Abstract
BACKGROUND:
Targeting insect-specific genes through post-transcriptional gene silencing with RNA interference (RNAi) is a new strategy for insect pest management. However, lepidopterans are recalcitrant to RNAi, which prevents the application of novel RNAi technology to many notorious pests, including Ostrinia nubilalis (ECB). Strategies for enhancing RNAi efficiency, including large doses of double-stranded RNA (dsRNA), nuclease inhibitors, transfection reagents, and nanoparticles, have proved useful in other insects exhibiting substantial dsRNA degradation, a major mechanism limiting RNAi efficacy. To determine if similar strategies can enhance RNAi efficiency in ECB, various reagents were tested for their ability to enhance dsRNA stability in ECB tissues, then compared for their effectiveness in whole ECB.
RESULTS:
Ex vivo incubation experiments revealed that Metafectene Pro, EDTA, chitosan-based dsRNA nanoparticles, and Zn2+ enhanced dsRNA stability in ECB hemolymph and gut content extracts, compared to uncoated dsRNA. Despite these positive results, the reagents used in this study were ineffective at enhancing RNAi efficiency in ECB in vivo. To reduce assay time and required dsRNA, midguts were dissected and incubated in tissue culture medium containing dsRNA with and without reagents. These experiments showed that RNAi efficiency varied between target genes, and nuclease inhibitors improved RNAi efficiency for only a portion of the refractory target genes investigated ex vivo.
CONCLUSION:
These results indicate that enhancing dsRNA stability is insufficient to improve RNAi efficiency in ECB and suggests the existence of additional, complex mechanisms contributing to low RNAi efficiency in ECB.
Keywords: RNAi, European corn borer, double-stranded RNA stability, feeding, injection, tissue culture
1. Introduction
Targeting insect-specific genes through post-transcriptional gene silencing with RNA interference (RNAi) is a new strategy for insect pest management.1 RNAi-mediated crop protection strategies, including transgenic plants, baits, and sprays, are valuable approaches for sustainable agriculture and food security because they provide new approaches to address yield loss,2 they offer a novel mechanism of action to combat pesticide resistance,3 and they provide greater species-specificity to reduce off-target environmental effects.4 Unfortunately, RNAi technology is currently hindered by inefficient and highly variable results when different insects are targeted, especially lepidopterans.5,6 Given that lepidopterans are among the most devastating pests of crops, forests, and stored products, basic research is needed to elucidate and overcome factors limiting RNAi efficiency in agriculturally relevant lepidopteran pests, such as the European corn borer (ECB), Ostrinia nubilalis (Hübner).7,8
The ECB is a small, light brown, crambid moth native to Europe that was first introduced into the United States in the early 1900s and is now established in North America.9,10 ECB is one of the most damaging pests of corn in North America and Europe,10 and a variety of methods have been employed to limit crop loss, including genetic modification of corn and conventional insecticides. Corn plants have been genetically engineered to resist ECB feeding through recombinant expression of Bt Cry toxins from Bacillus thuringiensis.9,11 Unfortunately, resistance to these methods of control has already been documented.12,13 Therefore, new approaches that exploit novel modes of action are badly needed for ECB to combat insecticide resistance and prolong the use of Bt Cry toxins. However, before RNAi-based pest management strategies can be developed for ECB, the molecular mechanisms limiting RNAi efficiency in ECB and other lepidopteran pests must be understood and overcome.
Investigations of molecular mechanisms influencing RNAi efficiency reveals five main factors limiting RNAi efficiency in insects: instability of dsRNA in body fluids, inadequate internalization of dsRNA, deficient core RNAi machinery, insufficient systemic spreading of the silencing signal, and refractory target genes.1,14 To combat these limitations in RNAi efficiency, there is a growing suite of strategies and technologies for dsRNA delivery to enhance RNAi efficiency in RNAi-refractory insects, including Drosophila suzukii,15 Spodoptera exigua,16,17 Hyphantria cunea,18 and Anopheles gambiae.19 These strategies include large doses of double-stranded RNA (dsRNA), repeated exposure to dsRNA, cationic liposomes, and dsRNA-nanoparticles, among others.1,14 Thus, it may be possible to enhance RNAi efficiency (i.e., suppression at a given dose of dsRNA) in ECB by optimizing dsRNA delivery procedures and reagents. However, strategies for enhancing RNAi efficiency in ECB have not yet been evaluated.
In the present study, various strategies, procedures, and reagents were tested for their ability to enhance dsRNA stability and RNAi efficiency in ECB. Ex vivo incubation experiments were used to identify transfection reagents, nanoparticles, and nuclease inhibitors that enhanced dsRNA stability in ECB gut contents (GC) and hemolymph (HE). These reagents, as well as a peritrophic matrix (PM) disrupting reagent, were then tested on a variety of ECB developmental stages to determine if they could enhance oral or injected RNAi efficiency in vivo. An assay using midgut tissue culture was also used for the rapid comparison of RNAi efficiency among different target genes. Together, these findings support the hypothesis that RNAi efficiency in ECB is limited by multiple mechanisms and cannot be enhanced using individual strategies or reagents.
2. Materials and Methods
2.1. Insect rearing
The ECB used in this study originated from French Agricultural Research (Lumberton, MN, USA) and were continuously reared in the laboratory at Kansas State University, Manhattan, KS, as described in Cooper et al.20
2.2. Preparation of dsRNA and reagents
To obtain dsRNA for stability enhancement experiments and RNAi assays, dsRNA targeting either an endogenous gene encoding lethal giant larvae protein (OnLgl; MT467568) from the ECB or an exogenous gene encoding enhanced green fluorescent protein (GFP; LC336974.1) were synthesized, purified, and assessed as described in Cooper et al.20 The primer sequences for dsRNA synthesis are listed in Table S-1. OnLgl was selected as the target gene because it is a single copy gene with an essential role in cell proliferation,21 and it is expressed in all developmental stages and tissues of ECB. In addition, RNAi of TcLgl led to efficient and rapid suppression of transcript levels that lasted over 10 days and resulted in molting defects and mortality in all developmental stages of Tribolium castaneum tested.21 GFP was selected as a non-target control to account for non-specific changes in gene expression associated with dsRNA exposure.22 To form chitosan-based dsRNA nanoparticles (NP dsRNA), ethanol precipitated dsRNA was first purified with the MEGAclear Transcription Clean-Up Kit (Invitrogen, Carlsbad, CA) and eluted in nuclease-free water. Eight micrograms of dsRNA (in 8 μl water) were then combined with 2 μl (8 μg/μl) proprietary chitosan-based polymer in 1% acetic acid solution, and 198 μl of 1 % acetic acid. This solution was incubated for three hours at room temperature to allow NP dsRNA to form. After incubation, newly formed NP dsRNA was pelleted out of solution by centrifuging at 16,000 × g for 30 min. The supernatant was then transferred to another tube for quantification. Unincorporated dsRNA remaining in the supernatant was quantified with a NanoPhotometer (Implen, Westlake Village, CA, USA), and subtracted from the starting quantity, to obtain the amount of NP dsRNA in the pellet. The dsRNA incorporation efficiency was then calculated, and water added to the NP dsRNA pellet to reach the desired final concentration. The NP dsRNA solution was stored at 4°C, and a hand-held tissue homogenizer was used to fully resuspend the solution prior to use.
To form Lipofectamine RNAiMax dsRNA lipoplexes (Lipo dsRNA), two mixes were created and then combined. Mix 1 contained 1 μl of Lipofectamine RNAiMax reagent (Invitrogen) and 7 μl of buffered sucrose. Mix 2 contained 32 μg of dsRNA and enough buffered sucrose to bring the final volume to 8 μl. Mix 1 and 2 were combined together, vortexed, spun down and incubated at room temperature for 30 min to allow Lipo dsRNA lipoplexes to form. Buffered sucrose solution contained 2% sucrose, 10 mM Tris, (pH 7.5), and 0.5 mM spermidine.15 Lipo dsRNA solution was stored at 4°C.
To form Metafectine Pro dsRNA lipoplexes (Meta dsRNA), 3.5 μl of Metafectene Pro (Biontex Laboratories GmbH, Munchen, Germany) was mixed with 3.5 μl of nuclease-free water containing 14 μg of dsRNA. The solution was then vortexed, spun down, and incubated at room temperature for 30 min to allow Meta dsRNA liposomes to form. Meta dsRNA solution was stored at 4°C.
Nuclease inhibitor solutions were prepared by dissolving powdered ethylenediaminetetraacetic acid (EDTA), Zn2+, Mn2+, or Co2+ in 1X phosphate-buffered saline (PBS) to create 25–100 mM (w/v) stock solutions for use in subsequent assays.
2.3. dsRNA-stability assays
To assess which of the candidate reagents could enhance dsRNA stability in ECB tissues, ex vivo incubation experiments were performed using hemolymph (HE) and gut contents (GC) harvested from fifth-instar larvae following the detailed protocols described in Cooper et al.20 In brief, after harvesting tissues from the ECB larvae, PBS was used to normalize the total protein content between biological replicates (each containing tissue pooled from 15 individuals). One microliter containing 1 μg of coated or uncoated 300 bp dsGFP (i.e., naked dsGFP, Lipo dsGFP, Meta dsGFP, or NP dsGFP) was then incubated with 2.7 μl of the normalized tissue extracts (or PBS for control incubations), and enough PBS and/or nuclease inhibitor (i.e., EDTA, Zn2+, Mn2+, or Co2+) to reach a final volume of 14 μl. After incubating at room temp for 30 min, the reactions were quenched by either adding 2.8 μl of 50 mM EDTA or by heating to 65°C for 10 min. The remaining dsRNA in each sample was then converted to cDNA and quantified with RT-PCR (see Table S-1 for primers used). Cycle threshold (Ct) values were converted to the amount of dsGFP in nanograms based on standard curves (Fig S-1) as previously described.20 Experiments followed a two-way factorial treatment design to allow for assessment of significant effects on dsRNA stability due to reagent (present or absent), incubation fluid (tissue extract or PBS) and/or interaction between both factors. Fold-increases in dsRNA stability due to each reagent were calculated by taking the mean amount of dsRNA in the “dsRNA plus reagent in tissue extract” treatment divided by the mean amount of dsRNA in the “dsRNA in tissue extract” treatment.
