Abstract
Objectives:
Low-density granulocytes (LDGs) are a distinct subset of pro-inflammatory and vasculopathic neutrophils expanded in systemic lupus erythematosus (SLE). Neutrophil trafficking and immune function are intimately linked to cellular biophysical properties. This study used proteomic, biomechanical, and functional analyses to further define neutrophil heterogeneity in the context of SLE.
Methods:
Proteomic/phosphoproteomic analyses were performed in healthy control (HC) normal density neutrophils (NDNs), SLE NDNs and autologous SLE LDGs. The biophysical properties of these neutrophil subsets were analyzed by real-time deformability cytometry (RT-DC) and lattice light-sheet microscopy. A two-dimensional endothelial flow system and a three-dimensional microfluidic microvasculature mimetic (MMM) were used to decouple the contributions of cell surface mediators and biophysical properties to neutrophil trafficking, respectively.
Results:
Proteomic and phosphoproteomic differences were detected between HC and SLE neutrophils and between SLE NDNs and LDGs. Increased abundance of type 1 interferon-regulated proteins and differential phosphorylation of proteins associated with cytoskeletal organization were identified in SLE LDGs relative to SLE NDNs. The cell surface of SLE LDGs was rougher than in SLE and HC NDNs, suggesting membrane perturbances. While SLE LDGs did not display increased binding to endothelial cells in the two-dimensional assay, they were increasingly retained/trapped in the narrow channels of the lung MMM.
Conclusions:
Modulation of the neutrophil proteome and distinct changes in biophysical properties are observed alongside differences in neutrophil trafficking. SLE LDGs may be increasingly retained in microvasculature networks, which has important pathogenic implications in the context of lupus organ damage and small vessel vasculopathy.
Keywords: lupus, neutrophils, cell traffic, proteomics
Introduction
Neutrophil dysregulation may play critical roles in SLE pathogenesis1. Enhanced release of neutrophil extracellular traps (NETs)- the externalization of oxidized nucleic acids and granule proteins- promotes immune dysregulation, vasculopathy and organ damage associated with SLE2–5.
We previously identified a subset of SLE proinflammatory neutrophils (low density granulocytes, LDGs), purified from the peripheral blood mononuclear cell (PBMC) layer 6. In contrast to normal dense neutrophils (NDNs), LDGs spontaneously form proinflammatory NETs 7,8 induce endothelial damage 6, and associate with in vivo vascular inflammation, coronary atherosclerosis 5,9–11, and T cell activation 12, suggesting they play important roles in SLE pathogenesis.
Previous LDG studies focused on transcriptomic analysis, with little known about proteome modulation and protein function 8,9,13–16. Proteomic analyses comparing SLE LDGs to SLE and healthy control (HC) NDNs, identified differential phosphorylation of proteins associated with cytoskeletal organization. Using real-time deformability cytometry (RT-DC) and a polydimethylsiloxane (PDMS) device mimicking neutrophil trafficking through the pulmonary microvasculature, we determined SLE LDGs are biophysically distinct from other neutrophil subsets, which may affect their ability to traffic through small blood vessels.
Methods
Results
Differential protein profiles of lupus and HC neutrophils
Proteomic/phosphoproteomic analyses were performed in SLE LDGs and NDNs, and HC NDNs (n=5/group; Supplementary Tables 1, 2). As controls, HC NDNs were also analyzed following priming with N-formylmethionine leucyl-phenylalanine (fMLF), given that priming decreases HC NDN density 17. Neutrophil preparations used identical protocols optimized to minimize biophysical or functional disruption of cells from their unstimulated state in whole blood (Supplementary Figure 1).
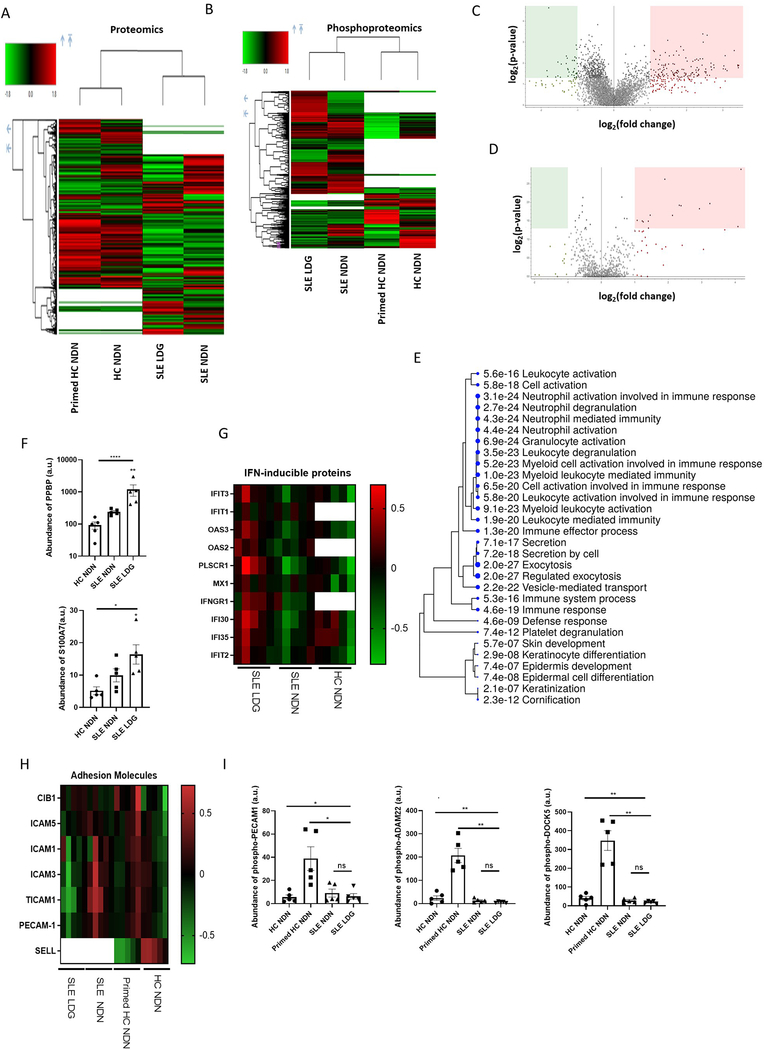
Neutrophil mass spectrometry analysis identified 4109 proteins (Figure 1A), of which 601 (14.6%) and 685 (16.6%) were identified only in HC or SLE neutrophils, respectively (Supplementary Figure 2A–B). This is comparable to the most robust neutrophil proteomic analysis previously reported 18. Results were aligned with SLE LDG, NDN, and HC NDN transcriptomics (GEO GSE139358) 16 to identify proteins not present at the mRNA level that may be of exogenous source (Supplementary Figure 3). SLE LDGs and NDNs showed complete proteome overlap, albeit with considerable variation in protein abundance. Indeed, 9.4% of proteins expressed by SLE neutrophils were differentially abundant in SLE LDGs vs NDNs, with 270 more abundant and 60 less abundant (ratio cut-off >1.5 or <0.5 in at least 4/5 matched samples; Figure 1C). Of the 2823 proteins common to both SLE and HC NDNs, 304 (10.7%) showed differential abundance. FMLF-primed and unstimulated HC NDNs showed complete proteome overlap with little variation in protein abundance, except for decreased abundance of L-selectin in primed HC NDNs, suggesting protein shedding from in vitro activation 19 (Supplementary Figure 2D–H). Overall, many proteins were uniquely present in either HC or SLE neutrophils and protein abundances varied between subsets, indicating neutrophil proteome heterogeneity.
Figure 1. SLE NDNs differ in their proteome and phosphoproteome compared to HC NDNs.
