Abstract
Among mammalian species, skeletal structures vary greatly in their size and shapes, leading to the dramatic variety of body sizes and proportions. How different bones grow to different lengths, whether among different species, different individuals of the same species, or even in different anatomical parts of our own bodies, have always been a fascinating subject of research in biology and physiology. In the current review, we will focus on some of the recent advances in the field and discuss how these recent findings provided important new insights into the mechanisms regulating bone lengths and skeletal proportions.
Keywords: Body size, growth plate, senescence, skeletal growth, chondrocyte
Introduction
Bones come in a wide variety of sizes and shapes. The bones found in places like the skull and the pelvis are called flat bones because they appear as broad and flat plates, whose principal functions are to provide broad surfaces for muscular attachment and to protect our internal organs. In contrast, bones found in our upper and lower extremities like our legs and finger bones are called long bones because they are longer than they are wide. The principal functions of long bones are to provide support for our body weight and to facilitate movement. Importantly, elongation of long bones, or longitudinal bone growth, also represents perhaps the most important determinant of overall body growth and body size.
One longstanding mystery in biology is how different long bones grow to different lengths. Each of the long bones initially forms in the embryo as cartilage rudiment of similar sizes but subsequently diverges with differential regulation of growth. For example, human femurs in the thighs are approximately 20 times longer than the phalanges in the fingers and toes. Interestingly, mechanisms controlling differential bone growth also underlies the differences in body proportions between different mammals, as signified by the long necks in giraffes and the long fingers in bats, and even more strikingly, contributes to the vast differences in adult body sizes, for example, when comparing a mouse to a buffalo or an elephant.
Bone Elongation: Endochondral Ossification
Longitudinal bone growth proceeds through a process called endochondral ossification that happens at the growth plate, a cartilaginous structure located at the ends of long bones. The growth plate consists mainly of chondrocytes, which are arranged into three separate layers: the resting, proliferative, and hypertrophic zones. Closest to the bone end lies the resting zone which consists of chondrocytes that behave as progenitor cells that give rise to columnar clones of rapidly proliferating chondrocytes, thereby generating the proliferative zone of the growth plate (Abad et al., 2002; Newton et al., 2019; Lui, 2020). As longitudinal bone growth continues, proliferative chondrocytes farthest from the epiphysis undergo terminal differentiation into hypertrophic zone chondrocytes. During this process, the chondrocytes stop proliferating and start enlarging. These cells then either transdifferentiate into bone-forming osteoblasts (Ono et al., 2014; Yang et al., 2014; Zhou et al., 2014) or generate a cartilage matrix scaffold that is subsequently remodeled into bone. These processes ultimately give rise to new bone formation.
The final length of any given bone can be thought to be a product of three factors: the rate of growth at the growth plate, the duration by which growth happens at the growth plate, and the efficiency by which the cartilage matrix is turned into bone matrix. The growth rate at the growth plate is in turn a product of the rate of chondrocyte proliferation (in the proliferative zone) and the height of each terminally differentiated hypertrophic chondrocyte (Kember, 1993), which are regulated by numerous factors including genetic, nutritional, and environmental factors that can act independently or in conjunction to modify the rate and duration of bone elongation (Gat-Yablonski et al., 2009; Lui et al., 2014; Yakar & Isaksson, 2016). Some of these factors that regulates the growth plate act systemically, such as hormonal and nutritional factors, while some others tend to act locally and/or intracellularly, such as paracrine factors, transcription factors, and epigenetic modifications.
Femurs versus Phalanges: differential aging underlying differences in bone length
The growth rates between different growth plates could differ dramatically in a single animal at any particular age (Wilsman et al., 2008), thus contributing to the eventual difference in lengths of bones from different anatomical locations. Several studies have demonstrated that such difference in growth rates between different growth plates were highly correlated with hypertrophic chondrocyte volume (Breur et al., 1991; Kuhn et al., 1996). However, it is worth noting that chondrocyte hypertrophy is only part of the regulatory mechanisms underlying the differential growth rate between different long bones. We recently showed that the previously described differences in chondrocyte hypertrophy in different bones is due in part to a much broader phenomenon, in which the growth plates in different bones were aging differently (Lui et al., 2018).
The idea of growth plate aging, or growth plate senescence, is not particularly new. It was used to describe a gradual decline in growth plate functions with age, including a decrease in chondrocyte proliferation rate, decrease in chondrocyte hypertrophy, and the associated structural and molecular changes in the growth plate (Weise et al., 2001) preceding epiphyseal fusion, when longitudinal bone growth comes to a complete stop. Interestingly, brilliantly designed growth plate-transplantation experiments previously showed that the growth rate of the transplanted growth plate depends on the age of the donor rather than the recipient (Stevens et al., 1999), suggesting that the age of the growth plate appears not dependent on the systemic environment, but rather on the intrinsic property of the growth plate itself.
In our recent study, we compared multiple senescent changes in the growth plates of mouse tibia, femur, metacarpal and phalanx (Lui et al., 2018). We found that the gradual decline in chondrocyte proliferation, column height, and hypertrophic cell size were declining with age in all different growth plates studied, which is expected. But interestingly, the time course for these senescent changes in the shorter bones closely paralleled longer bones but appeared to be left-shifted or down-shifted, which meant that these changes were advanced in the shorter bones. Furthermore, the final stage of growth plate senescence, which is epiphyseal fusion, also occurred earlier in shorter bones than in longer bones. Consequently, the longer bones were allowed to grow at a high rate and for a longer period of time, which contributed to their greater final bone length (Figure 1). Importantly, our findings to a certain extent is consistent with prior studies that attributed the differential growth of different bones primarily to hypertrophic chondrocyte size (Breur et al., 1991; Kuhn et al., 1996). Because the ability of chondrocytes to undergo hypertrophy gradually diminish during growth plate senescence, accelerated senescence in shorter bones would also be manifested as a difference in hypertrophic size when the observations were made at any specific age.
Figure 1. Comparing the change in body length between different long bones in mice.
