Abstract
A subset of human prostate cancer exhibits increased de novo synthesis of fatty acids, but the molecular driver(s) of this metabolic abnormality remains obscure. This study demonstrates a novel metabolic function of c-Myc (Myc) in regulation of fatty acid synthesis. The role of Myc in regulation of fatty acid synthesis was investigated by: (a) interrogation of the prostate cancer The Cancer Genome Atlas (TCGA) dataset, (b) chromatin immunoprecipitation, and (c) determination of the expression of fatty acid synthesis enzymes and targeted metabolomics using a mouse model and human specimens. The expression of MYC was positively associated with that of key fatty acid synthesis genes including ACLY, ACC1, and FASN in prostate cancer TCGA dataset. Chromatin immunoprecipitation revealed Myc occupancy at the promoters of ACLY, ACC1, and FASN. Prostate-specific overexpression of Myc in Hi-Myc transgenic mice resulted in overexpression of ACLY, ACC1, and FASN proteins in neoplastic lesions and increased circulating levels of total free fatty acids. Targeted metabolomics confirmed increased circulating levels of individual fatty acids in the plasma of Hi-Myc mice and human subjects when compared to corresponding controls. Immunohistochemistry also revealed a positive and statistically significant association in expression of Myc with that of ACC1 in human prostate adenocarcinoma specimens. We propose that Myc-regulated fatty acid synthesis is a valid target for therapy and/or prevention of prostate cancer.
Keywords: Prostate cancer, Myc, lipogenesis
Introduction
Molecular heterogeneity of human prostate cancer, a leading cause of cancer-related lethality in American men, is characterized by genomic alterations including DNA copy number changes, gene fusion, deletion, and point mutations [1–5]. For example, deletion on chromosome 8p21 for NKX3.1, amplification of chromosome 8q24 mapping Myc oncogene, and homozygous deletion of tumor suppressor phosphatase and tensin homolog (PTEN) on chromosome 10q23 are some of the most frequently observed genomic alterations in human prostate cancers [6–8]. The oncogenic role of c-Myc (Myc) in the pathogenesis of prostate cancer is substantiated by both rodent studies and analysis of human prostate cancer specimens. Prostate-specific overexpression of Myc in male transgenic mice (Hi-Myc mice) results in prostatic intraepithelial neoplasia (PIN) by 2–4 weeks of age [9]. The PIN lesions in Hi-Myc mice progress to invasive prostate adenocarcinoma by 6 months of age [9]. Analysis of the surgical specimens of prostate adenocarcinoma (n=9) and benign hypertrophied prostate (n=19) revealed substantially high Myc transcript level in 67% of prostate adenocarcinoma specimens, but not in benign tissues [10]. Gene amplification, rearrangement, and overexpression of Myc was reported in a human prostate cancer cell line (LNCaP) derived from a lymph node metastasis of a human prostate cancer patient [11]. Amplification of Myc together with other oncogenes (e.g., epidermal growth factor receptor 1 gene) was associated with emergence of hormone-resistant (castration-resistant) prostate cancer [12]. Studies have also indicated that increased nuclear Myc protein level is an early event in the pathogenesis of human prostate cancer and evident in luminal cells of PIN lesions [13]. Moreover, Myc overexpression was observed in roughly 70% of early stage prostate cancer [14]. Nuclear Myc protein expression was associated with prostate cancer progression and it was suggested to be a possible predictor of two-year overall survival [15].
At the biochemical level, deregulated metabolism is one of the hallmarks of different cancers including prostate adenocarcinoma [16–18]. Prostate cancers frequently exhibit increased de novo synthesis of fatty acids, which is very low in normal prostate gland [17, 18]. In prostate cancer, citrate from tricarboxylic acid cycle is utilized for the synthesis of acetyl-CoA that is eventually converted to saturated fatty acids (e.g., C16:0 palmitic acid) and further metabolized to unsaturated fatty acids (e.g., oleic acid, an omega-9 monounsaturated fatty acid) through a series of catalytic reactions facilitated by ATP citrate lyase (ACLY), acetyl-CoA carboxylase 1 (a rate limiting enzyme; ACC1 or ACACA), fatty acid synthase (FASN) complex, elongases, and desaturases [17, 18]. Studies have suggested that β-oxidation of fatty acids may be the primary bioenergetics pathway for ATP production in human prostate cancers [19]. In addition to ATP production, the fatty acids are utilized for post-translational modification of oncogenic proteins as well as maintenance of membrane structure in hyper-proliferative cancer cells [17, 18]. Circulating levels of saturated fatty acids and monounsaturated fatty acids were shown to be markers of de novo fatty acid synthesis as well as risk factors for prostate cancer [20]. Collectively, these studies indicate that deregulated fatty acid synthesis is a druggable target for therapy and/or prevention of human prostate cancer.
Integrated metabolic profiling of RWPE-1 normal prostate epithelial cell line stably transfected to overexpress Myc, ventral prostate lobes from 12–13-week old Lo-Myc transgenic mice, and human prostate cancer specimens indicated that Myc expression was associated with deregulated lipid metabolism [21]. However, the mechanism by which Myc regulates fatty acid synthesis in prostate cancer is still elusive. In this study, we have systematically investigated the role of Myc in regulation of fatty acid synthesis by analysis of the prostate cancer The Cancer Genome Atlas (TCGA) RNA-Seq data, and by using prostate cancer specimens and plasma from a transgenic mouse model (Hi-Myc mice) as well as human subjects.
Results
Analyses of TCGA RNA-Seq data and chromatin immunoprecipitation (ChIP) assay
Figure 1a summarizes steps in biochemical conversion of citrate to saturated fatty acids (chemical structure of only C16:0 palmitic acid is shown) and the associated enzymes of this pathway. In the prostate cancer TCGA RNA-Seq dataset, expression of MYC (c-Myc) was positively associated with that of ACLY, ACC1 (also known as ACACA), and FASN (Figure 1b). The MYCN (n-Myc) expression was inversely associated with that of ACLY, ACC1, and FASN in the prostate cancer TCGA (Supplementary Figure S1a). The ovarian tumor TCGA dataset indicated an inverse association for MYC expression with that of ACLY (Supplementary Figure S1b). Collectively, these data implicated Myc in regulation of fatty acid synthesis in prostate cancer, but not in ovarian cancer.
Figure 1.

MYC expression is positively correlated with that of key genes involved in fatty acid synthesis in prostate cancer TCGA. a A simplified diagram showing intermediates of fatty acid synthesis and the enzymes associated with this metabolic pathway. b Correlation of MYC expression with that of key fatty acid synthesis genes in prostate tumors from TCGA (n = 497). Pearson test was used to determine the correlation coefficient and statistical significance. c ChIP assay showing recruitment of Myc on the ACLY, ACC1, and FASN promoters in 22Rv1 cells. Results shown are mean ± SD (n = 3). Statistical significance was determined by Student’s t-test.