(A & B) A total of 4109 proteins (A) and 875 phosphoproteins (B) were identified by mass spectrometry in LDGs and NDNs from SLE subjects (n=5) and unstimulated and primed NDNs from HC volunteers (n=5). Volcano plots depict differences between SLE NDN and LDG proteomes (C) and phosphoproteomes (D). The upregulated (red) and downregulated (green) proteomes are in LDGs, while NDNs are the reference proteome. (E) Gene ontology biological process analysis highlighting biological networks associated with proteins more abundant in at least 4/5 SLE NDN samples relative to HC NDNs. Proteins with abundance ratios greater than 1.5 were included and significance was established by false discovery rate (FDR). (F) Proteins responsible for upregulation of networks associated with neutrophil activation in SLE NDNs relative to HC NDNs in arbitrary units. Significance was established by Kruskal-Wallis test with post hoc Dunn’s tests for multiple comparisons (3 comparisons) or by Mann-Whitney U test (2 comparisons). (G) Relative abundance of IFN inducible proteins in SLE NDNs and HC NDNs relative to SLE LDGs. SLE NDNs were compared to autologous SLE LDGs. HC NDNs were compared to the mean protein abundance in SLE LDGs. Open boxes in heatmaps indicate the given protein was not identified in the sample. (H) Abundance of cell integrins and adhesion-related proteins in SLE NDNs and HC NDNs relative to autologous SLE LDGs and autologous primed HC NDNs respectively. (I) Abundance of phosphoproteins associated with regulation of neutrophil-endothelial interactions in all neutrophil subsets, in arbitrary units. Significance was established by Kruskal-Wallis test with post hoc Dunn’s tests for multiple comparisons. All results are mean ± SEM and significance was set at *p ≤ 0.05, **p≤ 0.01, ns=not significant.
We identified 875 proteins phosphorylated on serine, threonine, and/or tyrosine residues in neutrophils (Figure 1B). Some proteins were phosphorylated at multiple sites (Supplementary Table 3). Of these phosphoproteins, 48 (5.4%) and 366 (41.8%) were only identified in HC NDNs and SLE neutrophils, respectively. The same phosphoproteins were identified in HC unstimulated and fMLF-primed NDNs, and one phosphoprotein was uniquely identified in SLE LDGs (round spermatid basic protein 1-like protein, pRSBN1L; Supplementary Figure 2C). When comparing SLE LDGs and NDNs, 95 phosphoproteins (11.5%) were differentially abundant, with 11 less and 84 more abundant in SLE LDGs (Figure 1D). Of the 509 phosphoproteins co-expressed in fMLF-primed and unstimulated HC NDNs, 167 (32.8%) were differentially abundant (Supplementary Figure 2E). Of the 460 phosphoproteins common to all neutrophils, 100 (21.7%) were differentially abundant between HC and SLE NDNs (Supplementary Figure 2G). These data support neutrophil phosphoproteome heterogeneity.
The LDG proteome displays a distinct profile
Using ShinyGO 20 and MetaScape 21, we mapped proteins differentially abundant in at least 4/5 samples to known gene-ontology biological processes. Proteins more abundant in SLE NDNs relative to HC NDNs mapped to neutrophil activation networks, including proteins facilitating migration to inflammatory sites and release from bone marrow 22–24. Some proteins associated with neutrophil activation were most abundant in SLE LDGs (Figure 1E–F).
SLE subjects express elevated type 1 IFN-stimulated genes (ISGs) in various organs and cells, including NDNs and LDGs 16,25. While ISG-encoded proteins were not uniformly upregulated in SLE NDNs vs HC NDNs, many were upregulated in SLE LDGs relative to SLE or HC NDNs (Figure 1G). ISG transcription is mediated by phosphorylation of signal transducer and activator of transcription (STAT) molecules 26 but we did not detect phospho-STATs, possibly because pTyr residues are less abundant than pSer 27. Limited LDG numbers prevented immunoprecipitation of pTyr residues alongside phosphopeptide enrichment. Collectively, the SLE neutrophil proteome suggests an activated status, while the IFN-associated protein signature is distinct to SLE LDGs.
Neutrophil priming/activation facilitates interactions with the endothelium 28. There were no differences in adhesion molecule or integrin expression among neutrophil subsets. However, phosphoproteins regulating neutrophil-endothelial interactions were more abundant in fMLF-primed HC NDNs than other neutrophil subsets (Figures 1H–I). This suggests differences between SLE LDGs/NDNs and fMLF-primed HC NDNs.
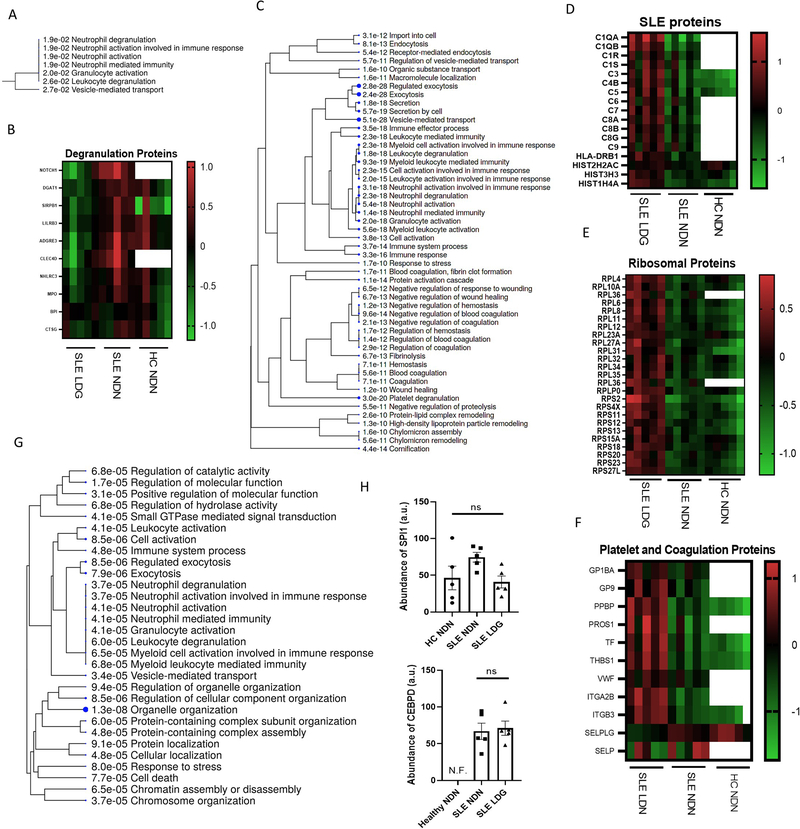
Proteins with differential phosphorylation in SLE NDNs vs HC NDNs were associated with organelle organization and actin cytoskeletal organization, including phospho-coronin 1A (pCORO1A) and phospho-heat shock protein 90AA1 (pHSP90AA1) (Supplementary Figure 2H). Some proteins less abundant in SLE LDGs vs SLE NDNs associated with neutrophil degranulation but key granule proteins, including myeloperoxidase and cathepsin-G, were not decreased (Figures 2A–B). Rather, lower abundance of membrane proteins, particularly ficolin-1-rich granule membrane proteins, accounted for downregulated degranulation-associated networks in SLE LDGs. Differences in degranulation capabilities did not explain changes in the neutrophil proteome between neutrophil subsets.
Figure 2. SLE LDGs have a distinct proteomic profile characterized by enhanced pathways associated to translational activity, intracellular trafficking and type I IFN-induced protein pathways.
