Abstract
Actin filament crosslinking, bundling and molecular motor proteins are necessary for the assembly of epithelial projections such as microvilli, stereocilia, hairs, and bristles. Mutations in such proteins cause defects in the shape, structure, and function of these actin-based protrusions. One protein necessary for stereocilia formation, Myosin VIIA, is an actin-based motor protein conserved throughout phylogeny. In Drosophila melanogaster, severe mutations in the MyoVIIA homologue crinkled (ck) are “semi-lethal” with only a very small percentage of flies surviving to adulthood. Such survivors show morphological defects related to actin bundling in hairs and bristles. To better understand ck/MyoVIIA’s function in bundled-actin structures, we used dominant female sterile approaches to analyze the loss of maternal and zygotic (M/Z) ck/MyoVIIA in the morphogenesis of denticles, small actin-based projections on the ventral epidermis of Drosophila embryos. M/Z ck mutants displayed severe defects in denticle morphology-actin filaments initiated in the correct location, but failed to elongate and bundle to form normal projections. Using deletion mutant constructs, we demonstrated that both of the C-terminal MyTH4 and FERM domains are necessary for proper denticle formation. Furthermore, we show that ck/MyoVIIA interacts genetically with dusky-like (dyl), a member of the ZPD family of proteins that links the extracellular matrix to the plasma membrane, and when mutated also disrupts normal denticle formation. Loss of either protein alone does not alter the localization of the other; however, loss of the two proteins together dramatically enhances the defects in denticle shape observed when either protein alone was absent. Our data indicate that ck/MyoVIIA plays a key role in the formation and/or organization of actin filament bundles, which drive proper shape of cellular projections.
Keywords: Drosophila, denticles, actin cytoskeleton, Myosin VIIA, actin-based projections
Introduction
During embryogenesis cells undergo extensive modifications in their shape and their location within the embryo. Changes in cell migration and three-dimensional morphology depend on the function(s) of the dynamic cytoskeleton (Pollard and Cooper, 2009; Tang and Gerlach, 2017; Schaks et al., 2019). Actin-based extensions from the apical surfaces of epithelial cells are common and occur in numerous cell types including intestinal epithelial cells (microvilli), inner ear hair cells (stereocilia), and cells in developing embryos (filopodia and lamellipodia, DeRosier and Tilney, 2000). Drosophila epithelial cells display planar cell polarity and actin bundle organization similar to their vertebrate counterparts (DeRosier and Tilney, 2000). Moreover, many proteins responsible for organizing networks of actin filaments in vertebrates are conserved across evolution and have orthologs in flies, including but not limited to Fascin (Singed), Espin (Forked), Villin (Quail), Scaffold protein containing ankyrin repeats and SAM domain referred to as SANS (Sans), and Protocadherin 15 (Cad99C,Mahajan-Miklos and Cooley, 1994; Wulfkuhle et al., 1998; D’Alterio et al., 2005; Demontis and Dahmann, 2009). Evidence suggests that many, if not all, of the molecular functions and mechanisms of these proteins are also conserved.
Previous studies used bristle and hair formation on the Drosophila thorax and wing as model systems to examine the ontogeny of actin-based cellular protrusions (Tilney et al., 1996; Turner and Adler, 1998; Tilney et al., 2000a; Tilney et al., 2000b). The formation of denticles, actin-based projections on the ventral epidermis of the Drosophila embryo, is also an excellent model system for understanding protrusion morphogenesis and the developmental cues that specify it (Nusslein-Volhard and Wieschaus, 1980; Dickinson and Thatcher, 1997; Price et al., 2006; Bejsovec and Chao, 2012; Spencer et al., 2017). Moreover, this system has an added benefit of allowing for the investigation of homozygous recessive alleles that fail to survive beyond early developmental stages. With the use of germline clones, the effect of early protein requirement due to maternally loaded products in the early embryos can be investigated.
The thorax and abdomen of Drosophila embryos are segmented along the anterior-posterior axis into 11 repeating pairs. Each pair consists of an anterior region that is decorated with denticles and a posterior region that is smooth or naked (Dickinson and Thatcher, 1997). Each anterior hemi-segment contains 6 columns of cells which produce uniquely patterned denticles that can be recognized by their variations in size, shape, number, and hook orientation (Fig. 1A; Nusslein-Volhard and Wieschaus, 1980; Bejsovec and Wieschaus, 1993; Martinez Arias, 1993). Due to the denticles’ highly reproducible pattern, this system has extensively been used to investigate the signaling pathways that are responsible for embryonic patterning and planar cell polarity (Nusslein-Volhard and Wieschaus, 1980; DiNardo et al., 1994; Hatini and DiNardo, 2001; Payre, 2004; Dilks and DiNardo, 2010). We and others use the formation of denticles as a model system to investigate the molecular mechanisms that specify how cytoskeletal proteins orchestrate the formation of three dimensional structures of bundled F-actin with unique architectures (Chanut-Delalande et al., 2006; Price et al., 2006; Bejsovec and Chao, 2012). Denticles form during the middle stages of embryogenesis (stages 14–16, approximately 10–13 hours after egg lay, AEL, at 25°C). At a rudimentary level, the cellular structures that form denticles like those that form bristles and hairs are structurally similar to vertebrate microvilli and stereocilia; growth of the structure begins with a small microtubule-rich base and core, and cortical actin nucleation in a small patch beneath the plasma membrane (DeRosier and Tilney, 2000; Price et al., 2006). During 12–13 hours AEL, these condensations become more organized and the actin filaments bundle and elongate in the posterior direction while the plasma membrane is reshaped around the growing extensions until mature F-actin rich denticles are formed (Dickinson and Thatcher, 1997; Price et al., 2006)). In denticles, the overall organization of actin filaments is orchestrated by several actin-associated proteins such as Diaphanous, Actin-related protein 3 (Arp3), and Enabled (Price et al., 2006), which are also important in vertebrate actin organization. Mutations in additional cytoskeleton remodeling components Singed, Forked and Multiple Wing Hair also show defects in denticle size and shape (Dickinson and Thatcher, 1997; Chanut-Delalande et al., 2006).
Figure 1:
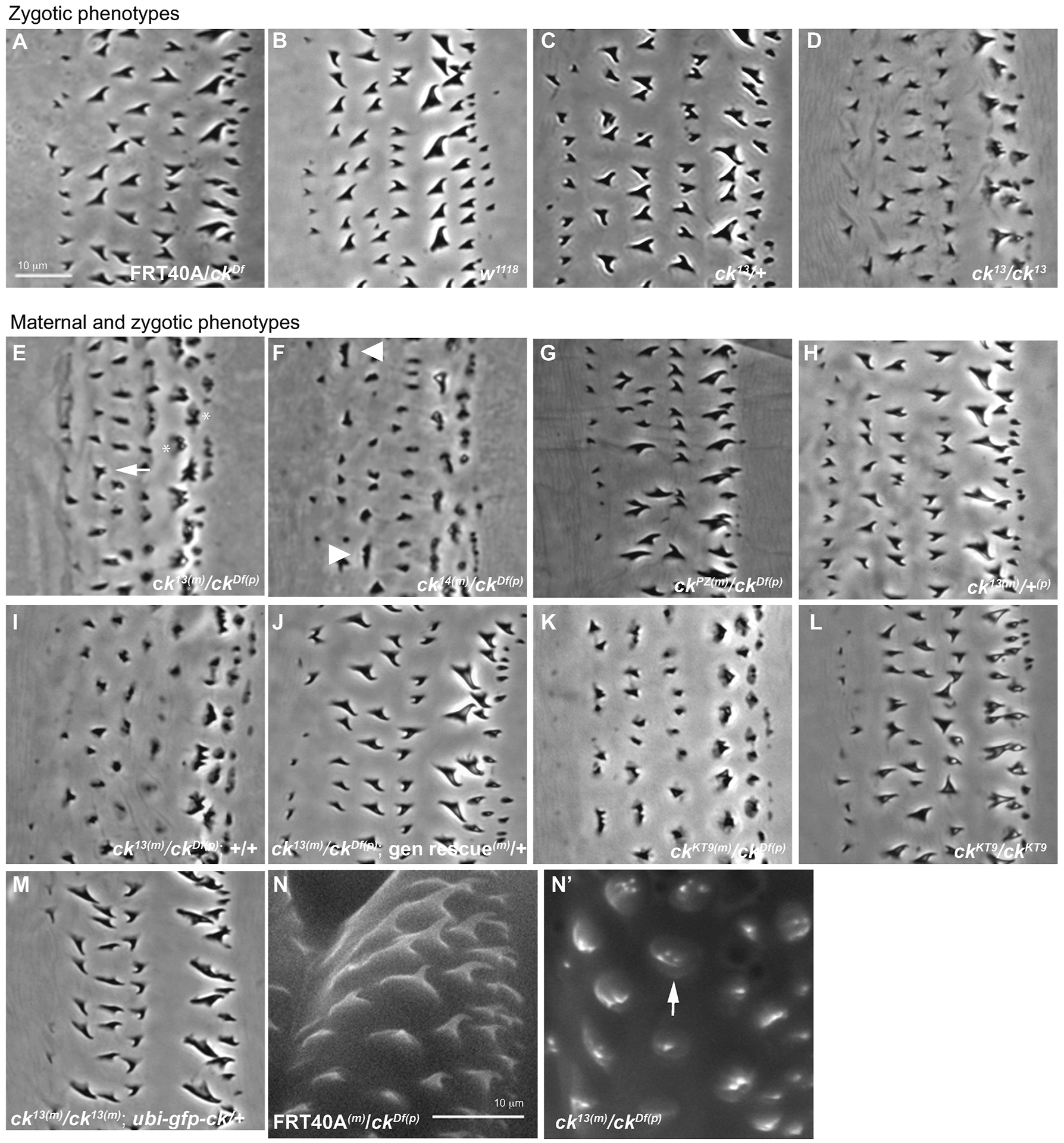
Loss of ck/MyoVIIA perturbs denticle morphology. (m) marks maternal chromosome and (p) marks paternal chromosome. (A, B, C) Phase contrast image of cuticle preps of control (FRT40A(m)/ckDf(p), w1118 (wildtype), and zygotic ck13(m)/+) denticles show normal size, structure and hooking. (D) Homozygous zygotic ck13/ck13 mutants display mild defects in height and structure. (E–G) M/Z ck allelic mutant embryos show defects in denticle morphology (ck13(m)/ckDf(p), ck14(m)/ckDf(p), ckPZ(m)/ckDf(p)) compared to controls (ck13(m)/+, FRT40A(m)/ckDf(p)). (H) maternal ck13(m)/+(p) heterozygote embryos displays no defects. (I) M/Z ck13(m)/ckDf(p) mutant denticles compared to (J) sibling denticles rescued with a genomic promoter driven cDNA transgene of ck/MyoVIIA. (K) M/Z ckKT9(m)/ckDf(p) show a similar phenotype as our ck13 allele. (L) Homozygous zygotic ckKT9/ckKT9 denticles have a milder phenotype than germ line clones. (M) The mutant denticle phenotype is rescued by expression of a ubiquitin-driven GFP-tagged ck cDNA in the M/Z ck13(m)/ck13(p) mutant background. All images are of the second abdominal belt. (N, N’) Environmental scanning electron microscopy of first instar larvae denticles in control (FRT40A(m)/ckDf(p)) and ck/MyoVIIA null organisms (M/Z ck13(m)/ckDf(p)). Anterior position is left in all figures. Each scale bar is 10 μm. The scale bar in A applies to panels A–M, while the scale bar in N applies to panels N and N’.
Between 13–15 hours AEL the epidermal cells secrete cuticle, a hardened mixture of the polysaccharide chitin, extracellular matrix (ECM) proteins and lipids. Cuticle protects the larvae from the environment (Payre, 2004; Moussian et al., 2006). The plasma membrane is anchored to the ECM by zona-pellucida domain (ZPD) proteins, a family of single pass transmembrane proteins with small cytoplasmic tails (Plaza et al., 2010). Different ZPD proteins accumulate and organize specific sub-apical regions of the forming denticles. Loss of these proteins results in defects in the interactions between membrane proteins and ECM proteins of the cuticle as well as defects in denticle shape (Fernandes et al., 2010). Once deposited, the cuticle lies tightly opposed to the plasma membrane and it’s underlying cortex and thereby creates a permanent record of the mature denticle’s shape. Ultimately, the cellular projections retract such that denticles in larvae are composed only of cuticle (Payre, 2004). What specifies the hooking (thorn-like shape) that is seen in the final cuticle but is not present in the F-actin networks (cone shape) of the denticles remains unknown.
One protein necessary for actin-based protrusion formation in vertebrates is the unconventional myosin motor, Myosin VIIA (MyoVIIA). MyoVIIA localizes to vertebrate microvilli and stereocilia and is necessary for the structural organization of, and vesicle transport within, stereocilia (Hasson et al., 1997; Self et al., 1998; Wolfrum et al., 1998). Humans homozygous and transheterozygous for mutations in MyoVIIA suffer from congenital deafness and blindness, clinically known as human Usher syndrome 1B (Weil et al., 1995) and non-syndromic deafness DFNB2 and DFNA11 (Liu et al., 1997a; Liu et al., 1997b; Weil et al., 1997; El-Amraoui and Petit, 2005). In addition, mice lacking MyoVIIA are also deaf and show severe structural defects in stereocilia (Gibson et al., 1995; Self et al., 1998).
Drosophila crinkled (ck) encodes MyoVIIA. It’s motor domain is 67% identical to human Myo VIIA’s motor domain, and overall the protein has a 62% identity and 73% similarity to human Myo VIIA (Kiehart et al., 2004; Orme et al., 2016). Severe mutations in ck/MyoVIIA are “semi-lethal,” with only a small fraction of homozygous flies (0.5–5% of Mendelian expectation) reaching adulthood (Kiehart et al., 2004). Such escapers are deaf, infertile, and show defects related to actin bundling in hair and bristle formation (Kiehart et al., 2004; Todi et al., 2005; Todi et al., 2008; Singh, 2012). Thus, ck/Myosin VIIA has a similar, crucial role in organizing actin bundles in Drosophila.
