Summary:
Cancer immunotherapy shows limited efficacy against many solid tumors that originate from epithelial tissues, including triple-negative breast cancer (TNBC). We identify the SOX4 transcription factor as an important resistance mechanism to T cell-mediated cytotoxicity for TNBC cells. Mechanistic studies demonstrate that inactivation of SOX4 in tumor cells increases the expression of genes in a number of innate and adaptive immune pathways important for protective tumor immunity. Expression of SOX4 is regulated by the integrin αvβ6 receptor on the surface of tumor cells which activates TGFβ from a latent precursor. An integrin αvβ6/8 blocking monoclonal antibody (mAb) inhibits SOX4 expression and sensitizes TNBC cells to cytotoxic T cells. This integrin mAb induces a substantial survival benefit in highly metastatic murine TNBC models poorly responsive to PD-1 blockade. Targeting of the integrin αvβ6 – TGFβ – SOX4 pathway therefore provides therapeutic opportunities for TNBC and other highly aggressive human cancers of epithelial origin.
eTOC: Context and Significance
Bagati et al. show that the SOX4 transcription factor induces tumor cell resistance to cytotoxic T cells. Integrin αvβ6 on the surface of epithelial cancer cells activates TGFβ from a latent precursor to induce SOX4 expression, and antibody-mediated inhibition of integrin αvβ6 induces T cell-mediated immunity in immunotherapy-resistant tumor models.
Graphical Abstract
Introduction
Triple-negative breast cancer (TNBC) has a high propensity for metastatic dissemination, and the prognosis is poor for patients who fail to respond to chemotherapy (Denkert et al., 2017). Recent evidence suggests a role for T cell-mediated immunity in TNBC. The presence of tumor infiltrating lymphocytes (TIL) is both predictive of response to neoadjuvant chemotherapy and is associated with improved survival in TNBC (Adams et al., 2014; Denkert et al., 2018). Survival benefit is associated with a higher density of infiltrating CD8+ T cells and a higher CD8/FoxP3 ratio in pre-treatment biopsies (Adams et al., 2014; Miyashita et al., 2015). In addition, the Impassion130 phase 3 clinical trial demonstrated that the combination of a PD-L1 antibody (atezolizumab) with nab-paclitaxel increased progression-free survival in patients with previously untreated metastatic TNBC compared to nab-paclitaxel (Schmid et al., 2018). This drug combination represents a significant advance for the treatment of TNBC, but the majority of patients still fail to benefit from immunotherapy.
Published studies in melanoma demonstrated that a lack of CD8+ T cell infiltration can be caused by an overactive β-catenin pathway or inactivation of the PTEN tumor suppressor gene (Peng et al., 2016; Spranger et al., 2015). Recent genetic screens performed in human and murine melanoma cell lines have highlighted the importance of genes related to the MHC class I and IFNγ signaling pathways in T cell-mediated immunity (Manguso et al., 2017; Pan et al., 2018; Patel et al., 2017). Loss of function mutations in MHC class I and IFNγ signaling pathways have also been identified in melanoma patients as mechanisms of secondary resistance to checkpoint blockade (Gide et al., 2018; Zaretsky et al., 2016). However, the tumor-intrinsic mechanisms of resistance to immunotherapy remain poorly understood in TNBC and many other human cancers of epithelial origin.
We recently performed a genome-scale CRISPR knockout screen to identify genes that render tumor cells resistant to cytotoxic T cells (Pan et al., 2018). A total of 313 genes were identified as hits in the primary screen, and our prior study focused on three genes that encoded subunits of the SWI/SNF complex. In the present study, we focused on the transcription factor SOX4 because it is associated with cancer cell invasion (Tiwari et al., 2013; Zhang et al., 2012). High expression of SOX4 is associated with a poor prognosis in many human cancers, in particular TNBC (Chen et al., 2016; Hazelbag et al., 2007; Song et al., 2015; Tavazoie et al., 2008; Vervoort et al., 2013b; Zhang et al., 2012). We hypothesized that Itgav (encoding the integrin αv protein) also identified in the screen could be connected to the SOX4 pathway because integrin αv proteins activate TGFβ (Aluwihare et al., 2009; Henderson and Sheppard, 2013; Munger et al., 1999), and SOX4 is a TGFβ target gene (Peng et al., 2017; Vervoort et al., 2013a; Zhang et al., 2012). The integrin αvβ6 heterodimer (encoded by ITGAV and ITGB6) is overexpressed by many human epithelial cancers, and this integrin activates TGFβ from a latent form deposited on the extracellular matrix (Dong et al., 2017; Munger et al., 1999). We therefore tested the hypothesis that the integrin αvβ6 – TGFβ – SOX4 pathway represents a major mechanism for resistance of TNBC to T cell-mediated immunity.
Results
SOX4 and ITGAV genes confer tumor cell resistance to T cell-mediated cytotoxicity
We initially evaluated the functional significance of the integrin αvβ6 – TGFβ – SOX4 pathway in a co-culture assay of human BT549 TNBC cells and CD8+ T cells. These TNBC cells express HLA-A2 and the NY-ESO-1 antigen (Fig. S1A, B) which enabled T cell cytotoxicity assays with human CD8+ T cells transduced with a T cell receptor (TCR) specific for a HLA-A2 bound NY-ESO-1 peptide. Tumor cells were edited by electroporation with ribonucleoprotein complexes (RNPs) composed of Cas9 protein and bound gRNAs. Indeed, editing of BT549 cells with SOX4, ITGAV or ITGB6 gRNAs significantly increased T cell-mediated cytotoxicity compared to editing with a control gRNA (Fig. 1A–C, S1C). Re-expression of SOX4 in SOX4−/− cells restored resistance to T cell-mediated killing (Fig. S1D–G).
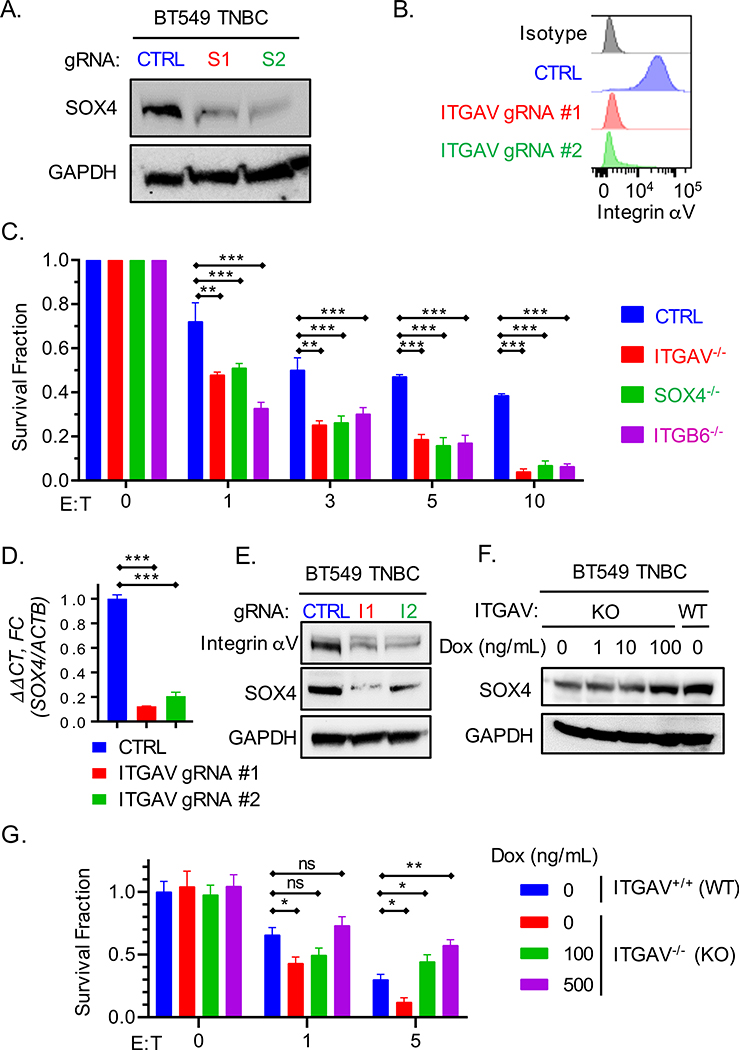
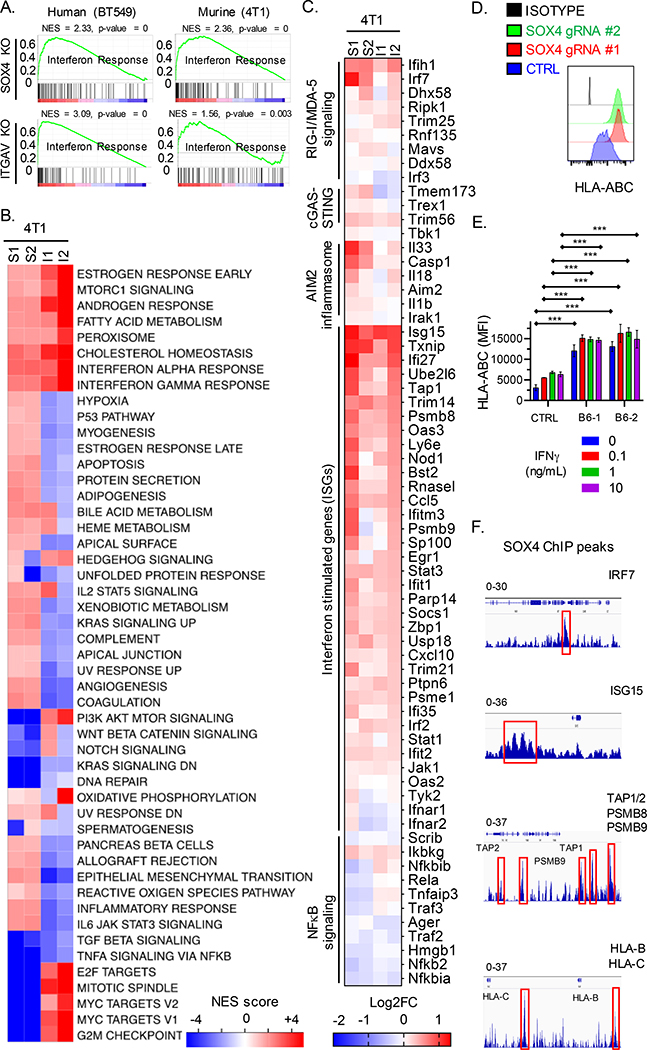
Fig. 1. Inactivation of SOX4 or ITGAV genes sensitizes tumor cells to cytotoxic T cells.
(A) Immunoblot showing SOX4 protein expression by human BT549 TNBC cells edited using two SOX4 gRNAs (S1, S2) or a control gRNA (CTRL). (B) Cell surface expression of integrin αv in BT549 TNBC cells edited with two ITGAV gRNAs (ITGAV gRNA#1 and #2) or a control gRNA (CTRL). (C) T cell cytotoxicity assay with human BT549 TNBC cells edited with SOX4, ITGAV, ITGB6 or control gRNAs. Human T cells expressing a NY-ESO-1 specific TCR were co-cultured for 24 h with tumor cells at the indicated E:T ratios. Data represent the mean of surviving tumor cell fraction after 24 h of co-culture for two independent gRNAs +/− SEM; data are shown relative to condition without T cells (E:T = 0). (D) RT-qPCR analysis of SOX4 mRNA levels in BT549 cells edited with ITGAV or control gRNAs represented as mean ± S.E.M. (E) Immunoblot showing SOX4 protein levels in human BT549 TNBC cells edited with two ITGAV targeting gRNAs (I1, I2) or a control gRNA (CTRL). (F-G) Impact of doxycycline-inducible SOX4 expression in ITGAV KO BT549 tumor cells on resistance to cytotoxic T cells. (F) Immunoblot showing levels of SOX4 and GAPDH proteins in ITGAV+/+ (WT) or ITGAV−/− (KO) BT549 human TNBC cells containing a doxycycline (DOX) inducible SOX4 cDNA construct. Cells were treated with the indicated concentration of doxycycline for 48 h. (G) T cell cytotoxicity assay with ITGAV WT or KO BT549 TNBC cells co-cultured with human NY-ESO-1 specific CD8+ T cells at indicated E:T ratios following pre-treatment with the indicated concentrations of doxycycline for 48 h. Data in (C, D, and G) are representative of at least two independent experiments with technical triplicates and summarized as mean ± S.E.M. Data in [A, B, E, F] were repeated at least three times with consistent results. To determine statistical significance, a two way ANOVA with Dunnett’s [C] or Tukey’s [G] post hoc test or an unpaired Student t-test [D] was used. ***P < 0.001; **P < 0.01; *P < 0.05; n.s., not significant. See also Figure S1.
TGFβ has been reported to induce SOX4 expression (Peng et al., 2017; Vervoort et al., 2013a; Zhang et al., 2012), and we therefore investigated whether inactivation of the ITGAV gene would impact SOX4 expression in TNBC cells. We found that inactivation of the ITGAV gene significantly reduced SOX4 mRNA and protein levels in human BT549 and Hs578T TNBC cells (Fig. 1D, E, S1H, I) as well as murine 4T1 and Py8119 TNBC cells (Fig. S1J–M). We further examined this pathway by restoring SOX4 expression in ITGAV−/− BT549 TNBC cells to levels observed in ITGAV+/+ cells using a doxycycline-inducible promoter. Doxycycline-induced re-expression of SOX4 indeed restored resistance of ITGAV−/− cells to T cell-mediated killing (Fig. 1F, G). However, inactivation of SOX4 did not affect in vitro proliferation of human or murine TNBC cell lines (Fig. S1N, O).