2.4. RNAi bioassays
To compare strategies for enhancing RNAi efficiency in ECB, dsRNA, in combination with candidate RNAi-efficiency enhancing reagents, were fed or injected into numerous ECB developmental stages. DsRNAs (500 bp) targeting either the endogenous OnLgl gene or the exogenous GFP gene were used for all bioassays. Experiments were generally designed in a two-way factorial treatment structure so that significant effects on relative gene expression, weight, and survivorship due to dsRNA (e.g., dsOnLgl, dsGFP, water), reagent (e.g., 0, 10, 20 mM Zn2+), or interaction between both factors could be investigated. Optimization experiments (data not shown) were preformed using the reagents alone to determine the highest dose the insects could tolerate without negative effects on body weight and survivorship.
Injection bioassays consisted of a single, high-dose injection of dsRNA, so comparisons of the enhancement of RNAi efficiency could be made using each of the dsRNA stabilizing reagents, as well as among developmental stages. First instar neonate larvae proved challenging to inject, so they were only used for oral RNAi. However, both delivery methods were tested on second instar larvae. To reduce the amount of dsRNA required for the assays, RNAi efficiency was compared among developmental stages using injection, rather than ingestion. Injection of dsRNA was performed on second-, third-, fifth-instar larvae, pupae, and adults using a Nanoinject II system (Drummond Scientific, PA, USA) coupled with a SYS-Micro4 controller (World Precision Instruments, Sarasota, FL, USA). Each 125-nl injection contained 500 ng of dsRNA with and without reagent delivered at the intersegmental membrane of abdominal segments A5–A6. An equal volume of nuclease-free water or PBS (pH 7.0) was used for buffer-only control injections. Twenty to twenty-five individuals were injected per replicate and either placed on artificial diet inside of 37-ml clear plastic cups sealed with oversnap caps (Frontier Agricultural Sciences, Newark, DE, USA) or placed inside of a covered 1-L glass beaker containing a 90 ×15 mm plastic petri dish with a cotton ball soaked in 20% sucrose/water solution. Three biological replicates, each containing 20–25 individuals, were injected 24–48 h apart. Insects were maintained under rearing conditions until sampling.
Oral RNAi bioassays for ECB were designed based on those used by Khajuria et al.,7,23 for young larvae continuously fed on a high dose of dsRNA. Ten micrograms of dsRNA with and without each reagent were applied to artificial diet. An equal volume of nuclease-free water or PBS (pH 8.0) was used for buffer-only controls. Optimization experiments were performed to determine how much diet 25 neonate larvae consume on average in the first 3 d of life, and then the next 3 d of life, to ensure that the full dose of dsRNA would be consumed. Nitrogen gas was used to dry dsRNA/reagent solutions (10 μg/larvae) onto 20 mg squares of artificial diet inside individual Bio-Assay Tray cells (Frontier Agricultural Sciences). Twenty-five larvae were then transferred into each prepared cell using a fine-point paintbrush and sealed inside with Bio-Assay Tray Lids (Frontier Agricultural Sciences). Three biological replicates of every treatment, each containing 25 individuals, were started 12–24 h apart. Larvae were allowed to feed under rearing conditions for three days. On the third day (once all the diet had been consumed), the larvae were transferred to a new cell containing a dsRNA/reagent-treated 40 mg diet square (10 μg/larvae), and then allowed to feed for another three days (until all diet was again consumed). Thus, about 20 μg of dsRNA was fed to each larva over a period of six days. To avoid negative effects on larvae due to starvation, additional untreated diet was added as needed if the larvae finished the treated diet before the 3 or 6 d period was over. Larvae were maintained under rearing conditions until sampling.
Sampling was performed 3 and 6 d after the start of dsRNA exposure as well as at other time points as needed to assess phenotypic effects. The average body weight was determined by weighing all the individuals from each replicate on a laboratory balance and then dividing by the total number of larvae weighed. Percent survivorship was calculated as the number of live individuals divided by the starting number of larvae times one hundred. For expression analysis, three individuals (the largest, the smallest, and an intermediate sized individual) from each replicate of each treatment were pooled at each time point and homogenized in TRIzol reagent (Invitrogen) and frozen at −80°C until further processing. The effects on transcript levels were calculated as described below.
2.5. RNAi tissue culture assays
To determine how RNAi efficiency for OnLgl compares to the RNAi efficiency for other target genes, a protocol for incubating excised ECB midguts in tissue culture media containing dsRNA was developed, and used to compare the RNAi efficiency of five additional target genes, encoding clathrin heavy chain (Chc), clathrin coat adaptor protein (AP50), ADP ribosylation factor 72A (ARF72A), V-type proton ATPase 16 kD proteolipid subunit (Vha16), and V-type proton ATPase subunit H (VhaSFD). To determine if enhanced stability of dsRNA in the ECB midgut is sufficient to enhance RNAi efficiency, midgut tissue culture assays were performed with and without the addition of Zn2+, Mn2+, and Co2+ in the medium. Only the divalent cations were tested on ECB midgut cultures because they were the most promising candidates, as EDTA and transfection reagents were toxic to larvae.
Whole guts were dissected from fifth instar ECB larvae in PBS buffer (pH 8.0) and trimmed to remove the foregut and hindgut. Fifth-instar larvae were selected for this assay due to their large size because they were easiest to dissect and three pooled midguts provided ample tissue for RNA extractions. Excised midguts were gently massaged with forceps to remove the food bolus, and directly transferred to a 96 well tissue culture plate containing 50 μl aliquots of prepared insect medium. Prepared insect medium was comprised of 1 X Gibco Grace’s Insect Medium (Fisher Scientific, Huston, TX, USA) containing 1% penicillin-streptomycin, nuclease inhibitor (i.e., Zn2+, Mn2+, or Co2+), and various concentrations of dsRNA (50, 100, or 150 ng/μl) for either the target gene or the GFP-control, all diluted with 1X PBS (pH 8.0). After dissections were completed, the 96-well plate was sealed with Parafilm and incubated at 25°C. After 24 h, three midguts were pooled four times from each treatment, homogenized in TRIzol (Invitrogen) and frozen at −80°C until further processing for reverse transcription quantitative PCR (RT-qPCR).
2.6. Analysis of transcript levels
To determine if RNAi-mediated suppression of the target gene occurred, relative expression analysis was performed with RT-qPCR. Total RNA was extracted from three pooled individuals (whole bodies) following the manufacturer’s instructions and resuspended in DEPC-treated water. The quality and quantity of total RNA were assessed using a NanoPhotometer (Implen). One microgram of total RNA from each sample was treated with DNase I (Thermo Fisher Scientific, Waltham, MA, USA) to remove genomic DNA, and then converted to cDNA using the EasyScript cDNA synthesis kit (Applied Biological Materials, Richmond, Canada) following the manufactures instructions. The resulting cDNA was diluted 5-fold with nuclease-free water for use as template for RT-qPCR.
RT-qPCR was performed in accordance with MIQE guidelines24 on a CFX Connect Real-Time System (Bio-Rad, Hercules, CA, USA) using BrightGreen 2x qPCR master mix (Applied Biological Materials) in a 20-μl reaction volume with the following program: 95°C for 3 min, 39 cycles at 95°C for 30 s, 60°C for 30 s and 72°C for 30 s, followed by melt curve analysis to confirm amplicon specificity using a temperature range from 65 to 95°C. Primer sequences for RT-qPCR are included in Table S-1. The primer efficiency [E=10^(−1/slope)] and percent primer efficiency [%E=(E-1)*100] were calculated using the standard curve method for each primer pair. The cycle threshold (Ct) was plotted against concentration for each primer set, and a regression line was generated to determine the slope and correlation confidence (i.e., R2 value). All RT-qPCR reactions were performed on at least three biological replicates, each with two technical replicates. Technical replicates that had Ct values more than 0.5 apart were repeated. In each run, non-template (water only) control reactions were used as negative controls for each primer set.
Ct values were analyzed following the ΔΔCT method.25 First, the mean Ct values of all technical replicates were normalized to the geometric mean of ribosomal protein S3 gene (Rps3, DQ988989) and elongation factor-1 alpha gene (Ef1a, AF173392) to calculate ΔCt. Then ΔCt values were normalized to the mean of the calibrator treatment (i.e., either the dsGFP or buffer-only control). Finally, fold change for each biological replicate was calculated, subjected to statistical analysis, and the mean and standard error of each treatment graphed. Ct values over 32 were considered non-detectible, and a fold change of zero was used for analysis. NormFinder,26 geNorm,27 BestKeper,28 and RefFinder (http://150.216.56.64/referencegene.php) were used to verify the stability of reference genes across ECB tissues and developmental stages.20 Percent suppression of the target gene was calculated as [(control-target)/control)*100].
