Abstract
Aberrant expression of certain genes and microRNAs (miRNAs) has been shown to drive cancer development and progression, thus the modification of aberrant gene and miRNA expression presents an opportunity for therapeutic targeting. Ectopic modulation of a single dysregulated miRNA has the potential to revert therapeutically unfavorable gene expression in cancer cells by targeting multiple genes simultaneously. Although the use of noncoding RNA-based cancer therapy is a promising approach, the lack of a feasible delivery platform for small noncoding RNAs has hindered the development of this therapeutic modality. Recently, however, there has been an evolution in RNA nanotechnology, in which small noncoding RNA is loaded onto nanoparticles derived from the pRNA-3WJ viral RNA motif of the bacteriophage phi29. Preclinical studies have shown the capacity of this technology to specifically target tumor cells by conjugating these nanoparticles with ligands specific for cancer cells and resulting in the endocytic delivery of siRNA and miRNA inhibitors directly into the cell. Here we provide a systematic review of the various strategies, which have been utilized for miRNA delivery with a specific focus on the preclinical evaluation of promising RNA nanoparticles for glioblastoma (GBM) targeted therapy.
Keywords: RNA nanotechnology, RNA nanoparticle, microRNA, siRNA, glioblastoma
1. Introduction
Although great strides have been made in cancer therapy, cancer is still a leading cause of death in modern world. Over the past century, a major focus of cancer research has been limited to understanding genetic and epigenetic alterations of protein coding genes. However, decades of research on noncoding RNA (ncRNA) has clearly shown that the sole analysis of molecular changes in protein coding genes is not sufficient to understand the whole disease. Indeed, cumulative data clearly indicates that ncRNAs play a critical role in cancer, not merely as diagnostic or prognostic markers but also as therapeutically druggable targets. NcRNAs are endogenously transcribed functional RNA moieties which do not undergo protein translation, as they lack an open reading frame (ORF) [1]. In general, ncRNAs are categorized into two groups depending on size: long noncoding RNAs (lncRNAs) with over 200 nucleotides and short noncoding RNAs (sncRNAs) which are shorter than 200 nucleotides.
MicroRNAs (miRNAs), the smallest sncRNAs, are approximately 22 nucleotides long and have been characterized as some of the key regulators in the translation of messenger RNAs (mRNAs) into proteins [2]. MiRNAs were first discovered in Caenorhabditis elegans (C. elegans) [3]. Victor Ambros and his colleagues found that a 22 nucleotide-long piece of RNA derived from abnormal cell LINeage gene (Lin-4) inhibited the translation of the protein coding gene Lin-14 through its direct interaction with the 3’ untranslated region (3’ UTR) of the Lin-14 mRNA [3, 4]. A similar small RNA regulator, Let-7, was also found in C. elegans [5], prompting further research towards the identification of miRNAs in other organisms, including humans. It was discovered that miRNAs are regulators of gene expression in both normal cells as well as diseased. In human cancers, the first pathological evidence that miRNAs are directly involved in cancer development and progression was reported in 2002 by Carlo Croce’s laboratory [6]. In a landmark study, authors identified a frequently deleted miR-15/16 cluster of genes in chronic lymphocytic leukemia (CLL) patient tissues. It was discovered that miR-15/16 is a tumor suppressor which is deleted in CLL, and normally functions to directly inhibit the expression of the oncogene B-cell lymphoma gene 2 (BCL2) [6]. Since this seminal finding, numerous additional studies have revealed that miRNAs take on various roles in many aspects of cancer initiation, progression, and metastasis [7], such as cell proliferation and apoptosis [8], differentiation [9], cancer cell metabolism [10] and cell cycle [11]. The aberrant dysregulation of miRNAs in cancer cells results in a unique expression profile and led to the identification of “signature miRNAs”, which have been proposed to serve as diagnostic or prognostic markers. Many of these signature miRNAs have also been explored as potential tools in the development of cancer therapies. However, a lack of proper strategies to ensure efficient targeting and delivery of miRNAs into cancer cells has hampered the therapeutic potential of miRNAs in the clinical setting. Recent advances in RNA nanotechnology have enabled new strategies for targeted delivery of therapeutically useful sncRNAs. This review will describe the current progress of RNA nanotechnology-based targeted delivery of sncRNAs for glioblastoma (GBM), highlighting an important translational implication for future clinical trials.
2. MiRNAs in human cancers
Global transcriptome analysis has predicted that 70% of miRNAs are transcribed from protein coding host genes, while the other 30% of miRNAs are found in intergenic areas [2, 12, 13]. The long primary transcript of miRNA (pri-miR) is transcribed by RNA polymerase II [14], and then processed into a shorter hairpin-shaped precursor miRNA (pre-miR) by the class 2 nuclear RNase III Drosha complexed with DGCR8 [15]. The pre-miR translocates through the nuclear membrane channel protein Exportin-5 (EXPO5) into the cytoplasm [16], where it is further cleaved by the cytoplasmic RNase III Dicer to produce a 22 nucleotide-long mature miRNA [17]. The double-stranded mature form of miRNA binds to Argonaute 2 (Ago2) along with the transactivation-responsive RNA-binding protein (TRBP) to form the RNA-induced silencing complex (RISC), in which the active miRNA strand remains accessible to bind the 3’UTR of target mRNAs [13]. Unlike small interfering RNAs (siRNAs), which require perfectly complementary sequences against their target sequences, miRNAs can bind to their target mRNAs through partial complementarity between its “seed sequences”, which span from the second to the eighth nucleotide, and the 3’ UTR of its targeted mRNA. The association of miRNA with the 3’UTR of mRNA physically inhibits the interaction between mRNA with ribosomal complexes, abolishing successful protein synthesis [2, 12], and eventually, miRNA-bound mRNAs are deadenylated and degraded in the cytoplasm [18, 19].
As noted earlier, aberrant dysregulation of miRNA in human cancers has been widely identified through global gene expression profiling, such as hybridization-based microarrays and RNA sequencing. As such, cancer-specific miRNA signatures have been proposed as useful biomarkers for diagnostic and prognostic purposes. Importantly, preclinical studies have shown that restoration of tumor suppressive miRNAs may be an attractive therapeutic option for many types of cancers. For example, restoration of miR-15a and miR-16 downregulates BCL2 expression and results in an induction of apoptosis in CLL cells [6, 20, 21]. In lung cancer, miR-29 is frequently down-regulated, leading to an up-regulation of DNA methyltransferase (DNMT)-3A and 3B, and a worse prognosis for patients [22]. Another extensively studied tumor suppressive miRNA, miR-34a, has been found to be significantly down-regulated in many types of cancers due to a CpG island methylation of its host gene [23]. However, expression of miR-34a can be induced by the tumor suppressor p53 [24], and its restoration in cancer cells can result in cell cycle arrest or cancer cell apoptosis through its direct targeting of proteins specific to each function: c-MET, c-Myc, CCND1, and CDK6 [25–27], or Notch1 and Bcl2 [28, 29], respectively. Interestingly, survivin, another direct target of miR-34a, was also found to be an inhibitor of p53, thus forming a positive feedback loop [30].