The mechanisms by which MyoVIIA promotes the formation and maintenance of actin-based protrusions are still unclear. Its N-terminus contains a head with a motor domain, which binds to actin, hydrolyzes ATP, and generates motor function (Yang et al., 2006). Its C-terminal tail contains a tandem repeat of MyTH4 and FERM domains (a motif shared by the highly conserved band 4.1 family proteins that link actin to trans-membrane proteins (reviewed in Baines et al., 2014)) separated by an SH3 domain (Kiehart et al., 2004). The tail participates in auto regulation of MyoVIIA function through intra-molecular interactions (Umeki et al., 2009; Yang et al., 2009), and facilitates transporting cargo and targeting ck/MyoVIIA to different intracellular locations (Sato et al., 2017; Yu et al., 2017). For example, C-terminal binding partners of vertebrate MyoVIIA are necessary for retinal melanosome transport (MyRIP) and for targeting proteins to lateral adhesions between stereocilia (Harmonin B, Protocadherin 15, and Sans,Todorov et al., 2001; Boëda et al., 2002; El-Amraoui et al., 2002; Adato et al., 2005; Senften et al., 2006; Etournay et al., 2007). More recent evidence suggests that both recombinant mammalian and Drosophila MyoVIIA, previously proposed to exist as dimers, are instead predominantly monomeric (Yang et al., 2009; Haithcock et al., 2011). However, this does not discount the possibility that multiple MyoVIIAs may function collectively to transport cargo or to bind to partners.
Here we employed classical and molecular genetic approaches in flies to investigate the role of ck/MyoVIIA in the ontogeny of denticles. Using an allelic series ranging from mild to complete loss-of-function alleles, we constructed organisms with reduced or absent ck/MyoVIIA protein and showed that reduced levels of ck/MyoVIIA alter the actin-filament arrays that underlie denticle structure. We generated a series of C-terminal domain-deletion constructs of GFP-ck/MyoVIIA and examined their role in protein localization and function in actin-filament organization. Lastly, we examined genetic interactions between dusky-like (dyl), a member of the ZPD family of proteins, and ck/MyoVIIA and found they interact synergistically to effect normal denticle morphology, likely through separate pathways.
Materials and Methods
Fly stocks
Drosophila melanogaster stocks and crosses were maintained using standard methods (Roberts, 1996). w1118 embryos were used as a wild type control because many of the transgenic stocks were mutant for w at the endogenous locus for white. GFP-ABD-Moe (sequences encoding GFP fused to sequences encoding the F-Actin binding domain of Moesin) were under control of the spaghetti squash promoter (Edwards et al., 1997; Kiehart et al., 2000) in the sGMCA line. Other stocks were obtained from the Bloomington Drosophila Stock Center (Bloomington, IN): a) Df(2L)PZ07130-mr9, dpov1 b1 cn1/Cyo, b) P[ovoD1–18]2La P[ovoD1–18]2Lb P[neoFRT]40A/Dp(?;2)bwD, S1 wgSp-1 Ms(2)M1 bwD/CyO, c) w, hs-FLP; Sco/CyO, d) da-GAL4III, and e) sqh-GAL4III. Flies null for the ZPD protein Zye, zyeex72, and deficiency spanning Dyl, dylΔ26, were kindly provided by Serge Plaza (Université de Toulouse, France). Chromosomes that were doubly mutant for ck (ck13) and ZPD proteins (zyeex72, dylΔ26) and had an engineered [neoFRT]40A site were generated using meiotic recombination. The ckKT9 allele of ck was kindly provided by Amy Besjovec (Duke University) and was also recombined into the [neoFRT]40A background using meiotic recombination. Flies mutant for sans245 (data not shown) were a kind gift from Christian Dahmann (University of Dresden).
ck/MyoVIIA alleles and Truncation Mutants
pUASt-GFP-ck and pUASt-GFP-ckΔhead (ck/MyoVIIA-tail) were described previously (Todi et al., 2008). hGFP-ck/MyoVIIA was cloned downstream of the ubiquitin promoter in the modified pCaSper 3Up2 RHX poly A vector (to which BamH1, Not1, Acc651 and Kpn1 enzyme cut sites were added to the multiple cloning sequence, a gift from Rick Fehon, University of Chicago). hGFP was amplified from the UAS-hGFP-ck/MyoVIIA template (Todi et al., 2005) and cloned into the EcoR1 and Not1 cut pCaSper 3Up2 RHX poly A modified vector. The cDNA coding for the ck/MyoVIIA ORF (Todi et al., 2005) was amplified with a 5’ primer that included a Not1 and 3’ primer that included a Spe1 restriction site. The Not1 and Spe1 cut ck/MyoVIIA was cloned into Not1 and Xba1 cut hGFP-pCaSper 3Up2 RHX poly A (modified) plasmid. The hGFP and ck ORF are linked by a sequence (AAG CGG CCG CCG ACG AAC ACG ACG AAC) introduced by the 5’ Not1 primer and creates the amino acids KRPPTNTTN.
UAS GFP-ck/MyoVIIA deletion alleles
The first MyTH4 FERM1,2 domains, SH3 domain in the tail and the second MyTH4 FERM1,2 domains were deleted from the UAS-GFP-ck/MyoVIIA rescue construct (Todi et al., 2005) using a SOEing PCR strategy (Horton, 1995). The fused fragment with the required domains deleted was digested with appropriate enzymes and cloned into the pUAST-GFP-ck/MyoVIIA rescue plasmid replacing the “wild type” fragment. In order to compromise MyTH4-FERM function we removed the MyTH4-FERM1,2 domains. Our expectation is that the MyTH7 domain (also called the FERM3 domain) could not function on its own.
RKAA mutant
As described in Yang et al. (2009), the constitutively active mutant GFP-ckRKAA was generated in the UAS-GFP-ck/MyoVIIA rescue construct (Todi et al., 2005) using a SOEing PCR mutagenesis strategy. The double mutation of the R2129 and K2132 amino acids to alanine abolishes the inhibitory, bent conformation of myosin. Please note that the highly conserved R2129 and K2132 amino acids were incorrectly numbered as R2140 and K2143 in (Yang et al., 2009).
Germline clones
Flies with the following genotypes were used to make germline clones: ck13[neoFRT]40A/Twist Gal4 GFP Cyo (TGC); sGMCA/TM3Sb, ck14[neoFRT]40A/TGC; sGMCA/TM6BTb, and ckPZ[neoFRT]40A/TGC;sGMCA/sGMCA (Kiehart et al., 2000; Kiehart et al., 2004). ck germline clone embryos were generated using the FLP/DFS system (Chou et al., 1993) by crossing hs-flp/y; p[ovoD1–18]2L FRT40A/CyO males to ckallele FRT40A/Twist GAL4 Cyo (TGC); sGMCA virgin females. Resulting first and second-instar larvae were heat shocked on two consecutive days for 1 hour at 37°C to induce Flp/FRT-mediated recombination as previously described (Chou and Perrimon, 1996). The resulting hs-flp/w; ckallele FRT40A/p[ovoD1–18]2L FRT40A; SGMCA/+ virgin females were then crossed to Df(2L)PZ07130-mr9, dpov1 b1 cn1/TGC males. In addition, germline clones were made with ck13FRT40A/Cyo (krupple-GAL4 UAS-GFP); dylΔ26/TM6B Sb Tb Hu dfd-eYFP, and ck13FRT40A/KGC; zyeex72/TM6B Sb Tb Hu dfd-eYFP flies. Embryos were manually sorted using a Zeiss Discovery V12 SteREO microscope (Carl Zeiss, Thornwood, NY) equipped to detect green fluorescence to distinguish homozygous mutant embryos from those carrying a fluorescently tagged balancer.
Immunolocalization and cuticle preps
10–12 hour old embryos were dechorionated in 50% bleach, fixed in 1:1 heptane: 4% paraformaldehyde/PBS, and devitellinized using 85% ethanol. PBS was made without divalent cations and used at a pH of 7.4. Embryos were incubated in blocking solution, PBT (PBS/0.1% Tx-100)+ 20% NGS (normal goat serum) for 1 hour. Primary antibodies were incubated overnight in PBT + 5% goat serum at 4°C. Antibody concentrations were as follows: guinea pig anti-Ck 1:2000 (kindly provided by D. Godt, Glowinski et al., 2014), mouse monoclonal E7 anti-β-tubulin 1:500 (DSHB), rabbit anti-Zipper 1:1000, rabbit anti-Dyl 1:300 (from Serge Plaza, Fernandes et al., 2010), mouse anti-Actin 1:3000 (Chemicon, Temecula, CA), mouse anti-Singed 1:50 (DSHB, Iowa City, IA), goat anti-Arp2 (Santa Cruz Biotechnology, Dallas, TX), and rabbit anti-GFP 1:1000 (A-11122, ThermoScientific, Waltham, MA). Secondary antibodies (1:2000) were diluted in PBT +5% goat serum and incubated with embryos at room temperature for 2–3 hours. Secondary antibodies were goat anti-rabbit Alexa 568, goat anti-mouse Alexa 488 (Molecular Probes, Eugene, OR) or goat anti-mouse Cy3 (Molecular Probes, Eugune, OR), and goat anti-guinea pig Cy2 (Sigma, St. Louis, MO). To examine embryonic and larval cuticles, 10–12 hour old embryos were dechorionated, transferred to an agar slab and allowed to develop. Embryos that failed to hatch were transferred to coverslips and mounted on a slide with 1:1 mixture of Hoyer’s media and lactic acid, weighted with 6 pennies, and baked at 65°C overnight. First instar larvae preps were prepared as described previously (Alexandre, 2008).
Imaging
Time-lapse microscopy:
Embryos were dechorionated and mounted in chambers for imaging as previously described (Kiehart et al., 2006). Images were obtained with a Zeiss Axiovert 200 M microscope (Carl Zeiss, Thornwood, NY) with a Yokogawa CSU-10 spinning disk confocal head (Yokogawa Corp., Sugarland, TX) and a Hamamatsu Orca-ER CCD camera (Hamamatsu Corp., Hamamatsu City, Japan) for time-lapse capture using a 100x/1.3 NA oil-immersion objective. For general denticle analysis, every 2–3 minutes a stack of 5–10 Z-planes covering an volume 1–3 μm in depth were acquired and z-projections were analyzed with Metamorph acquisition software (Molecular Devices, San Jose, CA).
Cuticle preps:
Samples were imaged with phase contrast optics on a Zeiss Axio Imager M2m microscope (Carl Zeiss, Thornwood, NY) through a Yokogawa CSU-10 spinning disk confocal head (Yokogawa Corp., Sugarland, TX) using μManager software (Edelstein et al., 2014) and a 100x/1.3 NA oil-immersion objective.
Immunofluorescence:
Fixed samples were imaged on a Zeiss Axio Observer LSM510 confocal microscope with LSM v4.2 acquisition software using a 100x/1.4 NA oil-immersion objective. 3D image reconstruction was done with Imaris (Bitplane Scientific Software, Zurich, Switzerland).
Scanning electron microscopy:
Newly hatched first instar larvae were fixed (4% PFA in PBS) overnight, rinsed with PBS, then distilled water and mounted on agar slab to retain hydration. Slabs were placed on EM stubs and imaged using an environmental scanning electron microscope (FEI XL30 ESEM (FEI Company, Hillsboro, OR) with Bruker XFlash 4010 EDS Detector (Bruker, Billerica, MA)).
Widefield fluorescence intensity:
Live embryos were prepared as described below and imaged on a Nikon Eclipse Ts2R microscope (Nikon Instruments, Melville, NY) with a 4X/0.13 NA objective and X-CITE 120LED Boost High-Power LED light system (Excelitas Technologies, Waltham, MA) kept at constant power and exposure across experiments. Images were obtained with a Nikon DS-QI2 CMOS camera and analyzed using Nikon Elements software.
All images were adjusted for contrast and brightness using Adobe Photoshop or Fiji. 3D reconstruction of the Z-stack of UAS-GFP-ck/MyoVIIA was denoised using Nikon Denoise and blur was removed using Nikon Clarify in Nikon Elements.
Denticle Morphology Quantification
Images of denticles were taken as stated above. They were oriented with anterior to the left and posterior to the right and cropped to a 4-inch square. Eight to twelve independent A2 denticle belts were examined for each genotype. Within this random field, denticle morphologies were separated into 7 groups (see text). Denticles were divided into rows (1–6) and values were displayed as a percentage of the number of denticles in the row across all embryos.
Fluorescence Intensity Quantification
To quantify fluorescence of GFP-tagged ck domain deletion constructs, UAS lines were crossed to daughterless-GAL4 flies. Embryos were collected in a 2-hour window and then aged for 10–12 hours. Embryos were dechorionated as described above and all genotypes were glued to a single coverslip and incubated in PBS during imaging. Two independent experiments were conducted with a total of 19–26 embryos examined for each genotype. Background fluorescence was subtracted from all values. Embryos were detected using Nikon Elements binary function, binary values were converted to ROI, and each embryo generated fluorescent intensity values from zero-16,383. Mean fluorescence intensity and a standard deviations of fluorescence intensity were calculated for each embryo. The average mean fluorescent intensities were calculated for each genotype and error bars shown are the average of the standard deviations found for each embryo within a genotype allowing us to see the deviation in intensity within each genotype.
