Abstract
Glia shape the development and function of the C. elegans nervous system, especially its sense organs and central neuropil (nerve ring). Cell-type-specific promoters allow investigators to label or manipulate individual glial cell types, and therefore provide a key tool for deciphering glial function. In this technical resource, we compare the specificity, brightness, and consistency of cell-type-specific promoters for C. elegans glia. We identify a set of promoters for the study of seven glial cell types (F16F9.3, amphid and phasmid sheath glia; F11C7.2, amphid sheath glia only; grl-2, amphid and phasmid socket glia; hlh-17, cephalic (CEP) sheath glia; and grl-18, inner labial (IL) socket glia) as well as a pan-glial promoter (mir-228). We compare these promoters to promoters that are expressed more variably in combinations of glial cell types (delm-1 and itx-1). We note that the expression of some promoters depends on external conditions or the internal state of the organism, such as developmental stage, suggesting glial plasticity. Finally, we demonstrate an approach for prospectively identifying cell-type-specific glial promoters using existing single-cell sequencing data, and we use this approach to identify two novel promoters specific to IL socket glia (col-53 and col-177).
INTRODUCTION
Cell-type-specific promoters are the key to studying any individual cell in C. elegans. With such a promoter in hand, one can visualize the cell in live animals by expressing a soluble fluorescent protein; probe cell biology in real time by expressing fluorescent constructs that label subcellular structures or serve as biosensors of cellular activity; disrupt cell function by genetic ablation or cell-specific depletion of single proteins; and determine cell-autonomy of gene function by expressing a rescuing transgene in specific cells in an otherwise mutant background.
C. elegans glia illustrate the power of cell-specific promoters to open up a new area of study. The powerful approaches listed above have enabled the study of several glial cell types. In particular, the amphid sheath (AMsh) and cephalic sheath (CEPsh) glia have been studied in detail over the last two decades, while more recent work has begun to examine the amphid socket (AMso), phasmid socket (PHso1 and PHso2), and inner labial socket (ILso) glia (Singhvi and Shaham, 2019). In contrast, other glial cell types, for which no cell-type-specific promoters have been identified, remain largely unexplored.
In this technical resource, we summarize and compare the promoters that have been used to study distinct glial cell types in C. elegans. We evaluate a set of specific, bright, and consistent promoters for the study of seven glial cell types as well as a pan-glial driver. We note that some promoters exhibit interesting dynamics in their expression patterns that are suggestive of glial plasticity. Finally, we demonstrate an approach to identify new cell-type-specific glial promoters by taking advantage of recent single-cell sequencing experiments, and we use this approach to identify col-53 and col-177 as novel cell-specific promoters for ILso glia.
RESULTS AND DISCUSSION
Survey of glial promoters used in the literature
We performed a survey of cell-specific promoters that have been used to label and manipulate C. elegans glia (Table 1). All C. elegans glia are associated with sense organs, mostly in the head and tail (Ward et al., 1975). Hermaphrodites have 24 sense organs: 18 in the head (two amphid, AM; four cephalic, CEP; six inner labial, IL; six outer labial, OL), two in the anterior midbody (anterior deirid, ADE); two in the posterior midbody (posterior deirid, PDE), and two in the tail (phasmid, PH) (White et al., 1986). Each sense organ comprises one or more ciliated sensory neurons; one sheath glial cell; and, in most cases, one socket glial cell (the phasmid has two socket glia). The distal endings of the sheath and socket glia are arranged to form a tube-shaped epithelium that is continuous with the skin (Fig. 1) (Low et al., 2019; Ward et al., 1975). The proximal portion of this tube is formed by the ending of the sheath glial cell, while the distal portion of the tube is formed by the ending of the socket glial cell. In each sense organ, sensory neurons protrude through this glial tube to directly access the environment (Fig. 1). The sheath (sh) and socket (so) glia of most sense organs appear to represent distinct cell types. For example, the AMsh glia are a different cell type than the AMso glia or CEPsh glia based on morphology, molecular profile, and function (Mizeracka and Heiman, 2015). As discussed in detail below, each of these glial cell types expresses a distinct transcriptional reporter. The functional relevance of these striking transcriptional differences has not been fully established, but these transcriptional reporters serve as powerful tools to study and manipulate individual glial cell types. In addition to the glial cell types described above, there are also sex-specific glia in copulatory sense organs in the male tail, and glial-like functions can be performed by mesoderm-derived GLR cells and by the skin (Singhvi and Shaham, 2019; Yang and Chien, 2019). Here, we will focus exclusively on sheath and socket glia of sense organs that are present in both sexes.
Table 1. Glial promoters used in the literature.
Promoters with glial-specific expression were identified in the literature. Within each group, promoters are listed in order from most to least frequently used.
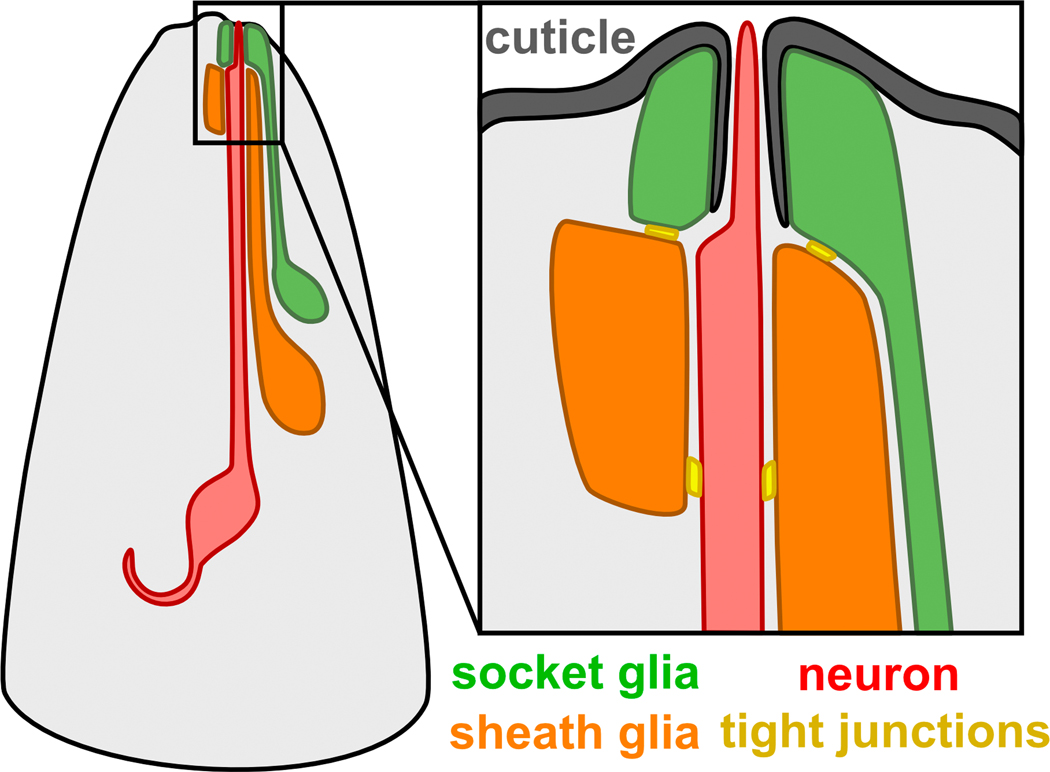
Fig. 1. Schematic of a C. elegans sense organ in the head.
A typical C. elegans sense organ contains one or more sensory neurons (red) and two glial cells, called the sheath (orange) and socket (green). Most sensory neurons extend a ciliated dendritic ending through a tube-shaped pore formed by the sheath and socket glia. The socket glia secretes cuticle (gray) that forms an open pore through which chemosensory dendrite endings protrude, as shown, or a closed sheet into which mechanosensory dendrite endings are embedded (not shown). Tight junctions (yellow) are present between the neuron and sheath glia, the sheath and socket glia, and the socket glia and skin.