(A) Gene ontology biological process analysis highlighting biological networks associated with proteins less abundant in SLE LDGs (n=5) relative to SLE NDNs (n=5). Proteins with abundance ratios less than 0.5 in at least 4/5 matched samples were included and significance was established by false discovery rate (FDR). (B) Relative abundance of degranulation network-associated proteins in SLE NDNs and HC NDNs relative to SLE LDGs. SLE NDNs were compared to autologous SLE LDGs. HC NDNs were compared to the mean protein abundance in SLE LDGs. (C) Gene ontology biological process analysis highlighting biological networks associated with proteins more abundant in at least 4/5 SLE LDG samples relative to SLE NDNs. Proteins with abundance ratios greater than 1.5 were included and significance was established by false discovery rate (FDR). (D) Relative abundance of SLE network-associated proteins in SLE NDNs and HC NDNs relative to SLE LDGs. SLE NDNs were compared to autologous SLE LDGs. HC NDNs were compared to the mean protein abundance in SLE LDGs. (E) Relative abundance of eukaryotic translation network-associated proteins in SLE NDNs and HC NDNs relative to SLE LDGs. SLE NDNs were compared to autologous SLE LDGs. HC NDNs were compared to the mean protein abundance in SLE LDGs. Open boxes in heatmaps indicate the given protein was not identified in the sample. (F) Relative abundance of coagulation and platelet network-associated proteins in SLE NDNs and HC NDNs relative to SLE LDGs. SLE NDNs were compared to autologous SLE LDGs. HC NDNs were compared to the mean protein abundance in SLE LDGs. (G) Gene ontology biological process analysis highlighting biological networks associated with phosphoproteins differentially abundant in SLE LDGs and NDNs. Phosphoproteins with abundance ratios less than 0.5 or greater than 1.5 in at least 4/5 matched samples were included and significance was established by false discovery rate (FDR). (H) Abundance of transcription factors CEBPD and SPI1 in arbitrary units. CEBPD not identified in HC NDNs. Results are mean ± SEM, with comparisons between autologous SLE LDG and NDNs. Significance established by Kruskal-Wallis test with post hoc Dunn’s tests for multiple comparisons and set at *p ≤ 0.05, ns=not significant. N.F. = not found.
Abundant proteins in SLE LDGs vs SLE NDNs clustered in neutrophil activation, coagulation, platelet, and intracellular trafficking networks (Figure 2C). The SLE biological network (False Discovery Rate (FDR) = 10−11.119) was upregulated in SLE LDGs vs autologous NDNs, primarily driven by complement proteins (Figure 2D). Immunoglobin chains and apolipoproteins were more abundant in SLE LDGs vs other neutrophils. Differential phosphorylation in SLE LDGs vs NDNs also associated with neutrophil activation and intracellular trafficking. In addition, SLE LDGs expressed higher abundances of ribosomal proteins (Figures 2E–G).
SLE LDGs are a heterogeneous group comprising of CD10− (immature, less abundant) and CD10+ (intermediate-mature, most abundant) subsets. CD10− LDGs have decreased CEBPD and SPI1 transcripts relative to SLE NDNs and CD10+ LDGs 16. Proteomic analysis was completed on unfractionated SLE LDGs and displayed similar SPI1 protein abundance across neutrophil subsets. CEBPD was not identified in HC NDNs but was similar in SLE LDGs and NDNs (Figure 2H). Most SLE LDGs had multi-lobulated nuclei (Supplementary Figure 1C), confirming intermediate-mature cells represent the most abundant LDG subset 16.
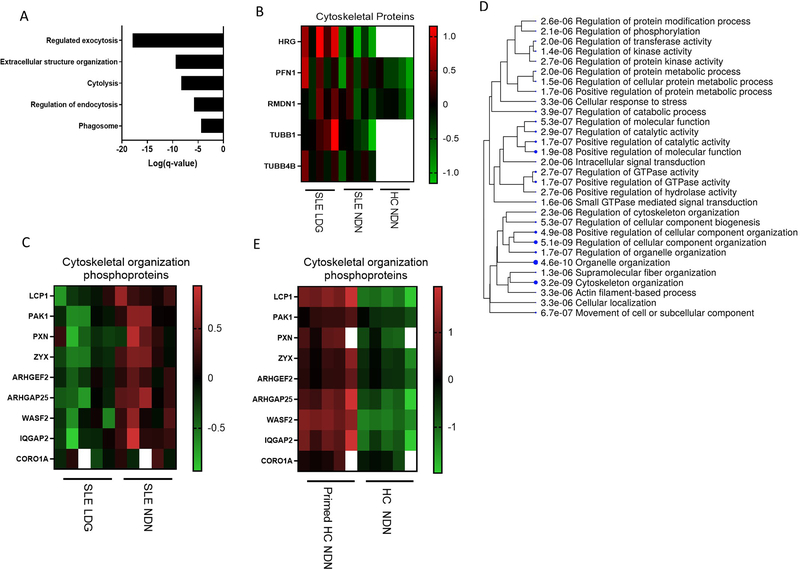
SLE LDGs and NDNs differ in expression of cytoskeleton-associated proteins
Consistent with evidence that cytoskeleton-associated transcriptional networks are enhanced in SLE LDGs 8, we found upregulation at the protein level when compared to autologous NDNs (Figure 3A). Many proteins differentially expressed and/or phosphorylated in LDGs regulate intracellular trafficking (Figure 3B–C)29–33. In addition, we assessed modulation of the HC NDN phosphoproteome by fMLF 17,34,35. Many proteins differentially phosphorylated in fMLF-primed HC NDNs were associated with cytoskeletal organization (Figure 3D–E). Upregulation of cytoskeleton-associated networks in the SLE LDG proteome, alongside phosphoproteomic findings suggestive of differential cytoskeletal reorganization among neutrophil subsets, prompted investigation of neutrophil biomechanical properties.
Figure 3. Proteomic and phosphoproteomic analyses indicate differential expression of proteins associated to cytoskeleton function between SLE NDNs and LDGs.
(A) Gene ontology biological process analysis highlighting biological networks related to the cytoskeleton and associated with proteins more abundant in at least 4/5 SLE LDG samples relative to their matched SLE NDNs. Proteins with abundance ratios greater than 1.5 were included and significance was established by false discovery rate (FDR). (B) Abundance of cytoskeleton-associated proteins in SLE NDNs and HC NDNs relative to SLE LDGs. SLE NDNs were compared to autologous SLE LDGs. HC NDNs were compared to the mean protein abundance in SLE LDGs. Open boxes in heatmaps indicate the given protein was not identified in one of the two autologous samples being compared. (C) Abundance of cytoskeleton network-associated phosphoproteins in SLE LDGs relative to abundance in autologous SLE NDNs. (D) Gene ontology biological process analysis highlighting biological networks associated with phosphoproteins differentially abundant in primed HC NDNs (n=5) and unstimulated HC NDNs (n=5). Proteins with abundance ratios greater than 1.5 or less than 0.5 in at least 4/5 matched samples were included and significance was established by false discovery rate (FDR). (E) Abundance of phosphoproteins regulating the cytoskeleton in primed HC NDNs relative to autologous HC NDNs.
Neutrophil biomechanical properties are altered in clinically active SLE.
