Abstract
INTRODUCTION:
Periprosthetic joint infection (PJI) data elements are contained in both structured and unstructured documents in electronic health records and require manual data collection. The goal of this study was to develop a natural language processing (NLP) algorithm to replicate manual chart review for PJI data elements.
METHODS:
PJI were identified among all TJA procedures performed at a single academic institution between 2000 and 2017. Data elements that comprise the Musculoskeletal Infection Society (MSIS) criteria were manually extracted and used as the gold standard for validation. A training sample of 1197 TJA surgeries (170 PJI cases) was randomly selected to develop the prototype NLP algorithms and an additional 1179 surgeries (150 PJI cases) were randomly selected as the test sample. The algorithms were applied to all consultation notes, operative notes, pathology reports and microbiology reports to predict the correct status of PJI based on MSIS criteria.
RESULTS:
The algorithm --which identified patients with PJI based on MSIS criteria--achieved an f1-score (harmonic mean of precision and recall) of 0.911. Algorithm performance in extracting the presence of sinus tract, purulence, pathological documentation of inflammation, and growth of cultured organisms from the involved TJA achieved f1-scores ranged from 0.771 to 0.982, sensitivity ranged from 0.730 to 1.000, and specificity ranged from 0.947 to 1.000.
CONCLUSION:
NLP-enabled algorithms have the potential to automate data collection for PJI diagnostic elements, which could directly improve patient care and augment cohort surveillance and research efforts. Further validation is needed in other hospital settings.
Level of Evidence:
Level III, Diagnostic
Keywords: total joint arthroplasty, periprosthetic joint infection, natural language processing, electronic health records, informatics, artificial intelligence, data science, machine learning
INTRODUCTION
Periprosthetic joint infections (PJI) following total joint arthroplasty (TJA) are associated with significant morbidity, mortality and economic burden (1, 2). In the clinical setting, diagnosing PJI remains a major challenge as there are no singular, conclusive diagnostic tests. Most patients present with joint pain as the main symptom, which carries a broad differential diagnosis. PJI diagnosis is typically based on a combination of clinical findings, laboratory results from peripheral blood and synovial fluid, microbiological culture, histological evaluation of periprosthetic tissue and intraoperative findings, as defined by the Musculoskeletal Infection Society (MSIS) and the Infectious Diseases Society of America (3, 4). These definitions, although relatively new and subject to periodic refinement and scrutiny, are widely adopted in the orthopedic and infectious diseases communities. Since their creation, evidence-based criteria have significantly improved clinical decision-making and research by allowing for consistency across studies, thus enhancing the potential for collaboration. Yet, data elements that are included in these definitions are recorded in multiple sections of electronic health records (EHR), which leads to a cumbersome process for physicians caring for patients with suspected PJI and is an even more daunting challenge for patient surveillance and research efforts. Furthermore, although diagnostic tests for PJI continue to evolve, timely, consistent, and actionable diagnosis of PJI remains elusive in the clinical setting. Similarly, in the research setting, large administrative databases and surveillance programs (i.e., U.S. National Healthcare Safety Network) offer unique opportunities for evidence generation in large cohorts; yet distinguishing the type of surgical site infections (superficial infections involving the skin and soft tissues beneath the skin versus PJI involving deeper tissues and indwelling orthopedic hardware) remains a methodological challenge that prevents comparisons across studies. Manual abstraction of PJI data elements for research purposes is also time-intensive even for trained and experienced nurse abstractors.
As described by our group and others, natural language processing (NLP) methods are increasingly used for both clinical and research purposes and offer an opportunity to efficiently extract data elements that are embedded in the unstructured text of the EHR (5–7). Several groups also described application of NLP methods for identification of surgical site infections (8–11). Most recently, Thirukumaran and colleagues developed an orthopedic-specific NLP algorithm to retrospectively identify 172 surgical site infections in a cohort of 1,407 patients who underwent various orthopedic procedures (12). Yet, the algorithm was not specific to TJA (for which deep infections are more devastating than outside of a joint) and did not distinguish the type of surgical site infections (superficial versus deep versus PJI). In partnership with orthopedic surgeons, infectious disease physicians and data scientists, we developed a PJI-specific NLP algorithm to replicate manual chart review for specific PJI data elements as well as PJI case detection based on MSIS criteria. We evaluated the accuracy of the algorithm by comparing it against the gold standard of manual chart review by trained registry specialists.