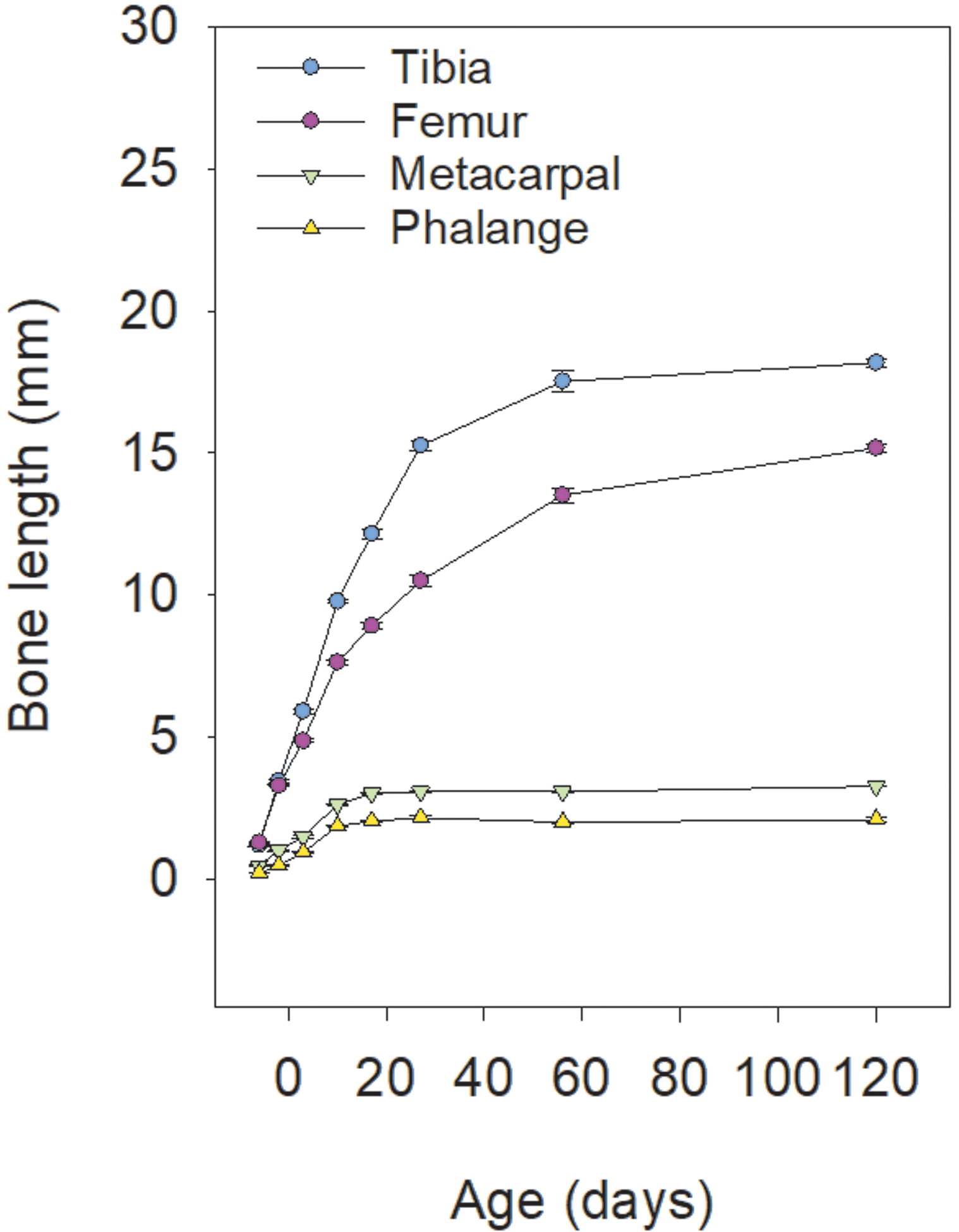
The slowing of growth, due in part to growth plate senescence, happens more slowly in longer bones like tibias and femurs, compared with metacarpals and phalanges, thus allowing a longer period of bone growth and greater final bone length.
Digging deeper, we used laser capture microdissection followed by RNA-Seq to elucidate the underlying molecular mechanisms that contribute to the accelerated growth plate senescence in the shorter bones. We found that several key paracrine signaling pathways that modulate growth plate functions were differentially regulated. For example, Insulin-like growth factors (IGF) signaling is crucial for chondrocyte proliferation and hypertrophy (Hunziker et al., 1994; Wang et al., 1999; Lui et al., 2019), and we found that IGF signaling in the longer bones appeared to be intrinsically more robust than in the shorter bones (Lui et al., 2018). Compared with the phalanges and metacarpals, chondrocytes from tibias and femurs proliferate faster and expressed higher endogenous levels of Igf2 in culture. Importantly, administration of exogenous Igf1 were able to increase proliferation in phalanges/metacarpals chondrocytes to a rate comparable to untreated tibias/femurs but were unable to further increase chondrocyte proliferation from the tibias/femurs, indicating that IGF signaling is already optimal in tibia/femoral chondrocytes. Similarly, bone morphogenetic proteins (BMPs), another important paracrine signaling pathway for chondrocyte function (De Luca et al., 2001; Yoon et al., 2006; Garrison et al., 2017), appeared to be differentially regulated in different bones. The proximal tibial growth plate expressed higher levels of BMP ligand and lower levels of BMP antagonist and when challenged with exogenous BMP antagonist, exhibited much greater inhibition of chondrocyte hypertrophy compared with that in the metacarpal. Our observations therefore suggest an intrinsically more robust BMP signaling in the longer bones, which in turn contribute to more prominent chondrocyte hypertrophy in the tibias and femurs. Taken together, our recent findings suggest that the size disparities between different long bones in our body arise in part from intrinsic differences in paracrine signaling such as IGFs and BMPs, which determines the differential modulation of growth plate senescence.
Bats, Jerboas, and Giraffes: differences in skeletal proportions
Evolutionary modulation of bone growth and bone length also play an important role in achieving different skeletal proportions in different species to facilitate adaptations to their environment. For example, wing skeletons of bats (or chiropterans), which are essentially their forelimbs, demonstrate a musculoskeletal specialization unique among mammals, including extremely elongated metacarpals and phalanges (Swartz & Middleton, 2008). Interestingly, quantitative analysis comparing the growth plates in distal elements of forelimbs (the manus) and most strikingly that of hindlimbs (the pes) showed that hypertrophic zones were vastly expanded and hypertrophic chondrocytes were dramatically elongated only in the manus (up to 60 μm paralleling the direction of growth), far exceeding what has been reported previously in any species (Farnum et al., 2008). A separate study compared gene expression between the forelimbs and hindlimbs of bats and mice and showed using immunohistochemistry that Bmp2 and BMP signaling were up-regulated specifically in the bats forelimbs (Sears et al., 2006). Taken together, these studies suggest chondrocyte hypertrophy driven by up-regulation of BMP signaling could be a major factor underlying the evolutionary modulation of forelimb elongation in chiropterans.
Another interesting study (Cooper et al., 2013) examined the Egyptian jerboas, a small bipedal rodent similar to the size of a mouse but with vastly elongated hindlimbs (Moore et al., 2015). Notably, the metatarsal length in jerboa is about two to three times the relative proportion of mouse metatarsals. Similar to the manus of bats, the hypertrophic chondrocytes in the jerboa metatarsals were dramatically enlarged compared to the mouse. But most interestingly, this study also elucidated the cellular mechanisms by which this unique cellular enlargement is achieved. The authors used quantitative phase microscopy to show that mammalian chondrocytes undergo three distinct phases of volume increase, with an initial phase of true hypertrophy (proportionate increase in dry mass and fluid uptake), followed by massive cell swelling in which the cellular dry mass is significantly diluted, and a final phase of slow and proportional increase in dry mass and fluid volume at this new lower dry mass density (Cooper et al., 2013). Remarkably, this third and final phase of chondrocyte hypertrophy was prolonged in the metatarsals of the jerboa while truncated in that of the mouse. Furthermore, the authors showed that IGF signaling is important for volume enlargement and continued production of dry mass in chondrocytes. Their findings therefore provided novel insights not only into the evolutionary modulation in jerboas but also provided a framework for understanding how cellular enlargement and skeletal sizes are regulated more generally.
