Abstract
Chromatin is a dynamic structure composed of DNA, RNA, and proteins, regulating storage and expression of the genetic material in the nucleus. Heterochromatin plays a crucial role in driving the three-dimensional arrangement of the interphase genome, and in preserving genome stability by maintaining a subset of the genome in a silent state. Spatial genome organization contributes to normal patterns of gene function and expression, and is therefore of broad interest. Mammalian heterochromatin, the focus of this review, mainly localizes at the nuclear periphery, forming Lamina-associated domains (LADs), and at the nucleolar periphery, forming Nucleolus-associated domains (NADs). Together, these regions comprise approximately one-half of mammalian genomes, and most but not all loci within these domains are stochastically placed at either of these two locations after exit from mitosis at each cell cycle. Excitement about the role of these heterochromatic domains in early development has recently been heightened by the discovery that LADs appear at some loci in the preimplantation mouse embryo prior to other chromosomal features like compartmental identity and topologically-associated domains (TADs). While LADs have been extensively studied and mapped during cellular differentiation and early embryonic development, NADs have been less thoroughly studied. Here, we summarize pioneering studies of NADs and LADs, more recent advances in our understanding of cis/trans-acting factors that mediate these localizations, and discuss the functional significance of these associations.
2. Introduction: Heterochromatin
The terms euchromatin and heterochromatin were coined by Emil Heitz in 1928, when he observed that some chromosome regions were poorly stained with acetocarmine after telophase (euchromatin), while other regions were strongly stained throughout the cell cycle (heterochromatin) [1]. Currently, we more commonly differentiate between euchromatin and heterochromatin using DNA dyes such as 4′,6-diamidino-2-phenylindole (DAPI), which has a higher affinity for AT-rich heterochromatic regions [2]. Although the terms “euchromatin” and “heterochromatin” were coined based on the microscopy described above, “heterochromatin” has come to be defined as loci that are condensed and transcriptionally less active, whereas “euchromatic” regions are decondensed and display more active gene expression. These classifications generally correlate with several other characteristics. For example, euchromatic regions are generally accessible to DNase I cleavage, while heterochromatic regions are more resistant [3]. Likewise, euchromatic regions replicate in early S-phase, and heterochromatin regions replicate in mid to late S-phase [4,5]. Heterochromatin in general is also characterized by global hypoacetylation, which contributes to its condensation [6]. Covalent addition of an acetyl group to the terminal ε-amino group on lysine side chains neutralizes that amino acid’s positive charge. For histone residues, this reaction eliminates electrostatic bonds between histones and DNA, reducing the strength of these interactions (reviewed in [7,8]). This can result in increased accessibility for transcription factors and RNA polymerases [8]. Additionally, bromodomains of transcription-promoting factors and nucleosome remodeling complexes can specifically recognize and bind acetylated histone lysines [7,8]. Conversely, removal of acetyl groups by histone deacetylases (HDACs) facilitates decreased chromatin accessibility, as found in heterochromatic regions [7,9]. Also, lysine deacetylation can do more than restore the positive charge on lysine. For example, deacetylation of histone H3 lysine 9 (H3K9) is a prerequisite for methylation of this residue, which then can serve as a binding site for the chromodomain of heterochromatin protein 1 (HP1), a key protein for the formation, maintenance and propagation of heterochromatin [10]. HDACs also regulate heterochromatin via other histone residues, such as H3K14 [11,12], (reviewed in [13]).
Heterochromatin is largely divided into two subcategories: constitutive and facultative. Constitutive heterochromatin is generally unchanged in its genomic location across the cell cycle or during differentiation [14], generally adopts characteristic sub-cellular localizations [15,16], tends to include repeat-rich loci, and is generally gene-poor [6]. Constitutive heterochromatin is mainly found at telomeres, centromeres, and adjacent silent regions (subtelomeres and pericentric regions, respectively) [13]. These regions are highly condensed, highly repetitive, constitutively repressed and enriched in repressive H3K9me2/3 and H4K20me2/3 histone modifications, cytosine methylation at CpG dinucleotides, and are often bound by HP1 [2]. HP1 directly binds to histone H3 methylated at lysine 9 [17–19], providing the foundation of constitutive heterochromatin organization [6,20].
In contrast to constitutive heterochromatin, facultative heterochromatin is defined as transcriptionally inactive loci that are less condensed [21] and become transcriptionally active at specific developmental stages, or during nuclear relocalization, or in a heritable context such as monoallelic gene expression [22]. Polycomb group (PcG) proteins play important role in facultative heterochromatin formation, and the H3K27me3 modification created by the Ezh2 methyltransferase subunit of the Polycomb Repressive Complex 2 (PRC2) is the signature mark of facultative heterochromatin [22].
Heterochromatin is spatially concentrated for the most part in two regions in the nucleus (reviewed in [23,24]: 1) loci enriched at the perinucleolar region are termed nucleolus-associated domains (NADs) [25,26]; and 2) loci enriched at the nuclear periphery are termed lamina-associated domains (LADs) [27–30]. We will review these spatial domains separately, with a special focus on NADs.
3. Chromatin at the nuclear lamina: LADs
LADs are genomic regions that frequently associate with the nuclear lamina (NL) [31]. LADs have been mapped in several species, including Caenorhabditis elegans, Drosophila melanogaster, mouse, and human cells [27–30,32,33]. Similarly, a recent study in the plant Arabidopsis thaliana identified interactions between lamin-like protein CRWN1 and chromatin-associated protein PWO1 at the nuclear periphery, which regulate nuclear morphology and expression of target genes [34]. Localization of heterochromatin to the nuclear periphery is also observed in budding and fission yeasts [35,36], and therefore is widely observed throughout eukaryotes. However, in the absence of nuclear lamins in yeast, the silencing at their nuclear periphery is executed through the action of the inner nuclear membrane proteins [37,38].
Many studies of LADs were performed using DNA adenine methyltransferase identification (DamID). DamID utilizes bacterial adenine methyltransferase (Dam) fused to a protein of interest, in this case a NL protein such as lamin B. This results in genomic regions that interact with the NL protein becoming methylated on adenines, which is a mappable modification normally absent in metazoan cells [27]. This method provided genome-wide maps of LADs, as well as tools to visualize LADs by microscopy via the use of a GFP-m6A-Tracer protein that binds adenine-methylated regions [39]. Additionally, LADs have been mapped by ChIP-seq protocols using anti-lamin antibodies [40–42].
Mammalian LADs are typically 10 kb-10 Mb long, with median size ~0.5 Mb, covering ~40% of the genome [28,30]. LADs generally display low gene density and expression, and are mostly late-replicating [28,30,43]. LADs include pericentric heterochromatin and a subset of telomeric regions [28] and are enriched for heterochromatic marks H3K9me2/3 and H3K27me3 [28,32,44,45]. Studies of mouse ESC differentiation led to the identification of constitutive LADs (cLADs), which remain invariant during differentiation, in contrast to cell type-specific facultative LADs (fLADs), and to constitutive interLADs (ciLADs), which are regions outside of LADs in all cell types investigated [30,46]. cLADs are the most gene-poor subset of LADs, and are enriched in LINE transposable elements and AT-rich DNA [46]. Comparing mouse and human syntenic regions, cLAD genomic positions and sizes are strongly conserved [46]. Therefore, cLADs are suggested to be a “structural backbone” of chromatin organization and folding in interphase nuclei [47].
DamID methods have been extended to single cells in order to detect heterogeneity that is masked in studies of cell populations [32]. Comparison of data from hundreds of individual cells allowed genome-wide calculation of contact frequencies (CF), a measure of the fraction of cells in a population in which a locus contacts the NL [32]. About 15% of genomic regions exhibited high CF (>80%), whereas approximately 30% of genomic regions showed intermediate (~20–80%) CFs. Consistent with population-based DamID data, single-cell DamID results showed that cLADs tended to have high CFs, and fLADs mostly had intermediate CFs [32]. Regions with high CFs exhibited about 20-fold lower gene densities compared to regions with no NL contact, and single-cell mRNA expression levels negatively correlated with CF values [32]. CF values positively correlated with the presence of H3K9me3, but not H3K27me3, consistent with the idea that high CF regions are composed predominantly of constitutive heterochromatin [32]. Additionally, the mean CF value across LADs positively correlated with length (plateauing above ~6 Mb), suggesting that LADs are attached to NL via multivalent interactions distributed across them [32]. Supporting this idea, individual cell data included greater than expected frequencies of long, contiguous stretches of both NL-contacting and non-contacting sequences, suggesting coordinated switching between bound and unbound states. Thus, these single cell experiments allowed detection of intrachromosomal coordination of NL association that could not be detected in population experiments.
NL is generally a repressive nuclear compartment [47,48]. During the differentiation of mESCs to neural precursor cells (NPCs) and astrocytes, many genes detach from the NL, frequently accompanied by gene activation; conversely, regions that increase their frequency of NL interactions often display decreased gene expression [30]. Likewise, the NL-tethering of a hygromycin resistance-LacO reporter gene in mouse fibroblasts using LacI-emerin, an inner nuclear membrane (INM) protein, resulted in decreased H4 acetylation levels and transcriptional repression [49]. A more recent study from the van Steensel laboratory showed that LADs are generally repressive but can be heterogeneous with respect to transcription [50]. Leemans et al. demonstrated that although many promoters become silenced when they were inserted into LAD regions, a subset of promoters were less sensitive to the repressive effect of LADs. These escaper promoters were locally detached from the NL despite being located inside LADs, but they did not display significant differences in their histone modification patterns compared to promoters that were repressed in LADs. The authors speculate that the insensitivity of these escaper promoters could be due to their recruitment of transcription factors (TFs) that are more efficient in resisting LAD repression [50]. Notably, the authors demonstrated that escaper enhancers displayed enriched association with pioneer factors, which are defined as TFs capable of activating promoters in regions with high nucleosome occupancy [51]. The authors also suggested that TFs that bind escaper promoters are likely to function in a combinatorial manner, where they act synergistically to counteract the silencing effect of NL tethering [50]. Importantly, compared to earlier studies [52,53] that analyzed a small number integration sites for NL localization arrays, the recent studies [50,54] tested numerous integration sites. Thus, these recent studies favor the view that LADs generally are repressive but can in some cases be overcome by strong TFs.
There are several models of how LADs can promote repression. LADs could inactivate genes by the virtue of repressive enzyme activities in the NL, e.g. histone deacetylase 3 (HDAC3) [47] (See Figure 1). Emerin, an integral INM protein that is found to be attached to lamins, binds and catalytically activates HDAC3, which then facilitates gene repression at the nuclear periphery [55]. Another study showed that HDAC3, complexed with transcriptional repressor cKrox and INM protein Lap2β, binds to lamina-associated sequences (LASs; 4–6 kb DNA sequences that are sufficient to tether chromatin to the NL) and promotes NL localization and gene repression [56]. Thus, LAD repression appears to include spatial regulation of histone modification.
Figure 1. Molecular interactions at the Inner Nuclear Membrane (INM).
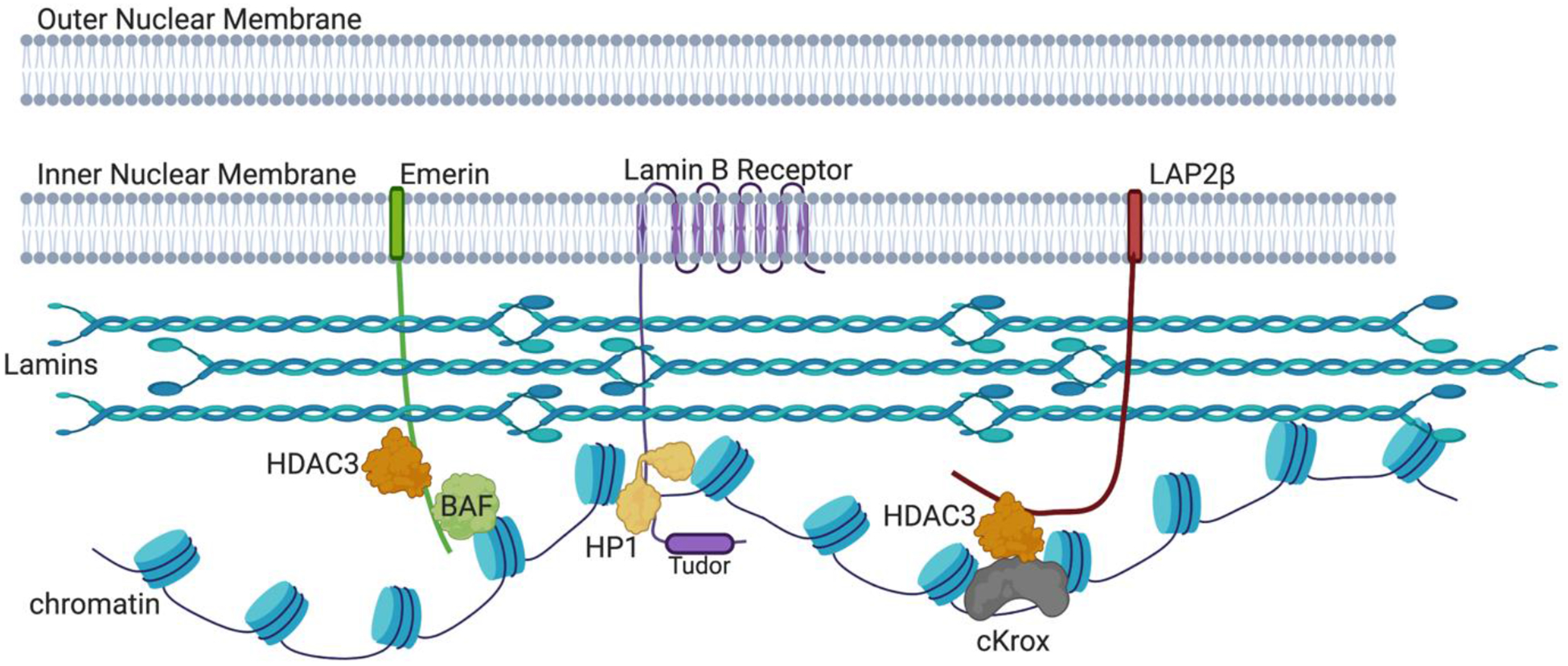
We emphasize here molecules discussed in this review, omitting many interactions at the nuclear lamina for clarity, including lamin-DNA interactions [72,75,79]. Individual lamin protein molecules form dimeric coiled coils, which in turn form polar head-to-tail polymers. These polymers make antiparallel interactions to form a network of intermediate filaments [80–82]. The INM proteins Emerin and LAP2β [83] both have “LEM” (LAP2-Emerin-MAN1) domains (not depicted), and bind lamins and HDAC3 (histone deacetylase 3). Emerin also contacts chromatin via its interaction with Barrier-to-Autointegration Factor (BAF) [84]. LAP2β binds to HDAC3 together with cKrox, a DNA-binding protein that contacts LAS elements [56]. The Lamin B receptor (LBR) binds not only to Lamin B, but also to DNA, histones H3/H4 and Heterochromatin Protein 1 (HP1) [85], which binds methylated histone H3K9 residues [17–19]. Additionally, the LBR Tudor domain binds H4K20me3-marked heterochromatin [86]. The contributions of H3K9 methylation by Suv3–9 enzymes and H3K27 methylation by Ezh2 are discussed in the text but not depicted here. Created with BioRender.com.
An alternative model, not exclusive with the one above, suggests that genes in LADs are inactive because they are shielded from transcriptionally active “A”-type nuclear compartments [47]. Nuclear “A” and “B” compartments were identified in early Hi-C studies. Hi-C measures physical interactions of genomic loci based on frequency of ligation events that follow nuclear fixation and restriction enzyme digestion, detected via deep sequencing [57,58]. Loci within a compartment have higher contact frequencies with regions of the same compartment type (A-A, B-B), and lower contact frequencies with regions of a different compartment type (A-B) [58,59]. The A compartment corresponds to active chromatin, and is characterized by chromatin accessibility, transcriptional activity and H3K36me3 enrichment. In contrast, the B compartment is associated with an inactive chromatin; it is less transcriptionally active and more densely packed than compartment A, and enriched for heterochromatic marks H3K9me3 and H3K27me3 [58,60,61]. A study from the Zheng group integrated Hi-C and DamID data and demonstrated that LADs correspond to an inactive B compartment, and showed that lamin depletion leads to relocalization of a subset of LADs from compartment B to A [62]. Those data are also consistent with other recent studies of the self-association of heterochromatin in the B compartment described in section 6c (“Phase Separation”). Together, these data suggest that physical segregation of LADs from the more active A-compartment contributes to limiting TF access to LADs [63].
Major questions remain regarding the hierarchy and kinetics of forming different levels of chromosome organization, including chromosomal compartments, LADs and topologically-associated domains (TADs). TADs are self-associating domains on the megabase size scale, which were discovered in Hi-C data as regions which display more interactions in cis within each domain and significantly fewer interactions with outside loci [64,65]. TADs are significant because they represent regions permissive for local promoter-enhancer interactions, which form much less frequently between loci found in different TADs [64,66,67].
A recent study from the Torres-Padilla and Kind groups has explored the kinetics of genome organization during early mouse embryonic development. Specifically, they measured compartment identity, LADs and TADs, from prior to fertilization through the early pre-implantation cell divisions [33]. They found that LADs were already established in murine zygotes after fertilization but prior to the first cell division; in contrast, the boundaries of (TADs) were not well-defined at this stage [33]. Additionally, in some genomic regions, 2-cell stage LADs predated the formation of B-compartments. Thus, these results indicate that LAD establishment can precede TAD formation and full establishment of compartmental identity. These studies suggest that chromatin attachment at the NL is a foundational event in shaping a mammalian genome, instructing the formation of multiple aspects of higher-order chromosome organization during early embryonic development [33]. Therefore, future studies of the molecular mechanisms of LAD establishment in zygotic cells and the 2-cell embryo will be of particular importance for understanding development of mammalian gene expression programs.
4. Cis and trans-acting factors mediating the NL-LAD interactions
4a. Lamin proteins.
Several studies have investigated the proteins and genomic sequences that are responsible for NL-LAD interactions. The NL is a meshwork of proteins associated with the inner nuclear membrane (INM) that is attached to chromatin (See Figure 1). NL is mainly composed of type V intermediate filaments that consist of nuclear lamin proteins [68]. Mammalian lamins include lamin B1 and B2, encoded by the LMNB1 and LMNB2 genes, respectively, and lamins A and C, derived from a single gene, LMNA, by alternative splicing [69].
There are multiple interactions between the lamin network and chromatin. First, the C-terminal tail domain of lamins interacts with N-and C-terminal tail domains of core histones [70–74]. Additionally, it has been reported that lamins can bind directly to DNA sequences [75–77] (reviewed in [68,78]).