As can be seen in Supplementary Figure S2, putative Myc occupancy sequence CATGTG or CACGTG was observed in the promoters of ACLY (2 sites), ACC1 (one site), and FASN (2 sites). The ChIP assay revealed recruitment of Myc at both sites of ACLY promoter and one site each of ACC1 and FASN promoters (Figure 1c). These results suggested direct transcriptional regulation of ACLY, ACC1, and FASN by Myc in prostate cancer.
Expression of fatty acid synthesis proteins in the prostate of WT and Hi-Myc mice
Age-matched prostate sections from WT (normal mice) and Hi-Myc transgenic mice (5, 10, and 26 weeks of age) were used for immunohistochemical analyses of ACLY (Figure 2a), ACC1 (Figure 2b), and FASN (Figure 2c) protein expression to further test the in vivo validity of the gene expression correlations from the prostate cancer TCGA RNA-Seq data (Figure 1b). The incidence of PIN at 5-, 10- and 26-week old Hi-Myc mice varied between 91–100%. The incidence of adenocarcinoma (ADC) in Hi-Myc mice at 5-, 10-, and 26-weeks of age was 43%, 69%, and 73%, respectively. Even at 5-weeks of age with earliest onset of PIN and ADC, the expression of ACC1 (P = 0.04) and FASN (P = 0.01) were significantly higher in the prostate of Hi-Myc mice when compared with age-matched WT mice (Figure 2d). Increased protein levels of ACLY, ACC1, and FASN were also observed in the prostate of Hi-Myc mice at 10 and 26 weeks of age in comparison with corresponding WT mice (Figure 2d). These results indicated upregulation of ACLY, ACC1, and FASN in Myc-driven prostate cancer in a mouse model.
Figure 2.
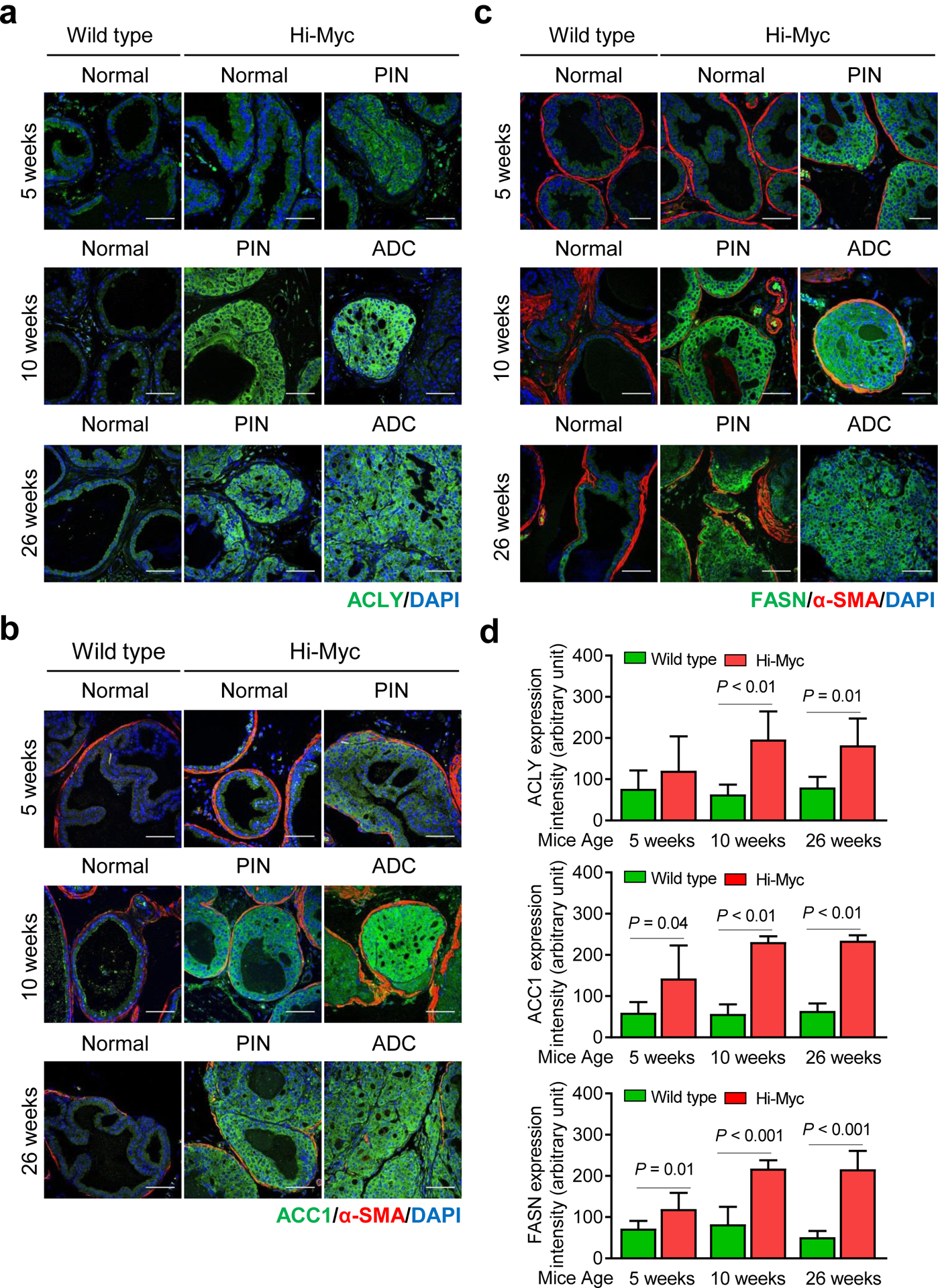
Expression of proteins responsible for fatty acid synthesis is increased in prostatic intraepithelial neoplasia (PIN) and adenocarcinoma (ADC) by prostate-specific transgenic expression of Myc in Hi-Myc mice. Representative immunohistochemical images for (a) ATP citrate lyase (ACLY), (b) acetyl-CoA carboxylase 1 (ACC1), and (c) fatty acid synthase (FASN) in normal prostate, PIN, and ADC of wild-type (WT) or Hi-Myc mice (40× oil objective magnification; scale bar- 50 μm). Sections were stained with a stromal marker α-smooth muscle actin (SMA) protein. Nuclei were visualized by staining with 4′,6-diamidino-2-phenylindole (DAPI), which is a dye that strongly binds to adenine-thymine rich regions in DNA. d Bar graphs showing quantitation of ACLY, ACC1, and FASN protein expression by ImageJ software. Results shown are mean ± SD (n = 6). Statistical significance was determined by Student’s t-test.
Analysis of fatty acids and their intermediaries in the plasma of WT and Hi-Myc mice
The circulating levels of acetyl-CoA, malonyl-CoA, and total free fatty acids were higher in Hi-Myc mice when compared to WT mice at 10 and 26 weeks of age, except that the difference from WT group was not significant for malonyl-CoA at 26 weeks of age possibly due to large data scatter in the Hi-Myc group (Figure 3). Similar increase in Hi-Myc groups compared to corresponding WT mice were also seen for total phospholipids and cholesterol, which are also derived from the fatty acid metabolism pathway at 10 and 26 weeks of age.