We also performed T cell cytotoxicity assays with murine 4T1 TNBC cells in which Itgav, Sox4 or Itgav plus Sox4 were inactivated. We found that addition of active TGFβ1 induced significant resistance to CD8+ T cell-mediated cytotoxicity in control and Itgav deficient tumor cells; addition of active TGFβ to Itgav deficient tumor cells bypassed the requirement for integrin αvβ6-mediated activation of latent TGFβ (Fig. S1P, Q). Importantly, Sox4 or Sox4/Itgav deficient 4T1 tumor cells remained sensitive to T cell-mediated cytotoxicity even when treated with active TGFβ, indicating that SOX4 was the major TGFβ target gene responsible for this phenotype. These data indicated that SOX4 was a major TGFβ effector gene in conferring resistance to killing by CTLs. Also, integrin αv and SOX4 were part of the same resistance pathway to T cell-mediated cytotoxicity in TNBC cells.
Targeting of SOX4 with an integrin αvβ6/8 specific mAb
We therefore investigated whether an integrin αvβ6 blocking antibody could sensitize TNBC cells to cytotoxic T cells. Expression of the integrin β6 subunit (ITGB6 gene) is restricted to epithelial cells. Importantly, integrin αvβ6 expression is low on healthy epithelial cells but upregulated in many cancers of epithelial origin, including breast, gastric, pancreatic, colorectal, lung and ovarian cancers (Bandyopadhyay and Raghavan, 2009; Niu and Li, 2017).
The previously reported 264RAD mAb binds with high affinity to human and murine integrin αvβ6 heterodimers and blocks integrin-mediated activation of TGFβ (Eberlein et al., 2013). This mAb also binds with lower affinity to the integrin αvβ8 heterodimer expressed by regulatory T cells (Tregs) and dendritic cells (Paidassi et al., 2011; Worthington et al., 2015). The 264RAD mAb has a human IgG1 Fc region which binds to activating Fc receptors; antibody-dependent cellular cytotoxicity (ADCC) mediated by NK cells and macrophages may therefore have contributed to its anti-tumor activity. We introduced two mutations into the Fc region to prevent binding to activating Fc receptors, thus limiting the activity of this mAb to its blocking function (designated here as integrin αvβ6/8 mAb). Treatment of human and murine TNBC cells with this mAb reduced SOX4 protein levels and TGFβ signaling (phospho-SMAD2) (Fig. 2A, Fig. S2A). It also reduced levels of active TGFβ in co-cultures of human TNBC cell lines with a TGFβ reporter cell line (Fig. 2B). Importantly, pre-treatment of human and murine TNBC cell lines with this integrin αvβ6/8 mAb sensitized them to killing by CD8+ T cells (Fig. 2C–D); treatment with both integrin αvβ6/8 and PD-1 mAbs further enhanced killing of human TNBC cells by cytotoxic T cells (Fig. 2D).
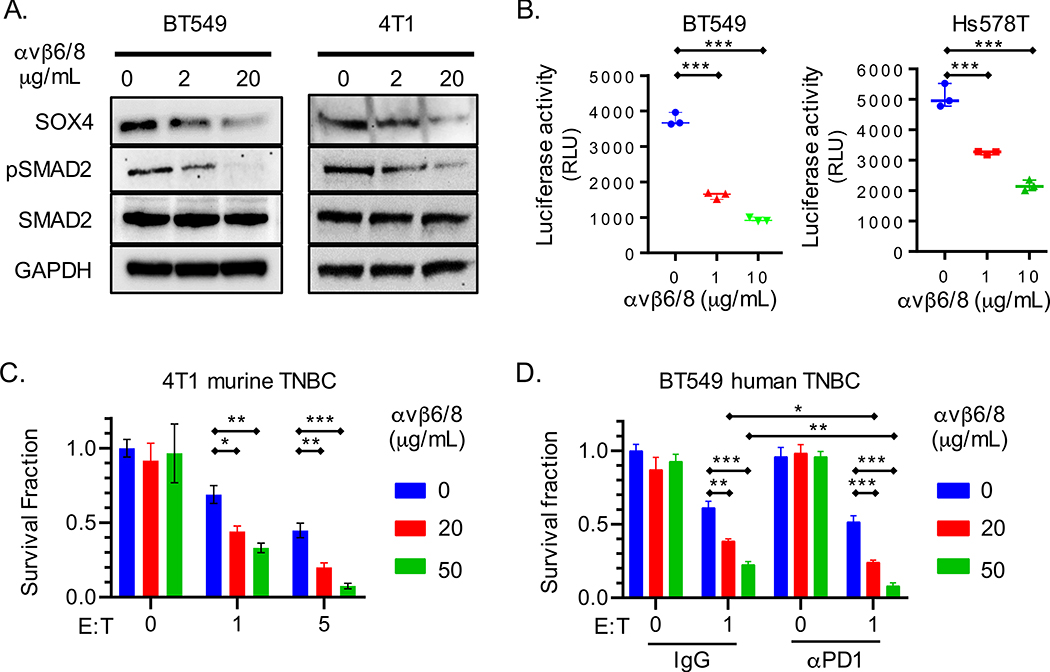
Fig. 2. An integrin αvβ6/8 mAb inhibits SOX4 expression and sensitizes TNBC cells to cytotoxic T cells.
(A) Immunoblot for indicated proteins in human (BT549) or murine (4T1) TNBC cells lines treated with integrin αvβ6/8 blocking mAb for 72 h. (B) Luciferase-based TGFβ reporter assay with human BT549 and Hs578T TNBC cell lines. HepG2-TGFβ reporter cells were co-cultured for 24 h with human TNBC cells that had been pre-treated with indicated concentrations of integrin αvβ6/8 mAb for 72 h. Data are represented as relative luciferase units (RLU). (C) T cell cytotoxicity assay with GFP+ murine 4T1 TNBC cells. Tumor cells were co-cultured for 48 h with GFP-specific CD8+ T cells (JEDI T cells) at indicated E:T ratios. Tumor cells were pre-treated with indicated concentrations of integrin αvβ6/8 mAb for 72 h prior to co-culture. (D) T cell cytotoxicity assay with human BT549 TNBC and human CD8+ T cells that expressed a NY-ESO-1 TCR. Tumor cells were pre-treated with indicated concentrations of control IgG or integrin αvβ6/8 mAb for 72 h; control IgG or PD-1 mAbs were added to co-cultures (20 μg/ml). Y-axis shows number of surviving tumor cells after 24 h of co-culture. Data are summarized as mean ± S.E.M and are representative of at least two independent experiments with technical triplicates. A one-way [B] or two-way [C and D] ANOVA with Dunnett’s post hoc test was used to determine statistical significance, ***P < 0.001; **P < 0.01; *P < 0.05. See also Figure S2.
Addition of active TGFβ1 reversed the effect of the integrin αvβ6/8 mAb and rendered tumor cells resistant to CD8+ T cells (Fig. S2B, C), consistent with the role of this integrin in activating TGFβ from a latent form. We also used a small molecule inhibitor of TGFβ receptor signaling (Galunisertib, LY2157299) to examine whether it recapitulated the effects of the integrin αvβ6/8 mAb. We found that both approaches to inhibiting the TGFβ pathway significantly decreased SOX4 protein levels and increased tumor cell sensitivity to killing by CD8+ T cells (Fig. S2D, E). While the integrin αVvβ6/8 mAb only modestly decreased PD-L1 levels on the surface of human and murine TNBC cells (Fig. S2F), inactivation of Itgav and Sox4 genes significantly reduced levels of PD-L1 (Fig. S2G, H). Conversely, ectopic expression of Sox4 or addition of active TGFβ1 significantly increased PD-L1 while reducing MHC-I protein levels (Fig. S2F, I, J).
In vivo efficacy of targeting the integrin αv – SOX4 pathway
High expression of integrin αv and SOX4 is associated with reduced patient survival and progression of several aggressive human cancers, particularly breast cancer (Fig. S3A) (Desai et al., 2016; Song et al., 2015; Tavazoie et al., 2008; Zhang et al., 2012). We hypothesized that targeting of SOX4 with this integrin αvβ6/8 mAb could simultaneously reduce tumor cell invasiveness and sensitize tumor cells to T cell-mediated immunity. We therefore investigated the efficacy of the integrin αvβ6/8 mAb in two highly metastatic murine models of TNBC (4T1 and Py8119) that are poorly responsive to PD-1 blockade. We observed that monotherapy with the integrin αvβ6/8 mAb substantially reduced primary tumor burden and resulted in a substantial survival benefit in both TNBC models compared to control IgG treated mice (Fig. 3A–D, Fig. S3B–D). Combination therapy with integrin αvβ6/8 and PD-1 mAbs further enhanced therapeutic benefit and significantly enhanced survival compared to monotherapy with the integrin αvβ6/8 mAb (Fig. 3A–D, Fig. S3B–D).
Fig. 3. Efficacy of integrin αvβ6/8 mAb in metastatic murine TNBC models resistant to PD-1 blockade.
(A) Py8119GFP+ TNBC (n=10 mice/group) or (B) 4T1 TNBC (n=12 mice/group) primary tumor volume shown at indicated time points. Mice with similar tumor burden were treated with indicated antibodies (IP, 0.2 mg/dose, twice weekly) until tumor volume in any group reached 1000 mm3. To deplete CD8+ T cells, mice were treated with anti-CD8β antibodies (0.1 mg/dose) on days −1, 1, and weekly thereafter. (C-D) Kaplan-Meier analysis of survival for mice described in (A) and (B), respectively. (E) Primary tumor volume shown at indicated time points for Py8119 model following monotherapy with integrin αvβ6 or isotype control mAbs; in the indicated groups CD8+ T cells (CD8β mAb) and/or NK cells (NK1.1 mAb) were also depleted by administration of the respective antibodies (0.1 mg/dose) on days −1, 1, and weekly thereafter. (F) Number of 4T1 lung surface metastases in mice treated as described in (A) following staining with picric acid for 24 h. (G) Representative images of 4T1 lung surface metastases on day 24 following tumor inoculation. Data are summarized as mean ± SD of tumor volume and are an average of two independent experiments. To determine statistical significance, a two way [A, B, and E] or a one-way [F] ANOVA with Dunnett’s post hoc test and Kaplan-Meier log-rank (Mantel-Cox) test [C and D] were used. ***P < 0.001; **P < 0.01; *P < 0.05; n.s., not significant. See also Figure S3.
Next, we addressed whether CD8+ T cells and/or NK cells contributed to the efficacy of integrin αvβ6/8 Ab treatment (Fig. S3E). Depletion of CD8+ T cells resulted in loss of anti-tumor efficacy by monotherapy with the integrin αvβ6/8 mAb in the Py8119 model (Fig. 3E) while the effect of NK cell depletion (NK1.1 mAb) on primary tumor growth was modest (Fig. 3E, S3E, F). In the 4T1 tumor model, we found that CD8+ T cell depletion abrogated the protective effect of integrin αvβ6/8 plus PD-1 mAb combination therapy on lung metastases and reduced the therapeutic effect of this combination on primary tumor growth (Fig. 3B, 3F, S3B). In vitro studies demonstrated that inactivation of the Sox4 gene also increased the invasiveness of 4T1 TNBC cells (Fig. S3G) which may also have contributed to the efficacy of the integrin αvβ6/8 mAb. Collectively, these data indicated that CD8+ T cells play an important role in the efficacy of integrin αvβ6/8 mAb therapy. Other cell populations such as NK cells could have more modest contributions.
Integrin αvβ6/8 mAb monotherapy also greatly reduced lung metastatic burden (lung surface metastases, 4T1 model) (Fig. 3F, G, Fig. S3C). PD-1 blockade did not reduce the number of lung metastases, but enhanced the effect of the integrin αvβ6/8 mAb on lung metastases (Fig. 3F, S3C). Given that we did not surgically remove primary tumors prior to initiation of therapy, the reduced number of metastases may be explained by treatment effects on primary tumors and metastatic lesions. These data demonstrated that an integrin αvβ6/8 mAb resulted in a substantial therapeutic benefit in aggressive models of TNBC.
Remodeling of the tumor microenvironment by integrin αvβ6/8 mAb treatment
In human cancers, resistance to checkpoint blockade is frequently associated with poor CD8+ T cell infiltration (also referred to as ‘cold’ tumors) (Denkert et al., 2017), and both TNBC models were poorly infiltrated by CD8+ T cells in the absence of treatment. Flow cytometric analysis demonstrated that integrin αvβ6/8 mAb monotherapy significantly enhanced the number of infiltrating CD8+ T cells in 4T1 and Py8119 TNBC tumors, a finding that was confirmed by immunofluorescence analysis of tissue sections (Fig. 4A–C, Fig. S4A). Also, a smaller percentage of tumor-infiltrating CD8+ T cells from integrin αvβ6/8 compared to control mAb treated mice were positive for the PD-1 inhibitory receptor (Fig. S4B–C).
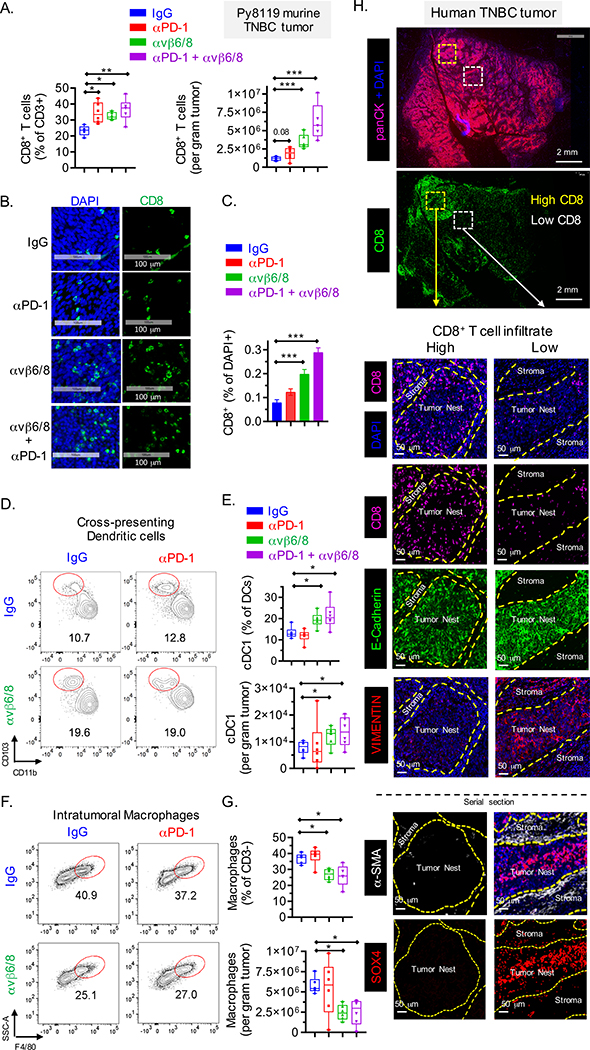
Fig. 4. Analysis of tumor microenvironment in murine and human TNBC.