Lamins are important for NL-tethering throughout multicellular animals including Drosophila and C. elegans [87,88]. In mammalian cells, there is a division of labor among lamin proteins. In murine fibroblasts, depletion of both the lamin A/C isoforms, but not lamin A alone, led to the loss of NL association with LAS-containing regions that are normally targeted to nuclear periphery [45]. These findings have been supported by a recent study, in which microscopy of individual mouse fibroblasts detected reduced LAD attachments to the NL in cells specifically depleted of lamin C, but not lamin A or lamin B1 [89]. Notably, lamin C was particularly important for the normal kinetics of reforming NL-LAD associations after exit from mitosis, and for self-association of both LAD and non-LAD clusters. Remarkably, lamin C preferentially associates with euchromatin and not LAD regions prior to NL association in early G1, disfavoring a simple model in which lamin C would serve as a guide that brings heterochromatin to the NL [89]. Instead, the authors propose that lamin C governs self-association properties of different genomic subsets. This could be part of a complex set of phase-separation-based associations (see section 7c).
In addition to somatic cell studies, several experiments have explored the roles for lamins in mouse embryonic stem cells (mESCs). In one study, triple knockout of lamin B1, lamin B2 and lamin A/C in mESCs did not cause dramatic changes in LADs, as measured by DamID analysis of emerin [90]. A different study of triple lamin knockout mESCs showed that the overall TAD structure was largely similar to that observed in wild-type cells, and many LADs remained unperturbed [62]. However, at specific loci the more recent study detected changes in inter-TAD interactions, decondensation of cLADs, and detachment of fLADs from the NL in lamin-null mESCs [62]. This study extended previous Hidden Markov Model-based analyses combining lamin B1-DamID, histone modification, and linker and core histone occupancy data to subclassify the genome into six histone-lamin landscapes (“HiLands”), including two distinct LAD states [91]. Specifically, the HiLands-P LAD subset covers longer stretches of chromatin with higher lamin B1-DamID values, but lower abundance of H3K27me3-modified regions, and thus generally corresponds to constitutive LADs [91]. In contrast, the HiLands-B subset generally corresponds to facultative LADs that display reduced NL interaction (measured by emerin-DamID) upon lamin B deletion [62,91]. Therefore, the general consensus is that lamins make important contributions to NL associations, and that distinct subsets of LADs can have different characteristics. Likewise, distinct subclasses of nucleolar-associated domains will be discussed below in section 5.
4b. Lamin B receptor.
Lamin B receptor (LBR) is an integral membrane protein found in the INM, which together with lamin A/C mediates chromatin attachment to the nuclear periphery (Figure 1) [15]. Solovei et al. demonstrated that the absence of both LBR and lamin A/C in mouse cells results in inverted nuclei, where the heterochromatin cannot bind to the nuclear lamina but maintains its self-association within the nuclear interior [15]. Indeed, a more recent study combined Hi-C, microscopy and polymer simulations to suggest a model in which the interactions between lamina and heterochromatin are necessary to obtain the conventional nuclear organization, i.e. when heterochromatin segregates at the nuclear periphery [16]. Together, these results are consistent with the view that lamins, LBR, emerin and possibly other INM proteins act redundantly to promote chromatin attachment to the NL [47].
4c. Histone modifications.
H3K9 methylation is a central component of heterochromatin (reviewed in [6,14,20]). H3K9me2-enriched domains largely overlap with LADs in mammalian cells [39,44]. Several groups investigated whether H3K9me2 contributes to recruitment of chromatin to the NL. Indeed, depletion of H3K9 methyltransferase G9a resulted in decreased frequencies of NL-LAD interactions [39], or in some cases dissociation of LADs from the NL [45]. In C. elegans, H3K9me1/2 promotes the NL attachment of repeat-rich chromosome arms, whereas H3K9me3 reinforces this attachment and establishes silencing at these regions [92]. In mouse fibroblasts, a prominent study of H3K9 methylation involved integrated bacterial artificial chromosomes (BACs) containing human β-globin (HBB) [93]. Suv39H1/2 knockdown (i.e. H3K9me3 depletion) decreased BAC association with the NL, and G9a knockdown (i.e. H3K9me2 depletion) resulted in the lamina dissociation of the LAD region adjacent to the HBB locus. However, only combined G9a and Suv39H1/H2 knockdown led to the detachment of the entire HBB BAC with the adjacent LAD region [93]. Together, these investigations indicated overlapping roles for multiple H3K9 methylation states in NL association in diverse eukaryotes.
H3K27me3 marks are enriched at LAD borders [28]. The Reddy group showed that inhibition of H3K27me3 either via RNAi knockdown or chemical inhibition of the Ezh2 histone methyltransferase led to relocalization of ectopically integrated LASs and endogenous LADs away from the lamina [45]. In section 7a we will explain that H3K27me3 is important not only for NL localization but also for perinucleolar localization of heterochromatin.
4d. Cis-acting DNA sequences.
Lamina-associating sequences (LASs) were defined as genomic sequences within LADs that conferred NL association when inserted ectopically [56]. The LASs analyzed were enriched for the GAGA motif, which promotes NL attachment via binding a complex composed of transcription factor cKrox, nuclear membrane protein Lap2β and histone deacetylase HDAC3 (Figure 1) [56]. However, cKrox knockdown in mouse fibroblasts did not alter the peripheral localization of an integrated HBB-BAC transgene that included GAGA motif clusters adjacent to HBB [93]. The Belmont group defined peripheral targeting regions (PTRs) that can confer peripheral targeting to the HBB-BAC, whereas the same sequence confers pericentric heterochromatin targeting to another BAC, suggesting that PTR targeting is likely to be epigenetic [93]. Together, these types of data suggest that multiple factors can contribute to localization of loci. It will be of great interest to determine how cis-acting targeting sequences might in some cases act in a cell-type or context-specific manner.
5. Chromatin at the nucleolar periphery: NADs
NADs are genomic regions enriched at the nucleolar periphery in interphase cells [25,26]. The nucleolus is the largest nuclear substructure, best known as the site of rRNA synthesis and ribosome assembly. Ribosomal DNA (rDNA) tandem repeats (also known as 45S rDNA) are the templates for formation of nucleolus organizer regions (NORs), which cluster and initiate assembly of the nucleolus [94,95]. rDNA repeats each contain a ~14 kb RNA Pol I-transcribed region that encodes the 47S pre-rRNA; these are separated by ~30 kb intergenic spacers (IGS) [96,97]. 47S pre-rRNAs are processed into 18S, 5.8S and 28S rRNAs, which, together with the RNA pol III-transcribed 5S rRNA that is encoded at separate loci, constitute the RNA backbone of the ribosome [96]. 45S rDNA and 5S rDNA occur in tandem repeats in several hundred copies per genome, with the distinction that 45S rDNA repeats are found on the acrocentric ends of five NOR-bearing chromosomes, whereas 5S rDNA repeats are mostly arrayed at a single locus [98].
The nucleolus lacks a membrane and is composed of three layers: a large granular component (GC), with one or a few dense fibrillar components (DFC), each of which possesses a fibrillar center (FC) [99] (Figure 2). Transcription of 47S pre-rRNA occurs in the FC or at the border between FC and DFC, whereas processing of rRNA happens in the DFC, and the first steps of ribosome assembly occur in the GC [100]. The rDNA outside of the nucleolus is usually inactive, whereas the active and poised rDNA genes are generally found within the nucleolus, close to the DFC and FC [101]. However, recent fluorescence in situ hybridization (FISH) experiments combined with super-resolution microscopy showed that inactive rDNA can also be found inside the nucleolus, where they co-localized with HP1 staining [102]. Another recent super-resolution microscopy study of nucleolar structure revealed that transcriptionally active rDNA form ring-shaped loops, with each active rDNA unit consisting of one or two transcribed rRNA genes [103]. Importantly, all these structures are regulated during the cell cycle: nucleoli disassemble when cells enter mitosis and begin reassembling in early telophase [104].
Figure 2: Nucleolar tripartite structure.
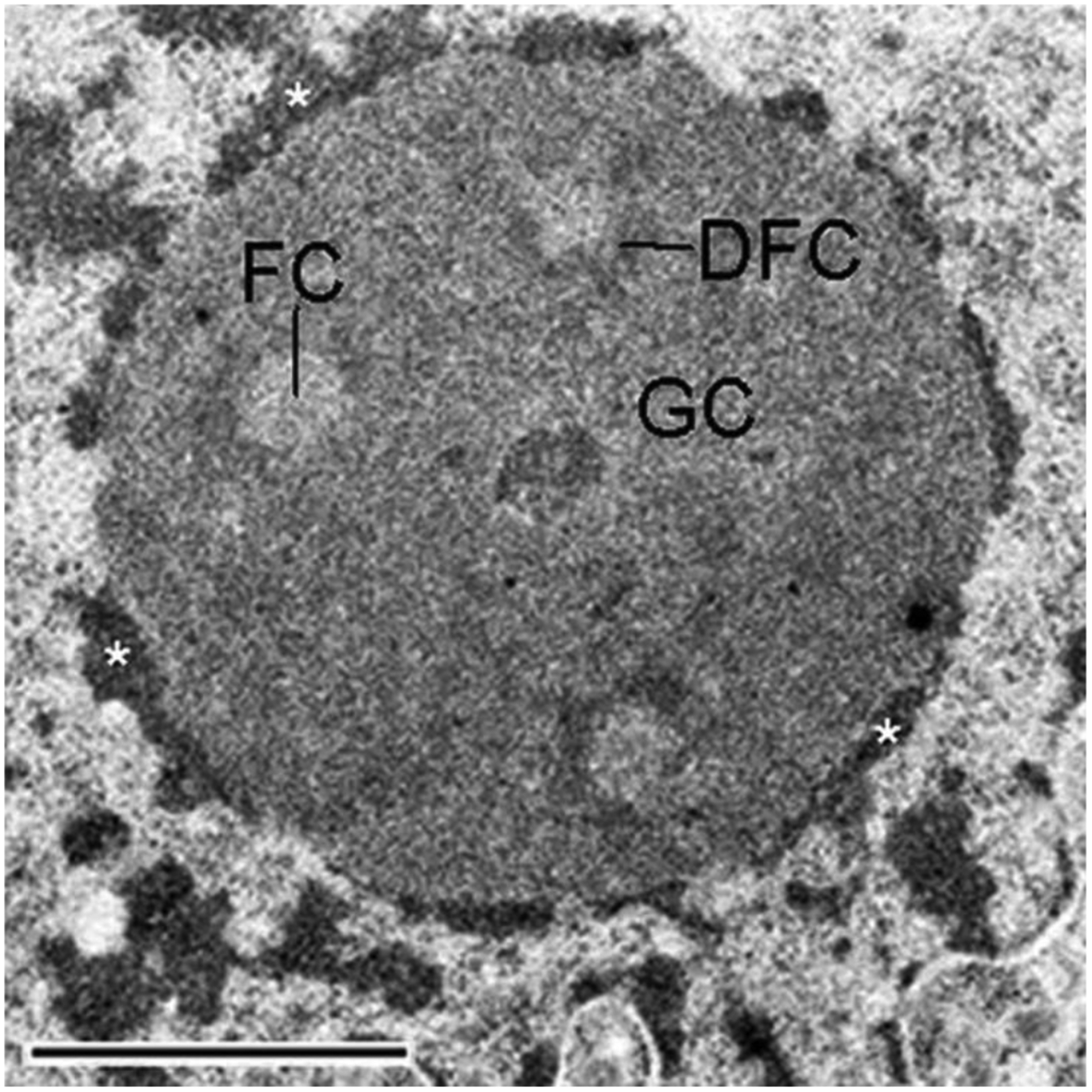
Shown is an electron microscopy image of a nucleolus. Nucleoli possess a tripartite structure consisting of Fibrillar Center (FC), Dense Fibrillar Component (DFC) and Granular Component (GC). The FC appears as a less-well labeled region surrounded by the more darkly stained DFC. The GC comprises the area outside of the DFC. Ribosomal DNA transcription occurs in the FC or at the interface between FC and DFC, this is where the rRNA-encoding Nucleolar Organizer Regions (NORs) are located. Processing of ribosomal RNA takes place in the DFC, and ribosomal subunit assembly occurs in the GC [105]. On the periphery of this nucleolus is darkly stained heterochromatin, illustrated by asterisks. These regions are where NAD loci are located. This image is from Schöfer and Weipoltshammer, Histochemistry and Cell Biology 150:209–225 (2018) [106], under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/).
Due to the proximity of centromeres and rDNA repeats, centromeres of NOR-bearing chromosomes often cluster near nucleoli [107]. Additionally, centromeres of non-NOR chromosomes have also been shown to frequently localize near nucleoli [108,109]. Thus, perinucleolar heterochromatin is enriched in centromeres and pericentric heterochromatin, and includes other regions both from NOR and non-NOR bearing chromosomes (reviewed in [2,110]). Prior to the generation of genome-wide NAD maps, some genomic regions besides rDNA repeat-containing NORs were found to associate with the nucleolar periphery (reviewed in [111]), including centromeres of human chromosomes 1 and 9 [112], transfer RNA (tRNA) gene families in Saccharomyces cerevisiae [113], and the mammalian inactive X chromosome (Xi) [114]. In some cases, such associations were shown to limit the mobility of the loci involved. For example, telomeric repeats in S. cerevisiae were shown via Chromosome Conformation Capture-based (3C) methods to associate frequently with rDNA, and this has been suggested to limit the movement of chromosomes and define the nuclear chromatin architecture [115]. Similarly, chromatin in human cells was reported to be limited in mobility at nucleolar and nuclear peripheries based on live cell tracking of GFP-tagged genomic regions [116].
5a. Methods for NAD identification.
NADs have been analyzed in multiple eukaryotic species via several distinct methods (recently reviewed in [117]). NADs in mammalian cells have primarily been identified via deep sequencing of DNA associated with biochemically-purified nucleoli (NAD-seq). These experiments depended on sonication of whole cell or nuclear extracts, in order to break covalent connections between the nucleolus-associated DNA and the non-associated portion of chromosomes [25,26,118–121]. Two main variations of this technique have been employed. Some experiments used formaldehyde to crosslink nucleic acids and proteins, followed by sonication of whole cells to release nucleoli [25,118–120], a method developed by the McStay group [122]. Alternatively, a non-crosslinked method was originally developed by Muramatsu et al. [123] and optimized by the Lamond group [124]. In the non-crosslinked [26,119,121] experiments, cells were first swollen in hypotonic buffer, plasma membranes were broken with a Dounce homogenizer, nuclei were isolated by centrifugation and further cleaned away from cytoplasmic material on sucrose cushion [124,125], and nucleoli were then released from nuclei by sonication. Both methods rely on the fact that nucleoli are the fastest-sedimenting components of nuclei, and thus can be separated via centrifugation in a high-density sucrose medium [122,126]. The sonication buffers contain Mg2+ ions, which promote formation of compact nucleolar structures that are resistant to disruption during sonication, which disintegrates the nuclear membranes [123]. These techniques therefore require the optimization of sonication burst intensities and durations, solution volumes, and Mg2+ concentrations for each specific cell type, as these parameters affect the purity and integrity of isolated nucleoli [123,124,126,127]. Differences in sonication equipment, or the age of sonication tips could also contribute to experimental differences among experiments.
Our laboratory compared mouse embryonic fibroblast (MEF) NADs generated by both crosslinked and non-crosslinked nucleoli isolation methods [119]. One of the possible advantages of non-crosslinked NAD identification is that nuclei are purified first before isolating nucleoli, possibly resulting in cleaner and more intact nucleoli. It has been suggested that purifying nucleoli from whole cells without isolating nuclei might result in nucleoli contacting cytoplasmic or lysosomal degrading enzymes [128]. However, we observed that the genome coverages of crosslinked and non-crosslinked NADs in MEFs were 41% and 30%, respectively, and almost all peaks in the non-crosslinked data set were present in the crosslinked data set, suggesting that crosslinking improves the recovery of some loci that may associate transiently or less avidly. One of the concerns about crosslinking is whether it generates false-positive DNA-nucleolus contacts. However, 3D immuno-FISH assays in MEFs and F121–9 ESCs validated the NADs predicted by NAD-seq in these cells. Nevertheless, the classical DNA-FISH assay using BAC probes is labor-intensive, thereby limiting the number of probes that can be tested. To this end, innovative, high-throughput live-cell imaging of chromosomal regions has been accomplished using fluorescent CRISPR/Cas9-based systems [129–133]. Alternatively, repetitive hybridization-based approaches, e.g. Oligopaint [134,135] have revolutionized the analysis of chromosome structures in cells. Such approaches have been recently reviewed elsewhere [136–139].
Another variable among the different NAD-seq studies has been the bioinformatic analysis of the data. Most studies identified NAD peaks by separately sequencing total genomic and nucleolar DNA, and determining regions with high nucleolar/genomic DNA ratios [26,118–120]. Dillinger et al. [118] successfully used two-state hidden Markov model (HMM)-based analyses to assign regions as either NADs or interNADs, as had been done in previous LAD analyses [32,39,46,140]. Our group developed software named NADfinder that uses nucleolar/genomic DNA ratio and local background correction to identify NADs, increasing the sensitivity for weak peaks far from centromeres (discussed in [119,120], see also section 5d). A different normalization sample was used by Lu et al., who sequenced nucleolar DNA from cells treated with nucleolus-disrupting transcription inhibitors (ActD and DRB) as samples for normalization [121], followed by peak analysis using MACS [121]. The differences in both the sample preparation and bioinformatics tools likely contribute to the large differences between the percentage of the genome identified in mouse ESC NADs in ESCs in the Lu et al. study (7.5%) [121] and our study (31%) [120] (see also section 5d). Comparison of these data sets revealed that some NADs confirmed by immuno-FISH analysis of F121–9 mESCs [120] were not designated as NADs in the non-crosslinked experiment [121].
In contrast to the methods used for mammalian cells involving purification of nucleoli via sedimentation, NADs were mapped in transgenic Arabidobsis thaliana [141] plants with the aid of a genetically-encoded yellow fluorescent protein (YFP) tag on fibrillarin (FBL), a nucleolar protein found in the DFC layer of the nucleolus [142]. A. thaliana leaves were fixed with formaldehyde and homogenized, and nuclei containing FBL-YFP were then purified by FACS sorting. The purified nuclei were either used in their entirety to obtain genomic DNA as the input for normalization, or sonicated to release nucleoli, which were isolated via an additional round FACS-based sorting [141].
Other techniques for NAD analyses have included innovative genomic analyses. The Guttman lab pioneered the Split-Pool Recognition of Interactions by Tag Extension (SPRITE) method [143], in which crosslinked extracts are subjected to repeated rounds of split-pool barcoding, in which material is tagged with oligonucleotides followed by dilution and subdivision into separate populations. After many rounds, deep sequencing identifies DNA [143] and RNA species [144] that co-associated over the course of the experiment. The resulting data provide Hi-C-like maps of genome-scale DNA-DNA contacts. In addition, this technique can detect multiple tagged nucleic acids per complex and is therefore not limited to the detection of pair-wise interactions. Therefore, this method has provided detailed maps of DNA interactions at some nuclear bodies, including the transcription-associated nuclear speckles, the clustered histone gene loci, and at the nucleolar surface [143]. Conclusions from these studies are discussed in section 6a.