A recent study examining the tallest terrestrial animal, the giraffe, has provided intriguing insights into the genes contributing to its unique features: its long neck and long legs (Agaba et al., 2016). The giraffe’s closest relative is the okapi, which is also a member of the Giraffidae family, but does not exhibit the unique characteristics of a giraffe. By comparative analysis of the genomes of the giraffe and okapi, this study found that two-thirds of the genes that diverged most significantly in giraffes played roles in physiological development. This study identified unique genetic changes in genes of the HOX, NOTCH, and FGF signaling pathways, which likely contributes to distinctive skeletal proportions in the giraffe. Significant divergences from other mammals were found in the homeobox genes HOXB3, CDX4, and NOTO. These genes are speculated to be required for restricting differential growth to the cervical vertebrae and legs in the giraffe, as homeobox genes function to identify specific body regions. Additionally, seven unique substitutions, or variants, were found in FGFRL1, a decoy FGF receptor. Notably, FGF signaling is known to regulate proper skeletal development and somite size (Dubrulle & Pourquié, 2004; Hung et al., 2007; Liu et al., 2007), and FGFRL1 is likely an inhibitor of FGF signaling associated with human overgrowth (Matoso et al., 2014). Thus, these findings indicate that FGFRL1 is a strong candidate for regulating bone growth and contributing to the unique skeletal proportions in giraffes.
Small dogs and big dogs
Body sizes also vary dramatically among different mammalian species. The mechanisms regulating not only bone sizes and lengths, but also overall body size among mammals, have remained fascinating yet elusive. Recent genome wide association studies (GWAS) on human stature (Wood et al., 2014; Marouli et al., 2017) represents an important first step toward understanding size regulation within a species. Perhaps expectedly, most of these height-associated variants commonly found in the human population have very small effect sizes, contributing to less than 1 centimeter of height gain, and hundreds if not thousands of such genetic variants - many of which located in genes expressed and function at the growth plate (Lui et al., 2012) - collectively determine the genetic potential of final adult body height among individuals.
The size and height variations within the human species is rather unremarkable compared with that found in the domestic dog, exceeding 50-fold differences in mature body weights across different breed and exhibiting the greatest physiological disparity in growth rate out of any other terrestrial vertebrate (Tryfonidou et al., 2010). The domestic dog therefore provides an excellent model for study of body size regulation within a species. One study conducted a genome-wide scan and identified a locus on chromosome 15 modulating body size variation. At this locus, a single Igf1 single-nucleotide polymorphism haplotype was present in small dog breeds but absent in large breeds, suggesting that this variant is an important contributor to body size in small dog breeds (Sutter et al., 2007).
Although virtually all small dog breeds exhibit the same fixed Igf1 haplotype, within small dogs, there still exists substantial variation in body sizes. In Portuguese water dogs, a breed that exhibit large variation in body size, the Igf1 haplotype could explain 15% of the size variation within this single breed, providing further support for the significant role Igf1 plays in contributing to body size (Sutter et al., 2007). Another study identified a nonsynonymous variant in the Igf1 receptor (IGF1R) that is associated with dog size (Hoopes et al., 2012). This variant alters a highly conserved arginine at position 204 to histidine in the IGF1R, which is predicted to inhibit hydrogen bond formation within the ligand binding subunit. Out of 13 tiny dog breeds, nine carried this genetic variant. Moreover, the allelic frequency increases further within the Poodle and Dachshund dog breeds as the size becomes smaller. These findings suggest that at least in some small dog breeds, combinations of Igf1 haplotype and Igf1r variant may contribute to regulation of body sizes.
As one might expect, the growth rates among different dog breeds also vary greatly (Parcher & Williams, 1970). By comparing a small dog breed Miniature Poodle (MP) with a large-dog breed Great Dane (GD), a study established that significant differences in IGF signaling along with differential expression of key components of the vitamin D pathway and PTHrP-IHH feedback loop at the growth plate notably contribute to the disparities in growth rates seen between small and large dog breeds (Tryfonidou et al., 2010). Interestingly, the increased longitudinal bone growth rate seen in the GD compared to the MP is in part due to the significantly reduced levels of Igf1 binding proteins (igfbp-2, −4, −6). Since Igfbps regulates the actions of Igf1 and Igf1 upregulates growth plate chondrocyte proliferation, the downregulation of Igfbps allow for increased levels of free Igf1, thereby leading to a higher rate of bone growth in the GD. Collectively, these results further signify the importance of the Igf1 signaling pathway in regulating body size variation across dog breeds.
Recent studies have used companion dogs as animal models for the study of aging as their disease profile is similar to humans (Hoffman et al., 2020). Dogs have a unique life history pattern that allows them to provide important information regarding body size and longevity. The weights of dogs were used as an approximation of longevity as a 50-fold variation in body mass is negatively associated with a 2-fold range in lifespan (Fleming et al., 2011; O’Neill et al., 2013). A particular study conducted a large-scale global metabolomic profiling of dogs ranging in size and age (Hoffman et al., 2020) and found that tryptophan was strongly associate with dog weight and that smaller dogs had significantly higher levels of tryptophan pathway metabolites, supporting that tryptophan metabolism may play an essential role in influencing aging and longevity. Given that tryptophan metabolism is strongly implicated in regulation of appetite and nutritional statues (Le Floc’h et al., 2011), it could provide another potential mechanism contributing to the regulation of adult body sizes among different dog breeds.
How about mice versus elephants?
Even more so than among different breeds of dogs, body and skeletal sizes range dramatically across different mammalian species. Interestingly, what is common among all mammalian species is that body growth and bone growth occur rapidly during early development, then slowly tapers off when approaching adulthood. However, in small mammals, the decline of body growth may occur within weeks, and in larger mammals, like humans, body growth suppression occurs gradually over years, which could help explain the larger adult body sizes (Lui & Baron, 2011). In the human fetus, body length increases rapidly at a rate of 100cm/year but declines to approximately 5cm/year by mid-childhood (Tanner & Davies, 1985). This suppression of body growth is in part driven by an intrinsic developmental genetic program that commonly occurs in multiple different organs (Lui et al., 2010). This genetic program consists of a collection of growth-promoting genes, such as Igf2, that were highly expressed during early life to allow rapid body growth but were subsequently down-regulated with age. Interestingly, because the downregulation of these growth-promoting genes were being driven by growth itself, it forms a negative feedback loop where growth gradually slow and halt when more and more growth were fulfilled as adult body size was attained (Lui et al., 2008; Lui et al., 2010).