(A) Quantification of tumor infiltrating CD8+ T cells, represented as percentage of CD3+ cells (left) and per gram of tumor (right) in Py8119 TNBC tumors (n=6) treated with indicated mAbs (FACS analysis 22 days following tumor inoculation). (B-C) Representative images showing CD8+ T cell infiltration into Py8119 TNBC tumors (n=6) (B) and quantification of CD8+ cells as percentage of DAPI+ cells (C). (D-E) Contour plot showing migratory cross-presenting DCs, defined as CD45+/CD3−/F4/80−/CD11c+/MHC-IIhigh/CD103+/CD11b− cells (D) and quantification of these cells as percentage of DCs (CD45+/CD3−/F4/80−/CD11c+/MHC-IIhigh) and total count (E). (F-G) Contour plot showing intra-tumoral F4/80+ macrophages, defined as CD45+/CD3−/Gr1−/CD11b+/MHC-II+/F4/80+ cells (F) and quantification of these cells as percentage of CD45+ CD3− cells (top) and per gram of tumor (bottom) (G). (H) Human TNBC tumor sections stained with indicated markers. Serial sections were stained with DAPI and antibodies specific for CD8, E-cadherin and vimentin (panel #1) and DAPI, SOX4 and αSMA (panel #2). Data are summarized as mean ± SD. Data in [B, C] is an average of two independent experiments and data in [A, D, E, F and G] are representative of at least two independent experiments. For box plots, dots denote all individual values, horizontal lines denote median values, boxes extend from 25th - 75th percentile of each group’s distribution, and no data points were excluded. An unpaired Student’s t-test was used to determine statistical significance, ***P < 0.001; **P < 0.01; *P < 0.05. See also Figure S4 and S5.
Tumors from integrin αvβ6/8 compared to control mAb treated mice also contained significantly larger numbers of CD4+ T cells but a smaller percentage of CD4+ T cells were Foxp3+ regulatory T cells (Fig. S4D, E). Cross-presenting DCs are critical for induction of tumor immunity mediated by CD8+ T cells, and the percentage of DCs with this phenotype (CD11c+/MHC−IIhi/CD103+/CD11b−) was higher in 4T1 and Py8119 tumors following treatment with integrin αvβ6/8 compared to control mAb (Fig. 4D, E, S4F, G), while the percentage of F4/80+ macrophages was reduced in tumors from integrin αvβ6/8 compared to control mAb treated mice (Fig. 4F, G, S4H, I). This result is relevant because macrophages have been shown to promote tumor growth and suppress T cell-mediated tumor immunity (Goplen et al., 2019; Peranzoni et al., 2018).
Multi-color immunofluorescence analysis of serial sections from five human TNBC archival specimens showed either regional intra-tumoral heterogeneity in the degree of CD8+ T cell infiltration or poor T cell infiltration (Fig. 4H, S5A, S5B). Tumor cell nests and stromal regions were identified by labeling with antibodies specific for E-cadherin and α-smooth muscle actin (αSMA), respectively. Tumor nests from areas with poor infiltration by CD8+ cells tended to show higher SOX4 labeling whereas tumor nests with higher infiltration by CD8+ cells showed limited labeling with a SOX4 antibody (Fig. 4H, S5A and S5B). Tumor nests with strong SOX4 labeling also tended to be labeled by a vimentin specific antibody (a marker of EMT) (Fig. 4H, S5B). Tumor nests devoid of any CD8 infiltrate tended to express high levels of SOX4 and vimentin and were surrounded by stroma with high levels of αSMA and vimentin (Fig. 4H, S5B). EMT represents a gradient ranging from fully epithelial to highly mesenchymal cells. Analysis of human tumors has shown substantial regional heterogeneity in EMT and identified partial EMT phenotypes, for example at the invasive margin (Grigore et al., 2016; Puram et al., 2018). The observation that these SOX4 and vimentin positive tumor nests retained E-cadherin labeling is consistent with such a partial EMT phenotype.
We also further investigated the in vivo relevance of SOX4 to the efficacy integrin αvβ6/8 mAb treatment by generating 4T1 murine TNBC cells with a doxycycline-inducible Sox4 cDNA construct. Doxycycline treatment resulted in increased SOX4 protein levels and a dose-dependent increase in resistance to CD8+ T cell-mediated cytotoxicity (Fig. 5A, B). Notably, doxycycline pre-treatment of tumor cells diminished the sensitizing effect of the integrin αvβ6/8 mAb on T cell-mediated cytotoxicity (Fig. 5C). We also implanted these 4T1Sox4-Dox cells into the mammary fat pads of Balb/c mice. Once tumors were palpable, mice with similar tumor burden received either a regular or doxycycline-containing diet (625ppm, Envigo Teklad) as well as monotherapy with integrin αvβ6/8 or isotype control mAbs. Integrin αvβ6/8 mAb treatment only reduced primary and metastatic tumor burden for mice on a regular diet, but not the doxycycline-containing diet (Fig. 5D, E). Moreover, significantly lower numbers of CD8+ T cells were present in tumors following integrin αvβ6/8 monotherapy for mice on the doxycycline-containing compared to the control diet (Fig. 5F). We also tested the level of SOX4 protein in whole tumor lysates from mice treated with integrin αvβ6/8 or isotype control IgG mAbs. The doxycycline-containing diet resulted in higher SOX4 protein levels in tumors compared to the regular diet, even when mice received the integrin αvβ6/8 mAb (Fig. 5G).
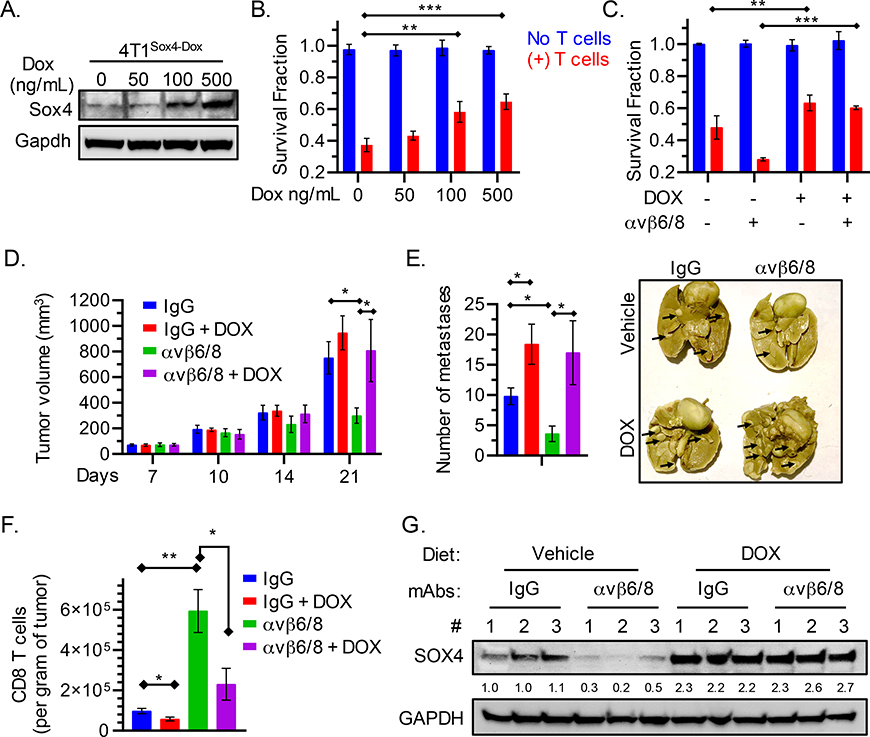
Fig. 5. Relevance of SOX4 to the efficacy of integrin αvβ6/8 mAb treatment.
(A) Immunoblot showing levels of SOX4 and GAPDH proteins in GFP+ 4T1 murine TNBC cells containing a doxycycline (DOX) inducible Sox4 cDNA construct (4T1Sox4-Dox). Cells were treated with the indicated concentrations of doxycycline for 48 h. (B) Cells from (A) were co-cultured with murine GFP-specific CD8+ T cells (red) (E:T = 1:1), and the fraction of surviving cells was quantified (Y-axis) after 24 h of co-culture. (C) 4T1Sox4-Dox tumor cells were treated with either DOX (500 ng/mL) alone or in combination with integrin αvβ6/8 mAb for 48 h followed by co-culture with murine GFP specific CD8+ T cells (red) for 18 h. (D) 4T1Sox4-Dox TNBC (n=10 mice/group) primary tumor volume shown at indicated time points. Mice with similar tumor burden were fed either a regular diet or a doxycycline-containing diet (625 ppm, Envigo Teklad) starting on day 7 to induce the expression of SOX4 in tumor cells. Mice receiving either diet also received monotherapy with integrin αvβ6/8 or isotype control mAbs (IP, 0.25 mg/dose, twice weekly) until tumor volume in any group reached 1000 mm3. Data are summarized as mean ± SD of tumor volume. (E) Number of lung surface metastases in mice treated as described in (D) following staining with picric acid for 24 h. Summary of number of lung surface metastases (left) and representative images (right) on day 21 following tumor inoculation. (F) Quantification of tumor-infiltrating CD8+ T cells per gram of tumor (right) following treatment as described in (D) on day 21 following tumor inoculation. (G) Immunoblot showing levels of SOX4 and GAPDH proteins in 4T1Sox4-Dox tumors (n=3 per group) derived from mice treated as described in (D) on day 21. Numbers represent relative quantification of SOX4 to GAPDH, normalized to the average expression in vehicle and IgG treated controls. Data in [A, B, C and G] are representative of at least two independent experiments with technical triplicates and summarized as mean ± SEM [B, C]. Data in [D, E, F] are an average of two independent experiments and summarized as mean ± SD. To determine statistical significance, a one way ANOVA with Dunnett’s [B-E] post hoc test or an unpaired Student t-test [F] was used. ***P < 0.001; **P < 0.01; *P < 0.05; n.s., not significant.
SOX4 regulates multiple pathways relevant for T cell-mediated tumor immunity
GSEA analysis of RNA-seq data showed that the ‘interferon response’ represented one of the major pathways for genes upregulated in SOX4 or ITGAV edited compared to control edited human BT549 and murine 4T1 TNBC cells (Fig. 6A, B, Fig. S6A–F, Table S1). In contrast, gene sets associated with TGFβ and TNFα/NF-κB were negatively enriched in both Sox4 and Itgav edited 4T1 TNBC cells (Fig. 6B). Further analysis of RNA-seq data showed that Sox4 or Itgav edited 4T1 TNBC cells contained higher mRNA levels of many interferon-stimulated genes (ISGs), including genes associated with important innate immune pathways such as RIG-I/MDA-5, cGAS – STING and the AIM2 inflammasome (Fig. 6C). It is important to note that these RNA-seq experiments were performed in the absence of T cells or added interferons. We confirmed increased mRNA and protein levels for selected genes in human BT549 and murine 4T1 TNBC cells edited with SOX4 or control gRNAs (Fig. S6C–E).
Fig. 6. SOX4 regulates multiple innate and adaptive immune genes to inhibit T cell-mediated tumor immunity.
(A) GSEA analysis for gene sets associated with an interferon response in human (BT549, left) and murine (4T1, right) TNBC cells edited with SOX4 (top) or ITGAV (bottom) gRNAs compared to control edited cells. (B) GSEA results are summarized as the normalized enrichment score (NES) in Sox4 (S1, S2) or Itgav (I1, I2) deficient 4T1 TNBC cells as compared to control edited counterparts. (C) Heat map showing RNA-seq data for indicated genes from 4T1 TNBC cells edited with either two Sox4 (S1, S2) or two Itgav (I1, I2) gRNAs. Gene expression is shown relative to control edited 4T1 cells (log2FC, color scale). (D) Surface HLA-ABC protein levels on BT549 cells edited with two different SOX4 gRNAs or a control gRNA (CTRL); isotype control antibody staining is shown in black. (E) Surface HLA-ABC protein levels on BT549 cells edited with two different ITGB6 (B6–1, B6–2) or control gRNAs, followed by stimulation with indicated concentrations of IFNγ for 24 h. (F) ChIP-seq with SOX4 mAb in human BT549 TNBC cells. SOX4 specific peaks (red boxes) at the indicated gene loci relative to reference genome Hg19 analyzed using the IGV viewer (IGV, Broad Institute). The data range is shown on top for each indicated gene. Data in [A-C] represent an average of triplicates of two independent gRNAs for each gene knockout. Data in [D, E] are representative of at least two independent experiments with technical triplicates. Data in [F] was assessed using biological triplicates each composed of technical duplicates. A two-way ANOVA with Dunnett’s post hoc test was used to determine significance in [D and E]. Data are summarized as mean ± S.E.M, *** P < 0.001; **P < 0.01; *P < 0.05; n.s., not significant. See also Figure S6, Table S1, and S2.
SOX4 or ITGB6 edited human BT549 TNBC cells had higher surface levels of MHC class I proteins (HLA-ABC) compared to control edited cells in the absence and presence of IFNγ stimulation (Fig. 6D, E). Consistent with these findings, RNA-seq data showed higher level expression of a number of genes in the MHC class I pathway (including HLA-A, HLA-B and TAP1) in SOX4 knockout compared to control BT549 cells (Fig. S6B, C).