Another alternative view of nucleolar interactions starting with crosslinked samples came from the Lemos laboratory [145,146]. They mined Hi-C data sets for paired reads that contained covalent links between rDNA and the rest of the genome (results discussed in section 7b; we will also discuss a 4C-based analysis of rDNA contacts in section 6c). It has been noted that not all NADs necessarily overlap with rDNA-contacting genomic regions [117], another example of interactions detected being affected by the experimental method.
Additional alternative NAD mapping methods exist, including Dam-ID, which relies on a genetically-encoded Dam methylase fusion to the marker protein gene [27], or tyramide signal amplification sequencing (TSA-seq), an antibody-based proximity labeling method [147,148]. Indeed, DamID analysis of nucleoli was recently obtained by fusing Dam to a nucleolus-targeting peptide repeat [149]. Results from these studies are discussed in section 6a. Because DamID and TSA-seq do not rely on the biochemical isolation of nucleoli, they could serve as a powerful orthogonal set of experiments to verify NAD peaks identified via NAD-seq. For these techniques, the marker protein in question must be very specifically localized to nucleoli, whereas proteins that shuttle among multiple nuclear locations would be expected to mark DNA in non-nucleolar sites, thereby obscuring nucleolar-specific signals. A strong advantage of these techniques is that they could in principle be extended to single-cell based analyses, which would not be feasible for biochemistry-based methods.
5b. Human NADs
Two seminal studies identified the first genome-wide map of NADs in human cells [25,26]. The Längst group used formaldehyde-crosslinked HeLa cells, whereas the Lamond group utilized non-crosslinked HT1080 fibrosarcoma cells as the source for biochemical purification of nucleolus-associated DNA. Nucleolar enrichments were then measured via DNA microarrays [25] or DNA sequencing [26]. Both studies reported significant nucleolar enrichment of gene-poor and transcriptionally silent regions, including satellite repeats. Additionally, many classes of repeats enriched in NADs could not be localized to specific regions because of the ambiguity inherent in their mapping [26]. Both studies showed that genes encoding zinc finger proteins, olfactory receptors, immunoglobulins and 5S rRNA were highly enriched in NADs. NADs were found to be enriched for repressive histone marks H3K9me3, H3K27me3 and H4K20me3 [25]. Although these early studies identified some regions in common with NADs, the datasets diverged in many cases, e.g. regarding whether tRNA genes were present and the degree of overlap with LADs. In retrospect, most of these differences likely arose because of the relatively sparse nature of the data from that era.
Subsequent to the initial papers identifying NADs, a more recent study from the Nemeth group analyzed human IMR90 embryonic fibroblasts, a diploid human primary cell line, obtaining higher-resolution microarray-based maps of NADs in young proliferating and old senescent cells [118]. In this study, approximately 38% of human non-repetitive genome was designated as NAD regions in the proliferating cells. The median length of IMR90 NADs was 361 kb. The identified NADs were enriched in heterochromatic features including late replication, high levels of DNA methylation, low DNaseI accessibility, low gene density, and low gene expression. The authors noted that the great majority of NADs are in the heterochromatic type “B” chromosome compartment. They also compared their data to subcompartments first defined in earlier Hi-C experiments [61]. The B1 subcompartment contains regions resembling facultative heterochromatin, in that they correlate positively with H3K27me3 marks and negatively with H3K36me3, and replicate at the end of mid-S phase [61]. In contrast, the B2 and B3 subcompartments resemble constitutive heterochromatin: they are not enriched for either H3K27me3 or H3K36me3 and replicate in late S-phase [61]. Furthermore, the B2 subcompartment is enriched at the nuclear lamina and nucleolar periphery, whereas the B3 subcompartment is more specifically localized to the nuclear lamina [61]. In the IMR90 cells, 74% of the identified NADs are in B2/B3-subcompartment constitutive heterochromatin, and 15% of NADs overlap B1-subcompartment facultative heterochromatin. Therefore, nucleolar-associated domains are largely, but not exclusively, constitutive heterochromatin.
Surprisingly, in this study the senescent IMR90 cells yielded NAD maps highly similar to young IMR90 cells. These results were unexpected because of the documented effects of senescence on nucleolar morphology: senescent cells generally exhibit one large nucleolus, whereas non-senescent cells have several smaller nucleoli [150]. Additionally, according to the rDNA theory of aging proposed by Kobayashi (2008), rDNA is the region most sensitive to genome instability and its instability is the driving force of cellular senescence [151] (reviewed in [152]). Another model suggests that chromatin relocalization during DNA repair promotes senescence [153]. Thus, rDNA instability in senescent cells might be expected to cause large-scale chromatin alterations [154]. However, in contrast to expectations of large differences between NADs in young and senescent IMR90 cells, the observed changes had a median size less than 20 kb, and they usually reflected altered borders of otherwise conserved NADs [118]. However, these restricted alterations did correlate with functional consequences. For example, the NAD regions unique to either young or senescent cells were enriched in protein-coding genes. Furthermore, the loss of nucleolar association in either young or senescent cells correlated with higher gene expression; conversely, the gain of NAD status correlated with decreased gene expression. Additionally, 3D immunoFISH experiments showed that the satellite repeats at centromeric and pericentric regions displayed decreased association with nucleolar periphery, accompanied by decreased H3K9me3 signal intensity at perinucleolar regions in senescent cells. Therefore, the authors speculated that the nucleolus safeguards the maintenance of NADs and 3D genome organization during senescence [118].
Another functional consequence of nucleolar association has been discovered in a new study from the Cheeseman group, which revealed that nucleolus-centromere interactions repress the RNA pol II-mediated transcription of α-satellite repeats [155]. Specifically, in a variety of human cell lines the number of nucleolus-associated centromeres was inversely related to the number of α-satellite RNA foci. The functionality of nucleolar association was established upon demonstration that inhibition of RNA pol I or inducible deletion of genes encoding the nucleolar proteins Ki-67 or FBL increased transcription of α-satellite repeats. Therefore, these studies demonstrate that nucleolar association negatively regulates centromeric transcription. Because the satellite transcripts are not stably associated with centromeres, the authors hypothesize that the act of transcription itself is the important regulatory event, perhaps affecting centromeric protein dynamics [155].
5c. Plant NADs
As mentioned above, NADs were also sequenced in plant Arabidopsis thaliana [141]. As in mammalian cells, these were shown to be enriched in pseudogenes, tRNA genes, and heterochromatic marks H3K27me3 and H3K9me2, whereas they were depleted of actively transcribed genes [141]. However, human and mouse NADs (see section 5d below), cover approximately one third of their genomes and are distributed widely across all chromosomes [118–120]. In contrast, NADs in A. thaliana are found only on the short arm of NOR-bearing chromosome 4, and at telomeric regions from all five pairs of chromosomes, representing approximately 4% of the genome [141]. As we learn more about the mechanisms governing perinucleolar localization of chromatin (see section 7 below), it will be of interest to discover which of these might explain the distinct NAD distributions between plants and mammals.
5d. Mouse NADs
The first genome-wide maps of NADs in mouse cells have been published by our group [119]. Of note, mouse chromosomes are acrocentric, i.e. the centromeres are found at one telomeric end of chromosomes [156], and these experiments detected a gradient of nucleolar enrichment across the length of most mouse chromosomes. Our group developed software called NADfinder that uses local background correction during the identification of NAD peaks ((https://bioconductor.org/packages/release/bioc/html/NADfinder.html). This aspect of NADfinder helped in identifying peaks at chromosome ends distal from the centromere [119].
However, a more problematic issue was whether valleys in the nucleolus-association scores (termed “NAD-splitting regions”) [119] represent NAD regions or not (Figure 3). It was clear that such valleys had distinct functional characteristics (e.g. higher levels of gene expression than the surrounding strong NAD peaks that were largely transcriptionally silent). However, DNA-FISH analyses suggested that these valleys indeed can be associated with nucleoli, even in regions where so few reads were obtained that they were not identified as nucleolus-associated regions by a variety of software packages [120] (Figure 3). One possibility consistent with these data is that some of these “valleys” represent loops of transcriptionally active, nucleolus-associated chromatin that are sensitive to the sonication steps inherent in the biochemical method used here to isolated nucleoli (Fig. 3B).
Figure 3: Genomic locations of BACs that correspond to NAD Splitting Regions (NSRs).
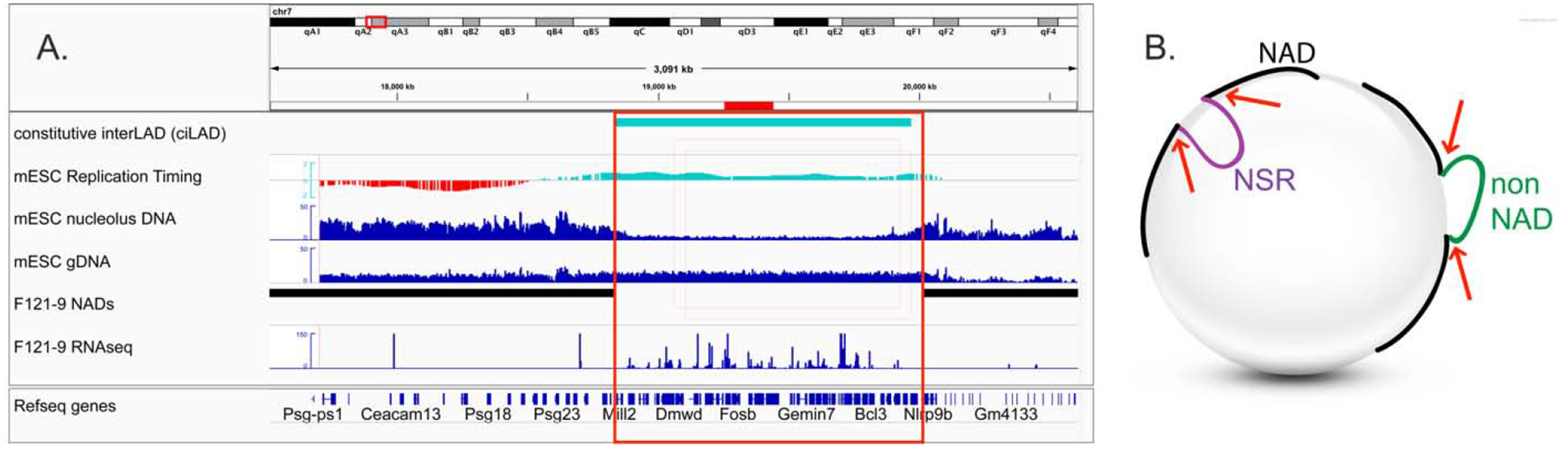
“NAD-splitting regions” (NSRs) have distinct attributes: early replication timing, increased transcriptional activity, and low numbers of reads in nucleolus DNA samples from NAD-seq experiments. These features led us to speculate that NSRs are not associated with nucleoli [119]. However, one example of an NSR which is nucleolus-associated in F121–9 mESCs in immuno-FISH assays is illustrated here [120]. These data suggested that NSRs can be systematically lost during the sonication-based purification during the NADseq protocol.
(A) A genome browser view of the NSR region around BAC probe pPK1007. This overlaps a constitutive interLAD (ciLAD, cyan), which is early-replicating (cyan portion of replication timing track) and displays elevated transcript levels (RNA-seq) compared to the neighboring NADs (black). The region between the two NADs was designated an NSR based on the low read numbers in the nucleolus DNA sample. The BAC location is indicated with a red horizontal bar above the top track, and the red box higlights the region between the neighboring NADs. ciLAD and mESC LAD tracks are from [30]. mESC DNA replication timing data are from [157]. mESC NAD-seq analyses of nucleolus and genomic DNA (gDNA), and also RNA-seq, are from [120].
(B) Graphical hypothesis. NSRs (purple) and non-NAD regions (green) are released from nucleoli during sonication steps of NAD-seq protocols (sonication-mediated breaks indicated by red arrows). However, in intact cells, NADs (black) and NSRs, but not non-NADs, are closely associated with the nucleolar periphery.
As a whole, MEF NADs exhibited heterochromatic features: they often overlapped with MEF LADs [30] and late-replicating regions [157], were enriched in heterochromatic marks H3K9me3 and H3K27me3, and displayed low gene expression levels. However, there were distinct subsets within the NADs: Type I NADs that associated with both nucleolar and nuclear peripheries, and Type II NADs that associated with nucleolar periphery, but not the nuclear periphery [119]. Type II NADs displayed higher abundance of facultative heterochromatic mark H3K27me3 and lower levels of constitutive heterochromatic mark H3K9me3 than do Type I NADs. Additionally, Type II NADs were often early-replicating, with higher gene density and expression levels than Type I NADs [119].
For comparison to the mouse fibroblast NADs, we recently generated a high-resolution map of NADs in formaldehyde-crosslinked F121–9 hybrid mouse ESCs [120]. NADs comprised 31% of the genome in F121–9 mESCs, less than observed in crosslinked MEFs (41%) [119], consistent with the presence of less condensed chromatin and a higher ratio of euchromatin to heterochromatin in stem cells compared to differentiated cells [157–159] (reviewed in [38,160]. As was observed in MEFs [119], the large Type I subset of NADs in mESCs exhibited features of constitutive heterochromatin: low gene expression and gene density and late replication timing. In contrast, Type II NADs, which are found only in the nucleolar periphery and not at the nuclear lamina, were a smaller subset of total NADs in F121–9 mESCs compared to MEFs (Figure 4A). Additionally, F121–9 NADs were less enriched for H3K27me3-modified regions [120] than were NADs in MEFs [119] (Figure 4B–C). These data suggest that Polycomb Repressive Complex 2 (PRC2)-mediated formation of facultative heterochromatin during stem cell differentiation is reflected in Type II NAD expansion.
Figure 4. Comparison of NADs in mouse embryonic stem cells (mESCs) and embryonic fibroblasts (MEFs).
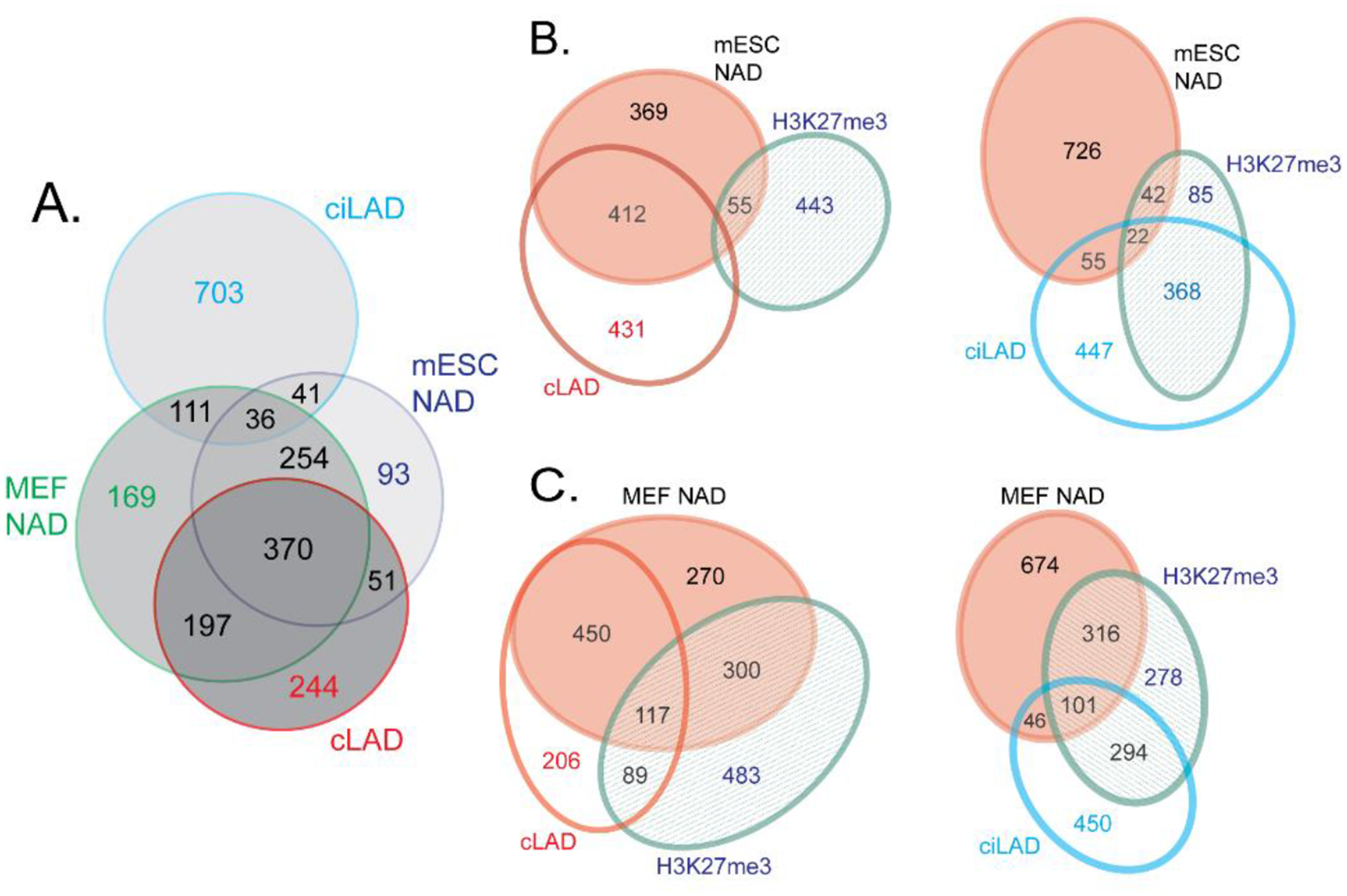
Numbers indicate the size of regions in Mb.
A) Venn diagram showing the overlaps between mESC NADs [120], MEF NADs [119] and constitutive LADs (cLADs) and constitutive interLADs (ciLADs) [30].
B) Venn diagrams illustrating the overlaps among mESC NADs [120], cLADs, ciLADs [30] and mESC H3K27me3-enriched domains [161].
C) As in B, except here the indicated overlaps are between crosslinked MEF NADs [119] and MEF H3K27me3-enriched domains [162]. Note that the overlap between H3K27me3-rich loci is much larger with the NADs from MEFs (panel C) than with the NADs from mESCs (panel B).
License 4882511241435. Reprinted by permission from [the Licensor]: [Springer Nature] [Chromosoma] (Distinct features of nucleolus-associated domains in mouse embryonic stem cells. Bizhanova, Yan, Yu, Zhu, Kaufman) [120] [Copyright] (2020)
As observed for NAD-localized genes in general, genes within conserved and cell type-specific NADs display low levels of gene expression. For example, NADs conserved in both F121–9 mESCs and MEFs were highly enriched for olfactory and vomeronasal receptor genes [119,120], genes that are not expressed in stem cells and fibroblasts. Similarly, developmentally-regulated genes tend to be nucleolus-associated when poorly expressed. For example, chemotactic cytokines are frequently in NADs in the F121–9 ESCs, and genes responsible for differentiation along the anterior-posterior axis are enriched in MEF NADs [119,120].
