Abstract
There has been rapid growth in the use of transcriptomic analyses to study the interplay between gene expression and behavior. Experience can modify gene expression in the brain, leading to changes in internal state and behavioral displays, while gene expression variation between species is thought to specify many innate behavior differences. However, providing a causal association between a gene and a given behavior remains challenging as it is difficult to determine when and where a gene contributes to the function of a behaviourally-relevant neuronal population. Moreover, given that there are fewer genetic tools available for non-traditional model organisms, transcriptomic approaches have been largely limited to profiling of bulk tissue, which can obscure the contributions of subcortical brain regions implicated in multiple behaviors. Here, we discuss how emerging single cell technologies combined with methods offering additional spatial and connectivity information can give us insight about the genetic profile of specific cells involved in the neural circuit of target social behaviors. We also emphasize how these techniques are broadly adaptable to non-traditional model organisms. We propose that, ultimately, a combination of these approaches applied throughout development will be key to discerning how genes shape the formation of social behavior circuits.
Identification of genes associated with behavior
As described by Robinson, Fernald, and Clayton, two principal lines of investigation in the field of sociogenomics are 1) understanding how social experiences modify gene expression to produce lasting effects on brain function and 2) understanding how information encoded in the genome leads to a unique repertoire of social behaviors (Robinson, Fernald, and Clayton 2008). A central challenge in addressing these questions is identifying neuronal populations in which expression of a given candidate gene shows some kind of causality for or effect from the behavior of interest. Moreover, most genetic gain- or loss-of function experiments, have implicated brain regions in behavior rather than genetically-defined cell types, leading to studies focusing on gene expression in bulk tissue dissections. This has been the case for quantitative trait locus (QTL) analyses, which have revealed many loci that associate with a behavioral trait, but cannot address how specific neural substrates mediate such traits. Given that brain regions are highly heterogeneous, it is likely that only a subset of cells in a given region are uniquely contributing to the behavior of interest, and the differential gene expression signal from these cells may be lost in the noise from their neighbors, making it difficult to identify genes involved in specific behaviors.
An example of this idea was shown in the mouse arcuate–median eminence complex, where fasting behavior was shown to significantly affect gene expression in subsets of pro-opiomelanocortin (POMC)-expressing neurons and Agouti-related protein (AGRP)-expressing neurons, contrary to previous bulk tissue analyses that had not reported these changes as significant (Campbell et al. 2017). Indeed, characterizing specific cell types involved in target behaviors within heterogeneous brain regions would be the first step to identifying gene candidates associated with the organization and display of social behaviors.
In this review, we will discuss recent technical advances that can be applied to study neuronal populations associated with specific neural pathways or behaviors, schematized in Figure 1. These include methods to capture active neuronal populations and gene expression programs that are active during or as a consequence of specific behaviors, as well as approaches that can help characterize the development of specific cell types and their connectivity patterns. Notably, many of these approaches do not require germline genetic modification, making them suitable for a variety of interesting species with diverse behavioral repertoires. We describe isolation of RNA from acutely activated neurons, projection-specific RNA-seq, and single-cell methodologies that provide cellular and spatial resolution. We conclude by suggesting that the field consider a developmental approach to understanding species differences in behavior, based on the hypothesis that the neural pathways that drive such differences are specified in early life. That is to say, in order to fully understand how social behavior neural circuits function, gene programs dictating the organization and activation of such neural pathways must be equally considered, as articulated by Tinbergen’s questions about ontogeny and mechanism for understanding animal behavior (Tinbergen 2010; Bateson and Laland 2013).
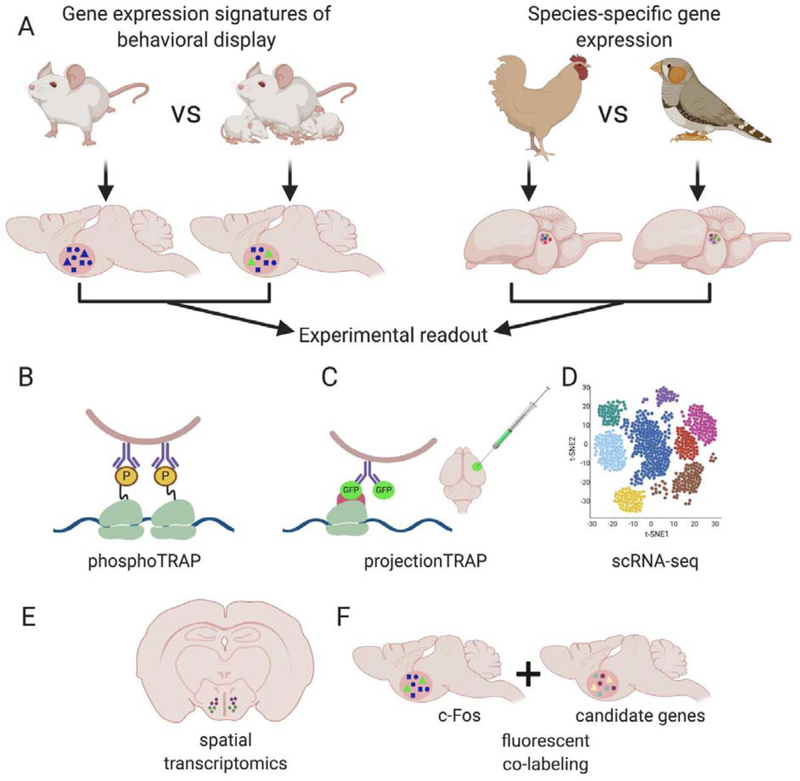
Figure 1. Strategies for pinpointing behaviorally-relevant genes.
(A) Schematic of behavioral paradigms that can be used to discern the genetic and molecular signatures of social behaviors. On the left, a mouse is shown with pups and no pups. Since it is known that the medial preoptic area (mPOA) is involved in parental behavior in mice, eliciting this behavioral response can be used to find cells in the mPOA that participate in the neural circuit involved in parental behavior display. This in turn would allow assessment of gene expression changes in specific cells that are relevant to parenting behavior. On the right, two species of birds with innate differences in behavior are shown; a vocal-learner songbird and a non-vocal learner bird (chicken). In this case, it is known that the medial portion of the dorsolateral thalamus (DLM) is one of the regions involved in vocal learning in birds (Goldberg and Fee 2011). Comparing both species would help identify key differences in the genetic profile of cells in the DLM of vocal learners and non-vocal learners. It could be the case, for example, that there are different quantities of a certain cell type in one species, similar cell types with different connectivity patterns, and/or specific genes with differential gene expression patterns.
(B-F) A summary of molecular approaches that can be employed to study social behaviors are shown.
(B) Phosphorylated Translating Ribosome Affinity Purification (phosphoTRAP) can be used to select for transcripts of genes being expressed during a behavioral response.
(C) ProjectionTRAP can be employed to capture transcripts of cells with specific projection patterns within a neural circuit, which is ideal for comparing projection pattern differences between species.
(D) Single cell RNA-seq (scRNA-seq) can be used in combination with TRAP methods to genetically profile relevant cells to a social behavior’s neural circuit.
(E,F) Information obtained from scRNA-seq can be further used for spatial transcriptomics
(E) and candidate gene co-labeling with c-Fos (F), to obtain an integral understanding of relevant genes being expressed in their native context. Figure created with BioRender.com.