In contrast to tumor suppressive miRNAs, many miRNAs are significantly overexpressed in human cancers. Often, up-regulated miRNAs have oncogenic roles and are thus referred to as onco-miRs. The first discovered onco-miR is miR-155 and it was found as a noncoding transcript of B cell integration cluster (BIC) in human B-cell lymphomas [31]. Later, miR-155 was found to be an onco-miR in many other types of cancers as well including pediatric Burkitt’s lymphoma, Hodgkin’s disease, primary mediastinal non-Hodgkin’s lymphoma, CLL, AML, lung cancer, and breast cancer [32]. In a transgenic mouse model, specific overexpression of miR-155 in B cells resulted in polyclonal pre-leukemic B cell proliferation and ultimately generated a high level of B cell malignancy [33]. Later, Src homology 2 domain-containing inositol-5-phosphatase (SHIP) and CCAAT enhancer-binding protein beta (C/EBPbeta) were identified as the direct targets of miR-155 [34]. To date, the most frequently up-regulated miRNA in both hematopoietic and solid tumors is miR-21 [35]. Its validated targets include many tumor suppressor genes, such as Phosphatase and tensin homolog (PTEN) [36], Programmed cell death protein 4 [37], and B-cell translocation gene 2 (BTG2) [38]. Suppression of onco-miRs has been shown to be a promising therapeutic strategy for cancer treatment: down-regulation of these onco-miRs results in an induction of apoptosis, cell cycle arrest, and inhibition of cancer cell invasion and metastasis sufficient to prevent tumor initiation and progression.
In cancer cell populations, some miRNAs have been shown to be involved in the formation and maintenance of cancer stem cells (CSCs). CSCs have been proposed to initiate tumor formation and induce cancer recurrence, and therefore CSC-specific miRNAs may also become attractive therapeutic targets as well as diagnostic or prognostic markers. For example, miR-200b was found to be a CSC-specific miRNA that is down-regulated in breast cancers [39]. The direct target of miR-200b, SUZ12, epigenetically regulates polycomb-mediated repression of E-cadherin. Additionally, tumor suppressive Let-7 is down-regulated in breast CSCs and its loss results in the over-expression of oncogenes, H-RAS and HMGA2, prolonging stem cell-like self-renewal and reduced cellular differentiation [40].
In human glioblastoma (GBM), many miRNAs have been identified as either oncogenic or tumor suppressive from a number of human patient samples and functionally validated in established GBM cell lines, which has been well described recently by Rezaei et al [41]. For examples, miR-21 suppresses various tumor suppressors in GBM, such as PTEN, PDCD4, or EGFR, to increase cell proliferation, metastasis and drug resistance [37, 42–45]. MiR-221/222 targets protein tyrosine phosphatase mu (PTPmu) to induce invasiveness of GBM cells [46]. On the other hands, tumor suppressive miRNAs, such as miR-181, miR-34a, miR-29b or miR-133, are downregulated in GBM by suppressing oncogenes.
Although ectopic modulation of these dysregulated miRNAs in cancer cells has been proposed to show therapeutic potential [47], only limited studies have been advanced to clinical trials due to the lack of proper and efficient modulation strategies.
3. Delivery strategies of miRNAs into cancer tissue
Since the discovery of miRNAs over the last two decades, many attempts have been made to develop therapeutic strategies to return dysregulated miRNA expression levels back to normal with the hope of ceasing cancer progression and ultimately curing it. Despite promising results in many preclinical studies, only a few miRNA-based therapies have reached clinical trials and their outcomes are still dismal. For instance, a clinical trial testing the safety of delivering miR-34a formulated in a liposome-based carrier called Miramax (MRX34) (NCT01829971) was terminated early due to elevated cytokine-related adverse events in patients [48]. This was attributed to a lack of specificity. Aside from specificity, though, delivery strategies also need to ensure the stability of cargo miRNA, since naked, foreign RNA species will be cleared or degraded within 30 minutes of entering the blood stream by way of self-hydrolysis or RNase-mediated enzymatic degradation. Traditional efforts to achieve optimal miRNA delivery have been focused on three strategies: 1) modification of naked nucleic acids, 2) optimizing viral vector systems, and 3) optimizing non-viral vector systems.
A variety of chemically modified nucleotide analogs have been used to generate anti-miRNA oligonucleotides (AMO) by replacing the 2’-OH group with a chemically inert group, such as 2′-O-methyl-(2′-O-Me)-oligonucleotides, 2′-O-methoxyethyl-oligonucleotides (2′-O-MOE), or 2’ fluoride derivatives (2′-F) [49–51]. For example, 2’-O-Me AMO duplexes against miR-122 sequences injected through the mouse tail vein at 240 mg/kg/day successfully degraded liver abundant miR-122, and the protein expression levels of its known direct targets, Aldo-A, Ndrg3, Tmed3 and Hfe2, were increased up to 6.8 fold as compared to negative control mice [52]. Locked nucleic acid (LNA), an RNA analogue with a chemical lock between the 2′-oxygen and 4′-carbon on ribose, has been widely used in miRNA delivery due to its superior serum stability: up to 15 hours compared to 1.5 hours for unmodified oligonucleotides [53]. Lipid nanocapsules containing LNA sequences against miR-21 were shown to reduce miR-21 expression in U87MG cells and sensitize cells to radiation-induced death [54]. MiRNA sponge or target mimic concept, which uses an artificial competitive inhibitor, is another approach to modulate an intrinsic expression level of certain miRNAs of interest [55, 56]. For examples, an oncogenic long noncoding RNA, called HOTAIR, serves as a miRNA sponge against miR-34a-5p to increase cancer progression, drug resistance or metastasis [57–59].
In the case of restoring tumor suppressive miRNAs in cancer cells, viral-induced overexpression has been examined by encoding miRNA sequences into retroviruses, lentiviruses, adenoviruses, and adeno-associated viruses (AAVs) under a strong promoter system [60]. For GBM, overexpression of miR-128 by a lentivirus construct in the U87MG cell line resulted in cell growth inhibition and increased sensitivity to radiation therapy [61]. However, there are some concerns with viral delivery, such as the potential activation of oncogenes, systemic toxicity, and generation of an immunogenic response, which have arisen following the use of systemic administration [62].
In an effort to avoid these viral delivery-related concerns, non-viral delivery strategies of miRNAs artificially formulated with polymers, lipids, or inorganic nanoparticles have been extensively studied. For example, lipid-based nanoparticles formulated with cationic lipids to accommodate negatively charged RNAs have been commonly used [63] and polymer nanogels made with a polyglycerol-scaffold used to deliver tumor suppressive miR-34a systemically, resulted in tumor growth inhibition in mice [64]. Biodegradable polymeric nanoparticles were used to deliver miR-7 into GBM and endothelial cells resulting in reduction of its target gene and a functional inhibition of angiogenesis and tumor growth [65]. Transferrin-targeting lipopolyplex nanoparticles were used to specifically deliver miR-1 into GBM cells, and resulted in a significant reduction of target genes, MET and EGFR [66]. In another study, anti-miR-21 oligonucleotides were delivered via a chlorotxin-coupled lipid particle in order to specifically target brain tumors in mice [67]. Silencing of miR-21 in this model resulted in an increased expression of RhoB and decreased tumor cell proliferation and tumor size. In another study, systemic injection of antagomiR-21 and antagomiR-10b encapsulated in cRGD-tagged PEG-PLGA nanoparticles rescued expression of tumor suppressive and apoptotic genes including PTEN, PDCD4, HOXD10, P53 and CASP3, and sensitized GBM bearing mice to temozolomide (TMZ) [68]. Although nanoparticles are relatively simple and easy to generate, their penetration into brain tumors has been challenging by systemic administration due to the blood brain barrier (BBB) [69]. The BBB regulates brain homeostasis and the transport of endogenous and exogenous compounds by controlling their selective and specific uptake, efflux, and metabolism in the brain. While the BBB is meant to protect the brain from noxious agents, it also significantly hinders the delivery of therapeutics to brain tumors [70–75]. Although it is still far beyond the satisfaction for a successful clinical application, many attempts have been or still being made to overcome the natural barrier. It includes use of polypeptide-bearing amphiphilic block-copolymers or peptide amphiphiles (PAs) [76] or chitosan adsorption [77] to improve the penetrability through the BBB.