Chromatin immunoprecipitation (ChIP) experiments in human BT549 TNBC cells with a SOX4 versus control IgG antibody identified SOX4 specific peaks in the regulatory regions of interferon pathway genes (such as IRF7 and ISG15) and MHC-I pathway genes (including TAP1, TAP2, PSMB9, HLA-B and HLA-C) (Fig. 6F, Table S2). These data are consistent with the hypothesis that SOX4 regulates the expression of multiple genes in innate and adaptive immune pathways in tumor cells.
Inhibition of the SOX4 pathway prevents the emergence of MHC-Ilow tumor cells resistant to CD8+ T cells
Tumor cell escape from cytotoxic T cells is frequently mediated by inactivation of MHC-I pathway genes by mutational or transcriptional mechanisms (Gide et al., 2018; Zaretsky et al., 2016). When BT549 TNBC cells were co-cultured with CD8+ T cells for 24 hours, we observed the emergence of a substantial population of tumor cells with low/absent MHC-I cell surface protein (HLAlow) (Fig. 7A, Fig. S7A). Notably, inactivation of the SOX4, ITGAV or ITGB6 genes substantially reduced the percentage of these HLAlow BT549 tumor cells (Fig. 7A, B). Similarly, pre-treatment of tumor cells with integrin αvβ6/8 but not a control mAb inhibited the emergence of these HLAlow tumor cells (Fig. 7C, D).
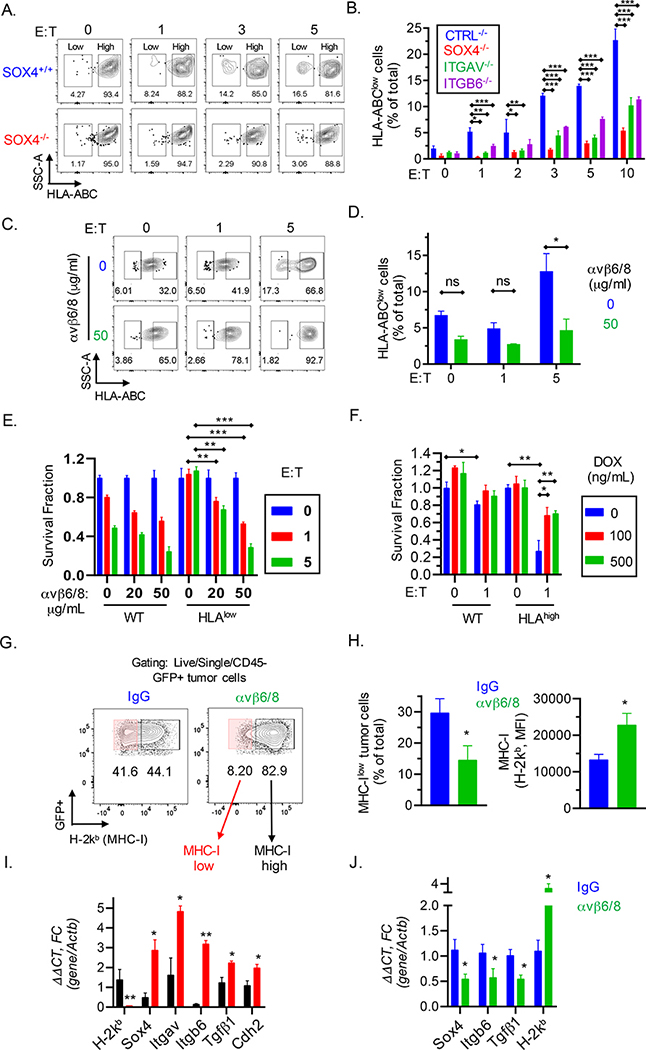
Fig. 7. Inactivation of SOX4 gene inhibits emergence of MHC class I deficient TNBC cells during selection by cytotoxic T cells.
(A) Contour plots showing enrichment of HLA-ABC low/negative populations following a 24 h co-culture of SOX4+/+ or SOX4−/− BT549 human TNBC cells with human T cells expressing a NY-ESO-1 TCR at the indicated E:T ratios. (B) Quantification of HLA-ABC low/negative cells for indicated gene edited BT549 tumor populations following co-culture with CD8+ T cells as described in (A). Data are summarized as mean ± S.E.M. (C-D) BT549 tumor cells were pretreated with indicated concentrations of integrin αvβ6/8 mAb for 72 h and then co-cultured with CD8+ T cells. Contour (C) and summary (D) plots of HLA-ABC low/negative BT549 TNBC cells following co-culture with CD8+ T cells for 24 h at indicated E:T ratios. Isotype control Ab was used to define MHC-I negative populations. (E) Human BT549 TNBC cells expressing wild-type (WT) or low levels of HLA-ABC (HLAlow) were sorted and then pre-treated with indicated concentrations of integrin αvβ6/8 mAb followed by co-culture with NY-ESO-1 specific CD8+ T cells at the indicated E:T ratios. (F) BT549 TNBC cells were transduced with a doxycycline inducible SOX4 cDNA construct followed by FACS-based enrichment of HLA-ABChigh cells; tumor cells were then pre-treated for 48 h with the indicated concentrations (ng/ml) of doxycycline (DOX). Numbers of surviving wild-type (WT) or HLA-ABChigh tumor cells were quantified after co-culture with CD8+ T cells for 24 h. (G-J) Characterization of emergence of MHC class I deficient TNBC cells in vivo. (G) Contour plots showing expression of MHC-I (H-2Kb) in Py8119 tumors derived from mice treated with either control IgG Abs or integrin αvβ6/8 mAbs. (H) Quantification of MHC-Ilow (H-2Kb) cells shown in (G), represented as a percentage of total cells (left) or as MFI (right). (I) mRNA levels of indicated genes relative to β-actin in sorted MHC-Ihigh (black) and MHC-Ilow (red) murine TNBC cells derived from isotype control IgG treated tumors as shown in (G) or (J) in whole tumors from mice treated as described in (G). Data in [A-D, G-J] are representative of at least two independent experiments. Data in [E, F] are representative of three independent experiments. A two-way ANOVA with Dunnett’s post hoc test [B, D, E, and F] and an unpaired Student t-test [H-J] were used to determine significance, ***P < 0.001; **P < 0.01; *P < 0.05; n.s., not significant. See also Figure S7.
We further investigated the molecular mechanism by sorting HLAlow and HLAhigh BT549 tumor cells after co-culture with CD8+ T cells for 48 hours (Fig. S7B, C). Interestingly, HLAlow tumor cells had significantly higher mRNA levels of SOX4, ITGAV and ITGB6 as well as lower mRNA levels of HLA-A and HLA-B compared to non-sorted BT549 TNBC cells (Fig. S7D, E). Consistent with these RT-qPCR data, HLAlow tumor cells had substantially higher levels of integrin β6 protein compared to non-sorted BT549 cells (Fig. S7F). In contrast, HLAhigh tumor cells expressed lower mRNA levels of SOX4, ITGAV, and ITGB6 genes (Fig. S7G). Importantly, pre-treatment of MHC-Ilow cells with integrin αvβ6/8 mAb re-sensitized them to cytotoxic T cells while control mAb treated MHC-Ilow cells were highly resistant to T cells (Fig. 7E). Conversely, doxycycline-induced overexpression of SOX4 in MHC-Ihigh cells conferred resistance to CD8+ T cells (Fig. 7F).
To investigate the in vivo relevance of these findings, we evaluated the expression of MHC-I (H2-Kb) on Py8119 tumor cells from mice treated with integrin αvβ6/8 or isotype control mAbs. We found that integrin αvβ6/8 monotherapy significantly decreased the number of MHC-Ilow tumor cells (Fig. 7G, H). Similar to the in vitro studies described above, we utilized FACS to enrich MHC-Ilow and MHC-Ihigh tumor cells followed by RT-qPCR analysis of key genes. Notably, MHC-Ilow cells had significantly increased expression of genes belonging to the integrin αVβ6 – SOX4 (Itgav, Itgb6, and Sox4), TGFβ (Tgfb1) and EMT (N-cadherin, cdh2) pathways compared to MHC-Ihigh tumor cells (Fig. 7I). Consistent with these findings, we observed an increase in MHC-I (H2-kb) and a decrease in Sox4, Itgb6, and Tgfβ1 mRNA levels in whole tumors following integrin αvβ6/8 versus isotype control mAb monotherapy (Fig. 7J). These data demonstrate that targeting of the integrin αv – SOX4 pathway can reduce the emergence of MHC-I deficient TNBC cells during selection by cytotoxic T cells, both in vitro and in vivo.
Discussion
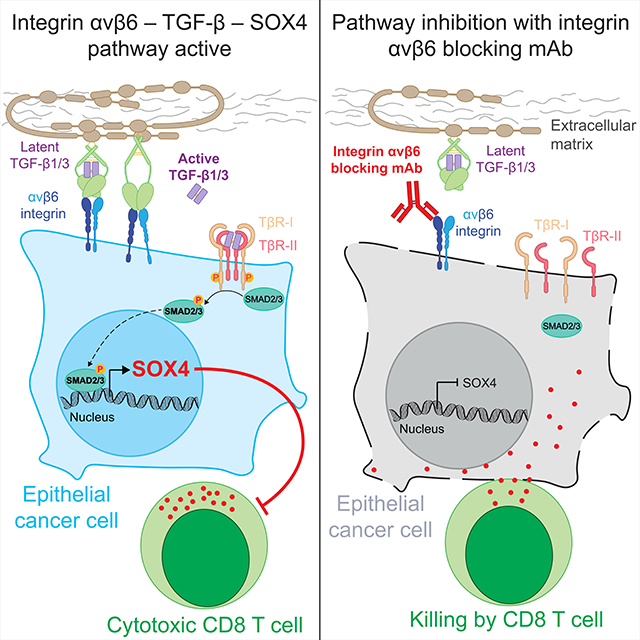
Expression of the SOX4 transcription factor has been associated with EMT and a poor prognosis in many human cancers but its role in promoting immune evasion was previously not known. Here we show that SOX4 promotes resistance of human and murine TNBC cells to cytotoxic T cells. Mechanistically, SOX4 regulates several important innate and adaptive immune pathways in tumor cells. In SOX4 KO tumor cells, expression of many type 1 interferon-inducible genes is upregulated, including genes in the RIG-I/MDA-5, cGAS – STING and AIM2 inflammasome pathways. Inactivation of SOX4 also increases expression of genes in the MHC class I pathway while reducing expression of PD-L1. Importantly, targeting of SOX4 with an integrin αvβ6 mAb inhibits the emergence of resistant tumor cells with low MHC class I levels during selection by cytotoxic T cells.
What is the relationship between the SOX4-mediated immune evasion program and EMT? Several recent studies in murine models and human cancers have proposed that EMT is associated with impaired tumor immunity (Chockley and Keshamouni, 2016; Dongre et al., 2017), but the molecular mechanisms were not fully defined. We propose that these two biological processes are interconnected, but also partially distinct. The fundamental connection between these two cellular programs is that both are induced by TGFβ, a cytokine that serves a fundamental role in tissue homeostasis by promoting repair and suppressing adaptive immunity (Morikawa et al., 2016). A second connection between EMT and immune evasion is the SOX4 transcription factor. SOX4 expression is directly induced by TGFβ signaling and contributes to the EMT program (Lourenco and Coffer, 2017), although other transcription factors may arguably play a more central role in the cellular programs leading to EMT. A third connection between EMT, SOX4 and immune evasion relates to the differentiation state of epithelial cells. In breast cancer, EMT has been associated with a less differentiated state of tumor cells (Ye et al., 2015). In several human cancers, SOX4 is associated with a stem-like state that correlates with poor survival outcomes (Ikushima et al., 2011; Peng et al., 2017; Zhang et al., 2012). SOX4 may therefore contribute to immune evasion by less differentiated tumor cells in cancers of epithelial origin.
The integrin αvβ6 heterodimer is expressed at low levels by healthy epithelial cells. Infection and transformation induce upregulation of integrin αvβ6 on the surface of epithelial cells, thus enhancing activation of TGFβ deposited on the extracellular matrix (Munger et al., 1999). In many human cancers of epithelial origin, integrin αvβ6 expression has been associated with a poor prognosis (Niu and Li, 2017). Also, recent studies have implicated TGFβ in resistance to checkpoint blockade. For example, in patients with metastatic bladder cancer who received a PD-L1 blocking mAb (atezolizumab), a TGFβ gene expression signature in tumor RNA-seq data was associated with a poor treatment response (Mariathasan et al., 2018). Thus, a series of clinical studies have separately demonstrated an association of integrin αvβ6, TGFβ or SOX4 with poor survival and/or response to therapy. This study demonstrates that these three molecules form an important immune evasion pathway which confers tumor cell resistance to cytotoxic T cells.
Multiple lines of experimental evidence indicate that integrin αvβ6 serves as a key regulator of this SOX4-driven immune evasion pathway. Specifically, we show that the SOX4 transcription factor can be therapeutically targeted with an integrin αvβ6/8 mAb which inhibits activation of TGFβ from a latent precursor. Our findings suggest that SOX4 plays a dual role in promoting progression of TNBC and other human cancers: it promotes invasion/metastasis and inhibits T cell-mediated immunity against invasive cancer cells. Therefore, reduced invasion/metastasis and enhanced T cell-mediated immunity are likely to contribute to the therapeutic efficacy of the integrin αvβ6/8 mAb, in particular against metastases.