Figure 3.
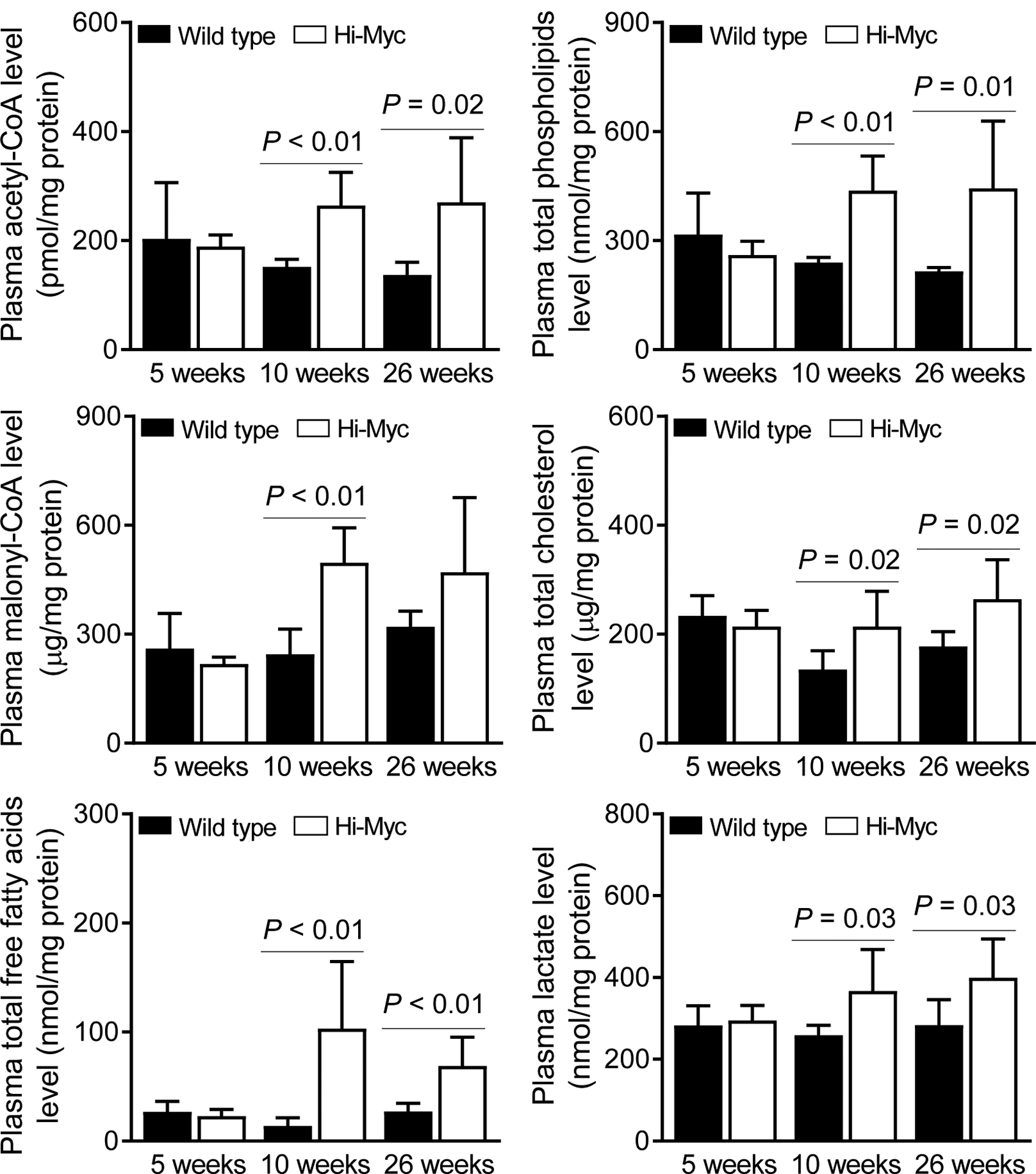
Circulating levels of total free fatty acids and their metabolic intermediaries is increased by prostate-specific transgenic expression of Myc in Hi-Myc mice. Levels of acetyl-CoA, malonyl-CoA, total free fatty acids, total phospholipids, total cholesterol, and lactate in the plasma of Hi-Myc mice and age-matched wild-type (WT) mice. Results shown are mean ± SD (n = 6). Statistical significance was determined by Student’s t-test.
Increased glycolysis in cancer cells (the Warburg effect), a concept first introduced by Dr. Otto Warburg in the fifties, [22] is another common metabolic deregulation. Because Myc is known to regulate expression of certain glycolysis-related genes (e.g., lactate dehydrogenase A), [23, 24] we also measured lactate levels in the plasma of WT and Hi-Myc mice. Plasma level of lactate, the end-product of glycolysis, was also higher in Hi-Myc mice when compared to WT mice at 10 and 26 weeks of age (Figure 3). Collectively, these results indicated that Myc-driven prostate cancer in Hi-Myc mice resulted in elevated circulating levels of total free fatty acids as well as lactate.
The levels of individual fatty acids were also determined in the plasma of 5- and 10-week old WT and Hi-Myc mice by targeted metabolomics, and the results are summarized in Table 1. The levels of two of the three saturated fatty acids (palmitic acid and stearic acid) were higher in the plasma of 10-week old Hi-Myc mice when compared to age-matched WT mice (Table 1). The circulating levels of oleic acid (18:1 cis-9 monounsaturated fatty acid) and linoleic acid (C18:2 cis,cis-9,12-octadecadienoic acid) were also higher in the plasma of Hi-Myc mice in comparison with control (Table 1).
Table 1.