Results
Loss of maternal and zygotic ck produces severe denticle defects
Based on the bristle and wing hair morphology defects observed in crinkled mutant escaper flies (Kiehart et al., 2004), we examined actin-based protrusions in embryos homozygous (allele/allele) and hemizygous (allele/deficiency) for an allelic series of ck/MyoVIIA mutations (strong loss of function ck13 and ck14, to weak loss of function ckPZ). These embryos displayed mild defects in denticle morphologies including occasional forked or multiple distal tips and loss of denticle hooking (compare various controls in Figure 1A–C to ck13/ck13 in Figure 1D and Table 1 for ck13-data not shown for other alleles).
Table 1.
Summary of genotypes and phenotypes of different of flies. Ooplasm protein represents whether maternal protein deposited in the eggs was wild type or mutant. Nuclear genotype explains which gene the mother and father contributed.
| Ooplasm protein | Maternal nucleus | Sperm nucleus | Phenotype | Figure Reference |
|---|---|---|---|---|
| WT | WT | WT | wild type (Control) | Figure 1B |
| WT | WT | ckDf | wild type (ckDf is recessive) | Figure 1A |
| WT | ck13 | ck13 | mild mutant (maternal protein can compensate) | Figure 1D |
| WT | ck13 | WT | wild type (paternal rescue) | Figure 1C |
| ck13 | ck13 | ckDf | severe mutant | Figure 1E, I |
| ck13 | ck13 | WT | wild type (paternal Rescue) | Figure 1H |
| ck13, (ckwt-transgene) | ck13, (ckwt-transgene) | ckDf | wild type (transgenic Rescue) | Figure 1J |
| ckKT9 | ckKT9 | ckDf | severe mutant(KT9 fails to rescue) | Figure 1K |
| wt | ckKT9 | ckKT9 | mild mutant (maternal protein can compensate) | Figure 1L |
| ck13, ubi-GFP-ck | ck13, ubi-GFP-ck | ckDf | Mild mutant (hooking defect) | Figure 1M |
ck/MyoVIIA is a maternally loaded protein (Kiehart et al., 2004) so we investigated whether the removal of the maternal contribution would enhance the denticle phenotype. The Flp-FRT/dominant female sterile technique (Chou et al., 1993) was used to generate germline clones in which only female germ cells containing two mutant copies of the gene could produce mature eggs. Such mature eggs include only negligible amounts (if any) of wild type Ck protein and have a haploid genotype that corresponds to the mutant ck allele contributed by the mother, so embryos derived from these eggs cannot express wild type Ck zygotically from the allele donated by the mother. Germline clone females were fertilized by males heterozygous for ck (ckDf/Balancer, which contains a wild type copy of ck/MyoVIIA donated by the balancer). This approach enabled us to analyze denticle development in the absence of both maternal and zygotic contributions of ck/MyoVIIA (hereafter referred to as M/Z ck; Figure 1E, F) or in the absence of just maternally contributed ck/MyoVIIA but in the presence of a wild type ck/MyoVIIA allele contributed paternally. Cuticles from embryos that lacked both maternal and zygotic ck/MyoVIIA showed severe defects in the size and shape of the denticles compared to controls (Figure 1E, F, compared to Figure 1A,Table 1, detailed below). The denticle phenotypes caused by severe ck alleles were substantially more expressive and penetrant than denticles from embryos that were homozygous or hemizgous for severe ck alleles zygotically (see above). In contrast, even in the absence of a maternal contribution of wild type Ck gene product, when the male parent contributed a wild type copy of ck/MyoVIIA, the mutant denticle phenotypes were rescued and embryos survived to adult stages of development (Figure 1H, Table 1). At least to a first approximation, full rescue of the ck phenotypes by paternally contributed ck indicates that ck function is not required for the production of normal eggs and early development of the zygote.
Two observations confirmed that the mutant phenotype in our alleles was due to a mutation in the ck gene and not at a secondary site. First, a genomic transgene of ck/MyoVIIA, included in the maternal background so that it can contribute maternally loaded ck gene product, rescued the denticle phenotypes that resulted from ck13 hemizygous germlines (compare Figure 1J, to 1I). Moreover, the genomic transgene rescued the overall lethality of the flies (data not shown), further demonstrating that the phenotype was a direct result of loss of ck/MyoVIIA function and that the ck13 chromosome carries no other lesions that affect cuticle formation. Second, germline clones were derived from an independently generated nonsense allele of ck, ckKT9FRT40A (a stop at residue Q445, Bejsovec and Chao, 2012). M/Z ckKT9(m)/ckKT9(p) hemizgyous larvae displayed a similar denticle phenotype as the ck13 allele, consistent with the phenotype being due to loss of ck/MyoVIIA (Figure 1K).
We further characterized the denticle phenotype of M/Z ck mutant embryos as follows. M/Z ck mutant denticles due to severe, nonsense mutations (e.g., ck13(m)/ckDf(p), Figure 1E) or missense mutations (e.g., ck14(m)/ckDf(p), Figure 1F) are shorter, more stout, more triangularly shaped and lack characteristic hooks when compared to denticles from controls (wild type or appropriate transheterozygotes, see Figure 1A, B, H, Table 1). Germline clones generated with a substantially weaker, hypomorphic allele, ckPZ, which has a P-element insert in 5’ UTR (Kiehart et al., 2004), and displayed a much less severe phenotype, sometimes indistinguishable from wild type (Figure 1G).
Embryos homozygous for each of the ck alleles examined displayed a range of denticle phenotypes. Occasionally mutant denticles had a split distal end with two or more tips (arrow, Figure 1E). We often saw several small denticles (arrowheads, Figure 1F) clustered around a site normally characterized by a single denticle in controls. In the two most posterior rows of denticles (rows 5 and 6), the bases of the denticle created a wide and uneven footprint not seen in other rows (asterisk, Figure 1E). Increased magnification of denticles in M/Z ck13(m)/ckDf(p) embryos using environmental scanning electron microscopy (ESEM) confirmed multiple small protrusions at the site of the denticles, suggesting that smaller bundles of actin filaments failed to coalesce into the larger protrusion (compare arrow in Figure 1N’ to control denticles in Figure 1N). These data are consistent with the small denticles observed in cuticle preps (arrowheads, Figure 1F). Despite the strong denticle phenotypes observed in M/Z ck mutant embryos, denticle fields were normal in several respects. The embryos showed normal segmentation patterns and normal distribution of denticles (6 rows of denticles in the anterior end of each segment and naked cuticle in the posterior half of each segment). Moreover, there was no significant difference in denticle number between mutant and control animals, suggesting that the nucleation of actin filaments, a first step in denticle formation, is not affected by the absence of ck (data not shown). In sum, disruption of MyoVIIA gene function alters F-actin bundle organization resulting in denticles with split tips and broad bases but does not alter epidermal pattern formation.
Ck/MyoVIIA localizes to the periphery and base of developing denticles
We used two experimental approaches to evaluate the distribution of ck/MyoVIIA in the cuticle forming epidermis of developing embryos: fixation with indirect immunofluorescence of endogenous protein and live time-lapse imaging of GFP-tagged ck/MyoVIIA. Antibody staining of 12–13 hour AEL embryos revealed ck/MyoVIIA localized along the periphery of the denticles and was enriched at the denticle base (Figure 2A, B) where it co-localized with F - actin (Figure 2A”, 2B” arrows). Analysis of Z-slices that extend from the tip to the base of the denticle (Figure 2A”’, 2B”’) and their 3D reconstruction (Figure 2C, C’) indicates that ck/MyoVIIA does not directly co-localize with actin filaments in the center of the forming denticle. In order to test whether artifacts due to fixation and staining were the reason that ck/MyoVIIA was not in the center of the growing denticle, we used live-cell imaging to examine the localization patterns of GFP-tagged ck/MyoVIIA encoded by an appropriately expressed construct composed of sequences encoding GFP fused to the N-terminus of a wild type ck cDNA transgene that can rescue the mutant phenotype in the M/Z ck13(m)/Df(p) genetic background (Figure 1M, see below). A series of confocal Z-slices through denticles expressing UAS-GFP-ck/MyoVIIA driven with a ubiquitous driver (sqh-GAL4) confirmed that GFP-ck does not localize to the center of the denticle but instead was confined to the periphery of the denticle near the plasma membrane (Figure 2D–D”, E, E’, Supplemental Movie 3). Based on this localization pattern, we hypothesize that ck/MyoVIIA functions to stabilize actin filaments along the plasma membrane interface like it does in the vertebrate system (Hasson et al., 1997; Boëda et al., 2002; Adato et al., 2005; El-Amraoui and Petit, 2005; Senften et al., 2006).
Figure 2:
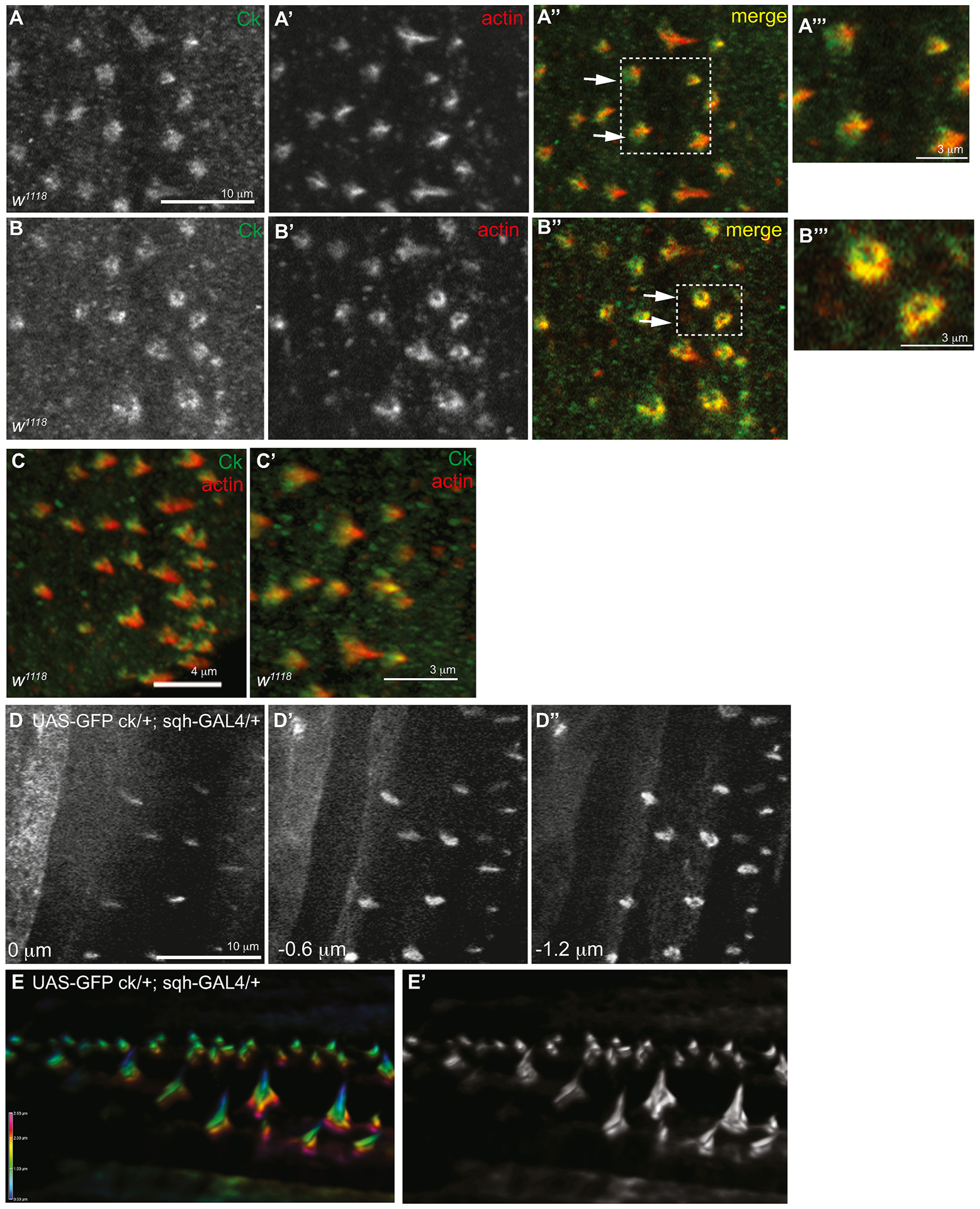
ck/MyoVIIA localizes to the periphery of denticles and not the central region of the actin protrusion. w1118 embryos were stained with antibodies to ck/MyoVIIA (green, single channel A, B) and Actin (red, single channel in A’, B’). (A) Maximum projection of 3 Z-slices. (B) Single Z-slice at the base of the denticle. A”’, B”’ are enlarged from the box in A” and B”. (C, C’) 3D reconstruction of confocal Z-stack. (D–D”) Z-series through the denticles of a late stage embryo expressing UAS-GFP-ck/MyoVIIA driven with sqh-GAL4 (UAS-GFP-ck/MyoVIIA/+; sqh-GAL4/+). Slices are 0.6 μm apart and 0 represents the apex of the denticles. (E, E’) Sideview of 3D reconstruction of Z-series of denticles expressing UAS-GFP-ck/MyoVIIA in black and white and color-coded depth scale. The scale bars in panels A–A”, B–B”, and D–D” are 10 μm. The scale bar for panel C is 4 μm and for panels A”’, B”’, and C’ is 3 μm.