Interestingly, when comparing the progression of this growth-limiting genetic program between the three mammalian species: mouse, rat, and sheep, we found a strong evolutionary conservation of this genetic program between the three species, and yet the progression of the program is played out much more gradually in the sheep, persisting beyond 31 weeks postfertilization, than in mice, which was largely completed by 11 weeks postfertilization (Delaney et al., 2014). Our findings therefore suggest that different mammals may use similar molecular mechanisms to control adult body sizes, but importantly, the modulation of the progression of these similar mechanisms could contribute to dramatic disparity in final body sizes. This same concept might be applicable to the regulation of longitudinal bone growth. As mentioned above, the slowing of growth in long bones are driven by growth plate senescence, followed by growth plate fusion (or closure) shortly after sexual maturity. Therefore, one may hypothesize that in larger mammals, the long bones are allowed to grow longer because they have a more gradual and paced out growth plate senescence, or that the growth plate remains open for longer or never fuses. Indeed, the decline of bone growth rate is slower in the rat compared to the mouse (Figure 2), and we predict that in even larger mammals, like in the sheep or the elephant, the decline of bone growth rate would happen yet even more slowly. Interestingly though, while in most species, epiphyseal fusion occurs shortly after the growth potential is exhausted in the growth plate (Weise et al., 2001), it does not seem to occur in mice and rats (Kilborn et al., 2002; Martin et al., 2003), which therefore argues against the notion that growth plate closure (or the lack thereof) per se is a likely explanation for interspecies size disparity. It would be interesting to see if the changes of gene expression that drives growth plate senescence progress more gradually in larger mammals, and if so, what are the molecular mechanisms that allow differentially modulation of growth plate senescence between mammals of dramatically different sizes and lengths.
Figure 2. Comparing the pace of growth plate senescence between mice and rats.
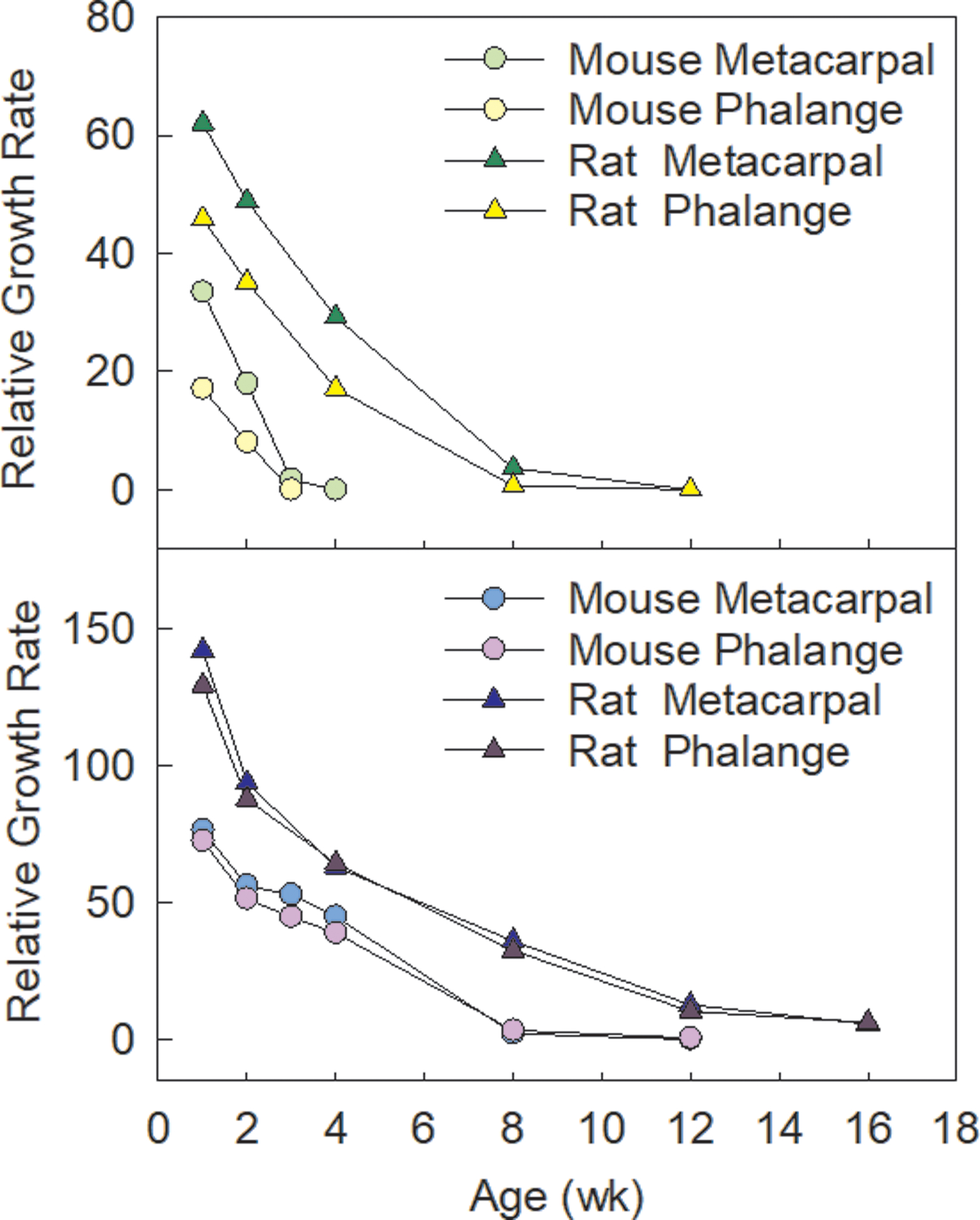
The decline of bone growth rate is slower in the rat compared to the mouse as shown in metacarpals, phalanges, tibias and femurs, which may contribute to the generally greater final bone length in rats.
Concluding Remarks
Advances in high throughput sequencing and comparative genomics in the last decade has greatly improved our understanding to the fascinating question of how body sizes and shapes are regulated. Many of the identified variants or differential expression between different bones or species were understandably implicated in signaling pathways that are important in the growth plate, such as IGFs, FGFs, and BMPs. However, modulating the IGF signaling pathway alone, for example, would likely not turn a mouse into the size of an elephant, and one must feel that something bigger, some holistic regulatory mechanisms seem to be missing. The answer to that might go well beyond the coding region in the mammalian genome. Identification of more powerful underlying regulatory elements, genetic or genomic network that can integrate and modulate gene expression more globally to exert the control over the evolution of skeletal size and proportion represents the next major challenge in the field.