Levels of individual fatty acids in the plasma of 5- and 10-week old Hi-Myc mice and corresponding age-matched wild-type (WT) mice.
| Relative amount to internal control (mean ± SD) | ||||||
|---|---|---|---|---|---|---|
| Metabolites | 5 weeks | 10 weeks | ||||
| WT (n=4~6) | Hi-Myc (n=6) | P value | WT (n=6) | Hi-Myc (n=6) | P value | |
| Palmitic acid | 424.4 ± 26.6 | 524.2 ± 87.8 | 0.06 | 454.9 ± 50.1 | 697.2 ± 145.6 | <0.01 |
| Stearic acid | 197.3 ± 41.47 | 220.6 ± 45.27 | 0.40 | 193.5 ± 23.4 | 309.9 ± 86.9 | 0.01 |
| Palmitoleic acid | 33.6 ± 8.1 | 47.9 ± 6.7 | 0.02 | 44.5 ± 20.0 | 47.1 ± 16.8 | 0.81 |
| Oleic acid | 206.4 ± 14.1 | 240.6 ± 26.4 | <0.05 | 175.7 ± 44.6 | 254.7 ± 60.3 | 0.03 |
| Linoleic acid | 287 ± 42.34 | 338.9 ± 43.5 | 0.10 | 376 ± 70.2 | 522.7 ± 79.5 | <0.01 |
| γ-Linolenic acid | 14.7 ± 3.4 | 16.6 ± 4.7 | 0.50 | 26.2 ± 9.8 | 24.1 ± 8.7 | 0.70 |
| Arachidonic acid | 57.4 ± 17.2 | 75.6 ± 23.1 | 0.18 | 86.5 ± 8.6 | 156.4 ± 80.9 | 0.06 |
| Dihomo-γ-Linolenic acid | 13.1 ± 3.3 | 16.0 ± 3.8 | 0.22 | 14.4 ± 3.0 | 22.3 ± 12.3 | 0.15 |
| Myristic acid | 22.9 ± 18.2 | 26.0 ± 10.4 | 0.72 | 20.2 ± 9.4 | 16.4 ± 10.7 | 0.53 |
| Eicosadienoic acid | 2.8 ± 0.5 | 3.73 ± 0.6 | 0.03 | 3.4 ± 0.8 | 3.4 ± 1.3 | 0.89 |
| Eicosapentaenoic acid | 11.1 ± 3.8 | 11.0 ± 3.5 | 0.98 | 4.1 ± 2.0 | 5.6 ± 2.2 | 0.24 |
| Docosapentaenoic acid | 4.2 ± 1.3 | 5.2 ± 1.6 | 0.31 | 3.9 ± 0.6 | 3.9 ± 1.7 | 0.93 |
| Docosahexaenoic acid | 75.1 ± 24.5 | 94.7 ± 31.1 | 0.28 | 82.3 ± 10.0 | 77.3 ± 43.8 | 0.79 |
| SCD 18/18:1 | 1.2 ± 0.3 | 1.1 ± 0.2 | 0.38 | 0.9 ± 0.2 | 0.9 ± 0.2 | 0.70 |
| SCD 16/16:1 | 0.1 ± 0.03 | 0.1 ± 0.03 | 0.86 | 0.1 ± 0.04 | 0.1 ± 0.02 | 0.12 |
Statistical significance was determined by two-sided Student’s t test. Abbreviations: SCD, Stearoyl-CoA desaturase-1
Expression of ACLY and ACC1 in human PIN and prostate adenocarcinoma
A previous study showed elevated expression of FASN protein in human PIN and prostate adenocarcinoma in comparison with normal prostate tissue [25]. In the present study, we extended these findings and determined the expression of ACLY and ACC1 proteins using tissue microarray consisting of normal prostate, PIN, and prostate adenocarcinoma. Figure 4a shows immunohistochemical staining for ACLY and ACC1 proteins. The mean H-score for ACLY protein was higher by about 2-fold in the prostate adenocarcinoma when compared to PIN (Figure 4b). The expression of ACC1 protein was also higher in prostate adenocarcinoma in comparison with normal prostate tissue (P < 0.001) or PIN (P < 0.001) (Figure 4b). Consistent with these results, the circulating levels of total free fatty acids (Figure 4c) and total phospholipids (Figure 4d) were higher in the plasma of prostate cancer patients than those of patients with PIN and/or normal prostate.
Figure 4.

Expression of fatty acid synthesis proteins in normal and cancerous human prostate. a Representative immunohistochemical images for ACLY and ACC1 protein expression in human normal prostate, prostatic intraepithelial neoplasia (PIN), and adenocarcinoma (ADC) (20× objective magnification, scale bar- 100 μm). b Quantitation of ACLY and ACC1 protein expression (H-score). Results shown are mean ± SD (n = 20 for normal; n = 42 for PIN; n = 66 for ADC). Statistical significance was determined by one-way ANOVA followed by Bonferroni’s test. Plasma levels of (c) total free fatty acids and (d) total phospholipids in men with normal or cancerous prostate. Results shown are mean ± SD (n = 20 for normal and PIN and n = 18 for ADC). Statistical significance was determined by one-way ANOVA followed by Bonferroni’s test.
Association of Myc protein expression with that of ACLY and ACC1
Details of patients whose normal or cancerous prostate specimens were used for co-staining of Myc and ACLY or Myc and ACC1 are included in Supplementary Table S1. Figure 5a shows co-staining for Myc and ACLY or Myc and ACC1 proteins in normal human prostate tissue and in human PIN or prostate adenocarcinoma. Quantitative data for expression intensity of each individual protein are summarized in Figure 5b. A positive association between expression of ACC1 and Myc protein was discernible for human prostate adenocarcinoma specimens (Figure 5c).
Figure 5.
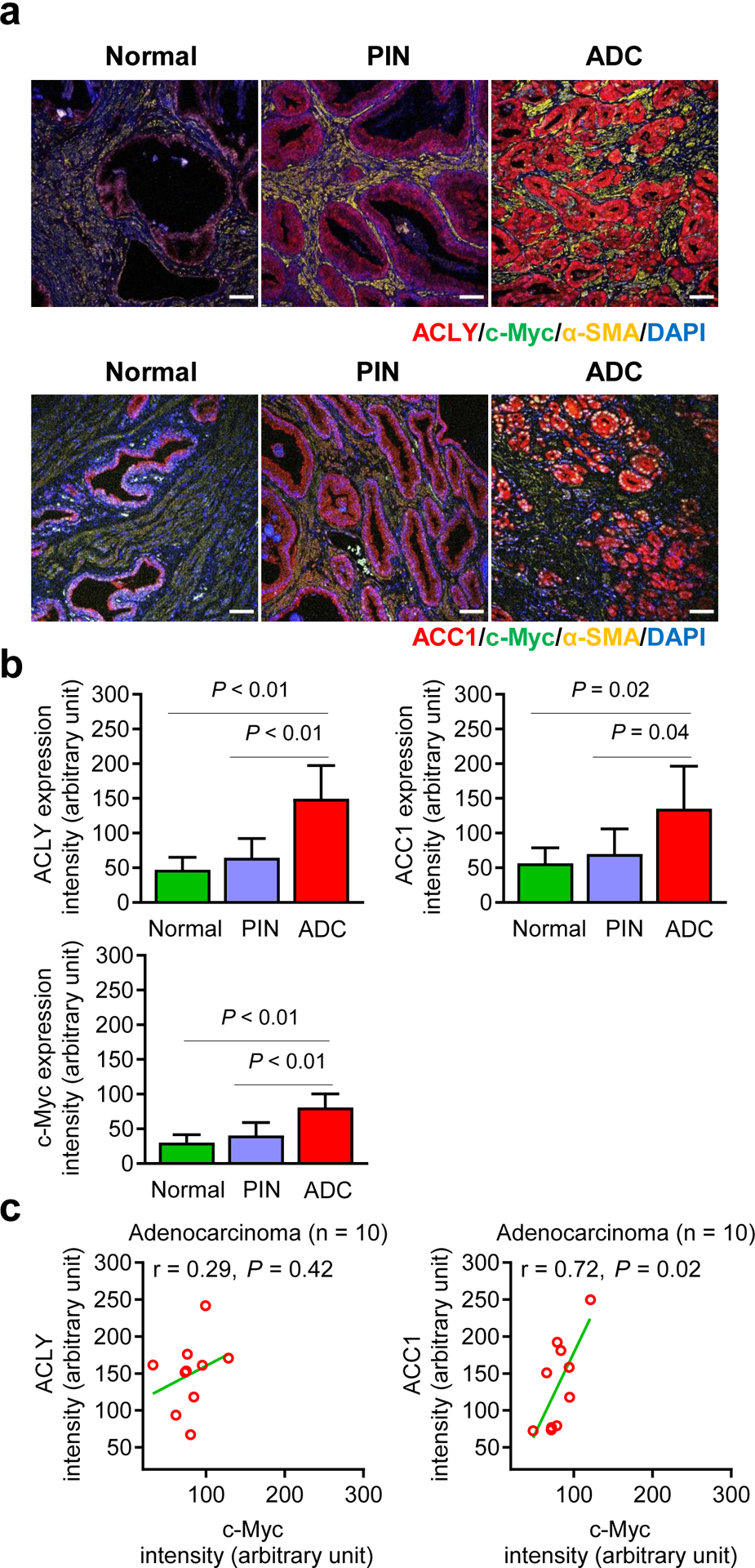
Association of Myc protein expression with that of ACLY or ACC1 in normal and cancerous human prostate. a Representative immunohistochemical images for ACLY, ACC1, c-Myc, and α-smooth muscle actin (α-SMA) in normal and cancerous prostate tissues (20× objective magnification; scale bar- 50 μm). Nuclei were visualized by staining with 4′,6-diamidino-2-phenylindole (DAPI). b Bar graphs show quantitation of ACLY, ACC1, and c-Myc protein expression by ImageJ software. Results shown are mean ± SD [n = 5 for normal prostate, n = 7 for PIN, and n = 10 for adenocarcinoma (ADC)]. Statistical significance was determined by one-way ANOVA followed by Bonferroni’s test. c Correlation of Myc protein expression with that of ACLY or ACC1 protein in prostate ADC (n = 10). Pearson test was used to determine the correlation coefficient and statistical significance.