METHODS
Study Setting
This study was approved by the institutional review board. The study cohort comprised 48,962 primary total hip and knee arthroplasty procedures performed by 35 orthopedic surgeons at a single academic institution between 2000 and 2017. During this time frame, the EHR in our institution was an in-house system based on general electric (GE) Centricity, an EHR system developed by GE Healthcare. All infectious disease consultation notes, operative notes, pathology reports and microbiology reports present in the EHR since the date of TJA were evaluated. Our institution maintains a total joint arthroplasty registry as part of routine care of all patients. Registry data collection is performed in a comprehensive fashion on all aspects related to TJA outcomes through manual chart review of EHRs by trained registry personnel, including the use of standardized definitions for TJA-specific data elements and PJI. All MSIS criteria (3, 4) data elements were manually abstracted and recorded. Therefore, the gold standard data for validation was readily available for all PJI events. In this cohort, we defined positive cases as a PJI (hip or knee) infection found anytime within 12 months after the TJA procedures performed between 2000 and 2017. Of note, restricting PJI cases to those diagnosed within 12 months after TJA was for logistical reasons to ensure all data elements were available. Negative controls without PJI were defined as patients who had TJA between 2000 and 2017 without prior or subsequent PJI (hip or knee) infection at any time after the surgery.
Study Design
PJI cases were sampled from primary TJA procedures at Mayo Clinic Rochester. Controls were matched on age, sex, and year of surgery. We then randomly split the study sample (total 2387) into 50% training and 50% test datasets, ensuring training and test datasets contained an equal number of cases and controls. The training dataset comprised 170 PJI cases and 1027 matched controls with a mean age of 64 (±15) years and women comprised 50%. The test dataset comprised 150 PJI cases and 929 matched controls with a mean age of 65 (±15) years and women comprised 48%.
The PJI data elements were searched within the twelve months’ time window after index surgery and included (a) presence of a sinus tract communicating with the prosthesis, (b) two or more intraoperative cultures or a combination of preoperative aspiration and intraoperative cultures that yield the same organism, (c) presence of elevated laboratory results for erythrocyte sedimentation rate (ESR >29mm/h), Creactive protein (CRP >8 mg/L), (d) synovial leukocyte count (>3000 cells/uL) and synovial neutrophil percentage >80%, (e) presence of purulence without another known etiology surrounding the prosthesis, (f) presence of acute inflammation on histopathologic examination (i.e., greater than five neutrophils per high-power field in five high-power fields observed from histologic analysis of periprosthetic tissue at ×400 magnification).