Defects in actin elongation and filament organization contribute to mutant denticle shapes
To further test the role of ck/MyoVIIA in the organization of actin filaments in bundled F-actin structures, we used time-lapse approaches to evaluate the assembly of the F-actin structures that drive denticle formation. We analyzed denticles from embryos which were maternally and zygotically null for ck/MyoVIIA (maternal ck13 embryos fertilized with a male ckDf/Balancer fly: M/Z ck13(m)/ckDf(p)) because they were the most severe compared to other ck alleles tested. Indistinguishable results were seen when maternal ck13 embryos were fertilized with male ck13/Balancer flies (M/Z ck13(m)/ck13(p)) indicating that there is no difference in denticle phenotypes between the two M/Z genotypes (allele vs Df), thus supporting the notion that ck13 is functionally a genetic null. To follow F-actin dynamics, M/Z ck13(m)/ckDf(p)mutants were created in a genetic background containing the F-actin binding domain of Drosophila moesin fused to GFP (GFP-ABD-moesin previously referred to as GFP-moe, see Edwards et al., 1997; Kiehart et al., 2000). Time-lapse movies (Figure 3, and supplemental movies M1 and M2) beginning at approximately 10–12 hours AEL showed control (FRT40A(m)/ckDf(p)) embryo denticles started as condensations of F-actin at the apical surface near the posterior edge of epithelial cells, extending the indirect immunofluorescence observations previously published (Price et al., 2006). The actin protrusions elongated posteriorly over neighboring cells and the actin bundled to form a single denticle (wild type denticles are fully formed in ~ 100 min, Figure 3A–A”, C). In M/Z ck13(m)/ckDf(p) embryos, the F-actin condensations formed at the apical surface along the posterior edge of the epithelial cells in a similar manner as the controls but did not develop into persistent, elongated actin protrusions, even after 3 hours (Figure 3B–B”, C’, C”). Instead, the bases of the denticles widened with time and did not elongate, suggesting that ck/MyoVIIA may play a key role in directly drawing nascent actin filaments, or small bundles of actin filaments, together or indirectly by recruiting an F-actin bundling protein to the denticle. Occasionally, the wide bases that characterized M/Z ck mutant embryos displayed a small concentration of elongated filaments that emanated from the center of the denticle but these elongations were transient (Figure 3C’ arrows) suggesting that ck/MyoVIIA also plays a role in stabilizing forming denticles. Several denticles had multiple small bundles of F-actin projecting from the apical surface, which we surmise are responsible for the multiple distal tips phenotype we observed in cuticle preparations (Figure 3C’, 3C” solid arrowheads) and again point to a possible role for ck/MyoVIIA in drawing actin bundles together either directly or indirectly via binding partners. Ultimately, mutants failed to bundle actin into discrete protrusions resulting in denticles with multiple pointed tips (Figure 3C” arrowhead) or denticles that formed broad lamella-like structures (Figure 3C” asterisk).
Figure 3.
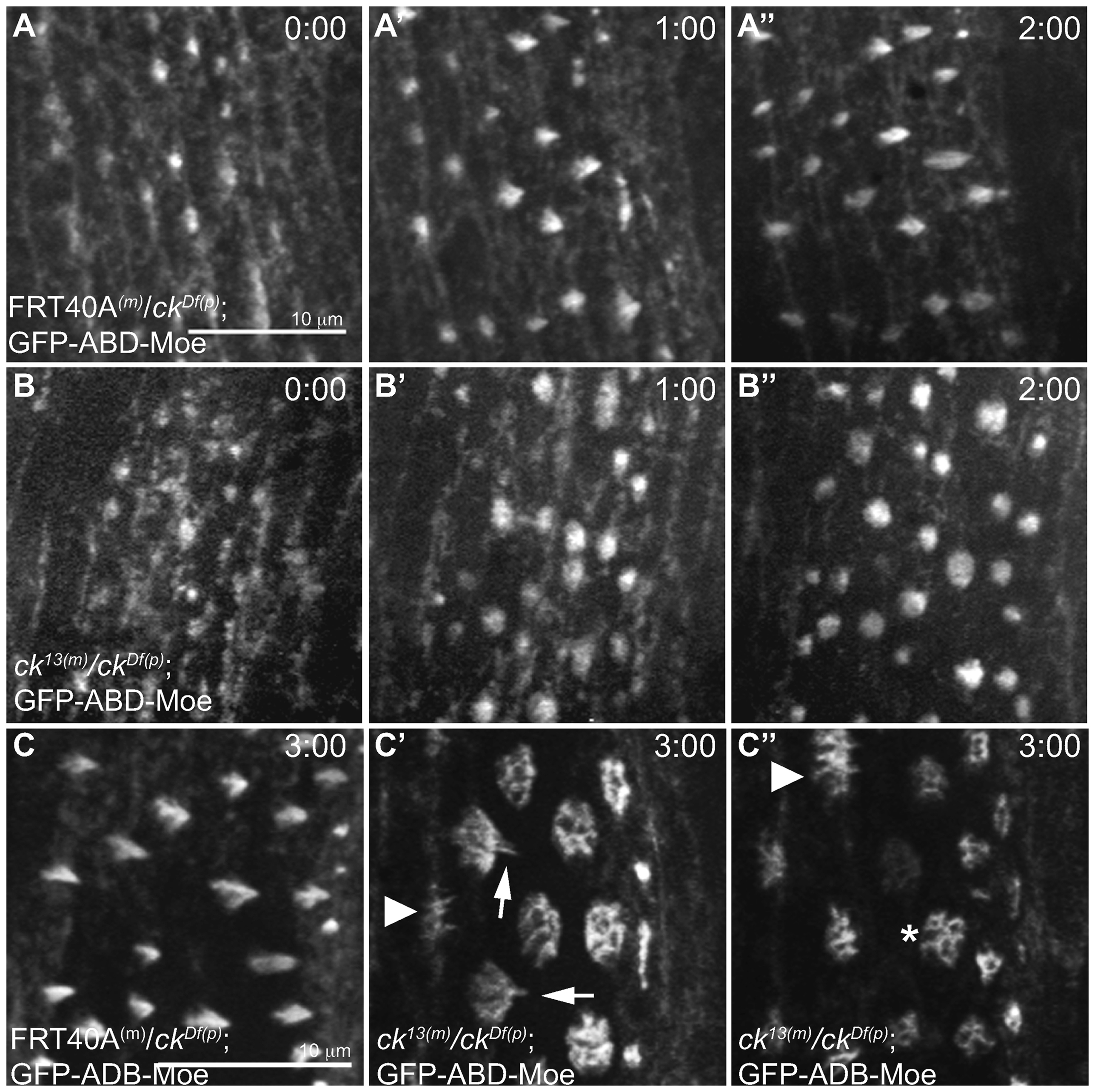
ck/MyoVIIA mutant denticles fail to coalesce actin filaments and elongate into persistent structures. (A) Time-lapse images of GFP-ABD-Moe under the control of the spaghetti squash promoter (sGMCA) in control (FRT40A(m)/ckDf(p)) or (B) ck null (M/Z ck13(m)/ckDf(p)) backgrounds. Initiation of the denticle starts with the formation of apical condensations of actin at the posterior edge of the epidermal cells (0:00 hr) and continues to elongate (1:00, 2:00 hrs). (C–C”) 3 hours after initiation of denticle formation, control denticles were fully formed and elongated (C) while ck null (M/Z ck13(m)/ckDf(p)) embryos failed to elongate (C” asterisk) and instead coalesce into a stubby splotch of short projections (arrow and arrowhead in C’, C”). The scale bars are 10μm. The scale bar in A applies to all panels in A and B, while the scale bar in C applies all panels in C.
Loss of MyoVIIA does not alter the localization of several cytoskeleton proteins
Because defects in denticle morphology appear to be related to defects in actin filament elongation and bundling, we examined the location of other actin associated proteins known to localize to the developing denticle. Previous work has shown that zipper/MyoII and ck/MyoVIIA act antagonistically in planar cell polarity and wing hair development (Franke et al., 2010) and ck/MyoVIIA synergistically interacts with non-muscle myosin II regulatory light chain protein encoded by sqh in Johnston’s Organ function (Todi et al., 2008). When examined by immunohistochemistry and confocal microscopy, zipper/MyoII heavy chain was localized normally in MZ ck13(m)/ckDf(p) denticles compared to controls (Figure 4A, B). Singed (Fascin) is known to typically bundle F-actin into ~ 5 filament diameter bundles with a maximum bundle diameter of 20 filaments (Claessens et al., 2008), supporting a role for Singed in organizing actin filaments in denticles. Staining embryos for Singed revealed that the protein is still present in the M/Z ck13(m)/ckDf(p) denticles, although due to the altered morphology of the mutant denticle the localization looks slightly different than controls (Figure 4 C, D). This suggests that ck/MyoVIIA is not needed to recruit Singed to denticles, and that Singed is involved in bunding the small actin filaments in the mutant denticles (filaments seen in Figure 3C’, C”). There is a possible indirect interaction between ck/MyoVIIA and Singed in forming larger F-actin bundles but this was not tested. Arp2, an actin nucleating protein, was also found to correctly localize at the apical subdomain of growing M/Z ck13(m)/ckDf(p) denticles (Fig. 4C–F, Price et al., 2006). Together this data shows that the cytoskeleton organizing proteins that we investigated did not require ck/MyoVIIA in order to be recruited to the developing denticles.
Figure 4.
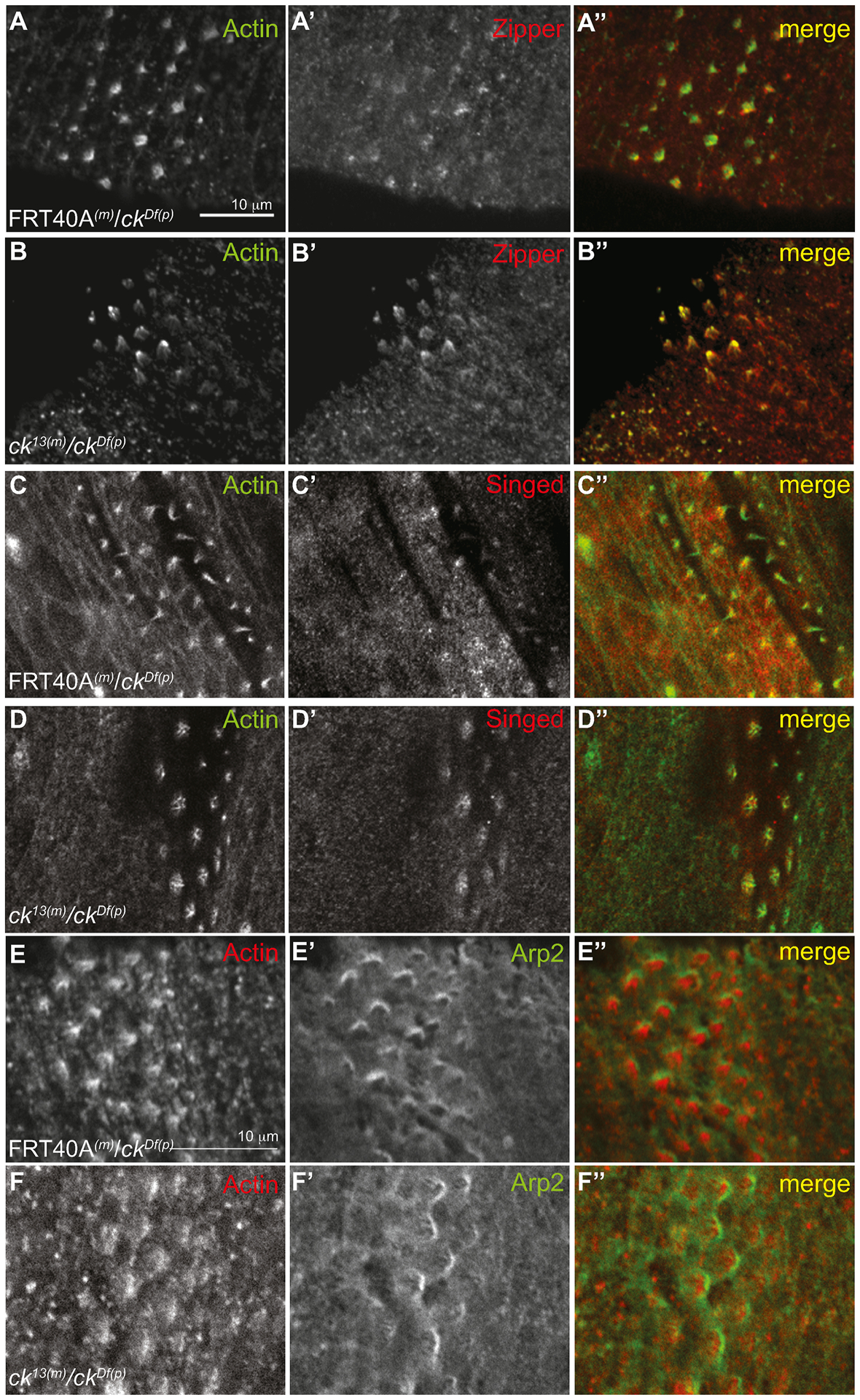
Localization of several cytoskeletal proteins is not altered in M/Z ck null embryos. Control (FRT40A(m)/ckDf(p)) and ck/MyoVIIA null (M/Z ck13(m)/ckDf(p)) embryos were stained with actin and zipper (myoII) (A, B), singed (C, D), or Arp2 (E, F) antibodies. Anterior is to the left. The scale bars are 10μm. The scale bar in A applies to panels in A–D, while the scale bar in D applies to panels in D and E.