Circulating levels of fatty acids in human subjects with normal or cancerous prostate
Circulating levels of individual fatty acids were also measured in subjects with normal prostate or with PIN or ADC in the prostate. Details of the patients whose blood were used for targeted metabolomics are included in Supplementary Table S2. The levels of myristic acid and stearic acid were higher in the plasma of prostate ADC patients and/or those with PIN than in the plasma of human subjects with normal prostate (Supplementary Table S3). The level of palmitic acid was higher in the plasma of prostate ADC patients when compared to those with normal prostate, but the difference did not reach significance (Supplementary Table S3). We also observed increased circulating levels of certain other complex lipids, including oleic acid, palmitoleic acid (a monounsaturated fatty acid; 9Z-hexadec-9-enoic acid), and linolenic acid (all-cis-9,12,15-octadecatrienoic acid) and so forth, in prostate ADC patients and/or those with PIN when compared to normal subjects (Supplementary Table S3).
Discussion
Sterol regulatory element binding protein 1 is considered the master regulator of fatty acid synthesis in normal tissues [26]. The present study reveals that Myc is another novel regulator of fatty acid synthesis especially in prostate cancer. Interrogation of the prostate cancer TCGA RNA-Seq data reveals a positive association between expression of Myc with that of ACLY, ACC1, and FASN, and this association seems unique for this malignancy. In ovarian cancer TCGA dataset, a positive association is not seen between Myc expression with that of ACLY, ACC1, or FASN. The molecular basis for prostate tumor-specific function of Myc in regulation of fatty acid synthesis is unclear, but this may be related to androgen receptor (AR). The AR is a well-known driver of human prostate cancer [27]. Both Myc and AR genes are amplified in human prostate cancers [28]. Studies have also shown that Myc is a ligand-independent AR target gene [29]. A strong correlation of Myc expression with that of AR is observed in tumors from patients with castration-resistant prostate cancer [29]. Exposure of androgen-responsive LNCaP human prostate cancer cell line to endogenous androgens (testosterone and dihydrotestosterone) as well as synthetic androgens (R1881 and mibolerone) results in accumulation of neutral lipid droplets [30]. Neutral lipid droplet accumulation after androgen addition was not observed in DU145 and PC-3 human prostate cancer cells that lack AR expression [30]. Interestingly, Myc has been shown to cooperate with sterol regulatory element binding protein 1 to promote lipogenesis in hepatocellular carcinoma, T-cell acute lymphoblastic leukemia, and renal cell carcinoma [31]. Whether a similar cooperation exists between Myc and sterol regulatory element binding protein 1 in prostate cancer is yet to be determined.
The present study reveals that neoplastic transformation induced by prostate-specific overexpression of Myc in Hi-Myc mice leads to increased expression of ACLY, ACC1, and FASN proteins in PIN as well as adenocarcinoma and elevated circulating levels of total free fatty acids and certain individual fatty acids. We also show recruitment of Myc at the promoters of ACLY, ACC1, and FASN by ChIP assay suggesting direct transcriptional regulation of these genes by Myc. Prior work has implicated ACLY, ACC1, and FASN in prostate carcinogenesis. For example, the gene for ACLY protein, which catalyzes the conversion of citrate to acetyl-CoA, is overexpressed in prostate cancer as judged by analysis of the GSE103512 dataset [32]. Consistent with gene expression results, analysis of the Human Protein Atlas also revealed upregulation of ACLY protein in prostate cancer when compared to normal prostate [32]. Chemical inactivation of ACLY by a natural product (Cucurbitacin B) resulted in inhibition of prostate cancer cell growth [33]. Inhibition of ACC1, which catalyzes the rate limiting step in fatty acid synthesis and converts acetyl-CoA to malonyl-CoA, decreases the matrigel invasion ability of PC-3 cells that can be restored by lipid supplementation [34]. Similarly, extensive literature exists on oncogenic role of FASN in prostate cancer. In one such study, the expression of FASN protein was observed in 59% of prostate cancer tissues [35]. Pharmacological inhibition and RNA interference of FASN in LNCaP cells resulted in growth inhibition in vitro and in vivo [36]. Prostate-specific expression of FASN is sufficient to induce PIN lesions in mice [37].