NLP Algorithm Development
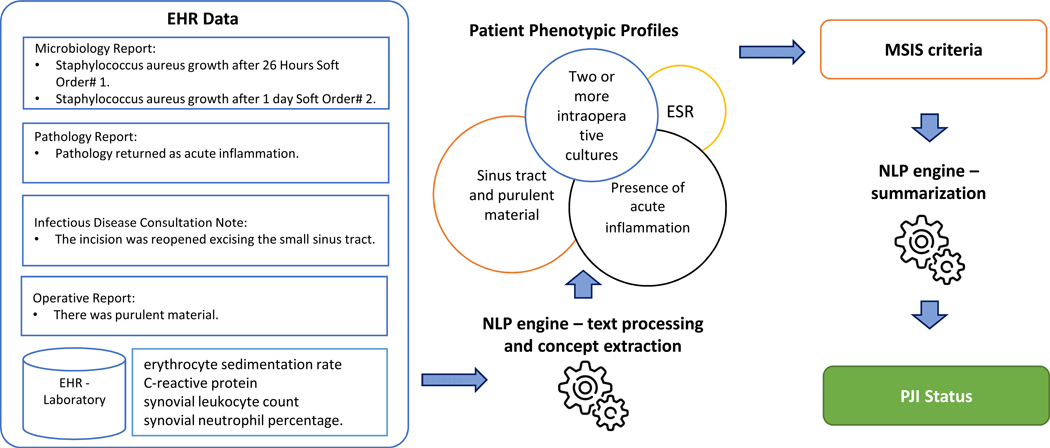
The NLP algorithm for each MSIS criteria data element was developed on a training dataset and validated on a blinded test dataset. Our NLP algorithm was based on expert rules—target “textual markers” (i.e., keywords related to PJI) that were specified in the clinical narratives defined by orthopedic surgeons or infectious diseases specialists. The NLP algorithm had three main components: text processing, concept extraction, and classification. The key components of the text processing pipeline were sentence segmentation, assertion identification, and temporal extraction. Assertion of each concept includes certainty (i.e., positive, possible, and negative) along with experiencer (i.e., patient or family member), while temporality determining whether the event is historical or present. For example, the sentence “Postoperative diagnosis: draining sinus tract on patient’s right knee was not found” will be processed into assertion status “negative”, temporality “present”, and experiencer “associated with the patient”. Concept extraction is a knowledge-driven annotation and indexing process to identify phrases referring to concepts of interest in unstructured text (13). In the previous example, “draining sinus tract” would be extracted as a concept associated with sinus tract. After concepts are extracted, they are normalized to a patient phenotypic profile. Non-negated and present findings from a patient phenotypic profile are summarized into final PJI status based on MSIS criteria. Figure 1 shows the process for extracting and classifying PJI status.
Figure 1.
Process for Extracting and Classifying PJI Status
The development of the NLP algorithm was an iterative process involving informatics frameworks, cross-functional expert knowledge, and logic. The algorithm was first applied to the training data. Error cases (falsely classified) were manually reviewed by an orthopedic surgeon or an infectious diseases specialist. Keywords were manually curated through an iteratively refining process until all issues were resolved.
The NLP algorithm was implemented using the institutional NLP-as-a-service infrastructure (14) which utilizes big data platforms to support high-throughput NLP. The infrastructure contains an open-source NLP pipeline MedTaggerIE resource-driven open-source with an Unstructured Information Management Architecture (UIMA) (15)-based IE framework. The solution separates domain-specific NLP knowledge engineering from the generic NLP process, which enables words and phrases containing clinical information to be directly coded by subject matter experts. The full list of concepts, keywords, modifiers, and rules are listed in Table 1.
Table 1.
PJI Keywords and Rules for Concept Extraction.
| Confirmation keywords | Rules | Data sources |
|---|---|---|
|
Purulent Material: purulence; purulent; purulently; purulent-appearing; Drain: abscess drained; abscessed drained; abscesses drained; drain abscesses; drained abscess; drained abscesses; draining abscesses; Fluid: fluid cloudy; turbid fluid; yellowish fluid; infected fluid collection; cloudy looking fluid; brown yellow discharge; cloudy serous fluid; serosanguineous fluid; greenish fluid; draining cloudy fluid; cloudy fluid; aspiration cloudy Negation: minor amount of; slightly; nothing |
Positive purulent material: • Mention of Purulent Material • Mention of Drain • Mention of Fluid AND NOT Negation (within 3-word distance) |
Operative report; ID consultation notes |
|
Acute inflammation: acute inflammation; acute inflammatory cells; acute inflammatory debris Negation: looking for |
Positive acute inflammation: • Mention of Acute Inflammation (within 8-word distance) |
Pathology Report |
|