In two of the bioassays, additional samples were collected at the 3, 6, 12, and 24 h time points to investigate short-term transcriptional responses of the target gene to dsRNA exposure. To provide better visualization of fluctuations in gene expression over the time points analyzed, relative expression was calculated as the expression of OnLgl relative to the geometric mean of the reference genes (i.e., ΔCt rather than ΔΔCt).
2.7. Statistical analysis
Statistical differences between treatment means were assessed in Minitab 18 or 19 (Minitab, State College, PA, USA) with either a two-sample t-test or a one-way ANOVA followed by the Tukey post hoc test procedure depending on the number of groups that were compared. The significance level (α) was set at 0.05 for the entire family of comparisons. All data sets were subjected to the Anderson-Darling normality test and/or the Levene’s test for equal variance to verify statistical assumptions. Data that did not meet the assumptions were subjected to either a Mann-Whitney U test, or a Kruskal-Wallis one-way ANOVA on ranks test, followed by multiple Wilcoxon signed-rank tests for each desired pairwise comparison. The Bonferroni correction was used to control for type I error occurring from the use of multiple parametric statistical tests. In all experiments, treatments were replicated at least three times.
3. Results
3.1. Putative dsRNA stability-enhancing reagents
Comparison of dsRNA stability in ECB tissue extracts, or PBS as a control, indicated that NP dsGFP was not significantly degraded in HE compared to the control incubations, despite significant degradation of naked dsGFP in HE [F(1,20)=6.11, p=0.023*] (Fig. 1A). Conversely, NP dsGFP was significantly degraded in GC compared to the PBS control incubations, but NP dsGFP incubated in GC was still significantly more stable than dsGFP incubated in GC [F(1,8)=5.66, p=0.045*] (Fig. 1B). Specifically, NP enhanced the stability of dsRNA in ECB HE and GC by 16.6- and 36.1-fold, respectively (Table 1). However, there was still 2.2-fold less dsRNA in the NP dsGFP in GC treatment than in the NP dsGFP in PBS control treatment, indicating that NP could not completely protect dsRNA from degradation in ECB GC but could completely protect dsRNA from degradation in ECB HE.
Figure 1. Enhancement of dsRNA stability in A) hemolymph (HE) and B) gut contents (GC) due to chitosan-based dsRNA nanoparticles (NP).
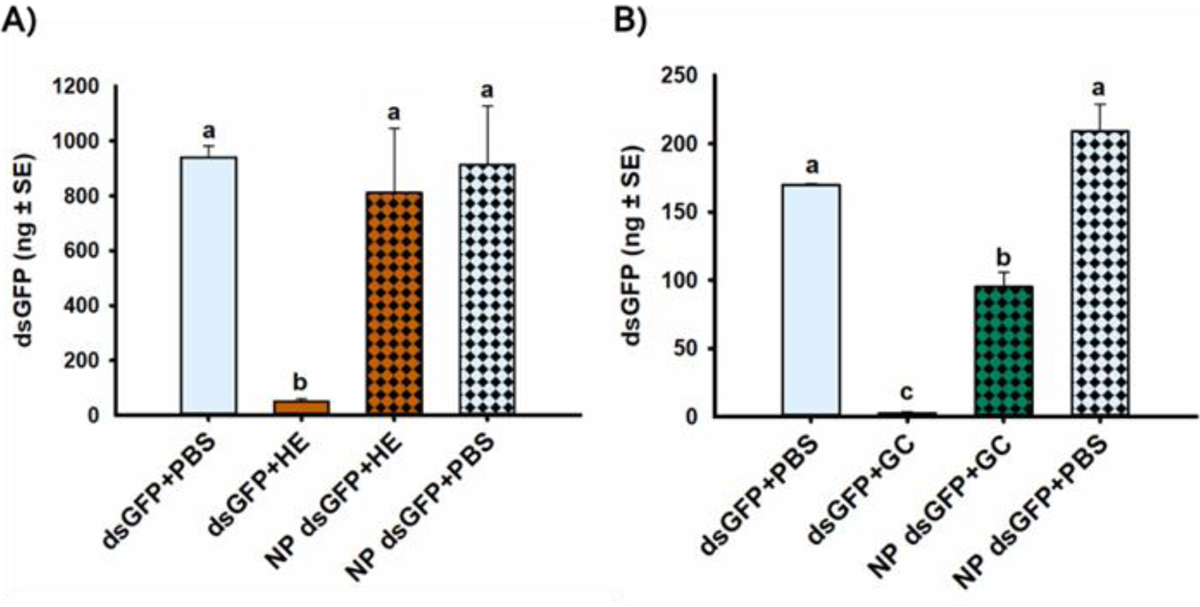
The mean amount of dsGFP (in ng) recovered after 30 min incubation in PBS buffer (control) or tissue extracts harvested from 15 fifth-instar ECB larvae. Means that do not share a letter are significantly different.
Table 1. Comparison of fold-increase in stability of dsRNA in ECB tissue extracts due to candidate RNAi-enhancing reagents.
The mean amounts (nanograms plus and minus standard error) of 300 bp dsGFP that was recovered after incubation in ECB tissue extracts with and without each reagent are presented. Significant values are indicated with an asterisk.
| Enhancement of dsRNA stability in ECB | ||||||
|---|---|---|---|---|---|---|
| Hemolymph (HE) | Gut contents (GC) | |||||
| Reagent | dsGFP +HE [control] |
Reagent dsGFP + HE | Fold increase | dsGFP +GC [control] |
Reagent dsGFP +GC | Fold increase |
| Lipo | 30.2±11.0 | 24.1±8.8 | 0.8 | 0.0013±0.0011 | 0.005±0.002 | 3.7 |
| Meta | 128.5±54.0 | 717.9±166.1* | 5.6 | -- | -- | n/a |
| NP | 49.0±10.3 | 811.7±234.3* | 16.6 | 2.6±0.9 | 95.0±10.6* | 36.1 |
| EDTA | 0.079±0.007 | 148.9±20.9* | 6 mM: 1,874.8 | 0.0013± 0.0011 | 182.7±27.9* | 6 mM: 135,947.2 |
| Zn2+ | 38.0±12.3 | 165.4±12.3* | 10 mM: 4.4 | 0.098±0.038 | 77.1±26.5 | 10 mM: 790.6 |
| Mn2+ | 38.0±16.6 | 96.8±20.6 | 10 mM: 2.5 | 0.40±0.29 | 3.3±1.8 | 10 mM: 8.2 |
| Co2+ | 18.7±8.2 | 61.7±23.2 | 10 mM: 3.3 | 0.093±0.048 | 8.9±1.8 | 10 mM: 95.6 |
Abbreviations: mM, millimolar, Lipo, Lipofectamine RNAiMax; Meta, Metafectene Pro; EDTA; ethylenediaminetetraacetic acid.
Similarly, Meta (Fig. S-2), EDTA (Fig. S-3), and Zn2+ (Fig. S-4A, S-5A) all significantly enhanced dsRNA stability and completely protected dsRNA from degradation in ECB tissue extracts (Table 1). However, Lipo (Fig. S-6), Mn2+ (Fig. S-4B, S-5B), and Co2+ (Fig. S-4C, S-5C) did not significantly enhance dsRNA stability in ECB GC or HE (Table 1).
3.2. RNAi efficiency in ECB
Feeding of dsOnLgl to neonate ECB larvae did not significantly affect the relative gene expression of OnLgl in four separate bioassays, even when candidate RNAi-enhancing reagents such as Lipo, 1% fluorescent brightener 28 (FB28), 50 mM Zn2+, and 6 mM EDTA were employed (Table 2). However, when a similar oral RNAi experiment was performed with 0, 10, and 20 mM Zn2+ on second-instar larvae, there was a significant effect on the relative expression of OnLgl due to dsRNA after six days of dsRNA feeding (Fig S-9A). Specifically, relative expression of OnLgl was suppressed by 68% or 3.2-fold after six days of feeding on dsOnLgl compared to larvae that fed on dsGFP, regardless of Zn2+ concentrations examined [F(2,12)=9.93, p=0.003*] (Fig. 2). No phenotypes were observed due to the suppression of OnLgl at any of the time points investigated (Table 2, Fig S-9B, C).
Table 2. Comparison of RNAi efficiency among ECB developmental stages when various delivery methods and RNAi-enhancing reagents were employed.