A major remaining question is whether silencing is the cause or consequence of nucleolar association. A recent study from the van Steensel group showed that forced activation of genes within LADs leads to their local detachment from the NL; vice versa, inactivation of genes found in fLADs resulted in their increased association with the lamina [54]. Similar studies are needed to investigate whether transcription has a similar causative effect on nucleolar localization of genomic regions.
In sum, recent years have established maps of murine NADs in fibroblasts and ES cells. These will provide the baseline for future exploration of developmental dynamics of these genomic regions. This will be particularly important in the murine system, because it has been shown that perturbation of perinucleolar chromatin disrupts early development ([163]; see also Conclusion section).
6. Functional significance of NADs
6a. Silencing hub and chromatin organization.
Genome-wide maps of NADs [25,26,118–120,141] and studies of trans-acting factor-mediated localizations at the nucleolar periphery [114,164–168] suggest that the perinucleolar space is a heterochromatic region. Comparison of NADs with Hi-C data revealed that NADs mostly overlap B2/B3-type constitutive heterochromatin and B1-type facultative heterochromatin subcompartments [61,118]. A study based on the SPRITE technique (introduced in section 5a) from the Guttman group also implicated the nucleolar periphery as a silencing hub [143]. This study determined that both mouse and human cells contain two prominent, distinct hubs of frequent genomic interactions. The “active” hub was enriched for transcriptionally active genes and localized around nuclear speckles, whereas the “silencing” hub was enriched for inactive genes and located around the nucleolus. Interestingly, genomic DNA showed preference either for nucleolar periphery, or nuclear speckles, i.e. regions found in these two hubs were mutually exclusive [143].
A newly developed computational method named SPIN (Spatial Position Inferences of the Nuclear genome) was developed to integrate different types of data for enhanced analysis of three-dimensional genome organization, in order to derive domain compartmentalization patters [149]. Here, SPIN was used to combine data from Hi-C experiments, DamID analysis of lamina and nucleoli, and TSA-seq analysis of lamina and transcription-related nuclear speckles. In this case, 10 spatial compartmentalization states were described in human K562 leukemia cells [149]. In addition to regions adjacent to two nuclear landmarks (Speckles and Lamina), eight other states were recognized: Interior Active 1, 2, 3; Interior Repressive 1, 2; Near Lamina 1, 2; and Lamina-like. These SPIN states correlated with genomic functions such as transcriptional activity, replication timing and histone modifications more accurately than subcompartments that are based solely on Hi-C data [61]. Interestingly, DamID analysis of a nucleolus-localized peptide repeat suggested that several SPIN states displayed elevated nucleolar localization. As expected because of the high degree of LAD and NAD overlap, these included the Near Lamina 1, 2, Lamina-like and Lamina states. However, the Interior Repressive 2 state also had high nucleolus DamID scores yet had low lamin B-DamID and lamin B TSA-seq signals compared to the SPIN states near the NL [149]. Interior Repressive 2 SPIN state was also highly enriched for H3K27me3 marks compared with all other SPIN states. Hence, Interior Repressive 2 resembles nucleolar-specific Type II NADs [119,120]. As a wider variety of genomic data becomes available for additional nuclear bodies, the SPIN classification software will become ever more useful in the identification of functionally distinct genomic subclasses.
A recent study from the Dekker group used a “liquid Hi-C” technique to determine the strength of chromatin interactions in a genome-wide manner [169]. This technique involves extensive chromatin fragmentation via restriction enzyme digestion in situ in the nucleus, followed by fixation and Hi-C analysis to identify the kinetics of chromatin dissociation. Regions around the nucleoli exhibited one of the most stable interactions based on half-life of chromatin interactions upon digestion, with heterochromatin in general showing the most stable chromatin associations in the genome [169]. This is in agreement with the hypothesis that stable interactions in heterochromatin drive the A-B compartmentalization [16].
Together, the SPRITE, SPIN and liquid Hi-C data support the notion that the nucleolar periphery serves as heterochromatin docking site, thereby limiting chromatin movement and influencing chromatin organization, as it has been shown in human and yeast cells [115,116].
6b. Developmental regulation.
The existence of cell type-specific NADs in F121–9 mESCs and MEFs suggests that NADs are likely to be involved in developmental regulation of genomic regions. For example, the Pcsk6 gene, important for anterior-posterior axis establishment in early embryogenesis [170], is part of a NAD in MEFs, but not in F121–9 cells [119,120]. Correspondingly, this gene has higher transcriptional activity in F121–9 mESCs (FPKM value 22.2; [120]) than in MEFs (FPKM value 6.6; [162]); thus, reinforcing nucleolar localization as a possible repressive mechanism in developmental regulation.
A landmark paper from the Torres-Padilla group revealed the importance of nucleolar localization in early embryonic development [163]. Instead of tripartite nucleoli, mammalian oocytes and early preimplantation embryos contain transcriptionally inactive nucleolus precursor bodies (NPBs) that consist of homogenous fibrillar material (reviewed in [171]). Pericentric heterochromatin forms a ring-like structure around these NPBs. This configuration is maintained until the late two-cell stage, after which pericentric heterochromatic regions (major satellites in mouse cells) cluster together and form chromocenters, the characteristic dense DNA structures also observed in adult mouse cells [172,173]. Jachowicz et al. artificially tethered pericentric heterochromatic regions to the NL immediately after fertilization and analyzed the resulting two-cell stage embryos [150]. NL tethering caused high transcriptional activity at major satellite repeats and decreased staining for H3K27me3 and Polycomb Repressive Complex 1 (PRC1) subunit Ring1b. Strikingly, about half of the embryos with NL-tethered major satellites halted their development between the two- and eight-cell stages [163]. These results suggest that the nucleolar precursors play a crucial role in generating a repressive environment for major satellite repeats at these critical early cell divisions, that this repression is important for normal development, and this role cannot be substituted by the environment at the nuclear periphery.
6c. Roles for nucleoli and NADs in health and disease.
Studies in several species have linked nucleolar biology to organismal health and longevity. For example, small nucleolar size was correlated with longevity in C. elegans, D. melanogaster and mice, and long-lived C. elegans displayed reduced FBL expression, ribosomal protein synthesis, and rRNA levels, in a manner dependent on the nucleolar protein nucleolin [174]. Furthermore, muscle biopsies from humans on calorie restriction diets in conjunction with increased exercise regimes also exhibited reduced nucleolar size [174]. Conversely, Buchwalter et al. observed a positive correlation between human aging and increased nucleolar size and activity [175]. These authors also detected enlarged nucleoli in fibroblasts from Hutchinson-Gilford progeria syndrome (HGPS) patients, in which protein translation, ribosome synthesis, and rRNA transcription were elevated and rDNA methylation was decreased. Remarkably, induced expression of progerin, a mutant form of lamin A that causes HGPS [176], led to increased nucleolar size [175]. Levels of lamin A appeared to be pivotal, because depletion of this protein also resulted in nucleolar expansion. Together, these data indicate that lamin A is important not only for NL organization, but also affects nucleolar size and function. This role may be related to its localization within the nucleoplasm in addition to its canonical placement at the lamina [41,42], (reviewed in [63]). Evidence linking rDNA epigenetics to aging came from the Lemos group, who determined that rDNA CpG methylation increases with aging in mice, canids and humans, leading to the proposition that rDNA methylation can serve as an evolutionarily conserved clock of aging [177]. In sum, data from multiple organisms indicates that nucleolar physiology and epigenetics correlate with aging phenotypes. Exploration of the molecular mechanisms involved will therefore be a high priority, and it will be of much interest to determine if formation and silencing of perinucleolar heterochromatin contributes to these processes in aging and HGPS, extending existing NAD studies in senescent cells (see section 5b) [118].
A recent study by the Hannan and Poortinga groups showed that genomic regions change their interaction with rDNA repeats during Myc-driven malignant transformation of mouse pre-B cells [178]. The transition from pre-malignant to malignant pre-B cells was accompanied by UBF-mediated switching of a significant fraction of pseudo-silenced, i.e. hypomethylated but transcriptionally inactive rDNA promoters [179] to active, i.e. transcriptionally competent and hypomethylated rDNA promoters. In contrast, the transition from wild-type to pre-malignant cells did not exhibit similar phenomenon [178].
Psoralen more readily incorporates into accessible DNA [180], and thus Diesch et al. were able to distinguish active and inactive rDNA via psoralen crosslinking. The authors termed changes in rDNA chromatin during malignant transformation as rDNA class switching, however, this switching was not associated with increased rRNA transcription [178]. Diesch et al. performed 4C-seq, a modification of 3C and Hi-C technique [181,182], to map the genomic regions that interact with rDNA, which the authors equated to NADs [178]. The major changes in genome-rDNA interactions occurred between pre-malignant and malignant cells, concomitant with rDNA class switching, and these perturbations were accompanied by changes in gene expression levels. Increased rDNA interactions correlated with decreased expression of rDNA-interacting genes, and vice versa [178]. Gene Ontology analysis revealed that genes which were found in regions exhibiting increased rDNA interaction and decreased transcript levels were enriched for B-cell differentiation and lineage specification genes [178], which are often dysregulated in hematological cancers [183]. Genes located in regions that showed diminished rDNA interaction and increased expression were enriched for genes in ribosome function, RNA processing and mitochondrial function pathways [178]. Indeed, it has been shown that Myc drives increased rRNA transcription RNA processing and mitochondrial biogenesis [184] (reviewed in [111,185]). The authors suggested that since Myc drives many human cancers, rDNA class switching might be a common occurrence in human malignant transformations [178]. It would be of special interest to determine whether changes in NADs are common in diseases where perturbations in nucleolar morphology or rDNA repeat number or chromatin state occur. More intriguingly, investigation of mechanisms by which NAD alterations might possibly contribute to human malignancies is an exciting and unexplored avenue in the field of genome organization.
7. Factors and mechanisms regulating nucleolar association
7a. Protein factors
There are multiple studies that implicate various cis and trans-acting factors in tethering genomic regions to the perinucleolar space. The breadth of these factors suggest multiple mechanisms for perinucleolar association exist, and in some cases the mechanisms provide for aspects of locus specificity. We also note that many of these factors have other cellular functions besides the nucleolar tethering.
CTCF
CTCF is a DNA-binding protein that binds insulator elements in vertebrates and aids in blocking an enhancer of one gene from activating a promoter of another gene [186]. This insulating activity of CTCF is achieved through loop formation at topologically-associated domain (TAD) boundaries [61,64,65]. Regarding its role at the nucleolus, the Felsenfeld group showed that human cell CTCF co-purifies with nucleophosmin (NPM-1); furthermore, CTCF- and NPM-1-bound insulators on transgenes preferentially localized to nucleolar periphery in a CTCF binding site-dependent manner [187].
Nucleophosmin
Nucleophosmin (NPM-1) is a histone chaperone enriched in the GC layer of the nucleolus [104]. NPM-1 plays important roles in various cellular processes, including ribosome biogenesis, chromatin remodeling, and DNA damage responses [188]. Its Drosophila homolog, nucleoplasmin-like protein (NLP), along with CTCF and Modulo (Drosophila homolog of the nucleolar protein nucleolin) mediate centromere clustering around the nucleolar periphery [189]. Depletion of either NLP, or Modulo, or CTCF causes de-clustering of centromeres and relocalization of heterochromatin away from the nucleolar periphery. This was accompanied by derepression of centromeric repeats and mitotic defects, such as lagging chromosomes and anaphase bridges [189]. In human and mouse cells, depletion of NPM-1 resulted in altered nucleolar morphology and decreased levels of H3K9me3 and HP1γ foci at perinucleolar regions [190], suggesting that NPM-1 is important for tethering HP1γ to the nucleolus. These studies highlight the challenges in the analysis of nucleolar association, as depletion of proteins such as nucleolin and NPM-1 lead to changes in nucleolar morphology, heterochromatin organization and mitotic defects, confounding the direct and indirect functions of these factors.
CAF-1
CAF-1 (chromatin assembly factor-1) is a histone chaperone complex that deposits newly synthesized H3/H4 tetramers onto replicating DNA [191]. Depletion of CAF-1 subunit p150 in human cells causes partial dispersal of several nucleolar proteins: NPM-1, Ki-67, nucleolar phosphoprotein 140 (Nopp140), upstream binding factor (UBF), transcription termination factor 1 (TTF1), and nucleolin [192]. Additionally, depletion of CAF-1 p150 reduces nucleolar association of NADs, as demonstrated for the chromosome 10q telomere, the 5S rDNA array, and centromeric α-satellite DNA. The activity that localizes nucleolar proteins and NADs was mapped to a p150 domain that is not required for the histone deposition activity of CAF-1. Specifically, a Sumo-interaction motif (SIM) within this domain was essential [192]. Notably, this p150 domain also regulates the extent of recruitment of Ki-67 to the periphery of mitotic chromosomes [193].
One possible explanation for the role of p150 in NAD localization is its effect on cell proliferation marker Ki-67. This is because depletion of Ki-67 results in decreased nucleolar association of the centromeric histone CENP-A, reduced nucleolar staining of H3K9me3 and H4K20me3 in human and mouse cells [194], and leads to decreased association of α-satellite DNA with the nucleolar periphery [193] and elevated α-satellite transcript levels [155]. Ki-67 is not only required for NAD interactions in interphase cells, but it is also essential for recruitment of proteins to the surface of the mitotic chromosome [195] (reviewed in [196,197]). Furthermore, Ki-67 shapes the morphology of the mitotic chromosome [198,199] and is important for avoiding checkpoint-mediated cell cycle delays at the G1/S boundary of the cell cycle [200]. Therefore, Ki-67 is of high interest as a chromosome architectural protein across the cell cycle.
A recent study implicated a ribonucleoprotein complex termed MiCEE in nucleolar tethering and silencing of genes expressed from bidirectionally-oriented promoters [168]. MiCEE contains Mirlet7d, a let-7 miRNA family member, and protein subunits of both PRC2 and the nucleolytic exosome. Many of Mirlet7d targets are non-coding RNAs (ncRNAs) encoded near promoters of adjacent coding RNAs (cRNAs) [168]. Of these Mirlet7d target ncRNA and cRNAs, 20% are bidirectionally transcribed from shared promoter regions. MiCEE is targeted to these bidirectionally-transcribed loci by Mirlet7d-based recognition of locally-transcribed ncRNAs, triggering exosome-mediated degradation of the ncRNAs and PRC2-mediated transcriptional silencing of adjacent cRNAs. The loss of Mirlet7d, the exosome complex, or PRC2 subunits (EZH2, SUZ12, or EED), led to derepression of Mirlet7d targets in mouse and human cells. Furthermore, the effects of deleting one Mirlet7d ncRNA target locus was tested. Remarkably, this resulted in the relocalization of a Mirlet7d target cRNA-encoding locus away from the nucleolar periphery to the nuclear periphery [168]. Therefore, this study illustrates how ncRNAs can initiate nuclear body-specific tethering and silencing of specific loci. Furthermore, MiCEE supports localization of a locus to the perinucleolar chromatin, preventing what appears to be a default localization among LADs. It will be of great interest to see how many such mechanisms exist across the genome.
Additional studies confirm that PRC2 contributes broadly to NAD localization, because small molecule Ezh2 inhibitors led to decreased nucleolar association of Type I & Type II NADs [119]. However, PRC2 is not a nucleolus-specific trans-acting factor, because Ezh2 inhibition also led to decreased nuclear lamina association of LADs and Type I NADs [45,119]. These data are consistent with a role for H3K27me3 in heterochromatin association with both lamina and nucleoli. High resolution microscopy studies have indicated that H3K27me3-rich heterochromatin is spatially distinct from and has a different density than euchromatin or H3K9me3-rich chromatin [21]. Therefore, defining the factors that generate this distinct identity is of major interest to chromatin researchers.
TIP5
TIP5 (TTF-1-interacting protein-5), together with the ATPase SNF2h, are the protein subunits of the nucleolar remodeling complex (NoRC) [201,202]. NoRC represses rDNA transcription through its interactions with DNA methyltransferases DNMT1 and DNMT3b, histone deacetylase 1 (HDAC1), and the small RNA transcribed from the rDNA Intragenic Spacer (IGS) region, known as promoter-associated RNA (pRNA) [201–205]. In MEFs, TIP5 depletion also led to coalescence of CENP-A-bound centromeres and DAPI-intense heterochromatin into a smaller number of more-intensely stained nucleolar foci [206]. Notably, electron microscopy revealed the disappearance of densely-staining perinucleolar heterochromatin upon TIP5 depletion. These changes were accompanied by reduced levels of DNA methylation and gene silencing at rDNA repeats, and reduced heterochromatic marks H3K9me3 and H4K20me3 at centromeric minor satellite and pericentric major satellite repeats [207,208], accompanied by a shift from late to early DNA replication at these satellites and rDNA [206]. Additionally, TIP5 depletion reduced the DNA copy number of rDNA and major and minor satellite repeats, suggesting increased recombination [206]. Together, these data indicate TIP5 is a master regulator of heterochromatin at rDNA and satellite repeats, which impacts genome stability at these loci. Furthermore, given its role in silencing of satellite repeats, it is tempting to speculate that TIP5 may also be important for silencing of these repeats at the perinucleolar region during early mouse embryogenesis. This is of critical importance because in the absence of efficient silencing there are defects in later development [163].
A crucial partner of TIP5 is the short, structured pRNA molecule [204,205]. Normally, mESCs do not express mature pRNA, however, ectopic expression of pRNA in stem cells resulted in the formation of electron-dense perinucleolar heterochromatin-like structures, as well as increased occurrence of H3K9me2/3 marks at repetitive regions within mESCs, similar to the nuclear heterochromatin observed in NPCs, which were differentiated from these mESCs [209]. Conversely, depletion of TIP5 impaired mESC differentiation [209]. Therefore, NoRC-mediated rRNA silencing seems to promote not only nucleolar, but also nuclear heterochromatin condensation that is critical for exit from pluripotency.
7b. Nucleic acid factors
Unlike the nucleolar arrays of RNA Pol I-transcribed rDNA genes, the genes encoding ribosomal 5S rRNA are transcribed by RNA polymerase III and are found in distinct 5S rDNA arrays. These frequently localize to the nucleolar periphery, as do 5S rRNA transgenes, accompanied by transcriptional silencing [166]. Surprisingly, Hi-C maps did not reveal direct 5S-45S rDNA interactions, although they shared many common interaction sites [145,146]. Yu and Lemos [146] proposed a competitive exclusion model in which these anchoring sites are bound either by 5S or 45S arrays at any given time. Alternatively, 5S and 45S rDNA might interact within large protein complexes but fail to be captured via crosslinking as directly interacting entities in Hi-C experiments, which involve short formaldehyde-based crosslinks [146].