Sinus Tract: sinu tract; sinus tract; sinus tracts; sinus track; draining sinus; fistulization tract; fistulizing tract; fistulous tract; fistulous tracts; sinus drain; sinus draining; sinus-draining; sinus drainage; draining chronic sinus; Communication: communicated; communication; communicate; tracked down; tracking all the way; pinhole leaking; coming from deep Joint and Tissue-related: calf; cavity; joint; deep; tissue; periprosthetic; fracture; hip; knee; femur; arthroplasty Fascia: fascia Size of Defect: cm; -cm; 1-cm; 2-cm Surgical Complication: rent; defect; dehiscence; exposing; large hole; not well sealed Negation: completely sealed; no further fluid; low threshold to open; well-sealed; closed |
Positive sinus tract: • Mention of Sinus Tract • Mention of (Fluid AND Communication AND Joint and Tissue-related AND NOT Negation) within 5-word distance • Mention of Fascia AND Size of Defect AND NOT Negation • Mention of Surgical Complication AND Fascia AND NOT Negation |
Operative report, ID consultation notes |
|
Bacteria: streptococcus agalactiae; staphylococcus epidermidis; staphylococcus aureus; pseudomonas aeruginosa; proteus mirabilis; enterococcus sp; staphylococcus coagulase negative; actinomyces neuii; finegoldia magna; clostridium perfringens; clostridium bifermentans; klebsiella pneumoniae complex; escherichia coli; streptococcus beta hemolytic group b; small nonsporeforming gpb res coryne sp not c jeikeium; corynebacterium striatum; peptoniphilus sp; helcococcus sueciensis; bacillus; propionibacterium acnes; enterococcus faecalis; candida albicans; basidiomycete; peptostreptococcus sp; peptostreptococcus magnus; lelliottia enterobacter amnigena; lelliottia (enterobacter) amnigena Anatomic: leg; hip; joint; knee; femur; femoral; synovial; cartilage; acetabulum; trochanter; pelvis; buttock; left aspirate; right aspirate Soft Order Number: retrieve from laboratory |
One positive culture: • Bacteria AND Anatomic AND (One unique) Soft Order Number |
Microbiology Report |
All findings need to be within 180 days after the TJA surgery; Generic negation status from MedTaggerIE needs to be applied to all findings.
Statistical Analysis
The performance of each NLP algorithm was assessed using the gold standard manually abstracted data from the institutional total joint registry. Performance was assessed through sensitivity (recall), specificity, positive predictive value (PPV or precision), negative predictive value (NPV) and f1-score (weighted harmonic mean of precision and recall and calculated as 2* [ (precision*recall)/(precision+recall)] (13). The error analysis was performed by an orthopedic surgeon through manually reviewing falsely predicted cases from EHRs.
RESULTS
Among the 48,962 primary TJA procedures at our institution, 338 PJI cases (occurring within 12 months of TJA) were randomly sampled. 2049 controls were matched on age, sex, and year of surgery. Age and date of surgery between cases and controls were similar with mean of 0 (0.60) years and 0 (0.23) years, respectively. 95% of controls were within 1 year of the cases on age and 0.57 years on surgery date. Among the 2387 cases and controls, 45% were primary total knee replacement and 55% were primary total hip replacement patients. Of the 338 PJI cases, 43% were diagnosed within the first month after surgery, and 66% were diagnosed within three months after surgery (cumulative). None of the PJI cases had infection in more than on joint.
The data element specific NLP algorithms were able to identify individual data elements very well except for the presence of sinus tract (Table 2). The performance of extracting the presence of sinus tract achieved f1-score of 0.771, sensitivity 0.887 and specificity 0.991. For presence of purulence, pathological documentation of inflammation and growth of cultured organisms, f1-scores ranged from 0.909 to 0.982, sensitivity ranged from 0.833 to 1.000, and specificity ranged from 0.947 to 1.000. These results demonstrated a good feasibility of an automated PJI algorithm. The final PJI algorithm that combined the four data elements to identify patients with PJI based on MSIS criteria achieved the f1-score, sensitivity, specificity, PPV and NPV of 0.911, 0.887, 0.991, 0.937, and 0.984, respectively (Table 3).
Table 2.