The results of our study suggest that inhibition of fatty acid synthesis by targeting Myc-ACLY/ACC1/FASN axis may be a viable strategy for prevention and/or therapy of prostate cancer. In this context, we have reported previously that in vivo prevention of prostate cancer by sulforaphane administration in TRAMP mice and inhibition of 22Rv1 prostate cancer xenograft growth in nude mice by leelamine administration is associated with downregulation of ACLY, ACC1, and/or FASN proteins as well as suppression of tumor and plasma levels of total free fatty acids in TRAMP model by sulforaphane [38, 39]. Effect of leelamine on Myc is yet to be determined, but sulforaphane downregulates Myc protein level in prostate cancer cells [40].
Comparative analysis of prostate tissues from WT and Hi-Myc mice in the present study reveals upregulation of ACLY, ACC1, and FASN proteins in both PIN lesions as well as in prostate adenocarcinoma. These results suggest that elevated fatty acid synthesis may be an early metabolic deregulation in Myc-driven prostate cancer in mice. Overexpression of FASN has been documented in human PIN and prostate adenocarcinoma in comparison with normal tissue [25]. In the present study, elevated expression of ACLY and ACC1 is seen in the human prostate adenocarcinoma, but not in PIN relative to normal prostate. These differences in results between mouse versus human specimens may be due to Myc expression status that was not available for the tissue microarrays used in the present study. We tried to explore this possibility by dual staining of Myc and ACLY or ACC1. We found a strong positive and significant correlation between Myc expression and that of ACC1 at least in human prostate adenocarcinoma. A trend for a similar positive correlation is also evident for Myc and ACLY protein expression in prostate adenocarcinoma, although the correlation is not statistically significant possibly due to small sample size. Additional work is necessary to determine if there is positive correlation between Myc expression and that of ACLY and/or ACC1 in human PIN, which has been reported for FASN [25].
In conclusion, the present study reveals, for the first time, that expression of key fatty acid synthesis enzyme proteins in prostate cancer is regulated by Myc. Therefore, inhibition of Myc-ACLY/ACC1/FASN axis to suppress fatty acid synthesis represents an attractive target for prevention and/or treatment of human prostate cancer.
Materials and methods
Ethics statement and study design
The male transgenic Hi-Myc [FVB-Tg (ARR2/Pbsn-MYC)] mice were obtained from the National Cancer Institute Mouse Repository and were crossed with WT female FVB/N mice to obtain male WT and Hi-Myc mice. Plasma and prostate tumor sections from the male WT and male transgenic Hi-Myc mice (n=6 for both groups) were used to determine the expression of fatty acid metabolism proteins as well as metabolomics. Because the goal of this study was to compare plasma and/or ADC levels of fatty acid synthesis intermediates or tumor level of fatty acid synthesis enzyme proteins in Hi-Myc transgenic mice and age-matched WT mice, power calculation or randomization was not used. All samples were used for analysis and thus there was no pre-determined exclusion/inclusion criteria. The investigator was not blinded to the group allocation during experiment or data analysis. Specific pathogen free housing was used for up to four mice per cage. The animals were monitored once a day and body weight of mice were recorded at the end of experiment and no adverse events were observed. The mice were euthanized by cervical dislocation. Both age-matched WT and Hi-Myc mice at 5, 10, and 26 weeks of age were sacrificed. Male WT and Hi-Myc mice were used to test the hypothesis that prostate specific overexpression of Myc increases fatty acid synthesis. The primary end-point for this study was plasma levels of total free fatty acids. The secondary end points included circulating levels of fatty acid synthesis intermediates (e.g., acetyl-CoA and malonyl-CoA) and expression of fatty acid synthesis enzyme proteins in the normal and cancerous prostate of WT and Hi-Myc mice.
Clinical specimens
Plasma (Figure 4c and 4d) and paraffin-embedded normal prostate tissue, prostate tissue with histological evidence of PIN, and prostate ADC (Figure 5b and 5c) were provided by the University of Pittsburgh Biospecimen Core Facility under three different requisition numbers, including (#1498 for plasma), and (#1534 and #1681 for tissue specimens). The PIN samples were collected by radical retropubic prostatectomy or cystoprostatectomy, whereas all the adenocarcinoma samples were from radical prostatectomy. Most of the samples from project #1498 were from consented patients, except for IB570 and TP11-S1333 that were excess/nonconsented collections. For Project #1534 and #1681, the request was from the clinical archive so they were not consented to a specific research study. The acquisition of these specimens for the work presented in this article was approved by the University of Pittsburgh Institutional Review Board (protocol # PRO18080058 and 0506140).
Reagents
Fetal bovine serum, phosphate-buffered saline (PBS), antibiotic mixture, and other cell culture reagents were purchased from Life Technologies-Thermo Fisher Scientific (Waltham, MA). RPMI1640 media was purchased from Mediatech (a Corning subsidiary, Manassas, VA). The antibodies were purchased from the following vendors: anti-ACC1 antibody (cat. #21923-AP) was from Proteintech (Rosemont, IL); anti-FASN antibody (cat. #3180) was from Cell Signaling Technology (Danvers, MA); anti-c-Myc antibody (cat. #39688) and anti-ACLY antibody (cat. #ab40793) were from Abcam (Cambridge, MA); and anti-α-smooth muscle actin antibody (α-SMA; cat. #sc-32251) was from Santa Cruz Biotechnology (Dallas, TX). The kits for measurement of metabolites were purchased from the following companies: kits for acetyl-CoA (cat. #K317–100), cholesterol (cat. # K603–100), and lactate (cat. # K607–100) were from BioVision (Milpitas, CA); the kits for total free fatty acids (cat. #MAK044) and total phospholipids (cat. #MAK122) were from Sigma-Aldrich (St. Louis, MO). Tissue microarrays (cat. # PR806 and cat. # BB19012) containing normal prostate, PIN, and prostate ADC were purchased from US Biomax (Derwood, MD).
RNA-Seq analysis from prostate cancer TCGA dataset
Association of MYC or MYCN expression with that of ACLY, ACACA (ACC1), FASN or ACACB (ACC2) in prostate and ovarian cancers were determined from the RNA-Seq data from TCGA. University of California Santa Cruz Xena Browser (http://xena.ucsc.edu/public-hubs/) was used for these analyses.
The ChIP assay
The 22Rv1 human prostate cancer cell line, which was purchased from the American Type Culture Collection (Manassas, VA) and maintained as recommended by the provider, was used for chromatin immunoprecipitation (ChIP) assay. This cell line was last authenticated by us in March of 2017 by short tandem repeat profiling, and found to be of human origin. The ChIP assay was performed using a commercially available Pierce™ Magnetic ChIP kit (Thermo Fisher Scientific, Waltham, MA; cat. #26157) according to the instructions provided by the supplier. In brief, cross-linking was done with 1% formaldehyde for 15 minutes at room temperature and the reaction was stopped by treatment with 125 mM glycine for 5 minutes. Fixed cells were washed with PBS and lysed in immunoprecipitation buffer. Lysate was sonicated to generate 200- to 600-bp DNA fragments and the lysate was cleared by centrifugation. The supernatant was immunoprecipitated with anti-c-Myc antibody (Santa Cruz Biotechnology) or normal IgG (as a negative control) and then reverse cross-linked for DNA isolation. The putative c-Myc binding sites at the ACLY, ACACA (ACC1), and FASN promoters were amplified by the real time-polymerase chain reaction.