Cell-type-specific promoters for C. elegans glia have been identified through three approaches: reverse genetics, forward genetics, and transcriptional profiling. In reverse genetics, the molecular identity of a gene motivates investigation of its expression pattern. The study of molecularly interesting gene families led to the first identification of promoters expressed specifically in glia, including the basic helix-loop-helix transcription factor gene hlh-17 (McMiller and Johnson, 2005; Yoshimura et al., 2008); the inositol triphosphate receptor gene itr-1 (Gower et al., 2001); the neurexin gene itx-1 (Haklai-Topper et al., 2011); the VEGF receptor gene ver-1 (Popovici et al., 2002); the venom allergen protein gene vap-1 (Bacaj et al., 2008); the ion channel genes delm-1 and delm-2 (Han et al., 2013); the microRNA mir-228 (Pierce et al., 2008); and a group of nematode-specific genes with distant similarity to Hedgehog genes, including the “groundhog” (grd) and “ground-like” (grl) genes (Hao et al., 2006). In a typical experiment, a putative promoter fragment from the gene of interest is inserted upstream of GFP in a transgene and the resulting expression pattern is examined. Expression in the AMsh (vap-1, ver-1) or CEPsh (hlh-17) glia is readily recognized due to the symmetry and distinctive morphology of these cells (Table 1). However, expression in the ILsh, ILso, OLsh, and OLso glia is harder to recognize due to the variable cell body positions and similar symmetry and morphology of these glial types (Bargmann and Avery, 1995). Thus, most promoters that label combinations of these cells, including itx-1 and the delm, grl, and grd gene families, have not had their expression patterns precisely defined (Table 1).
As a second approach, forward genetics has led to the discovery of glial-expressed genes through the identification of mutants that disrupt neuronal or glial function. These include daf-6 and lit-1, which affect glial morphogenesis and thus sensory neuron function (Oikonomou et al., 2011; Perens and Shaham, 2005); ttx-1 and ztf-16, which affect a glial-specific temperature response (Procko et al., 2012, 2011); and kcc-3, which affects the microenvironment that glia create around sensory neuron endings (Singhvi et al., 2016; Yoshida et al., 2016) (Table 1). These genes came under study due to their roles in AMsh glia, but several are also expressed in other classes of glia.
Finally, using transcriptional profiling approaches, existing glial markers have been used to “bootstrap” the way to new markers, typically by using fluorescence-activated cell sorting to purify a glial population and then subjecting it to RNA profiling. This approach led to the identification of numerous AMsh markers, including fig-1, F16F9.3, T02B11.3, and others (Bacaj et al., 2008; Wallace et al., 2016) (Table 1). The expression of these genes is highly specific to the AMsh, but they were missed by other approaches because they are not members of a conserved gene family and (with the exception of fig-1) have not been found to cause phenotypes when disrupted. The AMsh and PHsh are highly similar, and many AMsh markers are also expressed in the PHsh.
Recommended promoters for specific glial cell types
There is currently no consensus on which promoters are most appropriate for studying specific glia types. Therefore, we collected glial promoters from the literature and reexamined the specificity, brightness, and consistency of their expression patterns. Specificity requires that expression is restricted to individual glial cell types for easy visualization and for reducing off-target effects of genetic manipulations. Brightness allows fine structural details of glia to be resolved, which provides a tool for examining glia-neuron interactions and an efficient way to screen for mutants with glial morphology defects. Last, consistency assesses whether the promoter reliably labels the same number of cells across individuals. Based on these criteria, we selected five promoters to target seven glial cell types: the AMsh and PHsh; CEPsh; the AMso, PHso1 and PHso2; and ILso glia (Fig. 2, Table 2). However, it is important to note that brightness and consistency are also affected by the structure of the transgene, including copy number and whether it is genomically integrated, as well as the fluorescent reporter used.
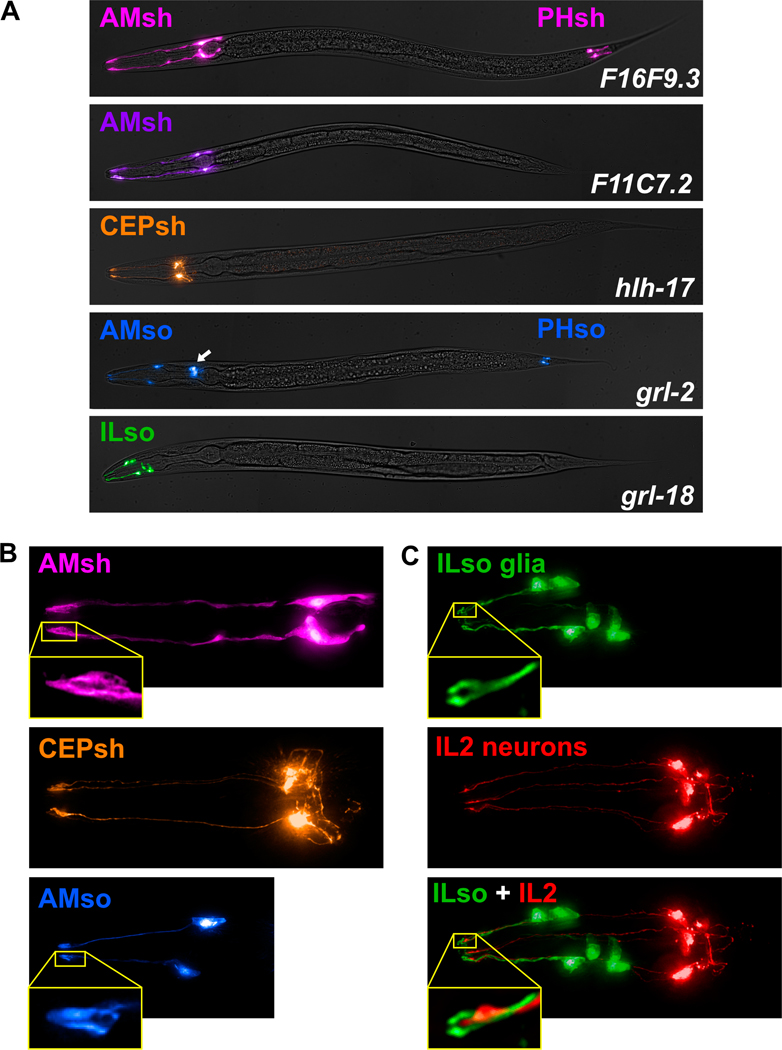
Fig. 2. Cell-type-specific promoters for sheath and socket glia.
(A) Merged brightfield images and pseudo-colored fluorescence projections of animals expressing fluorescent proteins under control of promoters selected for brightness, specificity, and consistency. F16F9.3 (pink, AMsh and PHsh); F11C7.2 (purple, AMsh only); hlh-17 (orange, CEPsh); grl-2 (blue, AMso, PHso1, and PHso2; also expressed in excretory duct and pore cells, white arrow); grl-18 (green, ILso). (B) Magnified images of AMsh (pink, F16F9.3), CEPsh (orange, hlh-17), and AMso (blue, grl-2), showing that their brightness is sufficient to resolve fine structural details including the tube-like pores of the AMsh and AMso glia and the branch-like posterior processes of the CEPsh glia. (C) Head of an animal expressing grl-18pro:GFP (green, ILso glia) and klp-6pro:mCherry (red, IL2 neurons), demonstrating that the grl-18 promoter labels the six ILso glia, identified by their processes which each form a pore for the ciliated dendritic ending of an IL2 sensory neuron.
Table 2. Recommended cell-type-specific glial promoters.