4. Development of RNA nanoparticle by RNA nanotechnology for cancer cell targeting
RNA interference (RNAi) has a great potential of modulating gene expressions since its discovery in 1998, therefore its application for disease therapy has been extensively studied through many clinical trials as well reviewed previously [78, 79]. Despite the numerous efforts over the last two decades, the first siRNA-based therapeutics, Onpattro (patisiran) by Alnylam Pharmaceuticals, was only approved by FDA in 2018 for the treatment of peripheral nerve disease, polyneuropathy, through siRNA against hereditary transthyretin-mediated amyloidosis (hATTR) [80, 81]. A year later in 2019, the second siRNA-based drug from the same manufacturer, Givlaari, was approved to lower the expression of aminolevulinic acid synthase 1 (ALAS1) for the treatment of acute hepatic porphyria (AHP) [82]. Main reason behind the serious delay of the clinical applications may lie in the lack of proper strategy for efficient and safe delivery, nor even tissue-specific targeting if possible, which still requires new methodologies for the successful application of RNAi technique.
Over the years, RNA nanotechnology has evolved from conventional RNA studies describing their relationship between sequences and folding structures to the engineering of RNA molecules in order to generate therapeutically promising nanomedicine. Overcoming the sensitive nature of RNAs to RNases in serum by the previously mentioned chemical modifications has been a game-changer. The pioneering work of RNA nanotechnology emerged in 1998 by Peixuan Guo and colleagues, when they engineered bacteriophage phi29 viral RNA into an artificial hexameric RNA ring [83]. Exploiting the unique stability of the packaging RNA (pRNA) from the phi29 DNA packaging motor system, the first pRNA-based siRNA delivery vector platform for cancer cell targeting emerged [84]. Further research on these pRNA sequences revealed that the three-way junction (3WJ) region of the pRNA can be assembled from three RNA molecules, denoted as a3WJ, b3WJ and c3WJ, maintaining an intact folding structure for RNA nanoparticles (RNPs) [85]. The size of these RNPs was determined to be approximately 5 nm based on dynamic light scattering (DLS) analysis. With the use of chemically modified 2’-F uridine or cytosine, the artificially constructed pRNA-3WJ RNPs exhibited comparable enzymatic, thermodynamic, and chemical stability with resistance to serum for up to 36 hours, a Tm value of 58°C, and stability up to 8 M urea [85–88]. Based on this, the individual RNA oligos (a3WJ, b3WJ and c3WJ) were further modified to harbor functional moieties including fluorescent dyes for tracking, cancer cell-specific ligands for selective targeting, and siRNAs/miRNAs as therapeutic cargo. Systemic injection of the pRNA-3WJ RNPs showed their specific accumulation in tumors [85] and pharmacological profiling of the pRNA-3WJ based RNP platform revealed little to no side effects or toxicity [89]. Chemically synthesized 2’-F-modified RNPs administered systemically demonstrated a favorable pharmacokinetic profile in mice: 5–10 hours half-life, <0.13 L/kg/hour clearance rate, and 1.2 L/kg volume of distribution [89]. In addition, there was no detectable toxicity as measured by interferon (IFN) and there was little to no inflammatory cytokine response. For specific targeting, pRNA-3WJ RNPs were conjugated with folate (FA) as most cancer cells overexpress folate receptors (FRs) to support DNA replication [90, 91]. The FA-conjugated pRNA-3WJ RNP platform has been extensively tested for tumor targeting in various cancer types [92–95] and non-cancer diseases [96].
5. Ligand specific targeting to GBM by RNA nanoparticles
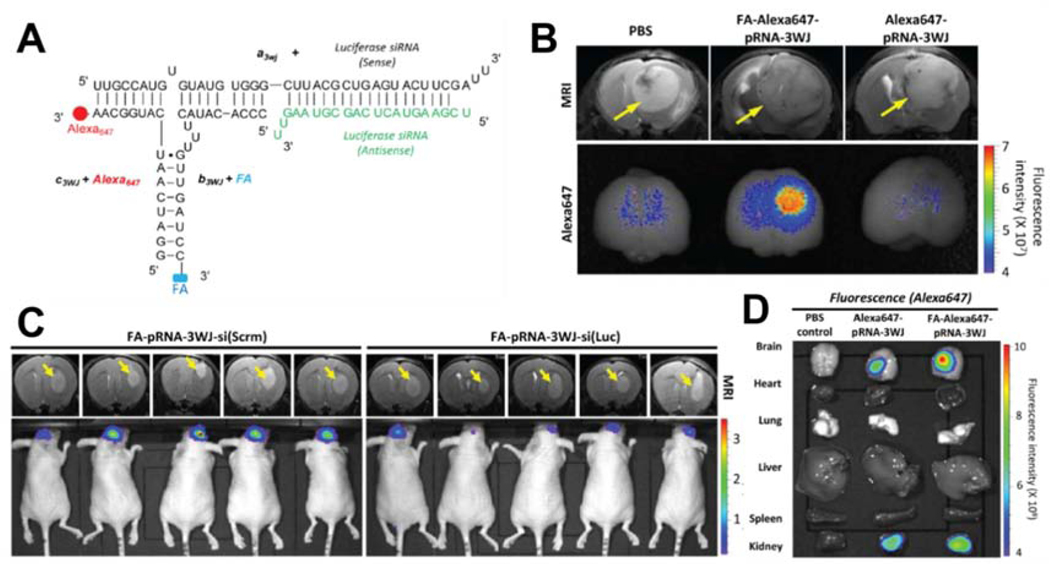
Tissue specific targeting is a critical factor to consider during systemic application of any therapeutic in order to reduce fatal or unnecessary side effects caused by non-specific targeting of healthy normal cells. This can be achieved by adding cell receptor recognizing agents, such as ligands, antibodies, or aptamers [97]. As described earlier, FA conjugation onto pRNA-3WJ RNPs enabled cancer cell specific targeting. In GBM, FRs are found to be highly overexpressed in tumor cells and not in adjacent normal brain cells [98]. Using FA conjugation, a new pRNA-3WJ RNP was constructed to test GBM targeting in orthotopic brain tumor mouse models [93]. Each of the three RNA modules (a3WJ, b3WJ and c3WJ) was modified to carry a functional moiety: 1) FA for FR targeting, 2) Alexa647 fluorophore for tracking, and 3) siRNA against luciferase as the gene silencing functional moiety (or scrambled RNA as a negative control). As expected, one step self-assembly of the three modified RNAs produced uniformed RNPs, denoted FA-pRNA-3WJ-si(Luc) RNPs (Fig. 1A), when analyzed for zeta potential and atomic force microscopy (AFM) imaging clearly revealed homogeneous three way-branched structure of the RNPs. An in vitro binding test in the cultured glioma cell line U87EGFRvIII showed that FA-pRNA-3WJ-si(Luc) RNPs bound to cells at higher levels than FA-free negative control RNPs. When U87EGFRvIII cells were pre-incubated with free folate-containing culture media to mask the FRs on their cell surface, the binding of FA-pRNA-3WJ-si(Luc) RNP to the cells was abolished. These data indicated that U87EGFRvIII cell targeting by FA-pRNA-3WJ-si(Luc) RNP is specific to the presence of FA on the RNPs. For the preclinical evaluation of the FA-mediated specific targeting, Alexa647-labeled FA-pRNA-3WJ RNPs were systemically injected into mice bearing intracranial brain tumor xenografts. While the brain tumor sizes were similar among the tested mice as confirmed by MRI, a stronger Alexa647 fluorescence signal overlapped with the brain tumor region in the mice injected with FA-Alexa647-pRNA-3WJ (mean fluorescence intensity 2.052±0.416, s.e.m.) compared to mice injected with control Alexa647-pRNA-3WJ (mean fluorescence intensity 1.014±0.279, s.e.m.) which was comparable to the signal obtained by PBS-injected control mice (mean fluorescence intensity 1.000±0.298, s.e.m.) (Fig. 1B). In addition to the appropriate targeting of RNPs to tumor cells, delivery of cargo RNA must also be functionally intact in order to actively repress its target mRNAs. To evaluate the functionality of cargo RNA, mice bearing intracranial GBM xenografts expressing luciferase were administered FA-pRNA-3WJ RNPs containing siRNA cargo directed against luciferase (FA-pRNA-3WJ-si(Luc)). Systemic injection of the FA-pRNA-3WJ-si(Luc) RNPs resulted in a significant reduction of bioluminescence, up to 5 times lower compared to mice treated with the control RNP, FA-pRNA-3WJ-si(Scrm) (Fig. 1C). Together these data successfully demonstrated that FA-pRNA-3WJ RNPs can specifically target GBM cells, internalize, and deliver cargo siRNAs. More importantly, the delivered siRNAs remained functionally intact, reducing target gene expression in tumor cells. A biodistribution profile of the mice injected with the Alexa647-labeled FA-pRNA-3WJ RNPs revealed that, aside from kidney, there was no significant fluorescence signal detected in major internal organs, such as heart, lung, liver, or spleen (Fig. 1D). This observation reduces the safety concern of unbound FA-pRNA-3WJ RNPs circulating in the blood stream. Pharmacological study showed that the FA-pRNA-3WJ RNPs are sufficiently large enough not to be accumulated in kidneys and the fluorescently labeled FA-pRNA-3WJ RNPs were found from urines immediately after tail vein injection, indicating they were readily cleared from the mouse through their kidneys [89]. This was the first report demonstrating the suitability of FA-pRNA-3WJ RNPs for the targeted delivery of small noncoding RNAs into GBM xenografts [93, 99].