Unfed larvae were starved for 24 h prior to dsRNA delivery. Fed larvae were allowed to feed normally (i.e., not starved) prior to dsRNA delivery. Non-feeding stages do not feed as part of their natural developmental cycle.
| RNAi Bioassays | ||||||
|---|---|---|---|---|---|---|
| Delivery method | Stage | Feeding status | Reagent | Significant suppression of OnLgl | Phenotype(s) | Results |
| Oral | First-instar larvae | Unfed | 50 mM Zn2+ | No | None | Fig. S-10 |
| 6 mM EDTA | No | EDTA reduced weight by 1.7-fold at 6 d (6 mM vs 0 mM) | Figs. S-7, S-8 | |||
| 1% FB28 | No | FB28 reduced weight by 2.7-fold at 6 d (1% vs 0%) | Figs. S-12, S-13 | |||
| Lipo | No | None | Fig. S-11 | |||
| Second-instar larvae | Fed | 10 & 20 mM Zn2+ | 68%, or 3.2-fold decrease at 6 d (dsOnLgl vs dsGFP) | None | Figs. 2, S-9 | |
| Injection | Second-instar larvae | Fed | 10 & 20 mM Zn2+ | No | None | Fig. S-21 |
| Third-instar larvae | Fed | 25 mM Zn2+ | 32%, or 1.5-fold decrease at 3 d (Zn2+ dsOnLgl vs Zn2+ dsGFP) | None | Fig. 4, S-19 | |
| 50 mM Zn2+ | No | None | Fig. S-20 | |||
| Meta | 57%, or 2.3-fold decrease at 6 d (naked dsOnLgl vs naked dsGFP) | Meta decrease survivorship by 1.25-fold & weight by 1.24-fold at 6 d (Meta dsRNA vs naked dsRNA) | Figs. 3, S-14 | |||
| Wandering (fifth-instar) larvae | Nonfeeding | Meta | No* | None | Fig. S-15 | |
| Pupae | Nonfeeding | Lipo | No | None | Fig. S-18 | |
| Meta | No | None | Fig. S-16 | |||
| Adults | Unfed | Meta | No, 1.4-fold increase at 3 d (dsOnLgl vs dsGFP) | None | Fig. S-17 | |
Significant differences were detected between some treatment groups but could not be meaningfully interpreted. Abbreviations: mM, millimolar; Lipo, Lipofectamine RNAiMax; Meta, Metafectene Pro; EDTA; ethylenediaminetetraacetic acid; Zn2+, nuclease inhibitor; FB28, fluorescent brightener 28; h, hour; d, day.
Figure 2. Significant effects on RNAi efficiency in 48-h old ECB larvae six days after oral delivery of dsRNA, regardless of nuclease inhibitor (Zn2+) concentration.
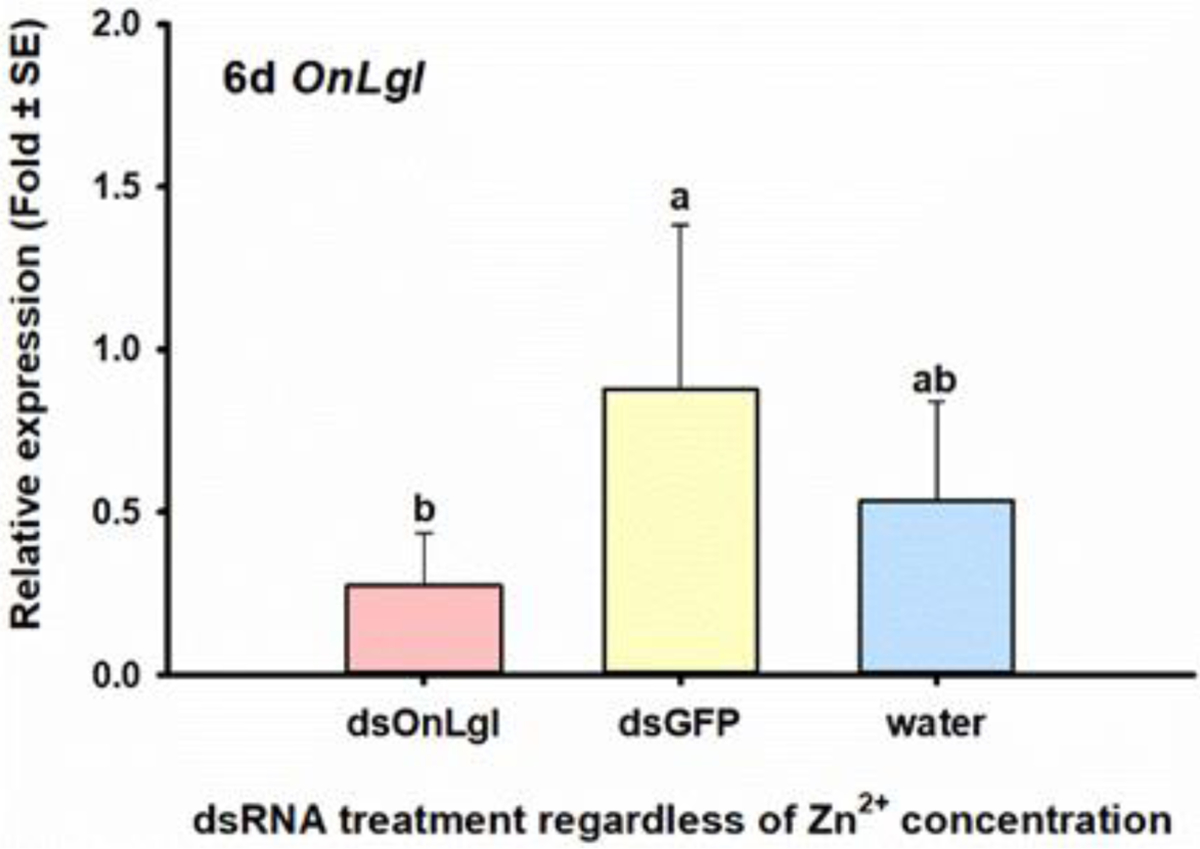
Relative expression of the OnLgl six days after feeding of dsOnLgl or dsGFP incorporated into diet, with 10 or 20 mM Zn2+. Means that do not share a letter are significantly different.
Injection of dsOnLgl into third-instar ECB larvae resulted in RNAi-mediated suppression of OnLgl in two out of three bioassays (Table 2). However, injection of dsOnLgl into second instar larvae, wandering fifth-instar larvae, and pupae did not significantly affect the relative expression of OnLgl, even when RNAi enhancing reagents such as Zn2+, Meta, and Lipo were used (Table 2). In adults, the relative expression of OnLgl increased by 1.4-fold three days after injection [F(1,8)=5.13, p=0.053*] (Table 2, Fig S-17).
Specifically, third-instar ECB larvae injected with dsOnLgl in one bioassay had 2.3-fold lower relative expression of OnLgl, or 57% gene suppression, by day 6 compared to larvae injected with dsGFP, regardless of whether dsRNA was encapsulated in Meta or not [F(1,8)=6.16, p=0.038*] (Fig. 3A). There was no significant effect of reagent on the relative expression of the target gene in this experiment, indicating that Meta dsRNA did not enhance RNAi efficiency when dsOnLgl was injected into third-instar ECB larvae (Fig S-14A). The Meta reagent significantly reduced survivorship by 1.25-fold [F(1,8)=13.50, p=0.006*] (Fig. 3B) and body weight by 1.24-fold [F(1,8)=11.95, p=0.009*] (Fig. 3C) during this bioassay, but had no effect in other bioassays testing Meta on other developmental stages (Table 2).
Figure 3. Significant effects on RNAi efficiency in third-instar ECB larvae after injection of dsRNA with and without Metafectene Pro (Meta) lipoplexes.
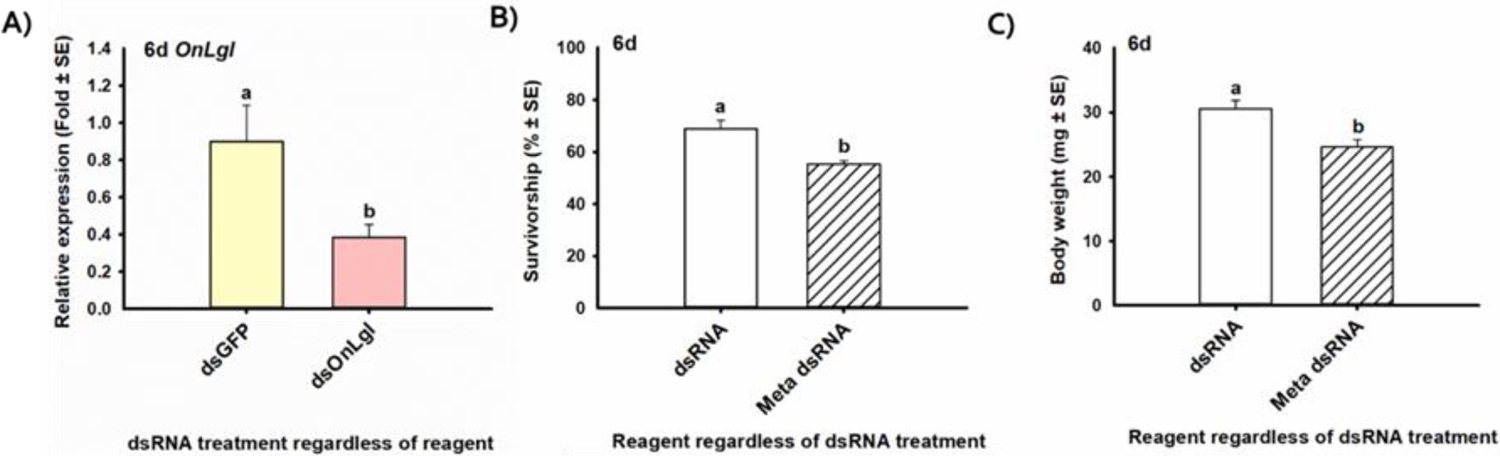
The mean A) relative expression of the OnLgl target gene, B) percent of injected larvae to survive, and C) body weight, six days after microinjection of dsRNA targeting either OnLgl or GFP, with and without encapsulation of dsRNA in Meta lipoplexes. Means that do not share a letter are significantly different.
In another bioassay, third-instar ECB larvae injected with dsOnLgl in the presence of 25 mM Zn2+ had 1.5-fold or 32% suppression of OnLgl three days after injection [t(4)=3.69, p=0.021*] (Fig. 4), which is less suppression than observed for feeding of second instars (68%; Fig. 2) and injection of third instars in the Meta bioassay (57%; Fig. 3A), suggesting that 25 mM Zn2+ did not enhance RNAi efficiency in ECB (Table 2). No phenotypes were observed due to the suppression of OnLgl at any of the time points investigated (Table 2, Fig S-19B, C).