Logical insights from flies and worms
Associating genes with behavior necessitates identification of the anatomic location and developmental time in which a gene product acts in the brain to facilitate said behavior. Previous studies using chemo- and opto-genetic approaches have shown necessity and sufficiency of genetically-defined cell populations in the display of behaviors. Testing the requirement for an individual gene in a distinct population of neurons has only recently become possible with the advent of a CRISPR activation or inhibition (CRISPRa/i) approach. CRISPRa uses a single guide RNA to direct a nuclease-deficient Cas9 (dCas9) fused with a transcriptional activator to regulatory elements of a gene in order to increase its expression, and has been useful to confirm the role of specific gene candidates in behavioral circuits (Matharu et al. 2019; Carpenter et al. 2020).
Work in invertebrate model organisms has historically had an advantage in providing this connection due to the relative ease of genetic labeling to fluorescently tag neurons that express a particular gene. As in the field of behavioral neuroendocrinology, reproductive behaviors provide a robust readout for experimental manipulation in Drosophila; males engage in a stereotyped, yet complex, courtship routine that plays a crucial role in species-recognition (Greenspan and Ferveur 2000). This sexual behavior is thought to have been recruited by sexual selection operating on pre-existing neuronal pathways, thus leading to similar courtship behavior across Drosophila species. Most initial studies looking into the molecular and genetic basis of courtship behavior in this genus, however, were done in Drosophila melanogaster. For example, the fruitless gene (fru) was first identified as having a role in Drosophila melanogaster male courtship behaviors using linkage mapping (Gailey and Hall 1989). With further studies, it was found that fru yielded sex-specific splice variants, with the male variant producing transcription factor FruM, which is required for male-specific courtship behavior (Baker, Taylor, and Hall 2001). Genetic tagging of the fru locus allowed visualization of Fru-expressing cells, and revealed neural pathways relevant to courtship, including sex dimorphisms (Demir and Dickson 2005; Dornan, Gailey, and Goodwin 2005; Kimura et al. 2005; Manoli et al. 2005) and, more recently, species differences between Drosophila melanogaster and Drosophila simulans (Seeholzer et al. 2018).
Caenorhabditis elegans is also an ideal organism in which to study behavioral genetics due to the tractable size of its nervous system, ease of genetic approaches, and availability of diverse naturally-occurring strains. de Bono and Bargmann leveraged these qualities to characterize the genetics of one of the most complex behaviors in this species: aggregation around limited food sources, which closely resembles social activity (de Bono and Bargmann 1998). They found that a natural polymorphism in the gene for the neuropeptide receptor npr-1 confers sociality across multiple strains. Importantly, insertion of green fluorescent protein (GFP) into the npr-1 locus revealed that this gene is expressed in a discrete subset of neurons. These findings demonstrate the function of a neuropeptide receptor in aggregation, a behavior that often underlies population dynamics across taxa.
Another neuropeptide receptor that regulates complex social-like behaviors in C. elegans is NTR-1, which binds nematocin, the worm analog of oxytocin/vasopressin (OT/AVP) (Beets et al. 2012). GFP labeling of NTR-1 and nematocin-expressing cells, allowed Beets et al to determine the role of this signaling pathway in a specific learned behavior: chemotaxis to salt. Studying nematocin, although seemingly not related to sociality at first, does demonstrate the role of homologous neuropeptides in sensory processing throughout evolution, which later allowed more complex social behaviors to arise. For example, AVP has been implicated in parental behaviors in certain frog and Peromyscus mice species, and the receptor for AVP, V1aR, has been found to have a role in pair bonding in prairie voles (Fischer et al. 2019; Bester-Meredith and Marler 2012; Bendesky et al. 2017; Lim et al. 2004). Additionally, neuromodulators have been suggested to have an extensive role in controlling the development of behavioral circuits across species by modulating the synaptic strength and excitability of neurons, resulting in the activation or repression of circuit states (Tosches 2017).
The logic of these approaches in flies and worms can be applied to vertebrate systems to achieve a similar level of circuit-specificity via selective transcriptomic profiling of behavior-associated neuronal populations. However, similar advances in vertebrates have been restricted to lab mice as genetic labelling hasn’t been extensively developed for other model species. Nonetheless, there are alternative approaches that can be used to initially capture relevant cell populations across species.
Capturing active neuronal populations
One way to identify neuronal populations potentially associated with behaviors is to look for cells that become active during a sensory experience or behavioral display. Researchers have long used neural activity regulated immediate early genes (IEG) such as FBJ osteosarcoma oncogene (Fos), activity regulated cytoskeletal-associated protein (Arc), and early growth response 1 (Egr1) to identify brain regions that are active during specific behaviors. Cellular resolution can be obtained by co-labeling with individual cell-type markers using immunostaining or fluorescent ISH (FISH). The cellular compartment analysis of temporal activity by FISH (catFISH) assay takes advantage of the kinetics of IEG transcription and splicing to determine if the same cells are active while the animal is performing two sequential behaviors (Guzowski et al. 1999). One RNA probe is for the immature nuclear transcript, while another labels the mature, cytoplasmic transcript, allowing for distinct labeling of both cellular compartments with an approximately 30 minute separation between stimuli. Hashikawa et al employed cFos catFISH to demonstrate that cells involved in mating and aggression in the ventromedial hypothalamus show distinct topographical organization (Hashikawa et al. 2017). Similarly, the Xu lab found that males exposed to females or pups activated largely separate populations of neurons in the medial pre-optic area (mPOA) (Wei et al. 2018), a region implicated in multiple innate behaviors including parental care, mating, temperature regulation, and sleep. Thus, IEGs are a valuable tool for obtaining population specificity within heterogeneous subcortical brain regions (See Fig. 1A).
In 2012, the phosphoTRAP (TRAP: translating ribosome affinity purification) method was developed to obtain the transcriptome directly from activated neurons, making it possible to determine the identity of neurons responding to distinct stimuli (see Fig. 1B). PhosphoTRAP takes advantage of ribosomal protein S6 (RPS6) phosphorylation downstream of neural activation in response to multiple signaling cascades including protein kinase A (PKA), mitogen-activated protein kinase (MAPK), and Phosphoinositide 3-kinase/mechanistic target of rapamycin kinase (PI3K/MTOR) (Knight et al. 2012). Phosphorylated ribosomes are immunoprecipitated, enriching for mRNA selectively translating in active neurons, which is then sequenced and characterized. The sequenced genes are not specific to activity-regulated transcripts, but rather capture all translating transcripts within an active cell. The Knight lab has, for example, applied this approach to the POA, identifying a subset of neurons that respond to ambient temperature changes (Tan et al. 2016). However, there are certain considerations to keep in mind when working with data generated with this method. Baran and Streelman discuss these, emphasizing that PhosphoTRAP has a lower gene detection rate when compared to traditional RNA-seq, possibly due to a low RNA concentration or because those genes were not expressed in cells active during the behavior of interest (Baran and Streelman 2020). They discuss that there is also a tendency for higher variance in PhosphoTRAP compared to RNA-seq data, likely because of PCR amplification bias or because the genes are mostly expressed in active neurons. This reflects on the statistical analysis of PhosphoTRAP data, given that the ratio of fold-enrichment value for each gene is calculated as the immunoprecipitated (IP) sample read count divided by the input sample read count (which is obtained via traditional RNA-seq). Thus, Baran and Streelman suggest performing the data analyses considering differences in batch, species, condition and sample type (IP vs input).