The enrichment of centromeric repeats within NADs suggests that the repeats have important functional roles in nucleolar association [25,210]. For example, human centromeric RNA has been implicated in the nucleolar association of centromeric proteins INCENP and CENP-C1 during interphase, prior to their release into the nucleoplasm for kinetochore assembly in mitosis [211].
A study from the Shen group suggests that L1 repeat RNAs, encoded by L1 non-long terminal repeat (non-LTR) retrotransposons, can facilitate localization of L1-enriched genes at nuclear and nucleolar peripheries [121]. Specifically, depletion of L1 RNA transcripts resulted in relocalization of L1-rich DNA sequences away from these regions. Indeed L1 RNA is a hub for structural interactions, because it mediates the binding of nucleolar protein nucleolin to rDNA [121,212]. Conversely, nucleolin preferentially binds to L1 repeats relative to other classes of repeats [121], indicating the importance of nucleolin-L1 RNA interactions for tethering nucleolus-associated genomic regions. These interactions are functionally important, because depletion of either L1 RNA transcripts or nucleolin led to upregulation of L1-enriched genes [121].
The nucleolar association of Xi is facilitated by the activity of long non-coding RNAs (lncRNAs, defined as ncRNAs longer than 200 nucleotides). During random X chromosome inactivation (XCI) in early female embryos, the X chromosome-linked X inactivation center (Xic) locus produces the lncRNA Xist (X-inactive specific transcript), which binds in cis to the X chromosome of origin and initiates silencing, generating the Xi [213–215]. Xist also targets Xi to the perinucleolar space during S-phase in mouse cells [114]. Loss of Xist causes detachment of Xi from the nucleolar periphery, reduced levels of H3K27me3, and some gene derepression on the Xi, suggesting that the Xi’s nucleolar association helps maintain its repressed state. One possibility to explain how this localization relates to silencing is based on the observation that the nucleolar periphery is enriched for Snf2h [114]. This is the catalytic subunit of the ACF1-ISWI chromatin remodeling enzyme complex that is needed for efficient DNA replication fork progression through heterochromatin [216]. These observations suggest that the nucleolar periphery could contribute to silencing in part by maintaining the fidelity of heterochromatin replication [114].
Another X-linked lncRNA named Firre escapes XCI and is encoded far (54 Mb) from the Xic [217]. Of note, CTCF binds Firre lncRNA, and together Firre and CTCF contribute to the nucleolar association and H3K27me3 enrichment of the Xi. However, depletion of Firre does not reactivate X-linked genes, consistent with overlapping mechanisms that prevent their reactivation [217,218]. Firre and Ctcf knockdown also led to the reduced nucleolar association of another X-linked lncRNA Dxz4 [167], but Dxz4 deletion did not result in changes in nucleolar association of Xi, or its H3K27me3 enrichment [219]. However, Firre depletion did not affect Xist lncRNA levels, or the shape of Xist RNA clouds [218], suggesting that interactions between CTCF and Firre lncRNA contribute to nucleolar association of the Xi, independent of the role Xist lncRNA plays in tethering Xi to the nucleolus.
LncRNA Kcnq1ot1 regulates the ~1 Mb Kcnq1-imprinted domain on mouse chromosome 7 by being paternally expressed and silencing neighboring genes in cis [220–222]. Insertion of a silencing domain found at the 5’ end of the Kcnq1ot1 transcript into an episomal vector led to the nucleolar association of this vector and silencing of a flanking reporter gene [164]. This silencing occurs with some tissue specificity, because the Kcnq1ot1 domain was more enriched for H3K9me3 and H3K27me3, as well as for histone methyltransferases G9a and PRC2, in mouse placenta than in fetal liver cells [165]. This correlated with increased nucleolar association of Kcnq1 in placenta, but not in liver cells [164]. However, a study by Magnuson group showed that mouse trophoblast stem cells deleted for PRC2 subunit Eed still exhibited frequent nucleolar association of Kcnq1ot1 lncRNA, despite derepression of Kcnq1-imprinted domain genes [223]. Hence, the nucleolar localization can be maintained even when transcriptional repression and PRC2 are lost, at least in specific cell types investigated. These results reinforce the idea that multiple pathways can contribute to nucleolar association and gene silencing.
7c. Phase separation
Recent studies have revealed that phase separation is an important mechanism regarding the formation and function of membraneless cellular bodies. Perhaps most importantly for this discussion, nucleoli form as a result of liquid-liquid phase separation [224], and NPM-1 and FBL form immiscible phases within nucleoli in vitro, recapitulating the distinct tripartite structure of nucleoli in vivo [225]. FBL and NPM-1 are proteins with intrinsically disordered regions (IDRs) that drive their liquid droplet formation, whereas RNA binding to these low complexity proteins drives them to their respective subcompartments [225,226]. Another study used temperature dependence and reversibility parameters to discern two distinct mechanisms of nucleolar protein assembly: IDR-driven phase separation of FBL and Nopp140, and active recruitment of nucleolin homolog Modulo and Nucleostemin 1 via rDNA [227].
Therefore, it is tempting to speculate that genomic regions might also be driven to associate with nucleolar periphery through combinations of phase separation and active recruitment. Examples might include the CTCF-NPM-1-Firre complex or the 5S rDNA arrays discussed above. Another possible phase separation-related example includes Alu repeat-derived RNAs, which contribute to nucleolar structure and perinucleolar associations through their specific interactions with nucleolin and NPM-1 [228]. Consistent with a role for phase separation, hexanediol treatment, which disrupts liquid-liquid phase separation, reduces the nucleolar association levels of NADs in mouse fibroblasts [119].
8. Overlap between NADs and LADs
Genome-wide mapping of NADs in human and mouse cells revealed extensive overlap between NADs and LADs [25,26,118–120]. One of these early studies also included a key experiment demonstrating that NADs labeled via a photoactivatable fluorescently-tagged histone protein would either be distributed around the nuclear periphery or the nucleolar periphery in the next interphase after progression through mitosis [26]. These results, consistent with experiments following the fate of LADs after mitosis [39], led to the conclusion that the bulk of NAD and LAD sequences are stochastically distributed to either the nuclear or nucleolar periphery at each mitotic exit. One proposed mechanism is that lamin A found at both the nuclear and nucleolar peripheries could tether these interactions [229]. Additionally, lamin B2 localizes at the nucleolar border in close proximity to nucleolin, and depletion of lamin B2 leads to disruption of both nuclear and nucleolar morphologies [230]. Together, these observations suggest that lamins are central to multiple heterochromatin localizations.
Apart from reshuffling during mitosis, evidence for plasticity in nuclear localization of mammalian heterochromatin comes from nucleolus perturbation studies in human lymphoblastoid cells [109]. This study focused on late-replicating regions, which are distributed among the NL, pericentric regions, and the nucleolar periphery. In general, the late-replicating regions on large chromosomes were frequently found at the nuclear periphery, whereas those on small chromosomes were more often associated with nucleoli. Treatment of cells with actinomycin D, which inhibits RNA Pol I and disrupts nucleoli, drastically decreased association of heterochromatic regions with the nucleolar periphery, but increased association of these regions with the NL [109]. Similarly, our group also showed that Ki-67-depleted cells exhibited reduced Xi association with the nucleolar periphery, and increased association of Xi with the nuclear periphery [200]. Hence, when one of the heterochromatic nuclear subcompartments is disrupted, the heterochromatic regions tend to relocalize to other intact subcompartments. This suggests that some common mechanisms operate at each location.
Indeed, the majority of human and mouse NADs overlap with LADs [118–120]. Both nuclear and nucleolar peripheries are enriched for centromeric and pericentric heterochromatin [231], and for repetitive elements such as LINEs and LTR retrotransposons [118,121]. However, the overlap between LADs and NADs is incomplete. About one third of human NADs are non-overlapping with LADs [118], and likewise there is a subset of mouse NADs found only at the nucleolar periphery, i.e. Type II NADs [119,120]. As discussed above, a full understanding of the mechanisms and functional significance of each type of nuclear localization remains to be elucidated. For example, decoding the biology of nucleolar localization will require systematic investigation of the molecular determinants and examining whether silencing is a cause or consequence of nucleolar tethering.
Conclusion
Interest in the nucleolar periphery as a heterochromatin hub was sparked ten years ago with the initial sequencing of human NAD DNA [25,26]. This interest has only increased with subsequent, more detailed analyses in animal and plant cells [118–121,141] and the advent of novel genomic techniques, such as Hi-C [58,61]; SPRITE [143] and SPIN [149], which reinforced the importance of the nucleolar periphery as a site for heterochromatin localization. Many insights into the biology of perinucleolar heterochromatin have been made by comparisons to nuclear lamina-associated domains (LADs), which have been more extensively characterized. The similarity among NADs and LADs arises because many constitutive heterochromatic loci adopt a nucleolar or laminar localization stochastically each time cells exit mitosis [26,39]. Self-interaction mechanisms including phase separation will be important for understanding the localizations of these loci [16].
However, some heterochromatic regions appear to be localized by less general mechanisms. For example, because Type II NADs are found only in the perinucleolar space, but not at the nuclear periphery [119,120], it is tempting to speculate that specific trans-acting factors will be required for such localizations. Indeed, positioning of some perinucleolar chromatin is known to be disturbed in the absence of the MiCEE and NoRC complexes [168,209]. Because both of those complexes contain small RNAs (let7d and pRNA, respectively) required for targeting, it will be of great interest to determine how general RNA-based mechanisms for nucleolar targeting will be.
It is clear that progress in our understanding of NAD biology requires the dissection of the different mechanisms of nucleolar association. Future studies of the dynamics and cell-to-cell variability of nucleolar association could be particularly aided by methodologies that can be extended to single-cell analyses, such as DamID and TSA-seq, as well as high-throughput imaging. Finally, it will be of great interest to extend to techniques to understanding the mechanisms underlying perinucleolar chromatin localization during early mammalian embryogenesis. This is of particular importance because perturbation of this localization by dragging mouse pericentric heterochromatin to the lamina via a zinc finger-emerin fusion protein not only impaired silencing of satellite repeats, but also greatly impaired formation of blastocysts [163]. Therefore, how perinucleolar chromatin is formed in the embryo, and how these mechanisms might differ across development, are major questions in mammalian developmental biology.
Highlights.
We review the spatial organization of eukaryotic genomes, with particular emphasis on heterochromatin in mammalian cells.
Heterochromatin is largely localized to the nuclear and nucleolar peripheries.
Localization to these regions occurs by a variety of molecular mechanisms, some being region specific.
Mechanisms of heterochromatin localization are important for normal patterns of gene expression, cellular differentiation, and organismal development.
We review multiple technologies being applied to the study of chromatin localization.
Acknowledgements
This work was supported by NIH grants U01 CA260669 and R35 GM127035 to PDK.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Author Agreement
The authors, Aizhan Bizhanova and Paul D. Kaufman, certify that all authors have seen and approved the final version of the manuscript being submitted. They warrant that the article is the authors’ original work, hasn’t received prior publication, and isn’t under consideration for publication elsewhere.
References
- [1].Heitz E, Das Heterochromatin der Moose., Jahrbücher Für Wissenschaftliche Bot. (1928). [Google Scholar]
- [2].Politz JCR, Scalzo D, Groudine M, Something Silent This Way Forms: The Functional Organization of the Repressive Nuclear Compartment, Annu. Rev. Cell Dev. Biol 29 (2013) 241–270. 10.1146/annurev-cellbio-101512-122317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Weintraub H, Groudine M, Chromosomal subunits in active genes have an altered conformation, Science (80-.). 193 (1976) 848–856. 10.1126/science.948749. [DOI] [PubMed] [Google Scholar]
- [4].O’Keefe RT, Henderson SC, Spector DL, Dynamic organization of DNA replication in mammalian cell nuclei: spatially and temporally defined replication of chromosome-specific alpha-satellite DNA sequences, J. Cell Biol 116 (1992) 1095–1110. 10.1083/jcb.116.5.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Wu R, Terry AV, Singh PB, Gilbert DM, Differential Subnuclear Localization and Replication Timing of Histone H3 Lysine 9 Methylation States, Mol. Biol. Cell 16 (2005) 2872–2881. 10.1091/mbc.e04-11-0997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Saksouk N, Simboeck E, Déjardin J, Constitutive heterochromatin formation and transcription in mammals, Epigenetics and Chromatin. 8 (2015). 10.1186/1756-8935-8-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Kurdistani SK, Grunstein M, Histone acetylation and deacetylation in yeast, Nat. Rev. Mol. Cell Biol 4 (2003) 276–284. 10.1038/nrm1075. [DOI] [PubMed] [Google Scholar]
- [8].Shahbazian MD, Grunstein M, Functions of Site-Specific histone acetylation and deacetylation, Annu. Rev. Biochem 76 (2007) 75–100. 10.1146/annurev.biochem.76.052705.162114. [DOI] [PubMed] [Google Scholar]
- [9].Richards EJ, Elgin SCR, Epigenetic codes for heterochromatin formation and silencing: Rounding up the usual suspects, Cell. 108 (2002) 489–500. 10.1016/S0092-8674(02)00644-X. [DOI] [PubMed] [Google Scholar]
- [10].Taddei A, Maison C, Roche D, Almouzni G, Reversible disruption of pericentric heterochromatin and centromere function by inhibiting deacetylases, Nat. Cell Biol 3 (2001) 114–120. 10.1038/35055010. [DOI] [PubMed] [Google Scholar]
- [11].Yamada T, Fischle W, Sugiyama T, Allis CD, Grewal SIS, The nucleation and maintenance of heterochromatin by a histone deacetylase in fission yeast, Mol. Cell 20 (2005) 173–185. 10.1016/j.molcel.2005.10.002. [DOI] [PubMed] [Google Scholar]
- [12].Alper BJ, Job G, Yadav RK, Shanker S, Lowe BR, Partridge JF, Sir2 is required for Clr4 to initiate centromeric heterochromatin assembly in fission yeast, EMBO J. 32 (2013) 2321–2335. 10.1038/emboj.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Janssen A, Colmenares SU, Karpen GH, Heterochromatin: Guardian of the Genome, Annu. Rev. Cell Dev. Biol 34 (2018) 265–288. 10.1146/annurev-cellbio-100617-062653. [DOI] [PubMed] [Google Scholar]
- [14].Allshire RC, Madhani HD, Ten principles of heterochromatin formation and function, Nat. Rev. Mol. Cell Biol 19 (2018) 229–244. 10.1038/nrm.2017.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Solovei I, Wang AS, Thanisch K, Schmidt CS, Krebs S, Zwerger M, Cohen TV, Devys D, Foisner R, Peichl L, Herrmann H, Blum H, Engelkamp D, Stewart CL, Leonhardt H, Joffe B, LBR and lamin A/C sequentially tether peripheral heterochromatin and inversely regulate differentiation, Cell. 152 (2013) 584–598. 10.1016/j.cell.2013.01.009. [DOI] [PubMed] [Google Scholar]
- [16].Falk M, Feodorova Y, Naumova N, Imakaev M, Lajoie BR, Leonhardt H, Joffe B, Dekker J, Fudenberg G, Solovei I, Mirny LA, Heterochromatin drives compartmentalization of inverted and conventional nuclei, Nature. 570 (2019) 395–399. 10.1038/s41586-019-1275-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T, Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain, Nature. 410 (2001) 120–124. 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- [18].Jacobs SA, Khorasanizadeh S, Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail, Science (80-.). 295 (2002) 2080–2083. 10.1126/science.1069473. [DOI] [PubMed] [Google Scholar]
- [19].Nielsen PR, Nietlispach D, Mott HR, Callaghan J, Bannister A, Kouzarides T, Murzin AG, Murzina NV, Laue ED, Structure of the HP1 chromodomain bound to histone H3 methylated at lysine 9, Nature. 416 (2002) 103–107. 10.1038/nature722. [DOI] [PubMed] [Google Scholar]
- [20].Nishibuchi G, Déjardin J, The molecular basis of the organization of repetitive DNA-containing constitutive heterochromatin in mammals, Chromosom. Res 25 (2017) 77–87. 10.1007/s10577-016-9547-3. [DOI] [PubMed] [Google Scholar]