For each marker, the promoter column indicates the size (bp) of the genomic DNA fragment that is taken directly upstream of the gene translation start site, except hlh-17 for which the 3’ end of the promoter fragment is 118 bp upstream of the translation start site.
| Gene | Promoter (bp) | Glia marked | Other cells marked | References |
|---|---|---|---|---|
| F16F9.3 | 2000 | AMsh, PHsh | none | (Bacaj et al., 2008) |
| F11C7.2 | 350 | AMsh | none | (Bacaj et al., 2008) |
| hlh-17 | 2500 | CEPsh | none | (McMiller and Johnson, 2005; Yoshimura et al., 2008) |
| grl-2 | 2981 | AMso, PHso1, PHso2 | excretory duct, pore | (Hao et al., 2006) |
| grl-18 | 2962 | ILso | vulval epithelial cells | (Cebul et al., 2020; Hao et al., 2006) |
| mir-228 | 2200 | all (or nearly all) glia | seam cells, excretory cell | (Pierce et al., 2008) |
A pair of AMsh glia are part of the bilateral amphid sense organs in the head (Ward et al., 1975). Each AMsh is associated with 12 sensory neurons that respond to diverse stimuli including chemical and mechanical cues, temperature, osmolarity, and pheromones (Bargmann, 2006). AMsh glia are the most well-studied C. elegans glia, with many highly specific markers identified (Bacaj et al., 2008; Wallace et al., 2016) (Table 1). We selected F16F9.3 and F11C7.2 as AMsh promoters that offer different advantages (Fig. 2A). F16F9.3 is reliably expressed in AMsh glia as well as the PHsh glia, their functional counterparts in the tail (24/25 animals with an integrated F16F9.3pro:mCherry transgene had 2/2 AMsh and 2/2 PHsh glia marked; 1/25 animals had 1/2 AMsh and 2/2 PHsh glia marked) (Fig. 2A, Supp. Table S2). The strong expression of F16F9.3 allows us to clearly visualize the tube-shaped pores in the sheath glia through which the sensory dendrite endings of the neurons protrude (Fig. 2B). Other promoters, including F53F4.13 and T02B11.3, also offer strong specific labeling of AMsh and PHsh. By comparison, because F11C7.2 expression is restricted to the AMsh glia (Fig. 2A), it offers the advantage of genetically manipulating AMsh glia without affecting PHsh glia (16/23 animals with an F11C7.2pro:GFP extrachromosomal array had 2/2 AMsh glia and no PHsh glia marked, whereas 7/23 animals had 1/2 AMsh glia and no PHsh glia marked, Supp. Table S2). In summary, F16F9.3 has higher overall expression, while F11C7.2 has higher specificity for AMsh glia.
Each of four CEPsh glia is associated with the four-fold symmetric cephalic sense organs in the head. Each CEPsh glial cell wraps around the dendrite of a mechanosensory neuron (CEP) and, in males, an additional pheromone-detecting neuron (CEM) (Ward et al., 1975). We selected hlh-17 as a cell-specific promoter for CEPsh glia (Fig. 2A). We evaluated the expression pattern of a transcriptional reporter for hlh-17 (McMiller and Johnson, 2005; Yoshimura et al., 2008) and observed that it is restricted to CEPsh glia and is consistently expressed in all four cells (22/22 animals with an integrated hlh-17pro:GFP transgene had 4/4 CEPsh glia marked, Supp. Table S2). Though the transcriptional reporter we examined uses a ~2.5 kb promoter fragment and appears to be highly specific to CEPsh glia, it is important to note that reporters containing a different promoter fragment of the hlh-17 gene are expressed in additional cells (Yoshimura et al., 2008). The marker typically becomes brighter as animals reach adulthood but the extended anterior and posterior processes of the glia are visible at all larval stages. We can clearly observe the thin, branch-like posterior processes that wrap around the nerve ring – a feature that distinguishes CEPsh glia from other glia of C. elegans (Fig. 2B). The ability to visualize these fine details makes it possible to investigate the role of CEPsh glia in shaping neuronal connections in the central neuropil, or nerve ring (Colón-Ramos et al., 2007; Ji et al., 2019; Rapti et al., 2017; Shao et al., 2013).
The two AMso glia wrap around the distal tips of a subset of amphid sensory neurons and guide them towards pores in the overlying cuticle so that they can access the external environment (Ward et al., 1975). We selected grl-2 to label and target AMso glia (Fig. 2A). It is consistently expressed in AMso glia as well as the PHso glia in the tail (23/23 animals with an integrated grl-2pro:YFP transgene have 2/2 AMso glia, 2/2 PHso1, and 2/2 PHso2 glia marked, Supp. Table S2). However, the marker is also expressed brightly in the excretory duct and pore cells (Hao et al., 2006) (Fig. 2A, arrow), which makes it problematic to use grl-2 for genetic perturbations, as disrupting the excretory cells can lead to lethality or defects in osmoregulation (Nelson and Riddle, 1984; Sundaram and Buechner, 2016). Although grl-2 is less cell-type-specific than the AMsh or CEPsh promoters, it is bright (Fig. 2B) and consistent and exhibits a more restricted expression pattern than other AMso glia markers that have been identified to date (Table 1). As previous work has shown, we found that lin-48 offers more restricted expression in the phasmid glia (PHso1 but not PHso2) but is also expressed in the posterior intestine and additional cells in the head, including the excretory duct and unidentified neurons (Johnson et al., 2001). It will be important to identify new AMso glia markers whose expression is excluded from the excretory cells so that they can be used as tools to elucidate AMso glia biology and function in future studies.
Last, we selected the grl-18 promoter for the six ILso glia that are part of the six-fold symmetric inner labial sense organs of the head (Fig. 2A). In a previous study (Hao et al., 2006), grl-18 was tentatively identified as a potential marker for ILso or the OLso quadrant (dorsal and ventral, but not lateral) glia based on position and morphology. We find that grl-18 is a highly specific marker for ILso glia. Each ILso glia wraps around the dendrite endings of two sensory neurons, called the IL1 and IL2 neurons (Ward et al., 1975). Therefore, we examined animals co-expressing grl-18pro:GFP and the IL2 neuron-specific marker klp-6pro:mCherry (Fig. 2C). We observed the ciliated endings of the IL2 dendrites protruding through the tube-like pores formed by the glia expressing grl-18pro:GFP, demonstrating that grl-18 labels ILso glia. This is consistent with our previous observation that grl-18pro:GFP labels the lateral ILso glial cells that form specialized attachments to the BAG and URX neurons (Cebul et al., 2020). To assess the quality of this marker, we examined the expression pattern more carefully. We found that its expression is highly consistent (28/28 animals with an integrated grl-18pro:GFP transgene had 6/6 ILso glia marked, Supp. Table S2). It is mostly specific to the ILso glia, however after animals enter the fourth larval (L4) stage, grl-18pro:GFP is also expressed in vulval epithelial cells. The marker is very bright, so it can be used to visualize the pore structure of the glia (Fig. 2C) and screen for mutants with defects in morphology to investigate ILso glia development. It is also suitable for structured illumination microscopy, which requires greater brightness than conventional imaging (Cebul et al., 2020). An inherent limitation is that the ILso cell body positions are variable (Bargmann and Avery, 1995) and often appear to overlap in a two-dimensional projection, making it difficult to show all six cells clearly in a single image. Overall, we have demonstrated that grl-18 specifically labels ILso glia, and this opens up opportunities for studying ILso glia, which remain largely unexplored.
Based on their specificity, brightness, and consistency, the markers listed in Table 2 are especially useful for labeling and genetically manipulating the AMsh, PHsh, CEPsh, AMso, PHso1, PHso2, and ILso glia. We are not aware of promoters that specifically label the CEPso, OLsh, OLso, ADEsh, ADEso, PDEsh, or PDEso. In fact, little is known about these glia at all. Currently, the best approach for studying them may be to use promoters that are expressed in combinations of multiple glial cell types, as described further below.