Immunohistochemistry
Prostate sections (4–5 μm thick) were de-paraffinized, hydrated, washed with PBS, and incubated in citrate retrieval buffer solution (pH 6.0) for 20 minutes at 100°C followed by treatment with 0.3% hydrogen peroxide in 100% methanol for 20 minutes at room temperature. Immunohistochemistry was performed essentially as described by us previously [38, 39, 41]. Stained sections were examined under Nikon A1 confocal microscope or Leica microscope equipped with DFC 450C digital camera. Quantitation of immunofluorescence for ACLY, ACC1, and FASN protein expression intensity was performed using ImageJ software. For human prostate tumor tissue array, the H-score was analyzed by positive pixel count V9 algorithm using Aperio ImageScope software.
Quantitation of metabolites levels
Levels of acetyl-CoA, malonyl-CoA, total free fatty acids (C8 and longer), total phospholipids, cholesterol, and lactate in the plasma of age-matched WT and Hi-Myc mice and human subjects were determined using commercially available colorimetric/fluorometric assay kits as described by us previously [38, 39].
Targeted metabolomics by liquid chromatography-high resolution mass spectrometry
Individual fatty acids were analyzed by liquid chromatography-high resolution mass spectrometry (LC-HRMS) as described by Li and Franke [42]. Briefly,10 μL of a deuterated fatty acid internal standard mixture (50 μg/mL) was spiked into 50 μL plasma and the sample was brought to 500 μL with PBS before extraction using a modified Folch method. After centrifugation, the organic phase was moved to a new tube and dried under N2 for derivatization. To each sample, 200 μL of oxalyl chloride (2 M in dichloromethane) was added and the mixture was incubated for 5 minutes at 65°C. Samples were dried under N2 and 150 μL of 3-picolylamine (1% v/v in acetonitrile) was added for a 5-minute incubation at room temperature to form the 3-picolylamide ester. Samples were dried a final time under N2 and reconstituted in 1 mL methanol and 5 μL of this sample was injected onto the Vanquish-Q Exactive LC-HRMS system. (Thermo Fisher Scientific). Fatty acid-3-picolylamide ester derivatives were separated on a reversed phase C8 column (Phenomenex, 2.1 × 150 mm, 5 μm) using H2O + 0.1% acetic acid (Solvent A) and acetonitrile + 0.1% acetic acid (Solvent B). The gradient started at 65% B and increased linearly to 85% B at 10 minutes and held for 1 minute before ramping to 100% B for 2 minutes. Finally, the gradient returned to 65% B for a 2-minute equilibration. Total run time was 15 minutes. Samples were analyzed using positive electrospray ionization in full scan at a resolution of 70K. Relative amounts were obtained by taking the area under the peak of the fatty acid-3-picolylamide ester analyte and normalizing to its corresponding deuterated internal standard.
Statistical analysis
Statistical tests were performed using GraphPad Prism software (version 7.02). One-way analysis of variance (ANOVA) followed by Bonferroni’s multiple comparisons test or Kruskal-Wallis test or Student’s t-test were used for statistical tests. Pearson test was used to determine the correlation coefficient and statistical significance for the TCGA data analyses.
Supplementary Material
Funding:
This study was supported by the National Cancer Institute grant R01 CA225716 (to S.V.S.). This study used the Animal Facility and the Tissue and Research Pathology Facility partly supported by the National Cancer Institute grant P30 CA047904. The funders had no role in the design of the study, data collection, analysis or interpretation of the data, manuscript preparation or decision to submit the manuscript for publication.
Footnotes
Competing interests: The authors don’t declare any competing interests.
Data availability: The data and material generated for this study are available upon request to the corresponding author.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin 2020; 70: 7–30. [DOI] [PubMed] [Google Scholar]
- 2.Sun J, Liu W, Adams TS, Sun J, Li X, Turner AR et al. DNA copy number alterations in prostate cancers: a combined analysis of published CGH studies. Prostate 2007; 67: 692–700. [DOI] [PubMed] [Google Scholar]
- 3.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS et al. Integrative genomic profiling of human prostate cancer. Cancer Cell 2010; 18: 11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grasso CS, Wu YM, Robinson DR, Cao X, Dhanasekaran SM, Khan AP et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature 2012; 487: 239–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rubin MA, Maher CA, Chinnaiyan AM. Common gene rearrangements in prostate cancer. J Clin Oncol 2011; 29: 3659–3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.He WW, Sciavolino PJ, Wing J, Augustus M, Hudson P, Meissner PS et al. A novel human prostate-specific, androgen-regulated homeobox gene (NKX3.1) that maps to 8p21, a region frequently deleted in prostate cancer. Genomics 1997; 43: 69–77. [DOI] [PubMed] [Google Scholar]
- 7.Jenkins RB, Qian J, Lieber MM, Bostwick DG. Detection of c-myc oncogene amplification and chromosomal anomalies in metastatic prostatic carcinoma by fluorescence in situ hybridization. Cancer Res 1997; 57: 524–531. [PubMed] [Google Scholar]
- 8.Wang SI, Parsons R, Ittmann M. Homozygous deletion of the PTEN tumor suppressor gene in a subset of prostate adenocarcinomas. Clin Cancer Res 1998; 4: 811–815. [PubMed] [Google Scholar]
- 9.Ellwood-Yen K, Graeber TG, Wongvipat J, Iruela-Arispe ML, Zhang J, Matusik R et al. Myc-driven murine prostate cancer shares molecular features with human prostate tumors. Cancer Cell 2003; 4: 223–238. [DOI] [PubMed] [Google Scholar]
- 10.Buttyan R, Sawczuk IS, Benson MC, Siegal JD, Olsson CA. Enhanced expression of the c-myc protooncogene in high-grade human prostate cancers. Prostate 1987; 11: 327–337. [DOI] [PubMed] [Google Scholar]
- 11.Nag A, Smith RG. Amplification, rearrangement, and elevated expression of c-myc in the human prostatic carcinoma cell line LNCaP. Prostate 1989; 15: 115–122. [DOI] [PubMed] [Google Scholar]
- 12.Edwards J, Krishna NS, Witton CJ, Bartlett JM. Gene amplifications associated with the development of hormone-resistant prostate cancer. Clin Cancer Res 2003; 9: 5271–5281. [PubMed] [Google Scholar]