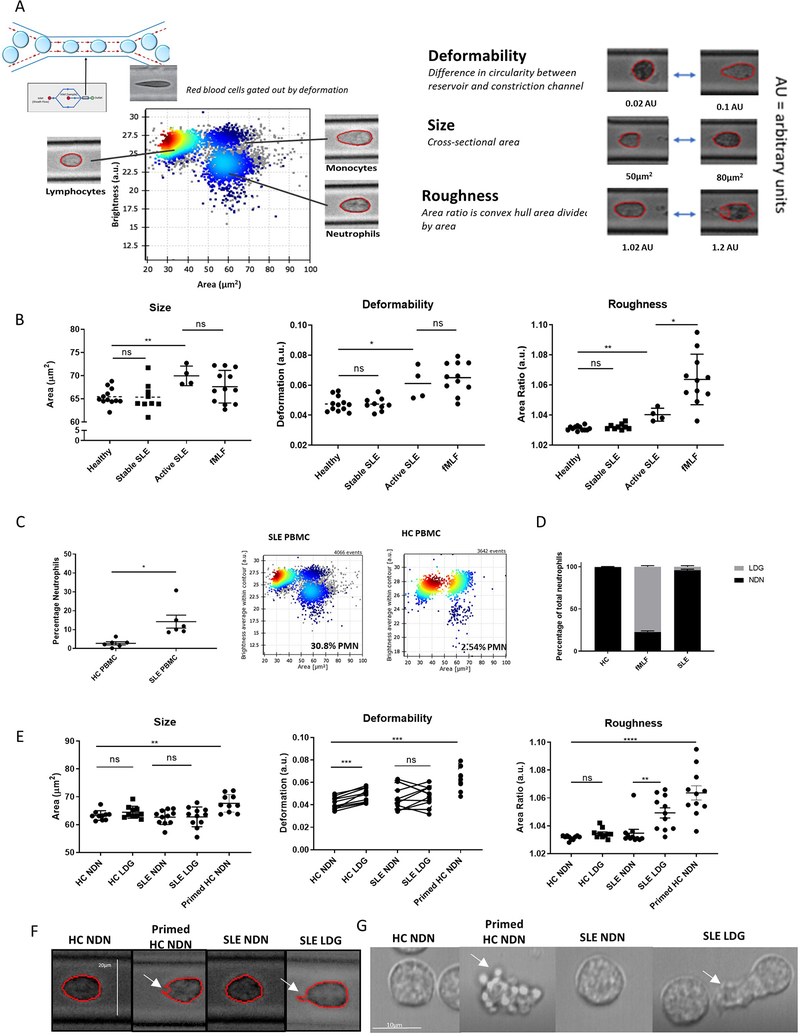
RT-DC is a high-throughput technique that analyzes biomechanical properties of thousands of cells in suspension 36. An inverted microscope with a high-speed camera captures images of individual cells moving through a narrow constriction channel within a PDMS microfluidic chip, where cells are deformed by hydrodynamic shear stress. Cell tracing algorithms generate biomechanical profiles per cell, including cell size (cross-sectional area), roughness (cell surface perturbations quantified by dividing the convex hull area by the cross-sectional area), and deformability (one minus the value of circularity within a constriction channel). Cell populations are identified in blood by size and brightness (Figure 4A) 37. These measurements were obtained in peripheral blood from HC (N=11), clinically quiescent (N=11) or clinically active SLE (N=4). In some experiments, fMLF was added directly to HC peripheral blood to prime neutrophils prior to analysis. Neutrophils from HC and clinically quiescent SLE were biomechanically identical, while active SLE neutrophils had larger areas, enhanced deformability and roughness. FMLF-primed HC neutrophils were also larger and more deformable than unstimulated neutrophils and significantly rougher than any unstimulated neutrophil subsets (Figure 4B). Overall, neutrophils from active SLE subjects displayed altered biomechanical properties and cell membrane perturbations.
Figure 4. SLE LDGs are biomechanically rougher than other neutrophil subsets by real-time deformability cytometry.
(A) RT-DC was used to biomechanically characterize the shape, size, and deformability of neutrophil subsets. (B) Biomechanical profiling of neutrophils in blood samples obtained from healthy volunteers (n=12), clinically quiescent SLE patients (n=9), and active SLE patients (n=4) by RT-DC. In some instances, 100nM fMLF was used to prime neutrophils within the healthy blood. (C) Percentage of LDGs identified in SLE (n=6) and HC PBMCs (n=6) by RT-DC. (D) LDGs as a percentage of total neutrophils in SLE patients (n=6), HC volunteers (n=6), and fMLF-primed HC blood (n=6). (E) Biomechanical profiling of isolated NDNs and LDGs from HC volunteers (n=11) and clinically quiescent SLE patients (n=11) by RT-DC. Some HC NDNs were primed with fMLF prior to isolation. For each sample analyzed by RT-DC, the median measurement of over 500 neutrophils is graphed and the mean ± SEM for each neutrophil subset is depicted. Autologous unstimulated/primed HC NDNs or autologous SLE NDN/LDGs were compared and significance was established by Wilcoxon matched-pairs signed rank tests. In other comparisons, significance was established by Mann-Whitney U-tests. Significance was set at *p ≤ 0.05, **p≤ 0.01, ns=not significant. (F) Images of NDNs and LDGs captured during RT-DC, representative of >500 images of each neutrophil subset from n=11 SLE patients and n=11 HC volunteers, 10x objective. White arrows identify a concave cell surface feature common in primed HC NDNs and an irregular protrusion in the cell surface common in SLE LDGs. (G) Brightfield microscopy of NDNs and LDGs (n=3). White arrows identify rounded, protruded, membrane features observed in nearly 100% of primed HC NDNs and an irregular protrusion observed in ~30% of SLE LDGs. See supplementary movie files.
SLE LDGs and NDNs are biomechanically distinct
Biomechanical properties of purified SLE LDGs/NDNs from clinically quiescent subjects and HC NDNs were quantified, using optimized purification strategies to avoid disruption of biomechanical properties (Supplementary Figure 1). Gating strategies allowed for identification of neutrophils, monocytes, lymphocytes and eosinophils in mixed cell fractions (Figure 4A) 37. Biomechanical properties did not differ in lymphocytes and monocytes between SLE and HC subjects (Supplementary Figure 4). In contrast, SLE LDGs displayed distinct biomechanical features relative to other neutrophil subsets (Figure 4E). While HC and SLE NDNs were round and smooth, SLE LDGs had significantly rougher cell surfaces that correlated with age but not with other clinical/demographic characteristics (Supplementary Figure 5). HC NDNs incubated for various time-points with Sm/RNP immune complexes 7 and/or recombinant IFN-α displayed no changes in neutrophil roughness (Supplementary Figure 6). Overall, SLE LDGs display distinct biomechanical properties seemingly unrelated to exposure to immune complexes or type I IFNs.
Neutrophil percentages were higher in SLE than HC PBMC fractions (Figure 4C–D), consistent with higher LDG numbers 9. It is unclear whether LDGs are present in small numbers in healthy individuals but expanded in SLE 38. We compared biomechanical properties of SLE LDGs to the small population of HC LDGs. Like autologous HC NDNs, and not consistent with the SLE LDG biomechanical phenotype, HC LDGs displayed smooth, non-polarized surfaces. HC LDGs were also more deformable than HC NDNs (Figure 5C). These differences in biomechanical properties support SLE LDGs do not represent expansion of a minor LDG population found in HCs.
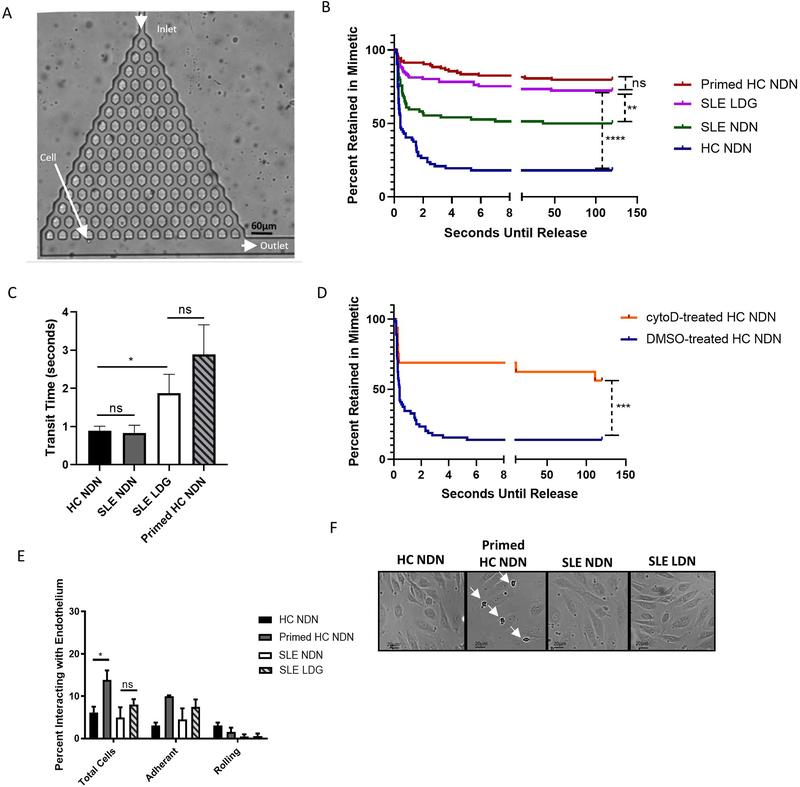
Figure 5. SLE LDGs are increasingly retained within a three-dimensional pulmonary microvasculature mimetic, but do not display enhanced adherence to endothelial cells in a two-dimensional system of flow.