Promoters that label multiple glial cell types
Pan-neuronal promoters, such as rab-3pro (Nonet et al., 1997), have proven vital for the study of neuronal function and, similarly, the ability to drive expression in all glial cells is important for studying glial function. Several promoters that drive expression in many glial types have been identified (Table 1). We selected mir-228 as a pan-glial promoter, because it drives robust expression in most if not all glia (Fig. 3A, C) (Pierce et al., 2008). However, it is important to note that it is also expressed in some non-glial cells, including the hypodermal seam cells and the excretory cell.
Fig. 3. Promoters expressed in combinations of glia.
(A) Images of head expression of GFP under control of the pan-glial promoter mir-228 and the variable glial promoters delm-1 and itx-1. (B) Quantification of the number of GFP-expressing glia observed with delm-1 (n=20) and itx-1 (n=24) extrachromosomal reporters. Error bars indicate standard deviation. p<0.0001, unpaired t-test with Welch’s correction. (C) Merged brightfield and fluorescence projections of a whole animal expressing pan-glial mir-228pro:GFP.
Promoters expressed in other combinations of glia are also useful for studying glial biology. Curiously, some of these promoters exhibit more variable expression across individuals than the highly consistent cell-type-specific promoters described above. Two such promoters are delm-1 and itx-1, both of which are expressed in combinations of glia in the head (Fig. 3A) (Haklai-Topper et al., 2011; Han et al., 2013). While the identity of some of these glia is known (see Table 1), the expression patterns of these promoters have not been fully characterized. Although there is some overlap in their expression, as described previously (Han et al., 2013), it is clear that delm-1 and itx-1 are expressed in different combinations of glia (Fig. 3A). In contrast to the highly consistent cell-type-specific promoters described above, delm-1pro:GFP and itx-1pro:GFP extrachromosomal arrays exhibit greater variability from animal to animal, although each promoter shows a consistent range of expression (Fig. 3B). delm-1 is expressed in fewer glia and has less variability in expression than itx-1 (Fig. 3B, Supp. Table S2). A high degree of variability in GFP intensity between different glia within the same animal was often observed, particularly for itx-1. It is possible that differences in the threshold of detection introduced more variability into the number of GFP-expressing glia that could be discerned. Despite this variability, these promoters provide a useful tool for driving expression in combinations of glia that cannot be accessed with other promoters.
Plasticity in expression of glial promoters
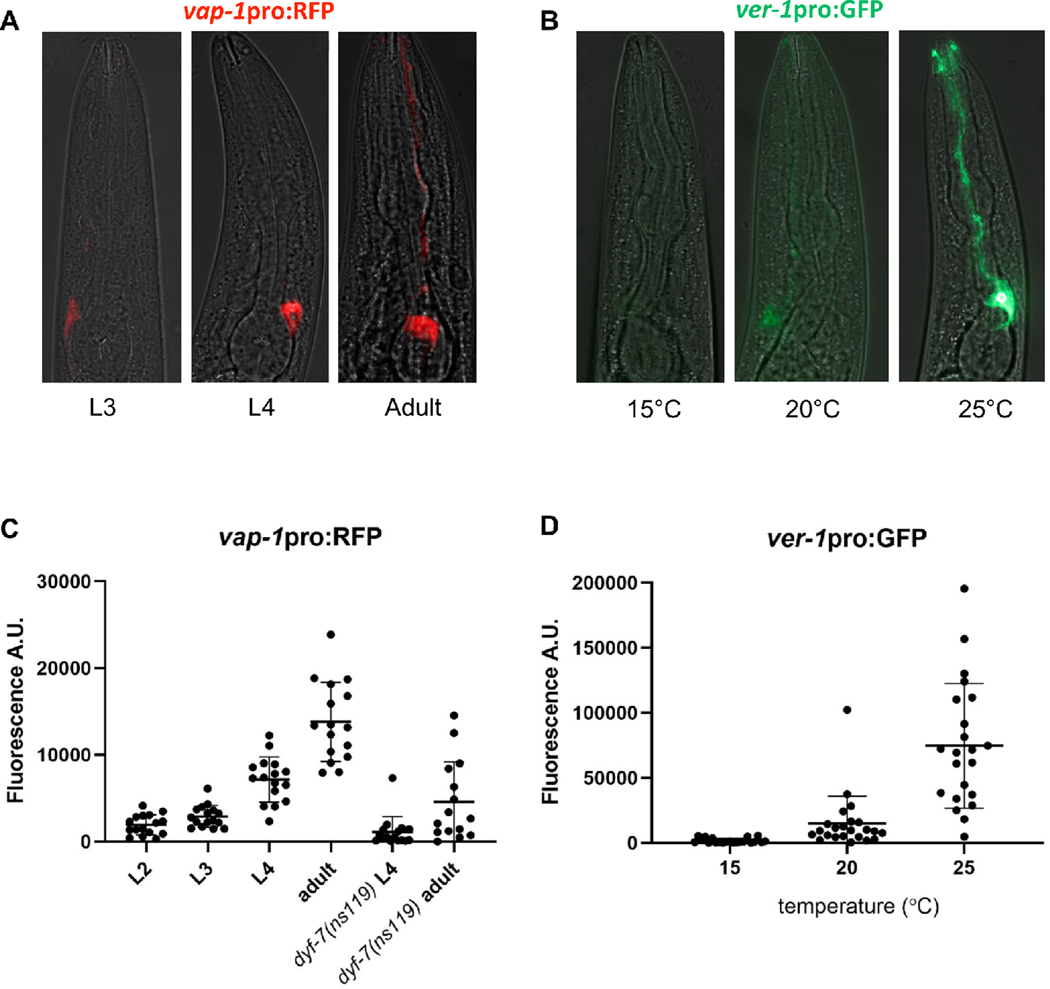
The variability in expression of these promoters may reflect transgene artifacts or true biological differences across individuals. Indeed, the expression of some glial promoters has been shown to be dynamic across life stages, suggesting that glia exhibit surprising plasticity in response to environmental conditions and the internal state of the animal. We noted that the expression of vap-1 in the AMsh is dependent upon developmental stage. Using a 2797 bp promoter, an integrated vap-1pro:RFP transgene exhibits absent or very faint expression in the AMsh prior to the L4 stage, but it then becomes robust in L4 animals and is upregulated further in adulthood (Fig. 4A, C). These developmental changes in expression suggest that changes in glial function may occur during late larval development and maturation. In addition to developmental stage, vap-1 expression is subject to further modulation through activity of amphid sensory neurons. DYF-7 is required to anchor the dendrite endings of sensory neurons to the tip of the nose, and loss of function of this protein results in shortened dendrites that diminish the function of these sensory neurons (Heiman and Shaham, 2009; Low et al., 2019). Loss of DYF-7 function due to the null allele dyf-7(ns119) causes severe defects in vap-1 expression in AMsh glia, suggesting that input from the external environment may modulate AMsh gene expression (Fig. 4C). Interestingly, while vap-1pro:RFP expression at the L4 stage is nearly lost in dyf-7(ns119) mutants, it is still upregulated to some degree in adults, albeit to a much lesser extent than in wild type (Fig. 4C). These observations suggest that there may be multiple layers of plasticity in glial gene expression, both through developmental stage and sensory activity.
Fig. 4. Plasticity in glial gene expression.
(A) vap-1pro:RFP expression in AMsh glia in animals at the third larval (L3), fourth larval (L4), and 1-day adult life stages, showing developmental regulation of gene expression. (B) ver-1pro:GFP expression in AMsh glia in L4 animals grown at 15°C, 20°C, or 25°C, showing thermoregulation of gene expression as previously described (Procko et al., 2012, 2011). (C) Quantification of vap-1pro:RFP fluorescence in AMsh glia from second larval (L2) stage to 1-day adults and in dyf-7(ns119) L4 and adult animals, showing that sensory defects alter gene expression. n=15–16 per group. p<0.05, dyf-7 L4 vs dyf-7 adults; p<0.0001, dyf-7 adults vs wild-type adults; unpaired t-test with Welch’s correction (D) Quantification of ver-1pro:GFP fluorescence in the AMsh glia in L4 animals from 15°C to 25°C. n=21–23 per group. p<0.05, 20°C vs 15°C; p<0.0001, 20°C vs 25°C; Brown-Forsyth and Welch one-way ANOVA with Dunnett’s multiple comparisons test.