Targeting of the integrin αvβ6 – TGFβ – SOX4 pathway may be relevant for many other human epithelial cancers, in addition to TNBC. These findings could be rapidly advanced to clinical trials because high-affinity blocking antibodies and small molecule inhibitors for integrin αvβ6/8 are already available (Raab-Westphal et al., 2017). It is worth noting that a phase II clinical trial with an integrin αvβ6 blocking antibody (BG00011) in patients with pulmonary fibrosis was terminated due to safety concerns. This antibody had a human IgG1 Fc region that binds with high affinity to activating Fc receptors expressed by NK cells and myeloid cells (Raghu et al., 2018). It is therefore possible that antibody-dependent cellular toxicity (ADCC) contributed to the side effects observed with this antibody. In contrast, an integrin αv blocking mAb (abituzumab) with a human IgG2 Fc region was found to be well tolerated in phase I and II clinical trials in patients with prostate and colon cancer (Elez et al., 2015; Uhl et al., 2014). The IgG2 Fc region binds only with low affinity to some but not all activating Fc receptors (Vidarsson et al., 2014), and abituzumab may therefore not induce a significant level of ADCC. Taken together, these findings provide the rationale for therapeutic targeting of the integrin αvβ6 – TGFβ – SOX4 immune evasion pathway to promote tumor immunity in TNBC and other aggressive human cancers of epithelial origin.
STAR METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Kai Wucherpfennig (Kai_wucherpfeenig@dfci.harvard.edu)
Materials Availability
This study did not generate new materials or mouse models.
Data and Code Availability
The accession number for the raw RNA-seq and ChIP-seq data reported in this paper is GEO: GSE144014.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mouse strains
4–6-week-old mice female Balb/c (JAX stock #000651) or C57Bl/6J (JAX stock #000664) mice were purchased from The Jackson Laboratory. Pmel transgenic mice (B6.Cg-Thy1a/Cy Tg(TcraTcrb) 8Rest/J, JAX stock #005023) were also purchased from JAX labs. CD8+ T cells from these mice express a TCR specific for a peptide of pmel-17 which is expressed by melanocytes and melanoma cells including the B16F10 cell line. Experiments in murine models were performed in accordance with protocols approved by the Institutional Animal Care and Use Committee (IACUC) at DFCI.
Cell lines
Murine TNBC cells (4T1, Py8119), human TNBC cells (BT549, Hs578T), murine melanoma cells (B16F10), and human HEK293T cells were obtained from ATCC. TGFβ luciferase reporter cell line HepG2 was purchased from Signosis (SL-0016-NP).
METHOD DETAILS
Reagents
The following cytokines were used: Human IL-2 (BioLegend 589104), murine IL-2 (BioLegend 575406), murine TGFβ1 (BioLegend 763104), murine IFNγ (Abcam #Ab9922), human IFNγ (PeproTech #300–02). In vivo experiments were performed with the following mAbs: inVivoMAb mouse anti-PD1 (BioXcell RMPI-14 clone), inVivoMAb rat IgG2a, isotype control (BioXcell 2A3 clone) and CD8α depletion antibody (BioXcell, 2.43 clone). Other reagents included Doxycycline hyclate (Sigma-Aldrich #D9891), collagenase type IV (Sigma-Aldrich #C5138), DNAse type IV (Sigma-Aldrich #D5205), Hyaluronidase Type V (Sigma-Aldrich #H6254), ACK lysis buffer (Life Technologies #A1049201), Percoll density gradient media (Sigma-Aldrich #P1644) and TGFβ receptor I inhibitor Galunisertib, LY2157299 (Selleck #S2230).
Plasmids
pINDUCER21-SOX4 was a gift from George Daley (Addgene plasmid # 51304, http://n2t.net/addgene:51304; RRID: Addgene_51304). The 1539 bp ORF of human SOX4 containing C-terminal FLAG and His tags (pENTER-CMV-SOX4) was purchased from Vigene Biosciences (CH830603). Following PCR amplification using FWD primer 5’-AAAAAAGCTAGCGCCGCCAC CATGGTGCAG CAAACCAACA ATGCCGAGAA-3’ containing a 5’ NheI restriction enzyme (RE) site and REV primer TTTTTTGGATCCTTAGT GGTGGTGGTG GTGGTGCTCGAC containing a 3’ BamHI RE site, the SOX4 ORF was cloned into the pHAGE-ZsGreen lentiviral plasmid cut with NheI and BamHI RE enzymes (SOX4-FLAG).
The 1320 bp murine Sox4 cDNA equipped with a Myc-DDK tags in a pCMV6 vector was purchased from Origene (Cat #MR207005). Following PCR amplification using FWD primer 5’-CATACTAGTATGGTACAACAGACCA-3’ containing a 5’ SpeI restriction enzyme (RE) site and REV primer AAAAAACTCGAGTCAGTAGGTGAAGACCAGGTT containing a 3’ PspXI RE site, the Sox4 cDNA was cloned into the pINDUCER21-ORF-EG (Addgene Plasmid #46948) plasmid cut with SpeI and PspXI RE enzymes (Sox4-DOX).
Expression of integrin αvβ6/8 mAb in CHO cells
The 264RAD mAb binds with high affinity to both human and murine integrin αvβ6 proteins and inhibits integrin αvβ6-mediated TGFβ activation (29). We expressed this antibody in CHO cells and introduced two mutations into mouse IgG2b Fc region (D265A and N297A) to prevent antibody binding to activating Fc receptors (Shields et al., 2001). This approach thus limited the activity of this antibody to its blocking function and prevented a contribution of antibody-mediated cellular cytotoxicity (ADCC) to in vivo efficacy. The cDNAs encoding the mAb heavy and light chains were cloned into the UCOE Hu-P vector (EMD Millipore). The two cDNAs were separated by viral 2A skip sequence which enabled stoichiometric expression from a single plasmid for efficient antibody expression. Selection of transfected cells was performed with puromycin (InvivoGen) at concentrations up to 50 μg/ml. Expression was scaled up in Freestyle CHO medium supplemented with 40 ml GlutaMAX and 10 ml anti-clumping agent (Life Technologies) per liter. Cells were split to 0.25×106/ml in 5 L Optimum Growth shaker flasks (Thompson Scientific) and incubated in a Multitron incubation shaker (Infors HT) at 37°C, 8% CO2, 120 rpm. Supernatant containing the antibody was collected after 8–10 days and purified using Protein G Sepharose affinity columns (GE Healthcare). Size-exclusion chromatography was performed using a Superose 6 HPLC column (GE Biosciences). Expression of stable clones was 25–100 mg per liter. Antibody was concentrated using Amicon spin columns (Millipore) using PBS as the final buffer and sterile filtered prior to in vivo experiments.
Culture media
Tumor cells were cultured in RPMI 1640 media (+ L-glutamine) supplemented with 10% Fetal Bovine Serum (FBS), 2 mM L-Glutamine (Glu), 100 IU/ml Penicillin/Streptomycin (Pen/Strep).
Human T cells were cultured in human T cell media (hTCM): RPMI 1640 (+ L-glutamine) containing HEPES (5mM), Glutamax (2 mM), Pen/Strep (50ug/mL), non-essential amino acids (NEAA, 5 mM), sodium pyruvate (5 mM), fetal bovine serum (9 %), human serum (1 %), and beta-mercaptoethanol (50 μM). Media was replenished with fresh human IL-2 (20 ng/mL) every 2–3 days. Murine T cells were cultured in murine T cell media (mTCM): RPMI 1640 (+ L-glutamine) containing HEPES (5 mM), Glutamax (2 mM), Pen/Strep (50 μg/mL), non-essential amino acids (NEAA, 5 mM), sodium pyruvate (5mM), fetal bovine serum (10%), and beta-mercaptoethanol (50 μM). Media was replenished with fresh human IL-2 (20 ng/mL) every 2–3 days.
CRISPR/Cas9 editing
Editing of tumor cell lines was performed using ribonuclear protein complexes (RNP) of Cas9 protein with bound gRNAs. As a first step in the assembly of RNPs, 100 μM of tracrRNA (IDT) was mixed with the appropriate crRNA (100 μM) at a 1:1 ratio, incubated at 95 °C for 5min followed by cooling to room temperature. Cas9 protein (20 μM, Macrolab) was then added and RNPs were incubated for 15min at 37 °C.
RNPs were introduced into cells by electroporation. Cells were electroporated using Lonza 4D Nucleofector Core Unit (Lonza #AAF-1002B) with 100μM of assembled RNPs in SF buffer using SF Cell Line 96-well Nucleofector™ Kit (#V4SC-2096) and program number DJ-110, as per manufacturer’s instructions. See Table S3 for full list of crRNA sequences. Editing efficiency was determined by DNA sequencing, immunoblot analysis and/or flow cytometry, depending on the targeted gene.
Immunoblotting
Cells were washed with PBS and lysed using RIPA cell lysis buffer (Thermofisher Scientific #89900) supplemented with protease inhibitor cocktail (Roche, complete mini, EDTA free protease inhibitor tablets, #11836170001) and phosphatase inhibitors (Thermo Scientific, Halt Phosphatase Inhibitor Cocktail, #78427). Total protein concentration was determined using BCA Protein Assay Kit (ThermoFisher Pierce, 23225). Proteins were separated using NuPAGE Novex 4–12% Bis-Tris gels using 1X MOPS SDS running buffer and transferred to PVDF membranes. Blots were blocked in PBS containing 4% milk powder and 0.2% Tween and then incubated overnight with primary antibodies followed by washes and exposure to secondary antibody for 2 hours at room temperature. Western blots were then incubated in luminol-based substrate for HRP-catalyzed detection (Perkin Elmer #NEL104001EA) and luminescence was captured on ChemiDoc MP Imaging System (Bio-Rad #12003154). See Table S4 for a detailed list of antibodies used in immunoblotting.
Flow cytometry
Single cell suspensions were stained with primary antibodies at 4°C for 20min in FACS buffer (2% FBS, 2mM EDTA) following blockade of Fc receptors in PBS for 10min. For intracellular staining, cells were first labeled with antibodies specific for investigated surface markers, fixed in Fix/Perm buffer (eBioscience) for 15 min, washed twice with permeabilization buffer (eBioscience) and stained with primary antibodies targeting intracellular proteins in permeabilization buffer for 30 min at 4°C. Cells were analyzed on a BD Biosciences Fortessa instrument or sorted on a BD Biosciences Aria III instrument. Data analysis was performed using FlowJo 10. See Table S4 for a detailed list of antibodies used in flow cytometry.
RT-PCR
Total RNA was isolated from cells using the RNeasy Mini Kit (QIAGEN, Valencia, CA). cDNAs were synthesized from 1 μg of total RNA using the PrimeScript RT reagent Kit (Takara) and were amplified by SYBR Premix Ex Taq II (Takara) using the CFX96 Real-Time PCR System (Bio-Rad) according to the manufacturer’s protocols. RT-qPCR was performed using 7900HT Fast Real-Time PCR System (Applied Biosystems, Carlsbad, CA) using SYBr GreenMaster Mix (Invitrogen). For a detailed list of human and murine RT-qPCR primer sequences, see Table S3.
TGFβ reporter assay
SMAD2/3 responsive luciferase reporter HepG2 cells (hygromycin resistant) stably expressed a firefly luciferase reporter gene under the control of the SMAD2/3 response element (Signosis, SL-0016-NP). Cell lines were co-cultured with this reporter cell line for 24–48 hours followed by detection of luciferase activity using Promega Luciferase Assay System® (Glo Lysis Buffer (#E2661), Cell Culture Lysis Reagent (#E1531), Passive Lysis Buffer (#E1941) and Reporter Lysis Buffer (#E3971) as per manufacturer’s instructions.
Isolation and propagation of primary murine CD8+ T cells
Murine T cells that expressed a gp100 (Pmel-1) or GFP specific (JEDI) TCR were cultured in murine T cell media (mTCM): RPMI (+ L-glutamine) containing HEPES (5 mM), Glutamax (2 mM), Pen/Strep (50 μg/mL), NEAA (5 mM), sodium pyruvate (5mM), FBS (10%), beta-mercaptoethanol (50 μM). Cells were cultured at 37°C under an atmosphere of 5% carbon dioxide. Cell line were recently authenticated and verified for being mycoplasma-free using the MycoAlert mycoplasma detection kit (Lonza #LT07–118).
Pmel-1 TCR transgenic mice were purchased from Jackson Laboratory (stock # 005023). This transgenic strain carries a TCR transgene specific for the mouse homologue (pmel-17) of human pre-melanosome protein (PMEL, or gp100). JEDI mice which carry a GFP-specific TCR transgene on the Balb/c background were kindly provided by Dr. Judith Agudo (Agudo et al., 2015). Murine CD8+ T cells were isolated from spleens using a CD8+ T cell isolation kit (STEMCELL #19753) according to the manufacturer’s protocol. Freshly isolated CD8+ T cells were stimulated with αCD3/αCD28 Dynabeads (Life Technologies #11453D) at a bead to cell ratio of 1:2. On day 3, recombinant mouse IL-2 (Biolegend #575406) was added to the culture at 20 ng/ml.
Isolation and generation of primary human CD8+ T cells expressing NY-ESO-1 specific TCR
Peripheral blood mononuclear cells (PBMCs) were isolated by Ficoll density gradient centrifugation from leukopheresis collars from healthy donors (Brigham and Women’s Hospital Blood Bank). CD8+ T cells were purified from PBMCs using CD8 Dynabeads (StemCell # 19053) following the manufacturer’s instructions. Isolated CD8 cells were activated for 48 hours with αCD3/αCD28 beads (Life Technologies #11132D, 1:2 ratio of beads to T cells) and grown in the presence of 30U/mL of human IL-2 for one week.
Expanded CD8+ T cells were transduced with the lentivirus by spin infection to introduce the NY-ESO-1 TCR. A non-tissue culture treated 24 well plate was coated with 0.8 ml of 15 μg/ml Retronectin (Takara; Kyoto, Japan) overnight at 4°C. Wells were blocked with sterile 2% BSA for 15 minutes at room temperature and gently washed once with PBS. Next, lentivirus was added to wells of the retronectin-coated plate at a multiplicity of infection (MOI) of 15, and plates were spun for 2.5 hours at 2,000 × g, 32°C. The supernatant in the wells was then carefully decanted, and wells were gently washed with 0.5 ml of PBS. 0.5 × 106 T cells were transferred to wells containing 10 μg/ml protamine sulfate (Sigma-Aldrich) in RPMI-1640 media containing 30 U/ml IL-2 and cultured for three days. NY-ESO-1 TCR+ T cells were isolated to >90% purity by FACS and expanded with Dynabeads and IL-2 (30 U/ml).