Figure 1. Preclinical Glioblastoma Targeting by FA-pRNA-3WJ-si(Luc) RNPs.
(A) Schematic diagram showing sequence map of FA-pRNA-3WJ-si(Luc) RNP harboring FA (targeting ligand), Alexa647 (imaging module), and siRNA against luciferase (therapeutic module). (B) Glioblastoma targeting by FA-Alexa647-pRNA-3WJ RNP in U87EGFRvIII-induced intracranial brain tumor mice models. Tumor location and size were determined by MRI, while the specific targeting of RNPs were visualized by fluorescence intensity. (C) Representative bioluminescence intensity from intracranial glioblastoma bearing mice showing reduction of luciferase activity by systemic injection of FA-pRNA-3WJ-siRNA(Luc). (D) Biodistribution profile of FA-Alexa647-pRNA-3WJ RNP obtained from major internal organs collected from intracranial brain tumor bearing mice 15 hours after systemic injection through tail vein. All figures were adapted from Lee et al., 2015 [68].
6. Targeted inhibition of onco-miRs in GBM by RNA nanoparticles
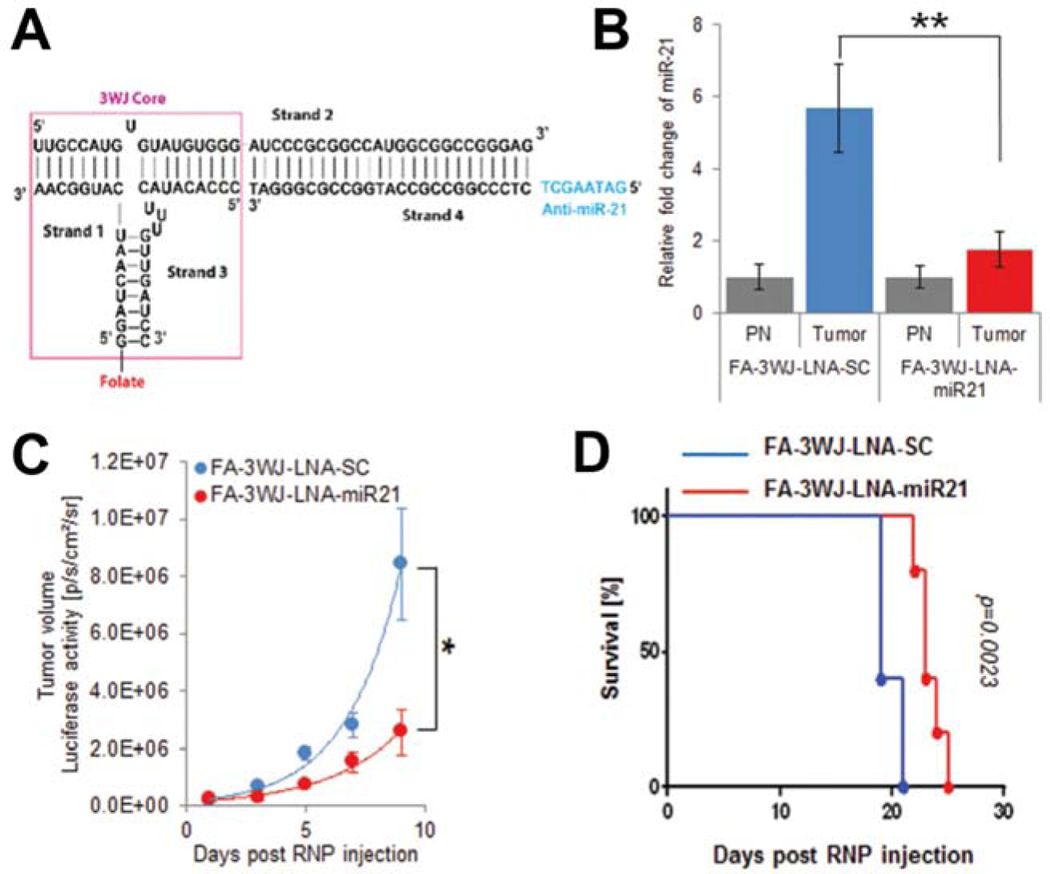
Inhibition of aberrantly overexpressed onco-miRs has been proposed as a promising therapeutic strategy to prevent tumor progression [7]. MiR-21 is one of the most frequently overexpressed onco-miRs in many type of cancers and plays a role in the oncogenesis of GBM by suppressing important tumor suppressors, such as PTEN, PDCD4, and Caspase-3, suggesting miR-21 as a promising therapeutic target [35, 37, 100–102]. The above mentioned pRNA-3WJ RNPs have been previously shown to specifically deliver anti-miR-21 LNA into triple-negative breast cancer (TNBC) cells using a 39-nucleotide long epidermal growth factor receptor (EGFR) targeting aptamer [94]. For targeted therapy of GBM using an anti-miR-21 LNA, a new RNP, named FA-3WJ-LNA-miR21 (Fig. 2A), was generated [95]. The newly synthesized FA-3WJ-LNA-miR21 was directed towards FR expressing cells and was shown to decrease the expression level of miR-21 in GBM cells (Fig. 2B). Glioma cell killing in vitro was significantly increased when U87EGFRvIII-Luc cells were incubated with FA-3WJ-LNA-miR21 RNPs compared to cells incubated with the negative control RNPs (FA-3WJ-LNA-SC). Additionally, inhibition of miR-21 in GBM cells with FA-3WJ-LNA-miR21 resulted in the rescue of multiple tumor suppressors, such as PTEN and PDCD4, that were otherwise suppressed. Importantly, this led to an increase in caspase/PARP activation and ultimately tumor cell apoptotic death. Systemic treatment of mice bearing intracranial GBM xenografts with FA-3WJ-LNA-miR21 RNPs reduced tumor growth (Fig. 2C) and increased animal survival (median survival 23 days compared to 19 days in negative control RNP-treated mice group) (Fig. 2D). These results demonstrated the preclinical potential of FA-targeted delivery of anti-miR-21 LNA via pRNA-3WJ RNPs for GBM.