The expression of ver-1 in AMsh glia has been previously reported to be modulated by temperature, increasing steadily from undetectable expression at 15°C to very strong expression at 25°C (Procko et al., 2012, 2011). We were able to recapitulate these results (Fig. 4B, D). Additionally, ver-1 expression has also been shown to be induced by entry into dauer, a long-lived stress-resistant life stage, concomitant with remodeling of glia (Procko et al., 2011).
Other examples of dynamic gene expression in glia include hmit-1.2, which is upregulated in AMsh glia in response to osmotic stress (Kage-Nakadai et al., 2011), and daf-6, which is downregulated in AMsh glia during dauer (Moussaif and Sze, 2009). While these data provide evidence for glial plasticity, the functional significance of these changes in gene expression for AMsh function and its interactions with neurons remain unclear. While all of these examples involve AMsh glia, it is likely that other glia may exhibit similar plasticity. Dynamic expression of promoters in other glia has yet to be identified and the full extent of glial plasticity in response to different environmental and state changes remains unknown.
Single-cell transcriptional profiling identifies new cell-type-specific promoters for glia
Discovery of glial cell-type-specific promoters has often relied on serendipity, and has been limited to study of one glial cell type at a time. The recent revolution in single-cell transcriptional profiling raises the possibility that promoters specific to each glial cell type can be identified systematically. Single-cell transcriptional profiling of embryos identified progenitor cells that give rise to eight major groups of tissues, including a group that contains glia and excretory cells. Packer et al. used existing markers from the literature to propose identities for clusters of cells corresponding to nine glial subtypes (Packer et al., 2019).
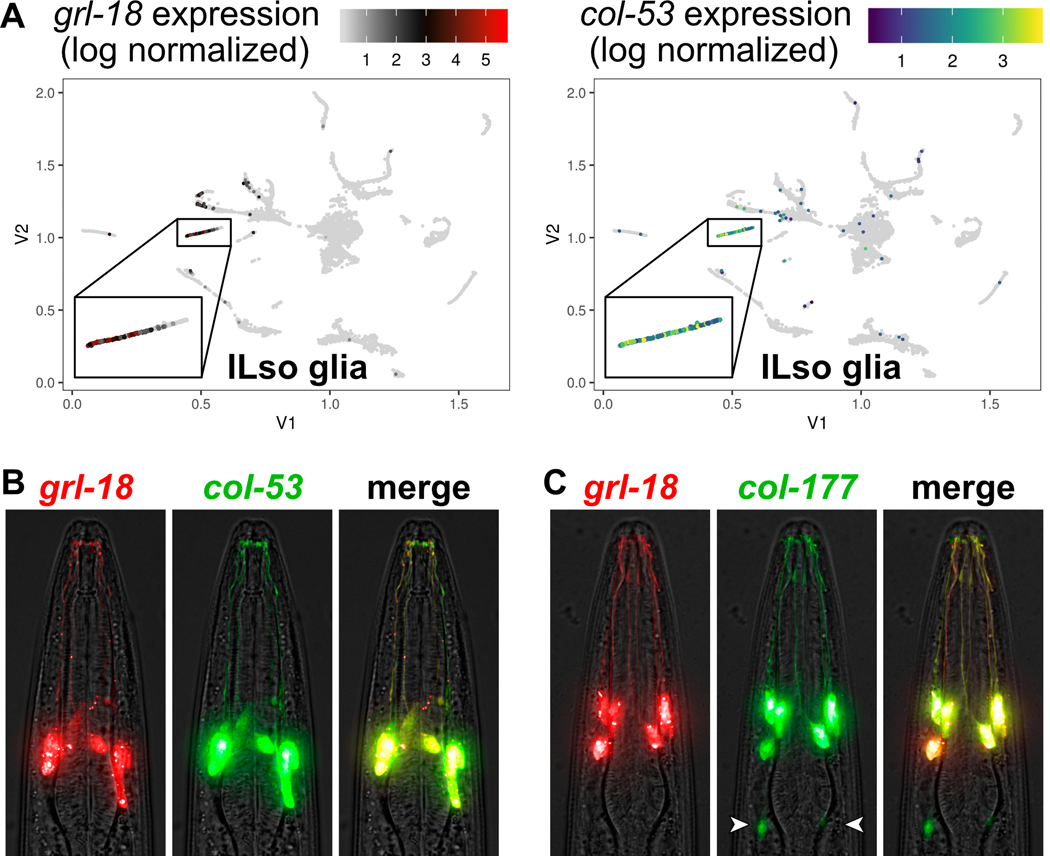
In order to test whether this dataset could be used to prospectively identify novel cell-type-specific glial markers, we attempted to identify a novel marker for ILso glia. The ILso glia cluster is characterized by its enrichment in grl-18 expression (Fig. 5A). Using the VisCello data explorer (http://github.com/qinzhu/VisCello.celegans), we generated a list of additional genes that are highly enriched in the ILso glia cluster compared to the global dataset (see Methods). From the list, we selected the collagen genes, col-53 and col-177, to characterize further. They are strong candidates to be markers for ILso glia, because they are highly expressed in the ILso glia cluster (Fig. 5A, Supp. Fig. S1A), like grl-18, and because socket glia secrete cuticle, which consists of collagen and other extracellular matrix proteins.
Fig. 5. Prospective identification of novel cell-type-specific promoters for ILso glia.
(A) Cell cluster plots illustrating the expression of grl-18 and col-53 in embryonic glia and excretory cells from Packer et al., 2019. Each point represents an individual cell. The color indicates the relative expression level of each gene. For grl-18, low expression is black and high expression is red. For col-53, low is blue and high is green/yellow. Gray indicates no detected transcripts for the gene of interest. Inset box, the cell cluster predicted to include ILso glia. Cell cluster plot for col-177 is in Supp. Fig. S1A. (B, C) Merged brightfield and fluorescence images of an animal expressing grl-18pro:mApple together with (B) col-53pro:GFP or (C) col-177pro:GFP, showing that these markers are expressed in the same cells. col-177 is also faintly expressed in cells with a glial morphology, tentatively identified as OLL socket glia in the head (arrowheads, see Supp. Fig. S1B) and ADE and PDE socket glia in the body.
We created col-53 and col-177 reporters consisting of 733 bp or 2.75 kb promoter regions upstream of their respective translation start sites fused to GFP. We generated animals with a col-53pro:GFP or col-177pro:GFP extrachromosomal array and an integrated grl-18pro:mApple transgene. We found that col-53 and col-177 are each expressed in the same six glial cells as grl-18 (Fig. 5B, C), demonstrating that they are novel cell-type-specific promoters for ILso glia. For col-177, we found that it is also expressed in two additional cells in the head that sit posteriorly to the ILso glia and extend processes to the tip of the nose. Based on morphology, position, symmetry, and the association of OLL neuron sensory dendrites with their distal endings (Supp. Fig. S1B), we have tentatively identified these cells as the lateral OLL/Rso glia, for which there is currently no marker. We also observe faint expression in cells in the body whose morphology, position, and symmetry is consistent with the anterior and posterior deirid (ADE, PDE) socket glia.