Both MyTH4-FERM1,2 domains are necessary for denticle formation
Our rescue experiments demonstrated that expression of a full-length ck/MyoVIIA transgene can rescue proper denticle formation (Figure 1J, M). Because ck/MyoVIIA consists of several domains (Kiehart et al., 2004), each of which presumably contributes to one or more of its functions, we sought to determine which domain(s) are essential in denticle development. We generated four GFP-tagged domain deletion constructs (in order of largest protein product to smallest protein product) that removed sequences encoding either the C-terminal SH3 domain (ckΔSH3), the first MyTH4-FERM domain (ckΔM1F1), the second MyTH4-FERM domain (ckΔM2F2), or the head domain (ckΔhead) from the full length ck/MyoVIIA cDNA (ckFL; Figure 5A). In addition, we created a full-length GFP-ck/MyoVIIA construct that had two point mutations in the second MyTH7 subdomain of the second FERM domain (R2129A, K2132A, referred to as ckRKAA). In vitro, this combination of amino acid substitutions prevents the tail from binding back on the head, maintaining MyoVIIA in the open and active conformation (Figure 5B, 6G, Yang et al., 2009). The expression of these fusion proteins was driven by the bipartite GAL4-UAS system in a homozygous null germline clone background with sqh-GAL4, an essentially ubiquitous driver (M/Z ck13(m)/ck13(p); ckdeletion(m)/sqh-GAL4). Because each construct was incorporated into the genome at random locations, we determined relative expression levels of GFP fluorescence for each construct in approximately 15 hour AEL embryos. Full-length GFP-ck/MyoVIIA was the strongest expressor by almost double and the GFP-ckΔhead was the lowest expressor (Figure 5B). However, the remaining four constructs had overlapping mean fluorescent intensities and standard deviations suggesting that there was no substantial difference in their expression values (Figure 5B). In addition, all proteins exhibited similar localization to the denticle as full-length GFP-ck/MyoVIIA (Figure 5C–H).
Figure 5.
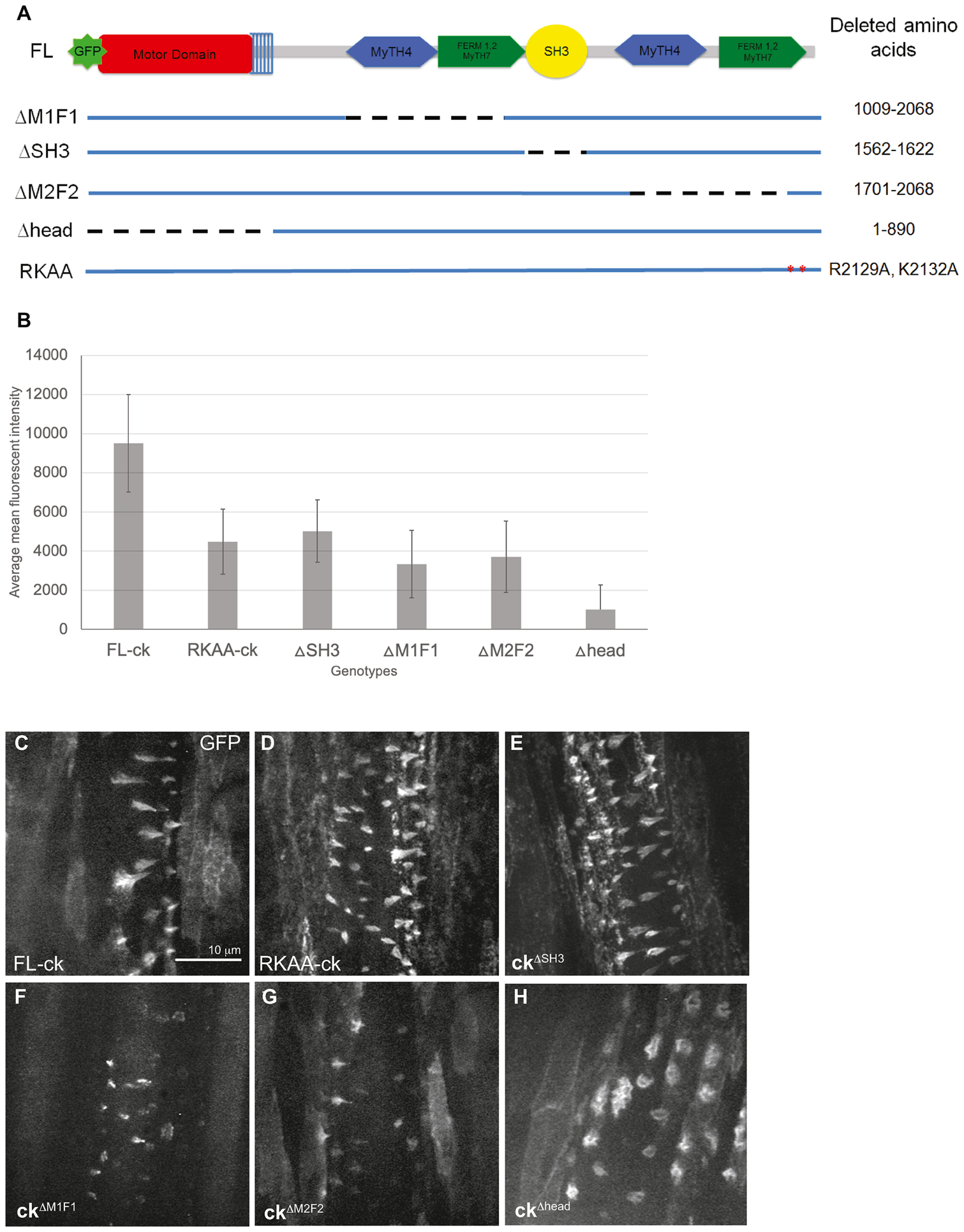
GFP-mutant ck proteins are expressed and localized to denticles. (A) Cartoon of deletion mutants with dashed lines corresponding to amino acid deletions. (B) The average mean fluorescence instenity of UAS-GFP-ck constructs in live 12–14 hours AEL embryos (n=19–26 embryos depending on genotype). Error bars are the average standard deviations of fluorescence intensity from each embryo within a genotype. GFP-ckΔhead only has the top error bar due to the lower error bar extending below zero. (C–H) Images of live embryos expressing UAS-GFP-ck mutants driven by sqh-GAL4 in the M/Z homozygous null background (M/Z ck13(m)/ck13(p); UAS-ckmutant/sqh-GAL4). The scale bar is 10 μm and can be applied to all panels.
Figure 6.
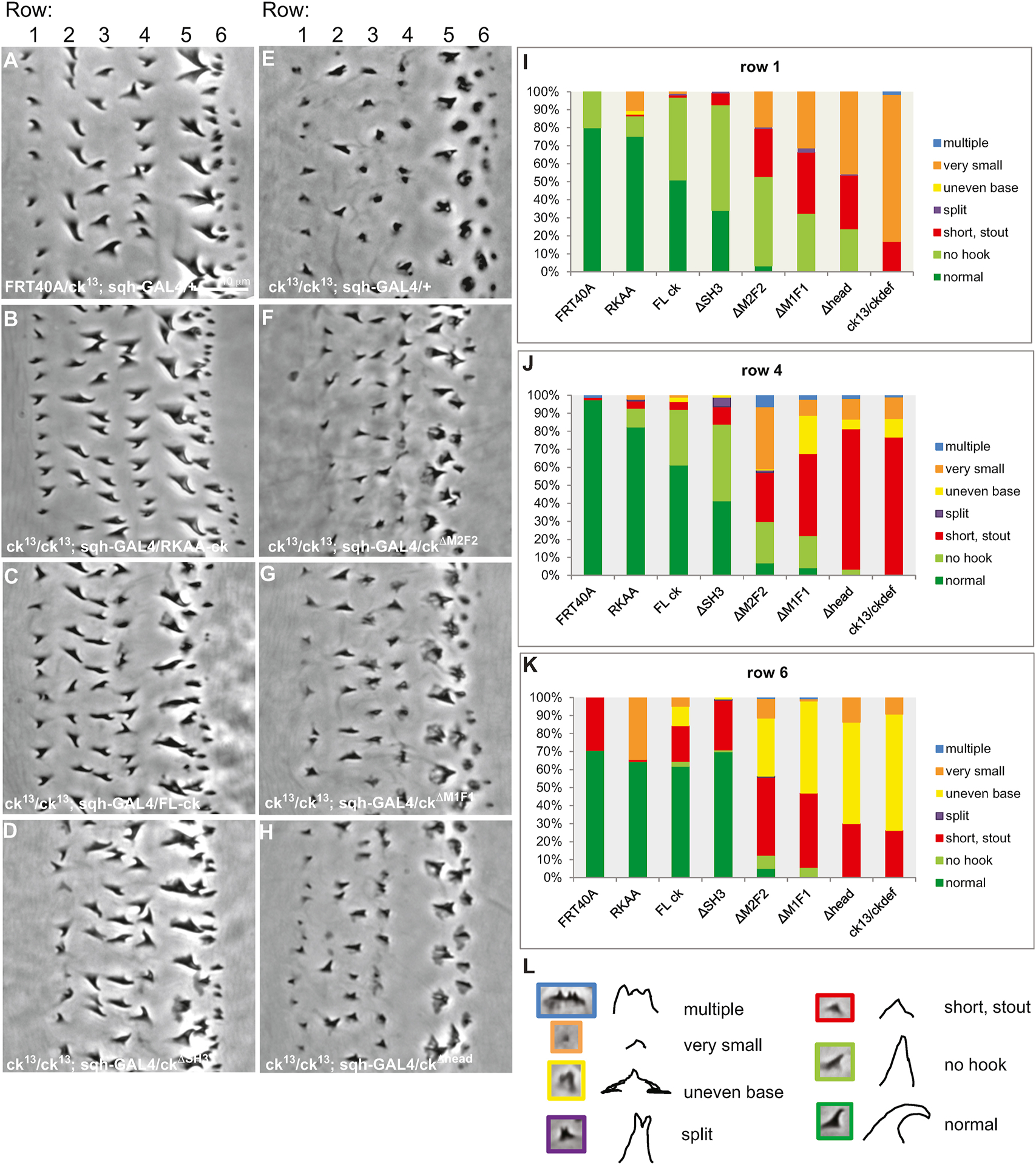
The MyTH4-FERM1,2 and head domains of ck/MyoVIIA are necessary for proper denticle formation. Cuticle preps of control embryos (A) FRT40A/ck13; sqh-GAL4/+ show normal denticle phenotypes. (B–H) Homozygous M/Z null embryos expressing sqh-driven deletion constructs (ck13(m)/ck13(p); UAS-ckmutant/sqh-GAL4) display varying levels of rescue. (B–D) UAS-GFP-RKAA-ck, UAS-GFP-FL-ck, and UAS-GFP-ckΔSH3 show significant rescue of the mutant denticle phenotypes compared to the null denticles (M/Z ck13(m)/ck13(p)) (E). (F–H) Constructs with deletions of either of the MyTH4-FERM1,2 domains or of the motor domain do not rescue the null phenotype (E). All images are of the second abdominal belt. (I–K) The degree of phenotype rescue is quantified for denticle rows 1, 4 and 6 in FRT40A/ck13; sqh-GAL4/+, ck13(m)/ck13(p); UAS-ckmutant/sqh-GAL4, or ck13(m)/ckDf(p) embryos. (L) Representative images and cartoons of each phenotype are color coded to match the charts. Scale bar is 10 μm and can be applied to all panels.
In order to assess the domain functions in final denticle morphologies, cuticle preps were made of the denticle belts in embryos expressing the constructs and we categorized the denticle phenotypes into 7 groups: 1) normal, 2) no hook (but the appropriate size), 3) short and stout, 4) split denticle, 5) multiple distal tips, 6) very small, and 7) uneven base (usually referring to the melted appearance prominent in the 5th and 6th rows, Figure 6E, L). In embryos expressing the full length MyoVIIA (GFP-ckFL) or the constitutively active mutant (GFP-ckRKAA) proteins, the denticle morphology showed complete or nearly complete rescue (Figure 6A–C, I – K). In GFP-ckΔSH3 embryos, the majority of denticles were also rescued to a normal size although there was a failure to hook in approximately half of the denticles in columns 1 and 4 (Figure 6D, I – J). Live images of the denticles expressing the GFP constructs at 13 hours AEL showed that GFP-ckFL, GFP-ckRKAA, and GFP-ckΔSH3 proteins produced normal cone-shaped denticles that elongated over the posterior edge of cells (Figure 5C–E).
In contrast, GFP-ckΔM1F1and GFP-ckΔM2F2 expressing embryos did not show complete rescue (Figure 6E–G, I – K). The majority of mutant embryos had smaller denticles lacking the characteristic hook shape. Denticle height in columns 1 and 4 was partially rescued but the uneven and wide base phenotype remained in columns 5 and 6. Live images of GFP-ckΔM1F1 and GFP-ckΔM2F2 embryos showed several denticles where some F-actin bundling and elongation occurred and a tapered structure was formed but denticles appeared smaller than controls (Figure 5F, G). In addition, cuticle preparations of GFP-ckΔhead expressing embryos did not show rescue of the mutant phenotypes (Figure 6H–K), despite GFP-ckΔhead localizing to the denticles (Figure 5H). While this observation is consistent with a role for the ck/MyoVIIA motor domain in denticle formation, because the GFP-ckΔhead protein was produced at < 50% of the other constructs, the observation cannot be unambiguously interpreted and GFP-ckΔhead was not investigated further. In addition, no significant change in the number of denticles was seen in these genotypes (data not shown). Expression of these deletion constructs in a wild type background occasionally produced mild dominant negative effects of smaller, less hooked denticles, but the phenotypes were not very penetrant affecting only a few denticles in each belt (data not shown).
Dyl and ck collaborate in denticle formation
Lastly, we examined other denticle-associated proteins that might influence the ability of ck/MyoVIIA to participate in denticle formation. The ZPD family of proteins is associated with control of the shape of the apical domains of epithelial cells (Fernandes et al., 2010). In addition, these proteins are targets of the transcription factor ovo/shavenbaby (svb) and previous work has suggested that ck/MyoVIIA acts in collaboration with certain svb targets to regulate denticle shape (Bejsovec and Chao, 2012). Here we extended these studies and examined the relationship between MyoVIIA and two of the ZPD genes which are downstream targets of svb, dyl (dusky-like) and zye. Dyl localizes to the tips of the bundled F-actin and Zye accumulates at the base of the extensions (Fernandes et al., 2010), both locations where we have observed ck/MyoVIIA localization (Figure 2). Homozygous dyl deficiency (dylΔ26/dylΔ26) denticles are small, unhooked and often have split tips (Fig. 7A, Fernandes et al., 2010), a phenotype similar to that caused by ck/MyoVIIA (M/Z) mutants (Figure 1). Homozygous ck13; dylΔ26(M/Z for ck13 only) double mutants displayed defects in denticle morphology that were substantially more severe than either mutant alone (compare Figure 7A, B to 7C)-denticles were smaller in size and in some cases nonexistent. Despite smaller denticles in the cuticle preps, phallodin staining in the double mutants showed the actin cytoskeleton resembled that of the M/Z ck13(m)/ck13(p) embryos (Figure 7G, H) indicating that the increase in severity of the phenotype may be due to defects in cuticle deposition and not in actin cytoskeleton organization.