Figure 4. Significant effects on RNAi efficiency following injection of third-instar ECB larvae with dsRNA and 25 mM nuclease inhibitor (Zn2+).
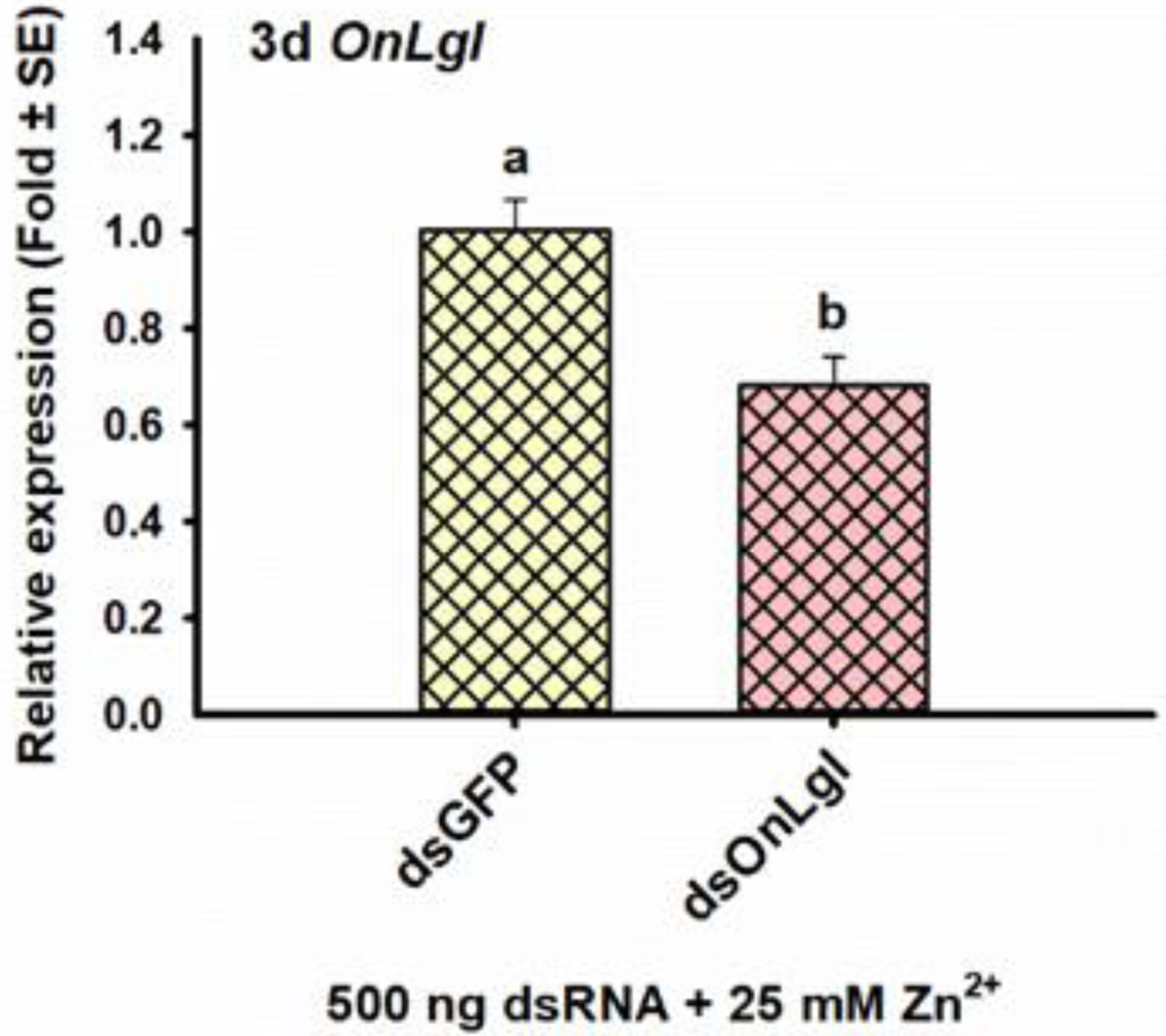
Relative expression of the OnLgl target three days after injection of dsOnLgl or dsGFP in conjunction with 25 mM Zn2+. Means that do not share a letter are significantly different.
To determine if the expression of OnLgl was affected at earlier time points than those investigated in the majority of these bioassays, transcriptional responses of OnLgl to dsRNA exposure were investigated at earlier time points after dsRNA feeding and injection in the two bioassays conducted on second instar larvae (Table 2). However, OnLgl expression was unaffected at 3, 6, 12, and 24 h after dsRNA feeding (Fig. S-22) or injection (Fig. S-23), even when various concentrations of the Zn2+ nuclease inhibitor were used.
3.3. Comparison of multiple target genes in tissue culture
When excised midguts were incubated with dsGFP or various concentrations of dsOnLgl, there were no significant differences in the relative expression of OnLgl between treatments, indicating that suppression of OnLgl did not occur at a measurable level (Fig. S-24A). Similar findings were obtained for dsRNA targeting OnChc, OnArf72A, and OnAP50 (Fig. S-24; Table 3). Conversely, excised midguts incubated with dsOnVha16 had significantly lower relative expression of OnVha16 transcripts compared to midguts incubated with dsGFP, regardless of dsOnVha16 concentrations tested [F(3,17)=7.20, p=0.003*] (Fig. 5A). Similar results were observed for OnVhaSFD [H(4)=17.20, p=0.002*] (Fig. 5B). Expression of OnVha16 was suppressed by 54.8% on average, and OnVhaSFD was suppressed by 46.2% on average (Table 3).
Table 3. Comparison of enhancement of RNAi efficiency in ECB tissue culture for multiple target genes due to nuclease inhibitor Zn2+.
The relative expression (in fold change plus and minus standard error) of each target gene after incubation of excised midguts in media containing 100 ng/ml of dsGFP or dsRNA specific to each target gene are presented.
| RNAi Efficiency in ECB Midgut Cultures | ||||||
|---|---|---|---|---|---|---|
| Without Zn2+ | With Zn2+ | |||||
| Target gene | dsGFP | dsTarget | Significant suppression | dsGFP | dsTarget | Significant suppression |
| OnLgl | 1.00±0.06 | 0.83±0.20 | No | 1.01±0.10 | 0.95±0.21 | No |
| OnArf72A | 1.08±0.23 | 0.78±0.17 | No | 1.14±0.33 | 0.08±0.03 | 92% |
| OnChc | 1.03±0.13 | 1.42±0.10 | No | 1.04±0.16 | 0.69±0.07 | 33% |
| OnAP50 | 1.02±0.11 | 1.19±0.29 | No | 1.14±0.33 | 0.08±0.03 | No |
| OnVha16 | 1.04±0.16 | 0.38±0.04 | 64% | 1.06±0.20 | 0.44±0.14 | 59% |
| OnVhaSFD | 1.10±0.21 | 0.68±0.10 | 38% | 1.04±0.16 | 0.69±0.07 | 33% * |
Italicized averages are trends (not statistically significant). Abbreviations: On, Ostrinia nubilalis; Lgl, lethal giant larvae; Chc, clathrin heavy chain; AP50, clathrin coat adaptor protein AP50; Arf72A, ADP ribosylation factor 72A; Vha16, V-type proton ATPase 16 kD proteolipid subunit; VhaSFD, V-type proton ATPase subunit H.
Figure 5. RNAi efficiency of two alternative target genes in ECB midgut tissue cultures.
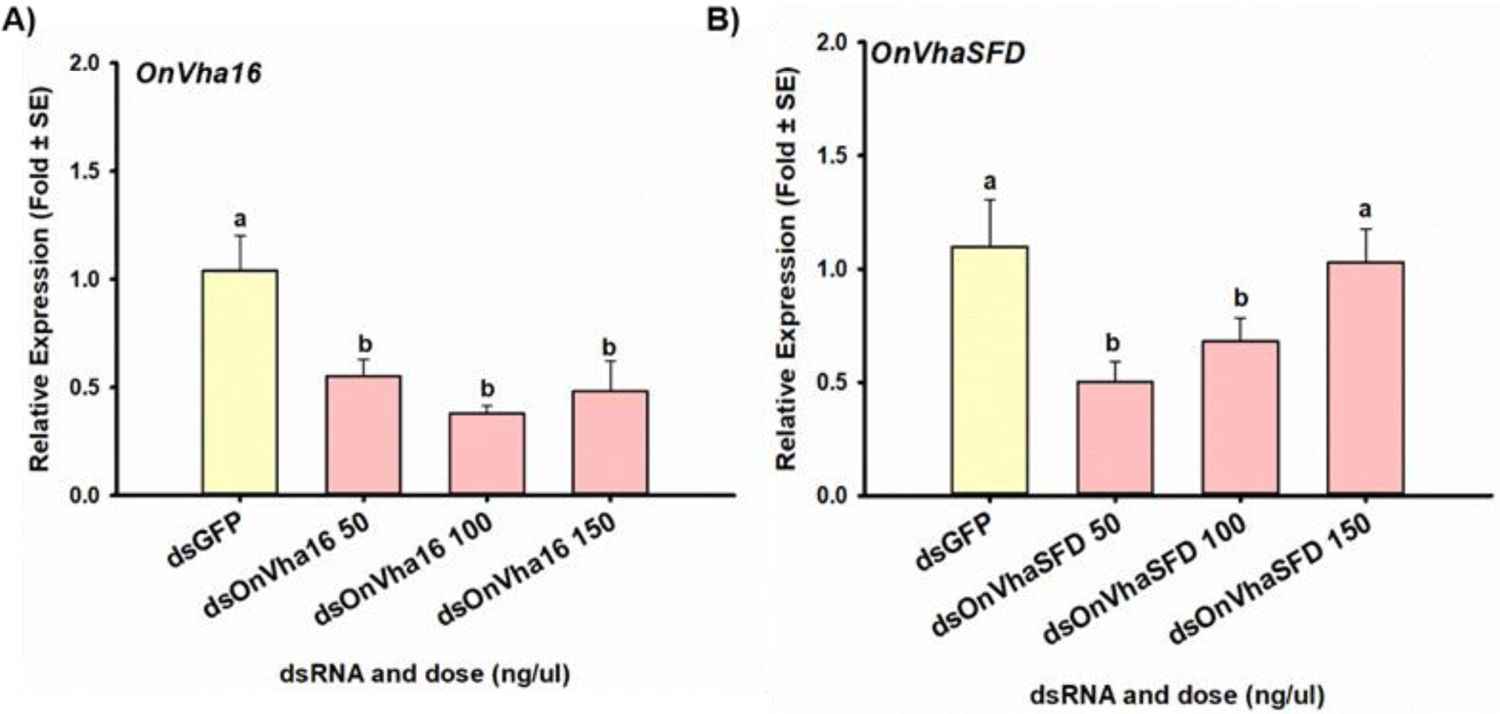
The mean relative expression of A) the V-type proton ATPase 16 kD proteolipid subunit (OnVha16), and B) the V-type proton ATPase subunit H (OnVhaSDF) target genes after incubation of excised midguts from fifth instar ECB larvae in media containing various concentrations (50, 100, and 150 ng/μl) of either dsOnVha16, dsOnVhaSFD, or dsGFP. Means that do not share a letter are significantly different.