Our findings provide proof-of-principle that single-cell transcript profiling enables prospective identification of novel cell-type-specific glial promoters. Indeed, we have also found that this approach can be used to identify promoters that are expressed in combinations of glia (mam-5, Supp. Fig. S2A, B) or that are likely to be expressed in other discrete glial types (AMso, Supp. Fig. S2C). Markers for the remaining unexplored glia will be essential for studies that systematically examine the roles of glia in the diverse sense organs of C. elegans. Furthermore, it will be extremely valuable to identify glial promoters that are expressed early in development or whose expression changes with environmental context, such as neuronal activity. Glial markers expressed in embryos will allow us to visualize and track the birth, migration, and morphogenesis of glia during nervous system assembly (Rapti et al., 2017). Meanwhile, glial markers whose expression is regulated by the environment will provide insight on how glia adapt and potentially alter interactions with their associated neurons to drive appropriate behaviors. These rich datasets and tools will open up new avenues for discovery in the field of glial biology.
MATERIALS AND METHODS
Strains and plasmids
All strains were grown at 20ºC on nematode growth media (NGM) with E. coli OP50 bacteria (Brenner, 1974). Transgenic strains generated in this study were constructed in an N2 background. N2 animals were injected with 25–75 ng/μL per plasmid at a final concentration of 100 ng/μL DNA. Additional information on strains and plasmids used in this study are in Supplemental Table S1.
Fluorescence microscopy and image processing
L2 to L3 animals were selected based on size and the extent of vulva development on the day of imaging. Animals were washed and immobilized in M9 solution containing 50 mM sodium azide and mounted on 2% agarose pads. Image stacks were collected on a DeltaVision Core imaging system (Applied Precision) with a UApo 40x/1.35 NA, PlanApo 60x/1.42 NA, or UPlanSApo 100x/1.40 NA oil immersion objective and a CoolSnap HQ2 camera. Images were subsequently deconvolved using Softworx (Applied Precision) and maximum intensity projections were generated in ImageJ. The brightness and contrast of each projection were linearly adjusted in Affinity Photo 1.7.3. Fluorescent signals were pseudo-colored, and merged images were generated using the Screen layer mode in Affinity Photo 1.7.3. When the whole animal is shown, a composite of multiple fields was manually stitched together. The maximum intensity projections of the stitched images were generated with the same optical stacks across the whole animal, except for F16F9.3 and grl-2, for which different optical stacks were used to generate the maximum intensity projections of the head and tail, because the head and tail glia are in different planes. Additionally, for grl-2, the head was split into two regions and different optical stacks were used to generate the maximum intensity projections to resolve both the AMso glia and the excretory cells separately. All figures were assembled in Affinity Designer 1.7.3. For quantification of cell fluorescence expression, FIJI software (Schindelin et al., 2012) was used to measure the integrated density of GFP or RFP in the cell body and the average of three background regions in the head was subtracted.
Assessment of glial marker consistency
To assess the consistency of each glial marker, L2 to L3 transgenic animals were washed and immobilized in M9 solution containing 50 mM sodium azide and mounted on 2% agarose pads. Fluorescently labeled glial cells were scored visually across optical stacks with a Deltavision Core imaging system (Applied Precision) and a PlanApo 60x/1.42 NA oil immersion objective.
Analysis of single-cell RNA sequencing data
To identify novel markers for ILso glia, we used the “Differential Expression” feature on the VisCello data explorer (Packer et al., 2019). We generated a list of genes that are highly enriched in the predicted ILso glia cluster compared to all other cell types in the single-cell RNA sequencing dataset. This was achieved by first selecting the following: Global dataset (Choose Sample), Cell type/subtype (Meta Class), and ILso (Group 1). Group 2 was left blank. Next, downsample cells was performed with the default settings (0.05 FDR cutoff, 200 Max Cell in subset, and 1000 Max Cell in background). Finally, differential expression (DE) analysis was run to generate a table of differentially expressed genes for the ILso glia cluster. A list of genes that are enriched in other glial types, including AMso glia (Supp. Fig. S2C), can be generated by replacing ILso with the glial type of interest (Group 1).
The graphical plots of grl-18 and col-53 or col-177 expression (Fig. 5A, Supp. Fig. S1A) were generated using the online VisCello data explorer (https://cello.shinyapps.io/celegans_explorer/). The “Cell Type” explorer was used to select the glia and excretory cell subset, and data was displayed using the “UMAP-2D [Paper]” projection colored by cell type/subtype. Separate searches for grl-18 and col-53 or col-177 were used to generate a plot of the glia and excretory cell subset showing the relative expression levels of each gene using the black_red palette for grl-18 and viridis palette for col-53 or col-177. Inset boxes in Fig. 5A and Supp. Fig. S1A are magnifications of the predicted ILso cluster, which contains 163 cells.
Supplementary Material
ACKNOWLEDGMENTS
We thank Shai Shaham and members of the Shaham laboratory, Meera Sundaram, and John Murray for sharing information and advice on glial-specific promoters and for providing strains and plasmids, and John Murray for help using the VisCello data explorer. We thank members of the Heiman laboratory for unpublished work further characterizing candidate promoters, including Ian McLachlan (itx-1), Elizabeth Cebul (grl-18, delm-1, delm-2), and Karolina Mizeracka (grl-2, lin-48). Some strains were provided by the CGC, which is funded by National Institutes of Health Office of Research Infrastructure Programs (P40 OD010440). This work was supported by NIH R01NS112343 to M.G.H.
REFERENCES
- Bacaj T, Tevlin M, Lu Y, Shaham S, 2008. Glia are essential for sensory organ function in C. elegans. Science 322, 744–747. 10.1126/science.1163074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargmann CI, 2006. Chemosensation in C. elegans. WormBook Online Rev. C Elegans Biol 1–29. 10.1895/wormbook.1.123.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargmann CI, Avery L, 1995. Laser killing of cells in Caenorhabditis elegans. Methods Cell Biol. 48, 225–250. 10.1016/s0091-679x(08)61390-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunreiter K, Hamlin S, Lyman-Gingerich J, 2014. Identification and characterization of a novel allele of Caenorhabditis elegans bbs-7. PloS One 9, e113737. 10.1371/journal.pone.0113737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cebul ER, McLachlan IG, Heiman MG, 2020. Dendrites with specialized glial attachments develop by retrograde extension using SAX-7 and GRDN-1. Dev. Camb. Engl 147 10.1242/dev.180448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cianciulli A, Yoslov L, Buscemi K, Sullivan N, Vance RT, Janton F, Szurgot MR, Buerkert T, Li E, Nelson MD, 2019. Interneurons Regulate Locomotion Quiescence via Cyclic Adenosine Monophosphate Signaling During Stress-Induced Sleep in Caenorhabditis elegans. Genetics 213, 267–279. 10.1534/genetics.119.302293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colón-Ramos DA, Margeta MA, Shen K, 2007. Glia promote local synaptogenesis through UNC-6 (netrin) signaling in C. elegans. Science 318, 103–106. 