- [21].Boettiger AN, Bintu B, Moffitt JR, Wang S, Beliveau BJ, Fudenberg G, Imakaev M, Mirny LA, Wu CT, Zhuang X, Super-resolution imaging reveals distinct chromatin folding for different epigenetic states, Nature. 529 (2016) 418–422. 10.1038/nature16496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Trojer P, Reinberg D, Facultative Heterochromatin: Is There a Distinctive Molecular Signature?, Mol. Cell 28 (2007) 1–13. 10.1016/J.MOLCEL.2007.09.011. [DOI] [PubMed] [Google Scholar]
- [23].Pombo A, Dillon N, Three-dimensional genome architecture: Players and mechanisms, Nat. Rev. Mol. Cell Biol 16 (2015) 245–257. 10.1038/nrm3965. [DOI] [PubMed] [Google Scholar]
- [24].Politz JCR, Scalzo D, Groudine M, The redundancy of the mammalian heterochromatic compartment, Curr. Opin. Genet. Dev 37 (2016) 1–8. 10.1016/j.gde.2015.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Németh A, Conesa A, Santoyo-Lopez J, Medina I, Montaner D, Péterfia B, Solovei I, Cremer T, Dopazo J, Längst G, Initial genomics of the human nucleolus, PLoS Genet. 6 (2010). 10.1371/journal.pgen.1000889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].van Koningsbruggen S, Gierlinski M, Schofield P, Martin D, Barton GJ, Ariyurek Y, den Dunnen JT, Lamond AI, High-resolution whole-genome sequencing reveals that specific chromatin domains from most human chromosomes associate with nucleoli., Mol. Biol. Cell 21 (2010) 3735–48. 10.1091/mbc.E10-06-0508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Pickersgill H, Kalverda B, de Wit E, Talhout W, Fornerod M, van Steensel B, Characterization of the Drosophila melanogaster genome at the nuclear lamina, Nat. Genet 38 (2006) 1005–1014. 10.1038/ng1852. [DOI] [PubMed] [Google Scholar]
- [28].Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W, Eussen BH, de Klein A, Wessels L, de Laat W, van Steensel B, Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions, Nature. 453 (2008) 948–951. 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- [29].Ikegami K, Egelhofer TA, Strome S, Lieb JD, Caenorhabditis elegans chromosome arms are anchored to the nuclear membrane via discontinuous association with LEM-2, Genome Biol. 11 (2010) R120 10.1186/gb-2010-11-12-r120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Peric-Hupkes D, Meuleman W, Pagie L, Bruggeman SWM, Solovei I, Brugman W, Gräf S, Flicek P, Kerkhoven RM, van Lohuizen M, Reinders M, Wessels L, van Steensel B, Molecular Maps of the Reorganization of Genome-Nuclear Lamina Interactions during Differentiation, Mol. Cell 38 (2010) 603–613. 10.1016/j.molcel.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Kind J, van Steensel B, Genome–nuclear lamina interactions and gene regulation, Curr. Opin. Cell Biol 22 (2010) 320–325. 10.1016/j.ceb.2010.04.002. [DOI] [PubMed] [Google Scholar]
- [32].Kind J, Pagie L, de Vries SS, Nahidiazar L, Dey SS, Bienko M, Zhan Y, Lajoie B, de Graaf CA, Amendola M, Fudenberg G, Imakaev M, Mirny LA, Jalink K, Dekker J, van Oudenaarden A, van Steensel B, Genome-wide Maps of Nuclear Lamina Interactions in Single Human Cells, Cell. 163 (2015) 134–147. 10.1016/J.CELL.2015.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Borsos M, Perricone SM, Schauer T, Pontabry J, de Luca KL, de Vries SS, Ruiz-Morales ER, Torres-Padilla M-E, Kind J, Genome-lamina interactions are established de novo in the early mouse embryo., Nature. 569 (2019) 729–733. 10.1038/s41586-019-1233-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Mikulski P, Hohenstatt ML, Farrona S, Smaczniak C, Stahl Y, Kalyanikrishna, Kaufmann K, Angenent G, Schubert D, The chromatin-associated protein pwo1 interacts with plant nuclear lamin-like components to regulate nuclear size, Plant Cell. 31 (2019) 1141–1154. 10.1105/tpc.18.00663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Holla S, Dhakshnamoorthy J, Folco HD, Balachandran V, Xiao H, ling Sun L, Wheeler D, Zofall M, Grewal SIS, Positioning Heterochromatin at the Nuclear Periphery Suppresses Histone Turnover to Promote Epigenetic Inheritance, Cell. 180 (2020) 150–164.e15. 10.1016/j.cell.2019.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Akhtar A, Gasser SM, The nuclear envelope and transcriptional control, Nat. Rev. Genet 8 (2007) 507–517. 10.1038/nrg2122. [DOI] [PubMed] [Google Scholar]
- [37].Towbin BD, Meister P, Gasser SM, The nuclear envelope - a scaffold for silencing?, Curr. Opin. Genet. Dev 19 (2009) 180–186. 10.1016/j.gde.2009.01.006. [DOI] [PubMed] [Google Scholar]
- [38].Mattout A, Cabianca DS, Gasser SM, Chromatin states and nuclear organization in development — a view from the nuclear lamina, Genome Biol. 16 (2015) 174 10.1186/s13059-015-0747-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Kind J, Pagie L, Ortabozkoyun H, Boyle S, De Vries SS, Janssen H, Amendola M, Nolen LD, Bickmore WA, Van Steensel B, Single-cell dynamics of genome-nuclear lamina interactions, Cell. 153 (2013) 178–192. 10.1016/j.cell.2013.02.028. [DOI] [PubMed] [Google Scholar]
- [40].Shah PP, Donahue G, Otte GL, Capell BC, Nelson DM, Cao K, Aggarwala V, Cruickshanks HA, Rai TS, McBryan T, Gregory BD, Adams PD, Berger SL, Lamin B1 depletion in senescent cells triggers large-scale changes in gene expression and the chromatin landscape, Genes Dev. 27 (2013) 1787–1799. 10.1101/gad.223834.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Lund EG, Duband-Goulet I, Oldenburg A, Buendia B, Collas P, Distinct features of lamin A-interacting chromatin domains mapped by Chip-sequencing from sonicated or micrococcal nuclease-digested chromatin, Nucleus. 6 (2015) 30–39. 10.4161/19491034.2014.990855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Gesson K, Rescheneder P, Skoruppa MP, Von Haeseler A, Dechat T, Foisner R, A-type Lamins bind both hetero- and euchromatin, the latter being regulated by lamina-associated polypeptide 2 alpha, Genome Res. 26 (2016) 462–473. 10.1101/gr.196220.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Pope BD, Ryba T, Dileep V, Yue F, Wu W, Denas O, Vera DL, Wang Y, Hansen RS, Canfield TK, Thurman RE, Cheng Y, Gülsoy G, Dennis JH, Snyder MP, Stamatoyannopoulos JA, Taylor J, Hardison RC, Kahveci T, Ren B, Gilbert DM, Topologically associating domains are stable units of replication-timing regulation, Nature. 515 (2014) 402–405. 10.1038/nature13986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Wen B, Wu H, Shinkai Y, Irizarry RA, Feinberg AP, Large histone H3 lysine 9 dimethylated chromatin blocks distinguish differentiated from embryonic stem cells, Nat. Genet 41 (2009) 246–250. 10.1038/ng.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Harr JC, Luperchio TR, Wong X, Cohen E, Wheelan SJ, Reddy KL, Directed targeting of chromatin to the nuclear lamina is mediated by chromatin state and A-type lamins, J. Cell Biol 208 (2015) 33–52. 10.1083/jcb.201405110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Meuleman W, Peric-Hupkes D, Kind J, Beaudry J-B, Pagie L, Kellis M, Reinders M, Wessels L, van Steensel B, Constitutive nuclear lamina-genome interactions are highly conserved and associated with A/T-rich sequence., Genome Res. 23 (2013) 270–80. 10.1101/gr.141028.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].van Steensel B, Belmont AS, Lamina-Associated Domains: Links with Chromosome Architecture, Heterochromatin, and Gene Repression., Cell. 169 (2017) 780–791. 10.1016/j.cell.2017.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Lochs SJA, Kefalopoulou S, Kind J, Lamina Associated Domains and Gene Regulation in Development and Cancer., Cells. 8 (2019). 10.3390/cells8030271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Reddy KL, Zullo JM, Bertolino E, Singh H, Transcriptional repression mediated by repositioning of genes to the nuclear lamina, Nature. 452 (2008) 243–247. 10.1038/nature06727. [DOI] [PubMed] [Google Scholar]
- [50].Leemans C, van der Zwalm MCH, Brueckner L, Comoglio F, van Schaik T, Pagie L, van Arensbergen J, van Steensel B, Promoter-Intrinsic and Local Chromatin Features Determine Gene Repression in LADs, Cell. 177 (2019) 852–864.e14. 10.1016/J.CELL.2019.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Iwafuchi-Doi M, Zaret KS, Pioneer transcription factors in cell reprogramming, Genes Dev. 28 (2014) 2679–2692. 10.1101/gad.253443.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Kumaran RI, Spector DL, A genetic locus targeted to the nuclear periphery in living cells maintains its transcriptional competence, J. Cell Biol 180 (2008) 51–65. 10.1083/jcb.200706060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Finlan LE, Sproul D, Thomson I, Boyle S, Kerr E, Perry P, Ylstra B, Chubb JR, Bickmore WA, Recruitment to the nuclear periphery can alter expression of genes in human cells, PLoS Genet. 4 (2008). 10.1371/journal.pgen.1000039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Brueckner L, Zhao PA, Schaik T, Leemans C, Sima J, Peric-Hupkes D, Gilbert DM, Steensel B, Local rewiring of genome–nuclear lamina interactions by transcription, EMBO J. 39 (2020). 10.15252/embj.2019103159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Demmerle J, Koch AJ, Holaska JM, The Nuclear Envelope Protein Emerin Binds Directly to Histone Deacetylase 3 (HDAC3) and Activates HDAC3 Activity, J. Biol. Chem 287 (2012) 22080 10.1074/JBC.M111.325308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Zullo JM, Demarco IA, Piqué-Regi R, Gaffney DJ, Epstein CB, Spooner CJ, Luperchio TR, Bernstein BE, Pritchard JK, Reddy KL, Singh H, DNA sequence-dependent compartmentalization and silencing of chromatin at the nuclear lamina, Cell. 149 (2012) 1474–1487. 10.1016/j.cell.2012.04.035. [DOI] [PubMed] [Google Scholar]
- [57].Dekker J, Rippe K, Dekker M, Kleckner N, Capturing chromosome conformation., Science. 295 (2002) 1306–11. 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- [58].Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, Sandstrom R, Bernstein B, Bender MA, Groudine M, Gnirke A, Stamatoyannopoulos J, Mirny LA, Lander ES, Dekker J, Comprehensive mapping of long-range interactions reveals folding principles of the human genome., Science. 326 (2009) 289–93. 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Zhang Y, McCord RP, Ho YJ, Lajoie BR, Hildebrand DG, Simon AC, Becker MS, Alt FW, Dekker J, Spatial organization of the mouse genome and its role in recurrent chromosomal translocations, Cell. 148 (2012) 908–921. 10.1016/j.cell.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Gibcus JH, Dekker J, The Hierarchy of the 3D Genome, Mol. Cell 49 (2013) 773–782. 10.1016/j.molcel.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Rao SSP, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, Aiden EL, A 3D Map of the Human Genome at Kilobase Resolution Reveals Principles of Chromatin Looping, Cell. 159 (2014) 1665–1680. 10.1016/J.CELL.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Zheng X, Hu J, Yue S, Kristiani L, Kim M, Sauria M, Taylor J, Kim Y, Zheng Y, Lamins Organize the Global Three-Dimensional Genome from the Nuclear Periphery., Mol. Cell 71 (2018) 802–815.e7. 10.1016/j.molcel.2018.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Buchwalter A, Kaneshiro JM, Hetzer MW, Coaching from the sidelines: the nuclear periphery in genome regulation, Nat. Rev. Genet 20 (2019) 39–50. 10.1038/s41576-018-0063-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, Piolot T, van Berkum NL, Meisig J, Sedat J, Gribnau J, Barillot E, Blüthgen N, Dekker J, Heard E, Spatial partitioning of the regulatory landscape of the X-inactivation centre, Nature. 485 (2012) 381–385. 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B, Topological domains in mammalian genomes identified by analysis of chromatin interactions, Nature. 485 (2012) 376–380. 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Symmons O, Uslu VV, Tsujimura T, Ruf S, Nassari S, Schwarzer W, Ettwiller L, Spitz F, Functional and topological characteristics of mammalian regulatory domains, Genome Res. 24 (2014) 390–400. 10.1101/gr.163519.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Mifsud B, Tavares-Cadete F, Young AN, Sugar R, Schoenfelder S, Ferreira L, Wingett SW, Andrews S, Grey W, Ewels PA, Herman B, Happe S, Higgs A, LeProust E, Follows GA, Fraser P, Luscombe NM, Osborne CS, Mapping long-range promoter contacts in human cells with high-resolution capture Hi-C, Nat. Genet 47 (2015) 598–606. 10.1038/ng.3286. [DOI] [PubMed] [Google Scholar]
- [68].Dechat T, Pfleghaar K, Sengupta K, Shimi T, Shumaker DK, Solimando L, Goldman RD, Nuclear lamins: major factors in the structural organization and function of the nucleus and chromatin, Genes Dev. 22 (2008) 832–853. 10.1101/gad.1652708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Burke B, Stewart CL, The nuclear lamins: flexibility in function, Nat. Rev. Mol. Cell Biol 14 (2013) 13–24. 10.1038/nrm3488. [DOI] [PubMed] [Google Scholar]
- [70].Yuan J, Simos G, Blobel G, Georgatos SD, Binding of lamin A to polynucleosomes, J. Biol. Chem 266 (1991) 9211–9215. [PubMed] [Google Scholar]
- [71].Höger TH, Krohne G, Kleinschmidt JA, Interaction of Xenopus lamins A and LII with chromatin in vitro mediated by a sequence element in the carboxyterminal domain, Exp. Cell Res 197 (1991) 280–289. 10.1016/0014-4827(91)90434-V. [DOI] [PubMed] [Google Scholar]
- [72].Taniura H, Glass C, Gerace L, A chromatin binding site in the tail domain of nuclear lamins that interacts with core histones, J. Cell Biol 131 (1995) 33–44. 10.1083/jcb.131.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Goldberg M, Harel A, Brandeis M, Rechsteiner T, Richmond TJ, Weiss AM, Gruenbaum Y, The tail domain of lamin Dm0 binds histones H2A and H2B, Proc. Natl. Acad. Sci. U. S. A 96 (1999) 2852–2857. 10.1073/pnas.96.6.2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Mattout A, Goldberg M, Tzur Y, Margalit A, Gruenbaum Y, Specific and conserved sequences in D. melanogaster and C. elegans lamins and histone H2A mediate the attachment of lamins to chromosomes, J. Cell Sci 120 (2007) 77–85. 10.1242/jcs.03325. [DOI] [PubMed] [Google Scholar]
- [75].Shoeman RL, Traub P, The in Vitro DNA-binding Properties of Purified Nuclear Lamin Proteins and Vimentin, J. Biol. Chem 265 (1990) 9055–9061. [PubMed] [Google Scholar]
- [76].Rzepecki R, Bogachev SS, Kokoza E, Stuurman N, Fisher PA, In vivo association of lamins with nucleic acids in Drosophila melanogaster, J. Cell Sci 111 (1998) 121–129. [DOI] [PubMed] [Google Scholar]
- [77].Stierlé V, Couprie J, Östlund C, Krimm I, Zinn-Justin S, Hossenlopp P, Worman HJ, Courvalin JC, Duband-Goulet I, The carboxyl-terminal region common to lamins A and C contains a DNA binding domain, Biochemistry. 42 (2003) 4819–4828. 10.1021/bi020704g. [DOI] [PubMed] [Google Scholar]
- [78].Gruenbaum Y, Foisner R, Lamins: Nuclear Intermediate Filament Proteins with Fundamental Functions in Nuclear Mechanics and Genome Regulation, Annu. Rev. Biochem 84 (2015) 131–164. 10.1146/annurev-biochem-060614-034115. [DOI] [PubMed] [Google Scholar]
- [79].Bruston F, Delbarre E, Östlund C, Worman HJ, Buendia B, Duband-Goulet I, Loss of a DNA binding site within the tail of prelamin A contributes to altered heterochromatin anchorage by progerin, FEBS Lett. 584 (2010) 2999–3004. 10.1016/j.febslet.2010.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Dobrzynska A, Gonzalo S, Shanahan C, Askjaer P, The nuclear lamina in health and disease, Nucleus. 7 (2016) 233–248. 10.1080/19491034.2016.1183848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Ho CY, Lammerding J, Lamins at a glance, J. Cell Sci 125 (2012) 2087–2093. 10.1242/jcs.087288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Dittmer T, Misteli T, The lamin protein family, Genome Biol. 12 (2011). 10.1186/gb-2011-12-5-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Barton LJ, Soshnev AA, Geyer PK, Networking in the nucleus: A spotlight on LEM-domain proteins, Curr. Opin. Cell Biol 34 (2015) 1–8. 10.1016/j.ceb.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Berk JM, Simon DN, Jenkins-Houk CR, Westerbeck JW, Wilson KL, Carlson CR, The molecular basis of emerin-emerin and emerin-BAF interactions, J. Cell Sci 127 (2014) 3956–3969. 10.1242/jcs.148247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Nikolakaki E, Mylonis I, Giannakouros T, Lamin B Receptor: Interplay between Structure, Function and Localization, Cells. 6 (2017) 28 10.3390/cells6030028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Hirano Y, Hizume K, Kimura H, Takeyasu K, Haraguchi T, Hiraoka Y, Lamin B receptor recognizes specific modifications of histone H4 in heterochromatin formation, J. Biol. Chem 287 (2012) 42654–42663. 10.1074/jbc.M112.397950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Kohwi M, Lupton JR, Lai S-L, Miller MR, Doe CQ, Developmentally regulated subnuclear genome reorganization restricts neural progenitor competence in Drosophila., Cell. 152 (2013) 97–108. 10.1016/j.cell.2012.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Mattout A, Pike BL, Towbin BD, Bank EM, Gonzalez-Sandoval A, Stadler MB, Meister P, Gruenbaum Y, Gasser SM, An EDMD Mutation in C. elegans Lamin Blocks Muscle-Specific Gene Relocation and Compromises Muscle Integrity, Curr. Biol 21 (2011) 1603–1614. 10.1016/j.cub.2011.08.030. [DOI] [PubMed] [Google Scholar]
- [89].Wong X, Hoskins VE, Harr JC, Gordon M, Reddy K, Lamin C regulates genome organization after mitosis, BioRxiv. (2020) 2020.07.28.213884. 10.1101/2020.07.28.213884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Amendola M, van Steensel B, Nuclear lamins are not required for lamina-associated domain organization in mouse embryonic stem cells, EMBO Rep. 16 (2015) 610–617. 10.15252/embr.201439789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Zheng X, Kim Y, Zheng Y, Identification of lamin B-regulated chromatin regions based on chromatin landscapes, Mol. Biol. Cell 26 (2015) 2685–2697. 10.1091/mbc.E15-04-0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Towbin BD, González-Aguilera C, Sack R, Gaidatzis D, Kalck V, Meister P, Askjaer P, Gasser SM, Step-Wise Methylation of Histone H3K9 Positions Heterochromatin at the Nuclear Periphery, Cell. 150 (2012) 934–947. 10.1016/j.cell.2012.06.051. [DOI] [PubMed] [Google Scholar]
