. Sci Rep, 5, 11126. doi: 10.1038/srep11126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S, Tucker-Burden C, Zhang C, Moberg K, Read R, Hadjipanayis C, & Brat DJ (2016). Drosophila Brat and Human Ortholog TRIM3 Maintain Stem Cell Equilibrium and Suppress Brain Tumorigenesis by Attenuating Notch Nuclear Transport. Cancer Res, 76(8), 2443–2452. doi: 10.1158/0008-5472.CAN-15-2299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muraro NI, Weston AJ, Gerber AP, Luschnig S, Moffat KG, & Baines RA (2008). Pumilio binds para mRNA and requires Nanos and Brat to regulate sodium current in Drosophila motoneurons. J Neurosci, 28(9), 2099–2109. doi:28/9/2099 [pii] 10.1523/JNEUROSCI.5092-07.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumuller RA, Betschinger J, Fischer A, Bushati N, Poernbacher I, Mechtler K, … Knoblich JA (2008). Mei-P26 regulates microRNAs and cell growth in the Drosophila ovarian stem cell lineage. Nature, 454(7201), 241–245. doi: 10.1038/nature07014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumuller RA, Gross T, Samsonova AA, Vinayagam A, Buckner M, Founk K, … Perrimon N (2013). Conserved regulators of nucleolar size revealed by global phenotypic analyses. Sci Signal, 6(289), ra70. doi: 10.1126/scisignal.2004145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumuller RA, Richter C, Fischer A, Novatchkova M, Neumuller KG, & Knoblich JA (2011). Genome-wide analysis of self-renewal in Drosophila neural stem cells by transgenic RNAi. Cell Stem Cell, 8(5), 580–593. doi: 10.1016/j.stem.2011.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton FG, Harris RE, Sutcliffe C, & Ashe HL (2015). Coordinate post-transcriptional repression of Dpp-dependent transcription factors attenuates signal range during development. Development, 142(19), 3362–3373. doi: 10.1242/dev.123273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen DTT, Richter D, Michel G, Mitschka S, Kolanus W, Cuevas E, & Wulczyn FG (2017). The ubiquitin ligase LIN41/TRIM71 targets p53 to antagonize cell death and differentiation pathways during stem cell differentiation. Cell Death Differ, 24(6), 1063–1078. doi: 10.1038/cdd.2017.54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlstein B, & McKearin D (1997). Ectopic expression of the Drosophila Bam protein eliminates oogenic germline stem cells. Development, 124(18), 3651–3662. [DOI] [PubMed] [Google Scholar]
- Oskarsson T, & Trumpp A (2005). The Myc trilogy: lord of RNA polymerases. Nat Cell Biol, 7(3), 215–217. doi: 10.1038/ncb0305-215 [DOI] [PubMed] [Google Scholar]
- Ozato K, Shin DM, Chang TH, & Morse HC 3rd. (2008). TRIM family proteins and their emerging roles in innate immunity. Nat Rev Immunol, 8(11), 849–860. doi: 10.1038/nri2413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page SL, McKim KS, Deneen B, Van Hook TL, & Hawley RS (2000). Genetic studies of mei-P26 reveal a link between the processes that control germ cell proliferation in both sexes and those that control meiotic exchange in Drosophila. Genetics, 155(4), 1757–1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichardt I, Bonnay F, Steinmann V, Loedige I, Burkard TR, Meister G, & Knoblich JA (2018). The tumor suppressor Brat controls neuronal stem cell lineages by inhibiting Deadpan and Zelda. EMBO Rep, 19(1), 102–117. doi: 10.15252/embr.201744188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond A, Meroni G, Fantozzi A, Merla G, Cairo S, Luzi L, … Ballabio A (2001). The tripartite motif family identifies cell compartments. Embo J, 20(9), 2140–2151. doi: 10.1093/emboj/20.9.2140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybak A, Fuchs H, Smirnova L, Brandt C, Pohl EE, Nitsch R, & Wulczyn FG (2008). A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat Cell Biol, 10(8), 987–993. doi: 10.1038/ncb1759 [DOI] [PubMed] [Google Scholar]
- San-Juan BP, & Baonza A (2011). The bHLH factor deadpan is a direct target of Notch signaling and regulates neuroblast self-renewal in Drosophila. Dev Biol, 352(1), 70–82. doi: 10.1016/j.ydbio.2011.01.019 [DOI] [PubMed] [Google Scholar]