Figure 2. Targeted Glioblastoma Therapy by FA-3WJ-LNA-miR21 RNPs.
(A) Schematic diagram showing sequence map of FA-3WJ-LNA-miR21 RNP constructed by mixing four module strands containing FA (targeting ligand), Alexa647 (imaging module), and anti-miR-21 LNA (therapeutic module). (B) Knockdown of onco-miR, miR-21, expression in glioblastoma tumor regions of mice treated with FA-3WJ-LNA-miR21 RNPs. (C) Glioblastoma tumor growth suppression in mice by repeated systemic administration of FA-3WJ-LNA-miR21 RNPs through tail vein injection. (D) Kaplan-Meier survival curves of mice systemically treated with 3WJ-LNA-miR21 RNPs. All figures were adapted under permission from Lee et al., 2017 [70].
7. Perspective
The therapeutic potential of siRNAs or miRNAs has been demonstrated for years by a number of preclinical studies. Compared to the extensive clinical application of synthetic drugs and protein-based biopharmaceuticals however, RNA-based cancer therapy remains at an early stage and this is mainly due to the difficulty of specific and safe delivery into cancer cells. Early studies investigating sncRNA delivery platforms resulted in the development of pRNA-3WJ RNPs derived from the viral RNA of bacteriophage phi29 [85]. Since the successful construction of this delivery platform, pRNA-3WJ RNPs have shown the capacity for specific cell targeting, cell internalization, and delivery of functional siRNA and miRNA inhibitors in both breast cancer or brain tumor model systems [93–95, 99]. These achievements successfully demonstrate the potential of pRNA-3WJ RNPs in the development of drug delivery for targeted cancer therapy showing the following: 1) pRNA-3WJ RNPs can be charged with a cancer cell targeting agent, FA; 2) pRNA-3WJ RNPs can gain access to brain tumor cells in mice; 3) pRNA-3WJ RNPs can deliver sncRNAs, such as siRNA and miRNA inhibitors, into brain tumor cells without losing their functionality; and 4) this targeted therapy avoids collateral damage in normal cells. Additional research on RNP design, including ligand selection, may further improve the therapeutic potential of the pRNA-3WJ RNPs, ultimately enhancing their suitability for cancer treatment in the clinic.
Acknowledgement:
Special thanks to Amy Thorne for helpful proofreading of this manuscript. This work was supported in part by Cancer Prevention and Research Institute of Texas (CPRIT) [RP200615] to T.J.L.; Research Scholar Grants (RSG-19–185-01-MPC) from the American Cancer Society to J.Y.Y; National Institutes of Health (NIH) National Cancer Institute (NCI) (R01 CA150153 and P01 CA163205) to B.K.; and NIH National Institute of Neurological Disorders and Stroke (NINDS) (R61 NS112410) to B.K.
Footnotes
Conflicts of interest: The authors declare no conflicts of interest.
Declaration of interests
☒ The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Cech TR, Steitz JA, The noncoding RNA revolution-trashing old rules to forge new ones, Cell, 157 (2014) 77–94. [DOI] [PubMed] [Google Scholar]
- [2].Bartel DP, Metazoan MicroRNAs, Cell, 173 (2018) 20–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Lee RC, Feinbaum RL, Ambros V, The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14, Cell, 75 (1993) 843–854. [DOI] [PubMed] [Google Scholar]
- [4].Wightman B, Ha I, Ruvkun G, Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans, Cell, 75 (1993) 855–862. [DOI] [PubMed] [Google Scholar]
- [5].Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G, The 21-nucleotide let7 RNA regulates developmental timing in Caenorhabditis elegans, Nature, 403 (2000) 901–906. [DOI] [PubMed] [Google Scholar]
- [6].Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, Rassenti L, Kipps T, Negrini M, Bullrich F, Croce CM, Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia, Proceedings of the National Academy of Sciences of the United States of America, 99 (2002) 15524–15529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Calin GA, Croce CM, MicroRNA signatures in human cancers, Nat Rev Cancer, 6 (2006) 857–866. [DOI] [PubMed] [Google Scholar]
- [8].Lynam-Lennon N, Maher SG, Reynolds JV, The roles of microRNA in cancer and apoptosis, Biol Rev Camb Philos Soc, 84 (2009) 55–71. [DOI] [PubMed] [Google Scholar]
- [9].Chen CZ, Li L, Lodish HF, Bartel DP, MicroRNAs modulate hematopoietic lineage differentiation, Science (New York, N.Y.), 303 (2004) 83–86. [DOI] [PubMed] [Google Scholar]
- [10].Chen B, Li H, Zeng X, Yang P, Liu X, Zhao X, Liang S, Roles of microRNA on cancer cell metabolism, J Transl Med, 10 (2012) 228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Singh DK, Bose S, Kumar S, Role of microRNA in regulating cell signaling pathways, cell cycle, and apoptosis in non-small cell lung cancer, Curr Mol Med, (2016). [PubMed] [Google Scholar]
- [12].Bartel DP, MicroRNAs: target recognition and regulatory functions, Cell, 136 (2009) 215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Ha M, Kim VN, Regulation of microRNA biogenesis, Nat Rev Mol Cell Biol, 15 (2014) 509–524. [DOI] [PubMed] [Google Scholar]
- [14].Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN, MicroRNA genes are transcribed by RNA polymerase II, The EMBO journal, 23 (2004) 4051–4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, Kim VN, The nuclear RNase III Drosha initiates microRNA processing, Nature, 425 (2003) 415–419. [DOI] [PubMed] [Google Scholar]
- [16].Yi R, Qin Y, Macara IG, Cullen BR, Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs, Genes & development, 17 (2003) 3011–3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD, A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA, Science (New York, N.Y.), 293 (2001) 834–838. [DOI] [PubMed] [Google Scholar]
- [18].Bagga S, Bracht J, Hunter S, Massirer K, Holtz J, Eachus R, Pasquinelli AE, Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation, Cell, 122 (2005) 553–563. [DOI] [PubMed] [Google Scholar]
- [19].Eichhorn SW, Guo H, McGeary SE, Rodriguez-Mias RA, Shin C, Baek D, Hsu SH, Ghoshal K, Villen J, Bartel DP, mRNA destabilization is the dominant effect of mammalian microRNAs by the time substantial repression ensues, Molecular cell, 56 (2014) 104–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Calin GA, Liu CG, Sevignani C, Ferracin M, Felli N, Dumitru CD, Shimizu M, Cimmino A, Zupo S, Dono M, Dell’Aquila ML, Alder H, Rassenti L, Kipps TJ, Bullrich F, Negrini M, Croce CM, MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias, Proceedings of the National Academy of Sciences of the United States of America, 101 (2004) 11755–11760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE, Iorio MV, Visone R, Sever NI, Fabbri M, Iuliano R, Palumbo T, Pichiorri F, Roldo C, Garzon R, Sevignani C, Rassenti L, Alder H, Volinia S, Liu CG, Kipps TJ, Negrini M, Croce CM, A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia, The New England journal of medicine, 353 (2005) 1793–1801. [DOI] [PubMed] [Google Scholar]