Concordance in PJI Status between NLP and gold standard
| PJI Status / Data Element | F1-score | Sensitivity | Specificity | PPV | NPV |
|---|---|---|---|---|---|
| Sinus Tract | 0.771 | 0.730 | 0.951 | 0.818 | 0.921 |
| Purulence | 0.946 | 0.940 | 0.947 | 0.951 | 0.935 |
| Pathology Inflammation | 0.909 | 0.833 | 1.000 | 1.000 | 0.944 |
| Growth of Cultured Organisms | 0.982 | 1.000 | 0.998 | 0.965 | 1.000 |
| PJI (n = 1179) | 0.911 | 0.887 | 0.991 | 0.937 | 0.984 |
PPV positive predictive value, NPV negative predictive value
Table 3.
Confusion Matrix for PJI detection.
| Gold Standard → NLP ↓ | Yes | No | Total | |
|---|---|---|---|---|
| Yes | 133 | 9 | 142 | Positive predictive value (Precision) 133/142=0.937 |
| No | 17 | 1020 | 1037 | Negative predictive value 1020/1037=0.984 |
| Total | 150 | 1029 | 1179 | F1 score=0.911 |
| Sensitivity (recall) 133/150=0.887 | Specificity 1020/1029=0.991 |
DISCUSSION
The systematic identification of patients with PJI from EHRs can drastically improve the effectiveness and efficiency of chart review for clinical quality improvement, clinical research, and registry development. In our study, we developed and evaluated an NLP algorithm that identified patients with PJI from EHRs. The evaluation statistics showed a high performance, validating the proof-of-concept for this application.
The combination of multiple EHR sources and comprehensive MSIS criteria enhances the high stability of the PJI phenotyping algorithm described in this study. The PJI algorithm was developed using four different clinical report types (infectious disease consultation notes, operative notes, pathology reports, and microbiology reports) and seven MSIS criteria. These individual features such as laboratory values, documentation of a sinus tract communicating with the arthroplasty, pathologic evidence of inflammation, and the presence of purulent materials are then aggregated to generate a positive or negative determination. This aggregation minimizes the variation caused by any inherent characteristics of individual features and allows the algorithm to remain robust.
Although the overall performance of the PJI algorithm was robust (Table 2), we found it challenging to extract some of the concepts, particularly the first MSIS major criteria – presence of sinus tract communicating with the joint. This was due to high variation in description of sinus tract in clinical and surgical notes. There are many different ways to express this finding in clinical documentation. For example, a positive indication can be expressed as “fluid tracking all the way to the joint.” Similarly, it can also be expressed as “there was a rent in the fascia.” Both sentences share the same semantic meaning but different syntactic structures. Our iterative chart review and rule refining process helped capture the majority of the cases. However, around 25% of expressions were still missed. We plan to address the challenge through leveraging statistical machine learning, a method that can learn patterns without explicit programming through learning the association of input data and labeled outputs (16, 17). We also identified that not all data elements were systematically documented for every patient. For example, orthopedic surgeons or infectious diseases specialists do not strictly follow all MSIS criteria to make diagnostic decisions. In addition, we found that some cases have minor data quality issues including abstraction errors from the registry and missing laboratory results.
Our study has potential limitations. First, despite the fact that we limited the search to a specific time range, inaccurate information from the heterogeneous EHR may still be copied and used. Furthermore, cases were restricted to those diagnosed within 12 months after index TJA. This time frame was chosen for convenience. The algorithm can theoretically be applied to other times frames both prospective and retrospectively and even as a real time screening tool. It should be noted that applying the algorithm to a longer time frame may pose additional complexity because a patient may experience multiple different procedures that makes it difficult to correctly associate a given TJA with a corresponding PJI. Second, despite the high feasibility of detecting PJI from EHR, the performances of the algorithm are limited by the number of positive cases. Additional data are required to have a comprehensive evaluation of the system. Third, the algorithms were only evaluated using datasets from one institution, and therefore, the generalizability of the systems may be limited. In future studies, we plan to validate and refine the algorithm in other health care systems.