- Sardiello M, Cairo S, Fontanella B, Ballabio A, & Meroni G (2008). Genomic analysis of the TRIM family reveals two groups of genes with distinct evolutionary properties. BMC Evol Biol, 8, 225. doi: 10.1186/1471-2148-8-225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulman BR, Esquela-Kerscher A, & Slack FJ (2005). Reciprocal expression of lin-41 and the microRNAs let-7 and mir-125 during mouse embryogenesis. Dev Dyn, 234(4), 1046–1054. doi: 10.1002/dvdy.20599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekelsky JJ, McKim KS, Messina L, French RL, Hurley WD, Arbel T, … Hawley RS (1999). Identification of novel Drosophila meiotic genes recovered in a P-element screen. Genetics, 152(2), 529–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sgromo A, Raisch T, Backhaus C, Keskeny C, Alva V, Weichenrieder O, & Izaurralde E (2018). Drosophila Bag-of-marbles directly interacts with the CAF40 subunit of the CCR4-NOT complex to elicit repression of mRNA targets. Rna, 24(3), 381–395. doi: 10.1261/rna.064584.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sgromo A, Raisch T, Bawankar P, Bhandari D, Chen Y, Kuzuoglu-Ozturk D, … Izaurralde E (2017). A CAF40-binding motif facilitates recruitment of the CCR4-NOT complex to mRNAs targeted by Drosophila Roquin. Nat Commun, 8, 14307. doi: 10.1038/ncomms14307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Short KM, & Cox TC (2006). Subclassification of the RBCC/TRIM superfamily reveals a novel motif necessary for microtubule binding. J Biol Chem, 281(13), 8970–8980. doi: 10.1074/jbc.M512755200 [DOI] [PubMed] [Google Scholar]
- Slack FJ, & Ruvkun G (1998). A novel repeat domain that is often associated with RING finger and B-box motifs. Trends in Biochemical Sciences, 23(12), 474–475. doi: 10.1016/S0968-0004(98)01299-7 [DOI] [PubMed] [Google Scholar]
- Slaidina M, & Lehmann R (2014). Translational control in germline stem cell development. J Cell Biol, 207(1), 13–21. doi: 10.1083/jcb.201407102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, & Lu B (2011). Regulation of cell growth by Notch signaling and its differential requirement in normal vs. tumor-forming stem cells in Drosophila. Genes Dev, 25(24), 2644–2658. doi: 10.1101/gad.171959.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda J, & Wharton RP (2001). Drosophila Brain Tumor is a translational repressor. Genes Dev, 15(6), 762–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling AC, Stern D, Beaton A, Rhem EJ, Laverty T, Mozden N, … Rubin GM (1999). The Berkeley Drosophila Genome Project gene disruption project: Single P-element insertions mutating 25% of vital Drosophila genes. Genetics, 153(1), 135–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stathakis DG, Pentz ES, Freeman ME, Kullman J, Hankins GR, Pearlson NJ, & Wright TR (1995). The genetic and molecular organization of the Dopa decarboxylase gene cluster of Drosophila melanogaster. Genetics, 141(2), 629–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subtelny AO, Eichhorn SW, Chen GR, Sive H, & Bartel DP (2014). Poly(A)-tail profiling reveals an embryonic switch in translational control. Nature, 508(7494), 66–71. doi: 10.1038/nature13007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadros W, & Lipshitz HD (2009). The maternal-to-zygotic transition: a play in two acts. Development, 136(18), 3033–3042. doi: 10.1242/dev.033183 [DOI] [PubMed] [Google Scholar]
- Temme C, Simonelig M, & Wahle E (2014). Deadenylation of mRNA by the CCR4-NOT complex in Drosophila: molecular and developmental aspects. Front Genet, 5, 143. doi: 10.3389/fgene.2014.00143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temme C, Zaessinger S, Meyer S, Simonelig M, & Wahle E (2004). A complex containing the CCR4 and CAF1 proteins is involved in mRNA deadenylation in Drosophila. Embo J, 23(14), 2862–2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temme C, Zhang L, Kremmer E, Ihling C, Chartier A, Sinz A, … Wahle E (2010). Subunits of the Drosophila CCR4-NOT complex and their roles in mRNA deadenylation. Rna, 16(7), 1356–1370. doi:rna.2145110 [pii] 10.1261/rna.2145110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas. TCGA Research Network: https://www.cancer.gov/tcga.
- The Human Protein Atlas. TRIM71/tissue/testis, available from v19.3.proteinatlas.org <https://www.proteinatlas.org/ENSG00000206557-TRIM71/tissue/testis>.