In vitro cytotoxicity assays
All in vitro cytotoxicity assays were performed in human or murine T cell media (without addition of IL-2). Cells were co-cultured on collagen I coated 96 well plates (ThermoFisher #A1142803) or Corning® 96 Well Black Polystyrene Microplates (Corning #3603) coated with Collagen I (5μg/cm2) according to the manufacturer’s instructions (ThermoFisher #A1048301).
Human cytotoxicity assay
BT549 human TNBC cells are HLA-A02*01 positive (Figure S1A) and endogenously express the NY-ESO-1 antigen (Fig. S1B), allowing recognition by T cells specific for a NY-ESO-1 peptide presented by HLA-A2*01 (Zhao et al., 2005). BT549 cells were co-cultured with human CD8+ T cells that expressed a NY-ESO-1 TCR (generated as described above) at increasing effector to target (E:T) ratios for 12–72 hours. Cytotoxicity was determined using flow cytometry.
Murine cytotoxicity assays
4T1 murine TNBC cells expressing GFP (4T1GFP) were generated as described previously (Agudo et al., 2015). 4T1GFP cells were co-cultured with murine CD8+ T cells derived from JEDI mice that recognized a GFP peptide presented by H2-Kd (Agudo et al., 2015). B16F10 murine melanoma cells which endogenously expressed the Pmel antigen (Pmel-17) were pre-treated for 24 hours with 0.1–10 ng/mL of murine IFNγ to induce surface expression of MHC class I protein (Zhou, 2009). The melanoma cells were then co-cultured with CD8+ T cells from Pmel-1 transgenic mice to study T cell-mediated cytotoxicity. The number of tumor cells that were seeded remained fixed (5–10×103 per well depending upon the tumor cell line) and CD8+ T cells were added at increasing effector to target ratios. The total number of surviving tumor cells was quantified 12–72 hours after initiation of co-cultures by either flow cytometry or image cytometry (Celigo, Nexcelom Bioscience).
Celigo Image Cytometer instrumentation
The Celigo Image Cytometer is designed to perform plate-based image cytometric analysis and was used here to quantify the number of surviving fluorescent tumor cells in the presence of cytotoxic T cells. It is equipped with one bright-field (BF) and four fluorescence (FL) imaging channels: Blue (EX: 377/50 nm, EM: 470/22 nm), Green (EX: 483/32 nm, EM: 536/40 nm), Red (EX: 531/40 nm, EM: 629/53 nm), and Far Red (EX: 628/40 nm, EM: 688/31 nm). The image cytometer allows auto-focusing in the well based on image contrast or the thickness of the bottom surface. The Celigo software application “Target 1 + 2” was used to identify and count the number of GFP+ tumor cells (Green channel). The Celigo instrument was set up to acquire images including brightfield (Target 1) and Green fluorescent (Target 2) channels with an exposure time of 10,000 μs. Next, hardware-based autofocus (HWAF) was used to focus in the BF channel, and the focus offset was applied to the Green (+26 μm) channel. GFP+ target cells above an intensity threshold were counted, and the data were analyzed using GraphPad Prism software (GraphPad Software Inc, San Diego, CA).
Treatment of cells with integrin αvβ6/8 and PD-1 mAbs
Human BT549, murine 4T1 or murine B16F10 cells were pre-treated with 2–50 μg/mL of integrin αvβ6/8 or control IgG mAb for 24–72 hours before co-culture with CD8+ T cells. The exact conditions are described for each experiment in the Fig. legends. Anti-human PD-1 (20 μg/mL, Bioxcell clone J116, BE0188), control IgG (20μg/mL, Bioxcell, clone MOPC-21 #BE0083), anti-murine PD-1 (20 ug/mL, Bioxcell, clone RMP1–14 #BE0146) or rat IgG2a isotype control, anti-trinitrophenol (Bioxcell, clone 2A3 #BE0089) were added when co-cultures were set up.
Animal Experiments
Female BALB/c (Jackson Laboratory #000651) or C57Bl/6J (Jackson Laboratory #000664) mice of 4–6 weeks of age were purchased from The Jackson Laboratory. 4T1 (2 × 105) or Py8119 (5 × 105) TNBC cells were injected in 50 μl of PBS orthotopically into the mammary fat pads of syngeneic mice (BALB/c mice for 4T1 cells, C57Bl/6J mice for Py8119 cells). When tumors reached approximately 50mm3, mice carrying similar tumor burden were randomized into treatment groups and treated as described below.
Treatment with integrin αvβ6/8 antibodies
Mice received biweekly intraperitoneal (IP) injections of 0.2 mg of integrin αvβ6/8 or IgG2b control mAbs in 100μL of PBS solution for 3–8 weeks depending upon the experimental endpoint. The specific endpoint for each experiment is indicated in the Fig. legend.
Treatment with PD-1 antibodies
Mice received biweekly intraperitoneal (IP) injections of 0.2 mg of PD-1 mAb (rat IgG2a, RMP1–14 clone) or rat IgG2a control mAb in 100 μL of PBS for 3–8 weeks depending upon the experimental endpoint. The specific endpoint for each experiment is indicated in the Fig. legend.
Depletion of CD8+ T cells using anti-CD8β antibodies
The depletion of CD8+ T cells in BALB/c and C57BL/6J mice was achieved by IP injection of 0.1 mg of CD8β mAb (BioXCell, Clone 53–5.8 #BE0223) in 100 μL of PBS on days −1, 0, 7 and 14 relative to tumor inoculation. Mice receiving an isotype control mAb (Bio × cell, clone HRPN #BE0088) at the same dose in PBS were used as controls. CD8+ T cell depletion was confirmed by labeling of CD8+ T cells from spleens with a CD8 mAb (Biolegend #100741) followed by flow cytometric analysis (BD Fortessa, BD Biosciences). CD8+ T cells were significantly depleted within 24 hours of administration of CD8β antibodies and at the experimental endpoint.
Induction of SOX4 expression in vivo.
To induce expression of SOX4 in 4T1Sox4-Dox cells in vivo, BALB/c mice were fed a doxycycline containing diet (625ppm, Envigo Teklad) until the experimental endpoint. Mice receiving a regular feed were used as controls. Intra-tumoral induction of SOX4 was confirmed by immunoblotting at experimental endpoint (2–3 weeks after initiation of the DOX diet).
Endpoints
Primary tumor volumes were determined using calipers to measure dimensions and calculated using the formula: Volume (mm3) = 0.5 × Length (mm) × (Width (mm))2 At the experimental endpoint, mice were euthanized followed by surgical excision of tumors, tumor draining lymph nodes (TdLN), spleens and/or lung tissue for downstream analyses. The experimental endpoint for individual mice was either a tumor volume >1000 mm3, tumor ulceration, interference of tumors with movement, a moribund state or conclusion of the experiment. For the purpose of Kaplan-Meier survival curves, mice were considered dead when tumor volumes exceeded 1000 mm3. All tumor experiments were performed in accordance with the protocols approved by the Institutional Animal Care and Use Committee (IACUC) at DFCI.
FACS analysis of tumor-infiltrating immune cells
Tumors were excised when the majority of tumors in any experimental group reached the endpoint (tumor size of ~1000 mm3), approximately 3–5 weeks following tumor inoculation. The tumors were cut into small pieces using sterile scalpels in serum free RPMI 1640 media (ThermoFisher #11875093). Samples were dissociated in 1 mg/ml Collagenase Type IV (Sigma-Aldrich #C5138), 20 units/ml DNAse Type IV (Sigma-Aldrich #D5205), 0.1 mg/ml Hyaluronidase Type V (Sigma-Aldrich #H6254) using GentleMACS C or M tubes using Gentle MACS m_impTumor04 program in the gentleMACS™ Dissociator (Miltenyibiotech #130–093-235) followed by incubation at 37°C. The suspension was passed through a 70 μm filter and pelleted by centrifugation at 300 × g for 5min. To remove red blood cells, ACK lysis buffer (3x by volume) was added for 45–60 seconds followed by 2 volumes of RPMI to stop red cell lysis. Crude bulk removal of tumor cells was performed by centrifugation at a low g force (50 × g) for 5min, maximum acceleration and deceleration. Pelleted cells from pooled supernatants (>300 × g or 1500 rpm, 5 min) were resuspended in the appropriate buffer for flow cytometric analysis of tumors.
Tumor draining lymph nodes, spleens and lungs were physically dissociated using 70μm strainers (Miltenyi Biotec) and 3mL syringe handles. Cells were washed with RPMI 1640 medium. Red blood cells were lysed with ammonium chloride solution for 5min on ice (Stemcell), washed with RPMI 1640 medium and resuspended in the appropriate buffer for flow cytometry. Single cell suspensions were stained with 5μg/mL Fc receptor blocking anti-mouse CD16/CD32 antibody (clone 2.4G2, BD PharMingen) at 4°C for 5 min before surface staining with an antibody cocktail at 4°C for 30min in 100 μL. Cells were then washed twice with PBS, stained with LIVE/DEAD Fixable Dead Cell Stain Kit (Molecular Probes) at 4°C for 15min and washed twice with staining buffer (PBS supplemented with 1% BSA and 2 mM EDTA). Finally, cells were fixed by incubation in BD Cytofix Fixation Buffer (BD Biosciences) at 4°C for 30min. Samples were then analyzed using a BD LSR Fortessa X-20 cell analyzer and BD FACSDiva Software version 8.0. For intracellular staining, cells were stained with surface markers, fixed in Fix/Perm buffer (eBioscience) for 15min, washed in permeabilization buffer (eBioscience) twice and stained with primary antibodies targeting intracellular proteins in permeabilization buffer for 30 min at 4°C. Cells were sorted using a BD Biosciences Aria III or analyzed using BD Biosciences Fortessa instruments, and data analysis was performed on FlowJo 10. See Table S4 for a detailed list of antibodies used in flow cytometry.
RNA-seq
Total RNA was extracted from control, SOX4 or ITGAV edited human BT549 and murine 4T1 cells cultured in complete RPMI media in biological triplicates. RNA extraction was performed using the RNeasy Plus Mini Kit (Qiagen # 74134) following the manufacturer’s protocol. Total RNA was quality-checked using an Agilent BioAnalyzer 2000 instrument. RNA with an integrity number of greater than 9.5 was used for subsequent analyses. Total RNA was submitted to GeneWiz for RNA-seq analysis. Libraries were prepared with TruSeq RNA Sample Prep Kit v2 (Illumina). Library concentrations were quantified by Qubit (Invitrogen) and mixed equally for single-end 75 bp sequencing using an Illumina NextSeq 500 instrument. Statistics for differentially expressed genes were calculated using DESeq2 (version 3.5) (Love et. al) and Cufflinks (Trapnel et. al). Differential gene expression was analyzed using the DESeq2 (1.8.1) package in R using default settings. Principal component analyses were generated using the prcomp function in R and plotted with ggplot2. Human and mouse gene homologues were matched using the Mouse Genome Informatics annotation. Heatmaps were generated using the heatmap.2 function in R. RNA-seq. data have been deposited at the Gene Expression Omnibus under accession number GSE144014.
Gene sets enrichment analysis (GSEA)
For gene set identification, the hypergeometric overlap statistic tool (http://software.broadinstitute.org/gsea/msigdb/annotate.jsp) was used to calculate the overlap between a gene list and pathways in MSIgDB (Broad Institute, Molecular signature database). GSEA on gene expression data was performed by loading cufflink count table for each comparison into the GSEA package.
ChIP-Seq
Generally, chromatin from 10×106 cells was used for each ChIP. Nuclei/cells were fixed with 2 mM DSG (Pierce) for 45 min at RT (shaking) prior to formaldehyde fixation for 10 min at RT. The reaction was quenched with glycine (0.125 M). Nuclei/cells were then washed twice with ice-cold PBS, lysed in ChIP sonication buffer (50 mM HEPES pH7.9, 140 mM NaCl, 1 mM EDTA, 1 % Triton X-100, 0.1 % sodium deoxycholate, 0.2 % SDS) supplemented with protease inhibitors, and were subjected to sonication to obtain DNA fragments of 300–800 bp. Immunoprecipitation was done using 5 μl of antibody and 40 μg of chromatin. The soluble chromatin (40 μg) was immunoprecipitated with 10 μg of SOX4 antibody (ab86809 Abcam). ChIP-seq libraries were constructed using Accel-NGS 2S DNA library kit from Swift Biosciences. Fragments of the desired size were enriched using AMPure XP beads (Beckman Coulter). 36-bp paired-end reads were sequenced on a Nextseq instrument (Illumina). The raw data are deposited at the Gene Expression Omnibus (GEO) under the entry GSE144014.
Raw reads were aligned to hg19 using bwa. The resulting sam files were converted to bam with samtools. MACS2 was used to call peaks on the bam files. The bedGraph files containing signal per million reads produced from MACS2 were converted to bigwig files with ucsctool kit. ChIP-seq signals were extracted with bwtool from bigwig files and visualized in R. A peak catalog consisting of all possible peak intervals in ChIP-seq was produced. ChIP-seq signals were extracted with bwtool from bigwig files and then visualized in R. IGV viewer was used to visualize enriched peak relative to hg19 reference genome.