New Findings:
1. What is the topic of this review?
Mechanisms regulating bone length and skeletal proportions
2. What advances does it highlight?
Recent advances being highlighted included study on differential bone length between leg and finger bones (Lui, Plos Biol 2018), metatarsals of the Egyptian jerboa (Cooper, Nature 2013) and genomic analysis of Giraffes (Agaba, Nat Comm 2016).
Source of Funding:
This work was supported by the Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development, NIH
Biography
Julian Lui is a staff scientist in the Section on Growth and Development, National Institute of Child Health and Human Development, NIH. He received his PhD in biochemistry from the Chinese University of Hong Kong in 2006 and joined his current group after his graduation. In the past 10 years, Dr. Julian Lui focuses on understanding the genetics and molecular pathophysiology of childhood growth disorders, and developing new treatment for skeletal diseases. He has published more than 50 peer-reviewed research article, reviews and book chapters in the field.
Footnotes
Conflict of Interest: None declared
Reference
- Abad V, Meyers JL, Weise M, Gafni RI, Barnes KM, Nilsson O, Bacher JD & Baron J (2002). The role of the resting zone in growth plate chondrogenesis. Endocrinology 143, 1851–1857. [DOI] [PubMed] [Google Scholar]
- Agaba M, Ishengoma E, Miller WC, McGrath BC, Hudson CN, Bedoya Reina OC, Ratan A, Burhans R, Chikhi R, Medvedev P, Praul CA, Wu-Cavener L, Wood B, Robertson H, Penfold L & Cavener DR (2016). Giraffe genome sequence reveals clues to its unique morphology and physiology. Nat Commun 7, 11519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breur GJ, VanEnkevort BA, Farnum CE & Wilsman NJ (1991). Linear relationship between the volume of hypertrophic chondrocytes and the rate of longitudinal bone growth in growth plates. J Orthop Res 9, 348–359. [DOI] [PubMed] [Google Scholar]
- Cooper KL, Oh S, Sung Y, Dasari RR, Kirschner MW & Tabin CJ (2013). Multiple phases of chondrocyte enlargement underlie differences in skeletal proportions. Nature 495, 375–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Luca F, Barnes KM, Uyeda JA, De-Levi S, Abad V, Palese T, Mericq V & Baron J (2001). Regulation of growth plate chondrogenesis by bone morphogenetic protein-2. Endocrinology 142, 430–436. [DOI] [PubMed] [Google Scholar]
- Delaney A, Padmanabhan V, Rezvani G, Chen W, Forcinito P, Cheung CS, Baron J & Lui JC (2014). Evolutionary conservation and modulation of a juvenile growth-regulating genetic program. J Mol Endocrinol 52, 269–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubrulle J & Pourquié O (2004). fgf8 mRNA decay establishes a gradient that couples axial elongation to patterning in the vertebrate embryo. Nature 427, 419–422. [DOI] [PubMed] [Google Scholar]
- Farnum CE, Tinsley M & Hermanson JW (2008). Forelimb versus hindlimb skeletal development in the big brown bat, Eptesicus fuscus: functional divergence is reflected in chondrocytic performance in Autopodial growth plates. Cells Tissues Organs 187, 35–47. [DOI] [PubMed] [Google Scholar]
- Fleming JM, Creevy KE & Promislow DE (2011). Mortality in north american dogs from 1984 to 2004: an investigation into age-, size-, and breed-related causes of death. J Vet Intern Med 25, 187–198. [DOI] [PubMed] [Google Scholar]
- Garrison P, Yue S, Hanson J, Baron J & Lui JC (2017). Spatial regulation of bone morphogenetic proteins (BMPs) in postnatal articular and growth plate cartilage. PLoS One 12, e0176752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gat-Yablonski G, Yackobovitch-Gavan M & Phillip M (2009). Nutrition and bone growth in pediatrics. Endocrinol Metab Clin North Am 38, 565–586. [DOI] [PubMed] [Google Scholar]
- Hoffman JM, Kiklevich JV, Austad M, Tran V, Jones DP, Royal A, Henry C & Austad SN (2020). Tryptophan metabolism is differently regulated between large and small dogs. Geroscience 42, 881–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoopes BC, Rimbault M, Liebers D, Ostrander EA & Sutter NB (2012). The insulin-like growth factor 1 receptor (IGF1R) contributes to reduced size in dogs. Mamm Genome 23, 780–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung IH, Yu K, Lavine KJ & Ornitz DM (2007). FGF9 regulates early hypertrophic chondrocyte differentiation and skeletal vascularization in the developing stylopod. Dev Biol 307, 300–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunziker EB, Wagner J & Zapf J (1994). Differential effects of insulin-like growth factor I and growth hormone on developmental stages of rat growth plate chondrocytes in vivo. J Clin Invest 93, 1078–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kember NF (1993). Cell kinetics and the control of bone growth. Acta Paediatr Suppl 82 Suppl 391, 61–65. [DOI] [PubMed] [Google Scholar]
- Kilborn SH, Trudel G & Uhthoff H (2002). Review of growth plate closure compared with age at sexual maturity and lifespan in laboratory animals. Contemp Top Lab Anim Sci 41, 21–26. [PubMed] [Google Scholar]
- Kuhn JL, DeLacey JH & Leenellett EE (1996). Relationship between bone growth rate and hypertrophic chondrocyte volume in New Zealand white rabbits of varying ages. J Orthop Res 14, 706–711. [DOI] [PubMed] [Google Scholar]
- Le Floc’h N, Otten W & Merlot E (2011). Tryptophan metabolism, from nutrition to potential therapeutic applications. Amino Acids 41, 1195–1205. [DOI] [PubMed] [Google Scholar]