- [22].Fabbri M, Garzon R, Cimmino A, Liu Z, Zanesi N, Callegari E, Liu S, Alder H, Costinean S, Fernandez-Cymering C, Volinia S, Guler G, Morrison CD, Chan KK, Marcucci G, Calin GA, Huebner K, Croce CM, MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B, Proceedings of the National Academy of Sciences of the United States of America, 104 (2007) 15805–15810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Lodygin D, Tarasov V, Epanchintsev A, Berking C, Knyazeva T, Korner H, Knyazev P, Diebold J, Hermeking H, Inactivation of miR-34a by aberrant CpG methylation in multiple types of cancer, Cell Cycle, 7 (2008) 2591–2600. [DOI] [PubMed] [Google Scholar]
- [24].He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, Jackson AL, Linsley PS, Chen C, Lowe SW, Cleary MA, Hannon GJ, A microRNA component of the p53 tumour suppressor network, Nature, 447 (2007) 1130–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Yamamura S, Saini S, Majid S, Hirata H, Ueno K, Chang I, Tanaka Y, Gupta A, Dahiya R, MicroRNA-34a suppresses malignant transformation by targeting c-Myc transcriptional complexes in human renal cell carcinoma, Carcinogenesis, 33 (2012) 294–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Li N, Fu H, Tie Y, Hu Z, Kong W, Wu Y, Zheng X, miR-34a inhibits migration and invasion by down-regulation of c-Met expression in human hepatocellular carcinoma cells, Cancer Lett, 275 (2009) 44–53. [DOI] [PubMed] [Google Scholar]
- [27].Sun F, Fu H, Liu Q, Tie Y, Zhu J, Xing R, Sun Z, Zheng X, Downregulation of CCND1 and CDK6 by miR-34a induces cell cycle arrest, FEBS Lett, 582 (2008) 1564–1568. [DOI] [PubMed] [Google Scholar]
- [28].Li L, Yuan L, Luo J, Gao J, Guo J, Xie X, MiR-34a inhibits proliferation and migration of breast cancer through down-regulation of Bcl-2 and SIRT1, Clin Exp Med, 13 (2013) 109–117. [DOI] [PubMed] [Google Scholar]
- [29].Pang RT, Leung CO, Ye TM, Liu W, Chiu PC, Lam KK, Lee KF, Yeung WS, MicroRNA-34a suppresses invasion through downregulation of Notch1 and Jagged1 in cervical carcinoma and choriocarcinoma cells, Carcinogenesis, 31 (2010) 1037–1044. [DOI] [PubMed] [Google Scholar]
- [30].Yamakuchi M, Ferlito M, Lowenstein CJ, miR-34a repression of SIRT1 regulates apoptosis, Proc Natl Acad Sci U S A, 105 (2008) 13421–13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Eis PS, Tam W, Sun L, Chadburn A, Li Z, Gomez MF, Lund E, Dahlberg JE, Accumulation of miR-155 and BIC RNA in human B cell lymphomas, Proceedings of the National Academy of Sciences of the United States of America, 102 (2005) 3627–3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Jurkovicova D, Magyerkova M, Kulcsar L, Krivjanska M, Krivjansky V, Gibadulinova A, Oveckova I, Chovanec M, miR-155 as a diagnostic and prognostic marker in hematological and solid malignancies, Neoplasma, 61 (2014) 241–251. [DOI] [PubMed] [Google Scholar]
- [33].Costinean S, Zanesi N, Pekarsky Y, Tili E, Volinia S, Heerema N, Croce CM, Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice, Proceedings of the National Academy of Sciences of the United States of America, 103 (2006) 7024–7029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Costinean S, Sandhu SK, Pedersen IM, Tili E, Trotta R, Perrotti D, Ciarlariello D, Neviani P, Harb J, Kauffman LR, Shidham A, Croce CM, Src homology 2 domain-containing inositol-5-phosphatase and CCAAT enhancer-binding protein beta are targeted by miR-155 in B cells of Emicro-MiR-155 transgenic mice, Blood, 114 (2009) 1374–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Pfeffer SR, Yang CH, Pfeffer LM, The Role of miR-21 in Cancer, Drug Dev Res, 76 (2015) 270–277. [DOI] [PubMed] [Google Scholar]
- [36].Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T, MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer, Gastroenterology, 133 (2007) 647–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Gaur AB, Holbeck SL, Colburn NH, Israel MA, Downregulation of Pdcd4 by mir-21 facilitates glioblastoma proliferation in vivo, Neuro-oncology, 13 (2011) 580–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Liu M, Wu H, Liu T, Li Y, Wang F, Wan H, Li X, Tang H, Regulation of the cell cycle gene, BTG2, by miR-21 in human laryngeal carcinoma, Cell Res, 19 (2009) 828–837. [DOI] [PubMed] [Google Scholar]
- [39].Iliopoulos D, Lindahl-Allen M, Polytarchou C, Hirsch HA, Tsichlis PN, Struhl K, Loss of miR-200 inhibition of Suz12 leads to polycomb-mediated repression required for the formation and maintenance of cancer stem cells, Molecular cell, 39 (2010) 761–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong C, Huang Y, Hu X, Su F, Lieberman J, Song E, Let-7 Regulates Self Renewal and Tumorigenicity of Breast Cancer Cells, Cell, 131 (2007) 1109–1123. [DOI] [PubMed] [Google Scholar]
- [41].Rezaei O, Honarmand K, Nateghinia S, Taheri M, Ghafouri-Fard S, miRNA signature in glioblastoma: Potential biomarkers and therapeutic targets, Exp Mol Pathol, 117 (2020) 104550. [DOI] [PubMed] [Google Scholar]
- [42].Ren Y, Kang CS, Yuan XB, Zhou X, Xu P, Han L, Wang GX, Jia Z, Zhong Y, Yu S, Sheng J, Pu PY, Co-delivery of as-miR-21 and 5-FU by poly(amidoamine) dendrimer attenuates human glioma cell growth in vitro, J Biomater Sci Polym Ed, 21 (2010) 303–314. [DOI] [PubMed] [Google Scholar]
- [43].Shi L, Chen J, Yang J, Pan T, Zhang S, Wang Z, MiR-21 protected human glioblastoma U87MG cells from chemotherapeutic drug temozolomide induced apoptosis by decreasing Bax/Bcl-2 ratio and caspase-3 activity, Brain Res, 1352 (2010) 255–264. [DOI] [PubMed] [Google Scholar]
- [44].Zhang S, Wan Y, Pan T, Gu X, Qian C, Sun G, Sun L, Xiang Y, Wang Z, Shi L, MicroRNA-21 inhibitor sensitizes human glioblastoma U251 stem cells to chemotherapeutic drug temozolomide, J Mol Neurosci, 47 (2012) 346–356. [DOI] [PubMed] [Google Scholar]
- [45].Costa PM, Cardoso AL, Nobrega C, Pereira de Almeida LF, Bruce JN, Canoll P, Pedroso de Lima MC, MicroRNA-21 silencing enhances the cytotoxic effect of the antiangiogenic drug sunitinib in glioblastoma, Hum Mol Genet, 22 (2013) 904–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Quintavalle C, Garofalo M, Zanca C, Romano G, Iaboni M, del Basso De Caro M, Martinez-Montero JC, Incoronato M, Nuovo G, Croce CM, Condorelli G, miR-221/222 overexpession in human glioblastoma increases invasiveness by targeting the protein phosphate PTPmu, Oncogene, 31 (2012) 858–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Hanahan D, Weinberg RA, Hallmarks of cancer: the next generation, Cell, 144 (2011) 646–674. [DOI] [PubMed] [Google Scholar]