- [93].Bian Q, Khanna N, Alvikas J, Belmont AS, β-Globin cis-elements determine differential nuclear targeting through epigenetic modifications, J. Cell Biol 203 (2013) 767–783. 10.1083/jcb.201305027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Heitz E, Die Ursache der gesetzmässigen Zahl, Lage, Form und Grösse pflanzlicher Nukleolen, Planta. 12 (1931) 775–844. 10.1007/BF01912443. [DOI] [Google Scholar]
- [95].McClintock B, The relation of a particular chromosomal element to the development of the nucleoli in Zea mays, Zeitschrift Für Zellforsch. Und Mikroskopische Anat. 21 (1934) 294–326. 10.1007/BF00374060. [DOI] [Google Scholar]
- [96].Grummt I, Wisely chosen paths--regulation of rRNA synthesis: delivered on 30 June 2010 at the 35th FEBS Congress in Gothenburg, Sweden., FEBS J. 277 (2010) 4626–39. 10.1111/j.1742-4658.2010.07892.x. [DOI] [PubMed] [Google Scholar]
- [97].Henras AK, Plisson-Chastang C, O’Donohue MF, Chakraborty A, Gleizes PE, An overview of pre-ribosomal RNA processing in eukaryotes, Wiley Interdiscip. Rev. RNA 6 (2015) 225–242. 10.1002/wrna.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Potapova TA, Gerton JL, Ribosomal DNA and the nucleolus in the context of genome organization, Chromosom. Res 27 (2019) 109–127. 10.1007/s10577-018-9600-5. [DOI] [PubMed] [Google Scholar]
- [99].Pederson T, The nucleolus., Cold Spring Harb. Perspect. Biol 3 (2011). 10.1101/cshperspect.a000638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Grob A, McStay B, Construction of synthetic nucleoli and what it tells us about propagation of sub-nuclear domains through cell division, Cell Cycle. 13 (2014) 2501–2508. 10.4161/15384101.2014.949124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Yao RW, Xu G, Wang Y, Shan L, Luan PF, Wang Y, Wu M, Yang LZ, Xing YH, Yang L, Chen LL, Nascent Pre-rRNA Sorting via Phase Separation Drives the Assembly of Dense Fibrillar Components in the Human Nucleolus, Mol. Cell 76 (2019) 767–783.e11. 10.1016/j.molcel.2019.08.014. [DOI] [PubMed] [Google Scholar]
- [102].Tatavosian R, Kent S, Brown K, Yao T, Duc HN, Huynh TN, Zhen CY, Ma B, Wang H, Ren X, Nuclear condensates of the Polycomb protein chromobox 2 (CBX2) assemble through phase separation., J. Biol. Chem 294 (2019) 1451–1463. 10.1074/jbc.RA118.006620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Maiser A, Dillinger S, Längst G, Schermelleh L, Leonhardt H, Németh A, Super-resolution in situ analysis of active ribosomal DNA chromatin organization in the nucleolus, Sci. Rep 10 (2020) 1–11. 10.1038/s41598-020-64589-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Boisvert FM, Van Koningsbruggen S, Navascués J, Lamond AI, The multifunctional nucleolus, Nat. Rev. Mol. Cell Biol 8 (2007) 574–585. 10.1038/nrm2184. [DOI] [PubMed] [Google Scholar]
- [105].Raška I, Shaw PJ, Cmarko D, Structure and function of the nucleolus in the spotlight, Curr. Opin. Cell Biol 18 (2006) 325–334. 10.1016/j.ceb.2006.04.008. [DOI] [PubMed] [Google Scholar]
- [106].Schöfer C, Weipoltshammer K, Nucleolus and chromatin, Histochem. Cell Biol 150 (2018) 209–225. 10.1007/s00418-018-1696-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].McStay B, Nucleolar organizer regions: Genomic ‘dark matter’ requiring illumination, Genes Dev. 30 (2016) 1598–1610. 10.1101/gad.283838.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Carvalho C, Pereira HM, Ferreira J, Pina C, Mendonça D, Rosa AC, Carmo-Fonseca M, Chromosomal G-dark Bands Determine the Spatial Organization of Centromeric Heterochromatin in the Nucleus, Mol. Biol. Cell 12 (2001) 3563–3572. 10.1091/mbc.12.11.3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Ragoczy T, Telling A, Scalzo D, Kooperberg C, Groudine M, Functional redundancy in the nuclear compartmentalization of the Late-Replicating genome, Nucleus. 5 (2014) 626–635. 10.4161/19491034.2014.990863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].McStay B, Grummt I, The Epigenetics of rRNA Genes: From Molecular to Chromosome Biology, Annu. Rev. Cell Dev. Biol 24 (2008) 131–157. 10.1146/annurev.cellbio.24.110707.175259. [DOI] [PubMed] [Google Scholar]
- [111].V Cerqueira A, Lemos B, Ribosomal DNA and the Nucleolus as Keystones of Nuclear Architecture, Organization, and Function., Trends Genet. 0 (2019). 10.1016/j.tig.2019.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Stahl A, Hartung M, Vagner-Capodano AM, Fouet C, Chromosomal constitution of nucleolus-associated chromatin in man, Hum. Genet 35 (1976) 27–34. 10.1007/BF00295616. [DOI] [PubMed] [Google Scholar]
- [113].Thompson M, Haeusler RA, Good PD, Engelke DR, Nucleolar Clustering of Dispersed tRNA Genes, Science (80-.). 302 (2003) 1399–1401. 10.1126/science.1089814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Zhang LF, Huynh KD, Lee JT, Perinucleolar Targeting of the Inactive X during S Phase: Evidence for a Role in the Maintenance of Silencing, Cell. 129 (2007) 693–706. 10.1016/j.cell.2007.03.036. [DOI] [PubMed] [Google Scholar]
- [115].O’Sullivan JM, Sontam DM, Grierson R, Jones B, Repeated elements coordinate the spatial organization of the yeast genome, Yeast. 26 (2006) 125–138. 10.1002/yea.1657. [DOI] [PubMed] [Google Scholar]
- [116].Chubb JR, Boyle S, Perry P, Bickmore WA, Chromatin motion is constrained by association with nuclear compartments in human cells., Curr. Biol 12 (2002) 439–45. 10.1016/s0960-9822(02)00695-4. [DOI] [PubMed] [Google Scholar]
- [117].Gupta S, Santoro R, Regulation and Roles of the Nucleolus in Embryonic Stem Cells: From Ribosome Biogenesis to Genome Organization, Stem Cell Reports. (2020). 10.1016/j.stemcr.2020.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Dillinger S, Straub T, Nemeth A, Nucleolus association of chromosomal domains is largely maintained in cellular senescence despite massive nuclear reorganisation, PLoS One. 12 (2017) 1–28. 10.1371/journal.pone.0178821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Vertii A, Ou J, Yu J, Yan A, Pagès H, Liu H, Zhu LJ, Kaufman PD, Two contrasting classes of nucleolus-associated domains in mouse fibroblast heterochromatin, Genome Res. (2019) gr.247072.118. 10.1101/gr.247072.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Bizhanova A, Yan A, Yu J, Zhu LJ, Kaufman PD, Distinct features of nucleolus-associated domains in mouse embryonic stem cells, Chromosoma. (2020). 10.1007/s00412-020-00734-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Lu JY, Shao W, Chang L, Yin Y, Li T, Zhang H, Hong Y, Percharde M, Guo L, Wu Z, Liu L, Liu W, Yan P, Ramalho-Santos M, Sun Y, Shen X, Genomic Repeats Categorize Genes with Distinct Functions for Orchestrated Regulation, Cell Rep. 30 (2020) 3296–3311.e5. 10.1016/j.celrep.2020.02.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Sullivan GJ, Bridger JM, Cuthbert AP, Newbold RF, Bickmore WA, McStay B, Human acrocentric chromosomes with transcriptionally silent nucleolar organizer regions associate with nucleoli, EMBO J. 20 (2001) 2867–2877. 10.1093/emboj/20.11.2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Muramatsu M, Smetana K, Busch H, Quantitative Aspects of Isolation of Nucleoli of the Walker Carcinosarcoma and Liver of the Rat, Cancer Res. 23 (1963) 510–518. [PubMed] [Google Scholar]
- [124].Lam Y, Lamond A, Isolation of Nucleoli, in: Cell Biol, Elsevier, 2006: pp. 103–107. 10.1016/b978-012164730-8/50087-3. [DOI] [Google Scholar]
- [125].Busch H, Muramatsu M, Adams H, Steele WJ, Liau MC, Smetana K, Isolation of nucleoli, Exp. Cell Res 9 (1963) 150–163. 10.1016/0014-4827(63)90255-6. [DOI] [PubMed] [Google Scholar]
- [126].Dillinger S, Garea AV, Deutzmann R, Németh A, Analysis of histone posttranslational modifications from nucleolus-associated chromatin by mass spectrometry, Methods Mol. Biol 1094 (2014) 277–293. 10.1007/978-1-62703-706-8_22. [DOI] [PubMed] [Google Scholar]
- [127].Muramatsu M, Onishi T, Isolation and Purification of Nucleoli and Nucleolar Chromatin from Mammalian Cells, Methods Cell Biol. 17 (1978) 141–161. 10.1016/S0091-679X(08)61142-5. [DOI] [PubMed] [Google Scholar]
- [128].Muramatsu M, Onishi T, Isolation and Purification of Nucleoli and Nucleolar Chromatin from Mammalian Cells, Methods Cell Biol. 17 (1978) 141–161. 10.1016/S0091-679X(08)61142-5. [DOI] [PubMed] [Google Scholar]
- [129].Chen B, Gilbert LA, Cimini BA, Schnitzbauer J, Zhang W, Li GW, Park J, Blackburn EH, Weissman JS, Qi LS, Huang B, Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system, Cell. 155 (2013) 1479–1491. 10.1016/j.cell.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Ma H, Tu LC, Naseri A, Huisman M, Zhang S, Grunwald D, Pederson T, Multiplexed labeling of genomic loci with dCas9 and engineered sgRNAs using CRISPRainbow, Nat. Biotechnol 34 (2016) 528–530. 10.1038/nbt.3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Ma H, Tu LC, Naseri A, Chung YC, Grunwald D, Zhang S, Pederson T, CRISPR-Sirius: RNA scaffolds for signal amplification in genome imaging, Nat. Methods 15 (2018) 928–931. 10.1038/s41592-018-0174-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Ma H, Naseri A, Reyes-Gutierrez P, Wolfe SA, Zhang S, Pederson T, Multicolor CRISPR labeling of chromosomal loci in human cells, Proc. Natl. Acad. Sci. U. S. A 112 (2015) 3002–3007. 10.1073/pnas.1420024112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Zhou Y, Wang P, Tian F, Gao G, Huang L, Wei W, Xie S, Painting a specific chromosome with CRISPR/Cas9 for live-cell imaging, Cell Res. 27 (2017) 298–301. 10.1038/cr.2017.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Beliveau BJ, Joyce EF, Apostolopoulos N, Yilmaz F, Fonseka CY, McCole RB, Chang Y, Li JB, Senaratne TN, Williams BR, Rouillard JM, Wu CT, Versatile design and synthesis platform for visualizing genomes with Oligopaint FISH probes, Proc. Natl. Acad. Sci. U. S. A 109 (2012) 21301–21306. 10.1073/pnas.1213818110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Beliveau BJ, Boettiger AN, Avendaño MS, Jungmann R, McCole RB, Joyce EF, Kim-Kiselak C, Bantignies F, Fonseka CY, Erceg J, Hannan MA, Hoang HG, Colognori D, Lee JT, Shih WM, Yin P, Zhuang X, Wu CT, Single-molecule super-resolution imaging of chromosomes and in situ haplotype visualization using Oligopaint FISH probes, Nat. Commun 6 (2015) 1–13. 10.1038/ncomms8147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Lakadamyali M, Cosma MP, Visualizing the genome in high resolution challenges our textbook understanding, Nat. Methods 17 (2020) 371–379. 10.1038/s41592-020-0758-3. [DOI] [PubMed] [Google Scholar]
- [137].Knight SC, Tjian R, Doudna JA, Genomes in Focus: Development and Applications of CRISPR-Cas9 Imaging Technologies, Angew. Chemie - Int. Ed 57 (2018) 4329–4337. 10.1002/anie.201709201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Eykelenboom JK, Tanaka TU, Zooming in on chromosome dynamics, Cell Cycle. 19 (2020) 1422–1432. 10.1080/15384101.2020.1757242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Shaban HA, Seeber A, Monitoring the spatio-temporal organization and dynamics of the genome, Nucleic Acids Res. 48 (2020) 3423–3434. 10.1093/nar/gkaa135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Filion GJ, van Steensel B, Reassessing the abundance of H3K9me2 chromatin domains in embryonic stem cells, Nat. Genet 42 (2010) 4–4. 10.1038/ng0110-4. [DOI] [PubMed] [Google Scholar]
- [141].Pontvianne F, Carpentier MC, Durut N, Pavlištová V, Jaške K, Schořová Š, Parrinello H, Rohmer M, Pikaard CS, Fojtová M, Fajkus J, Sáez-Vásquez J, Identification of Nucleolus-Associated Chromatin Domains Reveals a Role for the Nucleolus in 3D Organization of the A. thaliana Genome, Cell Rep. 16 (2016) 1574–1587. 10.1016/j.celrep.2016.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Boisvert F-M, van Koningsbruggen S, Navascués J, Lamond AI, The multifunctional nucleolus, Nat. Rev. Mol. Cell Biol 8 (2007) 574–585. 10.1038/nrm2184. [DOI] [PubMed] [Google Scholar]
- [143].Quinodoz SA, Ollikainen N, Tabak B, Palla A, Schmidt JM, Detmar E, Lai MM, Shishkin AA, Bhat P, Takei Y, Trinh V, Aznauryan E, Russell P, Cheng C, Jovanovic M, Chow A, Cai L, McDonel P, Garber M, Guttman M, Higher-Order Inter-chromosomal Hubs Shape 3D Genome Organization in the Nucleus, Cell. 174 (2018) 744–757.e24. 10.1016/j.cell.2018.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Quinodoz SA, Bhat P, Ollikainen N, Jachowicz JW, Banerjee AK, Chovanec P, Blanco MR, Chow A, Markaki Y, Plath K, Guttman M, RNA promotes the formation of spatial compartments in the nucleus, BioRxiv. (2020) 2020.08.25.267435. 10.1101/2020.08.25.267435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Yu S, Lemos B, A portrait of ribosomal DNA contacts with Hi-C reveals 5S and 45S rDNA anchoring points in the folded human genome, Genome Biol. Evol 8 (2016) 3545–3558. 10.1093/gbe/evw257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Yu S, Lemos B, The long-range interaction map of ribosomal DNA arrays, PLOS Genet. 14 (2018) e1007258 10.1371/journal.pgen.1007258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Zhang L, Zhang Y, Chen Y, Gholamalamdari O, Wang Y, Ma J, Belmont A, TSA-Seq reveals a largely “hardwired” genome organization relative to nuclear speckles with small position changes tightly correlated with gene expression changes, BioRxiv. (2019) 824433 10.1101/824433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Chen Y, Zhang Y, Wang Y, Zhang L, Brinkman EK, Adam SA, Goldman R, Van Steensel B, Ma J, Belmont AS, Mapping 3D genome organization relative to nuclear compartments using TSA-Seq as a cytological ruler, J. Cell Biol 217 (2018) 4025–4048. 10.1083/jcb.201807108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Wang Y, Zhang Y, Zhang R, van Schaik T, Zhang L, Sasaki T, Hupkes DP, Chen Y, Gilbert D, van Steensel B, Belmont A, Ma J, SPIN reveals genome-wide landscape of nuclear compartmentalization, BioRxiv. (2020) 2020.03.09.982967. 10.1101/2020.03.09.982967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Bemiller PM, Lee LH, Nucleolar changes in senescing WI-38 cells, Mech. Ageing Dev 8 (1978) 417–427. 10.1016/0047-6374(78)90041-6. [DOI] [PubMed] [Google Scholar]
- [151].Kobayashi T, A new role of the rDNA and nucleolus in the nucleus - RDNA instability maintains genome integrity, BioEssays. 30 (2008) 267–272. 10.1002/bies.20723. [DOI] [PubMed] [Google Scholar]
- [152].Tiku V, Antebi A, Nucleolar Function in Lifespan Regulation, Trends Cell Biol. 28 (2018) 662–672. 10.1016/j.tcb.2018.03.007. [DOI] [PubMed] [Google Scholar]
- [153].Oberdoerffer P, Sinclair DA, The role of nuclear architecture in genomic instability and ageing, Nat. Rev. Mol. Cell Biol 8 (2007) 692–702. 10.1038/nrm2238. [DOI] [PubMed] [Google Scholar]
- [154].Ganley ARD, Kobayashi T, Ribosomal DNA and cellular senescence: New evidence supporting the connection between rDNA and aging, FEMS Yeast Res. 14 (2014) 49–59. 10.1111/1567-1364.12133. [DOI] [PubMed] [Google Scholar]
- [155].Bury L, Moodie B, McKay LS, Miga KH, Cheeseman IM, Alpha-satellite RNA transcripts are repressed by centromere-nucleolus associations, BioRxiv. (2020) 2020.04.14.040766. 10.1101/2020.04.14.040766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Silver LM, Karyotypes, Chromosomes, and Translocations, in: Mouse Genet. Concepts Appl, Oxford: Oxford University Press, 1995: pp. 83–92. [Google Scholar]
- [157].Hiratani I, Ryba T, Itoh M, Rathjen J, Kulik M, Papp B, Fussner E, Bazett-Jones DP, Plath K, Dalton S, Rathjen PD, Gilbert DM, Genome-wide dynamics of replication timing revealed by in vitro models of mouse embryogenesis., Genome Res. 20 (2010) 155–69. 10.1101/gr.099796.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Meshorer E, Yellajoshula D, George E, Scambler PJ, Brown DT, Misteli T, Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells, Dev. Cell (2006). 10.1016/j.devcel.2005.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Ahmed K, Dehghani H, Rugg-Gunn P, Fussner E, Rossant J, Bazett-Jones DP, Global Chromatin Architecture Reflects Pluripotency and Lineage Commitment in the Early Mouse Embryo, PLoS One. 5 (2010) e10531 10.1371/journal.pone.0010531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Gaspar-Maia A, Alajem A, Meshorer E, Ramalho-Santos M, Open chromatin in pluripotency and reprogramming, Nat. Rev. Mol. Cell Biol 12 (2011) 36–47. 10.1038/nrm3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Cruz-Molina S, Respuela P, Tebartz C, Kolovos P, Nikolic M, Fueyo R, van Ijcken WFJ, Grosveld F, Frommolt P, Bazzi H, Rada-Iglesias A, PRC2 Facilitates the Regulatory Topology Required for Poised Enhancer Function during Pluripotent Stem Cell Differentiation, Cell Stem Cell. 20 (2017) 689–705.e9. 10.1016/j.stem.2017.02.004. [DOI] [PubMed] [Google Scholar]