- Tocchini C, & Ciosk R (2015). TRIM-NHL proteins in development and disease. Semin Cell Dev Biol, 47–48, 52–59. doi: 10.1016/j.semcdb.2015.10.017 [DOI] [PubMed] [Google Scholar]
- Torres-Fernandez LA, Jux B, Bille M, Port Y, Schneider K, Geyer M, … Kolanus W (2019). The mRNA repressor TRIM71 cooperates with Nonsense-Mediated Decay factors to destabilize the mRNA of CDKN1A/p21. Nucleic Acids Res, 47(22), 11861–11879. doi: 10.1093/nar/gkz1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trendel J, Schwarzl T, Horos R, Prakash A, Bateman A, Hentze MW, & Krijgsveld J (2019). The Human RNA-Binding Proteome and Its Dynamics during Translational Arrest. Cell, 176(1–2), 391–403 e319. doi: 10.1016/j.cell.2018.11.004 [DOI] [PubMed] [Google Scholar]
- Tsukamoto T, Gearhart MD, Spike CA, Huelgas-Morales G, Mews M, Boag PR, … Greenstein D (2017). LIN-41 and OMA Ribonucleoprotein Complexes Mediate a Translational Repression-to-Activation Switch Controlling Oocyte Meiotic Maturation and the Oocyte-to-Embryo Transition in Caenorhabditis elegans. Genetics, 206(4), 2007–2039. doi: 10.1534/genetics.117.203174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vastenhouw NL, Cao WX, & Lipshitz HD (2019). The maternal-to-zygotic transition revisited. Development, 146(11). doi: 10.1242/dev.161471 [DOI] [PubMed] [Google Scholar]
- Wang H, Somers GW, Bashirullah A, Heberlein U, Yu F, & Chia W (2006). Aurora-A acts as a tumor suppressor and regulates self-renewal of Drosophila neuroblasts. Genes Dev, 20(24), 3453–3463. doi: 10.1101/gad.1487506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster MW, Chen YH, Stowell JAW, Alhusaini N, Sweet T, Graveley BR, … Passmore LA (2018). mRNA Deadenylation Is Coupled to Translation Rates by the Differential Activities of Ccr4-Not Nucleases. Mol Cell, 70(6), 1089–1100 e1088. doi: 10.1016/j.molcel.2018.05.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welte T, Tuck AC, Papasaikas P, Carl SH, Flemr M, Knuckles P, … Grosshans H (2019). The RNA hairpin binder TRIM71 modulates alternative splicing by repressing MBNL1. Genes Dev, 33(17–18), 1221–1235. doi: 10.1101/gad.328492.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams FP, Haubrich K, Perez-Borrajero C, & Hennig J (2019). Emerging RNA-binding roles in the TRIM family of ubiquitin ligases. Biol Chem, 400(11), 1443–1464. doi: 10.1515/hsz-2019-0158 [DOI] [PubMed] [Google Scholar]
- Wissel S, Harzer H, Bonnay F, Burkard TR, Neumuller RA, & Knoblich JA (2018). Time-resolved transcriptomics in neural stem cells identifies a v-ATPase/Notch regulatory loop. J Cell Biol, 217(9), 3285–3300. doi: 10.1083/jcb.201711167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodhouse E, Hersperger E, & Shearn A (1998). Growth, metastasis, and invasiveness of Drosophila tumors caused by mutations in specific tumor suppressor genes. Dev Genes Evol, 207(8), 542–550. [DOI] [PubMed] [Google Scholar]
- Worringer KA, Rand TA, Hayashi Y, Sami S, Takahashi K, Tanabe K, … Yamanaka S (2014). The let-7/LIN-41 pathway regulates reprogramming to human induced pluripotent stem cells by controlling expression of prodifferentiation genes. Cell Stem Cell, 14(1), 40–52. doi: 10.1016/j.stem.2013.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wreden C, Verrotti AC, Schisa JA, Lieberfarb ME, & Strickland S (1997). Nanos and pumilio establish embryonic polarity in Drosophila by promoting posterior deadenylation of hunchback mRNA. Development, 124(15), 3015–3023. [DOI] [PubMed] [Google Scholar]
- Wright TR, Beermann W, Marsh JL, Bishop CP, Steward R, Black BC, … Wright EY (1981). The genetics of dopa decarboxylase in Drosophila melanogaster. IV. The genetics and cytology of the 37B10–37D1 region. Chromosoma, 83(1), 45–58. doi: 10.1007/bf00286015 [DOI] [PubMed] [Google Scholar]
- Wright TR, Hodgetts RB, & Sherald AF (1976). The genetics of dopa decarboxylase in Drosophila melanogaster. I. Isolation and characterization of deficiencies that delete the dopa-decarboxylase-dosage-sensitive region and the alpha-methyl-dopa-hypersensitive locus. Genetics, 84(2), 267–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Q, Komori H, & Lee CY (2012). klumpfuss distinguishes stem cells from progenitor cells during asymmetric neuroblast division. Development, 139(15), 2670–2680. doi: 10.1242/dev.081687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, & Min J (2011). Structure and function of WD40 domain proteins. Protein Cell, 2(3), 202–214. doi: 10.1007/s13238-011-1018-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi H, Park J, Ha M, Lim J, Chang H, & Kim VN (2018). PABP Cooperates with the CCR4-NOT Complex to Promote mRNA Deadenylation and Block Precocious Decay. Mol Cell, 70(6), 1081–1088 e1085. doi: 10.1016/j.molcel.2018.05.009 [DOI] [PubMed] [Google Scholar]
- Yi YH, Ma TH, Lee LW, Chiou PT, Chen PH, Lee CM, … Lo SJ (2015). A Genetic Cascade of let-7-ncl-1-fib-1 Modulates Nucleolar Size and rRNA Pool in Caenorhabditis elegans. PLoS Genet, 11(10), e1005580. doi: 10.1371/journal.pgen.1005580 [DOI] [PMC free article] [PubMed] [Google Scholar]