Notably, phosphoTRAP can be applied to non-Mus musculus species, as only the antibody to the conserved pS6 residue is required to achieve specificity. As an example, phosphoTRAP was used to study the molecular basis of parental behaviors in frogs (Fischer et al. 2019). Three species of frogs known to display either male uniparental (Dendrobates tinctorius), female uniparental (Oophaga sylvatica) or biparental behaviors (Ranitomeya imitator) were compared in order to identify sex and species independent brain regions involved in tadpole transport. By quantifying the number of RPS6+ cells, neural activity increases during tadpole transport were found in the POA and medial pallium (Mp) across all frog species, implicating these brain regions in the behavioral display. Transcripts with significant expression changes during tadpole transport were also identified, with a special focus on active neurons in the POA and Mp. Moreover, co-labelling of RPS6 and galanin revealed an increase of galanin neurons quantity and activity exclusively in the biparental frog species, which is also monogamous, as opposed to the uniparental species. Similarly, phosphoTRAP and RPS6 immunostaining have been used for comparing neural activity in two species of cichlid fish with differences in aggressive behaviors (Baran and Streelman 2020). Although this study used whole brain samples for the phosphoTRAP experiments, it revealed genes with selectively enriched expression in an aggressive species following mirror-elicited aggression, including neuropeptide receptor Y (Npy) and insulin receptor. Thus, approaches that isolate neurons active during a behavioral response are available to improve our understanding of the gene expression of neuronal populations associated with behavior in diverse species. Moreover, in vertebrates, the aforementioned approaches can be combined with methods that allow us to characterize the transcriptomic profile of cells based on their connectivity patterns. This can be highly informative because cells active during a behavior could project to different brain regions, and looking at their specific transcriptomic profiles can indicate how these connections are formed and maintained.
Projection-specific transcriptomics
Another way to isolate cells participating in an individual behavior is by considering their connectivity patterns. That is to say, when the brain regions and neuronal populations active during a behavior are known, they can then go on to be profiled based on their projection targets. Species and sex differences in behavior may result from cells receiving different inputs by having formed different neuronal connections throughout development. In Drosophila, differences in weighting of excitatory and inhibitory inputs to courtship-promoting neurons cause the same female pheromonal cues to have different meaning to males of different species (Seeholzer et al. 2018). Likewise, differences in distribution of AVP receptors throughout various developmental stages and adulthood have been found between monogamous and promiscuous vole species, suggesting a role for AVP receptors in modulating the development of the pair bonding circuitry in these species (Lim et al. 2004; Z. Wang et al. 1996). There are many other examples of species-specific neuronal circuitry underlying differences in social behavior well summarized in previous reviews (P. S. Katz and Harris-Warrick 1999; Paul S. Katz 2011; Godwin and Thompson 2012).
Projection-specific viral TRAP allows the systematic profiling of presynaptic neurons based on their projection target (See Fig. 1C). It consists of having an anti-GFP nanobody fused to a ribosomal protein, which can then bind GFP intracellularly. The ribosomes are then captured by immunoprecipitation. Injection of a GFP-expressing retrograde virus allows for enrichment of RNA specifically expressed in cell bodies of neurons that project to the targeted region (Ekstrand et al. 2014). The initial version of this method was used to profile dopaminergic cells of the ventral tegmental area (VTA) that specifically project to the nucleus accumbens (NAc). More recently Cre-dependent viral TRAP has identified markers for cells within the prelimbic cortex that uniquely project to the amygdala, VTA, or NAc (Murugan et al. 2017). Although this approach has only been published in Mus musculus, it could be adapted for use in other rodents in which viral approaches have been developed, e.g. prairie voles (Scribner et al. 2020), Peromyscus californicus (Duque-Wilckens et al. 2020), rhesus macaques (Stauffer et al. 2016), zebrafish (Tabor et al. 2019), and zebra finches (Hisey, Kearney, and Mooney 2018; Zhao et al. 2019; Duque-Wilckens et al. 2020), which will permit comparative transcriptomic analyses of the same functional projection in multiple species.
Comparing gene expression across species
One approach that has been employed to study how candidate genes affect social behaviors in non-traditional model species, is to compare gene sequence variation amongst members of the same species, to assess how these differences contribute to varying degrees of behavioral display. For example, sequence variation in potential regulatory regions for OT and AVP receptors resulting in varying degrees of gene expression in specific brain regions, were linked to differences in social attachment and sexual fidelity in prairie voles (Okhovat et al. 2015; King et al. 2016). This approach has been useful to begin understanding how gene variation in outbred species affects behavioral display. Several groups have also performed RNA-seq in multiple species to find the molecular basis of differing social behaviors, which has yielded clues as to potential behavior-modulating genes. Recent papers of note in vertebrates include (Pfenning et al. 2014; Bendesky et al. 2017; York et al. 2018; Young et al. 2019). Bendesky et al compared two species of Peromyscus, P. maniculatus and P. polionotus, to find genetic variants associated with paternal behavior and nest building. Through quantitative trait locus (QTL) analysis and transcriptomics, they found that increased levels of AVP gene expression is linked with less nest building in P. maniculatus. Pharmacological studies involving broad intracerebroventricular administration of AVP into P. polionotus inhibited nest building. Previous work in another Peromyscus species, however, reported that higher levels of AVP in the bed nucleus of the stria terminalis (BNST) of biparental P. californicus females correlate with more nest building (Bester-Meredith and Marler 2012). These results highlight the importance of assessing the location and identity of cells expressing a gene candidate, such as AVP, to further identify its role in a target behavior. Moreover, determining whether the quantity of AVP receptors and the cell types expressing them vary across species, or if cells expressing the AVP receptor receive different inputs will be possible by employing approaches that allow single cell resolution.
A prominent example of comparative transcriptomics can also be seen in the study of vocal learning in birds. Pfenning et al. sought to make specific brain transcriptome comparisons, initially sampling from vocal-learner birds, vocal non-learner birds, humans, and macaques followed by RNA-seq on matched laser-dissected brain regions from motor cortex and striatum. Regions of interest included striatal Area X, the robust nucleus arcopallium (RA), and the lateral magnocellular nucleus of the nidopallium (LMAN) in birds, the laryngeal motor cortex (LMC) in humans, and primary motor cortex in primates. After candidate genes were identified, those with the most notable differential expression between vocal learners and vocal non-learners were chosen for ISH confirmation. One major finding was that Slit1 was consistently downregulated in the RA of vocal learning birds, while the gene for its regulator, the transcription factor FOXP2, was downregulated in the LMC of humans, potentially both having a role in vocal learning. Slit1 was also found to be downregulated in the primary cortex of rhesus macaques, but neurons from the macaque ventral premotor region, which is hypothesized to be a precursor of LMC, and which indirectly projects to vocal motor neurons in the brainstem, showed no downregulation of SLIT1. Conversely, parrots, which also have an RA analog with neurons from the RA core projecting to vocal motor neurons, showed Slit1 downregulation in this particular RA core portion. In fact, Slit1 encodes for an axon guidance ligand, and differential expression patterns were later assessed with in-situ hybridization in vocal learner and vocal non-learner birds, further suggesting a potential role for Slit1 in regulating the development of the vocal learning circuit (R. Wang et al. 2015). As for FOXP2, a previous study had already reported that human FOXP2 more differentially up-regulates SLIT1 compared to chimpanzee FOXP2, alluding to how the downregulation of FOXP2 would affect SLIT1 expression in human LMC. The authors suggested that species with convergent vocal learning circuits likely underwent convergent molecular changes of multiple genes. Additional insight will likely arise as a consequence of The Avian Phylogenomics Project, which has the target of sequencing all ~10,500 bird genomes. Comparison of additional genomes and transcriptomes will permit a better understanding of the evolution of vocal learning circuitry (Jarvis 2016). Similar broad genome sequencing efforts will be key to studying the genetic underpinnings of a wealth of complex behavioral traits (see the Vertebrate Genomes Project https://vertebrategenomesproject.org). Notably, finding and studying gene expression programs that control behaviors in non-traditional model species will require incorporating different types of analyses that will yield high specificity of cell populations, such as those that are currently employed in Mus musculus.