- Liu Z, Lavine KJ, Hung IH & Ornitz DM (2007). FGF18 is required for early chondrocyte proliferation, hypertrophy and vascular invasion of the growth plate. Dev Biol 302, 80–91. [DOI] [PubMed] [Google Scholar]
- Lui JC (2020). Home for a rest: stem cell niche of the postnatal growth plate. J Endocrinol 246, R1–r11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui JC & Baron J (2011). Mechanisms limiting body growth in mammals. Endocr Rev 32, 422–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui JC, Colbert M, Cheung CSF, Ad M, Lee A, Zhu Z, Barnes KM, Dimitrov DS & Baron J (2019). Cartilage-Targeted IGF-1 Treatment to Promote Longitudinal Bone Growth. Mol Ther 27, 673–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui JC, Finkielstain GP, Barnes KM & Baron J (2008). An imprinted gene network that controls mammalian somatic growth is down-regulated during postnatal growth deceleration in multiple organs. Am J Physiol Regul Integr Comp Physiol 295, R189–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui JC, Forcinito P, Chang M, Chen W, Barnes KM & Baron J (2010). Coordinated postnatal down-regulation of multiple growth-promoting genes: evidence for a genetic program limiting organ growth. Faseb j 24, 3083–3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui JC, Jee YH, Garrison P, Iben JR, Yue S, Ad M, Nguyen Q, Kikani B, Wakabayashi Y & Baron J (2018). Differential aging of growth plate cartilage underlies differences in bone length and thus helps determine skeletal proportions. PLoS Biol 16, e2005263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui JC, Nilsson O & Baron J (2014). Recent research on the growth plate: Recent insights into the regulation of the growth plate. J Mol Endocrinol 53, T1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui JC, Nilsson O, Chan Y, Palmer CD, Andrade AC, Hirschhorn JN & Baron J (2012). Synthesizing genome-wide association studies and expression microarray reveals novel genes that act in the human growth plate to modulate height. Hum Mol Genet 21, 5193–5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marouli E, Graff M, Medina-Gomez C, Lo KS, Wood AR, Kjaer TR, Fine RS, Lu Y, Schurmann C, Highland HM, Rüeger S, Thorleifsson G, Justice AE, Lamparter D, Stirrups KE, Turcot V, Young KL, Winkler TW, Esko T, Karaderi T, Locke AE, Masca NG, Ng MC, Mudgal P, Rivas MA, Vedantam S, Mahajan A, Guo X, Abecasis G, Aben KK, Adair LS, Alam DS, Albrecht E, Allin KH, Allison M, Amouyel P, Appel EV, Arveiler D, Asselbergs FW, Auer PL, Balkau B, Banas B, Bang LE, Benn M, Bergmann S, Bielak LF, Blüher M, Boeing H, Boerwinkle E, Böger CA, Bonnycastle LL, Bork-Jensen J, Bots ML, Bottinger EP, Bowden DW, Brandslund I, Breen G, Brilliant MH, Broer L, Burt AA, Butterworth AS, Carey DJ, Caulfield MJ, Chambers JC, Chasman DI, Chen YI, Chowdhury R, Christensen C, Chu AY, Cocca M, Collins FS, Cook JP, Corley J, Galbany JC, Cox AJ, Cuellar-Partida G, Danesh J, Davies G, de Bakker PI, de Borst GJ, de Denus S, de Groot MC, de Mutsert R, Deary IJ, Dedoussis G, Demerath EW, den Hollander AI, Dennis JG, Di Angelantonio E, Drenos F, Du M, Dunning AM, Easton DF, Ebeling T, Edwards TL, Ellinor PT, Elliott P, Evangelou E, Farmaki AE, Faul JD, Feitosa MF, Feng S, Ferrannini E, Ferrario MM, Ferrieres J, Florez JC, Ford I, Fornage M, Franks PW, Frikke-Schmidt R, Galesloot TE, Gan W, Gandin I, Gasparini P, Giedraitis V, Giri A, Girotto G, Gordon SD, Gordon-Larsen P, Gorski M, Grarup N, Grove ML, Gudnason V, Gustafsson S, Hansen T, Harris KM, Harris TB, Hattersley AT, Hayward C, He L, Heid IM, Heikkilä K, Helgeland Ø, Hernesniemi J, Hewitt AW, Hocking LJ, Hollensted M, Holmen OL, Hovingh GK, Howson JM, Hoyng CB, Huang PL, Hveem K, Ikram MA, Ingelsson E, Jackson AU, Jansson JH, Jarvik GP, Jensen GB, Jhun MA, Jia Y, Jiang X, Johansson S, Jørgensen ME, Jørgensen T, Jousilahti P, Jukema JW, Kahali B, Kahn RS, Kähönen M, Kamstrup PR, Kanoni S, Kaprio J, Karaleftheri M, Kardia SL, Karpe F, Kee F, Keeman R, Kiemeney LA, Kitajima H, Kluivers KB, Kocher T, Komulainen P, Kontto J, Kooner JS, Kooperberg C, Kovacs P, Kriebel J, Kuivaniemi H, Küry S, Kuusisto J, La Bianca M, Laakso M, Lakka TA, Lange EM, Lange LA, Langefeld CD, Langenberg C, Larson EB, Lee IT, Lehtimäki T, Lewis CE, Li H, Li J, Li-Gao R, Lin H, Lin LA, Lin X, Lind L, Lindström J, Linneberg A, Liu Y, Liu Y, Lophatananon A, Luan J, Lubitz SA, Lyytikäinen LP, Mackey DA, Madden PA, Manning AK, Männistö S, Marenne G, Marten J, Martin NG, Mazul AL, Meidtner K, Metspalu A, Mitchell P, Mohlke KL, Mook-Kanamori DO, Morgan A, Morris AD, Morris AP, Müller-Nurasyid M, Munroe PB, Nalls MA, Nauck M, Nelson CP, Neville M, Nielsen SF, Nikus K, Njølstad PR, Nordestgaard BG, Ntalla I, O’Connel JR, Oksa H, Loohuis LM, Ophoff RA, Owen KR, Packard CJ, Padmanabhan S, Palmer CN, Pasterkamp G, Patel AP, Pattie A, Pedersen O, Peissig PL, Peloso GM, Pennell CE, Perola M, Perry JA, Perry JR, Person TN, Pirie A, Polasek O, Posthuma D, Raitakari OT, Rasheed A, Rauramaa R, Reilly DF, Reiner AP, Renström F, Ridker PM, Rioux JD, Robertson N, Robino A, Rolandsson O, Rudan I, Ruth KS, Saleheen D, Salomaa V, Samani NJ, Sandow K, Sapkota Y, Sattar N, Schmidt MK, Schreiner PJ, Schulze MB, Scott RA, Segura-Lepe MP, Shah S, Sim X, Sivapalaratnam S, Small KS, Smith AV, Smith JA, Southam L, Spector TD, Speliotes EK, Starr JM, Steinthorsdottir V, Stringham HM, Stumvoll M, Surendran P, t Hart LM, Tansey KE, Tardif JC, Taylor KD, Teumer A, Thompson DJ, Thorsteinsdottir U, Thuesen BH, Tönjes A, Tromp G, Trompet