10.1126/science.1143762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding G, Zou W, Zhang H, Xue Y, Cai Y, Huang G, Chen L, Duan S, Kang L, 2015. In vivo tactile stimulation-evoked responses in Caenorhabditis elegans amphid sheath glia. PloS One 10, e0117114 10.1371/journal.pone.0117114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenk LA, de Bono M, 2015. Environmental CO2 inhibits Caenorhabditis elegans egg-laying by modulating olfactory neurons and evokes widespread changes in neural activity. Proc. Natl. Acad. Sci. U. S. A 112, E3525–3534. 10.1073/pnas.1423808112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson CL, Balbona JT, Niedzwiecki A, Rodriguez P, Nguyen KCQ, Hall DH, Blakely RD, 2018. Glial loss of the metallo β-lactamase domain containing protein, SWIP-10, induces age- and glutamate-signaling dependent, dopamine neuron degeneration. PLoS Genet. 14, e1007269. 10.1371/journal.pgen.1007269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gower NJ, Temple GR, Schein JE, Marra M, Walker DS, Baylis HA, 2001. Dissection of the promoter region of the inositol 1,4,5-trisphosphate receptor gene, itr-1, in C. elegans: a molecular basis for cell-specific expression of IP3R isoforms. J. Mol. Biol 306, 145–157. 10.1006/jmbi.2000.4388 [DOI] [PubMed] [Google Scholar]
- Grant J, Matthewman C, Bianchi L, 2015. A Novel Mechanism of pH Buffering in C. elegans Glia: Bicarbonate Transport via the Voltage-Gated ClC Cl- Channel CLH-1. J. Neurosci. Off. J. Soc. Neurosci 35, 16377–16397. 10.1523/JNEUROSCI.3237-15.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haklai-Topper L, Soutschek J, Sabanay H, Scheel J, Hobert O, Peles E, 2011. The neurexin superfamily of Caenorhabditis elegans. Gene Expr. Patterns GEP 11, 144–150. 10.1016/j.gep.2010.10.008 [DOI] [PubMed] [Google Scholar]
- Han L, Wang Y, Sangaletti R, D’Urso G, Lu Y, Shaham S, Bianchi L, 2013. Two novel DEG/ENaC channel subunits expressed in glia are needed for nose-touch sensitivity in Caenorhabditis elegans. J. Neurosci. Off. J. Soc. Neurosci 33, 936–949. 10.1523/JNEUROSCI.2749-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao L, Johnsen R, Lauter G, Baillie D, Bürglin TR, 2006. Comprehensive analysis of gene expression patterns of hedgehog-related genes. BMC Genomics 7, 280 10.1186/1471-2164-7-280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardaway JA, Sturgeon SM, Snarrenberg CL, Li Z, Xu XZS, Bermingham DP, Odiase P, Spencer WC, Miller DM, Carvelli L, Hardie SL, Blakely RD, 2015. Glial Expression of the Caenorhabditis elegans Gene swip-10 Supports Glutamate Dependent Control of Extrasynaptic Dopamine Signaling. J. Neurosci. Off. J. Soc. Neurosci 35, 9409–9423. 10.1523/JNEUROSCI.0800-15.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiman MG, Shaham S, 2009. DEX-1 and DYF-7 establish sensory dendrite length by anchoring dendritic tips during cell migration. Cell 137, 344–355. 10.1016/j.cell.2009.01.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt-Newbury R, Viveiros R, Johnsen R, Mah A, Anastas D, Fang L, Halfnight E, Lee D, Lin J, Lorch A, McKay S, Okada HM, Pan J, Schulz AK, Tu D, Wong K, Zhao Z, Alexeyenko A, Burglin T, Sonnhammer E, Schnabel R, Jones SJ, Marra MA, Baillie DL, Moerman DG, 2007. High-throughput in vivo analysis of gene expression in Caenorhabditis elegans. PLoS Biol. 5, e237 10.1371/journal.pbio.0050237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji T, Wang K, Fan J, Huang J, Wang M, Dong X, Shi Y, Manning L, Zhang X, Shao Z, Colón-Ramos DA, 2019. ADAMTS-family protease MIG-17 regulates synaptic allometry by modifying the extracellular matrix and modulating glia morphology during growth (preprint). Neuroscience. 10.1101/734830 [DOI] [Google Scholar]
- Johnson AD, Fitzsimmons D, Hagman J, Chamberlin HM, 2001. EGL-38 Pax regulates the ovo-related gene lin-48 during Caenorhabditis elegans organ development. Dev. Camb. Engl 128, 2857–2865. [DOI] [PubMed] [Google Scholar]
- Kage-Nakadai E, Ohta A, Ujisawa T, Sun S, Nishikawa Y, Kuhara A, Mitani S, 2016. Caenorhabditis elegans homologue of Prox1/Prospero is expressed in the glia and is required for sensory behavior and cold tolerance. Genes Cells Devoted Mol. Cell. Mech 21, 936–948. 10.1111/gtc.12394 [DOI] [PubMed] [Google Scholar]
- Kage-Nakadai E, Uehara T, Mitani S, 2011. H+/myo-inositol transporter genes, hmit-1.1 and hmit-1.2, have roles in the osmoprotective response in Caenorhabditis elegans. Biochem. Biophys. Res. Commun 410, 471–477. 10.1016/j.bbrc.2011.06.001 [DOI] [PubMed] [Google Scholar]
- Katz M, Corson F, Iwanir S, Biron D, Shaham S, 2018. Glia Modulate a Neuronal Circuit for Locomotion Suppression during Sleep in C. elegans. Cell Rep. 22, 2575–2583. 10.1016/j.celrep.2018.02.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low IIC, Williams CR, Chong MK, McLachlan IG, Wierbowski BM, Kolotuev I, Heiman MG, 2019. Morphogenesis of neurons and glia within an epithelium. Dev. Camb. Engl 146 10.1242/dev.171124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMiller TL, Johnson CM, 2005. Molecular characterization of HLH-17, a C. elegans bHLH protein required for normal larval development. Gene 356, 1–10. 10.1016/j.gene.2005.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuary PR, Liao C-Y, Chang JT, Kumsta C, She X, Davis A, Chu C-C, Gelino S, Gomez-Amaro RL, Petrascheck M, Brill LM, Ladiges WC, Kennedy BK, Hansen M, 2016. C. elegans S6K Mutants Require a Creatine-Kinase-like Effector for Lifespan Extension. Cell Rep. 14, 2059–2067. 10.1016/j.celrep.2016.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizeracka K, Heiman MG, 2015. The many glia of a tiny nematode: studying glial diversity using Caenorhabditis elegans. Wiley Interdiscip. Rev. Dev. Biol 4, 151–160. 10.1002/wdev.171 [DOI] [PubMed] [Google Scholar]
- Mizeracka K, Rogers JM, Shaham S, Bulyk ML, Heiman MG, 2019. Lineage-specific control of convergent cell identity by a Forkhead repressor (preprint). Developmental Biology. 10.1101/758508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina-García L, Cook SJ, Kim B, Bonnington R, Sammut M, O’Shea J, Elliott DJ, Hall DH, Emmons SW, Barrios A, Poole RJ, 2018. A direct glia-to-neuron natural transdifferentiation ensures nimble sensory-motor coordination of male mating behaviour (preprint). Neuroscience. 10.1101/285320 [DOI] [Google Scholar]
- Moussaif M, Sze JY, 2009. Intraflagellar transport/Hedgehog-related signaling components couple sensory cilium morphology and serotonin biosynthesis in Caenorhabditis elegans. J. Neurosci. Off. J. Soc. Neurosci 29, 4065–4075. 10.1523/JNEUROSCI.0044-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson FK, Riddle DL, 1984. Functional study of the Caenorhabditis elegans secretory-excretory system using laser microsurgery. J. Exp. Zool 231, 45–56. 10.1002/jez.1402310107 [DOI] [PubMed] [Google Scholar]
- Nonet ML, Staunton JE, Kilgard MP, Fergestad T, Hartwieg E, Horvitz HR, Jorgensen EM, Meyer BJ, 1997. Caenorhabditis elegans rab-3 Mutant Synapses Exhibit Impaired Function and Are Partially Depleted of Vesicles. J. Neurosci 17, 8061–8073. 10.1523/JNEUROSCI.17-21-08061.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkura K, Bürglin TR, 2011. Dye-filling of the amphid sheath glia: implications for the functional relationship between sensory neurons and glia in Caenorhabditis elegans. Biochem. Biophys. Res. Commun 406, 188–193. 10.1016/j.bbrc.2011.02.003 [DOI] [PubMed] [Google Scholar]