- [162].Delbarre E, Ivanauskiene K, Spirkoski J, Shah A, Vekterud K, Moskaug JØ, Bøe SO, Wong LH, Küntziger T, Collas P, PML protein organizes heterochromatin domains where it regulates histone H3.3 deposition by ATRX/DAXX, Genome Res. 27 (2017) 913–921. 10.1101/gr.215830.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Jachowicz JW, Santenard A, Bender A, Muller J, Torres MEP, Heterochromatin establishment at pericentromeres depends on nuclear position, Genes Dev. 27 (2013) 2427–2432. 10.1101/gad.224550.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Mohammad F, Pandey RR, Nagano T, Chakalova L, Mondal T, Fraser P, Kanduri C, Kcnq1ot1/Lit1 Noncoding RNA Mediates Transcriptional Silencing by Targeting to the Perinucleolar Region, Mol. Cell. Biol 28 (2008) 3713–3728. 10.1128/MCB.02263-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Pandey RR, Mondal T, Mohammad F, Enroth S, Redrup L, Komorowski J, Nagano T, Mancini-DiNardo D, Kanduri C, Kcnq1ot1 Antisense Noncoding RNA Mediates Lineage-Specific Transcriptional Silencing through Chromatin-Level Regulation, Mol. Cell 32 (2008) 232–246. 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- [166].Fedoriw AM, Starmer J, Yee D, Magnuson T, Nucleolar association and transcriptional inhibition through 5S rDNA in mammals, PLoS Genet. 8 (2012). 10.1371/journal.pgen.1002468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Yang F, Deng X, Ma W, Berletch JB, Rabaia N, Wei G, Moore JM, Filippova GN, Xu J, Liu Y, Noble WS, Shendure J, Disteche CM, The lncRNA Firre anchors the inactive X chromosome to the nucleolus by binding CTCF and maintains H3K27me3 methylation, Genome Biol. 16 (2015) 52 10.1186/s13059-015-0618-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Singh I, Contreras A, Cordero J, Rubio K, Dobersch S, Günther S, Jeratsch S, Mehta A, Krüger M, Graumann J, Seeger W, Dobreva G, Braun T, Barreto G, MiCEE is a ncRNA-protein complex that mediates epigenetic silencing and nucleolar organization, Nat. Genet 50 (2018) 990–1001. 10.1038/s41588-018-0139-3. [DOI] [PubMed] [Google Scholar]
- [169].Belaghzal H, Borrman T, Stephens AD, Lafontaine DL, Venev SV, Weng Z, Marko JF, Dekker J, Compartment-dependent chromatin interaction dynamics revealed by liquid chromatin Hi-C, BioRxiv. (2019) 704957 10.1101/704957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Constam DB, Robertson EJ, SPC4/PACE4 regulates a TGFβ signaling network during axis formation, Genes Dev. 14 (2000) 1146–1155. 10.1101/gad.14.9.1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Fulka H, Aoki F, Nucleolus Precursor Bodies and Ribosome Biogenesis in Early Mammalian Embryos: Old Theories and New Discoveries1, Biol. Reprod 94 (2016). 10.1095/biolreprod.115.136093. [DOI] [PubMed] [Google Scholar]
- [172].Martin C, Beaujean N, Brochard V, Audouard C, Zink D, Debey P, Genome restructuring in mouse embryos during reprogramming and early development, Dev. Biol 292 (2006) 317–332. 10.1016/j.ydbio.2006.01.009. [DOI] [PubMed] [Google Scholar]
- [173].Probst AV, Santos F, Reik W, Almouzni G, Dean W, Structural differences in centromeric heterochromatin are spatially reconciled on fertilisation in the mouse zygote, Chromosoma. 116 (2007) 403–415. 10.1007/s00412-007-0106-8. [DOI] [PubMed] [Google Scholar]
- [174].Tiku V, Jain C, Raz Y, Nakamura S, Heestand B, Liu W, Späth M, Suchiman HED, Müller RU, Slagboom PE, Partridge L, Antebi A, Small nucleoli are a cellular hallmark of longevity, Nat. Commun 8 (2017) 16083 10.1038/ncomms16083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Buchwalter A, Hetzer MW, Nucleolar expansion and elevated protein translation in premature aging, Nat. Commun 8 (2017). 10.1038/s41467-017-00322-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Eriksson M, Brown WT, Gordon LB, Glynn MW, Singer J, Scott L, Erdos MR, Robbins CM, Moses TY, Berglund P, Dutra A, Pak E, Durkin S, Csoka AB, Boehnke M, Glover TW, Collins FS, Recurrent de novo point mutations in lamin A cause Hutchinson-Gilford progeria syndrome, Nature. 423 (2003) 293–298. 10.1038/nature01629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Wang M, Lemos B, Ribosomal DNA harbors an evolutionarily conserved clock of biological aging, Genome Res. 29 (2019) 325–333. 10.1101/gr.241745.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Diesch J, Bywater MJ, Sanij E, Cameron DP, Schierding W, Brajanovski N, Son J, Sornkom J, Hein N, Evers M, Pearson RB, McArthur GA, Ganley ARD, O’Sullivan JM, Hannan RD, Poortinga G, Changes in long-range rDNA-genomic interactions associate with altered RNA polymerase II gene programs during malignant transformation, Commun. Biol 2 (2019) 1–14. 10.1038/s42003-019-0284-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Sanij E, Hannan RD, The role of UBF in regulating the structure and dynamics of transcriptionally active rDNA chromatin, Epigenetics. 4 (2009) 374–382. 10.4161/epi.4.6.9449. [DOI] [PubMed] [Google Scholar]
- [180].Conconi A, Widmer RM, Koller T, Sogo JM, Two different chromatin structures coexist in ribosomal RNA genes throughout the cell cycle, Cell. 57 (1989) 753–761. 10.1016/0092-8674(89)90790-3. [DOI] [PubMed] [Google Scholar]
- [181].Zhao Z, Tavoosidana G, Sjölinder M, Göndör A, Mariano P, Wang S, Kanduri C, Lezcano M, Sandhu KS, Singh U, Pant V, Tiwari V, Kurukuti S, Ohlsson R, Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions, Nat. Genet 38 (2006) 1341–1347. 10.1038/ng1891. [DOI] [PubMed] [Google Scholar]
- [182].Simonis M, Klous P, Splinter E, Moshkin Y, Willemsen R, De Wit E, Van Steensel B, De Laat W, Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C), Nat. Genet 38 (2006) 1348–1354. 10.1038/ng1896. [DOI] [PubMed] [Google Scholar]
- [183].Mullighan CG, Goorha S, Radtke I, Miller CB, Coustan-Smith E, Dalton JD, Girtman K, Mathew S, Ma J, Pounds SB, Su X, Pui CH, Relling MV, Evans WE, Shurtleff SA, Downing JR, Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia, Nature. 446 (2007) 758–764. 10.1038/nature05690. [DOI] [PubMed] [Google Scholar]
- [184].Grandori C, Gomez-Roman N, Felton-Edkins ZA, Ngouenet C, Galloway DA, Eisenman RN, White RJ, c-Myc binds to human ribosomal DNA and stimulates transcription of rRNA genes by RNA polymerase I, Nat. Cell Biol 7 (2005) 311–318. 10.1038/ncb1224. [DOI] [PubMed] [Google Scholar]
- [185].Dang CV, MYC on the path to cancer, Cell. 149 (2012) 22–35. 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [186].West AG, Gaszner M, Felsenfeld G, Insulators: many functions, many mechanisms, Genes Dev. 16 (2002) 271–288. 10.1101/gad.954702. [DOI] [PubMed] [Google Scholar]
- [187].Yusufzai TM, Tagami H, Nakatani Y, Felsenfeld G, CTCF Tethers an Insulator to Subnuclear Sites, Suggesting Shared Insulator Mechanisms across Species, Mol. Cell 13 (2004) 291–298. 10.1016/S1097-2765(04)00029-2. [DOI] [PubMed] [Google Scholar]
- [188].Box JK, Paquet N, Adams MN, Boucher D, Bolderson E, O’Byrne KJ, Richard DJ, Nucleophosmin: from structure and function to disease development, BMC Mol. Biol 17 (2016). 10.1186/S12867-016-0073-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [189].Padeken J, Mendiburo MJ, Chlamydas S, Schwarz H-J, Kremmer E, Heun P, The Nucleoplasmin Homolog NLP Mediates Centromere Clustering and Anchoring to the Nucleolus, Mol. Cell 50 (2013) 236–249. 10.1016/j.molcel.2013.03.002. [DOI] [PubMed] [Google Scholar]
- [190].Holmberg Olausson K, Nistér M, Lindström MS, Loss of Nucleolar Histone Chaperone NPM1 Triggers Rearrangement of Heterochromatin and Synergizes with a Deficiency in DNA Methyltransferase DNMT3A to Drive Ribosomal DNA Transcription, J. Biol. Chem 289 (2014) 34601–34619. 10.1074/jbc.M114.569244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [191].Kaufman PD, Kobayashi R, Kessler N, Stillman B, The p150 and p60 subunits of chromatin assembly factor I: a molecular link between newly synthesized histones and DNA replication., Cell. 81 (1995) 1105–14. 10.1016/s0092-8674(05)80015-7. [DOI] [PubMed] [Google Scholar]
- [192].Smith CL, Matheson TD, Trombly DJ, Sun X, Campeau E, Han X, Yates JR, Kaufman PD, A separable domain of the p150 subunit of human chromatin assembly factor-1 promotes protein and chromosome associations with nucleoli, Mol. Biol. Cell 25 (2014) 2866–2881. 10.1091/mbc.e14-05-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [193].Matheson TD, Kaufman PD, The p150N domain of chromatin assembly factor-1 regulates Ki-67 accumulation on the mitotic perichromosomal layer, Mol. Biol. Cell 28 (2017) 21–29. 10.1091/mbc.e16-09-0659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [194].Sobecki M, Mrouj K, Camasses A, Parisis N, Nicolas E, Llères D, Gerbe F, Prieto S, Krasinska L, David A, Eguren M, Birling M-C, Urbach S, Hem S, Déjardin J, Malumbres M, Jay P, Dulic V, Lafontaine DL, Feil R, Fisher D, The cell proliferation antigen Ki-67 organises heterochromatin, Elife. 5 (2016) e13722 10.7554/eLife.13722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [195].Booth DG, Takagi M, Sanchez-Pulido L, Petfalski E, Vargiu G, Samejima K, Imamoto N, Ponting CP, Tollervey D, Earnshaw WC, Vagnarelli P, Ki-67 is a PP1-interacting protein that organises the mitotic chromosome periphery, Elife. 2014 (2014). 10.7554/eLife.01641.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [196].Matheson TD, Kaufman PD, Grabbing the genome by the NADs, Chromosoma. 125 (2016) 361–371. 10.1007/s00412-015-0527-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [197].Booth DG, Earnshaw WC, Ki-67 and the Chromosome Periphery Compartment in Mitosis, Trends Cell Biol. 27 (2017) 906–916. 10.1016/j.tcb.2017.08.001. [DOI] [PubMed] [Google Scholar]
- [198].Cuylen S, Blaukopf C, Politi AZ, Muller-Reichert T, Neumann B, Poser I, Ellenberg J, Hyman AA, Gerlich DW, Ki-67 acts as a biological surfactant to disperse mitotic chromosomes, Nature. 535 (2016) 308–312. 10.1038/nature18610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [199].Takagi M, Natsume T, Kanemaki MT, Imamoto N, Perichromosomal protein Ki67 supports mitotic chromosome architecture, Genes to Cells. 21 (2016) 1113–1124. 10.1111/gtc.12420. [DOI] [PubMed] [Google Scholar]
- [200].Sun X, Bizhanova A, Matheson TD, Yu J, Zhu LJ, Kaufman PD, Ki-67 Contributes to Normal Cell Cycle Progression and Inactive X Heterochromatin in p21 Checkpoint-Proficient Human Cells, Mol. Cell. Biol 37 (2017). 10.1128/mcb.00569-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [201].Strohner R, Nemeth A, Jansa P, Hofmann-Rohrer U, Santoro R, Längst G, Grummt I, NoRC - A novel member of mammalian ISWI-containing chromatin remodeling machines, EMBO J. 20 (2001) 4892–4900. 10.1093/emboj/20.17.4892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [202].Santoro R, Li J, Grummt I, The nucleolar remodeling complex NoRC mediates heterochromatin formation and silencing of ribosomal gene transcription, Nat. Genet 32 (2002) 393–396. 10.1038/ng1010. [DOI] [PubMed] [Google Scholar]
- [203].Zhou Y, Santoro R, Grummt I, The chromatin remodeling complex NoRC targets HDAC1 to the ribosomal gene promoter and represses RNA polymerase I transcription, EMBO J. 21 (2002) 4632–4640. 10.1093/emboj/cdf460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [204].Mayer C, Schmitz K-M, Li J, Grummt I, Santoro R, Intergenic Transcripts Regulate the Epigenetic State of rRNA Genes, Mol. Cell 22 (2006) 351–361. 10.1016/j.molcel.2006.03.028. [DOI] [PubMed] [Google Scholar]
- [205].Mayer C, Neubert M, Grummt I, The structure of NoRC-associated RNA is crucial for targeting the chromatin remodelling complex NoRC to the nucleolus, EMBO Rep. 9 (2008) 774–780. 10.1038/embor.2008.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [206].Guetg C, Lienemann P, Sirri V, Grummt I, Hernandez-Verdun D, Hottiger MO, Fussenegger M, Santoro R, The NoRC complex mediates the heterochromatin formation and stability of silent rRNA genes and centromeric repeats, EMBO J. 29 (2010) 2135–2146. 10.1038/emboj.2010.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [207].Vissel B, Choo KH, Mouse major (gamma) satellite DNA is highly conserved and organized into extremely long tandem arrays: implications for recombination between nonhomologous chromosomes., Genomics. 5 (1989) 407–14. 10.1016/0888-7543(89)90003-7. [DOI] [PubMed] [Google Scholar]
- [208].Kipling D, Ackford HE, Taylor BA, Cooke HJ, Mouse minor satellite DNA genetically maps to the centromere and is physically linked to the proximal telomere., Genomics. 11 (1991) 235–41. 10.1016/0888-7543(91)90128-2. [DOI] [PubMed] [Google Scholar]
- [209].Savić N, Bär D, Leone S, Frommel SC, Weber FA, Vollenweider E, Ferrari E, Ziegler U, Kaech A, Shakhova O, Cinelli P, Santoro R, lncRNA Maturation to Initiate Heterochromatin Formation in the Nucleolus Is Required for Exit from Pluripotency in ESCs, Cell Stem Cell. 15 (2014) 720–734. 10.1016/j.stem.2014.10.005. [DOI] [PubMed] [Google Scholar]
- [210].Németh A, Längst G, Genome organization in and around the nucleolus, Trends Genet. 27 (2011) 149–156. 10.1016/j.tig.2011.01.002. [DOI] [PubMed] [Google Scholar]
- [211].Wong LH, Brettingham-Moore KH, Chan L, Quach JM, Anderson MA, Northrop EL, Hannan R, Saffery R, Shaw ML, Williams E, Choo KHA, Centromere RNA is a key component for the assembly of nucleoproteins at the nucleolus and centromere, Genome Res. 17 (2007) 1146–1160. 10.1101/gr.6022807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [212].Percharde M, Lin CJ, Yin Y, Guan J, Peixoto GA, Bulut-Karslioglu A, Biechele S, Huang B, Shen X, Ramalho-Santos M, A LINE1-Nucleolin Partnership Regulates Early Development and ESC Identity, Cell. 174 (2018) 391–405.e19. 10.1016/j.cell.2018.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [213].Brown CJ, Ballabio A, Rupert JL, Lafreniere RG, Grompe M, Tonlorenzi R, Willard HF, A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome, Nature. 349 (1991) 38–44. 10.1038/349038a0. [DOI] [PubMed] [Google Scholar]
- [214].Brockdorff N, Ashworth A, Kay GF, Cooper P, Smith S, McCabe VM, Norris DP, Penny GD, Patel D, Rastan S, Conservation of position and exclusive expression of mouse Xist from the inactive X chromosome, Nature. 351 (1991) 329–331. 10.1038/351329a0. [DOI] [PubMed] [Google Scholar]
- [215].Clemson CM, McNeil JA, Willard HF, Lawrence JB, XIST RNA paints the inactive X chromosome at interphase: evidence for a novel RNA involved in nuclear/chromosome structure, J. Cell Biol 132 (1996) 259–275. 10.1083/jcb.132.3.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [216].Collins N, Poot RA, Kukimoto I, García-Jiménez C, Dellaire G, Varga-Weisz PD, An ACF1–ISWI chromatin-remodeling complex is required for DNA replication through heterochromatin, Nat. Genet 32 (2002) 627–632. 10.1038/ng1046. [DOI] [PubMed] [Google Scholar]
- [217].Yang F, Babak T, Shendure J, Disteche CM, Global survey of escape from X inactivation by RNA-sequencing in mouse, Genome Res. 20 (2010) 614–622. 10.1101/gr.103200.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [218].Csankovszki G, Nagy A, Jaenisch R, Synergism of Xist RNA, DNA methylation, and histone hypoacetylation in maintaining X chromosome inactivation, J. Cell Biol 153 (2001) 773–783. 10.1083/jcb.153.4.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [219].Bonora G, Deng X, Fang H, Ramani V, Qiu R, Berletch JB, Filippova GN, Duan Z, Shendure J, Noble WS, Disteche CM, Orientation-dependent Dxz4 contacts shape the 3D structure of the inactive X chromosome, Nat. Commun 9 (2018) 1445 10.1038/s41467-018-03694-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [220].Fitzpatrick GV, Soloway PD, Higgins MJ, Regional loss of imprinting and growth deficiency in mice with a targeted deletion of KvDMR1, Nat. Genet 32 (2002) 426–431. 10.1038/ng988. [DOI] [PubMed] [Google Scholar]
- [221].Thakur N, Tiwari VK, Thomassin H, Pandey RR, Kanduri M, Gondor A, Grange T, Ohlsson R, Kanduri C, An Antisense RNA Regulates the Bidirectional Silencing Property of the Kcnq1 Imprinting Control Region, Mol. Cell. Biol 24 (2004) 7855–7862. 10.1128/MCB.24.18.7855-7862.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [222].Mancini-DiNardo D, Steele SJS, Levorse JM, Ingram RS, Tilghman SM, Elongation of the Kcnq1ot1 transcript is required for genomic imprinting of neighboring genes, Genes Dev. 20 (2006) 1268–1282. 10.1101/gad.1416906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [223].Fedoriw AM, Calabrese JM, Mu W, Yee D, Magnuson T, Differentiation-driven nucleolar association of the mouse imprinted Kcnq1 locus., G3 (Bethesda). 2 (2012) 1521–8. 10.1534/g3.112.004226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [224].Brangwynne CP, Mitchison TJ, Hyman AA, Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes, Proc. Natl. Acad. Sci 108 (2011) 4334–4339. 10.1073/pnas.1017150108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [225].Feric M, Vaidya N, Harmon TS, Mitrea DM, Zhu L, Richardson TM, Kriwacki RW, Pappu RV, Brangwynne CP, Coexisting Liquid Phases Underlie Nucleolar Subcompartments, Cell. 165 (2016) 1686–1697. 10.1016/j.cell.2016.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [226].Mitrea DM, Cika JA, Guy CS, Ban D, Banerjee PR, Stanley CB, Nourse A, Deniz AA, Kriwacki RW, Nucleophosmin integrates within the nucleolus via multi-modal interactions with proteins displaying R-rich linear motifs and rRNA, Elife. 5 (2016). 10.7554/eLife.13571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [227].Falahati H, Wieschaus E, Independent active and thermodynamic processes govern the nucleolus assembly in vivo., Proc. Natl. Acad. Sci. U. S. A 114 (2017) 1335–1340. 10.1073/pnas.1615395114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [228].Caudron-Herger M, Pankert T, Seiler J, Nemeth A, Voit R, Grummt I, Rippe K, Alu element-containing RNAs maintain nucleolar structure and function, EMBO J. 34 (2015) 2758–2774. 10.15252/embj.201591458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [229].Kind J, van Steensel B, Stochastic genome-nuclear lamina interactions, Nucleus. 5 (2014) 124–130. 10.4161/nucl.28825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [230].Sen Gupta A, Sengupta K, Lamin B2 Modulates Nucleolar Morphology, Dynamics, and Function, Mol. Cell. Biol 37 (2017). 10.1128/mcb.00274-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [231].Solovei I, Schermelleh L, Düring K, Engelhardt A, Stein S, Cremer C, Cremer T, Differences in centromere positioning of cycling and postmitotic human cell types, Chromosoma. 112 (2004) 410–423. 10.1007/s00412-004-0287-3. [DOI] [PubMed] [Google Scholar]