Multiplex Immunofluorescence of TNBC sections
Paraffin embedded archival treatment naïve human TNBC tumor specimens were stained for multiplex immunofluorescence analysis sequentially on the Leica Bond automated staining platform using the Leica Biosystems Refine Detection Kit with citrate antigen retrieval (Leica Biosystems, DS9800). The BOND Polymer Refine Detection utilizes controlled polymerization technology to prepare polymeric HRP-linker antibody conjugates. The detection system avoids the use of streptavidin and biotin, and therefore eliminates non-specific staining as a result of endogenous biotin. The tissue specimens were incubated with hydrogen peroxide to quench the endogenous peroxidase activity followed by staining with a specific primary antibody. A post primary IgG linker reagent was used to localize mouse antibodies. And a Poly-HRP IgG reagent was used to localize rabbit antibodies. The substrate chromogen, 3,3’-Diaminobenzidine tetrahydrochloride hydrate (DAB), stains the complex and is visualized as a brown precipitate. DAPI (blue) counterstaining was used to visualize nuclei. The BOND Polymer Refine Detection kit was used in combination with the BOND automated system to reduce human error and variability resulting from individual reagent dilution, manual pipetting or reagent application.
SOX4 antibody (Abcam #86809) was used at a 1:100 dilution and labeled with Alexa Fluor 594 (Thermo #40957). An E-cadherin antibody (CST3195S, Clone 24E10) was used at a 1:100 dilution and labeled with Alexa Fluor 488 (Thermo # 40953). A CD8 antibody (Dako, M7103, Clone C8/144B) was used at a 1:100 dilution and labeled with Alexa Fluor 647 (Thermo # B40958). A Vimentin antibody (Dako, 0725, Clone V9) was used at a 1:400 dilution and labelled with Alexa Fluor 488 (Thermo # 40953). An αSMA antibody (Abcam, ab5694) was run at a 1:400 dilution and labeled with Alexa Fluor 594 (Thermo #40957). Whole-slide digital image acquisition was performed using the Aperio ScanScope CS System (Aperio Technologies, USA) at a 20x objective. Quality control of the scanned images and analysis were performed using ImageScope software (Aperio, V10.2.1.2315, Nussloch, Germany).
Quantification and Statistical Analyses
Statistical analyses were performed using GraphPad Prism 8 software. Comparisons between two groups were made using an unpaired two-tailed Student’s t-test. For multiple comparisons, analysis of variance (ANOVA) followed by Dunnett’s or Tukey’s post hoc test were used. For nonparametric data, Kruskal-Wallis or Mann Whitney U test followed by Dunn’s test were used. For animal studies, sample size was determined as a function of effect size ((difference in means)/(SD) = 2.0) for a two-sample t-test comparison assuming a significance level of 5%, a power of 90%, and a two-sided t-test. Normal distribution was confirmed using normal probability plot (GraphPad Prism 8.0, GraphPad Software, San Diego, CA), variance was assessed within and between groups. The exact number of mice (n) is listed in the Fig. legend for each experiment. The growth of primary tumors over time was analyzed using two-way ANOVA with multiple comparisons. For comparing mouse survival curves, a log-rank (Mantel-Cox) test was used. For ChIP-seq and RNA-seq data, all statistical analysis was performed with R (version 3.4.0) unless otherwise specified. All p-values are two-sided, and statistical significance was evaluated at the 0.05 level.
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| SOX4 | Abcam | ab86809 |
| SOX4 | Diagenode | CS-129-100 |
| ITGAV | CST | 4711S |
| GAPDH | CST | 8884 |
| pSMAD2 | CST | 18338 |
| CD51 | ThermoFisherScientific | 14051282 |
| ITGB6 | ThermoFisherScientific | PAS-47588 |
| CD45 | BioLegend | 103112 |
| CD3 | BioLegend | 100222 |
| CD4 | BioLegend | 100552 |
| CD8 | BioLegend | 100742 |
| PD1 | BioLegend | 135225 |
| IFNγ | BioLegend | 505836 |
| F4/80 | BioLegend | 123132 |
| Gr1 | Biolegend | 108407 |
| CD11b | BioLegend | 101216 |
| CD11c | BioLegend | 117328 |
| MHC-II | BioLegend | 107606 |
| H-2Kb | BioLegend | 116518 |
| E-cadherin | BioLegend | 147304 |
| HLA-ABC | BioLegend | 311410 |
| HLA-A0201 | BioLegend | 343306 |
| Mouse anti-PD1 | inVivoMAb | BioXcell RMPI-14 clone |
| Rat IgG2a isotype control | inVivoMAb | BioXcell 2A3 clone |
| CD8 depletion antibody | inVivoMAb | BioXcell, 2.43 clone |
| Alexa Fluor 594 | Thermo | #40957 |
| E-cadherin antibody | CST | CST3195S, Clone 24E10 |
| Alexa Fluor 488 | Thermo | #40953 |
| CD8 antibody | Dako | M7103, Clone C8/144B |
| Alexa Fluor 647 | Thermo | # B40958 |
| Vimentin antibody | Dako | 0725, Clone V9 |
| αSMA antibody | Abcam | ab5694 |
| Anti-human PD-1 | Bioxcell | clone J116, BE0188 |
| IgG control | Bioxcell | clone MOPC-21 #BE0083 |
| anti-murine PD-1 | Bioxcell | clone RMP1-14 #BE0146 |
| anti-trinitrophenol rat IgG2a isotype control | Bioxcell | clone 2A3 #BE0089 |
| CD16/CD32 antibody | PharMingen | clone 2.4G2, BD |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Human IL2 | BioLegend | 589104 |
| Murine IL2 | BioLegend | 575406 |
| Human IFNγ | PeproTech | #300-02 |
| Murine IFNγ | Abcam | #Ab9922 |
| collagenase type IV | Sigma-Aldrich | #C5138 |
| DNAse type IV | Sigma-Aldrich | #D5205 |
| Hyaluronidase Type V | Sigma-Aldrich | #H6254 |
| ACK lysis buffer | Life Technologies | #A1049201 |
| Percoll density gradient media | Sigma-Aldrich | #P1644 |
| puromycin | Takara Bio | 631306 |
| GlutaMAX | Life Technologies | 35050061 |
| Cas9 protein (20 μM) | Macrolab | CAS9-200 |
| Protease inhibitor cocktail | Roche | 11836170001 |
| Halt Phosphatase Inhibitor Cocktail | Thermo Scientific | #78427 |
| HRP-catalyzed detection | Perkin Elmer | #NEL104001EA |
| Cell Culture Lysis Reagent | Promega | #E1531 |
| Passive Lysis Buffer | Promega | #E1941 |
| Reporter Lysis Buffer | Promega | #E3971 |
| Collagen I | ThermoFisher | #A1048301 |
| Fibronectin | Corning | 356008 |
| protamine sulfate | Sigma-Aldrich | P3369 |
| Retronectin | Takara Bio | T100B |
| αCD3/αCD28 beads | Life Technologies | #11132D |
| CD8 Dynabeads | StemCell | #19053 |
| CD3/CD28 Dynabeads | Life Technologies | #11453D |
| RPMI 1640 media | ThermoFisher | #11875093 |
| Fix/Perm buffer | eBioscience | #00-5521-00 |
| Hyaluronidase Type V | Sigma-Aldrich | #H6254 |
| DNAse Type IV | Sigma-Aldrich | #D5205 |
| Collagenase Type IV | Sigma-Aldrich | #C5138 |
| Critical Commercial Assays | ||
| TSA Fluorescein kit | Perkin-Elmer | #NEL701001KT |
| TSA Biotin kit | Perkin-Elmer | #NEL700001KT |
| BOND Polymer Refine Detection kit | Leica | #DS9800 |
| BOND Polymer Refine Red Detection kit | Leica | #DS9390 |
| BOND Intense R Detection kit | Leica | #DS9263 |
| Cytofix kit | BD Bioscience | #554655 |
| Transcription Factor Staining kit | eBioscience | #00-5523-00 |
| cDNA synthesis kit | Applied Biosystems, Carlsbad, CA | 4368813 |
| CD8 T cell isolation kit | STEMCELL | #19753 |
| MycoAlert mycoplasma detection kit | Lonza | #LT07-118 |
| Pierce BCA Protein Assay Kit | ThermoFisher Pierce | 23225 |
| PrimeScript RT reagent kit | Takara | RR037B |
| SYBR Premix Ex Taq II | Takara | RR820B |
| RNeasy Plus Mini Kit | Qiagen | (# 74134 |
| TruSeq RNA Sample Prep Kit v2 | Illumina | RS-122-2001 |
| Luciferase Assay System® (Glo Lysis Buffer | Promega | #E2661 |
| LIVE/DEAD Fixable Dead Cell Stain Kit | Molecular Probes | L23105 |
| gentleMACS™ Dissociator | Miltenyibiotech | #130-093-235 |
| CFX96 Real-Time PCR System | Bio-Rad | 1855196 |
| 7900HT Fast Real-Time PCR System | Applied Biosystems, Carlsbad, CA | 4351405 |
| The BOND Polymer Refine Detection kit | Leica Biosystems | DS9800 |
| Aperio ScanScope CS System | Aperio Technologies | NA |
| Lonza 4D Nucleofector Core Unit | Lonza | #AAF-1002B |
| SF Cell Line 96-well NucleofectorTM Kit | Lonza | #V4SC-2096 |
| ChemiDoc MP Imaging System | Bio-Rad | #12003154 |
| FACSDiva Software version 8.0. | BD | 330798 |
| LSR Fortessa X-20 cell analyzer | BD | 658222R1 |
| Deposited Data | ||
| RNA Seq. raw reads and processed data | this paper | GEO: GSE144014 |
| ChIP Seq. raw reads and processed data | this paper | GEO: GSE144014 |
| Experimental Models: Cell Lines | ||
| 4T1 | ATCC | CRL-2539 |
| Py8119 | ATCC | CRL-3278 |
| BT549 | ATCC | HTB-122 |
| Hs578T | ATCC | HTB-126 |
| B16F10 | ATCC | CRL-6475 |
| HEK293T | ATCC | CRL-11268 |
| HepG2-Luc | Signosis | SL-0016-NP |
| Experimental Models: Organisms/Strains | ||
| Balb/c | JAX | #000651 |
| C57BI/6J | JAX | #000664 |
| B6.Cg-Thy1a/Cy Tg(TcraTcrb) 8Rest/J | JAX | #005023 |
| Oligonucleotides | ||
| Primers for RT-qPCR, see Table S3 | NA | NA |
| crRNA sequences, see Table S3 | NA | NA |
| Recombinant DNA | ||
| pINDUCER21-SOX4 | Gift from George Daley | Addgene plasmid # 51304 |
| pENTER-CMV-SOX4 | Vigene Biosciences | CH830603 |
| Software and Algorithms | ||
| Flowjo v10.5 | Flowjo, L.L.C. | RRID: SCR_008520 |
| Prism v8.0.1 | Graphpad | RRID: SCR_002798 |
| Fiji v2.0.0 | ImageJ | RRID: SCR_002285 |
| cBioportal v2.2.0 | MSK Center for Mol Onc | https://www.cbioportal.org/ |
| ssGSEA v2.0 | Broad Institute | http://software.broadinstitute.org/gsea/index.jsp |
| FACSDiva | BD Biosciences | RRID: SCR_001456 |
| MSIgDB (Molecular signature database) | Broad Institute | https://www.gsea-msigdb.org/gsea/msigdb |
| ImageScope software | Aperio Technologies | Aperio, V10.2.1.2315 |
| GraphPad Prism | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| Other | ||
| Optimum Growth shaker flasks | Thompson Scientific | 507516355 |
| Multitron incubation shaker | Infors HT | IN3001 |
| Protein G Sepharose affinity columns | GE Healthcare | 17061802 |
| Superose 6 HPLC column | GE Biosciences | 29091596 |
| Amicon spin columns | Millipore | UFC900396 |
| 96 Well Black Polystyrene Microplates | Corning | #3603 |
| Collagen I coated 96 well plates | ThermoFisher | #A1142803 |
Highlights.
The SOX4 transcription factor induces tumor cell resistance to cytotoxic T cells
SOX4 inhibits expression of genes in innate and adaptive immune pathways
SOX4 expression can be inhibited with an integrin αvβ6 blocking antibody
Antibody treatment results in a substantial survival benefit in models of TNBC
Acknowledgments
This work was supported by the Ludwig Center at Harvard Medical School, NIH grants R01CA251599, R01CA238039, and P01CA163222 (to K.W.W.), T32CA207021 (to A.B. and K.W.W.), fellowships from AACR (to S.K. and N.D.M) and the Cancer Research Institute (CRI, to A.N.R.C.) as well as the DF/HCC Breast SPORE 1P50CA168504.
We thank the Specialized Histopathology Core of the Dana-Farber/Harvard Cancer Center for performing immunofluorescence studies on human and murine TNBC specimens. Dana-Farber/Harvard Cancer Center is supported in part by an NCI Cancer Center Support Grant # NIH 5 P30CA06516. We also thank the Center for Functional Cancer Epigenetics for performing the SOX4 ChIP-seq experiment.
Footnotes
Declaration of interests
K.W.W. serves on the scientific advisory board of TCR2 Therapeutics, T-Scan Therapeutics, SQZ Biotech and Nextechinvest, and he receives sponsored research funding from Novartis. He is a co-founder of Immunitas Therapeutics, a biotech company. D.D. consults for Novartis and is on the advisory board for Oncology Analytics, Inc.