- Oikonomou G, Perens EA, Lu Y, Shaham S, 2012. Some, but not all, retromer components promote morphogenesis of C. elegans sensory compartments. Dev. Biol 362, 42–49. 10.1016/j.ydbio.2011.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikonomou G, Perens EA, Lu Y, Watanabe S, Jorgensen EM, Shaham S, 2011. Opposing activities of LIT-1/NLK and DAF-6/patched-related direct sensory compartment morphogenesis in C. elegans. PLoS Biol. 9, e1001121. 10.1371/journal.pbio.1001121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packer JS, Zhu Q, Huynh C, Sivaramakrishnan P, Preston E, Dueck H, Stefanik D, Tan K, Trapnell C, Kim J, Waterston RH, Murray JI, 2019. A lineage-resolved molecular atlas of C. elegans embryogenesis at single-cell resolution. Science 365 10.1126/science.aax1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perens EA, Shaham S, 2005. C. elegans daf-6 encodes a patched-related protein required for lumen formation. Dev. Cell 8, 893–906. 10.1016/j.devcel.2005.03.009 [DOI] [PubMed] [Google Scholar]
- Pierce ML, Weston MD, Fritzsch B, Gabel HW, Ruvkun G, Soukup GA, 2008. MicroRNA-183 family conservation and ciliated neurosensory organ expression. Evol. Dev 10, 106–113. 10.1111/j.1525-142X.2007.00217.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popovici C, Isnardon D, Birnbaum D, Roubin R, 2002. Caenorhabditis elegans receptors related to mammalian vascular endothelial growth factor receptors are expressed in neural cells. Neurosci. Lett 329, 116–120. 10.1016/s0304-3940(02)00595-5 [DOI] [PubMed] [Google Scholar]
- Procko C, Lu Y, Shaham S, 2012. Sensory organ remodeling in Caenorhabditis elegans requires the zinc-finger protein ZTF-16. Genetics 190, 1405–1415. 10.1534/genetics.111.137786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Procko C, Lu Y, Shaham S, 2011. Glia delimit shape changes of sensory neuron receptive endings in C. elegans. Dev. Camb. Engl 138, 1371–1381. 10.1242/dev.058305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapti G, Li C, Shan A, Lu Y, Shaham S, 2017. Glia initiate brain assembly through noncanonical Chimaerin-Furin axon guidance in C. elegans. Nat. Neurosci 20, 1350–1360. 10.1038/nn.4630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sammut M, Cook SJ, Nguyen KCQ, Felton T, Hall DH, Emmons SW, Poole RJ, Barrios A, 2015. Glia-derived neurons are required for sex-specific learning in C. elegans. Nature 526, 385–390. 10.1038/nature15700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez J-Y, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A, 2012. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Z, Watanabe S, Christensen R, Jorgensen EM, Colón-Ramos DA, 2013. Synapse location during growth depends on glia location. Cell 154, 337–350. 10.1016/j.cell.2013.06.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhvi A, Liu B, Friedman CJ, Fong J, Lu Y, Huang X-Y, Shaham S, 2016. A Glial K/Cl Transporter Controls Neuronal Receptive Ending Shape by Chloride Inhibition of an rGC. Cell 165, 936–948. 10.1016/j.cell.2016.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhvi A, Shaham S, 2019. Glia-Neuron Interactions in Caenorhabditis elegans. Annu. Rev. Neurosci 42, 149–168. 10.1146/annurev-neuro-070918-050314 [DOI] [PubMed] [Google Scholar]
- Stout RF, Parpura V, 2011. Voltage-gated calcium channel types in cultured C. elegans CEPsh glial cells. Cell Calcium 50, 98–108. 10.1016/j.ceca.2011.05.016 [DOI] [PubMed] [Google Scholar]
- Stringham E, Pujol N, Vandekerckhove J, Bogaert T, 2002. unc-53 controls longitudinal migration in C. elegans. Dev. Camb. Engl 129, 3367–3379. [DOI] [PubMed] [Google Scholar]
- Sundaram MV, Buechner M, 2016. The Caenorhabditis elegans Excretory System: A Model for Tubulogenesis, Cell Fate Specification, and Plasticity. Genetics 203, 35–63. 10.1534/genetics.116.189357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanis JE, Bellemer A, Moresco JJ, Forbush B, Koelle MR, 2009. The potassium chloride cotransporter KCC-2 coordinates development of inhibitory neurotransmission and synapse structure in Caenorhabditis elegans. J. Neurosci. Off. J. Soc. Neurosci 29, 9943–9954. 10.1523/JNEUROSCI.1989-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timbers TA, Garland SJ, Mohan S, Flibotte S, Edgley M, Muncaster Q, Au V, Li-Leger E, Rosell FI, Cai J, Rademakers S, Jansen G, Moerman DG, Leroux MR, 2016. Accelerating Gene Discovery by Phenotyping Whole-Genome Sequenced Multi-mutation Strains and Using the Sequence Kernel Association Test (SKAT). PLoS Genet. 12, e1006235. 10.1371/journal.pgen.1006235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker M, Sieber M, Morphew M, Han M, 2005. The Caenorhabditis elegans aristaless orthologue, alr-1, is required for maintaining the functional and structural integrity of the amphid sensory organs. Mol. Biol. Cell 16, 4695–4704. 10.1091/mbc.e05-03-0205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace SW, Singhvi A, Liang Y, Lu Y, Shaham S, 2016. PROS-1/Prospero Is a Major Regulator of the Glia-Specific Secretome Controlling Sensory-Neuron Shape and Function in C. elegans. Cell Rep. 15, 550–562. 10.1016/j.celrep.2016.03.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Perens EA, Oikonomou G, Wallace SW, Lu Y, Shaham S, 2017. IGDB-2, an Ig/FNIII protein, binds the ion channel LGC-34 and controls sensory compartment morphogenesis in C. elegans. Dev. Biol 430, 105–112. 10.1016/j.ydbio.2017.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Apicella A, Lee S-K, Ezcurra M, Slone RD, Goldmit M, Schafer WR, Shaham S, Driscoll M, Bianchi L, 2008. A glial DEG/ENaC channel functions with neuronal channel DEG-1 to mediate specific sensory functions in C. elegans. EMBO J. 27, 2388–2399. 10.1038/emboj.2008.161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, D’Urso G, Bianchi L, 2012. Knockout of glial channel ACD-1 exacerbates sensory deficits in a C. elegans mutant by regulating calcium levels of sensory neurons. J. Neurophysiol 107, 148–158. 10.1152/jn.00299.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward S, Thomson N, White JG, Brenner S, 1975. Electron microscopical reconstruction of the anterior sensory anatomy of the nematode Caenorhabditis elegans.?2UU. J. Comp. Neurol 160, 313–337. 10.1002/cne.901600305 [DOI] [PubMed] [Google Scholar]
- White JG, Southgate E, Thomson JN, Brenner S, 1986. The structure of the nervous system of the nematode Caenorhabditis elegans. Philos. Trans. R. Soc. Lond. B. Biol. Sci 314, 1–340. 10.1098/rstb.1986.0056 [DOI] [PubMed] [Google Scholar]
- Yang W-K, Chien C-T, 2019. Beyond being innervated: the epidermis actively shapes sensory dendritic patterning. Open Biol. 9, 180257 10.1098/rsob.180257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J-A, Gao G, Liu X-J, Hao Z-Q, Li K, Kang X-L, Li H, Shan Y-H, Hu W-L, Li H-P, Cai S-Q, 2017. Genetic variation in glia-neuron signalling modulates ageing rate. Nature 551, 198–203. 10.1038/nature24463 [DOI] [PubMed] [Google Scholar]
- Yip ZC, Heiman MG, 2018. Ordered arrangement of dendrites within a C. elegans sensory nerve bundle. eLife 7 10.7554/eLife.35825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida A, Nakano S, Suzuki T, Ihara K, Higashiyama T, Mori I, 2016. A glial K(+) /Cl(−) cotransporter modifies temperature-evoked dynamics in Caenorhabditis elegans sensory neurons. Genes Brain Behav. 15, 429–440. 10.1111/gbb.12260 [DOI] [PubMed] [Google Scholar]
- Yoshimura S, Murray JI, Lu Y, Waterston RH, Shaham S, 2008. mls-2 and vab-3 Control glia development, hlh-17/Olig expression and glia-dependent neurite extension in C. elegans. Dev. Camb. Engl 135, 2263–2275. 10.1242/dev.019547 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.