- 13.Gurel B, Iwata T, Koh CM, Jenkins RB, Lan F, Van Dang C et al. Nuclear MYC protein overexpression is an early alteration in human prostate carcinogenesis. Mod Pathol 2008; 21: 1156–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fox SB, Persad RA, Royds J, Kore RN, Silcocks PB, Collins CC. p53 and c-myc expression in stage A1 prostatic adenocarcinoma: useful prognostic determinants? J Urol 1993; 150: 490–494. [DOI] [PubMed] [Google Scholar]
- 15.Zeng W, Sun H, Meng F, Liu Z, Xiong J, Zhou S et al. Nuclear C-MYC expression level is associated with disease progression and potentially predictive of two year overall survival in prostate cancer. Int J Clin Exp Pathol 2015; 8: 1878–1888. [PMC free article] [PubMed] [Google Scholar]
- 16.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011; 144: 646–674. [DOI] [PubMed] [Google Scholar]
- 17.Wu X, Daniels G, Lee P, Monaco ME. Lipid metabolism in prostate cancer. Am J Clin Exp Urol 2014; 2: 111–120. [PMC free article] [PubMed] [Google Scholar]
- 18.Suburu J, Chen YQ. Lipids and prostate cancer. Prostaglandins Other Lipid Mediat 2012; 98: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Y Fatty acid oxidation is a dominant bioenergetic pathway in prostate cancer. Prostate Cancer Prostatic Dis 2006; 9: 230–234. [DOI] [PubMed] [Google Scholar]
- 20.Chavarro JE, Kenfield SA, Stampfer MJ, Loda M, Campos H, Sesso HD et al. Blood levels of saturated and monounsaturated fatty acids as markers of de novo lipogenesis and risk of prostate cancer. Am J Epidemiol 2013; 178: 1246–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Priolo C, Pyne S, Rose J, Regan ER, Zadra G, Photopoulos C et al. AKT1 and MYC induce distinctive metabolic fingerprints in human prostate cancer. Cancer Res 2014; 74: 7198–7204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Warburg O On respiratory impairment in cancer cells. Science 1956; 124: 269–270. [PubMed] [Google Scholar]
- 23.Shim H, Dolde C, Lewis BC, Wu CS, Dang G, Jungmann RA et al. c-Myc transactivation of LDH-A: implications for tumor metabolism and growth. Proc Natl Acad Sci U S A 1997; 94: 6658–6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stine ZE, Walton ZE, Altman BJ, Hsieh AL, Dang CV. MYC, Metabolism, and Cancer. Cancer Discov 2015; 5: 1024–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rossi S, Graner E, Febbo P, Weinstein L, Bhattacharya N, Onody T et al. Fatty acid synthase expression defines distinct molecular signatures in prostate cancer. Mol Cancer Res 2003; 1: 707–715. [PubMed] [Google Scholar]
- 26.Shimano H Sterol regulatory element-binding proteins (SREBPs): transcriptional regulators of lipid synthetic genes. Prog Lipid Res 2001; 40: 439–452. [DOI] [PubMed] [Google Scholar]
- 27.Schmidt LJ, Tindall DJ. Androgen receptor: past, present and future. Curr Drug Targets 2013; 14: 401–407. [DOI] [PubMed] [Google Scholar]
- 28.Miyoshi Y, Uemura H, Fujinami K, Mikata K, Harada M, Kitamura H et al. Fluorescence in situ hybridization evaluation of c-myc and androgen receptor gene amplification and chromosomal anomalies in prostate cancer in Japanese patients. Prostate 2000; 43: 225–232. [DOI] [PubMed] [Google Scholar]
- 29.Gao L, Schwartzman J, Gibbs A, Lisac R, Kleinschmidt R, Wilmot B et al. Androgen receptor promotes ligand-independent prostate cancer progression through c-Myc upregulation. PLoS One 2013; 8: e63563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Swinnen JV, Van Veldhoven PP, Esquenet M, Heyns W, Verhoeven G. Androgens markedly stimulate the accumulation of neutral lipids in the human prostatic adenocarcinoma cell line LNCaP. Endocrinology 1996; 137: 4468–4474. [DOI] [PubMed] [Google Scholar]
- 31.Gouw AM, Margulis K, Liu NS, Raman SJ, Mancuso A, Toal GG et al. The MYC oncogene cooperates with sterol-regulated element-binding protein to regulate lipogenesis essential for neoplastic growth. Cell Metab 2019; 30: 556–572 e555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Q, Yin X, Pan Z, Cao Y, Han S, Gao G et al. Identification of potential diagnostic and prognostic biomarkers for prostate cancer. Oncol Lett 2019; 18: 4237–4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gao Y, Islam MS, Tian J, Lui VW, Xiao D. Inactivation of ATP citrate lyase by Cucurbitacin B: A bioactive compound from cucumber, inhibits prostate cancer growth. Cancer Lett 2014; 349: 15–25. [DOI] [PubMed] [Google Scholar]
- 34.Scott KE, Wheeler FB, Davis AL, Thomas MJ, Ntambi JM, Seals DF et al. Metabolic regulation of invadopodia and invasion by acetyl-CoA carboxylase 1 and de novo lipogenesis. PLoS One 2012; 7: e29761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shah US, Dhir R, Gollin SM, Chandran UR, Lewis D, Acquafondata M et al. Fatty acid synthase gene overexpression and copy number gain in prostate adenocarcinoma. Hum Pathol 2006; 37: 401–409. [DOI] [PubMed] [Google Scholar]
- 36.Chen HW, Chang YF, Chuang HY, Tai WT, Hwang JJ. Targeted therapy with fatty acid synthase inhibitors in a human prostate carcinoma LNCaP/tk-luc-bearing animal model. Prostate Cancer Prostatic Dis 2012; 15: 260–264. [DOI] [PubMed] [Google Scholar]
- 37.Migita T, Ruiz S, Fornari A, Fiorentino M, Priolo C, Zadra G et al. Fatty acid synthase: a metabolic enzyme and candidate oncogene in prostate cancer. J Natl Cancer Inst 2009; 101: 519–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Singh KB, Kim SH, Hahm ER, Pore SK, Jacobs BL, Singh SV. Prostate cancer chemoprevention by sulforaphane in a preclinical mouse model is associated with inhibition of fatty acid metabolism. Carcinogenesis 2018; 39: 826–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Singh KB, Hahm ER, Pore SK, Singh SV. Leelamine is a novel lipogenesis inhibitor in prostate cancer cells in vitro and in vivo. Mol Cancer Ther 2019; 18: 1800–1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vyas AR, Moura MB, Hahm ER, Singh KB, Singh SV. Sulforaphane inhibits c-Myc-mediated prostate cancer stem-like traits. J Cell Biochem 2016; 117: 2482–2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Singh KB, Hahm ER, Alumkal JJ, Foley LM, Hitchens TK, Shiva SS et al. Reversal of the Warburg phenomenon in chemoprevention of prostate cancer by sulforaphane. Carcinogenesis 2019; 40: 1545–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li X, Franke AA. Improved LC-MS method for the determination of fatty acids in red blood cells by LC-orbitrap MS. Anal Chem 2011; 83: 3192–3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