Single-cell methodologies
As mentioned previously, subcortical brain regions such as the POA and ventromedial hypothalamus (VMH) are highly heterogeneous; one region contains multiple sub-nuclei, which could be participating in different behavioral circuits. Several parameters can be used collectively to characterize different neuronal types, which include spatial context, morphology, connectivity, gene expression patterns, and evolutionary history (Arendt et al. 2016; Mukamel and Ngai 2019). Gene products are especially key players in shaping the development and governing the function of cells participating in behavioral circuits. With advancements in sequencing technologies, we can now look at individual cells within brain regions of interest to characterize neuronal types based on their gene expression patterns (Zeng and Sanes 2017). This has led to the emergence of unparalleled possibilities, such as cross-species comparisons of cell types within homologous brain regions (See Fig. 1A and 1D).
The Laurent lab employed Drop-seq (Macosko et al. 2015), a droplet-based single cell RNA sequencing (scRNA-seq) method, in the pallium of turtles and lizards to assess the evolutionary conservation of inhibitory neuron classes (Tosches et al. 2018). Krienen et al compared the molecular identities of telencephalic interneurons from humans, macaques, marmosets, and mice (Krienen et al. 2019). The authors found that conserved interneuron types have undergone evolutionary changes in density, distribution and genetic programs, especially across mice and primates. Similar approaches could be applied to assess how behavioral circuits have evolved at a molecular level across species.
Spatial information can help reveal the function of different cell types identified with scRNA-seq, especially in heterogeneous sub-cortical brain regions that control innate behaviors. Recent approaches combine scRNA-seq with in-situ hybridization (ISH) to obtain spatial resolution of cells using target gene markers (see Fig. 1D and 1E). One prominent example integrated scRNA-seq combined with a single cell transcriptome imaging method called multiplexed error robust fluorescence ISH (MERFISH) to assess the molecular organization of cells in the mouse POA (Moffitt et al. 2018). MERFISH works by assigning barcodes to individual RNA species and labelling them combinatorially with oligonucleotide probes containing a representation of those barcodes. The barcodes can then be read sequentially by single or multi colored single molecule FISH imaging (Moffitt et al. 2016). Thus, RNA probes for gene products identified with scRNA-seq as strong cell type markers can be designed and used with MERFISH. For the purposes of finding cell types associated with mating, parenting, and aggressive behaviors in the POA, the study by Moffitt et al. initially included neuropeptide receptors, such as OXT receptors, as cell type markers. This was followed by c-Fos labelling during the aforementioned behaviors, and comparison with MERFISH results to find which cell-type clusters were associated with target behaviors (Moffitt et al. 2018). An interesting finding of this study was that spatial proximity between cell types is a possible indicator of their function, thus supporting the idea that integrating spatial and genetic information of cell types is crucial for understanding their function in neural circuits.
A similar approach was employed by Kim et al to characterize cell types in the ventrolateral VMH (VMHvl) of mice, by combining scRNA-seq and sequential in-situ hybridization (seqFISH), a technique similar to MERFISH in which RNA is barcoded as a color sequence instead of a binary string (Kim et al. 2019). They found that neurons in this region could be characterized by their expression patterns of three different genes: estrogen receptor 1 alpha (Esr1), special AT-rich sequence binding protein 2 (Satb2) and delta like non-canonical Notch ligand 2 (Dlk2). Kim et al. also incorporated retrograde labeling in known target brain regions to combine gene expression, spatial and projection data in the characterization of VMHvl cells. Similar to Moffitt et al., Fos expression was used as a readout for neuronal activity following mating, social fear, and aggressive behaviors. The results showed that most cells did not display a clear relationship between gene expression, projections, and behavioral relevance, with a few exceptions. Notably, VMH neurons that express the nucleoporin 62 C-terminal like gene (Nup62cl) preferentially project to the lateral periaquaductal grey, and are activated by social fear.
Moreover, in order to study how connectivity patterns are established in different cell types in an experience-dependent manner, single cell analysis after a behavioral stimulus will be useful in revealing activity-dependent gene programs that might control learning. Notably, activity-regulated IEGs such as Fos are transcription factors (TFs) that initiate cell-type specific cascades of gene expression to regulate neural development, synaptic plasticity, and learning and memory (Yap and Greenberg 2018). Hrvatin et al. employed scRNA-seq to identify cell type-specific, activity-induced transcriptional programs in visual cortex. (Hrvatin et al. 2018). For example, they found that corticotropin releasing hormone (Crh) is induced 1 hr after light stimulation specifically within Vip+ interneurons, while its receptor Crhr1 is induced in excitatory pyramidal cells, and the secreted negative inhibitor Crhbp is uniquely in Sst+ interneurons. Thus, single-cell analyses can reveal microcircuits that respond to sensory inputs and is effective for identification of genes that mediate transient or long-lasting effects of other stimuli, such as a social interaction, on the brain.
Importantly, combining spatial and scRNA-seq information has been explored in rats. One such study combined scRNA-seq with immunocytochemistry to study cell types within the pituitary gland (Fletcher et al. 2019). Therefore, scRNA-seq combined with spatial approaches can potentially be applied to non-traditional model organisms, and will be key for isolating cell types specifically associated with target behaviors within heterogeneous brain regions implicated in such behaviors. Moreover, it will be key to finding gene targets in such specific cell types, for the development of genetic tools in vertebrates to study complex behaviors in similar fashion as how neural circuits have been studied in invertebrate species.
Genetic labeling to enhance scRNA-seq
Various studies have sought to determine which genes are the best classifiers of cell type and have found that transcription factors (TFs) are the most robust discriminators of neuronal types and their progenitors throughout embryonic development (Shimogori et al. 2010; Arber 2012; Hobert 2016; Li et al. 2017; Paul et al. 2017; Huisman et al. 2019). Neuromodulator receptors, although weakly expressed in comparison to TFs, can also be used to define cell types, and can give clues as to which cells in different brain regions are involved in specific behaviors (Moffitt et al. 2018; Kim et al. 2019). In lab mice, genetic labeling of specific classes of neurons allows purification before scRNA-seq, so that only the cells of interest are sequenced. This enrichment permits deeper sequencing of desired populations, revealing more genes per sequenced cell.