S, Tsafantakis E, Tuomilehto J, Tybjaerg-Hansen A, Tyrer JP, Uher R, Uitterlinden AG, Ulivi S, van der Laan SW, Van Der Leij AR, van Duijn CM, van Schoor NM, van Setten J, Varbo A, Varga TV, Varma R, Edwards DR, Vermeulen SH, Vestergaard H, Vitart V, Vogt TF, Vozzi D, Walker M, Wang F, Wang CA, Wang S, Wang Y, Wareham NJ, Warren HR, Wessel J, Willems SM, Wilson JG, Witte DR, Woods MO, Wu Y, Yaghootkar H, Yao J, Yao P, Yerges-Armstrong LM, Young R, Zeggini E, Zhan X, Zhang W, Zhao JH, Zhao W, Zhao W, Zheng H, Zhou W, Rotter JI, Boehnke M, Kathiresan S, McCarthy MI, Willer CJ, Stefansson K, Borecki IB, Liu DJ, North KE, Heard-Costa NL, Pers TH, Lindgren CM, Oxvig C, Kutalik Z, Rivadeneira F, Loos RJ, Frayling TM, Hirschhorn JN, Deloukas P & Lettre G (2017). Rare and low-frequency coding variants alter human adult height. Nature 542, 186–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin EA, Ritman EL & Turner RT (2003). Time course of epiphyseal growth plate fusion in rat tibiae. Bone 32, 261–267. [DOI] [PubMed] [Google Scholar]
- Matoso E, Ramos F, Ferrão J, Pires LM, Mascarenhas A, Melo JB & Carreira IM (2014). Interstitial 287 kb deletion of 4p16.3 including FGFRL1 gene associated with language impairment and overgrowth. Mol Cytogenet 7, 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore TY, Organ CL, Edwards SV, Biewener AA, Tabin CJ, Jenkins FA Jr. & Cooper KL (2015). Multiple phylogenetically distinct events shaped the evolution of limb skeletal morphologies associated with bipedalism in the jerboas. Curr Biol 25, 2785–2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton PT, Li L, Zhou B, Schweingruber C, Hovorakova M, Xie M, Sun X, Sandhow L, Artemov AV, Ivashkin E, Suter S, Dyachuk V, El Shahawy M, Gritli-Linde A, Bouderlique T, Petersen J, Mollbrink A, Lundeberg J, Enikolopov G, Qian H, Fried K, Kasper M, Hedlund E, Adameyko I, Sävendahl L & Chagin AS (2019). A radical switch in clonality reveals a stem cell niche in the epiphyseal growth plate. Nature 567, 234–238. [DOI] [PubMed] [Google Scholar]
- O’Neill DG, Church DB, McGreevy PD, Thomson PC & Brodbelt DC (2013). Longevity and mortality of owned dogs in England. Vet J 198, 638–643. [DOI] [PubMed] [Google Scholar]
- Ono N, Ono W, Nagasawa T & Kronenberg HM (2014). A subset of chondrogenic cells provides early mesenchymal progenitors in growing bones. Nat Cell Biol 16, 1157–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parcher JW & Williams JR (1970). Skeletal system: ossification In The Beagle as an Experimental Dog, vol. 9 ed. Andersen AC, pp. 149–225. Iowa State Univ. Press, Ames, IA. [Google Scholar]
- Sears KE, Behringer RR, Rasweiler JJt & Niswander LA (2006). Development of bat flight: morphologic and molecular evolution of bat wing digits. Proc Natl Acad Sci U S A 103, 6581–6586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens DG, Boyer MI & Bowen CV (1999). Transplantation of epiphyseal plate allografts between animals of different ages. J Pediatr Orthop 19, 398–403. [PubMed] [Google Scholar]
- Sutter NB, Bustamante CD, Chase K, Gray MM, Zhao K, Zhu L, Padhukasahasram B, Karlins E, Davis S, Jones PG, Quignon P, Johnson GS, Parker HG, Fretwell N, Mosher DS, Lawler DF, Satyaraj E, Nordborg M, Lark KG, Wayne RK & Ostrander EA (2007). A single IGF1 allele is a major determinant of small size in dogs. Science 316, 112–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swartz SM & Middleton KM (2008). Biomechanics of the bat limb skeleton: scaling, material properties and mechanics. Cells Tissues Organs 187, 59–84. [DOI] [PubMed] [Google Scholar]
- Tanner JM & Davies PS (1985). Clinical longitudinal standards for height and height velocity for North American children. J Pediatr 107, 317–329. [DOI] [PubMed] [Google Scholar]
- Tryfonidou MA, Hazewinkel HA, Riemers FM, Brinkhof B, Penning LC & Karperien M (2010). Intraspecies disparity in growth rate is associated with differences in expression of local growth plate regulators. Am J Physiol Endocrinol Metab 299, E1044–1052. [DOI] [PubMed] [Google Scholar]
- Wang J, Zhou J & Bondy CA (1999). Igf1 promotes longitudinal bone growth by insulin-like actions augmenting chondrocyte hypertrophy. Faseb j 13, 1985–1990. [DOI] [PubMed] [Google Scholar]
- Weise M, De-Levi S, Barnes KM, Gafni RI, Abad V & Baron J (2001). Effects of estrogen on growth plate senescence and epiphyseal fusion. Proc Natl Acad Sci U S A 98, 6871–6876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilsman NJ, Bernardini ES, Leiferman E, Noonan K & Farnum CE (2008). Age and pattern of the onset of differential growth among growth plates in rats. J Orthop Res 26, 1457–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood AR, Esko T, Yang J, Vedantam S, Pers TH, Gustafsson S, Chu AY, Estrada K, Luan J, Kutalik Z, Amin N, Buchkovich ML, Croteau-Chonka DC, Day FR, Duan Y, Fall T, Fehrmann R, Ferreira T, Jackson AU, Karjalainen J, Lo KS, Locke AE, Mägi R, Mihailov E, Porcu E, Randall JC, Scherag A, Vinkhuyzen AA, Westra HJ, Winkler TW, Workalemahu T, Zhao JH, Absher D, Albrecht E, Anderson D, Baron J, Beekman M, Demirkan A, Ehret GB, Feenstra B, Feitosa MF, Fischer K, Fraser RM, Goel A, Gong J, Justice AE, Kanoni S, Kleber ME, Kristiansson K, Lim U, Lotay V, Lui JC, Mangino M, Mateo Leach I, Medina-Gomez C, Nalls MA, Nyholt DR, Palmer CD, Pasko D, Pechlivanis S, Prokopenko I, Ried JS, Ripke S, Shungin D, Stancáková A, Strawbridge RJ, Sung YJ, Tanaka T, Teumer A, Trompet S, van der Laan SW, van Setten J, Van