(A) A branching microfluidic mimetic of the pulmonary microvasculature was designed. Arrows indicate the inlet, outlet, and a cell navigating the network, 10x objective. (B) Retention of NDNs and LDGs in the microvasculature mimetic (n>150 neutrophils per subset from n=6 HC volunteers and n=6 SLE patients). Seconds until release refers to the amount of time each neutrophil was retained within the mimetic, measured from entry at the inlet to exit at an outlet. Seconds until release was recorded as >120 seconds if cells did not exit the mimetic within the two-minute video. Times were determined manually with a timer superimposed on the video during data collection. Significance was determined by log-rank test. (C) Transit times through the microvasculature mimetic for all NDNs and LDGs navigating the entire device. Significance assessed by Mann-Whitney U-test. (D) Retention of HC NDNs treated with DMSO or cytochalasin D in the microvasculature mimetic (n>50 neutrophils from n=3 HC volunteers). Significance was determined by log-rank test. (E) Percentage of neutrophils interacting with endothelium under 0.4mL/min flow. Significance was determined by Kruskal-Wallis test with post hoc Dunn’s tests for multiple comparisons. (F) Light microscopy of neutrophil binding to endothelium post-flow assay. Arrows show enhanced binding of primed NDNs. Images representative of n=4 images obtained for each neutrophil subset isolated from n=6 SLE patients or n=6 HC volunteers. All results are mean ± SEM with significance was set at *p ≤ 0.05, **p≤ 0.01, ****p≤ 0.0001, ns=not significant.
While fMLF-primed HC NDNs are morphologically rougher 17 and localize to the PBMC interphase on density gradients (Figure 4D) 17, they were consistently larger than autologous unprimed NDNs. This contrasts with SLE LDGs, which were similar in size to autologous SLE NDNs (Figure 4E), supporting primed HC NDNs and SLE LDGs are biomechanically distinct. These biomechanical differences were confirmed by brightfield (Figure 4F–G) and lattice light-sheet fluorescence microscopy (Supplementary Figure 7). The cell surface of fMLF-primed HC NDNs appeared to ruffle, with small membrane perturbations moving inwards and outwards. In contrast, SLE LDGs’ cell surface was smooth, except for sections of dramatic protrusions, which were irregularly shaped (Figure 4F–G). This suggests SLE LDGs have distinct biophysical properties not consistent with acutely primed phenotypes.
SLE LDGs are retained in a microfluidic microvasculature mimetic (MMM)
Neutrophil biomechanical properties can modulate transit through the pulmonary microvasculature. Primed neutrophils are retained in pulmonary capillary beds 39, possibly due to enhanced cell stiffness and/or irregular cell shape 40. To mimic trafficking through the pulmonary microvasculature, we developed an MMM formed of a branched pyramidal network within a PDMS chip (Figure 5A). Neutrophils flowed through this network at physiologically relevant pressures (10 and 50mbar, or 10.2 and 51.0 cmH2O, respectively) 40 without impact on viability (Supplemental Figure 8). As previously reported 39,40, fMLF-primed HC NDNs were increasingly retained in the MMM, with >80% unable to fully navigate it. In contrast, >80% unprimed HC NDNs navigated the MMM within three seconds. Trafficking patterns of SLE LDGs resembled those of primed HC NDNs, with >75% SLE LDGs retained in the MMM vs approximately 50% SLE NDNs (Figure 5B). Of neutrophils transiting the entire MMM, SLE and unprimed HC NDNs averaged a transit time of <0.9 seconds, while SLE LDGs and primed HC NDNs averaged transit times of 1.87 and 2.89 seconds, respectively (Figure 5C).
HC NDNs treated with cytochalasin D, which disassembles filament actin and decreases neutrophil deformability 36, were increasingly retained in the MMM (>75% retention vs. <20% in vehicle-treated HC NDNs (Figure 5D)), supporting biomechanical modulation alters neutrophil trafficking. Like primed HC NDNs 39, SLE LDGs may be preferentially retained in microvasculature due to biomechanical property differences.
The MMM evaluated effects of cellular biomechanical properties on trafficking but not the putative role of neutrophil-endothelial interactions. By decoupling effects of biophysical properties and cell-surface markers on neutrophil trafficking, we evaluated their independent contributions. A two-dimensional assay evaluated neutrophil interactions-rolling alongside or adherence to microvascular endothelium- in a circulatory flow system mimicking physiological conditions (flow rate 0.4 mL/min). Over three minutes, 15% and 60% primed HC NDNs interacted with unstimulated or stimulated endothelium, respectively. In contrast, <10% and <20% HC NDNs, SLE LDGs and NDNs interacted with unstimulated and stimulated endothelium, respectively (p<0.01 compared to primed HC NDNs and p>0.05 comparing other neutrophil subsets; Figure 5E–F; Supplementary Figure 9). These observations suggest that, while enhanced neutrophil-endothelium interactions may contribute to microvasculature retention of primed HC NDNs, they do not explain differences in microvasculature trafficking observed between SLE LDGs and NDNs. Overall, SLE LDGs may be retained in microvasculature networks 39, by intrinsic changes in cellular biomechanical properties rather than by specific neutrophil-endothelium interactions.
Discussion
We identified significant differences between the SLE and healthy control neutrophil proteomes as well as heterogeneity in the proteome of SLE neutrophils including proteins involved in formation/rearrangement of the cytoskeleton. In addition, we identified biomechanical differences in SLE LDGs with implications for neutrophil trafficking in the microvasculature.
Consistent with the proteomics data and previous transcriptomic analyses reporting differential gene expression associated with the actin cytoskeleton in SLE LDGs 8, we found SLE LDGs are biomechanically distinct and showed cell membrane perturbations differing from fMLF-primed neutrophils 17,41. While mechanisms promoting enhanced SLE LDG cytoskeletal changes remain unclear, differential abundance of proteins associated with extracellular structure organization and cytolysis may be implicated. For example, PFN1 modulates actin/microtubule dynamics 30,42 and actin polymerization 43, while HRG induces neutrophil morphologic changes 29 implicated in neutrophil retention in microvasculature 29,44. Furthermore, the enhanced ability of LDGs to form NETs may contribute to cytoskeleton perturbations and disruptions in cell membrane integrity 15,45–47. Although SLE LDGs did not morphologically resemble HC neutrophils treated with Sm/RNP immune complexes or PMA to induce NETosis (Supplementary Figures 6, 7), differences in spontaneous LDG NET formation and PMA-induced NET formation have been reported 48,7,8. The potential link between NET formation, neutrophil proteome and biomechanical properties of LDGs should be studied further.
Previous studies indicate rougher, primed neutrophils are retained in the lungs 29,40. Our MMM data suggests LDG roughness may similarly hinder LDGs’ ability to traffic through narrow capillaries and biophysical properties, not enhanced binding to endothelium 40. Increased retention in microvasculature networks could have pathogenic implications in lung or kidney damage, and in development of small vessel vasculopathy. SLE lung manifestations are associated with blood vessel damage triggered by neutrophils 49–51. Circulating immune complexes can activate neutrophils, promote endothelial cell barrier dysfunction and perturbed vascular permeability 52,53. While distinct biomechanical properties of SLE LDGs did not align with preferential binding to microvascular endothelial cells, LDGs have potent deleterious effects on endothelium through NET formation 7,54,55. Accordingly, we propose a model where slow LDG microvasculature transit, coupled to enhanced NETosis, promotes vasculopathy. Future studies should assess mechanisms of enhanced SLE LDG roughness and in vivo significance of its effect on LDG trafficking.