Figure 7.
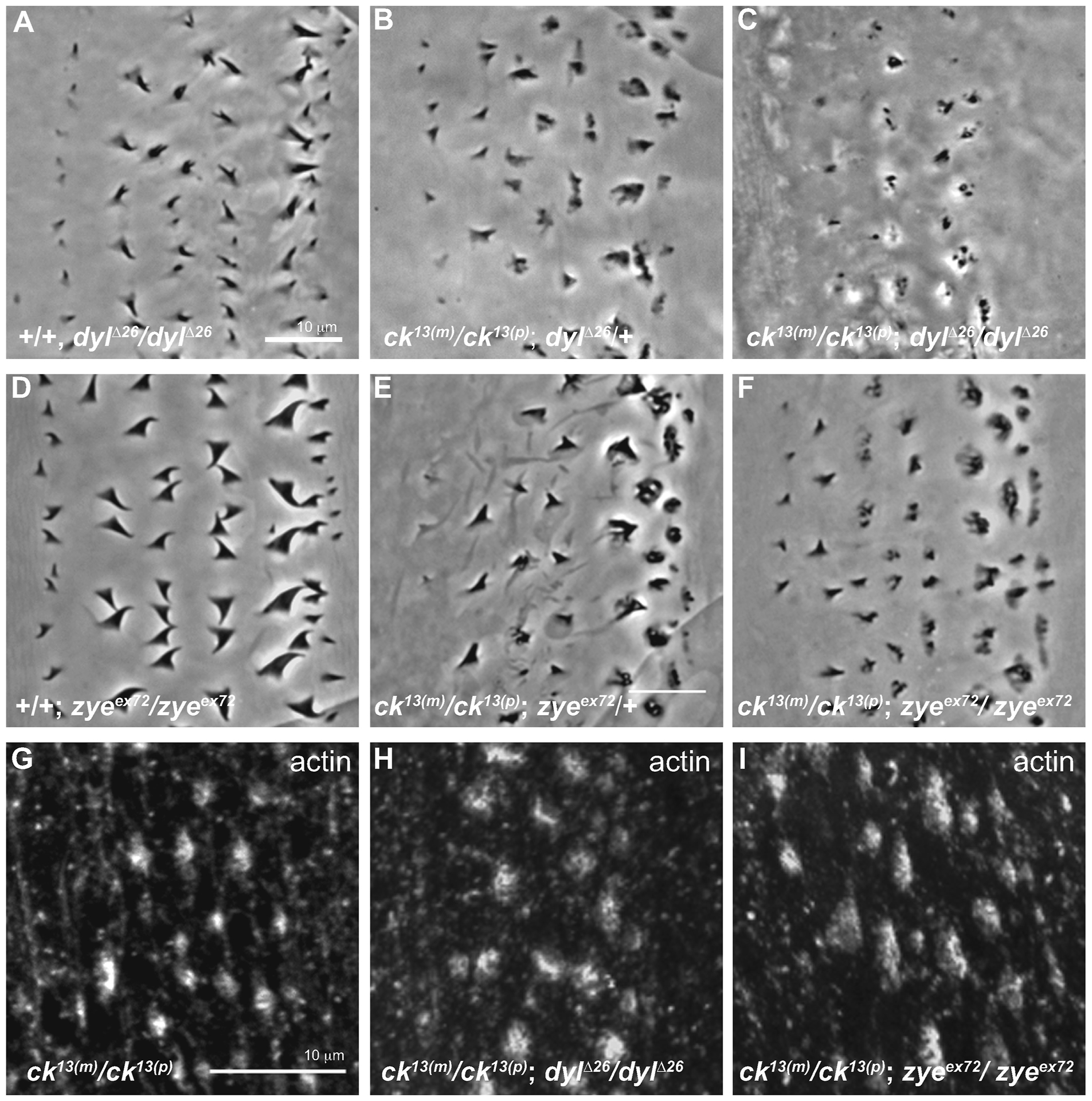
Mutations in dusky-like and ck/MyoVIIA genetically interact to produce severe defects in denticle morphology. Cuticle preps are from the second abdominal belt of embryos that failed to hatch after 48 hours. (A) Flies homozygous for the dyl deficiency (dylΔ26/dylΔ26) produce thinner denticles that were often split at the top. (B) Homozygous M/Z ck13(m)/ck13(p) mutants expressing a single copy of the deficiency for the dyl (dylΔ26/+) gene show the typical M/Z ck13(m)/ck13(p) phenotype. (C) Homozygous double mutants M/Z ck13(m)/ck13(p); dylΔ26/dylΔ26 display an additive effect on denticle morphology with very small, punctate remnants of denticles. (D) Homozygous zye mutants (zyeex72/zyeex72) produce denticles with wide bases. (E) Homozygous M/Z ck13(m)/ck13(p) mutants heterozygous for zye (zyeex72/+) show the typical M/Z ck13(m)/ck13(p) phenotype. (F) Homozygous double mutants of M/Z ck13(m)/ck13(p); zyeex72/zyeex72 do not show a different phenotype compared to D. (G–I) Actin staining of 12–16 hour old double mutants (H, I) shows no difference in actin structures compared to single ck13 homozygous mutants (G). Note: B, C, E, F, G, H, I are M/Z ck13(m)/ck13(p)mutants while A, D are zygotic mutants. Anterior is left in all figures. Each scale bar is 10 μm. The scale bar in A applies to A–F, while the scale bar in G applies to G–I.
Homozygous zye null denticles (zyeex72/zyeex72) are characterized by a thin and twisted apical region (Fig. 7D, Fernandes et al., 2010), with denticle defects that are less severe than in dylΔ26/dylΔ26 embryos. Anterior (rows 1, 2) denticles from homozygous M/Z ck13; zyeex72 double mutants showed a similar phenotype to the M/Z ck13(m)/ck13(p); zyeex72/+ (with one copy of wild type zye) mutants but posterior rows had decidedly more severe denticle defects (compare Figure 7E to Figure 7F). Indeed, homozygous M/Z ck13; zyeex72 double mutant cuticles look similar to M/Z ck13 cuticles (compare Figure 7F to Figure 6E, 1I) further indicating both that the M/Z ck13 homozygotes show a range of phenotypes and the genetic interaction between ck and zye is less severe than that between ck and dyl (compare Figure 7C to 7F). Actin staining was similar to M/Z ck13(m)/ck13(p) embryos (Figure 7I).
To determine if the loss of these genes affects the localization of each other we used indirect immunofluorescence to examine protein localization in the mutant backgrounds. Endogenous Ck localization was not altered in dylΔ26 homozygous mutants (Figure 8A, B) and despite the altered structure of the ck/MyoVIIA null denticles, Dyl still localized to the apical region of the denticles (Figure 8 C, D). Live images showed localization of ubi-GFP-ck/MyoVIIA (GFP-ck driven with a ubiquitin promoter) in zyeex72 and dylΔ26 homozygous mutants was also unaffected (Figure 8E, F) supporting the endogenous protein data in the dylΔ26 mutants (Figure 8B). These data indicate that both ck/MyoVIIA and Dyl synergistically interact and are together necessary for proper denticle formation but do not directly specify each other’s location.
Figure 8.
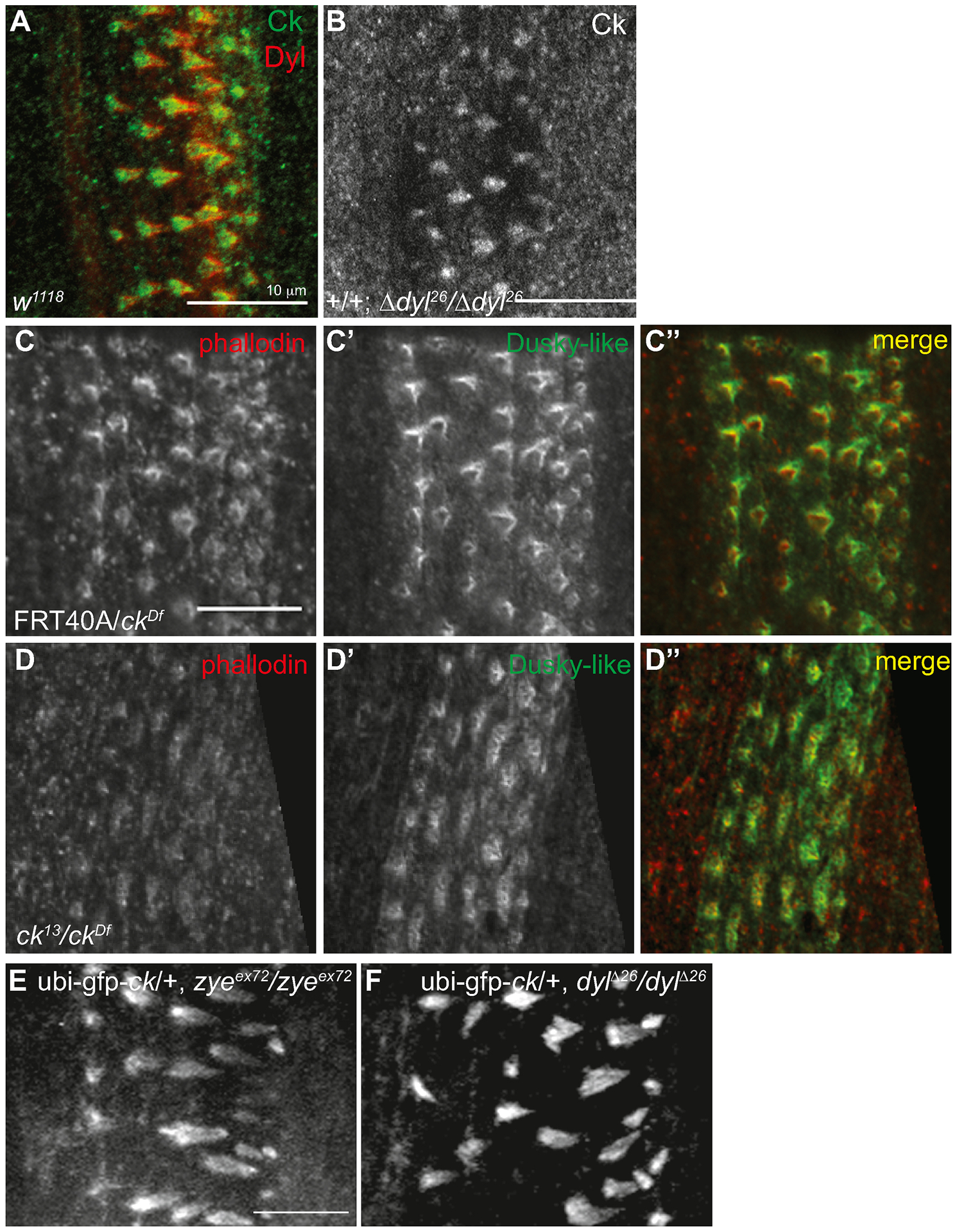
ck/MyoVIIA and Dyl are not dependent on each other for their localization. (A) w1118 embryos were stained for endogenous Ck and Dyl. (B) Homozygous zygotic dylΔ26/dylΔ26 embryos were stained for endogenous Ck. (C,D) Control (FRT40A(m)/ckDf(p)) and ck/MyoVIIA null (M/Z ck13(m)/ckDf(p)) embryos were stained with phallodin (C, D) and dusky-like antibody (C’, D’) and merge (C”, D”) and show accumulation at the tip of the actin filaments despite the altered morphology in the mutant denticles. (E, F) GFP-ck driven with the ubiquitin promoter (ubi-GFP-ck) was expressed in homozygous zygotic zyeex72/zyeex72 and dylΔ26/dylΔ26 embryos and localized correctly to the growing denticles. Anterior is left in all figures. Each scale bar is 10μm. The scale bar in C applies to all C and D panels while the scale bar in E applies to F as well.
Discussion
Here we demonstrate that loss of both maternal and zygotic ck/MyoVIIA in Drosophila melanogaster results in severe defects in denticles, actin-based protrusions found on the ventral surface of the embryo. Loss of ck/MyoVIIA did not alter the number or the location of the denticles in the epidermal cells but the actin-based denticles displayed reduced height and failed to organize into tapered actin structures suggesting a major defect in cytoskeletal organization. Our data are consistent with several, not mutually exclusive, hypotheses regarding the function of ck/MyoVIIA in denticles: 1) ck/MyoVIIA aids in bundling actin filaments into the larger structures necessary for elongation, 2) ck/MyoVIIA links the plasma membrane and actin filaments through interactions at this interface, allowing for stabilization and elongation of the actin filaments, and 3) ck/MyoVIIA transports bundling and/or stabilizing proteins to the growing denticles.