Vliet-Ostaptchouk JV, Wang Z, Yengo L, Zhang W, Afzal U, Arnlöv J, Arscott GM, Bandinelli S, Barrett A, Bellis C, Bennett AJ, Berne C, Blüher M, Bolton JL, Böttcher Y, Boyd HA, Bruinenberg M, Buckley BM, Buyske S, Caspersen IH, Chines PS, Clarke R, Claudi-Boehm S, Cooper M, Daw EW, De Jong PA, Deelen J, Delgado G, Denny JC, Dhonukshe-Rutten R, Dimitriou M, Doney AS, Dörr M, Eklund N, Eury E, Folkersen L, Garcia ME, Geller F, Giedraitis V, Go AS, Grallert H, Grammer TB, Gräßler J, Grönberg H, de Groot LC, Groves CJ, Haessler J, Hall P, Haller T, Hallmans G, Hannemann A, Hartman CA, Hassinen M, Hayward C, Heard-Costa NL, Helmer Q, Hemani G, Henders AK, Hillege HL, Hlatky MA, Hoffmann W, Hoffmann P, Holmen O, Houwing-Duistermaat JJ, Illig T, Isaacs A, James AL, Jeff J, Johansen B, Johansson Å, Jolley J, Juliusdottir T, Junttila J, Kho AN, Kinnunen L, Klopp N, Kocher T, Kratzer W, Lichtner P, Lind L, Lindström J, Lobbens S, Lorentzon M, Lu Y, Lyssenko V, Magnusson PK, Mahajan A, Maillard M, McArdle WL, McKenzie CA, McLachlan S, McLaren PJ, Menni C, Merger S, Milani L, Moayyeri A, Monda KL, Morken MA, Müller G, Müller-Nurasyid M, Musk AW, Narisu N, Nauck M, Nolte IM, Nöthen MM, Oozageer L, Pilz S, Rayner NW, Renstrom F, Robertson NR, Rose LM, Roussel R, Sanna S, Scharnagl H, Scholtens S, Schumacher FR, Schunkert H, Scott RA, Sehmi J, Seufferlein T, Shi J, Silventoinen K, Smit JH, Smith AV, Smolonska J, Stanton AV, Stirrups K, Stott DJ, Stringham HM, Sundström J, Swertz MA, Syvänen AC, Tayo BO, Thorleifsson G, Tyrer JP, van Dijk S, van Schoor NM, van der Velde N, van Heemst D, van Oort FV, Vermeulen SH, Verweij N, Vonk JM, Waite LL, Waldenberger M, Wennauer R, Wilkens LR, Willenborg C, Wilsgaard T, Wojczynski MK, Wong A, Wright AF, Zhang Q, Arveiler D, Bakker SJ, Beilby J, Bergman RN, Bergmann S, Biffar R, Blangero J, Boomsma DI, Bornstein SR, Bovet P, Brambilla P, Brown MJ, Campbell H, Caulfield MJ, Chakravarti A, Collins R, Collins FS, Crawford DC, Cupples LA, Danesh J, de Faire U, den Ruijter HM, Erbel R, Erdmann J, Eriksson JG, Farrall M, Ferrannini E, Ferrières J, Ford I, Forouhi NG, Forrester T, Gansevoort RT, Gejman PV, Gieger C, Golay A, Gottesman O, Gudnason V, Gyllensten U, Haas DW, Hall AS, Harris TB, Hattersley AT, Heath AC, Hengstenberg C, Hicks AA, Hindorff LA, Hingorani AD, Hofman A, Hovingh GK, Humphries SE, Hunt SC, Hypponen E, Jacobs KB, Jarvelin MR, Jousilahti P, Jula AM, Kaprio J, Kastelein JJ, Kayser M, Kee F, Keinanen-Kiukaanniemi SM, Kiemeney LA, Kooner JS, Kooperberg C, Koskinen S, Kovacs P, Kraja AT, Kumari M, Kuusisto J, Lakka TA, Langenberg C, Le Marchand L, Lehtimäki T, Lupoli S, Madden PA, Männistö S, Manunta P, Marette A, Matise TC, McKnight B, Meitinger T, Moll FL, Montgomery GW, Morris AD, Morris AP, Murray JC, Nelis M, Ohlsson C, Oldehinkel AJ, Ong KK, Ouwehand WH, Pasterkamp G, Peters A, Pramstaller PP, Price JF, Qi L, Raitakari OT, Rankinen T, Rao DC, Rice TK, Ritchie M, Rudan I, Salomaa V, Samani NJ, Saramies J, Sarzynski MA, Schwarz PE, Sebert S, Sever P, Shuldiner AR, Sinisalo J, Steinthorsdottir V, Stolk RP, Tardif JC, Tönjes A, Tremblay A, Tremoli E, Virtamo J, Vohl MC, Amouyel P, Asselbergs FW, Assimes TL, Bochud M, Boehm BO, Boerwinkle E, Bottinger EP, Bouchard C, Cauchi S, Chambers JC, Chanock SJ, Cooper RS, de Bakker PI, Dedoussis G, Ferrucci L, Franks PW, Froguel P, Groop LC, Haiman CA, Hamsten A, Hayes MG, Hui J, Hunter DJ, Hveem K, Jukema JW, Kaplan RC, Kivimaki M, Kuh D, Laakso M, Liu Y, Martin NG, März W, Melbye M, Moebus S, Munroe PB, Njølstad I, Oostra BA, Palmer CN, Pedersen NL, Perola M, Pérusse L, Peters U, Powell JE, Power C, Quertermous T, Rauramaa R, Reinmaa E, Ridker PM, Rivadeneira F, Rotter JI, Saaristo TE, Saleheen D, Schlessinger D, Slagboom PE, Snieder H, Spector TD, Strauch K, Stumvoll M, Tuomilehto J, Uusitupa M, van der Harst P, Völzke H, Walker M, Wareham NJ, Watkins H, Wichmann HE, Wilson JF, Zanen P, Deloukas P, Heid IM, Lindgren CM, Mohlke KL, Speliotes EK, Thorsteinsdottir U, Barroso I, Fox CS, North KE, Strachan DP, Beckmann JS, Berndt SI, Boehnke M, Borecki IB, McCarthy MI, Metspalu A, Stefansson K, Uitterlinden AG, van Duijn CM, Franke L, Willer CJ, Price AL, Lettre G, Loos RJ, Weedon MN, Ingelsson E, O’Connell JR, Abecasis GR, Chasman DI, Goddard ME, Visscher PM, Hirschhorn JN & Frayling TM (2014). Defining the role of common variation in the genomic and biological architecture of adult human height. Nat Genet 46, 1173–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakar S & Isaksson O (2016). Regulation of skeletal growth and mineral acquisition by the GH/IGF-1 axis: Lessons from mouse models. Growth Horm IGF Res 28, 26–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Tsang KY, Tang HC, Chan D & Cheah KS (2014). Hypertrophic chondrocytes can become osteoblasts and osteocytes in endochondral bone formation. Proc Natl Acad Sci U S A 111, 12097–12102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon BS, Pogue R, Ovchinnikov DA, Yoshii I, Mishina Y, Behringer RR & Lyons KM (2006). BMPs regulate multiple aspects of growth-plate chondrogenesis through opposing actions on FGF pathways. Development 133, 4667–4678. [DOI] [PubMed] [Google Scholar]
- Zhou X, von der Mark K, Henry S, Norton W, Adams H & de Crombrugghe B (2014). Chondrocytes transdifferentiate into osteoblasts in endochondral bone during development, postnatal growth and fracture healing in mice. PLoS Genet 10, e1004820. [DOI] [PMC free article] [PubMed] [Google Scholar]