The Correa lab sought to identify the distinct populations of VMH Esr1 cells that contribute to physical activity and thermogenesis in female mice. They began by targeting cells from the Steroidogenic factor 1 (Sf1) lineage via a Cre-dependent reporter, which is unique to the VMH, and performed scRNA-seq in juvenile females and males (van Veen et al. 2020). Two subpopulations were identified, distinguished by differential expression of the reprimo (Rprm) and tachykinin-1 (Tac1) genes in the two sexes. Further functional studies, which involved ablation of Rprm expression, demonstrated the importance of Esr1+ Rprm+ neurons in temperature regulation, while ablating Tac1 expression showed the role of Esr1+ Tac1+ cells in movement coordination. Thus, genetic profiling of cells expressing a specific TF allowed the identification of a neuromodulatory gene product involved in a specific behavioral response. Indeed, plasticity in neuromodulatory systems is thought to be a key driver of the evolution of social behavior (Donaldson and Young 2008; Paul S. Katz 2011; Garrison et al. 2012; Kelly and Vitousek 2017; Phelps, Okhovat, and Berrio 2017; Edsinger and Dölen 2018; Bendesky and Bargmann 2011; Goodson, Wilson, and Schrock 2012; Kocher et al. 2018; Lim et al. 2004; Wu et al. 2014; Okhovat et al. 2015; Fischer and O’Connell 2020; Niepoth and Bendesky 2020) In the future, genetic labeling of cells expressing specific neuromodulators identified via these types of studies will be key to understanding via single-cell transcriptomic profiling how neural circuits involved in complex behaviors differ between species, sexes, and potentially how they might be modified by environmental inputs.
Development and circuit specification
Differential gene expression signatures likely reflect differences in internal state, neuronal activity patterns, or in the case of species comparisons, composition of cell types. While the approaches described above may be used to pinpoint functional populations that are modified by experience, in our view identification of gene programs responsible for the organization of specific behavioral circuits requires a neurodevelopmental approach. As previously stated, an integrative understanding of behavior requires us to address questions about genes orchestrating embryonic and postnatal development, behavioral display, and evolutionary history (Tinbergen 2010; Bateson and Laland 2013). Genes that show increased expression in one or multiple species which possess a behavior of interest are not necessarily the genes that specify the formation of the unique neural pathways making that behavior possible. Finding such genes would be facilitated by performing transcriptomic experiments in the developing brain, as wiring of relevant circuitry is occurring. However, to the best of our knowledge, all such comparative screens to date have been performed in adult animals e.g. (Pfenning et al. 2014; Baran and Streelman 2020; Fischer and O’Connell 2020; Fischer et al. 2019). Even when causal species differences in gene expression are well-established, such as the expression of Oxtr and Avpr1a in different vole species, when these differences arise during development and how they relate to wiring of behavioral circuitry remains an open question (Insel and Shapiro 1992; Z. Wang et al. 1997).
Innate behavioral circuits can be considered analogous to morphological traits, in that their developmental programs are encoded in the genome. In fact, the same mechanistic properties that govern the evolution of morphological traits through alterations to developmental processes, have been described to contribute to behavioral variation (Hoke et al. 2019) Understanding the evolution of such traits, particularly limb development, has required knowledge of when species-specific factors are expressed. Examples include decreased paired like homeodomain 1 (Pitx1) expression leading to hindlimb reduction in stickleback fish (Shapiro et al. 2004), or loss of an E26 transformation-specific (ETS) TF binding site in the Zone of Polarising Activity (ZPA) Regulatory Sequence (ZRS) enhancer causing loss of all limbs in snakes (Kvon et al. 2016). Notably the drivers of evolution in these examples are TFs. Although many markers of social behavior diversity identified to date are neuropeptides, including AVP, TAC1, or NPY, we expect that the principal drivers of diversity are the TFs that define identity and connectivity of cells that express such neuropeptides and their receptors. Accordingly, recent analysis of both gene expression and chromatin accessibility throughout cortical development revealed specific TFs and regulatory elements that shape the unique identity of cells from upper or lower cortical layers (Heavner et al. 2020). Application of similar approaches across species is likely to reveal TF-directed gene regulatory networks that direct the development of social behavior circuitry (Baran, McGrath, and Streelman 2017). With this “evo-devo” framework as a guide, we anticipate that the adoption of the techniques described in this review, combined with the ever-expanding repertoire of model organisms, will provide a powerful approach to identify conserved principles of sociality.
Highlights.
Increased specificity is particularly important for characterization of heterogeneous sub-cortical brain regions that regulate innate behaviors
Capturing active neuronal populations can improve transcriptomic specificity
Single-cell approaches can reveal cell types that differ across species
Acknowledgments
Funding information
This work was supported by the NIH (R01 MH113628 to JT and T-32 2T32GM065094 to J R-O) the Pershing Square Innovation Fund, and the Stanley Family Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arber Silvia. 2012. “Motor Circuits in Action: Specification, Connectivity, and Function.” Neuron 74 (6): 975–89. [DOI] [PubMed] [Google Scholar]
- Arendt Detlev, Musser Jacob M., Baker Clare V. H., Bergman Aviv, Cepko Connie, Erwin Douglas H., Pavlicev Mihaela, et al. 2016. “The Origin and Evolution of Cell Types.” Nature Reviews. Genetics 17 (12): 744–57. [DOI] [PubMed] [Google Scholar]
- Baker BS, Taylor BJ, and Hall JC. 2001. “Are Complex Behaviors Specified by Dedicated Regulatory Genes? Reasoning from Drosophila.” Cell 105 (1): 13–24. [DOI] [PubMed] [Google Scholar]
- Baran Nicole M., McGrath Patrick T., and Streelman J. Todd. 2017. “Applying Gene Regulatory Network Logic to the Evolution of Social Behavior.” Proceedings of the National Academy of Sciences of the United States of America 114 (23): 5886–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baran Nicole M., and Streelman J. Todd. 2020. “Ecotype Differences in Aggression, Neural Activity and Behaviorally Relevant Gene Expression in Cichlid Fish.” Genes, Brain, and Behavior, April, e12657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateson Patrick, and Laland Kevin N.. 2013. “Tinbergen’s Four Questions: An Appreciation and an Update.” Trends in Ecology & Evolution 28 (12): 712–18. [DOI] [PubMed] [Google Scholar]