When excised midguts were incubated with dsGFP or dsOnARF72A in the presence of various nuclease inhibitors (i.e.., Zn2+, Mn2+, Co2+), significant differences in target transcript levels relative to control were observed for Zn2+ [U(4)=10.00, p=0.030*], but not Mn2+ or Co2+ (Fig. 6). These findings suggest that Zn2+ enhanced RNAi efficiency in ECB midgut cultures. However, testing of Zn2+ on additional target genes revealed that RNAi-efficiency was only enhanced for two of the six target genes (Table 3). In the presence of Zn2+, OnARF72A, OnChc, OnVha16, and OnVhaSFD were suppressed by 92, 33, 59, and 33% respectively, but no suppression was detected for OnLgl or OnAP50 (Figs. 6A, S-25; Table 3). Compared to the previous results for these target genes without Zn2+, it appears that Zn2+ enhanced the RNAi efficiency of OnARF72A and OnChc, but not of OnVha16 or VhaSFD, which were suppressed at similar levels with and without Zn2+ (Table 3).
Figure 6. Enhancement of RNAi efficiency in ECB midgut tissue cultures due to inclusion of nuclease inhibitor Zn2+, but not Mn2+ and Co2+.
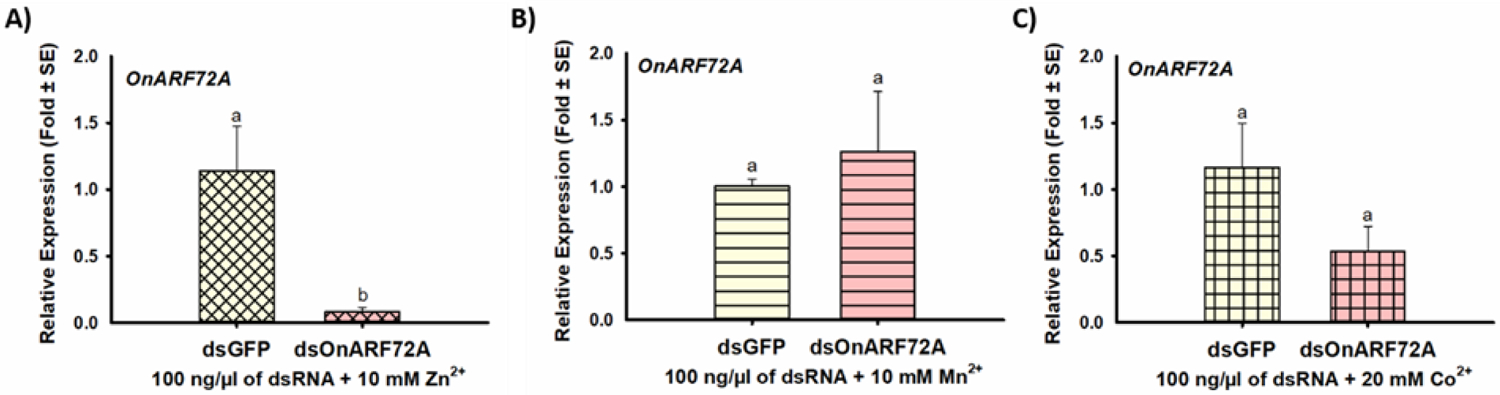
The mean relative expression of the ADP ribosylation factor 72A (OnArf72A) target gene after incubation of excised midguts from fifth instar ECB larvae in media containing either dsOnARF72A or dsGFP along with A) 10 mM Zn2+, B) 10 mM Mn2+, or C) 20 mM Co2+. Means that do not share a letter are significantly different.
4. Discussion
In the present study, we developed oral and injection RNAi procedures for multiple developmental stages of ECB and used them to test seven reagents for their ability to enhance dsRNA stability in ECB tissue extracts, as well as their ability to improve RNAi efficiency in ECB. A total of 13 RNAi bioassays were performed, but RNAi-mediated suppression of OnLgl was only observed in one of five oral RNAi experiments, and two out of eight injection experiments (Table 2). In these experiments where suppression was observed, the relative expression of OnLgl was suppressed by 68, 32, and 57%, respectively. Nonetheless, the suppression of OnLgl was only observed in 23% of all bioassays presented here (Table 2). These findings confirm that ECB is refractory to RNAi and support the need for additional strategies and reagents that can be used to enhance RNAi efficiency in ECB.
Liposomal transfection reagents are cationic lipids used to facilitate the delivery of nucleic acids into cells. Lipofectamine 2000 (Invitogen) and Metafectene Pro (Biontex) are two propriety liposomal formulations developed for the transfection of nucleic acids into eukaryotic cells. Previously, these reagents enhanced oral RNAi efficiency in D. suzukii larvae and adults15 in Euschistus heros nymphs,29 and in Chilo suppressalis.30 Metafectene Pro (Meta) dsRNA lipoplexes were previously used to achieve successful RNAi with injection in H. cunea pupae,18 and S. exigua larvae.17,16 Enhanced RNAi efficiency associated with transfection reagents was originally attributed to enhanced dsRNA uptake,15,30 but evidence suggests that both Lipofectaimine 2000 and Meta may also enhance dsRNA stability.29,30
In our experiments, Lipofectamine RNAiMax (Lipo), an RNAi-specific cationic lipid formulation designed explicitly for high transfection efficiency of siRNA and miRNA into a variety of cell types, and Meta were tested on ECB. Unfortunately, encapsulation of dsRNA in Lipo or Meta did not enhance RNAi efficiency when fed or injected into any of the ECB developmental stages tested (Table 2), even though Meta increased the stability of dsRNA in ECB hemolymph ex vivo (Table 1). These results conflict with previous reports of enhanced dsRNA-stability and/or RNAi efficiency when transfection reagents were used in lepidopteran,16–18,30 hemipteran,29 and dipteran15 insects, but agree with a report that Lipofectamine 2000 did not enhance oral RNAi efficiency in an orthopteran.31 Together these results suggest that transfection reagents, such as Lipo and Meta, are not suitable for enhancing RNAi in ECB, despite the success found in other species.
Chitosan/dsRNA nanoparticles were first developed to facilitate oral RNAi in Anopheles gambiae larvae.19 The nanoparticles have been used to facilitate oral RNAi in Aedes aegypti larvae,32,33 Chilio suppressalis larvae,30 Chironomus tentans larvae,34 Penaeus vannamei shrimp,35 and Caenorhabditis elegans nematodes.36 In C. suppressalis, chitosan/dsRNA nanoparticles enhanced dsRNA stability and uptake, resulting in enhanced RNAi efficiency.30 In addition, evidence from C. elegans indicates that uptake of chitosan/dsRNA nanoparticles occurs through clathrin-mediated endocytosis.36 Initial testing of chitosan/dsRNA nanoparticles on ECB larvae indicated that oral RNAi efficiency was not enhanced (unpublished). Therefore, a new version of proprietary chitosan-based/dsRNA nanoparticle (NP) was developed for testing on ECB. Excitingly, NP completely protected dsRNA from degradation in ECB HE and enhanced dsRNA stability in GC (Fig. 1; Table 1). Unfortunately, the NP dsRNA formulation proved challenging to assay as it was problematic to solubilize, difficult to resuspend into a homogenous solution, and would not migrate through 1% agarose gels. Nonetheless, the ex vivo data presented here demonstrate that NP are promising candidates for enhancing RNAi in ECB, as a result of enhanced dsRNA stability, but more testing is needed to ascertain effectiveness in vivo.