In contrast to SLE LDGs, fMLF-primed NDNs showed enhanced adherence to endothelium and higher abundance of phosphoproteins linked to cell adhesion 56–59. Actin-regulatory proteins are de-phosphorylated in LDGs but phosphorylated in primed neutrophils, suggesting both actin depolymerization and polymerization may induce biophysical changes affecting trafficking. Indeed, imaging showed primed NDNs with contracted cortical actin rings while LDGs appeared irregularly shaped with incomplete actin rings (Supplementary Figure 7, Movies 1–4). Overall, SLE LDGs differ from acutely primed neutrophils and interact with the vasculature differently.
The type I IFN pathway is linked to SLE pathogenesis and neutrophils responding to these cytokines exhibit proinflammatory responses 4. ISG-encoded proteins were higher in SLE LDGs, consistent with transcriptome reports 16. Why SLE LDGs express higher ISG-encoded proteins than autologous SLE NDNs, exposed to similar levels of cytokines in vivo, could be related to differences in JAK-STAT activity or to differences in activation status 16. SLE LDGs have enhanced ISG hypomethylation relative to HC neutrophils, perhaps modulating the protein response 60. Future studies should address how enhanced IFN responses modulate pathogenic differences linked to LDGs’ ability to NET and damage vasculature.
The SLE LDG proteome contained increased acute phase response proteins associated with complement and coagulation 61. Corroborating our findings, LDGs display significantly enhanced transcription of several complement components (Supplementary Figure 3). Some complement proteins identified by proteomics were not identified by transcriptomics. These proteins, including C6-C9, may bind to circulating neutrophils. This aligns with findings of C6-C9 contributing to formation of MAC-induced lytic pores in rheumatoid arthritis neutrophils62. Additionally, activated HC NDNs upregulate C3 transcription (Supplementary Figure 10), suggesting activated LDGs may behave in a similar manner. This LDG-complement relationship should be investigated further.
Some proteins associated to platelet biology were more abundant in SLE LDGs, similar to descriptions in psoriasis LDGs63. This was confirmed by fluorescence microscopy (Supplementary Figure 1) and suggests commonalities in the proteome of LDGs across inflammatory diseases associated with enhanced vascular damage. Platelet-neutrophil interactions can drive inflammation and thrombosis 64; thus, increased platelet presence in LDG samples may contribute to their upregulation of coagulation and some neutrophil activation-associated proteins relative to NDNs. Ultimately, LDG-platelet interactions may play distinct pathophysiologic roles in vasculopathy development.
Alongside the proteomics, the biomechanical profile and trafficking pattern of SLE LDGs support reports that LDGs represent a distinct neutrophil subset rather than expansion of immature/primed neutrophils present in healthy subjects 15,65,66. SLE LDGs have a distinct proteomic signature and specific biomechanical features impacting transit through the microvasculature. This study adds to the understanding of neutrophil heterogeneity in the context of blood vessel trafficking, with important implications for development of small vessel vasculopathy and organ damage and development of therapeutics modulating neutrophil biomechanical properties 67.
Supplementary Material
Key messages:
What is already known about this subject? Low density granulocytes (LDGs) are a subset of neutrophils expanded in SLE. These cells have been shown to have a pathogenic role through their enhanced ability to form neutrophil extracellular traps, promote type I IFN responses and damage the vasculature. Their levels and gene signature associate with enhanced vasculopathy and atherosclerosis in lupus patients.
What does this study add? The findings from this study indicate that lupus LDGs display distinct proteomic and biomechanical properties that may impact their ability to travel through the vasculature, interact with the endothelium and enhance their trapping in the small vessels of various organs.
How might this impact on clinical practice? Increased retention of lupus LDGs in microvasculature could have pathogenic implications in lung or kidney damage, and in development of small vessel vasculopathy. These results suggest that development of therapeutics modulating neutrophil biomechanical properties could modulate deleterious responses in lupus and other autoimmune diseases.
Acknowledgements:
We thank Jane Hollis, Cecelia Matara, and Frederica Mescla for their assistance in drawing clinical blood samples.
Funding Information: This research was supported by the Intramural Research programs at NIAMS (ZIA-AR041199), NHLBI and NIBIB. ACM is supported by a Clinical Research Career Development Fellowship from the Wellcome Trust (WT 2055214/Z/16/Z). CS is funded by Medical Research Council, Wellcome Trust, British Heart Foundation, Glaxo Smith Kline, Astra Zeneca and NIHR Cambridge Biomedical Research Centre. ERC receives funding from MRC, Wellcome Trust, GlaxoSmithKline, the NHLI Foundation and the NIHR Imperial Biomedical Research Centre. AJTW was supported by a Gates Cambridge Scholarship. KRB was also supported by a National Institutes of Health OxCam Scholarship.
Footnotes
Completing Interests: The authors declare no competing interests exist.
Ethical Approval Information: All studies were approved by site-specific IRBs: the Cambridge Local Research Ethics Committee (REC reference 06/Q0108/281) and NIAMS/NIDDK IRB (NIH 94-AR-0066). All subjects signed informed consent.
Data Sharing: The raw proteomics data are being uploaded into the PRIDE database.
Patient and public involvement: Patients and/or the public were not involved in the design, or conduct, or reporting, or dissemination plans of this research.
References
- 1.Cusick MF, Libbey JE, Fujinami RS. Molecular mimicry as a mechanism of autoimmune disease. Clin Rev Allergy Immunol 2012;42(1):102–11. doi: 10.1007/s12016-011-8294-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Obermoser G, Pascual V. The interferon-α signature of systemic lupus erythematosus. Lupus 2010;19(9):1012–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dema B, Charles N. Advances in mechanisms of systemic lupus erythematosus. Discov Med 2014;17(95):247–55. [published Online First: 2014/06/03] [PubMed] [Google Scholar]
- 4.Garcia-Romo GS, Caielli S, Vega B, et al. Netting neutrophils are major inducers of type I IFN production in pediatric systemic lupus erythematosus. Sci Transl Med 2011;3(73):73ra20–73ra20. doi: 10.1126/scitranslmed.3001201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carmona-Rivera C, Zhao W, Yalavarthi S, et al. Neutrophil extracellular traps induce endothelial dysfunction in systemic lupus erythematosus through the activation of matrix metalloproteinase-2. Annals of the rheumatic diseases 2015;74(7):1417–24. doi: 10.1136/annrheumdis-2013-204837 [published Online First: 2014/02/25] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaplan MJ. Neutrophils in the pathogenesis and manifestations of SLE. Nature reviews Rheumatology 2011;7(12):691–99. doi: 10.1038/nrrheum.2011.132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lood C, Blanco LP, Purmalek MM, et al. Neutrophil extracellular traps enriched in oxidized mitochondrial DNA are interferogenic and contribute to lupus-like disease. Nature medicine 2016;22(2):146–53. doi: 10.1038/nm.4027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Villanueva E, Yalavarthi S, Berthier CC, et al. Netting neutrophils induce endothelial damage, infiltrate tissues and expose immunostimulatory molecules in systemic lupus erythematosus. J Immunol 2011;187(1):538–52. doi: 10.4049/jimmunol.1100450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Denny