- Beets Isabel, Janssen Tom, Meelkop Ellen, Temmerman Liesbet, Suetens Nick, Rademakers Suzanne, Jansen Gert, and Schoofs Liliane. 2012. “Vasopressin/Oxytocin-Related Signaling Regulates Gustatory Associative Learning in C. Elegans.” Science 338 (6106): 543–45. [DOI] [PubMed] [Google Scholar]
- Bendesky Andres, and Bargmann Cornelia I.. 2011. “Genetic Contributions to Behavioural Diversity at the Gene-Environment Interface.” Nature Reviews. Genetics 12 (12): 809–20. [DOI] [PubMed] [Google Scholar]
- Bendesky Andres, Kwon Young-Mi, Lassance Jean-Marc, Lewarch Caitlin L., Yao Shenqin, Peterson Brant K., He Meng Xiao, Dulac Catherine, and Hoekstra Hopi E.. 2017. “The Genetic Basis of Parental Care Evolution in Monogamous Mice.” Nature 544 (7651): 434–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bester-Meredith Janet K., and Marler Catherine A.. 2012. “Naturally Occurring Variation in Vasopressin Immunoreactivity Is Associated with Maternal Behavior in Female Peromyscus Mice.” Brain, Behavior and Evolution 80 (4): 244–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bono M. de, and Bargmann CI. 1998. “Natural Variation in a Neuropeptide Y Receptor Homolog Modifies Social Behavior and Food Response in C. Elegans.” Cell 94 (5): 679–89. [DOI] [PubMed] [Google Scholar]
- Campbell John N., Macosko Evan Z., Fenselau Henning, Pers Tune H., Lyubetskaya Anna, Tenen Danielle, Goldman Melissa, et al. 2017. “A Molecular Census of Arcuate Hypothalamus and Median Eminence Cell Types.” Nature Neuroscience 20 (3): 484–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter Marco D., Hu Qiwen, Bond Allison M., Lombroso Sonia I., Czarnecki Kyle S., Lim Carissa J., Song Hongjun, Wimmer Mathieu E., Pierce R. Christopher, and Heller Elizabeth A.. 2020. “Nr4a1 Suppresses Cocaine-Induced Behavior via Epigenetic Regulation of Homeostatic Target Genes.” Nature Communications 11 (1): 504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demir Ebru, and Dickson Barry J.. 2005. “Fruitless Splicing Specifies Male Courtship Behavior in Drosophila.” Cell 121 (5): 785–94. [DOI] [PubMed] [Google Scholar]
- Donaldson Zoe R., and Young Larry J.. 2008. “Oxytocin, Vasopressin, and the Neurogenetics of Sociality.” Science 322 (5903): 900–904. [DOI] [PubMed] [Google Scholar]
- Dornan Anthony J., Gailey Donald A., and Goodwin Stephen F.. 2005. “GAL4 Enhancer Trap Targeting of the Drosophila Sex Determination Gene Fruitless.” Genesis 42 (4): 236–46. [DOI] [PubMed] [Google Scholar]
- Duque-Wilckens Natalia, Torres Lisette Y., Yokoyama Sae, Minie Vanessa A., Tran Amy M., Petkova Stela P., Hao Rebecca, et al. 2020. “Extrahypothalamic Oxytocin Neurons Drive Stress-Induced Social Vigilance and Avoidance.” Proceedings of the National Academy of Sciences of the United States of America, October 10.1073/pnas.2011890117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edsinger Eric, and Dölen Gül. 2018. “A Conserved Role for Serotonergic Neurotransmission in Mediating Social Behavior in Octopus.” Current Biology: CB 28 (19): 3136–3142.e4. [DOI] [PubMed] [Google Scholar]
- Ekstrand Mats I., Nectow Alexander R., Knight Zachary A., Latcha Kaamashri N., Pomeranz Lisa E., and Friedman Jeffrey M.. 2014. “Molecular Profiling of Neurons Based on Connectivity.” Cell 157 (5): 1230–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer Eva K., and O’Connell Lauren A.. 2020. “Hormonal and Neural Correlates of Care in Active versus Observing Poison Frog Parents.” Hormones and Behavior 120 (April): 104696. [DOI] [PubMed] [Google Scholar]
- Fischer Eva K., Roland Alexandre B., Moskowitz Nora A., Tapia Elicio E., Summers Kyle, Coloma Luis A., and O’Connell Lauren A.. 2019. “The Neural Basis of Tadpole Transport in Poison Frogs.” Proceedings. Biological Sciences / The Royal Society 286 (1907): 20191084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher Patrick A., Smiljanic Kosara, Prévide Rafael Maso, Iben James R., Li Tianwei, Rokic Milos B., Sherman Arthur, Coon Steven L., and Stojilkovic Stanko S.. 2019. “Cell Type- and Sex-Dependent Transcriptome Profiles of Rat Anterior Pituitary Cells.” Frontiers in Endocrinology 10 (September): 623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gailey DA, and Hall JC. 1989. “Behavior and Cytogenetics of Fruitless in Drosophila Melanogaster: Different Courtship Defects Caused by Separate, Closely Linked Lesions.” Genetics 121 (4): 773–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrison JL, Macosko EZ, Bernstein S, Pokala N, Albrecht DR, and Bargmann CI. 2012. “Oxytocin/Vasopressin-Related Peptides Have an Ancient Role in Reproductive Behavior.” Science 338 (6106): 540–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godwin John, and Thompson Richmond. 2012. “Nonapeptides and Social Behavior in Fishes.” Hormones and Behavior 61 (3): 230–38. [DOI] [PubMed] [Google Scholar]
- Goldberg Jesse H., and Fee Michale S.. 2011. “Vocal Babbling in Songbirds Requires the Basal Ganglia-Recipient Motor Thalamus but Not the Basal Ganglia.” Journal of Neurophysiology 105 (6): 2729–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodson James L., Wilson Leah C., and Schrock Sara E.. 2012. “To Flock or Fight: Neurochemical Signatures of Divergent Life Histories in Sparrows.” Proceedings of the National Academy of Sciences of the United States of America 109 Suppl 1 (June): 10685–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenspan RJ, and Ferveur JF. 2000. “Courtship in Drosophila.” Annual Review of Genetics 34: 205–32. [DOI] [PubMed] [Google Scholar]
- Guzowski JF, McNaughton BL, Barnes C. a., and Worley PF. 1999. “Environment-Specific Expression of the Immediate-Early Gene Arc in Hippocampal Neuronal Ensembles.” Nature Neuroscience 2 (12): 1120–24. [DOI] [PubMed] [Google Scholar]
- Hashikawa Koichi, Hashikawa Yoshiko, Tremblay Robin, Zhang Jiaxing, Feng James E., Sabol Alexander, Piper Walter T., Lee Hyosang, Rudy Bernardo, and Lin Dayu. 2017. “Esr1+ Cells in the Ventromedial Hypothalamus Control Female Aggression.” Nature Neuroscience 20 (11): 1580–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heavner Whitney E., Ji Shaoyi, Notwell James H., Dyer Ethan S., Tseng Alex M., Birgmeier Johannes, Yoo Boyoung, Bejerano Gill, and McConnell Susan K.. 2020. “Transcription Factor Expression Defines Subclasses of Developing Projection Neurons Highly Similar to Single-Cell RNA-Seq Subtypes.” Proceedings of the National Academy of Sciences of the United States of America 117 (40): 25074–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hisey Erin, Kearney Matthew Gene, and Mooney Richard. 2018. “A Common Neural Circuit Mechanism for Internally Guided and Externally Reinforced Forms of Motor Learning.” Nature Neuroscience 21 (4): 589–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobert Oliver. 2016. “Terminal Selectors of Neuronal Identity.” Current Topics in Developmental Biology 116 (January): 455–75. [DOI] [PubMed] [Google Scholar]