Actin polymerization can produce or transmit forces that overcome the resistance of the actin-rich cortex and the overlying plasma membrane to allow for the extension of actin-based protrusions from the surface of the cell (Peskin et al., 1993; Mogilner and Rubinstein, 2005). While individual actin filaments are flexible, the force they produce or transmit can be increased dramatically when filaments are bundled, a process which serves to increase the rigidity of the structure (Mogilner and Rubinstein, 2005). Thus, large diameter bundles are able to grow but small F-actin bundles do not generate enough force to make protrusions (Mogilner and Rubinstein, 2005). In the M/Z ck null background we saw nucleation of actin filaments (with GFP-Moe, Figure 3B) indicating that nucleation is ck/MyoVIIA independent. However, we failed to see long and thick F-actin structures. Instead, small bundles of F-actin were short lived and failed to organize into normal length protrusions (Figure 3C’, C”). The majority of M/Z ck denticles created ridges of actin that resulted in a “melted” denticle phenotype. Our interpretation is that ck/Myosin VIIA is not necessary for the nucleation of short actin filaments or for bundling small groups of filaments. Instead, ck/MyoVIIA either directly or indirectly drives coalescence of actin filaments into large filament bundles thereby increasing the stability of the protrusion and increasing the force exerted on the plasma membrane from the growing F-actin tips. This allows for tapered, full length denticles to develop. The multiple denticle phenotype and environmental scanning electron microscopy (ESEM) data in the M/Z ck null embryos supports this bundling hypothesis: M/Z ck null denticles show several small, short projections at denticle sites suggesting that individual filaments, or small bundles of filaments fail to bundle into a larger structure (Figure 1N’). ck/MyoVIIA may have innate bundling activity, but none has been reported thus far and bundling activity could not be directly tested due to the inability to purify full length ck/MyoVIIA for in-vitro actin sedimentation experiments. Another possibility is that ck/MyoVIIA may coordinate the function of known actin “bundlers”. For example, in vertebrates, Harmonin may act as an actin bundler in stereocilia cells (fly does not have an identifiable Harmonin homolog, Boëda et al., 2002). It is interesting to note that in fly, zygotic mutations in other known bundling proteins such as forked, singed (Dickinson and Thatcher, 1997), and quail (our unpublished results) show significantly milder denticle phenotypes compared to maternal and zygotic mutations in ck/MyoVIIA. Singed and quail are maternally loaded proteins (Fisher et al., 2012) and therefore, one possibility is that wild type maternal load of these proteins reduces the severity of phenotypes. This observation also suggests that either an as yet unidentified bundler is essential for making large actin filament bundles during denticle formation or that ck/MyoVIIA is responsible for facilitating bundling directly or indirectly, through a multi-protein complex, which affects protein transport or mediates direct interactions with the plasma membrane.
Indeed, our data suggest that ck/MyoVIIA may be required for anchoring membrane-bound elements to the actin bundle, similar to its role in mammalian stereocilia (Peskin et al., 1993; Kros et al., 2002; El-Amraoui and Petit, 2005; Mogilner and Rubinstein, 2005). Recall that ck/MyoVIIA is localized along the length of the denticle but is absent from the F-actin at the center of the actin bundle in the forming denticle (Figure 2). Thus its localization is similar to vertebrate MyoVIIA, which is localized around the periphery of stereocilia (at the actin-plasma membrane interface) and microvilli (Hasson et al., 1997; Grati and Kachar, 2011; Rzadzinska et al., 2004). In addition, the FERM domains of the vertebrate MyoVIIA tail can bind to integral membrane proteins either directly or indirectly making it plausible that ck/MyoVIIA may link the plasma membrane to the actin bundle (Küssel-Andermann et al., 2000; Boëda et al., 2002; Bahloul et al., 2010; Liu et al., 2014; Yu et al., 2017). Further, transient state kinetic analysis of the purified Drosophila MyoVIIA motor domain alone suggests that it can exert and hold tension on actin filaments, consistent with a role for MyoVIIA protein in tethering the plasma membrane to the denticles’ actin-rich cytoskeleton (Yang et al., 2006).
Myosin VIIA is believed to be targeted to intracellular locations by actin-binding through the motor domain and by binding of the C-terminal tail domains to specific protein binding partners. Work in vertebrates demonstrate that MyosinVIIA’s localization at the tip link of the stereocilia is via interactions with Harmonin (via the 2nd MyTH4-FERM domain) and Sans (strongly, via the first MyTH4-FERM domain and more weakly with the2nd MyTH4-FERM domain), cadherin 23, and protocadherin15 (unspecified domains, Boëda et al., 2002; Adato et al., 2005; Senften et al., 2006; Grati and Kachar, 2011). Indeed, we saw the tail-only construct (ckΔhead) correctly localized to forming denticles supporting that the C-terminal domain alone is sufficient to recruit ck/MyoVIIA to cellular locations via it’s interactors. However, the low expression level (less than half of other constructs) of the ckΔhead construct makes unambiguous interpretation of these data impossible-the failure to rescue the phenotype could be due to low expression levels, the lack of the motor domain, or a combination of both. In addition, loss of either of the MyTH4-FERM domains of ck/MyoVIIA inhibits normal denticle formation (Figure 6F, G) but allows for some organization of the cytoskeleton (Figure 5F, G). This supports the MyTH4-FERM domains play a similar role in Drosophila as they do in vertebrates. Because some organization of the actin cytoskeleton occurs (Figure 5F, G) the remaining domains provide partial MyoVIIA function probably through binding of a subset of interacting proteins or binding to the plasma membrane via the FERM domains.
We used a candidate approach to try to identify potential protein interactors with ck/MyoVIIA through localization studies of actin-binding proteins in control and M/Z ck embryos. Arp2, Singed, and Zipper remained in M/Z ck deficient denticles indicating that their recruitment to the forming denticle is not dependent on ck/MyoVIIA. Although there is no known fly ortholog for Usher Type 1 protein Harmonin or Cadherin 23, the Sans and Protocadherin 15 proteins have fly orthologs (Sans and Cad99C; Schlichting et al., 2006; Demontis and Dahmann, 2009). Cad99C binds to ck/MyoVIIA in oocytes (Glowinski et al., 2014) however, homozygous mutant Cad99C oocytes are not viable (D’Alterio et al., 2005) and so the role of Cad99C in denticle formation is not easily tested. We have examined loss of Sans (Demontis and Dahmann, 2009) in denticle formation and found that there was mild to no phenotype (Sarah LaCoppola and Jennifer Sallee, unpublished observation) in homozygous null embryos, suggesting no role for Sans in the formation of these actin-based protrusions. Further research is needed to identify potential genetic interactors as well as protein binding partners to provide more insight into the mechanics behind how MyoVIIA regulates the actin cytoskeleton in denticles. Advances in both super resolution microscopy and biochemical purification will hopefully aid in these pursuits.
Data has shown that ck/MyoVIIA genetically interacts with the svb target Miniature in a wingless background and ck/MyoVIIA may help distribute products of the svb target genes to aid in the final morphology of denticles (Bejsovec and Chao, 2012). Miniature is a member of the family of single pass transmembrane ZPD proteins and links extracellular matrix proteins to the plasma membrane (Fernandes et al., 2010). Therefore, we examined other members of the ZPD family, which are also targets of svb, and found that dyl genetically interacts with ck/MyoVIIA in our maternal null background. Our data suggest that these two proteins are working in two separate yet converging pathways. Previous work on fly bristles and wing hairs (Nagaraj and Adler, 2012; Adler et al., 2013) shows that knockdown of Dyl does not affect the growth of those actin-based protrusions but instead perturbs chitin deposition. Our data show that in homozygous dyl embryos the actin staining of denticles displays a normal phenotype (data not shown) and in embryos doubly homozygous for ck and dyl, actin staining demonstrates defects in denticle morphology which phenocopy defects due to mutations in ck/MyoVIIA alone (Figure 7G, H). Together this suggests that Dyl also affects denticle morphology through its role in chitin deposition and not by affecting the actin cytoskeleton. From these data, we propose a model where ck/MyoVIIA may link actin to the plasma membrane via its FERM domains and unidentified binding partners, while Dyl anchors the cuticle and membrane to F-actin via a complex associated with the membrane (Fernandes et al., 2010). If correct, this would explain why when both Dyl and ck/MyoVIIA are lost, actin filaments are shorter, fail to form large bundles, and the cuticle is improperly linked to the membrane and the cytoskeleton. The consequence is extremely small and punctate denticles. This model suggests that key, as yet un-identified proteins function to bridge these two pathways.
In summary, loss of M/Z ck/MyoVIIA disrupts the normal activity of actin cytoskeletal architecture that forms the basis of denticle morphology. ck/MyoVIIA likely forms a key complex(es) with other proteins and it is loss of this complex(es) that disrupts denticle morphogenesis. Studying the formation of denticles, instead of wing hairs and bristles, will allow researchers to examine the in vivo function of actin-based protrusion genes where mutants do not survive to adulthood. This model system will aid in the unraveling of the complicated networks of proteins that remodel these embryonic epithelial cells, will generate further understanding of the players involved in the formation of actin-based protrusions in general, and will inform future work on deafness and/or microvillus function in disease.
Supplementary Material
Highlights:
Loss of maternal and zygotic ck/MyoVIIA disrupts denticle morphology
ck/MyoVIIA mutant denticles fail to elongate and have multiple apices
MyTH4-FERM domains necessary for actin-based protrusion formation
Loss of ck/MyoVIIA and Dyl results in severe abnormalities in denticle morphology
Acknowledgements
We thank the DPK lab for thoughtful discussions and reading of the manuscript, and S. Plaza, C. Dahman, A. Bejsovec, and D. Godt for reagents and flies. We appreciate Chandreyee Mitra’s help with the analysis of the fluorescent intensity data, Sarah LaCoppola’s work characterizing the sans245 mutant embryos, and Michael Kerber’s help with 3D image rendering.
Funding
Funded by the National Institutes of Health [RO1 NIDCD007673, R01 GM033830 and R35 GM127059 to DPK and NRSA F32 GM093592 to JLS]
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing Interests
Authors declare no competing financial interests.
References
- Adato A, Michel V, Kikkawa Y, Reiners J, Alagramam KN, Weil D, Yonekawa H, Wolfrum U, El-Amraoui A and Petit C (2005). Interactions in the network of Usher syndrome type 1 proteins. Hum. Mol. Genet 14, 347–356. [DOI] [PubMed] [Google Scholar]
- Adler PN, Sobala LF, Thom D and Nagaraj R (2013). dusky-like is required to maintain the integrity and planar cell polarity of hairs during the development of the Drosophila wing. Developmental Biology 379, 76–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandre C (2008). Cuticle Preparation of Drosophila Embryos and Larvae In Drosophila (ed. Dahmann C), pp. 197–205: Humana Press. [DOI] [PubMed] [Google Scholar]
- Bahloul A, Michel V, Hardelin J-P, Nouaille S, Hoos S, Houdusse A, England P and Petit C (2010). Cadherin-23, myosin VIIa and harmonin, encoded by Usher syndrome type I genes, form a ternary complex and interact with membrane phospholipids. Human Molecular Genetics 19, 3557–3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baines A, Lu H and Bennett P (2014). The Protein 4.1 family: hub proteins in animals for organizing membrane proteins. Biochim Biophys Acta. 1838, 605–619. [DOI] [PubMed] [Google Scholar]
- Bejsovec A and Chao AT (2012). crinkled reveals a new role for Wingless signaling in Drosophila denticle formation. Development 139, 690–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejsovec A and Wieschaus E (1993). Segment polarity gene interactions modulate epidermal patterning in Drosophila embryos. Development 119, 501–517. [DOI] [PubMed] [Google Scholar]
- Boëda B, El-Amraoui A, Bahloul A, Goodyear R, Daviet L, Blanchard S, Perfettini I, Fath K, Shorte S, Reiners J, et al. (2002). Myosin VIIa, harmonin and cadherin 23, three Usher I gene products that cooperate to shape the sensory hair cell bundle. EMBO Journal 21, 6689–6699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanut-Delalande H, Fernandes I, Roch F, Payre F and Plaza S (2006). Shavenbaby Couples Patterning to Epidermal Cell Shape Control. PLoS Biol 4, e290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou TB, Noll E and Perrimon N (1993). Autosomal P[ovoD1] dominant female-sterile insertions in Drosophila and their use in generating germ-line chimeras. Development 119, 1359–1369. [DOI] [PubMed] [Google Scholar]
- Chou TB and Perrimon N (1996). The Autosomal FLP-DFS Technique for Generating Germline Mosaics in Drosophila melanogaster. Genetics 144, 1673–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claessens M, Semmrich C, Ramos L and Bausch A (2008). Helical twist controls the thickness of F-actin bundles. . Proceedings of the National Academy of Sciences 105, 8819–8822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Alterio C, Tran DDD, Yeung MWYA, Hwang MSH, Li MA, Arana CJ, Mulligan VK, Kubesh M, Sharma P, Chase M, et al. (2005). Drosophila melanogaster Cad99C, the orthologue of human Usher cadherin PCDH15, regulates the length of microvilli. J. Cell Biol 171, 549–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demontis F and Dahmann C (2009). Characterization of the Drosophila Ortholog of the Human Usher Syndrome Type 1G Protein Sans. PLoS ONE 4, e4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRosier DJ and Tilney LG (2000). F-actin bundles are derivatives of microvilli:What does this tell us about how bundles might form? J Cell Biol 148, 1–6. [PMC free article] [PubMed] [Google Scholar]
- Dickinson WJ and Thatcher JW (1997). Morphogenesis of Denticles and Hairs in Drosophila Embryos: Involement of Actin-Associated Proteins that Also Affect Adult Structures. Cell Motility and the Cytoskeleton 38, 9–21. [DOI] [PubMed] [Google Scholar]
- Dilks SA and DiNardo S (2010). Non-cell-autonomous control of denticle diversity in the Drosophila embryo. Development 137, 1395–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiNardo S, Heemskerk J, Dougan S and O’Farrell PH (1994). The making of a maggot: patterning the Drosophila embryonic epidermis. Curr Opin Genet Dev 4, 529–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelstein AD, Tsuchida MA, Amodai N, Pinkard H, Vale RD and Stuurman N (2014). Advanced Methods of microscope control using microManager software. Journal of Biological Methods 1, e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards KA, Demsky M, Montague RA, Weymouth N and Kiehart DP (1997). GFP-Moesin Illuminates Actin Cytoskeleton Dynamics in Living Tissue and Demonstrates Cell Shape Changes during Morphogenesis inDrosophila. Developmental Biology 191, 103–117. [DOI] [PubMed] [Google Scholar]
- El-Amraoui A and Petit C (2005). Usher I syndrome: unravelling the mechanisms that underlie the cohesion of the growing hair bundle in inner ear sensory cells. J Cell Sci 118, 4593–4603. [DOI] [PubMed] [Google Scholar]
- El-Amraoui A, Schonn J, Küssel-Andermann P, Blanchard S, Desnos C, Henry J, Wolfrum U, Darchen F and Petit C (2002). MyRIP, a novel Rab effector, enables myosin VIIa recruitment to retinal melanosomes. EMBO reports 3, 463–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etournay R, Zwaenepoel I, Perfettini I, Legrain P, Petit C and El-Amraoui A (2007). Shroom2, a myosin-VIIa- and actin-binding protein, directly interacts with ZO-1 at tight junctions. J Cell Sci 120, 2838–2850. [DOI] [PubMed] [Google Scholar]
- Fernandes I, Chanut-Delalande H, Ferrer P, Latapie Y, Waltzer L, Affolter M, Payre F and Plaza S (2010). Zona Pellucida Domain Proteins Remodel the Apical Compartment for Localized Cell Shape Changes. Developmental cell 18, 64–76. [DOI] [PubMed] [Google Scholar]
- Fisher B, Weiszmann R, Frise E, Hammonds A, Tomancak P, Beaton A, Berman B, Quan E, Shu S, Lewis S, et al. (2012). BDGP insitu homepage.