EDTA is a chelating agent that sequesters di- and trivalent metal ions. EDTA is often used as a nuclease inhibitor because many enzymes use metal ions as inorganic cofactors, which are required for catalytic activity. A final concentration of 10–20 mM EDTA was previously reported to enhance dsRNA stability in tissue extracts from Manduca sexta,37 T. castaneum,38 Acyrthosiphon pisum,38 and ECB.20 In addition, 3% (w/v) EDTA was reported to enhance dsRNA stability in saliva from nymphs as well as oral RNAi efficiency in Euschistus heros.29 Ex vivo incubation in ECB tissue extracts indicated that 6 mM EDTA (the highest concentration that was consumed without detrimental effects in optimization experiments) completely protected dsRNA in ECB tissue extracts (Table 1). Despite this, oral delivery of dsRNA with and without EDTA did not enhance RNAi efficiency in unfed neonate ECB larvae (Table 2). However, the consumption of EDTA for six days significantly reduced larval body weight.
Zn2+, Mn2+, and Co2+ are three divalent ions that inhibited the activity of purified BmdsRNase from Bombyx mori.39 Ex vivo incubation assays indicated that 10–20 mM Zn2+ was able to completely protect dsRNA from degradation in ECB HE and GC, whereas Mn2+ and Co2+ did not (Table 1). Injection of 25 mM Zn2+ in third instar ECB larvae resulted in 32% suppression of the OnLgl target gene (Table 2, Fig. 4). Two additional bioassays were then performed on second instar larvae using feeding and injection of dsRNA in combination with 0, 10, and 20 mM Zn2+ so the effects of dsRNA and Zn2+ on relative gene expression could be investigated for the target gene. Relative expression of OnLgl was not significantly affected at any of the early time points examined after dsRNA feeding or injection (Figs. S-22, S-23). In addition, significant suppression was only observed at the 6-d time point of the oral RNAi bioassay, but not at the 3-d time point for either delivery method (Table 2). In Ostrinia furnacalis, the transcriptional response of numerous genes was evaluated after dsRNA exposure, and most of the transcripts that were unaffected or upregulated six hours after dsRNA exposure were shown to be refractory to RNAi, whereas genes that were downregulated within six hours were effective RNAi targets.22 Based on this report, and the short-term transcriptional response observed for OnLgl after dsRNA exposure, there may be other target genes that are more sensitive to RNAi in ECB than OnLgl. Further testing of Zn2+ is needed to understand how it enhances RNAi efficiency ex vivo, and testing on additional insects is recommended.
To determine how sensitivity of OnLgl to RNAi compares to other target genes in ECB, a midgut tissue culture system was used to compare RNAi efficiency between six target genes (Table 3). This investigation was performed using ECB tissue cultures to reduce the assay time, as well as the amount of dsRNA required to screen multiple target genes. Only two of the six target genes (OnVha16 and OnSFD) exhibited RNAi-mediated suppression after 24 h of incubation in tissue culture media containing dsRNA (Table 3; Figs. 5, S-24). Surprisingly, suppression in ECB midgut cultures did not appear to be dose-dependent (Fig. 5). However, RNAi-mediated suppression was also not dose-dependent in A. aegypti,40 and A. pisum41 possibly due to insufficient activation of core RNAi pathway components.
Comparison of Zn2+, Mn2+ and Co2+ on RNAi efficiency in ECB midgut tissue cultures indicated that 20 mM Zn2+ significantly enhanced RNAi efficiency of the OnArf72A target gene (Fig. 6). Therefore, tissue culture assays using 20 mM Zn2+ were performed on the other five target genes. Surprisingly, RNAi efficiency was only enhanced by Zn2+ in two of the six genes assayed (OnArf72A and OnChc) (Table 3). Based on these results, it appears that nucleases may impact RNAi efficiency differently for different genes in ECB, possibly suggesting that nuclease activity is sequence-specific, or another unknown mechanism is present. However, our previous investigation of dsRNA stability in ECB found no evidence for sequence-specific degradation of dsRNA in ECB GC.20 Therefore, additional research is needed to understand why inhibition of nuclease activity enhances RNAi efficiency for some but not all genes in ECB midgut tissue cultures.
Taken together, the results presented here indicate that instability of dsRNA due to nuclease activity does contribute to low RNAi efficiency in ECB (at least for some target genes in ECB midgut cultures), but enhancing dsRNA stability alone may not improve RNAi efficiency in ECB. Further, RNAi efficiency in ECB did not appear to be higher in early developmental stages or non-feeding stages of ECB (Table 2), as previously observed in other insects, including many lepidopterans.5,42 Therefore, RNAi efficiency in ECB appears to be more refractory than in other species, perhaps due to the involvement of multiple mechanisms. Instability of dsRNA20 and refractory target genes (Table 3) contribute to limited RNAi efficiency in ECB, and there is evidence that deficient activation of the core RNAi pathway components may limit RNAi efficiency in ECB as well (unpublished). While incomplete internalization of dsRNA and systemic spreading have not been investigated in ECB yet, these mechanisms are known to contribute to low RNAi efficiency in other lepidopterans and may be at play in ECB. Thus, multiple reagents and strategies may need to be combined in order to enhance RNAi efficiency in ECB. In addition, it is important to select a wide range of candidate genes for screening both ex vivo and in vivo, as not all genes exhibiting specific transcript depletion will necessarily cause mortality or phenotypic defects, as was the case here for OnLgl (Table 2).
Despite the complex and challenging nature of enhancing RNAi efficiency in lepidopterans, RNAi represents a novel mode of action to combat insecticide resistance in many devastating pests, such as ECB. The reagents evaluated here that enhance dsRNA stability in ECB HE and GC are good candidates for testing in other insect species, including orthopterans and hemipterans, which often exhibit low RNAi efficiency due to rapid degradation of dsRNA.20 Furthermore, enhancing dsRNA stability could possibly reduce the amount of dsRNA needed for pest control by improving the environmental stability of dsRNA and/or the susceptibility of the pest to dsRNA (data not shown). Enhanced stability and lower doses would be especially beneficial, given the high price of producing dsRNA for pest control.43
5. Conclusion
This investigation demonstrated that RNAi efficiency in ECB was very low, and highly variable, with both feeding and injection of dsRNA targeting OnLgl, as well as incubation of midgut tissue cultures with dsRNA targeting six genes. Of the 13 RNAi bioassays and 12 tissue culture assays, gene suppression was only observed in 23 and 50% of the assays, respectively. In addition, four of the six stabilizing reagents tested enhanced the stability of dsRNA in ECB tissue extracts, ex vivo, but this was insufficient to improve RNAi efficiency in ECB, in vivo. However, enhanced dsRNA stability translated to improved RNAi efficiency in tissue culture for two of the four refractory target genes tested. These findings indicate that multiple mechanisms are contributing to low RNAi efficiency in ECB, suggesting that conventional strategies for enhancing RNAi efficiency in insects may be insufficient to increase RNAi efficiency in ECB. This knowledge will facilitate the development of novel approaches that simultaneously overcome multiple RNAi efficiency limitations in lepidopteran insects.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Wanlun Cai, Dr. Matt Heerman, Dr. Young Ho Kim, Dr. Hao Zhang, Mylah Knight, Kyle Ismert, Lina Parks, and Miriam Reyanldo for their help conducting and analyzing experiments. This research was partially supported by U.S. Department of Agriculture (USDA/NIFA 2014-67013-21714) to KYZ, and by the KSU Research and Extension Experiences for Undergraduates Program (funded by USDA/NIFA 2019-67032) to MB. Research reported in this publication was, in part, supported by the National Institute of General Medical Sciences of the National Institutes of Health (under award number NIH R01GM128659) to DHH. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. This manuscript is contribution 20-332-J from the Kansas Agricultural Experiment Station, Kansas State University, Manhattan, KS, USA.
Footnotes
CONFLICT OF INTEREST DECLARATION
The authors have no conflicts of interest to declare.