- [48].Beg MS, Brenner AJ, Sachdev J, Borad M, Kang YK, Stoudemire J, Smith S, Bader AG, Kim S, Hong DS, Phase I study of MRX34, a liposomal miR-34a mimic, administered twice weekly in patients with advanced solid tumors, Invest New Drugs, 35 (2017) 180–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Lennox KA, Behlke MA, Chemical modification and design of anti-miRNA oligonucleotides, Gene Ther, 18 (2011) 1111–1120. [DOI] [PubMed] [Google Scholar]
- [50].Prakash TP, Bhat B, 2’-Modified oligonucleotides for antisense therapeutics, Curr Top Med Chem, 7 (2007) 641–649. [DOI] [PubMed] [Google Scholar]
- [51].Prakash TP, An overview of sugar-modified oligonucleotides for antisense therapeutics, Chem Biodivers, 8 (2011) 1616–1641. [DOI] [PubMed] [Google Scholar]
- [52].Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M, Silencing of microRNAs in vivo with ‘antagomirs’, Nature, 438 (2005) 685–689. [DOI] [PubMed] [Google Scholar]
- [53].Kurreck J, Wyszko E, Gillen C, Erdmann VA, Design of antisense oligonucleotides stabilized by locked nucleic acids, Nucleic Acids Res, 30 (2002) 1911–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Griveau A, Bejaud J, Anthiya S, Avril S, Autret D, Garcion E, Silencing of miR-21 by locked nucleic acid-lipid nanocapsule complexes sensitize human glioblastoma cells to radiation-induced cell death, Int J Pharm, 454 (2013) 765–774. [DOI] [PubMed] [Google Scholar]
- [55].Ebert MS, Neilson JR, Sharp PA, MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells, Nat Methods, 4 (2007) 721–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Franco-Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, Rubio-Somoza I, Leyva A, Weigel D, Garcia JA, Paz-Ares J, Target mimicry provides a new mechanism for regulation of microRNA activity, Nat Genet, 39 (2007) 1033–1037. [DOI] [PubMed] [Google Scholar]
- [57].Duan Y, Chen J, Yang Y, Qu Z, Lu Y, Sun D, LncRNA HOTAIR contributes Taxol-resistance of hepatocellular carcinoma cells via activating AKT phosphorylation by down-regulating miR-34a, Biosci Rep, 40 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Peng CL, Zhao XJ, Wei CC, Wu JW, LncRNA HOTAIR promotes colon cancer development by down-regulating miRNA34a, Eur Rev Med Pharmacol Sci, 23 (2019) 5752–5761. [DOI] [PubMed] [Google Scholar]
- [59].Zheng F, Li J, Ma C, Tang X, Tang Q, Wu J, Chai X, Xie J, Yang XB, Hann SS, Novel regulation of miR-34a-5p and HOTAIR by the combination of berberine and gefitinib leading to inhibition of EMT in human lung cancer, J Cell Mol Med, 24 (2020) 5578–5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Davidson BL, Harper SQ, Viral delivery of recombinant short hairpin RNAs, Methods Enzymol, 392 (2005) 145–173. [DOI] [PubMed] [Google Scholar]
- [61].Peruzzi P, Bronisz A, Nowicki MO, Wang Y, Ogawa D, Price R, Nakano I, Kwon CH, Hayes J, Lawler SE, Ostrowski MC, Chiocca EA, Godlewski J, MicroRNA-128 coordinately targets Polycomb Repressor Complexes in glioma stem cells, Neuro-oncology, 15 (2013) 1212–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Penaud-Budloo M, Francois A, Clement N, Ayuso E, Pharmacology of Recombinant Adeno-associated Virus Production, Mol Ther Methods Clin Dev, 8 (2018) 166–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Campani V, Salzano G, Lusa S, De Rosa G, Lipid Nanovectors to Deliver RNA Oligonucleotides in Cancer, Nanomaterials (Basel), 6 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Shatsberg Z, Zhang X, Ofek P, Malhotra S, Krivitsky A, Scomparin A, Tiram G, Calderon M, Haag R, Satchi-Fainaro R, Functionalized nanogels carrying an anticancer microRNA for glioblastoma therapy, J Control Release, 239 (2016) 159–168. [DOI] [PubMed] [Google Scholar]
- [65].Babae N, Bourajjaj M, Liu Y, Van Beijnum JR, Cerisoli F, Scaria PV, Verheul M, Van Berkel MP, Pieters EH, Van Haastert RJ, Yousefi A, Mastrobattista E, Storm G, Berezikov E, Cuppen E, Woodle M, Schaapveld RQ, Prevost GP, Griffioen AW, Van Noort PI, Schiffelers RM, Systemic miRNA-7 delivery inhibits tumor angiogenesis and growth in murine xenograft glioblastoma, Oncotarget, 5 (2014) 6687–6700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Wang X, Huang X, Yang Z, Gallego-Perez D, Ma J, Zhao X, Xie J, Nakano I, Lee LJ, Targeted delivery of tumor suppressor microRNA-1 by transferrin-conjugated lipopolyplex nanoparticles to patient-derived glioblastoma stem cells, Curr Pharm Biotechnol, 15 (2014) 839–846. [DOI] [PubMed] [Google Scholar]
- [67].Costa PM, Cardoso AL, Custodia C, Cunha P, Pereira de Almeida L, Pedroso de Lima MC, MiRNA-21 silencing mediated by tumor-targeted nanoparticles combined with sunitinib: A new multimodal gene therapy approach for glioblastoma, J Control Release, 207 (2015) 31–39. [DOI] [PubMed] [Google Scholar]
- [68].Malhotra M, Sekar TV, Ananta JS, Devulapally R, Afjei R, Babikir HA, Paulmurugan R, Massoud TF, Targeted nanoparticle delivery of therapeutic antisense microRNAs presensitizes glioblastoma cells to lower effective doses of temozolomide in vitro and in a mouse model, Oncotarget, 9 (2018) 21478–21494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Pardridge WM, The blood-brain barrier: bottleneck in brain drug development, NeuroRx, 2 (2005) 3–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Da Ros M, De Gregorio V, Iorio AL, Giunti L, Guidi M, de Martino M, Genitori L, Sardi I, Glioblastoma Chemoresistance: The Double Play by Microenvironment and Blood-Brain Barrier, Int J Mol Sci, 19 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Harder BG, Blomquist MR, Wang J, Kim AJ, Woodworth GF, Winkles JA, Loftus JC, Tran NL, Developments in Blood-Brain Barrier Penetrance and Drug Repurposing for Improved Treatment of Glioblastoma, Front Oncol, 8 (2018) 462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Jain KK, A Critical Overview of Targeted Therapies for Glioblastoma, Front Oncol, 8 (2018) 419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Sarkaria JN, Hu LS, Parney IF, Pafundi DH, Brinkmann DH, Laack NN, Giannini C, Burns TC, Kizilbash SH, Laramy JK, Swanson KR, Kaufmann TJ, Brown PD, Agar NYR, Galanis E, Buckner JC, Elmquist WF, Is the blood-brain barrier really disrupted in all glioblastomas? A critical assessment of existing clinical data, Neuro Oncol, 20 (2018) 184–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Shergalis A, Bankhead A 3rd, Luesakul U, Muangsin N, Neamati N, Current Challenges and Opportunities in Treating Glioblastoma, Pharmacol Rev, 70 (2018) 412–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Huang R, Boltze J, Li S, Strategies for Improved Intra-arterial Treatments Targeting Brain Tumors: a Systematic Review, Front Oncol, 10 (2020) 1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Duro-Castano A, Leite DM, Forth J, Deng Y, Matias D, Jesus CN, Battaglia G, Designing peptide nanoparticles for efficient brain delivery, Adv Drug Deliv Rev, (2020). [DOI] [PubMed] [Google Scholar]