MF, Yalavarthi S, Zhao W, et al. A distinct subset of proinflammatory neutrophils isolated from patients with systemic lupus erythematosus induces vascular damage and synthesizes type I IFNs. J Immunol 2010;184(6):3284–97. doi: 10.4049/jimmunol.0902199 [published Online First: 2010/02/19] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carlucci PM, Purmalek MM, Dey AK, et al. Neutrophil subsets and their gene signature associate with vascular inflammation and coronary atherosclerosis in lupus. JCI Insight 2018;3(8) doi: 10.1172/jci.insight.99276 [published Online First: 2018/04/20] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Denny MF, Thacker S, Mehta H, et al. Interferon-alpha promotes abnormal vasculogenesis in lupus: a potential pathway for premature atherosclerosis. Blood 2007;110(8):2907–15. doi: 10.1182/blood-2007-05-089086 [published Online First: 2007/07/16] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rahman S, Sagar D, Hanna RN, et al. Low-density granulocytes activate T cells and demonstrate a non-suppressive role in systemic lupus erythematosus. Ann Rheum Dis 2019;78(7):957–66. doi: 10.1136/annrheumdis-2018-214620 [published Online First: 2019/05/02] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mistry P, Carmona-Rivera C, Ombrello AK, et al. Dysregulated neutrophil responses and neutrophil extracellular trap formation and degradation in PAPA syndrome. Ann Rheum Dis 2018;77(12):1825–33. doi: 10.1136/annrheumdis-2018-213746 [published Online First: 2018/08/23] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wright HL, Makki FA, Moots RJ, et al. Low-density granulocytes: functionally distinct, immature neutrophils in rheumatoid arthritis with altered properties and defective TNF signalling. J Leukoc Biol 2017;101(2):599–611. doi: 10.1189/jlb.5A0116-022R [published Online First: 2016/09/08] [DOI] [PubMed] [Google Scholar]
- 15.Carmona-Rivera C, Kaplan MJ. Low density granulocytes: a distinct class of neutrophils in systemic autoimmunity. Seminars in immunopathology 2013;35(4):455–63. doi: 10.1007/s00281-013-0375-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mistry P, Nakabo S, O’Neil L, et al. Transcriptomic, epigenetic, and functional analyses implicate neutrophil diversity in the pathogenesis of systemic lupus erythematosus. Proceedings of the National Academy of Sciences 2019;116(50):25222–28. doi: 10.1073/pnas.1908576116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bashant KR, Vassallo A, Herold C, et al. Real-time deformability cytometry reveals sequential contraction and expansion during neutrophil priming. Journal of Leukocyte Biology 2019;105(6):1143–53. doi: 10.1002/JLB.MA0718-295RR [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wood AJT, Vassallo AM, Ruchaud-Sparagano M-H, et al. C5a impairs phagosomal maturation in the neutrophil through phosphoproteomic remodeling. JCI Insight 2020;5(15) doi: 10.1172/jci.insight.137029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Amulic B, Cazalet C, Hayes GL, et al. Neutrophil Function: From Mechanisms to Disease. Annual Review of Immunology 2012;30(1):459–89. doi: 10.1146/annurev-immunol-020711-074942 [DOI] [PubMed] [Google Scholar]
- 20.Ge SX, Jung D, Yao R. ShinyGO: a graphical gene-set enrichment tool for animals and plants. Bioinformatics 2019. doi: 10.1093/bioinformatics/btz931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou Y, Zhou B, Pache L, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nature Communications 2019;10(1):1523. doi: 10.1038/s41467-019-09234-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reutershan J, Morris MA, Burcin TL, et al. Critical role of endothelial CXCR2 in LPS-induced neutrophil migration into the lung. J Clin Invest 2006;116(3):695–702. doi: 10.1172/JCI27009 [published Online First: 2006/02/16] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stillie R, Farooq SM, Gordon JR, et al. The functional significance behind expressing two IL–8 receptor types on PMN. Journal of Leukocyte Biology 2009;86(3):529–43. doi: 10.1189/jlb.0208125 [DOI] [PubMed] [Google Scholar]
- 24.Smith NLD, Bromley MJ, Denning DW, et al. Elevated Levels of the Neutrophil Chemoattractant Pro–Platelet Basic Protein in Macrophages From Individuals With Chronic and Allergic Aspergillosis. The Journal of Infectious Diseases 2014;211(4):651–60. doi: 10.1093/infdis/jiu490 [DOI] [PubMed] [Google Scholar]
- 25.Tucci M, Quatraro C, Lombardi L, et al. Glomerular accumulation of plasmacytoid dendritic cells in active lupus nephritis: Role of interleukin-18. Arthritis & Rheumatism 2008;58(1):251–62. doi: 10.1002/art.23186 [DOI] [PubMed] [Google Scholar]
- 26.Leonard WJ, O’Shea JJ. JAKS AND STATS: Biological Implications. Annual Review of Immunology 1998;16(1):293–322. doi: 10.1146/annurev.immunol.16.1.293 [DOI] [PubMed] [Google Scholar]
- 27.Lombardi B, Rendell N, Edwards M, et al. Evaluation of phosphopeptide enrichment strategies for quantitative TMT analysis of complex network dynamics in cancer-associated cell signalling. EuPA Open Proteomics 2015;6:10–15. doi: 10.1016/j.euprot.2015.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Akk A, Springer LE, Yang L, et al. Complement activation on neutrophils initiates endothelial adhesion and extravasation. Mol Immunol 2019;114:629–42. doi: 10.1016/j.molimm.2019.09.011 [published Online First: 2019/09/23] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nishibori M, Wake H, Mori S, et al. Histidine-rich glycoprotein prevents septic lethality through neutrophil regulation. Crit Care 2014;18(Suppl 2):P23–P23. doi: 10.1186/cc14026 [published Online First: 2014/12/03] [DOI] [Google Scholar]
- 30.Pinto-Costa R, Sousa MM. Profilin as a dual regulator of actin and microtubule dynamics. Cytoskeleton 2020;77(3–4):76–83. doi: 10.1002/cm.21586 [DOI] [PubMed] [Google Scholar]
- 31.Schenk LK, Möller-Kerutt A, Klosowski R, et al. Angiotensin II regulates phosphorylation of actin-associated proteins in human podocytes. The FASEB Journal 2017;31(11):5019–35. doi: 10.1096/fj.201700142R [DOI] [PubMed] [Google Scholar]
- 32.Föger N, Jenckel A, Orinska Z, et al. Differential regulation of mast cell degranulation versus cytokine secretion by the actin regulatory proteins Coronin1a and Coronin1b. J Exp Med 2011;208(9):1777–87. doi: 10.1084/jem.20101757 [published Online First: 2011/08/15] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sandí M-J, Marshall CB, Balan M, et al. MARK3-mediated phosphorylation of ARHGEF2 couples microtubules to the actin cytoskeleton to establish cell polarity. Science Signaling 2017;10(503):eaan3286. doi: 10.1126/scisignal.aan3286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pai A, Sundd P, Tees DFJ. In situ Microrheological Determination of Neutrophil Stiffening Following Adhesion in a Model Capillary. Annals of Biomedical Engineering 2008;36(4):596–603. doi: 10.1007/s10439-008-9437-8 [DOI] [PubMed] [Google Scholar]
- 35.Worthen GS, Schwab B, Elson EL, et al. Mechanics of stimulated neutrophils: cell stiffening induces retention in capillaries. Science 1989;245(4914):183. [DOI] [PubMed] [Google Scholar]
- 36.Otto O, Rosendahl P, Mietke A, et al. Real-time deformability cytometry: on-the-fly cell mechanical phenotyping. Nature Methods 2015;12:199. doi: 10.1038/nmeth.3281 https://www.nature.com/articles/nmeth.3281#supplementary-information [DOI] [PubMed] [Google Scholar]
- 37.Toepfner N, Herold C, Otto O, et al. Detection of human disease conditions by single-cell morpho-rheological phenotyping of blood. eLife 2018;7:e29213. doi: 10.7554/eLife.29213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kegerreis BJ, Catalina MD, Geraci NS, et al. Genomic Identification of Low-Density Granulocytes and Analysis of Their Role in the Pathogenesis of Systemic Lupus Erythematosus. The Journal of Immunology 2019;202(11):3309–17. doi: 10.4049/jimmunol.1801512 [DOI] [PubMed] [Google Scholar]