- Hoke Kim L., Elizabeth Adkins-Regan Andrew H. Bass, McCune Amy R., and Wolfner Mariana F.. 2019. “Co-Opting Evo-Devo Concepts for New Insights into Mechanisms of Behavioural Diversity.” The Journal of Experimental Biology 222 (Pt 8). 10.1242/jeb.190058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrvatin Sinisa, Hochbaum Daniel R., Nagy M. Aurel, Cicconet Marcelo, Robertson Keiramarie, Cheadle Lucas, Zilionis Rapolas, et al. 2018. “Single-Cell Analysis of Experience-Dependent Transcriptomic States in the Mouse Visual Cortex.” Nature Neuroscience 21 (1): 120–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huisman Christian, Cho Hyeyoung, Brock Olivier, Su Jeong Lim Sung Min Youn, Park Younjung, Kim Sangsoo, Lee Soo-Kyung, Delogu Alessio, and Lee Jae W.. 2019. “Single Cell Transcriptome Analysis of Developing Arcuate Nucleus Neurons Uncovers Their Key Developmental Regulators.” Nature Communications 10 (1): 3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insel TR, and Shapiro LE. 1992. “Oxytocin Receptor Distribution Reflects Social Organization in Monogamous and Polygamous Voles.” Proceedings of the National Academy of Sciences of the United States of America 89 (July): 5981–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis Erich D. 2016. “Perspectives from the Avian Phylogenomics Project: Questions That Can Be Answered with Sequencing All Genomes of a Vertebrate Class.” Annual Review of Animal Biosciences 4: 45–59. [DOI] [PubMed] [Google Scholar]
- Katz PS, and Harris-Warrick RM. 1999. “The Evolution of Neuronal Circuits Underlying Species-Specific Behavior.” Current Opinion in Neurobiology 9 (5): 628–33. [DOI] [PubMed] [Google Scholar]
- Katz Paul S. 2011. “Neural Mechanisms Underlying the Evolvability of Behaviour.” Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences 366 (1574): 2086–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly Aubrey M., and Vitousek Maren N.. 2017. “Dynamic Modulation of Sociality and Aggression: An Examination of Plasticity within Endocrine and Neuroendocrine Systems.” Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences 372 (1727). 10.1098/rstb.2016.0243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Dong-Wook, Yao Zizhen, Graybuck Lucas T., Kim Tae Kyung, Nguyen Thuc Nghi, Smith Kimberly A., Fong Olivia, et al. 2019. “Multimodal Analysis of Cell Types in a Hypothalamic Node Controlling Social Behavior.” Cell 179 (3): 713–728.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura Ken-Ichi, Ote Manabu, Tazawa Tatsunori, and Yamamoto Daisuke. 2005. “Fruitless Specifies Sexually Dimorphic Neural Circuitry in the Drosophila Brain.” Nature 438 (7065): 229–33. [DOI] [PubMed] [Google Scholar]
- King Lanikea B., Walum Hasse, Inoue Kiyoshi, Eyrich Nicholas W., and Young Larry J.. 2016. “Variation in the Oxytocin Receptor Gene Predicts Brain Region–Specific Expression and Social Attachment.” Biological Psychiatry 80 (2): 160–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight Zachary A., Tan Keith, Birsoy Kivanc, Schmidt Sarah, Garrison Jennifer L., Wysocki Robert W., Emiliano Ana, Ekstrand Mats I., and Friedman Jeffrey M.. 2012. “Molecular Profiling of Activated Neurons by Phosphorylated Ribosome Capture.” Cell 151 (5): 1126–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocher Sarah D., Mallarino Ricardo, Rubin Benjamin E. R., Yu Douglas W., Hoekstra Hopi E., and Pierce Naomi E.. 2018. “The Genetic Basis of a Social Polymorphism in Halictid Bees.” Nature Communications 9 (1): 4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krienen FM, Goldman M, Zhang Q, and del Rosario R. 2019. “Innovations in Primate Interneuron Repertoire.” BioRxiv. https://www.biorxiv.org/content/10.1101/709501v1.abstract.
- Kvon Evgeny Z., Kamneva Olga K., Melo S, Dickel Diane E., Melo S, Barozzi Iros, Osterwalder Marco, et al. 2016. “Progressive Loss of Function in a Limb Enhancer during Snake Evolution.” Cell, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Hongjie, Horns Felix, Wu Bing, Xie Qijing, Li Jiefu, Li Tongchao, Luginbuhl David J., Quake Stephen R., and Luo Liqun. 2017. “Classifying Drosophila Olfactory Projection Neuron Subtypes by Single-Cell RNA Sequencing.” Cell 171 (5): 1206–1220.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim Miranda M., Wang Zuoxin, Olazábal Daniel E., Ren Xianghui, Terwilliger Ernest F., and Young Larry J.. 2004. “Enhanced Partner Preference in a Promiscuous Species by Manipulating the Expression of a Single Gene.” Nature 429 (6993): 754–57. [DOI] [PubMed] [Google Scholar]
- Macosko Evan Z., Basu Anindita, Satija Rahul, Nemesh James, Shekhar Karthik, Goldman Melissa, Tirosh Itay, et al. 2015. “Highly Parallel Genome-Wide Expression Profiling of Individual Cells Using Nanoliter Droplets.” Cell 161 (5): 1202–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manoli Devanand S., Foss Margit, Villella Adriana, Taylor Barbara J., Hall Jeffrey C., and Baker Bruce S.. 2005. “Male-Specific Fruitless Specifies the Neural Substrates of Drosophila Courtship Behaviour.” Nature 436 (7049): 395–400. [DOI] [PubMed] [Google Scholar]
- Matharu Navneet, Rattanasopha Sawitree, Tamura Serena, Maliskova Lenka, Wang Yi, Bernard Adelaide, Hardin Aaron, Eckalbar Walter L., Vaisse Christian, and Ahituv Nadav. 2019. “CRISPR-Mediated Activation of a Promoter or Enhancer Rescues Obesity Caused by Haploinsufficiency.” Science 363 (6424). 10.1126/science.aau0629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffitt Jeffrey R., Bambah-Mukku Dhananjay, Eichhorn Stephen W., Vaughn Eric, Shekhar Karthik, Perez Julio D., Rubinstein Nimrod D., et al. 2018. “Molecular, Spatial, and Functional Single-Cell Profiling of the Hypothalamic Preoptic Region.” Science 362 (6416). 10.1126/science.aau5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffitt Jeffrey R., Hao Junjie, Bambah-Mukku Dhananjay, Lu Tian, Dulac Catherine, and Zhuang Xiaowei. 2016. “High-Performance Multiplexed Fluorescence in Situ Hybridization in Culture and Tissue with Matrix Imprinting and Clearing.” Proceedings of the National Academy of Sciences of the United States of America 113 (50): 14456–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukamel Eran A., and Ngai John. 2019. “Perspectives on Defining Cell Types in the Brain.” Current Opinion in Neurobiology 56 (June): 61–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murugan Malavika, Jang Hee Jae, Park Michelle, Miller Ellia M., Cox Julia, Taliaferro Joshua P., Parker Nathan F., et al. 2017. “Combined Social and Spatial Coding in a Descending Projection from the Prefrontal Cortex.” Cell 171 (7): 1663–1677.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niepoth Natalie, and Bendesky Andres. 2020. “How Natural Genetic Variation Shapes Behavior.” Annual Review of Genomics and Human Genetics 21 (August): 437–63. [DOI] [PubMed] [Google Scholar]