In conclusion, PJI is a common complication following TJA, and our results indicate that it is feasible to ascertain both structured and unstructured PJI data elements in an automated fashion using rule-based natural language processing algorithms. These algorithms offer great potential to augment data collection capabilities for clinical and research purposes.
Supplementary Material
Acknowledgments
Funding: Supported by the National Institutes of Health (NIH) grants R01AR73147 and P30AR76312.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Kurtz SM, Lau E, Watson H, Schmier JK, Parvizi JJTJoa. Economic burden of periprosthetic joint infection in the United States. 2012;27(8):61–5. e1. [DOI] [PubMed] [Google Scholar]
- 2.Yao JJ, Kremers HM, Abdel MP, Larson DR, Ransom JE, Berry DJ, et al. Long-term mortality after revision THA. 2018;476(2):420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Osmon DR, Berbari EF, Berendt AR, Lew D, Zimmerli W, Steckelberg JM, et al. Diagnosis and management of prosthetic joint infection: clinical practice guidelines by the Infectious Diseases Society of America. 2013;56(1):e1–e25. [DOI] [PubMed] [Google Scholar]
- 4.Parvizi J, Gehrke TJTJoa. Definition of periprosthetic joint infection. 2014;29(7):1331. [DOI] [PubMed] [Google Scholar]
- 5.Wyles CC, Tibbo ME, Fu S, Wang Y, Sohn S, Kremers WK, et al. Use of natural language processing algorithms to identify common data elements in operative notes for total hip arthroplasty. 2019;101(21):1931–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tibbo ME, Wyles CC, Fu S, Sohn S, Lewallen DG, Berry DJ, et al. Use of natural language processing tools to identify and classify periprosthetic femur fractures. 2019;34(10):2216–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murff HJ, FitzHenry F, Matheny ME, Gentry N, Kotter KL, Crimin K, et al. Automated identification of postoperative complications within an electronic medical record using natural language processing. 2011;306(8):848–55. [DOI] [PubMed] [Google Scholar]
- 8.FitzHenry F, Murff HJ, Matheny ME, Gentry N, Fielstein EM, Brown SH, et al. Exploring the frontier of electronic health record surveillance: the case of post-operative complications. 2013;51(6):509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sohn S, Larson DW, Habermann EB, Naessens JM, Alabbad JY, Liu HJJoSR Detection of clinically important colorectal surgical site infection using Bayesian network. 2017;209:168–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chapman AB, Mowery DL, Swords DS, Chapman WW, Bucher BT, editors. Detecting evidence of intra-abdominal surgical site infections from radiology reports using natural language processing. AMIA Annual Symposium Proceedings; 2017: American Medical Informatics Association. [PMC free article] [PubMed] [Google Scholar]
- 11.Shen F, Larson DW, Naessens JM, Habermann EB, Liu H, Sohn SJJohir. Detection of surgical site infection utilizing automated feature generation in clinical notes. 2019;3(3):267–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thirukumaran CP, Zaman A, Rubery PT, Calabria C, Li Y, Ricciardi BF, et al. Natural Language Processing for the Identification of Surgical Site Infections in Orthopaedics. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Manning CD, Schütze H. Foundations of statistical natural language processing. MIT press; 1999. [Google Scholar]
- 14.Wen A, Fu S, Moon S, El Wazir M, Rosenbaum A, Kaggal VC, et al. Desiderata for delivering NLP to accelerate healthcare AI advancement and a Mayo Clinic NLP-as-a-service implementation. 2019;2(1):1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferrucci D, Lally A. UIMA: an architectural approach to unstructured information processing in the corporate research environment. Nat Lang Eng. 2004;10 (3–4):327–48. [Google Scholar]
- 16.Sebastiani F Machine learning in automated text categorization. ACM computing surveys (CSUR). 2002;34(1):1–47. [Google Scholar]
- 17.Freitag D Machine learning for information extraction in informal domains. Machine learning. 2000;39(2–3):169–202. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.