- [77].Cortes H, Alcala-Alcala S, Caballero-Floran IH, Bernal-Chavez SA, Avalos-Fuentes A, Gonzalez-Torres M, Gonzalez-Del Carmen M, Figueroa-Gonzalez G, Reyes-Hernandez OD, Floran B, Del Prado-Audelo ML, Leyva-Gomez G, A Reevaluation of Chitosan-Decorated Nanoparticles to Cross the Blood-Brain Barrier, Membranes (Basel), 10 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Hu B, Zhong L, Weng Y, Peng L, Huang Y, Zhao Y, Liang XJ, Therapeutic siRNA: state of the art, Signal Transduct Target Ther, 5 (2020) 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Zuckerman JE, Davis ME, Clinical experiences with systemically administered siRNA-based therapeutics in cancer, Nat Rev Drug Discov, 14 (2015) 843–856. [DOI] [PubMed] [Google Scholar]
- [80].Wood H, FDA approves patisiran to treat hereditary transthyretin amyloidosis, Nat Rev Neurol, 14 (2018) 570. [DOI] [PubMed] [Google Scholar]
- [81].Adams D, Gonzalez-Duarte A, O’Riordan WD, Yang CC, Ueda M, Kristen AV, Tournev I, Schmidt HH, Coelho T, Berk JL, Lin KP, Vita G, Attarian S, Plante-Bordeneuve V, Mezei MM, Campistol JM, Buades J, Brannagan TH 3rd, Kim BJ, Oh J, Parman Y, Sekijima Y, Hawkins PN, Solomon SD, Polydefkis M, Dyck PJ, Gandhi PJ, Goyal S, Chen J, Strahs AL, Nochur SV, Sweetser MT, Garg PP, Vaishnaw AK, Gollob JA, Suhr OB, Patisiran, an RNAi Therapeutic, for Hereditary Transthyretin Amyloidosis, N Engl J Med, 379 (2018) 11–21. [DOI] [PubMed] [Google Scholar]
- [82].Gonzalez-Aseguinolaza G, Givosiran - Running RNA Interference to Fight Porphyria Attacks, N Engl J Med, 382 (2020) 2366–2367. [DOI] [PubMed] [Google Scholar]
- [83].Guo P, Zhang C, Chen C, Garver K, Trottier M, Inter-RNA interaction of phage phi29 pRNA to form a hexameric complex for viral DNA transportation, Molecular cell, 2 (1998) 149–155. [DOI] [PubMed] [Google Scholar]
- [84].Guo S, Tschammer N, Mohammed S, Guo P, Specific delivery of therapeutic RNAs to cancer cells via the dimerization mechanism of phi29 motor pRNA, Hum Gene Ther, 16 (2005) 1097–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Shu D, Shu Y, Haque F, Abdelmawla S, Guo P, Thermodynamically stable RNA three-way junction for constructing multifunctional nanoparticles for delivery of therapeutics, Nature nanotechnology, 6 (2011) 658–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Zhang H, Endrizzi JA, Shu Y, Haque F, Sauter C, Shlyakhtenko LS, Lyubchenko Y, Guo P, Chi YI, Crystal structure of 3WJ core revealing divalent ion-promoted thermostability and assembly of the Phi29 hexameric motor pRNA, RNA, 19 (2013) 1226–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Binzel DW, Khisamutdinov EF, Guo P, Entropy-driven one-step formation of Phi29 pRNA 3WJ from three RNA fragments, Biochemistry, 53 (2014) 2221–2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Shu D, Khisamutdinov EF, Zhang L, Guo P, Programmable folding of fusion RNA in vivo and in vitro driven by pRNA 3WJ motif of phi29 DNA packaging motor, Nucleic Acids Res, 42 (2014) e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Abdelmawla S, Guo S, Zhang L, Pulukuri SM, Patankar P, Conley P, Trebley J, Guo P, Li QX, Pharmacological characterization of chemically synthesized monomeric phi29 pRNA nanoparticles for systemic delivery, Molecular therapy : the journal of the American Society of Gene Therapy, 19 (2011) 1312–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Xia W, Low PS, Folate-targeted therapies for cancer, Journal of medicinal chemistry, 53 (2010) 6811–6824. [DOI] [PubMed] [Google Scholar]
- [91].Low PS, Antony AC, Folate receptor-targeted drugs for cancer and inflammatory diseases, Advanced Drug Delivery Reviews, 56 (2004) 1055–1058. [DOI] [PubMed] [Google Scholar]
- [92].Rychahou P, Haque F, Shu Y, Zaytseva Y, Weiss HL, Lee EY, Mustain W, Valentino J, Guo P, Evers BM, Delivery of RNA nanoparticles into colorectal cancer metastases following systemic administration, ACS Nano, 9 (2015) 1108–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Lee TJ, Haque F, Shu D, Yoo JY, Li H, Yokel RA, Horbinski C, Kim TH, Kim SH, Kwon CH, Nakano I, Kaur B, Guo P, Croce CM, RNA nanoparticle as a vector for targeted siRNA delivery into glioblastoma mouse model, Oncotarget, 6 (2015) 14766–14776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Shu D, Li H, Shu Y, Xiong G, Carson WE 3rd, Haque F, Xu R, Guo P, Systemic Delivery of Anti-miRNA for Suppression of Triple Negative Breast Cancer Utilizing RNA Nanotechnology, ACS nano, 9 (2015) 9731–9740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Lee TJ, Yoo JY, Shu D, Li H, Zhang J, Yu JG, Jaime-Ramirez AC, Acunzo M, Romano G, Cui R, Sun HL, Luo Z, Old M, Kaur B, Guo P, Croce CM, RNA Nanoparticle-Based Targeted Therapy for Glioblastoma through Inhibition of Oncogenic miR-21, Mol Ther, 25 (2017) 1544–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Shi Z, Li SK, Charoenputtakun P, Liu CY, Jasinski D, Guo P, RNA nanoparticle distribution and clearance in the eye after subconjunctival injection with and without thermosensitive hydrogels, J Control Release, 270 (2018) 14–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Paulmurugan R, Malhotra M, Massoud TF, The protean world of non-coding RNAs in glioblastoma, J Mol Med (Berl), 97 (2019) 909–925. [DOI] [PubMed] [Google Scholar]
- [98].Parker N, Turk MJ, Westrick E, Lewis JD, Low PS, Leamon CP, Folate receptor expression in carcinomas and normal tissues determined by a quantitative radioligand binding assay, Analytical Biochemistry, 338 (2005) 284–293. [DOI] [PubMed] [Google Scholar]
- [99].Lee TJ, Haque F, Vieweger M, Yoo JY, Kaur B, Guo P, Croce CM, Functional assays for specific targeting and delivery of RNA nanoparticles to brain tumor, Methods in molecular biology (Clifton, N.J.), 1297 (2015) 137–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Krichevsky AM, Gabriely G, miR-21: a small multi-faceted RNA, Journal of Cellular and Molecular Medicine, 13 (2009) 39–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Zhou X, Ren Y, Moore L, Mei M, You Y, Xu P, Wang B, Wang G, Jia Z, Pu P, Zhang W, Kang C, Downregulation of miR-21 inhibits EGFR pathway and suppresses the growth of human glioblastoma cells independent of PTEN status, Laboratory investigation; a journal of technical methods and pathology, 90 (2010) 144–155. [DOI] [PubMed] [Google Scholar]
- [102].Shi L, Chen J, Yang J, Pan T, Zhang S, Wang Z, MiR-21 protected human glioblastoma U87MG cells from chemotherapeutic drug temozolomide induced apoptosis by decreasing Bax/Bcl-2 ratio and caspase-3 activity, Brain research, 1352 (2010) 255–264. [DOI] [PubMed] [Google Scholar]