- Okhovat Mariam, Berrio Alejandro, Wallace Gerard, Ophir Alexander G., and Phelps Steven M.. 2015. “Sexual Fidelity Trade-Offs Promote Regulatory Variation in the Prairie Vole Brain.” Science 350 (6266): 1371–74. [DOI] [PubMed] [Google Scholar]
- Paul Anirban, Crow Megan, Raudales Ricardo, He Miao, Gillis Jesse, and Huang Z. Josh. 2017. “Transcriptional Architecture of Synaptic Communication Delineates GABAergic Neuron Identity.” Cell 171 (3): 522–539.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfenning Andreas R., Hara Erina, Whitney Osceola, Rivas Miriam V., Wang Rui, Roulhac Petra L., Howard Jason T., et al. 2014. “Convergent Transcriptional Specializations in the Brains of Humans and Song-Learning Birds.” Science 346 (6215): 1256846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelps Steven M., Okhovat Mariam, and Berrio Alejandro. 2017. “Individual Differences in Social Behavior and Cortical Vasopressin Receptor: Genetics, Epigenetics, and Evolution.” Frontiers in Neuroscience 11 (October): 537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson Gene E., Fernald Russell D., and Clayton David F.. 2008. “Genes and Social Behavior.” Science 322 (5903): 896–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scribner Jennifer L., Vance Eric A., Protter David S. W., Sheeran William M., Saslow Elliott, Cameron Ryan T., Klein Eric M., Jimenez Jessica C., Kheirbek Mazen A., and Donaldson Zoe R.. 2020. “A Neuronal Signature for Monogamous Reunion.” Proceedings of the National Academy of Sciences of the United States of America 117 (20): 11076–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeholzer Laura F., Seppo Max, Stern David L., and Ruta Vanessa. 2018. “Evolution of a Central Neural Circuit Underlies Drosophila Mate Preferences.” Nature 559 (7715): 564–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro Michael D., Marks Melissa E., Peichel Catherine L., Blackman Benjamin K., Nereng Kirsten S., Jónsson Bjarni, Schluter Dolph, and Kingsley David M.. 2004. “Genetic and Developmental Basis of Evolutionary Pelvic Reduction in Threespine Sticklebacks.” Nature 428 (6984): 717–23. [DOI] [PubMed] [Google Scholar]
- Shimogori Tomomi, Lee Daniel A., Miranda-Angulo Ana, Yang Yanqin, Wang Hong, Jiang Lizhi, Yoshida Aya C., et al. 2010. “A Genomic Atlas of Mouse Hypothalamic Development.” Nature Neuroscience 13 (6): 767–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stauffer William R., Lak Armin, Yang Aimei, Borel Melodie, Paulsen Ole, Boyden Edward S., and Schultz Wolfram. 2016. “Dopamine Neuron-Specific Optogenetic Stimulation in Rhesus Macaques.” Cell 166 (6): 1564–1571.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor Kathryn M., Marquart Gregory D., Hurt Christopher, Smith Trevor S., Geoca Alexandra K., Bhandiwad Ashwin A., Subedi Abhignya, et al. 2019. “Brain-Wide Cellular Resolution Imaging of Cre Transgenic Zebrafish Lines for Functional Circuit-Mapping.” ELife 8 (February). 10.7554/eLife.42687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Chan Lek, Cooke Elizabeth K., Leib David E., Lin Yen Chu, Daly Gwendolyn E., Zimmerman Christopher A., and Knight Zachary A.. 2016. “Warm-Sensitive Neurons That Control Body Temperature.” Cell 167 (1): 47–59.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinbergen N 2010. “On Aims and Methods of Ethology.” Zeitschrift Für Tierpsychologie 20 (4): 410–33. [Google Scholar]
- Tosches Maria Antonietta. 2017. “Developmental and Genetic Mechanisms of Neural Circuit Evolution.” Developmental Biology 431 (1): 16–25. [DOI] [PubMed] [Google Scholar]
- Tosches Maria Antonietta, Yamawaki Tracy M., Naumann Robert K., Jacobi Ariel A., Tushev Georgi, and Laurent Gilles. 2018. “Evolution of Pallium, Hippocampus, and Cortical Cell Types Revealed by Single-Cell Transcriptomics in Reptiles.” Science 360 (6391): 881–88. [DOI] [PubMed] [Google Scholar]
- Veen J. Edward van, Kammel Laura G., Bunda Patricia C., Shum Michael, Reid Michelle S., Massa Megan G., Arneson Douglas V., et al. 2020. “Hypothalamic Oestrogen Receptor Alpha Establishes a Sexually Dimorphic Regulatory Node of Energy Expenditure.” Nature Metabolism 2 (4): 351–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Rui, Chen Chun-Chun, Hara Erina, Rivas Miriam V., Roulhac Petra L., Howard Jason T., Chakraborty Mukta, Audet Jean-Nicolas, and Jarvis Erich D.. 2015. “Convergent Differential Regulation of SLIT-ROBO Axon Guidance Genes in the Brains of Vocal Learners.” The Journal of Comparative Neurology 523 (6): 892–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Liu Y, Young LJ, and Insel TR. 1997. “Developmental Changes in Forebrain Vasopressin Receptor Binding in Prairie Voles (Microtus Ochrogaster) and Montane Voles (Microtus Montanus).” Annals of the New York Academy of Sciences 807 (January): 510–13. [DOI] [PubMed] [Google Scholar]
- Wang Z, Zhou L, Hulihan TJ, and Insel TR. 1996. “Immunoreactivity of Central Vasopressin and Oxytocin Pathways in Microtine Rodents: A Quantitative Comparative Study.” The Journal of Comparative Neurology 366 (4): 726–37. [DOI] [PubMed] [Google Scholar]
- Wei Yi-Chao, Wang Shao-Ran, Jiao Zhuo-Lei, Zhang Wen, Lin Jun-Kai, Li Xing-Yu, Li Shuai-Shuai, Zhang Xin, and Xu Xiao-Hong. 2018. “Medial Preoptic Area in Mice Is Capable of Mediating Sexually Dimorphic Behaviors Regardless of Gender.” Nature Communications 9 (1): 279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Zheng, Autry Anita E., Bergan Joseph F., Watabe-Uchida Mitsuko, and Dulac Catherine G.. 2014. “Galanin Neurons in the Medial Preoptic Area Govern Parental Behaviour.” Nature 509 (7500): 325–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap Ee-Lynn, and Greenberg Michael E.. 2018. “Activity-Regulated Transcription: Bridging the Gap between Neural Activity and Behavior.” Neuron 100 (2): 330–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- York Ryan A., Patil Chinar, Abdilleh Kawther, Johnson Zachary V., Conte Matthew A., Genner Martin J., McGrath Patrick T., Fraser Hunter B., Fernald Russell D., and Streelman J. Todd. 2018. “Behavior-Dependent Cis Regulation Reveals Genes and Pathways Associated with Bower Building in Cichlid Fishes.” Proceedings of the National Academy of Sciences of the United States of America 115 (47): E11081–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young Rebecca L., Ferkin Michael H., Ockendon-Powell Nina F., Orr Veronica N., Phelps Steven M., Pogány Ákos, Richards-Zawacki Corinne L., et al. 2019. “Conserved Transcriptomic Profiles Underpin Monogamy across Vertebrates.” Proceedings of the National Academy of Sciences of the United States of America 116 (4): 1331–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Hongkui, and Sanes Joshua R.. 2017. “Neuronal Cell-Type Classification: Challenges, Opportunities and the Path Forward.” Nature Reviews. Neuroscience 18 (9): 530–46. [DOI] [PubMed] [Google Scholar]
- Zhao Wenchan, Garcia-Oscos Francisco, Dinh Daniel, and Roberts Todd F.. 2019. “Inception of Memories That Guide Vocal Learning in the Songbird.” Science 366 (6461): 83–89. [DOI] [PMC free article] [PubMed] [Google Scholar]