- Franke JD, Montague RA and Kiehart DP (2010). Nonmuscle myosin II is required for cell proliferation, cell sheet adhesion and wing hair morphology during wing morphogenesis. Developmental Biology 345, 117–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson F, Walsh J, Mburu P, Varela A, Brown K, Antonio M, Beisel K, Steel K and Brown S (1995). A type VII myosin encoded by the mouse deafness gene shaker-1. Nature 374, 62–64. [DOI] [PubMed] [Google Scholar]
- Glowinski C, Liu R-HS, Chen X, Darabie A and Godt D (2014). Myosin VIIA regulates microvillus morphogenesis and interacts with cadherin Cad99C in Drosophila oogenesis. Journal of Cell Science 127, 4821–4832. [DOI] [PubMed] [Google Scholar]
- Grati M. h. and Kachar B (2011). Myosin VIIa and sans localization at stereocilia upper tip-link density implicates these Usher syndrome proteins in mechanotransduction. Proceedings of the National Academy of Sciences 108, 11476–11481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haithcock J, Billington N, Choi K, Fordham J, Sellers JR, Stafford WF, White H and Forgacs E (2011). The Kinetic Mechanism of Mouse Myosin VIIA. Journal of Biological Chemistry 286, 8819–8828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasson T, Gillespie PG, Garcia JA, MacDonald RB, Zhao Y. d., Yee AG, Mooseker MS and Corey DP (1997). Unconventional Myosins in Inner-Ear Sensory Epithelia. J. Cell Biol 137, 1287–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatini V and DiNardo S (2001). Divide and conquer: pattern formation in Drosophila embryonic epidermis. Trends in Genetics 17, 574–579. [DOI] [PubMed] [Google Scholar]
- Horton R (1995). PCR-mediated Recombination and Mutagenesis. SOEing Together Tailor-made Genes. Molecular Biotechnology 3, 93–99. [DOI] [PubMed] [Google Scholar]
- Kiehart D, Franke J, Chee M, Montague R, Chen T, Roote J and Ashburner M (2004). Drosophila crinkled, mutations of which disrupt morphogenesis and cause lethality, encodes fly myosin VIIA. Genetics 168, 1337–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiehart DP, Galbraith CG, Edwards KA, Rickoll WL and Montague RA (2000). Multiple Forces Contribute to Cell Sheet Morphogenesis for Dorsal Closure in Drosophila. J Cell Biol 149, 471–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiehart DP, Tokutake Y, Chang MS, Huston MS, Wiemann JM, Peralta XG, Toyama Y, Wells AR, Rodriguez A and Edwards GS (2006). Ultraviolet Laser Microbeam for Dissections of Drosophila Embryos In Cell Biology: A Laboratory Handbook. (ed. Cells JE), pp. 87–103: Academic Press. [Google Scholar]
- Kros C, Marcotti W, van Netten S, Self T, Libby R, Brown S, Richardson G and Steel K (2002). Reduced climbing and increased slipping adaptation in cochlear hair cells of mice with Myo7a mutations. Nat Neurosci 5, 41–47. [DOI] [PubMed] [Google Scholar]
- Küssel-Andermann P, El-Amraoui A, Safieddine S, Nouaille S, Perfettini I, Lecuit M, Cossart P, Wolfrum U and Petit C (2000). Vezatin, a novel transmembrane protein, bridges myosin VIIA to the cadherin-catenins complex. EMBO Journal 19, 6020–6029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Walsh J, Mburu P, Kendrick-Jones J, Cope M, Steel K and Brown S (1997a). Mutations in the myosin VIIA gene cause non-syndromic recessive deafness. Nature Genetics 16, 188–190. [DOI] [PubMed] [Google Scholar]
- Liu X. z., Newton VE, Steel KP and Brown SDM (1997b). Identification of a new mutation of the myosin VII head region in Usher syndrome type 1. Human Mutation 10, 168–170. [DOI] [PubMed] [Google Scholar]
- Liu Y, Guan L, Zhan J, Lu D, Wan J and Zhang H (2014). FERM domain-containing unconventional myosin VIIA interacts with integrin β5 subunit and regulates αvβ5-mediated cell adhesion and migration. FEBS Letters 588, 2859–2866. [DOI] [PubMed] [Google Scholar]
- Mahajan-Miklos S and Cooley L (1994). The villin-like protein encoded by the Drosophila quail gene is required for actin bundle assembly during oogenesis. Cell 78, 291–301. [DOI] [PubMed] [Google Scholar]
- Martinez Arias A (1993). Development and patterning of the larval epidermis of Drosophila. The development of Drosophila melanogaster 1, 517–608. [Google Scholar]
- Mogilner A and Rubinstein B (2005). The Physics of Filopodial Protrusion. Biophysical Journal 89, 782–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moussian B, Seifarth C, Müller U, Berger J and Schwarz H (2006). Cuticle differentiation during Drosophila embryogenesis. Arthropod Structure & Development 35, 137–152. [DOI] [PubMed] [Google Scholar]
- Nagaraj R and Adler PN (2012). Dusky-like functions as a Rab11 effector for the deposition of cuticle during Drosophila bristle development. Development 139, 906–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusslein-Volhard C and Wieschaus E (1980). Mutations affecting segment number and polarity in Drosophila. Nature 287, 795–801. [DOI] [PubMed] [Google Scholar]
- Orme MH, Liccardi G, Moderau N, Feltham R, Wicky-John S, Tenev T, Aram L, Wilson R, Bianchi K, Morris O, et al. (2016). The Unconventional Myosin CRINKLED and Its Mammalian Orthologue Myo7A Regulates Caspases in Their Signalling Roles. Nature Communication 7, 10972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payre F (2004). Genetic control of epidermis differentiation in Drosophila. The International Journal of Developmental Biology 48, 207–215. [PubMed] [Google Scholar]
- Peskin CS, Odell GM and Oster GF (1993). Cellular motions and thermal fluctuations: the Brownian ratchet. Biophysical Journal 65, 316–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaza S, Chanut-Delalande H, Fernandes I, Wassarman PM and Payre F (2010). From A to Z: apical structures and zona pellucida-domain proteins. Trends in Cell Biology 20, 524–532. [DOI] [PubMed] [Google Scholar]
- Pollard TD and Cooper JA (2009). Actin, a Central Player in Cell Shape and Movement. Science 326, 1208–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MH, Roberts DM, McCartney BM, Jezuit E and Peifer M (2006). Cytoskeletal dynamics and cell signaling during planar polarity establishment in the Drosophila embryonic denticle. Journal of Cell Science 119, 403–415. [DOI] [PubMed] [Google Scholar]
- Roberts DB (1996). Basic Drosophila care and techniques In Drosophila a practical approach (ed. Roberts DB), pp. 1–38. Oxford: IRL Press. [Google Scholar]
- Sato O, Komatsu S, Sakai T, Tsukasaki Y, Tanaka R, Mizutani T, Watanabe TM, Ikebe R and Ikebe M (2017). Human Myosin VIIa is a Very Slow Progessive Motor Protein on Various Cellular Actin Structures. J Biol Chem 292, 10950–10960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaks M, Giannone G and Rottner K (2019). Actin Dynamics in Cell Migration. Essays in Biochemistry 63, 483–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlichting K, Wilsch-Brauninger M, Demontis F and Dahmann C (2006). Cadherin Cad99C is required for normal microvilli morphology in Drosophila follicle cells. J Cell Sci 119, 1184–1195. [DOI] [PubMed] [Google Scholar]
- Self T, Mahony M, Fleming J, Walsh J, Brown SD and Steel KP (1998). Shaker-1 mutations reveal roles for myosin VIIA in both development and function of cochlear hair cells. Development 125, 557–566. [DOI] [PubMed] [Google Scholar]
- Senften M, Schwander M, Kazmierczak P, Lillo C, Shin J-B, Hasson T, Geleoc GSG, Gillespie PG, Williams D, Holt JR, et al. (2006). Physical and Functional Interaction between Protocadherin 15 and Myosin VIIa in Mechanosensory Hair Cells. J. Neurosci 26, 2060–2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh V (2012). Analysis of crinkled Function in Drosophila melanogaster Hair and Bristle Morphogenesis. Dissertation Duke University, Print. [Google Scholar]
- Spencer AK, Schaumberg AJ and Zallen JA (2017). Scaling of Cytoskeletal Organization with Cell Size in Drosophila Molecular Biology of the Cell 28, 1519–1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang DD and Gerlach BD (2017). The Roles and Regulation of the Actin Cytoskeleton, Intermediate Filaments and Microtubules in Smooth Muscle Cell Migration. Respiratory Research 18, 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilney LG, Connelly PS and Guild GM (1996). F-actin bundles in Drosophila bristles are assembled from modules composed of short filaments. J Cell Biol 135, 1291–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilney LG, Connelly PS, Vranich KA, Shaw MK and Guild GM (2000a). Actin filaments and microtubules play different roles during bristle elongation in Drosophila. Journal of Cell Science 113, 1255–1265. [DOI] [PubMed] [Google Scholar]
- Tilney LG, Connelly PS, Vranich KA, Shaw MK and Guild GM (2000b). Regulation of actin filaments cross-linking and bundle shape in Drosophila bristles. J Cell Biol 148, 87–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todi S, Sivan-Loukianova E, Jacobs J, Kiehart D and Eberl D (2008). Myosin VIIA, important for human auditory function, is necessary for Drosophila auditory organ development. PLoS ONE 3, 2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todi SV, Franke JD, Kiehart DP and Eberl DF (2005). Myosin VIIA Defects, which Underlie the Usher 1B Syndrome in Humans, Lead to Deafness in Drosophila. 15, 862–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todorov PT, Hardisty RE and Brown SD (2001). Myosin VIIA is specifically associated with calmodulin and microtubule-associated protein-2B (MAP-2B). Biochem. J 354, 267–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner CM and Adler PN (1998). Distinct roles for the actin and microtubule cytoskeletons in the morphogenesis of epidermal hairs during wing development in Drosophila. Mech Dev 70, 181–192. [DOI] [PubMed] [Google Scholar]
- Umeki N, Jung HS, Watanabe S, Sakai T, Li X. d., Ikebe R, Craig R and Ikebe M (2009). The tail binds to the head-neck domain, inhibiting ATPase activity of myosin VIIA. Proceedings of the National Academy of Sciences 106, 8483–8488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weil D, Blanchard S, Kaplan J, Guilford P, Gibson F, Walsh J, Mburu P, Varela A, Levilliers J, Weston MD, et al. (1995). Defective myosin VIIA gene responsible for Usher syndrome type 1B. Nature 374, 60–61. [DOI] [PubMed] [Google Scholar]
- Weil D, Küssel P, Blanchard S, Lévy G, Levi-Acobas F, Drira M, Ayadi H and Petit C (1997). The autosomal recessive isolated deafness, DFNB2, and the Usher 1B syndrome are allelic defects of the myosin-VIIA gene. Nature Genetics 16, 191–193. [DOI] [PubMed] [Google Scholar]
- Wolfrum U, Liu X, Schmitt A, Udovichenko IP and Williams DS (1998). Myosin VIIa as a common component of cilia and microvilli. Cell Motility and the Cytoskeleton 40, 261–271. [DOI] [PubMed] [Google Scholar]
- Wulfkuhle JD, Petersen NS and Otto JJ (1998). Changes in the F-actin cytoskeleton during neurosensory bristle development in Drosophila: The role of singed and forked proteins. Cell Motility and the Cytoskeleton 40, 119–132. [DOI] [PubMed] [Google Scholar]
- Yang Y, Baboolal TG, Siththanandan V, Chen M, Walker ML, Knight PJ, Peckham M and Sellers JR (2009). A FERM domain autoregulates Drosophila myosin 7a activity. Proceedings of the National Academy of Sciences 106, 4189–4194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Kovacs M, Sakamoto T, Zhang F, Kiehart DP and Sellers JR (2006). Dimerized Drosophila myosin VIIa: A processive motor. 103, 5746–5751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu I-M, Planelles-Herrero VJ, Sourigues Y, Moussaoui H, Sirkia H, Kikuti C, Stroebel D, Titus MA and Houdusse A (2017). Myosin 7 and Its Adaptors Link Cadherins to Actin. Nature Communication 8, 15864. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