- 39.Summers C, Singh NR, White JF, et al. Pulmonary retention of primed neutrophils: a novel protective host response, which is impaired in the acute respiratory distress syndrome. Thorax 2014;69(7):623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ekpenyong AE, Toepfner N, Fiddler C, et al. Mechanical deformation induces depolarization of neutrophils. Science Advances 2017;3(6) doi: 10.1126/sciadv.1602536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Doodnauth SA, Grinstein S, Maxson ME. Constitutive and stimulated macropinocytosis in macrophages: roles in immunity and in the pathogenesis of atherosclerosis. Philos Trans R Soc Lond B Biol Sci 2019;374(1765):20180147. doi: 10.1098/rstb.2018.0147 [published Online First: 2019/04/11] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nejedla M, Sadi S, Sulimenko V, et al. Profilin connects actin assembly with microtubule dynamics. Mol Biol Cell 2016;27(15):2381–93. doi: 10.1091/mbc.E15-11-0799 [published Online First: 2016/06/17] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alkam D, Feldman EZ, Singh A, et al. Profilin1 biology and its mutation, actin(g) in disease. Cell Mol Life Sci 2017;74(6):967–81. doi: 10.1007/s00018-016-2372-1 [published Online First: 2016/09/28] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Terao K, Wake H, Adachi N, et al. Histidine-Rich Glycoprotein Suppresses Hyperinflammatory Responses of Lung in a Severe Acute Pancreatitis Mouse Model. Pancreas 2018;47(9):1156–64. doi: 10.1097/mpa.0000000000001153 [published Online First: 2018/09/08] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thiam HR, Wong SL, Qiu R, et al. NETosis proceeds by cytoskeleton and endomembrane disassembly and PAD4-mediated chromatin decondensation and nuclear envelope rupture. Proceedings of the National Academy of Sciences 2020;117(13):7326. doi: 10.1073/pnas.1909546117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Metzler KD, Goosmann C, Lubojemska A, et al. A myeloperoxidase-containing complex regulates neutrophil elastase release and actin dynamics during NETosis. Cell Rep 2014;8(3):883–96. doi: 10.1016/j.celrep.2014.06.044 [published Online First: 2014/07/30] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Neubert E, Meyer D, Rocca F, et al. Chromatin swelling drives neutrophil extracellular trap release. Nature Communications 2018;9(1):3767. doi: 10.1038/s41467-018-06263-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gupta S, Chan DW, Zaal KJ, et al. A High-Throughput Real-Time Imaging Technique To Quantify NETosis and Distinguish Mechanisms of Cell Death in Human Neutrophils. J Immunol 2018;200(2):869–79. doi: 10.4049/jimmunol.1700905 [published Online First: 2017/12/03] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shahane A Pulmonary hypertension in rheumatic diseases: epidemiology and pathogenesis. Rheumatol Int 2013;33(7):1655–67. doi: 10.1007/s00296-012-2659-y [published Online First: 2013/01/22] [DOI] [PubMed] [Google Scholar]
- 50.Carreira PE. Pulmonary hypertension in autoimmune rheumatic diseases. Autoimmunity Reviews 2004;3(4):313–20. doi: 10.1016/j.autrev.2003.11.004 [DOI] [PubMed] [Google Scholar]
- 51.Keane MP, Lynch JP. Pleuropulmonary manifestations of systemic lupus erythematosus. Thorax 2000;55(2):159. doi: 10.1136/thorax.55.2.159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xia Y, Herlitz LC, Gindea S, et al. Deficiency of fibroblast growth factor-inducible 14 (Fn14) preserves the filtration barrier and ameliorates lupus nephritis. J Am Soc Nephrol 2015;26(5):1053–70. doi: 10.1681/ASN.2014030233 [published Online First: 2014/09/30] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Burg N, Swendeman S, Worgall S, et al. Sphingosine 1-Phosphate Receptor 1 Signaling Maintains Endothelial Cell Barrier Function and Protects Against Immune Complex-Induced Vascular Injury. Arthritis Rheumatol 2018;70(11):1879–89. doi: 10.1002/art.40558 [published Online First: 2018/05/22] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.King KR, Aguirre AD, Ye Y-X, et al. IRF3 and type I interferons fuel a fatal response to myocardial infarction. Nature Medicine 2017;23(12):1481–87. doi: 10.1038/nm.4428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Knight JS, Luo W, O’Dell AA, et al. Peptidylarginine deiminase inhibition reduces vascular damage and modulates innate immune responses in murine models of atherosclerosis. Circ Res 2014;114(6):947–56. doi: 10.1161/CIRCRESAHA.114.303312 [published Online First: 2014/01/14] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lertkiatmongkol P, Liao D, Mei H, et al. Endothelial functions of platelet/endothelial cell adhesion molecule-1 (CD31). Curr Opin Hematol 2016;23(3):253–59. doi: 10.1097/MOH.0000000000000239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhu P, Sang Y, Xu H, et al. ADAM22 plays an important role in cell adhesion and spreading with the assistance of 14–3-3. Biochemical and Biophysical Research Communications 2005;331(4):938–46. doi: 10.1016/j.bbrc.2005.03.229 [DOI] [PubMed] [Google Scholar]
- 58.Frank SR, Köllmann CP, van Lidth de Jeude JF, et al. The focal adhesion-associated proteins DOCK5 and GIT2 comprise a rheostat in control of epithelial invasion. Oncogene 2017;36(13):1816–28. doi: 10.1038/onc.2016.345 [published Online First: 2016/09/26] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Albarran-Juarez J, Iring A, Wang S, et al. Piezo1 and Gq/G11 promote endothelial inflammation depending on flow pattern and integrin activation. J Exp Med 2018;215(10):2655–72. doi: 10.1084/jem.20180483 [published Online First: 2018/09/09] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Coit P, Yalavarthi S, Ognenovski M, et al. Epigenome profiling reveals significant DNA demethylation of interferon signature genes in lupus neutrophils. J Autoimmun 2015;58:59–66. doi: 10.1016/j.jaut.2015.01.004 [published Online First: 2015/01/28] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.de Bont CM, Boelens WC, Pruijn GJM. NETosis, complement, and coagulation: a triangular relationship. Cellular & Molecular Immunology 2019;16(1):19–27. doi: 10.1038/s41423-018-0024-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Romero V, Fert-Bober J, Nigrovic PA, et al. Immune-mediated pore-forming pathways induce cellular hypercitrullination and generate citrullinated autoantigens in rheumatoid arthritis. Sci Transl Med 2013;5(209):209ra150. doi: 10.1126/scitranslmed.3006869 [published Online First: 2013/11/01] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Teague HL, Varghese NJ, Tsoi LC, et al. Neutrophil Subsets, Platelets, and Vascular Disease in Psoriasis. JACC: Basic to Translational Science 2019;4(1):1. doi: 10.1016/j.jacbts.2018.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lisman T Platelet-neutrophil interactions as drivers of inflammatory and thrombotic disease. Cell Tissue Res 2018;371(3):567–76. doi: 10.1007/s00441-017-2727-4 [published Online First: 2017/11/25] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Singh N, Traisak P, Martin KA, et al. Genomic alterations in abnormal neutrophils isolated from adult patients with systemic lupus erythematosus. Arthritis Res Ther 2014;16(4):R165. doi: 10.1186/ar4681 [published Online First: 2014/08/12] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hassani M, Hellebrekers P, Chen N, et al. On the origin of low-density neutrophils. Journal of Leukocyte Biology 2020;107(5):809–18. doi: 10.1002/JLB.5HR0120-459R [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Craciun EM, Altfelder F, Kuss N, et al. Anti-inflammatory effects of selected drugs on activated neonatal and adult neutrophils. Scandinavian Journal of Clinical and Laboratory Investigation 2013;73(5):407–13. doi: 10.3109/00365513.2013.796591 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.