SUPPORTING INFORMATION
Cooper et al., RNAi efficiency in Ostrinia nubilalis
REFERENCES
- 1.Zhu KY, Palli SR, 2020. Mechanisms, applications, and challenges of insect RNA interference. Annu Rev Entomol 65, 293–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Godfray HCJ, Beddington JR, Crute IR, Haddad L, Lawrence D, Muir JF, et al. , 2010. Food security: the challenge of feeding 9 billion people. Sci 327, 812–818. [DOI] [PubMed] [Google Scholar]
- 3.Khajuria C, Ivashuta S, Wiggins E, Flagel L, Mora W, Pleau M, et al. , 2018. Development and characterization of the first dsRNA-resistant insect population from western corn rootworm, Diabrotica virgifera virgifera LeConte: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Whyard S, Singh AD, Wong S, 2009. Ingested double-stranded RNAs can act as species-specific insecticides. Insect Biochem Mol Biol 39, 824–32. [DOI] [PubMed] [Google Scholar]
- 5.Terenius O, Papanicolaou A, Garbutt JS, Eleftherianos I, Huvenne H, Sriramana K, et al. , 2011. RNA interference in Lepidoptera: An overview of successful and unsucessful studies and implications for experimental design. J Insect Physiol 57, 231–245. [DOI] [PubMed] [Google Scholar]
- 6.Kolliopoulou A, Swevers L, 2014. Recent progress in RNAi research in Lepidoptera: intracellular machinery, antiviral immune response and prospects for insect pest control. Curr Opin Insect Sci 6, 28–34. [DOI] [PubMed] [Google Scholar]
- 7.Khajuria C, Buschman LL, Chen M-S, Siegfried BD, Zhu KY, 2011. Identification of a novel aminopeptidase P-like gene (OnAPP) possibly involved in Bt toxicity and resistance in a major corn pest (Ostrinia nubilalis). PLoS One 6, e23983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smagghe G, Swevers L, 2014. Editorial overview: Pests and resistance — RNAi research in insects. Curr Opin Insect Sci 6, iv–v. [DOI] [PubMed] [Google Scholar]
- 9.Capinera JL, 2000. Featured Creatures: European corn borer (Web: http://entnemdept.ufl.edu/creatures/field/e_corn_borer.htm). Entomology and Nematollogy Department. Univeristy Florida: EENY-156 (2017). [Google Scholar]
- 10.Thieme TGM, Buuk C, Gloyna K, Ortego F, Farinós GP, 2018. Ten years of MON 810 resistance monitoring of field populations of Ostrinia nubilalis in Europe. J Appl Entomol 192–200. [Google Scholar]
- 11.Whitworth RJ, Michaud JP, Davis HN, 2014. Corn Insect Management 2014. Kansas State University Agricultural Experiment Station and Cooperative Extension Service, Manhattan KS. [Google Scholar]
- 12.Siegwart M, Thibord JB, Olivares J, Hirn C, Elias J, Maugin S, et al. 2017. Biochemical and molecular mechanisms associated with the resistance of the European corn borer (Lepidoptera: Crambidae) to Lambda-Cyuhalothrin and first monitoring tool. J Econ Entomol 110: 598–606. [DOI] [PubMed] [Google Scholar]
- 13.Yu T, Li X, Coates BS, Zhang Q, Siegfried BD, Zhou X, 2018. microRNA profiling between Bacillus thuringiensis Cry1Ab-suceptile and resistant European corn borer, Ostrinia nubilalis (Hubner). Insect Mol Biol 27: 279–294. [DOI] [PubMed] [Google Scholar]
- 14.Cooper AMW, Silver K, Zhang J, Park Y, Zhu KY, 2019. Molecular mechanisms influencing efficiency of RNA interference in insects. Pest Manag Sci 75, 18–28. [DOI] [PubMed] [Google Scholar]
- 15.Taning CNT, Christiaens O, Berkvens N, Casteels H, Maes M, Smagghe G, 2016. Oral RNAi to control Drosophila suzukii: laboratory testing against larval and adult stages. J Pest Sci 89:803–814. [Google Scholar]
- 16.Park Y, Kim Y, 2013. RNA interference of cadherin gene expression in Spodoptera exigua reveals its significance as a specific Bt target. J Invertebr Pathol 114, 285–291. [DOI] [PubMed] [Google Scholar]
- 17.Choi B-G., Hepat R, Kim Y, 2014. RNA interference of a heat shock protein, Hsp70, loses its protection role in indirect chilling injury to the beet armyworm, Spodoptera exigua. Comp Biochem Physiol Part A 168, 90–95. [DOI] [PubMed] [Google Scholar]
- 18.Yang F, Kamruzzaman ASM, Hiragaki S, Takeda M, 2016. The involvement of circadian clock gene period in photoperiodic time measurement of the fall webworm, Hyphantria cunea. Glob J Adv Res 3, 985–991. [Google Scholar]
- 19.Zhang X, Zhang J, Zhu KY, 2010. Chitosan/double-stranded RNA nanoparticle-mediated RNA interference to silence chitin synthase genes through larval feeding in the African malaria mosquito (Anopheles gambiae). Insect Mol Biol 19, 683–693. [DOI] [PubMed] [Google Scholar]
- 20.Cooper AMW, Yu Z, Biondi M, Song H, Silver K, Zhang J, Zhu KY, Submitted. Stability of double-stranded RNA in gut contents and hemolymph of Ostrinia nubilalis larvae. Pest Biochem Physiol XX, XXX–XXX. [DOI] [PubMed] [Google Scholar]
- 21.Xiao D, Liang X, Gao X, Yao J, Zhu KY, 2014. The lethal giant larvae gene in Tribolium castaneum: Molecular properties and roles in larval and pupal development as revealed by RNA interference. Int J Mol Sci 15, 6880–6896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guan RB, Li HC, Miao XX, 2018. Prediction of effective RNA interference targets and pathway-related genes in lepidopteran insects by RNA sequencing analysis. Insect Sci 3, 356–367. [DOI] [PubMed] [Google Scholar]
- 23.Khajuria C, Buschman LL, Chen M-S, Muthukrishnan S, Zhu KY, 2010. A gut-specific chitinase gene essential for regulation of chitin content of peritrophic matrix and growth of Ostrinia nubilalis larvae. Insect Biochem Mol Biol 40, 621–629. [DOI] [PubMed] [Google Scholar]
- 24.Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, et al. , 2009. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55, 611–622. [DOI] [PubMed] [Google Scholar]
- 25.Livak KJ, Schmittgen TD, 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(Delta Delta C(T)) Method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- 26.Andersen CL, Jensen JL, Ørntoft TF, 2004. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res 64, 5245–5250. [DOI] [PubMed] [Google Scholar]
- 27.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, et al. , 2002. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3, 34–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP, 2004. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper--Excel-based tool using pairwise correlations. Biotechnol Lett 26, 509–515. [DOI] [PubMed] [Google Scholar]
- 29.Castellanos NL, Smagghe G, Sharma R, Oliveira EE, Christiaens O, 2019. Liposome encapsulation and EDTA formulation of dsRNA targeting essential genes increase oral RNAi-caused mortality in the Neotropical stink bug Euschistus heros. Pest Manag Sci 75, 537–548. [DOI] [PubMed] [Google Scholar]
- 30.Wang K, Peng Y, Chen J, Peng Y, Wang X, Shen Z, et al. , 2019. Comparison of efficacy of RNAi mediated by various nanoparticles in the rice striped stem borer (Chilo suppressalis). Pest Biochem Physiol In Press, 104467. [DOI] [PubMed] [Google Scholar]
- 31.Luo Y, Wang X, Yu D, Chen B, Kang L, 2013. Differential responses of migratory locusts to systemic RNA interference via double-stranded RNA injection and feeding. Insect Mol Biol 22, 574–583. [DOI] [PubMed] [Google Scholar]
- 32.Das S, Debnath N, Cui Y, Unrine J, Palli S, 2015. Chitosan, carbon quantum dot, and silica nanoparticle mediated dsRNA delivery for gene silencing in Aedes aegypti: A comparative analysis. ACS Appl Mater Interfaces 7, 19530–19535. [DOI] [PubMed] [Google Scholar]
- 33.Kumar DR, Kumar PS, Gandhi MR, Al-Dhabi NA, Paulraj MG, Ignacimuthu S, 2016. Delivery of chitosan/dsRNA nanoparticles for silencing of wing development vestigial (vg) gene in Aedes aegypti mosquitoes. Int J Biol Macromol 86, 89–95. [DOI] [PubMed] [Google Scholar]
- 34.Tang G, Yao J, Li D, He Y, Zhu Y-C, Zhang X, et al. , 2017. Cytochrome P450 genes from the aquatic midge Chironomus tentans: Atrazine-induced up-regulation of CtCYP6EX3 enhanced the toxicity of chlorpyrifos. Chemosphere 186, 68e77. [DOI] [PubMed] [Google Scholar]
- 35.Ufaz S, Balter A, Tzror C, Einbender S, Koshet O, Shainsky-Roitman J, et al. , 2018. Anti-viral RNAi nanoparticles protect shrimp against white spot disease. Mol Syst Des Eng 3, 38–48. [Google Scholar]
- 36.Lichtenberg SS, Tsyusko OV, Palli SR, Unrine JM, 2019. Uptake and bioactivity of chitosan/double-stranded RNA polyplex nanoparticles in Caenorhabditis elegans. Environ Sci Technol 53, 3832–3840. [DOI] [PubMed] [Google Scholar]
- 37.Garbutt JS, Bellés X, Richards EH, Reynolds SE, 2013. Persistence of double-stranded RNA in insect hemolymph as a potential determiner of RNA interference success: evidence from Manduca sexta and Blattella germanica. J Insect Physiol 59, 171–178. [DOI] [PubMed] [Google Scholar]
- 38.Cao M, Gatehouse JA, Fitches EC, 2018. A systematic study of RNAi effects and dsRNA stability in Tribolium castaneum and Acyrthosiphon pisum, following injection and ingestion of analogous dsRNAs. Int J Mol Sci 19, 29–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arimatsu Y, Kotani E, Sugimura Y, Furusawa T, 2007. Molecular characterization of a cDNA encoding extracellular dsRNase and its expression in the silkworm, Bombyx mori. Insect Biochem Mol Biol 37, 176–183. [DOI] [PubMed] [Google Scholar]
- 40.Lopez SBG, Guimaraes-Ribeiro V, Rodriguez JVG, Dorand FAPS, Salles TS, Sa-Guimaraes TE, et al. , 2019. RNAi-based bioinsecticide for Aedes mosquito control. Sci Rep 9, 4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ye C, An X, Jiang Y-D, Ding B-Y, Shang F, Christiaens O, et al. , 2019. Induction of RNAi core machinery’s gene expression by exogenous dsRNA and the effects of pre-exposure to dsRNA on the gene silencing efficiency in the pea aphid (Acyrthosiphon pisum). Front Physiol 9, 1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Belles X, 2010. Beyond Drosophila: RNAi in vivo and functional genomics in insects. Annu Rev Entomol 55, 111–128. [DOI] [PubMed] [Google Scholar]
- 43.Mitter N, Worrall EA, Robinson KE, Xu ZP, Carroll BJ, 2017. Induction of virus resistance by exogenous application of double-stranded RNA. Curr Opin Virol 26, 49–55. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