Data and materials availability: The accession number for the raw RNA-seq and ChIP-seq data reported in this paper is GSE144014.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams S, Gray RJ, Demaria S, Goldstein L, Perez EA, Shulman LN, Martino S, Wang M, Jones VE, Saphner TJ, et al. (2014). Prognostic value of tumor-infiltrating lymphocytes in triple-negative breast cancers from two phase III randomized adjuvant breast cancer trials: ECOG 2197 and ECOG 1199. J Clin Oncol 32, 2959–2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agudo J, Ruzo A, Park ES, Sweeney R, Kana V, Wu M, Zhao Y, Egli D, Merad M, and Brown BD (2015). GFP-specific CD8 T cells enable targeted cell depletion and visualization of T-cell interactions. Nat Biotechnol 33, 1287–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aluwihare P, Mu Z, Zhao Z, Yu D, Weinreb PH, Horan GS, Violette SM, and Munger JS (2009). Mice that lack activity of alphavbeta6- and alphavbeta8-integrins reproduce the abnormalities of Tgfb1- and Tgfb3-null mice. Journal of cell science 122, 227–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay A, and Raghavan S (2009). Defining the role of integrin alphavbeta6 in cancer. Curr Drug Targets 10, 645–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Ju HL, Yuan XY, Wang TJ, and Lai BQ (2016). SOX4 is a potential prognostic factor in human cancers: a systematic review and meta-analysis. Clin Transl Oncol 18, 65–72. [DOI] [PubMed] [Google Scholar]
- Chockley PJ, and Keshamouni VG (2016). Immunological Consequences of Epithelial-Mesenchymal Transition in Tumor Progression. J Immunol 197, 691–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C et al. , Differential gene and transcript expression analysis of RNA-seq experiments ith TopHat and Cufflinks. Nature protocols 7, 562–578 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denkert C, Liedtke C, Tutt A, and von Minckwitz G (2017). Molecular alterations in triple-negative breast cancer-the road to new treatment strategies. Lancet 389, 2430–2442. [DOI] [PubMed] [Google Scholar]
- Denkert C, von Minckwitz G, Darb-Esfahani S, Lederer B, Heppner BI, Weber KE, Budczies J, Huober J, Klauschen F, Furlanetto J, et al. (2018). Tumour-infiltrating lymphocytes and prognosis in different subtypes of breast cancer: a pooled analysis of 3771 patients treated with neoadjuvant therapy. The Lancet Oncology 19, 40–50. [DOI] [PubMed] [Google Scholar]
- Desai K, Nair MG, Prabhu JS, Vinod A, Korlimarla A, Rajarajan S, Aiyappa R, Kaluve RS, Alexander A, Hari PS, et al. (2016). High expression of integrin β6 in association with the Rho-Rac pathway identifies a poor prognostic subgroup within HER2 amplified breast cancers. Cancer Med 5, 2000–2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X, Zhao B, Iacob RE, Zhu J, Koksal AC, Lu C, Engen JR, and Springer TA (2017). Force interacts with macromolecular structure in activation of TGF-β. Nature 542, 55–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dongre A, Rashidian M, Reinhardt F, Bagnato A, Keckesova Z, Ploegh HL, and Weinberg RA (2017). Epithelial-to-Mesenchymal Transition Contributes to Immunosuppression in Breast Carcinomas. Cancer research 77, 3982–3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberlein C, Kendrew J, McDaid K, Alfred A, Kang JS, Jacobs VN, Ross SJ, Rooney C, Smith NR, Rinkenberger J, et al. (2013). A human monoclonal antibody 264RAD targeting alphavbeta6 integrin reduces tumour growth and metastasis, and modulates key biomarkers in vivo. Oncogene 32, 4406–4416. [DOI] [PubMed] [Google Scholar]
- Elez E, Kocakova I, Hohler T, Martens UM, Bokemeyer C, Van Cutsem E, Melichar B, Smakal M, Csoszi T, Topuzov E, et al. (2015). Abituzumab combined with cetuximab plus irinotecan versus cetuximab plus irinotecan alone for patients with KRAS wild-type metastatic colorectal cancer: the randomised phase I/II POSEIDON trial. Ann Oncol 26, 132–140. [DOI] [PubMed] [Google Scholar]
- Gide TN, Wilmott JS, Scolyer RA, and Long GV (2018). Primary and Acquired Resistance to Immune Checkpoint Inhibitors in Metastatic Melanoma. Clinical Cancer Research 24, 1260. [DOI] [PubMed] [Google Scholar]
- Goplen NP, Huang S, Zhu B, Cheon IS, Son YM, Wang Z, Li C, Dai Q, Jiang L, and Sun J (2019). Tissue-Resident Macrophages Limit Pulmonary CD8 Resident Memory T Cell Establishment. Frontiers in Immunology 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigore AD, Jolly MK, Jia D, Farach-Carson MC, and Levine H (2016). Tumor Budding: The Name is EMT. Partial EMT . J Clin Med 5, 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazelbag S, Kenter GG, Gorter A, Dreef EJ, Koopman LA, Violette SM, Weinreb PH, and Fleuren GJ (2007). Overexpression of the alpha v beta 6 integrin in cervical squamous cell carcinoma is a prognostic factor for decreased survival. J Pathol 212, 316–324. [DOI] [PubMed] [Google Scholar]
- Henderson NC, and Sheppard D (2013). Integrin-mediated regulation of TGFβ in fibrosis. Biochimica et biophysica acta 1832, 891–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikushima H, Todo T, Ino Y, Takahashi M, Saito N, Miyazawa K, and Miyazono K (2011). Glioma-initiating cells retain their tumorigenicity through integration of the Sox axis and Oct4 protein. J Biol Chem 286, 41434–41441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lourenco AR, and Coffer PJ (2017). SOX4: Joining the Master Regulators of Epithelial-to-Mesenchymal Transition? Trends Cancer 3, 571–582. [DOI] [PubMed] [Google Scholar]
- Manguso RT, Pope HW, Zimmer MD, Brown FD, Yates KB, Miller BC, Collins NB, Bi K, LaFleur MW, Juneja VR, et al. (2017). In vivo CRISPR screening identifies Ptpn2 as a cancer immunotherapy target. Nature 547, 413–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariathasan S, Turley SJ, Nickles D, Castiglioni A, Yuen K, Wang Y, Kadel EE III, Koeppen H, Astarita JL, Cubas R, et al. (2018). TGFbeta attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature 554, 544–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashita M, Sasano H, Tamaki K, Hirakawa H, Takahashi Y, Nakagawa S, Watanabe G, Tada H, Suzuki A, Ohuchi N, and Ishida T (2015). Prognostic significance of tumor-infiltrating CD8+ and FOXP3+ lymphocytes in residual tumors and alterations in these parameters after neoadjuvant chemotherapy in triple-negative breast cancer: a retrospective multicenter study. Breast Cancer Res 17, 124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morikawa M, Derynck R, and Miyazono K (2016). TGF-beta and the TGF-beta Family: Context-Dependent Roles in Cell and Tissue Physiology. Cold Spring Harbor perspectives in biology 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munger JS, Huang X, Kawakatsu H, Griffiths MJ, Dalton SL, Wu J, Pittet JF, Kaminski N, Garat C, Matthay MA, et al. (1999). The integrin alpha v beta 6 binds and activates latent TGF beta 1: a mechanism for regulating pulmonary inflammation and fibrosis. Cell 96, 319–328. [DOI] [PubMed] [Google Scholar]
- Niu J, and Li Z (2017). The roles of integrin alphavbeta6 in cancer. Cancer Lett 403, 128–137. [DOI] [PubMed] [Google Scholar]
- Paidassi H, Acharya M, Zhang A, Mukhopadhyay S, Kwon M, Chow C, Stuart LM, Savill J, and Lacy-Hulbert A (2011). Preferential expression of integrin alphavbeta8 promotes generation of regulatory T cells by mouse CD103+ dendritic cells. Gastroenterology 141, 1813–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D, Kobayashi A, Jiang P, Ferrari de Andrade L, Tay RE, Luoma AM, Tsoucas D, Qiu X, Lim K, Rao P, et al. (2018). A major chromatin regulator determines resistance of tumor cells to T cell-mediated killing. Science 359, 770–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel SJ, Sanjana NE, Kishton RJ, Eidizadeh A, Vodnala SK, Cam M, Gartner JJ, Jia L, Steinberg SM, Yamamoto TN, et al. (2017). Identification of essential genes for cancer immunotherapy. Nature 548, 537–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng W, Chen JQ, Liu C, Malu S, Creasy C, Tetzlaff MT, Xu C, McKenzie JA, Zhang C, Liang X, et al. (2016). Loss of PTEN Promotes Resistance to T Cell-Mediated Immunotherapy. Cancer discovery 6, 202–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng X, Liu G, Peng H, Chen A, Zha L, and Wang Z (2017). SOX4 contributes to TGF-β-induced epithelial-mesenchymal transition and stem cell characteristics of gastric cancer cells. Genes Dis 5, 49–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peranzoni E, Lemoine J, Vimeux L, Feuillet V, Barrin S, Kantari-Mimoun C, Bercovici N, Guérin M, Biton J, Ouakrim H, et al. (2018). Macrophages impede CD8 T cells from reaching tumor cells and limit the efficacy of anti–PD-1 treatment. Proceedings of the National Academy of Sciences 115, E4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puram SV, Parikh AS, and Tirosh I (2018). Single cell RNA-seq highlights a role for a partial EMT in head and neck cancer. Molecular & cellular oncology 5, e1448244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raab-Westphal S, Marshall JF, and Goodman SL (2017). Integrins as Therapeutic Targets: Successes and Cancers. Cancers (Basel) 9, 110–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghu G, Mouded M, Culver DA, Hamblin MJ, Golden JA, Veeraraghavan S, Enelow RI, Lancaster LH, Goldberg HJ, Frost AE, et al. (2018). Randomized, Double-Blind, Placebo-Controlled, Multiple Dose, DoseEscalation Study of BG00011 (Formerly STX-100) in Patients with Idiopathic Pulmonary Fibrosis (IPF). Am J Respir Crit Care Med 197, A7785. [Google Scholar]
- Schmid P, Adams S, Rugo HS, Schneeweiss A, Barrios CH, Iwata H, Dieras V, Hegg R, Im SA, Shaw Wright G, et al. (2018). Atezolizumab and Nab-Paclitaxel in Advanced Triple-Negative Breast Cancer. N Engl J Med 379, 2108–2121. [DOI] [PubMed] [Google Scholar]
- Shields RL, Namenuk AK, Hong K, Meng YG, Rae J, Briggs J, Xie D, Lai J, Stadlen A, Li B, et al. (2001). High resolution mapping of the binding site on human IgG1 for Fc gamma RI, Fc gamma RII, Fc gamma RIII, and FcRn and design of IgG1 variants with improved binding to the Fc gamma R. J Biol Chem 276, 6591–6604. [DOI] [PubMed] [Google Scholar]
- Song GD, Sun Y, Shen H, and Li W (2015). SOX4 overexpression is a novel biomarker of malignant status and poor prognosis in breast cancer patients. Tumour Biol 36, 4167–4173. [DOI] [PubMed] [Google Scholar]
- Spranger S, Bao R, and Gajewski TF (2015). Melanoma-intrinsic β-catenin signalling prevents anti-tumour immunity. Nature 523, 231–235. [DOI] [PubMed] [Google Scholar]
- Stone RC, Pastar I, Ojeh N, Chen V, Liu S, Garzon KI, and Tomic-Canic M (2016). Epithelial-mesenchymal transition in tissue repair and fibrosis. Cell Tissue Res 365, 495–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavazoie SF, Alarcon C, Oskarsson T, Padua D, Wang Q, Bos PD, Gerald WL, and Massague J (2008). Endogenous human microRNAs that suppress breast cancer metastasis. Nature 451, 147–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari N, Tiwari VK, Waldmeier L, Balwierz PJ, Arnold P, Pachkov M, Meyer-Schaller N, Schubeler D, van Nimwegen E, and Christofori G (2013). Sox4 is a master regulator of epithelial-mesenchymal transition by controlling Ezh2 expression and epigenetic reprogramming. Cancer cell 23, 768–783. [DOI] [PubMed] [Google Scholar]
- Uhl W, Zuhlsdorf M, Koernicke T, Forssmann U, and Kovar A (2014). Safety, tolerability, and pharmacokinetics of the novel alphav-integrin antibody EMD 525797 (DI17E6) in healthy subjects after ascending single intravenous doses. Invest New Drugs 32, 347–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vervoort SJ, Lourenço AR, van Boxtel R, and Coffer PJ (2013a). SOX4 mediates TGF-β-induced expression of mesenchymal markers during mammary cell epithelial to mesenchymal transition. PloS one 8, e53238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vervoort SJ, van Boxtel R, and Coffer PJ (2013b). The role of SRY-related HMG box transcription factor 4 (SOX4) in tumorigenesis and metastasis: friend or foe? Oncogene 32, 3397–3409. [DOI] [PubMed] [Google Scholar]
- Vidarsson G, Dekkers G, and Rispens T (2014). IgG subclasses and allotypes: from structure to effector functions. Front Immunol 5, 10.3389/fimmu.2014.00520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worthington JJ, Kelly A, Smedley C, Bauche D, Campbell S, Marie JC, and Travis MA (2015). Integrin alphavbeta8-Mediated TGF-beta Activation by Effector Regulatory T Cells Is Essential for Suppression of T-Cell-Mediated Inflammation. Immunity 42, 903–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X, Tam WL, Shibue T, Kaygusuz Y, Reinhardt F, Ng Eaton E, and Weinberg RA (2015). Distinct EMT programs control normal mammary stem cells and tumour-initiating cells. Nature 525, 256–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaretsky JM, Garcia-Diaz A, Shin DS, Escuin-Ordinas H, Hugo W, Hu-Lieskovan S, Torrejon DY, Abril-Rodriguez G, Sandoval S, Barthly L, et al. (2016). Mutations Associated with Acquired Resistance to PD-1 Blockade in Melanoma. N Engl J Med 375, 819–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Liang Q, Lei Y, Yao M, Li L, Gao X, Feng J, Zhang Y, Gao H, Liu DX, et al. (2012). SOX4 induces epithelial-mesenchymal transition and contributes to breast cancer progression. Cancer Res 72, 4597–4608. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Zheng Z, Robbins PF, Khong HT, Rosenberg SA, and Morgan RA (2005). Primary human lymphocytes transduced with NY-ESO-1 antigen-specific TCR genes recognize and kill diverse human tumor cell lines. J Immunol 174, 4415–4423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F (2009). Molecular mechanisms of IFN-gamma to up-regulate MHC class I antigen processing and presentation. Int Rev Immunol 28, 239–260. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for the raw RNA-seq and ChIP-seq data reported in this paper is GEO: GSE144014.